Abstract
There are considerable interindividual variations in drug absorption, distribution, metabolism and excretion (ADME) in humans, which may lead to undesired drug effects in pharmacotherapy. Some of the mechanistic causes are known, e.g., genetic polymorphism, inhibition and induction of ADME enzymes and transporters, while others such as posttranscriptional regulation of ADME genes are under active study. MicroRNAs (miRNAs) are a large group of small, noncoding RNAs that control posttranscriptional expression of target genes. More than 1000 miRNAs have been identified in the human genome, which may regulate thousands of protein-coding genes. Some miRNAs directly or indirectly control the expression of xenobiotic-metabolizing cytochrome P450 enzymes, ATP-binding cassette or solute carrier transporters and/or nuclear receptors. Consequently, intervention of miRNA epigenetic signaling may alter ADME gene expression, change the capacity of drug metabolism and transport, and influence the sensitivity of cells to xenobiotics. In addition, the expression of some ADME regulatory miRNAs is significantly changed in cells following the exposure to a given drug, and the consequent changes in ADME gene expression might result in distinct ADME properties and drug response. In this review, we summarized recent findings on the role of noncoding miRNAs in epigenetic regulation of ADME genes and discussed the potential impact on pharmacokinetics and pharmacodynamics.
Keywords: MicroRNA, Epigenetics, Gene regulation, Pharmacokinetics, Multidrug resistance, Cancer
1. Introduction
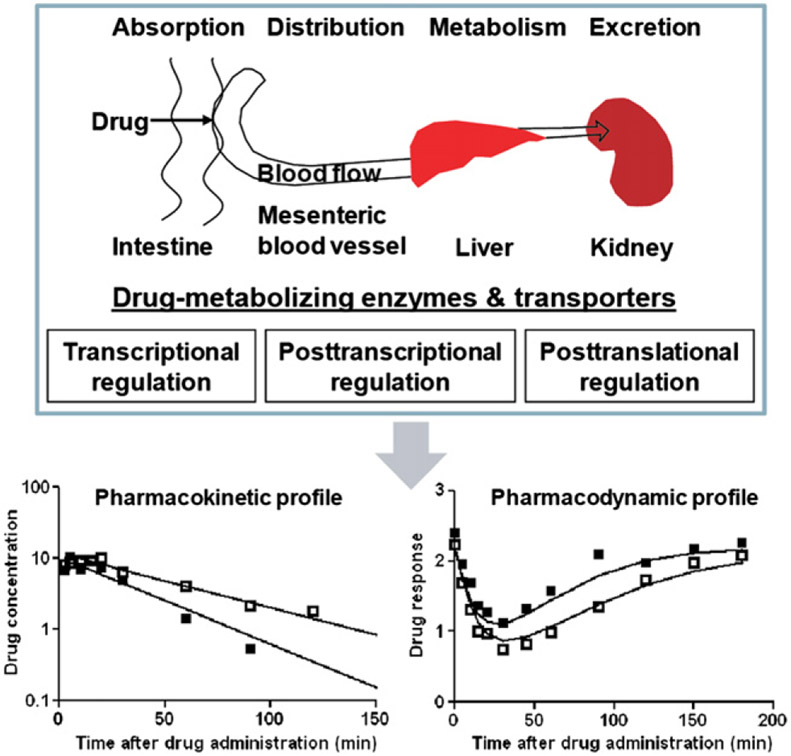
Drugs administered orally undergo a series of processes, namely absorption, distribution, metabolism and excretion (ADME). The ADME of drugs are mediated by drug-metabolizing enzymes and transporters expressed in different tissues including small intestine, liver and kidney. In particular, xenobiotic-metabolizing enzymes such as cytochrome P450 (CYP or P450) isoforms play a critical role in metabolic elimination of drugs1, and transporters such as ATP binding cassette (ABC) and solute carrier (SLC) transporters have high impact on drug absorption, distribution and excretion2. The interactions between drugs and enzymes/transporters ultimately determine the pharmacokinetic properties and subsequently influence the pharmacodynamics (Fig. 1).
Figure 1.
Xenobiotic-metabolizing enzymes and transporters underlying drug absorption, distribution, metabolism and excretion processes are regulated at the transcriptional, posttranscriptional and posttranslational levels. Different extents of ADME gene expression may result in considerable variations in pharmacokinetics and pharmacodynamics.
There are considerable variations in the ADME of xenobiotics in humans, which may lead to a reduced drug efficacy or an adverse drug reaction in pharmacotherapy. It is noteworthy that adverse drug reaction is one of the leading causes of morbidity and mortality, and imposes an enormous financial burden on health care3,4. Some of the mechanistic causes of interindividual variability in ADME are known, e.g., genetic polymorphism, induction and inhibition of drug-metabolizing enzymes and/or transporters5,6. For instance, genetic polymorphism of CYP2D6 is a well-known cause of the loss, reduction or enhancement of drug elimination. Thus knowing a patient’s CYP2D6 genotype or phenotype would allow clinicians to practice individualized medication, i.e. to achieve a desired drug response and avoid the adverse drug events by prescribing an alternative drug or adjusting the dose7-9. As another example, unwanted drug effects may occur when one drug alters the exposure to a concurrent drug through the inhibition or induction of drug-metabolizing enzyme or transporter. Given the risk of adverse drug–drug interactions, one is advised to avoid concomitant use of the drugs10. Therefore, a mechanistic understanding of the risk of interpatient variability in ADME can help clinicians to develop appropriate approaches to prevent potential adverse drug events and achieve the desired drug efficacy.
Expression of ADME genes is tightly regulated at the transcriptional and translational levels, as well as through posttranslational modification, membrane trafficking, subcellular organization and some signal transduction pathways11-14. The important role of xenobiotic receptors in transcriptional regulation of ADME genes has been recognized well. Drugs can activate or deactivate a nuclear transcriptional factor to different extents and cause considerable variation in ADME (Fig. 1). Attention is drawn to the epigenetic regulation of ADME genes, including DNA methylation, histone modification and noncoding RNA (ncRNA) mediated posttranscriptional regulation15-23. DNA methylation is the addition of methyl group to DNA, predominantly at cytosine residues of the dinucleotide sequence CpG, mediated by the DNA methyltransferase (DNMT). Methylation of CpG islands is a known cause of gene silencing found in human diseases. Histone modifications, such as acetylation, phosphorylation and ubiquitination of histone code, are strongly associated with the de novo methylation of DNA underlying gene silencing. Some ADME genes are subject to DNA methylation and histone modifications, leading to an altered gene expression in cells24-29. In addition, there are increased studies on microRNA (miRNA) controlled posttranscriptional regulation of ADME genes (Table 1), which binds to the complementary sequences of mRNA targets and reduces gene expression. In this review, we focus on the discussion miRNA-controlled regulation of ADME genes.
Table 1.
Drug-metabolizing enzymes, transporters and nuclear receptors shown to be targeted by miRNAs.
| Name | MicroRNA | Reference | |
|---|---|---|---|
| Enzymes | CYP1B1 | miR-27b | 50 |
| CYP2A3 | miR-126*, miR-34 | 60 | |
| CYP2E1 | miR-378 | 53 | |
| CYP3A4 | miR-27b, mmu-miR-298 | 54 | |
| svRNAb | 57 | ||
| CYP7A1 | miR-122a, miR-422a | 58 | |
| CYP24A1 | miR-125b | 59 | |
| Transporters | ABCB1/P-gp | miR-451 | 61,63 |
| miR-27a and miR-451 | 62,64 | ||
| ABCG2/BCRP | miR-520h | 65,66,70 | |
| miR-519c | 65,68,69 | ||
| miR-328 | 65,67 | ||
| ABCA1 | miR-101, miR-135 | 74 | |
| miR-33a, miR-33b | 94-96 | ||
| miR-758 | 97 | ||
| miR-106b | 98 | ||
| ABCC1 | miR-134 | 72 | |
| miR-326 | 71 | ||
| miR-199a, miR-199b, miR-296 | 74 | ||
| ABCC2 | miR-379 | 73 | |
| ABCC3 | mmu-miR-665 | 99 | |
| miR-9* | 100 | ||
| ABCC4 | miR-125a, miR-125b | 74 | |
| ABCC5 | miR-101, miR-125a, Let-7a | 74 | |
| miR-128 | 101 | ||
| ABCC6 | miR-9* | 100 | |
| ABCC7 | hsa-miR-145, hsa-miR-494 | 102 | |
| ABCC10 | Let-7a, Let-7e | 74 | |
| ABCE1 | miR-26a, miR-135b, miR-145 | 74 | |
| SLC6A4 | miR-16 | 76 | |
| SLC15A1 | miR-92b | 75 | |
| SLC12A2 | hsa-miR-384, hsa-miR-494 and | 102 | |
| hsa-miR-1246 | |||
| SLC16A1 | miR-124 | 103 | |
| Nuclear receptors | NR1I2/PXR | miR-148a | 78 |
| NR3C1/GR | miR-18 and miR-124a | 87 | |
| miR-130b | 88 | ||
| NR2B1/RXRα | miR-27a/b | 77 | |
| NR1I1/VDR | miR-27b and mmu-miR-298 | 54 | |
| miR-125b | 104 | ||
| NR1C1/PPARα | miR-10b | 83 | |
| miR-506 | 82 | ||
| miR-21 | 81,105 | ||
| miR-27b | 81 | ||
| NR1C3/PPARγ | miR-130 | 106 | |
| miR-27b | 107,108 | ||
| miR-27a | 109 | ||
| NR1C2/PPARβ | miR-15a | 110 | |
| NR1H3/LXRα | miR-613 | 84 | |
| NR2A1/HNF4α | miR-24, miR-34a | 79,80 | |
| NR3A1/ERα | miR-221, miR-222 | 85,111 | |
| Let-7 | 86 | ||
| miR-130a | 112 | ||
| miR-22 | 113 |
2. MicroRNAs in posttranscriptional gene regulation
A large portion (~98%) of human genome is comprised of non-coding DNAs, which do not code for proteins, but are proceeded to produce functional RNAs30. These non-coding RNAs (ncRNAs) include ribosomal RNAs, transfer RNAs, miRNAs, small nucleolar RNAs (snoRNAs) and small nuclear RNAs (snRNAs), which have critical roles in gene regulation and biological processes fundamental to physiological and pathophysiological conditions such as diseases. Study on the function of ncRNAs is rapidly expanding towards the elucidation of their roles along with protein-coding genes in human diseases as well as development of therapeutic strategies. Among them the miRNAs, a class of small (18–25 nt in length) ncRNAs in control of posttranscriptional regulation of target genes31-33, represent one of the most intensively studied groups of ncRNAs. Since the discovery of the first miRNA lin-4 from C. elegans in 199334, over 1000 miRNAs have been identified in human genome, which are predicted to regulate 30–60% of protein-coding genes critical for essentially all biological processes35,36.
The biogenesis of canonical miRNAs consists of multiple steps37. The primary miRNA transcript (pri-miRNA) is transcribed by RNA polymerase II or III, and cleaved by the microprocessor complex Drosha-DGCR8 (Pasha) to release hairpin structured miRNA precursor (pre-miRNA) in the nucleus. Besides the Drosh-DGCR8-dependent pathway, some pre-miRNAs are generated from intronic miRNAs (Mirtrons) via the mRNA precursor splicing pathway. Pre-miRNA is transported from nucleus to cytoplasm by exportin-5-Ran-GTP and processed by the complex of RNase Dicer and double-stranded RNA-binding protein TRBP into a short double stranded miRNA duplex. After the duplex is unwound, the resultant guide strand mature miRNA is preferentially assembled into the RNA-induced silencing complex (RISC).
Mature miRNA incorporated in the RISC complex binds to the 3′ untranslated region (3′UTR) of target mRNAs through complementary Watson–Crick base pairings, leading to an enhanced mRNA cleavage, translational repression or dead-enylation38. Despite the fact that miRNA-recognition elements (MREs) are often present within the 3′UTR of target mRNAs, some sites may reside in the protein-coding regions of the target genes. The extent of gene suppression by miRNA may be related to the miRNA and mRNA levels, number of MRE sites, strength of contiguous base pairings and target site accessibility. Given the fact that a miRNA may act on multiple targets including the transcriptional factors of a given gene, knowing the miRNA signaling pathways shall help better understand miRNA regulatory mechanisms.
MicroRNAs are named using the prefix “miR” followed by a dash and a number (e.g., miR-27), according to the standard nomenclature guidelines39,40. Capital and lowercase “R” are used to annotate mature miRNA (e.g., miR-27) and pre-miRNA (mir-27), respectively. Another three-letter prefix is employed to identify the species of origin. For instance, homo sapiens miR-27 is named hsa-miR-27, and Mus musculus miR-27 is called mmu-miR-27. MicroRNAs with very similar sequences within a species are given the same number, with an additional lower case letters as suffixes to distinguish their difference in 1–2 nucleotides (e.g., hsa-miR-27b versus hsa-miR-27a). In case that the double stranded miRNA duplex is processed to two mature miRNAs, the resultant sense and antisense are denoted with −5p and −3p suffix (e.g., hsa-miR-27b-5p and hsa-miR-27b-3p), respectively. Rather, a number of early discovered miRNAs are still denoted with trivial names (e.g., let-7).
MicroRNAs play important roles in control of biological processes through posttranscriptional regulation of target genes. For instance, neuronal miR-124 has been shown to promote neuronal differentiation via the regulation of a complex network of nervous system-specific alternative pre-mRNA splicing41. Liver-specific miR-122 regulates cholesterol and lipid metabolism by targeting those enzymes and transporters42,43. The evolutionarily conserved, pancreatic islet-specific miR-375 is revealed to regulate the expression of myotrophin and thus control insulin secretion44. In addition, a number of miRNAs have been identified as oncogenes or tumor suppressors to promote or inhibit tumor initiation, progression and metastasis45-49. Therefore, miRNAs constitute a group of novel pharmaceutical targets for the treatment of various human diseases.
3. MicroRNAs in control of ADME gene expression
There is also increasing evidence indicating that miRNAs are involved in posttranscriptional regulation of ADME genes19,20, which in turn is anticipated to affect pharmacokinetics and pharmacodynamics (Fig. 1). More and more studies have demonstrated that miRNAs are able to target the 3′UTR of ADME genes, including P450 enzymes, ABC and SLC transporters, and xenobiotic receptors (Table 1). Change of ADME gene expression by miRNAs may lead to an altered capacity of drug metabolism and disposition, as well as a different response to xenobiotics.
3.1. MicroRNAs regulate the expression of P450 enzymes
CYP1B1 is an extrahepatic P450 enzyme that plays an important role in procarcinogen activation. miR-27b was shown to suppress CYP1B1 protein expression in human cell lines via direct action on the miR-27b MRE site near the poly(A) tail50. Modulation of miR-27b expression/function altered CYP1B1 enzymatic activity, as indicated by the change in metabolism of various procarcinogens and 4-hydroxylation of 17β-estradiol. Interestingly, the 5′UTR of CYP1B1 was found to interact with the miR-27b MRE within the 3′UTR of CYP1B1 mRNA and almost completely block the translation of CYP1B151, indicating the importance of 3′UTR in CYP1B1 gene expression.
Accounting for about 7% of total hepatic P450 enzymes in human liver, CYP2E1 contributes to the metabolism of many toxicologically important chemicals (e.g., ethanol, acetone, and benzene) and therapeutic drugs (e.g., acetaminophen). CYP2E1 is readily induced by ethanol through a posttranslational regulatory mechanism52. The 3′UTR of human CYP2E1 was found to contain a MRE site for miR-378 53. Overexpression of miR-378 caused a decrease in CYP2E1 protein expression, likely through a translation inhibition mechanism. Furthermore, overexpression of miR-378 significantly reduced chlorzoxazone 6-hydroxylase activity in cells, suggesting a potential impact of miR-378 on drug metabolism.
CYP3A4 is the most abundant P450 enzyme in human liver and small intestine, and metabolizes over 50% of pharmaceutical drugs such as benzodiazepines, antivirals and steroids. It was revealed that miR-27b and mmu-miR-298 acted directly on the 3′UTR of CYP3A4 (Fig. 2A), and modulated CYP3A4 expression in human cancer cell lines following the induction with vitamin D54. The involvement of miRNAs in regulation of CYP3A4 might provide helpful explanation for the developmental expression of CYP3A455,56. Moreover, CYP3A4 was identified as a major target for the small, non-coding vault RNA (svRNA), svRNAb, which was derived from vault RNA by a pathway different from miRNA biogenesis57. svRNAb was shown to down-regulate CYP3A4 3′UTR-luciferase reporter activity. This novel finding on svRNA in regulation of CYP3A4 may not only expand the repertoire of small RNAs in gene regulation but also provide mechanistic understanding of the association between vault particles and drug resistance.
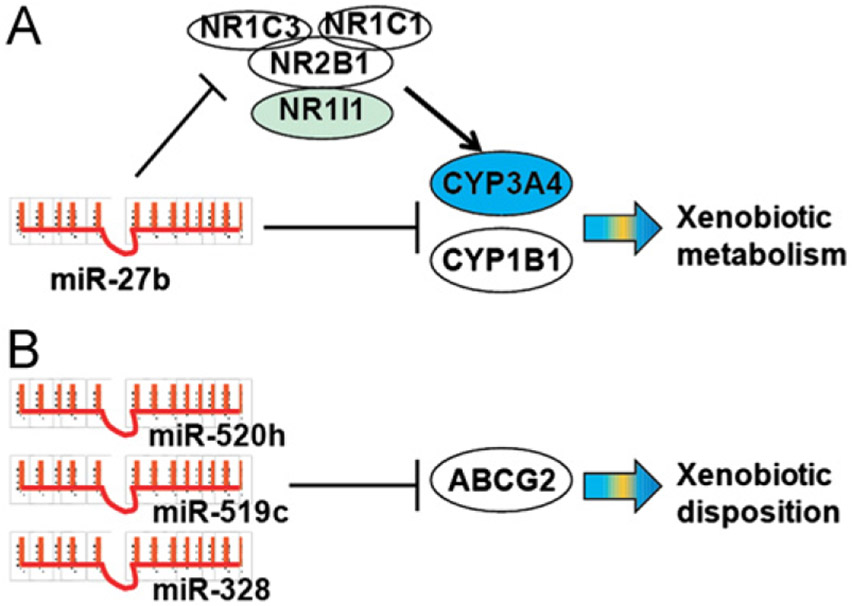
Figure 2.
(A) One microRNA (e.g., miR-27b) may regulate the expression of an ADME gene via direct targeting the gene (e.g., CYP3A4) and/or “indirect” targeting its regulatory factors (e.g., PPARα/NR1C1, PPARγ/NR1C3, VDR/NR1I1 and RXRα/NR2B1). (B) Multiple microRNAs (e.g., miR-520h, −519c and −328) can target the same ADME gene (e.g., BCRP/ABCG2).
Some other P450 genes were also targeted by miRNAs (Table 1), including the regulation of CYP7A1 by miR-122a and −422a58, CYP24A1 by miR-125b59, and CYP2A3 by miR-126*60. With an increased research on miRNA-controlled epigenetic regulation of drug-metabolizing enzymes, one would expect to have an improved understanding of the significance of miRNAs in drug metabolism.
3.2. MicroRNAs regulate the expression of drug transporters
Human P-glycoprotein (P-gp/ABCB1) is expressed at high levels on the apical membrane of enterocytes, biliary surface of hepatocytes, and luminal (apical) side of kidney proximal tubule cells, and P-gp has high impact on drug absorption, distribution and excretion. Regulation of P-gp by miR-451 and −27a in human cancer cells was defined by different groups61-64. Following the characterization of distinct miRNA expression profiles between parental MCF-7 cells and doxorubicin (DOX)-resistant MCF-7 (MCF-7/DOX) cells, Kovalchuk et al.61 identified a miR-451 MRE within ABCB1 3′UTR and further verified the MRE site using luciferase reporter assays. ABCB1 mRNA and protein expression was altered in cells transfected with miR-451 precursor or inhibitor. Moreover, the transfection of MCF-7/DOX cells with miR-451 precursors sensitized the cells to DOX. Meanwhile, Zhu et al.62 found that miR-451 and −27a levels were elevated more than 2-fold in drug-resistant cancer cells than parental cells. Likewise, miRNA antagomirs or mimics changed ABCB1 expression and altered the intracellular accumulation of DOX and rhodamine 123 (RH-123). These findings demonstrate the impact of miRNAs on cellular drug disposition and chemosensitivity through epigenetic regulation of membrane transporter gene expression.
Breast cancer resistance protein (BCRP/ABCG2) plays an important role in cellular transport of anticancer drugs (e.g., mitoxantrone, doxorubicin and topotecan). Regulation of ABCG2 at the 3′UTR by miRNAs (Fig. 2B) was studied by multiple groups65-70. Using luciferase reporter assay, Liao et al.66 showed that 3′UTR of ABCG2 might be targeted by miR-520h. After defining the distinct ABCG2 mRNA expression patterns in parent S1 and drug-resistant S1MI80 cells, To et al.68,69 found that miR-519c controlled posttranscriptional expression of ABCG2. Meanwhile, Pan et al.67 demonstrated the regulation of ABCG2 protein expression by miR-328, after observing an inverse relationship between miR-328 and ABCG2 expression levels in drug-sensitive MCF-7 and drug-resistant MCF-7/MX100 cancer cells. Further comparative studies65 revealed that miR-519c and −328 had stronger effects than miR-520h in regulation of ABCG2 expression in human breast cancer cells, and these ABCG2-regulatory miRNAs were down-regulated in stem-like cancer cells. Nevertheless, miR-520h was shown to target ABCG2 in PANC-1 cells and inhibit cell migration, invasion and side population70, suggesting that miRNA-controlled regulation of ABCG2 could be cell selective.
Multidrug resistance-associated proteins (MRP/ABCC) are ubiquitously expressed in human tissues and these ABCC transporters are able to transport many endobiotics and xenobiotics. Liang et al.71 found that miR-326 down-regulated MRP1/ABCC1 protein expression in drug-resistant, MRP1-overexpressing MCF-7/VP cells. Consequently, overexpression of miR-326 could reverse the multidrug resistance phenotype, and sensitize the cells to DOX. Meanwhile, Guo et al.72 showed that miR-134 inhibited ABCC1 expression, and down-regulation of ABCC1 protein expression was largely associated with the increased levels of miR-134 in H69AR cells. In addition, Haenisch et al.73 found that miR-379 could target the 3′UTR of ABCC2 and suppress ABCC2 gene expression at the posttranscriptional level, providing a mechanistic understanding of nuclear receptor-independent induction of ABCC2 in HepG2 cells by rifampicin.
A very recent study reported by Borel et al.74 demonstrated that a large number of ABC transporters (e.g., ABCA2, ABCB1, ABCB6, ABCC1, ABCC2, ABCC3, ABCC4, ABCC5, ABCC10, ABCC11, ABCC12 and ABCE1) were up-regulated in hepatocellular carcinoma prior to chemotherapy. Up-regulation of ABC transporters was associated with down-regulation of various miRNAs. Following computational prediction of miRNA target sites in ABC genes, a total of 13 miRNAs were shown to act on the 3′UTRs of ABCA1, ABCC1, ABCC5, ABCC10 and ABCE1 by luciferase reporter assays. These findings support the potential role of miRNAs in epigenetic regulation of ADME genes, as well as the intervention of miRNA signaling as a novel means to overcome multidrug resistance.
Besides the regulation of ABC transporters, some miRNAs were found to control the expression of SLC family transporters (Table 1). Human peptide transporter 1 (PEPT1/SLC15A1) is expressed specifically on the brush border membrane of intestinal epithelial cells, and mediates cellular uptake of various di- or tripeptides and peptidomimetic drugs such as β-lactam antibiotics. miR-92b was shown to target the 3′UTR of PEPT1, suppress PEPT1 expression at both mRNA and protein levels, and subsequently reduce PEPT1-mediated drug transport activity75. As another example, miR-16 was revealed to target serotonin transporter (SERT/SLC6A4)76, the pharmacological target of selective serotonin reuptake inhibitor antidepressants. Chronic administration of fluoxetine increased miR-16 levels in serotonergic raphe nuclei and subsequently reduced SERT expression in mice, which offers a novel view on the pharmacological actions of fluoxetine.
3.3. MicroRNAs regulate the expression of nuclear receptors
MicroRNAs may regulate the expression of ADME genes not only via direct targeting the 3′UTRs of enzymes or transporters but also via “indirect” targeting the 3′UTRs of nuclear receptors (Fig. 2A) (Table 1). For instance, vitamin D receptor (VDR/NR1I1) recruits retinoid X receptor α (RXRα/NR2B1) to form functional heterodimer and thus controls the transcriptional expression of many critical ADME genes including CYP3A4. In addition to a direct targeting the 3′UTR of CYP3A4, miR-27b was found to regulate VDR54 and RXRα77 gene expression. The actions of miR-27b on VDR/RXRα presumably enhanced its impact on CYP3A4 gene expression (Fig. 2A). Another miRNA, miR-148a, was shown to regulate the expression of pregnane X receptor (PXR/NR1I2) and subsequently influence the expression of PXR targeted ADME genes such as CYP3A478. In addition, the 3′UTR of hepatocyte nuclear factor 4α (HNF4α/NR2A1) was targeted by a number of miRNAs79,80. Among them the MRE for miR-34a was demolished by the single nucleotide polymorphism (SNP rs11574744), which might affect the regulation of ADME processes as well as the etiology of diabetes or renal cell carcinoma80.
Peroxisome proliferator-activated receptor alpha (PPARα/NR1C1) is another important transcriptional factor that regulates the expression of genes encoding endo/xenobiotic-metabolizing enzymes. Kida et al.81 found that miR-21 and miR-27b could target PPARα. The overexpression and inhibition of miR-21 or miR-27b in HuH-7 cells significantly decreased and increased the PPARα protein level, respectively. Further, Tong et al.82 showed that miR-506 was overexpressed in the hydroxycamptothecin (HCPT)-resistant human colon cancer cell line, which conferred the resistance to HCPT through the suppression of PPARα expression. In addition, Zheng et al.83 validated the MRE for miR-10b within the 3′UTR of PPARα, and demonstrated the modulation of PPARα protein expression by miR-10b in steatotic L02 cells. These findings suggest that miRNAs may regulate PPARα expression and modulate drug transport.
The liver X receptor alpha (LXRα/NR1H3) is a nuclear receptor closely related to RXR and PPAR in regulation of cholesterol, lipid, bile acid, and steroid metabolism and homeostasis. hsa-miR-613 directly targeted the 3′UTR of LXRα84. Interestingly, the activation of LXRα led to an increased expression of hsa-miR-613. The auto-regulation of LXRα was mediated through the action of sterol regulatory element binding protein (SREBP)-1c, a known LXR target gene, on the SREBP response element within the promoter of hsa-miR-613 gene. This feedback loop might be important for a tight regulation of LXRα critical for endobiotic/xenobiotic metabolism and transport.
Estrogen receptor alpha (ERα/NR3A1) and glucocorticoid receptor (GR/NR3C1), known to control transcriptional regulation of some ADME genes (e.g., CYP3A4 and ABCG2), were also targeted by various miRNAs (Table 1). miR-221/222 was found to negatively regulate ERα protein expression85. Human breast cancer cells transfected with miR-221/222 became resistant to tamoxifen, and knockdown of miR-221/222 sensitized the cells to tamoxifen treatment, suggesting the importance of miR-221/222 in tamoxifen chemotherapy. In addition, miRNA let-7 was shown to induce cell sensitivity to tamoxifen through a negative regulation of ERα signaling in breast cancer cells86, and miR-18, −124a and −130b were found to down-regulate GR protein expression and consequently modulate GR signaling87,88. While the influence of GR/ERα regulatory miRNAs on ADME gene expression has not been investigated, one may not underestimate their importance in ADME of xenobiotics.
4. Modulation of microRNA expression/maturation by small molecule drugs
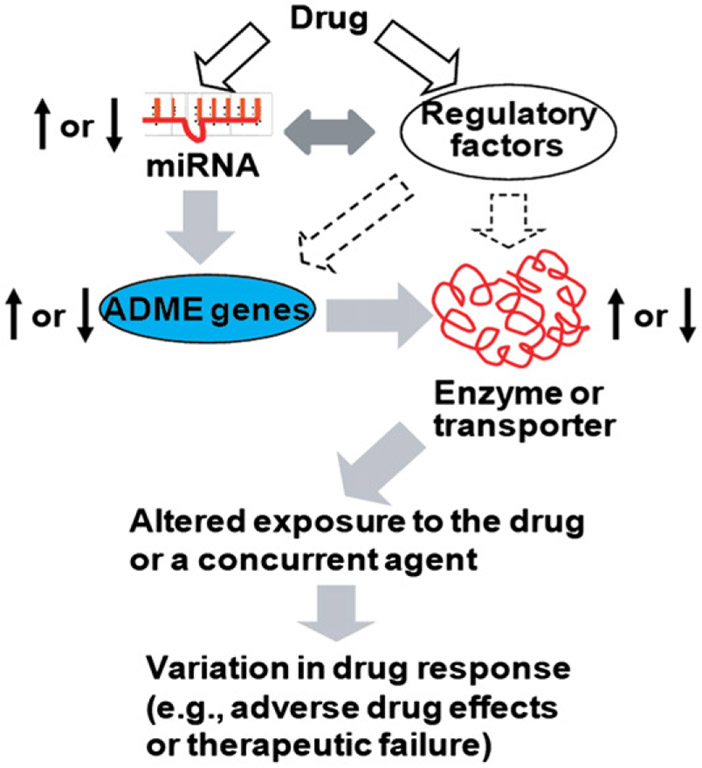
The exposure to small molecule compounds such as medications, drugs of abuse, toxins or hormones may alter the expression of miRNAs in cells through cis- or trans-regulatory mechanisms (reviews19,89). Change in the expression or maturation of miRNAs in regulation of ADME genes would help facilitate the elimination or disposition of the drug or a concomitant agent (Fig. 3). This process presumably represents a feedback mechanism in cellular defense against xenobiotics, while it might cause considerable variations in pharmacokinetics and dynamics.
Figure 3.
Change in the expression/maturation of ADME gene regulatory miRNAs by a drug may alter the expression of drug-metabolizing enzymes and/or transporters, resulting in a variable drug exposure and drug response.
As mentioned above, fluoxetine treatment increased miR-16 expression in mouse serotonergic raphe nuclei76, and the activation of LXRα upregulated hsa-miR-613 expression in primary human hepatocytes as well as hepatocarcinoma cells84. Recently, we examined the impact of 19 drugs (e.g. dexamethasone, vinblastine, bilobalide and cocaine) on the expression of ten miRNAs (e.g., miR-18a, −27b, −124a, −148a, −328, −451 and −519c) in MCF-7, Caco-2, SH-SY5Y and BE(2)-M17 cell systems90. Our data indicated that miRNAs were differentially expressed in human cell lines and the change in miRNA expression was dependent on multiple factors including the nature of drug, type of cells and exposure time. It was noted that dexamethasone and vinblastine, inducers of drug-metabolizing enzymes and transporters, suppressed the expression of miR-27b, −148a and −451 that were shown to down-regulate ADME genes. These observations are helpful for understanding the variable pharmacokinetics, multidrug resistance, drug–drug interactions that are independent upon nuclear receptor-mediated transcriptional regulation.
Because miRNAs are generated from pre-miRNAs derived from the human genome or through mRNA splicing pathways, change of miRNA expression may be attributed to the actions of drugs on cis- or trans-regulatory factors of miRNA genes. For instance, estrogen was found to alter the maturation of a large number of miRNAs through the activation of ERα/NR3A191, and GW3965 was revealed to up-regulate miR-613 through the action of SREBP-1c following the activation of LXRα84. On the other hand, drugs may affect miRNA expression or maturation via targeting the proteins within miRNA processing machinery. As an example, enoxacin was found to enhance the RNA-binding protein TRBP in promoting RNA interference and miRNA processing92,93. Identifying how the expression of ADME regulatory miRNAs is altered by drugs and the underlying mechanisms would help understand drug actions and predict potential variation in ADME.
5. Conclusions
MicroRNAs have been recognized as critical factors in posttranscriptional regulation of target genes in humans. Many miRNAs are revealed to act on the 3′UTRs of drug-metabolizing enzymes, transporters and nuclear receptors underlying ADME processes. The same miRNA (e.g., miR-27b) may control the expression of an ADME gene (e.g., CYP3A4) via direct and/or indirect targeting. On the other hand, multiple miRNAs (e.g., miR-520h, −519c and −328) can regulate the expression of the same ADME gene (e.g., BCRP/ABCG2) to different extents. Change in ADME regulatory miRNA signaling, which can be driven by the drug via various mechanisms, may lead to an altered drug metabolism and transport, as well as variable pharmacological or toxicological responses. With the increase of studies on the relationship between miRNAs as well as other epigenetic factors and pharmacological properties, pharmacoepigenetics and pharmacoepigenomics17,22 are expected to provide an improved understanding of interindividual variations in pharmacotherapy towards rational drug therapy.
Acknowledgment
A.M. Yu would like to thank financial support (award number R01DA021172) from the National Institute On Drug Abuse, National Institutes of Health (NIH).
References
- 1.Guengerich FP. Cytochrome P450s and other enzymes in drug metabolism and toxicity. AAPS J 2006;8:E101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, Chu X, et al. Membrane transporters in drug development. Nat Rev Drug Discov 2010;9:215–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett CL, Nebeker JR, Lyons EA, Samore MH, Feldman MD, McKoy JM, et al. The research on adverse drug events and reports (RADAR) project. J Am Med Assoc 2005;293:2131–40. [DOI] [PubMed] [Google Scholar]
- 4.Gurwitz JH, Field TS, Harrold LR, Rothschild J, Debellis K, Seger AC, et al. Incidence and preventability of adverse drug events among older persons in the ambulatory setting. J Am Med Assoc 2003;289:1107–16. [DOI] [PubMed] [Google Scholar]
- 5.Haga SB, Thummel KE, Burke W. Adding pharmacogenetics information to drug labels: lessons learned. Pharmacogenet Genomics 2006;16:847–54. [DOI] [PubMed] [Google Scholar]
- 6.Thummel KE, Wilkinson GR. In vitro and in vivo drug interactions involving human CYP3A. Annu Rev Pharmacol Toxicol 1998;38:389–430. [DOI] [PubMed] [Google Scholar]
- 7.Ma Q, Lu AY. Pharmacogenetics pharmacogenomics, and individualized medicine. Pharmacol Rev 2011;63:437–59. [DOI] [PubMed] [Google Scholar]
- 8.Nebert DW, Zhang G, Vesell ES. From human genetics and genomics to pharmacogenetics and pharmacogenomics: past lessons, future directions. Drug Metab Rev 2008;40:187–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu AM, Idle JR, Gonzalez FJ. Polymorphic cytochrome P450 2D6: humanized mouse model and endogenous substrates. Drug Metab Rev 2004;36:243–77. [DOI] [PubMed] [Google Scholar]
- 10.Astrand B. Avoiding drug–drug interactions. Chemotherapy 2009;55:215–20. [DOI] [PubMed] [Google Scholar]
- 11.Morgan ET. Impact of infectious and inflammatory disease on cytochrome P450-mediated drug metabolism and pharmacokinetics. Clin Pharmacol Ther 2009;85:434–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muntane J. Regulation of drug metabolism and transporters. Curr Drug Metab 2009;10:932–45. [DOI] [PubMed] [Google Scholar]
- 13.Willson TM, Kliewer SA. PXR CAR and drug metabolism. Nat Rev Drug Discov 2002;1:259–66. [DOI] [PubMed] [Google Scholar]
- 14.Correia MA, Sadeghi S, Mundo-Paredes E. Cytochrome P450 ubiquitination: branding for the proteolytic slaughter? Annu Rev Pharmacol Toxicol 2005;45:439–64. [DOI] [PubMed] [Google Scholar]
- 15.Choudhuri S, Cui Y, Klaassen CD. Molecular targets of epigenetic regulation and effectors of environmental influences. Toxicol Appl Pharmacol 2010;245:378–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez-Antona C, Gomez A, Karlgren M, Sim SC, Ingelman-Sundberg M. Molecular genetics and epigenetics of the cytochrome P450 gene family and its relevance for cancer risk and treatment. Hum Genet 2010;127:1–17. [DOI] [PubMed] [Google Scholar]
- 17.Gomez A, Ingelman-Sundberg M. Pharmacoepigenetics: its role in interindividual differences in drug response. Clin Pharmacol Ther 2009;85:426–30. [DOI] [PubMed] [Google Scholar]
- 18.Gomez A, Ingelman-Sundberg M. Epigenetic and microRNA-dependent control of cytochrome P450 expression: a gap between DNA and protein. Pharmacogenomics 2009;10:1067–76. [DOI] [PubMed] [Google Scholar]
- 19.Yu AM. Role of microRNAs in the regulation of drug metabolism and disposition. Expert Opin Drug Metab Toxicol 2009;5:1513–28. [DOI] [PubMed] [Google Scholar]
- 20.Nakajima M, Yokoi T. MicroRNAs from biology to future pharmacotherapy: regulation of cytochrome P450s and nuclear receptors. Pharmacol Ther 2011;131:330–7. [DOI] [PubMed] [Google Scholar]
- 21.Ingelman-Sundberg M, Gomez A. The past, present and future of pharmacoepigenomics. Pharmacogenomics 2010;11:625–7. [DOI] [PubMed] [Google Scholar]
- 22.Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther 2007;116:496–526. [DOI] [PubMed] [Google Scholar]
- 23.Yu AM. Small interfering RNA in drug metabolism and transport. Curr Drug Metab 2007;8:700–8. [DOI] [PubMed] [Google Scholar]
- 24.Anttila S, Hakkola J, Tuominen P, Elovaara E, Husgafvel-Pursiainen K, Karjalainen A, et al. Methylation of cytochrome P4501A1 promoter in the lung is associated with tobacco smoking. Cancer Res 2003;63:8623–8. [PubMed] [Google Scholar]
- 25.Tokizane T, Shiina H, Igawa M, Enokida H, Urakami S, Kawakami T, et al. Cytochrome P450 1B1 is overexpressed and regulated by hypomethylation in prostate cancer. Clin Cancer Res 2005;11:5793–801. [DOI] [PubMed] [Google Scholar]
- 26.Gomez A, Karlgren M, Edler D, Bernal ML, Mkrtchian S, Ingelman-Sundberg M. Expression of CYP2W1 in colon tumors: regulation by gene methylation. Pharmacogenomics 2007;8:1315–25. [DOI] [PubMed] [Google Scholar]
- 27.Schnekenburger M, Peng L, Puga A. HDAC1 bound to the Cyp1a1 promoter blocks histone acetylation associated with Ah receptor-mediated trans-activation. Biochim Biophys Acta 2007;1769:569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin B, Ryu DY. Regulation of CYP1A2 by histone deacetylase inhibitors in mouse hepatocytes. J Biochem Mol Toxicol 2004;18:131–2. [DOI] [PubMed] [Google Scholar]
- 29.Yang H, Nie Y, Li Y, Wan YJ. Histone modification-mediated CYP2E1 gene expression and apoptosis of HepG2 cells. Exp Biol Med (Maywood) 2010;235:32–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattick JS. RNA regulation: a new genetics?. Nat Rev Genet 2004;5:316–23. [DOI] [PubMed] [Google Scholar]
- 31.Ambros V The functions of animal microRNAs. Nature 2004;431:350–5. [DOI] [PubMed] [Google Scholar]
- 32.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hobert O Gene regulation by transcription factors and microRNAs. Science 2008;319:1785–6. [DOI] [PubMed] [Google Scholar]
- 34.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843–54. [DOI] [PubMed] [Google Scholar]
- 35.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 2005;37:766–70. [DOI] [PubMed] [Google Scholar]
- 36.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 2009;11:228–34. [DOI] [PubMed] [Google Scholar]
- 38.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science 2011;331:550–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, et al. A uniform system for microRNA annotation. RNA 2003;9:277–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006;34:D140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol cell 2007;27:435–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005;438:685–9. [DOI] [PubMed] [Google Scholar]
- 43.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006;3:87–98. [DOI] [PubMed] [Google Scholar]
- 44.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004;432:226–30. [DOI] [PubMed] [Google Scholar]
- 45.Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature 2012;481:190–4. [DOI] [PubMed] [Google Scholar]
- 46.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008;451:147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature 2005;435:828–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007;315:1576–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 2006;124:1169–81. [DOI] [PubMed] [Google Scholar]
- 50.Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res 2006;66:9090–8. [DOI] [PubMed] [Google Scholar]
- 51.Devlin AH, Thompson P, Robson T, McKeown SR. Cytochrome P450 1B1 mRNA untranslated regions interact to inhibit protein translation. Mol Carcinog 2010;49:190–9. [DOI] [PubMed] [Google Scholar]
- 52.Song BJ, Veech RL, Park SS, Gelboin HV, Gonzalez FJ. Induction of rat hepatic N-nitrosodimethylamine demethylase by acetone is due to protein stabilization. J Biol Chem 1989;264:3568–72. [PubMed] [Google Scholar]
- 53.Mohri T, Nakajima M, Fukami T, Takamiya M, Aoki Y, Yokoi T. Human CYP2E1 is regulated by miR-378. Biochem Pharmacol 2010;79:1045–52. [DOI] [PubMed] [Google Scholar]
- 54.Pan YZ, Gao W, Yu AM. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos 2009;37:2112–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Felmlee MA, Lon HK, Gonzalez FJ, Yu AM. Cytochrome P450 expression and regulation in CYP3A4/CYP2D6 double transgenic humanized mice. Drug Metab Dispos 2008;36:435–41. [DOI] [PubMed] [Google Scholar]
- 56.Yu AM, Fukamachi K, Krausz KW, Cheung C, Gonzalez FJ. Potential role for human cytochrome P450 3A4 in estradiol homeostasis. Endocrinology 2005;146:2911–9. [DOI] [PubMed] [Google Scholar]
- 57.Persson H, Kvist A, Vallon-Christersson J, Medstrand P, Borg A, Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol 2009;11:1268–71. [DOI] [PubMed] [Google Scholar]
- 58.Song KH, Li T, Owsley E, Chiang JY. A putative role of micro RNA in regulation of cholesterol 7alpha-hydroxylase expression in human hepatocytes. J Lipid Res 2010;51:2223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Komagata S, Nakajima M, Takagi S, Mohri T, Taniya T, Yokoi T. Human CYP24 catalyzing the inactivation of calcitriol is post-transcriptionally regulated by miR-125b. Mol Pharmacol 2009;76:702–9. [DOI] [PubMed] [Google Scholar]
- 60.Kalscheuer S, Zhang X, Zeng Y, Upadhyaya P. Differential expression of microRNAs in early-stage neoplastic transformation in the lungs of F344 rats chronically treated with the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis 2008;29:2394–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, et al. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther 2008;7:2152–9. [DOI] [PubMed] [Google Scholar]
- 62.Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu CG, et al. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol 2008;76:582–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bitarte N, Bandres E, Boni V, Zarate R, Rodriguez J, Gonzalez-Huarriz M, et al. MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cells 2011;29:1661–71. [DOI] [PubMed] [Google Scholar]
- 64.Li Z, Hu S, Wang J, Cai J, Xiao L, Yu L, et al. MiR-27a modulates MDR1/P-glycoprotein expression by targeting HIPK2 in human ovarian cancer cells. Gynecol Oncol 2010. [DOI] [PubMed] [Google Scholar]
- 65.Li X, Pan YZ, Seigel GM, Hu ZH, Huang M, Yu AM. Breast cancer resistance protein BCRP/ABCG2 regulatory microRNAs (hsa-miR-328, −519c and −520h) and their differential expression in stem-like ABCG2+ cancer cells. Biochem Pharmacol 2011;81:783–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao R, Sun J, Zhang L, Lou G, Chen M, Zhou D, et al. MicroRNAs play a role in the development of human hematopoietic stem cells. J Cell Biochem 2008;104:805–17. [DOI] [PubMed] [Google Scholar]
- 67.Pan YZ, Morris ME, Yu AM. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol 2009;75:1374–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.To KK, Robey RW, Knutsen T, Zhan Z, Ried T, Bates SE. Escape from hsa-miR-519c enables drug-resistant cells to maintain high expression of ABCG2. Mol Cancer Ther 2009;8:2959–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.To KK, Zhan Z, Litman T, Bates SE. Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Mol Cell Biol 2008;28:5147–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang F, Xue X, Wei J, An Y, Yao J, Cai H, et al. hsa-miR-520h downregulates ABCG2 in pancreatic cancer cells to inhibit migration, invasion, and side populations. Br J Cancer 2010;103:567–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liang Z, Wu H, Xia J, Li Y, Zhang Y, Huang K, et al. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol 2010;79:817–24. [DOI] [PubMed] [Google Scholar]
- 72.Guo L, Liu Y, Bai Y, Sun Y, Xiao F, Guo Y. Gene expression profiling of drug-resistant small cell lung cancer cells by combining microRNA and cDNA expression analysis. Eur J Cancer 2010;46:1692–702. [DOI] [PubMed] [Google Scholar]
- 73.Haenisch S, Laechelt S, Bruckmueller H, Werk A, Noack A, Bruhn O, et al. Down-regulation of ABCC2 protein expression in HepG2 cells after rifampicin treatment is mediated by microRNA-379. Mol Pharmacol 2011;80:314–20. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 74.Borel F, Han R, Visser A, Petry H, van Deventer SJ, Jansen PL, et al. ATP-binding cassette transporter genes up-regulation in untreated hepatocellular carcinoma is mediated by cellular microRNAs. Hepatology 2011 10.1002/hep.24682. [DOI] [PubMed] [Google Scholar]
- 75.Dalmasso G, Nguyen HT, Yan Y, Laroui H, Charania MA, Obertone TS, et al. MicroRNA-92b regulates expression of the oligopeptide transporter PepT1 in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 2011;300:G52–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science 2010;329:1537–41. [DOI] [PubMed] [Google Scholar]
- 77.Ji J, Zhang J, Huang G, Qian J, Wang X, Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett 2009;583:759–66. [DOI] [PubMed] [Google Scholar]
- 78.Takagi S, Nakajima M, Mohri T, Yokoi T. Post-transcriptional regulation of human pregnane X receptor by microRNA affects the expression of cytochrome P450 3A4. J Biol Chem 2008;283:9674–80. [DOI] [PubMed] [Google Scholar]
- 79.Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem 2010;285:4415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wirsing A, Senkel S, Klein-Hitpass L, Ryffel GU. A systematic analysis of the 3′UTR of HNF4A mRNA reveals an interplay of regulatory elements including miRNA target sites. PLoS One 2011;6:e27438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kida K, Nakajima M, Mohri T, Oda Y, Takagi S, Fukami T, et al. PPARalpha is regulated by miR-21 and miR-27b in human liver. Pharm Res 2011;28:2467–76. [DOI] [PubMed] [Google Scholar]
- 82.Tong JL, Zhang CP, Nie F, Xu XT, Zhu MM, Xiao SD, et al. MicroRNA 506 regulates expression of PPAR alpha in hydroxycamptothecin-resistant human colon cancer cells. FEBS Lett 2011;585:3560–8. [DOI] [PubMed] [Google Scholar]
- 83.Zheng L, Lv GC, Sheng J, Yang YD. Effect of miRNA-10b in regulating cellular steatosis level by targeting PPAR-alpha expression, a novel mechanism for the pathogenesis of NAFLD. J Gastroenterol Hepatol 2010;25:156–63. [DOI] [PubMed] [Google Scholar]
- 84.Ou Z, Wada T, Gramignoli R, Li S, Strom SC, Huang M, et al. MicroRNA hsa-miR-613 targets the human LXRalpha gene and mediates a feedback loop of LXRalpha autoregulation. Mol Endocrinol 2011;25:584–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem 2008;283:31079–86. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86.Zhao Y, Deng C, Lu W, Xiao J, Ma D, Guo M, et al. Let-7 microRNAs induce tamoxifen sensitivity by down-regulation of estrogen receptor alpha signaling in breast cancer. Mol Med 2011. doi: 10.2119/molmed.2010.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vreugdenhil E, Verissimo CS, Mariman R, Kamphorst JT, Barbosa JS, Zweers T, et al. MicroRNAs miR-18 and miR-124a downregulate the glucocorticoid receptor: implications for glucocorticoid responsiveness in the brain. Endocrinology 2009;150:2220–8. [DOI] [PubMed] [Google Scholar]
- 88.Tessel MA, Benham AL, Krett NL, Rosen ST, Gunaratne PH. Role for microRNAs in regulating glucocorticoid response and resistance in multiple myeloma. Horm Cancer 2011;2:182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li Y, Kong D, Wang Z, Sarkar FH. Regulation of microRNAs by natural agents: an emerging field in chemoprevention and chemotherapy research. Pharm Res 2010;27:1027–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rodrigues AC, Li X, Radecki L, Pan YZ, Winter JC, Huang M, et al. MicroRNA expression is differentially altered by xenobiotic drugs in different human cell lines. Biopharm Drug Dispos 2011;32:355–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, et al. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol cell 2009;36:340–7. [DOI] [PubMed] [Google Scholar]
- 92.Shan G, Li Y, Zhang J, Li W, Szulwach KE, Duan R, et al. A small molecule enhances RNA interference and promotes microRNA processing. Nat Biotechnol 2008;26:933–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Melo S, Villanueva A, Moutinho C, Davalos V, Spizzo R, Ivan C, et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc Natl Acad Sci USA 2011;108:4394–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010;328:1570–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci USA 2010;107:12228–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010;328:1566–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ramirez CM, Dávalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D, et al. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler Thromb Vasc Biol 2011;31:2707–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim J, Yoon H, Ramirez CM, Lee SM, Hoe HS, Fernandez-Hernando C. miR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp Neurol 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dalmasso G, Nguyen HT, Yan Y, Laroui H, Charania MA, Ayyadurai S, et al. Microbiota modulate host gene expression via microRNAs. PLoS One 2011;6:e19293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jeon HM, Sohn YW, Oh SY, Kim SH, Beck S, Kim S, et al. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res 2011;71:3410–21. [DOI] [PubMed] [Google Scholar]
- 101.Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao H, et al. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin Cancer Res 2011;17:7105–15. [DOI] [PubMed] [Google Scholar]
- 102.Gillen AE, Gosalia N, Leir SH, Harris A. MicroRNA regulation of expression of the cystic fibrosis transmembrane conductance regulator gene. Biochem J 2011;438:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li KK, Pang JC, Ching AK, Wong CK, Kong X, Wang Y, et al. miR-124 is frequently down-regulated in medulloblastoma and is a negative regulator of SLC16A1. Hum Pathol 2009;40:1234–43. [DOI] [PubMed] [Google Scholar]
- 104.Mohri T, Nakajima M, Takagi S, Komagata S, Yokoi T. MicroRNA regulates human vitamin D receptor. Int J Cancer 2009;125:1328–33. [DOI] [PubMed] [Google Scholar]
- 105.Zhou J, Wang KC, Wu W, Subramaniam S, Shyy JY, Chiu JJ, et al. MicroRNA-21 targets peroxisome proliferators-activated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc Nat Acad Sci USA 2011;108:10355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, Srikantan S, et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol 2011;31:626–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jennewein C, von Knethen A, Schmid T, Brune B. MicroRNA-27b contributes to lipopolysaccharide-mediated peroxisome proliferator-activated receptor gamma (PPARgamma) mRNA destabilization. J Biol Chem 2010;285:11846–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Karbiener M, Fischer C, Nowitsch S, Opriessnig P, Papak C, Ailhaud G, et al. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem Biophys Res Commun 2009;390:247–51. [DOI] [PubMed] [Google Scholar]
- 109.Kim SY, Kim AY, Lee HW, Son YH, Lee GY, Lee JW, et al. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem Biophys Res Commun 2010;392:323–8. [DOI] [PubMed] [Google Scholar]
- 110.Yin KJ, Deng Z, Hamblin M, Xiang Y, Huang H, Zhang J, et al. Peroxisome proliferator-activated receptor delta regulation of miR-15a in ischemia-induced cerebral vascular endothelial injury. J Neurosci 2010;30:6398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guttilla IK, Phoenix KN, Hong X, Tirnauer JS, Claffey KP, White BA. Prolonged mammosphere culture of MCF-7 cells induces an EMT and repression of the estrogen receptor by microRNAs. Breast Cancer Res Treat 2012;132:75–85. [DOI] [PubMed] [Google Scholar]
- 112.Tang L, Pu Y, Wong DK, Liu T, Tang H, Xiang T, et al. The hepatitis B virus-associated estrogen receptor alpha (ERalpha) was regulated by microRNA-130a in HepG2.2.15 human hepatocellular carcinoma cells. Acta Biochim Biophys Sin (Shanghai) 2011;43:640–6. [DOI] [PubMed] [Google Scholar]
- 113.Xiong J, Yu D, Wei N, Fu H, Cai T, Huang Y, et al. An estrogen receptor alpha suppressor, microRNA-22, is downregulated in estrogen receptor alpha-positive human breast cancer cell lines and clinical samples. FEBS J 2010;277:1684–94. [DOI] [PubMed] [Google Scholar]