Graphical Abstract
Keywords: colloidal crystals, azobenzene, light-responsive, DNA nanoparticle superlattices, optical patterning, Colloidal Crystal Engineering
TOC entry
Light-responsive colloidal crystals are constructed with azobenzene-tethered DNA to undergo reversible structural change between assembled and disassembled states under isothermal conditions. Such dynamic control in bonding between nanoparticles allows one to combine light-responsive colloidal crystals with a photopatterning technique to enable printing of crystalline DNA-nanoparticle thin films into pre-designed shapes.
A novel method for synthesizing and photopatterning colloidal crystals via light-responsive DNA has been developed. These crystals are comprised of 10-30 nm gold nanoparticles interconnected with azobenzene-modified DNA strands. The photoisomerization of the azobenzene molecules leads to reversible assembly and disassembly of the bcc and fcc crystalline nanoparticle lattices. In addition, UV light is used as a trigger to selectively remove nanoparticles on centimeter-scale thin-films of colloidal crystals, allowing one to photopattern them into pre-conceived shapes. The design of the azobenzene-modified linking DNA is critical and involves complementary strands, with azobenzene moieties deliberately staggered between the bases that define the complementary code. This results in a tunable wavelength-dependent melting temperature (Tm) window (4.5-15 °C) and one suitable for effecting the desired transformations. In addition to the isomeric state of the azobenzene groups, the size of the particles can be used to modulate the Tm window over which these structures are light-responsive.
Colloidal crystals are the basis for functional materials such as photonic lattices,[1–3] electronic device components,[4–5] advanced catalysts,[6–8] and magnetic storage devices.[9–11] Therefore, a variety of particle assembly techniques (e.g., interface/template-assisted, field-induced, diffusion-controlled, and ligand-directed strategies)[12–15] have been developed to engineer their formation. Colloidal crystal engineering with DNA,[16–19] which relies on DNA-grafted nanoparticles as “programmable atom equivalents” (PAEs) to generate crystalline nanoparticle superlattices (i.e. soft colloidal crystals), offers advantages over other assembly techniques, because the size, shape, and composition of the core nanoparticle “atom” can be tuned independent of the length, sequence, and density of the DNA “bonds”. Moreover, stimuli-responsive DNA “bonds” render PAEs sensitive to external cues such as pH,[20] enzymes,[21] dielectric media,[22] and small molecules;[23–24] this can yield crystals that reconfigure their structures based upon the introduction of a trigger. As a stimulus, light is widely used in chemical and biological systems to effect structural changes and regulate important processes. Indeed, light provides high spatial and temporal resolution, remote control, and tunability in terms of wavelength.[25–48] We hypothesized that by using a crystal engineering approach with DNA, we can build off of early reports involving amorphous structures, which show particle-tethered azobenzene-DNA can be used in conjunction with light to control duplex stability,[49–51] to design and synthesize light-responsive colloidal crystals.
To integrate colloidal crystals into device structures, a variety of different lithography techniques have been developed, including nano-imprinting,[52] direct-write 3D printing,[53] and e-beam lithography (EBL).[54] Although quite useful for many purposes, nanoimprinting often causes structural deformation to the structures, direct write 3D printing is limited with respect to the size and composition of the crystal building blocks and the types of symmetries that can be accessed, and EBL is time-consuming, labor-intensive, and difficult to implement over large scales. On the other hand, photopatterning is easily scalable and could be used, in principle, to customize light-responsive colloidal materials into predesigned shapes without introducing chemical contaminants into the system. The ability to photo-direct the generation and disassembly of colloidal crystals on surfaces might provide a general way to pattern and integrate colloidal structures with surfaces and device components over large areas. In addition, by combining such capabilities with particle-based lattices engineered from DNA, one could effect changes within nanoscale building blocks with almost unlimited access to crystal symmetry and related parameters.
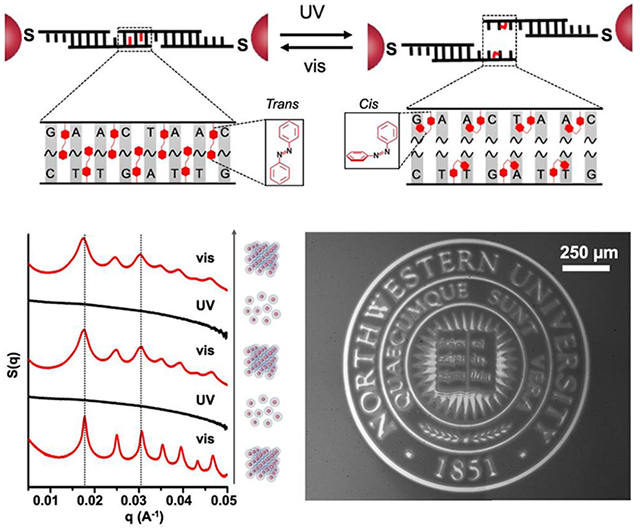
Herein, we report the proof-of-concept synthesis of light-responsive DNA-interconnected colloidal crystals (Figure 1A) and describe two separate light-directed processes – (i) light-directed assembly and disassembly of crystalline nanoparticle superlattices, respectively (Figure 1B), and (ii) light-directed patterning of colloidal crystals (Figure 1C). Light-responsive PAEs were synthesized by using azobenzene-modified DNA (azoDNA) as bonding elements such that the configurational change of azobenzene molecules (between trans- and cis-states) effects DNA hybridization or dehybridization, causing the bonding between, or dissociation of, PAEs, respectively. The azoDNA linkers explored herein were designed to exhibit large wavelength-dependent Tm windows, substantially greater than designs previously explored for effecting simple aggregation/dissociation of amorphous DNA-nanoparticle assemblies.[51] Unlike previous systems in which particle-tethered azoDNA included tracts of 10-base complementary strands to effect aggregation of nanoparticles,[51] the designs reported herein, involve strands with short “sticky ends”, typically between 6 to 8 nucleotide bases and with 7-10 azobenzene moieties incorporated within the duplex that results from the hybridization of the sticky ends. This design enables the reorganization of PAEs even after they have bonded to one another to form a crystalline lattice[55] and provides a wavelength-dependent Tm window suitable for cleanly controlling crystallization or subsequent disassembly.
Figure 1.
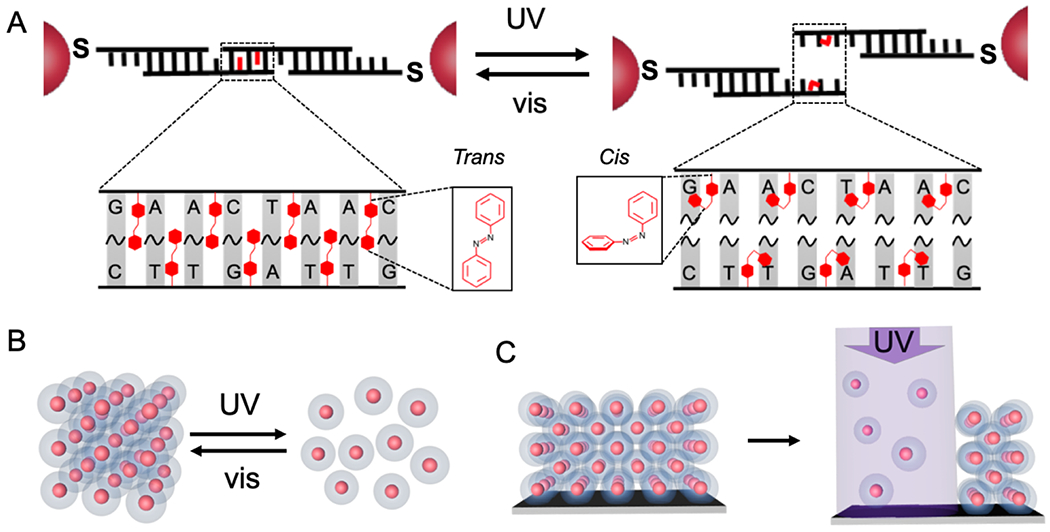
(A) Schematic shows how azobenzene residues are incorporated in the DNA sticky end, such that the trans (left) and cis (right) states of the azobenzene molecules can be toggled via visible or UV light, respectively. Under visible light (trans-form), the DNA sticky ends remain hybridized, but under UV light, the cis-form azobenzene molecules induce dehybridization of the DNA sticky ends. (B) Under isothermal conditions, light is applied to trigger a reversible change in PAE crystals between assembled and disassembled states. (C) UV-light-directed disassembly of surface-grown, crystalline PAE thin films can be used to selectively remove nanoparticles.
In a proof-of-concept experiment, PAEs were synthesized by densely coating gold nanoparticles with a monolayer of “anchor” DNA strands that contain 3’ thiol moieties, and then hybridizing complementary “linker” DNA strands to the “anchor” strands, exposing short, single-stranded “sticky ends” at the outer-surface of PAEs for particle binding (see schematic in Figure S1). Upon annealing, two PAEs of the same size but with complementary “sticky ends” form a bcc crystal, while those with self-complementary “sticky ends” form an fcc crystal.[55] Short “sticky ends” favor particle reorganization and higher crystal quality,[55] but with these systems, the sequences must be long enough to include an adequate number of azobenzene molecules to induce light-responsive particle assembly and dissociation. We tested the design and synthesis of light-responsive PAEs with both complementary and self-complementary sticky ends by incorporating azobenzene moieties in a staggered arrangement (see sequences in Tables S1–S2). For example, 15 nm gold nanoparticle cores were assembled with complementary azoDNA linkers, which had a total of 7 staggered azobenzene groups located at sticky ends that were 8 bases in length (4 and 3 azobenzene moieties on particle type A and B, respectively). After annealing with a slow cooling method, these azoDNA-containing PAEs formed pellets (Figure 2A). Both scanning electron microscopy (SEM) and small angle X-ray scattering (SAXS) of the assembled structures confirms that the nanoparticles are arranged into faceted bcc crystals in the form of the expected rhombic dodecahedron[56] (Figures 2B, 2C, S7A, and S8). Similar results are obtained regardless of gold particle core size (10 – 30 nm in diameter), as long as the same linkers are used. In the case of linkers with self-complementary sticky ends (comprising of a total of 10 azobenzene moieties within 6 pairs of nucleotide bases), the anticipated fcc structures form, as evidenced by SAXS and SEM (Figures S7–S10). Collectively, these experiments and data show that incorporating staggered azobenzene residues in short sticky ends does not hinder the PAE crystallization process.
Figure 2.
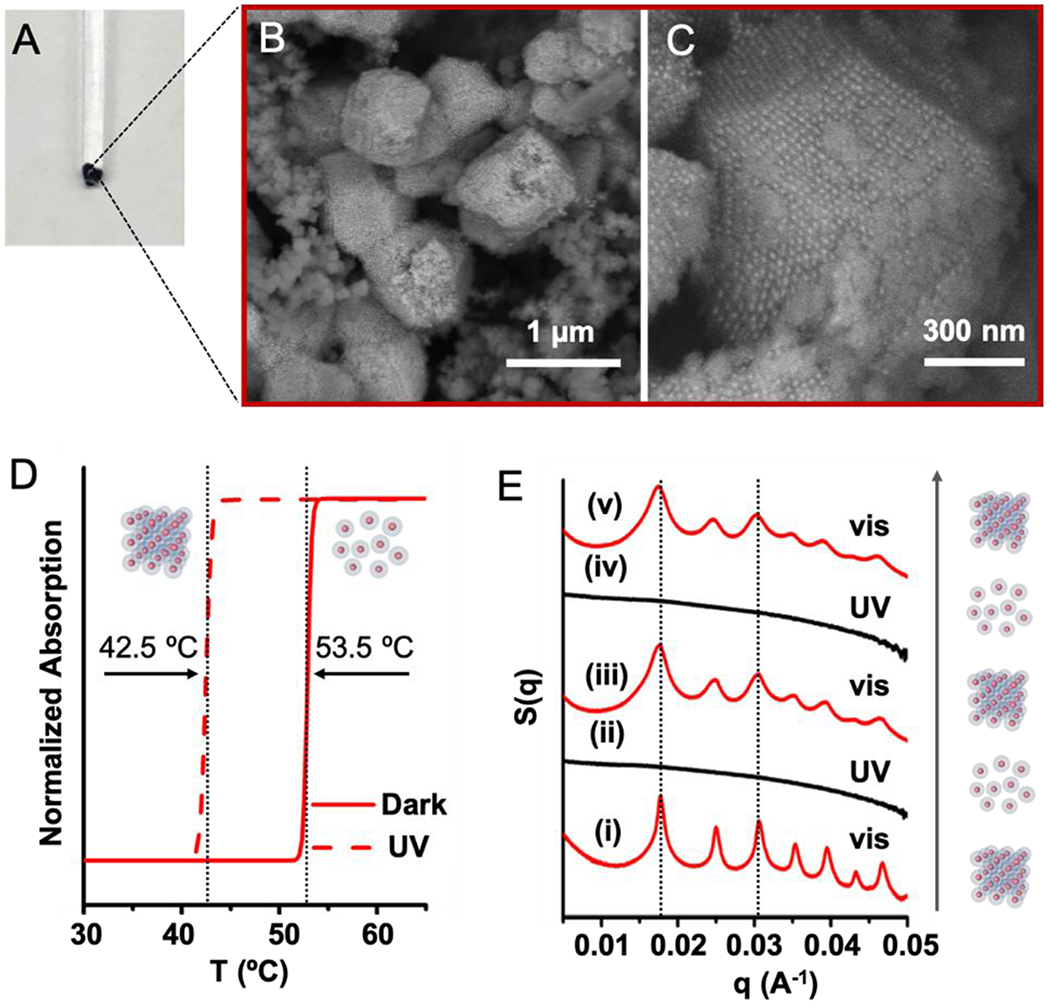
Light-responsive PAEs functionalized with azoDNA linkers successfully assembled into ordered crystals in aqueous solution. (A) Aggregated PAEs were annealed via a slow-cool method, and (B, C) SEM images confirm the formation of ordered PAE crystals. (D) Melting curves of light-responsive PAEs show two different melting temperatures, one in the dark and the other under UV light. (E) SAXS data confirm the formation of bcc crystals and also a dynamic and reversible wavelength-dependent change between assembled (visible light) and disassembled states (UV light). UV light (365 nm, 5 mW cm−2, 30 min irradiation) and visible light (405 nm, 10 mW cm−2, 30 min irradiation) were utilized to effect the reversible structural changes at 53 °C. SAXS measurements were performed within 2 min of removing the crystals from the heating environment. The cooling process prior to the SAXS measurement induces negligible changes to the crystal structure.
To determine the light-dependent characteristics of these structures, samples containing PAEs with either complementary or self-complementary sequences, respectively, were studied as a function of UV-irradiation (Figure S2). UV irradiation (300 - 400 nm) effects a trans-to-cis isomerization,[49–50] which destabilizes the duplexes that form from PAE hybridization. Therefore, crystals formed from PAEs (15 nm core and azoDNA linkers) with complementary sticky ends, including seven azobenzene moieties, exhibit a remarkable eleven-degree difference in Tm in the dark (53.5 °C), as compared to under UV light (42.5 °C). It is important to note that the number of azobenzene molecules within the sticky ends as well as the size of the core nanoparticle dictate the breadth of the Tm window (Figures S11–S13). For example, 4 azobenzene moieties in the sticky ends on 15 nm particle cores result in a Tm window of 4.5 °C (Figures S11B, S12B and S12C), while if the number of azobenzene groups is kept constant (7) and core size is systematically increased from 10 to 30 nm, the Tm window decreases from 15 °C to 9.5 °C. This difference in Tm window is likely due to the facts that smaller PAEs form fewer DNA bonds and a comparable amount of isomerization results in a greater degree of crystal destabilization.[57] Similarly, a particle-size dependent decrease in the wavelength-dependent Tm window was also observed for the fcc crystals formed from PAEs with self-complementary sticky ends (Figure S14). Finally, no melting temperature differences were observed with crystals formed from unmodified DNA sticky ends, confirming that the azobenzene moieties, and not the Watson-Crick base pairs, are responsible for the UV light-dependent Tms (Figures S11A, S12A and S12C).
The ability to tune the melting temperatures for PAE crystals enables one to predictably control the assembly and disassembly and even reversibly toggle between the two states. It is important to note that indeed, azoDNA bonds should exhibit light-induced hybridization and dehybridization at any temperature within the Tm window between 42.5 °C and 53.5 °C for bcc crystals, Figure 2D). To test this hypothesis, we set the temperature at 53 °C, a temperature slightly lower than the Tm for the crystals measured in the dark (i.e. azobenzene molecules are predominately in the trans-form), to maximize the thermal energy for crystallization. Under visible light (405 nm), the PAEs assemble into bcc crystals, as evidenced by SAXS (Figures 2E (i) and S7). Upon switching to UV irradiation (365 nm), the PAEs that defined the crystals dissociated under these isothermal conditions into a colloidal dispersion, due to the light-induced dehybridization of azoDNA (Figure 2E (ii)). Once the UV was replaced by visible light (405 nm), the PAEs again formed bcc crystals with the same lattice constant as that of the original lattice, confirmed by the re-appearance of SAXS peaks at the same locations (Figure 2E (iii)). This reversible change was repeated for two full cycles without any washing steps, while keeping the same set of samples at 53 °C (Figures 2E and S16). Note that the reassembly of colloidal crystals was observed in the dark, albeit at a slower rate (i.e., over a couple of hours) than under visible light. This indicates that the assembly/disassembly process is a consequence of the photoisomerization of azobenzene molecules and that visible light irradiation accelerates the cis-to-trans isomerization. It is important to note that this light-directed assembly and disassembly of PAE crystals does not require external denaturing conditions for dissociating the lattices, such as excess Urea or temperatures above 70 °C, which were used in a previous study that relied on light-induced ligation of DNA bonds in DNA-nanoparticle superlattices.[32]
While PAE crystals in-solution are suitable for studying reversible changes, PAEs grown on a substrate layer-by-layer are appropriate for photopatterning (Figure 1C). Centimeter scale, light-responsive PAE thin films (5 layers) were synthesized via a stepwise growth method[58] (Figures 3A and S17). SEM images as well as grazing-incidence small angle X-ray scattering (GISAXS) measurements confirmed that PAEs with azoDNA-sticky ends form ordered bcc lattices on a substrate (Figures 3B, 3C, 3D and S18). Surface-grown PAE crystals also responded to a change in wavelength; PAEs remained intact in a crystalline thin film under visible light, while exposure to UV dissociated all PAEs within 30 minutes (Figure S19A).
Figure 3.
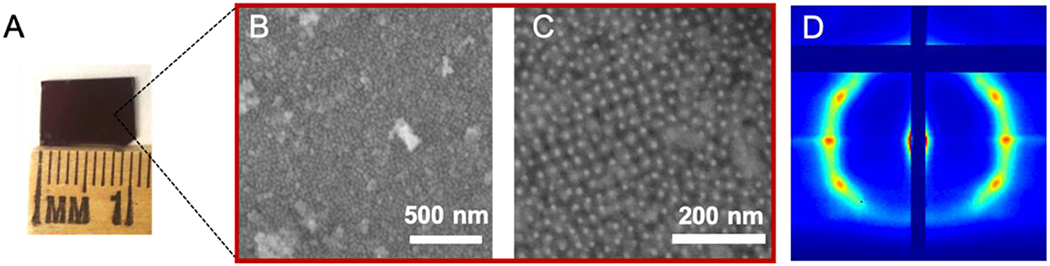
(A) Centimeter-scale, light-responsive PAE thin films on a substrate synthesized via a layer-by-layer process. (B, C) SEM images and (D) GISAXS measurement confirm the formation of ordered PAE thin films.
Based on the previous results, we hypothesized that soft colloidal crystal thin films could be photopatterned into designed shapes. To evaluate possibilities, three different optical lithography approaches were studied. First, a custom-built photomask was applied, in which centimeter scale patterns were hand-crafted and used to mask the desired region above the crystalline PAE thin film (Figures 4A, 4B and S3). These custom-built photomasks are low resolution, due to the gap between the photomask and thin film, which allows light to scatter within the gap and effect crystal dissolution. However, a gap is necessary when using a photomask to pattern colloidal crystals because the soft DNA-mediated colloidal crystal thin films are easily damaged by external forces such as direct contact with a photomask. In order to achieve higher resolution, a direct UV laser writing technique was utilized to make micron-size features from crystalline PAE thin films (Figures S4 and S20). Although effective, laser writing is difficult to scale up because the UV laser spot size is small (15 to 30 microns), and therefore photopatterning large areas becomes time-consuming. In order to achieve high resolution over large scales, a TERA-fab E series with a digital micromirror device (DMD) was used in cantilever-free, scanning probe lithography mode to print micron sized features over centimeter scale areas (Figures 4C, 4D, 4E (middle), S5, S21 and S22). SEM images of the patterned regions confirmed that the majority of light-responsive PAEs were removed, while non-irradiated areas exhibited intact crystals (Figures 4E (left and right)). To our knowledge, this is the first report to show that photopatterning techniques can be used to customize colloidal crystals on surfaces, and as such, these findings mark a significant step towards using colloidal crystals to prepare functional materials that could be integrated with existing microelectronic and optical circuitry. Future studies will explore the potential for incorporating anisotropic nanoparticles of different compositions to build novel stimuli-responsive optical devices or enzymes for constructing thin-film biological sensors.
Figure 4.
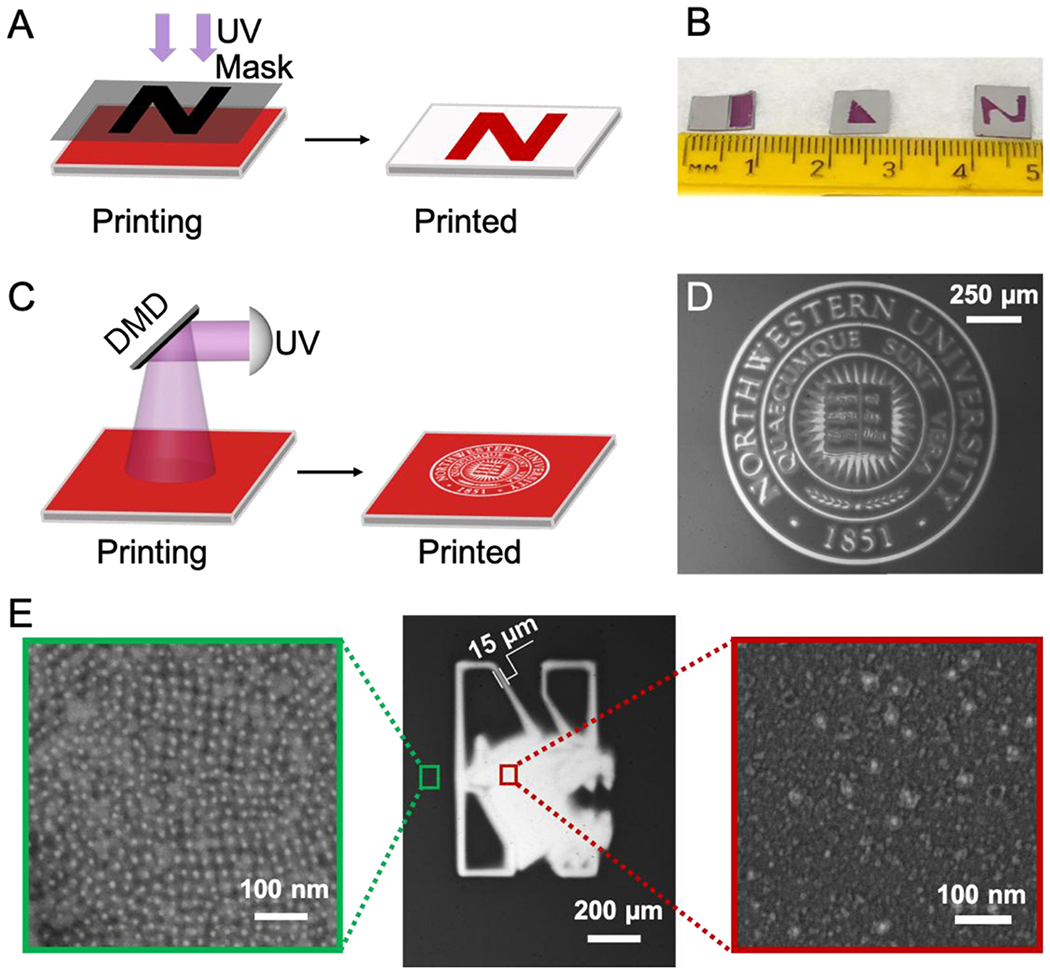
(A) A custom-made photomask can be used to pattern crystalline PAE thin films into desired shapes. (B) Photograph showing photomask-patterned samples of three different shapes (rectangle, triangle, and the letter “N”). (C) A digital micromirror device (DMD) is used with a cantilever-free scanning probe lithography instrument to photopattern crystalline PAE thin films. (D) Optical microscope image confirms successful photopatterning of a predesigned image. (E) Optical microscope image (middle), with a zoomed-in SEM image confirming that PAEs remain intact and ordered on the area that was not exposed to UV light (left) and another zoomed-in SEM image showing sparsely distributed nanoparticles on the UV-exposed region and demonstrating that the majority of light-responsive PAEs are successfully removed from the substrate (right).
Experimental Section
Light-Responsive Assembly and Disassembly of 3D Colloidal Crystals:
Light-responsive PAEs that contain complementary azoDNA-containing linkers were initially synthesized and then assembled into bcc crystals via a slow-cool method from 60 °C to 25 °C at a rate of 0.1 °C/10 min in 0.2 M NaCl buffer solution. Light-triggered disassembly and re-assembly of 3D PAE crystals were performed at a temperature within the light-dependent Tm window of PAEs. UV light (centered at 365 nm, 5 mW cm−2, 30 min) and visible light (centered at 405 nm, 10 mW cm−2, 30 min) were utilized to effect the reversible structural changes under an isothermal condition. SAXS measurements were performed within 2 min of removing the crystals from the heating environment, where the cooling process within 2 min induces negligible changes to the crystal structures.[18, 55] DNA design, sequences, and PAE assembly protocols are described in the Supporting Information.
Photopatterning of Colloidal Crystal Thin Films:
Light-responsive PAEs were layered on gold-coated Si wafer via a stepwise method and then annealed into ordered superlattice thin films. Photomask patterning was performed with a custom-made apparatus where PAE crystal thin films were covered by a photomask under UV light (centered at 365 nm, 0.5 mW cm−2, 30 min). Photopatterning with a digital micromirror device was performed on TERA fab E-series printer with UV light (centered at 365 nm, 1.1 mW cm−2, 15 min). Preparation procedures including functionalization of the substrate, layer-by-layer growth of PAEs, and photopatterning protocols are described in the Supporting Information.
Supplementary Material
Acknowledgements
The authors acknowledge Dr. Katherine Bujold for helpful discussions, and Dr. Andrey Ivankin and Dr. William Hutson for assistance with photopatterning using the TERA fab E-series instrument. This material is based upon work supported by the following awards: Air Force Office of Scientific Research FA9550-17-1-0348 (DNA-nanoparticle functionalization and assembly) and FA9550-18-1-0493 (patterning from colloidal crystals); Vannevar Bush Faculty Fellowship program sponsored by Basic Research Office of Assistant Secretary of Defense for Research and Engineering and funded by Office of Naval Research N00014-15-1-0043 (DNA synthesis); Center for Bio-Inspired Energy Science, an Energy Frontier Research Center funded by U.S. Department of Energy, Office of Science, Basic Energy Sciences DE-SC0000989 (SEM characterization); Sherman Fairchild Foundation, Inc. (patterning experimental design); and National Cancer Institute of National Institutes of Health U54CA199091 (DNA purification). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This work utilized the Northwestern University Micro/Nano Fabrication Facility (NUFAB), which is partially supported by the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-1542205), the Materials Research Science and Engineering Center (NSF DMR-1720139), the State of Illinois, and Northwestern University.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Jinghan Zhu, Department of Materials Science and Engineering, Northwestern University, 2220 Campus Drive, Evanston, IL 60208, USA; International Institute for Nanotechnology, 2190 Campus Drive, Evanston, IL 60208, USA.
Haixin Lin, Department of Chemistry, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208, USA; International Institute for Nanotechnology, 2190 Campus Drive, Evanston, IL 60208, USA.
Youngeun Kim, Department of Materials Science and Engineering, Northwestern University, 2220 Campus Drive, Evanston, IL 60208, USA; International Institute for Nanotechnology, 2190 Campus Drive, Evanston, IL 60208, USA.
Muwen Yang, Department of Chemistry, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208, USA; International Institute for Nanotechnology, 2190 Campus Drive, Evanston, IL 60208, USA.
Kacper Skakuj, Department of Chemistry, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208, USA; International Institute for Nanotechnology, 2190 Campus Drive, Evanston, IL 60208, USA.
Jingshan S. Du, Department of Materials Science and Engineering, Northwestern University, 2220 Campus Drive, Evanston, IL 60208, USA; International Institute for Nanotechnology, 2190 Campus Drive, Evanston, IL 60208, USA
Byeongdu Lee, X-ray Science Division, Argonne National Laboratory, 9700 S. Cass Ave., Argonne, IL 60439, USA.
George C. Schatz, Department of Chemistry, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208, USA; International Institute for Nanotechnology, 2190 Campus Drive, Evanston, IL 60208, USA
Richard P. Van Duyne, Department of Chemistry, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208, USA; International Institute for Nanotechnology, 2190 Campus Drive, Evanston, IL 60208, USA.
Chad A. Mirkin, Department of Materials Science and Engineering, Northwestern University, 2220 Campus Drive, Evanston, IL 60208, USA; Department of Chemistry, Northwestern University, 2145 Sheridan Road, Evanston, IL 60208, USA; International Institute for Nanotechnology, 2190 Campus Drive, Evanston, IL 60208, USA
References
- [1].Sun L, Lin H, Kohlstedt KL, Schatz GC, Mirkin CA, Proc. Natl. Acad. Sci 2018, 115, 7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].von Freymann G, Kitaev V, Lotsch BV, Ozin GA, Chem. Soc. Rev 2013, 42, 2528. [DOI] [PubMed] [Google Scholar]
- [3].Teyssier J, Saenko SV, van der Marel D, Milinkovitch MC, Nat. Commun 2015, 6, 6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Urban JJ, Talapin DV, Shevchenko EV, Kagan CR, Murray CB, Nat. Mater 2007, 6, 115. [DOI] [PubMed] [Google Scholar]
- [5].Lee J-S, Kovalenko MV, Huang J, Chung DS, Talapin DV, Nat. Nano 2011, 6, 348. [DOI] [PubMed] [Google Scholar]
- [6].Auyeung E, Morris W, Mondloch JE, Hupp JT, Farha OK, Mirkin CA, J. Am. Chem. Soc 2015, 137, 1658. [DOI] [PubMed] [Google Scholar]
- [7].Chen Q-X, Liu Y-H, Qi X-Z, Liu J-W, Jiang H-J, Wang J-L, He Z, Ren X-F, Hou Z-H, Yu S-H, J. Am. Chem. Soc 2019, 141, 10729. [DOI] [PubMed] [Google Scholar]
- [8].Begley MR, Gianola DS, Ray TR, Science 2019, 364, 1250. [DOI] [PubMed] [Google Scholar]
- [9].Chen J, Dong A, Cai J, Ye X, Kang Y, Kikkawa JM, Murray CB, Nano Lett. 2010, 10, 5103. [DOI] [PubMed] [Google Scholar]
- [10].Zeng H, Li J, Liu JP, Wang ZL, Sun S, Nature 2002, 420, 395. [DOI] [PubMed] [Google Scholar]
- [11].Black CT, Murray CB, Sandstrom RL, Sun S, Science 2000, 290, 1131. [DOI] [PubMed] [Google Scholar]
- [12].Grzelczak M, Vermant J, Furst EM, Liz-Marzán LM, ACS Nano 2010, 4, 3591. [DOI] [PubMed] [Google Scholar]
- [13].Boles MA, Engel M, Talapin DV, Chem. Rev 2016, 116, 11220. [DOI] [PubMed] [Google Scholar]
- [14].Yan C, Wang T, Chem. Soc. Rev 2017, 46, 1483. [DOI] [PubMed] [Google Scholar]
- [15].Whitesides GM, Grzybowski B, Science 2002, 295, 2418. [DOI] [PubMed] [Google Scholar]
- [16].Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ, Nature 1996, 382, 607. [DOI] [PubMed] [Google Scholar]
- [17].Nykypanchuk D, Maye MM, van der Lelie D, Gang O, Nature 2008, 451, 549. [DOI] [PubMed] [Google Scholar]
- [18].Park SY, Lytton-Jean AKR, Lee B, Weigand S, Schatz GC, Mirkin CA, Nature 2008, 451, 553. [DOI] [PubMed] [Google Scholar]
- [19].Jones MR, Seeman NC, Mirkin CA, Science 2015, 347, 1260901. [DOI] [PubMed] [Google Scholar]
- [20].Zhu JH, Kim Y, Lin HX, Wang SZ, Mirkin CA, J. Am. Chem. Soc 2018, 140, 5061. [DOI] [PubMed] [Google Scholar]
- [21].Barnaby SN, Ross MB, Thaner RV, Lee B, Schatz GC, Mirkin CA, Nano Lett. 2016, 16, 5114. [DOI] [PubMed] [Google Scholar]
- [22].Mason JA, Laramy CR, Lai C-T, O’Brien MN, Lin Q-Y, Dravid VP, Schatz GC, Mirkin CA, J. Am. Chem. Soc 2016, 138, 8722. [DOI] [PubMed] [Google Scholar]
- [23].Pal S, Zhang Y, Kumar SK, Gang O, J. Am. Chem. Soc 2015, 137, 4030. [DOI] [PubMed] [Google Scholar]
- [24].Kim Y, Macfarlane RJ, Jones MR, Mirkin CA, Science 2016, 351, 579. [DOI] [PubMed] [Google Scholar]
- [25].Kuzyk A, Yang YY, Duan X, Stoll S, Govorov AO, Sugiyama H, Endo M, Liu N, Nat. Commun 2016, 7, 10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Stoychev G, Kirillova A, Ionov L, Adv. Opt. Mater 2019, 7, 1900067. [Google Scholar]
- [27].Klajn R, Wesson PJ, Bishop KJM, Grzybowski BA, Angew. Chem., Int. Ed 2009, 48, 7035. [DOI] [PubMed] [Google Scholar]
- [28].Kim Y, Shah AA, Solomon MJ, Nat. Commun 2014, 5, 3676. [DOI] [PubMed] [Google Scholar]
- [29].Feng L, Romulus J, Li MF, Sha RJ, Royer J, Wu KT, Xu Q, Seeman NC, Weck M, Chaikin P, Nat. Mater 2013, 12, 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Klajn R, Bishop KJM, Grzybowski BA, Proc. Natl. Acad. Sci 2007, 104, 10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Vialetto J, Anyfantakis M, Rudiuk S, Morel M, Baigl D, Angew. Chem., Int. Ed 2019, 58, 9145. [DOI] [PubMed] [Google Scholar]
- [32].De Fazio AF, El-Sagheer AH, Kahn JS, Nandhakumar I, Burton MR, Brown T, Muskens OL, Gang O, Kanaras AG, ACS Nano 2019, 13, 5771. [DOI] [PubMed] [Google Scholar]
- [33].Wang Q, Li D, Xiao J, Guo F, Qi L, Nano Res. 2019, 12, 1563. [Google Scholar]
- [34].Chen Y, Wang Z, He Y, Yoon YJ, Jung J, Zhang G, Lin Z, Proc. Natl. Acad. Sci 2018, 115, E1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Stricker L, Fritz E-C, Peterlechner M, Doltsinis NL, Ravoo BJ, J. Am. Chem. Soc 2016, 138, 4547. [DOI] [PubMed] [Google Scholar]
- [36].Kundu PK, Das S, Ahrens J, Klajn R, Nanoscale 2016, 8, 19280. [DOI] [PubMed] [Google Scholar]
- [37].He H, Feng M, Chen Q, Zhang X, Zhan H, Angew. Chem., Int. Ed 2016, 55, 936. [DOI] [PubMed] [Google Scholar]
- [38].Zhao H, Sen S, Udayabhaskararao T, Sawczyk M, Kučanda K, Manna D, Kundu PK, Lee J-W, Král P, Klajn R, Nat. Nano 2015, 11, 82. [DOI] [PubMed] [Google Scholar]
- [39].Zhang L, Dai L, Rong Y, Liu Z, Tong D, Huang Y, Chen T, Langmuir 2015, 31, 1164. [DOI] [PubMed] [Google Scholar]
- [40].Manna D, Udayabhaskararao T, Zhao H, Klajn R, Angew. Chem., Int. Ed 2015, 54, 12394. [DOI] [PubMed] [Google Scholar]
- [41].Kundu PK, Samanta D, Leizrowice R, Margulis B, Zhao H, Börner M, Udayabhaskararao T, Manna D, Klajn R, Nat. Chem 2015, 7, 646. [DOI] [PubMed] [Google Scholar]
- [42].Shiraishi Y, Shirakawa E, Tanaka K, Sakamoto H, Ichikawa S, Hirai T, ACS Appl. Mater. Interfaces 2014, 6, 7554. [DOI] [PubMed] [Google Scholar]
- [43].Köhntopp A, Dabrowski A, Malicki M, Temps F, Chem. Commun 2014, 50, 10105. [DOI] [PubMed] [Google Scholar]
- [44].Han H, Lee JY, Lu X, Chem. Commun 2013, 49, 6122. [DOI] [PubMed] [Google Scholar]
- [45].Das S, Ranjan P, Maiti PS, Singh G, Leitus G, Klajn R, Adv. Mater 2013, 25, 422. [DOI] [PubMed] [Google Scholar]
- [46].Chovnik O, Balgley R, Goldman JR, Klajn R, J. Am. Chem. Soc 2012, 134, 19564. [DOI] [PubMed] [Google Scholar]
- [47].Liu D, Chen W, Sun K, Deng K, Zhang W, Wang Z, Jiang X, Angew. Chem., Int. Ed 2011, 50, 4103. [DOI] [PubMed] [Google Scholar]
- [48].Fava D, Winnik MA, Kumacheva E, Chem. Commun 2009, 2571. [DOI] [PubMed] [Google Scholar]
- [49].Kamiya Y, Asanuma H, Acc. Chem. Res 2014, 47, 1663. [DOI] [PubMed] [Google Scholar]
- [50].Asanuma H, Liang X, Nishioka H, Matsunaga D, Liu M, Komiyama M, Nat. Protoc 2007, 2, 203. [DOI] [PubMed] [Google Scholar]
- [51].Yan YQ, Chen JIL, Ginger DS, Nano Lett. 2012, 12, 2530. [DOI] [PubMed] [Google Scholar]
- [52].Paik T, Yun H, Fleury B, Hong SH, Jo PS, Wu YT, Oh SJ, Cargnello M, Yang H, Murray CB, Kagan CR, Nano Lett. 2017, 17, 1387. [DOI] [PubMed] [Google Scholar]
- [53].Tan ATL, Beroz J, Kolle M, Hart AJ, Adv. Mater 2018, 30, 1803620. [DOI] [PubMed] [Google Scholar]
- [54].Wang MX, Seo SE, Gabrys PA, Fleischman D, Lee B, Kim Y, Atwater HA, Macfarlane RJ, Mirkin CA, ACS Nano 2017, 11, 180. [DOI] [PubMed] [Google Scholar]
- [55].Macfarlane RJ, Lee B, Jones MR, Harris N, Schatz GC, Mirkin CA, Science 2011, 334, 204. [DOI] [PubMed] [Google Scholar]
- [56].Auyeung E, Li TING, Senesi AJ, Schmucker AL, Pals BC, de la Cruz MO, Mirkin CA, Nature 2014, 505, 73. [DOI] [PubMed] [Google Scholar]
- [57].Lee J-S, Stoeva SI, Mirkin CA, J. Am. Chem. Soc 2006, 128, 8899. [DOI] [PubMed] [Google Scholar]
- [58].Senesi AJ, Eichelsdoerfer DJ, Macfarlane RJ, Jones MR, Auyeung E, Lee B, Mirkin CA, Angew. Chem., Int. Ed 2013, 52, 6624. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


