Abstract
Chemoresistance is a leading cause of tumor relapse and treatment failure in colorectal cancer (CRC) patients and is correlated with epithelial-mesenchymal transition (EMT). This study was aimed to explore the mechanism of EMT in chemoresistant CRC. Bioinformatic method was used to screen differentially expressed genes between 5-FU sensitive and resistant CRC cells. Immunohistochemistry staining was utilized to analyze the expression of FLNA in CRC tissues. The roles of FLNA in chemoresistance were validated via loss-of-function and gain-of-function experiments in vitro and in an orthotopic CRC animal model. The regulation of c-Met signaling by FLNA was explored via Co-Immunoprecipitation and luciferase reporter assays. Our results suggested FLNA directly regulated the metastasis and EMT of chemoresistant CRC cells. Moreover, c-Met-AKT mediated ser2152 phosphorylation of FLNA was demonstrated to be correlated with EMT. In turn, FLNA enhanced c-Met promoter activity by its interaction with smad2. Clinically, the expression of FLNA was significantly associated with c-Met protein levels in CRC tissues. These data established that FLNA could be a novel and reliable CRC marker and a potential therapeutic target against CRC.
Keywords: Colorectal cancer, chemoresistance, FLNA, smad2, EMT
Introduction
Colorectal cancer (CRC) is the third most common cancer and the fourth leading cause of cancer-related mortality worldwide [1]. Although chemotherapy is well demonstrated to reduce tumor burden and prolong survival, it remains a palliative treatment since most CRC patients eventually exhibit drug-resistance [2]. Currently, 5-fluorouracil (5-FU) is widely used as one of the classic and basic drugs in adjuvant chemotherapy and palliative chemotherapy of CRC patients. 5-FU resistance is often coupled with local recurrence and distant metastasis, which always contribute to failure of anticancer therapy [3]. Thus, it is necessary to reveal the potential mechanisms of 5-Fu resistance in CRC patients. Epithelial-mesenchymal transition (EMT), characterized by cytokeratin cytoskeletal transformation and decreased expression of cell adhesion molecules (such as E-cadherin), exhibited important roles in embryonic development, chronic inflammation, tissue remodeling, cancer metastasis as well as fibrosis [4]. Tumor cells could obtain aggressive phenotypes including migration, invasion, anti-apoptosis, and degradation of extracellular matrix through EMT [4]. Recent evidences indicated that EMT might be responsible for the aggressive phenotype in chemotherapy-resistant cancer cells. EMT can be observed in adriamycin-resistant gastric cancer, cisplatin-resistant neuroblastoma and folate-resistant lung cancer [5-7]. Several studies revealed that EMT is correlated with 5-FU resistance in CRC [8,9]. However, the molecular mechanism that regulates EMT in 5-FU resistant colorectal cancer is still elusive and needs further investigation.
Filamin A (FLNA), the most abundant and widely distributed member of the filamin family (Filamin A, Filamin B and Filamin C), was first identified as a non-muscle actin filament cross-linking protein in 1975 [10]. FLNA crosslinks actin filaments into an orthogonal network, which contributes to stabilize the cytoskeleton network and supports cell integrity [11,12]. Interestingly, several studies showed that FLNA associates with multiple functional non-cytoskeletal proteins and is involved in several unrelated pathways that regulate cell migration and adhesion [13]. FLNA was generally revealed as a cancer promoting protein in multiple human malignancies including metastatic melanoma, lung cancer and hepatocellular carcinoma [14-16]. However, Xu et al. reported that FLNA expression was decreased in breast cancer tissues and was negatively correlated with lymph node metastasis. Their biological studies indicated that knockdown of FLNA could promoted cell migration and invasion [17]. These studies revealed controversial roles of FLNA in human malignancies. Accumulating evidences showed that FLNA ser2152 phosphorylation was critical for its biological functions [18]. Woo et al. first reported that N-terminal kinase domain of RSK could phosphorylate FLNA on Ser2152 in response to mitogens [19]. Later studies revealed that epidermal growth factor (EGF) stimulation promoted α5β1 integrin activation of human cancer cell lines through p90RSK-dependent FLNA phosphorylation [20]. The carcinogenic effect of FLNA ser2152 phosphorylation was also well demonstrated in breast cancer. Ravid et al. reported that caveolin-1 up-regulated FLNA ser2152 phosphorylation via Akt pathway, thereby mediating breast cancer cell migration [21]. Cyclin D1 interacted with FLNA and induced its ser2152 phosphorylation in breast cancer cell adhesion and motility [22]. Therefore, it seemed that the role of FLNA expression level and phosphorylation status in tumor progression should be discussed separately according to the types of tumors.
The clinical expression and biological function of FLNA in colorectal cancer seemed to be controversial. Tian et al. reported that FLNA was lower expressed in 46 colorectal adenocarcinoma tissues compared with normal mucosa [23]. However, in another study, FLNA overexpression and ser2152 phosphorylation were demonstrated to be correlated with snail-induced EMT and cell adhesion [24]. In current study, we further analyzed the expression and clinical significance of FLNA in CRC clinical samples, and initially elucidated the potential role of FLNA in 5-FU resistant CRC cells. Developing drugs that target FLNA or inhibit FLNA ser2152 phosphorylation might contribute to overcome 5-FU resistance of CRC.
Materials and methods
Biological information analysis
Colorectal cancer cell lines and 5-FU treatment information were obtained from the CCLE database and the CTRP database. The differentially expressed genes were compared by ‘limma’ package of R. Functional association network of the differentially expressed genes were constructed by Cytoscape software. RNA-seq data of CRC tissues in TCGA database and the matched clinical data were downloaded. The functional enrichment analysis of FLNA was performed using the ‘clusterProfiler’ package of R, and the bubble map was drawn using the “ggplot2” package of R.
Clinical specimens
The clinical research protocol was approved by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University (Zhengzhou, China). All patients provided written informed consent. A total of 152 patients were enrolled in this study. Paired tumor and adjacent non-tumor CRC tissues were collected from patients who underwent surgical resection in The First Affiliated Hospital of Zhengzhou University between January 2011 and December 2018. The specimens were immediately frozen in liquid nitrogen or fixed in 10% formalin and paraffin embedded. No patient had received any anti-tumor treatment preoperatively. Immunohistochemistry staining and scoring of each slide were performed as previously described [25]. Slides were examined by two researchers (Yang and Zhao) independently. FLNA expression status was determined according to the staining intensity and percentage of positive cells. Percentage of positive cells was determined as 0 (<10%), 1 (10%-25%), 2 (26-50%) and 3 (>50%). Staining intensity was determined as no staining (0), weak staining (1), moderate staining (2) and strong staining (3). The final score of each case was determined by multiplying the score of intensity and score of positive cells. Therefore, cases were considered negative if the final score was 0-3 and positive if the final score was 4-9.
Cell culture conditions and reagents
Human CRC cell lines, HCT116 and HT29, were purchased from the American Type Culture Collection (Manassas, VA, USA). 5-FU resistant HCT116 and HT29 (HCT116R and HT29R) cell lines were established by continuous culture in media containing increasingly higher concentrations of 5-FU (Selleckchem, Houston, TX, USA). HCT116 and HT29 cell lines were cultured in RPMI 1640 (Corning Cellgro®, Manassas, VA, USA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA). HCT116R and HT29R cell lines were cultured in the same medium containing 1 μg/mL of 5-FU. All cells were maintained in a humidified incubator at 37°C with 5% CO2. Crizotinib (c-Met inhibitor), LY294002 (PI3K/AKT pathway inhibitor) and LY2109761 (smad2 signaling inhibitor) were purchased from Selleckchem (Houston, TX, USA) and used according to manufacturer’s instructions.
Short hairpin RNA (shRNA) and constructs
The short hairpin RNA (shRNA) targeting FLNA were established using the following targets: sequence 1, 5’-GGGCTGACAACAGTGTGGTGC-3’ (targeting nt 3323-3343); sequence 2, 5’-GGTGCTGCCTACTCATGATGC-3’ (targeting nt 4663-4683). Stably transfected cells were selected with 5 μg/ml puromycin and tested regularly by immunoblotting to ensure downregulation of FLNA. pGL3-c-Met and pcDNA3-smad2 plasmids were obtained from Sangon Biotech (Shanghai, China) and amplified by PureLink HiPure Plasmid Filter Midiprep Kit (Invitrogen). pcDNA3-myc expression vectors coding for wild-type and S2152A FLNA were obtained from Addgene (Cambridge MA, USA). Three µg of empty vector pcDNA3 (control) or the vector expressing pcDNA3-myc-FLNA (WT) or mutant pcDNA3-myc-FLNA (S2152A) were transfected into HCT116 or HT29 cells using Lipofectamine 2000 (Invitrogen). Stable clones were obtained by G418 (500 μg/ml) selection. All plasmids transfection was performed according to the manufacturer’s instruction.
Cell proliferation and colony formation assay
Cells were seeded in a 96-well plate overnight at a density of 5 × 103 cells per well and treated with indicated doses of 5-FU (2, 4, 6, 8, 16, 32, 64, 128, 256, 512 μM) for 48 h. Then, cells were incubated with 10 μL CCK-8 for 60 min at 37°C, 5% CO2. The absorption value was detected at 450 nm with a spectrophotometer. Each assay was conducted in triplicate and repeated three times. For colony formation assay, 1 × 103 treated cells were seeded into 6-well plates. The drug-containing medium was changed every 3 days. At 15th day, these plates were washed with phosphate buffered saline (PBS) twice, fixed by methanol for 10 min and stained with 0.1% crystal violet solution for further analysis.
Flow cytometry analysis
Apoptosis was examined by flow cytometric analysis. Cells were collected, washed with PBS and incubated with Annexin V and propidium iodide (BD Biosciences, San Jose, CA, USA) for 15 min. Then cell apoptosis was analyzed by flow cytometry (FACS Calibur, Becton Dickinson, Sparks, MD) according to the manufacturer’s instruction. All the assays were performed in triplicate.
Cell migration, invasion and adhesion assay
Cell migration/invasion assays were performed to analyze the migration and invasiveness of CRC cells according to a previously described protocol [25]. The adhesion of CRC cells to ECM components was evaluated using a CytoSelect 48-Well Cell Adhesion Assay Kit (Cell Biolabs, San Diego, CA, USA) following the manufacturer’s instructions. Firstly, cells were seeded onto the coated substrate and the adherent cells were captured. After unbound cells were washed away, the adherent cells were fixed and stained. Finally, the stain is extracted and quantified colorimetrically.
Quantitative reverse transcription-PCR
Total RNA was extracted from CRC cells and subjected to qRT-PCR as previously described [25]. Primers for qRT-PCR were listed in Supplementary Table 1.
Immunoblotting and Co-Immunoprecipitation assay
Immunoblotting and Co-Immunoprecipitation (Co-IP) assays were performed as previously described [25]. The primary antibodies were: anti-GAPDH and anti-HGF from Abcam (Cambridge, MA, USA); anti-FLNA from Santa Cruz (Santa Cruz Biotechnology, Santa Cruz, CA); anti-phospho-FLNA, anti-phospho-AKT, anti-smad2, anti-phospho-smad2, anti-c-Met, anti-vimentin, anti-N-cadherin and anti-E-cadherin from Cell Signaling Technology (Beverly, MA, USA).
Immunofluorescence assay
Cells were cultured on confocal dishes, fixed in methyl alcohol, permeabilized with 0.5% Triton X-100, blocked with 5% BSA (in TBST), and incubated with anti-smad2 (1:100) at 4°C overnight. Cells were then incubated with secondary antibody (Cell Signaling Technology) at room temperature for 30 min. Immunofluorescence (IF) images were obtained with confocal microscopy (Olympus, Japan).
Nude mouse tumor transplantation model
All animal procedures were performed with the approval of the Local Medical Experimental Animal Care Commission of the First Affiliated Hospital of Zhengzhou University. Mice were randomly grouped (5 mice per group). Four-week-old male nude BALB/C mice were subcutaneously injected with HCT116/shFLNA or control cells (1 × 106). When tumor volumes reached approximately 0.2 cm3, mice were subcutaneously injected with control vehicle (DMSO) or 5-FU (20 mg/kg every week). Body weights, tumor length (L) and width (W) were measured every three days. Tumor volumes were calculated by the following formula: V=0.5 × LW2. Thirty days later, mice were sacrificed. Subcutaneous tumor grafts were excised and weighed. For lung metastasis assay, 2 × 106 cells re-suspended in 100 μl PBS were injected into caudal vena. On the 60th day, mice were killed by cervical decapitation. The pulmonary metastatic masses were removed and fixed with 10% buffered formalin for hematoxylin-eosin (HE) staining.
Luciferase assay
The c-Met promoter fragments were amplified from human genomic DNA, and were inserted into pGL3-basic vector. The smad2/FLNA (WT)/FLNA (S2152A) overexpressing or control cells were co-transfected with pGL3-c-Met-promotor constructs. Luciferase assays were performed 24 h after transfection using Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. The ratios of luminescence intensities were measured relative to that of the pGL3 empty vector.
Statistical analysis
The statistical analysis was performed using SPSS version 25.0 for Windows (SPSS, Chicago, USA). All data were presented as the mean ± SD and repeated from at least three independent experiments. Statistical difference between two groups was examined by Student’s t-test. Multiple comparisons were conducted by one-way analysis of variance. P value less than 0.05 was considered statistically significant.
Results
Identification of FLNA as a potential target of 5-FU resistance in CRC
To explore potential target of 5-FU resistance in CRC, we first extracted the 5-FU treatment information of CRC cell lines by using the CCLE database and the CTRP database. As shown in Supplementary Table 2, CRC cell lines were divided into 3 groups (Low IC50, Moderate IC50, High IC50) according to different sensitivities to 5-FU. The differentially expressed genes between low IC50 group and high IC50 group were compared by “limma” package in R, and 254 genes (fold change >2, P<0.05) were obtained (176 of which were up-regulated in High IC50 group, 77 were down-regulated, Figure 1A and Supplementary Table 3). We next used the above genes to build a functional association network, and the most relevant 6 genes (ACTN1, COL6A1, EGFR, FLNA, FN1 and ITGA1) were used for further analysis (Figure 1B). We further analyzed the prognostic significance of these genes in patients receiving 5-FU based chemotherapy. As the result, only FLNA was up-regulated in High IC50 group and correlated with poorer survival status of CRC patients (Figure 1C). Thus, we selected FLNA for further characterization.
Figure 1.
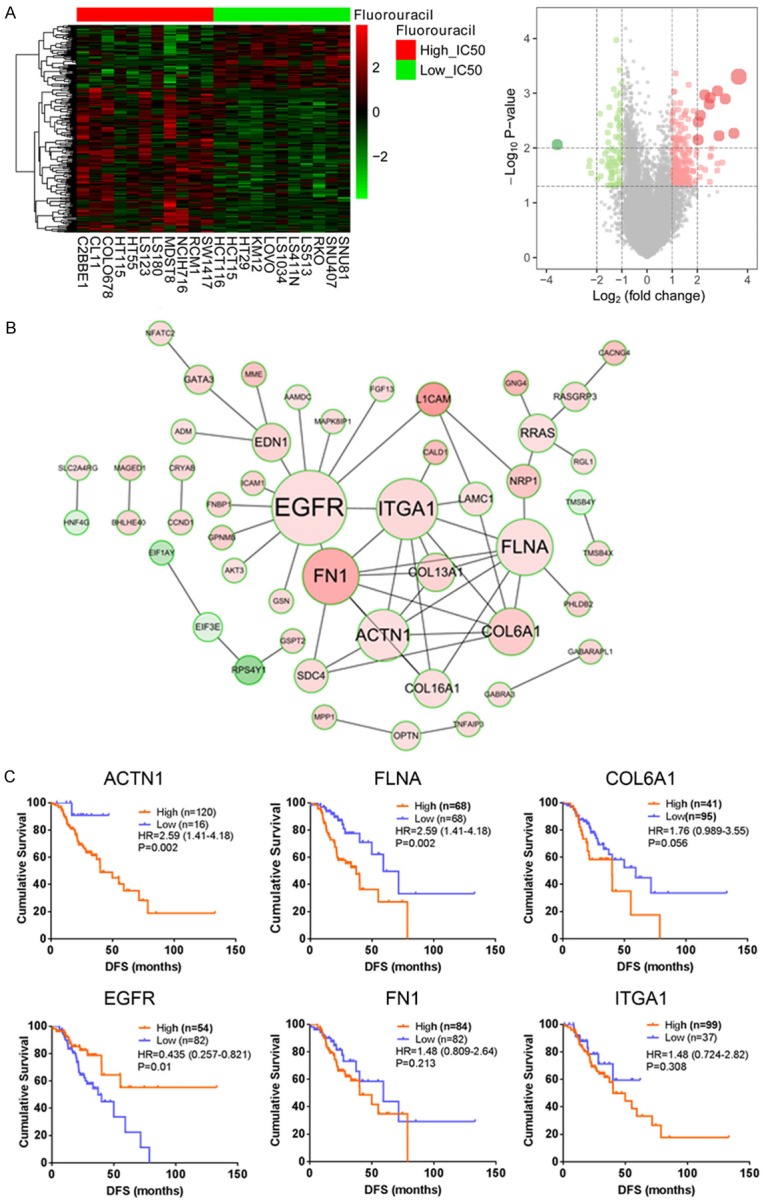
Comprehensive analysis of FLNA mediated 5-FU resistance in CRC. A. The heatmap and volcano plot of differentially expressed genes in 5-FU sensitive and resistant CRC cells. B. Functional association network of the differentially expressed genes. C. Prognostic significance of ACTN1, COL6A1, EGFR, FLNA, FN1 and ITGA1 in patients receiving 5-FU based chemotherapy.
High FLNA expression correlates with poor survival of CRC patients
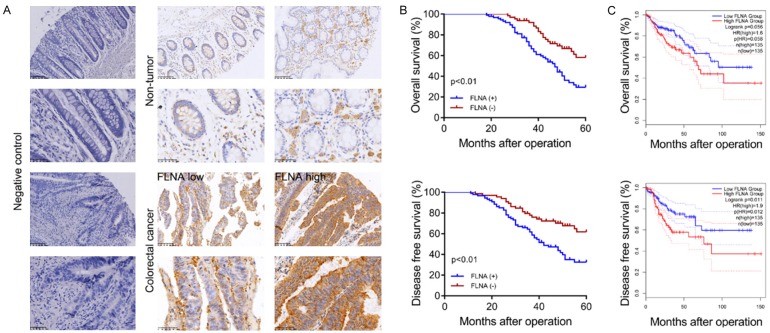
To clarify whether FLNA expression is aberrantly elevated in CRC tissues, we first detected the mRNA levels of FLNA in 40 paired CRC tissues. As shown in Supplementary Figure 1A, FLNA mRNA levels were highly expressed in CRC tissues compared with normal tissues. The clinical significance of FLNA was further analyzed by IHC assay of FLNA protein expression in 152 paired CRC tissues (Figure 2A). High levels of FLNA expression were found in 86 of 152 (56.6%) CRC samples. To investigate the clinical significance of FLNA in CRC, we analyzed the association between FLNA expression and the clinicopathologic characteristics of CRC patients. As shown in Supplementary Table 4, FLNA expression was significantly associated with tumor size (P=0.048), extent of invasion (P=0.022) and lymphatic metastasis (P=0.009), while there was no significant association between FLNA expression and age, gender, location, tumor histology and CEA level. Additionally, FLNA high expression was associated with short Overall Survival (OS, P<0.01) and Disease Free Survival (DFS, P<0.01, Figure 2B) time. We next assessed the prognostic significance of FLNA in a TCGA pan-cancer data set obtained from Gene Expression Profiling Interactive Analysis (GEPIA) online database (http://gepia.cancer-pku.cn). The result indicated that higher FLNA expression was correlated with shorter Disease Free Survival (HR=1.9, P=0.012) but not Overall Survival (HR=1.6, P=0.058) of CRC patients (Figure 2C).
Figure 2.
FLNA is highly expressed in CRC and associated with poor prognosis. A. The typical IHC images of FLNA expression in CRC tissues. B. Kaplan-Meier analysis of OS and DFS of 152 CRC patients. C. OS and DFS curves of CRC patients from TCGA database.
FLNA regulates the sensitivity of CRC cells to 5-FU treatment
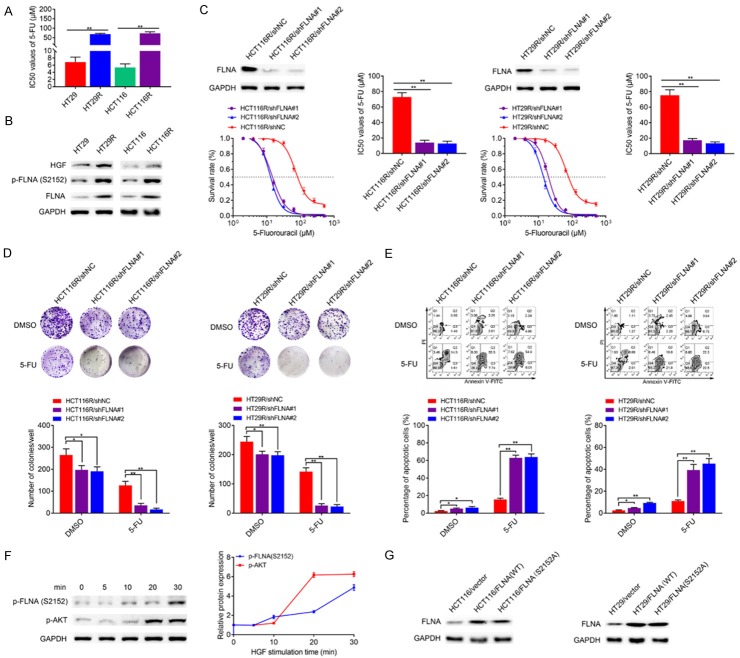
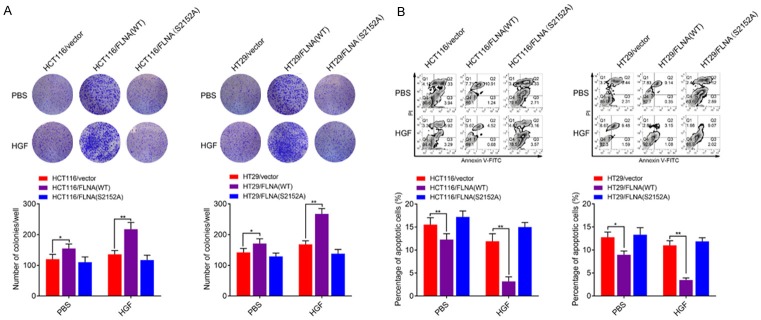
Two 5-FU resistant cell models (HT29R and HCT116R) were established from the human CRC cell line HT29 and HCT116 through serial 5-FU induction. We evaluated the IC50 values of HT29, HT29R, HCT116 and HCT116R cells under 5-FU treatment. HT29R and HCT116R cells exhibited significantly resistance to 5-FU treatment in vitro compared with parental HT29 and HCT116 cells (Figure 3A). As to immunoblotting assay, protein levels of FLNA and its phosphorylated form p-FLNA (Ser2152) were up-regulated in HT29R/HCT116R cells compared with the parental cells (Figure 3B). We investigated the role of FLNA in chemoresistance by knocking down its expression in the two acquired 5-FU resistant cells. As indicated in Figure 3C, both shRNAs targeting FLNA suppressed FLNA expression in HCT116R or HT29R cells compared with the mock shRNA. Meanwhile, FLNA knockdown in HCT116R or HT29R cells resulted in a fifty percent decrease (P<0.01) in its sensitivity to 5-FU. Engineered cell lines were then treated with 5-FU for colony formation. shRNA HCT116R and shRNA HT29R cells had relatively lower colony survival rates compared to mock cells under 5-FU treatment (Figure 3D). To examine whether FLNA mediated 5-FU resistance through the apoptosis, we performed flow cytometry analysis by staining cells with annexin V-FITC and PI. As the result, knockdown of FLNA only slightly promoted the apoptotic rate of HCT116R or HT29R cells without 5-FU treatment, while knockdown of FLNA in HCT116R or HT29R cells significantly promoted apoptosis with 5-FU treatment (Figure 3E). Since AKT could phosphorylate FLNA at Ser2152. We then stimulated the parental HCT116 cells with different concentrations of HGF to activate c-Met-AKT signaling pathway. Immunoblotting analysis of HCT116 cell lysates showed that Ser2152 phosphorylation of FLNA began 5-10 min following HGF stimulation and was increased by 3-fold within 30 min (Figure 3F). To address the role of FLNA overexpression and phosphorylation in the regulation of 5-FU resistance, HCT116 or HT29 cell lines that expressed either FLNA (WT) or the non-phosphorylatable mutant FLNA (S2152A) were constructed (Figure 3G). In the presence of HGF, stable cells expressing FLNA (WT) exhibited higher colony survival rates and lower apoptosis rates, while cells expressing FLNA (S2152A) showed no difference compared with the control cells. Following the addition of HGF, cells expressing FLNA (WT) exhibited significantly higher colony survival rates (Figure 4A) and lower apoptosis rates (Figure 4B) than other groups.
Figure 3.
FLNA is critical for 5-FU resistance of CRC cells. A. IC50 dose of acquired 5-FU resistant cells and the parental cells. B. Immunoblotting analysis shows 5-FU resistant cells exhibit higher FLNA, p-FLNA (Ser2152) and HGF expression than parental cells. C. Protein of HCT116R/shNC, HCT116R/shFLNA#1, HCT116R/shFLNA#2, HT29R/shNC, HT29R/shFLNA#1 and HT29R/shFLNA#2 were extracted and the knockdown efficacy of FLNA were verified by immunoblotting. Statistical chart showed IC50 dose of 5-FU calculated from measurement of cell viability in different concentrations of 5-FU. D. Representative images of colony formation of transfected CRC cells after 5-FU treatment. E. Representative images of apoptosis of transfected CRC cells after 5-FU treatment. F. HCT116 cells were stimulated with HGF (50 ng/ml) at the indicated times. Cell lysates were immunoblotted using anti-p-FLNA and anti-p-AKT. Bands for p-FLNA and p-AKT were quantified and normalized to GAPDH. G. Lysates from control cells (HCT116/vector, HT29/vector), clonal cell lines expressing either wild-type FLNA (HCT116/FLNA (WT), HT29/FLNA (WT)) or the mutant FLNA (HCT116/FLNA (S2152A), HT29/FLNA (S2152A)) were immunoblotted to verify FLNA expression. All data are presented as the mean ± SD of three experiments. *P<0.05, **P<0.01.
Figure 4.
Clonal cell lines expressing either wild-type FLNA or the mutant FLNA were pre-treated with HGF (50 ng/ml) or PBS for 30 min. The colony formation (A) and apoptosis (B) were compared after cultured with 5-FU (10 μM) for 48 h. All data are presented as the mean ± SD of three experiments. *P<0.05, **P<0.01.
FLNA maintains 5-FU resistance of CRC cells through inducing EMT
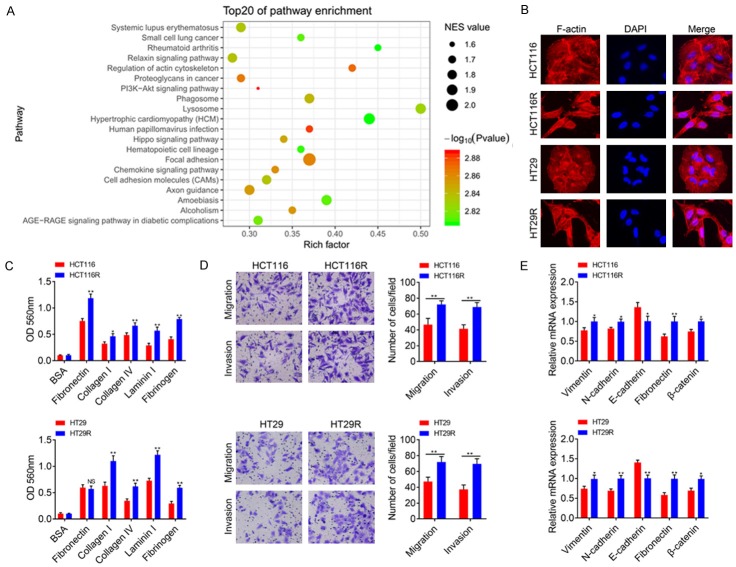
To clarify the mechanism of FLNA in 5-FU resistant CRC cells, we analyzed FLNA related signaling pathways by GSEA analysis. Among the top 20 enriched pathways, FLNA was significantly correlated with regulation of actin cytoskeleton, cell adhesion (focal adhesion and cell adhesion molecules) and PI3K-AKT signaling pathway (Figure 5A). Since EMT of cancer cells is tightly correlated with drug resistance and is always characterized as enhanced migration and invasion ability, anti-apoptosis, extracellular matrix remodeling as well as cytoskeletal reconstruction, we suspected that FLNA might maintain 5-FU resistance of CRC cells through inducing EMT. We first compared cell motility between the chemoresistant and parental CRC cells. As shown in Figure 5B, the parental HCT116 and HT29 cells exhibited a flat epithelial phenotype with extensive cell-cell contacts, whereas HCT116R and HT29R cells exhibited spindle-shaped morphology with thin pseudopods and reduced cell-cell contacts. Since distinct morphology is associated with cell invasiveness and typical EMT characteristics, we compared the adhesion, migration and invasion ability of 5-FU resistant cells and parental cells. As expected, HCT116R and HT29R cells exhibited higher adhesion rates to fibronectin, collagen I, collagen IV, Laminin I and fibrinogen compared with their parental cells, respectively (Figure 5C). Additionally, transwell assays showed that migration and invasion capacities of resistant cells were higher than parental cells (Figure 5D). Epithelial marker protein E-cadherin levels were reduced, while the levels of mesenchymal marker proteins vimentin, N-cadherin, fibronectin and β-catenin were increased in acquired 5-FU resistant cells (Figure 5E). These results suggested that migration and invasion ability of CRC cells were elevated upon acquired 5-FU resistance. To confirm the necessity of FLNA in EMT process of 5-FU resistance, cell adhesion, migration and invasion assays were performed to detect alterations after shFLNA or shNC transfection. The cells exhibited lower adhesion rates to fibronectin, collagen I, collagen IV, Laminin I and fibrinogen when FLNA was down-regulated (Figure 6A). Transwell assays showed that invasion and migration capacities of the resistant-shFLNA cells were lower than that of resistant-shNC cells (Figure 6B). We also found that the levels of E-cadherin were increased, whereas the levels of vimentin, N-cadherin, fibronectin and β-catenin were reduced in cells transfected with shFLNA (Figure 6C). Using the nude mice model, we noticed that knockdown of FLNA significantly inhibited lung metastases of HCT116R cells (Figure 6D).
Figure 5.
FLNA is associated with metastasis and EMT of chemoresistant CRC. A. Bubble chart of top 20 pathways enriched from TCGA data according to FLNA expression. B. Phalloidin staining (red) in 5-FU resistant CRC cells and parental cells. C. HCT116, HCT116R, HT29 and HT29R cells that adhered to plates coated with different ECM components after 30 min of incubation were quantified at OD 560 nm after 30 min incubation. D. Representative figures and data from the transwell assay of HCT116, HCT116R, HT29 and HT29R cells. E. qRT-PCR analysis for the expression of EMT-associated markers in HCT116, HCT116R, HT29 and HT29R cells. All data are presented as the mean ± SD of three experiments. *P<0.05, **P<0.01.
Figure 6.
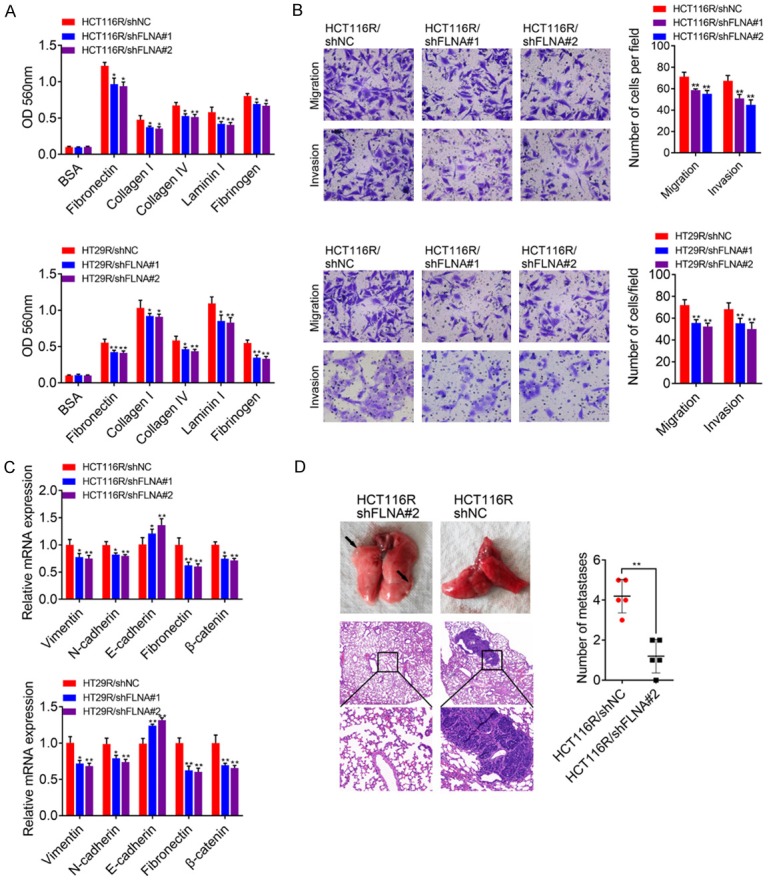
A. Adhesion assay of CRC cells in which FLNA was knocked down. B. Transwell analysis of HCT116R and HT29R cells transfected with shNC, shFLNA#1 or shFLNA#2. Five randomly selected fields were assessed under a microscope. C. qRT-PCR analysis of EMT-associated markers in HCT116R and HT29R cells transfected with shNC, shFLNA#1 or shFLNA#2. D. 2 × 106 HCT116R/shFLNA#2 or HCT116R/shNC cells were injected into nude mice through the tail vein for 60 days to evaluate the lung metastasis ability. Representative figures for lung metastatic tumor were presented by HE staining.
c-Met-AKT mediated ser2152 phosphorylation of FLNA is essential for its function
We then conducted FLNA gain-of-function assay in parental HCT116 and HT29 cells. In cell adhesion assays, overexpression of FLNA (WT) but not FLNA (S2152A) promoted the adhesion ability compared with control cells (Figure 7A). FLNA (WT) expressing cells but not FLNA (S2152A) expressing cells exhibited enhanced cell migration and invasion ability compared with control cells (Figure 7B). In addition, the level of E-cadherin was decreased while the levels of vimentin, N-cadherin, fibronectin and β-catenin were increased in FLNA (WT) group but not FLNA (S2152A) group compared with control group (Figure 7C). Since mutation of FLNA ser2152 attenuated its oncogenic roles, we attempted to determine whether inhibition of c-Met-AKT pathway, a well-established regulator of FLNA Ser2152 phosphorylation, influences the downstream biological processes. In HGF stimulated HCT116/FLNA (WT) or HT29/FLNA (WT) cells, Crizotinib (c-Met inhibitor) or LY294002 (PI3K-AKT inhibitor) treatment significantly decreased FLNA ser2152 phosphorylation (Figure 7D). Crizotinib or LY294002 treatment also attenuated the adhesion/migration/invasion ability of HCT116/FLNA (WT) or HT29/FLNA (WT) cells (Figure 7E, 7F). In addition, Crizotinib or LY294002 treatment also decreased the expression of vimentin, N-cadherin, fibronectin, β-catenin and increased the expression of E-cadherin of HCT116/FLNA (WT) or HT29/FLNA (WT) cells (Figure 7G).
Figure 7.
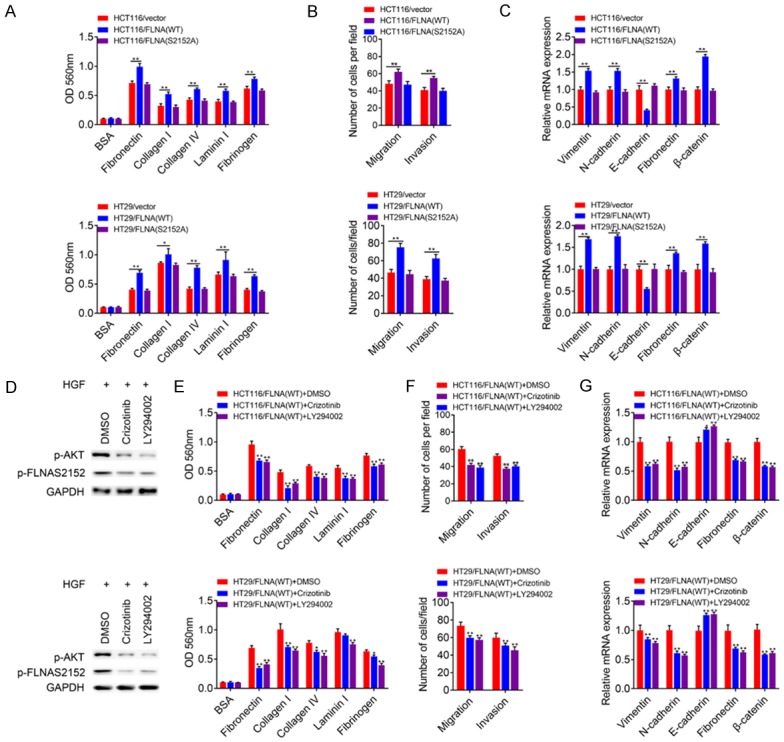
c-Met-AKT mediated ser2152 phosphorylation of FLNA was essential for EMT. (A) HCT116 or HT29 cells were transfected with empty vector, wild-type FLNA, or the mutant S2152A. Then, clonal cell lines expressing either WT-FLNA or the mutant S2152A were compared in ECM-based adhesion assay (A) and transwell assay (B). (C) RNA were extracted from clonal cells expressing empty vector, or clonal cell lines expressing either WT-FLNA or the S2152A mutant to determine the mRNA levels of EMT-associated markers. (D-G) HCT116 or HT29 cells expressing WT-FLNA were pre-treated with HGF (50 ng/ml) for 30 min. Then, cells were cultured with DMSO, Crizotinib (2 μM) or LY294002 (5 μM) for 48 h to evaluate the adhesion, migration, invasion ability and the mRNA levels of EMT-associated markers. All data are presented as the mean ± SD of three experiments. *P<0.05, **P<0.01.
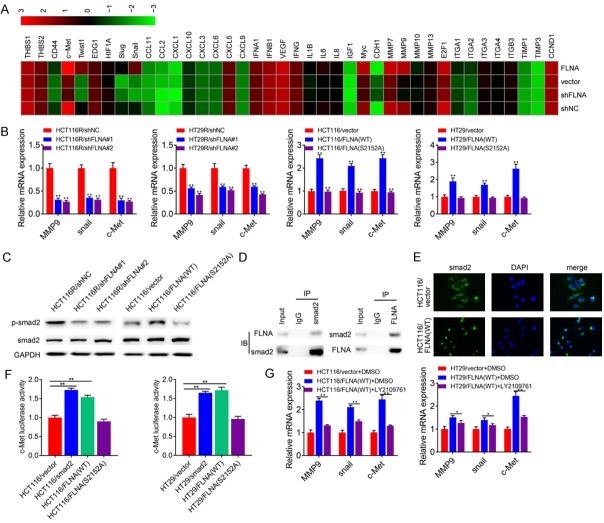
FLNA interacts with smad2 to promote EMT
To further clarify the molecular mechanisms of FLNA induced EMT, we performed a signal transduction RT2 Profiler PCR Array to profile differentially expressed genes between each group (HCT116R/shNC, HCT116R/shFLNA#2; HCT116/vector, HCT116/FLNA (WT)). The heatmap produced by this array shows gene expression levels relative to GAPDH on a log2 scale (Figure 8A). As the result, seven genes were significantly (fold change >2) down-regulated and one gene was up-regulated in HCT116R/shFLNA#2 cells compared with HCT116R/shNC cells. Eight genes were up-regulated and one gene was down-regulated in HCT116/FLNA (WT) cells compared with HCT116/vector cells. Among these genes, MMP9, snail and c-Met were well-reported targets of smad2. Thus, we suspected that FLNA might be involved in smad2 signaling. We first confirmed the results of PCR array by qRT-PCR assay. As shown in Figure 8B, knockdown of FLNA in HCT116R or HT29R cells significantly suppressed the expression of all the three genes, while overexpression of FLNA (WT) in HCT116 or HT29 cells promoted the expression of these genes. Correlation between FLNA and smad2 downstream genes prompted us to examine whether FLNA could regulate smad2 signaling. As indicated in Figure 8C, knockdown of FLNA decreased the levels of p-smad2, whereas overexpression of FLNA promoted the levels of p-smad2. To elucidate how FLNA regulated smad2 signaling, we analyzed the relationship between FLNA and smad2. Co-IP assay indicated that FLNA could interact with smad2 in HCT116 cells (Figure 8D). Immunofluorescence assay demonstrated that HCT116/vector cells mainly exhibited cytoplasmic smad2 staining, while smad2 mainly accumulated in the nucleus in HCT116/FLNA (WT) cells (Figure 8E). The promoter activity of c-MET in HCT116 cells was significantly increased after transfection with plasmid vectors driving either smad2 or FLNA (Figure 8F). To clarify FLNA-mediated function/chemoresistance was dependent on FLNA, we blocked smad2 signaling by a specific inhibitor LY2109761 in CRC cells expressing FLNA (WT). As shown in Figure 8G, LY2109761 treatment suppressed MMP8, snail and c-Met expression in HCT116/FLNA (WT) and HT29/FLNA (WT) cells. Blockage of smad2 signaling decreased the IC50 value of HCT116/FLNA (WT) and HT29/FLNA (WT) cells (Supplementary Figure 1A). Moreover, Blockage of smad2 signaling also significantly inhibited the migration and invasion ability of HCT116/FLNA (WT) and HT29/FLNA (WT) cells (Supplementary Figure 1B).
Figure 8.
FLNA regulates EMT through smad2 signaling. A. A total of 40 genes are quantified using a human signal transduction PCR array. The color scheme represents gene expression changes on a log2 scale. B. Three smad2 target genes (MMP9, snail and c-Met) reached the cutoff value (fold change >2 or <0.5) and their expression levels were confirmed in both two groups of cells. C. Expression of smad2 and p-smad2 in the indicated cell lines were determined by immunoblotting. D. Co-IP of smad2 and FLNA. HCT116 cells were immunoprecipitated using smad2 or FLNA Abs and immunoblotted with FLNA or smad2 Abs, respectively. E. Smad2 expression and its cellular location were detected by immunostaining with anti-smad2 antibody. DAPI stains nuclei. F. Promoter activity of c-Met in clonal cell lines expressing empty vector, smad2, WT-FLNA or the mutant S2152A were analyzed by luciferase reporter assay. G. The mRNA expression levels of MMP9, snail and c-Met in HCT116/vector cells and HCT116/FLNA (WT) cells treated with LY2109761. All data are presented as the mean ± SD of three experiments. *P<0.05, **P<0.01.
Clinical significance of FLNA and c-Met expression in CRC specimens
Given the regulatory relationship between FLNA and c-Met in CRC cells, we investigated the clinical significance of FLNA and c-Met in CRC specimens. Representative images of FLNA and c-Met staining were shown in Figure 9A. Pearson’s correlation analysis indicated that FLNA expression was positively correlated with c-Met expression (Figure 9B, P<0.01, r=0.462) in this cohort. We then performed Kaplan-Meier analyses to evaluate whether combination of FLNA and c-Met could better predict the prognosis of CRC patients. As illustrated in Figure 9C, the FLNA(+)/c-Met(+) group had the shortest 5-year overal survival and disease-free survival, and the FLNA(-)/c-Met(-) group had the longest survival (P<0.01); there was no significant difference of survival between the FLNA(+)/c-Met(-) group and FLNA(-)/c-Met(+) group (P>0.05).
Figure 9.
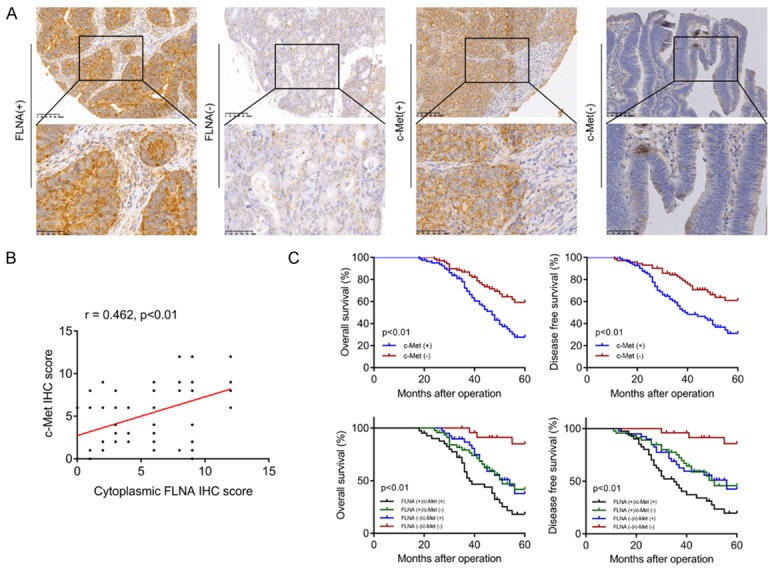
Clinical significance of FLNA and c-Met expression in CRC patients. A. Microphotographs of FLNA and c-Met protein expression in CRC specimens. B. The correlation between FLNA and c-Met expression was analyzed in a cohort of 152 CRC tissues (Pearson’s correlation, P<0.01, r = 0.462). C. Kaplan-Meier curves showing OS and DFS based on c-Met protein expression or FLNA combined with c-Met.
Knockdown of FLNA promotes chemosensitivity in a xenograft model
A xenograft mouse model was employed to further validate whether silencing FLNA could promote chemosensitivity in vivo. The tumor weight and growth rates were lower in HCT116/shFLNA group compared to HCT116/shNC group. Moreover, xenografts derived from HCT116/shFLNA cells exhibited decreased volume and weight than that from controls in response to 5-FU (Figure 10A, 10B). Decreased ki67 signals were observed in the shFLNA group compared to the shNC group. Correspondingly, TUNEL assay results indicated that knockdown of FLNA could promote apoptosis in vivo (Figure 10C). Furthermore, three specimens were randomly selected from each group for IHC and immunoblotting assay. Knockdown of FLNA could inhibit EMT in vivo, as demonstrated by the up-regulation of E-cadherin and the down-regulation of vimentin and N-cadherin. Moreover, Knockdown of FLNA could suppress the c-Met-AKT signaling pathway in vivo (Figure 10D, 10E). Altogether, the schematic diagram of our study is shown in Figure 10F.
Figure 10.
Knockdown of FLNA promotes tumor 5-FU sensitivity in a xenograft model. Nude mice were injected with HCT116R/shNC or HCT116R/shFLNA cells, followed by 5-FU or vehicle treatment. A. Tumor growth rate in each group. B. Average weight of subcutaneous xenografts. C. Representative immunohistochemical staining of TUNEL and ki67 from tumor samples in each group. D. Representative immunohistochemical staining of FLNA, p-AKT, c-Met, E-cadherin, N-cadherin and vimentin from tumor samples in each non-treated group. E. Expression of FLNA, p-AKT, c-Met, E-cadherin, N-cadherin and vimentin were determined using Immunoblotting. F. Schematic illustration of the mechanism by which FLNA promotes 5-FU resistance of CRC cells.
Discussion
Acquisition of 5-FU resistance is a major clinical obstacle for successful treatment of CRC. Although multiple mechanisms of 5-FU resistance had been reported, we focused on FLNA since bioinformatics analysis revealed that FLNA was up-regulated in a series of primary 5-FU resistant CRC cell lines. Moreover, Kaplan-Meier survival analysis of public database and our cohort showed a negative correlation between FLNA expression and disease free survival of CRC patients. Thus, FLNA may be a potential chemotherapeutic drug sensitivity marker for CRC patients. We then used common CRC cell lines that were selected over time for acquired resistance to 5-FU. After weeks of exposure to increasing concentrations of 5-FU, we observed remarkably elevated FLNA levels in two acquired 5-FU-resistant cells HCT116R and HT29R compared to parental cells, which prompted us to further analyze the exact roles of FLNA in chemoresistance.
Our study highlights the function and mechanisms of FLNA in regulating chemosensitivity of CRC. Though FLNA has been demonstrated to participate in several cancer-related processes, little is known about its role in regulation of drug resistance. In the present study, FLNA knockdown sensitized the CRC cells to 5-FU while FLNA overexpression promoted 5-FU resistance. Further functional studies confirmed that FLNA down or up-regulation altered the colony formation ability or apoptotic rate under 5-FU treatment. To be noticed, we also found that p-FLNA (ser2152) was up-regulated in acquired 5-FU resistant HCT116R and HT29R cells compared with the parental cells. Transfection of wild-type FLNA into HCT116 or HT29 cells promoted 5-FU resistance while transfection of mutant FLNA (S2152A) didn’t alter 5-FU sensitivity. Thus, FLNA mediated 5-FU resistance might be dependent on ser2152 phosphorylation, at least partly.
Chemoresistance is closely related to EMT during malignant tumor progression. In lung adenocarcinoma specimens, Uramoto et al. found that sensitive tumors exhibited positive expression of epithelial markers [26]. Biological studies have linked EMT to the emergence of drug resistance such as gefitinib, adriamycin, and cisplatin [5,27,28]. In CRC, there are multiple known factors responsible for EMT induced 5-FU resistance. Sun et al. reported that HES1 promotes 5-FU resistance of colorectal cancer cells (RKO and HCT8, LOVO) by prompting EMT and several ABC transporter genes such as ABCC1, ABCC2 and P-gp1 [9]. In comparison with parental HT29 cells, 5-FU-resistant HT29R cells exhibited enhanced migration, mesenchymal-like morphology, increased expression of mesenchymal marker fibronectin and EMT-inducing transcription factors Twist, Zeb1, and Zeb2 [8]. In accordance with the findings of the previous studies, our established 5-FU resistant HT29R cells were observed obvious acquisition of the mesenchymal properties. Moreover, another 5-FU-resistant cell line HCT116R also acquired mesenchymal properties compared with the parental HCT116 cells. FLNA is well recognized to regulate EMT-like phenotypes such as cell adhesion, migration and cytoskeletal remodeling [16,18]. In our research, knockdown of FLNA in HT29R and HCT116R cells suppressed EMT while overexpression of FLNA (WT) in HT29 and HCT116 cells promoted EMT, which demonstrated FLNA was essential for EMT in CRC cells. However, overexpression of FLNA (S2152A) in HT29 and HCT116 cells had no effect on EMT, suggesting FLNA ser2152 phosphorylation is essential to regulate EMT. There are several known signaling pathways responsible for chemoresistance as well as EMT, including PI3K/AKT, MAPK/ERK and Hippo/YAP [29-31]. Among these pathways, AKT activation was well reported to promote FLNA ser2152 phosphorylation [19]. Our results indicated HGF stimulation promoted phosphorylation of AKT and FLNA (ser2152), while blockage of AKT signaling by c-Met inhibitor Crizotinib or PI3K inhibitor LY294002 significantly decreased FLNA ser2152 phosphorylation. Furthermore, Crizotinib or LY294002 treatment also suppressed the adhesion/migration/invasion ability of HCT116/FLNA (WT) and HT29/FLNA (WT) cells. Based on these findings, we concluded that FLNA dephosphorylation with the c-Met inhibitor or PI3K inhibitor resulted in decreased invasiveness of CRC cells.
Dissection of the downstream signaling responsible for FLNA-mediated 5-FU resistance and EMT is another challenge. Firstly, we used a PCR array to profile differentially expressed signal transduction-related genes in two groups (HCT116R/shNC vs HCT116R/shFLNA#2 cells, HCT116/vector vs HCT116/FLNA). Among these differential genes, snail and MMP9 are well recognized targets of smad2. Snail is a well-known EMT-promoting transcription factor; MMP9 is mainly involved in the degradation of extracellular matrix to promote tumor invasion and metastasis. Thus, our results suggested smad2 signaling might play a crucial role in FLNA mediated EMT. To date, the most well recognized mechanism for smad2 mediated function is the phosphorylation and nuclear translocation. In our results, knockdown of FLNA in HCT116R cells led to a significant decrease of smad2 phosphorylation. On the contrary, overexpression of FLNA (WT) but not FLNA (S2152A) in HCT116 cells drastically promoted the protein level of p-smad2. Immunofluorescence assay indicated smad2 mainly exhibited cytoplasmic staining in HCT116/vector cells and nuclear staining in HCT116/FLNA (WT) cells, suggesting FLNA led to an accumulation of smad2 in the nucleus. Intriguingly, c-Met was also identified as a target of FLNA-smad2 signaling. On one hand, c-Met is a tyrosine kinase receptor that can participate in the activation of PI3K/AKT, MAPK and other signaling pathways when stimulated by the ligand HGF. On the other hand, c-Met-AKT pathway could induce FLNA ser2152 phosphorylation. Thus, our results highlighted a novel mechanism that c-Met-AKT-FLNA formed a positive feedback loop to regulate EMT. We then investigated whether smad2 activation induced by FLNA was essential for the expression of downstream effectors. LY2109761 was used to block smad2 signaling in HCT116/FLNA (WT) cells and HT29/FLNA (WT) cells. qRT-PCR assay suggested LY2109761 treatment suppressed the expression of c-Met, snail and MMP9, which meant an attenuation of FLNA mediated EMT. Clinically, IHC analysis revealed a positive correlation between FLNA and c-Met expression in a cohort of CRC tissues. Furthermore, combination of FLNA and c-Met was a better prognostic marker for CRC patients.
A limitation of our current work is that cell motility could be influenced by various pathways, such as NF-kB, Wnt and Notch signaling, which also play important roles in EMT induced chemoresistance. Despite the chemoresistant CRC model used in our study exhibited enhanced adhesion/migration/invasion ability, we could not definitively conclude that increased cell motility was driven by EMT. Actually, several recent studies show that EMT might be necessary for chemoresistance but may not be essential for distant metastasis [32,33]. Therefore, whether changes in cell motility and EMT in chemoresistant CRC cells occur in parallel or have a cause-and-effect relationship still need further study. Secondly, despite FLNA could interact with smad2 to promote smad2 phosphorylation, the mechanism seemed to be complicated. Zhou et al. reported that FLNA interacted with smad2 but didn’t affect its stability [34]. Several researchers speculated that FLNA might serve as an anchor protein to control localization of smad proteins near the cell surface receptors, but still lack of powerful evidences [35,36]. In CRC cells, we confirmed the result that FLNA showed no effect on ubiquitin-dependent smad2 degradation (data not shown). However, we cannot elucidate deeper molecular mechanisms except that FLNA has an effect on smad2 phosphorylation. Thus, how does FLNA work in the phosphorylation of smad2 also need further investigation.
In conclusion, we elucidated the potential roles of FLNA overexpression and ser2152 phosphorylation in acquired 5-FU resistance of CRC for the first time. We provided evidences that c-Met-AKT-FLNA formed a positive feedback loop to enhance EMT, which contributes to 5-FU resistance of CRC cells. In the future, disruption of the functionally relevant FLNA-smad2 interaction and c-Met-AKT mediated FLNA ser2152 phosphorylation might contribute to development of biomarkers and treatment strategies for CRC patients.
Acknowledgements
The authors want to thank Xueshan Yin for excellent technical assistance.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 2.Salonga D, Danenberg KD, Johnson M, Metzger R, Groshen S, Tsao-Wei DD, Lenz HJ, Leichman CG, Leichman L, Diasio RB, Danenberg PV. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res. 2000;6:1322–1327. [PubMed] [Google Scholar]
- 3.Longley DB, Allen WL, Johnston PG. Drug resistance, predictive markers and pharmacogenomics in colorectal cancer. Biochim Biophys Acta. 2006;1766:184–196. doi: 10.1016/j.bbcan.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang B, Yang Y, Shi X, Liao W, Chen M, Cheng AS, Yan H, Fang C, Zhang S, Xu G, Shen S, Huang S, Chen G, Lv Y, Ling T, Zhang X, Wang L, Zhuge Y, Zou X. Proton pump inhibitor pantoprazole abrogates adriamycin-resistant gastric cancer cell invasiveness via suppression of Akt/GSK-beta/beta-catenin signaling and epithelial-mesenchymal transition. Cancer Lett. 2015;356:704–712. doi: 10.1016/j.canlet.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 6.Piskareva O, Harvey H, Nolan J, Conlon R, Alcock L, Buckley P, Dowling P, Henry M, O’Sullivan F, Bray I, Stallings RL. The development of cisplatin resistance in neuroblastoma is accompanied by epithelial to mesenchymal transition in vitro. Cancer Lett. 2015;364:142–155. doi: 10.1016/j.canlet.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Liang SQ, Marti TM, Dorn P, Froment L, Hall SR, Berezowska S, Kocher G, Schmid RA, Peng RW. Blocking the epithelial-to-mesenchymal transition pathway abrogates resistance to anti-folate chemotherapy in lung cancer. Cell Death Dis. 2015;6:e1824. doi: 10.1038/cddis.2015.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim AY, Kwak JH, Je NK, Lee YH, Jung YS. Epithelial-mesenchymal transition is associated with acquired resistance to 5-fluorocuracil in HT-29 colon cancer cells. Toxicol Res. 2015;31:151–156. doi: 10.5487/TR.2015.31.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L, Ke J, He Z, Chen Z, Huang Q, Ai W, Wang G, Wei Y, Zou X, Zhang S, Lan P, Hong C. HES1 promotes colorectal cancer cell resistance to 5-Fu by inducing Of EMT and ABC transporter proteins. J Cancer. 2017;8:2802–2808. doi: 10.7150/jca.19142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartwig JH, Stossel TP. Isolation and properties of actin, myosin, and a new actinbinding protein in rabbit alveolar macrophages. J Biol Chem. 1975;250:5696–5705. [PubMed] [Google Scholar]
- 11.Tseng Y, An KM, Esue O, Wirtz D. The bimodal role of filamin in controlling the architecture and mechanics of F-actin networks. J Biol Chem. 2004;279:1819–1826. doi: 10.1074/jbc.M306090200. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adh Migr. 2011;5:160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Boren J, Akyurek LM. Filamins in cardiovascular development. Trends Cardiovasc Med. 2007;17:222–229. doi: 10.1016/j.tcm.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Zhang K, Zhu T, Gao D, Zhang Y, Zhao Q, Liu S, Su T, Bernier M, Zhao R. Filamin A expression correlates with proliferation and invasive properties of human metastatic melanoma tumors: implications for survival in patients. J Cancer Res Clin Oncol. 2014;140:1913–1926. doi: 10.1007/s00432-014-1722-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uramoto H, Akyurek LM, Hanagiri T. A positive relationship between filamin and VEGF in patients with lung cancer. Anticancer Res. 2010;30:3939–3944. [PubMed] [Google Scholar]
- 16.Kircher P, Hermanns C, Nossek M, Drexler MK, Grosse R, Fischer M, Sarikas A, Penkava J, Lewis T, Prywes R, Gudermann T, Muehlich S. Filamin A interacts with the coactivator MKL1 to promote the activity of the transcription factor SRF and cell migration. Sci Signal. 2015;8:ra112. doi: 10.1126/scisignal.aad2959. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Bismar TA, Su J, Xu B, Kristiansen G, Varga Z, Teng L, Ingber DE, Mammoto A, Kumar R, Alaoui-Jamali MA. Filamin A regulates focal adhesion disassembly and suppresses breast cancer cell migration and invasion. J Exp Med. 2010;207:2421–2437. doi: 10.1084/jem.20100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savoy RM, Ghosh PM. The dual role of filamin A in cancer: can’t live with (too much of) it, can’t live without it. Endocr Relat Cancer. 2013;20:R341–356. doi: 10.1530/ERC-13-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo MS, Ohta Y, Rabinovitz I, Stossel TP, Blenis J. Ribosomal S6 kinase (RSK) regulates phosphorylation of filamin A on an important regulatory site. Mol Cell Biol. 2004;24:3025–3035. doi: 10.1128/MCB.24.7.3025-3035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vial D, McKeown-Longo PJ. Epidermal growth factor (EGF) regulates alpha5beta1 integrin activation state in human cancer cell lines through the p90RSK-dependent phosphorylation of filamin A. J Biol Chem. 2012;287:40371–40380. doi: 10.1074/jbc.M112.389577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravid D, Chuderland D, Landsman L, Lavie Y, Reich R, Liscovitch M. Filamin A is a novel caveolin-1-dependent target in IGF-I-stimulated cancer cell migration. Exp Cell Res. 2008;314:2762–2773. doi: 10.1016/j.yexcr.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Zhong Z, Yeow WS, Zou C, Wassell R, Wang C, Pestell RG, Quong JN, Quong AA. Cyclin D1/cyclin-dependent kinase 4 interacts with filamin A and affects the migration and invasion potential of breast cancer cells. Cancer Res. 2010;70:2105–2114. doi: 10.1158/0008-5472.CAN-08-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian ZQ, Shi JW, Wang XR, Li Z, Wang GY. New cancer suppressor gene for colorectal adenocarcinoma: filamin A. World J Gastroenterol. 2015;21:2199–2205. doi: 10.3748/wjg.v21.i7.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wieczorek K, Wiktorska M, Sacewicz-Hofman I, Boncela J, Lewinski A, Kowalska MA, Niewiarowska J. Filamin A upregulation correlates with Snail-induced epithelial to mesenchymal transition (EMT) and cell adhesion but its inhibition increases the migration of colon adenocarcinoma HT29 cells. Exp Cell Res. 2017;359:163–170. doi: 10.1016/j.yexcr.2017.07.035. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Zhang J, Liu L, Jiang Y, Ji J, Yan R, Zhu Z, Yu Y. Protein arginine methyltransferase 5-mediated epigenetic silencing of IRX1 contributes to tumorigenicity and metastasis of gastric cancer. Biochim Biophys Acta Mol Basis Dis. 2018;1864:2835–2844. doi: 10.1016/j.bbadis.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 26.Uramoto H, Iwata T, Onitsuka T, Shimokawa H, Hanagiri T, Oyama T. Epithelial-mesenchymal transition in EGFR-TKI acquired resistant lung adenocarcinoma. Anticancer Res. 2010;30:2513–2517. [PubMed] [Google Scholar]
- 27.Xie M, Zhang L, He CS, Xu F, Liu JL, Hu ZH, Zhao LP, Tian Y. Activation of Notch-1 enhances epithelial-mesenchymal transition in gefitinib-acquired resistant lung cancer cells. J Cell Biochem. 2012;113:1501–1513. doi: 10.1002/jcb.24019. [DOI] [PubMed] [Google Scholar]
- 28.Li S, Zhang X, Zhang R, Liang Z, Liao W, Du Z, Gao C, Liu F, Fan Y, Hong H. Hippo pathway contributes to cisplatin resistant-induced EMT in nasopharyngeal carcinoma cells. Cell Cycle. 2017;16:1601–1610. doi: 10.1080/15384101.2017.1356508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao M, Luo R, Liu Y, Gao L, Fu Z, Fu Q, Luo X, Chen Y, Deng X, Liang Z, Li X, Cheng C, Liu Z, Fang W. miR-3188 regulates nasopharyngeal carcinoma proliferation and chemosensitivity through a FOXO1-modulated positive feedback loop with mTOR-p-PI3K/AKT-c-JUN. Nat Commun. 2016;7:11309. doi: 10.1038/ncomms11309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JX, Xu Y, Gao Y, Chen C, Zheng ZS, Yun M, Weng HW, Xie D, Ye S. Decreased expression of miR-939 contributes to chemoresistance and metastasis of gastric cancer via dysregulation of SLC34A2 and Raf/MEK/ERK pathway. Mol Cancer. 2017;16:18. doi: 10.1186/s12943-017-0586-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Song R, Gu D, Zhang L, Zhang X, Yu B, Liu B, Xie J. Functional significance of Hippo/YAP signaling for drug resistance in colorectal cancer. Mol Carcinog. 2018;57:1608–1615. doi: 10.1002/mc.22883. [DOI] [PubMed] [Google Scholar]
- 32.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasaki A, Masuda Y, Ohta Y, Ikeda K, Watanabe K. Filamin associates with Smads and regulates transforming growth factor-β signaling. J Biol Chem. 2001;276:17871–17877. doi: 10.1074/jbc.M008422200. [DOI] [PubMed] [Google Scholar]
- 35.Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP. The small GTPase RalA targets filamin to induce filopodia. Proc Natl Acad Sci U S A. 1999;96:2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma CP, Ezzell RM, Arnaout MA. Direct interaction of filamin (ABP-280) with the beta 2-integrin subunit CD18. J Immunol. 1995;154:3461–3470. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.