Abstract
This study elaborated on the function of Low-density lipoprotein receptor-related protein 6 (LRP6), a critical component of Wnt signaling, in liver fibrosis. This study enrolled sixty-eight patients with liver fibrosis, with ten healthy liver tissue samples, served as the controls. A lentiviral vector expressing LRP6-CRISPR was constructed. Immortalized HSC-T6 cells were transfected with LRP6-CRISPR. A rat model of CCl4-induced liver fibrosis was established, and rats were injected with lentiviral vectors expressing LRP6-CRISPR. LRP6 expression and fibrosis biomarkers were examined by PCR, Western blot, and immunofluorescence assay, respectively. HSC growth and its ability of migration and invasion were evaluated by MTT and Transwell assay, separately. Wnt signaling activity was examined by Luciferase reporter assay. LRP6 was overexpressed in human fibrotic-liver tissues, and the expression of LRP6 was correlated with liver fibrosis stages. LRP6 knockout with CRISPR suppressed the Wnt signaling activities and consequently repressed HSC activation and relived liver injury in fibrotic-liver model rats. Our data revealed that the knockout of LRP6 weakens the binding of Wnt ligand with its cell surface receptors, the first step of Wnt transduction cascade, and consequently repressed HSC activation.
Keywords: Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), CRISPR-associated protein 9 (Cas9), single-guide RNA (sgRNA), Hepatic stellate cell (HSC), Low-density lipoprotein receptor-related protein 6 (LRP6)
Introduction
Liver fibrosis, mostly occurred in people with alcoholic liver disease, autoimmune and biliary diseases, or chronic HBV/HCV infections, is one of the significant life-threatening public health problems worldwide [1-4]. Although liver fibrosis, even at advanced stages, can now be reversed, there are still limited therapeutic options for this disease of high morbidity and mortality [4,5].
Thus, it is essential to understand the mechanism of liver fibrosis. In liver fibrosis, a pivotal cellular event is an activation of the hepatic stellate cells (HSC) [6]. Many signaling pathways involve in HSC activation [7]. Among which, Wnt signaling pathway, which has long been known as crucial to pathological conditions such as cancer and inflammatory diseases, significantly participates in HSC activation and liver fibrosis [8,9].
Low-density lipoprotein receptor-related protein 6 (LRP6) was first discovered by Schneider et al. in the mouse liver cDNA library [10]. LRP6 is a signal transmembrane protein. The interaction of Wnt ligand and their transmembrane receptor, Frizzled (FZD) family and LRP6, is the first step in the Wnt signal transduction cascade. Several reports indicate that LRP6 is associated with many diseases, such as metabolic syndrome, breast cancer, Alzheimer’s disease also osteoporosis [11]. Nevertheless, the role of LRP6 in liver fibrosis, especially in HSC activation, has not been well elucidated.
The clustered regularly interspaced short palindromic repeat/CRISPR-associated protein (CRISPR/Cas9) system has emerged as an accessible gene-editing technology in precision medicine [12]. This study aims to explore the functions and underlying mechanisms of LRP6 in the activation of the WNT pathway with CRISPR-based LRP6 gene knockout.
Materials and methods
Study design
This study investigated the role of LRP6 in liver fibrogenesis in vivo and in vitro (Figure 5). A total of sixty-eight patients with liver fibrosis were enrolled in this study, with ten healthy liver tissue samples served as the controls. Immortalized HSC-T6 cells were transfected with LRP6-CRISPR. A rat model of CCl4-induced liver fibrosis was established, and rats were injected with lentiviral vectors expressing LRP6-CRISPR. LRP6 expression and fibrosis biomarkers were examined by PCR, Western blot, and immunofluorescence assay, respectively. HSC growth and its ability of migration and invasion were evaluated by MTT and Transwell assay, separately. Wnt signaling activity was examined by Luciferase reporter assay.
Figure 5.
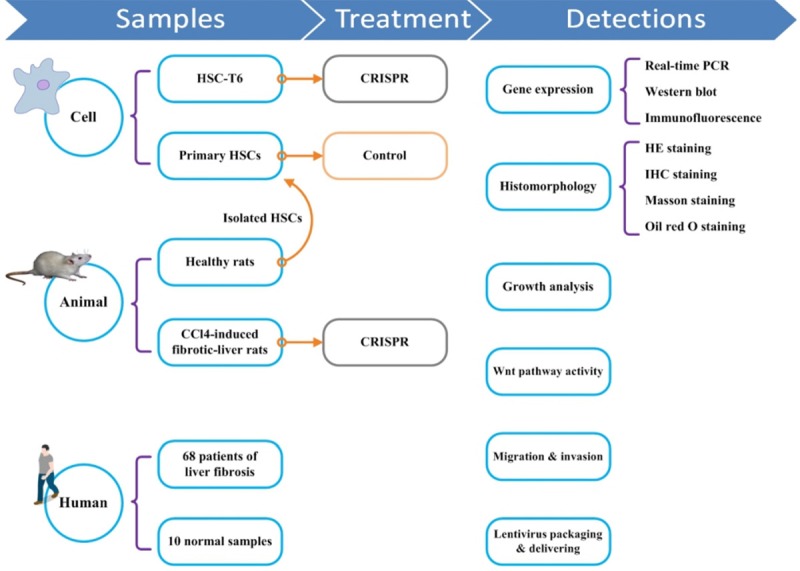
Study design. This study verified the role of LRP6 in liver fibrogenesis from various aspects, including cells, animals, and human. Lentivirus vector expressing LRP6-CRISPR was constructed and were then used for in vivo and in vitro experiments. A rat model of CCl4-induced liver fibrosis was established. The expression of fibrosis biomarkers and Wnt signaling activities were examined. HSCs proliferation, migration, and invasion were analyzed.
Patients and liver tissue samples
This study enrolled 68 patients with liver fibrosis from the First Affiliated Hospital of Jiaxing College, with written informed consent from each patient. Human fibrotic-liver tissue samples were collected, with ten healthy liver tissue samples served as the control. Tissue samples were preserved at the temperature of -80°C. The study was approved by the Ethics Committee of the First Affiliated Hospital of Jiaxing College (2015-039).
Liver fibrosis evaluations
Fibrotic stages of human liver tissues were evaluated by a modified Knodell scoring system [13-15]. The histology of liver samples was assessed by two experienced pathologists in a double-blinded manner. Results were graded in four categories: fibrous portal expansion (I), fibrous septa (II), bridging fibrosis (III), and cirrhosis (IV).
The animal model of liver fibrosis
Male Sprague-Dawley (SD) rats (8-10 weeks, 250-300 g) were purchased from Shanghai SLAC Laboratory Animal Co Ltd (Shanghai, China). Institutional and national guidelines for the care and use of laboratory animals were followed. The protocol of the experiments was approved by the Committee on the Ethics of Animal Experiments of Jiaxing College (JUMC2018-014).
The rat model of CCl4-induced hepatic fibrosis was induced by subcutaneous injection of 40% solution of CCl4 in olive oil at a dose of 5 ml/kg twice per week for four consecutive weeks. Forty male SD rats were randomly divided into two groups. The first group (n = 10) served as a normal control group, and the experimental group (n = 30) was liver fibrosis models.
Cell lines and culture
Immortalized rat (HSC-T6, # KCB200703YJ) cell line with characteristics of an activated HSCs phenotype and 293T cells (#KCB200744YJ) were purchased from Chinese Academy of Science Conservation Genetics CAS Kunming Cell Bank (Kunming, Yunnan, China).
Primary stellate cells were isolated from the livers of normal male SD rats (250-300 g of body weight), as previously described [16].
Plasmids
pLenti-CRISPR/Cas9-V2-GFP plasmid was purchased from Miaoling Bioscience & Technology (#P1289, Wuhan, China). pSpCas9 (BB)-2A-GFP (PX458) was a gift from Feng Zhang (Addgene plasmid #48138; http://n2t.net/addgene:48138; RRID: Addgene_48138). pMD2.G was a gift from Didier Trono (Addgene plasmid #12259; http://n2t.net/addgene:12259; RRID: Addgene_12259). psPAX2 was a gift from Didier Trono (Addgene plasmid #12260; http://n2t.net/addgene:12260; RRID: Addgene_12260). lenti dCAS-VP64_Blast was a gift from Feng Zhang (Addgene plasmid # 61425; http://n2t.net/addgene:61425; RRID: Addgene_61425). lenti MS2-P65-HSF1_Hygro was a gift from Feng Zhang (Addgene plasmid # 61426; http://n2t.net/addgene:61426; RRID: Addgene_61426). lenti sgRNA(MS2)_zeo backbone was a gift from Feng Zhang (Addgene plasmid #61427; http://n2t.net/addgene:61427; RRID: Addgene_61427). Four-plasmid lentiviral packaging system PG-p1-VSVG, PG-P2-REV, PG-P3-PRE was purchased from Genepharma (Shanghai, China).
sgRNA plasmid
Single-guide RNA (sgRNA) sequences targeting rat LRP6 (Gene ID: 312781) were designed using online software (http://crispr.mit.edu). This study selected three sgRNA target sequences (target 1, 2, and 3) for LRP6-CRISPR (Supplementary Table 1).
The LRP6-CRISPR expression vectors were constructed by ligating the cDNA expressing the sgRNA (target 1, 2, and 3) into the BmsBI-digested pLenti-CRISPR/Cas9-V2-GFP plasmid. The ligation products were transformed into Escherichia coli DB5 cells. Positive clones were screened by ampicillin and PCR and were further identified with sequencing. The ligation products were named as CRISPR-T1 (target 1), CRISPR-T2 (target 2), CRISPR-T3 (target 3), and CRISPR-NC (blank), respectively.
Screening of candidate sgRNAs
The LRP6-CRISPR expression vectors (CRISPR-T1 and CRISPR-T2) were transformed into 293T cells. Cells were harvested at 48 h, and the genomic DNA was extracted and purified using MiniBEST Universal Genomic DNA Extraction Kit (Takara Biotechnology, Dalian, China). The extracted DNA was amplified by PCR using the following primers: LRP6-F: CAGTACTTGTCGGCAGAGGA; LRP6-R: TGTCCCGCTTCCTTTGTACT (Supplementary Table 2). The PCR product was denatured at 95°C for 3 min, gradually cooled to room temperature, and then digested with T7E1 enzyme (New England Biolabs, Ipswich, MA, USA) according to manufacturer instructions. Results showed that CRISPR-T1 had the highest mutation rate (Supplementary Figure 2A). Results of sequencing indicated CRISPR-T1 vector containing the target sequence GCCAGTGCCAAGGCAACCGA (Supplementary Figure 2B). CRISPR-T1 was selected for the future study.
Lentiviral vectors construction and production
The packaging of recombinant lentivirus for CRISPR was performed by transfection of 293T cells with an LRP6-CRISPR expression vector (CRISPR-T1, CRISPR-NC), lentiviral envelope plasmid pMD.2G, and packaging vector psPAX2. Briefly, 2.5 × 106 293T cells were plated in 10 cm dish and cultured at 37°C, 50 mL/L CO2 for 20 h. Then the cells were cotransfected with 8 µg pMD.2G, 8 µg psPAX2, and 8 µg CRISPR-T1 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The supernatant containing the lentivirus (Lenti-CRISPR-T1, Lenti-CRISPR-NC) was harvested at 72 h, centrifuged at 4,000 rpm for 10 min, filtered through a 0.45 µm low protein-binding filter (Millipore, Bedford, MA), concentrated by Amicon Ultra-15 100KDa (Millipore, cat. no. UFC910024), and stored at -80°C. Lentiviral vector (Lenti-CRISPR-T1) titers were determined by fluorescence-activated cell sorting analysis, and the viral titer was 2.0 × 108 TU/ml (Supplementary Figure 2C).
In vitro transduction of lentivirus
HSC-T6 cells were co-cultured with a lentiviral vector (Lenti-CRISPR-T1 and Lenti-CRISPR-NC, MOI = 50). 48 h after infection, HSCs (HSC-T6 cells transfected with Lenti-CRISPR-NC served as negative controls, and non-transfected cells served as blank controls) were harvested for further study (Supplementary Figure 2D).
In vivo transduction of lentivirus
CCl4-induced liver-fibrotic rats were selected for the LRP6-CRISPR experiment. In vivo transduction of lentiviruses (Lenti-CRISPR-T1 and Lenti-CRISPR-NC) was achieved through in-situ injections of 30 uL of concentrated viral suspension with a viral titer of 1.0 × 108 TU/mL lentiviral particles in PBS; injections were conducted once. Three weeks after the dose, the animals (rats injected with Lenti-CRISPR-NC served as negative controls, and non-processed rats served as blank controls) were sacrificed by CO2 exposure, and liver tissues were harvested.
Statistical analysis
All statistical analyses were performed using R software (version 3.5.1). Data were presented as mean ± SEM. A value of P < 0.05 was considered to be statistically significant. A two-tailed Student’s t-test was employed to evaluate the differences between groups. For semi-quantitative analysis of histological staging, nonparametric tests (Mann-Whitney U Test or Kruskal-Wallis Rank Sum Test) were used.
Results
Overexpression of LRP6 was observed in hepatic tissues of patients with liver fibrosis
The expression of LRP6 protein in clinical samples of liver tissue was assessed by immunohistochemical staining (IHC). Liver tissue samples were judged positive if either cell nucleus or cytoplasm stained positive. A total of sixty-eight human fibrotic-liver tissue samples were examined with antibodies against human LRP6, with ten healthy liver tissue samples served as the controls. Results of immunostaining showed that the staining of LRP6 was positive in fibrotic-liver tissues, whereas it was negative in healthy liver tissues (Figure 1A).
Figure 1.
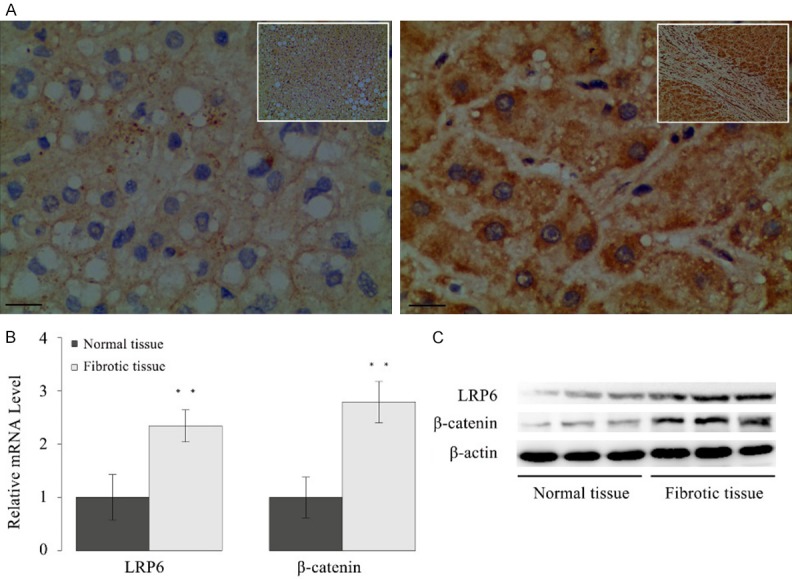
Overexpression of LRP6 was observed in human fibrotic-liver tissue. A. Representative pictures of immunostaining showed LRP6 expression was positive in liver tissues of patients with hepatic fibrosis, whereas it was negative in normal liver tissues (bar = 50 μm, magnification × 200/400). B. The mRNA level of LRP6 and β-catenin in human fibrotic-liver tissues (n = 40) were significantly higher than those of the normal liver tissues (n = 10). C. Expression of LRP6 and β-catenin were upregulated in liver tissues of patients with hepatic fibrosis. Data represent the mean of three independent experiments, and error bars are the standard deviation of means. *P < 0.05 compared with the control, **P < 0.01 compared with the control.
IHC results were further scored by the percentage of LRP6-stained cells as below: < 10% (score 0), 10%~50% (score 1), 51%~75% (score 2), and 76%~100% (score 3). Score zero and one were considered as the weak expression, whereas scores 2 and 3 were the strong expression. A strong expression of LRP6 (score 2: 20, score 3: 12) was found in 32 fibrotic-liver tissue samples, whereas a weak immunoreactivity of LRP6 (score 0: 16, score 1: 20) was identified in the remain fibrotic-liver tissue samples (Table 1). IHC results also indicated the expression of LRP6 in normal liver tissue samples (score 0: 7, score 1: 3) was significantly lower than that in the controls (Mann-Whitney U Test, P < 0.001).
Table 1.
Clinicopathological characteristics and LRP6 expression of liver fibrosis patients
| Characteristics | Patients number (n = 68) | LRP6-staining score | p-value | |||
|---|---|---|---|---|---|---|
|
| ||||||
| 0 | 1 | 2 | 3 | |||
| Age | 0.775 | |||||
| ≤ 50 | 24 | 5 | 7 | 8 | 4 | |
| > 50 | 44 | 11 | 13 | 12 | 8 | |
| Gender | 0.325 | |||||
| Male | 49 | 11 | 13 | 15 | 10 | |
| Female | 19 | 5 | 7 | 5 | 2 | |
| Residence | 0.148 | |||||
| Urban | 32 | 9 | 11 | 8 | 4 | |
| Rural | 36 | 7 | 9 | 12 | 8 | |
| Knodell score | 0.001 | |||||
| I | 12 | 7 | 4 | 1 | 0 | |
| II | 25 | 6 | 9 | 7 | 3 | |
| III | 23 | 3 | 6 | 8 | 6 | |
| IV | 8 | 0 | 1 | 4 | 3 | |
The mRNA level of LRP6 in the human liver tissues was further examined by real-time PCR (Figure 1B). Forty human fibrotic-liver tissue samples were randomly selected, with ten healthy liver tissue samples served as the controls. Results showed that the relative mRNA level of LRP6 was significantly higher in the fibrotic-liver tissues (P < 0.01) than that in the controls. The protein expression of LRP6 in human liver tissues was also measured by Western blot assay. Consistently, Western blot results showed that the protein expression of LRP6 increased in the fibrotic-liver tissues comparing with the controls (Figure 1C).
Since LRP6 involved in the Wnt pathway, this study also examined the level of β-catenin in human fibrotic-liver tissues. Results showed both the mRNA level (P < 0.01) and nuclear protein expression of β-catenin were significantly elevated in the fibrotic-liver tissue samples compared with the controls (Figure 1B, 1C).
A correlation between LRP6 expression and liver fibrosis stages was identified
The correlation between LRP6 expression and the clinicopathological characteristics of patients with liver fibrosis was analyzed (Table 1). Results showed there is no correlation between LRP6 expression and patients’ age (Mann-Whitney U Test, P = 0.775), gender (Mann-Whitney U Test, P = 0.325), and residence (Mann-Whitney U Test, P = 0.148). However, LRP6 expression was heavily correlated with the Knodell score for hepatic fibrosis sages [13-15] (Kruskal-Wallis Rank Sum Test, P = 0.001).
LRP6-CRISPR reversed HSC activation
This study tried to repress HSC activation, the center event of liver fibrogenesis, by knockout the LRP6 gene in the activated HSCs with the CRISPR system. Immortalized rat HSC cell line (HSC-T6), which presents with characteristics of an activated HSCs phenotype were transfected with a lentiviral vector expressing LRP6-CRISPR. HSC-T6 transfected with a lentiviral vector expressing CRISPR-NC served as the negative control, and those without treatment served as blank control. Upon transfected with LRP6-CRISPR, sgRNA complementary paired with the target sequence (coding region of LRP6 gene) and inhibited the expression of the LRP6 gene due to the frameshift mutation (Figure 2A). The mRNA levels of LRP6 and fibrosis biomarker (α-SMA and Collagen-I) in HSC-T6 cells transfected with LRP6-CRISP were significantly lower than those in the negative and blank control (P < 0.01 for LRP6, α-SMA, and collagen-I) (Figure 2B). Interestingly, the mRNA levels of LRP6, α-SMA, and collagen-I in HSC-T6 cells transfected with LRP6-CRISP tended to be close to those of the quiescent cells (freshly isolated HSC), which implied LRP6-CRISPR might reverse the HSC activation.
Figure 2.
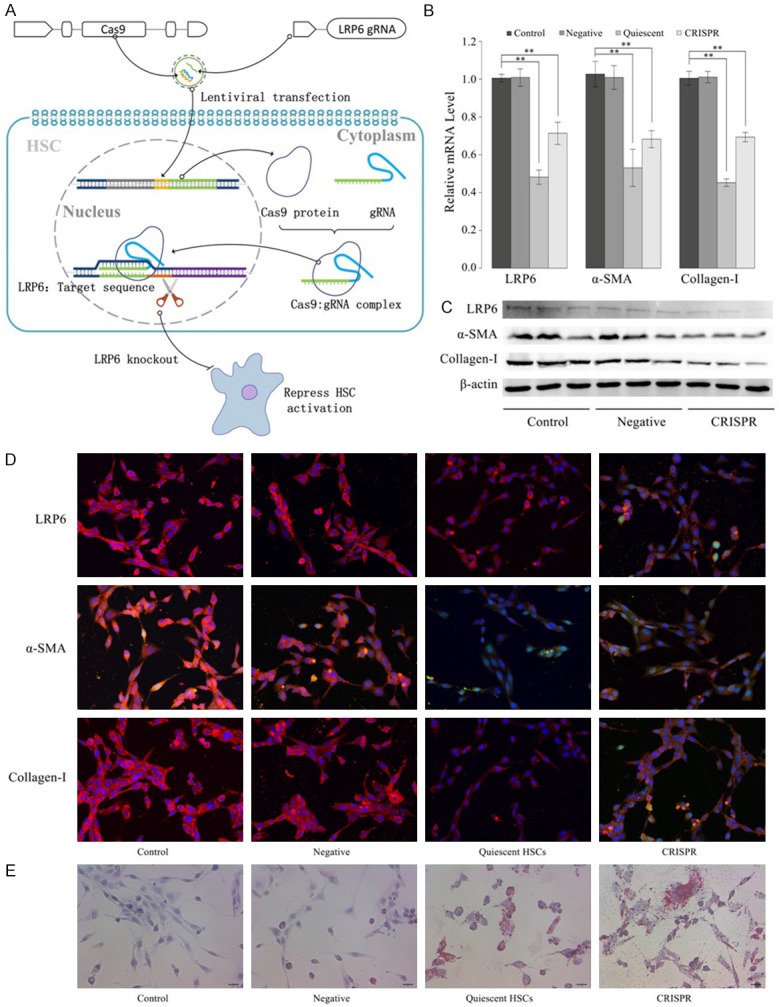
LRP6-CRISPR suppressed HSC activation. A. Editing the target sequence (LRP6) by CRISPR specificity is realized by the complementary pairing with the target sequence and double-stranded breaks at the target site. The repaired DNA (NHEJ or HDR) inhibits the expression of the LRP6 gene due to the frameshift mutation caused by the random insertion or deletion of the base, and consequently, repressed HSC activation. B. The mRNA level of LRP6, α-SMA, and collagen-I were significantly lower in HSC-T6 cells transfected with LRP6-CRISPR. C. The protein expression of LRP6, α-SMA, and collagen-I decreased in HSC-T6 cells treated with LRP6-CRISPR. D. Immunofluorescence results were consistent with those of Western blot assay. E. Oil red O staining demonstrated the lipid droplets increased in HSC-T6 cells transfected with LRP6-CRISPR. Data represent the mean of three independent experiments, and error bars are the standard deviation of means. *P < 0.05 compared with the blank control (HSC-T6 transfected with LRP6-NC served as the negative control, and those without treatment served as blank control), **P < 0.01 compared with the blank control.
The protein expressions of LRP6, α-SMA, and Collagen-I were analyzed by Western blot assay (Figure 2C). Consistent with the results of PCR, the protein expressions of LRP6, α-SMA, and Collagen-I were notably weakened in the HSC-T6 cells treated with LRP6-CRISPR. Similar results were also observed in the immunofluorescence test, the expressions of LRP6, α-SMA, and Collagen-I in HSC-T6 cells transfected with LRP6-CRISPR were significantly decreased compared with the negative and blank control. They were very close to those of the quiescent HSCs (Figure 2D). Consistently, oil red O staining results showed the cytoplasmic lipid droplets were regained in the activated HSCs treated with LRP6-CRISPR and were almost no different from those of the quiescent HSCs (Figure 2E). Together, these findings suggested HSCs transfected with LRP6-CRISPR present a trend towards the restoration to an inactive status.
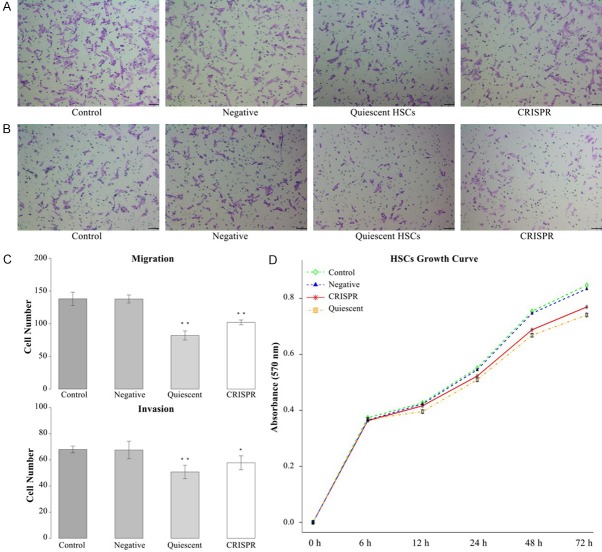
Activated HSC is characterized by the high ability of migration and invasion. This study examined HSCs’ ability of migration and invasion by Transwell assay. Results showed the migration ability of quiescent HSCs and HSC-T6 transfected with LRP6-CRISPR were significantly lower than the negative and blank control (P < 0.01 for both quiescent HSCs and HSC-T6 cells transfected with LRP6-CRISPR) (Figure 3A, 3C). Consistently, results of invasion assay indicated the invasion ability of quiescent HSCs and HSC-T6 treated with LRP6-CRISPR decreased significantly (P < 0.01 for quiescent HSCs, and P = 0.031 for HSC-T6 cells treated with LRP6-CRISPR) (Figure 3B, 3C). Results of proliferation assay (MTT) demonstrated the growth rate of HSC-T6 cells transfected with LRP6-CRISPR prominently down-regulated compared with the negative and blank control and was close to that of quiescent HSCs (Figure 3D).
Figure 3.
LRP6-CRISPR repressed HSCs’ ability of proliferation, migration, and invasion. A. Transwell assay showed LRP6-CRISPR weaken HSCs’ ability of migration. B. LRP6-CRISPR reduced HSCs’ ability of invasion. C. Cell numbers of both migration and invasion decreased significantly in HSC-T6 cells treated with LRP6-CRISPR. D. MTT assay showed LRP6-CRISPR down-regulated HSCs proliferation. Data represent the mean of three independent experiments, and error bars are the standard deviation of means. *P < 0.05 compared with the blank control (HSC-T6 transfected with LRP6-NC served as the negative control, and those without treatment served as blank control), **P < 0.01 compared with the blank control.
LRP6-CRISPR repressed WNT activation
As LRP6 participated in the Wnt pathway, WNT/β-catenin pathway activities in HSCs transfected with LRP6-CRISPR were analyzed. Real-time PCR showed that the relative mRNA level of β-catenin was significantly lower in HSC-T6 transfected with LRP6-CRISPR than that in the blank and the negative control (P < 0.01) (Supplementary Figure 1A). In line with the results discussed above, the mRNA level of β-catenin in HSC-T6 cells treated with LRP6-CRISPR approached that of quiescent HSCs. Accordingly, the result of Western blot assay indicated the protein expression of nuclear β-catenin weakened in HSC-T6 cells treated with LRP6-CRISP (Supplementary Figure 1B). Results of Luciferase reporter assay demonstrated TCF activities significantly dropped in HSCs treated with LRP6-CRISPR (P < 0.01 for TOPFlash/FOPFlash of both quiescent HSCs and HSC-T6 cells treated with LRP6-CRISPR) (Supplementary Figure 1C). Immunofluorescence results were consistent with the above findings that the expression of nuclear β-catenin decreased in HSC-T6 cells treated with LRP6-CRISPR and were close to those of quiescent HSCs (Supplementary Figure 1D). Together, these findings suggested the knockout of LRP6 by CRISPR repressed the Wnt pathway activities in activated HSCs.
LRP6-CRISPR relieved liver fibrogenesis in rats
To get a comprehensive understanding of the biological role of LRP6 in liver fibrogenesis, we established a rat model of CCl4-induced liver fibrosis for in vivo transduction of lentiviral vector expressing LRP6-CRISPR. The fibrotic-liver rats treated with a lentiviral vector expressing CRISPR-NC served as the negative control, and those without treatment served as the blank control. Liver tissues of fibrotic-liver rats were analyzed by Masson staining and HE staining, respectively. Masson staining revealed the LRP6-CRISPR might slow down the fibrogenesis in the liver tissues of CCl4-induced fibrotic-liver rats (Figure 4A). Consistently, HE staining showed the recovering of vacuoles degeneration in the liver tissues of CCl4-induced fibrotic-liver rats treated with LRP6-CRISPR, which indicate the knockout of LRP6 might relieve the liver tissue injury (Figure 4B). The expression of LRP6 protein in rats was evaluated by IHC. Results showed that the expression of LRP6 decreased in the fibrotic-liver rats treated with LRP6-CRISPR (Figure 4C). Consistent with the above findings, the results of PCR test showed the mRNA levels of LRP6, fibrosis biomarkers (α-SMA and collagen-I), and β-catenin in fibrotic-liver rats transfected with LRP6-CRISPR decreased significantly compared with the negative and blank control (P < 0.01 for LRP6, α-SMA, collagen-I, and β-catenin) (Figure 4D). Together, these findings suggested the LRP6-CRISPR might relief the liver fibrogenesis in rats with liver fibrosis.
Figure 4.
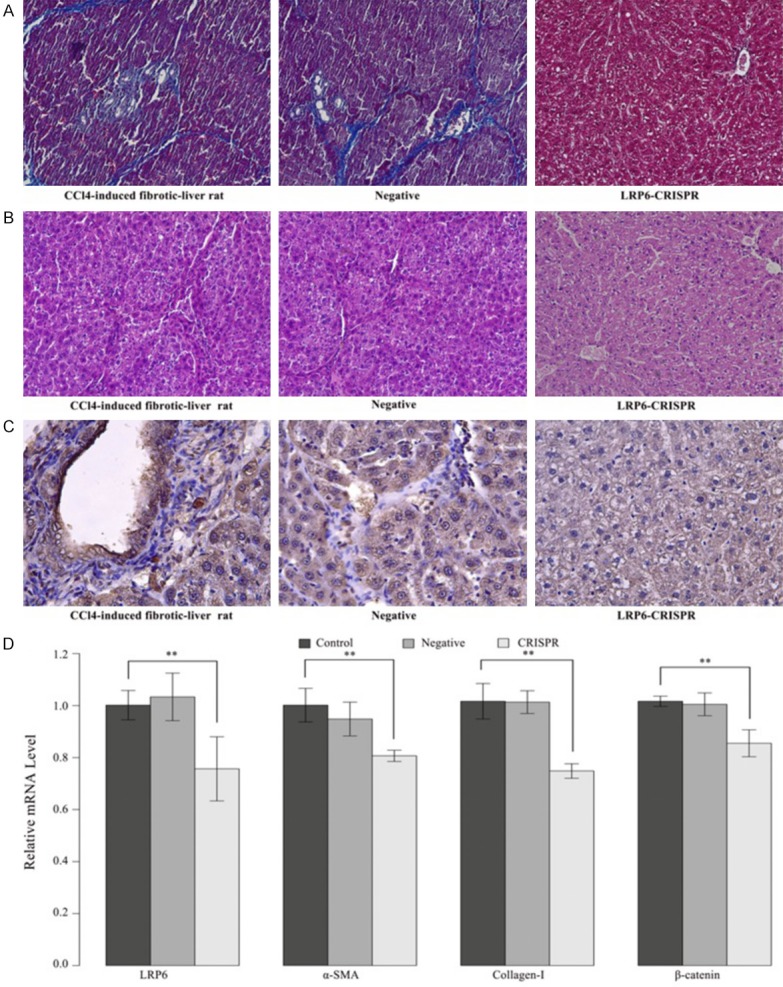
LRP6-CRISPR relieved hepatic fibrogenesis in a rat model of CCl4-induced liver fibrosis. A. Masson staining indicated the restoration of liver fibrosis in CCl4-induced fibrotic-liver rats transfected with LRP6-CRISPR. B. HE staining showed decreased vacuoles degeneration of hepatocytes in fibrotic-liver rats transfected with LRP6-CRISPR. C. IHC staining showed that the expression of LRP6 decreased in fibrotic-liver rats transfected with LRP6-CRISPR. D. the mRNA level of LRP6/α-SMA/collagen-I/β-catenin dropped significantly compared with the control. Data represent the mean of three independent experiments, and error bars are the standard deviation of means. *P < 0.05 compared with the blank control (CCl4-induced fibrotic-liver rats injected with CRISPR-NC served as the negative controls, and those non-processed rats served as the blank controls), **P < 0.01 compared with the blank control.
Discussion
Liver fibrosis is the most common pathological feature of many chronic liver diseases, and will gradually develop into liver cirrhosis and hepatocellular carcinoma [3,17]. Therefore, researchers have carried out a large number of studies on the early monitoring and treatment of liver fibrosis. Nevertheless, a number of detection and treatment of liver fibrosis were not highly targeted so far. In the present study, we identified that the expressions of LRP6 and nuclear β-catenin were significantly elevated in human fibrotic-liver tissue. The binding of LRP6 with its Wnt ligand is a crucial step in canonical Wnt/β-catenin signaling. Reports show LRP6 regulates cell fate, and the growth and repair of several tissues [18-20]. Our findings indicated that LRP6 might involve in the liver fibrogenesis.
By analyzing the LRP6 expression and the clinicopathological characteristics of patients with liver fibrosis, we found that LRP6 expression was correlated with hepatic fibrosis stages. Finding factors associated with the rapid development of liver fibrosis and sensitive biomarkers for evaluation of fibrosis progression has long been the unmet challenge for researchers [21]. The stages of liver fibrosis influence the doctor’s decision in treating and monitoring the disease complications [22]. Since liver biopsy has not considered as the “gold standard” in diagnosing liver fibrosis, our finding might provide new clues to explore efficient, safe, and noninvasive detection of liver fibrosis in personalized medicine [23].
The CRISPR-Cas9 system is an RNA-guided targeted gene-editing tool for cell lines and animals at the genomic DNA level [24]. The system requires two components, Cas9, the endonuclease, and a single-guide RNA (sgRNA), which accurately guides Cas9 to the location in the genome [25-28]. The effectiveness of the CRISPR system allows researchers to experiment in precision medicine nowadays. In this study, we knock out the LRP6 gene to inactivate HSCs with the CRISPR-Cas9 system. HSC-T6, an immortalized rat HSC cell line which presents with characteristics of an activated HSCs phenotype, were transfected with a lentiviral vector expressing LRP6-CRISPR. The expression of LRP6, and fibrosis biomarkers α-SMA and collagen-I in HSC-T6 cells transfected with LRP6-CRISPR decreased significantly and tended to be close to those of the quiescent cells. Accordingly, similar results were also observed in immunofluorescence experiments and oil red O staining. HSC activation companied with high ability of migration and invasion. Consistently, the results of the migration and invasion assay indicated that the migration and invasion ability of HSC-T6 treated with LRP6-CRISPR decreased significantly. Together, these findings implied the knockout of LRP6 suppressed HSC activation, the center event of liver fibrogenesis.
LRP6 is a “core” component of Wnt signaling. The interaction of Wnt proteins and LRP6 results in a series of downstream intracellular events [29]. LRP6 regulates the Wnt/β-catenin signaling, often referred to as the canonical pathway, which controls cell proliferation and differentiation [30,31]. Prior reports indicate that Wnt signaling was related to many human fibrosis diseases, such as pulmonary, renal, and liver fibrosis [2]. Furthermore, evidence showed that LRP6 might be more developmentally critical in the development and maintenance of disease than LRP5 [31]. Our data showed that the knockout of LRP6 with CRISPR system repressed Wnt signaling activities in the activated HSC-T6 cells and consequently weakened HSC-T6 activation.
To evaluate the viability of LRP6-CRISPR in the treatment of liver fibrosis, a rat model of CCl4-induced liver fibrosis was established for in vivo transduction of lentiviral vector expressing LRP6-CRISPR. Our results showed that LRP6-CRISPR relieve liver tissue injury. To our understanding, this is the first time CRISPR has been used in animals to treat liver fibrosis.
The above results open up a new strategy, LRP6-CRISPR, a DNA-level genetic medicine in inhibiting HSC activation and maintaining liver homeostasis to prevent fibrogenesis. Further studies are necessary to demonstrate the antifibrotic effects of LRP6-CRISPR on other mediators and signaling pathways.
Acknowledgements
This work was supported by grants from the Zhejiang Provincial Natural Science Fund (LY16H030016, LY17H030012), Center for gastroenterology and hepatology connecting with Shanghai. Jiaxing key medical discipline (gastroenterology and hepatology), Key medical discipline in Jiaxing-Infectious disease (2019-ZC-02). Key medical discipline in Jiaxing-Gastroenterology (2019-ZC-08).
Disclosure of conflict of interest
None.
Abbreviations
- LRP6
Low-density lipoprotein receptor-related protein 6
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- Cas9
CRISPR-associated protein 9
- sgRNA
single-guide RNA
- HSC
Hepatic stellate cell IHC, immunohistochemical staining
- SD
Sprague-Dawley
Supporting Information
References
- 1.Weersink RA, Bouma M, Burger DM, Drenth JP, Hunfeld NG, Kranenborg M, Monster-Simons MH, van Putten SA, Metselaar HJ, Taxis K, Borgsteede SD. Evaluating the safety and dosing of drugs in patients with liver cirrhosis by literature review and expert opinion. BMJ Open. 2016;6:e012991. doi: 10.1136/bmjopen-2016-012991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miao CG, Yang YY, He X, Huang C, Huang Y, Zhang L, Lv XW, Jin Y, Li J. Wnt signaling in liver fibrosis: progress, challenges and potential directions. Biochimie. 2013;95:2326–2335. doi: 10.1016/j.biochi.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Campana L, Iredale JP. Regression of liver fibrosis. Semin Liver Dis. 2017;37:1–10. doi: 10.1055/s-0036-1597816. [DOI] [PubMed] [Google Scholar]
- 4.Altamirano-Barrera A, Barranco-Fragoso B, Mendez-Sanchez N. Management strategies for liver fibrosis. Ann Hepatol. 2017;16:48–56. doi: 10.5604/16652681.1226814. [DOI] [PubMed] [Google Scholar]
- 5.Ren S, Johnson BG, Kida Y, Ip C, Davidson KC, Lin SL, Kobayashi A, Lang RA, Hadjantonakis AK, Moon RT, Duffield JS. LRP-6 is a coreceptor for multiple fibrogenic signaling pathways in pericytes and myofibroblasts that are inhibited by DKK-1. Proc Natl Acad Sci U S A. 2013;110:1440–1445. doi: 10.1073/pnas.1211179110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng JH, She H, Han YP, Wang J, Xiong S, Asahina K, Tsukamoto H. Wnt antagonism inhibits hepatic stellate cell activation and liver fibrosis. Am J Physiol Gastrointest Liver Physiol. 2008;294:G39–49. doi: 10.1152/ajpgi.00263.2007. [DOI] [PubMed] [Google Scholar]
- 7.Wang JN, Li L, Li LY, Yan Q, Li J, Xu T. Emerging role and therapeutic implication of Wnt signaling pathways in liver fibrosis. Gene. 2018;674:57–69. doi: 10.1016/j.gene.2018.06.053. [DOI] [PubMed] [Google Scholar]
- 8.Jiang J. CK1 in developmental signaling: hedgehog and wnt. Curr Top Dev Biol. 2017;123:303–329. doi: 10.1016/bs.ctdb.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson MD, Monga SP. WNT/beta-catenin signaling in liver health and disease. Hepatology. 2007;45:1298–1305. doi: 10.1002/hep.21651. [DOI] [PubMed] [Google Scholar]
- 10.Schneider WJ, Nimpf J, Brandes C, Drexler M. The low-density lipoprotein receptor family: genetics, function, and evolution. Curr Atheroscler Rep. 1999;1:115–122. doi: 10.1007/s11883-999-0007-9. [DOI] [PubMed] [Google Scholar]
- 11.Wang ZM, Luo JQ, Xu LY, Zhou HH, Zhang W. Harnessing low-density lipoprotein receptor protein 6 (LRP6) genetic variation and Wnt signaling for innovative diagnostics in complex diseases. Pharmacogenomics J. 2018;18:351–358. doi: 10.1038/tpj.2017.28. [DOI] [PubMed] [Google Scholar]
- 12.Aravalli RN, Steer CJ. CRISPR/Cas9 therapeutics for liver diseases. J Cell Biochem. 2018;119:4265–4278. doi: 10.1002/jcb.26627. [DOI] [PubMed] [Google Scholar]
- 13.Dufour JF, DeLellis R, Kaplan MM. Regression of hepatic fibrosis in hepatitis C with long-term interferon treatment. Dig Dis Sci. 1998;43:2573–2576. doi: 10.1023/a:1026601904609. [DOI] [PubMed] [Google Scholar]
- 14.Desmet VJ. Knodell RG, Ishak KG, Black WC, Chen TS, Craig R, Kaplowitz N, Kiernan TW, Wollman J. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis [Hepatology 1981;1:431-435] J Hepatol. 2003;38:382–386. doi: 10.1016/s0168-8278(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 15.Knodell RG, Ishak KG, Black WC, Chen TS, Craig R, Kaplowitz N, Kiernan TW, Wollman J. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology. 1981;1:431–435. doi: 10.1002/hep.1840010511. [DOI] [PubMed] [Google Scholar]
- 16.Ramm GA. Isolation and culture of rat hepatic stellate cells. J Gastroenterol Hepatol. 1998;13:846–851. doi: 10.1111/j.1440-1746.1998.tb00747.x. [DOI] [PubMed] [Google Scholar]
- 17.Shan L, Liu Z, Ci L, Shuai C, Lv X, Li J. Research progress on the anti-hepatic fibrosis action and mechanism of natural products. Int Immunopharmacol. 2019;75:105765. doi: 10.1016/j.intimp.2019.105765. [DOI] [PubMed] [Google Scholar]
- 18.Go GW. Low-density lipoprotein receptor-related protein 6 (LRP6) is a novel nutritional therapeutic target for hyperlipidemia, non-alcoholic fatty liver disease, and atherosclerosis. Nutrients. 2015;7:4453–4464. doi: 10.3390/nu7064453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janda CY, Dang LT, You C, Chang J, de Lau W, Zhong ZA, Yan KS, Marecic O, Siepe D, Li X, Moody JD, Williams BO, Clevers H, Piehler J, Baker D, Kuo CJ, Garcia KC. Surrogate Wnt agonists that phenocopy canonical Wnt and beta-catenin signalling. Nature. 2017;545:234–237. doi: 10.1038/nature22306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W, Singh R, Choi CS, Lee HY, Keramati AR, Samuel VT, Lifton RP, Shulman GI, Mani A. Low density lipoprotein (LDL) receptor-related protein 6 (LRP6) regulates body fat and glucose homeostasis by modulating nutrient sensing pathways and mitochondrial energy expenditure. J Biol Chem. 2012;287:7213–7223. doi: 10.1074/jbc.M111.286724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman SL. Evolving challenges in hepatic fibrosis. Nat Rev Gastroenterol Hepatol. 2010;7:425–436. doi: 10.1038/nrgastro.2010.97. [DOI] [PubMed] [Google Scholar]
- 22.Petitclerc L, Sebastiani G, Gilbert G, Cloutier G, Tang A. Liver fibrosis: review of current imaging and MRI quantification techniques. J Magn Reson Imaging. 2017;45:1276–1295. doi: 10.1002/jmri.25550. [DOI] [PubMed] [Google Scholar]
- 23.Li S, Sun X, Chen M, Ying Z, Wan Y, Pi L, Ren B, Cao Q. Liver fibrosis conventional and molecular imaging diagnosis update. J Liver. 2019;8 [PMC free article] [PubMed] [Google Scholar]
- 24.Maggio I, Stefanucci L, Janssen JM, Liu J, Chen X, Mouly V, Goncalves MA. Selection-free gene repair after adenoviral vector transduction of designer nucleases: rescue of dystrophin synthesis in DMD muscle cell populations. Nucleic Acids Res. 2016;44:1449–1470. doi: 10.1093/nar/gkv1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Yin M, Wang M, Wang Y. Phage AcrIIA2 DNA mimicry: structural basis of the CRISPR and anti-CRISPR arms race. Mol Cell. 2019;73:611–620. e613. doi: 10.1016/j.molcel.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Burmistrz M, Dudek B, Staniec D, Rodriguez Martinez JI, Bochtler M, Potempa J, Pyrc K. Functional analysis of porphyromonas gingivalis W83 CRISPR-Cas systems. J Bacteriol. 2015;197:2631–2641. doi: 10.1128/JB.00261-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karimian A, Azizian K, Parsian H, Rafieian S, Shafiei-Irannejad V, Kheyrollah M, Yousefi M, Majidinia M, Yousefi B. CRISPR/Cas9 technology as a potent molecular tool for gene therapy. J Cell Physiol. 2019;234:12267–12277. doi: 10.1002/jcp.27972. [DOI] [PubMed] [Google Scholar]
- 28.Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 30.Lara-Castillo N, Johnson ML. LRP receptor family member associated bone disease. Rev Endocr Metab Disord. 2015;16:141–148. doi: 10.1007/s11154-015-9315-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong Z, Baker JJ, Zylstra-Diegel CR, Williams BO. Lrp5 and Lrp6 play compensatory roles in mouse intestinal development. J Cell Biochem. 2012;113:31–38. doi: 10.1002/jcb.23324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.