Abstract
Circulating tumor cells (CTCs) are cells that are shed from the primary tumor and circulate in the blood, and their metastasis and formation of a secondary tumor are closely associated with cancer-related death. Therefore, regulating tumor metastasis through CTCs can be a novel strategy to fight cancer. It has been demonstrated that CTCs can reflect the profile of the primary tumor and provide valuable information about intratumoral heterogeneity and their evolution over time. Moreover, the revelation of the relationship between metastasis and CTCs suggests that CTC regulation represents a promising novel anticancer strategy. Above all, at the molecular level, genetic analysis might be vital in the new era of gene-targeted cancer therapies and contribute to personalized anti-metastasis tumor treatments. In this review, we will focus on the biological significance of CTCs in the peripheral blood and discuss their potential clinical implications in cancer management.
Keywords: Circulating tumor cells, enrichment technology, cancer metastasis, epithelial-mesenchymal transition, molecular analysis, clinical significance
Introduction
Circulating tumor cells (CTCs) were first defined by Asworth in 1869 and further elucidated in 1955 [1]. They can be free or clustered in the circulation or lodge themselves in new tissues. The presence of CTCs is common in most cancer patients, even in patients with localized disease and in patients at risk of recurrence after primary radical treatment [2]. However, CTCs are extraordinarily rare, and it is estimated that there is only one CTC in a million blood cells in the circulation of patients with advanced cancer. Therefore, a simple noninvasive blood sample is a uniquely accessible method.
CTCs are generally referred to as disseminated tumor cells (DTCs) in the bone marrow. In 2007, the American Society of Clinical Oncology first cited CTCs and DTCs in a suggestion about tumor markers [3]. Both CTCs and DCTs have the potential to predictive prognosis and are useful for monitoring therapeutic effects in cancer patients. However, sampling from the bone marrow is an invasive process that is not widely used clinically. The detection of CTCs in the peripheral blood seems more practical than detecting DTCs in the bone marrow [4]. A new technology called “liquid biopsy” provides real-time analysis of CTCs for patients with tumors [5]. As a blood test, the liquid biopsy is sufficiently sensitive to discover an individual tumor cell lurking in a billion ordinary hematopoietic cells and can be monitored in real time. Liquid biopsy results make important contributions in deciphering the genomic message related to tumor metastasis, providing comprehensive marker detection for targeted therapies and determining treatment resistance [6]. Liquid biopsy results can reflect information about different phases of metastasis and are helpful for the early diagnosis of malignant tumors and the accurate prediction of metastasis or recurrence [7].
With the development of CTC research, the simple enumeration of CTCs in the periphery cannot provide a comprehensive view of malignant tumors. In fact, CTCs provide the chance to assess the complexity of biological features of tumors and reflect genetic, epigenetic and proteomic features during progression, which can be very different from those of the primary tumor [8,9]. Therefore, collecting CTCs during disease progression reveal dynamically evolving results, reflecting the evolution of invasive tumor cells during treatment. With this information, mutations in tumor cells can be identified, and precision treatment can be used to treat the evolving cancer. The latest developed technologies to enrich and characterize CTCs have taken into account each step of metastasis and offer the opportunity to inhibit metastasis [10].
Collectively, the clinical significance of CTCs in malignant tumors warrants further investigation. In this review, we will discuss detection technologies based on liquid biopsy, concentrating on the characteristics of CTCs and the factors that impact CTC aggressiveness. In the future, the development of new CTC detection techniques and the further evaluation of CTCs will allow cancer research to enter a new era of clinical research.
Enrichment of CTCs
Several methods to capture CTCs from peripheral blood are based on their differential physical or biological properties [11,12] (Figure 1). Physical characteristics include size, density and electrical properties. These methods can separate CTCs without labeling. Compared with white blood cells (WBCs), CTCs are usually larger, and this characteristic has been utilized to capture CTCs with size-based filtration. On the basis of differences in sedimentation coefficients, density gradient centrifugation separates cells by centrifugal force. Furthermore, cells can be separated on the basis of differences in the charges of their membranes [8].
Figure 1.
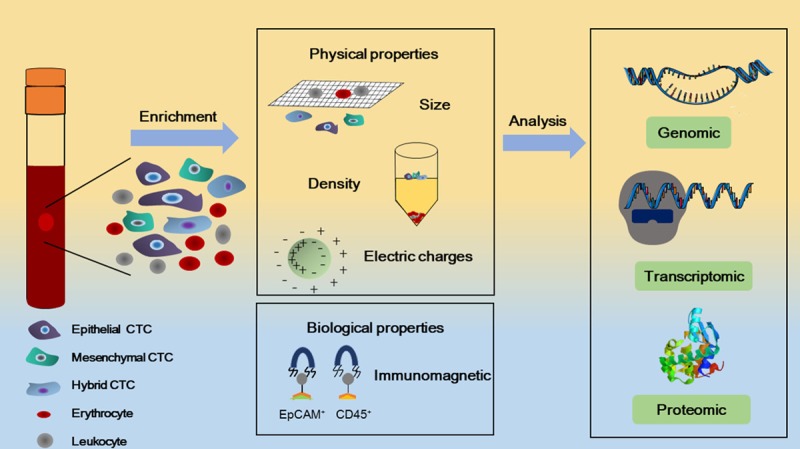
CTCs have physical properties and biological properties that can help separate them from normal peripheral blood cells. Physical properties include size, density and electrical charge. Biological properties are based on two principles: the expression of biomarkers (EpCAM for positive-selection, CD45 for negative-selection) and the separation of cells via immunomagnetic techniques. Further analysis of CTCs in terms of genomic, transcriptomic and proteomic characteristics distinguishes them from normal cells.
The majority of techniques based on biological properties are affinity-binding systems, which depend on the immunologic characteristics of CTCs. There are two methods that make use of this technique. The first approach is using cancer-special biomarkers to positively select the CTCs. According to the expression of surface biomarkers of epithelial-mesenchymal transition (EMT), CTCs are divided into three phenotypes: epithelial CTCs, mesenchymal CTCs or hybrid CTCs. The most successful detection technology has made use of EpCAM, which is commonly expressed on epithelial cells. Currently, the US Food and Drug Administration has approved only pass one validated and standardized assay, which is based on antibodies against EpCAM [13]. However, for aggressive triple negative breast cancers, gastrointestinal malignancies, lung cancers, and prostate cancers, positive selection based on EpCAM is limited [14-16]. In particular, some tumor cells may undergo EMT and have an EpCAM-negative biological phenotype. Researchers have found many biomarkers that can be used to detect EpCAM-negative tumor cells. One preferred method is to capture EpCAM-negative CTCs through the depletion of CD45-positive leukocytes (negative selection) [17]. Negative selection can remove red blood cells by size separation or chemical lysis. WBCs can also be removed by high-affinity antibodies and immunomagnetic separation [10]. Current strategies to capture hybrid CTCs (which express both epithelial and mesenchymal markers) combine antibodies against multiple cell surface markers [18].
With the advances in rearch, many devices have been used in capture CTCs. CTC-chip is a microfluidic platform, consisting of an array of anti-EpCAM antibody-coated microposts. The CTC-chip mechanism is based on physical biological properties and has been used to successfully isloate CTCs in metastatic lung, prostate, pancreatic, colon and breast cancer [19]. The source of the neoplasm is the key to isolating CTCs. Since the technique is not specific for isolating WBCs, it is possible to capture rare circulating epithelial cells and other nonimmune cells.
In other words, each technology has advantages and disadvantages. A combination of different technologies is likely to be beneficial for understanding the roles of CTCs in metastasis and holds great potential for new biological insights and clinical applications.
Migration of CTCs
It is accepted that CTCs are important in the hematogenous metastasis of malignant tumors. The process of CTC dissemination is complex and continuous and includes five steps: local invasion, intravasation, hematogenous survival and transport, extravasation and colonization [20,21]. Many factors may influence the diffusion and invasion of CTCs [22] (Figure 2).
Figure 2.

Metastasis includes five steps: local invasion, intravasation, hematogenous survival and transport, extravasation and colonization. EMT is a multistep transitional process that produces epithelial, mesenchymal and hybrid CTCs. Tumor cells may actively or passively enter the bloodstream via EMT or other biological events. In the circulation, CTCs may overcome several hurdles including shear forces, anoikis, and immune defense. After survival in the bloodstream, CTCs may move under the collisions with other blood cells and be recruited to the vascular walls, allowing them to transmigrate through the endothelium. At the distant site, CTCs may extravasate and undergo MET, which enables the colonization of CTCs and ultimately results in metastatic lesions.
EMT and MET
The transition between epithelial and mesenchymal states can include EMT and as mesenchymal-epithelial transition (MET). Both EMT and MET play a crucial role in embryonic development and have a key role in the tumorigenic process [23]. Emerging research suggests that EMT is likely a key event involved in the spread of epithelial cancer cells [24]. This process not only enables tumor cells to acquire migratory/invasive properties that facilitate infiltration of the vasculature system and production of CTCs but also promotes the ability of CTCs to survive in the blood and to penetrate the circulation and invade proximal tissues [25]. Apart from these roles, EMT has also been associated with drug resistance [26], genomic instability [27] and immune evasion [28].
EMT plays a crucial role during each step of tumor metastasis and is conducive to CTC generation, intravasation, survival in circulation and extravasation. Tumor cell invasion seems to start with the EMT program. In the process of EMT, tumor cells lose the junctions that connect them to other cells or with the extracellular matrix (EMC) [29]. This loss causes cells to become motile and aggressive, allowing them to transmigrate through the basement membrane and invade surrounding stromal tissues [25]. In a study of human hepatocellular carcinoma (HCC) tissues, HCC cells that had experienced EMT in cancer cell nests invaded the surrounding tumor stroma [30]. Once the tumor cells disperse into the bloodstream, they may face a formidable challenge from the vascular environment. They have to overcome intercellular collision and fluid shear forces caused by blood flow to survive [17]. The shearing forces significantly reduce the number of CTCs, and different shearing forces can change the movement path of CTCs. Studies have shown that CTCs with EMT phenotypes are more viable in the bloodstream. According via this pre-metastatic mechanism, EMT can also help tumor cells overcome anoikis [31]. After that, CTCs may move under the collisions with other blood cells and be recruited to the vascular walls. There, CTCs roll and adhere to the endothelium via interactions with adhesive factors, transmigrate through the endothelium and colonize the distant site [32]. Following metastatic spread and colonization, MET, the reverse biological process of EMT, converts the disseminated mesenchymal tumor cells back to the epithelial cell state which is a more differentiated state [23]. MET may result in the loss of the migratory capabilities of CTCs and the restoration of tight junctions between CTCs and other cells, which ultimately leads to formation of the metastatic lesion [33].
Host factors in CTC migration
New findings suggest that research concentrating on only the intrinsic features of tumor cells is insufficient, as host cells have coevolved with the tumor cells within the tumor microenvironment and thus also both play an important role in metastasis [34,35]. Therefore, the “premetastatic niche” as a new concept in recurrence and metastasis has attracted people’s attention [7]. The premetastatic niche is defined as a microenvironment that facilitates the invasion, proliferation and/or survival of metastatic tumor cells in the target metastatic organ [34]. Emerging evidence has shown that genetic activation and regulation of specific proteases and chemokines/cytokines may direct metastasis to a designated organ and form a premetastatic niche [36]. For instance, Minn et al. [37] and Kang et al. [38] have identified a unique set of genes that can predict lung or bone metastasis by examining subsets of breast tumor cells. These specific genes include VCAM1, matrix metalloproteinase 1 (MMP-1), CXCL1 and inhibitor of differentiation 1 (Id1) for lung metastasis, and these specific genes represent mediators and markers of tumor cell survival and growth [37]. Focus on the early molecular and cellular events in cancer dissemination will likely lead to new methods to detect and prevent metastasis in the initial stages of metastasis.
CTC clusters
Circulating tumor cell clusters are referred to as circulating tumor aggregates, circulating tumor microemboli or circulating micrometastases and are defined as groups of tumor cells travelling together in the bloodstream [39]. Various microfluidic devices have been developed to separate these clusters without compromising their integrity [40,41]. Recent studies have considered that CTC clusters have potential implications in the metastasis process and are relevant to clinical outcomes [42]. CTC clusters contain a number of tumor cells and may also contain platelets, immune cells, and cancer-associated fibroblasts. These nonmalignant components are conducive to the survival and metastasis of CTC clusters in different ways. For instance, the existence of heterotypic tumor-derived stromal cells within CTC clusters facilitated metastasis formation [43]. The presence of endothelial cells contributed to promoting angiogenesis and resulted in a larger size of metastases [44]. The CTC clusters have longer survival times than common CTCs in enduring hemodynamic shearing forces. However, the half-life of the CTCs (25-30 min) is longer than that of CTC clusters (estimated to be 6-10 min) [45].
CTC clusters are mainly composed of epithelial CTCs (which express biomarkers such as EpCAM and E-cadherin), but mesenchymal CTCs (which express biomarkers such as N-cadherin and EGFR) and hybrid CTCs are also observed [46-48]. The presence of two cell types and/or the ability to transfer between states are thought to be the reasons for their higher metastatic potential relative to individual CTCs, and the lack of proliferative biomarkers may explain why they are more resistant to chemotherapy [48-50]. Although it is widely believed that CTC clusters are relevant to the progression of tumor metastasis and that their presence is associated with poor clinical outcome, research on CTC clusters as biomarkers and therapeutic targets is limited by several factors [51]. The clinical significance of CTC clusters currently remains to be confirmed, and further efforts are needed to explore the potential of CTC clusters in clinical applications.
Molecular analysis of CTCs
The molecular analysis of CTCs has an impact on the molecular underpinnings of cancer in patients. It presents a valuable source for personalized anti-metastatic therapies and surveillance for drug resistance [52]. Advances in molecular technology allow molecular analysis at the single-cell level [53,54] (Table 1).
Table 1.
Analysis of CTCs
| Analysis | Techniques | Markers | References |
|---|---|---|---|
| Count | Stain | Antibodies against cytokeratins | [70] |
| DNA-based strategies | Sanger sequencing | PIK3CA and EGFR | [71] |
| Next-generation sequencing | The complete exome and genome | [72-74] | |
| Multiple displacement amplification | Single nucleotide polymorphism | [72] | |
| Comparative array genomic hybridization | Genomic profiling of CTCs | [75] | |
| mRNA-based strategies | Reverse-transcription PCR | CK18, CK19, CK20, MUC1, prostate-specific antigen, and carcinoembryonic antigen | [76] |
| Digital PCR | ATP | [77] | |
| RNA sequencing | Wnt2 | [78] | |
| Multicolor RNA in situ hybridization | Multiple gene targets | [79] | |
| Protein-based strategies | Mass cytometry | Phosphorylation states | [80-82] |
| CellSearch | CKs, CD45, and DAPI | [76] | |
| Epithelial ImmunoSPOT | Secreted, shed, or released proteins | [83] | |
| High-speed automated digital microscopy | Antibodies with fluorescent conjugates | [84] | |
| Microfluidic technologies | [85] | ||
| Mass spectrometry approaches | [82] |
Genomic and proteomic analyses can be used to provide clinical information on the DNA mutation status and evolution of cancer. Such analyses are critical for accurate identification and monitoring for the emergence of new mutations during the metastatic process [10]. For instance, in breast cancer, next-generation sequencing of CTCs has revealed significant inter- and intra-patient heterogeneity, which can be monitored over time. The emergence of estrogen receptor gene (ESR1) and fibroblast growth factor receptor gene (FGFR2) mutations revealed potential new therapeutic targets [55]. In addition, many genes associated with migration and adhesion such as CD44v6 and CD151, are strongly downregulated, which highlights the decreased invasion and adhesion of CTCs in the blood stream [56,57]. Detection of these mutations during the progression of the disease can direct follow-up treatment.
Transcriptomic analysis of CTCs provides great advantages for us to understand the metastatic process. In a transcriptomic-based assay, the relative change in the expression of epithelial and mesenchymal biomarkers during the treatment in 11 patients with metastatic breast cancer was studied by RNA in situ hybridization [46]. With the development of sequencing technologies, the mutation frequencies and expression profiles of CTCs can be provided. However, few technologies can be used to measure CTC protein expression, with the exception of antibody-staining approaches [10]. Proteomic-based assays can be used to detect the absence or presence of key signaling oncogenic aberrations [52]. For example, CD47 was found to be the only overexpressed protein on CTCs and can inhibit the toxicity and phagocytosis of immune cells [58]. Inhibition of the CD47 ligand and the receptor SIRPa can inhibit T cell and macrophage functions [59]. Therefore, CD47 is related to CTC immune- escape, and its upregulation permits survival of CTCs [57].
Molecular detection approaches such as polymerase chain reaction (PCR), reverse-transcription PCR (RT-PCR) and quantitative RT-PCR (RT-qPCR) can target nucleic acids to identify markers between tumor cells and nucleated hematopoietic cells. PCR uses genomic DNA as a starting material which is a high sensitivity approach. It can be used when the tumor cells have a specific mutation or when the mutation is already known [60]. However, its sensitivity is limited by the heterogeneity of CTCs. RT-PCR is used to detect tissue-specific mRNA. It is used to identify tumor-associated biomarkers such as EpCAM and stem cell biomarkers such as ALDH1. It can also identify EMT-associated transcripts such as PI3Kα and Twist1 [61]. Moreover, RT-qPCR can define a cut-off value for the marker transcript and compare it to the expression of the transcript in normal cells. However, the cut-off thresholds may not be applicable in individual patients; for example, it was not possible to set up a cut-off value to distinguish mRNA from illegitimate transcription in peripheral blood leukocytes and mRNA from tumor cells [62].
Clinical significance of CTCs
Methods for studying the spread of human tumors are very limited and human cancer metastasis is completely different from that in animal models. In such a setting, isolation and characterization of CTCs hold tremendous potential for new biological insights [63]. To date, many researchers have assessed the clinical value of CTC analyses and it has been proven that CTC analyses can provide significant prognostic information in breast cancer [13], prostate cancer [64] and colorectal cancer [65]. However, the primary challenge of CTC technologies is monitoring minimal residual disease in patients without signs of overt metastasis [66]. The analysis of higher blood volumes and more sensitive methods might be required to increase the reliability of CTC measurements.
The most exciting application of CTC detection technologies is the monitoring of systemic therapies. Molecular characterization of CTCs may be critical for the identification of therapeutic targets and for more “tailored” and personalized anti-metastatic therapies [17]. Such targeted therapies match the drug to the patient at different stages of disease progression [63]. Liquid biopsy, a method for analyzing CTCs in the peripheral blood, might become a less invasive and more cost-effective alternative to tissue biopsy [67]. It is a ‘real time’ noninvasive method that can be repeated during treatment to monitor the acquisition of new genetic abnormalities [63]. Metastatic tumor cells that spread to different organs can be used to construct CTC libraries and their analysis may provide helpful information on useful targets and resistance mechanisms related to systemic anticancer therapies [68].
In the new era of gene targeted cancer therapies, genetic analysis of tumor cells has become critical. Longitudinal monitoring of CTC-derived genotypes may provide an approach to identify drug- sensitivity and resistance-associated markers to guide treatment decisions [63]. A subgroup of NSCLC patients provided a powerful example of the application of CTC genotyping in targeted cancer therapy and showed a selective response to the EGFR tyrosine kinase inhibitor gefitinib. Most patients with NSCLC have no response to therapies targeting EGFR, and only 10 percent of patients have somatic activating mutations in EGFR. It is likely that patients can exhibit a response to selective EGFR kinase inhibitors [69].
In summary, the characterization of CTCs may have a significant influence in future clinical trials as a companion diagnostics strategy to new targeted therapies [68].
Conclusion
In the past few years, the field of CTCs has made great advances, and researchers are becoming increasingly aware of the importance of CTCs in cancer treatment, including recognizing the abilities of CTCs in cancer prediction, diagnosis and prognosis. Further elucidation of the characteristic of CTCs may provide vital insights into the mechanisms of metastasis and may contribute to the identification of novel targeted therapies. Based on the technology of the “liquid biopsy”, numerous targetable cellular markers of CTCs have been identified, which provides great advances for CTC characterization. Liquid biopsy can be an informative tool to provide comprehensive information for diagnosis through an optional and noninvasive procedure. These advances have revolutionized cancer detection and management. The information gained from these approaches can improve the selection of appropriate therapies and obtain further insights into the therapy-induced selection of tumor cells.
Analysis of the biological and molecular characteristics of CTCs might be vital to therapeutic decision and contribute to more personalized anti-metastatic therapies. It may provide an important understanding of tumor progression and can help to overcome the limitations of single biopsies, which cannot accurately describe the genomic landscape of cancer, and biopsy methods for multiple metastases [52].
In summary, if we can develop technology to detect the most aggressive subsets of CTCs by increasing assay analytical sensitivity, early detection and eradication of these CTCs may decrease cancer mortality. In addition, new therapy targets during the metastasis of CTCs can also be used to isolate and identify subpopulations of tumor cells. Focusing routine molecular research on rare circulating tumor cells, targeting cellular markers of CTCs and discovering new cellular markers may improve the management and prevention of metastatic disease in the near future.
Acknowledgements
This work was supported by the National Natural Science Foundation (81202115), Doctoral Innovation Fund of Shanghai Jiaotong University School of Medicine (No. BXJ201732).
Disclosure of conflict of interest
None.
References
- 1.ENGELL HC. Cancer cells in the circulating blood; a clinical study on the occurrence of cancer cells in the peripheral blood and in venous blood draining the tumour area at operation. Acta Chir Scand Suppl. 1955;201:1–70. [PubMed] [Google Scholar]
- 2.Hofman V, Bonnetaud C, Ilie MI, Vielh P, Vignaud JM, Fléjou JF, Lantuejoul S, Piaton E, Mourad N, Butori C, Selva E, Poudenx M, Sibon S, Kelhef S, Vénissac N, Jais JP, Mouroux J, Molina TJ, Hofman P. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res. 2011;17:827–835. doi: 10.1158/1078-0432.CCR-10-0445. [DOI] [PubMed] [Google Scholar]
- 3.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC Jr. American society of clinical oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J. Clin. Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 4.Cui L, Kwong J, Wang C. Prognostic value of circulating tumor cells and disseminated tumor cells in patients with ovarian cancer: a systematic review and meta-analysis. J Ovarian Res. 2015;8:38. doi: 10.1186/s13048-015-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pantel K, Alix-Panabières C. Liquid biopsy and minimal residual disease-latest advances and implications for cure. Nat Rev Clin Oncol. 2019;16:409–424. doi: 10.1038/s41571-019-0187-3. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Wu C, Lu C, Hsieh J, Wu D, Huang S, Lin S. Molecular detection of circulating tumor cells in the peripheral blood of patients with colorectal cancer using RT-PCR: significance of the prediction of postoperative metastasis. World J Surg. 2006;30:1007–1013. doi: 10.1007/s00268-005-0485-z. [DOI] [PubMed] [Google Scholar]
- 7.Masuda T, Hayashi N, Iguchi T, Ito S, Eguchi H, Mimori K. Clinical and biological significance of circulating tumor cells in cancer. Mol Oncol. 2016;10:408–417. doi: 10.1016/j.molonc.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salvianti F, Pazzagli M, Pinzani P. Single circulating tumor cell sequencing as an advanced tool in cancer management. Expert Rev Mol Diagn. 2016;16:51–63. doi: 10.1586/14737159.2016.1116942. [DOI] [PubMed] [Google Scholar]
- 9.Moon DH, Lindsay DP, Hong S, Wang AZ. Clinical indications for, and the future of, circulating tumor cells. Adv Drug Deliv Rev. 2018;125:143–150. doi: 10.1016/j.addr.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Micalizzi DS, Maheswaran S, Haber DA. A conduit to metastasis: circulating tumor cell biology. Genes Dev. 2017;31:1827–1840. doi: 10.1101/gad.305805.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Praharaj PP, Bhutia SK, Nagrath S, Bitting RL, Deep G. Circulating tumor cell-derived organoids: current challenges and promises in medical research and precision medicine. Biochim Biophys Acta Rev Cancer. 2018;1869:117–127. doi: 10.1016/j.bbcan.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen Z, Wu A, Chen X. Current detection technologies for circulating tumor cells. Chem Soc Rev. 2017;46:2038–2056. doi: 10.1039/c6cs00803h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cristofanilli M, Budd G, Ellis M, Stopeck A, Matera J, Miller M, Reuben J, Doyle G, Allard W, Terstappen L, Hayes D. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 14.Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11:R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beltran H, Jendrisak A, Landers M, Mosquera J, Kossai M, Louw J, Krupa R, Graf R, Schreiber N, Nanus D, Tagawa S, Marrinucci D, Dittamore R, Scher H. The initial detection and partial characterization of circulating tumor cells in neuroendocrine prostate cancer. Clin Cancer Res. 2016;22:1510–1519. doi: 10.1158/1078-0432.CCR-15-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poudineh M, Aldridge P, Ahmed S, Green B, Kermanshah L, Nguyen V, Tu C, Mohamadi R, Nam R, Hansen A, Sridhar S, Finelli A, Fleshner N, Joshua A, Sargent E, Kelley S. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat Nanotechnol. 2017;12:274–281. doi: 10.1038/nnano.2016.239. [DOI] [PubMed] [Google Scholar]
- 17.Joosse S, Gorges T, Pantel K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol Med. 2015;7:1–11. doi: 10.15252/emmm.201303698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satelli A, Mitra A, Brownlee Z, Xia X, Bellister S, Overman MJ, Kopetz S, Ellis LM, Meng QH, Li S. Epithelial-mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clin Cancer Res. 2015;21:899–906. doi: 10.1158/1078-0432.CCR-14-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaffer C, Weinberg R. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 21.Strilic B, Offermanns S. Intravascular survival and extravasation of tumor cells. Cancer Cell. 2017;32:282–293. doi: 10.1016/j.ccell.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaffer C, Thompson E, Williams E. Mesenchymal to epithelial transition in development and disease. Cells Tissues Organs (Print) 2007;185:7–19. doi: 10.1159/000101298. [DOI] [PubMed] [Google Scholar]
- 24.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Zhang X, Li J, Sun B, Qian H, Yin Z. The biological and clinical importance of epithelial-mesenchymal transition in circulating tumor cells. J Cancer Res Clin Oncol. 2015;141:189–201. doi: 10.1007/s00432-014-1752-x. [DOI] [PubMed] [Google Scholar]
- 26.Arumugam T, Ramachandran V, Fournier KF, Wang H, Marquis L, Abbruzzese JL, Gallick GE, Logsdon CD, McConkey DJ, Choi W. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 2009;69:5820–5828. doi: 10.1158/0008-5472.CAN-08-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comaills V, Kabeche L, Morris R, Buisson R, Yu M, Madden MW, LiCausi JA, Boukhali M, Tajima K, Pan S, Aceto N, Sil S, Zheng Y, Sundaresan T, Yae T, Jordan NV, Miyamoto DT, Ting DT, Ramaswamy S, Haas W, Zou L, Haber DA, Maheswaran S. Genomic instability is induced by persistent proliferation of cells undergoing epithelial-to-mesenchymal transition. Cell Rep. 2016;17:2632–2647. doi: 10.1016/j.celrep.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lou Y, Diao L, Cuentas ER, Denning WL, Chen L, Fan YH, Byers LA, Wang J, Papadimitrakopoulou VA, Behrens C, Rodriguez JC, Hwu P, Wistuba II, Heymach JV, Gibbons DL. Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin Cancer Res. 2016;22:3630–3642. doi: 10.1158/1078-0432.CCR-15-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grünert S, Jechlinger M, Beug H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol. 2003;4:657–665. doi: 10.1038/nrm1175. [DOI] [PubMed] [Google Scholar]
- 30.Sun B, Zhang X, Cheng X, Zhang Y, Chen L, Shi L, Liu Z, Qian H, Wu M, Yin Z. Intratumoral hepatic stellate cells as a poor prognostic marker and a new treatment target for hepatocellular carcinoma. PLoS One. 2013;8:e80212. doi: 10.1371/journal.pone.0080212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smit M, Geiger T, Song J, Gitelman I, Peeper D. A Twist-Snail axis critical for TrkB-induced epithelial-mesenchymal transition-like transformation, anoikis resistance, and metastasis. Mol Cell Biol. 2009;29:3722–3737. doi: 10.1128/MCB.01164-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rejniak K. Circulating tumor cells: when a solid tumor meets a fluid microenvironment. Adv Exp Med Biol. 2016;936:93–106. doi: 10.1007/978-3-319-42023-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hugo H, Ackland M, Blick T, Lawrence M, Clements J, Williams E, Thompson E. Epithelial--mesenchymal and mesenchymal--epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 34.Zoccoli A, Iuliani M, Pantano F, Imperatori M, Intagliata S, Vincenzi B, Marchetti P, Papapietro N, Denaro V, Tonini G, Santini D. Premetastatic niche: ready for new therapeutic interventions? Expert Opin Ther Targets. 2012;16(Suppl 2):S119–129. doi: 10.1517/14728222.2012.656092. [DOI] [PubMed] [Google Scholar]
- 35.Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, Psaila B, Kaplan RN, Bromberg JF, Kang Y, Bissell MJ, Cox TR, Giaccia AJ, Erler JT, Hiratsuka S, Ghajar CM, Lyden D. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer. 2017;17:302–317. doi: 10.1038/nrc.2017.6. [DOI] [PubMed] [Google Scholar]
- 36.Kaplan R, Rafii S, Lyden D. Preparing the “soil”: the premetastatic niche. Cancer Res. 2006;66:11089–11093. doi: 10.1158/0008-5472.CAN-06-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minn A, Gupta G, Siegel P, Bos P, Shu W, Giri D, Viale A, Olshen A, Gerald W, Massagué J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang Y, Siegel P, Shu W, Drobnjak M, Kakonen S, Cordón-Cardo C, Guise T, Massagué J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 39.Hou J, Krebs M, Lancashire L, Sloane R, Backen A, Swain R, Priest L, Greystoke A, Zhou C, Morris K, Ward T, Blackhall F, Dive C. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J. Clin. Oncol. 2012;30:525–532. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 40.Sarioglu AF, Aceto N, Kojic N, Donaldson MC, Zeinali M, Hamza B, Engstrom A, Zhu H, Sundaresan TK, Miyamoto DT, Luo X, Bardia A, Wittner BS, Ramaswamy S, Shioda T, Ting DT, Stott SL, Kapur R, Maheswaran S, Haber DA, Toner M. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat Methods. 2015;12:685–691. doi: 10.1038/nmeth.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Au SH, Edd J, Stoddard AE, Wong KHK, Fachin F, Maheswaran S, Haber DA, Stott SL, Kapur R, Toner M. Microfluidic isolation of circulating tumor cell clusters by size and asymmetry. Sci Rep. 2017;7:2433. doi: 10.1038/s41598-017-01150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabisiewicz A, Grzybowska E. CTC clusters in cancer progression and metastasis. Med Oncol. 2017;34:12. doi: 10.1007/s12032-016-0875-0. [DOI] [PubMed] [Google Scholar]
- 43.Duda D, Duyverman A, Kohno M, Snuderl M, Steller E, Fukumura D, Jain R. Malignant cells facilitate lung metastasis by bringing their own soil. Proc Natl Acad Sci U S A. 2010;107:21677–21682. doi: 10.1073/pnas.1016234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Upreti M, Jamshidi-Parsian A, Koonce N, Webber J, Sharma S, Asea A, Mader M, Griffin R. Tumor-endothelial cell three-dimensional spheroids: new aspects to enhance radiation and drug therapeutics. Transl Oncol. 2011;4:365–376. doi: 10.1593/tlo.11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aceto N, Bardia A, Miyamoto D, Donaldson M, Wittner B, Spencer J, Yu M, Pely A, Engstrom A, Zhu H, Brannigan B, Kapur R, Stott S, Shioda T, Ramaswamy S, Ting D, Lin C, Toner M, Haber D, Maheswaran S. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lecharpentier A, Vielh P, Perez-Moreno P, Planchard D, Soria JC, Farace F. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br J Cancer. 2011;105:1338–1341. doi: 10.1038/bjc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou JM, Krebs M, Ward T, Sloane R, Priest L, Hughes A, Clack G, Ranson M, Blackhall F, Dive C. Circulating tumor cells as a window on metastasis biology in lung cancer. Am J Pathol. 2011;178:989–996. doi: 10.1016/j.ajpath.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krebs MG, Hou JM, Sloane R, Lancashire L, Priest L, Nonaka D, Ward TH, Backen A, Clack G, Hughes A, Ranson M, Blackhall FH, Dive C. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol. 2012;7:306–315. doi: 10.1097/JTO.0b013e31823c5c16. [DOI] [PubMed] [Google Scholar]
- 50.Bithi SS, Vanapalli SA. Microfluidic cell isolation technology for drug testing of single tumor cells and their clusters. Sci Rep. 2017;7:41707. doi: 10.1038/srep41707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.May A, Crawford B, Nedelcu A. Model-systems to understand the biology and clinical significance of circulating tumor cell clusters. Front Oncol. 2018;8:63. doi: 10.3389/fonc.2018.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mateo J, Gerlinger M, Rodrigues DN, de Bono JS. The promise of circulating tumor cell analysis in cancer management. Genome Biol. 2014;15:448. doi: 10.1186/s13059-014-0448-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geigl JB, Speicher MR. Single-cell isolation from cell suspensions and whole genome amplification from single cells to provide templates for CGH analysis. Nat Protoc. 2007;2:3173–3184. doi: 10.1038/nprot.2007.476. [DOI] [PubMed] [Google Scholar]
- 54.Mathiesen RR, Fjelldal R, Liestøl K, Due EU, Geigl JB, Riethdorf S, Borgen E, Rye IH, Schneider IJ, Obenauf AC, Mauermann O, Nilsen G, Christian Lingjaerde O, Børresen-Dale AL, Pantel K, Speicher MR, Naume B, Baumbusch LO. High-resolution analyses of copy number changes in disseminated tumor cells of patients with breast cancer. Int J Cancer. 2012;131:E405–415. doi: 10.1002/ijc.26444. [DOI] [PubMed] [Google Scholar]
- 55.Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z, Wittner BS, Stojanov P, Brachtel E, Sgroi D, Kapur R, Shioda T, Ting DT, Ramaswamy S, Getz G, Iafrate AJ, Benes C, Toner M, Maheswaran S, Haber DA. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chien C, Lin S, Lai Y, Lin B, Lin S, Lee J, Tsai S. Regulation of CD151 by hypoxia controls cell adhesion and metastasis in colorectal cancer. Clin Cancer Res. 2008;14:8043–8051. doi: 10.1158/1078-0432.CCR-08-1651. [DOI] [PubMed] [Google Scholar]
- 57.Steinert G, Schölch S, Niemietz T, Iwata N, García S, Behrens B, Voigt A, Kloor M, Benner A, Bork U, Rahbari N, Büchler M, Stoecklein N, Weitz J, Koch M. Immune escape and survival mechanisms in circulating tumor cells of colorectal cancer. Cancer Res. 2014;74:1694–1704. doi: 10.1158/0008-5472.CAN-13-1885. [DOI] [PubMed] [Google Scholar]
- 58.Kim M, Lee J, Lee J, Kim S, Lee S, Park S, Sung M, Heo D. Association of CD47 with natural killer cell-mediated cytotoxicity of head-and-neck squamous cell carcinoma lines. Tumour Biol. 2008;29:28–34. doi: 10.1159/000132568. [DOI] [PubMed] [Google Scholar]
- 59.Avice M, Rubio M, Sergerie M, Delespesse G, Sarfati M. CD47 ligation selectively inhibits the development of human naive T cells into Th1 effectors. J Immunol. 2000;165:4624–4631. doi: 10.4049/jimmunol.165.8.4624. [DOI] [PubMed] [Google Scholar]
- 60.Zieglschmid V, Hollmann C, Böcher O. Detection of disseminated tumor cells in peripheral blood. Crit Rev Clin Lab Sci. 2005;42:155–196. doi: 10.1080/10408360590913696. [DOI] [PubMed] [Google Scholar]
- 61.Krawczyk N, Meier-Stiegen F, Banys M, Neubauer H, Ruckhaeberle E, Fehm T. Expression of stem cell and epithelial-mesenchymal transition markers in circulating tumor cells of breast cancer patients. Biomed Res Int. 2014;2014:415721. doi: 10.1155/2014/415721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schuster R, Max N, Mann B, Heufelder K, Thilo F, Gröne J, Rokos F, Buhr H, Thiel E, Keilholz U. Quantitative real-time RT-PCR for detection of disseminated tumor cells in peripheral blood of patients with colorectal cancer using different mRNA markers. Int J Cancer. 2004;108:219–227. doi: 10.1002/ijc.11547. [DOI] [PubMed] [Google Scholar]
- 63.Maheswaran S, Haber D. Circulating tumor cells: a window into cancer biology and metastasis. Curr Opin Genet Dev. 2010;20:96–99. doi: 10.1016/j.gde.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Bono J, Scher H, Montgomery R, Parker C, Miller M, Tissing H, Doyle G, Terstappen L, Pienta K, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 65.Cohen S, Punt C, Iannotti N, Saidman B, Sabbath K, Gabrail N, Picus J, Morse M, Mitchell E, Miller M, Doyle G, Tissing H, Terstappen L, Meropol N. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 66.Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16:398–406. doi: 10.1016/j.molmed.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 67.Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 68.Wan L, Pantel K, Kang Y. Tumor metastasis: moving new biological insights into the clinic. Nat Med. 2013;19:1450–1464. doi: 10.1038/nm.3391. [DOI] [PubMed] [Google Scholar]
- 69.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 70.Micalizzi DS, Maheswaran S, Haber DA. A conduit to metastasis: circulating tumor cell biology. Genes Dev. 2017;31:1827–1840. doi: 10.1101/gad.305805.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakaizawa K, Goto Y, Kiniwa Y, Uchiyama A, Harada K, Shimada S, Saida T, Ferrone S, Takata M, Uhara H, Okuyama R. Mutation analysis of BRAF and KIT in circulating melanoma cells at the single cell level. Br J Cancer. 2012;106:939–946. doi: 10.1038/bjc.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Costa A, Kieffer Y, Scholer-Dahirel A, Pelon F, Bourachot B, Cardon M, Sirven P, Magagna I, Fuhrmann L, Bernard C, Bonneau C, Kondratova M, Kuperstein I, Zinovyev A, Givel AM, Parrini MC, Soumelis V, Vincent-Salomon A, Mechta-Grigoriou F. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33:463–479. e410. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 73.Zahreddine H, Borden KL. Mechanisms and insights into drug resistance in cancer. Front Pharmacol. 2013;4:28. doi: 10.3389/fphar.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu J, Liang L, Jiao Y, Liu L U.S.-China Physical Sciences-Oncology Alliance. Enhanced invasion of metastatic cancer cells via extracellular matrix interface. PLoS One. 2015;10:e0118058. doi: 10.1371/journal.pone.0118058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salvianti F, Pazzagli M, Pinzani P. Single circulating tumor cell sequencing as an advanced tool in cancer management. Expert Rev Mol Diagn. 2016;16:51–63. doi: 10.1586/14737159.2016.1116942. [DOI] [PubMed] [Google Scholar]
- 76.Catherine AP, Klaus P. Circulating tumor cells: liquid biopsy of cancer. Clinical Chemistry. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 77.Cui CH, Chen RH, Zhai DY, Xie L, Qi J, Yu JL. Detection of FAM172A expressed in circulating tumor cells is a feasible method to predict high-risk subgroups of colorectal cancer. Tumour Biol. 2017;39:1010428317699126. doi: 10.1177/1010428317699126. [DOI] [PubMed] [Google Scholar]
- 78.Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, Ulman MJ, Xega K, Contino G, Alagesan B, Brannigan BW, Milos PM, Ryan DP, Sequist LV, Bardeesy N, Ramaswamy S, Toner M, Maheswaran S, Haber DA. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paolillo C, Mu Z, Rossi G, Schiewer MJ, Nguyen T, Austin L, Capoluongo E, Knudsen K, Cristofanilli M, Fortina P. Detection of activating estrogen receptor gene (ESR1) mutations in single circulating tumor cells. Clin Cancer Res. 2017;23:6086–6093. doi: 10.1158/1078-0432.CCR-17-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tellez-Gabriel M, Heymann MF, Heymann D. Circulating tumor cells as a tool for assessing tumor heterogeneity. Theranostics. 2019;9:4580–4594. doi: 10.7150/thno.34337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alix-Panabières C. EPISPOT assay: detection of viable DTCs/CTCs in solid tumor patients. Recent Results Cancer Res. 2012;195:69–76. doi: 10.1007/978-3-642-28160-0_6. [DOI] [PubMed] [Google Scholar]
- 84.Somlo G, Lau SK, Frankel P, Hsieh HB, Liu X, Yang L, Krivacic R, Bruce RH. Multiple biomarker expression on circulating tumor cells in comparison to tumor tissues from primary and metastatic sites in patients with locally advanced/inflammatory, and stage IV breast cancer, using a novel detection technology. Breast Cancer Res Treat. 2011;128:155–163. doi: 10.1007/s10549-011-1508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J, Arora KS, Desai N, Dahl DM, Sequist LV, Smith MR, Kapur R, Wu CL, Shioda T, Ramaswamy S, Ting DT, Toner M, Maheswaran S, Haber DA. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]


