Abstract
Oncogenic KRAS mutations are frequently found in non-small cell lung carcinoma (NSCLC) and cause constitutive activation of the MEK-ERK pathway. Many cancer types have been shown to overexpress PD-L1 to escape immune surveillance. FRA1 is a MEK/ERK-dependent oncogenic transcription factor and a member of the AP-1 transcriptional factor superfamily. This study assesses the hypothesis that KRAS mutation directly regulates PD-L1 expression through the MEK-ERK pathway mediated by FRA1. Premalignant human bronchial epithelial cell (HBEC) lines harboring the KRAS mutationV12, EGFR mutation, p53 knock-down, or both KRAS mutation and p53 knock-down were tested for levels of PD-L1, FRA1, and ERK activation (pERK). Our results showed that KRAS mutation alone, but not other genetic alterations, induced significantly higher expression of PD-L1 compared to its vector counterparts. The increased PD-L1 expression in the KRAS mutated cells was dramatically reduced by inhibition of ERK activation. Furthermore, the MEK-ERK pathway-dependent PD-L1 expression was markedly reduced by FRA1 silencing. Interestingly, FRA1 silencing led to inhibition of ERK activation, indicating that FRA1 plays a role in PD-L1 regulation via positive feedback of ERK activation. Correlation of PD-L1 and FRA1 mRNA expression was validated using human lung cancer specimens from The Cancer Genome Atlas (TCGA) and established NSCLC cell lines from Cancer Cell Line Encyclopedia (CCLE). FRA1 expression was significantly associated with PD-L1 expression, and high FRA1 expression was correlated with poor overall survival. Our findings suggest that oncogenic KRAS-driven PD-L1 expression is dependent on MEK-ERK and FRA1 in high risk, premalignant HBEC.
Keywords: FRA1, MEK-ERK pathway, PD-L1, KRAS, premalignant human bronchial epithelial cells
Introduction
KRAS oncogenic driver mutations occur in about 30% of patients with non-small cell lung cancer (NSCLC) and have been associated with poorer disease-free survival (DFS) and overall survival (OS) [1]. Programmed death ligand-1 (PD-L1) is overexpressed in many cancer types including lung cancer and plays a prominent role in immune resistance [2]. Recent clinical trials in NSCLC have shown promising results from immunotherapy by PD-1/PD-L1 checkpoint blockade [3]. Rizvi et al. reported that the clinical efficacy of PD-1/PD-L1 inhibition therapy is associated with a higher nonsynonymous tumor mutation burden (TMB) [4]. Interestingly, this study also found that tumors from 50% of patients with durable clinical benefit had KRAS mutation compared to only ~5% of tumors from patients with non-durable benefits. Although tumoral PD-L1 expression is a predictor of response to PD-1/PD-L1 blockade therapy [3], patients with PD-L1 positive tumors do not necessarily respond, and some responses occur in patients with PD-L1 weak or negative tumors [3,4]. The mechanistic relationship between TMB and PD-L1 expression that predicts clinical efficacy of PD-1/PD-L1 blockade therapy remain poorly understood [4]. Also, the use of tumoral PD-L1 positivity as a predictive biomarker for PD-1/PD-L1 blockade therapy is limited by the multitude of PD-L1 antibodies, assays, scoring systems, and varying clinical cutoffs of PD-L1 expression in clinical trials. As such, we are in need of pre-treatment biomarkers that can predict response to immunotherapy in oncology.
KRAS mutation has been known to activate the MEK-ERK pathway in many cancers [5]. FOS-related antigen 1 (FRA1) encoded by the FOS-like antigen-1 (FOSL1) gene is an oncogenic transcription factor and a member of the AP-1 family [6]. FRA1 is upregulated in many malignancies including lung cancer and plays an important role in lung carcinogenesis [7]. Under stimulated conditions, FRA1 is among the most highly upregulated transcriptional targets [8], and ERK activation is required for FRA1 accumulation [9]. Importantly, ectopic FRA1 expression in pulmonary malignant epithelial cell line was sufficient to enhance cell motility, invasion, and anchorage-independent growth as well as tumor growth and lung metastases [10].
Recent studies demonstrated that the MEK-ERK pathway was involved in the upregulation or posttranscriptional regulation of PD-L1 by KRAS [11-13]. However, the molecular basis of the upregulation remains largely unknown. Delineating the mechanisms by which PD-L1 expression is induced may lead to the identification of complementary biomarkers for increased efficacy to PD-1/PD-L1 blockade therapy. As such, our goal was to understand the mechanism of PD-L1 induction in NSCLC patients with KRAS mutation. We hypothesized that KRAS mutation could induce PD-L1 expression via MEK-ERK dependent oncogenic transcription factors, such as FRA1. In order to study this hypothesis and to avoid the multitude of tumor mutations coexistent in cancer cells, we used premalignant, high-risk human bronchial epithelial cell lines (HBEC) that expressed mutant KRAS, EGFR, or p53 knock-down [14,15]. These mutations in HBEC were not sufficient to confer a fully malignant phenotype [15]. Here, we report that oncogenic KRAS mutation-driven PD-L1 expression was dependent on FRA1-mediated ERK activation in HBEC, and KRAS mutation was also associated with increased PD-L1 expression in human NSCLC tissues and cell lines. Our findings suggest that KRAS mutation associated PD-L1 expression may be a mechanism that promotes tumor immune escape. Furthermore, our data support the concept of KRAS mutation directly driving PD-L1 expression via FRA1-mediated ERK activation rather than merely representing a surrogate marker of TMB or tumor antigenicity.
Materials and methods
Cells and culture conditions
All immortalized HBEC lines were provided by Dr. John D. Minna at the University of Texas, Southwestern Medical Center. The cells were immortalized without viral oncoproteins via ectopic expression of human telomerase (hTERT) and cyclin-dependent kinase 4 under control of puromycin and geneticin, respectively [16]. HBEC3 was subsequently manipulated to stably express the vector control (HBEC3 vector) or an activating point-mutation of the K-RAS proto-oncogene (K-RASV12; HBEC3 KRAS), alone or in combination with stable knockdown of the P53 tumor suppressor gene (HBEC3 KRAS/P53) [14]. HBEC3 cell line designated HBEC3/EGFR L858R and HBEC3/EGFR wild type [15] were also used. HBECs were cultured in Keratinocyte Serum-Free Media (Life Technologies) supplemented with 30 μg/mL Bovine Pituitary Extract and 0.2 ng/mL recombinant Epidermal Growth Factor (Life Technologies).
Flow cytometry
Cells were fixed with 2% paraformaldehyde in PBS for 15 min on ice and were washed with PBS and were stained with anti-human PD-L1 PE-conjugated monoclonal antibody or isotype control antibody listed in Table S1 for 30 min on ice and washed with PBS. Data were analyzed using a FACSCalibur™ and FlowJo software (Tree Star) and shown as normalized mean fluorescent intensity (MFI). MFI of PD-L1 stained sample was normalized to MFI of isotype control.
Western blot analysis
Preparation of total cell lysates and Western blotting were performed as described previously [17] using the primary and secondary antibodies as listed in Table S1. Primary antibodies were incubated overnight at 4°C. After incubating with primary antibodies, the membranes were washed with PBS containing 0.1% Tween 20 (PBST) three times. Then the membranes were incubated for 1 hour with IRDye800CW-conjugated goat anti-mouse IgG (1:15,000) and IRDye680RD-conjugated goat anti-rabbit IgG (1:15,000) secondary antibodies (LI-COR Biosciences, Lincoln, NE) diluted in PBST containing 5% BSA. The blots were then washed three times with PBST and rinsed with PBS. Proteins were visualized by scanning the membrane on an Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE) with both 700- and 800-nm channels. The blots were re-probed with the anti-GAPDH or anti-α-tubulin as internal loading controls. The relative densitometric values above the Figures were calculated using Image J software. The values were normalized to internal loading controls. Relative values were obtained using the values from the untreated groups.
Immunofluorescent staining
Immunofluorescent stainings were performed as previously described [18]. Antibodies used are listed in Table S1. For quantification, all images were acquired using the same photodetector gain and intensity to aid in quantitative comparisons of the relative fluorescence intensities (a measure of PD-L1 immunoreactivity) of different samples. All cells in the field of view for each experiment were scored. For the analysis of the fluorescence intensity, Image J (http://rsbweb.nih.gov/ij/) was used. Individual cells were selected by using a selection tool, followed by the use of the analyze menu to measure the area integrated intensity and mean grey value. This step was repeated for all cells in the field of view. Blank areas were selected, and the intensities used for data normalization. Quantified data are expressed as the mean ± SEM values. Significance testing was conducted via Student’s t-test. Calculated p-values are indicated in the Figure as follows: *: P ≤ 0.05, **: P ≤ 0.01; ***: P ≤ 0.001.
Quantitative real-time PCR
Quantitative real-time PCR was performed as previously described [19]. Total RNA was extracted using the Quick-RNA MiniPrep (Zymo Research, Irvine, CA) and reverse transcription reactions were performed using the High-Capacity RNA-to-cDNA (Applied Biosystems, Grand Island, NY) according to the manufacturer’s instructions. Transcript levels were measured using the Fast start universal SYBR green master mix (Roche, Pleasanton, CA) by iCycler thermal cycler (Bio-Rad, Hercules, CA). The primers are listed in Table S2. Data were normalized to β-actin levels in the samples in triplicates. mRNA expression is shown as 2-delta Ct calculated using the following equations: delta Ct (gene) = Ct (gene, sample) - Ct (gene, control).
Transient transfection of FRA1 siRNA transfection
Transient transfections were carried out on 50-60% confluent HBEC cells as previously described [18]. For each well of a 6-well plate, 100 nM final concentration of siRNA duplex in OPTIMEM (Invitrogen) was added. The transfection efficiency was evaluated by western blotting for FRA1. The FRA1 and control siRNAs are listed in Table S2.
TCGA and RNA seq analysis
Data from The Cancer Genome Atlas for Lung Adenocarcinoma (TCGA LUAD) were downloaded from the TCGA portal (http://tcga-data.nci.nih.gov/). Somatic mutations in KRAS were retrieved from cbioPortal (http://www.cbioportal.org/). RNA-seq analysis for 144 NSCLC cell lines from John D. Minna’s laboratory was performed as previously described [20]. The Spearman’s rank-order or Pearson’s correlation tests were applied to measure the strength of the association between PDL1 (CD274) and FRA1 (FOSL1) mRNA levels.
Immunohistochemistry
Lung tumor tissues removed from two patients, S11-21171 and S13-28321, who carried KRAS mutation with positive PD-L1 expression and KRAS wild type with PD-L1 negative expression, respectively, were embedded in paraffin, and micro-sectioned onto slides. Immunohistochemistry (IHC) was performed as previously described [19] using primary antibodies, PD-L1, and FRA1 listed in Table S1. The paraffin slides were placed in xylene to remove paraffin, followed by ethanol. Following a wash in tap water, the slides were incubated in 3% Hydrogen peroxide/methanol solution for 10 minutes and were baked for 1 hour at 65°C. Antigen retrieval was performed in a pressure cooker for 5 minutes with Tris-EDTA pH9 buffer and then cooled for 15 minutes at room temperature. Immunohistochemistry (IHC) was performed using a BOND III staining system (Leica Microsystems) programmed for primary antibodies. in Bond Antibody Diluent for 60 minutes, Polymer for 15 minutes, Peroxidase block for 5 minutes, DAB for 10 minutes, and Hematoxylin for 5 minutes followed by 0.5% cupric sulfate for 10 minutes with bond washes between steps. The Bond Refine Polymer Detection kit (DS9800) was utilized for all steps after primary antibody exposure. The staining results were digitally scanned at ×200 magnification using an Aperio CS2 whole slide scanner from Leica Biosystems. The images were visualized with ImageScope software and analyzed with the Aperio Image analysis toolkit (Leica Biosystems). Cellular expression of PD-L1 and FRA1 in tumor cells were analyzed using the Leica Image Analysis and membranous and cytoplasmic algorithms, respectively. The results were shown as the scored intensity of staining no (0), weak (1+), moderate (2+), or strong (3+), and the overall percentage of cell staining. Three regions of interest (ROI), representing average staining were evaluated in each tissue. The ROIs for each tissue totaled over 300,000 µm2. The ROIs were selected by a pathologist, blinded to patient data, and were areas free of necrosis with at least 90% tumor cells. Based on a side by side analysis, the ROIs were placed in the same location on each stain for each tissue.
Statistical analysis
Samples were plated/run in triplicate and all experiments were performed at least two or three times. The significance of the difference between groups was evaluated with Student’s t-test. P < 0.05 was considered statistically significant.
Results
Oncogenic KRAS mutation, but not EGFR mutation and p53 knock-down, induced PD-L1 expression in premalignant HBEC cell lines
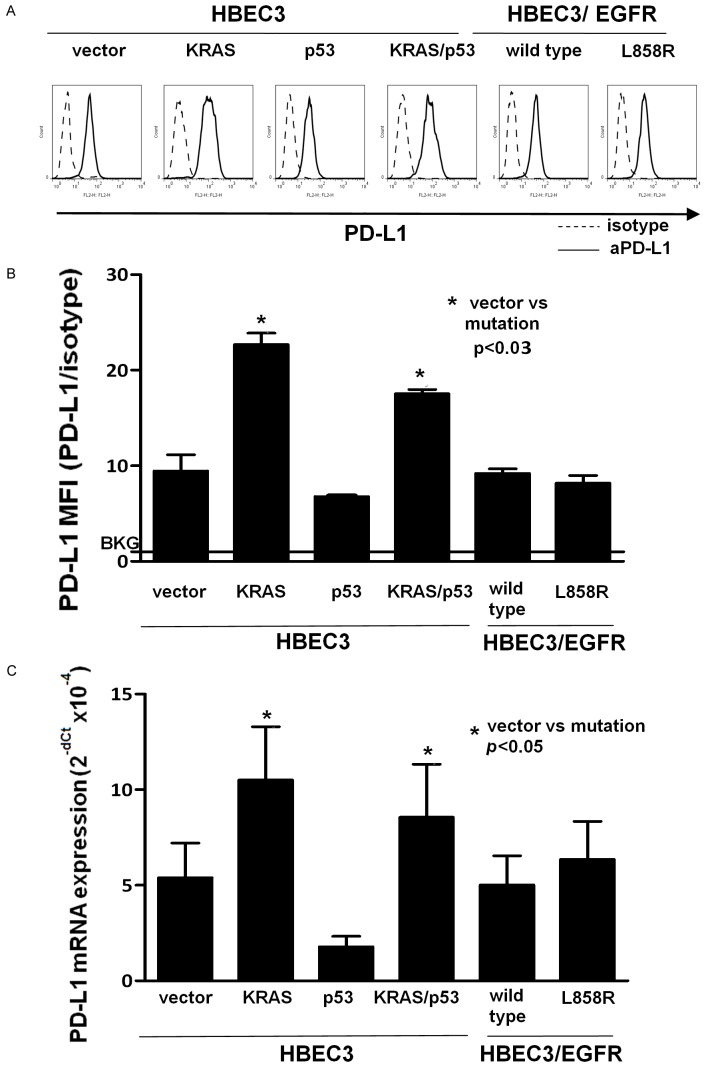
To evaluate the effect of common oncogenic driver mutations on PD-L1 expression, we examined PD-L1 expression in mutant KRASG12V (HBEC3/KRAS), knock-down of p53 (HBEC3/p53), KRAS mutation and knock-down of p53 (HBEC3/KRAS/p53), and mutant EGFR (HBEC3/L858R) HBEC3 cell lines. PD-L1 surface expression was determined by flow cytometry in all the HBEC cell lines (Figure 1A). There was a correlation between PD-L1 surface protein and mRNA expression levels in all the cell lines (Figure 1A-C). PD-L1 protein and mRNA expression were significantly increased by nearly 2-fold in HBEC3/KRAS and HBEC3/KRAS/p53 cells compared to wild type (HBEC3/vector) (Figure 1B and 1C). There was no significant increase in PD-L1 expression in the HBEC3/p53 and HBEC3/EGFR-L858R cell lines. Furthermore, PD-L1 expression levels in the HBEC3/KRAS and HBEC3/KRAS/p53 cell lines were comparable, indicating that knockdown of p53 did not alter increased PD-L1 expression induced by KRAS mutation (Figure 1A-C). These results highlight the predominant role of KRAS mutation over other oncogenic driver mutations in the induction of PD-L1 expression and implicate that KRAS mutation alone can induce PD-L1 expression in high risk, premalignant human bronchial epithelial cells.
Figure 1.
KRAS mutation alone induced PD-L1 expression in high risk, premalignant human bronchial epithelial cells. PD-L1 expression was examined in HBEC3 cell lines carrying the K-Rasv12 mutation (Kras), knock-down of p53 (p53) or both (Kras/p53), and EGFR mutation (L858R). PD-L1 surface expression was determined by flow cytometry and a representative histogram is shown (A). Mean fluorescence intensity (MFI) obtained from the histograms were normalized to an isotype control (B). A horizontal line at ratio 1 indicates the baseline (BKG). PD-L1 mRNA expression was determined by real-time qPCR. Data were shown as mean ± SEM from three independent experiments (C). Statistical analysis was done with Student’s t-test. BKG: background.
MEK-ERK pathway is a major regulator of constitutive and KRAS mutation-induced PD-L1 expression in HBEC cell line
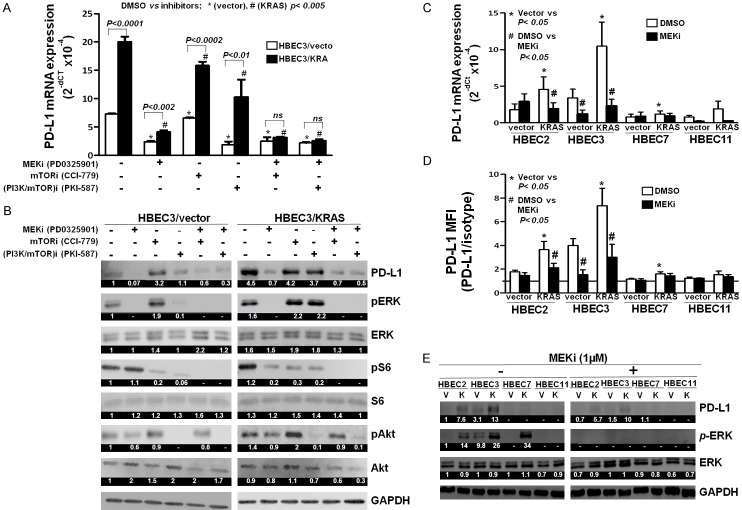
Oncogenic KRAS mutation stimulates a wide range of downstream signaling pathways, such as the RAF-MEK-ERK [5] and PI3K-Akt-mTOR pathways [21]. To examine the potential effects of these pathways on KRAS-induced PD-L1 expression, HBEC3/vector, and HBEC3/KRAS cells were treated with MEK inhibitor (MEKi), mTOR inhibitor (mTORi), and dual inhibitor of PI3K and mTOR (PI3K/mTOR)i, and analyzed for PD-L1 mRNA expression by RT-qPCR (Figure 2A). The efficacy of the inhibitors was also validated by western blot (Figure 2B). PD-L1 mRNA expression was significantly increased in HBEC3/KRAS cells compared to HBEC3/vector cells (Figure 2A), which was dramatically decreased (5-fold) by inhibition of MEK-ERK pathway (MEKi), while it was ~1.3-fold and ~2-fold decreased by inhibition of mTOR (mTORi) and PI3K/Akt/mTOR (PI3K/mTOR)i pathways, respectively (Figure 2A). These results indicate that KRAS-driven PD-L1 expression was mainly dependent on the MEK-ERK pathway. Combined inhibition of both MEK-ERK and mTOR pathways (MEKi+mTORi) or MEK-ERK and PI3K/Akt/mTOR pathways resulted in a significant decrease (P = 0.006 and P = 0.002) in KRAS-driven PD-L1 mRNA expression (Figure 2A), but not in protein levels (Figure 2B), when compared to MEKi alone. These results again support the finding of KRAS-driven PD-L1 expression was mainly dependent on the MEK-ERK pathway. We also found that MEKi treatment decreased constitutive PD-L1 mRNA expression by ~3-fold in HBEC3/vector cells (Figure 2A). However, there was only a slight reduction (1.1 fold) in PD-L1 mRNA expression by mTORi in HBEC3/vector cells, which was further significantly decreased by combination treatment with MEKi+mTORi (2.6-fold) compared to MEKi treatment alone (Figure 2A). There was ~3.5 fold decrease in the constitutive PD-L1 mRNA expression by treatment with (PI3K/mTOR)i in HBEC3/vector cells compared to mTORi alone or no treatment (Figure 2A). (PI3K/mTOR)i treatment alone led to almost complete inhibition of pERK, pAkt, and pS6 protein expression, relevant downstream mediators of PI3K/Akt/mTOR pathway (Figure 2B). There was a comparable reduction in PD-L1 expression by (PI3K/mTOR)i alone and by combination treatment of MEKi and (PI3K/mTOR)i in HBEC3/vector cells (Figure 2A and 2B). These findings suggest a possible mechanism of cross-talk between the MEK-ERK and PI3K/Akt/mTOR pathways in constitutive PD-L1 expression in HBEC3/vector cells but not in HBEC3/KRAS cells. Constitutive PD-L1 mRNA expression in HBEC3/vector cells was not significantly altered by the combined inhibition of MEK-ERK and mTOR pathways (MEKi+mTORi) or MEK-ERK and PI3K/Akt/mTOR pathways (MEKi+(PI3K/mTOR)i) compared to MEKi alone (Figure 2A). Collectively, these results demonstrate that the MEK-ERK pathway plays a major role in the regulation of oncogenic KRAS-driven PD-L1 expression in HBEC cells.
Figure 2.
KRAS-driven PD-L1 expression was inhibited by MEK inhibitor in multiple HBEC lines. HBECs (HBEC3/Vector and HBEC3/Kras) were treated with the inhibitors for MEK (PD0325901, 1 mM), mTOR (CCI779, 20 mM), and PI3K and mTOR (PKI-587, 3 mM) for 24 hours and the total RNA and cell lysates were collected to perform qPCR (A) and western blot (B) to measure PD-L1 mRNA expression and the efficacy of the inhibitors, respectively. A representative experiment was shown as mean ± SD from three independent experiments. Statistical analysis was done with Student’s t-test. P = 0.006 (HBEC3/KRAS, MEKi vs MEKi+mTORi), P = 0.002 ((HBEC3/KRAS, MEKi vs MEKi+(PI3K/mTOR)i) Four HBEC lines (HBEC2, HBEC3, HBEC7 and HBEC11) were treated with MEK inhibitor (PD0325901) at a final concentration of 1 µM for 24 hours. Vector and Kras were depicted as V and K, respectively. PD-L1 mRNA (C) and surface PD-L1 expression (D) were measured by qPCR and flow cytometry, respectively. Data were shown as mean ± SEM from three independent experiments. Statistical analysis was done with Student’s t-test. Western blot was performed to examine PD-L1 expression and ERK activation (E). The numeric values above the blots were obtained by densitometric analyses after normalized to internal loading controls (GAPADH).
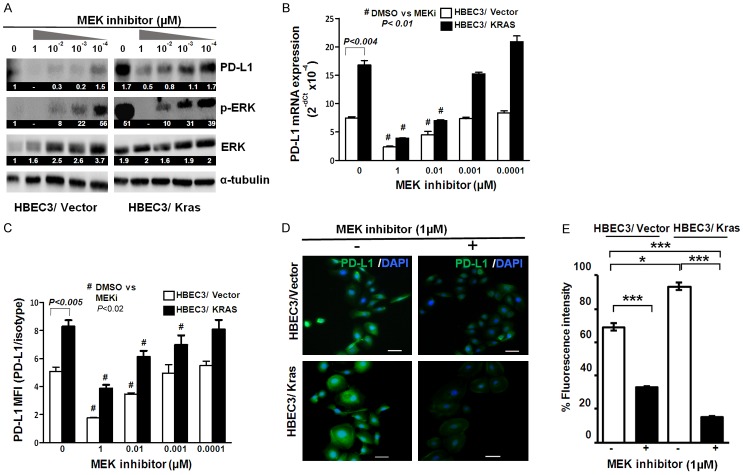
Next, we attempted to verify MEK-ERK dependent increase in PD-L1 expression in multiple HBEC cell lines with KRAS mutation. Of the four cell lines tested, three cell lines (HBEC2/KRAS, HBEC3/KRAS, and HBEC7/KRAS) showed significant increases in PD-L1 (mRNA, surface, and cytoplasmic protein) expression compared to respective wild type cell lines, which were significantly reduced by MEKi (Figure 2C and 2D). These findings further validate the concept of KRAS mediated PD-L1 expression in multiple high-risk HBEC lines. We also found that there was differential sensitivity to MEKi between cell lines. For example, the PD-L1 expressions in HBEC2/KRAS and HBEC3/KRAS were significantly reduced by MEKi, but not in HBEC7/KRAS and HBEC11/KRAS lines. Hence, we selected HBEC3 for further studies, in which KRAS-driven and MEK-ERK pathway-dependent PD-L1 expression were validated. Using HBEC3, we further confirmed that KRAS-driven PD-L1 protein and mRNA expression in HBEC3/KRAS were decreased by MEKi in a dose-dependent manner (Figure 3A-C). The reduced surface and intracellular PD-L1 protein expression by MEKi (1 µM) were also detected by immunofluorescence staining (Figure 3D).
Figure 3.
KRAS-driven PD-L1 expression was inhibited by MEK inhibitor in a dose-dependent manner in HBECs. HBECs (HBEC3/Vector and HBEC3/Kras) were treated with MEK inhibitor (PD0325901) at final concentrations of 10-4~1 µM for 24 hours. Western blot was performed to examine PD-L1 expression and ERK activation (pERK) (A). PD-L1 mRNA (B) and surface PD-L1 expression (C) were measured by qPCR and flow cytometry, respectively. Data were shown as mean ± SD. Immunofluorescent staining of PD-L1 expression and DAPI on HBEC3/Vector and HBEC3/Kras treated with MEK inhibitor (1 µM) for 24 hours (D). The relative percentage of PD-L1 expression was measured and expressed as fluorescence intensity under different experimental conditions (E). Scale bars, 50 µm, *: P ≤ 0.05, **: P ≤ 0.01; ***: P ≤ 0.001. The numeric values above the blots were obtained by densitometric analyses after normalized to internal loading controls (α-tubulin) (A).
FRA1 is upregulated in the KRAS mutant HBEC cell lines
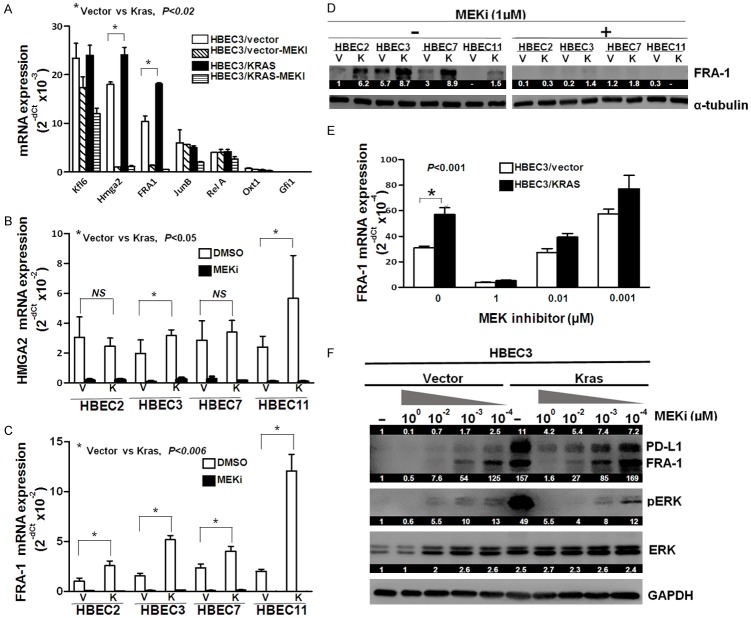
The MEK-ERK pathway regulates the activity of many different substrate proteins including the transcription of downstream target genes [22]. On the basis that altered expression of ERK-dependent transcription factors may contribute to oncogenic KRAS mutation mediated PD-L1 expression, we examined the expression of seven ERK-dependent transcription factors that were upregulated and strongly dependent on MEK-ERK signaling in KRAS oncogene transformed pre-neoplastic rat ovarian surface epithelial (ROSE) cells [23]. These transcription factors included Klf6, Hmga2, Fosl1 (FRA1), JunB, RelA, Otx1, and Gfi1. As expected, mRNA expression of Hmga2 (~1.3 fold) and FRA1 (~1.8 fold) were significantly increased, but not the other factors, in HBEC3/KRAS cells compared to HBEC3/vector cells (Figure 4A).
Figure 4.
FRA1 was upregulated in the KRAS mutant HBEC cell lines. Seven ERK-dependent transcription factors were tested for their mRNA expression levels in HBECs (HBEC3/Vector and HBEC3/Kras) (A). HMGA2 (B) and FRA1 (C) mRNA expression in HBECs were measured by qPCR after treatment with MEK inhibitor (PD0325901) at a final concentration of 1 µM for 24 hours. FRA1 protein expression levels were measured in multiple HBEC lines by western blot after treatment with or without 1 µM MEKi for 24 hours (D). FRA1 and PD-L1 mRNA and protein expression levels in HBECs (HBEC3/vector and HBEC3/Kras) were measured by qPCR and western blot after treated with MEK inhibitor (PD0325901) at final concentrations of 10-4~1 µM for 24 hours (E and F). The numeric values above the blots were obtained by densitometric analyses after normalized to internal loading controls (α-tubulin or GAPDH).
The concept of KRAS driven increased expression of Hmga2 and FRA1 was evaluated in multiple HBEC cell lines with KRAS mutation. All four HBEC/KRAS (HBEC2, 3, 7, and 11) cell lines tested showed significantly increased FRA1 mRNA and protein expression while two cell lines (HBEC3/KRAS and HBEC11/KRAS) exhibited moderate increase in Hmga2 mRNA expression (Figure 4B and 4C), which were dramatically and dose-dependently reduced by MEKi treatment (Figure 4B-E). The treatment of MEKi resulted in a parallel dose-dependent decrease in both PD-L1 and FRA1 protein expression with an associated marked reduction in pERK protein expression (Figure 4F). These results suggest that FRA1 may play a role in the regulation of MEK-ERK-dependent PD-L1 expression induced by KRAS mutation in HBEC cells.
FRA1 knock-down markedly suppresses KRAS mutation-induced and MEK-ERK-dependent PD-L1 expression and ERK activation in HBEC
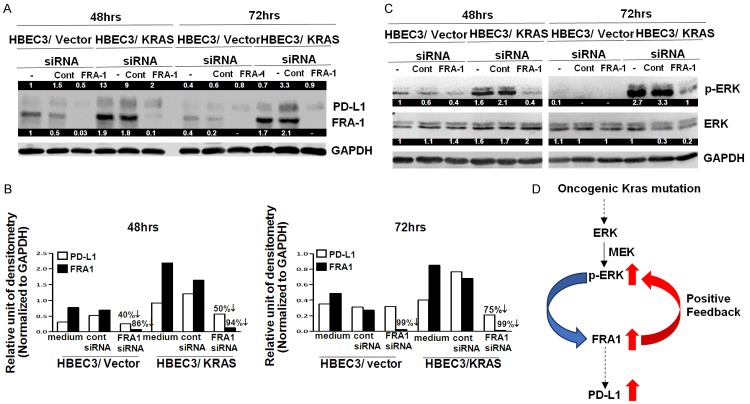
To test the functional role of FRA1 in regulating KRAS mutation-induced PD-L1 expression in HBEC, we used siRNA to inhibit FRA1 expression. At 48 hours siRNA transfection, FRA1 knockdown in both HBEC3/vector (86%) and HBEC3/KRAS (94%) cell lines was associated with a parallel 40% (HBEC3/vector) and 50% (HBEC3/KRAS) reduction of PD-L1 protein expression compared to non-targeting control siRNA transfection (Figure 5A and 5B). At 72 hours siRNA transfection, there was 99% knockdown by FRA1 siRNA and the PD-L1 protein expression further decreased (75%) compared to non-targeting control siRNA in HBEC3/KRAS cells. In contrast, there was also 99% knockdown by FRA1 siRNA in HBEC3/vector cells, but there was no associated further reduction in PD-L1 protein expression (Figure 5B). These findings suggest that FRA1 may regulate KRAS mutation-induced PD-L1 expression.
Figure 5.
The knockdown of FRA1 by siRNA led to the reduction of PD-L1 expression in HBECs. HBECs were treated with control (non-targeting) or FRA1 siRNA (a pool of 4 siRNAs) at a final concentration of 100 nM for 48 and 72 hours and western blot (A) and its densitometry analysis (B) using Image J were performed to measure PD-L1 and FRA1 expression. ERK activation (p-ERK) was also examined after FRA1 siRNA treatment by western blot (C). Schematic of a proposed mechanism for mutant KRAS-mediated PD-L1 upregulation through ERK pathway and FRA1 in premalignant, high risk human bronchial epithelial cells. This positive feedback loop between ERK activation and FRA1 up-regulation is a novel finding, particularly in a lung premalignancy model and sheds light on PD-L1 upregulation (D). The numeric values above the blots were obtained by densitometric analyses after normalized to internal loading controls (GAPDH).
To further evaluate if activation of ERK appears to be integral in oncogenic KRAS-mediated PD-L1 expression, we investigated whether ERK activation is impacted FRA1 knockdown. Surprisingly, we found that FRA1 knockdown led to a remarkable reduction in ERK activation, especially in HBEC3/KRAS cells (Figure 5C). These results suggest that FRA1 regulates mutant KRAS induced PD-L1 expression potentially through a positive feedback mechanism promoting sustained ERK activation (Figure 5D).
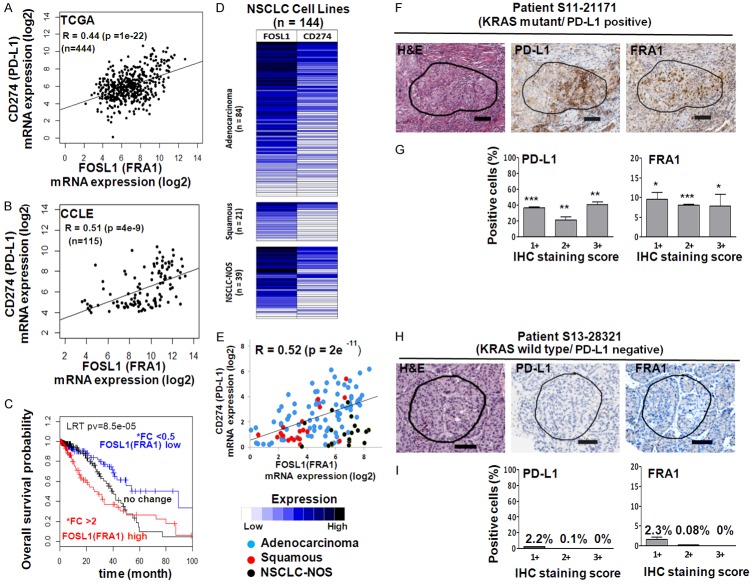
PD-L1 expression is significantly correlated with FRA1 expression in human lung cancer specimens (TCGA) and human NSCLC cell lines
To validate the correlation between PD-L1 and FRA1 mRNA expression in lung cancer, we analyzed TCGA samples [24] from 444 patients with NSCLC and 115 human lung cancer cell lines from CCLE [25]. As expected, there was a strong correlation between PD-L1 and FRA1 in the TCGA samples (R = 0.44, P = 1e-22) (Figure 6A) and CCLE lung cancer cell lines (R = 0.51, P = 4e-9) (Figure 6B). We also found that high levels of FRA1 expression independently predicted poor overall survival (OS) in NSCLC patients (Figure 6C). Additionally, RNA sequencing for PD-L1 (CD274) and FOSL1 (FRA1) were performed on NSCLC cell lines (n = 144) provided by John D. Minna’s lab. There was a significant correlation between PD-L1 and FRA1 expression in the NSCLC cell lines (R = 0.52, P = 2e-11) (Figure 6D and 6E).
Figure 6.
PD-L1 expression was significantly correlated with FRA1 expression in human lung cancer specimens (TCGA), human NSCLC cell lines and patient tumor tissues. Correlations between PD-L1 and FRA1 mRNA expression in samples from 444 patients with NSCLC from The Cancer Genome Atlas (TCGA) (A) from 115 NSCLC cell lines from Cancer Cell Line Encyclopedia (CCLE) (B) were evaluated. The Spearman’s rank-order correlation test was applied to measure the strength of the association between PD-L1 and FRA1 mRNA levels. Kaplan-Meier plots of overall survival in patients with lung adenocarcinoma (n = 444) according to the expression of FRA1 (C). *Fold-changes (FC) between tumors and healthy tissues; high (FC > 2), low (FC < 0.5). High expression in tumors is indicated in red, while low expression in tumors is shown in blue. A total of 144 NSCLC cell lines (84 adenocarcinomas, 21 squamous cell carcinomas, and 39 NSCLC-not otherwise specified (NSCLC-NOS) were provided by John D. Minna’s lab and were used to examine the correlation of PD-L1 and FRA1 mRNA expression by the Spearman’s rank-order correlation test (D, E). The log-rank test was used for comparisons. Hematoxylin and Eosin staining and immunohistochemical staining of expression of PD-L1 and FRA1 in NSCLC patients with KRAS mutant and positive expression of PD-L1 (F and G) and with KRAS wild type and negative expression of PD-L1 (H and I). IHC staining intensity was scored as 0 (negative), 1+ (weak), 2+ (moderate), and 3+ (strong) and the results were quantified using 3 ROIs from each tissue by Aperio Image analysis toolkit (Leica Biosystems) as described in the Methods. Results were shown as the average percentages of positive cells (G and I). One representative ROI was shown (F and H). *, P < 0.05; **, P < 0.005; ***, P < 10-5 (wild type KRAS vs mutant KRAS).
Given these findings that PD-L1 positively correlated with FRA1 mRNA expression from the analyses of 3 independent datasets, we further assessed this concept in patient tumor specimens using immunohistochemical analysis and found that FRA1 protein expression intensity was significantly higher in the KRAS mutant/PD-L1 positive tumor specimen (Figure 6F and 6G) than in KRAS wild type/PD-L1 negative tumor specimens (Figure 6H and 6I). These results further support the concept that tumor expression of PD-L1 is positively correlated with FRA1 expression in NSCLC patients at both mRNA and protein levels.
The prognostic significance of PD-L1 expression in human cancers including lung cancer is controversial and clinical outcomes are varied in studies [26-30]. To evaluate the impact of PD-L1 and FRA1 expression on overall survival (OS), we correlated the OS of patients with lung adenocarcinoma (n = 444) according to the expression of PD-L1 and FRA1. We found that patients with tumors that expressed high FRA1 and high PD-L1 levels had a trend toward lower OS than patients with low FRA1 and low PD-L1 expression (P > 0.05; Figure S1A). High FRA1 with low or high PD-L1 status did not correlate with a significant difference in OS (Figure S1B). Also, we did not find any significant association between KRAS mutation and mRNA expression of PD-L1 and FRA1 in the TCGA tumor samples and CCLE lung cancer cell lines (Figure S2A and S2B). These findings highlight the difficulty in studying specific driver mutations, such as KRAS, in patient samples (TCGA) or cancer cell lines (CCLE), and the role of potential biomarkers, such as PD-L1 and FRA1, in cancer specimens with clonal cancer subsets, varying mutational tumor burdens, concomitant other known and unknown mutations, and varied treatment exposures. As such, to understand the mechanism of KRAS driven PD-L1 expression, our studies were performed in high risk, premalignant HBEC cell lines to eliminate as much of the confounding variables, such as other driver mutations.
Discussion
KRAS mutation has remained an elusive target for cancer therapy [31]. There are currently no approved drugs that specifically target KRAS mutant tumors [31,32]. The presence of KRAS mutation has an attendant poorer prognosis, but it translates into little clinical utility [31,32]. Analyses have shown that KRAS mutations are more prevalent amongst former or active smokers compared to never-smokers suggesting that KRAS may merely reflect smoking status, tumor antigenicity, and TMB [31,33]. KRAS mutations are not mutually exclusive from EGFR mutations or ALK rearrangements [32]. Skoulidis et al. defined three major subgroups of KRAS mutant lung adenocarcinoma in which somatic genetic aberrations in STK11/LKB1, TP53, and CDKN2A/B can co-exist with KRAS mutation resulting in different phenotypes [32,34]. For instance, in the STK11/LKB1 alteration cohort, PD-L1 expression was lower compared to the other two subgroups [34]. As such, KRAS mutation in NSCLC is molecularly a diverse entity confounded by the presence of other driver mutations, which make elucidating the role of KRAS in NSCLC carcinogenesis and impact on immunotherapy difficult to interpret and study. On this basis and in an effort to avoid the multitude of coexistent tumor mutations in cancer, we used a premalignant, high-risk human bronchial epithelial cell line (HBEC) model that expressed mutant KRAS, EGFR, or p53 knock-down [14,15].
In checkpoint inhibition immunotherapy, total TMB and smoking status have been directly associated with better therapeutic efficacy. In lung cancer, there is wide variability in the frequency of somatic tumor mutations where tumors from smokers have relatively high TMB compared to tumors from nonsmokers [4]. The initial findings from Rizvi et al. of high TMB predicting the efficacy of PD-1/PD-L1 inhibition therapy [4] have been validated in CheckMate026 where patients with untreated advanced stage IV or recurrent NSCLC with ≥ 1% PD-L1 tumor expression were randomized to first-line nivolumab monotherapy or platinum-doublet chemotherapy [35]. In an exploratory analysis of CheckMate026, patients with high TMB had improved progression-free survival (PFS) and objective response rate with nivolumab compared to chemotherapy [35]. CheckMate026 was the first randomized phase 3 trials to demonstrate the concept of high TMB predicting efficacy to PD-1 inhibitor therapy. Dong et al. reported on clinical and mutational data of 34 NSCLC patients treated with pembrolizumab where they observed that TP53/KRAS mutation significantly increased PD-L1 expression and the TP53 or KRAS mutated tumors showed increased TMB [36].
Based on this body of evidence, the role of KRAS mutation in PD-L1 expression and TMB in NSCLC remains unclear [27,37]. It is controversial whether the presence of KRAS mutation is just a reflection of high TMB and smoking status or if there is a direct mechanism of KRAS mediated PD-L1 over-expression. As such, we hypothesized that oncogenic KRAS mutation induces PD-L1 upregulation via MEK-ERK dependent oncogenic transcription factor FRA1 (FOSL1). Here, our study demonstrated that oncogenic KRAS mutation mediated PD-L1 expression is driven by the MEK-ERK pathway in HBEC cell lines via FRA1, suggesting that KRAS can directly drive PD-L1 expression.
In an effort to validate our findings in cancer, we analyzed human NSCLC tissue specimens and cell lines and found a strong association between FRA1 and PD-L1 at mRNA and protein expression levels supporting our findings of FRA1 mediated PD-L1 expression in HBEC. Correlation between expression of FRA1 and PD-L1 was also confirmed in 33 NSCLC-PDX models (data not shown). Additionally, we found that oncogenic KRAS mutation (G12V) but not EGFR mutation (L858R), induced an increase in the surface and intracellular PD-L1 levels in HBEC carrying no other mutations. In comparison, D’Incecco et al. analyzed 125 NSCLC patients assessed PD-L1 and PD-1 protein expression by IHC in 56 EGFR mutated, 29 KRAS mutated, 10 ALK translocated, and 30 EGFR/KRAS/ALK wild type tumors [38]. PD-L1 expression was significantly associated with adenocarcinoma histology and EGFR mutations, whereas PD-1 expression was significantly associated with current smoking status and KRAS mutations [38]. In another study, Ji et al. evaluated 100 surgically resected lung adenocarcinoma specimens and assessed PD-1 and PD-L1 expression by IHC in relation to KRAS or EGFR mutational status [39]. In contrast, Ji et al. found a negative association between tumor PD-L1 expression and EGFR mutation, and also between tumor PD-1 expression and KRAS mutation [39]. These differences in studies that correlate PD-1/PD-L1 expression with driver mutations are likely explained by the phenotypically different subsets of KRAS mutations that co-existent with other driver mutations, and also the inherent variability in testing PD-1/PD-L1 protein expression by IHC.
Contrary to our results, Ma et al. reported a low expression of FRA1 from 118 NSCLC paraffin-embedded NSCLC tissue specimens (Beijing, China) correlated with advanced tumor stage and poor OS [40]. Notably, there was no mention of underlying driver mutations, such as EGFR status. A meta-analysis of clinical trials with Asians in 90 treatment arms revealed differences in OS and chemotherapy response rates [41]. As such, co-existent driver mutations, such as EGFR mutation, or ethnic differences in Asian subjects may account for the discrepancy reported by Ma et al. compared to our FRA1 results.
Correlation between PD-L1 expression and EGFR mutation remains inconclusive [42]. In contrast to our results, a previous study reported that ectopic expression of mutated EGFR, but not of mutated KRAS, caused an increase in PD-L1 levels in immortalized bronchial epithelial cells (BEAS2B) [43]. Although a more detailed investigation is necessary, the inconsistency in this finding with our result may be due to the differences in endogenous signaling contexts between the two cell lines. In fact, our results showed that there was no increase in PD-L1 expression in HBEC11/KRAS cell line, while there were significant increases in other HBEC/KRAS cell lines (HBEC2, 3, and 7). Furthermore, the enhanced PD-L1 expression by KRAS mutation was not impacted by the addition of p53 knockdown, indicating that there is no association between the level of PD-L1 expression and alterations of p53 signaling in HBEC.
Intrinsic tumor PD-L1 expression can be caused by the activation of oncogenic signaling pathways [44]. Oncogenic KRAS mutation results in the activation of mitogen-activated protein kinase (MAPK) signalling pathway (RAF-MEK-ERK) and PI3K pathway (PI3K-AKT-mTOR). Lastwika et al. showed that oncogenic and inducible PD-L1 expression was AKT-mTOR pathway-dependent in lung cancer [45]. In HBEC, we found that intrinsic and oncogenic KRAS activation-mediated PD-L1 expression was associated mainly with the MEK-ERK pathway whereas the mTOR pathway had a marginal effect on PD-L1 expression. In accordance with our results, Chen et al. revealed that PD-L1 upregulation was induced by Kras mutation through p-ERK and not p-AKT signaling [11]. Unexpectedly, we found that PD-L1 expression was independently associated with ERK activation in HBEC7 and HBEC11. PD-L2 is also a known ligand of PD-1 and was found to be expressed in the tumor microenvironment [46]. Although the current study did not address the inconsistency between the HBEC lines, our preliminary data showed that PD-L2 expression was more prevalent than PD-L1 in KRAS mutant HBEC7 and HBEC11, and the expression was dramatically reduced by MEK inhibitor (unpublished data). Thus, we speculate that there may be different mechanisms of selective or preferential expression between PD-L1 and PD-L2 in premalignant HBECs. Alternatively, differential sensitivity to the MEK inhibitor may also be present between HBECs, which is suggested by studies showing that RAS mutant cells did not demonstrate the same sensitivity to MEK inhibition despite the effective inhibition of p-ERK [47,48].
Interestingly, we found that PD-L1 inhibition by dual inhibitors of PI3K and mTOR in KRAS wild type, but not in KRAS mutant HBEC, was comparable to the inhibition by MEK inhibitor. This result is most likely due to a cross-talk between the MEK-ERK and PI3K-Akt-mTOR pathways [49] as ERK activation was nearly abolished by inhibition of PI3K and mTOR in the KRAS wild type HBEC while the activation was not affected in the KRAS mutant HBEC. Others have previously shown that MEK inhibition caused remarkably enhanced p-Akt levels under the RAS mutation, which resulted from the cross-regulation between the two signaling pathways and led to resistance to MEK inhibitor [49,50]. However, our study showed that the levels of p-Akt were not notably affected by MEK inhibition in both wild type and mutant KRAS HBECs.
The mechanisms of PD-L1 regulation in lung cancer remains poorly understood. Coelho et al. showed that Ras-MEK signaling elevated PD-L1 expression by modulating the stability of the transcript through tristetraprolin (TTP), an AU-rich element-binding protein [12], which may be an alternative FRA1 independent PD-L1 pathway in KRAS mutant cells. We elucidated in the current study that oncogenic KRAS mutation caused increased PD-L1 expression and its downstream MEK-ERK pathway was a major signaling pathway that mediated the upregulation of PD-L1 in HBEC. Importantly, we found that FRA1, a proto-oncogenic transcription factor, played an important role in the regulation of PD-L1 expression. The role of FRA1 in cancer progression is not clear. Our results represent the first evidence that FRA1 may promote cancer progression by facilitating immune evasion in high risk, premalignant bronchial epithelial cells. Specifically, we hypothesized that FRA1 might be pivotal in tumorigenesis via the regulation of immune checkpoint PD-L1 expression. Our speculation can be supported by the findings in this study that high FRA1 expression was a poor prognostic factor for NSCLC, and expression of FRA1 was strongly related to PD-L1 expression in lung cancer tissues and cell lines.
In conclusion, our study identified FRA-1 as one mechanism by which KRAS mutation resulted in direct regulation of PD-L1 expression in human premalignant, high-risk bronchial epithelium. As such, KRAS mutation may not merely represent TMB or tumor antigenicity. But rather, these findings suggest that MEK-ERK dependent FRA1 and PD-L1 status in NSCLC patients with KRAS mutation may serve as companion biomarkers that predict efficacy to PD-1/PD-L1 immune checkpoint blockade therapy or identify a patient population for cancer-prevention therapy. Additionally, there may be a role for targeting FRA-1 in potential combinational strategies with checkpoint inhibition in this patient population.
Acknowledgements
The authors thank all of the following funding sources. This work was supported by the Department of Surgery, David Geffen School of Medicine at UCLA, NIH/NCATS UL1TR001881, NCI 1K23 CA131577-01A1, Thoracic Surgery Foundation Research Award, STOP Cancer I.C.O.N./Natasha Girard Seed Grant, and NCI P50 CA07097.
Disclosure of conflict of interest
Dr. Jay M. Lee serves as an advisor to AstraZeneca, Genentech, Novartis, and Regeneron for clinical trials involving checkpoint inhibitors in early stage lung cancer. Dr. Jay M. Lee also receives research support (drug only) from Merck for an immunotherapy clinical trial.
Authors’ contribution
JML is the principal investigator of the study. MHL, JY, and JML conceived the idea, designed experiments. MHL and JML wrote the manuscript. MHL performed experiments. MHL, JY, and JML interpreted the results of experiments. MHL, BB, and JML edited and revised the manuscript. WDM contributed to the acquisition and analysis of the immunohistochemistry results. MKP contributed to the further analysis of the immune florescence staining results. LT, LG, BG, and JDM contributed to the acquisition and analyses of CCLE and TCGA data. TCW, EF, SJP, GZ, KK, and SMD assisted with manuscript writing. SMD and JML oversaw the study. All authors read and approved the final manuscript.
Abbreviations
- NSCLC
Non-small cell lung cancer
- PD-L1
Programmed death ligand-1
- TMB
tumor mutation burden
- IO
immunotherapy in oncology
- FRA1
FOS-related antigen 1
- FOSL1
FOS-like antigen-1
- HBEC
human bronchial epithelial cell lines
- ROI
regions of interest
- TCGA LUAD
The Cancer Genome Atlas for Lung Adenocarcinoma
- ROSE
rat ovarian surface epithelial
- OS
overall survival
- CCLE
Cancer Cell Line Encyclopedia
- PFS
progression-free survival
- BEAS2B
bronchial epithelial cells
- TTP
tristetraprolin
- MFI
Mean fluorescence intensity
Supporting Information
References
- 1.Mascaux C, Iannino N, Martin B, Paesmans M, Berghmans T, Dusart M, Haller A, Lothaire P, Meert AP, Noel S, Lafitte JJ, Sculier JP. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92:131–9. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, Waterhouse D, Ready N, Gainor J, Aren Frontera O, Havel L, Steins M, Garassino MC, Aerts JG, Domine M, Paz-Ares L, Reck M, Baudelet C, Harbison CT, Lestini B, Spigel DR. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123–35. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer. 2008;8:133–40. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- 6.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–68. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 7.Zhong G, Chen X, Fang X, Wang D, Xie M, Chen Q. Fra-1 is upregulated in lung cancer tissues and inhibits the apoptosis of lung cancer cells by the P53 signaling pathway. Oncol Rep. 2016;35:447–53. doi: 10.3892/or.2015.4395. [DOI] [PubMed] [Google Scholar]
- 8.Zuber J, Tchernitsa OI, Hinzmann B, Schmitz AC, Grips M, Hellriegel M, Sers C, Rosenthal A, Schafer R. A genome-wide survey of RAS transformation targets. Nat Genet. 2000;24:144–52. doi: 10.1038/72799. [DOI] [PubMed] [Google Scholar]
- 9.Casalino L, De Cesare D, Verde P. Accumulation of Fra-1 in ras-transformed cells depends on both transcriptional autoregulation and MEK-dependent posttranslational stabilization. Mol Cell Biol. 2003;23:4401–15. doi: 10.1128/MCB.23.12.4401-4415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adiseshaiah P, Vaz M, Machireddy N, Kalvakolanu DV, Reddy SP. A Fra-1-dependent, matrix metalloproteinase driven EGFR activation promotes human lung epithelial cell motility and invasion. J Cell Physiol. 2008;216:405–12. doi: 10.1002/jcp.21410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen N, Fang W, Lin Z, Peng P, Wang J, Zhan J, Hong S, Huang J, Liu L, Sheng J, Zhou T, Chen Y, Zhang H, Zhang L. KRAS mutation-induced upregulation of PD-L1 mediates immune escape in human lung adenocarcinoma. Cancer Immunol Immunother. 2017;66:1175–87. doi: 10.1007/s00262-017-2005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coelho MA, de Carne Trecesson S, Rana S, Zecchin D, Moore C, Molina-Arcas M, East P, Spencer-Dene B, Nye E, Barnouin K, Snijders AP, Lai WS, Blackshear PJ, Downward J. Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity. 2017;47:1083–99. e6. doi: 10.1016/j.immuni.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falk AT, Yazbeck N, Guibert N, Chamorey E, Paquet A, Ribeyre L, Bence C, Zahaf K, Leroy S, Marquette CH, Cohen C, Mograbi B, Mazieres J, Hofman V, Brest P, Hofman P, Ilie M. Effect of mutant variants of the KRAS gene on PD-L1 expression and on the immune microenvironment and association with clinical outcome in lung adenocarcinoma patients. Lung Cancer. 2018;121:70–75. doi: 10.1016/j.lungcan.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Sato M, Larsen JE, Lee W, Sun H, Shames DS, Dalvi MP, Ramirez RD, Tang H, DiMaio JM, Gao B, Xie Y, Wistuba II, Gazdar AF, Shay JW, Minna JD. Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Mol Cancer Res. 2013;11:638–50. doi: 10.1158/1541-7786.MCR-12-0634-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato M, Vaughan MB, Girard L, Peyton M, Lee W, Shames DS, Ramirez RD, Sunaga N, Gazdar AF, Shay JW, Minna JD. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–28. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 16.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM, Milchgrub S, Smith AL, Souza RF, Gilbey L, Zhang X, Gandia K, Vaughan MB, Wright WE, Gazdar AF, Shay JW, Minna JD. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–34. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 17.Lee MH, Kachroo P, Pagano PC, Yanagawa J, Wang G, Walser TC, Krysan K, Sharma S, John MS, Dubinett SM, Lee JM. Combination treatment with apricoxib and IL-27 enhances inhibition of epithelial-mesenchymal transition in human lung cancer cells through a STAT1 dominant pathway. J Cancer Sci Ther. 2014;6:468–77. doi: 10.4172/1948-5956.1000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kachroo P, Lee MH, Zhang L, Baratelli F, Lee G, Srivastava MK, Wang G, Walser TC, Krysan K, Sharma S, Dubinett SM, Lee JM. IL-27 inhibits epithelial-mesenchymal transition and angiogenic factor production in a STAT1-dominant pathway in human non-small cell lung cancer. J Exp Clin Cancer Res. 2013;32:97. doi: 10.1186/1756-9966-32-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JM, Lee MH, Garon E, Goldman JW, Salehi-Rad R, Baratelli FE, Schaue D, Wang G, Rosen F, Yanagawa J, Walser TC, Lin Y, Park SJ, Adams S, Marincola FM, Tumeh PC, Abtin F, Suh R, Reckamp KL, Lee G, Wallace WD, Lee S, Zeng G, Elashoff DA, Sharma S, Dubinett SM. Phase I trial of intratumoral injection of CCL21 gene-modified dendritic cells in lung cancer elicits tumor-specific immune responses and CD8(+) T-cell infiltration. Clin Cancer Res. 2017;23:4556–68. doi: 10.1158/1078-0432.CCR-16-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMillan EA, Ryu MJ, Diep CH, Mendiratta S, Clemenceau JR, Vaden RM, Kim JH, Motoyaji T, Covington KR, Peyton M, Huffman K, Wu X, Girard L, Sung Y, Chen PH, Mallipeddi PL, Lee JY, Hanson J, Voruganti S, Yu Y, Park S, Sudderth J, DeSevo C, Muzny DM, Doddapaneni H, Gazdar A, Gibbs RA, Hwang TH, Heymach JV, Wistuba I, Coombes KR, Williams NS, Wheeler DA, MacMillan JB, Deberardinis RJ, Roth MG, Posner BA, Minna JD, Kim HS, White MA. Chemistry-first approach for nomination of personalized treatment in lung cancer. Cell. 2018;173:864–78. e29. doi: 10.1016/j.cell.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castellano E, Downward J. RAS interaction with PI3K: more than just another effector pathway. Genes Cancer. 2011;2:261–74. doi: 10.1177/1947601911408079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stinchcombe TE, Johnson GL. MEK inhibition in non-small cell lung cancer. Lung Cancer. 2014;86:121–5. doi: 10.1016/j.lungcan.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stelniec-Klotz I, Legewie S, Tchernitsa O, Witzel F, Klinger B, Sers C, Herzel H, Bluthgen N, Schafer R. Reverse engineering a hierarchical regulatory network downstream of oncogenic KRAS. Mol Syst Biol. 2012;8:601. doi: 10.1038/msb.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorensen SF, Zhou W, Dolled-Filhart M, Georgsen JB, Wang Z, Emancipator K, Wu D, Busch-Sorensen M, Meldgaard P, Hager H. PD-L1 expression and survival among patients with advanced non-small cell lung cancer treated with chemotherapy. Transl Oncol. 2016;9:64–9. doi: 10.1016/j.tranon.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Wang L, Li Y, Pan Y, Wang R, Hu H, Li H, Luo X, Ye T, Sun Y, Chen H. Protein expression of programmed death 1 ligand 1 and ligand 2 independently predict poor prognosis in surgically resected lung adenocarcinoma. Onco Targets Ther. 2014;7:567–73. doi: 10.2147/OTT.S59959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massi D, Brusa D, Merelli B, Ciano M, Audrito V, Serra S, Buonincontri R, Baroni G, Nassini R, Minocci D, Cattaneo L, Tamborini E, Carobbio A, Rulli E, Deaglio S, Mandala M. PD-L1 marks a subset of melanomas with a shorter overall survival and distinct genetic and morphological characteristics. Ann Oncol. 2014;25:2433–42. doi: 10.1093/annonc/mdu452. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Kang S, Shen J, He J, Jiang L, Wang W, Guo Z, Peng G, Chen G, He J, Liang W. Prognostic significance of programmed cell death 1 (PD-1) or PD-1 ligand 1 (PD-L1) expression in epithelial-originated cancer: a meta-analysis. Medicine (Baltimore) 2015;94:e515. doi: 10.1097/MD.0000000000000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolazzo C, Raimondi C, Mancini M, Caponnetto S, Gradilone A, Gandini O, Mastromartino M, Del Bene G, Prete A, Longo F, Cortesi E, Gazzaniga P. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Sci Rep. 2016;6:31726. doi: 10.1038/srep31726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts PJ, Stinchcombe TE. KRAS mutation: should we test for it, and does it matter? J. Clin. Oncol. 2013;31:1112–21. doi: 10.1200/JCO.2012.43.0454. [DOI] [PubMed] [Google Scholar]
- 32.Matikas A, Mistriotis D, Georgoulias V, Kotsakis A. Targeting KRAS mutated non-small cell lung cancer: a history of failures and a future of hope for a diverse entity. Crit Rev Oncol Hematol. 2017;110:1–12. doi: 10.1016/j.critrevonc.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Calles A, Liao X, Sholl LM, Rodig SJ, Freeman GJ, Butaney M, Lydon C, Dahlberg SE, Hodi FS, Oxnard GR, Jackman DM, Jänne PA. Expression of PD-1 and its ligands, PD-L1 and PD-L2, in smokers and never smokers with KRAS-mutant lung cancer. J Thorac Oncol. 2015;10:1726–35. doi: 10.1097/JTO.0000000000000687. [DOI] [PubMed] [Google Scholar]
- 34.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, Behrens C, Kadara H, Parra ER, Canales JR, Zhang J, Giri U, Gudikote J, Cortez MA, Yang C, Fan Y, Peyton M, Girard L, Coombes KR, Toniatti C, Heffernan TP, Choi M, Frampton GM, Miller V, Weinstein JN, Herbst RS, Wong KK, Zhang J, Sharma P, Mills GB, Hong WK, Minna JD, Allison JP, Futreal A, Wang J, Wistuba II, Heymach JV. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–77. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, Felip E, van den Heuvel MM, Ciuleanu TE, Badin F, Ready N, Hiltermann TJN, Nair S, Juergens R, Peters S, Minenza E, Wrangle JM, Rodriguez-Abreu D, Borghaei H, Blumenschein GR Jr, Villaruz LC, Havel L, Krejci J, Corral Jaime J, Chang H, Geese WJ, Bhagavatheeswaran P, Chen AC, Socinski MA CheckMate 026 Investigators. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376:2415–26. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong ZY, Zhong W, Zhang XC, Su J, Xie Z, Liu SY, Tu HY, Chen HJ, Sun YL, Zhou Q, Yang J, Yang X, Lin JX, Yan H, Zhai HR, Yan LX, Liao RQ, Wu SP, Wu YL. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 37.Song Z, Yu X, Cheng G, Zhang Y. Programmed death-ligand 1 expression associated with molecular characteristics in surgically resected lung adenocarcinoma. J Transl Med. 2016;14:188. doi: 10.1186/s12967-016-0943-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D’Incecco A, Andreozzi M, Ludovini V, Rossi E, Capodanno A, Landi L, Tibaldi C, Minuti G, Salvini J, Coppi E, Chella A, Fontanini G, Filice ME, Tornillo L, Incensati RM, Sani S, Crinò L, Terracciano L, Cappuzzo F. PD-1 and PD-L1 expression in molecularly selected non-small-cell lung cancer patients. Br J Cancer. 2015;112:95–102. doi: 10.1038/bjc.2014.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji M, Liu Y, Li Q, Li X, Ning Z, Zhao W, Shi H, Jiang J, Wu C. PD-1/PD-L1 expression in non-small-cell lung cancer and its correlation with EGFR/KRAS mutations. Cancer Biol Ther. 2016;17:407–13. doi: 10.1080/15384047.2016.1156256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma K, Chang D, Gong M, Ding F, Luo A, Tian F, Liu Z, Wang T. Expression and significance of FRA-1 in non-small-cell lung cancer. Cancer Invest. 2009;27:353–9. doi: 10.1080/07357900802254008. [DOI] [PubMed] [Google Scholar]
- 41.Soo RA, Loh M, Mok TS, Ou SH, Cho BC, Yeo WL, Tenen DG, Soong R. Ethnic differences in survival outcome in patients with advanced stage non-small cell lung cancer: results of a meta-analysis of randomized controlled trials. J Thorac Oncol. 2011;6:1030–8. doi: 10.1097/JTO.0b013e3182199c03. [DOI] [PubMed] [Google Scholar]
- 42.Ji M, Liu Y, Li Q, Li XD, Zhao WQ, Zhang H, Zhang X, Jiang JT, Wu CP. PD-1/PD-L1 pathway in non-small-cell lung cancer and its relation with EGFR mutation. J Transl Med. 2015;13:5. doi: 10.1186/s12967-014-0373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, Mikse OR, Cherniack AD, Beauchamp EM, Pugh TJ, Wilkerson MD, Fecci PE, Butaney M, Reibel JB, Soucheray M, Cohoon TJ, Janne PA, Meyerson M, Hayes DN, Shapiro GI, Shimamura T, Sholl LM, Rodig SJ, Freeman GJ, Hammerman PS, Dranoff G, Wong KK. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013;3:1355–63. doi: 10.1158/2159-8290.CD-13-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, Jiang CC, Jin L, Zhang XD. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol. 2016;27:409–16. doi: 10.1093/annonc/mdv615. [DOI] [PubMed] [Google Scholar]
- 45.Lastwika KJ, Wilson W 3rd, Li QK, Norris J, Xu H, Ghazarian SR, Kitagawa H, Kawabata S, Taube JM, Yao S, Liu LN, Gills JJ, Dennis PA. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76:227–38. doi: 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- 46.Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, Iwai Y, Long AJ, Brown JA, Nunes R, Greenfield EA, Bourque K, Boussiotis VA, Carter LL, Carreno BM, Malenkovich N, Nishimura H, Okazaki T, Honjo T, Sharpe AH, Freeman GJ. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2:261–8. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 47.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, Golub TR, Sebolt-Leopold J, Sellers WR, Rosen N. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–62. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Irving J, Matheson E, Minto L, Blair H, Case M, Halsey C, Swidenbank I, Ponthan F, Kirschner-Schwabe R, Groeneveld-Krentz S, Hof J, Allan J, Harrison C, Vormoor J, von Stackelberg A, Eckert C. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood. 2014;124:3420–30. doi: 10.1182/blood-2014-04-531871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36:320–8. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Won JK, Yang HW, Shin SY, Lee JH, Heo WD, Cho KH. The crossregulation between ERK and PI3K signaling pathways determines the tumoricidal efficacy of MEK inhibitor. J Mol Cell Biol. 2012;4:153–63. doi: 10.1093/jmcb/mjs021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.