Abstract
PARP inhibitor (PARPi) therapies have been approved for treating multiple germline BRCA mutated (gBRCAm) advanced cancers including metastatic pancreatic cancer. Although significantly prolonged progression-free survival was observed in gBRCAm pancreatic cancer patients, there was no improved overall survival. The underlined resistant mechanism to PARPi therapy is worth pursuing. Here, we reported a patient with advanced pancreatic cancer harboring a germline deleterious BRCA2 V1804Kfs mutation as well as somatic mutations in KRAS, TP53 and PTEN. Stable disease was achieved with the combination therapy of cisplatin and PARPi olaparib, but the disease quickly progressed after 18 weeks of treatment. Next-generation sequencing (NGS)-based genomic profiling of the liver metastasis and liquid biopsy revealed four newly acquired BRCA2 indel mutations, including two reversion mutations that could potentially restore BRCA2 function in the PARPi-resistant tumor. Our case showed that although initial response to PARPi therapy can be achieved in advanced gBRCAm pancreatic cancer patient, the tumor rapidly evolved to acquire multiple secondary BRCA2 mutations to restore the integrity of DNA repair and confer drug resistance, which may contribute to the unimproved overall survival in pancreatic cancer patients.
Keywords: Pancreatic cancer, germline BRCA2 mutation, PARP inhibitor, reversion mutation
Introduction
The five-year survival rate for metastatic pancreatic cancer remains as low as 3% [1], and therefore it is important for these patients to consider treatment options outside of the standard therapy. As a central mediator in homologous recombination (HR), BRCA1/2 plays a crucial role in repairing damaged DNA and maintaining genome stability [2]. Patients with a germline BRCA mutation (gBRCAm) have been reported in various of cancers, such as breast cancer, ovarian cancer, and in a small subgroup of metastatic pancreatic cancer [3]. PARP inhibitors (PARPi), such as olaparib, rucaparib and talazoparib, have demonstrated clinical activity in BRCA-mutated cancers through synthetic lethality [4-6]. A recent study has shown that gBRCAm pancreatic cancer patients had a longer progression-free survival (PFS) with maintenance olaparib treatment [7]. However, the overall survival (OS) showed no difference between the olaparib and placebo groups. In this case report, we described a germline BRCA2 mutation carrier with metastatic pancreatic cancer, who acquired olaparib-resistance after 18 weeks of treatment, and discussed the potential association between secondary BRCA2 reversion mutations and olaparib-resistance. This study may provide a clue for treatment-decision making in gBRCAm pancreatic cancer patients.
Materials and methods
Genomic DNA and circulating tumor DNA were extracted from the tumor tissue and plasma sample of the patient, respectively, followed by sequencing library construction according to the published protocols [8]. Hybridization capture-based targeted next-generation sequencing (NGS) with a 422-cancer-relevant-gene panel was performed on Illumina Hiseq platform. Genomic alterations were analyzed as previously described [8].
For RT-PCR analysis, total RNA was extracted from tumor tissue with PureLink RNA Mini Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instruction. RNA concentration and quality were measured using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). SuperScript VILO MasterMix kit (Thermo Fisher Scientific, Waltham, MA, USA) was used for reverse transcription, followed by PCR amplification using AmpliTaq DNA Polymerase kit (Thermo Fisher Scientific, Waltham, MA, USA) with specific primer sets for different variants. Primers used for each variant are listed as follows: 5’-AGCAAAAAAATGGCTTAGAG-3’ and 5’-GCAGGTTTTGTTAAGAGTTT-3’ for c.5302_6841+2036del; 5’-GCAAAGACCCTAAAGTACAG-3’ and 5’-ACAGGTAATCGGCTCTAAAG-3’ for c.4897_6807del. 70 ng of each PCR product was subjected to library construction with a KAPA Hyper DNA LibraryPrep Kit (for Illumina) (KAPA Biosystems, Wilmington, MA, USA), followed by NGS sequencing on Illumina Hiseq platform. BRCA2 variants were analyzed and viewed with the Integrative Genomics Viewer (IGV) software.
Case report
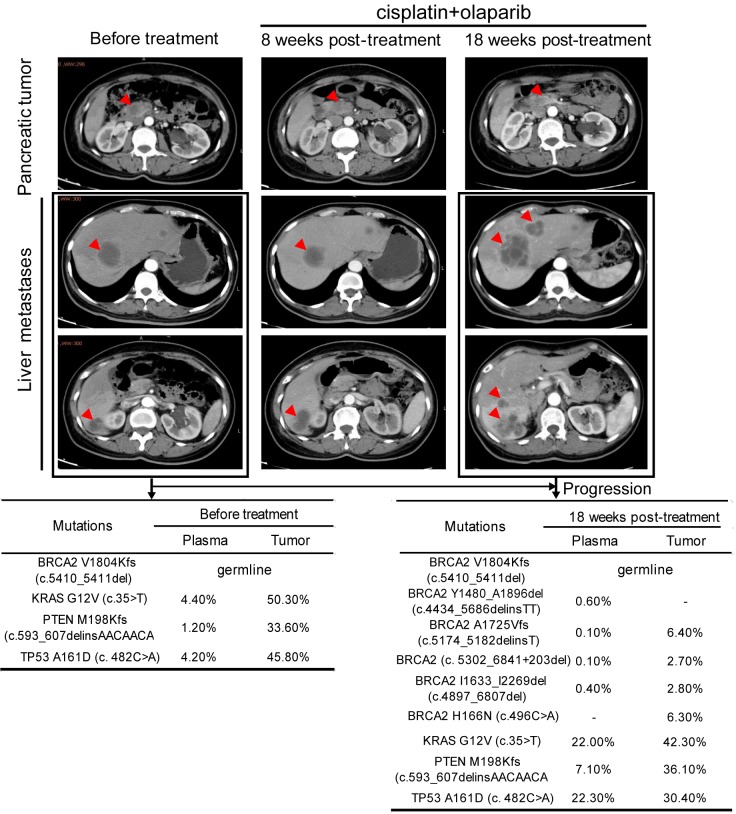
A 49-year-old female patient was diagnosed with stage IV pancreatic cancer accompanied by multiple liver metastases in January 2019 (Figure 1). To seek for more effective treatment strategy, the liver metastases tumor biopsy and plasma samples were subjected to genetic testing using targeted NGS for 422 cancer-related genes. A KRAS hotspot G12V mutation was revealed at a mutant allele frequency (MAF) of 50.3% in tumor sample, accompanied with a TP53 A161D mutation (45.8%) and a PTEN frameshift mutation M198Kfs (33.6%), which were also detected in the plasma sample at lower MAFs (Figure 1), and are all typical mutations frequently identified in pancreatic cancer. Although the clinical significance of KRAS G12V as an oncogenic driver is clear, no effective targeted drugs have been developed.
Figure 1.
Disease diagnosis and progression during treatment course with genetic testing results. Upper panel, CT images of the abdomen and pelvis showed the disease progression during treatment. Tumor lesions are indicated by red arrows in pancreas and liver. Lower panel, NGS-based genetic testing results for tissue and plasma biopsies collected before and 18 weeks after treatment. Mutant allele frequency (MAF) of each mutation was shown as percentage in the table. -: not detected.
Interestingly, a heterozygous BRCA2 frameshift c.5410_5411delGT (V1804Kfs) germline mutation that may result in a truncated and unfunctional BRCA2 protein was identified (Figures 1 and 2A), although no family history of cancers was reported. As a result, two cycles of cisplatin treatment (110 mg intravenously each cycle) with olaparib 300 mg orally twice a day were initiated in February 2019. When the patient was evaluated with computed tomography (CT) in March 2019, she achieved stable disease (SD) with a slight regression of the primary tumor and some lesions of the liver metastases, but other lesions in the liver continued to progress (Figure 1). Although the primary tumor in the pancreas was still stable at eighteen weeks post-treatment, aggressive progression was observed in the liver by CT scan (Figure 1). To figure out the underlying mechanisms of ineffective treatment in the metastatic areas, re-biopsy of the liver metastasis and plasma sample were collected and subjected to NGS-based genetic testing. Besides the previously detected KRAS, PTEN and TP53 mutations, four newly acquired somatic BRCA2 indels were observed, three of which were found both in the liver tumor and plasma sample, including c.5174_5182delinsT, c.4897_6807del and c.5302_6841+203del, whereas BRCA2 c.4434_5686delinsTT was only identified in the plasma sample with a relatively higher MAF level comparing to the three shared indels, suggesting that this mutation may come from the primary pancreatic tumor or other liver lesions (Figures 1 and 2A). Additionally, a liver tumor unique BRCA2 c.496C>A (H166N) with unknown significance was identified.
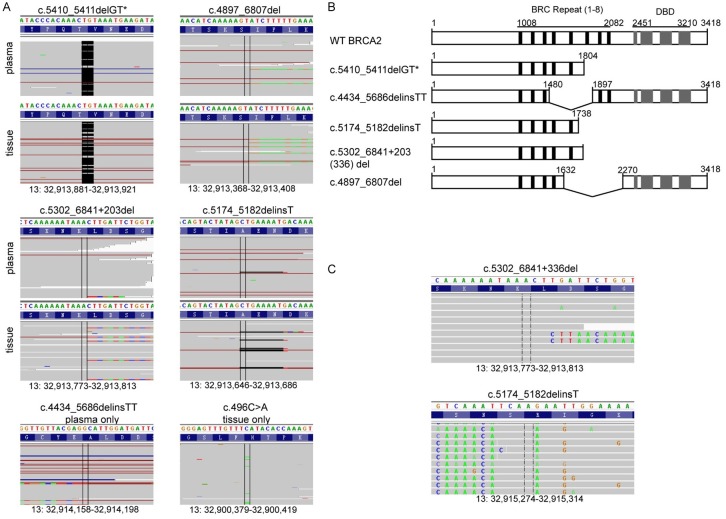
Figure 2.
Sequence analysis of BRCA2 mutants. A. Examination of the sequencing reads in Integrative Genomic Viewer (IGV) software revealed the germline and five newly acquired BRCA2 mutations in plasma and/or liver tumor biopsies. Bases that do not match to the reference genome due to point mutation or insertion are color coded, whereas deletions are indicated as a black dash (-). B. Diagram of BRCA2 protein domain structures caused by gBRCA2m and newly acquired BRCA2 mutations in treatment-resistance samples. C. NGS based RT-PCR analysis of the two long-range deletions were reviewed using IGV. Bases that do not match to the reference sequence are color coded. WT, wild type. *, germline mutation.
Within all these newly acquired BRCA2 mutations, two long-range deletions, the plasma-unique c.4434_5686delinsTT and the shared c.4897_6807del, have the potential to restore the open reading frame (ORF) of BRCA2 and express its c-terminal DNA-binding domain (DBD) (Figure 2B). c.4434_5686delinsTT (Y1480_A1896del) mutant produced a BRCA2 protein lacking a 417 amino-acid sequence that contains 3 BRC repeats (BRC 4-6). Similarly, c.4897_6807del (I1633_I2269del) mutant expressed a BRCA2 protein lacking 4 BRC repeats (BRC 5-8). These reversion mutations may contribute to the restoration of DNA repair through HR and potentially account for the resistance to olaparib and cisplatin treatment.
However, c.5174_5182delinsT and c.5302_6841+203del mutations could cause a truncated BRCA2 product similar to the germline mutation based on DNA-testing results. RT-PCR analysis followed by NGS of the resulted PCR products was used to confirm the effect of these BRCA2 mutations at RNA level. Indeed, the mRNA variation of c.4897_6807del was consistent with the prediction. However, c.5302_6841+203del mutation leaded to a c.5302_6841+336del at mRNA level (Figure 2C).
Discussion
BRCA mutation is widely reported in patients with breast and ovarian cancer, and may be a predictive biomarker for benefiting from DNA-damaging anticancer drugs such as PARP inhibitors [9-11]. The efficiency of PARPi in treating BRCA mutated advanced pancreatic cancer are currently under evaluation. A phase 3 clinical trial in metastatic pancreatic cancer patients who had a germline BRCA1/2 mutation demonstrated that olaparib-treated group has a significantly longer median PFS than the placebo group (7.4 months vs. 3.8 months) [7]. In our case, cisplatin plus olaparib treatment achieved a PFS of 4.5 months, shorter than the reported 7.4 months. We were aiming to identify the underlying mechanisms for the rapid olaparib resistance and tumor progression.
Several mechanisms account for PARPi resistance have been reported, including epigenetic regulation of BRCA expression [12], alternative mRNA splicing [13], regulation by miRNAs [14], and restoration of ORF to form a nearly full length BRCA by reversion mutations [15]. In our study, four newly acquired BRCA2 indel mutations were observed after PARPi and cisplatin resistance, indicating the heterogeneity and complexity of tumor genomic evolution during olaparib treatment. c.4434_5686delinsTT or c.4897_6807del could restore the ORF of BRCA2, and produce a BRCA2 protein lacking BRC repeats 4-6 or 5-8, respectively, but restoring the C-terminal DBD domain for DNA binding. BRC repeats of BRCA2 directly binds to RAD51, an important factor that catalyzes the recognition and strand exchange in DNA repair through HR [16,17]. However, studies have shown that each BRC repeat involved in different functions as only BRC 1-4 can enhance DNA strand exchange by RAD51 [18]. A previous in vitro study has shown that BRCA2 variants lacking BRC 6-8 were still competent in mediating PARPi resistance [19], suggesting that the c.4434_5686delinsTT (BRC4-6 loss) and c.4897_6807del (BRC 5-8 loss) reversion mutations identified in our study may restore certain BRCA2 function in order to confer PARPi resistance. More detailed functional investigation is needed for confirmation in future studies. Although c.4434_5686delinsTT variant was only detected in the plasma, it may be caused by the heterogeneity of cancer within different lesions, which cannot be fully captured by a single tumor biopsy sample.
c.5302_6841+203 (336) del and c.5174_5182delinsT mutations, which produced truncated BRCA2 proteins similar to the germline BRCA2 variant, were also identified at a similar or even higher MAF of c.4897_6807del reversion mutation in the treatment-resistant tumor. An additional BRCA2 H166N mutation with unknown significance was only identified in the relapsed liver tumor. The detailed function of these truncated mutations as well as the BRCA2 H166N mutation need to be further investigated in the future.
Conclusion
In summary, we reported an advanced pancreatic cancer patient with a BRCA2 germline mutation achieved SD on cisplatin plus olaparib treatment, but rapidly progressed after 4.5 months. Two ORF-restoring mutants lacking different BRC repeats and three mutants with unknown functions were newly acquired upon resistance. This report highlights a possible route to cisplatin and olaparib treatment failure for gBRCAm carriers, and demonstrated the heterogeneity and complexity of rapid tumor genomic evolution during treatment course of PARPi, which may provide a clue for treatment-decision making in gBRCAm pancreatic cancer patients.
Acknowledgements
The authors gratefully thank the patient and his family for their participation in this study. This work was supported by National Key R&D Program of China (No. 2017YFC097900/2017YFC0907904).
Disclosure of conflict of interest
Sisi Liu and Yang W. Shao are the employees of Nanjing Geneseeq Technology Inc.; Xue Wu is the employee of Geneseeq Technology Inc.; others claimed no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004;95:866–871. doi: 10.1111/j.1349-7006.2004.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2011;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spriggs DR, Longo DL. Progress in BRCA-mutated ovarian cancer. N Engl J Med. 2018;379:2567–2568. doi: 10.1056/NEJMe1812644. [DOI] [PubMed] [Google Scholar]
- 5.Ledermann JA. PARP inhibitors in ovarian cancer. Ann Oncol. 2016;27(Suppl 1):i40–i44. doi: 10.1093/annonc/mdw094. [DOI] [PubMed] [Google Scholar]
- 6.George A, Kaye S, Banerjee S. Delivering widespread BRCA testing and PARP inhibition to patients with ovarian cancer. Nat Rev Clin Oncol. 2017;14:284–296. doi: 10.1038/nrclinonc.2016.191. [DOI] [PubMed] [Google Scholar]
- 7.Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, Park JO, Hochhauser D, Arnold D, Oh DY, Reinacher-Schick A, Tortora G, Algul H, O’Reilly EM, McGuinness D, Cui KY, Schlienger K, Locker GY, Kindler HL. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z, Yang N, Ou Q, Xiang Y, Jiang T, Wu X, Bao H, Tong X, Wang X, Shao YW, Liu Y, Wang Y, Zhou C. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin Cancer Res. 2018;24:3097–3107. doi: 10.1158/1078-0432.CCR-17-2310. [DOI] [PubMed] [Google Scholar]
- 9.Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, Delaloge S, Li W, Tung N, Armstrong A, Wu W, Goessl C, Runswick S, Conte P. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 10.Papadimitriou M, Mountzios G, Papadimitriou CA. The role of PARP inhibition in triple-negative breast cancer: unraveling the wide spectrum of synthetic lethality. Cancer Treat Rev. 2018;67:34–44. doi: 10.1016/j.ctrv.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Tucker H, Charles Z, Robertson J, Adam J. NICE guidance on olaparib for maintenance treatment of patients with relapsed, platinum-sensitive, BRCA mutation-positive ovarian cancer. Lancet Oncol. 2016;17:277–278. doi: 10.1016/S1470-2045(16)00062-0. [DOI] [PubMed] [Google Scholar]
- 12.Ter Brugge P, Kristel P, van der Burg E, Boon U, de Maaker M, Lips E, Mulder L, de Ruiter J, Moutinho C, Gevensleben H, Marangoni E, Majewski I, Jozwiak K, Kloosterman W, van Roosmalen M, Duran K, Hogervorst F, Turner N, Esteller M, Cuppen E, Wesseling J, Jonkers J. Mechanisms of therapy resistance in patient-derived xenograft models of BRCA1-deficient breast cancer. J Natl Cancer Inst. 2016;108 doi: 10.1093/jnci/djw148. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Bernhardy AJ, Cruz C, Krais JJ, Nacson J, Nicolas E, Peri S, van der Gulden H, van der Heijden I, O’Brien SW, Zhang Y, Harrell MI, Johnson SF, Candido Dos Reis FJ, Pharoah PD, Karlan B, Gourley C, Lambrechts D, Chenevix-Trench G, Olsson H, Benitez JJ, Greene MH, Gore M, Nussbaum R, Sadetzki S, Gayther SA, Kjaer SK, kConFab I, D’Andrea AD, Shapiro GI, Wiest DL, Connolly DC, Daly MB, Swisher EM, Bouwman P, Jonkers J, Balmana J, Serra V, Johnson N. The BRCA1-Delta11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res. 2016;76:2778–2790. doi: 10.1158/0008-5472.CAN-16-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meghani K, Fuchs W, Detappe A, Drane P, Gogola E, Rottenberg S, Jonkers J, Matulonis U, Swisher EM, Konstantinopoulos PA, Chowdhury D. Multifaceted impact of microRNA 493-5p on genome-stabilizing pathways induces platinum and PARP inhibitor resistance in BRCA2-mutated carcinomas. Cell Rep. 2018;23:100–111. doi: 10.1016/j.celrep.2018.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pishvaian MJ, Biankin AV, Bailey P, Chang DK, Laheru D, Wolfgang CL, Brody JR. BRCA2 secondary mutation-mediated resistance to platinum and PARP inhibitor-based therapy in pancreatic cancer. Br J Cancer. 2017;116:1021–1026. doi: 10.1038/bjc.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Ma Z, Treszezamsky A, Powell SN. MDC1 interacts with Rad51 and facilitates homologous recombination. Nat Struct Mol Biol. 2005;12:902–909. doi: 10.1038/nsmb991. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez-Prieto R, Munoz-Cabello AM, Cabello-Lobato MJ, Prado F. Rad51 replication fork recruitment is required for DNA damage tolerance. EMBO J. 2013;32:1307–1321. doi: 10.1038/emboj.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carreira A, Kowalczykowski SC. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc Natl Acad Sci U S A. 2011;108:10448–10453. doi: 10.1073/pnas.1106971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]