Phalaenopsis orchids are popular potted ornamental plants around the world due to their beauty, floral diversity and long indoor blooming period. Orchids are also important in plant research, having tiny, dust‐like seeds, crassulacean acid metabolism photosynthesis, complex deletion of the genes encoding NADH dehydrogenase subunits and mycorrhizal symbiosis.
Previously, we investigated the orchid MADS gene family, which encodes DNA‐binding proteins that are highly expressed in floral organs and may be important for flower initiation and development (Lin et al., 2016). The Phalaenopsis MADS gene family includes more than 50 members (Chao et al., 2018). As it is challenging to obtain different combinations of mutants in perennial plants such as Phalaenopsis using traditional crosses, there is great interest in developing alternative techniques for gene family studies. Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR‐associated endonuclease (CRISPR/Cas) genome editing provides a convenient tool to obtain null and multiple mutants in nonmodel organisms, may prove useful for breeding and plant research (Li et al., 2019).
Here, we used two CRISPR/Cas strategies to generate multiple mutants in Phalaenopsis equestris MADS genes. First, using a vector containing a hygromycin selection marker (HPTII) and SpCas9 genes, we introduced three MADS target sites (MADS44, MADS36 and MADS8) together in one vector (pYLMADS8_36_44; Ma et al., 2015) or separately in individual vectors (P1300_MADS8, P1300_MADS36, P1300_MADS44; Lin et al., 2018) to produce single‐guide RNAs (sgRNAs). We used Agrobacterium tumefaciens‐mediated transformation (Hsing et al., 2016) to transfect explants with pYLMADS8_36_44 (the 3sg1C strategy) or a mixture of P1300_MADS8, P1300_MADS36 and P1300_MADS44 (the 3X1sg strategy). The transformed Agrobacteria were co‐cultured with Phalaenopsis explants (Figure 1a), and the explants were incubated on hygromycin medium for selection (Figure 1b, c). We numbered the antibiotic‐resistant explants (3sg1C#1 to 21 and 3X1sg#1 to 21) and incubated the regenerated transformants (e.g. 3sg1C#17‐1 to 3sg1C#17‐4) individually (Figure 1d). The genomic DNA of 3sg1C and 3X1sg transformants was isolated. Specific primers were used to amplify the target genes, SpCas9, sgRNA and HPTII.
Figure 1.
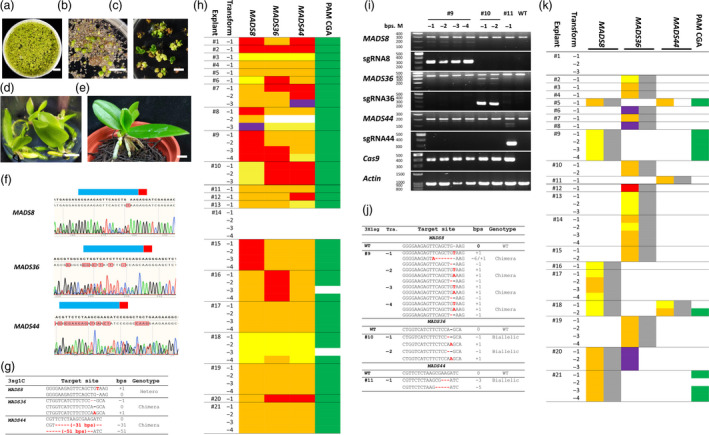
Strategies and results of CRISPR/Cas targeted mutagenesis of Phalaenopsis MADS genes. Homozygous, harbours the same edited sequences in both alleles; biallelic, both alleles were edited but the sequences were different; chimera, more than two alleles in the transformants; heterozygous, one wild type and one edited allele. In (g) and (j), letters in red, mutation; – in red, deletion. Numbers in parentheses indicate the number of bases deleted; ‘Hetero’, heterozygous. The coloured blocks in (h) and (k) indicate the genotypes of transformants that were edited in the genes shown above the column. Red, homozygous; orange, biallelic; yellow, chimera; purple, heterozygous and green, mutated in the sequences in MADS44 that are similar to the MADS8 target site. (a). One‐month‐old protocorms were incubated with 3sg1C Agrobacteria. Bar = 1 cm. (b). Transfected protocorms were incubated in hygromycin medium. The green protocorms are putative transformed explants. Bar = 1 cm. (c). Transformants with green true leaves. Bar = 1 cm. (d). Rooted 3sg1C#17‐1 to ‐4 incubated in hygromycin medium after 1‐month of subculture. Bar = 1 cm. (e). Two‐month‐old 3sg1C#13‐1 transformant. Bar = 0.5 cm. (f). In the 3sg1C#8‐3 transformant, three MADS gene target‐site regions were amplified and sequenced. Blue bar, target site; red bar, protospacer‐adjacent motif (PAM). Multiple peaks start from the sequences near the PAM indicating that there were mutated PCR products. (g). 3sg1C#8‐3 PCR products of three MADS gene target‐site regions were cloned, and eight clones from each construct were sequenced to determine the genotype. (h). MADS8, MADS36 and MADS44 target gene analysis in 3sg1C transformants. The explants are distinguished by lines (#1 to #21). Each row indicates one transformant (‐1 to ‐4). (i). DNA was isolated from each transformant derived from the 3X1sg strategy for PCR, including the target regions of MADS genes ( MADS8, MADS36 and MADS44); sgRNAs of each construct (sgRNA8, sgRNA36 and sgRNA44); Cas9 and actin as an internal control. The MADS gene PCR products were combined with wild‐type MADS DNA and tested using a T7 Endonuclease I assay. Cleavage of the PCR product indicates the presence of a mutation in this transformant. (j). These mutated MADS gene PCR products in (i) were cloned, and eight clones were sequenced for each construct. (k). MADS8, MADS36 and MADS44 target gene and sgRNA analysis in 3X1sg transformants. The coloured blocks indicate the genotype of transformants that were edited in the genes shown above the column.
The 51 3sg1C transformants were incubated in a growth chamber (Figure 1e). The DNAs of target gene regions were amplified and sequenced (Figure 1f, g, h). All except 3sg1C #14 (no sgRNA) contained the target MADS insertion/deletion (indel) mutation(s). Aside from 3sg1C#8‐2, 46 transformants derived from 20 explants contained triple mutants of all three target sites (97.9%, 46/47, Figure 1h). In comparison, a study in Dendrobium orchid produced an indel rate of only 10% (15/150) transformates in 15 target sites from five genes and only 33.3% target sites had mutations (Kui et al., 2017). Furthermore, 60.0% (12/20, Figure 1h) of explants were homozygous or biallelic for triple MADS gene mutations. Thus, all the transformants from these 12 explants were nonchimeric triple MADS‐null mutants. This feature makes the technique especially useful for gene editing of long‐juvenile‐phase, heterozygous and vegetative crops such as Phalaenopsis.
In the 3X1sg experiment, genotyping of 45 transformants derived from 21 explants indicated that except for 3X1sg#1 (no sgRNA) and 3X1sg#18, which carried two sgRNAs (MADS8 and MADS44), each carried only one sgRNA (Figure 1i, j, k). The numbers of transformants with one sgRNA in each of the indicated genes were 6, 12 and 1 for MADS8, MADS36 and MADS44, respectively (Figure 1k). The gene‐editing efficiencies of the sgRNAs were 100% (MADS8: 7/7; MADS36: 12/12 and MADS44: 2/2). The Cas9 codons in pYL‐derived and P1300‐derived transformants have been modified in different fashions—plant‐optimized in pYL‐derived (Ma et al., 2015) and human‐optimized in P1300‐derived (Lin et al., 2018)—but did not differ in their gene‐editing efficiencies in Phalaenopsis. These results indicate that multiple sgRNAs can be combined into a library and transformed to create an edited plant library. The mutants can be selected on the basis of phenotype, and the sgRNA sequenced to identify the gene(s) or DNA region(s) associated with the phenotype.
Compared with pYL‐derived (3sg1C) transformants, P1300‐derived (3X1sg) transformants had a higher proportion of chimerism (33.3%; 14/42, Figure 1k). This contrasts with the situation in protoplasts that were gene‐edited before cell division, which resulted mostly in homozygous or biallelic mutations. Therefore, high‐efficiency gene editing is important for vegetatively propagated crops for which there is no protoplast regeneration system. However, in some CRISPR/Cas9 systems, dicot plants are mutated less efficiently than monocots (Endo et al., 2019). Resolving the issue of chimerism in vegetatively propagated dicots will require increased efficiency of both CRISPR/Cas and protoplast regeneration.
Given current public concerns about genetically modified organisms (GMOs), methods to perform gene editing without transgenic gene integration are highly desirable. A recent Agrobacterium‐mediated transient expression experiment in tobacco (Nicotiana tabacum) using Cas9 and sgRNA yielded 8% nontransgenic gene‐editing mutants (Chen et al., 2018). The transformants from 3X1sg#20 had mutations in the target site of MADS36 without integration of MADS36 sgRNA and 3X1sg#5 had mutations in the target site of MADS44 without integration of MADS44 sgRNA into their genomes, indicating that nontransgenic targeted mutagenesis occurred in Phalaenopsis at an efficiency of 4.8% (2/42, Figure 1k).
The protospacer‐adjacent motif (PAM) neighbouring the target sequence is essential for CRISPR/Cas gene editing. The PAM sequence of SpCas9 is NRG. In Arabidopsis thaliana, the average efficiency was 52.7% when following an NGG PAM sequence but 0%–1.1% when following NGA, NGT or NGC (Ge et al., 2019). In rice (Oryza sativa), the same target site had a 65.5% efficiency when following a TGG PAM but 0% when following TGA, TGT or TGC (Endo et al., 2019). Notably, MADS44 contains a sequence similar to the MADS8 target sequence but with a CGA PAM. In our 3sg1C experiment, this PAM CGA target site was edited (Figure 1h). In the 3X1sg experiments, however, only the transformants with MADS8 sgRNA constructs had PAM CGA mutations in MADS44 (Figure 1k). Mutation to the PAM CGA target site in MADS44 created a sequence that could act as a MADS8 sgRNA.
In this study, single, double and triple Phalaenopsis mutants can be obtained by different sgRNA construction and transformation strategies, reducing the labour required for transformation. We obtained MADS‐null mutants of Phalaenopsis, a crop plant with a heterozygous genome and long juvenile period. This protocol has potential applications for gene family studies in other perennial plants.
Competing interests
The authors declare that they have no competing interests.
Author contributions
CGT performed Phalaenopsis transformation. FHW, YHY and YRC performed molecular biology experiments. CSL designed the experiments, interpreted the data and wrote the manuscript.
Acknowledgements
We thank Miranda Loney for English editing. This research was supported by Academia Sinica, Innovative Translational Agricultural Research Administrative Office (AS‐KPQ‐107‐ITAR‐10) and the Ministry of Science and Technology (105‐2313‐B‐001‐007 ‐MY3; 108‐2313‐B‐001 ‐011 ‐), Taiwan.
References
- Chao, Y.‐T. , Chen, W.‐C. , Chen, C.‐Y. , Ho, H.‐Y. , Yeh, C.‐H. , Kuo, Y.‐T. , Su, C.‐L. et al. (2018) Chromosome‐level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J. 16, 2027–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Li, W. , Katin‐Grazzini, L. , Ding, J. , Gu, X. , Li, Y. , Gu, T. et al. (2018) A method for the production and expedient screening of CRISPR/Cas9‐mediated non‐transgenic mutant plants. Hortic. Res. 5, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, M. , Mikami, M. , Endo, A. , Kaya, H. , Itoh, T. , Nishimasu, H. , Nureki, O. et al. (2019) Genome editing in plants by engineered CRISPR‐Cas9 recognizing NG PAM. Nat. Plants, 5, 14–17. [DOI] [PubMed] [Google Scholar]
- Ge, Z. , Zheng, L. , Zhao, Y. , Jiang, J. , Zhang, E.J. , Liu, T. , Gu, H. et al. (2019) Engineered xCas9 and SpCas9‐NG variants broaden PAM recognition sites to generate mutations in Arabidopsis plants. Plant Biotechnol. J. 17, 1865–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing, H.X. , Lin, Y.J. , Tong, C.G. , Li, M.J. , Chen, Y.J. and Ko, S.S. (2016) Efficient and heritable transformation of Phalaenopsis orchids. Bot. Stud. 57, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kui, L. , Chen, H. , Zhang, W. , He, S. , Xiong, Z. , Zhang, Y. , Yan, L. et al. (2017) Building a genetic manipulation tool box for orchid biology: identification of constitutive promoters and application of CRISPR/Cas9 in the orchid, Dendrobium officinale . Front. Plant Sci. 7, 2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. , Rui, H. , Li, Y. , Wang, Q. , Alariqi, M. , Qin, L. , Sun, L. et al. (2019) Robust CRISPR/Cpf1 (Cas12a)‐mediated genome editing in allotetraploid cotton (Gossypium hirsutum). Plant Biotechnol. J. 17, 1862–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.‐S. , Hsu, C.‐T. , Liao, D.‐C. , Chang, W.‐J. , Chou, M.‐L. , Huang, Y.‐T. , Chen, J.J.‐W. et al. (2016) Transcriptome‐wide analysis of the MADS‐box gene family in the orchid Erycina pusilla . Plant Biotechnol. J. 14, 284–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.‐S. , Hsu, C.‐T. , Yang, L.‐H. , Lee, L.‐Y. , Fu, J.‐Y. , Cheng, Q.‐W. , Wu, F.‐H. et al. (2018) Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single‐cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 16, 1295–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Zhang, Q. , Zhu, Q. , Liu, W. , Chen, Y. , Qiu, R. , Wang, B. , et al. (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 8, 1274–1284. [DOI] [PubMed] [Google Scholar]


