Summary
Oilseed rape (Brassica napus) is the third largest source of vegetable oil globally. In addition to food uses, there are industrial applications that exploit the ability of the species to accumulate the very‐long‐chain fatty acid (VLCFA) erucic acid in its seed oil, controlled by orthologues of FATTY ACID ELONGASE 1 (Bna.FAE1.A8 and Bna.FAE1.C3). The proportion of polyunsaturated fatty acids (PUFAs) in rapeseed oil is predicted to affect its thermal stability and is controlled by orthologues of FATTY ACID DESATURASE 2, particularly Bna.FAD2.C5. Our aim was to develop rapeseed lines combining high erucic and low PUFA characters and to assess the impact on thermal stability of the oil they produce. The new type of rapeseed oil (high erucic low polyunsaturate; HELP) contained a substantially greater proportion of erucic acid (54%) compared with high erucic rapeseed oil (46%). Although the total VLCFA content was greater in oil from HELP lines (64%) than from high erucic rapeseed (57%), analysis of triacylglycerol composition showed negligible incorporation of VLCFAs into the sn‐2 position. Rancimat analysis showed that the thermal stability of rapeseed oil was improved greatly as a consequence of reduction of PUFA content, from 3.8 and 4.2 h in conventional low erucic and high erucic rapeseed oils, respectively, to 11.3 and 16.4 h in high oleic low PUFA (HOLP) and HELP oils, respectively. Our results demonstrate that engineering of the lipid biosynthetic pathway of rapeseed, using traditional approaches, enables the production of renewable industrial oils with novel composition and properties.
Keywords: Brassica napus, rapeseed, oilseed rape, polyunsaturated fatty acids, thermal stability, erucic acid
Introduction
Oilseed rape (OSR) is the world’s third largest source of vegetable oil, after palm and soybean (FAO, 2018). It is a crop type of Brassica napus, which is a polyploid species (AACC, 2n = 38) formed by the spontaneous hybridization of B. rapa (AA, 2n = 20) and B. oleracea (CC, 2n = 18) (Nagaharu, 1935; Palmer et al., 1983; Parkin et al., 1995). Brassica is the crop genus most closely related to the widely used model plant species Arabidopsis thaliana (Yang et al., 1999), in which the lipid biosynthesis pathways leading to the complex suite of triacylglycerol (TAG) species found in storage oil have been extensively characterized (Li‐Beisson et al., 2013). An overview of key components of the pathway that are responsible for fatty acid synthesis and modification is provided in Figure 1. Rapeseed oil is an edible oil, but due to the presence of very‐long‐chain fatty acids (VLCFAs, carbon chain length ≥20), particularly erucic acid, it also has a wide range of industrial applications (Röbbelen, 1991; Zanetti et al., 2012). Feeding studies involving rats raised false concerns about adverse health effects of erucic acid (Beare et al., 1959; Charlton et al., 1975; Thomasson and Boldingh, 1955; de Wildt and Speijers, 1984), resulting in the breeding of low erucic acid rapeseed (LEAR) varieties for human consumption (Stefansson et al., 1961). High erucic acid rapeseed (HEAR) cultivars contain >45% erucic acid in their oil and are used as a ‘green feedstock’ for the oleochemical industry (Knutsen et al., 2016; Meakin, 2007). Erucic acid containing oils have high degree of lubricity and substantivity (ability to cling to the surfaces) than other oils (Piazza and Foglia, 2001). Erucic acid is derived into products such as erucamide, behenyl alcohol, brassylic acid, pelargonic acid, behenic acid, brassinolide and erucyl erucate (Caballero, 2006; Carlson et al., 1977; Molnar, 1974; Neischlag et al., 1967), which are used in the production of a wide range of industrial products, such as lubricants, slip additives, biofuels, pharmaceuticals, cosmetics, jet fuels, plasticizers, soaps, detergents, surfactants, textiles, recording material, rubber, nylon production and many more (Chang et al., 2013; Getachew et al., 2016; Iakovlieva et al., 2017; Johnson and Fritz, 1989; Neischlag et al., 1967; Nieschlag and Wolff, 1971; Pennick et al., 2012; Piazza and Foglia, 2001; Van Dyne and Blasé, 1991; Zanetti et al., 2012). Although other species have been evaluated for the production of erucic acid, the main source remains OSR (Hebard, 2016; Lalas et al., 2012; Sanyal et al., 2015).
Figure 1.
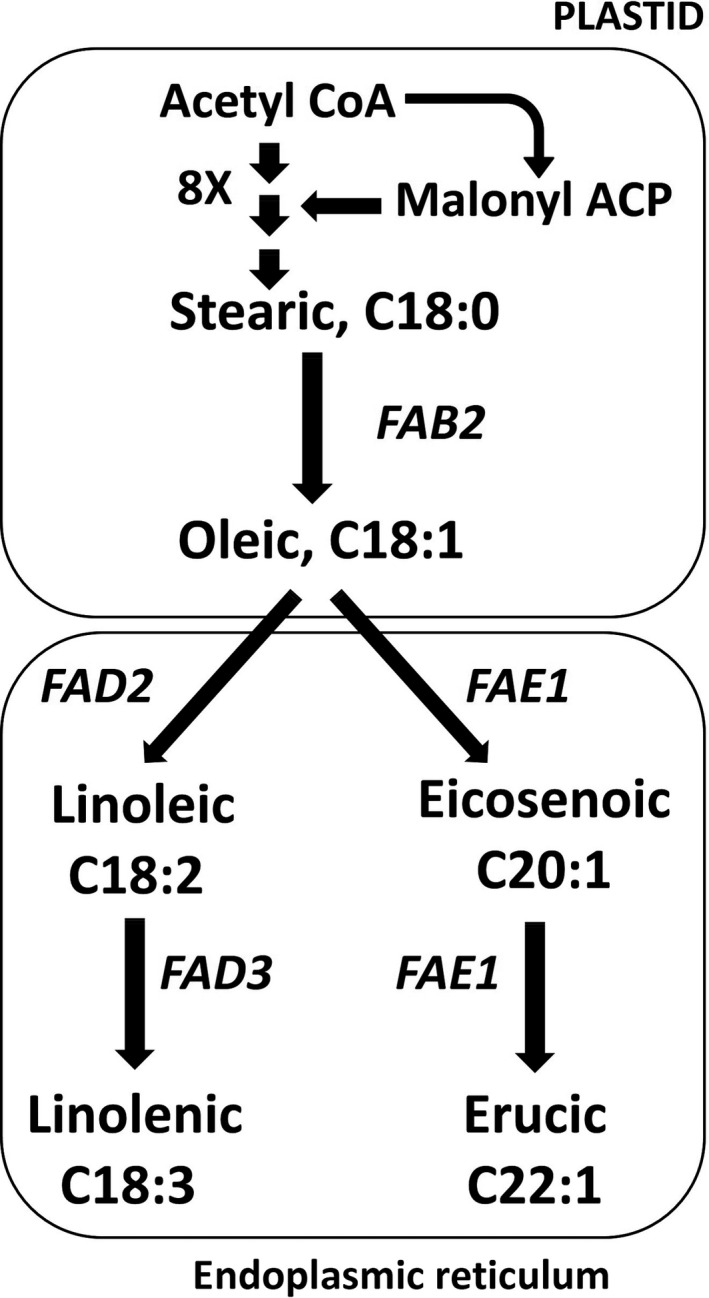
A simplified overview of the fatty acid biosynthesis pathway in A. thaliana seeds. In the Brassicaceae, the predominant product of fatty acid biosynthesis in the plastid is oleic acid (denoted C18:1 to indicate an acyl chain comprising 18 carbon atoms and containing 1 double bond). Further elaboration of acyl chains is catalysed by microsomal enzymes that either extend the acyl chains, for example to produce the very‐long‐chain fatty acids (VLCFAs) C20:1 and C22:1, or increase the extent of desaturation, that is the number bonds, for example to produce the polyunsaturated fatty acids (PUFAs) C18:2 and C18:3.
Studies in A. thaliana have shown the β‐ketoacyl‐CoA synthase encoded at the FATTY ACID ELONGASE1 (FAE1) locus to be rate limiting for the conversion of oleic acid (C18:1) to VLCFAs, and its orthologues have the same function in B. napus (Cassagne et al., 1987, 1994; James and Dooner, 1990; James et al., 1995; Kunst et al., 1992; Lemieux et al., 1990; Millar and Kunst, 1997; Roscoe et al., 2001; Stumpf and Pollard, 1983; Suneja et al., 1991). The same enzyme is required for two elongation steps: oleic to eicosenoic (C20:1) and eicosenoic to erucic acid (C22:1) (Kondra and Stefansson, 1965). B. napus has two orthologues of FAE1: Bna.FAE1.A8 and Bna.FAE1.C3. They act in an additive manner, with both being functional in HEAR varieties and nonfunctional in LEAR varieties (Ecke and Breeding, 1995; Harvey and Downey, 1964; Howell et al., 1996; Jourdren et al., 1996; Qiu et al., 2006; Thormann et al., 1996). The proportion of VLCFAs that can be accumulated in storage oil in B. napus is limited by the poor affinity of its native lysophosphatidic acid acyltransferase (LPAAT) for acyl groups with more than 18 carbon atoms, meaning that they are not incorporated into the sn‐2 position of the glycerol backbone during TAG biosynthesis (Frentzen, 1993; Katavic et al., 2001; Lassner et al., 1996). This results in a hypothetical limit of 66.7% VLCFAs. Up to 72% erucic acid has been achieved in B. napus by using a LPAAT transgene from Limnanthes douglasii to enable incorporation into the sn‐2 position (Nath, 2008; Nath et al., 2007).
Studies in A. thaliana have shown the oleate desaturase encoded at the FATTY ACID DESATURASE 2 (FAD2) locus to control the conversion of oleic acid (C18:1) to linoleic acid (C18:2) PUFAs (Miquel and Browse, 1992; Okuley et al., 1994). B. napus has four orthologues of FAD2: Bna.FAD2.A1, Bna.FAD2.A5, Bna.FAD2.C1 and Bna.FAD2.C5, corresponding to BnaA.FAD2.b, BnaA.FAD2.a, BnaC.FAD2.b and BnaC.FAD2.a, respectively, in previous studies (Hu et al., 2006; Scheffler et al., 1997; Schierholt et al., 2000; Smooker et al., 2011; Yang et al., 2012). A recent study (Wells et al., 2014) showed that, in rapeseed variety Cabriolet, only Bna.FAD2.C5 is functional. Thus, Bna.FAD2.C5 was targeted for chemical mutagenesis, resulting in population JBnaCAB_E, from which an allelic series of mutations of this gene was characterized. Several mutant lines produced seeds containing greatly reduced content of PUFAs, which was the aim of the study as polyunsaturation is anticipated to confer thermal instability (Browse et al., 1998; Durrett et al., 2008). However, this and a subsequent study (Bai et al., 2019) stopped short of confirming the predicted impact on thermal stability of rapeseed oil. An improvement in thermal stability improves the shelf life of edible oils (Kinney and Clemente, 2005) but would be transformative for industrial applications where greater thermal stability is required, such as feedstock for lubricants and hydraulic fluids, whilst minimizing environmental impact by retaining biodegradability (which mineral oil lacks).
In this study, we aimed primarily to characterize the impact on seed oil fatty acid composition of combining the low PUFA trait with the high erucic trait and thereby improving our understanding of the factors that limit erucic acid content. Having developed such rapeseed lines, we could address our secondary aim of assessing the impact of reducing PUFAs on the thermal stability of oil. Importantly, we used a strategy for development of the material that used genomics to accelerate ‘traditional’ mutation breeding, resulting in cultivars that are not considered genetically modified (GM) organisms.
Results
HELP development
We combined low PUFA and high erucic acid seed oil traits by crossing the low PUFA donors K0472 and K0047 (which had been generated by chemical mutagenesis of the low erucic acid variety Cabriolet as reported by Wells et al. (2014)) onto the current high erucic acid rapeseed cultivar Maplus. In F1 progeny (successful crosses assessed by Bna.FAD2 genotyping), two lines were randomly selected from each cross‐combination, grown and self‐pollinated to produce F2 progeny. Marker‐assisted selection was used in F2 generation to identify plants inheriting appropriate alleles at the two loci in this cross conferring the low PUFA trait (the inactive allele Bna.fad2.A5 and mutated versions of Bna.FAD2.C5 denoted either Bna.fad2.C5 K0472 or Bna.fad2.C5 K0047, depending on donor) along with the two alleles conferring the high erucic acid trait (Bna.FAE1.A8 and Bna.FAE1.C3). PCR primers and allele sequences are shown in Table S1, and sequence alignments of the amplicons of F2 plants are shown in Appendix [Link], [Link], [Link]. We had been unable to amplify the third orthologue of FAD2 reported previously as being required in inactive form for manifestation of the low PUFA trait (Bna.fad2.C1), and all individuals selected as inheriting low PUFA alleles Bna.fad2.A5 and Bna.fad2.C5 had inherited the low PUFA trait. From this, we deduce that variety Maplus already has the inactive (deleted) allele Bna.fad2.C1 as well as the inactive allele Bna.fad2.A1 that appears common in European rapeseed (Wells et al., 2014). Out of 192 F2 plants, one HELP plant ‘2‐91’ from cross Maplus x K0472 (K0472‐HE) and another HELP plant ‘4‐87’ from cross Maplus x K0047 (K0047‐HE) were selected. These were self‐pollinated up to F5 progeny, and the seeds were collected for fatty acid analysis. The summary of the development of HELP lines is shown in Figure S1.
Seed fatty acid composition of HELP lines
Fatty acid profile was analysed from F3 seeds of two HELP lines, 2‐91 (K0472‐HE) and 4‐87 (K0047‐HE), and high erucic acid control, Maplus as shown in Table 1. In comparison with Maplus, both HELP lines showed a slight increase in the very‐long‐chain fatty acids (sum of C20:1, C22:1 and C24:1) but showed a large decrease in the polyunsaturates (C18:2 and C18:3) percentages. As there were no biological replicates available for both HELP lines, F3 progeny was multiplied to F4 generation to assess the stability of fatty acid composition.
Table 1.
Seed fatty acid composition (% by weight, mean ± SD) of HELP F3 progeny and HEAR control, Maplus
| Line | Palmitic C16:0 |
Palmitoleic C16:1 |
Stearic C18:0 |
Oleic C18:1 |
Linoleic C18:2 |
Linolenic C18:3 |
Arachidic C20:0 |
Eicosenoic C20:1 |
Behenic C22:0 |
Erucic C22:1 |
Lignoceric C24:0 |
Nervonic C24:1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K0472‐HE (2‐91) | 2.2 ± 0.1 | 0.3 ± 0.1 | 0.7 ± 0.0 | 27.1 ± 0.5 | 1.7 ± 0.0 | 3.2 ± 0.1 | 0.6 ± 0.0 | 8.4 ± 0.1 | 0.4 ± 0.0 | 53.7 ± 1.0 | 0.1 ± 0.1 | 1.4 ± 0.9 |
| K0047‐HE (4‐87) | 2.3 ± 0.1 | 0.2 ± 0.0 | 0.7 ± 0.0 | 25.2 ± 0.1 | 2.9 ± 0.1 | 4.8 ± 0.0 | 0.6 ± 0.0 | 9.1 ± 0.0 | 0.4 ± 0.0 | 52.6 ± 0.5 | 0.1 ± 0.0 | 0.9 ± 0.3 |
| Maplus | 3.6 ± 0.1 | 0.2 ± 0.1 | 0.8 ± 0.0 | 12.3 ± 0.3 | 12.4 ± 0.0 | 8.2 ± 0.0 | 0.7 ± 0.0 | 8.7 ± 0.1 | 0.6 ± 0.0 | 51.1 ± 0.6 | 0.1 ± 0.0 | 0.8 ± 0.2 |
Each fatty acid value is the mean of three technical replicates.
Fatty acids were analysed for F4 seeds from 10 individual plants from each of the crosses, Maplus x K0472 and Maplus x K0047 (designated HELP lines K0472‐HE and K0047‐HE, respectively), along with parental lines as controls. The results are summarized in Table 2, and detailed fatty acid profiles for individual plants are provided in Appendix [Link], [Link], [Link]. Compared with Maplus, both HELP lines showed large increases in content of the main VLCFAs eicosenoic (C20:1), erucic (C22:1) and nervonic acid (C24:1) as shown in Figure 2. These increased from 55.7% of total fatty acids in Maplus to 63.3% in each of K0472‐HE and K0047‐HE HELP lines. Erucic acid increased from 46.1% in Maplus to 53.7% in K0047‐HE and 54.4% in K0472‐HE. One‐way ANOVA (analysis of variance) was calculated on five genotypes (having 10 replicates each)—Maplus, K0472, K0047, K0472‐HE and K0047‐HE. The P‐values for each of VLCFAs, PUFAs, OA and SAFAs were found to be <2 × 10− 16 (highly significant), and thus, these fatty acid groups differ significantly from each other. To know the difference in the individual genotypes, post hoc Tukey’s test was performed. The P‐values for the genotype groups, Maplus‐K0047 and Maplus‐K0472 were found to be highly significant. Thus, HELP lines differ significantly from HEAR, Maplus for these fatty acid groups. The relative proportions of the two most abundant VLCFAs (C20:1 and C22:1) varied between the individual plants representing the HELP lines and showed a strong inverse correlation (R 2 = 0.795), as shown in Figure S2. It shows that in some instances, the elongation from oleic acid (C18:1) goes up to eicosenoic acid and is not elongated to erucic acid, but the overall VLCFA composition remains constant.
Table 2.
Seed fatty acid composition (% by weight, mean ± SD) of HELP F4 progeny and parental controls
| Line | Mean fatty acid percentage composition of seed ± standard deviation | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Palmitic C16:0 |
Palmitoleic C16:1 |
Stearic C18:0 |
Oleic C18:1 |
Linoleic C18:2 |
Linolenic C18:3 |
Arachidic C20:0 |
Eicosenoic C20:1 |
Behenic C22:0 |
Erucic C22:1 |
Lignoceric C24:0 |
Nervonic C24:1 |
|
| K0472 | 3.4 ± 0.1 | 0.2 ± 0.2 | 1.2 ± 0.3 | 86.4 ± 1.4 | 1.8 ± 0.3 | 4.0 ± 0.5 | 0.6 ± 0.2 | 1.8 ± 0.2 | 0.2 ± 0.2 | 0.0 ± 0.1 | 0.2 ± 0.2 | 0.0 ± 0.1 |
| K0047 | 4.0 ± 0.3 | 0.2 ± 0.1 | 1.1 ± 0.2 | 82.0 ± 2.1 | 3.2 ± 0.6 | 6.5 ± 1.2 | 0.4 ± 0.2 | 1.8 ± 0.2 | 0.3 ± 0.2 | 0.0 ± 0.1 | 0.2 ± 0.1 | 0.1 ± 0.0 |
| Maplus | 4.2 ± 0.5 | 0.3 ± 0.1 | 0.8 ± 0.1 | 12.2 ± 1.8 | 16 ± 1.3 | 8.9 ± 1.0 | 0.7 ± 0.1 | 8.8 ± 1.3 | 0.6 ± 0.0 | 46.1 ± 2.3 | 0.2 ± 0.1 | 0.8 ± 0.2 |
| K0472‐HE (2‐91‐1 to 10) | 2.5 ± 0.4 | 0.2 ± 0.1 | 0.7 ± 0.1 | 26.8 ± 1.6 | 1.8 ± 0.4 | 3.6 ± 0.6 | 0.6 ± 0.1 | 8.1 ± 1.6 | 0.3 ± 0.1 | 54.4 ± 3.0 | 0.0 ± 0.0 | 0.8 ± 0.1 |
| K0047‐HE (4‐87‐1 to 10) | 2.2 ± 0.1 | 0.2 ± 0.0 | 0.8 ± 0.1 | 24.9 ± 1.3 | 2.7 ± 0.5 | 4.7 ± 0.5 | 0.7 ± 0.0 | 8.8 ± 1.2 | 0.4 ± 0.0 | 53.7 ± 1.7 | 0.1 ± 0.1 | 0.8 ± 0.0 |
Each fatty acid value is the mean of 10 biological replicates.
Figure 2.
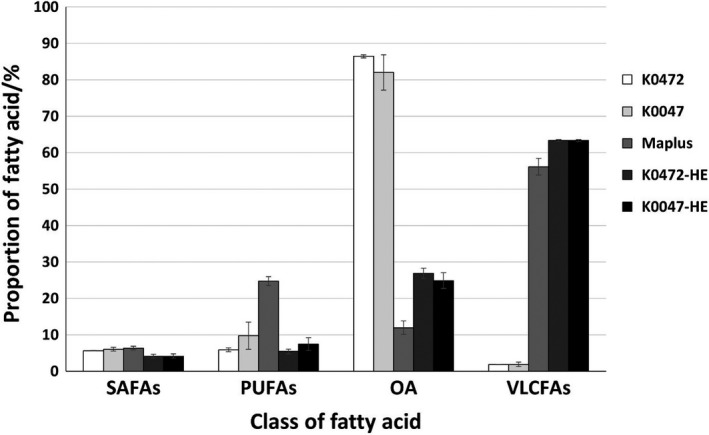
Seed fatty acid composition. Composition classified by fatty acid type: saturated fatty acids (SAFAs) = sum of C16:0, C18:0, C20:0, C22:0 and C24:0; polyunsaturated fatty acids (PUFAs) = sum of C18:2 and C18:3; oleic acid = C18:1; very‐long‐chain fatty acids (VLCFAs) = sum of C20:1, C22:1 and C24:1. Mean values plotted for 10 biological replicates each of K0472, K0047, Maplus, K0472‐HE and K0047‐HE, respectively. Standard deviation is shown as error bars.
In addition to the anticipated increase in the content of VLCFAs, the introduction of the low PUFA trait affected the content of other fatty acids as shown in Figure 2. As expected, compared with Maplus, both HELP lines showed large decreases in content of the main PUFA linoleic (C18:2) and linolenic acid (C18:3). These decreased from 24.9% of total fatty acids in Maplus to 5.4% in K0472‐HE and 7.4% in K0047‐HE. The proportion of saturated fatty acids (SAFAs) also reduced, with the total of all SAFAs reducing from 6.5% in Maplus to 4.1% in K0472‐HE and 4.2% in K0047‐HE. Both PUFAs and SAFAs are lower in the HELP lines than they are in the low erucic acid donors of the PUFA trait (6.6% and 7.0% SAFAs in K0472 and K0047, respectively; 7.0% and 8.9% PUFAs in K0472 and K0047, respectively).
In order to assess the stability of the HELP profile in further generations, 169 F4 plants were grown on, and selfed seed (F5 generation) was collected and analysed for fatty acid composition. The detailed fatty acid profiles for individual plants are provided in Appendix [Link], [Link], [Link]. These showed that the HELP profile is stably inherited. The relative proportions of erucic and eicosenoic acid were analysed. As illustrated in Figure S3, this showed correlation between the F5 and F4 generations. This suggests a heritable basis for the variation, such as a minor effect locus (the same alleles are present for the FAE1 orthologues, so the difference is not attributable to allelic variation in these).
Pattern of very‐long‐chain fatty acid incorporation into triacylglycerol
The increased proportion of VLCFAs in the HELP lines (63.3%) approaches the hypothetical limit for TAG in Brassica species (66.7%) imposed by the inability of Brassica LPAAT to incorporate such moieties at the sn‐2 position of the TAG molecule. We therefore tested whether, given the large proportion of VLCFAs that are synthesized and accumulated, incorporation had taken place at this position. Total lipids were extracted from HELP (K0472‐HE, F4 seeds) and its progenitors (Maplus and K0472) and TAGs analysed for fatty acids incorporated at the sn‐2 position (van Erp et al., 2010). The results, illustrated in Table S2, show negligible incorporation at the sn‐2 position, meaning that VLCFA occupation at the sn‐1 and sn‐3 positions can be calculated as ~95% (i.e. 63.3% observed of the 66.7% available).
Thermal stability of low polyunsaturated rapeseed oils
The reduced PUFA content of HELP rapeseed oil is predicted to improve thermal stability. To test this, we produced oil from field‐grown low PUFA lines K0472‐HE and K0472. The crude oil was subjected to Rancimat stability testing (Läubli et al., 1988), along with field‐grown conventional high and low erucic rapeseed oils (HEAR variety Maplus and LEAR variety Nikita, respectively). The fatty acid compositions of the crude oils are shown in Table S3, and the results of the stability test are shown in Table 3. As predicted, thermal stability improved greatly with the reduction in PUFA content. The stability of low erucic oil (variety Nikita) improved from 3.8 h for conventional LEAR rapeseed oil to 11.3 h for HOLP rapeseed oil. The stability of high erucic oil (variety Maplus) improved from 4.2 h for conventional HEAR to 16.4 h for HELP. The oxidative capacity of the oil types was assessed (shown in Table S4) and was not correlated with thermal stability.
Table 3.
Rancimat analysis of oil thermal stability
| Oil type | Source |
Induction time at 120 °C (h)† Mean ± SD |
|---|---|---|
| Low erucic acid rapeseed (LEAR) | LEAR (variety Nikita) | 3.8 ± 0.08 |
| High erucic acid rapeseed (HEAR) | HEAR (variety Maplus) | 4.2 ± 0.03 |
| High oleic low polyunsaturated (HOLP) | HOLP (line K0472) | 11.3 ± 0.17 |
| High erucic low polyunsaturated HELP) | HELP (line K0472‐HE) | 16.4 ± 0.24 |
Each value is mean of four technical replicates.
Discussion
Despite the opportunities for exploiting high erucic rapeseed oil as a renewable feedstock, growing demand from the oleochemical industry and its widespread cultivation, there has been relatively little progress increasing the proportion of erucic acid in oil produced commercially (Iakovlieva et al., 2017; Johnson and Fritz, 1989; Knutsen et al., 2016; Meakin, 2007; Nieschlag and Wolff, 1971; Röbbelen, 1991; Zanetti et al., 2012). Transgenic approaches have enabled exploration of one limitation in the accumulation of erucic acid in storage lipid, that imposed by the Brassica LPAAT enzyme, which cannot incorporate VLCFAs at the sn‐2 position of TAG (Bernerth and Frentzen, 1990; Brockerhoff, 1971; Cao et al., 1990; Frentzen, 1993; Katavic et al., 2001; Nath, 2008; Nath et al., 2007; Nath et al., 2009; Sasongko and Möllers, 2005). The analysis we undertook of FA composition at TAG sn‐2 positions (Table S2) confirmed expectations that even in the presence of additional substrate, VLCFAs are not incorporated at this position. Our studies do, however, provides insights into limitations imposed by the availability of monounsaturated 18‐carbon moieties for elongation to VLCFAs. Recapitulation of the combination of Bna.fad2 mutations shown previously to greatly restrict the conversion of oleic acid to PUFAs (Wells et al., 2014), but, in a high erucic acid background, increases the availability of C18:1 for extension by the Brassica elongases (controlled by the Bna.FAE1 loci). The observed impact is to increase the proportion of erucic acid in seed oil (by ~17% of its original value), demonstrating that the availability of C18:1 is, indeed, an important limitation for erucic acid content. To increase erucic acid content further, the introduction of an LPAAT capable of incorporation of VLCFAs at the sn‐2 position would be required. However, the pull through to VLCFAs already observed decreases the content of both saturated and polyunsaturated fatty acids (by ~38% and ~23%, respectively, of their values in the low PUFA donor line K0472), resulting in the oil produced by K0472‐HE containing ~90% monounsaturated fatty acids.
The same enzyme complex extends oleic (C18:1) to eicosenoic (C20:1) and eicosenoic to erucic acid (C22:1) (Kondra and Stefansson, 1965; Lassner et al., 1996). We observed variability in the content of eicosenoic, along with a negative correlation between contents of eicosenoic and erucic acids (Figure S2). Similar observations have been made previously in the progeny produced by crossing high erucic acid rapeseed with high oleic acid rapeseed (Sasongko and Möllers, 2005). It has been suggested that different alleles of B. napus orthologues of FAE1 have a different potential of producing erucic acid (Mahmood et al., 2003). This would not be unprecedented as, for example, the elongase encoded by the A. thaliana FAE1 locus uses oleic as a preferred substrate, resulting in the accumulation of eicosenoic rather than erucic acid (Katavic et al., 2001). In contrast, our results are consistent with the hypothesis that elongase activity is limiting the content of erucic acid in low PUFA lines, K0472‐HE and K0047‐HE. The observation of heritability to the F5 generation, as shown by correlation of the relative proportions of erucic and eicosenoic in F4 individuals and their progeny, indicates a genetic basis. As there is no variability for the active alleles of Bna.FAE1 in our experiments (these being inherited from Maplus), our observations can be attributed to the presence of ‘modifier’ loci for erucic acid content. An associative transcriptomics analysis of seed erucic acid content in a genetic diversity panel of B. napus indicated such a locus may be present on chromosome A5, with a gene encoding a fatty acid hydroxylase 1 identified as a candidate (Havlickova et al., 2018).
An increased content of erucic acid in HELP oil would undoubtedly be beneficial for industries producing compounds such as erucamide, for which rapeseed is the major source of erucic acid (Zanetti et al., 2012). HELP lines are expected to be superior to existing HEAR varieties because they contain a higher proportion of erucic acid in their oil (i.e. the yield of the primary product is greater) and due to anticipated reduction in processing costs. There is also an increasing global demand for more ‘environmental‐friendly’ alternatives to mineral oil. The key drawback of rapeseed oil (and, indeed for vegetable oils in general) is the thermal instability that is a consequence of their high content of PUFAs. To test our prediction that low PUFA rapeseed oil (both HOLP and HELP types) would have substantially greater thermal stability than conventional rapeseed oil, we tested this using an industry standard test (Rancimat; Läubli et al., 1988). The results confirmed a dramatic improvement compared with conventional rapeseed oil. For HOLP, the oil is suitable for edible uses such as repeated high‐temperature frying and retains the benefit of low saturate content. For HELP, the oil is a ‘non‐GM green feedstock’ for the production of large volumes of low‐cost, biodegradable products such as lubricants and hydraulic fluids, enabling more widespread use in sensitive environments. The development of non‐GM varieties even for industrial crops is important as the cost of deregulation (a requirement due to the release of the organisms into the environment) is prohibitive and many countries do not permit the cultivation of GM crops at all.
Experimental procedures
Plant material and growth conditions
Donors of the low PUFA trait were selected from the ethyl methane sulphonate‐mutagenized population JBnaCAB_E, derived from the winter oilseed rape variety Cabriolet (Wells et al., 2014). These lines carried mutations in Bna.FAD2.C5: K0472 and K0047. The donor of the high erucic acid trait was the current, open‐pollinated high erucic acid winter rapeseed variety Maplus (breeder: NPZ‐Lembke, Germany; https://www.npz.de/).
Seeds were sown in the medium‐grade compost (Scotts Levington F2 + S) and kept in the glasshouse under long‐day conditions of 16‐h photoperiod and temperatures of 20 °C/14 °C for day/night. At four‐leaf stage (after ~3 weeks of sowing), the plants were vernalized for 6 weeks with 8‐h photoperiod at 4 °C (vernalization being necessary as all lines are winter habit).
Development of HELP lines
K0472 and K0047 were crossed, as pollen donor, onto Maplus. F1 progeny were genotyped by PCR amplification and sequencing, with primers and alleles as indicated in Supplementary Table 1, to confirm the crosses. Two lines were randomly selected from each cross‐combination, grown and self‐pollinated. Ninety‐six F2 plants were grown for each and analysed for alleles of Bna.FAD2.A5, Bna.FAD2.C5, Bna.FAE1.A8 and Bna.FAE1.C3 by PCR amplification and sequencing, with primers and alleles as indicated in Table S1. Trace files were analysed using software Mutation Surveyor® (SoftGenetics LLC, State College, PA). One HELP plant ‘2‐91’ from cross Maplus x K0472 (K0472‐HE) and another HELP plant ‘4‐87’ from cross Maplus x K0047 (K0047‐HE) were confirmed to have inherited the required combination of homozygous alleles: Bna.FAE1.A8, Bna.FAE1.C3, Bna.fad2.A5 and Bna.fad2.C5. The sequence alignment files are provided in Appendix [Link], [Link], [Link]. The plants were self‐pollinated and the lines amplified to F4 for analysis.
DNA extraction and PCR amplification
One young leaf per plant was sampled at three‐leaf stage, and ‘DNeasy Plant 96 Qiagen Kit for 96 samples’ was used for the automated isolation of the DNA according to the manufacturer’s instructions (Qiagen, UK). The details of the primer pairs used for the amplification of Bna.FAD2.C5, Bna.FAD2.A5, Bna.FAE1.A8 and Bna.FAE1.C3 copies are shown in Table S1. DNA amplification was carried out in volumes of 25 μl with 50 ng of gDNA, 0.4 μM each of forward and reverse primers and 1× master mix (Thermo Scientific, Wilmington, DE). For Bna.FAD2 orthologues amplification, the PCR profile used was as follows: initial denaturation of 94 °C for 5 min; 35 cycles each with 94 °C for 30 s, 57 °C for 30 s and 72 °C for 1 min; final extension of 72 °C for 10 min. For Bna.FAE1 orthologues, the touchdown PCR profile was used: initial denaturation of 94 °C for 5 min; 15 cycles each with 94 °C for 30 s, 63 °C for 30 s (decrease by 1 °C every cycle) and 72 °C for 1 min; and 30 cycles each with 94 °C for 30 s, 53 °C for 30 s and 72 °C for 1 min; and final extension of 72 °C for 15 min. 1.5% agarose gel was used at 100 volts for 30 min for the separation of the PCR products, and these were observed under UV gel viewer.
PCR product purification, sequencing and analysis
PCR products were purified using the ‘Mag‐Bind® RXNPurePlus’ by following manufacturer’s protocol. Genewiz (https://www.genewiz.com) was used for the Sanger sequencing of the purified PCR products. Mutation surveyor® v5.0 software (SoftGenetics LLC, State College, PA) was used for the detection of the mutations from trace files.
Fatty acid composition analysis
Four milligrams of tripentadecanoin (Sigma‐Aldrich, Poole, Dorset, UK) internal standard was added to 30 mg of seeds, followed by the addition of 400 µL of cold hexane: isopropanol (3:2 v/v) solution. The seeds were ground using a Tissuelyser and incubated for 1 h on ice. The samples were centrifuged at ~9600 g for 5 min, and supernatant was transferred to a new tube. The pellet was washed another two times with 400 µL of cold hexane: isopropanol solution, vortexed, centrifuged and supernatants were pooled to the same tube. Then, 600 µL of 6.7% Na2SO4 was added, vortexed and centrifuged for 30 s. The upper phase was removed into a new tube and dried in the GenevacTM (EZ‐2.3 Elite model). These were reconstituted in 750 µL chloroform. 1/15th part of the sample (2–3 technical replicates) was transferred to a 2‐mL screw cap Supelco® glass vial. It was followed by the addition of 750 µL hexane and 500 µL 1N methanolic hydrochloric acid; incubation at 85 °C for 3 h; and addition of 250 µL of 0.9% KCl. The vials were vortexed, and upper hexane layer containing fatty acid methyl esters (FAMEs) was used the gas chromatography (GC) analysis. FAMEs were analysed on Thermo Scientific’s Trace GC Ultra‐FID with GC column (specifications: BP × forte 10 m × 0.1 mm ID × 0.2 m film thicknesses) with a run time of ~5 min per sample. Supelco® 37 component FAME mix (Sigma‐Aldrich) was used as an external standard, and hexane was used as a blank. Data were analysed with the Thermo Scientific’s ChromQuestTM software (version 4.2.34) platform.
Lipid extraction for TAG analysis
One millilitre of isopropanol containing butylated hydroxy toluene (BHT) was added to 4–6 seeds of each sample and heated in the oven at 85 °C for 15 min. Heat‐quenched samples were homogenized in a homogenizer; then, 2 mL of chloroform and 3 mL of methanol were added to each sample. It was followed by the addition of 1.6 mL of water, 2 mL of chloroform and 2 mL of 0.88% (w/v) KCl and gentle mixing. The tubes were centrifuged for 2 min at 400 g . The bottom phase was collected carefully in another tube and dried under N2. The lipid was resuspended in 500 µL of toluene containing 0.005% BHT for further analysis. FAMEs were derived using 10 µL of this sample, and total lipid content was calculated using GC results.
Positional distribution analysis of TAGs
Total lipid (1500 µL) was loaded on the thin‐layer chromatography (TLC) plate (20 × 20 cm, silica), and the mixture was separated using hexane: diethylether: acetic acid (70:30:1, v/v). TAGs were eluted from TLC silica twice by washing with 5 mL of chloroform: methanol (4:1, v/v). A phase separation was induced by adding 2 mL of methanol and 4 mL of 0.88% (w/v) potassium chloride. The chloroform phase was collected, and the aqueous phase was back extracted with 5 mL of chloroform. The chloroform phase was dried under N2 and re‐suspended in 500 L of toluene containing 0.005% BHT. TAGs were derivatized to FAMEs by using 10 µ of this sample and analysed by GC. Samples were dried down using N2, and 1 mg of TAG was resuspended in: 1 mL of diethylether, 0.8 mL of buffer (50 mM NaBr, 5 mM CaCl2, pH 7.6) and 200 µ of lipase (Rhizomucor miehei lipase; Sigma‐Aldrich). It was vortexed for 40 min, and the reaction was stopped by the addition of 2 mL of methanol: chloroform (1:1, v/v). The chloroform layer was collected, dried down and resuspended in 200 µ of chloroform. The mixture was separated by TLC using hexane: diethyl ether: acetic acid (35:70:1.5, v/v). 2‐monoacylglycerols (2‐MAGs) and TAGs were scrapped from the plate, and FAMEs were derived and analysed by GC.
Oxidative stability analysis
HOLP and HELP plants were grown in Moravian‐Silesian region of Czech Republic (49°56′N, 17°54′E), sown September 2017, harvested July 2018. Conventional low and high erucic rapeseed was sourced from the breeders (location grown not specified). Oil pressing was carried out using a small capacity Komet screw press (Model CA 59 G; IBG Monforts, Mönchengladbach, Germany), with a 6‐mm press nozzle die and a screw speed of 20 rpm. The oxidative stability of the pressed oils was determined using a ‘Metrohm Rancimat model 743’, according to AOCS Official Method Cd 12b‐92. Briefly, the induction times (n = 4) for portions of oil (3.0 g) was determined at 120 °C with 10 L/h air throughput. The induction point, as a measure of the oil stability index (OSI), is the time point of maximal change in the rate of oxidation.
Antioxidant properties of oil samples
The total antioxidant capacity (TAC) of oils was determined using a commercial kit (Sigma‐Aldrich, Catalogue number MAK187) to measure the concentration of the small molecule and protein antioxidants. This method is based on the detection of reduced Cu+ ion chelates with colorimetric probe at absorbance 570 nm, which is proportional to the total antioxidant capacity in Trolox equivalents (a water‐soluble vitamin E analogue used as an antioxidant standard). The absorbance at 570 nm was measured using spectrophotometric plate reader (Clariostar, BMG LABTECH Ltd. Oterberg, Germany). The concentrations of the samples were then calculated with the value obtained from trolox standards.
Conflict of interests
The authors declare no competing financial interests.
Author contributions
H.K., P.E. and I.B. conceived and designed the project. H.K., L.W., N.S., R.S and H.v.E. performed the experiments and analysed the results. H.K. and I.B wrote the manuscript. N.S., R.S. and P.E. revised the manuscript.
Supporting information
Figure S1 Summary of the development of HELP lines, K0472‐HE (2‐91) and K0047‐HE (4‐87).
Figure S2 Relationship between erucic and eicosenoic acid content of seeds of K0472‐HE and K0047‐HE.
Figure S3 Relationship between erucic / eicosenoic acid ratios in F4 plants and their F5 progeny.
Table S1 PCR primers used for selecting alleles required in HELP lines.
Table S2 Fatty acid composition at TAG sn‐2 position.
Table S3 Fatty acid composition of oils subjected to thermal stability testing.
Table S4 Total antioxidant capacity of oils subjected to thermal stability testing.
Appendix S1 Sequence alignment of amplicons of Bna.FAE1.A8, Bna.FAE1.C3, Bna.FAD2.A5 and Bna.FAD2.C5 copies.
Appendix S2 Detailed fatty acid composition of HELP F4 seeds. For each sample, the fatty acid percentage represents the mean of three technical replicates.
Appendix S3 Detailed fatty acid composition of HELP F5 seeds.
Acknowledgements
This work was supported by UK Biotechnology and Biological Sciences Research Council (BB/M028534/1 and BB/P012663/1) and an ICAR International Fellowship.
References
- Bai, S. , Engelen, S. , Denolf, P. , Wallis, J.G. , Lynch, K. , Bengtsson, J.D. , Van Thournout, M. et al.(2019) Identification, characterization and field testing of Brassica napus mutants producing high‐oleic oils. Plant J. 98, 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare, J.L. , Gregory, E.R.W. and Campbell, J.A. (1959) The effects of different varieties of rapeseed oil on weight gain, and of golden rapeseed oil on reproduction, of the rat. Can. J. Biochem. Physiol. 37, 1191–1195. [PubMed] [Google Scholar]
- Bernerth, R. and Frentzen, M. (1990) Utilization of erucoyl‐CoA by acyltransferases from developing seeds of Brassica napus (L.) involved in triacylglycerol biosynthesis. Plant Sci. 67, 21–28. [Google Scholar]
- Brockerhoff, H. (1971) Stereospecific analysis of triglycerides. Lipids, 6, 942–956. [DOI] [PubMed] [Google Scholar]
- Browse, J. , Spychalla, J. , Okuley, J. and Lightner, J. (1998) Altering the fatty acid composition of vegetable oils. In Plant Lipid Biosynthesis: Fundamentals and Agricultural Applications ( Harwood, J.L. , ed), pp. 131–153. New York, NY: Cambridge University Press (Seminar series). [Google Scholar]
- Caballero, A.G. (2006) DEFRAPLAST: New Polymer Plasticisers & Stabilisers from UK‐Grown Crambe. UK. Available at: http://sciencesearch.defra.gov.uk/Default.aspx?Menu=Menu&Module=More&Location=None&Completed=0&ProjectID=13368. [Google Scholar]
- Cao, Y.Z. , Oo, K.C. and Huang, A.H. (1990) Lysophosphatidate acyltransferase in the microsomes from maturing seeds of meadowfoam (Limnanthes alba). Plant Physiol. 94, 1199–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, K.D. , Sohns, V.E. , Perkins, R.B. and Huffman, E.L. (1977) Brassylic acid: chemical intermediate from high‐erucic oils. Indust. Eng. Chem. Prod. Res. Dev. 16, 95–101. [Google Scholar]
- Cassagne, C. , Lessire, R. , Bessoule, J.-J. and Moreau, P. (1987) Plant elongases. In The Metabolism, Structure, and Function of Plant Lipids ( Stumpf, P.K. , Mudd, J.B. and Nes, W.D. , eds), pp. 481–488. Boston, MA: Springer, New York. [Google Scholar]
- Cassagne, C. , Lessire, R. , Bessoule, J.J. , Moreau, P. , Creach, A. , Schneider, F. and Sturbois, B. (1994) Biosynthesis of very long chain fatty acids in higher plants. Prog. Lipid Res. 33, 55–69. [DOI] [PubMed] [Google Scholar]
- Chang, P. , Terbach, N. , Plant, N. , Chen, P.E. , Walker, M.C. and Williams, R.S.B. (2013) Seizure control by ketogenic diet‐associated medium chain fatty acids. Neuropharmacology, 69, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton, K.H. , Corner, A.H. , Davey, K. , Kramer, J.K. , Mahadevan, S. and Sauer, F.D. (1975) Cardiac lesions in rats fed rapeseed oils. Can. J. Comp. Med. 39, 261–269. [PMC free article] [PubMed] [Google Scholar]
- Durrett, T.P. , Benning, C. and Ohlrogge, J. (2008) Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 54, 593–607. [DOI] [PubMed] [Google Scholar]
- Ecke, W. and Breeding, P. (1995) Analysis of Synteny between Rapeseed (B. napus L.) and Arabidopsis thaliana’. p. 37075. [Google Scholar]
- van Erp, H. , Bates, P.D. , Burgal, J. , Shockey, J. and Browse, J. (2010) Castor phospholipid: diacylglycerol acyltransferase facilitates efficient metabolism of hydroxy fatty acids in transgenic arabidopsis. Plant Physiol. 155, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO . (2018) Food Outlook, Biannual Report on Global Food Markets. Available at: http://www.fao.org/3/CA0239EN/ca0239en.pdf [Google Scholar]
- Frentzen, M. (1993) Acyltransferases and Triacylglycerols. In Lipid metabolism in plants( Moore, T. S. ed.). Boca Raton, FL: CRC Press, Taylor & Francis Group. [Google Scholar]
- Getachew, P. , Getachew, M. , Joo, J. , Choi, Y.S. , Hwang, D.S. and Hong, Y.‐K. (2016) The slip agents oleamide and erucamide reduce biofouling by marine benthic organisms (diatoms, biofilms and abalones). Toxicol. Environ. Health Sci. 8, 341–348. [Google Scholar]
- Harvey, B.L. and Downey, R.K. (1964) The inheritance of erucic acid content in rapeseed (Brassica napus). Can. J. Plant Sci. 44, 104–111. [Google Scholar]
- Havlickova, L. , He, Z. , Wang, L. , Langer, S. , Harper, A.L. , Kaur, H. , Broadley, M.R. et al. (2018) Validation of an updated Associative Transcriptomics platform for the polyploid crop species Brassica napus by dissection of the genetic architecture of erucic acid and tocopherol isoform variation in seeds. Plant J. 93, 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebard, A. (2016) Successful commercialization of industrial oil Crops. In Indust Oil Crops (pp. 343–358). Champaign, IL: AOCS Press. [Google Scholar]
- Howell, P.M. , Lydiate, D.J. and Marshall, D.F. (1996) Towards developing intervarietal substitution lines in Brassica napus using marker‐assisted selection. Genome, 39, 348–358. [DOI] [PubMed] [Google Scholar]
- Hu, X. , Sullivan‐Gilbert, M. , Gupta, M. and Thompson, S.A. (2006) Mapping of the loci controlling oleic and linolenic acid contents and development of fad2 and fad3 allele‐specific markers in canola (Brassica napus L.). Theor. Appl. Genet. 113, 497–507. [DOI] [PubMed] [Google Scholar]
- Iakovlieva, A. , Boichenko, S. , Lejda, K. , Vovk, O. and Shkilniuk, I. (2017) ‘Vacuum distillation of rapeseed oil esters for production of jet fuel bio‐additives. Procedia Eng. 187, 363–370. [Google Scholar]
- James, D.W. and Dooner, H.K. (1990) Isolation of EMS‐induced mutants in Arabidopsis altered in seed fatty acid composition. Theor. Appl. Genet. 80, 241–245. [DOI] [PubMed] [Google Scholar]
- James, D.W. , Lim, E. , Keller, J. , Plooy, I. , Ralston, E. and Dooner, H.K. (1995) Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell, 7, 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.W. and Fritz, E. (1989) Fatty acids in industry: processes, properties, derivatives, applications. J. Dispers. Sci. Technol. 11, 433–434. [Google Scholar]
- Jourdren, C. , Barret, P. , Horvais, R. , Foisset, N. , Delourme, R. and Renard, M. (1996) Identification of RAPD markers linked to the loci controlling erucic acid level in rapeseed. Mol. Breeding, 2, 61–71. [Google Scholar]
- Katavic, V. , Friesen, W. , Barton, D.L. , Gossen, K.K. , Giblin, E. , Luciw, T. , An, J. et al. (2001) Improving erucic acid content in rapeseed through biotechnology: what can the arabidopsis FAE1 and the yeast SLC1‐1 genes contribute? Crop. Sci. 41, 739. [Google Scholar]
- Kinney, A.J. and Clemente, T.E. (2005) Modifying soybean oil for enhanced performance in biodiesel blends. Fuel. Proc. Technol. 86, 1137–1147. [Google Scholar]
- Knutsen, H.K. , Alexander, J. , Barregård, L. , Bignami, M. , Brüschweiler, B. , Ceccatelli, S. , Dinovi, M. et al. (2016) Erucic acid in feed and food. EFSA J. 14, 4593. [Google Scholar]
- Kondra, Z.P. and Stefansson, B.R. (1965) Inheritance of erucic and eicosenoic acid content of rapeseed oil (Brassica napus). Can. J. Genet. Cytol. 7, 505–510. [Google Scholar]
- Kunst, L. , Taylor, D.C. and Underhill, E.W. (1992) Fatty acid elongation in developing seeds of Arabidopsis thaliana . Plant Physiol. Biochem. 30(4), 425–434. [Google Scholar]
- Lalas, S. , Gortzi, O. , Athanasiadis, V. , Dourtoglou, E. and Dourtoglou, V. (2012) Full characterisation of crambe abyssinica hochst. Seed Oil. J. Am. Oil. Chemists Soc. 89, 2253–2258. [Google Scholar]
- Lassner, M.W. , Lardizabal, K. and Metz, J.G. (1996) A jojoba beta‐Ketoacyl‐CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant Cell, 8, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Läubli, M.W. , Bruttel, P.A. and Schalc, E. (1988) Bestimmung der Oxidationsstabilität von Fetten und Ölen – Vergleich zwischen der Active Oxygen method (AOCS Cd 12–57) und der Rancimat‐Methode. Lipid/Fett. 90, 56–58. [Google Scholar]
- Lemieux, B. , Miquel, M. , Somerville, C. and Browse, J. (1990) Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theoret. Appl. Genet. 80, 234–240. [DOI] [PubMed] [Google Scholar]
- Li‐Beisson, Y. , Shorrosh, B. , Beisson, F. , Andersson, M.X. , Arondel, V. , Bates, P.D. , Baud, S. et al. (2013) Acyl‐lipid metabolism. Arabidopsis Book, 11, e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood, T. , Ekuere, U. , Yeh, F. , Good, A.G. and Stringam, G.R . (2003) RFLP linkage analysis and mapping genes controlling the fatty acid profile of Brassica juncea using reciprocal DH populations. Theoret. Appl. Genet. 107, 283–290. [DOI] [PubMed] [Google Scholar]
- Meakin, S. (2007) High erucic acid rape (HEAR). In Crops for Industry: A Practical Guide to Non‐food and Oilseed Agriculture( Meakin, S. , ed.), pp. 94–110. Marlborough, UK: The Crowood Press Ltd. [Google Scholar]
- Millar, A.A. and Kunst, L. (1997) Very‐long‐chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 12, 121–131. [DOI] [PubMed] [Google Scholar]
- Miquel, M. and Browse, J. (1992) Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. Biochemical and genetic characterization of a plant oleoyl‐phosphatidylcholine desaturase. J. Biol. Chem. 267, 1502–1509. [PubMed] [Google Scholar]
- Molnar, N.M. (1974) Erucamide. J. Am. Oil Chem. Soc. 51, 84–87. [Google Scholar]
- Nagaharu, U. (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jap. J. Bot. 7, 389–452. [Google Scholar]
- Nath, U.K. (2008) Increasing erucic acid content in the seed oil of rapeseed (Brassica napus L.) by combining selection for natural variation and transgenic approaches. Georg‐August‐University Göttingen, Germany. [Google Scholar]
- Nath, U.K. , Becker, H.C. and Möllers, C. (2007) Increasing erucic acid content in rapeseed (Brassica napus L.). I(1997), 173–176. [DOI] [PubMed] [Google Scholar]
- Nath, U.K. , Goswami, G. , Clemens, R. , Becker, H.C. and Möllers, C. (2009) Inheritance and variation of erucic acid content in a transgenic rapeseed (Brassica napus L.) doubled haploid population. Mol. Breed. 23, 125–138. [Google Scholar]
- Neischlag, H.J. , Wolff, I.A. , Manley, T.C. and Holland, R.J. (1967) Brassylic acid from ozonolysis of erucic acid. Indust. Eng. Chem. Prod. Res. Dev. 6, 120–123. [Google Scholar]
- Nieschlag, H.J. and Wolff, I.A. (1971) Industrial uses of high erucic oils. J. Am. Oil Chem. Soc. 48, 723–727. [Google Scholar]
- Okuley, J. , Lightner, J. , Feldmann, K. , Yadav, N. , Lark, E. and Browse, J. (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell, 6, 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, J.D. , Shields, C.R. , Cohen, D.B. and Orton, T.J. (1983) Chloroplast DNA evolution and the origin of amphidiploid Brassica species. Theor. Appl. Genet. Theoretische und angewandte. 65, 181–189. [DOI] [PubMed] [Google Scholar]
- Parkin, I.A. , Sharpe, A.G. , Keith, D.J. and Lydiate, D.J. (1995) Identification of the A and C genomes of amphidiploid Brassica napus (Oilseed rape). Genome, 38, 1122–1131. [DOI] [PubMed] [Google Scholar]
- Pennick, G. , Chavan, B. , Summers, B. and Rawlings, A.V. (2012) The effect of an amphiphilic self‐assembled lipid lamellar phase on the relief of dry skin. Int. J. Cosmet. Sci. 34, 567–574. [DOI] [PubMed] [Google Scholar]
- Piazza, G.J. and Foglia, T.A. (2001) Rapeseed oil for oleochemical usage. Eur. J. Lipid Sci. Technol. 103, 450–454. [Google Scholar]
- Qiu, D. , Morgan, C. , Shi, J. , Long, Y. , Liu, J. , Li, R. , Zhuang, X. et al. (2006) A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theoret. Appl. Genet. 114(1), 67–80. [DOI] [PubMed] [Google Scholar]
- Röbbelen, G. (1991) Rapeseed in a changing world: plant production potential. In Proceedings of the 8th International Rapeseed Congress, Saskatoon, Canada, July 9–11. Saskatoon, Canada: Organizing Committee, Eighth International Rapeseed Congress, pp. 29–38. [Google Scholar]
- Roscoe, T.J. , Lessire, R. , Puyaubert, J. , Renard, M. and Delseny, M. (2001) Mutations in the fatty acid elongation 1 gene are associated with a loss of beta‐ketoacyl‐CoA synthase activity in low erucic acid rapeseed. FEBS Lett. 492, 107–111. [DOI] [PubMed] [Google Scholar]
- Sanyal, A. , Pinochet, X. , Merrien, A. , Laustriat, M., Decocq, G. and Fine, F. (2015) Rapeseed: Some examples of current French research. Erucic acid rapeseed: 1. Prospects of improvements. 22(3). [Google Scholar]
- Sasongko, N. and Möllers, C. (2005) Toward increasing erucic acid content in oilseed rape (Brassica napus L.) through the combination with genes for high oleic acid. J. Am. Oil Chem. Soc. 82, 445–449. [Google Scholar]
- Scheffler, J.A. , Sharpe, A.G. , Schmidt, H. , Sperling, P. , Parkin, I.A.P. , Lühs, W. , Lydiate, D.J. et al (1997) Desaturase multigene families of Brassica napus arose through genome duplication. Theoret. Appl. Genet. 94, 583–591. [Google Scholar]
- Schierholt, A. , Becker, H.C. and Ecke, W. (2000) Mapping a high oleic acid mutation in winter oilseed rape (Brassica napus L.). Theor. Appl. Genet. 101, 897–901. [Google Scholar]
- Smooker, A.M. , Wells, R. , Morgan, C. , Beaudoin, F. , Cho, K. , Fraser, F. and Bancroft, I. (2011) The identification and mapping of candidate genes and QTL involved in the fatty acid desaturation pathway in Brassica napus. Theor. Appl. Genet. 122, 1075–1090. [DOI] [PubMed] [Google Scholar]
- Stefansson, B.R. , Hougen, F.W. and Downey, R.K. (1961) Note on the isolation of rape plants with seed oil free from erucic acid. Can. J. Plant Sci. 41, 218–219. [Google Scholar]
- Stumpf, P.K. and Pollard, M. (1983) Pathways of fatty acid biosynthesis in higher plants with particular reference to developing rapeseed. In High and Low Erucic Acid Rapeseed Oils ( Kramer, J.K. , Sauer, F. and Pigden, W.K. , eds), pp. 131–141. New York: Academic Press. [Google Scholar]
- Suneja, S.K. , Nagi, M.N. , Cook, L. and Cinti, D.l. (1991) Decreased long‐chain fatty Acyl COA elongation activity in quaking and JIMPY mouse brain: deficiency in one enzyme or multiple enzyme activities? J. Neurochem. 57, 140–146. [DOI] [PubMed] [Google Scholar]
- Thomasson, H.J. and Boldingh, J. (1955) The biological value of oils and fatsii. The growth‐retarding substance in rapeseed oil: two figures. J. Nutri. 56, 469–475. [DOI] [PubMed] [Google Scholar]
- Thormann, C.E. , Romero, J. , Mantet, J. and Osborn, T.C. (1996) Mapping loci controlling the concentrations of erucic and linolenic acids in seed oil of Brassica napus L. Theoret. Appl. Genet. 93, 282–286. [DOI] [PubMed] [Google Scholar]
- Van Dyne, D.L. and Blasé, M. (1991) Commercialization of crops with high erucic acid for industrial uses. In Biotechnology of Plant Fats and Oils ( Rattray, J. , ed.), pp. 151–161. Champaign, IL: AOCS. [Google Scholar]
- Wells, R. , Trick, M. , Soumpourou, E. , Clissold, L. , Morgan, C. , Werner, P. , Gibbard, C. et al. (2014) The control of seed oil polyunsaturate content in the polyploid crop species Brassica napus . Mol. Breed. 33, 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wildt, D.J. and Speijers, G.J.A. (1984) Influence of dietary rapeseed oil and erucic acid upon myocardial performance and hemodynamics in rats. Toxicol. Appl. Pharmacol. 74, 99–108. [DOI] [PubMed] [Google Scholar]
- Yang, Y.W. , Lai, K.‐N. , Tai, P.‐Y. and Li, W.‐H. (1999) Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between brassica and other angiosperm lineages. J. Mol. Evol. 48, 597–604. [DOI] [PubMed] [Google Scholar]
- Yang, Q. , Fan, C. , Guo, Z. , Qin, J. , Wu, J. , Li, Q. , Fu, T. et al. (2012) Identification of FAD2 and FAD3 genes in Brassica napus genome and development of allele‐specific markers for high oleic and low linolenic acid contents. Theoret. Appl. Genet. 125, 715–729. [DOI] [PubMed] [Google Scholar]
- Zanetti, F. , Mosca, G. , Rampin, E. and Vamerali, T. (2012) Adaptability and sustainable management of high‐erucic brassicaceae in Mediterranean environment. In Oilseeds ( Akpan, U.G. ed.), pp. 99–116. Rome, Italy: InTech. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Summary of the development of HELP lines, K0472‐HE (2‐91) and K0047‐HE (4‐87).
Figure S2 Relationship between erucic and eicosenoic acid content of seeds of K0472‐HE and K0047‐HE.
Figure S3 Relationship between erucic / eicosenoic acid ratios in F4 plants and their F5 progeny.
Table S1 PCR primers used for selecting alleles required in HELP lines.
Table S2 Fatty acid composition at TAG sn‐2 position.
Table S3 Fatty acid composition of oils subjected to thermal stability testing.
Table S4 Total antioxidant capacity of oils subjected to thermal stability testing.
Appendix S1 Sequence alignment of amplicons of Bna.FAE1.A8, Bna.FAE1.C3, Bna.FAD2.A5 and Bna.FAD2.C5 copies.
Appendix S2 Detailed fatty acid composition of HELP F4 seeds. For each sample, the fatty acid percentage represents the mean of three technical replicates.
Appendix S3 Detailed fatty acid composition of HELP F5 seeds.


