Abstract
As the field of tissue engineering continues to advance rapidly, so too does the complexity of cell culture techniques used to generate in vitro tissue constructs, with the overall aim of mimicking the in vivo microenvironment. This complexity typically comes at a cost with regards to the size of the equipment required and associated expenses. We have developed a small, low-cost bioreactor system which overcomes some of the issues of typical bioreactor systems while retaining a suitable scale for the formation of complex tissues. Herein, we have tested this system with three cell populations/tissues: the culture of hepatocellular carcinoma cells, where an improved structure and basic metabolic function is seen; the culture of human pluripotent stem cells, in which the cultures can form more heterogeneous tissues resembling the in vivo teratoma and ex vivo liver tissue slices, in which improved maintenance of cellular viability is seen over the 3 days tested. This system has the flexibility to be used for a variety of further uses and has the potential to provide a more accessible alternative to current bioreactor technologies.
Keywords: bioreactor, three-dimensional, tissue culture, stem cells, liver slices
1. Introduction
Culturing cells and tissues in vitro is a complex undertaking with a wide range of factors to account for, from the composition of the growth medium to the mechanical properties of the culture surface or substrate. This has resulted in the development of a wide range of products which attempt to correctly recapitulate the desired in vivo conditions and thereby improve the viability, function and physiological relevance of cultured cells. Conventional two-dimensional (2D) cell culture is widely used and has many advantages, particularly its ease of use, high-throughput nature and scalability. However, it is severely limited by a number of factors, including enforced polarization through cellular flattening, the formation of unstirred layers and the absence of complex cell and tissue structures found in vivo [1].
To overcome the issues associated with 2D culture, a variety of more complex cell culture substrates can be employed. These fall broadly into several categories, including solid scaffolds, proteinaceous or polymer-based hydrogels, topographical surfaces and spheroid-based technologies [2,3]. These techniques are able to overcome some of the shortfalls of 2D culture through enabling the production of complex multi-layered cell structures or allowing for the patterning of cells in such a way to create a morphology more closely resembling that found in vivo while retaining a high level of user control over the tissue formation process [4–6].
While these methods allow for the recapitulation of the spatial arrangement of cells within the physiological environment, the introduction of improved structural organization alone is insufficient to mimic the in vivo microenvironment, and these techniques still cannot fully represent the complexity of dynamic biological systems. This is particularly important when in vitro systems are used to model human tissues and organs or maintain them ex vivo, as the vasculature and microcirculation have a crucial role in tissue formation and homeostasis [7,8]. This is due to the fact that vasculature not only maintains the viability of large tissue structures through oxygenation and the supply of nutrients, but also provides a route for the removal of waste and metabolites, preventing accumulation and the potential detrimental effects this leads to [9,10]. Introducing perfusion of medium into in vitro culture should promote the adoption of more physiological structures and functions, having previously been shown to lead to improved differentiation responses in a number of different cell populations including chondrocytes [11] and pluripotent stem cells [12–14].
Bioreactor systems have been developed to attempt to overcome some of the issues found in static culture conditions and so enhance cell structure and tissue viability. These typically introduce perfusion of the cell culture medium in order to replicate the dynamic in vivo environment and increase the diffusion rates close to the cell surface. Industrial bioreactors allow for large-scale culture and can allow for close monitoring of various in vitro parameters such as oxygen tension, glucose levels and metabolite production [2,15], but their applications are limited by the need for costly specialized equipment and the increased scale. This facilitates the culture of large homogeneous cell suspensions but is more limiting for the creation or ex vivo maintenance of complex heterogeneous tissues.
Microfluidic chip technology is also a popular choice for precision modelling of in vivo niches and environments while having the potential to reduce the size of cultures, allowing for higher throughput [16,17]. While these can be very successful, the small size that these cultures allow for requires technical expertise for assembling and maintaining the microfluidic systems. Other drawbacks of such systems are their inherent simplicity [15], limited capacity for synthesis and analysis owing to the small scale and inability to support the formation of appropriately sized tissue constructs with associated cellular complexity.
Because of the issues with creating appropriately sized tissue constructs in vitro alongside using a suitably scaled apparatus, there is demand for small and simple bioreactor systems which can create physiologically relevant tissue structures while retaining a small footprint. Other desirable properties of fluid flow systems, as with any cell culture technique, include low cost, reusability, ease of manipulation and routine use.
In this paper, we present a novel yet simple bench-top perfusion system, which can be used with standard cell culture equipment. The system has been fully designed and optimized with simplicity and accessibility in mind for analysis using a variety of techniques. We have investigated the physical properties of the system to demonstrate the fluid motions within and have tested it with the hepatocellular carcinoma cell line HepG2 and the pluripotent stem cell line TERA2.cl.SP12, as well as for the maintenance of ex vivo precision-cut liver tissue slices.
It is anticipated that this system can be used to create a more realistic and physiological microenvironment and, therefore, enhance the structure and functionality of cultures with a variety of applications across a wide range of tissue types. Potential applications for the system include basic research of cells in a dynamic environment, disease modelling with both in vitro models [18] and ex vivo tissues [19] owing to the ability to maintain large constructs, and more stringent analysis of the differentiation potential of pluripotent stem cell populations in an in vitro setting [20].
2. Material and methods
2.1. Bioreactor development
2.1.1. Design of the bioreactor
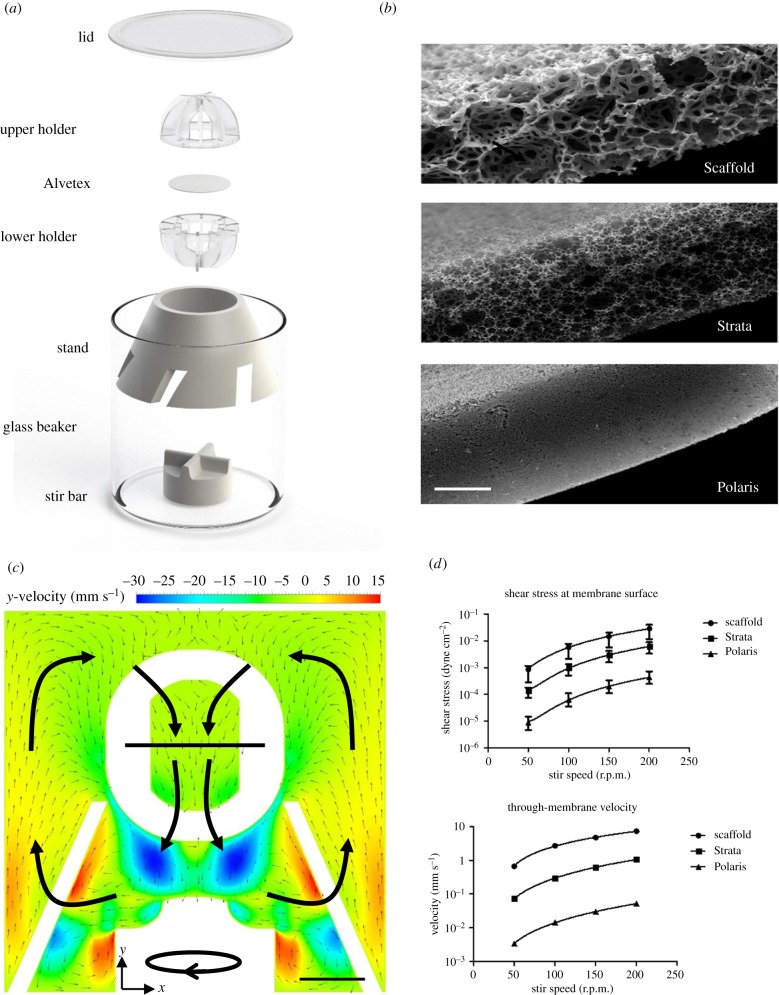
The bioreactor was designed to work using a magnetic stirrer to drive dynamic flow of the medium. A series of design properties were proposed for the system in order to achieve the desired conditions: minimal use of space, relatively low cost, ease of sterilization, flexibility to allow for use of different three-dimensional (3D) cell surfaces and the ability to tune the shear stress levels for different applications. A spherical polystyrene holder with a diameter of 28 mm was designed to house a porous membrane upon which cells would be grown or tissue slices maintained. This consists of a 5 mm wide central ring to clamp the membrane with ten 1 mm thick radially symmetric arches protruding outwards and meeting centrally to form the spherical shape. The design of this holder allows user control over the orientation of the culture in perfusion as well as making it easily transferrable between vessels and giving it the flexibility to be used in other applications. To support this holder, a vented cone was designed which acts as a baffle for the turbulence created by the stirrer, while the angled vents generate an effective medium recirculation pattern. The cone base diameter is 58 mm, which tapers to 32 mm at the top. It has a wall thickness of 2 mm. The vents cut into the base are 5 mm wide and 10 mm high and are installed at a 45° angle. This was made from polytetrafluoroethylene (PTFE), a highly inert and biocompatible polymer which has the benefit of being repeatedly autoclavable. This system is housed in a glass beaker (70 mm high, inner diameter 60 mm) with a loose-fitting polystyrene lid to enable adequate gas diffusion. To facilitate the stirring, a 25 mm magnetic stir bar was used and the apparatus was placed on a six-point magnetic stirrer (VELP Scientifica, Usmate, Italy) set to 80 r.p.m. Alvetex® membranes (Reprocell Europe, Sedgefield, UK) [21] were used in the system to support the cell culture owing to the highly porous nature of the membrane allowing adequate levels of flow of the medium through the culture.
2.1.2. Computational fluid dynamics
The system was analysed using transient 3D computational fluid dynamic (CFD) studies based on numerical solution of the Navier–Stokes equations. The geometry of the system was generated using Solidworks 2018 (Dassault Systèmes, Vélizy-Villacoublay, France). ANSYS Fluent (Ansys, Canonsburg, PA, USA) was used to carry out the meshing, fluid flow calculations and post processing. Patch-conforming tetrahedral meshing was used to create a mesh with 9.3 million elements. The fluid flow was simulated with an incompressible, isothermal fluid with a dynamic viscosity of 1.003 g m−1 s and a constant density of 998.2 kg m−3. A transient, pressure-based solver was used with the realizable k-epsilon turbulence model and scalable wall functions. The second-order special discretization was used throughout, and pressure–velocity coupling was undertaken using the pressure implicit with splitting of operator algorithm. The motion of the stir bar was modelled using a rotating zone with a frame motion in the y-axis centred on the origin at varying speeds from 50 to 200 r.p.m. Shear stress and through-membrane velocity were calculated as an average across a horizontal plane at the membrane surface and at the midpoint of the membrane, respectively, with minimum and maximum values as the limits.
2.2. Cell and tissue culture
2.2.1. HepG2 conventional cell culture
HepG2, a human hepatocellular carcinoma cell line, was maintained using minimum essential medium (MEM; Gibco, Invitrogen, Paisley, UK) supplemented with 10% fetal bovine serum (FBS; Gibco), 2 mM l-glutamine (Lonza, Basel, Switzerland) and 100 U ml−1 of penicillin/streptomycin (Gibco) in T75 and T175 flasks (Greiner Bio-One, Kremsmunster, Austria). Cells were passaged at 80% confluency using 0.25% trypsin/EDTA (Gibco) and split into new flasks at a ratio of 1 : 3. Cells were incubated and maintained at 37°C under 5% CO2.
2.2.2. HepG2 perfusion system set-up
Alvetex Strata inserts were prepared according to the manufacturer's instructions and placed in a six-well cell culture plate. HepG2 cells were detached from the cell culture flasks using 0.25% trypsin/EDTA, counted using the trypan blue exclusion assay to obtain accurate cell numbers and seeded at a density of 2 million cells per insert in 100 µl of medium. Five millilitres of medium was added after 1 h and the inserts were cultured for 2 days.
For perfusion experiments, the Alvetex Strata was unclipped from the inserts after 2 days and placed into the culture-holding device. A second prepared Alvetex Strata membrane was placed over the top of the culture before the device was clipped together and placed into the beaker of the perfusion system containing 110 ml of MEM prepared as previously described. The apparatus was placed in an incubator at 37°C on a magnetic stirrer set at 80 r.p.m. and cultured for a further 7 days. The same set-up method was used for the static controls, but with the magnetic stirrer set at 0 r.p.m.
2.2.3. TERA2.cl.SP12 conventional cell culture
The human embryonal carcinoma cell line TERA2.cl.SP12 was maintained using Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% heat-treated FBS (Gibco), 2 mM l-glutamine (Lonza) and 100 U ml−1 of penicillin/streptomycin (Gibco) in appropriate T25 and T75 flasks (BD Biosciences, Oxford, UK), with the medium being changed every 1–2 days as necessary. Cells were passaged on reaching 100% confluency by rolling acid-washed glass beads across the surface to dislodge cells, before transferring the cell suspension to new flasks. Cells were passaged into new flasks T75 at a ratio of 1 : 3 and were incubated and maintained at 37°C under 5% CO2.
2.2.4. TERA2.cl.SP12 perfusion system set-up
Cells were detached using the 0.25% trypsin/EDTA method and counted using the trypan blue exclusion assay to obtain accurate cell numbers. The appropriate cell volume for the number of inserts was then transferred to an Eppendorf tube, with the appropriate volume of growth factor-reduced Matrigel® matrix (Corning®, Fisher Scientific UK) to give a final concentration of 3 mg ml−1 in a defined volume. Alvetex Polaris inserts were prepared according to the manufacturer's instructions and placed in six-well plates ready for seeding, and the cell/matrix suspension was added to the top. Inserts were incubated at 37°C under 5% CO2 for at least 30 min to allow the Matrigel to set. After setting, medium was carefully added to the outside of the insert and left for 2 days for cells to adhere. To transfer to the perfusion system, perfusion devices were sterilized in 70% ethanol and washed in phosphate-buffered saline (PBS). Alvetex Polaris membranes were carefully unclipped from the holder and the disc was placed into the perfusion device, with a second Polaris disc added to the top. The device was then clipped together and placed in the vessel with 120 ml of medium and placed on the stirrer in the incubator for 7 days. Static control samples were generated by transferring the membranes to a six-well plate and changing the medium every 2–3 days. To compare these samples with in vivo tissue, samples from teratocarcinoma xenograft tumours were made available from previous work [22].
2.2.5. Rat precision-cut liver slice preparation and perfusion culture
Rat livers were harvested from Wistar rats killed by cervical dislocation after being anaesthetized with isoflurane. Livers were stored in ice-cold, oxygenated, Williams medium E (WME; Gibco) with 10% FBS (Gibco), 2 mM l-glutamine (Lonza), 100 U ml−1 penicillin/streptomycin (Gibco) and 1 mM sodium pyruvate (Gibco) immediately upon removal. Cores were cut from the livers using an 8 mm biopsy punch (Kai Medical, Solingen, Germany), and these were subsequently embedded in 4% low-gelling-temperature agarose (Sigma-Aldrich, UK) at 37°C. The embedded cores were sectioned at a thickness of 250 µm using a Leica VT1000 S vibratome (Leica Biosystems, Milton Keynes, UK). Sections were placed onto Alvetex Strata and clipped into the culture holders before being placed into 110 ml of oxygenated WME prepared as described previously in the perfusion apparatus. The beakers were sealed with parafilm and aluminium foil and placed into a 37°C incubator on a magnetic stirrer set to 80 r.p.m. and cultured for time periods of 24, 48 or 72 h. To measure the wet weight, the slices were placed onto filter paper to remove excess medium before being placed into a weighing boat and measured.
2.3. Analytical techniques
2.3.1. Haemotoxylin and eosin staining
Haematoxylin and eosin (H&E) staining was carried out according to the following protocol. Slides were deparaffinized in Histoclear II (National Diagnostics, USA) for 15 min, before rehydration through 100% ethanol for 2 min, 95% ethanol for 1 min, 70% ethanol for 1 min and demineralized water (dH2O) for 1 min. Slides were stained in Mayer's haematoxylin (Sigma-Aldrich) for 5 min, before washing in dH2O for 30 s, and incubating in alkaline alcohol for 30 s to blue the nuclei. Slides were dehydrated in 70% ethanol and 95% ethanol for 30 s each. Slides were stained in eosin for 1 min before two washes in 95% ethanol for 10 s. Slides were then dehydrated in 100% ethanol for 15 s, and then again in 100% ethanol for 30 s. Slides were cleared twice in Histoclear II for at least 5 min in each. To mount, excess Histoclear II was carefully removed from the slides, and a small amount of DPX (Fisher Scientific UK) or Omni-mount (National Diagnostics) placed on the slide, before a coverslip was added to the top. Slides were left to dry and set for at least 30 min before imaging using a Leica microscope.
2.3.2. Immunofluorescent staining
Slides were first deparaffinized in Histoclear II for at least 15 min, before rehydration for 5 min each in 100% ethanol, 95% ethanol, 70% ethanol and dH2O. Antigen retrieval was performed by incubation in citrate buffer at 95°C for 20 min, and slides were cooled by the addition of dH2O. Blocking buffer was prepared with either 20% normal goat serum (Sigma-Aldrich) or, for CGR8 teratoma slides, 20% newborn calf serum (Fisher Scientific UK) in 0.4% Triton-X-100 in PBS, and slides were blocked for at least 1 h at room temperature. Primary antibodies diluted in blocking buffer were added at the appropriate concentrations and incubated at 4°C overnight. The next day, slides were washed thrice in PBS before addition of appropriate fluorescently conjugated secondary antibodies and the nuclear stain Hoescht diluted in blocking buffer. Slides were incubated at room temperature for 1 h before washing thrice in PBS. Slides were mounted in Vectashield™ (Vector Labs, Peterborough, UK) and a coverslip placed on top, which was sealed around the edges using nail varnish. Slides were stored at 4°C until imaging.
2.3.3. MTT assay
Alvetex inserts were transferred to 12-well plates before washing in sterile PBS. PBS was then aspirated, thiazolyl blue tetrazolium bromide powder (MTT; Sigma-Aldrich) was dissolved in phenol-free DMEM (Gibco) at a concentration of 1 mg ml−1, and 1 ml was added to each well. Cells were incubated at 37°C under 5% CO2 for 60 min, before MTT solution was aspirated. One millilitre of acidified isopropanol was added to each well to lyse cells, which were placed in the dark on a shaker set at 80 r.p.m. for 10 min. Sample solution was then diluted appropriately in isopropanol to a total volume of 200 µl and placed in a flat-bottomed 96-well plate. Samples were then read at 570 nm on a microplate spectrophotometer.
2.4. Urea assay
Samples of medium were taken from the apparatus at days 1, 3, 5 and 7. Urea secretion was quantified using the QuantiChrom™ urea assay kit (BioAssay Systems, Haywood, CA, USA) according to the manufacturer's instructions for cell culture media with low urea levels. The resulting values were normalized to the total protein in the samples as measured with the Bradford assay.
2.5. Albumin assay
Albumin production was measured using the AssayMax human albumin enzyme-linked immunosorbent assay kit (AssayPro, St. Charles, MO, USA). The assay was carried out according to the manufacturer's instructions with all samples of medium diluted 20× in deionized water. Results were normalized to total protein in the samples as measured with the Bradford assay.
2.6. Bradford assay
The total protein in the samples was measured using a Bradford assay. Cells were lysed by placing in M-PER (Fisher Scientific, UK) at 4°C and sonicating for 30 min followed by freezing. Upon thawing, samples were centrifuged for 20 min at 13 000 r.p.m. and the supernatant was used for the assay. Five microlitres of sample was placed in each well of a 96-well plate with 250 µl of Bradford reagent and incubated at room temperature for 20 min. The absorbance at 570 nm was measured using a plate reader and values were quantified using a standard curve.
3. Results
3.1. Bench-top perfusion system
A system has been designed that has the following attributes: small yet relevant size, ease of use, low cost and ability to be re-used. As shown in figure 1a glass and PTFE were used for the larger components of the system, both of which are inert and autoclavable. The apparatus can be used to hold multiple formats of Alvetex membrane in support of cells and/or tissues (figure 1b). The addition of up to 120 ml of medium to the system allows the cultures to be carried out for up to two weeks without changing the medium, reducing rapid changes in culture conditions. Efficient stirring of the medium prevents the formation of unstirred layers and ensures nutrients and waste products in the medium are well distributed and exchanged effectively with the cells.
Figure 1.
Schematic, technical details and fluid flow of the bench-top bioreactor. (a) The system is composed of a glass beaker with an inner diameter of 60 mm and height of 70 mm with a volume of 198 ml. In the beaker sits a PTFE stand to secure the holder, into which the Alvetex membrane is placed. The stir bar is placed beneath the PTFE stand in order to generate the fluid flow. The 25 mm stir bar is commercially available (Fisher Scientific), while the rest of the apparatus was custom made by the Durham University mechanical workshops. (b) Scanning electron microscopy images of the three different forms of Alvetex membrane used in this study. Images courtesy of Reprocell Europe. Scale bar is 100 µm. (c) The CFD analysis of the system with an Alvetex Strata membrane inserted and the stirrer at 100 r.p.m. Fluid recirculation in the x–y plane is shown by the vectors, while the y-(vertical-) velocity component is shown by the heat map. Large arrows are added manually to show overall fluid recirculation pattern. Scale bar is 10 mm. (d) Shear stress and through-membrane velocity values were calculated in the system across a range of stir speeds. Values are mass weighted averages with error bars denoting the maximum and minimum values.
CFD analysis of fluid movement in the bioreactor shows the recirculation of medium throughout the system, which aids nutrient exchange (figure 1c). The use of different forms of Alvetex membranes allows control over the levels of shear stress experienced by cells (figure 1d) with only minor effects on the overall mixing of the medium. With control over the type of Alvetex as well as the speed of the stir bar the shear stress can be successfully tailored to mimic physiological conditions in a variety of tissues, allowing for great flexibility of use. The velocity through the membrane is also controlled in this manner. For example, for experiments on liver cells, Alvetex Strata was used and stirring performed at 80 r.p.m. This creates levels of shear stress below 10−3 dyne cm−2, which have been shown to have positive effects on hepatocyte metabolism [23,24] as well as being significantly below levels which reduce hepatocyte viability [25].
3.2. Formation of 3D complex structures by HepG2 cells
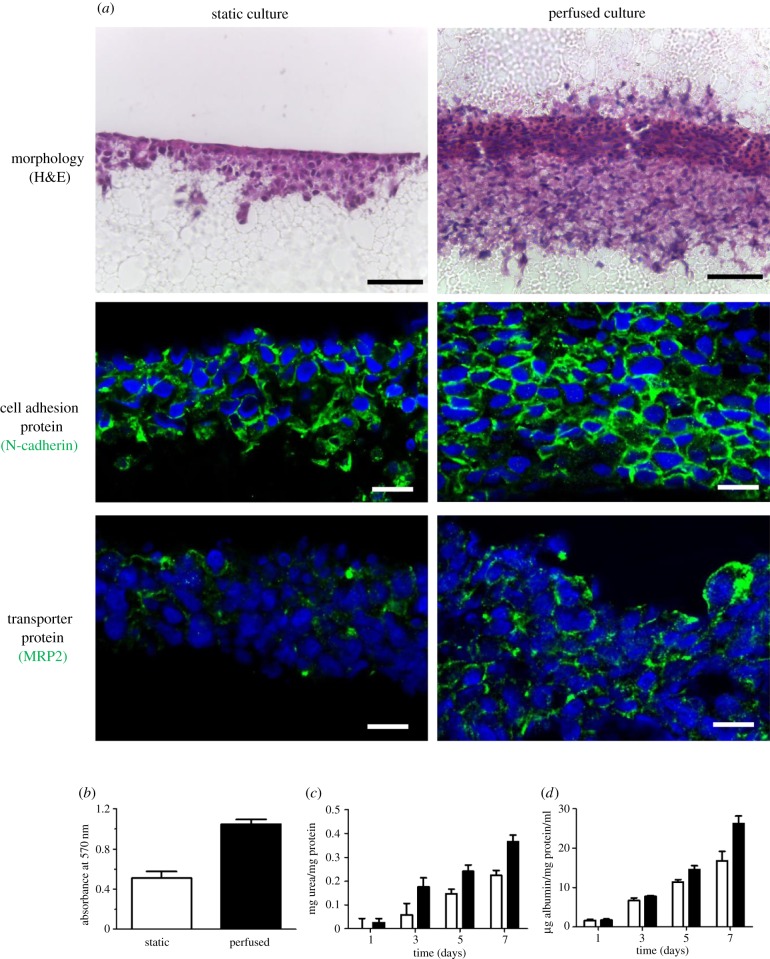
After 7 days in perfusion culture, HepG2 cells grow into a dense, multi-layered 3D structure, as shown in figure 2. The use of a sandwich of Alvetex membranes allowed for a more evenly distributed cell layer to be established with reduced clumping and island formation. Cells are anchored more effectively between membranes rather than on top of a single membrane. In perfusion culture, a significantly thicker cellular layer formed with greater levels of 3D cell–cell contacts (figure 2a). Enhanced levels of N-cadherin expression suggest increased cell–cell interactions. Apical cell markers such as MRP2 show more localized expression in perfusion cultures, indicating greater polarization of cells and the possible formation of bile canaliculi between adjacent cells (figure 2a). The observed increase in the number of viable cells present indicates that perfusion is having a beneficial effect on cell proliferation and/or survival (figure 2b). The results of the urea and albumin assays further demonstrate the enhanced metabolic capacity of the cells (figure 2c,d, respectively).
Figure 2.
Analysis of HepG2 cultures grown in perfusion compared with static culture. After 2 days in static culture, HepG2 cells were either kept in these conditions or made into a sandwich culture and grown in the perfusion system for a further 7 days. (a) H&E staining shows an increase in tissue thickness in perfusion alongside greater invasion into the Alvetex Strata, while immunofluorescent staining of N-cadherin shows a high level of cell–cell junctions and expression of MRP2 shows greater specificity and localization in perfused cultures compared with static. Scale bars are 50 µm for H&E and 10 µm for immunofluorescence images. (b) MTT assay indicates that the viability of cells is increased under perfused conditions. White columns are static cultures, while black columns are perfused cultures. (c,d) Urea and albumin assays, respectively, show increased activity when in perfusion, reflecting greater levels of functionality in dynamic 3D culture. Values are mean ± standard error of the mean, n = 3 for the urea and albumin assays while n = 6 for the MTT assay.
3.3. Pluripotent stem cells recapitulate aspects of teratocarcinoma structure in the perfused system
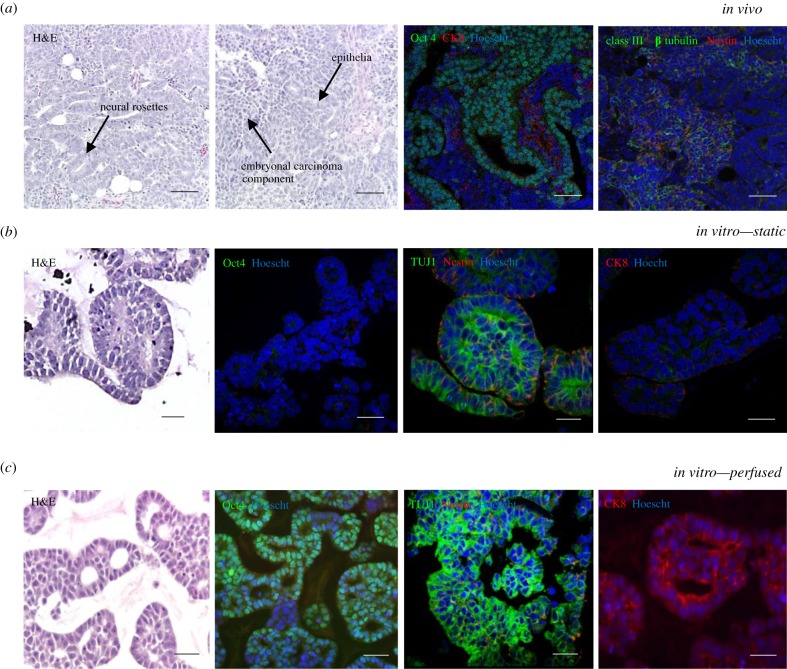
TERA2.cl.SP12 stem cells were seeded onto the Alvetex insert at a density of 2 million cells in Matrigel at a concentration of 3 mg ml−1 and cultured in the perfusion system for 7 days. The histology in figure 3 shows that cells in the perfused and static culture form organized morphologies. In the perfused system, cells produced an embryonal carcinoma phenotype with smaller, more rounded cells as well as cells showing a differentiated morphology, whereas in the static system cells produced more differentiated structures only. TERA2.cl.SP12 stem cells have a propensity to form neural derivatives; when compared with static cultures, the perfused samples showed an increase in differentiation marker class III β tubulin and a reduction in the early neural marker Nestin, indicating strong mature neural differentiation within the sample. In addition, there was evidence of ectodermal epithelial differentiation shown by positive staining for the epithelial marker cytokeratin 8. The expression of the pluripotency marker Oct4 indicates the maintenance of embryonal carcinoma stem cell populations and was only observed in the perfusion culture samples. The phenotype and morphology of the 3D tissue structures created in the perfused system bear some similarities to a teratocarcinoma xenograft tumour derived from the same cell lineage (figure 3a), with evidence of both pluripotent stem cells (embryonal carcinoma) and differentiated cells indicative of a teratoma (figure 3b,c).
Figure 3.
Perfusion of 3D culture enhances the phenotype of pluripotent stem cells in vitro, recapitulating aspects of the in vivo structure and phenotype of the teratocarcinoma. (a) Teratocarcinoma xenograft tumour derived from TERA2.cl.SP12 cells—teratocarcinomas derived from pluripotent stem cells display differentiated teratoma structures, neural rosette staining for Nestin and class III β tubulin (TUJ1) and cytokeratin 8-positive epithelial structures, and pluripotent embryonal carcinoma areas, which stain positively for Oct4. Scale bar is 100 µm. (b) Static culture—TERA2.cl.SP12 cells cultured in static conditions differentiate towards the neural lineage, but do not show the maintenance of any pluripotent areas. Scale bar is 50 µm. (c) Perfused culture—TERA2.cl.SP12 cells maintained in the perfused system showed both the maintenance of pluripotency and the formation of differentiated structures of both neural and epithelial identity, similar to the in vivo teratocarcinoma. This replicates both components of a germ cell tumour, notably aspects of the differentiated teratoma and undifferentiated stem cells in the embryonal carcinoma. Scale bar is 50 µm.
3.4. Enhanced viability of liver tissue slices maintained ex vivo
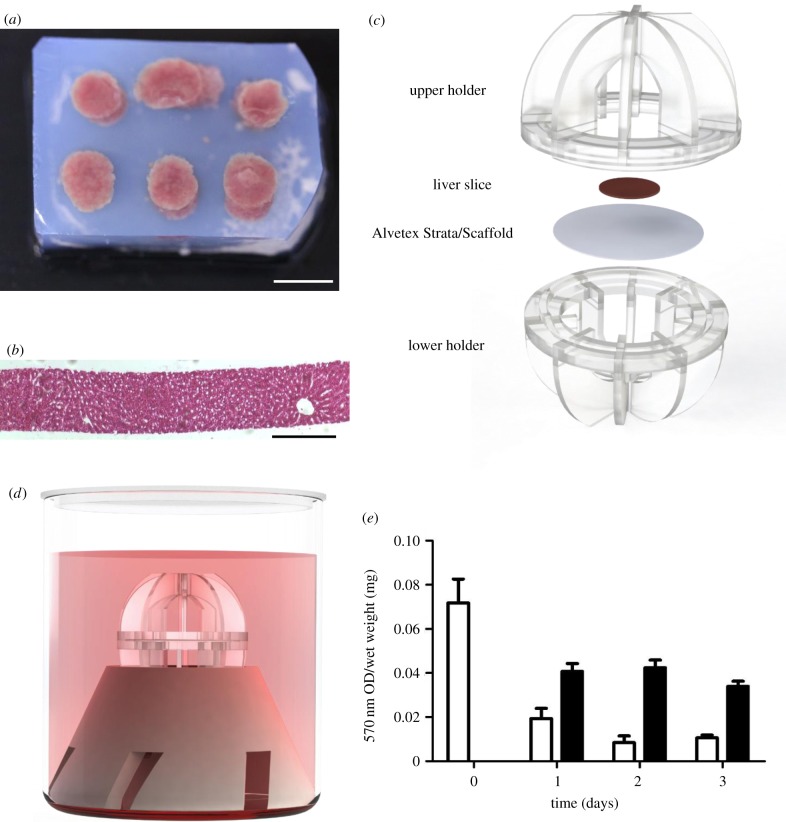
Precision-cut liver slices were produced with a thickness of 250 µm and cultured in the perfusion system with oxygenated medium supported on an Alvetex Strata membrane, as shown in figure 4. Histology of a liver slice from a tissue core (figure 4a) shows the uniformity of the tissue thickness and the presence of the lobular structure with features such as the central vein (figure 4b). A tissue slice was placed centrally on an Alvetex membrane and clipped together in the holder (figure 4c) and the bioreactor assembled (figure 4d).
Figure 4.
Culture of precision-cut liver slices in the bioreactor. (a) Rat livers were cut using an 8 mm biopsy punch and the resulting cores were embedded in low-gelling-temperature agarose. Scale bar is 10 mm. (b) H&E stain of a 250 µm liver slice sectioned using a Leica VT1000 S vibratome. Scale bar is 200 µm. (c) Precision-cut liver slices were placed in the culture holder on Alvetex Strata as shown. Slices were cultured for up to 3 days in the system. (d) Schematic of the fully assembled system used for the perfusion cultures. (e) MTT assay shows metabolic levels daily over 3 days of culture. White bars are for static culture, while black bars are perfused cultures. The MTT result was normalized to the wet weight of the slices to account for the small variation in slice size. Values are mean ± standard error of the mean, n = 3.
The MTT assay indicates a large initial drop after the first 24 h of culture in all samples, which is most likely due to damage incurred during the cutting process (figure 4e). The viability of the remaining tissue is sustained over the culture period in the perfused samples but decreases in static conditions.
4. Discussion
Microenvironmental signalling cues are crucial to cell structure and function, as well as having further roles in maintaining cell viability and homeostasis. Three-dimensional culture systems are able to recapitulate some of the in vivo spatial and topographical cues in an in vitro environment; however, the absence of microcirculatory elements to control gaseous exchange and metabolite removal from these culture systems ultimately limits their ability to support complex tissue constructs and effectively represent the physiological environment. In this study, we have developed a novel bioreactor system that enables flow of medium and cell perfusion of 3D cell cultures to enhance the growth and maintenance of tissue-like constructs in vitro.
Many of the current, more commonly used, techniques employed to introduce perfusion into culture systems are suitable for specific applications, such as single-cell studies and large-scale protein synthesis. These are often not representative of in vivo tissue architecture, which is particularly important for tissue-engineering applications and studies testing the efficacy and uptake of novel therapeutic compounds. The perfusion system presented herein was designed to allow for the creation of tissue constructs over a porous membrane with an area of 3 cm2, with a choice of three different pores and void sizes dependent on the application. Additional control is possible by adjusting the stir speed and hence the fluid flow dependent on the application. CFD analysis (figure 1c) confirms the recirculation route of the medium through the porous membrane, allowing for the disruption of unstirred layers surrounding the cells and tissues. This analysis also showed the adaptability of the system to provide different levels of shear stress to the cells as well as the variation in through-membrane velocity (figure 1d). By adjusting the parameters, it is possible to tailor the system for a variety of cell types and culture techniques.
Conventional 2D cultures of the HepG2 hepatocellular carcinoma cell line are commonly used to assess drug activity and compound testing as alternatives to primary hepatocytes owing to the difficulties encountered when attempting to use primary cells for in vitro liver studies [26]. While HepG2 cells are easy to handle and provide an infinite source of cells, their dissimilarity to primary hepatocytes is a key issue when interpreting data obtained from these cells, particularly in terms of their expression profile and metabolic activity [27,28]. Previous work has shown that growing these cells in static 3D culture has the capacity to improve their structure and function [5]. We have shown that HepG2 cells form 3D cultures and increase in proliferation and metabolic output when cultured in perfusion compared with the static medium controls. An increase in specificity of key structural markers suggests that the cells are adopting a more physiologically relevant phenotype. Improving the performance of these cells in applications such as drug toxicity assays is one of the potential uses of this technology, reducing the cost of the tests while pushing the cells to perform in a manner more closely resembling primary hepatocytes.
The embryonal carcinoma stem cell line TERA2.cl.SP12 has previously been used in a range of studies investigating aspects of human development and cellular pluripotency [29,30]. These cells provide an inexpensive yet robust option for the optimization of the culture of pluripotent stem cell populations in this system. When cultured in the perfused system, TERA2.cl.SP12 cells show an enhanced differentiated phenotype compared with the static system, with increased expression of mature neural marker class III β tubulin and evidence of ectodermal epithelial differentiation with the presence of cytokeratin 8-positive staining. Positive staining for the pluripotency marker Oct4 in the perfused samples indicates the maintenance of an embryonal carcinoma population of cells within the tissue which has formed, suggesting that the perfusion system has created an environment more similar to that seen in vivo. This has resulted in the creation of a tissue morphologically similar to a teratocarcinoma with both embryonal carcinoma and teratoma components observed. The main aim of these experiments was the optimization of this novel system using a well-characterized cell population, and these data provide proof-of-concept results for the use of pluripotent stem cells within the bioreactor. Further experiments can now be undertaken using more developmentally potent and relevant pluripotent stem cell populations, characterizing these and determining their developmental capacity. This is particularly relevant to the teratoma assay, where standardization or replacement of this in vivo technique is well recognized.
The maintenance of precision-cut tissue slices and subsequent analysis of test compound action ex vivo is a popular technique in pharmacology and toxicology studies [31]. However, the viability of these tissues in the in vitro environment severely limits the studies that can be conducted and the time scale over which these can be completed. Use of the perfusion system with ex vivo tissue slices such as the liver slices presented herein is another application in which this apparatus could be used to enhance cell and tissue viability over time. Perfusion allows for the mimicking of in vivo circulation as well as effective mixing of nutrients in order to maintain tissue slices for prolonged periods. Over the 3-day test period, we observed enhanced tissue viability in the liver slices in perfusion when compared with those in static culture. Further work will be carried out to examine the effect of longer time periods and the introduction of sustained oxygenation with the aim of achieving maintenance periods of over a week, bringing the effectiveness of this simple system closer to other more complex methods currently used [32,33]. This technology may allow for chronic tests to be carried out in applications such as fibrosis modelling, as well as enhancing data capture from precious tissue samples.
5. Conclusion
To summarize, we have developed a novel in vitro culture system, which uses a simple bioreactor to introduce fluid flow to support 3D cultures and intact tissue slices. The major components of the system are fully reusable, and the technology can be integrated into any routine cell culture facility. We have shown the maintenance of two very different cell lines within the system, with improvements in metabolic activity, structure and differentiated phenotype compared with static culture. The system can also be applied to the maintenance of precision-cut tissue slices, with a reduced loss of viability observed over the culture period, providing the potential for longer term studies. These improvements indicate the effectiveness and versatility of the system, offering the potential to introduce a key environmental factor into the in vitro culture and enhance the growth and function of 3D cultured cells and tissue models.
Supplementary Material
Acknowledgements
This work was supported by the technical staff of Durham University: Aaron Brown, Department of Chemistry Glassblowing Workshops; Neil Holmes and Paul White, Department of Chemistry Mechanical Workshop; Stephen Lishman and Malcolm Robertshaw, Department of Physics Mechanical Workshop; and Demi Minhinnett, Adele Kitching and Stacey Derivan, Life Science Support Unit (LSSU). The LSSU provided spare rat liver tissue from animals used for other experiments.
Data accessibility
All raw data are available in the electronic supplementary material.
Authors' contributions
H.W.H. and L.A.S. performed experiments. H.W.H., L.A.S. and S.A.P. wrote and revised the manuscript. R.J.W. provided advice with the CFD work and revised the article. S.A.P. had oversight of the project and approved the article for submission.
Competing interests
S.A.P. is the original inventor of Alvetex® technology now commercialized by Reprocell Europe Ltd. S.A.P. currently acts as a scientific consultant for Reprocell via a research collaboration with Durham University.
Funding
This work was supported by funding from the Engineering and Physical Sciences Research Council (EPSRC) and the Anatomical Society.
References
- 1.Yamada KM, Cukierman E. 2007. Modeling tissue morphogenesis and cancer in 3D. Cell 130, 601–610. ( 10.1016/j.cell.2007.08.006) [DOI] [PubMed] [Google Scholar]
- 2.Knight E, Murray B, Carnachan R, Przyborski S. 2011. Alvetex®: polystyrene scaffold technology for routine three dimensional cell culture. In 3D cell culture methods and protocols (ed. Haycock JW.), pp. 323–340. New York, NY: Humana Press. [DOI] [PubMed] [Google Scholar]
- 3.Mirbagheri M, Adibnia V, Hughes BR, Waldman SD, Banquy X, Hwang DK. 2019. Advanced cell culture platforms: a growing quest for emulating natural tissues. Mater. Horizons 6, 45–71. ( 10.1039/C8MH00803E) [DOI] [Google Scholar]
- 4.Knight E, Przyborski S. 2015. Advances in 3D cell culture technologies enabling tissue-like structures to be created in vitro. J. Anat. 227, 746–756. ( 10.1111/joa.12257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bokhari M, Carnachan RJ, Cameron NR, Przyborski SA. 2007. Culture of HepG2 liver cells on three dimensional polystyrene scaffolds enhances cell structure and function during toxicological challenge. J. Anat. 211, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross AM, Jiang Z, Bastmeyer M, Lahann J. 2012. Physical aspects of cell culture substrates: topography, roughness, and elasticity. Small 8, 336–355. ( 10.1002/smll.201100934) [DOI] [PubMed] [Google Scholar]
- 7.Rouwkema J, Rivron NC, van Blitterswijk CA. 2008. Vascularization in tissue engineering. Trends Biotechnol. 26, 434–441. ( 10.1016/j.tibtech.2008.04.009) [DOI] [PubMed] [Google Scholar]
- 8.Lovett M, Lee K, Edwards A, Kaplan DL. 2009. Vascularization strategies for tissue engineering. Tissue Eng. Part B Rev. 15, 353–370. ( 10.1089/ten.teb.2009.0085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelinsky M, Bernhardt A, Milan F. 2015. Bioreactors in tissue engineering: advances in stem cell culture and three-dimensional tissue constructs. Eng. Life Sci. 15, 670–677. ( 10.1002/elsc.201400216) [DOI] [Google Scholar]
- 10.Naing MW, Williams DJ. 2011. Three-dimensional culture and bioreactors for cellular therapies. Cytotherapy 13, 391–399. ( 10.3109/14653249.2011.556352) [DOI] [PubMed] [Google Scholar]
- 11.Pazzano D, Mercier KA, Moran JM, Fong SS, DiBiasio DD, Rulfs JX, Kohles SS, Bonassar LJ. 2000. Comparison of chondrogensis in static and perfused bioreactor culture. Biotechnol. Prog. 16, 893–896. ( 10.1021/bp000082v) [DOI] [PubMed] [Google Scholar]
- 12.Gerlach JC, et al. 2010. Dynamic 3D culture promotes spontaneous embryonic stem cell differentiation in vitro. Tissue Eng. Part C Methods 16, 115–121. ( 10.1089/ten.tec.2008.0654) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lei X, et al. 2014. Rotary suspension culture enhances mesendoderm differentiation of embryonic stem cells through modulation of Wnt/β-catenin pathway. Stem Cell Rev. Rep. 10, 526–538. ( 10.1007/s12015-014-9511-6) [DOI] [PubMed] [Google Scholar]
- 14.Stachelscheid H, et al. 2013. Teratoma formation of human embryonic stem cells in three-dimensional perfusion culture bioreactors. J. Tissue Eng. Regen. Med. 7, 729–741. ( 10.1002/term.1467) [DOI] [PubMed] [Google Scholar]
- 15.Stephenson M, Grayson W. 2018. Recent advances in bioreactors for cell-based therapies. F1000Research 7, 517 ( 10.12688/f1000research.12533.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coluccio ML, et al. 2019. Microfluidic platforms for cell cultures and investigations. Microelectron. Eng. 208, 14–28. ( 10.1016/j.mee.2019.01.004) [DOI] [Google Scholar]
- 17.Baudoin R, Corlu A, Griscom L, Legallais C, Leclerc E. 2007. Trends in the development of microfluidic cell biochips for in vitro hepatotoxicity. Toxicol. Vitr. 21, 535–544. ( 10.1016/j.tiv.2006.11.004) [DOI] [PubMed] [Google Scholar]
- 18.van Grunsven LA. 2017. 3D in vitro models of liver fibrosis. Adv. Drug Deliv. Rev. 121, 133–146. ( 10.1016/j.addr.2017.07.004) [DOI] [PubMed] [Google Scholar]
- 19.Palma E, Doornebal EJ, Chokshi S. 2019. Precision-cut liver slices: a versatile tool to advance liver research. Hepatol. Int. 13, 51–57. ( 10.1007/s12072-018-9913-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu M. 2013. Engineering stem cell niches in bioreactors. World J. Stem Cells 5, 124–135. ( 10.4252/wjsc.v5.i4.124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romo-Morales A, Knight E, Przyborski S. 2017. Alvetex, a highly porous polystyrene scaffold for routine three-dimensional cell culture. In Technology platforms for 3D cell culture: a user's guide (ed. Przyborski SA.), pp. 225–249. Hoboken, NJ: Wiley Blackwell. [Google Scholar]
- 22.Cooke MJ, Stojkovic M, Przyborski SA. 2006. Growth of teratomas derived from human pluripotent stem cells is influenced by the graft site. Stem Cells Dev. 15, 254–259. ( 10.1089/scd.2006.15.254) [DOI] [PubMed] [Google Scholar]
- 23.Leo HL, Xia L, Ng SS, Poh HJ, Zhang SF, Cheng TM, Xiao GF, Tuo XY, Yu H. 2009. Computational fluid dynamics investigation of the effect of the fluid-induced shear stress on hepatocyte sandwich perfusion culture. In Proc. 13th Int. Conf. on Biomedical Engineering, Singapore, 3–6 December 2008, vols 1–3, 23, 1405–1408. Heidelberg, Germany: Springer-Verlag. [Google Scholar]
- 24.Rashidi H, Alhaque S, Szkolnicka D, Flint O, Hay DC. 2016. Fluid shear stress modulation of hepatocyte-like cell function. Arch. Toxicol. 90, 1757–1761. ( 10.1007/s00204-016-1689-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasuda T, Obara H, Hsu H, Mizunuma H, Matsuno N, Enosawa S. 2015. Proposal of a novel evaluation index for the effects of shear stress and exposure time on hepatocyte damage. J. Artif. Organs 18, 236–242. ( 10.1007/s10047-015-0834-0) [DOI] [PubMed] [Google Scholar]
- 26.Gomez-Lechon M, Donato M, Lahoz A, Castell J. 2008. Cell lines: a tool for in vitro drug metabolism studies. Curr. Drug Metab. 9, 1–11. ( 10.2174/138920008783331086) [DOI] [PubMed] [Google Scholar]
- 27.Castell JV, Jover R, Martnez-Jimnez CP, Gmez-Lechn MJ. 2006. Hepatocyte cell lines: their use, scope and limitations in drug metabolism studies. Expert Opin. Drug Metab. Toxicol. 2, 183–212. ( 10.1517/17425255.2.2.183) [DOI] [PubMed] [Google Scholar]
- 28.Gerets HHJ, Tilmant K, Gerin B, Chanteux H, Depelchin BO, Dhalluin S, Atienzar FA. 2012. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol. Toxicol. 28, 69–87. ( 10.1007/s10565-011-9208-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Przyborski SA, Christie VB, Hayman MW, Stewart R, Horrocks GM. 2004. Human embryonal carcinoma stem cells: models of embryonic development in humans. Stem Cells Dev. 13, 400–408. ( 10.1089/scd.2004.13.400) [DOI] [PubMed] [Google Scholar]
- 30.Clarke KE, Tams DM, Henderson AP, Roger MF, Whiting A, Przyborski SA. 2017. A robust and reproducible human pluripotent stem cell derived model of neurite outgrowth in a three-dimensional culture system and its application to study neurite inhibition. Neurochem. Int. 106, 74–84. ( 10.1016/j.neuint.2016.12.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Bovenkamp M, Groothuis GMM, Draaisma AL, Merema MT, Bezuijen JI, van Gils MJ, Meijer DK, Friedman SL, Olinga P. 2005. Precision-cut liver slices as a new model to study toxicity-induced hepatic stellate cell activation in a physiologic milieu. Toxicol. Sci. 85, 632–638. ( 10.1093/toxsci/kfi127) [DOI] [PubMed] [Google Scholar]
- 32.Paish HL, et al. 2019. A bioreactor technology for modeling fibrosis in human and rodent precision-cut liver slices. Hepatology 70, 1377–1391. ( 10.1002/hep.30651) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, et al. 2018. Precision-cut human liver slice cultures as an immunological platform. J. Immunol. Methods. 455, 71–79. ( 10.1016/j.jim.2018.01.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data are available in the electronic supplementary material.