Abstract
The large family of KRAB zinc finger (KZNF) genes are transcription factors implicated in recognizing and repressing repetitive sequences such as transposable elements (TEs) in our genome. Through successive waves of retrotransposition-mediated insertions, various classes of TEs have invaded mammalian genomes at multiple timepoints throughout evolution. Even though most of the TE classes in our genome lost the capability to retrotranspose millions of years ago, it remains elusive why the KZNFs that evolved to repress them are still retained in our genome. One hypothesis is that KZNFs become repurposed for other regulatory roles. Here, we find evidence that evolutionary changes in KZNFs provide them not only with the ability to repress TEs, but also to bind to gene promoters independent of TEs. Using KZNF binding site data in conjunction with gene expression values from the Allen Brain Atlas, we show that KZNFs have the ability to regulate gene expression in the human brain in a region-specific manner. Our analysis shows that the expression of KZNFs shows correlation with the expression of their target genes, suggesting that KZNFs have a direct influence on gene expression in the developing human brain. The extent of this regulation and the impact it has on primate brain evolution are still to be determined, but our results imply that KZNFs have become widely integrated into neuronal gene regulatory networks. Our analysis predicts that gene expression networks have been repeatedly innovated throughout primate evolution, continuously gaining new layers of gene regulation mediated by both TEs and KZNFs in our genome.
This article is part of a discussion meeting issue ‘Crossroads between transposons and gene regulation’.
Keywords: KRAB zinc finger proteins, transposable elements, evolutionary arms race, co-option
1. Introduction
The family of KRAB zinc finger (KZNF) genes is a large and rapidly evolving gene family recently shown to be involved in the repression of transposable elements (TEs) in mammalian genomes. The presence of a large number of young, species-specific KZNFs alongside ancient, highly conserved KZNFs suggests that this gene family could have been guarding our genomes for a long time. For some KZNFs, the structural adaptations they underwent throughout evolution suggest that their main role is repressing the retrotransposition activity of TEs in an attempt to stop them from spreading throughout the genome. These KZNFs and the TEs they evolved to repress show clear signs of an evolutionary arms race that played out over the course of millions of years [1,2]. However, because many KZNFs are still retained in our genome long after the invasion of the TEs they evolved to repress has ceased, it is likely that KZNFs are co-opted for other regulatory roles. Indeed, for some KZNFs, their acquired ability to recognize TEs seems to have been repurposed for tissue-specific regulation of TE-derived gene regulatory elements [3]. It was recently shown that KZNFs bind to TEs in early embryonic stages and use TEs to modulate nearby gene expression. Importantly, it was shown that the regulatory influence of KZNFs on TEs becomes evident again at later stages of human brain development [3]. Whether KZNFs also have other regulatory functions independent of TEs remains elusive. Here, after reanalysis of KZNF binding site data [2,4,5], we find that evolutionary changes in KZNFs provide them not only with the ability to recognize and repress TEs, but also enabled some KZNFs to bind to a large number of gene promoters. The observation that, in the human brain, the expression of these KZNF genes shows widespread correlations with the expression of their target genes suggests that alongside their involvement in repressing and modulating the activity of TEs, some KZNFs integrate into existing gene expression networks and act as classical transcription factors (TFs) to establish novel gene regulatory networks.
2. Methods
(a). Analysis of MACS-peak data and KRAB zinc finger binding sites
The ChIP-seq and ChIP-exo data were generated by Schmitges et al. [5] (NCBI GEO database accession number GSE76496), Imbeault et al. [2] (NCBI GEO database accession number GSE78099), Najafabadi et al. [4] (NCBI GEO database accession number GSE52523) and Jacobs et al. [1] (NCBI GEO database accession number GSE60211). To visualize the data, raw fastq files were imported to the Galaxy US or EU servers [6]. Reads were processed, adaptor and illumina-specific sequences were removed using Trimmomatic (Galaxy v. 0.36.5) and mapped to the human genome (assembly GRCh37/hg19; [7]) using Bowtie2 (Galaxy v. 2.3.4.2) [8] with single-end, very sensitive end-to-end settings. BigWig files were generated using the bamCoverage tool (Galaxy v. 3.0.1.0). These were uploaded to the UCSC Genome Browser [9] to manually confirm that the reads were in concordance with the peak calling data.
A cut-off of 500 for the p-value score (10 × log10(p-value)) was decided after visual observation of the MACS-peaks using the UCSC genome browser. The script Match_final.py was used to compare remaining peaks with the location of gene promoter regions acquired from UCSC data hubs [10]. The region 1000 bp downstream and 5000 bp upstream of a gene transcription start site was taken and any gene promoter that overlapped with at least 50% of a MACS-peak was recorded using the match_prom() function. Further details on the function of all scripts can be found in the supplemental python files.
(b). Analysis of KRAB zinc finger expression data
KZNF expression data were obtained from the BrainSpan Gene Expression tool [11]. The function data_maker(brainarea) was used to extract the expression data for all the KZNFs with more than 50 binding sites on gene promoter regions. Heatmaps for whole-brain expression as well as expression in specific brain areas were generated with R [12] using the heatmap.2 function from the library gplots. For the specific brain regions, row order was determined by the clustering of the whole brain heatmap.
(c). Analysis of KRAB zinc finger expression correlation data
The api.py script contains the functions used in the following steps. Expression correlation values for all genes in the whole brain in relation to each zinc finger were downloaded from www.brainspan.org (2015 Allen Institute for Brain Science. Allen Brain Atlas API. Available from: brain-map.org/api/index.html) using their API. The correlation values for the genes directly bound by the KZNFs were extracted for each KZNF and the distribution of these correlation values was visualized in a histogram alongside the distribution of the expression correlation values of all genes relative to that KZNF. Histograms were made using Matplotlib [13]. For the histograms for specific brain regions, correlation values of 0.000 were removed.
Histograms were also generated showing the distribution of a random set of gene expression correlation values next to the distribution of expression values of all genes. The function rand_check_hist() selects at random an equal number of genes to the number of genes bound by the KZNF in question.
Heatmaps showing the distribution of the expression correlation values of the target genes of every KZNF in the whole brain and specific regions were generated using the function heatmap_exp_cor_dis() and the heatmap.2 function in R. The function assigns the segments of the heatmap based on the percentage of target genes that have a correlation value that falls within the range determined on the x-axis. If the bins 0.8–1.0 contained no values, they were removed. Row order for the specific tissues was based on the clustering for the dorsolateral prefrontal cortex (DFC). The heatmap.2 clustering algorithm was used to group the KZNFs into four categories. The skew and kurtosis of each distribution were calculated using the R package e1071. The skew and kurtosis of the groups were then tested for statistical difference using a Kruskal–Wallis test in R.
(d). Analysis of transcription factor data
Binding site data for TFs were obtained from the ENCODE database [14]. The functions match_prom_trans(tscf) and match_symbols_trans(tscf) in the Match_final.py script were used to generate lists of target gene promoters. This was then subjected to the same workflow as the ZNF gene expression correlations data to generate correlation histograms and heatmaps using the slightly altered heatmap_tscf_correlation() function.
3. Results
(a). KRAB zinc fingers bind to a large number of gene promoters
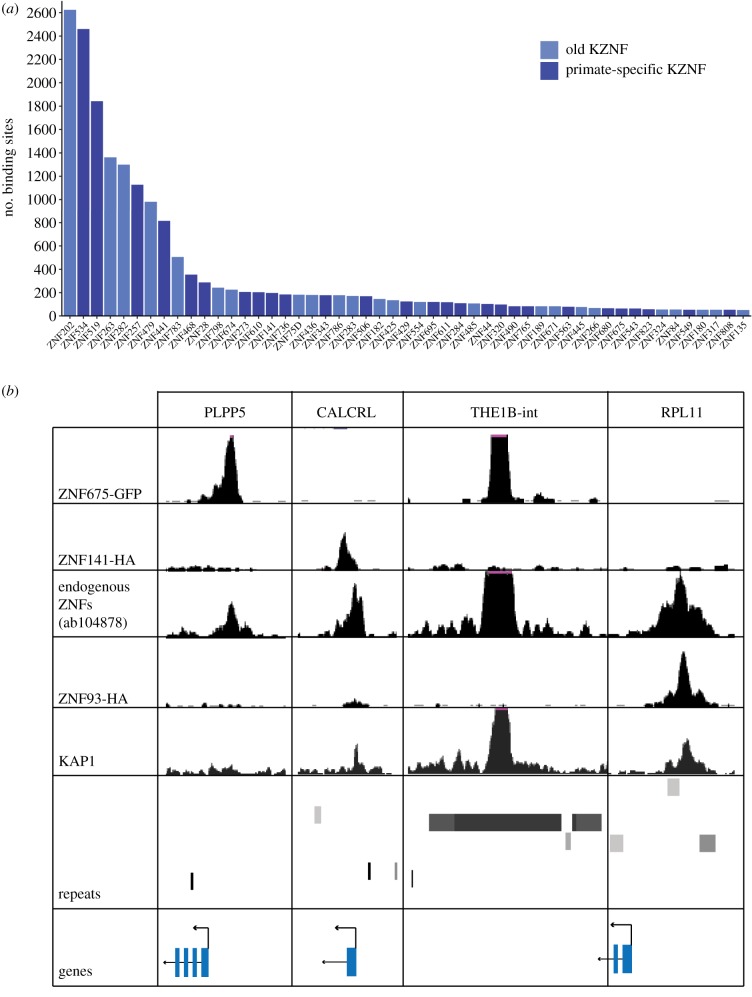
Previous ChIP-seq and ChIP-exo analyses have revealed widespread binding of KZNFs to TEs. To a lesser extent, some KZNFs were reported to bind to non-TE sequences as well, but the scope and the implications of this phenomenon were not thoroughly investigated. To explore the non-TE binding sites of KZNFs, we reanalysed the MACS-peak data for 197 KZNFs and focused on the binding sites that overlapped with gene promoters. Fifty-one of the KZNFs analysed were found to bind to at least 50 gene promoters (figure 1a). Even though the majority (45/51) of the KZNFs bind to a few hundred or fewer gene promoters genome-wide, some KZNFs (ZNF202, ZNF534, ZNF519, ZNF263, ZNF282, ZNF257) showed binding to more than a thousand promoters. Notably, there was not a big overlap of the set of promoters bound by each of the KZNFs, in line with the highly divergent DNA-binding domains of KZNFs: in total, 8888 genes showed binding of at least one KZNF in their promoter region, which corresponds to approximately 45% of the genes in the human genome. The ability of the KZNFs to bind to gene promoters was not restricted to very old KZNF genes, as almost half of the promoter-binding KZNFs were found to be primate-specific [15]. Taking this into account, approximately 26% of human gene promoters have a binding site for at least one primate-specific KZNF.
Figure 1.
Analysis of KZNF ChIP-seq data at gene promoters. (a) The number of gene promoters bound by KZNFs. Only KZNFs with more than 50 promoter targets are shown. Dark blue bars denote primate-specific KZNFs. Light blue bars indicate evolutionarily older, mammalian KZNFs. (b) ChIP-seq data for experiments using antibodies for KZNF-GFP and KZNF-HA fusion proteins and endogenous KZNFs showing peaks at gene promoters. All windows on the UCSC browser scaled to 50 except for the endogenous KZNF window, which was set to 20.
The ChIP-seq data used in this analysis were generated using the ectopic expression of KZNFs, raising the possibility that the ectopically expressed KZNF-GFP and KZNF-HA fusion proteins may bind to sites where the endogenous KZNF does not bind under physiological conditions. To assess if the observed promoter binding of KZNFs is a general artefact of the methodology used, we compared ChIP-seq signals of endogenous KZNFs with signals obtained from overexpression of these proteins (figure 1b). ChIP data from overexpressed ZNF675 and ZNF141 were compared to ChIP-seq data generated with an antibody that recognizes endogenous ZNF93 (ab104878; [1]), but also targets a small number of other ZNFs, including ZNF254, ZNF675 and ZNF141. In this comparison, we observe endogenous KZNF binding to the promoter regions of genes such as CALCRL and PLPP5 on the same location as where binding was observed for ectopically expressed ZNF141 and ZNF675 [2]. This validates that the binding pattern of ectopically expressed KZNFs can be used as a representation of the potential binding pattern of the endogenous KZNFs. This is further supported by previous observations where binding of ectopically expressed KZNFs was compared to endogenous TRIM28/KAP1 [2]. These data also confirmed that the binding pattern of ectopically expressed KZNFs closely resembles the binding pattern of endogenous KAP1, which is recruited to KZNFs through their KRAB domains [2]. Although this does not rule out that some of the KZNF binding can still be an artefact of ectopic expression, our analysis validates that promoter binding by endogenous ZNFs does occur under normal cellular conditions with endogenous levels of KZNF expression.
(b). KRAB zinc fingers show different expression patterns across time and space in the brain
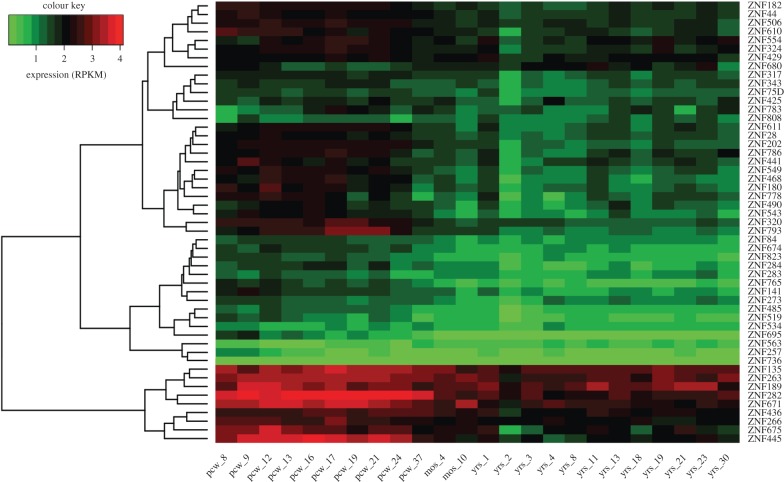
It was previously shown that KZNFs are highly expressed in the adult human brain [2]. To determine the expression dynamics of KZNFs throughout pre- and postnatal human brain development, we plotted the BrainSpan RNA-seq expression levels [11] of each of the 51 KZNFs across various neurodevelopmental timepoints (figure 2). Expression-based clustering shows some KZNFs have a uniform low or high expression across developmental time and some KZNFs are expressed at high levels in early development, followed by decreased expression in adult stages. Comparing the expression patterns between different brain regions revealed that the dynamics of KZNF expression is brain region specific. The developmental gene expression pattern of KZNFs in the whole brain (figure 2) was most similar to DFC but diverged from other brain regions, such as the cerebellum (CBC) (electronic supplementary material, figure S1).
Figure 2.
Heatmap showing whole-brain expression over time of all KZNFs that bind greater than 50 gene promoters. Brainspan expression values in reads per kilobase of transcript (RPKM), low expression in green, high expression denoted by red. Ages: postconceptional weeks (pcw), months (mos), years (yrs).
(c). Expression correlation of KRAB zinc fingers and target genes
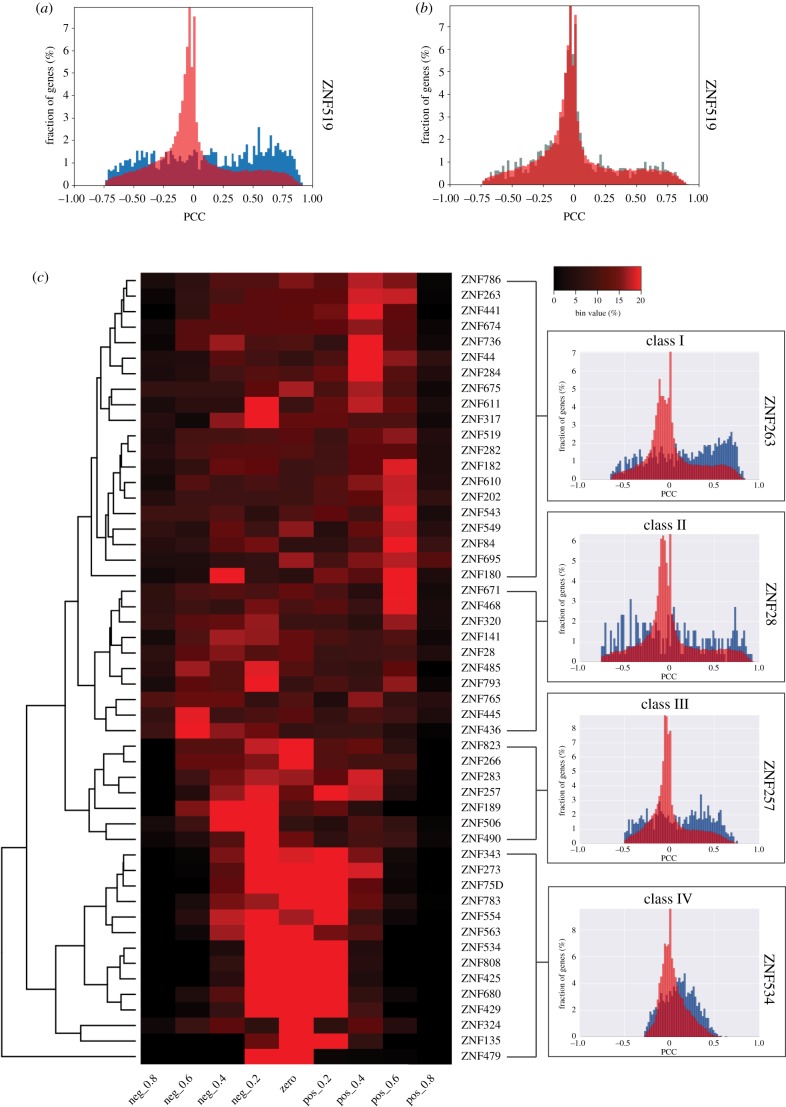
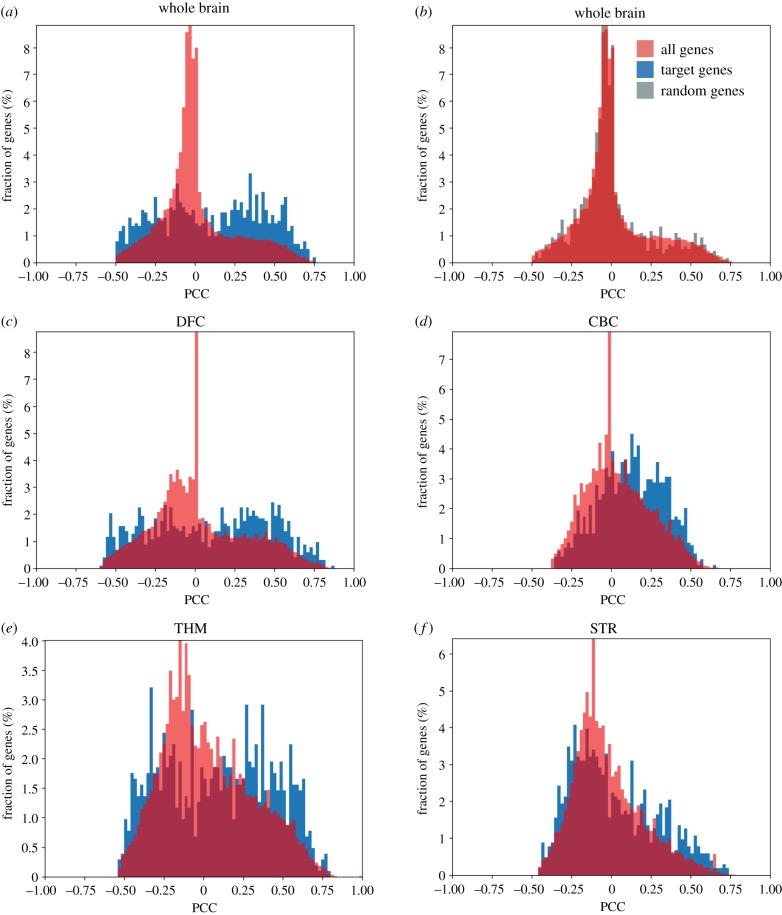
We next addressed whether we could find correlations between the expression of the KZNFs and their target genes, the set of genes where the KZNF binds the promoter (figure 3). For each of the selected 51 KZNFs, expression correlation values of all genes expressed in the brain were downloaded from the BrainSpan database [11]. The distribution of these correlation values was plotted in a histogram and, as expected, a normal distribution of the expression correlation values, clustered around 0, was observed (figure 3a; red histogram). Next, the correlation values for KZNF target genes in relation to the expression of their respective KZNF were extracted and plotted in a histogram. If no regulatory effect is present of the KZNF on its target promoters, a normal distribution of the expression correlation values would be expected as previously observed for all genes. Remarkably, for many KZNFs we investigated, the expression correlation values of the KZNF target genes were not normally distributed, and the KZNF target genes often showed strong correlation or anti-correlation to the expression pattern of the KZNF gene (figure 3a; blue histogram). As a control, we plotted an equal amount of randomly selected genes, and compared it to the distribution of all brain-expressed genes (figure 3b). Randomly selected genes show a normal distribution that closely matches the distribution of correlation values of all genes when correlated to the expression of the KZNF. Based on the distribution patterns of the correlation values, the KZNFs could be grouped in four classes (figure 3c): class I had distributions skewed towards positive correlation values, class II had uniform distributions, class III had distributions skewed towards negative correlation values and class IV followed the ‘normal’ distribution of the values for all genes (figure 3c; electronic supplementary material, figure S2). When compared using a Kruskal–Wallis test, measures of skew and kurtosis were significantly different between these classes (p-values < 0.001). These analyses support a regulatory relationship between 37/51 (72%) of the KZNFs and their target genes.
Figure 3.
Overview of KZNF target gene correlation. (a) Distribution of ZNF519 gene correlation values. Blue histogram indicates ZNF519 correlation with target genes. Red histogram indicates ZNF519 correlation with all genes. PCC, Pearson's correlation coefficient. (b) Distribution of ZNF519 gene correlation values using a random set of target genes (in grey) instead. (c) Heatmap showing the distribution of expression correlation values of genes bound by the ZNFs. Each square represents the percentage of target genes that fall within the range of correlation values covered by each bin. KZNFs clustered using the heatmap.2 dendrogram function; classes were defined using this unbiased clustering. On the right, representative histograms for each cluster showing in blue the distribution of target gene expression correlation values and in red the values of all genes relative to the zinc finger. Black–red gradient indicates the increasing percentage of genes per bin. Max value 20% (red), lowest value 0% (black).
(d). Based on gene expression correlations, KRAB zinc fingers act as regular transcription factors
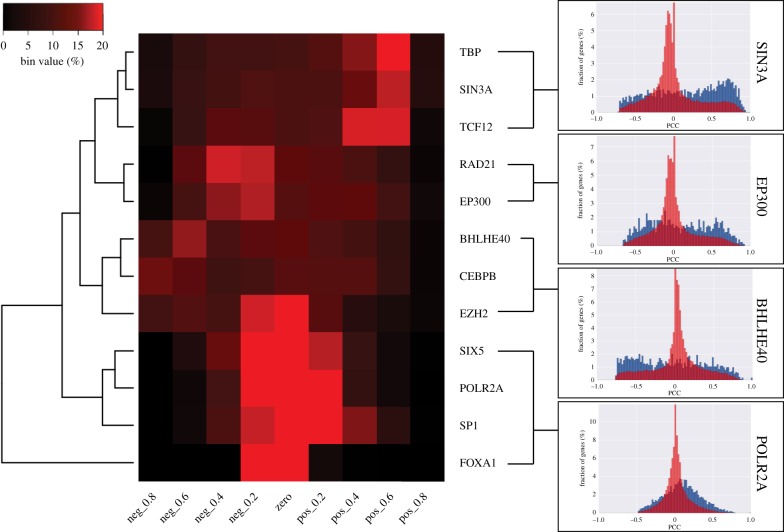
To compare the patterns of correlations between KZNFs and their target genes to other TFs and their target genes, the expression correlation analysis was repeated on a set of 12 well-documented TFs, part of the ENCODE consortium dataset [14] (figure 4). The distribution patterns of the correlation values for these TFs and their target genes showed a remarkably similar pattern to those observed for KZNFs. The described role of the TEs as activators or repressors does not seem to affect the distribution patterns. The known TFs also form similar clusters based on the distribution of the expression correlation values of the genes to which they bind. When choosing a random set of gene targets, we again observed a distribution mirroring the distribution of all genes instead. The similarities between the KZNF and TF correlation analyses indicate that certain KZNFs may have been repurposed to act as traditional TFs.
Figure 4.
Heatmap showing the distribution of expression correlation values of genes bound by 12 documented TFs. Generated with the same method as figure 3. To the right, representative histograms for each of the clusters. Black–red gradient indicates the increasing percentage of genes per bin. Max value 20% (red), lowest value 0% (black).
(e). The correlation between KRAB zinc finger and target genes is tissue-dependent
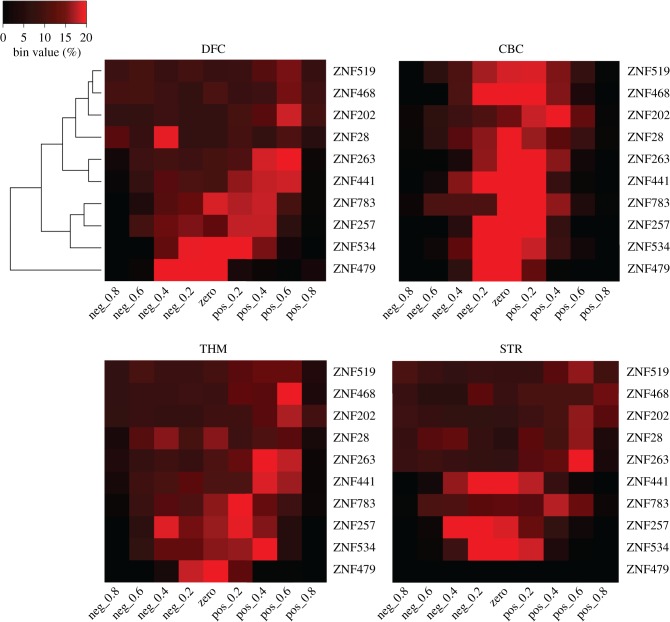
Our analysis revealed brain-region-specific expression of KZNFs during development. We therefore investigated whether there is region-specific regulatory influence of KZNFs on their target gene promoters (figure 5). For this analysis, we selected 10 KZNFs that bind to the highest number of gene promoters. In concordance with the differential gene expression dynamics of KZNFs in different brain regions, the expression correlation distributions are not consistent across the different brain regions. In the CBC, values seem to cluster around 0, suggesting that KZNFs do not have a major influence on gene expression in this region. The highest divergence from the normal distribution of all genes was observed for DFC, which indicates that in this tissue, KZNFs have the highest influence on the expression of their target genes (figure 6). The distributions of target genes and all genes for each brain region were compared using a Kolmogorov–Smirnov test. Each of the four regions had a p-value < 0.001 supporting the alternative hypothesis that the distributions of the target genes are different from those of all genes. Overall, our data suggest that the binding of KZNFs to gene promoters is associated with regulatory effects, and shows a brain-region-dependent influence on the expression of target genes.
Figure 5.
Heatmaps showing the distribution of expression correlation values for each of the top 10 promoter-binding KZNFs across different brain regions. Data from the dorsolateral frontal cortex (DFC), CBC, thalamus (THM) and striatum (STR). ZNF479 showed no expression or correlation values in the striatum and is depicted as a black line here. The order of ZNFs denoted by the clustering in the DFC. Black–red gradient indicates the increasing percentage of genes per bin. Max value 20% (red), lowest value 0% (black).
Figure 6.
Gene correlation histograms for ZNF257. (a,b) Distributions of expression correlation values of target genes (a) and a set of random genes (b) in the whole brain compared to all genes. All genes are shown in red, target genes in blue and random genes in grey. (c–f) Distribution of expression correlation values of target genes versus all genes in the different brain areas (c) DFC, (d) cerebellar cortex (CBC), (e) thalamus (THM) and (f) striatum (STR). For these regions, correlation values of 0.000 were removed owing to the large number of unexpressed genes in specific brain regions.
4. Discussion
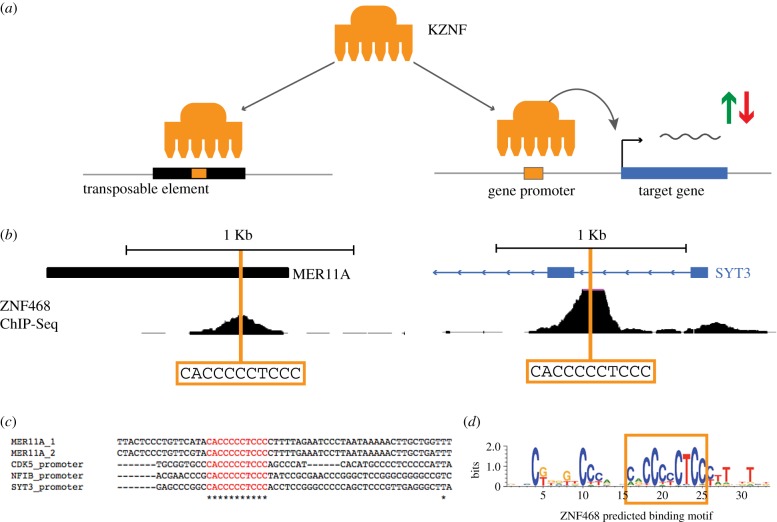
Our analysis of KZNF binding sites suggests that gene promoters have become regulatory targets of KZNFs, thereby forming a new layer of gene regulation as a consequence of the evolutionary arms race between KZNFs and TEs. This is supported by previous studies showing links between KZNFs and gene expression [16–18]. The question arises why and how these gene promoters became bound by the KZNFs. It was previously shown that some KZNFs and the TEs they repress are locked in an evolutionary arms race, each placing selective pressure on the other to evolve and fixate mutations in their DNA sequences. It is not unlikely that during the phase of structural evolution of the KZNF, when optimizations occur in the DNA-binding domains, the KZNF evolves to recognize and bind other non-TE sequences by chance (figure 7). Another scenario is that some KZNFs by default recognize GC-rich sequences, a feature often observed in both TEs and gene promoters [7]. There may be no possibility for a KZNF to bind and repress a GC-rich region in a TE without also binding and repressing GC-rich regions in gene promoters. In a third scenario, the binding of KZNFs to gene promoters may be explained by the strategy viruses use to maximize the production of viral components by the host cells. One of the most efficient ways of establishing this is having a long terminal repeat (LTR) region that contains sequences resembling the promoters of highly expressed genes. For the host, in its battle to control the retroviral activity and/or retrotransposon invasion, the most efficient strategy would be to repress the LTR region responsible for expression of these retroviral components. The consequence is that structural evolution of KZNFs to repress TEs results in the KZNF repressing the gene promoters that the original LTR was mimicking. Indeed, we find evidence for this scenario for some of the promoter-binding ZNFs, such as ZNF468, which clearly recognizes the same core 11 bp motif in MER11A elements (an HERVK-derived LTR) and ZN468-bound gene promoters that share no apparent homology to MER11A outside of the core motif (figure 7b–d). Furthermore, even though we observed many promoters that are bound by more than one KZNF, the binding sites of the KZNFs within the same promoter are usually not overlapping, showing a clear sequence specificity of KZNFs even in the repetitive parts of TEs and promoters. All of the scenarios above provide a good potential explanation for the fixation of KZNF genes after the TEs they evolved to repress have become inactive and neutralized. As such, each KZNF gene may have evolved via one of these scenarios, which may play on KZNF structural evolution as complementing evolutionary forces. Importantly, the non-TE-mediated binding of gene promoters by some KZNFs is complemented by other KZNFs that predominantly regulate gene expression by binding to TE-derived regulatory elements [3]. Together, they provide a picture of massive co-option of TEs, and also KZNFs, as two heavily interdependent layers of gene regulation.
Figure 7.
(a) Model showing a mechanism by which gene promoters may have become bound by KZNFs. (b) ChIP-seq data for ZNF468 at a MER11A element and the SYT3 promoter showing a shared binding motif in the core of the ZNF468 binding site. (c) Clustal-O multiple sequence alignment of ZNF468 binding at gene promoters and MER11A elements, the shared core binding site shown in red. (d) The predicted binding motif of ZNF468 based on protein sequence from http://zf.princeton.edu/ [19].
The role of KZNFs as gene regulators is supported by our analysis of expression correlation data. The expression of both KZNFs and TFs is correlated with the expression of the genes to which they bind. Out of the 10 KZNFs that bind the highest number of gene promoter regions, 7 have a correlation distribution skewed to the positive side, where the expression of bound genes increases with the expression of the KZNF. This is at odds with the well documented, repressive role of KZNFs but could be explained by the phenomena where KZNFs, especially ancient ones, lost their ability to recruit TRIM28 [2]. Indeed, a comparison of the promoter binding sites of the top 10 KZNFs with the binding sites of KAP1/TRIM28 shows very little overlap (electronic supplementary material, table S1). It remains unknown why KZNFs lose the ability to recruit TRIM28 and what happens after they lose this ability, but one possibility is that the loss of TRIM28 recruiting capacity may provide a mechanism in which KZNFs can be transformed into positive regulators of gene expression. This yields additional flexibility for KZNFs to integrate into gene regulatory networks.
For the gene promoters bound by KZNFs that retain the ability to recruit TRIM28, another question arises: what is the impact of this repressive machinery on gene expression? If important neurodevelopmental genes are bound, this could be negative for the organism. This, in turn, could lead to a selective pressure against binding of gene promoters by KZNFs and could explain why the majority of KZNFs bind less than 200 gene promoters. In combination with the KZNFs that lost their KRAB domain and repressive capabilities, this could exemplify how KZNFs form a complex gene regulatory network in the brain, consisting of a combination of repressive and promoting effects. The combined binding of multiple brain-expressed KZNFs to gene promoters may have subtle but significant effects on transcript levels of genes crucial to brain development. Taken together, our analysis suggests that both TEs and KZNFs may have contributed new primate-specific layers of gene regulation to our genome. This underlines how the intimate relation between TEs and KZNFs has the potential to repeatedly cause innovations in gene expression networks throughout primate evolution.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Michael Imbeault and members of the Jacobs lab for helpful discussions.
Data accessibility
This article has no additional data.
Authors' contributions
F.M.J.J. and G.A.L.: conceptualization; F.M.J.J., G.A.L., S.F.R., E.J.v.B. and G.F.: methodology; G.A.L., S.F.R., G.F.: investigation and validation, and writing—original draft and visualization; F.M.J.J., G.A.L, E.J.v.B. and G.F.: data curation, and writing—review and editing; F.M.J.J.: supervision, project administration and funding acquisition.
Competing interests
We declare we have no competing interests.
Funding
This project was supported by ERC-2016-StG-716035 (F.M.J.J.) and HFSP CDA00030/2016C (F.M.J.J.).
References
- 1.Jacobs FMJ, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. 2014. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516, 242–245. ( 10.1038/nature13760) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imbeault M, Helleboid P-Y, Trono D. 2017. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543, 550–554. ( 10.1038/nature21683) [DOI] [PubMed] [Google Scholar]
- 3.Pontis J. Planet E, Offner S, Turelli P, Duc J, Coudray A, Theunissen TW, Jaenisch R, Trono D. 2019. Hominoid-specific transposable elements and KZFPs facilitate human embryonic genome activation and control transcription in naive human ESCs. Cell Stem Cell 24, 724–735.e5. ( 10.1016/j.stem.2019.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Najafabadi HS, et al. 2015. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 33, 555–562. ( 10.1038/nbt.3128) [DOI] [PubMed] [Google Scholar]
- 5.Schmitges FW, et al. 2016. Multiparameter functional diversity of human C2H2 zinc finger proteins. Genome Res. 26, 1742–1752. ( 10.1101/gr.209643.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Afgan E, et al. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544. ( 10.1093/nar/gky379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409, 860–921. ( 10.1038/35057062) [DOI] [PubMed] [Google Scholar]
- 8.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. ( 10.1038/nmeth.1923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res. 12, 996–1006. ( 10.1101/gr.229102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raney BJ, et al. 2014. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics 30, 1003–1005. ( 10.1093/bioinformatics/btt637) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller JA, et al. 2014. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206. ( 10.1038/nature13185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.R Development Core Team. 2008. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org. [Google Scholar]
- 13.Hunter JD. 2007. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95. ( 10.1109/MCSE.2007.55) [DOI] [Google Scholar]
- 14.ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. ( 10.1038/nature11247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas JH, Schneider S. 2011. Coevolution of retroelements and tandem zinc finger genes. Genome Res. 21, 1800–1812. ( 10.1101/gr.121749.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W., et al. 2019. ZFP30 promotes adipogenesis through the KAP1-mediated activation of a retrotransposon-derived Pparg2 enhancer. Nat. Commun. 10, 1809 ( 10.1038/s41467-019-09803-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ecco G, et al. 2016. Transposable elements and their KRAB-ZFP controllers regulate gene expression in adult tissues. Dev. Cell 36, 611–623. ( 10.1016/j.devcel.2016.02.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang P, et al. 2017. A placental growth factor is silenced in mouse embryos by the zinc finger protein ZFP568. Science 356, 757–759. ( 10.1126/science.aah6895) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Persikov A, Singh M. . 2014. De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic Acids Res. 42, 97–108. Epub 3 October 2013 ( 10.1093/nar/gkt890) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.