Abstract
Transposable elements (TEs) are increasingly recognized as important contributors to mammalian regulatory systems. For instance, they have been shown to play a role in the human interferon response, but their involvement in other mechanisms of immune cell activation remains poorly understood. Here, we investigated the profile of accessible chromatin enhanced in stimulated human macrophages using ATAC-seq to assess the role of different TE subfamilies in regulating gene expression following an immune response. We found that both previously identified and new repeats belonging to the MER44, THE1, Tigger3 and MLT1 families provide 14 subfamilies that are enriched in differentially accessible chromatin and found near differentially expressed genes. These TEs also harbour binding motifs for several candidate transcription factors, including important immune regulators AP-1 and NF-κB, present in 96% of accessible MER44B and 83% of THE1C instances, respectively. To more directly assess their regulatory potential, we evaluated the presence of these TEs in regions putatively affecting gene expression, as defined by quantitative trait locus (QTL) analysis, and found that repeats are also contributing to accessible elements near QTLs. Together, these results suggest that a number of TE families have contributed to the regulation of gene expression in the context of the immune response to infection in humans.
This article is part of a discussion meeting issue ‘Crossroads between transposons and gene regulation’.
Keywords: transposable elements, infection, immune response, regulatory elements, ATAC-seq
1. Introduction
The regulatory networks responsible for immune function are particularly susceptible to evolutionary pressures as organisms adapt to withstand continuous assaults from a variety of infectious agents. Through their ability to move and replicate [1], be bound by transcription factors [2,3] and produce non-coding transcripts with regulatory potential [4,5], there is increasing evidence that transposable elements (TEs) have been co-opted throughout evolution and have contributed to the generation of regulatory diversity in mammalian genomes [6]. As such, they present interesting targets for the discovery of novel functional elements contributing to gene regulation in different human cell types, particularly immune cells. A notable example is the MER41 repeat and its associated subfamilies, which include an element in the AIM2 promoter with immune regulatory function confirmed through CRISPR-Cas9 knockouts [7]. In that analysis, Chuong et al. also described the enrichment of several other TE subfamilies within STAT1 and IRF1 transcription factor binding sites putatively involved in the interferon pathway of macrophages.
To gain a more comprehensive understanding of the TE-derived non-coding elements implicated in the immune response, we explored the landscape of accessible chromatin in macrophages during immune cell activation. More specifically, we investigated the association between repetitive elements and non-coding regions activated upon infection in the datasets published by Nédélec et al. [8], which evaluated population differences in the response to infection. From that study, we obtained assays for transposase-accessible chromatin (ATAC-seq) which were derived from macrophages challenged in vitro with Listeria monocytogenes and Salmonella typhimurium, two intracellular pathogens. The regulatory contribution of TEs has been previously described in immune cells [7], but analyses in that study were limited by targeting specific inflammatory pathways and transcription factor binding sites. In that respect, ATAC-seq data offer the advantage of targeting all regions of accessible chromatin genome-wide without the need for a pull-down experiment that would restrict the analysis to a pre-selected set of transcription factors [9]. Moreover, these data allowed us to explore the presence of TEs in the functional regions identified by expression quantitative trait locus (eQTL) analysis. Overall, we show that TEs contribute to non-coding sequences activated upon infection, as a set of subfamilies are overrepresented both in accessible chromatin and in proximity to differentially expressed genes (DEGs), harbour transcription factor motifs and are enriched near QTLs belonging to their haplotype block.
2. Results
(a). TEs contribute to accessible chromatin upon infection
We were interested in the contributions of TEs to the accessible chromatin that was specific to the immune response upon infection. For this purpose, we obtained ATAC-seq datasets from Nédélec et al. [8] and focused our analysis on regions of the genome where the chromatin becomes more accessible following bacterial exposure (see Material and methods). We processed the datasets separately for macrophages challenged with S. typhimurium (S12) and L. monocytogenes (L12), 12 h post-infection. By overlapping the peak summits with repetitive element annotations from the RepeatMasker track [10], we found that 25.6% (7995/31 256) and 23.6% (4678/19 788) of infection-induced accessible regions contain TEs in the S12 and L12 samples, respectively. That being said, given that nearly half of the genome is derived from repeats [6] and consistent with previous reports [11], we find that overall TEs are underrepresented in regions for which the chromatin becomes more accessible upon infection.
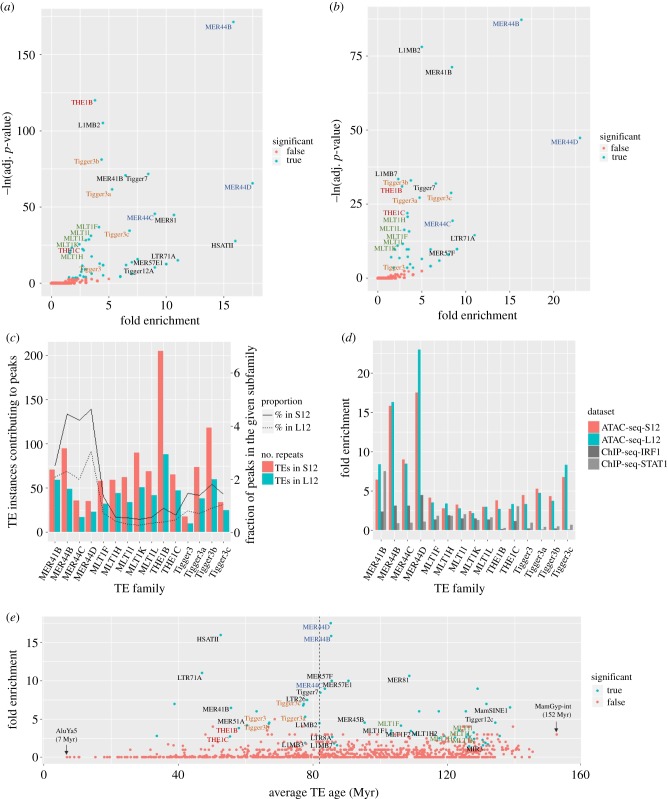
Next, we sought to identify specific TE families recurrently overrepresented in the S12 and L12 infected samples. We, therefore, compared the presence of peak-associated repeats (PARs) with their expected distribution and computed the statistical enrichment of TEs at three levels of repeat organization: individual subfamilies, families and the four main TE classes (LTR, DNA, LINE and SINE) (Material and methods). This analysis revealed 34 ‘immune’ subfamilies significantly enriched in both conditions (electronic supplementary material, tables S1 and S2), including the MER41B repeat previously shown to have a regulatory role in the immune activation of the AIM2 gene [7]. The most significant enrichment observed was for the MER44B subfamily, which was found 16 more times than expected in both conditions (p = 3.36 × 10−75 and p = 1.29 × 10−38 for S12 and L12, respectively) (figure 1a,b, electronic supplementary material, figure S1a). Notably, the related subfamilies MER44C and MER44D were also highly enriched, suggesting the potential presence of conserved activating elements within these related TEs. The THE1, Tigger3 and MLT1 subfamilies were also overrepresented, providing respectively two, four and nine related subfamilies enriched in at least one condition. However, the MER44B repeats were unique in their contribution, as they represent a small subfamily with only 2131 instances, but still provide as many PARs as the MLT1 K subfamily, which is nine times larger (figure 1c).
Figure 1.
TE enrichment in ATAC peaks by repeat subfamily in (a) S12 samples and (b) L12 samples. Significant families are represented by blue points (q-value < 0.05) and the remaining families by red. The coloured labels match the related families they belong to (blue for MER44, red for THE1, orange for Tigger3 and green for MLT1). adj., adjusted. (c) The absolute number of TE instances (bars) and the fraction of all instances in each subfamily (lines) contributing to accessible chromatin. (d) Comparison of TE enrichment results in our ATAC-seq peaks and the ChIP-Seq datasets published by Chuong et al. [7]. (e) Average age of TE instances for each repeat subfamily. Only the 34 immune families are labelled, with the coloured labels belonging to the 14 most interesting families. Dashed line marks the primate/rodent divergence time, 82 Ma [12].
From this analysis, we therefore identified 14 TE subfamilies belonging to four groups of related repeats belonging to the MER44, THE1, Tigger3 and MLT1 subfamilies, as well as the previously described MER41B subfamily (figure 1c–e). Although the above analysis has identified 34 enriched subfamilies, we are particularly interested in these 14 related repeats as they may share common features (see electronic supplementary material, figure S1b,c for results including all 34 immune subfamilies). A comparison with Chuong et al. [7] shows an overlapping set of TE subfamilies enriched in the STAT1 and IRF1 ChIP-Seq datasets, which map the transcription factor binding sites required for the regulation of interferon-stimulated genes in CD14+ cells. However, there are several notable differences between our findings and the previously described data. A number of subfamilies show increased enrichment in our ATAC-seq datasets, including MER81, MamSine1 and Tigger12c (electronic supplementary material, figure S1c). Moreover, the MER44, Tigger3 and THE1B repeats are several times more enriched in the overall landscape of accessible chromatin upon bacterial infection than in the previously defined STAT1 and IRF1 binding sites (figure 1d). Next, while these individual subfamilies appear to be contributing more than expected by chance to putative regulatory regions, the core TE classes they belong to, DNA and LTR, are comparatively poorly enriched overall, with a fold enrichment of only 1.25 and 1.03 (electronic supplementary material, figure S2a). However, the Tigger3 and MER44 subfamilies all belong to the larger TcMar-Tigger family, a subdivision of DNA transposons that is among the most highly enriched TE families in its category (electronic supplementary material, figure S2b).
Finally, we wanted to characterize the TE subfamilies according to their approximate age of insertion in the genome to determine whether the immune repeats have inserted early or late in evolution. We thus estimated the age of each repeat instance based on its similarity to the original sequence and compared the results between accessible and inaccessible repeats within each subfamily (Material and methods). Among the 14 related TEs, several subfamilies show a moderate difference in age between accessible and inaccessible instances (electronic supplementary material, table S3 and figure S3a,b), as is the case for Tigger3b (p = 6.60 × 10−05), and to a lesser extent, MER44B and THE1B (p = 0.028 and p = 0.035), where the accessible instances are slightly younger on average. Notably, we found that the four groups have inserted in the genome at different points in time (figure 1e). Where the younger THE1 and Tigger3 subfamilies have inserted on average 57.2 and 72.2 million years ago, the MER44 and MLT1 repeats have been present for 84.9 and 124 million years, respectively, before the estimated divergence time between rodents and primates, 82 million years [12]. As such, immune TEs may have contributed immune regulatory functions multiple times in the course of evolution.
(b). Selected TE subfamilies harbour motifs for master regulators of the immune response
While binding sites of the IRF1 and STAT1 transcription factors (TFs) were previously described in repeats in immune cells, [7] the use of ATAC-seq data enables the detection of multiple transcription factors by scanning for the presence of previously identified TF motifs within regions that become more open after infection using HOMER [13]. Overall, the most significantly enriched motifs in our ATAC-seq data belong to the AP-1 and NF-κB transcription factors, found, respectively, in 30.8 and 12.4% of all peaks enhanced upon infection (versus 6.6 and 4.0% genome-wide, p = 1 × 10−2112 and p = 1 × 10−325).
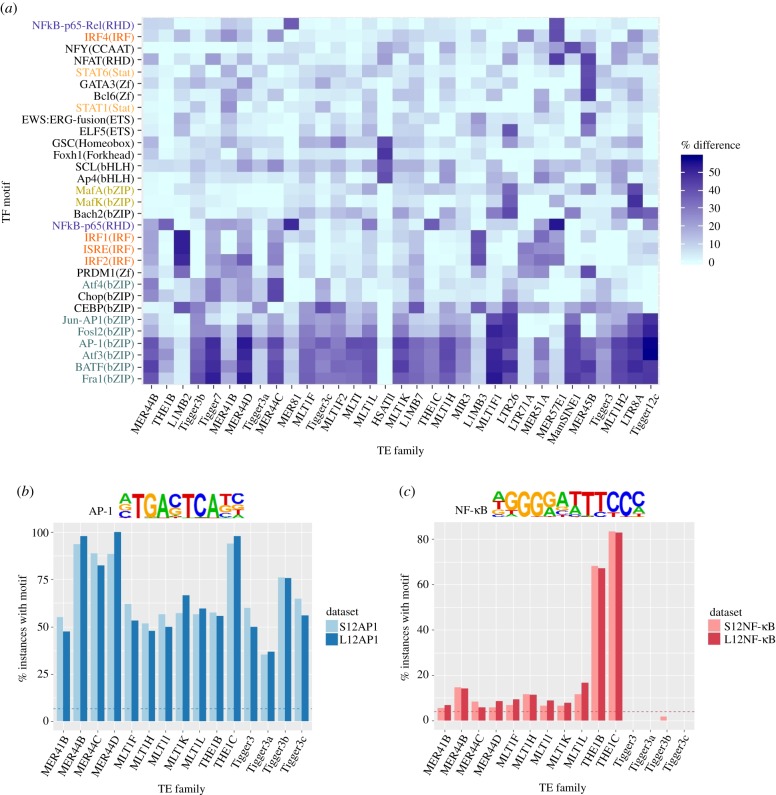
We specifically wanted to assess the presence of TF motifs within the immune TE subfamilies identified, comparing TF motifs found in repeat instances that are present in differentially accessible chromatin more often than in instances found in the rest of the genome (see Material and methods). We selected the top 30 motifs that were found in over one-third of accessible instances in at least one repeat subfamily and showed at least a 20% increase compared with inaccessible repeats (figure 2a and electronic supplementary material, figures S4 and S5). Several motifs showed more specific enrichment for a small set of subfamilies (e.g. Foxh1 in HSATII, and ELF5 in LTR26 and MER45B), while other motifs were found to be shared among multiple TE subfamilies (e.g. CEBP and AP-1 motifs). As expected, we find that the MER41B family is enriched with the STAT1 and IRF motifs (particularly IRF2), which are present in 87.7 and 57.7% of instances, respectively. However, these motifs are relatively depleted in many of the other TEs, including the THE1 and MLT1 subfamilies (electronic supplementary material, figure S6a,b). Instead, we observed that the motifs for the AP-1 family of transcription factors (including BATF, Fra1, Atf3 and JunB) are enriched in all of the 14 related subfamilies and in most of the 34 immune subfamilies (figure 2b and electronic supplementary material, figure S6c). Actually, 65.1% of accessible instances belonging to the former contain at least one of these motifs. It is comparatively depleted in activated repeats that do not belong to these 14 subfamilies, as it is found in only 48.5% of instances. The enrichment is particularly significant for MER44 and THE1, where the motifs are found in over 93% of MER44B and THE1C peaks, with less than 42 and 65% of non-accessible instances harbouring the motifs in each family, respectively. While the AP-1 motifs are present overall in accessible chromatin upon infection, their overrepresentation in these particular TE subfamilies suggests an association with immune-specific repeats. The second most significant enrichment is observed for the NF-κB motif, which is found almost exclusively in THE1 repeats (figure 2c), where it is present in 67 and 83% of THE1B and THE1C instances. It is also found in the smaller TE subfamilies MER81, MER57E1 and MER57F (electronic supplementary material, figure S6d). The enrichment of these motifs is reasonable overall within immune-specific accessible chromatin, as the significant role of these TFs in immunity is well described in the literature, both for AP-1 [14–16] and NF-κB [17–19]. Interestingly, functional NF-κB and AP-1 binding sites have both been previously validated in THE1B instances involved in gene reactivation of Hodgkin's lymphoma [20].
Figure 2.
(a) Absolute difference between the proportion of accessible repeats containing the top-most enriched TF motifs compared with their non-accessible counterparts within the same subfamily. The STAT1 motif was added as a control. The motif names are colour-coded according to the family of transcription factors they belong to: IRF in red, STAT in orange, AP-1 in turquoise, NF-κB in dark blue and Maf in yellow. The motifs in black are uniquely enriched. (b,c) Proportion of accessible repeats with the AP-1 (b) and NF-κB (c) motifs. Dashed lines show the proportion of background genome sequences containing the AP-1 (a) and (b) NF-κB motifs. See electronic supplementary material, figure S6 for results in all 34 immune subfamilies.
(c). A subset of TEs are enriched near DEGs
To clarify the role of TEs in the regulation of immunity, we are particularly interested in PARs found in proximity to genes that are up- or downregulated following infection. To determine the presence of PARs specifically near DEGs, we obtained lists of DEGs from Nédélec et al. [8] for both Salmonella and Listeria infected cells. We were able to confirm that accessible chromatin is enriched overall within 100 kb of DEGs (p = 4.94 × 10−324, see Material and methods). We then compared the presence of accessible and inaccessible repeats near DEGs and computed the enrichment of each TE subfamily contributing at least 10 instances to accessible chromatin. This analysis identified a new set of 85 and 28 subfamilies enriched in the S12 and L12 samples (electronic supplementary material, figure S7a–c and tables S4 and S5), with differential enrichment between the two conditions for many subfamilies, but also with 23 subfamilies significantly enriched in both conditions (electronic supplementary material, figure S8a,b). Notably, the previously identified THE1B repeat is the second most significantly enriched subfamily, with 57 instances near DEGs found in the S12 samples (we would have only expected 16, p-value = 3.77 × 10−16). The THE1C repeats are also observed, which further supports the possibility of a shared activating element within these TE subfamilies.
Similarly, the MLT1 repeats are also enriched near DEGs, with the MLT1 K family presenting as the most highly enriched subfamily (p = 8.06 × 10−5 and p = 6.38 × 10−4). For example, an interesting MLT1G instance, which is accessible in S12 but not in L12, can be found within 2.7 kb of the DAPP1 gene, which is upregulated only in S12 cells (electronic supplementary material, figure S8c). Another example is an MLT1 K instance 2 kb upstream of the TXN gene, which has been described as a player in redox reactions in stimulated macrophages [21,22]. As there are over 200 such PARs found within 100 kb of DEGs, this large group of repeats offers several other candidates for the contribution to genetic regulation (table 1 and electronic supplementary material, tables S6 and S7 for top PARs nearest DEGs).
Table 1.
Top 3 PARs nearest differentially expressed genes' transcription start sites for each TE group, present in both S12 and L12 samples. Distance is shown in base pairs. See electronic supplementary material, tables S6 and S7 for complete list.
| TE family | locus start | gene | distance to TSS (bp) | motif(s) present |
|---|---|---|---|---|
| MER44D | chr8: 90 794 231 | RIPK2 | 1915 | AP-1 |
| MER44B | chr8: 22 216 286 | SLC39A14 | 8207 | AP-1, NF-κB |
| MER44B | chr10: 26 970 664 | PDSS1 | 15 377 | AP-1 |
| MLT1H | chr1: 167 754 291 | MPZL1 | 1457 | — |
| MLT1 K | chr7: 89 786 435 | STEAP1 | 2665 | — |
| MLT1 K | chr6: 160 086 744 | SOD2 | 2973 | AP-1 |
| THE1B | chr2: 113 814 070 | IL36RN | 1776 | NF-κB |
| THE1B | chr2: 6 998 590 | CMPK2 | 2455 | AP-1, NF-κB |
| THE1B | chr12: 113 426 846 | OAS2 | 8848 | AP-1, NF-κB |
| Tigger3c | chr10: 90 595 119 | ANKRD22 | 12 561 | — |
| Tigger3b | chr9: 117 583 234 | TNFSF15 | 31 634 | AP-1 |
| Tigger3b | chr3: 172 279 540 | TNFSF10 | 44 395 | AP-1 |
Although THE1 and MLT1 subfamilies appear enriched both in accessible chromatin and near DEGs, it is surprising that the most significantly enriched TE subfamily, MER44B, is only moderately enriched near DEGs, with 19 and five notable instances within 100 kb of such genes for the two conditions. However, MER44 repeats may still be acting distally or contributing to gene regulation at earlier or later time points or through alternative mechanisms.
Finally, to explore the association between PARs and potential gene networks, we assessed gene ontology (GO) with GREAT [23], which is designed to assess possible biological functions of non-coding regions through the annotation of nearby genes (Material and methods). As a control, we evaluated all differentially accessible chromatin after infection against the background genome and obtained a majority of immune-related processes, as expected. Interestingly, the association with GO immune processes is not only preserved in PARs but actually enriched in the PARs belonging to the 34 ‘immune’ TEs when compared with all PARs, with biological processes including ‘regulation of innate immune response’ (p = 5.6 × 10−7), ‘cellular response to molecule of bacterial origin’ (p = 2.1 × 10−5) and ‘cellular response to peptidoglycan’ (p = 2.3 × 10−4) (electronic supplementary material, tables S8 and S9). Although this is a coarse analysis, it may further support a meaningful role for the enriched TE subfamilies we identified.
(d). A number of peak-associated repeats are found in QTL-regions
To further associate PARs with the genes they potentially regulate, we evaluated their presence in regions where genetic variation affects gene expression in response to infection, as defined by QTL analysis. These regions, termed reQTLs, were previously established by Nédélec et al. [8] through mapping of common SNP genotypes from 175 individuals to the magnitude of change in expression levels upon infection. We retrieved the most significant reQTLs for the S12 and L12 samples, 490 and 233, respectively. As SNPs in linkage disequilibrium (LD) are commonly grouped within haplotype blocks [24,25], we expanded each reQTL to encompass a larger interval based on its LD block. We used this method as an alternative to direct overlap with SNPs, as the latter returns no TEs overlapping the QTLs themselves, and only 53 and 15 TEs overlapping any proxy SNP in the corresponding LD block for S12 and L12, respectively (electronic supplementary material, tables S10 and S11). This is insufficient to perform statistical analysis. We therefore used the entire interval between the two most distal SNPs belonging to each LD block but extending no farther than 50 kb away from the original SNP, and we defined these as QTL-regions (see Material and methods). We first assessed the enrichment of all ATAC-seq peaks in these regions, as we expect accessible chromatin to localize to regions with a functional impact on gene expression. Only 645 (2.1%) and 225 (1.1%) of the accessible peaks in the S12 and L12 samples overlap a QTL-region, as is expected given the small number of QTLs identified. However, 310 (63.3%) and 129 (55.4%) of the QTL-regions do contain accessible peaks and the accessible chromatin does overlap the defined QTL-regions more than expected by chance (1.78- and 2.1-fold, p = 1.24 × 10−41 and p = 1.31 × 10−22 for S12 and L12, respectively). We then reproduced the analysis by limiting the accessible chromatin to repeat-associated peaks and confirmed that these elements are also enriched in the QTL-regions (2.37- and 2.85-fold, p = 5.66 × 10−23 and p = 9.63 × 10−12). Of the accessible peaks overlapping QTL-regions, 170 (26.4%) and 57 (25.3%) are PARs in the S12 and L12 samples, respectively.
As we have already defined above a set of 34 TE subfamilies enriched in accessible chromatin (electronic supplementary material, tables S1 and S2), we hypothesize that these immune-specific repeats are more likely contributing to regulatory networks and thus expect them to be overrepresented in QTL-regions. We, therefore, classified PARs by level of enrichment in accessible chromatin to create two distinct TE subgroups: immune (belonging to the 34 enriched subfamilies described above) and non-immune (belonging to the remaining subfamilies). We then compared the enrichment of PARs in QTL-regions separately for each subgroup (electronic supplementary material, figure S9). We observe a higher enrichment for immune TEs in the S12 sample (3.17-fold, p = 1.34 × 10−13) than for non-immune TEs (2.1-fold, p = 1.91 × 10−12). Conversely, there appears to be a slightly greater enrichment for non-immune TEs (2.93-fold, p = 8.52 × 10−10) than immune TEs (2.6-fold, p = 1.97 × 10−3) in the L12 sample, although the small number of immune TEs overlapping QTL-regions is insufficient to confirm the significance for this condition. To further quantify the association between significantly enriched TE families and their presence in the S12 QTL-regions, we performed a chi-square test to evaluate the correlation between the type of TE family (immune versus non-immune) and the type of peak (overlapping or not overlapping a QTL-region). We confirm a moderate association between TE category and peak type (p = 0.0239), which further supports that the immune-specific subfamilies we identified associate with reQTLs in their haplotype block in the S12 sample.
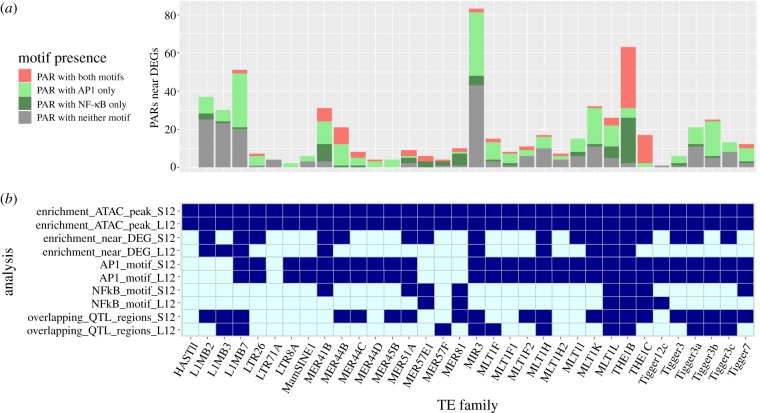
Finally, we observe interesting instances of PARs overlapping QTL-regions, including several of the 14 related TE subfamilies identified above from the MER44, THE1, Tigger3 and MLT1 subfamilies (figure 3, electronic supplementary material, tables S12 and S13). Notably, the most interesting MER44B instance, falling 15 kb upstream of PDSS1, also overlaps its corresponding QTL-region (electronic supplementary material, figure S10). We also observed more distal examples, such as a THE1B instance 150 kb downstream of BASP1, and an MLT1F2 instance 70 kb upstream of GADD45A, both repeats overlapping the corresponding QTL-region for those genes. Together, these results suggest that the significant TE subfamilies that we have identified are not only enriched in accessible chromatin but also contributing to regions that are affecting gene expression based on a QTL analysis.
Figure 3.
(a) Number of PARs within 100 kb of a DEG that overlap either only AP-1 motifs, the NF-κB motif, both or neither motif. Note that nearly all MER41B, MER44 and THE1 repeats near DEGs overlap at least one of the two motifs. (b) Summary of all results for the 34 immune TE subfamilies. Enrichment is indicated in dark blue. Enrichment of the AP-1 and NF-κB motifs is defined as p-value < 0.05 in HOMER, motif found in at least 30% of PARs and a difference of at least 20% in accessible versus inaccessible repeats. The bottom two rows indicate if the subfamily has at least one PAR overlapping a QTL-region.
3. Conclusion
In this study, we advanced our understanding of the TEs contributing to the accessible chromatin enhanced by an infection in macrophages stimulated in vitro. We specifically found a subset of families that are enriched in differentially accessible chromatin and near differentially accessible genes, with multiple related instances from the MER44, THE1, Tigger3 and MLT1 subfamilies. We identified binding motifs for the AP-1 immune transcription factor that were enriched in these subfamilies, and most particularly in the MER44 and THE1C subfamilies. NF-κB, another master regulator of immunity, also has enriched binding motifs specifically in the THE1B and THE1C subfamilies, which are the most highly enriched subfamilies near DEGs.
We also used a set of previously defined regulatory QTLs as a complementary analysis to define potential regions of influence on gene regulation. Although we do not have sufficient power to find statistically significant overlap between PARs and the QTLs themselves, using LD to define regions of influence flanking the QTLs shows that PARs contribute to putative regulatory regions more than expected. This also identifies a number of specific TE instances that would be interesting targets for biological validation through CRISPR-Cas9 deletions.
While the THE1B subfamily has a known role as an activated set of repeats in Hodgkin's lymphoma [20], it is interesting to note here that our most highly enriched subfamilies, MER44B, C and D, have not been previously identified. It would therefore be interesting to further explore these findings to determine if these small yet significant subfamilies have a regulatory role in other cell types and conditions.
4. Material and methods
(a). ATAC-seq dataset processing
We obtained raw ATAC-seq datasets from Nédélec et al. [8] with two replicates for each of three conditions: macrophages infected with L. monocytogenes (i), infected with S. typhimurium (ii) and non-infected (iii) at 12 h. The FASTQ files were processed using the MUGQIC ChIP-Seq pipeline v2.2.1 to obtain narrow peaks. Briefly, the pipeline pools the replicates for each condition and was adapted to retain reads longer than 37 bp for subsequent processing. Sequencing adaptors were trimmed using Trimmomatic v0.36 [26] before aligning to the hg19 human reference with BWA v0.7.12 [27]. Next, unique reads were filtered by mapping quality using Samtools v1.3.1 [28] and duplicates were marked with Picard v2.0.1 [29]. Narrow peaks were called with MACS2 v2.1.0 [30], using profiles from non-infected cells as background (input) to obtain differentially accessible peaks. Peaks with a q-value of less than 0.0005 and less than 1 kb in width were kept for downstream analyses.
(b). TE enrichment in accessible chromatin
Enrichment of each repeat family was computed within accessible chromatin. We obtained repeat annotations from the RepeatMasker track in the UCSC Table Browser and removed coordinates corresponding to transfer RNAs (tRNAs), simple repeats and tandem repeats. We intersected the remaining 4 506 876 repetitive sequences with S12 and L12 peak summits (centre 1 bp) using intersectBed from the BEDtools suite, with the –u option specified to avoid duplicates [31]. Strand information was not considered for any of the analyses, as it was unavailable for the peak calls.
We then computed the enrichment in accessible chromatin of each TE subfamily using a one-sided binomial test, comparing the number of TE instances overlapping peak summits with its expected counterpart. The expected distribution was obtained by shuffling the true peaks randomly across the genome for 1000 iterations, while maintaining a comparable distribution of peak locations. More specifically, the original peaks were annotated by categories based on their distance to RefSeq genes: 5′UTR, exon, intron, TSS (less than 1 kb upstream), promoter (1–5 kb upstream), proximal (5–10 kb upstream or less than 10 kb downstream), distal (10–100 kb upstream or downstream) and desert (greater than 100 kb upstream or downstream). We then separated the peaks based on their annotation and shuffled them separately with shuffleBed, using the –incl and –excl parameters to restrict the randomization within the corresponding genomic regions defined above. Each of the 1000 shuffled peak sets was overlapped with the RepeatMasker annotations and the number of peaks overlapping instances in each TE subfamily, large family and class was obtained.
The mean of the expected counts was taken and compared with the observed counts for each TE family using the binom.test() function in R and resulting p-values were adjusted for multiple testing using the Benjamini–Hochberg method with the p.adjust() function from the multtest R package [32]. TE families with a q-value of less than 0.05 were kept for further consideration.
(c). Estimation of TE age
We obtained an estimate of the age of each TE instance based on the sequence divergence (base mismatches in parts per thousand as defined by the milliDiv value from RepeatMasker). The milliDiv value of each instance was divided by 2.2 × 10−9, the substitution rate for the human genome, to obtain the final age [2,33]. Each TE instance in RepeatMasker was then classified as either accessible (overlapping ATAC-seq peaks) or inaccessible and the mean age was taken for each group and TE subfamily separately. The estimated divergence time between rodents and primates of 82 Myr was obtained from Meredith et al. [12].
(d). Transcription factor motif analysis
We scanned the PARs for known TF motifs using findMotifsGenome.pl from HOMER v4.9.1 [13]. This tool uses a hypergeometric test for each TF to compare the number of motifs found in a target set of genomic regions with that found in a specified set of background regions. We first compared the motifs detected in all ATAC-seq peaks with the STAT1 and IRF1 ChIP-Seq datasets published by Chuong et al. [7]. We restricted the width of all peaks to the centre 200 bp and separated them in two categories: (1) regions shared with the ChIP-Seq datasets (overlapping the STAT1 and IRF1 summits), and (2) regions that are specific to the ATAC-seq datasets. findMotifsGenome.pl was run twice using alternatively the shared regions and the ATAC-seq-specific regions as target and background sets, respectively, to detect the most overrepresented motifs in each dataset.
Next, we sought TF motifs that were enriched specifically in PARs belonging to the subset of 34 TE subfamilies enriched in accessible chromatin in both the S12 and L12 conditions. We defined the 34 target sets separately and ran findMotifsGenome.pl using a custom background set of all TEs in each subfamily not overlapping accessible chromatin. The –size parameter was set to ‘given’ to include the entire TE sequence for both the target and background regions for all analyses. Other parameters were left as default. We excluded the MER57F subfamily as HOMER returned no enriched motifs.
Finally, we used HOMER's scanMotifGenomeWide.pl to extract all loci containing the motifs for AP-1 and NF-κB, the most significant TFs detected, as well as the STAT1 and IRF2 motifs. The motifs were specified using HOMER's pre-defined motif files for AP-1(bZIP), Atf3(bZIP), BATF(bZIP), Fra1(bZIP), Fra2(bZIP), NF-κB-p65(RHD), STAT1(Stat) and IRF2(IRF). The motifs were subsequently overlapped with all repeats and PARs separately using intersectBED, with the –s option specified to intersect regions found on the same strand.
(e). TE enrichment near DEGs
The enrichment of TEs near DEGs was computed separately for each repeat subfamily. We defined DEGs as genes with a log2 fold-change in expression greater than 2 or less than −2. Lists of 1574 and 667 DEGs were thus obtained from the Nédélec et al. [8] dataset for S12 and L12 samples, respectively. For each TE subfamily, the number of PARs within 100 kb upstream or downstream of a DEG was obtained using the distanceToNearest() function from the GenomicRanges R package [34]. TE subfamilies with fewer than 10 instances in accessible chromatin were removed from the analysis. For each subfamily, a random set of the same size as the number of corresponding PARs was taken 1000 times from all TEs not overlapping peaks and was processed similarly to obtain an average expected number of TEs near DEGs. A binomial test was then performed comparing observed and expected counts, and p-values were adjusted as previously described. Individual repeat instances were visualized manually using the Integrative genomics viewer (IGV, v1.4.2) [35].
(f). GO analysis
We evaluated ontological associations for PARs using GREAT [23] v4.0.4. The software is built to evaluate associations with non-coding regions based on proximal and distal UCSC Known Genes and computes an adjusted p-value for biological processes, cellular components and molecular functions using a hypergeometric test. We selected the hg19 species assembly and applied the default association rule ‘basal + extension’, which includes 5000 bp upstream, 1000 bp downstream and 1 000 000 bp maximum extension, with curated regulatory domains included. First, we evaluated the GO processes for all differentially accessible chromatin, defining the ATAC-seq peaks as target sets and the entire genome as background. Second, we evaluated GO processes for ‘immune’ PARs, defining PARs belonging to the 34 immune subfamilies as target sets and all PARs as background. We ran the software directly in the Web browser and submitted two separate jobs for each condition (S12 and L12).
(g). Definition of QTL-regions and overlap with PARs
We defined QTL-regions for all reQTLs published by Nédélec et al. [8] for both S12 and L12 samples. The reQTLs were previously established through mapping of common SNP genotypes from 175 individuals to the magnitude of change in expression levels upon infection. We retrieved the most significant reQTLs for the S12 and L12 samples, 490 and 233, respectively. As SNPs in LD are commonly grouped within haplotype blocks, [24,25] we expanded each reQTL to encompass a larger interval based on its LD block using rAggr [36]. rAggr was adapted by Edlund et al. from the Haploview software [37] to provide proxy markers for SNPs in LD based on the 1000 Genomes (Phase 3) genotype data. The rsIDs for the 490 and 233 top SNPs were provided as input directly in the Web browser. The software was run with the CEU and YRI populations selected, a maximal distance of 50 kb and all other options left as default (minimum mean allele frequency = 0.001, r2 range of 0.8–1.0, maximum number of Mendelian errors = 1, HW p-value cut-off = 0 and minimum genotype percentage = 75). The interval between the resulting two most distant proxy SNPs was taken to define the QTL-regions for each original reQTL. The reQTLs with no proxy SNPs returned by rAggr were extended to encompass a 1 kb interval flanking each reQTL, thus creating intervals between 1 and 100 kb in width across all QTLs.
We then computed the enrichment of all ATAC-seq peaks in QTL-regions, as well as the subset of peaks overlapping TEs (corresponding to PARs). The statistical method is described above for the enrichment of repeats, comparing the true overlaps with an expected distribution obtained by shuffling the peaks across the genome while respecting their corresponding annotations. We also compared the categories of PARs according to the subfamilies they belong to (with immune-specific TEs belonging to the 34 families enriched in accessible chromatin) and the type of peak they overlap (peaks either found in or absent from the QTL-regions). The association between these groups was evaluated using the chisq.test() function in R.
Supplementary Material
Supplementary Material
Acknowledgements
We thank all members of the Bourque Lab for helpful comments and discussion, and David Venuto for advice on the peak enrichment analysis. We also thank Yohann Nédélec and Alain Pacis from the Barreiro Lab for providing access and support for the ATAC-seq data. Data analyses were enabled by computing and storage resources provided by Compute Canada and Calcul Québec.
Data accessibility
Data are available in GEO under GSE136566 and the code is available at: https://github.com/lubogdan/ImmuneTE.
Authors' contributions
Luc.B. participated in the design of the study, conceived and performed the computational analyses and drafted the manuscript. Lui.B. provided the data, suggested analyses and critically revised the manuscript. G.B. conceived, designed and directed the study, and edited the manuscript. All authors revised the paper and gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by funding from the Canadian Institute for Health Research (grant no. CIHR-MOP-115090) and Fonds de Recherche Santé Québec (grant no. FRSQ-25348 to G.B.).
References
- 1.Elbarbary RA, Lucas BA, Maquat LE. 2016. Retrotransposons as regulators of gene expression. Science 351, aac7247 ( 10.1126/science.aac7247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourque G, et al. 2008. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 18, 1752–1762. ( 10.1101/gr.080663.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. 2014. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 24, 1963–1976. ( 10.1101/gr.168872.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramsay L, Marchetto MC, Caron M, Chen SH, Busche S, Kwan T, Pastinen T, Gage FH, Bourque G. 2017. Conserved expression of transposon-derived non-coding transcripts in primate stem cells. BMC Genomics 18, 214 ( 10.1186/s12864-017-3568-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. 2013. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 9, e1003470 ( 10.1371/journal.pgen.1003470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 18, 71 ( 10.1038/nrg.2016.139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087. ( 10.1126/science.aad5497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nédélec Y, et al. 2016. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell 167, 657–669. ( 10.1016/j.cell.2016.09.025) [DOI] [PubMed] [Google Scholar]
- 9.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21–29. ( 10.1002/0471142727.mb2129s109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smit A, Hubley R, Green P. 1996–2012 RepeatMasker Open – 3.0, version 3.2.7. See http://www.repeatmasker.org/.
- 11.Jacques PE, Jeyakani J, Bourque G. 2013. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 9, e1003504 ( 10.1371/journal.pgen.1003504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meredith RW, et al. 2011. Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524. ( 10.1126/science.1211028) [DOI] [PubMed] [Google Scholar]
- 13.Heinz S, et al. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. ( 10.1016/j.molcel.2010.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foletta VC, Segal DH, Cohen DR. 1998. Transcriptional regulation in the immune system: all roads lead to AP-1. J. Leukoc. Biol. 63, 139–152. ( 10.1002/jlb.63.2.139) [DOI] [PubMed] [Google Scholar]
- 15.Schonthaler HB, Guinea-Viniegra J, Wagner EF. 2011. Targeting inflammation by modulating the Jun/AP-1 pathway. Ann. Rheum. Dis. 70(Suppl 1), i109–i112. ( 10.1136/ard.2010.140533) [DOI] [PubMed] [Google Scholar]
- 16.Glasmacher E, et al. 2012. A genomic regulatory element that directs assembly and function of immune-specific AP-1–IRF complexes. Science 338, 975–980. ( 10.1126/science.1228309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowe JM, Menendez D, Bushel PR, Shatz M, Kirk EL, Troester MA, Garantziotis S, Fessler MB, Resnick MA. 2014. p53 and NF-κB coregulate proinflammatory gene responses in human macrophages. Cancer Res. 74, 2182–2192. ( 10.1158/0008-5472.CAN-13-1070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker RG, Hayden MS, Ghosh S. 2011. NF-κB, inflammation, and metabolic disease. Cell Metab. 13, 11–22. ( 10.1016/j.cmet.2010.12.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oeckinghaus A, Hayden MS, Ghosh S. 2011. Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12, 695 ( 10.1038/ni.2065) [DOI] [PubMed] [Google Scholar]
- 20.Lamprecht B, et al. 2010. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat. Med. 16, 571–579. ( 10.1038/nm.2129) [DOI] [PubMed] [Google Scholar]
- 21.Carlson BA, Yoo MH, Conrad M, Gladyshev VN, Hatfield DL, Park JM. 2011. Protein kinase-regulated expression and immune function of thioredoxin reductase 1 in mouse macrophages. Mol. Immunol. 49, 311–316. ( 10.1016/j.molimm.2011.09.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schenk H, Klein M, Erdbrügger W, Dröge W, Schulze-Osthoff K. 1994. Distinct effects of thioredoxin and antioxidants on the activation of transcription factors NF-kappa B and AP-1. Proc. Natl Acad. Sci. USA 91, 1672–1676. ( 10.1073/pnas.91.5.1672) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495 ( 10.1038/nbt.1630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi F, Yanai K, Morii T, Ishinaga Y, Taniguchi-Yanai K, Nagano S, Kato N. 2005. Linkage disequilibrium grouping of single nucleotide polymorphisms (SNPs) reflecting haplotype phylogeny for efficient selection of tag SNPs. Genetics 170, 291–304. ( 10.1534/genetics.104.038232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. 2006. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat. Genet. 38, 82 ( 10.1038/ng1695) [DOI] [PubMed] [Google Scholar]
- 26.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler Transform. Bioinformatics 25, 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. ( 10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broad Institute. 2017. GitHub repository for Picard command-line tools. GitHub. See https://github.com/broadinstitute/picard (accessed 4 September 2017).
- 30.Zhang Y, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 ( 10.1186/gb-2008-9-9-r137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. ( 10.1093/bioinformatics/btq033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollard KS, Dudoit S, Laan MJ. 2005. Multiple testing procedures: R multtest package and applications to genomics. In Bioinformatics and computational biology solutions using R and Bioconductor (eds R Gentleman, V Carey, W Huber, R Irizarry, S Dudoit), pp. 249–272 Berlin, Germany: Springer. [Google Scholar]
- 33.International Human Genome Sequencing Consortium 2001. Initial sequencing and analysis of the human genome. Nature 409, 860–921. ( 10.1038/35057062) [DOI] [PubMed] [Google Scholar]
- 34.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 ( 10.1371/journal.pcbi.1003118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat. Biotechnol. 29, 24 ( 10.1038/nbt.1754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edlund CK, Conti DV, Van Den Berg DJ. 2017. rAggr. Los Angeles, CA: University of Southern California. See http://raggr.usc.edu/ (accessed 8 October 2018).
- 37.Barrett JC, Fry B, Maller JD, Daly MJ. 2004. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265. ( 10.1093/bioinformatics/bth457) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in GEO under GSE136566 and the code is available at: https://github.com/lubogdan/ImmuneTE.