Abstract
Transposable element (TE)-derived sequences comprise more than half of the human genome, and their presence has been documented to alter gene expression in a number of different ways, including the generation of alternatively spliced transcript isoforms. Alternative splicing has been associated with tumorigenesis for a number of different cancers. The objective of this study was to broadly characterize the role of human TEs in generating alternatively spliced transcript isoforms in cancer. To do so, we screened for the presence of TE-derived sequences co-located with alternative splice sites that are differentially used in normal versus cancer tissues. We analysed a comprehensive set of alternative splice variants characterized for 614 matched normal-tumour tissue pairs across 13 cancer types, resulting in the discovery of 4820 TE-generated alternative splice events distributed among 723 cancer-associated genes. Short interspersed nuclear elements (Alu) and long interspersed nuclear elements (L1) were found to contribute the majority of TE-generated alternative splice sites in cancer genes. A number of cancer-associated genes, including MYH11, WHSC1 and CANT1, were shown to have overexpressed TE-derived isoforms across a range of cancer types. TE-derived isoforms were also linked to cancer-specific fusion transcripts, suggesting a novel mechanism for the generation of transcriptome diversity via trans-splicing mediated by dispersed TE repeats.
This article is part of a discussion meeting issue ‘Crossroads between transposons and gene regulation'.
Keywords: transposable elements, alternative splicing, cancer, tumorigenesis, gene expression, gene regulation
1. Background
Half or more of the human genome is derived from transposable element (TE) sequences, remnants of formerly mobile genetic elements that can replicate to extremely high copy numbers over time [1,2]. TE sequences contribute to human gene regulation through a variety of distinct mechanisms [3–6]. Previous work from our own laboratory has documented the presence of TE-derived transcription factor binding sites [7–10], enhancers [11–15], chromatin insulators [16], microRNAs [17,18] and antisense RNAs [19] along with TE-derived alternative transcription initiation [20–22] and termination sites [23].
The provisioning of alternative splice sites is another way that TEs can contribute to the complexity of the human transcriptome [24–26]. A role for TEs in alternative splicing of human genes was discovered via classic studies on Alu elements in the early 2000s. Investigators from the laboratories of Gil Ast and Dan Graur uncovered evidence of Alu-derived splice sites, as well as the inclusion of Alu elements in alternatively spliced exons, for a number of human genes [27,28]. These studies suggested a potential role for TE-derived alternative splicing in disease, cancer in particular [29]. Nevertheless, compelling proof for such a connection has remained elusive.
The role of TEs in cancer has received substantial attention as of late [30–35], and alternative splicing has itself been widely associated with tumorigenesis [36–43]. As such, it seems reasonable to hypothesize that TE-derived alternative splicing could play an important role in cancer. Despite the seemingly obvious connections—among TEs, alternative splicing, and cancer—there has yet to be any systematic analysis on the contribution of TEs to alternative splicing events in tumour tissue. The goals of this study were to (i) survey the global landscape of TE-derived alternative splicing across a variety of cancer types, and (ii) identify individual cases where TE-derived splice sites are linked to splicing (isoform) alterations in cancer.
We analysed 614 matched normal-tumour samples pairs for 13 cancer types, characterized as part of The Cancer Genome Atlas (TCGA). Integrated analysis of RNA-seq data and genome annotations were used to generate a genome-wide atlas of TE-derived alternative splice sites, and differential expression analysis of alternative splice variants was used to identify ‘isoform switch' events, with TE-derived splice isoforms that show increased use in cancer samples. Our atlas of TE-derived alternative splice variants is made available to the research community via the UCSC Genome Browser. We go on to propose a potentially novel mechanism, whereby the dispersed repetitive nature of TE sequences facilitates the generation of fusion transcripts via trans-splicing events. Our TE trans-splicing mechanism is admittedly speculative at this time, and we suggest the kinds of tests that will need to be done to further interrogate our model.
2. Methods
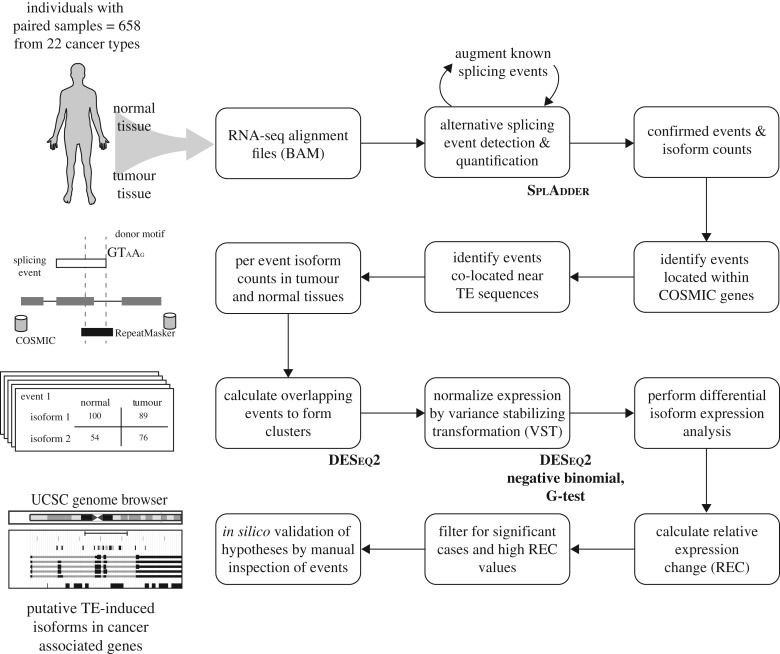
A schematic overview of the bioinformatics analysis pipeline used for this study can be seen in figure 1. A list of all data sources, programs and statistical methods used in the study can be seen in the electronic supplementary material, table S1.
Figure 1.
Bioinformatics analysis pipeline used for this study. RNA-seq datasets from 658 paired normal-tumour TCGA samples from 22 cancer types were analysed in this study. The schematic can be broadly divided into four stages: (row 1) detection of alternative splicing events and per-exon expression quantification, (row 2) identification of TE-derived alternative splicing events for cancer-associated genes, (row 3) statistical testing for differences in alternative splicing expression levels between matched normal and tumour tissues, and (row 4) evaluation of cases of interest to explore the potential functional impact of TE-derived alternative splicing on cancer.
(a). Genomic data
All analyses are based on the human genome reference sequence build hg19 (GRCh37). Genomic coordinates for NCBI RefSeq [44] and Ensembl transcript models, i.e. exon/intron boundaries, were taken from the UCSC Genome Browser [45]. Genomic coordinates for TE sequences were taken from RepeatMasker annotations [46]. Overlap analysis of gene, TE and alternative splice event coordinates were performed using the BEDTools program [47].
(b). Alternative splicing
The Catalogue Of Somatic Mutations In Cancer (COSMIC) Cancer Gene Census was used to identify cancer-associated genes—oncogenes, tumour suppressor genes and fusion genes—for subsequent alternative splicing analysis [48]. Transcriptome (RNA-seq) data for matched normal-tumour sample pairs of individual patients, across a variety of distinct cancer types, were taken from TCGA for alternative splice site analysis (electronic supplementary material, figure S1). RNA-seq data were mapped to the human reference genome sequence and processed using the program SplAdder, as previously described in [39], in order to characterize alternative splice events in cancer-associated genes. Four kinds of alternative splice events were analysed here: (i) intron retention, (ii) exon skipping, (iii) alternate 3′ splicing and (iv) alternate 5′ splicing (electronic supplementary material, figure S2). For all observed alternative splice events, two distinct isoforms were defined and quantified. Isoform 1 and isoform 2 are operationally defined as the shorter and longer isoforms, respectively. Thus, isoform 1 corresponds to the TE-derived isoform for exon skipping, whereas isoform 2 corresponds to the TE-derived isoform for intron retention, alternate 3′ splicing, and alternate 5′ splicing. The expected numbers of TE-derived isoforms for cancer-associated genes were calculated based on the total number TEs from each TE class within cancer genes:
Genomic coordinates for individual alternative splice sites and their corresponding isoforms are defined by the presence of overlapping RNA-seq reads for at least three individuals. Individual alternative splice events were characterized across all COSMIC genes, and each individual event was quantified as the number of reads mapping to the alternatively spliced exon. This was done for all genes from individual samples corresponding to each cancer type and its corresponding matched normal-tumour sample pair. Overlapping alternative splice event isoforms were clustered using single linkage clustering based on greater than or equal to 75% overlap of splice site genomic coordinates, and cluster coordinates were defined as the minimum and maximum start and stop sites for the individual constituent splice sites. Alternative splice site cluster counts for all isoforms were calculated as the average counts across all individual constituent splice sites within any given tissue type.
(c). Differential expression (splicing)
The program DESeq2 was used to normalize tissue-specific alternative splice site cluster counts using the variance stabilizing transformation technique [49]. Differential alternative splice isoform expression, between matched normal-tumour sample pairs, was measured using relative expression change (REC) and via a 2 × 2 contingency table with the G-test. For each alternative splice event, cluster average count values were computed across four conditions: (i) non-TE isoform normal, (ii) TE isoform normal, (iii) non-TE isoform tumour and (iv) TE isoform tumour. The REC value for individual alternative splice events are calculated as the normalized difference of the TE isoform in tumour versus normal tissue:
where, is the average normalized cluster count for the TE-derived isoform across all individuals in normal tissue. The statistical significance of normal-tumour differential expression (splicing), i.e. differences in average alternative splice site cluster counts, was evaluated using a 2 × 2 contingency table with the G-test:
| normal | tumour | ||
|---|---|---|---|
| isoform 1 | n1 | ||
| isoform 2 | n2 | ||
| nN | nT | N |
(d). Visualization
Individual cases TE-derived and differentially expressed alternative splice sites of interest were visualized using the UCSC Genome Browser. Locations of RNA-seq characterized alternative splice site clusters were compared to the locations of TE sequences and COSMIC gene exon/intron boundaries. Genomic coordinates of the TE-derived alternatively spliced exons characterized here are distributed as a UCSC Genome Brower Track hub.
3. Results and discussion
(a). Transposable element-derived alternative splice sites and cancer
We analysed RNA-seq data for matched normal-tumour sample pairs from individual patients in order to characterize the genomic landscape of alternative splicing in cancer. A total of 678 patient samples among 22 different cancer types were considered for preliminary analysis; cancer types with less than 10 patient samples were subsequently excluded, yielding a final dataset of 614 patients across 13 cancer types (table 1; electronic supplementary material, figure S1). We relied on a recently published approach to the characterization of alternative splicing in cancer, which has been shown to yield reliable results in terms of both characterizing and quantifying individual alternative splice sites and their corresponding isoforms [39]. We focused on four distinct types of alternative splicing events uncovered by the previous approach—(i) intron retention, (ii) exon skipping, (iii) alterative 3′ splicing and (iv) alterative 5′ splicing (electronic supplementary material, figure S2)—and modified the existing method to yield tissue-specific counts of alternative splice site isoforms for individual patients (see Methods).
Table 1.
TCGA patient samples analysed in this study.
| cancer type | TCGA abbreviations | number of samples | number of participants |
|---|---|---|---|
| breast invasive carcinoma | BRCA | 220 | 110 |
| kidney renal clear cell carcinoma | KIRC | 144 | 72 |
| thyroid carcinoma | THCA | 116 | 58 |
| lung adenocarcinoma | LUAD | 114 | 57 |
| prostate adenocarcinoma | PRAD | 104 | 52 |
| liver hepatocellular carcinoma | LIHC | 100 | 50 |
| lung squamous cell carcinoma | LUSC | 98 | 49 |
| head and neck squamous cell carcinoma | HNSC | 84 | 42 |
| kidney renal papillary cell carcinoma | KIRP | 62 | 31 |
| stomach adenocarcinoma | STAD | 54 | 27 |
| colon adenocarcinoma | COAD | 48 | 24 |
| kidney chromophobe | KICH | 46 | 23 |
| bladder urothelial carcinoma | BLCA | 38 | 19 |
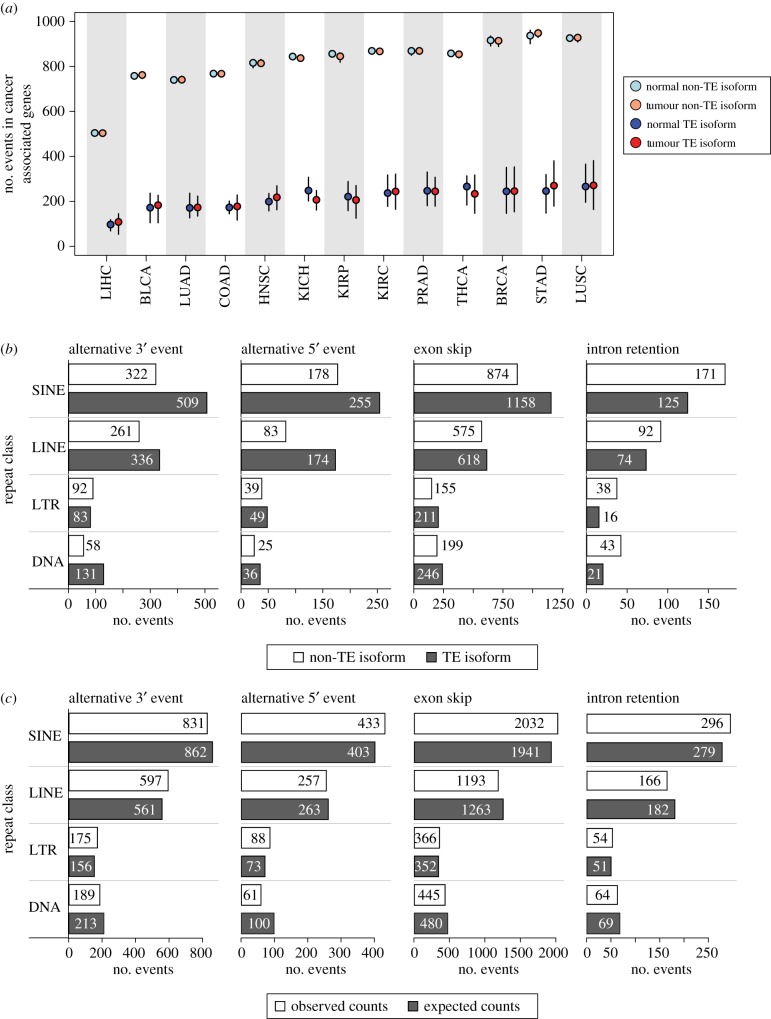
We then narrowed our analysis to a catalogue of 723 known cancer-associated genes and focused on the alternative splice sites in cancer genes that are derived from TE sequences. TE-derived splice sites were delineated by searching for canonical splice donor and acceptor site sequence motifs, located at 3′ and 5′ exon boundaries, that overlap with annotated TE sequences (electronic supplementary material, figure S3). Human TE sequences were divided into their four major classes – short interspersed nuclear elements (SINEs), long interspersed nuclear elements (LINEs), long terminal repeat elements (LTR) and DNA elements (DNA) (electronic supplementary material, figure S4)—and the overall extent of their contribution to alternative splicing in cancer was evaluated. TE sequences contribute 4820 distinct alternative splice sites genome-wide, ranging from 10.5% of alternative 5′ splice events to 14.0% of exon skipping events (electronic supplementary material, figure S5). TEs also contribute a substantial minority of the alternative splice sites to cancer-associated genes. Across the 13 cancer types, TE-derived isoforms are a consistent minority, and the numbers of alternative splice sites are more similar for TE- versus non-TE-derived isoforms, compared to the relatively small differences seen for normal versus cancer samples (figure 2a).
Figure 2.
Overall landscape of TE-derived alternative splicing in cancer. (a) Dot-and-whisker plot comparing the distributions of TE and non-TE isoforms in cancer-associated genes in normal (blue and light blue) and tumour (red and light-red) tissues across all samples within each cancer type. The median number of events are shown as dots and the outliers (defined classically as 1.5 × interquartile range) are shown as whiskers. Cancer tissue abbreviations are defined in table 1. (b) Counts of the total number of unique TE and non-TE isoforms in cancer-associated genes is shown by the splicing event type and TE class. (c) The observed counts of TE isoforms in cancer-associated genes for each event type and TE class is compared to expected counts.
At first glance, the overall landscape of TE-derived splicing isoforms in cancer-associated genes suggests the possibility that TE contributions to alternative splicing in cancer may not be very biologically significant. However, when alternative splicing events in cancer-associated genes are broken down by event type and TE class, the potential contribution of TEs becomes more apparent. This is because for any given splice site where a TE is present, the numbers of TE-derived splice isoforms tend to outnumber the non-TE-derived isoforms that do not show a splice site at the same TE (figure 2b). This holds true for three out of the four alternative splice event types; for intron retention, the non-TE-derived isoforms are more common. Finally, it is interesting to note that there is no particular enrichment for the contributions of any given TE class to any of the four kinds of alternative splice site events. The observed numbers of TEs from each class that contribute to these events are very similar to the expected numbers based on their background frequencies within cancer-associated genes (figure 2c).
(b). Differential expression of transposable element-derived splice sites
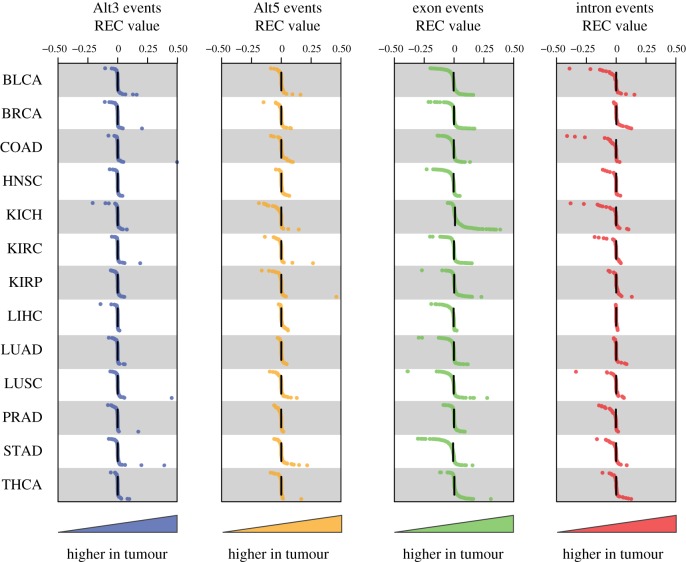
We analysed differences in the expression levels of alternative splice sites between matched normal-tumour sample pairs in an effort to evaluate the effects of individual TE-derived splice sites on cancer. The expression levels of individual alterative splice sites, and their corresponding isoforms, were quantified via normalized counts of mapped RNA-seq reads as detailed in the Methods section. For any given TE-derived splice site, there are four possible expression counts for an individual patient: (i) non-TE isoform normal, (ii) TE isoform normal, (iii) non-TE isoform tumour and (iv) TE isoform tumour. Expression counts for these four conditions can be averaged across individuals to measure the REC of TE-derived isoforms in tumour compared to normal tissue and to evaluate the significance of this difference (electronic supplementary material, figure S6). Distributions of REC values for the four types of TE-derived splice sites across the 13 cancer types are shown in figure 3. For the most part, these distributions are tightly clustered around the median value of 0, or no relative change, with sparsely populated tails that contain individual cases of potential interest. We evaluated a number of the outlier genes showing highly differentially expressed alternative splice isoforms between matched normal-cancer samples (table 2), in an effort to explore potential functional implications of TE-derived splice sites in cancer.
Figure 3.
Differential expression of TE-derived alternative splice isoforms in tumour versus normal samples. Distributions of the relative expression counts (REC) comparing TE-derived to non-TE-derived alternative splice isoforms in tumour versus normal samples. The formula for REC is described in the Methods and in the electronic supplementary material, figure S6. Data are shown for 13 cancer types and four alternative splice event types. Each dot represents an REC value derived from the average normalized expression counts of the TE- and non-TE-derived isoforms in normal and cancer samples. Higher expression (counts) of the TE-derived isoform in tumour are shown on the right side of the panels, whereas lower expression is shown to the left.
Table 2.
Candidate TE-derived isoform switching in cancer. (Examples are shown for individual TE-derived alternative splice events that are overexpressed in cancer compared to normal tissue.)
| clustera | gene | cancer type | %TEi-Nb | %TEi-Tc | event type |
|---|---|---|---|---|---|
| 467 | MYH11 | lung squamous cell carcinoma | 6.5 | 51.8 | Alt3 |
| 412 | CANT1 | stomach adenocarcinoma | 67.3 | 37.4 | exon |
| 132 | WHSC1 | stomach adenocarcinoma | 73.1 | 42.8 | exon |
| 412 | CANT1 | breast invasive carcinoma | 53.5 | 32.1 | exon |
| 154 | KMT2D | stomach adenocarcinoma | 21.3 | 31.8 | Alt5 |
| 397 | POLG | stomach adenocarcinoma | 34.8 | 54.6 | Alt3 |
| 397 | POLG | bladder urothelial carcinoma | 34.8 | 47.6 | Alt3 |
| 261 | PML | kidney renal papillary cell carcinoma | 57.2 | 70.3 | intron |
| 261 | PML | breast invasive carcinoma | 47.0 | 59.6 | intron |
| 261 | PML | kidney chromophobe | 65.5 | 75.8 | intron |
aCluster ID number corresponding to the distinct TE-derived alternative splicing event.
bRelative expression (percentage of total) for the TE-derived isoform in normal tissue.
cRelative expression (percentage of total) for the TE-derived isoform in tumour tissue.
(c). Potential functional implications of transposable element-derived splice sites in cancer
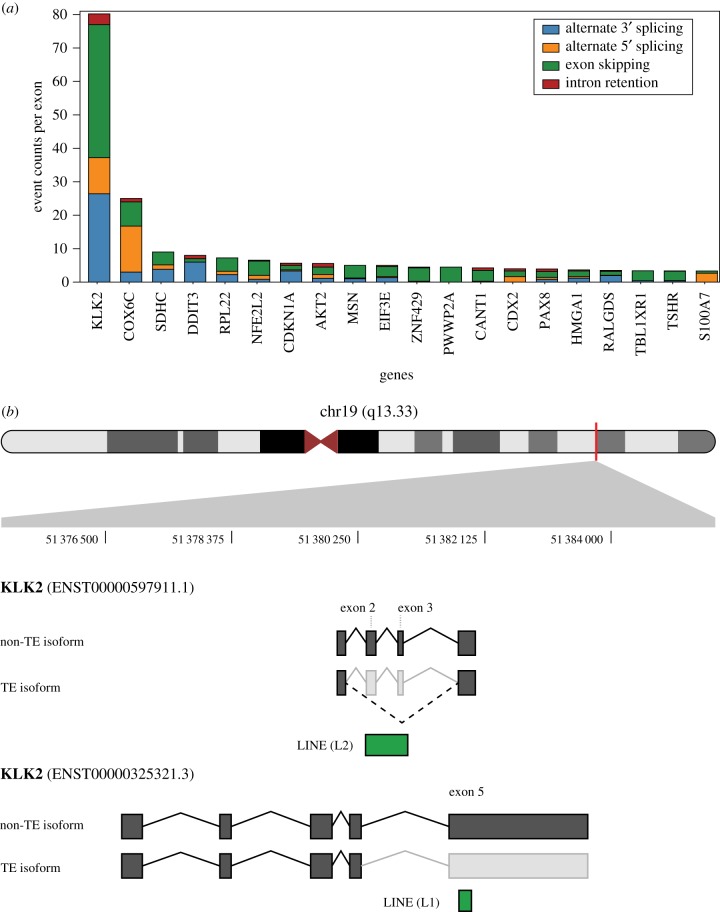
One particular result that stood out from this analysis was the observation that a few cancer-associated genes have extremely high counts of TE-derived alternative splicing events. The kallikrein-related peptidase 2 encoding gene KLK2 shows more than twice as many TE-derived alternative splice sites compared to the second rank gene on this list (figure 4a). There are a total of 297 TE-derived isoforms identified for this gene compared to 354 non-TE-derived isoforms. The KLK2 protein is primarily expressed in the prostate and has been shown to promote prostate cancer cell growth [50]. The connection between TE-derived alternative splicing and cancer is supported by the fact that all of the TE-derived isoforms observed here were identified in prostate adenocarcinoma samples. Examples of TE-derived isoforms for KLK2 are shown in figure 4b,c; these alternative splice events are predicted to induce frameshifts that would lead to truncated protein sequences. Alternative splicing of KLK2 results in fusions with the ETV1 and ETV4 genes in prostate cancer, and all of the known fusion transcripts for these genes are missing exon 3 of KLK2 [51,52]. Interestingly, exon skipping events are by far the most abundant TE-derived splice isoforms seen for this gene (figure 4). The large number of putative TE-derived alternative splicing events in KLK2, specifically exon skipping, suggests TEs could play an important role in the manifestation of KLK2 fusion transcripts and their contribution to prostate cancer. Given their dispersed repetitive nature, it is possible that TE sequences serve as hotspots for the generation of fusion transcripts in cancer. We further explore this potential model for transcriptome diversification by TE sequences in the Conclusion.
Figure 4.
Frequency of TE-derived alternative splice events for individual genes. (a) The total numbers of alternative splice counts per exon are shown for each cancer-associated gene, broken down by the four alternative splice event types. Genes with the highest counts of TE-derived alternative splice events across all cancer types are shown. (b) The location of KLK2 on the long arm of chromosome 19 is shown along with the locations of two TE-derived alternative splicing events. A LINE (L2) sequence generates an internal exon skipping event, and aLINE (L1) generates a terminal exon skipping event.
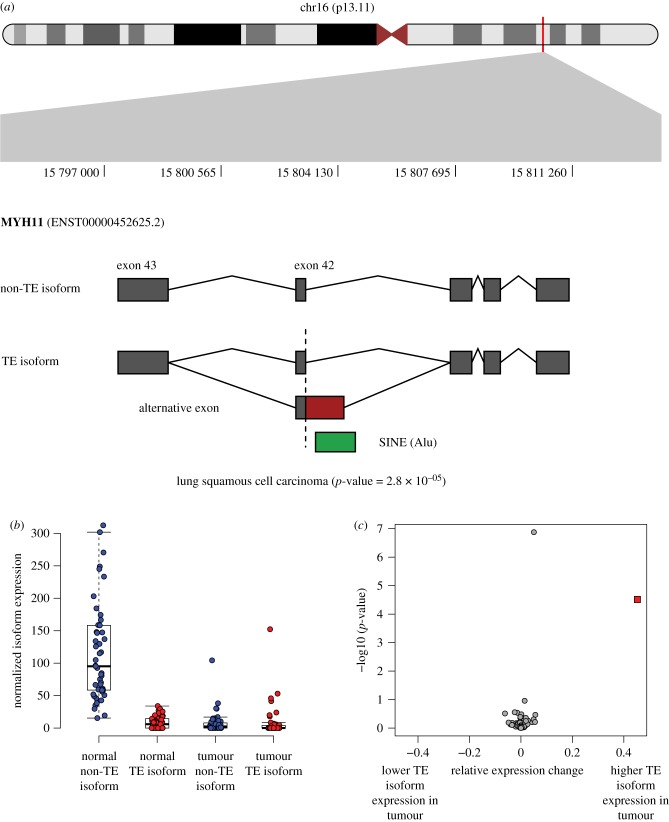
The Myosin Heavy Chain 11 gene MYH11 encodes part of a hexameric protein that functions as a major contractile complex, converting chemical energy into mechanical energy through the hydrolysis of ATP. MYH11 has been shown to contribute to tumorigenesis in both leukaemia and non-small cell lung cancer [53]. MYH11 undergoes alternative splicing, yielding isoforms that are differentially expressed in tumour samples [54]. MYH11 is also implicated in cancer-associated gene fusion events; for example, the CBFB-MYH11 gene fusion plays an important role in leukemogenesis [55–57]. Here, we observe differential isoform expression of MYH11 across 49 paired normal-tumour lung squamous cell carcinoma tissues, whereby an alternative 3′ splicing event within a SINE (Alu) yields a longer version of exon 41 (figure 5; electronic supplementary material, table S2). The longer SINE-derived isoform makes up 6.5% of the transcript population in normal samples compared to 51.8% in tumour samples. The SINE-derived isoform is predicted to result in a frameshift mutation and truncation of the MYH11 protein sequence.
Figure 5.
TE-derived alternative splicing in the MYH11 gene. (a) The location of MYH11 on the short arm of chromosome 16 is shown along with the specific location of its TE-derived alternative splicing event. A SINE (Alu) sequence provides an alternate 3′ splice site resulting in an extended exon 41. (b) Distributions of the non-TE (blue) and TE-derived (red) isoforms are shown for matched normal (left) and lung squamous cell carcinoma samples (right). (c) Relative expression change (REC) values are plotted against the corresponding G-test p-values (see Methods and the electronic supplementary material, figure S6) for the matched normal and lung squamous cell carcinoma samples. The MYH11 TE-derived isoform values are shown as a red square.
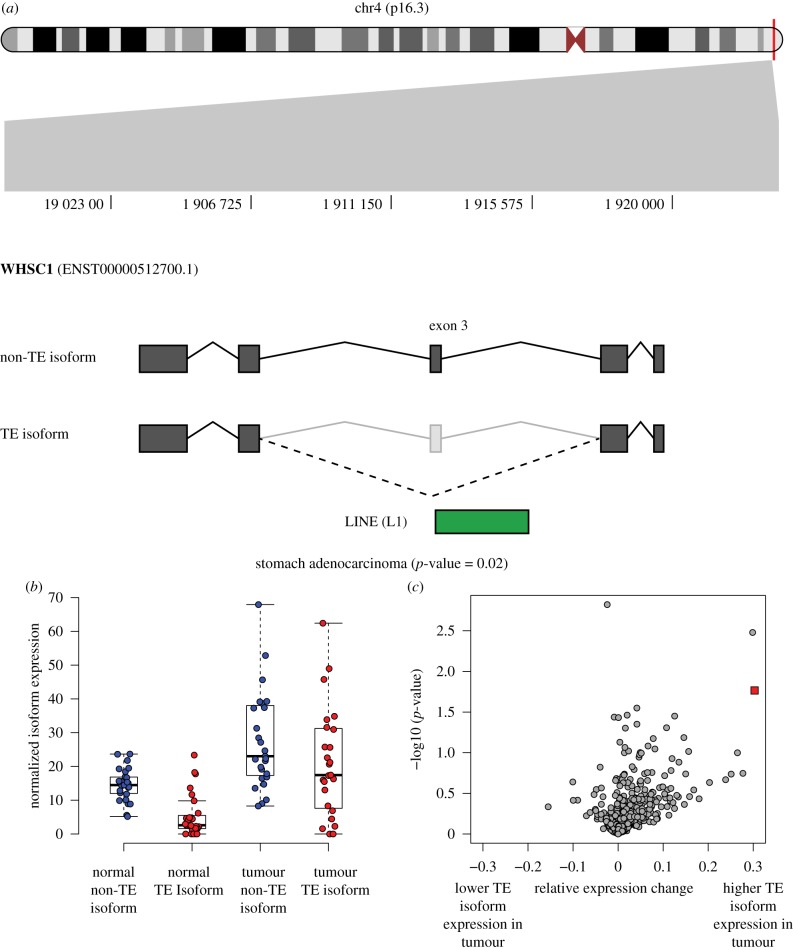
The Wolf-Hirschhorn Syndrome Candidate 1 Protein encoding gene WHSC1, also known as the Nuclear Receptor Binding SET Domain Protein 2 gene (NSD2), encodes a histone methyltransferase that catalyses the dimethylation of histone 3 lysine 36 (H3K36). WHSC1 expression is important for the epithelial-mesenchymal transition and metastasis in gastric cancer [58], and it is overexpressed in a number of different cancer types [59]. WHSC1 has been shown to undergo complex alternative splicing. Most of the primary transcripts of WHSC1 initiate from exon 3, which contains the canonical translation initiation site, although a small fraction of transcripts retain upstream non-coding sequences including exons 1 and 2 [60]. Here, we identified a LINE (L1) element apparently responsible for an exon skipping event in exon 3, which occurs much more frequently in stomach adenocarcinoma primary tumour tissues (57%) when compared to matched normal tissues (27%) (figure 6; electronic supplementary material, table S2). The L1 associated exon skipping event is predicted to cause a frameshift mutation and truncation of the WHSC1 protein sequence.
Figure 6.
TE-derived alternative splicing in the WHSC1 gene. (a) The location of WHSC1 on the short arm of chromosome 4 is shown along with the specific location of its TE-derived alternative splicing event. A LINE (L1) sequence generates an exon skipping event. (b) Distributions of the non-TE (blue) and TE-derived (red) isoforms are shown for matched normal (left) and stomach adenocarcinoma samples (right). (c) Relative expression change (REC) values are plotted against the corresponding G-test p-values (see Methods and the electronic supplementary material, figure S6) for the matched normal and stomach adenocarcinoma samples. The WHSC1 TE-derived isoform values are shown as a red square.
The calcium-activated nucleotidase 1 encoding gene CANT1 is overexpressed in prostate cancer and thought to be involved in proliferation, DNA synthesis, cell cycle and migration of prostate cancer cells [61]. CANT1 is known to undergo alternative splicing, with three well-defined isoforms. Here, we observe a novel exon skipping event, which includes both SINE and LINE elements and results in a differentially expressed isoform, found at 32.7% in normal samples and 62.6% in stomach adenocarcinoma tumour samples (electronic supplementary material, figure S7 and table S2). Interestingly, this particular TE-derived isoform does not lead to a change in the predicted protein sequence as exons 2 and 3 correspond to 5′ UTR sequence. Thus, TE-derived alternative splicing of CANT1 may have a regulatory as opposed to structural effect.
4. Conclusion
Our global survey of TE-derived alternative splicing in cancer revealed that TE sequences contribute to numerous alternative splice sites in cancer-associated genes, including cases where the TE isoforms are relatively overexpressed in tumour tissue. We hope that the landscape of TE-derived splice sites uncovered by our study can serve as a resource for further investigations into the role of TEs in tumorigenesis, and we have created a database of the TE-derived splice sites discovered here to facilitate follow-up studies on TE-derived alternative splicing. The data are distributed as a ‘Track data hub' [62] on the UCSC Genome Browser at: http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&hubUrl=http://jordan.biology.gatech.edu/teAs/hub.txt.
The tracks show the genomic locations of the TE-derived alternative splicing events, with a separate track for each of the four splicing event types. The tracks can be used for visual inspection of individual events of interest or for more large-scale studies via download with the Table Browser.
There are some important caveats to consider with respect to the overall contributions of TEs to the landscape of alternative splicing in cancer. For example, it should be noted that the majority of alternative splice sites in cancer are not TE-derived (electronic supplementary material, figure S5). Nevertheless, TE-derived splice sites are not rare events in cancer; TE sequences provide a substantial minority of alternative splice sites in cancer: 10.5–14.0% depending on the specific event type. Another point to consider is that the observed and expected counts of TE-derived splice isoforms are similar overall, suggesting that TEs' presence alone in gene bodies is enough to ensure that they will be recruited into splice variant isoforms (figure 2c). Thus, it is not clear whether there is an active mechanism by which the use of TE-derived splice sites is selected for in cancer. Finally, it must be emphasized that definitive proof for a functional role for TE-derived splice sites in cancer would require additional molecular biology work beyond the scope of this study.
One of the more intriguing results uncovered by our study was the potential connection between TE-derived alternative splicing and cancer fusion genes. Tumorigenesis is often characterized by large-scale genome rearrangements, and cancer fusion genes are thought to result from translocations, which bring genes that are normally far apart in the genome into close physical proximity. Our results showed numerous alternatively spliced exons that correspond to gene fusion junctions, particularly for the KLK2 gene that experiences both promiscuous alternative splicing and several gene fusion events, and these exons have previously been implicated in gene fusion events. We propose a model whereby apparent gene fusions actually occur at the transcript level via trans-splicing facilitated by TE sequences.
Pre-mRNA sequences destined for splicing are bound by heterogeneous ribonucleoprotein particle (hnRNP) proteins, which prevent the formation of short secondary structures caused by base pairing of complementary regions in the pre-mRNAs. In this way, the bound hnRNPs ensure that pre-mRNAs remain accessible for the assembly of the spliceosome. It occurs to us that hnRNP bound pre-mRNAs will also be open to trans interactions with pre-mRNAs from different loci, if they possess complementary sequences. Trans-splicing is the phenomenon whereby the splicing machinery joins splice donor and acceptor sites from different pre-mRNAs that are co-bound in the same spliceosome, yielding fused mature mRNAs. We propose that TE dispersed repeats provide complementary sequences for binding between pre-mRNAs from different loci, thereby serving as hot spots for trans-splicing. We envision this mechanism as an RNA level analogue of ectopic recombination between dispersed TE DNA sequences and a potential driver of transcriptome diversity.
It is important to note that our model of TE-derived trans-splicing for the generation of fusion transcripts is speculative and only suggested by our data. A number of additional analyses would need to be conducted to validate this model. DNA sequence analysis is needed to distinguish genome level rearrangements in cancer tissue from transcript fusions. TE homology (i.e. sequence complementarity) between transcript fusion partners, co-located with fusion junctions, would need to be confirmed. Explicit reconstruction of entire fusion transcript models, as opposed to individual alternative splice event analysis as was done here, needs to be performed to fully characterize observed gene fusions. Finally, it will be important to avoid RNA-seq experimental artefacts caused by template switching during the cDNA generation step, which could be done via single-molecule RNA-sequencing.
Supplementary Material
Acknowledgements
The results published here are in whole or part based upon data generated by The Cancer Genome Atlas managed by the NCI and NHGRI. Information about TCGA can be found at http://cancergenome.NIH.gov.
Data accessibility
TE-induced alternative splice variant data are distributed as a Track data hub on the UCSC Genome Browser at: http://genome.ucsc.edu/cgibin/hgTracks?db=hg19&hubUrl=http://jordan.biology.gatech.edu/teAs/hub.txt.
Authors' contributions
E.A.C., L.R., T.-C.H., S.G. and D.B. conducted all of the bioinformatics data analysis; E.A.C., L.R. and I.K.J. participated in the design of the study and drafted the manuscript; J.F.M. and I.K.J. conceived of the study, designed the study, coordinated the study and helped draft the manuscript. All authors gave final approval for publication and agree to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
E.C. was supported by the National Institutes of Health (T32, GM105490). L.R. and I.K.J. were supported by the IHRC-Georgia Tech Applied Bioinformatics Laboratory (ABiL). T.-C.H., S.G. and D.B. were supported by the Georgia Tech Bioinformatics Graduate Program. J.F.M. was supported by the Ovarian Cancer Institute (Atlanta). The funding bodies had no part in the design of the study, or collection, analysis, interpretation of data, or in writing the manuscript.
References
- 1.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7, e1002384 ( 10.1371/journal.pgen.1002384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409, 860–921. ( 10.1038/35057062) [DOI] [PubMed] [Google Scholar]
- 3.Jordan IK, Rogozin IB, Glazko GV, Koonin EV. 2003. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 19, 68–72. ( 10.1016/S0168-9525(02)00006-9) [DOI] [PubMed] [Google Scholar]
- 4.van de Lagemaat LN, Landry JR, Mager DL, Medstrand P. 2003. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 19, 530–536. ( 10.1016/j.tig.2003.08.004) [DOI] [PubMed] [Google Scholar]
- 5.Rebollo R, Romanish MT, Mager DL. 2012. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu. Rev. Genet. 46, 21–42. ( 10.1146/annurev-genet-110711-155621) [DOI] [PubMed] [Google Scholar]
- 6.Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 18, 71–86. ( 10.1038/nrg.2016.139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conley AB, Jordan IK. 2010. Identification of transcription factor binding sites derived from transposable element sequences using ChIP-seq. Methods Mol. Biol. 674, 225–240. ( 10.1007/978-1-60761-854-6_14) [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Bowen NJ, Marino-Ramirez L, Jordan IK. 2009. A c-Myc regulatory subnetwork from human transposable element sequences. Mol. Biosyst. 5, 1831–1839. ( 10.1039/b908494k) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polavarapu N, Marino-Ramirez L, Landsman D, McDonald JF, Jordan IK. 2008. Evolutionary rates and patterns for human transcription factor binding sites derived from repetitive DNA. BMC Genomics 9, 226 ( 10.1186/1471-2164-9-226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marino-Ramirez L, Lewis KC, Landsman D, Jordan IK. 2005. Transposable elements donate lineage-specific regulatory sequences to host genomes. Cytogenet Genome Res. 110, 333–341. ( 10.1159/000084965) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Norris ET, Jordan IK. 2017. Human retrotransposon insertion polymorphisms are associated with health and disease via gene regulatory phenotypes. Front. Microbiol. 8, 1418 ( 10.3389/fmicb.2017.01418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Rishishwar L, Marino-Ramirez L, Jordan IK. 2017. Human population-specific gene expression and transcriptional network modification with polymorphic transposable elements. Nucleic Acids Res. 45, 2318–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jjingo D, Conley AB, Wang J, Marino-Ramirez L, Lunyak VV, Jordan IK. 2014. Mammalian-wide interspersed repeat (MIR)-derived enhancers and the regulation of human gene expression. Mob. DNA 5, 14 ( 10.1186/1759-8753-5-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huda A, Tyagi E, Marino-Ramirez L, Bowen NJ, Jjingo D, Jordan IK. 2011. Prediction of transposable element derived enhancers using chromatin modification profiles. PLoS ONE 6, e27513 ( 10.1371/journal.pone.0027513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marino-Ramirez L, Jordan IK. 2006. Transposable element derived DNaseI-hypersensitive sites in the human genome. Biol. Direct. 1, 20 ( 10.1186/1745-6150-1-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, et al. 2015. MIR retrotransposon sequences provide insulators to the human genome. Proc. Natl Acad. Sci. USA 112, E4428–E4437. ( 10.1073/pnas.1507253112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piriyapongsa J, Marino-Ramirez L, Jordan IK. 2007. Origin and evolution of human microRNAs from transposable elements. Genetics 176, 1323–1337. ( 10.1534/genetics.107.072553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piriyapongsa J, Jordan IK. 2007. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS ONE 2, e203 ( 10.1371/journal.pone.0000203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conley AB, Miller WJ, Jordan IK. 2008. Human cis natural antisense transcripts initiated by transposable elements. Trends Genet. 24, 53–56. ( 10.1016/j.tig.2007.11.008) [DOI] [PubMed] [Google Scholar]
- 20.Huda A, Bowen NJ, Conley AB, Jordan IK. 2011. Epigenetic regulation of transposable element derived human gene promoters. Gene 475, 39–48. ( 10.1016/j.gene.2010.12.010) [DOI] [PubMed] [Google Scholar]
- 21.Huda A, Marino-Ramirez L, Landsman D, Jordan IK. 2009. Repetitive DNA elements, nucleosome binding and human gene expression. Gene 436, 12–22. ( 10.1016/j.gene.2009.01.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conley AB, Piriyapongsa J, Jordan IK. 2008. Retroviral promoters in the human genome. Bioinformatics 24, 1563–1567. ( 10.1093/bioinformatics/btn243) [DOI] [PubMed] [Google Scholar]
- 23.Conley AB, Jordan IK. 2012. Cell type-specific termination of transcription by transposable element sequences. Mob. DNA 3, 15 ( 10.1186/1759-8753-3-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowley M, Oakey RJ. 2013. Transposable elements re-wire and fine-tune the transcriptome. PLoS Genet. 9, e1003234 ( 10.1371/journal.pgen.1003234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelley DR, Hendrickson DG, Tenen D, Rinn JL. 2014. Transposable elements modulate human RNA abundance and splicing via specific RNA-protein interactions. Genome Biol. 15, 537 ( 10.1186/s13059-014-0537-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen S, Lin L, Cai JJ, Jiang P, Kenkel EJ, Stroik MR, Sato S, Davidson BL, Xing Y. 2011. Widespread establishment and regulatory impact of Alu exons in human genes. Proc. Natl Acad. Sci. USA 108, 2837–2842. ( 10.1073/pnas.1012834108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lev-Maor G, Sorek R, Shomron N, Ast G. 2003. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 300, 1288–1291. ( 10.1126/science.1082588) [DOI] [PubMed] [Google Scholar]
- 28.Sorek R, Ast G, Graur D. 2002. Alu-containing exons are alternatively spliced. Genome Res. 12, 1060–1067. ( 10.1101/gr.229302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mersch B, Sela N, Ast G, Suhai S, Hotz-Wagenblatt A. 2007. SERpredict: detection of tissue-or tumor-specific isoforms generated through exonization of transposable elements. BMC Genet. 8, 78 ( 10.1186/1471-2156-8-78) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anwar S, Wulaningsih W, Lehmann U. 2017. Transposable elements in human cancer: causes and consequences of deregulation. Int. J. Mol. Sci. 18, 974 ( 10.3390/ijms18050974) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burns KH. 2017. Transposable elements in cancer. Nat. Rev. Cancer 17, 415 ( 10.1038/nrc.2017.35) [DOI] [PubMed] [Google Scholar]
- 32.Carreira PE, Richardson SR, Faulkner GJ. 2014. L1 retrotransposons, cancer stem cells and oncogenesis. FEBS J. 281, 63–73. ( 10.1111/febs.12601) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clayton EA, Wang L, Rishishwar L, Wang J, McDonald JF, Jordan IK. 2016. Patterns of transposable element expression and insertion in cancer. Front. Mol. Biosci. 3, 76 ( 10.3389/fmolb.2016.00076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee E, et al. 2012. Landscape of somatic retrotransposition in human cancers. Science 337, 967–971. ( 10.1126/science.1222077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott EC, Gardner EJ, Masood A, Chuang NT, Vertino PM, Devine SE. 2016. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 26, 745–755. ( 10.1101/gr.201814.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El Marabti E, Younis I. 2018. The cancer spliceome: reprograming of alternative splicing in cancer. Front. Mol. Biosci. 5, 80 ( 10.3389/fmolb.2018.00080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Escobar-Hoyos L, Knorr K, Abdel-Wahab O. 2019. Aberrant RNA splicing in cancer. Annu. Rev. Cancer Biol. 3, 167–185. ( 10.1146/annurev-cancerbio-030617-050407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jayasinghe RG, et al. 2018. Systematic analysis of splice-site-creating mutations in cancer. Cell Rep. 23, 270–281.e273. ( 10.1016/j.celrep.2018.03.052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kahles A, et al. 2018. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 34, 211–224.e216. ( 10.1016/j.ccell.2018.07.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oltean S, Bates DO. 2013. Hallmarks of alternative splicing in cancer. Oncogene 33, 5311 ( 10.1038/onc.2013.533) [DOI] [PubMed] [Google Scholar]
- 41.Venables JP. 2004. Aberrant and alternative splicing in cancer. Cancer Res. 64, 7647–7654. ( 10.1158/0008-5472.CAN-04-1910) [DOI] [PubMed] [Google Scholar]
- 42.Venables JP, et al. 2009. Cancer-associated regulation of alternative splicing. Nat. Struct. Mol. Biol. 16, 670 ( 10.1038/nsmb.1608) [DOI] [PubMed] [Google Scholar]
- 43.Vitting-Seerup K, Sandelin A. 2017. The landscape of isoform switches in human cancers. Mol. Cancer Res. 15, 1206–1220. ( 10.1158/1541-7786.MCR-16-0459) [DOI] [PubMed] [Google Scholar]
- 44.O'Leary NA, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. ( 10.1093/nar/gkv1189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tyner C, et al. 2017. The UCSC genome browser database: 2017 update. Nucleic Acids Res. 45, D626–D634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smit A, Hubley R, Green P. 2015. RepeatMasker Open-4.0. 2013–2015. See http://www.repeatmasker.org.
- 47.Quinlan AR. 2014. BEDTools: the Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11.12.1–11.12.34. ( 10.1002/0471250953.bi1112s47) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. 2018. The COSMIC cancer gene census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 18, 696–705. ( 10.1038/s41568-018-0060-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 ( 10.1186/s13059-014-0550-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shang Z, Niu Y, Cai Q, Chen J, Tian J, Yeh S, Lai K-P, Chang C. 2014. Human kallikrein 2 (KLK2) promotes prostate cancer cell growth via function as a modulator to promote the ARA70-enhanced androgen receptor transactivation. Tumor Biol. 35, 1881–1890. ( 10.1007/s13277-013-1253-6) [DOI] [PubMed] [Google Scholar]
- 51.Adamopoulos PG, Kontos CK, Scorilas A. 2019. Discovery of novel transcripts of the human tissue kallikrein (KLK1) and kallikrein-related peptidase 2 (KLK2) in human cancer cells, exploiting next-generation sequencing technology. Genomics 111, 642–652. ( 10.1016/j.ygeno.2018.03.022) [DOI] [PubMed] [Google Scholar]
- 52.David A, et al. 2002. Unusual alternative splicing within the human kallikrein genes KLK2 and KLK3 gives rise to novel prostate-specific proteins. J. Biol. Chem. 277, 18 084–18 090. ( 10.1074/jbc.M102285200) [DOI] [PubMed] [Google Scholar]
- 53.Ma Q, et al. 2019. Identification and validation of key genes associated with non-small-cell lung cancer. J. Cell. Physiol. 234, 22 742–22 752. [DOI] [PubMed] [Google Scholar]
- 54.Sebestyén E, Zawisza M, Eyras E. 2015. Detection of recurrent alternative splicing switches in tumor samples reveals novel signatures of cancer. Nucleic Acids Res. 43, 1345–1356. ( 10.1093/nar/gku1392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castilla LH, et al. 1999. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat. Genet. 23, 144 ( 10.1038/13776) [DOI] [PubMed] [Google Scholar]
- 56.Liu PP, et al. 1996. Identification of the chimeric protein product of the CBFB-MYH11 fusion gene in inv (16) leukemia cells. Genes , Chromosomes Cancer 16, 77–87. () [DOI] [PubMed] [Google Scholar]
- 57.Castilla LH, et al. 1996. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB–MYH11. Cell 87, 687–696. ( 10.1016/S0092-8674(00)81388-4) [DOI] [PubMed] [Google Scholar]
- 58.Ezponda T, et al. 2013. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer. Oncogene 32, 2882–2890. ( 10.1038/onc.2012.297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hudlebusch HR, Santoni-Rugiu E, Simon R, Ralfkiær E, Rossing HH, Johansen JV, Jørgensen M, Sauter G, Helin K. 2011. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin. Cancer Res. 17, 2919–2933. ( 10.1158/1078-0432.CCR-10-1302) [DOI] [PubMed] [Google Scholar]
- 60.Keats JJ, et al. 2005. Overexpression of transcripts originating from the MMSET locus characterizes all t (4; 14)(p16; q32)-positive multiple myeloma patients. Blood 105, 4060–4069. ( 10.1182/blood-2004-09-3704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gerhardt J, et al. 2011. The androgen-regulated calcium-activated nucleotidase 1 (CANT1) is commonly overexpressed in prostate cancer and is tumor-biologically relevant in vitro. Am. J. Pathol. 178, 1847–1860. ( 10.1016/j.ajpath.2010.12.046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raney BJ, et al. 2014. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC genome browser. Bioinformatics 30, 1003–1005. ( 10.1093/bioinformatics/btt637) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TE-induced alternative splice variant data are distributed as a Track data hub on the UCSC Genome Browser at: http://genome.ucsc.edu/cgibin/hgTracks?db=hg19&hubUrl=http://jordan.biology.gatech.edu/teAs/hub.txt.