Abstract
Our perception of the role of the previously considered ‘selfish’ or ‘junk’ DNA has been dramatically altered in the past 20 years or so. A large proportion of this non-coding part of mammalian genomes is repetitive in nature, classified as either satellites or transposons. While repetitive elements can be termed selfish in terms of their amplification, such events have surely been co-opted by the host, suggesting by itself a likely altruistic function for the organism at the subject of such natural selection. Indeed numerous examples of transposons regulating the functional output of the host genome have been documented. Transposons provide a powerful framework for large-scale relatively rapid concerted regulatory activities with the ability to drive evolution. Mammalian totipotency has emerged as one key stage of development in which transposon-mediated regulation of gene expression has taken centre stage in the past few years. During this period, large-scale (epigenetic) reprogramming must be accomplished in order to activate the host genome. In mice and men, one particular element murine endogenous retrovirus with leucine tRNA primer (MERVL) (and its counterpart human ERVL (HERVL)) appears to have acquired roles as a key driving force in this process. Here, I will discuss and interpret the current knowledge and its implications regarding the role of transposons, particularly of long interspersed nuclear elements (LINE-1s) and endogenous retroviruses (ERVs), in the regulation of totipotency.
This article is part of a discussion meeting issue ‘Crossroads between transposons and gene regulation’.
Keywords: reprogramming, 2-cell-like cells, pluripotency, MERVL, LINE-1, transposable elements
1. Introduction
Our genomes are vastly populated by remnants of viruses and other genomic elements that have accumulated during mammalian evolution. In fact, in most common mammals, including mouse, cattle and humans, roughly half of the genome belongs to this category, commonly referred to as ‘repeats’ or repetitive elements [1]. Phylogenetically and from the point of view of their genome structure, repetitive elements can roughly be divided into transposable elements and satellite DNA. Satellite DNA is typically repeated in tandem at specific locations across the linear genome and can form clusters of up to several kilobases. The satellites forming the centromere of mouse and humans belong to this class. They are critical to the mitotic process, as they are the place for kinetochore loading. Their repetitive nature is thought to be important for the establishment of heterochromatin and consequent physical compaction of these loci, which is therefore essential for chromosome segregation and faithful chromosome inheritance [2].
Transposable elements are classified into DNA transposons and retrotransposons. The most abundant in rodent and human genomes are the retrotransposons. Generally speaking, retrotransposons are transcriptionally silenced in most somatic cells. This is achieved through the acquisition of constitutive heterochromatin signatures driven primarily by the catalysis of H3K9 tri-methylation by several enzymes, including Setdb1, Suv39h1/h2 and the downstream recruitment of heterochromatin protein 1 and Suv420h1/h2 and by DNA methylation. In the germline, young transposons are primarily silenced through the piRNA pathway (reviewed in [3]). Silencing of retrotransposons is thought to be tightly regulated, and orchestrated by a myriad of KRAB zinc-finger proteins and their interactor, Kap1/Trim28 (reviewed in [4]).
At the earliest developmental stages immediately after fertilization, in contrast, such heterochromatic signatures are thought to be largely absent or atypical [5]. In fact, the mouse zygote and 2-cell stage embryo actively transcribe many retrotransposons, and their transcriptional activity persists at least until the blastocyst stage, around the time of implantation [6–9]. The mechanisms regulating their transcriptional activation and repression are largely unknown. For example, the temporal dynamics of DNA de-methylation and re-methylation during pre-implantation development [10] are not sufficient to explain their changes in expression for the most part. Likewise, ChIP-seq analysis of H3K9me3 during pre-implantation development indicates that only a handful of retrotransposons from the long terminal repeat (LTR) family are enriched in H3K9me3 in the cleavage-stage embryos [11], and thus cannot explain by its simple presence/absence the transcriptional dynamics of most retrotransposons in the embryo.
While much remains to be done to understand how retrotransposons are regulated, recent work has started to identify key transcription factors (TF) involved in this process. These discoveries have emerged from the powerful combination of traditional in vivo (mouse embryo) and novel in vitro (cell culture) models. Here, I will briefly discuss both of these models, with emphasis on recent literature. However, before I go on and discuss these findings, I feel compelled to explain the definition of totipotency that I will use, since the word is used somewhat loosely and to refer to different things in the literature. The implications of this definition will be important when discussing the use of cellular models and the assays employed to address whether such models are indeed truly totipotent.
Totipotency is the ability of a single cell to give rise to a full organism by itself [12,13]. This contrasts to pluripotency, which in mammals refers to the ability of a cell to contribute and give rise to all three germ layers—including the germline—but cannot form a new organism by itself without supportive extra-embryonic tissues [14,15]. When I refer to totipotency, an important distinction must be made between chimeric contribution versus self-potential of a single cell. Indeed, while blastomeres of the 4-cell embryo can contribute to all lineages of the conceptus when transplanted in chimera, including embryonic and extraembryonic lineages, individually they are not able to form a new being. Thus, while they are extremely plastic given their ability to contribute to all lineages of the embryo, they are not totipotent, and instead totipotency in the mouse is restricted to the zygote and each of the blastomeres of the 2-cell stage embryo [16,17].
2. Changing cell fate
Work of Todd Macfarlan, Samuel Pfaff and colleagues in 2012 reported the existence of a subpopulation of cells in embryonic stem (ES) cell cultures, which highly resemble cells from the 2-cell stage embryo, based on their global transcriptional profile [18]. Remarkably, one of the main features of these ‘2-cell-like cells’ (2CLC) is that they robustly transcribe elements from an LTR family of endogenous retroviruses, specifically MERVL (murine endogenous retrovirus with leucine tRNA primer). Indeed, work by Barbara Knowles in the 2000s had reported that oocytes and early mouse embryos from cleavage stages can use sequences from MERVL (primarily Mt2_MM and MERVL_int) as alternative promoters of host genes, identifying several chimeric transcripts containing MERVL and single-copy genes sequences [6,8]. Thus, it would seem that the transcriptional activation of a retrotransposon could have a potential, direct role in changing cellular fate in cell culture, and in regulating the transcriptional programme of 2-cell stage embryos.
The discovery of 2CLC was ground-breaking for several reasons. First, it highlighted the general existence of additional heterogeneities in ES cell cultures in the mouse. This expanded our knowledge on the transient phenotypic features of embryonic stem cells (ESC) in culture, which already included the well-defined naive pluripotent cells (e.g. ‘Nanog-high’ typically enriched in ‘2i’ medium) as well as cells in a more intermediate pluripotency state (e.g. ‘Nanog-low’), but also cells that co-express epiblast and extraembryonic genes, similarly to morula stage blastomeres [19,20]. The latter were isolated as expressing the endoderm marker Hex-1 and shown to contribute to trophectoderm and inner cell mass (ICM) derivatives when aggregated in chimera. While these cells were referred to totipotent, I would argue that they are rather bipotent, and not totipotent, based on their ability to contribute to the two first lineages of the mouse blastocyst but not to a full organism by themselves, under the framework of the totipotency definition that I put forward. Of note, this capacity to contribute to ICM and trophectoderm derivatives has also recently been referred to as ‘expanded potential’ [21].
Second, and most importantly, the identification of 2CLC provided the community with a conceptual and experimental platform to seek to establish a biochemically tractable model to understand the molecular features of totipotency and the biology of the early mouse embryo. This has generated an enormous amount of interest in the past 5 years, and work aiming to thoroughly characterize 2CLC and identify the regulators of their emergence, primarily through the identification of factors that can regulate MERVL transcription, has flourished.
It is precisely this endeavour that led to the identification of the pioneer TF Dux as a key regulator of MERVL [22,23]. Dux (DUX4 in humans) is a double homeodomain TF, which is conserved in mammals and was originally identified as misregulated in facioscapulohumeral dystrophy in human biopsies [24]. The ectopic expression of Dux alone in mouse ESCs is sufficient to activate MERVL, along with a significant part of the ‘2C’ transcriptional programme, which corresponds in effect to the zygotic genome activation (ZGA) programme. Dux regulates MERVL expression through direct binding of the Dux-recognition motif within the LTR. DUX4, its human counterpart, leads to an equally robust increase in transcription of the human ERVL (HERVL) and of ZGA genes, which in humans reflects a ‘4C’ stage [22,23]. This is a remarkable observation, considering that the first homeodomains of Dux and DUX4 differ, and that the sequences of MERVL and HERVL are dissimilar, which indicates that Dux and DUX4 coevolved with their respective species-specific transposons, in an example of convergent evolution. Indeed, when expressed in mouse ESCs, mouse DUX can activate ZGA-related retrotransposons but DUX4 cannot [25]. This has led to the suggestion that ancestral DUX proteins emerged to regulate embryonic transcription, but their capacity to regulate the process of ZGA was multiplied through their acquisition of the ability to regulate retrotransposons [25]. This implies that MERVL has been co-opted to regulate the key developmental process of ZGA in mammals, which is supported by earlier findings of chimeric transcripts in early embryos as discussed above. This begs the question of how ‘2C’ genes became linked to MERVL elements and their LTRs through evolution.
Importantly however, while the overlap of genes regulated by Dux in mouse ESCs and the ‘2C’ genes activated during mouse ZGA is significant, it is not complete [22,23]. Similarly, DUX4 drives the expression of ZGA genes in human ESCs, but not all human ZGA genes become activated upon DUX4 expression. These findings established that although the Dux TF are key regulators of ZGA, they are clearly not the only players. In fact, only about 25% of the accessible chromatin regions specific to 2CLC are bound by Dux [23], again suggesting that a large, additional part of the ‘2C’ programme is regulated by other factors. It is therefore not surprising that a recent knock-out mouse model reported that Dux is not essential for development, since mice homozygous null for Dux are born [26,27]. However, Dux−/− are born at submendelian proportions and, as expected, only about 25% of minor ZGA genes were affected in embryos lacking Dux, although this phenotype appears to vary dependent on the genetic background.
These findings are perfectly in line with the concept that retrotransposons have been co-opted in many instances as gene regulatory regions during evolution [28,29]. Indeed, this hypothesis posits that MERVL amplification may have evolved to facilitate ZGA, perhaps by providing one single regulatory element across many genes to enable coordinated, fast and robust transcriptional activation at ZGA. However, the host-specific promoters remained intact and for the most part, functional, as seen by the differential promoter usage of some genes that are expressed during ZGA but also at later stages in development or in differentiated cells [8]. This notion would imply that, in case of mutation or deletion of the corresponding retrotransposon (or the TF regulating its expression), the host promoter would still be able to be activated, most likely explaining the minor changes in ZGA observed in the Dux−/− embryos. Analysis of the promoter usage of genes located in proximity to MERVL in these mutants would provide important insights on the impact of retrotransposon co-option on gene regulation. Thus, a direct relationship between MERVL and ZGA remains to be formally investigated.
Accordingly, additional TF for MERVL regulation and 2CLC emergence have been identified. Among them, Dppa2 and Dppa4 can activate ‘2C’ genes when overexpressed in mouse ESCs [30–32]. Like Dux, Dppa2 and Dppa4 are highly expressed in mouse embryos during ZGA, and highly upregulated in 2CLC. Dppa2/4 mRNA levels remain high through pre-implantation development. Dppa2/4 are thought to be regulated by DNA methylation [30], and depleting Dppa2 or Dppa4 reduces significantly, but did not fully eliminate, 2CLC induction [30,31]. Both Dppa2 and Dppa4 bind the Dux regulatory region and are necessary for Dux activation. However, whether Dppa2/4 act upstream and/or in parallel to Dux on MERVL activation is not yet fully clear, particularly considering that Dppa2/4 display broad binding to most active promoters. GATA2, has also been shown to regulate MERVL expression in mouse induced pluripotent stem (iPS) cells using a luciferase reporter, and when overexpressed in ESCs, it was found to bind MERVL by ChIP [33].
Additionally to searching for TF regulating MERVL—and the ‘2C’ programme—several groups have sought to identify chromatin modifiers involved in their regulation. Most of them have been mainly identified through screening strategies using a 2C-reporter as readout, consisting of the MERVL LTR driving expression of a fluorescent protein. The quest for identifying chromatin modifiers regulating 2CLC stems from at least two standpoints. First, chromatin regulators can enable cell fate conversions, acting as impediments or accelerators for reprogramming, but they can also stabilize cell fates. Second, work during the past years has established that the chromatin landscape of the totipotent cells of the early embryo differs significantly from chromatin in pluripotent and differentiated cells. Thus, identifying chromatin transitions in 2CLC has the potential to shed light on this very peculiar chromatin that characterizes the totipotent cells of the early mammalian embryo.
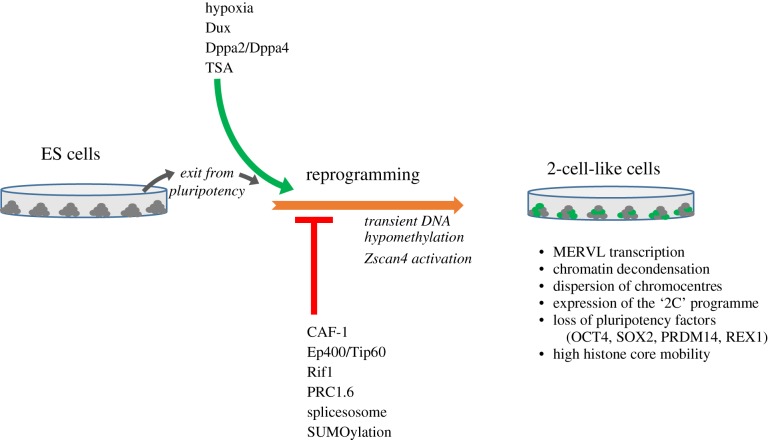
The original report by Macfarlan and colleagues [18] identified trichostatin A (TSA) as a 2CLC inducer, indicating that relaxing chromatin structure promotes 2CLC emergence. Indeed, subsequent MNase and ATACseq analyses revealed that 2CLC—and also 2-cell embryos—have a globally more open chromatin, compared to ESCs [23,34–36]. This is also in line with the increased histone mobility observed in 2CLC and 2-cell stage embryos [37,38]. Several chromatin modifiers have been identified as roadblocks for 2CLC reprogramming (figure 1). Our initial studies showed that chromatin assembly factor 1 (CAF-1), the complex responsible for depositing core H3/H4 tetramers in a replication-coupled manner, is a major regulator of such reprogramming [35]. Downregulating CAF-1 in ESCs led to a substantial increase in 2CLC and this effect was exclusively dependent on the ability of CAF-1 to assemble chromatin. Later work screened approximately 1400 chromatin modifiers using a siRNA approach [39]. We identified five main molecular pathways as major roadblocks for 2CLC reprogramming. These included replication and chromatin assembly, in line with our previous work, but also new regulators such as the Tip60/Ep400 complex, the splicesosome, the non-canonical PRC1 (polycomb repressor complex) complex PRC1.6 and various proteins related to Ubiquitin regulation. This study also suggested that several ‘epigenetic’ pathways can control 2CLC reprogramming, both directly and indirectly. A study published just a couple of months later using an shRNA screen also identified Rif1 (replication timing regulatory factor 1) as a regulator of 2CLC emergence [40]. An interesting observation from all of these findings is that induction of 2CLC emergence seems to be, at least partially, linked to the induction of some genes that are expressed in the germline. Components of the PRC1.6 complex had been identified as repressors of a germline transcriptional signature in ESCs [41]. Indeed, when Rybp, Pcgf6 and Max/Mga are downregulated, genes such as Stra8, otherwise exclusively expressed during meiosis, become activated. Max, which is a dimerization partner of Myc, was well known as a germ cell repressor [42,43]. Whether these specific changes pertaining to germline processes are important or necessary for acquiring a totipotent and/or a 2CLC identity remains to be determined. Notably, PRC1.6 binds directly to the Dux locus and is regulated by Sumoylation [44], and therefore the SUMO-pathway has emerged as a repressor of 2CLC fate. The regulation of 2CLC identity by SUMO occurs at least through two different mechanisms. The first one involves sumoylation of PRC1.6 components, which leads to the reduction of PRC1.6 occupancy at the Dux loci and eventual transcriptional activation of Dux [44]. The second one involves sumoylation of Dppa2 by the sumo ligase PIAS4, which renders Dppa2 inactive [32].
Figure 1.
Summary of the pathways shown so far to induce 2CLC in ESC cultures. Ectopic expression of the TF Dux and Dppa2/4 as well as culturing in hypoxia conditions or with TSA, increase the 2CLC population and therefore are positive regulators (green arrow). Downregulation of CAF-1, specific chromatin modifiers and components of the splicesosome also induces 2CLC, and therefore, these are negative regulators (red arrow). 2CLC emergence involves a transient DNA hypomethylation state, and a Zscan4-positive intermediate state. Global features of 2CLC are listed on the right.
More recently, another study using a wider dCas9 screening strategy revealed essentially the same factors identified in previous screenings [45]. Dnmt1 was also identified as promoting 2CLC maintenance. In a separate work, temporal changes in MERVL expression were correlated with DNA demethylation, upon addition of vitamin C to the culture [46]. Interestingly, the transition to a MERVL-expressing state had been previously shown to be accompanied by a global genome-wide DNA hypomethylation, including the decrease of DNA methylation at imprinted genes [34]. While this was associated with low levels of DNMT proteins in these cells, in particular of Dnmt1, the results indicated that reprogramming into a MERVL+ state was not a consequence of changes in DNA methylation. Indeed, triple-knock-out (TKO) of Dnmt1/3a/3b in ESCs, which results in loss of DNA methylation, does not affect 2CLC emergence [34]. In this regard, the RNA-binding protein PSPC1 inhibits expression of several MERVK, MERVL and mammalian apparent LTR-retrotransposon (MaLR) in mouse ESCs, presumably through physical interactions and recruitment of the Tet2 enzyme [47].
3. Cause or consequence?
In sum, the work above indicates that reprogramming towards 2CLC is regulated at multiple levels by different molecular pathways and chromatin regulators. It will be important to disentangle the direct and indirect regulators, and precise biochemical mechanisms and underlying regulatory actions. Concerning retrotransposon regulation, outstanding questions remain, including: (i) what provides specificity to transposon regulation, (ii) what other additional TF can regulate MERVL expression and repression and, most importantly, (iii) is retrotransposon expression sufficient to drive changes in cell fate, or is this just a consequence and/or a passenger?
Understanding the specificity in targeting each retrotransposon family and subfamilies will be crucial to understand how retrotransposons may help shape changes in cell fate. Such specificity is likely to be also cell type specific, and potentially achieved through KRAB-zinc-finger proteins [48]. Indeed, we know that depletion of the H3K9 methyltransferase Setdb1/ESET in ESCs results in derepression of class I (MLV) and class II (IAP and MusD) ERVs, but class III ERV (MTA) are unaffected [49,50]. This contrasts with fibroblasts, in which IAP and MusD are not derepressed upon Setdb1 loss, but MLV are. It is therefore possible that the ‘host’ chromatin state and/or cofactors, including TF, present in specific cell types aid in reaching specificity in retrotransposon regulation.
As discussed above, a key task would be to determine whether a ‘simple’ transcriptional activation of specific retrotransposons would be sufficient to drive cell fate changes. This question is now beginning to be addressed using different novel methodologies such as Crispr/Cas9-based genome editing methods and also TALE-mediated activation and CRISPRa strategies, whereby transcriptional activators or repressors are targeted directly to thousands of loci at their endogenous locations across the genome.
Is MERVL and its LTR-driven expression a driver for 2CLC reprogramming? Transcription from the MERVL LTR can also impact neighbouring genes by readthrough transcription and enhancer function. The fact that Dux can induce 2CLC emergence so efficiently suggests that transcription of MERVL may be sufficient for reprogramming [22,23]. However, because Dux can also bind additional single-copy genes, this possibility remains to be demonstrated. Activation of MERVL using Crispr/SAM did result in the induction of the handful of 2C genes analysed (11 genes in total) [47]. This indicates that activation of MERVL alone can potentially regulate—at least in part—the 2CLC programme.
4. Giving pluripotency away
The changes in cell fate that reprogramming ESCs to 2CLC entail also involve shutting down the pluripotency programme. The extent of this ‘off-switch’ is not yet very clear, but Macfarlan and colleagues documented that 2CLC lack detectable protein levels of OCT4 [18]. In fact, analysis of other TF of the pluripotency-gene regulatory network further demonstrated that this is not limited to OCT4, but that levels of SOX2, PRDM14, REX1 and AP2gamma proteins are also undetectable in 2CLC [39]. However, their mRNA levels are only slightly reduced or unchanged, indicating that shutting down the pluripotency network is regulated also at the translational level.
Importantly, analysis of an intermediate cellular state characterized by the expression of the TF Zscan4 but the absence of MERVL transcription, indicated that the pluripotency network is down-regulated before the transition into a 2CLC (MERVL+) state. These experiments led us to conclude that cells must exit pluripotency before committing to the 2CLC fate [39], potentially through translational regulation in Zscan4+ cells [51]. These results were recently confirmed by a later study [45]. In fact, neither Oct4, Rex1 or Nanog depletion affects MERVL or Zscan4 expression and, accordingly, their downregulation does not impact 2CLC emergence [39]. We also showed that it is the naïve ESCs that have the highest probability to reprogramme towards 2CLC [39]. At first sight, these conclusions would seem at odds with the observation that ESCs kept in a ‘ground’, naive state through the continuous presence of the ‘2i’ inhibitors cycle less often into 2CLC [39]. However, culturing ESCs in such pharmacological inhibitors is expected to ‘freeze’ ESCs in a ground fate, and therefore preventing spontaneous changes of cellular heterogeneities in such cultures.
Studying the natural progression from ESCs towards 2CLC reprogramming using single-cell expression analyses with 93 genes allowed us to establish the sequential changes in gene expression, including changes in pluripotency factors, that accompany 2CLC formation, thereby defining intermediate cellular states. Remarkably, computational modelling of single-cell RNAseq confirmed these transitions on a genome-wide level, and identified additional sets of genes differentially expressed at each of the transitions [39]. This indicates that the intermediate stages that we described reflect concrete and binary changes in the transcriptome of cells transiting towards the 2CLC. While informative, these analyses are highly correlative and do not really shed light on the actual molecular mechanisms that directly regulate changes in cell fate. Additional work, perhaps through the identification of specific TF and/or KRAB-ZFP will help us to better disentangle causal relationships and the role of retrotransposons in this process.
5. Can totipotency be reduced to a unique transcriptional state?
I believe that the answer is clearly no. Although phenotypically most of the molecular features that provide cell identity and function are a consequence of the genes transcribed, they are certainly not limited to that. For example, metabolic features and chromatin state or extrinsic factors need to be taken into account. Indeed, hypoxia reduces the percentage of 2CLC in ESC cultures [18]. Notwithstanding, a cell's potency should eventually be determined by its ability to ‘do something’ in its natural environment, when transplanted back into the respective native context.
The gold standard for testing whether cells in culture are pluripotent consists of introducing them into morula or blastocyst stage embryos and investigating their potential to contribute to the embryo, and most importantly, to the germline [52]. These chimera assays are relatively straightforward for ESCs with full pluripotency, since they readily contribute to all the lineages of the embryo proper, especially when more than one cell is injected per chimera. In analogy, therefore, totipotent cells should be tested in vivo. However, these experiments pose a conceptual problem, if one considers that totipotent cells are those that can give rise to a full organism without the need of carrier cells. Often however, totipotency is called positive when cells in culture are put back into late 8-cell stage embryos, and they are seen to occupy the trophectoderm layer of the blastocyst, and with more stringency, when cells are seen in the placenta of post-implantation embryos (figure 2). In the field, these experiments are generally done with various levels of stringency criteria and most of them lack evidence of proliferative trophoblast derivatives in the placenta, and are restricted to conclusions on contribution to yolk sac derivatives, which in fact is an ICM derivative, not a trophectoderm derivative. In addition, they have raised several reproducibility issues, which may be due to genetic background, mouse strains, timing, and many others, or simply to lack of robustness. Most importantly, however in my view these assays test for bipotentiality to contribute to the two first lineages of the blastocyst, but not for totipotency.
Figure 2.
Experimental approaches to investigate potential totipotency. In (a,b), typical chimera assays are shown. In (a), incorporation into 8-cell stage pre-implantation mouse embryos is done by aggregation, typically referred to as ‘morula aggregation’. In this assay, contribution to lineage is based on 3D position, but should be complemented with co-immunostaining to establish at least partial molecular identity. EGFP, enhanced green fluorescent protein; FACS, fluorescence activated cell sorting; H2B, histone H2B; NLS, nuclear localization signal. In (b), incorporation is achieved through microinjection of cells into the blastocoel of early blastocysts, followed by implantation and analysis of the conceptus, typically at embryonic day (E) 9.5. In this assay, contribution is based on expression of a fluorescent reporter in the placenta. These are difficult experiments, often hindered by the fact that the placenta is highly autofluorescent, and often it is not straightforward to distinguish between placenta and the yolk sac, which is an ICM derivative. These analyses should be accompanied by a stringent analysis through sections and molecular analysis of markers from the trophoblastic derivatives of the placenta. scRNAseq, single cell RNA sequencing. In (c), cell culture strategies are shown. In (c), as suggested by Baker and Pera [53], the ability of a single cell to give rise to stem cells from the three lineages of the mature blastocyst is depicted. XEN cells, primitive endoderm-derived stem cells; TS cells, trophoblast stem cells, derived from the trophectoderm. Molecular analysis of each of these cell types for the known relevant markers should be performed. (d) A potential design to promote self-aggregation of 2CLC, and derivation of cyst-containing structures also referred to as blastocyst-like. As in (c), a molecular analysis and exploration of lineage markers should be performed. In (e,f), nuclear transfer (NT) strategies are shown. (e) The rationale behind using somatic cell nuclear transfer as an assay to test cellular plasticity, based on the observations that nuclei derived from early embryos show a highest success in generating embryos and pups upon cloning, as opposed to pluripotent stem cells. Accordingly, 2CLC nuclei show a higher success in producing clone embryos upon NT [31]. SCNT, somatic cell nuclear transfer. In (f), the schematic of an NT experiment, including the potential outcomes and implications, is shown.
I will argue, therefore, that we currently do not have a correct assay to test for totipotency. It is also very improbable that one cell, for example, a 2CLC, can fulfill the geometrical needs and constraints of the in vivo embryo. Their diameter is at least 10 times smaller and thus their volume is 1000 times smaller than the 2-cell stage blastomeres ! Other differences beyond cell size, for example, in their metabolism [54] may also exist. However, aiming for strategies which can lead to the formation of blastuloids or blastocyst-like cyst structures will be a good starting point [55] (figure 2). In this regard, the current strategies based on self-assembly of embryonic and extra-ES cells into embryo-like structures will greatly facilitate the development of these approaches [56,57]. In addition, testing the capacity of a single cell to yield the three stem cell derivatives of the blastocysts in vitro, in cell culture, through the derivation of XEN (primitive endoderm-derived stem cells), ESC and TS cells (trophoblast stem cells, derived from the trophectoderm) has also been proposed as an assay to demonstrate totipotency [15]. Nuclear transfer efficiency is also an objective and robust experimental approach, which provides a clear metric of cell plasticity, considering that we have known for almost 50 years that differentiated cells are less likely to be fully reprogrammed upon cloning, compared to stem cells [58]. Likewise, ESCs donors exhibit a worse reprogramming efficiency upon nuclear transfer compared to totipotent mouse blastomeres [59]. It is therefore notable that 2CLC donors have a higher reprogrammability than ESCs [35].
6. Supporting developmental programmes
In all, work in the past years has highlighted that retrotransposons have the potential to support developmental programmes [4]. We are now also in the position to directly address whether retrotransposon expression is functionally important for developmental progression. For example, work in the Wysocka lab has used dCas9 and Crispr/Cas9 strategies to show that specific LTRs are in fact responsible for regulating full transcriptional induction of genes expressed in human ESCs [60]. Likewise, by manipulating the chromatin of long interspersed nuclear elements (LINE-1s) at their endogenous loci, our own work has indicated that their transcriptional activation regulates chromatin accessibility in the early mouse embryo [61].
Importantly, recent work has documented clear new insertions of LINE-1 in neurons of human beings, indicating current somatic transposition [62] and new roles for LINE-1 in nucleolar organization during development have emerged [63]. It is therefore very exciting and of utmost importance to fully dive into the consequences and mechanisms of retrotransposon regulation, function, and impact for developmental programmes, which in the longer term also dramatically impact the evolutionary path.
Acknowledgements
M.-E.T.-P. acknowledges past and current members of her laboratory for stimulating discussions and for building on the concepts laid down in this opinion over the past years and thanks A. Burton for critical reading of the manuscript, T. Nakatani for comments on the figures and J. Lin for providing the blastocyst-like image in figure 2.
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
Work in the Torres-Padilla laboratory is funded by the Helmholtz Association, the German Research Council (CRC 1064) and H2020 Marie-Curie Actions ITN EpiSystem and ITN ChromDesign.
References
- 1.Bourque G. 2009. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr. Opin. Genet. Dev. 19, 607–612. ( 10.1016/j.gde.2009.10.013) [DOI] [PubMed] [Google Scholar]
- 2.Fukagawa T, Earnshaw WC. 2014. The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell 30, 496–508. ( 10.1016/j.devcel.2014.08.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chuma S, Nakano T. 2013. piRNA and spermatogenesis in mice. Phil. Trans. R. Soc. B 368, 20110338 ( 10.1098/rstb.2011.0338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedli M, Trono D. 2015. The developmental control of transposable elements and the evolution of higher species. Annu. Rev. Cell Dev. Biol. 31, 429–451. ( 10.1146/annurev-cellbio-100814-125514) [DOI] [PubMed] [Google Scholar]
- 5.Burton A, Torres-Padilla ME. 2010. Epigenetic reprogramming and development: a unique heterochromatin organization in the preimplantation mouse embryo. Brief Funct. Genomics 9, 444–454. ( 10.1093/bfgp/elq027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evsikov AV, et al. 2004. Systems biology of the 2-cell mouse embryo. Cytogenet. Genome Res. 105, 240–250. ( 10.1159/000078195) [DOI] [PubMed] [Google Scholar]
- 7.Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E, Carninci P, Torres-Padilla M-E. 2013. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 20, 332–338. ( 10.1038/nsmb.2495) [DOI] [PubMed] [Google Scholar]
- 8.Peaston AE, Evsikov AV, Graber JH, De Vries WN, Holbrook AE, Solter D, Knowles BB. 2004. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7, 597–606. ( 10.1016/j.devcel.2004.09.004) [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez-Terrones D, Torres-Padilla ME. 2018. Nimble and ready to mingle: transposon outbursts of early development. Trends Genet. 34, 806–820. ( 10.1016/j.tig.2018.06.006) [DOI] [PubMed] [Google Scholar]
- 10.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. 2012. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339–344. ( 10.1038/nature10960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, et al. 2018. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 20, 620–631. ( 10.1038/s41556-018-0093-4) [DOI] [PubMed] [Google Scholar]
- 12.Ishiuchi T, Torres-Padilla ME. 2013. Towards an understanding of the regulatory mechanisms of totipotency. Curr. Opin Genet. Dev. 23, 512–518. ( 10.1016/j.gde.2013.06.006) [DOI] [PubMed] [Google Scholar]
- 13.Eckersley-Maslin MA, Alda-Catalinas C, Reik W. 2018. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 19, 436–450. ( 10.1038/s41580-018-0008-z) [DOI] [PubMed] [Google Scholar]
- 14.Arias AM, Nichols J, Schroter C. 2013. A molecular basis for developmental plasticity in early mammalian embryos. Development 140, 3499–3510. ( 10.1242/dev.091959) [DOI] [PubMed] [Google Scholar]
- 15.Baker CL, Pera MF. 2018. Capturing totipotent stem cells. Cell Stem Cell 22, 25–34. ( 10.1016/j.stem.2017.12.011) [DOI] [PubMed] [Google Scholar]
- 16.Casser E, Israel S, Witten A, Schulte K, Schlatt S, Nordhoff V, Boiani M. 2017. Totipotency segregates between the sister blastomeres of two-cell stage mouse embryos. Sci. Rep. 7, 8299 ( 10.1038/s41598-017-08266-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarkowski AK. 1959. Experiments on the development of isolated blastomeres of mouse eggs. Nature 184, 1286–1287. ( 10.1038/1841286a0) [DOI] [PubMed] [Google Scholar]
- 18.Macfarlan TS, et al. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63. ( 10.1038/nature11244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Festuccia N, Osorno R, Wilson V, Chambers I. 2013. The role of pluripotency gene regulatory network components in mediating transitions between pluripotent cell states. Curr. Opin Genet. Dev. 23, 504–511. ( 10.1016/j.gde.2013.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgani SM, Canham MA, Nichols J, Sharov AA, Migueles RP, Ko MS, Brickman JM. 2013. Totipotent embryonic stem cells arise in ground-state culture conditions. Cell Rep. 3, 1945–1957. ( 10.1016/j.celrep.2013.04.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, et al. 2017. Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell 169, 243–257. ( 10.1016/j.cell.2017.02.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. 2017. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 49, 941–945. ( 10.1038/ng.3858) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendrickson PG, et al. 2017. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 49, 925–934. ( 10.1038/ng.3844) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snider L, et al. 2010. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 6, e1001181 ( 10.1371/journal.pgen.1001181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whiddon JL, Langford AT, Wong C-J, Zhong JW, Tapscott SJ. 2017. Conservation and innovation in the DUX4-family gene network. Nat. Genet. 49, 935–940. ( 10.1038/ng.3846) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Z, Zhang Y. 2019. Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat. Genet. 51, 947–951. ( 10.1038/s41588-019-0418-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Iaco A, et al. 2019. DUX is a non-essential synchronizer of zygotic genome activation. bioRxiv.
- 28.Castro-Diaz N, Friedli M, Trono D. 2015. Drawing a fine line on endogenous retroelement activity. Mob. Genet. Elem. 5, 1–6. ( 10.1080/2159256X.2015.1006109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 18, 71–86. ( 10.1038/nrg.2016.139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckersley-Maslin M, Alda-Catalinas C, Blotenburg M, Kreibich E, Krueger C, Reik W. 2019. Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev. 33, 194–208. ( 10.1101/gad.321174.118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Iaco A, Coudray A, Duc J, Trono D. 2019. DPPA2 and DPPA4 are necessary to establish a 2C-like state in mouse embryonic stem cells. EMBO Rep. 20, e47382 ( 10.15252/embr.201847382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan YL, et al. 2019. DPPA2/4 and SUMO E3 ligase PIAS4 opposingly regulate zygotic transcriptional program. PLoS Biol. 17, e3000324 ( 10.1371/journal.pbio.3000324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi YJ, et al. 2017. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science 355, eaag1927 ( 10.1126/science.aag1927) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckersley-Maslin MA, et al. 2016. MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs. Cell Rep. 17, 179–192. ( 10.1016/j.celrep.2016.08.087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishiuchi T, Enriquez-Gasca R, Mizutani E, Bošković A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla M-E. 2015. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 22, 662–671. ( 10.1038/nsmb.3066) [DOI] [PubMed] [Google Scholar]
- 36.Wu J, et al. 2016. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657. ( 10.1038/nature18606) [DOI] [PubMed] [Google Scholar]
- 37.Boskovic A, Eid A, Pontabry J, Ishiuchi T, Spiegelhalter C, Raghu Ram EVS, Meshorer E, Torres-Padilla M-E. 2014. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev. 28, 1042–1047. ( 10.1101/gad.238881.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ooga M, Fulka H, Hashimoto S, Suzuki MG, Aoki F. 2016. Analysis of chromatin structure in mouse preimplantation embryos by fluorescent recovery after photobleaching. Epigenetics 11, 85–94. ( 10.1080/15592294.2015.1136774) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez-Terrones D, et al. 2018. A molecular roadmap for the emergence of early-embryonic-like cells in culture. Nat. Genet. 50, 106–119. ( 10.1038/s41588-017-0016-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li P, et al. 2017. Rif1 promotes a repressive chromatin state to safeguard against endogenous retrovirus activation. Nucleic Acids Res. 45, 12 723–12 738. ( 10.1093/nar/gkx884) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hisada K, Sanchez C, Endo TA, Endoh M, Roman-Trufero M, Sharif J, Koseki H, Vidal M. 2012. RYBP represses endogenous retroviruses and preimplantation- and germ line-specific genes in mouse embryonic stem cells. Mol. Cell. Biol. 32, 1139–1149. ( 10.1128/MCB.06441-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatsumi D, et al. 2018. DNMTs and SETDB1 function as co-repressors in MAX-mediated repression of germ cell-related genes in mouse embryonic stem cells. PLoS ONE 13, e0205969 ( 10.1371/journal.pone.0205969) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maeda I, et al. 2013. Max is a repressor of germ cell-related gene expression in mouse embryonic stem cells. Nat. Commun. 4, 1754 ( 10.1038/ncomms2780) [DOI] [PubMed] [Google Scholar]
- 44.Cossec JC, et al. 2018. SUMO safeguards somatic and pluripotent cell identities by enforcing distinct chromatin states. Cell Stem Cell 23, 742–757. ( 10.1016/j.stem.2018.10.001) [DOI] [PubMed] [Google Scholar]
- 45.Fu X, Wu X, Djekidel MN, Zhang Y. 2019. Myc and Dnmt1 impede the pluripotent to totipotent state transition in embryonic stem cells. Nat. Cell Biol. 21, 835–844. ( 10.1038/s41556-019-0343-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walter M, Teissandier A, Pérez-Palacios R, Bourc'his D. 2016. An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. Elife 5, e11418 ( 10.7554/eLife.11418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guallar D, et al. 2018. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat. Genet. 50, 443–451. ( 10.1038/s41588-018-0060-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ecco G, et al. 2016. Transposable elements and their KRAB-ZFP controllers regulate gene expression in adult tissues. Dev. Cell 36, 611–623. ( 10.1016/j.devcel.2016.02.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsui T, et al. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931. ( 10.1038/nature08858) [DOI] [PubMed] [Google Scholar]
- 50.Karimi MM, et al. 2011. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8, 676–687. ( 10.1016/j.stem.2011.04.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hung SS, Wong RCB, Sharov AA, Nakatake Y, Yu H, Ko MSH. 2013. Repression of global protein synthesis by Eif1a-like genes that are expressed specifically in the two-cell embryos and the transient Zscan4-positive state of embryonic stem cells. DNA Res. 20, 391–402. ( 10.1093/dnares/dst018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mascetti VL, Pedersen RA. 2016. Contributions of mammalian chimeras to pluripotent stem cell research. Cell Stem Cell 19, 163–175. ( 10.1016/j.stem.2016.07.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baker CL, Pera MF. 2018. Capturing totipotent stem cells. Cell Stem Cell 22, 25–34. ( 10.1016/j.stem.2017.12.011) [DOI] [PubMed] [Google Scholar]
- 54.Rodriguez-Terrones, et al. In Press.
- 55.Rivron NC, et al. 2018. Blastocyst-like structures generated solely from stem cells. Nature 557, 106–111. ( 10.1038/s41586-018-0051-0) [DOI] [PubMed] [Google Scholar]
- 56.Sozen B, et al. 2018. Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol. 20, 979–989. ( 10.1038/s41556-018-0147-7) [DOI] [PubMed] [Google Scholar]
- 57.Beccari L, et al. 2018. Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 562, 272–276. ( 10.1038/s41586-018-0578-0) [DOI] [PubMed] [Google Scholar]
- 58.Gurdon JB. 1992. The generation of diversity and pattern in animal development. Cell 68, 185–199. ( 10.1016/0092-8674(92)90465-O) [DOI] [PubMed] [Google Scholar]
- 59.McGrath J, Solter D. 1984. Inability of mouse blastomere nuclei transferred to enucleated zygotes to support development in vitro. Science 226, 1317–1319. ( 10.1126/science.6542249) [DOI] [PubMed] [Google Scholar]
- 60.Fuentes DR, Swigut T, Wysocka J. 2018. Systematic perturbation of retroviral LTRs reveals widespread long-range effects on human gene regulation. Elife 7, e35989 ( 10.7554/eLife.35989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jachowicz JW, Bing X, Pontabry J, Bošković A, Rando OJ, Torres-Padilla M-E. 2017. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 49, 1502–1510. ( 10.1038/ng.3945) [DOI] [PubMed] [Google Scholar]
- 62.Sanchez-Luque FJ, et al. 2019. LINE-1 evasion of epigenetic repression in humans. Mol. Cell 75, 590–604. ( 10.1016/j.molcel.2019.05.024) [DOI] [PubMed] [Google Scholar]
- 63.Percharde M, et al. 2018. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 174, 391–405. ( 10.1016/j.cell.2018.05.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.