Abstract
In disease-suppressive soils, microbiota protect plants from root infections. Bacterial members of this microbiota have been shown to produce specific molecules that mediate this phenotype. To date, however, studies have focused on individual suppressive soils and the degree of natural variability of soil suppressiveness remains unclear. Here, we screened a large collection of field soils for suppressiveness to Fusarium culmorum using wheat (Triticum aestivum) as a model host plant. A high variation of disease suppressiveness was observed, with 14% showing a clear suppressive phenotype. The microbiological basis of suppressiveness to F. culmorum was confirmed by gamma sterilization and soil transplantation. Amplicon sequencing revealed diverse bacterial taxonomic compositions and no specific taxa were found exclusively enriched in all suppressive soils. Nonetheless, co-occurrence network analysis revealed that two suppressive soils shared an overrepresented bacterial guild dominated by various Acidobacteria. In addition, our study revealed that volatile emission may contribute to suppression, but not for all suppressive soils. Our study raises new questions regarding the possible mechanistic variability of disease-suppressive phenotypes across physico-chemically different soils. Accordingly, we anticipate that larger-scale soil profiling, along with functional studies, will enable a deeper understanding of disease-suppressive microbiomes.
Keywords: Fusarium culmorum, rhizosphere microbiome, soil suppressiveness, soil volatiles
1. Introduction
The phenomenon of soil disease suppressiveness has been recognized for almost a century and was first defined by Cook & Baker [1] as soils where a particular soil-borne disease does not develop, despite the presence of the virulent pathogen, a susceptible host and favourable conditions for disease development. Physical and chemical properties of the soil can play a role in this phenomenon, but in many cases, disease suppressiveness is microbial in nature [2–5].
Two types of soil suppressiveness can be distinguished, namely, general and specific. General suppression is effective against a range of pathogens, whereas specific suppression operates against only one or a few of them. General soil suppressiveness is a result of the activity of the overall soil microbial community, whereas specific soil suppressiveness is due to the concerted action of specific microbial genera that interfere at some stage of the life cycle of the soil-borne pathogen. Specific soil suppressiveness can be eliminated by selective heat treatments and is transferable to a conducive soil by mixing in a small amount (1–10%) of suppressive soil (for review: [6,7]).
For many suppressive soils, the microorganisms and mechanisms have not been elucidated. Some of the best-studied suppressive soils to date are the take-all-decline (TAD) soils, where root disease of wheat or barley caused by the fungal pathogen Gaeumannomyces graminis var. tritici is suppressed [8–10]. Suppressiveness to take-all disease is, at least in part, due to the enrichment of populations of root-associated Pseudomonas spp. producing the antifungal polyketide 2,4-diacetylphlorogucinol (2,4-DAPG) [4,11–13]. For Fusarium wilt-suppressive soils, the microbes and mechanisms identified so far involve non-pathogenic Fusarium oxysporum and Pseudomonas species that, in a complementary manner, compete with pathogenic F. oxysporum for carbon and iron [14–16]. Recent studies on soils suppressive to Rhizoctonia damping-off disease of sugar beet revealed the involvement of multiple bacterial genera belonging to the Pseudomonadaceae, Streptomycetaceae and Burkholderiaceae. The suppression was linked to the production of antifungal lipopeptide thanamycin by Pseudomonas and to the production of volatile metabolites by Streptomyces spp. and Paraburkholderia graminis [10,17,18]. Volatiles are low-molecular-mass metabolites involved in long-distance interactions with potent antimicrobial activities [19–22]. For example, the effects of volatiles emitted from 50 agricultural soils on two soil-borne pathogenic fungi (F. oxysporum, R. solani) and a plant pathogenic oomycete (Pythium intermedium) have recently been studied [23] and revealed that most soils emit volatiles that inhibit hyphal growth, but the extent of growth inhibition per soil differed strongly for the three pathogens.
To date, most studies on suppressive soils have been limited to a single field soil. Except for the case of the TAD soils [24], it is currently unclear how widespread suppressiveness to specific pathogens is as a biological phenomenon. Furthermore, it is not established yet whether suppression is mediated by one or multiple taxa and mechanisms across various suppressive soils. Finally, the role of volatiles in soil suppressiveness in the presence of both the pathogen and the host plant is yet unexplored.
In this study, we screened a large collection of field soils for suppressiveness to Fusarium culmorum, a ubiquitous soil-borne fungus causing foot rot, root rot and Fusarium head blight of different cereals, in particular wheat and barley [25]. Using wheat (Triticum aestivum) as the host plant, we found F. culmorum suppressiveness in 4 out of 28 soils.
We hypothesized that (i) soils suppressive to F. culmorum have similar rhizobacterial community compositions with specific enriched taxa, (ii) volatile compounds contribute to disease suppression against F. culmorum and (iii) disease-suppressive soils have similar volatile profiles. We anticipated that specific rhizobacterial taxa and volatiles correlate with soil suppressiveness against F. culmorum.
However, our comparative analysis of bacterial rhizosphere microbiome composition of suppressive and non-suppressive (conducive) soils, along with volatile profiling, indicates that the phenomenon may be mediated by different rhizobacterial genera across these soils and that volatiles may contribute to the suppressive phenotype only for a limited number of soils.
2. Material and Methods
(a). Soil collection
In order to examine soil disease suppressiveness to F. culmorum, 28 sites in The Netherlands and Germany were chosen based on information on soil type and crop rotation. The sites included 25 arable fields, two pastures and one forest. The arable fields included sites with wheat present in crop rotation over the last three years and sites without wheat present in the available history of the field.
Soil samples were collected in the period from January to April 2017 from 3 m2 located in the middle of each agricultural field/pasture. In this area, top soil cores of approximately 30 cm deep were extracted. Samples were air-dried in room temperature, homogenized, sieved through 4 mm sieve and stored at 4°C. Heavy clay-type soils were additionally flaked using a jaw-crusher (Type BB-1, Retsch, Germany) after drying. Soil physical and chemical parameters were measured as described in electronic supplementary material, part I.
(b). Wheat growth conditions and pathogen inoculation
All the greenhouse experiments were performed in growth cabinets (MC 1750 VHO-EVD, Snijders Labs) in 20°C day and night, photoperiod 12 h day/12 h night and 60% relative humidity. In all the experiments, surface-sterilized and pre-germinated wheat seeds (JB Asano from Agrifirm, The Netherlands) were used. Plants were watered every second day and weekly supplemented with 0.5 Hoagland solution (1 ml per 80 cc of the soil, 0.5 M Ca(NO3)2·4H2O, 1 M KNO3, 1 M KH2PO4, 0.5 M MgSO4·7H2O and 98.6 mM ferric EDTA).
As a standard substrate, we used a dune soil collected near Bergharen, The Netherlands (BS) [26]. Before use, BS was air-dried, sieved and gamma-sterilized (Synergy Health Ede B.V., The Netherlands). Sub-samples of eight soils from the collection were gamma-sterilized as well in the same conditions for later use.
Prior to screening for disease suppressiveness, all soils were ‘activated’ in order to induce microbial activity in a so-called microbiome activation step. For this, soils hosted the growth of wheat for two weeks in the conditions indicated above. After this, plants, including their whole root system, were removed and the remaining soil with activated microbiome was mixed properly.
The soil-borne pathogen F. culmorum PV [27] was grown on 1/2 PDA media (Oxoid, The Netherlands) and maintained in continuous culture. For plant inoculation, the fungus was transferred to 1/4 PDA and grown for two weeks in 20°C. After the incubation, 6 mm plugs were cut from the border zone of Fusarium hyphae and mixed with the growth substrate (1 plug per 10 cc). Controls without pathogen were inoculated with sterile 1/4 PDA plugs. At the end of the experiments, disease symptoms were assessed and plants were used for rhizosphere DNA extraction and sequencing analysis (see electronic supplementary material, part I).
(c). Disease suppressiveness screening
Initial disease suppressiveness screening was performed in propagation trays containing 140 single pots (Teku, The Netherlands) each containing 38 cc of soil per pot. In order to minimize the physico-chemical differences between the 28 soils, after the activation step (described in the paragraph above) the natural soil was mixed 2 : 1 : 1 in volume with sterile BS soil and sterile vermiculite (Agra-vermiculite, The Netherlands). The soil was inoculated with Fusarium or mock inoculated with agar plugs, and one seedling was placed in the centre of each pot. Plants were grown in 10 replicates per treatment and, after three weeks, disease symptoms were assessed (see electronic supplementary material, part I). As a control, we used BS mixed with vermiculite 3 : 1 in volume.
(d). Confirmation of disease suppressiveness with soils sterilization
Four suppressive and four conducive soils were selected to confirm the previously observed levels of suppressiveness. The confirmation test was performed identically as the screening test but the system was scaled up to 380 cc soil per pot (7 × 7 × 8 cm, Teku, The Netherlands) with three plants in each. An additional treatment was included––sterilized soil inoculated with pathogen. Sterile BS and vermiculite mixture was used as a control. All the treatments had an additional control without pathogen.
(f). Transplantation assay
To assess whether soil suppressiveness is transferable, we mixed natural soils in proportions 1 : 9 and 3 : 7 in volume with a conducive substrate consisting of sterile BS and vermiculite. This experiment was also performed with 380 cc soil per pot (7 × 7 × 8 cm), each containing three plants, in treatments with and without the addition of the pathogen. Sterile BS and vermiculite mixture was used as a control.
(g). Sequencing and bioinformatics analysis
Rhizosphere DNA extraction, sequencing, 16S rRNA amplicon data processing and analysis are described in electronic supplementary material, part I. 16S rRNA raw amplicon sequencing data are available at EBI-PRJEB34717
(h). Effects of soil-emitted volatiles on fungal growth
Assays were performed as previously described by Garbeva et al. [22] with small modifications, see electronic supplementary material, part I.
(g). Effects of soil-emitted volatile compounds on disease suppression
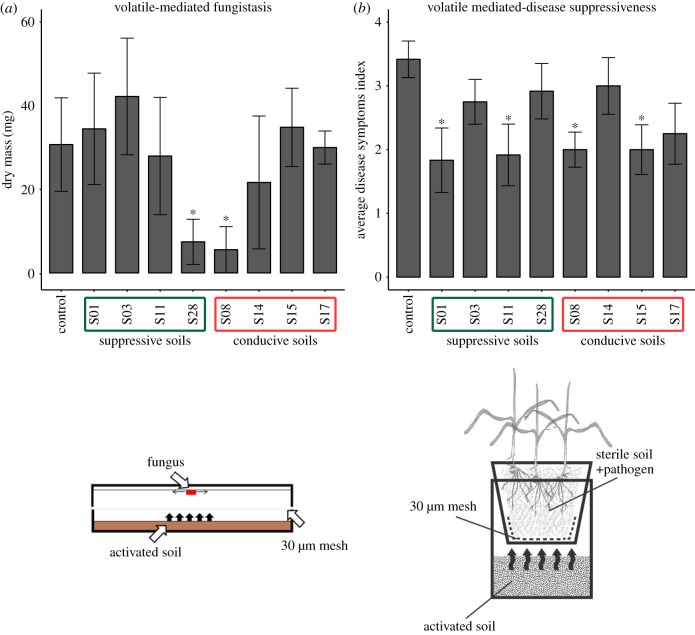
The eight soils selected in the suppressiveness screening were tested for their ability to induce soil suppressiveness via volatiles. For this, the modified method described in [28] was used. Briefly, wheat plants growing in conducive soil mixed with F. culmorum plugs were exposed to volatiles emitted by soil present in the compartment below the pot. To avoid any physical contact, compartments were separated and sealed with sterile nylon mesh (Sefar, Switzerland) and paper medical tape as described above. The scheme of the system is shown in figure 1b. Four pots per treatment with three plants per pot were grown for three weeks; afterwards, disease symptoms were assessed.
Figure 1.
The effect of the volatiles emitted by eight prioritized soils on F. culmorum growth (fungistasis) and disease suppression. (a) Average dry mass of the fungus with standard deviation, the statistically significant differences between treatments and control (based on ANOVA and Tuckey post-hoc test p < 0.05) are indicated by asterisk. (b) Average symptoms index with standard error. The statistically significant differences between treatments and control (based on Chi-square test p < 0.05) are indicated by asterisk. (Online version in colour.)
(i). Volatile trapping and GC-MS analysis
Volatile compounds were trapped using steel trap containing 150 mg Tenax TA and 150 mg Carbopack B (Markes International Ltd, Llantrisant, UK) and measured using GC-QTOF system. Statistical data analysis was performed using MetaboAnalyst 4.0 software (http://www.metaboanalyst.ca/MetaboAnalyst [29]). For details of Volatile trapping and GC-MS analysis, see electronic supplementary material, part I.
3. Results
(a). Identification of soils suppressive to F. culmorum
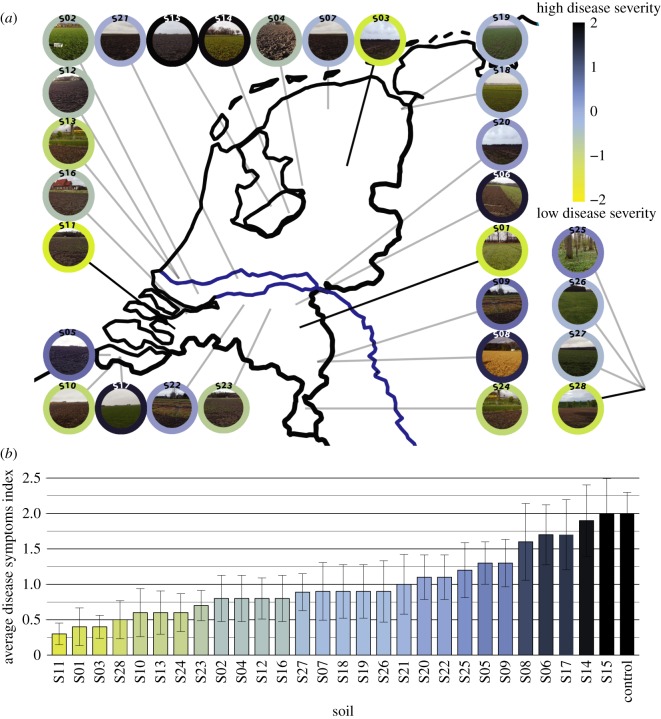
A collection of 28 soils of diverse geographical origins (figure 2a), soil types and agricultural histories (electronic supplementary material, table S1) was screened for disease suppressiveness against F. culmorum in greenhouse pot experiments. The disease symptoms of the plants grown in each of the soils were examined three weeks after pathogen inoculation. Overall, high variation in disease suppressiveness was observed between the 28 soils, while being largely consistent between replicates. The disease symptoms across the collection varied from mild or no infection to severe disease. Four soils (S01, S03, S11 and S28–yellow colour; figure 2) showed the lowest level of disease severity with an average score below 0.5 and were considered suppressive (figure 2). Four soils revealed high disease severity with average disease symptoms above 1.5 (S08, S14, S15 and S17––black colour; figure 2) and were considered conducive.
Figure 2.
Results of the phenotypic screening towards soil suppressiveness to F. culmorum in 28 soils. (a) Map showing locations of 28 sampling sites and the results of the screening. The colour of the outer circle around the photos represents the z-score transformed average disease symptoms value. Low disease severity indicates soil suppressiveness (yellow colour). (b) Bar graph showing average disease symptoms indexes with standard errors. (Online version in colour.)
(b). Disease suppression is independent of soil physico-chemical properties
In order to investigate the phenotypic variation across multiple soil types, our collection included soils from 25 arable fields, 2 pastures and 1 forest, representing different soil types, ranging from sands to heavy clays, with diverse pH (5.3–7.8) and C/N ratio (8.8–17.5). All physico-chemical parameters and field history are summarized in electronic supplementary material, table S1. No clear correlations between the level of disease suppressiveness and physico-chemical parameters and field history were found. Based on canonical correspondence analysis of the 28 soils, there was no separation between disease-suppressive and conducive soils (electronic supplementary material, figure S1, yellow and black dots accordingly). Only weak Pearson correlations were found between physico-chemical parameters and disease severity (electronic supplementary material, figure S2). Also, no relevant correlations were found between disease severity and soil pH and C/N ratio (0.24 and 0.25, respectively).
(c). Disease suppressiveness has a microbial basis
Based on the results from the initial screening, a set of eight soils was selected for confirmation assays and further analysis. This included the four soils with the highest level of disease suppressiveness (S01, S03, S11 and S28) and the four conducive soils showing the most contrasting phenotypes (S08, S14, S15 and S17). For the four suppressive soils, low average disease indices were again observed (figure 3a, red bars) but after gamma sterilization, disease indices increased significantly (figure 3a, blue bars). For the four conducive soils, except for S08, gamma sterilization did not substantially enhance disease severity. Furthermore, the results revealed that suppressiveness to F. culmorum is transferable (figure 3b). Mixing 10% or 30% of the suppressive soil (S01, S03, S11 and S28) in a conducive background soil transferred suppressiveness. There was no significant difference in the level of suppression between the transfer of 10 and 30% suppressive soil. Collectively, our data suggest that the microbial community contributes to the suppressive phenotype.
Figure 3.
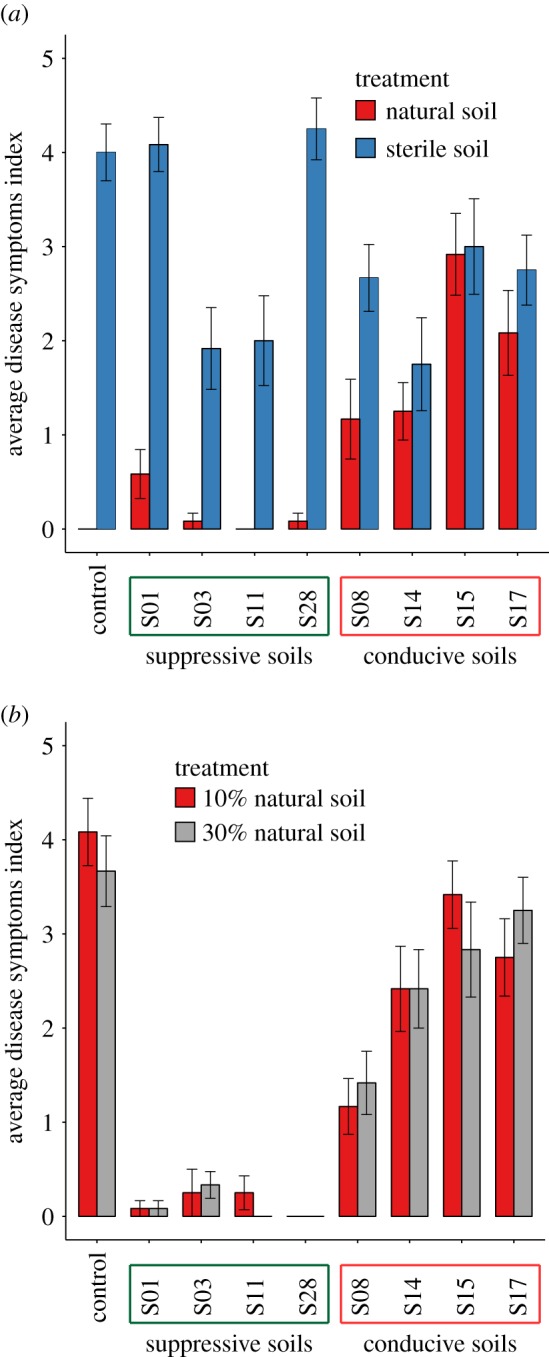
Disease symptoms observed in wheat inoculated with F. culmorum grown in eight prioritized soils. (a) Natural and gamma-sterilized soil with sterile BS soil/vermiculite mix as a control (b) 10% and 30% in volume of natural soil mixed with standardized sterile substrate or with sterile BS soil as a control. The bar indicates the average of the disease symptoms index, with the error bars representing the standard error. (Online version in colour.)
(d). Volatile-mediated inhibition of fungal growth and disease suppression is observed for suppressive and conducive soils
To determine the role of volatile compounds in disease suppression and antifungal activity, the eight selected soils were tested in two experimental systems: (i) hyphal growth of Fusarium on artificial media exposed to soil volatiles, and (ii) plants growing in conducive soil inoculated with the pathogen exposed to soil volatiles (figure 4a,b; electronic supplementary material, part I). The first assay revealed that only two soils (S28 suppressive and S08 conducive) emitted volatiles that significantly reduced growth of the fungus compared to the control. When plants were exposed to soil volatiles, disease suppression was observed for four out of eight soils (figure 4b). Again, these included both suppressive (S01, S11) and conducive (S08, S15) soils. Subsequent analysis of volatile profiles emitted by these eight soils did not reveal clear separation between suppressive and conducive soils (electronic supplementary material, figure S3). Interestingly, several suppressive and conducive soils (S03, S08, S11 and S14) revealed very similar volatile profiles. These results suggest that volatiles are not a common mechanism of soil suppressiveness to F. culmorum.
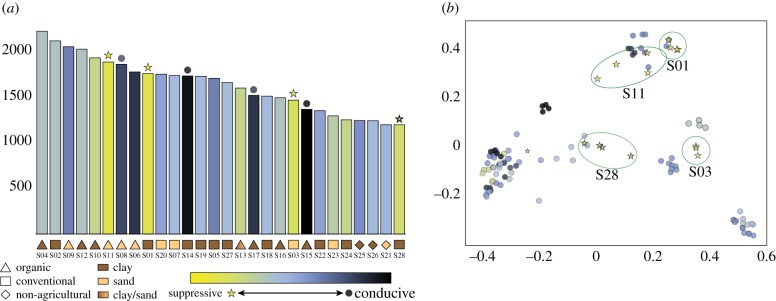
Figure 4.
Characteristics of rhizosphere bacterial communities across all 28 soils. (a) Bar plot representing the alpha diversity using the rarefied unique ASV counts. Samples are sorted according to their alpha diversity score and colour coded according to suppressive phenotype. (b) PCoA based on unweighted UniFrac distance between samples. Circles highlight the suppressive sample replicates clustered together. (Online version in colour.)
(e). Microbial profiling of F. culmorum-suppressive soils
To investigate possible links between the rhizobacterial community composition and the disease-suppressive phenotype, extensive 16S sequencing was performed for all 28 soils (see electronic supplementary material, part I). The rhizobacterial communities showed large variation between samples and even between replicates of the same samples, as can be seen from the inter- and intra-sample Jaccard similarity of 0.056 (s.d. = 0.033) and 0.346 (s.d. = 0.057), respectively. Based on the alpha- and beta-diversity, there were no significant community differences between suppressive and conducive soils. β-diversity for all sample pairs was calculated with unweighted UniFrac, which shows consistent grouping for soil sample replicates (figure 4b). Subsequent PCoA analysis of the rhizobacterial community composition of the suppressive and conducive soils indicated that the different suppressive soils have diverse taxonomic compositions and did not group together (figure 4b). This was also the case when calculating community diversity of the samples within specific taxonomic groups (electronic supplementary material, figures S4). As for alpha diversity, Wilcoxon rank-sum test showed no significant association between observed amplicon sequence variants (ASVs; p = 0.74) or Shannon diversity (p = 0.07) and soil suppressiveness. The suppressive soils included both the most and the least diverse communities. The soils from organic farms were associated with higher bacterial diversity compared to the soils from conventional farms and to the non-agricultural soils (figure 4a). Again, there was no consistent correlation between soil suppressiveness and alpha- or beta-diversity.
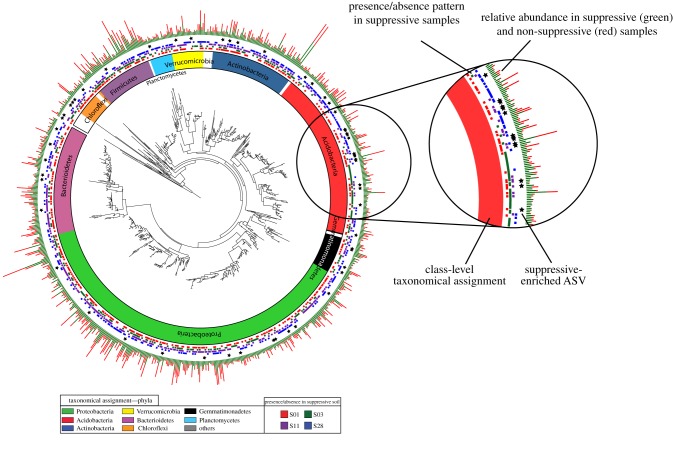
To establish whether the disease-suppressive phenotypes were mediated by specific rhizobacterial taxon or by multiple taxa, 16S amplicon data were analysed in more depth. To reveal if one or more individual abundant ASVs could be associated with disease suppressiveness, we inspected the presence–absence patterns of all ASVs that were significantly enriched in suppressive soils (figure 5). No individual ASV appeared to be exclusively present in all suppressive samples compared to the conducive samples (electronic supplementary material, figure S5). Random forest classifiers trained on their taxonomic profiles could not predict the suppressive soil phenotype in a leave-one-out analysis. The results were not significantly different when the same analysis was performed using OTU-level clustering of ASVs (at 97% identity).
Figure 5.
Phylogenetic tree of all ASVs consistently detected in one or more suppressive rhizosphere samples. The rings, from the inside to the outside, represent (1) the taxonomical assignment of the ASVs at the phylum level, (2) the presence/absence patterns of ASVs in the four different suppressive soils, (3) indications (with stars) of ASVs strongly enriched in suppressive samples and (4) the average cumulative normalized abundance of each ASV in suppressive (green) versus non-suppressive (red) samples. The results show that all ASVs enriched in suppressive samples are specific to a subset of the suppressive soils, while none of the ASVs that occur throughout all suppressive soils are significantly enriched in suppressive versus non-suppressive soils. (Online version in colour.)
(f). Co-occurrence network analysis indicates bacterial communities enriched in rhizosphere microbiomes of suppressive soils
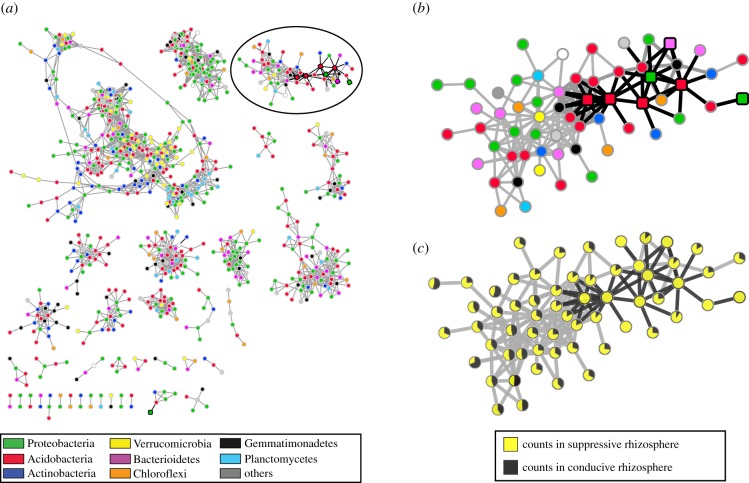
To investigate whether specific combinations of ASVs are associated with disease suppressiveness (in subsets of the four suppressive soils), we calculated the Spearman correlations between ASV occurrences across all 28 soils and constructed a co-occurrence network from the most highly correlating ASVs in the correlation matrix (see electronic supplementary material, part I). The resulting network (figure 6a) consists of 928 nodes (comprising 21% of all 4322 ASVs), distributed across 37 subnetworks. The ASVs in the network that were significantly enriched in suppressive soils are indicated by squares with black border in figure 6b. The subnetworks are taxonomically diverse, with no network being taxonomically homogeneous, even at the phylum level. Interestingly, one of the connected components was found to be particularly associated with the suppressive phenotype. Within this connected component, 7 out of the 163 ASVs were strongly enriched in suppressive soils and these were part of a more densely connected subcomponent comprising 60 ASVs that comprised 6 out of the 7 abovementioned ones. Taxonomic assignments of the suppression-associated subnetwork displayed an overrepresentation of ASVs belonging to different Acidobacteria, namely, Blastocatellales and members of subgroup 6, compared to the overall community composition (p = 0.0003, Fisher exact test; figure 6b). Moreover, several of these Acidobacteria displayed characteristics of being hub taxa in these subnetworks based on their high betweenness centrality. In addition, 11 of the 60 ASVs in the suppressive-associated network were exclusively present in two suppressive soils (figure 6c), 7 of which were taxonomically assigned to Acidobacteria.
Figure 6.
Co-occurrence network analysis of rhizosphere bacteria across samples. (a) Complete network of correlated ASVs using Spearman correlation. Nodes represent individual ASVs; edges link ASVs which had a correlation score above a given threshold. The node colour indicates the taxonomical annotation at the phylum level. Edges of ASVs that are significantly enriched in suppressive samples are highlighted black as the respective nodes. (b) Zoomed in view of a subnetwork of interest, which shows a number of ASVs associated with the suppressive phenotype indicated by squares with black border, along with its taxonomical distribution. (c) Normalized counts ratio between conducive (yellow) and suppressive (black) samples for ASVs from the network in (b). (Online version in colour.)
4. Discussion
Fusarium culmorum is an economically important fungal plant pathogen that causes disease in many cereal and non-cereal crops. However, little is known about the occurrence and distribution of soil suppressiveness to this pathogen. To our knowledge, this study is the first to screen for disease suppressiveness in a large collection of diverse soils. Although a high variation in the level of disease suppressiveness was observed between the soils, 14% of the tested soils revealed a clear suppressive phenotype. Interestingly, no physico-chemical parameter such as soil type, field history, pH, C/N ratio or content and concentration of bioavailable Fe, K, Mg, P and S correlated with the observed suppressive phenotype. In previous work, physico-chemical soil parameters such as soil type, moisture, pH, organic matter and microelements content have been more commonly associated with general disease suppressiveness [30,31].
In our study, the microbiological basis of specific suppressiveness to F. culmorum was revealed by two independent approaches: gamma sterilization and soil transplantation eliminated and conferred suppressiveness, respectively, and confirmed that suppressiveness to F. culmorum was not linked to soil physico-chemical parameters but rather to the soil (micro)biome.
Along with the soil, the plant itself is an important determinant of the structure of soil microbial communities and disease suppressiveness. Hence, the strength of disease suppressiveness is attributed to all players in the tripartite relationship of plant–soil–microorganisms [32]. Considering this, in the present study, all experiments included a ‘microbiome activation step’ by growing wheat for two weeks prior screening for disease suppressiveness. The application of plants provides substrates for growth via root exudates and space for the soil microbial community. Several studies have associated disease-suppressive phenotypes to individual bacterial groups based on their relative enrichment compared to a conducive phenotype [6,33,34]. However, rhizosphere microbiomes are highly diverse [35] and multiple differences may exist even between physico-chemical similar soils. In the present study, 28 different soils were examined and no individual bacterial taxon was found to be exclusively present or enriched in all suppressive soils. Bacterial taxonomic groups that were more prevalent in some of the suppressive soils were not prevalent in other suppressive soils.
Many studies have aimed to understand the relationship between microbial diversity and disease suppressiveness. However, both low [36] and high [32] community diversity have been associated with soil suppressiveness. Here, we found suppressive samples with both high and low community richness, which suggests community diversity is not the key driver of soil suppressiveness to F. culmorum. Community evenness is correlated with the phenotype but the strength of this association is not strong enough (p = 0.07) to warrant speculations on its role in disease suppressiveness.
To our surprise, no ASVs or OTU-level ASV clusters were found to be shared uniquely between the microbial communities of all four suppressive soils, or to be differentially abundant across all these four soils. The fact that random forest classifiers were unable to accurately predict suppressive phenotypes in a leave-one-out analysis of the soils suggests that (i) the suppressive phenotypes are mediated by different taxonomic groups across the different suppressive soils and/or (ii) that rhizobacteria do not play a major role in suppressiveness to F. culmorum. Many functional elements that are known to be able to drive disease suppression (such as biosynthetic gene clusters for secondary metabolites) are often strain-specific and are frequently transferred horizontally across species [37]. Hence, it is still possible that the same or similar functional elements could drive the suppressiveness across all four soils, while being undetectable by 16S sequencing due to its limited resolution or due to the elements being encoded in the genomes of diverse bacterial taxa.
Representatives of a range of bacterial groups can carry out functions that result in the suppression of soil-borne diseases. For example, several Bacillus, Pseudomonas, Streptomyces or Flavobacterium species are well known to play role in suppression of various soil-borne plant pathogens [5,33,38–40]. However, various previous studies indicate that many of these bacteria reveal antimicrobial activities only as results of interspecific interaction networks [19,41–43]. The correlation-based network analysis performed here revealed complex inter-sample connections between individual bacterial taxa that are likely interacting either directly or indirectly, based on their observed co-occurrence. One of the network components provided insights into a bacterial guild that is potentially associated with disease suppressiveness to F. culmorum. Multiple ASVs were found to be exclusively present in two distinct disease-suppressive microbiomes, while additionally being strongly correlated with another sub-community of ASVs that also consistently occurred in a conducive microbiome. Interestingly, most of these ASVs belonged to Acidobacteria. We observe an overrepresentation of Acidobacteria in the suppression-associated network when compared to the general community composition (Fisher exact test, p = 0.0003). This phylum has previously been associated with Rhizoctonia solani bare patch [34] and has been shown to harbour diverse species with a large specialized metabolic potential that could be involved in interactions with fungi [44]. Based on their specific enrichment, the identified bacterial guild might represent the suppressive core at the base of the phenotype for two of the four suppressive samples (soils S01 and S11). Further shotgun sequencing efforts as well as microbiological and biochemical analysis are needed to clarify the functional roles of these organisms. Furthermore, one should bear in mind that in addition to the bacterial community structure, the expression of functional genes conferring suppressiveness to soil-borne pathogens might be dependent on interactions with other soil microorganisms such as fungi, protists and viruses.
Frequently, the germination and growth of plant pathogenic fungi are negatively affected by direct or indirect contact with soils (without the presence of a host plant). This phenomenon is named soil fungistasis [19,45] and is often associated with general disease suppressiveness. Whereas abiotic soil factors can be involved, the major cause of fungistasis is biological since it is strongly reduced after soil sterilization, which is analogous to the soil disease suppressiveness. Recent studies revealed that microbial volatiles play important roles in both soil fungistasis as well as disease suppressiveness [23,46]. Our study did not reveal any congruence between volatile-mediated soil fungistasis and disease suppressiveness. Volatile-mediated soil suppressiveness was observed only with four soils, including both two suppressive and two conducive soils. This indicates that, even though volatile emission is one of the factors that may contribute to disease suppressiveness to F. culmorum, it is not a general one. Apparently, other (complementary) factors and mechanisms are needed to develop a full protective potential against F. culmorum. Alternatively, it might be that suppressiveness across the four soils is mediated by different mechanisms, involving volatiles in some cases but not in others. This could also explain the lack of commonly enriched taxa across the four suppressive soils.
In conclusion, we discovered several agricultural soils that protect wheat plants from F. culmorum infections through a microbial component. Moreover, these suppressive soils revealed different bacterial taxonomic patterns and diversity, as well as variable degrees of antifungal volatile emissions. These observations reject the hypothesis that specific rhizobacterial taxa and specific volatiles correlate with suppressiveness of F. culmorum.
Co-occurrence network analysis suggested that two of the suppressive samples share a similar microbial basis of the phenotype through a uniquely overrepresented bacterial guild dominated by Acidobacteria. Of course, taxonomic profiling alone cannot provide definitive answers on the actual biological mechanisms responsible for the suppressive phenotype. Accordingly, we anticipate our work to lay the foundation for a combination of functional metagenomics along with microbiological and biochemical analyses, in order to elucidate the functional mechanisms behind soil suppressiveness to F. culmorum in the near future.
Supplementary Material
Supplementary Material
Acknowledgements
This is publication 6904 of the NIOO-KNAW.
Data accessibility
Data available from the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.g884q70 [47].
Authors' contributions
A.O. designed and performed soil collection, soil analyses, greenhouse experiments and analysed the data; V.T. performed the analysis of amplicon data; V.T. and M.L.C.P. assisted with greenhouse experiments and soil collection; P.G., V.T., M.H.M. and A.O. wrote the manuscript; J.M.R., G.v.W. and M.L.C.P. critically revised the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by NWO ALWGR. 2015.1b grant.
References
- 1.Deacon JW. 1984. Managing disease. Nature 309, 732 ( 10.1038/309732a0) [DOI] [Google Scholar]
- 2.Andrade O, Campillo R, Peyrelongue A, Barrientos L. 2011. Soils suppressive against Gaeumannomyces graminis var. tritici identified under wheat crop monoculture in southern Chile. Cienc. Inv. Agr. 38, 345–356. ( 10.4067/S0718-16202011000300004) [DOI] [Google Scholar]
- 3.Wiseman BM, Neate SM, Keller KO, Smith SE. 1996. Suppression of Rhizoctonia solani anastomosis group 8 in Australia and its biological nature. Soil Biol. Biochem. 28, 727–732. ( 10.1016/0038-0717(95)00178-6) [DOI] [Google Scholar]
- 4.Weller DM, Raaijmakers JM, Gardener BB.M, Thomashow LS. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Ann. Rev. Phytopathol. 40, 309 ( 10.1146/annurev.phyto.40.030402.110010) [DOI] [PubMed] [Google Scholar]
- 5.Cha JY, et al. 2016. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. Isme J 10, 119–129. ( 10.1038/ismej.2015.95) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlatter D, Kinkel L, Thomashow L, Weller D, Paulitz T. 2017. Disease suppressive soils: new insights from the soil microbiome. Phytopathology 107, 1284–1297. ( 10.1094/Phyto-03-17-0111-Rvw) [DOI] [PubMed] [Google Scholar]
- 7.Mazzola M. 2002. Mechanisms of natural soil suppressiveness to soilborne diseases. Anton. Leeuw. 81, 557–564. ( 10.1023/A:1020557523557). [DOI] [PubMed] [Google Scholar]
- 8.Cook RJ, Rovira AD. 1976. Role of bacteria in biological control of Gaeumannomyces graminis by suppressive soils. Soil Biol. Biochem. 8, 269–273. ( 10.1016/0038-0717(76)90056-0) [DOI] [Google Scholar]
- 9.Hornby D. 1983. Suppressive soils. Ann. Rev. Phytopathol. 21, 65–85. ( 10.1146/annurev.py.21.090183.000433) [DOI] [Google Scholar]
- 10.Duran P, Jorquera M, Viscardi S, Carrion VJ, Mora MD, Pozo MJ. 2017. Screening and characterization of potentially suppressive soils against Gaeumannomyces graminis under extensive wheat cropping by Chilean Indigenous communities. Front. Microbiol. 8, 1552 ( 10.3389/fmicb.2017.01552). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raaijmakers JM, Bonsall RE, Weller DM. 1999. Effect of population density of Pseudomonas fluorescens on production of 2,4-diacetylphloroglucinol in the rhizosphere of wheat. Phytopathology 89, 470–475. ( 10.1094/Phyto.1999.89.6.470) [DOI] [PubMed] [Google Scholar]
- 12.Raaijmakers JM, Weller DM. 1998. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol. Plant Microbe Interact. 11, 144–152. ( 10.1094/Mpmi.1998.11.2.144) [DOI] [Google Scholar]
- 13.Kwak YS, Han S, Thomashow LS, Rice JT, Paulitz TC, Kim D, Weller DM. 2011. Saccharomyces cerevisiae genome-wide mutant screen for sensitivity to 2,4-diacetylphloroglucinol, an antibiotic produced by Pseudomonas fluorescens. Appl. Environ. Microb. 77, 1770–1776. ( 10.1128/Aem.02151-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alabouvette C. 1986. Fusarium-Wilt suppressive soils from the Chateaurenard region—review of a 10-year study. Agronomie 6, 273–284. ( 10.1051/agro:19860307) [DOI] [Google Scholar]
- 15.Siegel-Hertz K, Edel-Hermann V, Chapelle E, Terrat S, Raaijmakers JM, Steinberg C. 2018. Comparative microbiome analysis of a Fusarium wilt suppressive soil and a Fusarium wilt conducive soil from the Chateaurenard region. Front. Microbiol. 9, 568 ( 10.3389/fmicb.2018.00568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duijff BJ, Recorbet G, Bakker PAHM, Loper JE, Lemanceau P. 1999. Microbial antagonism at the root level is involved in the suppression of Fusarium wilt by the combination of nonpathogenic Fusarium oxysporum Fo47 and Pseudomonas putida WCS358. Phytopathology 89, 1073–1079. ( 10.1094/Phyto.1999.89.11.1073) [DOI] [PubMed] [Google Scholar]
- 17.Carrion VJ, Cordovez V, Tyc O, Etalo DW, de Bruijn I, de Jager VCL, Medema MH, Eberl L, Raaijmakers JM.. 2018. Involvement of Burkholderiaceae and sulfurous volatiles in disease-suppressive soils. Isme J. 12, 2307–2321. ( 10.1038/s41396-018-0186-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordovez V, Carrion VJ, Etalo DW, Mumm R, Zhu H, van Wezel GP, Raaijmakers JM.. 2015. Diversity and functions of volatile organic compounds produced by Streptomyces from a disease-suppressive soil. Front. Microbiol. 6, 1081 ( 10.5589/fmicb.2015.01081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garbeva P, Hol WHG, Termorshuizen AJ, Kowalchuk GA, de Boer W.. 2011. Fungistasis and general soil biostasis—a new synthesis. Soil Biol. Biochem. 43, 469–477. ( 10.1016/j.soilbio.2010.11.020) [DOI] [Google Scholar]
- 20.Cho G, Kim J, Park CG, Nislow C, Weller DM, Kwak YS. 2017. Caryolan-1-ol, an antifungal volatile produced by Streptomyces spp., inhibits the endomembrane system of fungi. Open Biol. 7, 170075 ( 10.1098/rsob.170075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ossowicki A, Jafra S, Garbeva P. 2017. The antimicrobial volatile power of the rhizospheric isolate Pseudomonas donghuensis P482. PLoS ONE 12, e174362 ( 10.1371/journal.pone.0174362) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garbeva P, Hordijk C, Gerards S, de Boer W.. 2014. Volatiles produced by the mycophagous soil bacterium Collimonas. Fems Microbiol. Ecol. 87, 639–649. ( 10.1111/1574-6941.12252) [DOI] [PubMed] [Google Scholar]
- 23.van Agtmaal M, Straathof AL, Termorshuizen A, Lievens B, Hoffland E, de Boer W.. 2018. Volatile-mediated suppression of plant pathogens is related to soil properties and microbial community composition. Soil Biol. Biochem. 117, 164–174. ( 10.1016/j.soilbio.2017.11.015) [DOI] [Google Scholar]
- 24.Landa BB, Mavrodi OV, Raaijmakers JM, Gardener BBM, Thomashow LS, Weller DM. 2002. Differential ability of genotypes of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens strains to colonize the roots of pea plants. Appl. Environ. Microb. 68, 3226–3237. ( 10.1128/Aem.68.7.3226-3237.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiese MV. 1987. Compendium of wheat diseases, 2nd edn St. Paul, MN: APS Press. [Google Scholar]
- 26.Schulz-Bohm K, Zweers H, de Boer W, Garbeva P.. 2015. A fragrant neighborhood: volatile mediated bacterial interactions in soil. Front. Microbiol. 6, 1212 ( 10.3389/fmicb.2015.01212). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blom D, Fabbri C, Eberl L, Weisskopf L. 2011. Volatile-mediated killing of Arabidopsis thaliana by bacteria is mainly due to hydrogen cyanide. Appl. Environ. Microb. 77, 1000–1008. ( 10.1128/Aem.01968-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park YS, Dutta S, Ann M, Raaijmakers JM, Park K. 2015. Promotion of plant growth by Pseudomonas fluorescens strain SS101 via novel volatile organic compounds. Biochem. Bioph. Res. Co. 461, 361–365. ( 10.1016/j.bbrc.2015.04.039) [DOI] [PubMed] [Google Scholar]
- 29.Chong J, Soufan O, Li C, Caraus I, Li SZ, Bourque G, Wishart DS, Xia JG. 2018. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–W494. ( 10.1093/nar/gky310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoper H, Alabouvette C. 1996. Importance of physical and chemical soil properties in the suppressiveness of soils to plant diseases. Eur. J. Soil Biol. 32, 41–58. [Google Scholar]
- 31.Janvier C, Villeneuve F, Alabouvette C, Edel-Hermann V, Mateille T, Steinberg C. 2007. Soil health through soil disease suppression: which strategy from descriptors to indicators? Soil Biol. Biochem. 39, 1–23. ( 10.1016/j.soilbio.2006.07.001) [DOI] [Google Scholar]
- 32.Garbeva P, van Veen JA, van Elsas JD.. 2004. Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Ann. Rev. Phytopathol. 42, 243–270. ( 10.1146/annurev.phyto.42.012604.135455) [DOI] [PubMed] [Google Scholar]
- 33.Donn S, Almario J, Mullerc D, Moenne-Loccoz Y, Gupta VV.S.R., Kirkegaard JA, Richardson AE. 2014. Rhizosphere microbial communities associated with Rhizoctonia damage at the field and disease patch scale. Appl. Soil Ecol. 78, 37–47. ( 10.1016/j.apsoil.2014.02.001) [DOI] [Google Scholar]
- 34.Yin CT, Hulbert SH, Schroeder KL, Mavrodi O, Mavrodi D, Dhingra A, Schillinger WF, Paulitz TC. 2013. Role of bacterial communities in the natural suppression of Rhizoctonia solani bare patch disease of wheat (Triticum aestivum L.). Appl. Environ. Microb. 79, 7428–7438. ( 10.1128/Aem.01610-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charlop-Powers Z, et al. 2015. Global biogeographic sampling of bacterial secondary metabolism. Elife 4, e5048 ( 10.7554/eLife.05048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mehrabi Z, McMillan VE, Clark IM, Canning G, Hammond-Kosack KE, Preston G, Hirsch PR, Mauchline TH. 2016. Pseudomonas spp. diversity is negatively associated with suppression of the wheat take-all pathogen. Sci. Rep-Uk 6, 29905 ( 10.1038/srep29905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medema MH, Cimermancic P, Sali A, Takano E, Fischbach MA. 2014. A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Comput. Biol. 10, e1004016 ( 10.1371/journal.pcbi.1004016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garbeva P, Postma J, van Veen JA, van Elsas JD.. 2006. Effect of above-ground plant species on soil microbial community structure and its impact on suppression of Rhizoctonia solani AG3. Environ. Microbiol. 8, 233–246. ( 10.1111/j.1462-2920.2005.00888.x) [DOI] [PubMed] [Google Scholar]
- 39.Chapelle E, Mendes R, Bakker PA, Raaijmakers JM. 2016. Fungal invasion of the rhizosphere microbiome. Isme J. 10, 265–268. ( 10.1038/ismej.2015.82) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwak MJ, et al. 2018. Rhizosphere microbiome structure alters to enable wilt resistance in tomato (vol 36, pg 1100, 2018). Nat. Biotechnol. 36, 1117 ( 10.1038/nbt1118-1117) [DOI] [PubMed] [Google Scholar]
- 41.Tyc O, et al. 2017. Exploring bacterial interspecific interactions for discovery of novel antimicrobial compounds. Microb. Biotechnol. 10, 910–925. ( 10.1111/1751-7915.12735). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyc O, van den Berg M, Gerards S, van Veen JA, Raaijmakers JM, de Boer W, Garbeva P. 2014. Impact of interspecific interactions on antimicrobial activity among soil bacteria. Front. Microbiol. 5, 567 ( 10.3389/fmicb.2014.00567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. 2013. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio. 4, pii. e00459-13 ( 10.1128/mBio.00459-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crits-Christoph A, Diamond S, Butterfield CN, Thomas BC, Banfield JF. 2018. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 558, 440–444. ( 10.1038/s41586-018-0207-y) [DOI] [PubMed] [Google Scholar]
- 45.Watson AG, Ford EJ. 1972. Soil fungistasis—a reappraisal. Ann. Rev. Phytopathol. 10, 327 ( 10.1146/annurev.py.10.090172.001551) [DOI] [Google Scholar]
- 46.Effmert U, Kalderas J, Warnke R, Piechulla B. 2012. Volatile mediated interactions between bacteria and fungi in the soil. J. Chem. Ecol. 38, 665–703. ( 10.1007/s10886-012-0135-5). [DOI] [PubMed] [Google Scholar]
- 47.Ossowicki A, Tracanna V, Petrus MLC, van Wezel G, Raaijmakers JM, Mederna MH, Garbeva P. 2020. Data from: Microbial and volatile profiling of soils suppressive to Fusarium culmorum of wheat Dryad Digital Repository. ( 10.5061/dryad.g884g70). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Ossowicki A, Tracanna V, Petrus MLC, van Wezel G, Raaijmakers JM, Mederna MH, Garbeva P. 2020. Data from: Microbial and volatile profiling of soils suppressive to Fusarium culmorum of wheat Dryad Digital Repository. ( 10.5061/dryad.g884g70). [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data available from the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.g884q70 [47].