Abstract
Numerous human disorders of the blood system would directly or indirectly benefit from therapeutic approaches that reconstitute the hematopoietic system. Hematopoietic stem cells (HSCs), either from matched donors or ex vivo manipulated autologous tissues, are the most used cellular source of cell therapy for a wide range of disorders. Due to the scarcity of matched donors and the difficulty of ex vivo expansion of HSCs, there is a growing interest in harnessing the potential of pluripotent stem cells (PSCs) as a de novo source of HSCs. PSCs make an ideal source of cells for regenerative medicine in general and for treating blood disorders in particular because they could expand indefinitely in culture and differentiate to any cell type in the body. However, advancement in deriving functional HSCs from PSCs has been slow. This is partly due to an incomplete understanding of the molecular mechanisms underlying normal hematopoiesis. In this review, we discuss the latest efforts to generate human PSC (hPSC)-derived HSCs capable of long-term engraftment. We review the regulation of the key transcription factors (TFs) in hematopoiesis and hematopoietic differentiation, the Homeobox (HOX) and GATA genes, and the interplay between them and microRNAs. We also propose that precise control of these master regulators during the course of hematopoietic differentiation is key to achieving functional hPSC-derived HSCs.
Keywords: Hematopoiesis, embryonic stem cell, induced pluripotent stem cell, differentiation, engraftment, HOX genes, GATA, transcription factors, microRNA, epigenetic regulation
1. INTRODUCTION
Hematopoiesis is a complex process through which hematopoietic stem cells (HSCs) generate all the cell types found in the blood. This originates during the early stages of embryonic development and continues in the bone marrow (BM) throughout adulthood to preserve homeostasis in the blood system [1, 2]. During hematopoiesis, HSCs undergo numerous changes at the epigenetic level and ultimately give rise to all the mature blood cells, such as T cells, macrophages, platelets, etc. [3, 4].
The interest in the expansion and production of HSCs has increased in recent years. This is partly because of the large number of blood diseases. Blood cancers, and more than 350 immune disorders that cause increased susceptibility to infections are associated with a high percentage of morbidity and mortality in the world [5]. In the past two decades, specific blood cell transfusion or HSC transplantation has been applied in the clinic. However, their maturation as standardized therapy is still an ongoing process [6]. Shortage of immunocompatible HSCs [7], poor ex vivo expansion of HSCs [8], and heterogeneity of response to therapy [5] are among the barriers that hinder full utilization of such promising cell therapies in clinics. Therefore, after the derivation of human embryonic stem cells (ESCs) [9] and the discovery of cellular reprogramming [10], much effort has been devoted to obtain HSCs and mature blood cells [11] from human pluripotent stem cells (PSCs). In this review, we discuss the process of hematopoietic differentiation of human PSCs, and we summarize recent reports of successful generation of HSC-like cells by the modulation of TFs and signaling pathways. We will specifically discuss the regulation of two sets of key TFs, HOX and GATA, as master regulators of hematopoiesis.
2. CLINICAL INTEREST IN PLURIPOTENT STEM CELLS IN HEMATOPOIESIS
When treating hematologic malignancies, chemotherapy and/or radiotherapy are used to eliminate malignant cells, but these treatments also harm healthy cells. Thus, HSC-based cell therapy is needed to replace blood cells, wherein successful hematopoietic reconstitution is of paramount importance [12]. Approximately 50,000 allogeneic transplants are performed every year worldwide [13]. Although the number of BM and umbilical cord blood HSC donations is adequate [14], Human Leukocyte Antigen (HLA) compatibility is still one of the most hindering obstacles [15]. Autologous transplantation of ex vivo expanded of HSCs is an option to circumvent the need for matched donors and avoid immune rejection and graft versus host disease [16-20]. Nevertheless, ex vivo expansion of HSCs remains challenging due to spontaneous differentiation and loss of self-renewal [8].
Due to the above, de novo generation of HSCs and functional blood cells from both human ESCs and induced PSCs (iPSCs) has become an attractive alternative to donation-based sources. PSCs have the capacity to self-renew indefinitely and give rise to all cell types found in the human body including the cells that make up the blood system. ESCs have been successfully differentiated into many types of cells in vitro since their first derivation [9]. However, ethical issues associated with ESC derivation and the risk of teratoma formation have hampered their advancement to clinical use [21]. Less than a decade later, Takahashi and Yamanaka, by the forced expression of a defined set of TFs (Oct4, Sox2, Klf4, and c-Myc (OSKM)), induced pluripotency in somatic cells and generated iPSCs that are indistinguishable from ESCs in their pluripotency, gene expression and self-renewal capacity. iPSCs are hailed as a promising source of cells for cellular therapy and regenerative medicine (Fig. 1), because they in principle evade the ethical concerns and risks of immune rejection that are associated with ESCs [10].
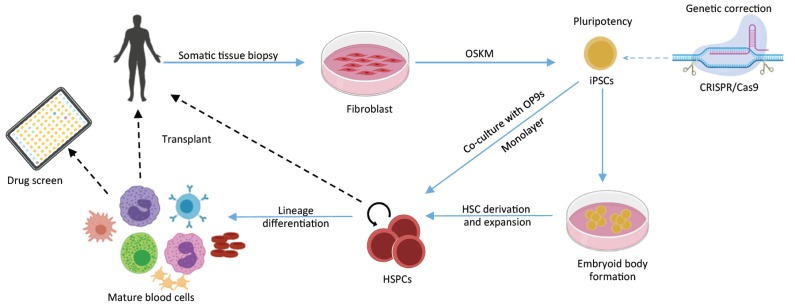
Fig. (1).
Hematopoietic cells engineering for autologous transplantation. Somatic cells are reprogrammed to pluripotency through modulation of OSKM expression (by TFs overexpression, small molecules in addition to reprogramming factors or integration-free methods). If needed, the genome of patient iPSCs could be edited to correct disease-causing mutations by the CRISPR/Cas9 system. Following differentiation, iPS-derived HSCs or terminally differentiated blood cells could then be transfused back to the patient to achieve the desired functional recovery, or used in high-throughput drug screens.
3. HEMATOPOIETIC DIFFERENTIATION OF HPSCS
Induction of pluripotency and iPSC generation offered valuable access to extensively study early human hematopoiesis. This is in addition to their significance in potentially being a versatile cell source for immunotherapies. Furthermore, differentiating patient-derived iPSC to generate HSCs could allow us to model heritable blood disorders and open unprecedented opportunities to investigate diseased mechanisms, novel therapeutics and high-throughput drug screening (Fig. 1).
Human PSC differentiation to blood cells has been accomplished using numerous strategies, which include conventional monolayer cultivation, three-dimensional cell aggregates, or Embryoid Bodies (EBs), or through feeder-dependent approaches that involve co-culturing.
A plethora of hematopoietic lineages were derived from hPSCs, including lymphoid cells, erythrocytes, megakaryocytes, platelets and macrophages [22]. These studies offered an insight that would ultimately help to complete our understanding of hemato-endothelial specification of human PSCs.
In the co-culturing method, hPSCs are cultured with murine BM-derived stromal cells using serum-containing medium [23]. Among the different murine BM stromal cells, the OP9 cell line has been shown to yield the most efficient hematopoietic differentiation [24]. OP9 co-culture was used to obtain multipotent hematopoietic progenitors and mature cells including T and B lymphocytes and megakaryocytes [25-29].
Although this method is attractive since differentiation can be achieved within 9 days without the use of cytokines, its clinical therapeutic potential is diminished due to the use of xenogenic material. In addition, factors like, OP9 cell density, size of hPSC cell aggregates and the quality of Fetal Bovine Serum (FBS) used, make the process variable, thus reducing its value as a method to study signaling pathways involved in the hemato-endothelial transition during the differentiation process.
Directed hematopoietic differentiation of hPSCs using chemically-defined media was also explored.
In this approach, cytokines, morphogens and/or small molecules are used to modulate signaling pathways in hPSCs-derived EBs. A common pattern in these protocols is the use of Wnt agonists and bone morphogenetic protein 4 (BMP4) to induce efficient mesoderm specification, Vascular Endothelial Growth Factor (VEGF) to promote angiogenesis, and hematopoietic cytokine cocktails to drive hematopoiesis [30, 31]. This approach was also done in monolayer protocols using extracellular matrix-coated plates. These 2D approaches counteract the variability associated with using EBs. Although these approaches are chemically defined, they have all failed to produce functional multipotent HSCs capable of multilineage long-term engraftment.
Cell identity is endowed by the presence of certain active gene regulatory networks governed by TFs. Since endothelial-hematopoietic transitions in Aorta-Gonad-Mesonephros (AGM) and the generation of HSCs is controlled by cell-specific set of TFs, numerous studies have utilized forced expression of key TFs to direct hPSCs to hematopoietic differentiation [32-36]. In 2013, Doulatov et al., have demonstrated that lineage-restricted CD34+CD45+ myeloid precursors derived from hPSCs could be reprogrammed into multilineage progenitors that can be expanded in vitro and engraft in vivo [32]. The TFs HOXA9, ERG, and RAR-related orphan receptor alpha (RORa) promoted self-renewal and multilineage potential in vitro. They also allowed in vitro maintenance of primitive CD34+CD38- cells. Additionally, the group performed transplantation-based cell screening and identified two additional TFs, SOX4 and MYB, that conferred engraftment ability. Hematopoietic cells generated with all five factors could short-term engraft and yield myeloid and erythroid lineages. Generated erythroid precursors silenced embryonic and activated adult globin expression.
In another study in the human system, it was demonstrated that the overexpression of TFs RUNX1, FOSB, SPI1, and GFI1 in human umbilical vein endothelial cells (HUVECs) followed by co-culture with AKT-activated endothelial cells, induced definitive hematopoietic development program [37].
Additionally, in another report, a gain-of-function screening revealed that overexpression of TFs ETV2 and GATA2 or GATA2 and TAL1 activated distinct hematopoietic programs, namely pan-myeloid and erythro-megakaryocytic, respectively [33].
Recently, de novo generation of engraftable HSCs has been achieved in the mouse system [38]. Lis, and colleagues reprogrammed mouse adult endothelial cells to match bone marrow HSCs in their transcriptome. Conditional expression of TFs FOSB, GFI1, RUNX1, and SPI1 (FGRS) in adult mouse endothelial cells co-cultured with vascular niche endothelial cells (VN-ECs) reprogrammed adult endothelial cells into engraftable HSCs (rEC-HSCs). During the conversion, mature endothelial cells transitioned through three subsequent stages. First, endothelial cells began to express Runx1 (induction phase, days 0-8). As a result of FGRS transduction, the cells were able to acquire hematopoietic morphology and markers (specification phase days 8-20). Lastly, the self-renewal and expansion stage included an essential direct cellular contact with vascular niche (expansion phase days 20-28). Interestingly, rEC-HSCs have the capability of long-term self-renewal and engraftment, and a gene expression profile that matched that of adult wild type HSCs. In addition, the same cells can be used for clonally derived engraftment and serial primary and secondary multi-lineage reconstitution, including antigen-dependent adaptive immune function [38].
In 2017, a milestone was reached when Sugimura and colleagues achieved long-term multilineage engraftment of human PSCs-derived hematopoietic progenitor cells [36]. Initially, hemogenic endothelium CD34+ cells were magnetically isolated from day 8 human PSCs-derived EBs and then cultured with hematopoietic cytokines. Even though, endothelial-hematopoietic transition was evident (a decrease in YAP, FOXC1, COUPTFII, and an increase in hematopoietic lineage gene expression RUNX1, MYB, GATA2, SCL), and CD34+CD45+ cells were emerging in culture, attempts to engraft irradiated immune-deficient recipient mice with these cells were not successful. Based on the assumption that TFs that govern hematopoiesis would be evolutionarily conserved, the group interrogated mouse and human databases of gene expression of fetal liver HSCs relative to hemogenic endothelium and, along with the previously identified factors, a library of 26 TFs was assembled and cloned individually into doxycycline-inducible lentivirus expression system. Three days following the magnetic isolation of hemogenic endothelium from EBs, cells were transduced with the library and 24 hours later transplanted in irradiated immunodeficient recipient mice. Chimerism (presence of human CD45+ cells) in peripheral blood of injected mice could be detected for up to 12 weeks following transplantation and doxycycline administration. Furthermore, the examination of bone marrow and thymus revealed a significant presence of human erythroid and myeloid cells and, B and T cells.
Using successfully engrafted human cells as a reference, (identifying what exogenous TFs they carried using PCR amplification), the group was able to identify the essential factors in the library that were indispensable for engraftment. A total of seven factors could be identified, ERG, HOXA5, HOXA9, HOXA10, LCOR, RUNX1, and SPI1. The use of only these seven factors resulted in long-term multilineage engraftment and serial primary and secondary multi-lineage reconstitution.
The significant presence of HOX family members in the seven factors is expected, given their previously identified role in hematopoiesis. HOXA9 is an essential factor in HSCs that promotes self-renewal during embryogenesis and is known to regulate transcription by interacting with numerous enhancers of genes important for hematopoiesis [39, 40]. HOXA5 is a direct transcriptional target of the Notch signaling pathway in T cell progenitors along with HOXA9 and HOXA10. This could be indicative of an essential role that these HOX genes may play in T-cell lymphopoiesis [41]. We will discuss HOX genes regulation in more detail in the following sections in this review.
4. REGULATION OF HOX AND GATA TRANSCRIPTION FACTORS AS MASTER REGULATORS OF HEMATOPOIESIS
Current protocols of hematopoietic differentiation of PSCs are based on mimicking in vivo signaling cascades and factors present during embryonic and adult hematopoiesis. However, a comprehensive understanding of the molecular mechanisms that govern hematopoietic differentiation and specification remains lacking. A large number of regulators work together to orchestrate epigenetic and transcription modifications to direct cellular differentiation [42, 43]. Such regulatory mechanisms are important for the proper temporal-spatial expression of master regulators including the HOX [44] and GATA [45] TFs that activate decisive pathways in the developmental process of hematopoietic specification. Furthermore, HOX and GATA factors can be indirectly regulated by other TFs, proteins, and miRNAs, which can also influence cell fate commitment (Fig. 2). Indeed, this is manifested in the most hematopoietic differentiation protocols where the HOX and GATA family of genes are frequently modulated (Table 1).
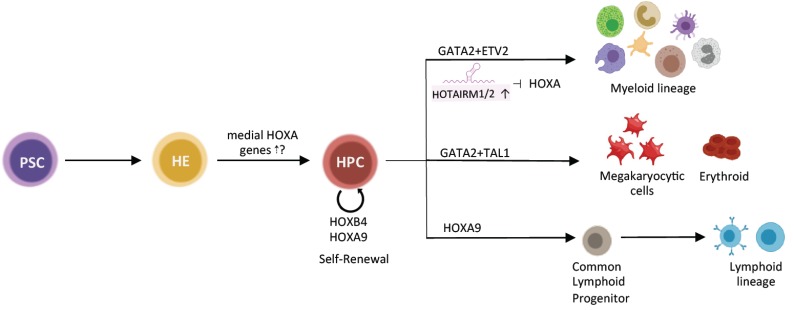
Fig. (2).
Regulators during hematopoietic differentiation of PSCs. Hematopoiesis is achieved through the interplay of transcription factors, signaling pathways, and lncRNAs. HOXB4 and HOXA9 promote self-renewal of HSCs through modulating cell cycle regulators. Medial HOXA genes have been shown to be expressed at higher levels in native HSCs compared to PSC-derived HSPCs. Combined upregulation of GATA2 and ETV2 confers pan-myeloid potential in hESC-derived HPCs. Whereas the upregulation of GATA2+TAL1 results in pan-erythroid/megakaryocytic potential. HOTAIRM1/2 upregulation also promotes myeloid differentiation potential by antagonizing HOXA genes. HOXA9 has been shown to be important for lymphoid differentiation.
Table 1. Summary of hematopoietic differentiation of human pluripotent cells.
| Cell Source | TF Cocktail | Growth Medium | Destination Cell Type | Notes | Refs. |
|---|---|---|---|---|---|
| ESCs | HOXB4 | SCF, IL-3, IL-6, Flt3 ligand, G-CSF, BMP4 | Hematopoietic Progenitor Cells (HPCs) with erythroid and myeloid commitment | - | [46, 47] |
| ESCs | ERG, HOXA9, RORA + SOX4 and MYB |
SCF, IL-3, G-CSF, Flt3 ligand, doxycycline. | Multipotent myeloid progenitor myeloid and erythroid engraftment |
- | [32] |
| iPSCs | None | In vivo teratoma | Erythroid, myeloid and lymphoid | Oncogenic risk | [48] |
| ESCs and iPSCs | GATA2, ETV2 or GATA2, TAL1 | SCF, bFGF, TPO | Pan-myeloid or limited to erythrocytes and megakaryocytes | - | [33] |
| ESCs | SCL/TAL1 | bFGF | Hematoendothelial progenitors | No in vivo reconstitution | [35] |
| ESCs | SCL/TAL1 | BMP4, VEGF, bFGF, SCF, TPO, Flt3 ligand | Megakaryocytic precursors, mature megakaryocytes (MKs), and platelets | Trichostatin A (TSA) and hydroxamic acid (SAHA), both mimic the SCL/TAL-induced effects | [49] |
| ESCs and iPSCs | GATA1, FLI1, TAL1 | TPO or EPO | Megakaryocytes and erythrocyte progenitors | - | [50] |
| ESCs and iPSCs | EKLF1 | bFGF, Y-27632 | Erythrocytes | Expressed in the AAVS1 locus | [11] |
| ESCs and iPSCs | None | rhSCF, rhFlt3 ligand, rh-IL7 | T-cells | Based on artificial thymic organoid | [51] |
| iPSCs | ERG, HOXA5, HOXA9, HOXA10, LCOR, RUNX1, SPI1 |
bFGF, VEGF, IL6, IGF-1, IL11, SCF | HSPCs capable of multilineage engraftment | Serially transplantable HSPCs | [36] |
| iPSCs | MLL-AF4 | BMP4, bFGF, VEGF, SB431542, SCF, IL3, TPO | Induced HSPCs (iHSPCs) | Reconstitute hematopoiesis without myeloid bias but prone to leukemic transformation | [52] |
| ESCs and iPSCs | None | Forskolin, cAMP, IBMX, SCF, IL-3, IL-5, GM-CSF and EPO. | Cells with HSC-like surface markers | cAMP induction decreased oxidative burden in hematopoietic cells | [53] |
| iPSCs | None, EB formation and OP-9 co-culture | BMP4, SCF, Flt3 ligand, IL-6, TPO, IL-3, G-CSF. | Neutrophils | Changes in expression of GATA-2, SCL/TAL1 and AML1/RUNX1 similar to normal hematopoiesis. | [54] |
| iPSCs | None, co-cultures or conditioned medium from OP-9 and WEHI-3 cells | Recombinant IL-2, IL-3, Flt3 ligand, and G-CSF | CD34+ hematopoietic cells (i CD34+ cells) | HLA-B4002-lacking i CD34+ cells escape cytotoxic T-cell attack | [55] |
4.1. Regulation of HOX Genes
The HOX genes encode a family of highly conserved TFs situated in four clusters–HOXA, HOXB, HOXC, and HOXD–in humans. HOX genes are master regulators of cellular differentiation during embryonic development and early hematopoiesis. Their spatio-temporal function suggests HOX genes as key players in hierarchical commitment of cell fate [44, 56].
Transcriptional activation of HOX genes is achieved in ESCs in the presence of Retinoic Acid (RA), an important extra- and intra-cellular metabolite involved in diverse biological processes including the stimulation of normal hematopoietic progenitors [57, 58]. Using murine ESCs lacking the RA receptor γ (RARγ) or in which the 3'-RA-responsive element of Hoxa1 has been deleted, Kashyap and colleagues have demonstrated that RARγ is essential for RA-induced HOX transcriptional activation. In addition, deletion of RARγ binding site in the HOXA1 enhancer diminished transcriptional and epigenetic activation of both the HOXA and HOXB gene clusters. Furthermore, RA/RARγ signaling was shown to be essential for the erasure of histone 3 lysine 27 trimethylation (H3K27me3), a repressive epigenetic mark, from activated HOX genes during ESC differentiation [59].
For function studies, overexpression of Hoxb4 was shown to enable definitive lymphoid-myeloid engraftment potential in mouse ESC-derived hematopoietic progenitors [60]. Additionally, hESCs that overexpress HOXB4 generate greater numbers of cells expressing hematopoietic cell surface markers than wild-type hESCs [46]. However, overexpression of HOXB4 in hESCs failed to confer engraftment on differentiated hematopoietic cells [61]. Multiple studies aimed to elucidate the mechanism by which Hoxb4 upregulation endowed definitive hematopoiesis and engraftment capacity to mESC-derived HSCs. Hoxb4 has been shown to promote the expansion of both adult and mESC-derived HSCs [62]. Its upregulation is associated with an increase in the expression of c-myc and cyclin-D1 [63]. Therefore, Hoxb4 may directly induce the expression of cell cycle regulators and drive cell division forward in adult and ESC-derived HSCs. Genome wide examination of potential targets of Hoxb4 revealed that Hoxb4 binds essential regulators of hematopoiesis including Runx1, stem cell leukemia/T-cell acute lymphocytic leukemia-1 (Scl/Tal1), and Gata2 [64]. Taken together, HOXB4 is likely to be the main regulator of hematopoiesis, which is further illustrated in studies demonstrating its direct interaction with other TFs and co-regulators [46, 65-67].
Hoxa9 is another HOX gene that is important in hematopoiesis. Although Hoxa9 knockout mice are viable, they exhibit smaller thymus and spleen and suffer from a reduction in myeloid cells, lymphocytes, and committed progenitors [68]. In addition, prior gene expression profile comparisons between hESC-derived HSCs and cord blood HSCs revealed HOXA9 to be the most differentially expressed (downregulated in hESCs) gene between the two [69]. HOXA9 overexpression was shown to augment hematopoietic differentiation of hESCs by specifically committing Hemogenic Endothelial (HE) precursor cells into primitive and total CD45+ blood cells [70], whereas, in the differentiation from HSCs to mature blood cells, HOXA9 is down-regulated [40]. HOXA9 is one of the most abundant HOX genes in HSCs that work together with HOXB4 to maintain stemness. In hematopoiesis, the progressive loss of expression of these genes sends signals to HSCs to proliferate and starts the downstream lymphoid and myeloid differentiation [40]. Medial HOXA genes including HOXA5, 7 and 9 are suppressed during hematopoietic differentiation of human PSCs [71]. However, simply overexpressing these medial HOXA genes is not sufficient to confer long-term engraftability in human PSC-derived hematopoietic progenitors [71]. These results suggest that the establishment of proper epigenetic state in the HOXA cluster and/or expression of additional regulatory factors are required for context-dependent regulation of HOXA target genes in bona fide HSCs (Fig. 2).
Interestingly, hematopoiesis differentiation is carried out by specific temporal regulation of HOX genes [72]. Many HOX genes (e.g. HOXA3, A7, A9 and A13, and HOXB2-B6) are up- and down-regulated during differentiation [73, 74]. In terms of down-regulation, the HOXA group has the task of activating the HOX gene repressors to gradually silence the HOX transcription according to the cellular commitment, which yields mature cells with minimal or no expression of HOXA genes [66, 67]. An example of a HOX gene regulator is the mix lineage leukemia (MLL) gene, a global epigenetic regulator characterized as a histone methyl transferase (HMT) that contains a DNA methyl transferase (DNMT) homologue domain CxxC that binds to unmethylated CpG residues in HOXA9 locus to protect them from potential DNA methylation [75]. Chromosomal translocation mediated fusion of MLL with TFs, such as MLL-AF4 and MLL-AF9, reinforces a hypomethylated state of CpG of HOXA9 and increases its expression in primitive hematopoiesis. However, the loss of MLL-CxxC domain allows hypermethylation, blocks transcription of HOXA9 gene and facilitates further differentiation [76].
HOX genes are additionally regulated by RNA-mediated mechanisms. Specific transcripts longer than 200 nucleotides called long non-coding RNAs (lncRNAs) can affect hematopoiesis through regulation of messenger RNA (mRNA) stability, chromatin structure or sequestration of regulatory molecules [77] (Fig. 2). One prominent example is the lncRNA HOTAIRM1 and its isoform HOTAIRM2, located between HOXA1 and HOXA2. HOTAIRM1 and 2 have important roles in ESC differentiation to HSCs, where they are rapidly up-regulated and recruited to HOXA1 and HOXB1 promoter regions to repress transcription and proceed with the proliferation and differentiation to HSCs and progenitors [78-80]. The same happens in native hematopoiesis, where HOTAIRM1 is up-regulated during myeloid and granulocyte differentiation with the same effect of blocking HOXA repression [79, 81]. HOXA transcript at the distal tip (HOTTIP), an additional lncRNA located in the active locus of HOXA13, acts on its target genes through chromosomal looping, which reduces the distance and facilitates the interaction [82]. Through its interaction with WD repeat domain 5 (WDR5) and MLL-1 which is a HMT, HOTTIP lncRNA drives histone H3 lysine 4 trimethylation (H3K4me3) and HOXA gene transcription [82].
4.2. Regulation of GATA Factors
The GATA TF family is composed of 6 members (GATA1-6), which are implicated in several biological processes. The discovery and genetic manipulation of GATA TFs have had a big impact on the understanding of hematopoiesis and development. GATA1 and GATA2 are important for hematopoietic regulation. Although GATA3 is also involved in this process, the molecular mechanisms are still poorly described [83]. GATA1 is a transcriptional activator expressed mainly in megakaryocytes and erythrocytes, but it also participates in the development of eosinophils, mast cells, and dendritic cells [84, 85]. GATA2 controls primitive and definitive hematopoiesis, as well as the maintenance of HSCs where it is highly expressed [86]. GATA3 is related to lymphoid lineages differentiation and has a less explored role in HSC quiescence [87].
However, in the context of hematopoiesis, the important roles of GATA1 and GATA2 have been elucidated.
In general, hematopoietic regulation by GATA activity can be viewed as a constant battle between GATA1 and GATA2 to gain chromatin occupancy, where less committed stem cells correlate with a greater presence of GATA2 in chromatin, whereas differentiation is associated with the gain of the territory of GATA1 that displaces GATA2 [85, 88, 89] (Fig. 2). GATA2 expression is essential for early embryonic development and early hematopoiesis because it is required for the maintenance and expansion of HSCs and/or multipotent progenitors [90-92]. Consequently, in early hematopoiesis, GATA2 is recruited to almost all GATA binding sites in chromatin, while GATA1 expression remains low [93, 94].
GATA activation can be affected by different factors that can change GATA gene activity in a coordinated fashion, in which positive regulators of GATA2 inhibit GATA1 expression and vice versa [92, 95]. The GATA proteins are composed of a common conserved C4 zinc finger domain that directly binds DNA and a transactivation domain in the N-terminus that can be positively or negatively regulated [96, 97]. Similar to HOX genes, RA exerts a big influence on GATA2 expression by binding with retinoic acid receptor alpha (RARα) expressed in hematopoietic cells. This is mediated by the interaction between the zinc fingers of GATA2 and the DNA-binding domain of RARα in the presence of RA, which allows active transcription in GATA2-bound chromatin sites [98]. On the other hand, the interaction between the N-terminal domain of GATA2 and transcriptional regulators, such as Scl/TAL1, LYL1, LMO2, ERG, FLI-1 or RUNX1, can affect GATA2 chromatin occupancy [97, 99]. Such interactions repress GATA2 binding by removing its binding domain from the DNA and vacating the chromatin sites for GATA1 occupancy [89].
Among the GATA1 positive regulators, Friend of GATA-1 (FOG-1) is one of the most important ones that are responsible for the activation and repression of most GATA1 genes. Although FOG-1 does not have a DNA-binding domain, the four FOG-1 zinc fingers can bind to the N-terminus of GATA1, which facilitates GATA1 occupancy at numerous genomic sites to proceed with “the GATA switch” (defined as the displacement of GATA2 and binding of GATA1) [100, 101]. FOG-1 recruits the nucleosome remodeling and deacetylation (NuRD) complex, which can be the mechanism through which GATA2 is displaced [102]. Disruption of the FOG1-NuRD association results in a reduction in mature erythroid and megakaryocyte colonies [102]. The histone acetyltransferases (HAT) CREB-binding protein (CBP) and E1A-associated protein p300 (p300) induce acetylation in GATA1, which reinforces its DNA binding to gain genomic occupancy [103]. Together, these co-factors help GATA factors to initiate megakaryocytic and erythroid specification [104].
PU.1 is recognized as an important regulator of cellular communication and immune differentiation by blocking erythroid commitment [105]. In early hematopoiesis, mutual inhibition between GATA1 and PU.1 occurs in progenitor cells, and regulates differentiation into erythroid or myeloid lineages respectively [106]. PU.1 regulates its own expression to drive the myeloid differentiation program through its binding to the upstream regulatory element [107, 108]. Targeted disruption of the upstream regulatory element (URE) results in a significant reduction in PU.1 levels and the development of acute myeloid leukemia (AML) [109]. To promote erythroid differentiation, GATA1 binds PU.1’s URE and together with DNMT1 mediates repression of PU.1 [106]. In addition, at the PU.1 promoter, histone 3 lysine 9 (H3K9) methylation and deacetylation as well as the H3K27 methylation occur to further inhibit its transcription [106, 110].
The Scl/TAL1 protein, which is involved in HSC maintenance and myeloid lineage differentiation, interacts as a multicomplex (Scl/TAL1:LMO:LDB1) with GATA proteins [49, 111, 112]. Interestingly, ChIP-sequencing analysis proposes that Scl/TAL1:LMO:LDB is recruited to GATA-bound active transcription sites, and suggests that GATA is a modulator of Scl/TAL1 transcription [111, 113, 114]. The previously described occupancy competition between GATA1 and GATA2 is responsible of the Scl/TAL1 specific function. This means that the co-occupancy with GATA2 promotes stem cell maintenance, while co-occupancy with GATA1 induces myeloid cell specification [114, 115]. Similar to the examples mentioned above, other molecules and TFs interacting with GATA, including GFI1b [116], IKZF1 [117], and RUNX [118], have co-regulator roles in hematopoietic commitment.
4.3. Regulation of HOX and GATA Genes by Small RNAs
Small non-coding RNA of around 22 nucleotides in length, called microRNA (miRNA), interferes in post-transcriptional gene regulation and therefore has a regulatory influence on developmental processes and diseases [119]. miRNA has been found in all cellular compartments including the nucleus, nucleolus, and mitochondria, where it plays different roles in transcription, translation, splicing, DNA repair, and even extracellular signaling events [120]. Around 40% of miRNAs can be found in introns and thus are usually regulated together with their host genes [120, 121]. In embryonic hematopoiesis, the miRNA-125 family members are highly expressed mainly in HSCs and progenitors, and their expression decreases with differentiation. Some studies suggest an important role of miRNA-125 in self-renewal of HSCs and its persistent expression in some myeloid and lymphoid leukemias confers resistance to apoptosis and halts differentiation [15, 122, 123].
The miRNA-196 family, which can be found between the HOXA9 and HOXA10 genes, supports the regulation of cell survival, proliferation, the undifferentiated state and long-term maintenance of HSCs by repressing genes involved in differentiation [124]. This has been demonstrated in HOXA7 and HOXB8, which contain complementary sequences to miRNA-196b and are transcriptionally suppressed in HSCs. Additionally, chromatin immunoprecipitation analysis revealed that the transcriptional repressor, Gfi1, binds directly to the miRNA-196b locus and downregulates its transcription level to permit myeloid differentiation [124, 125].
Many miRNAs are implicated in the differentiation of mature blood cells. For example, miRNAs-15, 24, 144, and 451 are required to silence the stemness program of HSCs [126-128]. As explained above, erythroid lineage differentiation is mainly mediated by GATA1 and GATA2, whose multiple regulatory pathways also involve miRNA-24 and miRNA-27a. The GATA2 protein binds the promoter region of the locus containing miRNA-27a and miRNA-24 (miR-27a≈24) and inhibits its transcription in immature erythroid progenitor cells [133]. During erythropoiesis, GATA1 displaces GATA2 from chromatin, including the promoter region of miR-27a≈24 and promotes its transcription. In a positive feedback loop, mature miR-27a and miR-24 bind the GATA2 mRNA and inhibit its translation, which further promotes erythroid maturation [129]. GATA1 also binds a distal upstream regulatory element of the common locus of miR-144 and miR-451. GATA1 binding recruits the transcription machinery and activates RNA polymerase II-mediated transcription of a common precursor RNA transcript (pre-miRNA) encoding both mature miRNAs. Inhibition of miR-451 in zebrafish embryo using antisense morpholinos yields erythroid progenitors incapable of developing into mature circulating red blood cells [127].
4.4. The Influence of Microenvironment on Signaling Pathways
We have so far focused on the discussion of epigenetic changes that modulate the activation or repression of master TFs during hematopoiesis. However, numerous factors (including the extracellular environment) can influence cell fate decisions. Another key element in hematopoietic differentiation of human PSCs is the signaling between cells and their microenvironment. Importantly, the interplay between components of the epigenetic machinery and the microenvironment in hematopoiesis may create context-dependent signaling events [130], which make the process more difficult to recreate in vitro.
Stem cell factor (SCF), an important hematopoietic factor, is one of the most used cytokines in the medium when HSCs and other hematopoietic progenitors are cultured in vitro. SCF promotes hematopoietic differentiation of hPSCs via interaction with its receptor c-KIT, a receptor tyrosine kinase under the control of both miR-221 and miR-222. The inhibition of SCF action during hematopoietic differentiation of human PSCs by using a c-KIT antagonist results in reduced multi-lineage potential of the obtained progenitors [131-133].
Bone morphogenetic protein (BMP) is an important protein that guides the blood formation process [134]. The BMP type 4 (BMP4) expression emerges in the early formation of blood cells in the human yolk sac [135]. In normal hematopoietic differentiation of ESCs, it has been showed that BMP4 first induces mesodermal lineages by activating nuclear Smad proteins. BMP4 signaling is linked to the Wnt signaling pathway, which regulates the caudal gene family (Cdx) and consequently has the capacity to activate transcription of posterior HOX genes [136-138]. In this linear process, BMP4-Wnt-Cdx-HOX is suggested to cause the activation of HOXA9 and HOXB1, resulting in an improved formation of myeloid colonies [138, 139]. This explains why induction protocols (Table 1) include BMP4 and molecules like CHIR99021, which modulate the Wnt/β-catenin pathway [30, 140].
Wnt signaling is known to be important for the definitive hematopoiesis potential of KDR+ mesodermal progenitors, which give rise to hemogenic endothelium from which HSCs emerge during development [12, 141]. Although many studies have elucidated the signaling requirements of the early stages of hematopoietic commitment during in vitro differentiation, the gene expression programs and signaling events important for subsequent specification and maturation of HSC from the early progenitors remain obscure. Interestingly, during mouse development β-catenin and Wnt target genes are expressed at higher levels in hemogenic endothelial precursors of HSCs, but are downregulated in HSC [142]. Therefore, it is important to understand the dynamic regulation of Wnt signaling during the specification of human hematopoietic stem/progenitor cells in the future.
5. CURRENT CHALLENGES
In the pursuit of de novo generation of functional HSCs from hPSCs for clinical applications, a number of obstacles remain to be overcome. Aside from the persisting challenge of differentiating hPSCs to HSCs that are capable of long-term engraftment, the safety of the generated blood stem cells needs to be determined. Precise control of the epigenetic changes during the course of hematopoietic differentiation is key in making the resulting cells safe for clinical use. Aberrant regulation of gene expression in hPSC-derived HSCs could lead to hematological malignancies [143]. For example, Enhancer of zeste homolog 2 (EZH2) is highly regulated in normal hematopoiesis in processes such as HSC self-renewal and B cell differentiation [144]. However, EZH2 overexpression or downregulation could lead to tumorigenesis [145]. This includes natural killer/T cell lymphoma [146], follicular lymphoma, T cell acute lymphoid leukemia and metastasis [147]. EZH2 works together with many transcription regulators and miRNAs to catalyze trimethylation of H3K27, which contributes to the silencing of multiple tumor suppressors such as INK4A/ARF and E-cadherin [147, 148]. Variability in the epigenome among iPSCs lines has been identified. Nishizawa and colleagues showed that transcriptomic and epigenetic variation contributes to the hematopoietic differentiation capacity (commitment and maturation) of 35 human iPSCs lines and 4 ESCs lines. They correlate the PSC commitment capacity with IGF2 levels that in turn depend on signaling-dependent chromatin accessibility at the mesoderm-related genes [149]. A recent report demonstrated that directed hematopoietic differentiation of hiPSCs using morphogens yields CD45+ cells that are epigenetically distinct from wild type human HSCs, when comparing their genome-wide DNA methylation patterns. Additionally, prolonged co-culture with stromal cells did not induce further epigenetic maturation of these iPSC-derived cells [150]. Taken together, with current differentiation protocols, iPSC-derived hematopoietic stem/progenitors seem to be both functionally and epigenetically immature, which brings into question their suitability for clinical application.
CONCLUSION AND FUTURE PERSPECTIVES
In addition to their unlimited, yet-to-be realized, therapeutic potential, human PSCs make a very useful tool that can be utilized to understand the signaling pathways involved in hematopoiesis. Recently, long term engraftment and multi-lineage differentiation of hPSCs-derived hematopoietic progenitor cells was achieved after screening large numbers of potential TFs that were activated in vivo following transplantation in mice [36]. Ideally, the generation of functional HSCs with the same capacity in vitro would allow deep interrogation of the differentiation process, and, eventually, the generation of therapeutic grade cells. In addition to using Cas9 as a nuclease for editing DNA sequences, the CRISPR/Cas9 technology can be used as a sequence-specific, non-mutagenic, gene regulation tool when fused with effector proteins (gene activators or repressors). This involves the usage of a deactivated Cas9 that harbors two mutations in its endonuclease domain, rendering it an RNA-guided DNA-binding protein unable to generate single or double DNA breaks [151]. This system can be easily engineered to be doxycycline-inducible, which could allow for multiplexed activation or repression of a large number of endogenous loci at carefully selected time points during the course of in vitro differentiation (Fig. 3). The ability to multiplex and to temporally control gene expression using endogenous loci, makes this system extremely valuable and appropriate to study the process of hPSC hematopoietic differentiation.
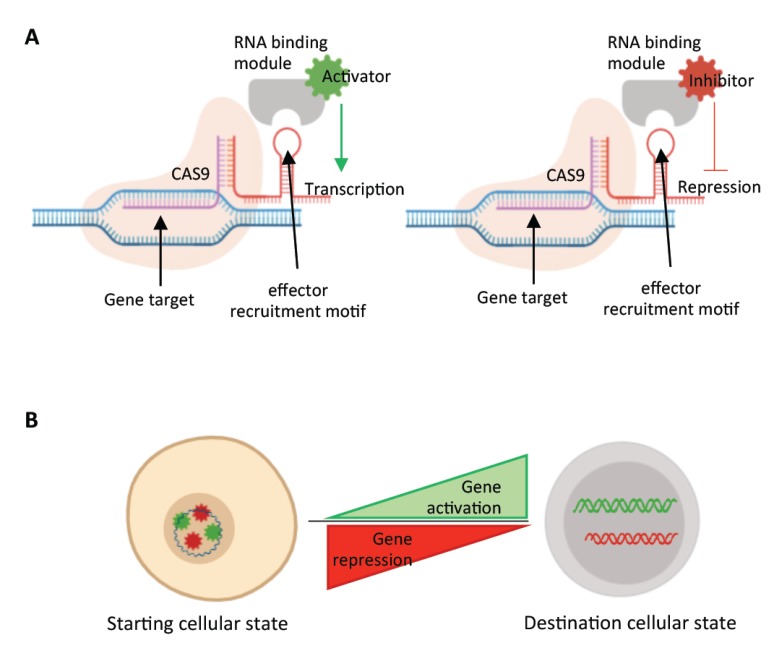
Fig. (3).
Multiplexed transcriptional gene activation or repression by CRISPR-dCAS9. A) Doxycycline-inducible dCas9 system could allow for precise temporal modulation of genes during the course of the differentiation process. Guide RNAs (gRNAs) of genes to be activated would be cloned into RNA scaffolds that harbor activator-binding motif, whereas gRNAs targeting genes to be repressed, would be cloned into scaffolds with repressor-recruiting motifs. B) The ability to temporally control multiple key TFs using this system could open new opportunities for large scale screenings to ultimately achieve functional HSCs in vitro.
Human PSCs represent a promising versatile source of cells for regenerative medicine. The fact that they could be derived from patients’ somatic cells and undergo clonal expansion in culture makes them ideal for the regeneration of the blood system. Their potential with regard to treating genetic blood disorders could be augmented when combined with genome editing techniques such as CRISPR/Cas9 [152-155].
However, two main hurdles limit the translation of iPSC-derived HSCs into cellular therapies. First, the generation of functional iPSC-derived HSCs capable of long-term engraftment and full reconstitution of the blood system. Second, the long-term safety of the generated cells. As mentioned above, HSCs derived from iPSCs remain transcriptionally and epigenetically distinct from cord blood HSCs. The impact of these differences on the safety and functionality of the generated HSCs is yet to be investigated.
After the maunuscript is accepeted for publication, the significant role of GATA2 as a master regulator of hematopoiesis was elucidated in a recent report. Castano and colleagues have demonstrated that GATA2 overexpression during the onset of mesodermal specification, generated hemogenic endothelial progenitors and promoted their further differentiation into hematopoietic progenitor cells. Interestingly, global RNA sequencing and chromatin immunoprecipitation coupled with DNA sequencing, demonstrated that GATA2 directly suppressed cardiac specification genes and activated master hematopoietic regulators, whereas its knockout diminished hematopoietic potential and improved cardiac differentation [156].
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
The study was funded by the KAUST Office of Sponsored Research (OSR), Awards No. BAS/1/1080-01.
ACKNOWLEDGEMENTS
We thank the staff of the Li laboratory for helpful discussions; Jinna Xu, Marie Krenz Y. Sicat and Xingxing Zhang for administrative support.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Daniel M.G., Pereira C.F., Lemischka I.R., Moore K.A. Making a hematopoietic stem cell. Trends Cell Biol. 2016;26(3):202–214. doi: 10.1016/j.tcb.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jagannathan-Bogdan M., Zon L.I. Hematopoiesis. Development. 2013;140(12):2463–2467. doi: 10.1242/dev.083147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deans C., Maggert K.A. What do you mean, “epigenetic”? Genetics. 2015;199(4):887–896. doi: 10.1534/genetics.114.173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinhold B. Epigenetics: The science of change. Environ. Health Perspect. 2006;114(3):A160–A167. doi: 10.1289/ehp.114-a160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos-Sanchez E., Martínez-Cano J., Del Pino Molina L., López-Granados E., Cobaleda C. Epigenetic deregulation in human primary immunodeficiencies. Trends Immunol. 2019;40(1):49–65. doi: 10.1016/j.it.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Siena S., Schiavo R., Pedrazzoli P., Carlo-Stella C. Therapeutic relevance of CD34 cell dose in blood cell transplantation for cancer therapy. J. Clin. Oncol. 2000;18(6):1360–1377. doi: 10.1200/JCO.2000.18.6.1360. [DOI] [PubMed] [Google Scholar]
- 7.Lim W.F., Inoue-Yokoo T., Tan K.S., Lai M.I., Sugiyama D. Hematopoietic cell differentiation from embryonic and induced pluripotent stem cells. Stem Cell Res. Ther. 2013;4(3):71. doi: 10.1186/scrt222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takizawa H., Schanz U., Manz M.G. Ex vivo expansion of hematopoietic stem cells: Mission accomplished? Swiss Med. Wkly. 2011;141:w13316. doi: 10.4414/smw.2011.13316. [DOI] [PubMed] [Google Scholar]
- 9.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Yang C.T., Ma R., Axton R.A., Jackson M., Taylor A.H., Fidanza A., Marenah L., Frayne J., Mountford J.C., Forrester L.M. Activation of KLF1 enhances the differentiation and maturation of red blood cells from human pluripotent stem cells. Stem Cells. 2017;35(4):886–897. doi: 10.1002/stem.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chivu-Economescu M., Rubach M. Hematopoietic stem cells therapies. Curr. Stem Cell Res. Ther. 2017;12(2):124–133. doi: 10.2174/1574888X10666151026114241. [DOI] [PubMed] [Google Scholar]
- 13.Liso A., Neri M., Maglietta F., La Russa R., Turillazzi E. Hematopoietic stem cell transplantation: A bioethical lens. Stem Cells Int. 2017;2017:1286246. doi: 10.1155/2017/1286246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Champlin R. Now everyone has a donor for HSCT. Blood. 2011;118(2):218. doi: 10.1182/blood-2011-05-352518. [DOI] [PubMed] [Google Scholar]
- 15.Guo S., Lu J., Schlanger R., Zhang H., Wang J.Y., Fox M.C., Purton L.E., Fleming H.H., Cobb B., Merkenschlager M., Golub T.R., Scadden D.T. MicroRNA miR-125a controls hematopoietic stem cell number. Proc. Natl. Acad. Sci. USA. 2010;107(32):14229–14234. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michallet M., Philip T., Philip I., Godinot H., Sebban C., Salles G., Thiebaut A., Biron P., Lopez F., Mazars P., Roubi N., Leemhuis T., Hanania E., Reading C., Fine G., Atkinson K., Juttner C., Coiffier B., Fière D., Archimbaud E. Transplantation with selected autologous peripheral blood CD34+Thy1+ hematopoietic stem cells (HSCs) in multiple myeloma: Impact of HSC dose on engraftment, safety, and immune reconstitution. Exp. Hematol. 2000;28(7):858–870. doi: 10.1016/S0301-472X(00)00169-7. [DOI] [PubMed] [Google Scholar]
- 17.Daikeler T., Tichelli A., Passweg J. Complications of autologous hematopoietic stem cell transplantation for patients with autoimmune diseases. Pediatr. Res. 2012;71(4 Pt 2):439–444. doi: 10.1038/pr.2011.57. [DOI] [PubMed] [Google Scholar]
- 18.Cioch M., Jawniak D., Wach M., Mańko J., Radomska K., Borowska H., Szczepanek A., Hus M. Autologous hematopoietic stem cell transplantation for adults with acute myeloid leukemia. Transplant. Proc. 2016;48(5):1814–1817. doi: 10.1016/j.transproceed.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 19.Kim A.R., Sankaran V.G. Development of autologous blood cell therapies. Exp. Hematol. 2016;44(10):887–894. doi: 10.1016/j.exphem.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watts M.J., Linch D.C. Optimisation and quality control of cell processing for autologous stem cell transplantation. Br. J. Haematol. 2016;175(5):771–783. doi: 10.1111/bjh.14378. [DOI] [PubMed] [Google Scholar]
- 21.King N.M., Perrin J. Ethical issues in stem cell research and therapy. Stem Cell Res. Ther. 2014;5(4):85. doi: 10.1186/scrt474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ackermann M., Liebhaber S., Klusmann J.H., Lachmann N. Lost in translation: Pluripotent stem cell-derived hematopoiesis. EMBO Mol. Med. 2015;7(11):1388–1402. doi: 10.15252/emmm.201505301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ledran M.H., Krassowska A., Armstrong L., Dimmick I., Renström J., Lang R., Yung S., Santibanez-Coref M., Dzierzak E., Stojkovic M., Oostendorp R.A., Forrester L., Lako M. Efficient hematopoietic differentiation of human embryonic stem cells on stromal cells derived from hematopoietic niches. Cell Stem Cell. 2008;3(1):85–98. doi: 10.1016/j.stem.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Slukvin I.I., Vodyanik M.A., Thomson J.A., Gumenyuk M.E., Choi K.D. Directed differentiation of human embryonic stem cells into functional dendritic cells through the myeloid pathway. J. Immunol. 2006;176(5):2924–2932. doi: 10.4049/jimmunol.176.5.2924. [DOI] [PubMed] [Google Scholar]
- 25.Vodyanik M.A., Thomson J.A., Slukvin I.I. Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood. 2006;108(6):2095–2105. doi: 10.1182/blood-2006-02-003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar A., Lee J.H., Suknuntha K., D’Souza S.S., Thakur A.S., Slukvin I.I. NOTCH activation at the hematovascular mesoderm stage facilitates efficient generation of T cells with high proliferation potential from human pluripotent stem cells. J. Immunol. 2019;202(3):770–776. doi: 10.4049/jimmunol.1801027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Timmermans F., Velghe I., Vanwalleghem L., De Smedt M., Van Coppernolle S., Taghon T., Moore H.D., Leclercq G., Langerak A.W., Kerre T., Plum J., Vandekerckhove B. Generation of T cells from human embryonic stem cell-derived hematopoietic zones. J. Immunol. 2009;182(11):6879–6888. doi: 10.4049/jimmunol.0803670. [DOI] [PubMed] [Google Scholar]
- 28.French A., Yang C.T., Taylor S., Watt S.M., Carpenter L. Human induced pluripotent stem cell-derived B lymphocytes express sIgM and can be generated via a hemogenic endothelium intermediate. Stem Cells Dev. 2015;24(9):1082–1095. doi: 10.1089/scd.2014.0318. [DOI] [PubMed] [Google Scholar]
- 29.Takayama N., Nishikii H., Usui J., Tsukui H., Sawaguchi A., Hiroyama T., Eto K., Nakauchi H. Generation of functional platelets from human embryonic stem cells in vitro via ES-sacs, VEGF-promoted structures that concentrate hematopoietic progenitors. Blood. 2008;111(11):5298–5306. doi: 10.1182/blood-2007-10-117622. [DOI] [PubMed] [Google Scholar]
- 30.Sturgeon C.M., Ditadi A., Awong G., Kennedy M., Keller G. Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat. Biotechnol. 2014;32(6):554–561. doi: 10.1038/nbt.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy M., Awong G., Sturgeon C.M., Ditadi A., LaMotte-Mohs R., Zúñiga-Pflücker J.C., Keller G. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep. 2012;2(6):1722–1735. doi: 10.1016/j.celrep.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Doulatov S., Vo L.T., Chou S.S., Kim P.G., Arora N., Li H., Hadland B.K., Bernstein I.D., Collins J.J., Zon L.I., Daley G.Q. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell. 2013;13(4):459–470. doi: 10.1016/j.stem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elcheva I., Brok-Volchanskaya V., Kumar A., Liu P., Lee J.H., Tong L., Vodyanik M., Swanson S., Stewart R., Kyba M., Yakubov E., Cooke J., Thomson J.A., Slukvin I. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nat. Commun. 2014;5:4372. doi: 10.1038/ncomms5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ran D., Shia W.J., Lo M.C., Fan J.B., Knorr D.A., Ferrell P.I., Ye Z., Yan M., Cheng L., Kaufman D.S., Zhang D.E. RUNX1a enhances hematopoietic lineage commitment from human embryonic stem cells and inducible pluripotent stem cells. Blood. 2013;121(15):2882–2890. doi: 10.1182/blood-2012-08-451641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Real P.J., Ligero G., Ayllon V., Ramos-Mejia V., Bueno C., Gutierrez-Aranda I., Navarro-Montero O., Lako M., Menendez P. SCL/TAL1 regulates hematopoietic specification from human embryonic stem cells. Mol. Ther. 2012;20(7):1443–1453. doi: 10.1038/mt.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugimura R., Jha D.K., Han A., Soria-Valles C., da Rocha E.L., Lu Y.F., Goettel J.A., Serrao E., Rowe R.G., Malleshaiah M., Wong I., Sousa P., Zhu T.N., Ditadi A., Keller G., Engelman A.N., Snapper S.B., Doulatov S., Daley G.Q. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature. 2017;545(7655):432–438. doi: 10.1038/nature22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandler V.M., Lis R., Liu Y., Kedem A., James D., Elemento O., Butler J.M., Scandura J.M., Rafii S. Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature. 2014;511(7509):312–318. doi: 10.1038/nature13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lis R., Karrasch C.C., Poulos M.G., Kunar B., Redmond D., Duran J.G.B., Badwe C.R., Schachterle W., Ginsberg M., Xiang J., Tabrizi A.R., Shido K., Rosenwaks Z., Elemento O., Speck N.A., Butler J.M., Scandura J.M., Rafii S. Conversion of adult endothelium to immunocompetent haematopoietic stem cells. Nature. 2017;545(7655):439–445. doi: 10.1038/nature22326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y., Sitwala K., Bronstein J., Sanders D., Dandekar M., Collins C., Robertson G., MacDonald J., Cezard T., Bilenky M., Thiessen N., Zhao Y., Zeng T., Hirst M., Hero A., Jones S., Hess J.L. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood. 2012;119(2):388–398. doi: 10.1182/blood-2011-03-341081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence H.J., Christensen J., Fong S., Hu Y.L., Weissman I., Sauvageau G., Humphries R.K., Largman C. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106(12):3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weerkamp F., Luis T.C., Naber B.A., Koster E.E., Jeannotte L., van Dongen J.J., Staal F.J. Identification of Notch target genes in uncommitted T-cell progenitors: No direct induction of a T-cell specific gene program. Leukemia. 2006;20(11):1967–1977. doi: 10.1038/sj.leu.2404396. [DOI] [PubMed] [Google Scholar]
- 42.Li M., Belmonte J.C. Ground rules of the pluripotency gene regulatory network. Nat. Rev. Genet. 2017;18(3):180–191. doi: 10.1038/nrg.2016.156. [DOI] [PubMed] [Google Scholar]
- 43.Li M., Liu G.H., Izpisua Belmonte J.C. Navigating the epigenetic landscape of pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 2012;13(8):524–535. doi: 10.1038/nrm3393. [DOI] [PubMed] [Google Scholar]
- 44.Argiropoulos B., Humphries R.K. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26(47):6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 45.Bresnick E.H., Katsumura K.R., Lee H.Y., Johnson K.D., Perkins A.S. Master regulatory GATA transcription factors: Mechanistic principles and emerging links to hematologic malignancies. Nucleic Acids Res. 2012;40(13):5819–5831. doi: 10.1093/nar/gks281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bowles K.M., Vallier L., Smith J.R., Alexander M.R., Pedersen R.A. HOXB4 overexpression promotes hematopoietic development by human embryonic stem cells. Stem Cells. 2006;24(5):1359–1369. doi: 10.1634/stemcells.2005-0210. [DOI] [PubMed] [Google Scholar]
- 47.Jackson M., Ma R., Taylor A.H., Axton R.A., Easterbrook J., Kydonaki M., Olivier E., Marenah L., Stanley E.G., Elefanty A.G., Mountford J.C., Forrester L.M. Enforced expression of hoxb4 in human embryonic stem cells enhances the production of hematopoietic progenitors but has no effect on the maturation of red blood cells. Stem Cells Transl. Med. 2016;5(8):981–990. doi: 10.5966/sctm.2015-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amabile G., Welner R.S., Nombela-Arrieta C., D’Alise A.M., Di Ruscio A., Ebralidze A.K., Kraytsberg Y., Ye M., Kocher O., Neuberg D.S., Khrapko K., Silberstein L.E., Tenen D.G. In vivo generation of transplantable human hematopoietic cells from induced pluripotent stem cells. Blood. 2013;121(8):1255–1264. doi: 10.1182/blood-2012-06-434407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toscano M.G., Navarro-Montero O., Ayllon V., Ramos-Mejia V., Guerrero-Carreno X., Bueno C., Romero T., Lamolda M., Cobo M., Martin F., Menendez P., Real P.J. SCL/TAL1-mediated transcriptional network enhances megakaryocytic specification of human embryonic stem cells. Mol. Ther. 2015;23(1):158–170. doi: 10.1038/mt.2014.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dalby A., Ballester-Beltrán J., Lincetto C., Mueller A., Foad N., Evans A., Baye J., Turro E., Moreau T., Tijssen M.R., Ghevaert C. Transcription factor levels after forward programming of human pluripotent stem cells with GATA1, FLI1, and TAL1 determine megakaryocyte versus erythroid cell fate decision. Stem Cell Reports. 2018;11(6):1462–1478. doi: 10.1016/j.stemcr.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amélie Montel-Hagen, C.S.S, Li, Suwen, S.L, Brent, C Yuhua, Z.; Patrick, C.; Steven, T.; Victoria, S.; Shawn, L.; Ho-Chung, C.; Chongbin, H.; Chee, J.C.; David, C.; Gay, M.C. Organoid-induced differentiation of conventional T cells from human pluripotent stem cells. Cell Stem Cell. 2019 doi: 10.1016/j.stem.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan Y.T., Ye L., Xie F., Beyer A.I., Muench M.O., Wang J., Chen Z., Liu H., Chen S.J., Kan Y.W. Respecifying human iPSC-derived blood cells into highly engraftable hematopoietic stem and progenitor cells with a single factor. Proc. Natl. Acad. Sci. USA. 2018;115(9):2180–2185. doi: 10.1073/pnas.1718446115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saxena S. Efficient production of human hematopoietic cells from pluripotent stem cells through cAMP induction. 2017 doi: 10.1038/protex.2017.012. [DOI] [Google Scholar]
- 54.Sweeney C.L., Teng R., Wang H., Merling R.K., Lee J., Choi U., Koontz S., Wright D.G., Malech H.L. Molecular analysis of neutrophil differentiation from human induced pluripotent stem cells delineates the kinetics of key regulators of hematopoiesis. Stem Cells. 2016;34(6):1513–1526. doi: 10.1002/stem.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Espinoza J.L., Elbadry M.I., Chonabayashi K., Yoshida Y., Katagiri T., Harada K., Nakagawa N., Zaimoku Y., Imi T., Takamatsu H., Ozawa T., Maruyama H., Hassanein H.A., Khalifa A Noreldin A., Takenaka K., Akashi K., Hamana H., Kishi H., Akatsuka Y., Nakao S. Hematopoiesis by iPSC-derived hematopoietic stem cells of aplastic anemia that escape cytotoxic T-cell attack. Blood Adv. 2018;2(4):390–400. doi: 10.1182/bloodadvances.2017013342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collins E.M., Thompson A. HOX genes in normal, engineered and malignant hematopoiesis. Int. J. Dev. Biol. 2018;62(11-12):847–856. doi: 10.1387/ijdb.180206at. [DOI] [PubMed] [Google Scholar]
- 57.Chanda B., Ditadi A., Iscove N.N., Keller G. Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell. 2013;155(1):215–227. doi: 10.1016/j.cell.2013.08.055. [DOI] [PubMed] [Google Scholar]
- 58.Cabezas-Wallscheid N. Vitamin A-retinoic acid signaling regulates hematopoietic stem cell dormancy. Cell. 2017;169(5):807. doi: 10.1016/j.cell.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 59.Kashyap V., Gudas L.J., Brenet F., Funk P., Viale A., Scandura J.M. Epigenomic reorganization of the clustered Hox genes in embryonic stem cells induced by retinoic acid. J. Biol. Chem. 2011;286(5):3250–3260. doi: 10.1074/jbc.M110.157545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kyba M., Perlingeiro R.C., Daley G.Q. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109(1):29–37. doi: 10.1016/S0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 61.Wang L., Menendez P., Shojaei F., Li L., Mazurier F., Dick J.E., Cerdan C., Levac K., Bhatia M. Generation of hematopoietic repopulating cells from human embryonic stem cells independent of ectopic HOXB4 expression. J. Exp. Med. 2005;201(10):1603–1614. doi: 10.1084/jem.20041888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schiedlmeier B., Santos A.C., Ribeiro A., Moncaut N., Lesinski D., Auer H., Kornacker K., Ostertag W., Baum C., Mallo M., Klump H. HOXB4's road map to stem cell expansion. Proc. Natl. Acad. Sci. USA. 2007;104(43):16952–16957. doi: 10.1073/pnas.0703082104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forrester L.M., Jackson M. Mechanism of action of HOXB4 on the hematopoietic differentiation of embryonic stem cells. Stem Cells. 2012;30(3):379–385. doi: 10.1002/stem.1036. [DOI] [PubMed] [Google Scholar]
- 64.Oshima M., Endoh M., Endo T.A., Toyoda T., Nakajima-Takagi Y., Sugiyama F., Koseki H., Kyba M., Iwama A., Osawa M. Genome-wide analysis of target genes regulated by HoxB4 in hematopoietic stem and progenitor cells developing from embryonic stem cells. Blood. 2011;117(15):e142–e150. doi: 10.1182/blood-2010-12-323212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fan R., Bonde S., Gao P., Sotomayor B., Chen C., Mouw T., Zavazava N., Tan K. Dynamic HoxB4-regulatory network during embryonic stem cell differentiation to hematopoietic cells. Blood. 2012;119(19):e139–e147. doi: 10.1182/blood-2011-12-396754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seifert A., Werheid D.F., Knapp S.M., Tobiasch E. Role of Hox genes in stem cell differentiation. World J. Stem Cells. 2015;7(3):583–595. doi: 10.4252/wjsc.v7.i3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zardo G., Cimino G., Nervi C. Epigenetic plasticity of chromatin in embryonic and hematopoietic stem/progenitor cells: Therapeutic potential of cell reprogramming. Leukemia. 2008;22(8):1503–1518. doi: 10.1038/leu.2008.141. [DOI] [PubMed] [Google Scholar]
- 68.Lawrence H.J., Helgason C.D., Sauvageau G., Fong S., Izon D.J., Humphries R.K., Largman C. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997;89(6):1922–1930. [PubMed] [Google Scholar]
- 69.Shojaei F., Menendez P. Molecular profiling of candidate human hematopoietic stem cells derived from human embryonic stem cells. Exp. Hematol. 2008;36(11):1436–1448. doi: 10.1016/j.exphem.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 70.Ramos-Mejía V., Navarro-Montero O., Ayllón V., Bueno C., Romero T., Real P.J., Menendez P. HOXA9 promotes hematopoietic commitment of human embryonic stem cells. Blood. 2014;124(20):3065–3075. doi: 10.1182/blood-2014-03-558825. [DOI] [PubMed] [Google Scholar]
- 71.Dou D.R., Calvanese V., Sierra M.I., Nguyen A.T., Minasian A., Saarikoski P., Sasidharan R., Ramirez C.M., Zack J.A., Crooks G.M., Galic Z., Mikkola H.K. Medial HOXA genes demarcate haematopoietic stem cell fate during human development. Nat. Cell Biol. 2016;18(6):595–606. doi: 10.1038/ncb3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He H., Hua X., Yan J. Epigenetic regulations in hematopoietic Hox code. Oncogene. 2011;30(4):379–388. doi: 10.1038/onc.2010.484. [DOI] [PubMed] [Google Scholar]
- 73.Bueno C., Montes R., Melen G.J., Ramos-Mejia V., Real P.J., Ayllón V., Sanchez L., Ligero G., Gutierrez-Aranda I., Fernández A.F., Fraga M.F., Moreno-Gimeno I., Burks D. Plaza-Calonge, Mdel.C.; Rodríguez-Manzaneque, J.C.; Menendez, P. A human ESC model for MLL-AF4 leukemic fusion gene reveals an impaired early hematopoietic-endothelial specification. Cell Res. 2012;22(6):986–1002. doi: 10.1038/cr.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chung N., Jee B.K., Chae S.W., Jeon Y.W., Lee K.H., Rha H.K. HOX gene analysis of endothelial cell differentiation in human bone marrow-derived mesenchymal stem cells. Mol. Biol. Rep. 2009;36(2):227–235. doi: 10.1007/s11033-007-9171-6. [DOI] [PubMed] [Google Scholar]
- 75.Slany R.K. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94(7):984–993. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Erfurth F.E., Popovic R., Grembecka J., Cierpicki T., Theisler C., Xia Z.B., Stuart T., Diaz M.O., Bushweller J.H., Zeleznik-Le N.J. MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression. Proc. Natl. Acad. Sci. USA. 2008;105(21):7517–7522. doi: 10.1073/pnas.0800090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alvarez-Dominguez J.R., Lodish H.F. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood. 2017;130(18):1965–1975. doi: 10.1182/blood-2017-06-788695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Kumar B., Parrish M.E., Slaughter B.D., Unruh J.R., Gogol M., Seidel C., Paulson A., Li H., Gaudenz K., Peak A., McDowell W., Fleharty B., Ahn Y., Lin C., Smith E., Shilatifard A., Krumlauf R. Analysis of dynamic changes in retinoid-induced transcription and epigenetic profiles of murine Hox clusters in ES cells. Genome Res. 2015;25(8):1229–1243. doi: 10.1101/gr.184978.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Z.H., Wang W.T., Huang W., Fang K., Sun Y.M., Liu S.R., Luo X.Q., Chen Y.Q. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017;24(2):212–224. doi: 10.1038/cdd.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhatlekar S., Fields J.Z., Boman B.M. Role of HOX genes in stem cell differentiation and cancer. Stem Cells Int. 2018;2018:3569493. doi: 10.1155/2018/3569493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang X., Lian Z., Padden C., Gerstein M.B., Rozowsky J., Snyder M., Gingeras T.R., Kapranov P., Weissman S.M., Newburger P.E. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113(11):2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A., Wysocka J., Lei M., Dekker J., Helms J.A., Chang H.Y. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weiss M.J., Orkin S.H. GATA transcription factors: Key regulators of hematopoiesis. Exp. Hematol. 1995;23(2):99–107. [PubMed] [Google Scholar]
- 84.Gutiérrez L., Nikolic T., van Dijk T.B., Hammad H., Vos N., Willart M., Grosveld F., Philipsen S., Lambrecht B.N. Gata1 regulates dendritic-cell development and survival. Blood. 2007;110(6):1933–1941. doi: 10.1182/blood-2006-09-048322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moriguchi T., Yamamoto M. A regulatory network governing Gata1 and Gata2 gene transcription orchestrates erythroid lineage differentiation. Int. J. Hematol. 2014;100(5):417–424. doi: 10.1007/s12185-014-1568-0. [DOI] [PubMed] [Google Scholar]
- 86.Lugus J.J., Chung Y.S., Mills J.C., Kim S.I., Grass J., Kyba M., Doherty J.M., Bresnick E.H., Choi K. GATA2 functions at multiple steps in hemangioblast development and differentiation. Development. 2007;134(2):393–405. doi: 10.1242/dev.02731. [DOI] [PubMed] [Google Scholar]
- 87.Hosoya T., Maillard I., Engel J.D. From the cradle to the grave: Activities of GATA-3 throughout T-cell development and differentiation. Immunol. Rev. 2010;238(1):110–125. doi: 10.1111/j.1600-065X.2010.00954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pevny L., Lin C.S., D’Agati V., Simon M.C., Orkin S.H., Costantini F. Development of hematopoietic cells lacking transcription factor GATA-1. Development. 1995;121(1):163–172. doi: 10.1242/dev.121.1.163. [DOI] [PubMed] [Google Scholar]
- 89.Huang K., Du J., Ma N., Liu J., Wu P., Dong X., Meng M., Wang W., Chen X., Shi X., Chen Q., Yang Z., Chen S., Zhang J., Li Y., Li W., Zheng Y., Cai J., Li P., Sun X., Wang J., Pei D., Pan G. GATA2(-/-) human ESCs undergo attenuated endothelial to hematopoietic transition and thereafter granulocyte commitment. Cell Regen. (Lond.) 2015;4(1):4. doi: 10.1186/s13619-015-0018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Frelin C., Herrington R., Janmohamed S., Barbara M., Tran G., Paige C.J., Benveniste P., Zuñiga-Pflücker J.C., Souabni A., Busslinger M., Iscove N.N. GATA-3 regulates the self-renewal of long-term hematopoietic stem cells. Nat. Immunol. 2013;14(10):1037–1044. doi: 10.1038/ni.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lim K.C., Hosoya T., Brandt W., Ku C.J., Hosoya-Ohmura S., Camper S.A., Yamamoto M., Engel J.D. Conditional Gata2 inactivation results in HSC loss and lymphatic mispatterning. J. Clin. Invest. 2012;122(10):3705–3717. doi: 10.1172/JCI61619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ohmori S., Moriguchi T., Noguchi Y., Ikeda M., Kobayashi K., Tomaru N., Ishijima Y., Ohneda O., Yamamoto M., Ohneda K. GATA2 is critical for the maintenance of cellular identity in differentiated mast cells derived from mouse bone marrow. Blood. 2015;125(21):3306–3315. doi: 10.1182/blood-2014-11-612465. [DOI] [PubMed] [Google Scholar]
- 93.Fujiwara T., O’Geen H., Keles S., Blahnik K., Linnemann A.K., Kang Y.A., Choi K., Farnham P.J., Bresnick E.H. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell. 2009;36(4):667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grass J.A., Boyer M.E., Pal S., Wu J., Weiss M.J., Bresnick E.H. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl. Acad. Sci. USA. 2003;100(15):8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kang H., Mesquitta W.T., Jung H.S., Moskvin O.V., Thomson J.A., Slukvin I.I. GATA2 is dispensable for specification of hemogenic endothelium but promotes endothelial-to-hematopoietic transition. Stem Cell Reports. 2018;11(1):197–211. doi: 10.1016/j.stemcr.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kaneko H., Kobayashi E., Yamamoto M., Shimizu R. N- and C-terminal transactivation domains of GATA1 protein coordinate hematopoietic program. J. Biol. Chem. 2012;287(25):21439–21449. doi: 10.1074/jbc.M112.370437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Matsunaga H., Sasaki S., Suzuki S., Matsushita A., Nakamura H., Nakamura H.M., Hirahara N., Kuroda G., Iwaki H., Ohba K., Morita H., Oki Y., Suda T. Essential role of GATA2 in the negative regulation of type 2 deiodinase gene by liganded thyroid hormone receptor β2 in thyrotroph. PLoS One. 2015;10(11):e0142400. doi: 10.1371/journal.pone.0142400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tsuzuki S., Kitajima K., Nakano T., Glasow A., Zelent A., Enver T. Cross talk between retinoic acid signaling and transcription factor GATA-2. Mol. Cell. Biol. 2004;24(15):6824–6836. doi: 10.1128/MCB.24.15.6824-6836.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilson N.K., Foster S.D., Wang X., Knezevic K., Schütte J., Kaimakis P., Chilarska P.M., Kinston S., Ouwehand W.H., Dzierzak E., Pimanda J.E., de Bruijn M.F., Göttgens B. Combinatorial transcriptional control in blood stem/progenitor cells: Genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7(4):532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 100.Pal S., Cantor A.B., Johnson K.D., Moran T.B., Boyer M.E., Orkin S.H., Bresnick E.H. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc. Natl. Acad. Sci. USA. 2004;101(4):980–985. doi: 10.1073/pnas.0307612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Katsumura K.R., Bresnick E.H., Group G.F.M. The GATA factor revolution in hematology. Blood. 2017;129(15):2092–2102. doi: 10.1182/blood-2016-09-687871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hong W., Nakazawa M., Chen Y.Y., Kori R., Vakoc C.R., Rakowski C., Blobel G.A. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 2005;24(13):2367–2378. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lamonica J.M., Vakoc C.R., Blobel G.A. Acetylation of GATA-1 is required for chromatin occupancy. Blood. 2006;108(12):3736–3738. doi: 10.1182/blood-2006-07-032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mancini E., Sanjuan-Pla A., Luciani L., Moore S., Grover A., Zay A., Rasmussen K.D., Luc S., Bilbao D., O’Carroll D., Jacobsen S.E., Nerlov C. FOG-1 and GATA-1 act sequentially to specify definitive megakaryocytic and erythroid progenitors. EMBO J. 2012;31(2):351–365. doi: 10.1038/emboj.2011.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Turkistany S.A., DeKoter R.P. The transcription factor PU.1 is a critical regulator of cellular communication in the immune system. Arch. Immunol. Ther. Exp. (Warsz.) 2011;59(6):431–440. doi: 10.1007/s00005-011-0147-9. [DOI] [PubMed] [Google Scholar]
- 106.Burda P., Vargova J., Curik N., Salek C., Papadopoulos G.L., Strouboulis J., Stopka T. GATA-1 Inhibits PU.1 Gene via DNA and histone H3K9 methylation of its distal enhancer in erythroleukemia. PLoS One. 2016;11(3):e0152234. doi: 10.1371/journal.pone.0152234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang Y., Li W., Laurent T., Ding S. Small molecules, big roles - the chemical manipulation of stem cell fate and somatic cell reprogramming. J. Cell Sci. 2012;125(Pt 23):5609–5620. doi: 10.1242/jcs.096032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Okuno Y., Huang G., Rosenbauer F., Evans E.K., Radomska H.S., Iwasaki H., Akashi K., Moreau-Gachelin F., Li Y., Zhang P., Göttgens B., Tenen D.G. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol. Cell. Biol. 2005;25(7):2832–2845. doi: 10.1128/MCB.25.7.2832-2845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rosenbauer F., Owens B.M., Yu L., Tumang J.R., Steidl U., Kutok J.L., Clayton L.K., Wagner K., Scheller M., Iwasaki H., Liu C., Hackanson B., Akashi K., Leutz A., Rothstein T.L., Plass C., Tenen D.G. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat. Genet. 2006;38(1):27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- 110.Burda P., Laslo P., Stopka T. The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis. Leukemia. 2010;24(7):1249–1257. doi: 10.1038/leu.2010.104. [DOI] [PubMed] [Google Scholar]
- 111.Tripic T., Deng W., Cheng Y., Zhang Y., Vakoc C.R., Gregory G.D., Hardison R.C., Blobel G.A. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2009;113(10):2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rodriguez P., Bonte E., Krijgsveld J., Kolodziej K.E., Guyot B., Heck A.J., Vyas P., de Boer E., Grosveld F., Strouboulis J. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 2005;24(13):2354–2366. doi: 10.1038/sj.emboj.7600702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Porcher C., Chagraoui H., Kristiansen M.S. SCL/TAL1: A multifaceted regulator from blood development to disease. Blood. 2017;129(15):2051–2060. doi: 10.1182/blood-2016-12-754051. [DOI] [PubMed] [Google Scholar]
- 114.Wu W., Morrissey C.S., Keller C.A., Mishra T., Pimkin M., Blobel G.A., Weiss M.J., Hardison R.C. Dynamic shifts in occupancy by TAL1 are guided by GATA factors and drive large-scale reprogramming of gene expression during hematopoiesis. Genome Res. 2014;24(12):1945–1962. doi: 10.1101/gr.164830.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hoang T., Lambert J.A., Martin R. SCL/TAL1 in hematopoiesis and cellular reprogramming. Curr. Top. Dev. Biol. 2016;118:163–204. doi: 10.1016/bs.ctdb.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 116.Gomes A.M. Cooperative transcription factor induction mediates hemogenic reprogramming. Cell Rep. 2018;25(10):2821–2835.:e7. doi: 10.1016/j.celrep.2018.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Malinge S., Thiollier C., Chlon T.M., Doré L.C., Diebold L., Bluteau O., Mabialah V., Vainchenker W., Dessen P., Winandy S., Mercher T., Crispino J.D. Ikaros inhibits megakaryopoiesis through functional interaction with GATA-1 and NOTCH signaling. Blood. 2013;121(13):2440–2451. doi: 10.1182/blood-2012-08-450627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rodrigues N.P., Tipping A.J., Wang Z., Enver T. GATA-2 mediated regulation of normal hematopoietic stem/progenitor cell function, myelodysplasia and myeloid leukemia. Int. J. Biochem. Cell Biol. 2012;44(3):457–460. doi: 10.1016/j.biocel.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bartel D.P. Metazoan microRNAs. Cell. 2018;173(1):20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rodriguez A., Griffiths-Jones S., Ashurst J.L., Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li Z., Huang H., Chen P., He M., Li Y., Arnovitz S., Jiang X., He C., Hyjek E., Zhang J., Zhang Z., Elkahloun A., Cao D., Shen C., Wunderlich M., Wang Y., Neilly M.B., Jin J., Wei M., Lu J., Valk P.J., Delwel R., Lowenberg B., Le Beau M.M., Vardiman J., Mulloy J.C., Zeleznik-Le N.J., Liu P.P., Zhang J., Chen J. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat. Commun. 2012;3:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gerrits A., Walasek M.A., Olthof S., Weersing E., Ritsema M., Zwart E., van Os R., Bystrykh L.V., de Haan G. Genetic screen identifies microRNA cluster 99b/let-7e/125a as a regulator of primitive hematopoietic cells. Blood. 2012;119(2):377–387. doi: 10.1182/blood-2011-01-331686. [DOI] [PubMed] [Google Scholar]
- 123.Ooi A.G., Sahoo D., Adorno M., Wang Y., Weissman I.L., Park C.Y. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc. Natl. Acad. Sci. USA. 2010;107(50):21505–21510. doi: 10.1073/pnas.1016218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yekta S., Shih I.H., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 125.Velu C.S., Baktula A.M., Grimes H.L. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. 2009;113(19):4720–4728. doi: 10.1182/blood-2008-11-190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao H., Kalota A., Jin S., Gewirtz A.M. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood. 2009;113(3):505–516. doi: 10.1182/blood-2008-01-136218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dore L.C., Amigo J.D., Dos Santos C.O., Zhang Z., Gai X., Tobias J.W., Yu D., Klein A.M., Dorman C., Wu W., Hardison R.C., Paw B.H., Weiss M.J.A. GATA-1-regulated microRNA locus essential for erythropoiesis. Proc. Natl. Acad. Sci. USA. 2008;105(9):3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kurkewich J.L., Hansen J., Klopfenstein N., Zhang H., Wood C., Boucher A., Hickman J., Muench D.E., Grimes H.L., Dahl R. The miR-23a~27a~24-2 microRNA cluster buffers transcription and signaling pathways during hematopoiesis. PLoS Genet. 2017;13(7):e1006887. doi: 10.1371/journal.pgen.1006887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang F., Zhu Y., Guo L., Dong L., Liu H., Yin H., Zhang Z., Li Y., Liu C., Ma Y., Song W., He A., Wang Q., Wang L., Zhang J., Li J., Yu J. A regulatory circuit comprising GATA1/2 switch and microRNA-27a/24 promotes erythropoiesis. Nucleic Acids Res. 2014;42(1):442–457. doi: 10.1093/nar/gkt848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Metcalf D. Hematopoietic cytokines. Blood. 2008;111(2):485–491. doi: 10.1182/blood-2007-03-079681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Felli N., Fontana L., Pelosi E., Botta R., Bonci D., Facchiano F., Liuzzi F., Lulli V., Morsilli O., Santoro S., Valtieri M., Calin G.A., Liu C.G., Sorrentino A., Croce C.M., Peschle C. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc. Natl. Acad. Sci. USA. 2005;102(50):18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lennartsson J., Rönnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol. Rev. 2012;92(4):1619–1649. doi: 10.1152/physrev.00046.2011. [DOI] [PubMed] [Google Scholar]
- 133.Lee J.Y., Kim M., Heo H.R., Ha K.S., Han E.T., Park W.S., Yang S.R., Hong S.H. Inhibition of MicroRNA-221 and 222 enhances hematopoietic differentiation from human pluripotent stem cells via c-KIT upregulation. Mol. Cells. 2018;41(11):971–978. doi: 10.14348/molcells.2018.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chadwick K., Wang L., Li L., Menendez P., Murdoch B., Rouleau A., Bhatia M. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood. 2003;102(3):906–915. doi: 10.1182/blood-2003-03-0832. [DOI] [PubMed] [Google Scholar]
- 135.Marshall C.J., Kinnon C., Thrasher A.J. Polarized expression of bone morphogenetic protein-4 in the human aorta-gonad-mesonephros region. Blood. 2000;96(4):1591–1593. [PubMed] [Google Scholar]
- 136.Sumi T., Tsuneyoshi N., Nakatsuji N., Suemori H. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development. 2008;135(17):2969–2979. doi: 10.1242/dev.021121. [DOI] [PubMed] [Google Scholar]
- 137.Verheyen E.M. Opposing effects of Wnt and MAPK on BMP/Smad signal duration. Dev. Cell. 2007;13(6):755–756. doi: 10.1016/j.devcel.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 138.Lengerke C., Schmitt S., Bowman T.V., Jang I.H., Maouche-Chretien L., McKinney-Freeman S., Davidson A.J., Hammerschmidt M., Rentzsch F., Green J.B., Zon L.I., Daley G.Q. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell. 2008;2(1):72–82. doi: 10.1016/j.stem.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 139.Nostro M.C., Cheng X., Keller G.M., Gadue P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell. 2008;2(1):60–71. doi: 10.1016/j.stem.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tran F.H., Zheng J.J. Modulating the wnt signaling pathway with small molecules. Protein Sci. 2017;26(4):650–661. doi: 10.1002/pro.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sturgeon C.M., Ditadi A., Awong G., Kennedy M., Keller G. Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat. Biotechnol. 2014;32(6):554–561. doi: 10.1038/nbt.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chanda B., Ditadi A., Iscove N.N., Keller G. Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell. 2013;155(1):215–227. doi: 10.1016/j.cell.2013.08.055. [DOI] [PubMed] [Google Scholar]
- 143.Rice K.L., Hormaeche I., Licht J.D. Epigenetic regulation of normal and malignant hematopoiesis. Oncogene. 2007;26(47):6697–6714. doi: 10.1038/sj.onc.1210755. [DOI] [PubMed] [Google Scholar]
- 144.Herviou L., Cavalli G., Cartron G., Klein B., Moreaux J. EZH2 in normal hematopoiesis and hematological malignancies. Oncotarget. 2016;7(3):2284–2296. doi: 10.18632/oncotarget.6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hu D., Shilatifard A. Epigenetics of hematopoiesis and hematological malignancies. Genes Dev. 2016;30(18):2021–2041. doi: 10.1101/gad.284109.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yan J., Ng S.B., Tay J.L., Lin B., Koh T.L., Tan J., Selvarajan V., Liu S.C., Bi C., Wang S., Choo S.N., Shimizu N., Huang G., Yu Q., Chng W.J. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood. 2013;121(22):4512–4520. doi: 10.1182/blood-2012-08-450494. [DOI] [PubMed] [Google Scholar]
- 147.Yamaguchi H., Hung M.C. Regulation and Role of EZH2 in Cancer. Cancer Res. Treat. 2014;46(3):209–222. doi: 10.4143/crt.2014.46.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Asangani I.A., Ateeq B., Cao Q., Dodson L., Pandhi M., Kunju L.P., Mehra R., Lonigro R.J., Siddiqui J., Palanisamy N., Wu Y.M., Cao X., Kim J.H., Zhao M., Qin Z.S., Iyer M.K., Maher C.A., Kumar-Sinha C., Varambally S., Chinnaiyan A.M. Characterization of the EZH2-MMSET histone methyltransferase regulatory axis in cancer. Mol. Cell. 2013;49(1):80–93. doi: 10.1016/j.molcel.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nishizawa M., Chonabayashi K., Nomura M., Tanaka A., Nakamura M., Inagaki A., Nishikawa M., Takei I., Oishi A., Tanabe K., Ohnuki M., Yokota H., Koyanagi-Aoi M., Okita K., Watanabe A., Takaori-Kondo A., Yamanaka S., Yoshida Y. Epigenetic variation between human induced pluripotent stem cell lines is an indicator of differentiation capacity. Cell Stem Cell. 2016;19(3):341–354. doi: 10.1016/j.stem.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 150.Cypris O., Frobel J., Rai S., Franzen J., Sontag S., Goetzke R., Szymanski de Toledo M.A., Zenke M., Wagner W. Tracking of epigenetic changes during hematopoietic differentiation of induced pluripotent stem cells. Clin. Epigenetics. 2019;11(1):19. doi: 10.1186/s13148-019-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Truong V.A., Hsu M.N., Kieu Nguyen N.T., Lin M.W., Shen C.C., Lin C.Y., Hu Y.C. CRISPRai for simultaneous gene activation and inhibition to promote stem cell chondrogenesis and calvarial bone regeneration. Nucleic Acids Res. 2019;47(13):e74. doi: 10.1093/nar/gkz267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li M., Suzuki K., Kim N.Y., Liu G.H., Izpisua Belmonte J.C. A cut above the rest: Targeted genome editing technologies in human pluripotent stem cells. J. Biol. Chem. 2014;289(8):4594–4599. doi: 10.1074/jbc.R113.488247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li M., Suzuki K., Qu J., Saini P., Dubova I., Yi F., Lee J., Sancho-Martinez I., Liu G.H., Izpisua Belmonte J.C. Efficient correction of hemoglobinopathy-causing mutations by homologous recombination in integration-free patient iPSCs. Cell Res. 2011;21(12):1740–1744. doi: 10.1038/cr.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Suzuki K., Yu C., Qu J., Li M., Yao X., Yuan T., Goebl A., Tang S., Ren R., Aizawa E., Zhang F., Xu X., Soligalla R.D., Chen F., Kim J., Kim N.Y., Liao H.K., Benner C., Esteban C.R., Jin Y., Liu G.H., Li Y., Izpisua Belmonte J.C. Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell. 2014;15(1):31–36. doi: 10.1016/j.stem.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu G.H., Suzuki K., Li M., Qu J., Montserrat N., Tarantino C., Gu Y., Yi F., Xu X., Zhang W., Ruiz S., Plongthongkum N., Zhang K., Masuda S., Nivet E., Tsunekawa Y., Soligalla R.D., Goebl A., Aizawa E., Kim N.Y., Kim J., Dubova I., Li Y., Ren R., Benner C., Del Sol A., Bueren J., Trujillo J.P., Surralles J., Cappelli E., Dufour C., Esteban C.R., Belmonte J.C.I. Modelling Fanconi anemia pathogenesis and therapeutics using integration-free patient-derived iPSCs. Nat. Commun. 2014;5:4330. doi: 10.1038/ncomms5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Castano J., Aranda S., Bueno C., Calero-Nieto F.J., Mejia-Ramirez E., Mosquera J.L. GATA2 promotes hematopoietic development and represses cardiac differentiation of human mesoderm. Stem Cell Reports. 13(3):515–529. doi: 10.1016/j.stemcr.2019.07.009. S2213- 6711(19)30261-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]