Abstract
Aging is a predominant risk factor for numerous chronic diseases that limit healthspan1. Mechanisms of aging are thus increasingly recognized as potential therapeutic targets. Blood from young mice reverses aspects of aging and disease across multiple tissues2–10, which supports a hypothesis that age-related molecular changes in blood could provide novel insights into age-related disease biology. We measured 2,925 plasma proteins from 4,263 young adults to nonagenarians (18–95 years old) and developed a novel bioinformatics approach, which uncovered marked non-linear alterations in the human plasma proteome with age. Waves of changes in the proteome in the fourth, seventh, and eighth decades of life reflected distinct biological pathways and revealed differential associations with the genome and proteome of age-related diseases and phenotypic traits. This new approach to the study of aging led to the identification of unexpected signatures and pathways, which might offer potential targets for age-related diseases.
Aging underlies declining organ function and is the primary risk factor for numerous diseases1. Thus, a deeper understanding of aging is likely to provide insight into mechanisms of disease and to facilitate the development of novel anti-aging therapeutics. A growing number of investigators have applied genomic, transcriptomic and proteomic assays (collectively referred to as omics) to studies of aging11. Human genetic studies have uncovered relatively few modifiers of aging, yet other omics modalities, which measure more dynamic gene modifications or products, have provided valuable insight. For example, the transcriptome varies greatly during aging across tissues and organisms12, pointing to evolutionarily conserved, fundamental roles of developmental and inflammatory pathways13. Protein composition of cells, bodily fluids, and tissues change similarly with age and provide insight into complex biological processes since proteins are often direct regulators of cellular pathways. In particular, blood, which contains proteins from nearly every cell and tissue, has been analyzed to discover biomarkers and gain insight into disease biology. Accordingly, organismal aging results in proteomic changes in blood that reflect aspects of aging of different cell types and tissues.
Perhaps the strongest evidence that blood can be used to study aging comes from experiments employing heterochronic parabiosis, a surgically induced state which connects the circulatory systems of young and old mice. These studies show that multiple tissues, including muscle, liver, heart, pancreas, kidney, bone, and brain can be rejuvenated in old mice2–10. Plasma (the soluble fraction of blood) from old mice is sufficient to accelerate brain aging following infusion into young mice9 and young plasma can reverse aspects of brain aging10,14. Together, these studies support the notion that the plasma proteome harbors key regulators of aging. Identifying such protein signatures may help understand mechanisms of organismal aging. However, plasma proteomic changes with age have not been thoroughly exploited and require novel tools to derive insight into the biology of aging. Here, we carried out a deep proteomic analysis of plasma from young adults to nonagenarians. Using novel analysis tools, we discovered changes in protein expression across the lifespan and link these changes to biological pathways and disease.
Results
Linear modeling links the plasma proteome to functional aging and identifies a conserved aging signature
We analyzed plasma isolated from ethylenediaminetetraacetic acid (EDTA)-treated blood acquired via venipuncture from 4,263 healthy individuals aged 18–95 years from the INTERVAL15 and LonGenity16 cohorts (Fig. 1a, Extended Data 1. Currently, one of the most advanced tools for the measurement of plasma proteins is the single‐stranded oligonucleotide known as aptamer17,18, which binds its target with high affinity and specificity. To generate a proteomic dataset of the human lifespan, we used the SomaScan aptamer technology, capable of quantifying thousands of proteins (Supplementary Tables 1 and 2) with high precision within and between runs19 (Supplementary Table 3). The INTERVAL and LonGenity datasets analyzed here can be interrogated with an interactive web interface (https://twc-stanford.shinyapps.io/aging_plasma_proteome/).
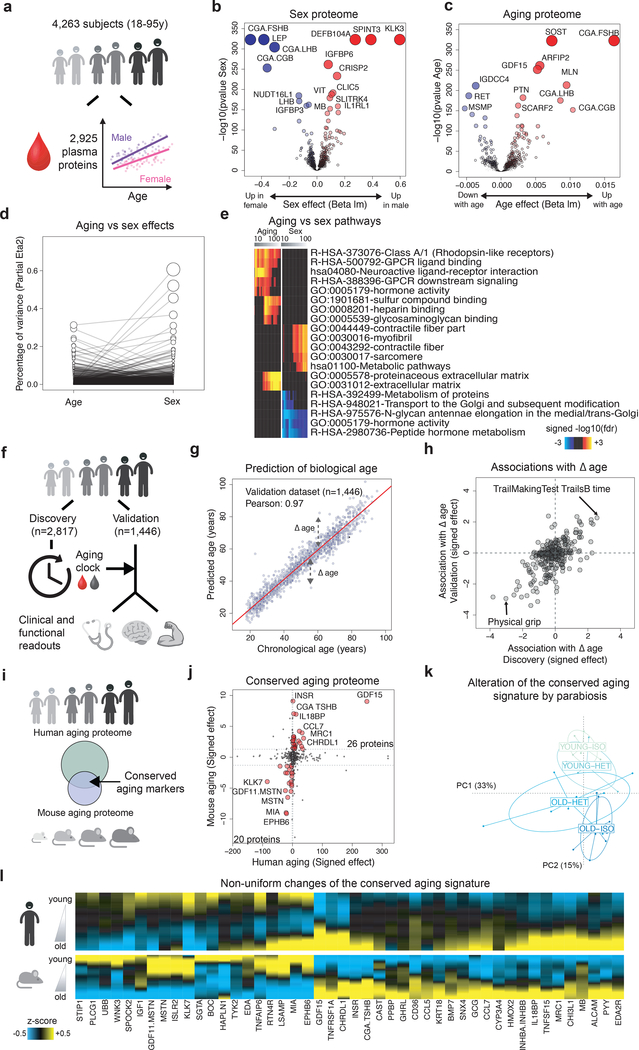
Figure 1: Linear modeling links the plasma proteome to functional aging and identifies a conserved aging signature.
(a) Schematic representation of analysis of the plasma proteome.
Volcano plots representing changes of the plasma proteome (n=4,263) with sex (b) and age (c). Linear models, adjusted for age, sex and subcohort were tested using F-test.
(d) Relative percentage of variance explained by age and sex. Values for each plasma protein are connected by edges.
(e) Pathways associated with sex and age identified by Sliding Enrichment Pathway Analysis (SEPA, n=4,263). Proteins upregulated and downregulated are analyzed separately. The top 10 pathways per condition are represented. Enrichment was tested using Fisher’s exact test (GO) and hypergeometric test (Reactome and KEGG).
(f) Schematic representation of the of biological age modeling using the plasma proteome.
(g) Prediction of age in the validation cohort (n=1,446) using 373 plasma proteins. Pearson correlation coefficient between chronological and predicted age is indicated.
(h) Association between delta age (difference between chronological age and chronological age) and functional readouts in old. Top associations in both Discovery and Validation datasets are represented.
(i) Schematic representation of the comparison between the human and mouse aging proteomes.
(j) Conserved markers of aging. Both human and mouse aging effects are signed by the beta age of their corresponding linear analysis. Forty-six plasma proteins are changing in the same direction in mouse and humans (red dots) and define a conserved aging signature.
(k) Alteration of the conserved aging signature by parabiosis. Normed principal component analysis was used to characterize changes of the conserved aging signature when mice are exposed to young or old blood.
(l) Age-related changes of the conserved aging signature. Plasma protein levels were z-scored and aging trajectories were estimated by locally estimated scatterplot smoothing.
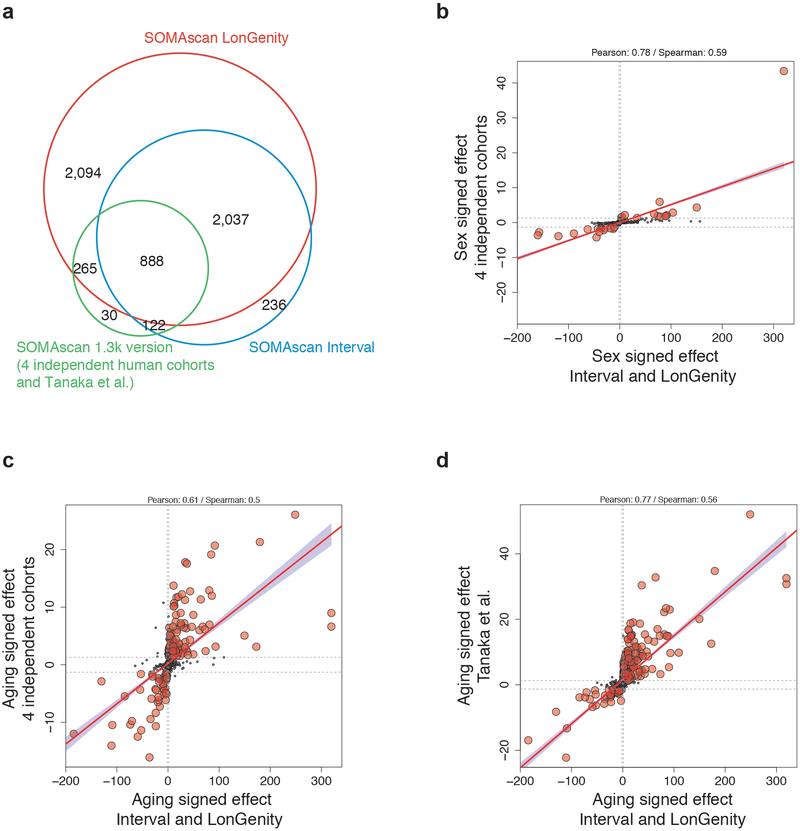
Since females have a longer average lifespan than males20, we assessed whether sex and aging proteomes are interconnected (Fig 1b–1d). Proteins most strongly changed with sex included well-known follicle stimulating hormone (CGA FSHB), human chorionic gonadotropin (CGA CGB), and prostate-specific antigen (KLK3). With age, the most prominent overall changes with respect to fold change and statistical significance included sclerostin (SOST), ADP Ribosylation Factor Interacting Protein 2 (ARFIP2), and growth differentiation factor 15 (GDF15), in addition to several sex proteins such as CGA FSHB. The proteins most strongly associated with age also changed significantly with sex, (Fig. 1d); 895 proteins out of the 1,379 proteins altered with age were significantly different between sexes (q<0.05, Supplementary Table 4). These results are aligned with a number of studies demonstrating that males and females age differently21. To determine whether these findings are representative of the general population, we compared changes identified in this study with findings from 4 independent cohorts from the US and Europe (n=171, age range 21–107y, Extended Data 1d) and with an independent study22. Although these independent cohorts used an older version of the SomaScan assay measuring only a subset of the current proteins (1,305 proteins; Supplementary Table 2), we observed high consistency of the aging and sex proteomes across cohorts (Extended Data 2).
To establish the biological relevance of these changes, we queried GO, KEGG, and Reactome databases and measured enrichment of proteins in pathways using a “Sliding Enrichment Pathway Analysis (SEPA)” (Supplementary Tables 5 and 6). The heatmaps produced by SEPA firstly illustrate the relationship between the top 100 proteins and the biological pathways they represent; secondly, the heatmaps emphasize how a restricted list of top aging proteins reveals biological pathways which would have escaped common pathway mining modalities (Fig. 1e). SEPA indicated that incremental lists of proteins are needed to determine biological functions of sex proteins, and pointed to expected differences in hormonal metabolism and activity. Conversely, an extensive list of aging proteins contained enrichment for blood-related pathways such as heparin and glycosaminoglycan binding, as recently reported22.
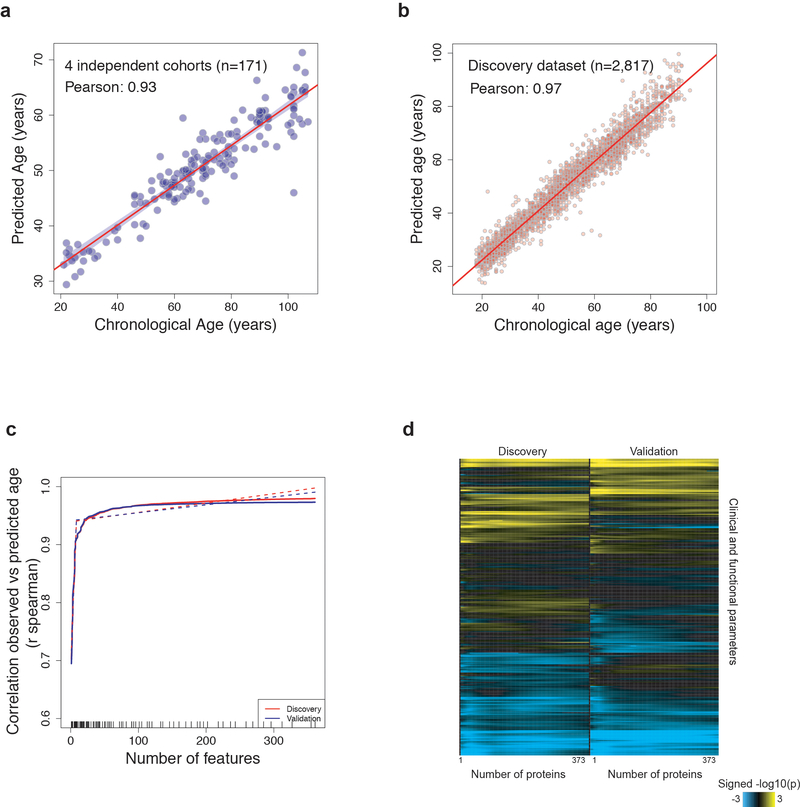
To determine whether the plasma proteome can predict biological age and serve as a “proteomic clock,” we used 2,817 randomly selected subjects to fine-tune a predictive model that was tested on the remaining 1,446 subjects (Fig. 1f). We identified a sex-independent plasma proteomic clock consisting of 373 proteins (Supplementary Table 7), which was highly accurate in predicting age in the discovery, validation, and the 4 independent cohorts (r=0.93–0.97, Fig. 1g and Extended Data 3a–b). Remarkably, subjects predicted younger than their chronologic age performed better on cognitive and physical tests (Fig. 1h and Supplementary Table 8). While a reduced model comprising only 9 proteins predicted age with good accuracy (Extended Data 3c and Supplementary Table 7), a combination of different sets of proteins may be required to model changes in a large set of clinical and functional parameters (Extended Data 3d).
Since most biological pathways that change with age are evolutionarily conserved23, we next identified aging proteins conserved between mice and humans. We analyzed mouse plasma (n=110, aged 1 month to 30 months) using SomaScan, which reliably measures hundreds of non-human proteins24 and has proven useful in mouse studies6,25 (Fig. 1i, Supplementary Table 9). In mice, 172 proteins changed with age (out of 1,305 measured, Supplementary Table 10, q<0.05) and 46 proteins overlapped with human aging proteins (Fig. 1j). Remarkably, many of these proteins were also modulated by heterochronic parabiosis: young mice exposed to old plasma (young-heterochronic mice) showed a relatively older plasma signature while aged mice exposed to young plasma (old-heterochronic mice) showed a younger signature (Fig. 1k). Altogether, standard linear modeling of the plasma proteome during the human lifespan revealed established aging pathways, possibly indicating accelerated and decelerated aging in humans and mice. Intriguingly, changes to the conserved aging proteins did not occur simultaneously (Fig. 1l). Thus, the chronology of aging in the plasma proteome requires further investigation.
Clustering protein trajectories reveals undulation of the aging plasma proteome
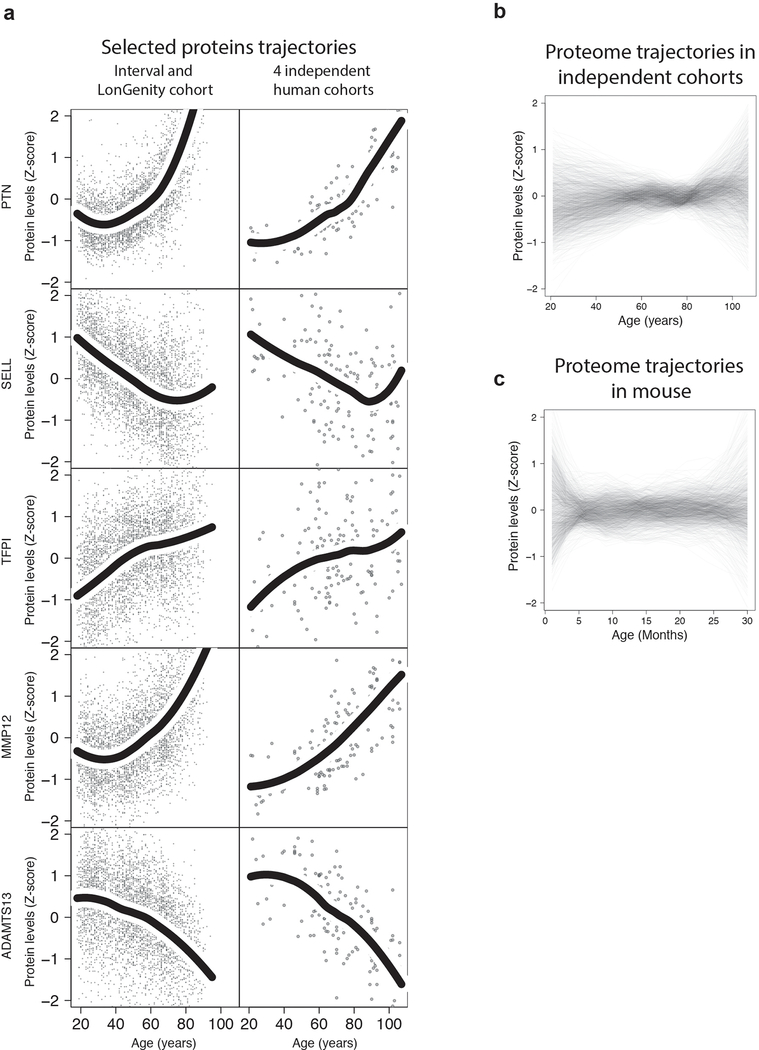
While standard linear modeling showed prominent changes in plasma protein composition, the undulating behavior of the 46 conserved proteins (Fig. 1l) and more globally by the 2,925 plasma proteins as a group when they were visualized as z-scored changes across the lifespan was striking (Fig. 2a–b). These undulating patterns were detected in independent human cohorts and in mice (Extended Data 4), suggesting they are robust and conserved.
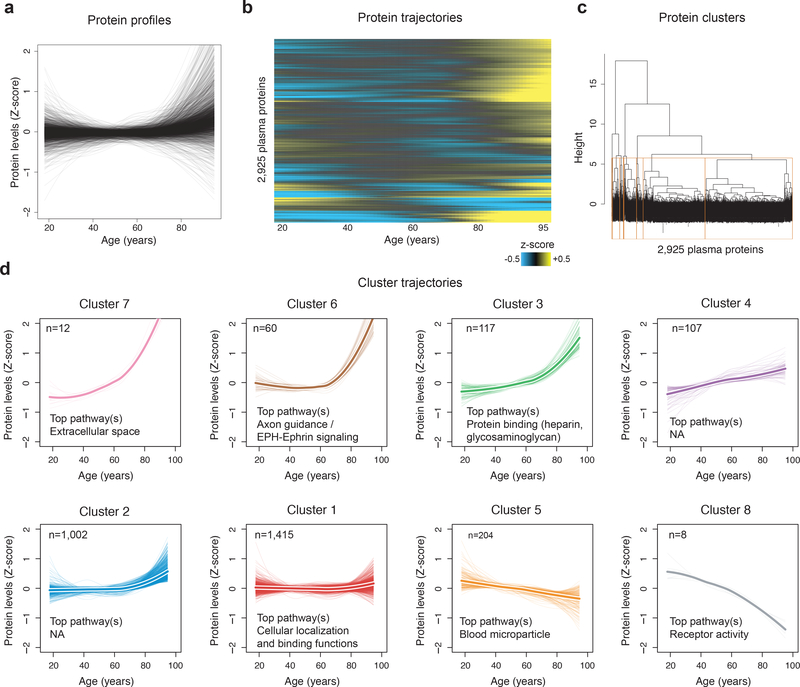
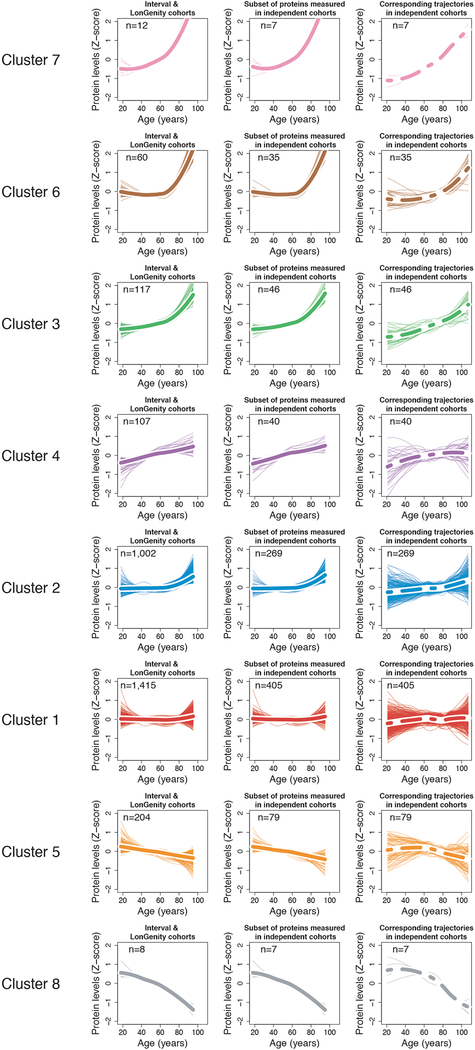
Figure 2: Clustering of protein trajectories identifies linear and non-linear changes during aging.
(a) Protein trajectories during aging. Plasma protein levels were z-scored and trajectories of the 2,925 plasma proteins were estimated by LOESS.
(b) Trajectories are represented in two dimensions by a heatmap and unsupervised hierarchical clustering was used to group plasma proteins with similar trajectories.
(c) Hierarchical clustering dendrogram. The 8 clusters identified are represented by orange boxes.
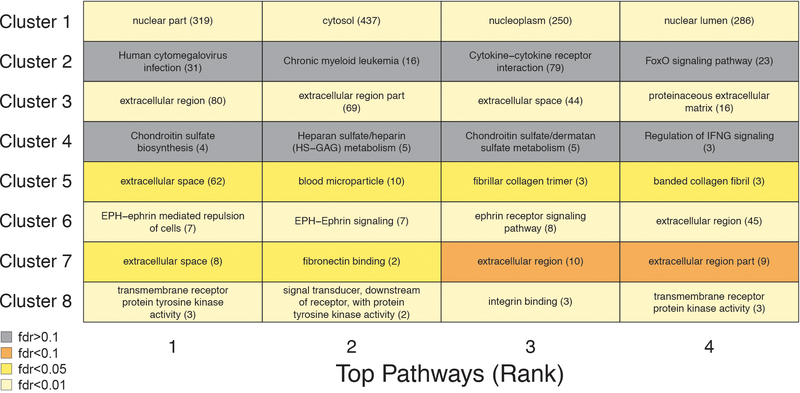
(d) Protein trajectories of the 8 identified clusters. Clusters are grouped by the similarity of global trajectories, the thicker lines representing the average trajectory for each cluster. The number of proteins and the most significant enriched pathways are represented for each cluster. Pathway enrichment was tested using GO, Reactome and KEGG databases. The top 20 pathways for each cluster are listed in Supplementary Table 12.
To reduce complexity of the proteome, we grouped proteins with similar trajectories using unsupervised hierarchical clustering (Fig. 2c) and identified 8 clusters of protein trajectories changing with age, which ranged in size from 8 to 1,415 proteins (Supplementary Table 11). In addition to linear patterns (clusters 1 and 5), several non-linear trajectories were evident, including stepwise, logarithmic, and exponential (clusters 2, 3, 4, 6, 7 and 8) (Fig. 2d). Importantly, these cluster trajectories were similarly detectable in independent cohorts (Extended Data 5). Out of the 8 clusters analyzed, 6 were enriched for specific biological pathways (q<0.05; Extended Data 6, Supplementary Table 12), suggesting distinct, yet orchestrated changes in biological processes during the lifespan. For example, proteins present in blood microparticles consistently decreased with age (cluster 5), other blood-related pathways such as heparin and glycosaminoglycan binding increased in a two-step manner (cluster 4); while levels of proteins involved in axon guidance and EPH-ephrin signaling remained constant until age 60 before rising exponentially (cluster 6) (Fig. 2d). Altogether, the majority of plasma proteome changes across the lifespan are non-linear.
Quantification of proteomic changes across the lifespan uncovers waves of aging proteins
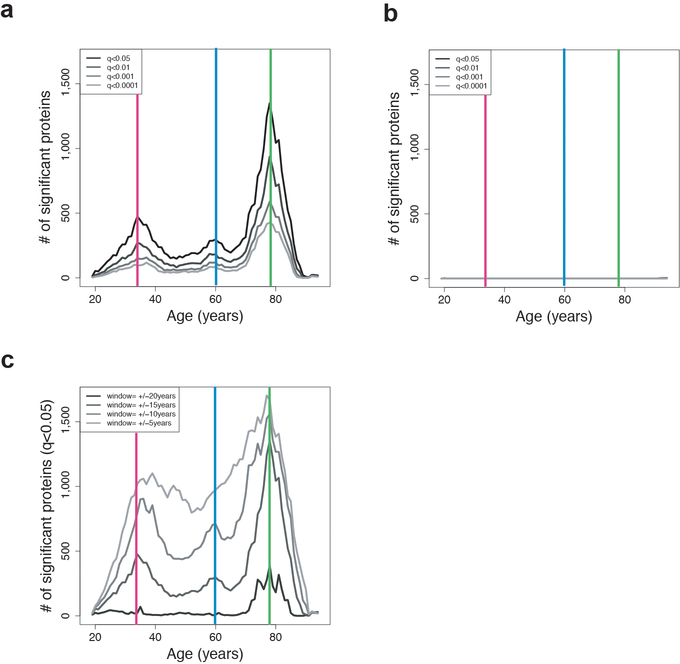
To quantitatively understand the proteomic changes occurring throughout life, we developed the software tool Differential Expression - Sliding Window ANalysis (DE-SWAN) (Fig. 3a). This algorithm analyzes protein levels within a window of 20 years and compares two groups in parcels of 10 years (e.g. 35–45y compared with 45–55y), while sliding the window in increments of 1 year from young to old. Using DE-SWAN, we detected changes at particular stages of life and determine the sequential effects of aging on the plasma proteome (while also controlling for the effect of confounding factors). This approach identified hundreds of proteins changing in waves throughout age (Fig. 3b). Summing the number of differentially expressed proteins at each age uncovered 3 crests at ages 34, 60, and 78 (Fig. 3c, Extended Data 7a and Supplementary Table 13). These crests disappeared when the ages of individuals were permutated (Extended Data 7b), but were still detectable using different statistical models (e.g. smaller/larger sliding windows) (Extended Data 7c, Supplementary Figure 1, Supplementary Table 13), indicating robustness of these age-related waves.
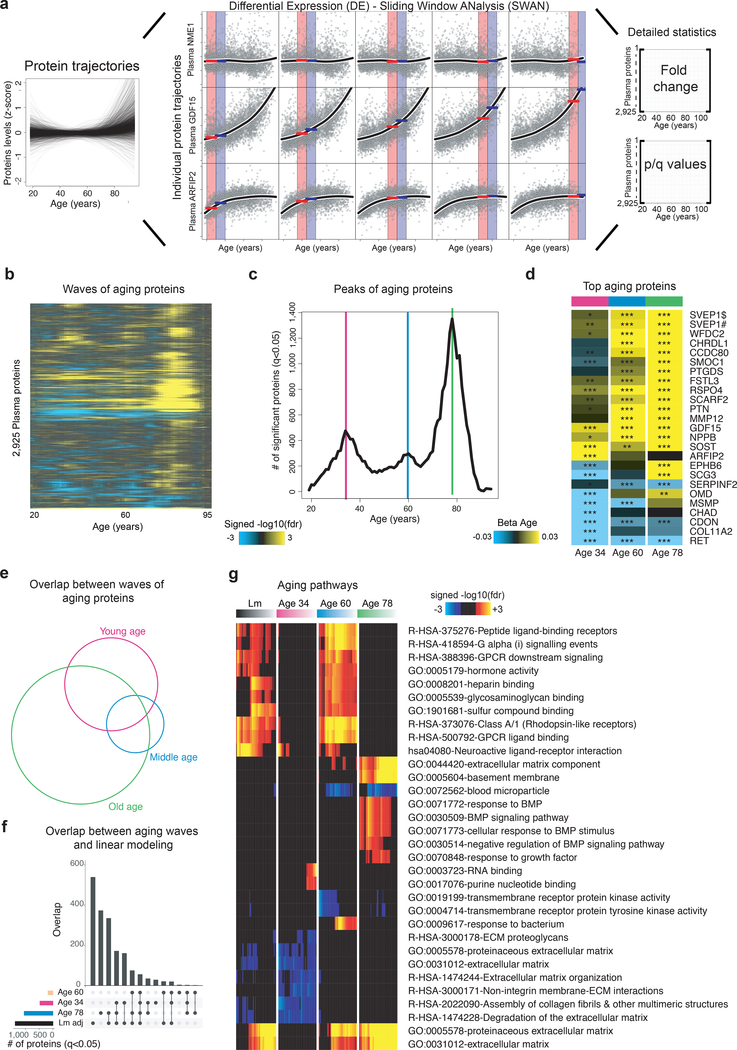
Figure 3: Sliding window analysis distinguishes waves of aging plasma factors.
(a) Differential Expression - Sliding Window ANalysis (DE-SWAN). DE-SWAN compares proteins levels between groups of individuals in parcels of 10 years, e.g. 30–40 compared with 40–50. DE-SWAN identifies linear and non-linear changes during aging. Examples of the DE-SWAN for 3 proteins and 5 age windows. Red and blue rectangles show the two parcels and the red and blue lines symbolize the mean within each parcel. DE-SWAN provides statistics for each age window and each plasma protein, allowing detailed analysis of plasma proteomic changes during aging.
(b) Waves of aging plasma proteins characterized by DE-SWAN (n=4,263). Within each window, −log10(p-values) and −log10(q-values) were estimated by linear modeling adjusted for age and sex and significance was tested using F-test. Local changes attributable to age were signed based on corresponding beta age.
(c) Number of plasma proteins differentially expressed during aging. Three local peaks at the ages of 34, 60 and 78 were identified by DE-SWAN.
(d) Top 10 plasma proteins identified by DE-SWAN at age 34, 60 and 78 (n=4,263). Linear models adjusted for age, sex and subcohort were used and significance was tested using F-test. Blue and yellow colors represent local decrease and increase, respectively. # and $ indicate different SOMAmers targeting the same protein. * q<0.05, ** q<0.01, *** q<0.001.
(e) Intersections between waves of aging proteins (n=4,263, q<0.05). Linear models adjusted for age, sex and subcohort were used and significance was tested using F-test.
(f) Intersections between linear modeling and the aging waves (n=4,263, q<0.05). Linear models adjusted for age, sex and subcohort were used and significance was tested using F-test.
(g) Visualization of pathways significantly enriched for aging proteins identified by linear modeling and DE-SWAN at age 34, 60 and 78 (n=4,263). Proteins upregulated and downregulated were analyzed separately. The top 10 pathways per condition are represented. Enrichment was tested using Fisher’s exact test (GO) and hypergeometric test (Reactome and KEGG).
Intriguingly, the 3 age-related crests were largely comprised of different proteins (Fig. 3d, Supplementary Table 14), but a few proteins were among the top 10 differentially expressed in each crest, such as GDF15, consistent with its pronounced increase across the lifespan (Fig. 3a). Other proteins, like chordin-like protein 1 (CHRDL1) or Matrix Metallopeptidase 12 (MMP12), were significantly changed only at the last two crests, reflecting their exponential increase with age. Overlap between proteins changing at age 34, 60, and 78 was statistically significant (p<0.05) but limited, (Fig. 3e) and most proteins changing in old age were not identified by linear modeling (Fig. 3f). This prompted us to use SEPA to determine whether these waves reflected distinct biological processes. Strikingly, we observed a prominent shift in multiple biological pathways with age (Fig. 3g). At young age (34 years), we observed a downregulation of proteins involved in structural pathways such as the extracellular matrix (ECM). These changes were reversed in middle and old age (60 and 78 years, respectively). At age 60, we found a prominent role of hormonal activity, binding functions, and blood pathways. At age 78, key processes still included blood pathways but also bone morphogenetic protein signaling, which is involved in numerous cellular functions26. Pathways changing with age by linear modeling overlapped most strongly with crests at age 34 and 60 (Fig. 3g), indicating that dramatic changes occurring in the elderly might be masked in linear modelling by more subtle changes at earlier ages. Altogether, these results show that aging is a dynamic, non-linear process characterized by waves of changes in plasma proteins that reflect complex shifts in biological processes.
Proteins linked to age-related diseases are enriched in distinct waves of aging
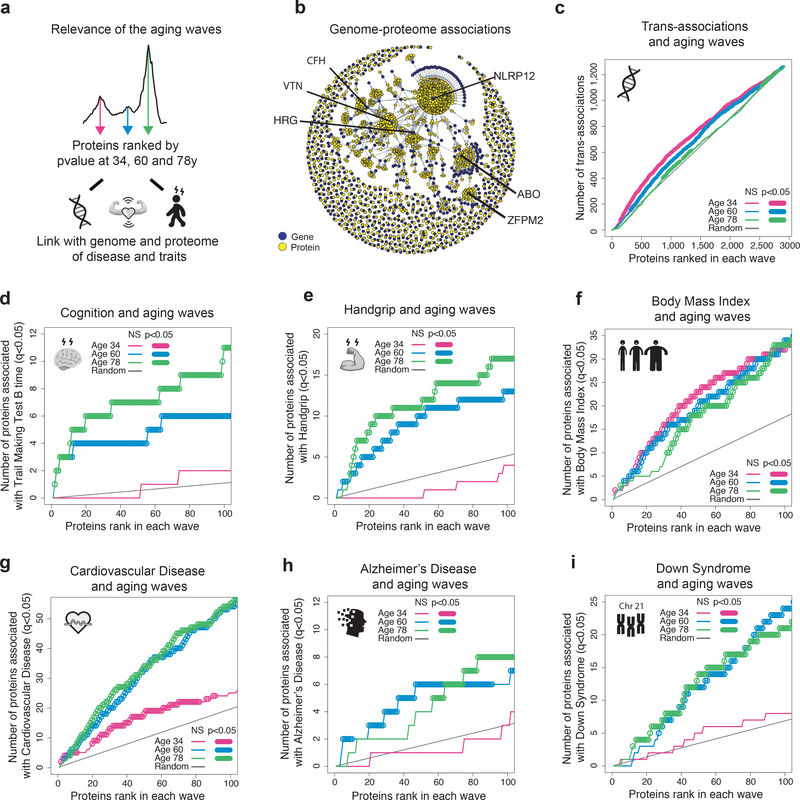
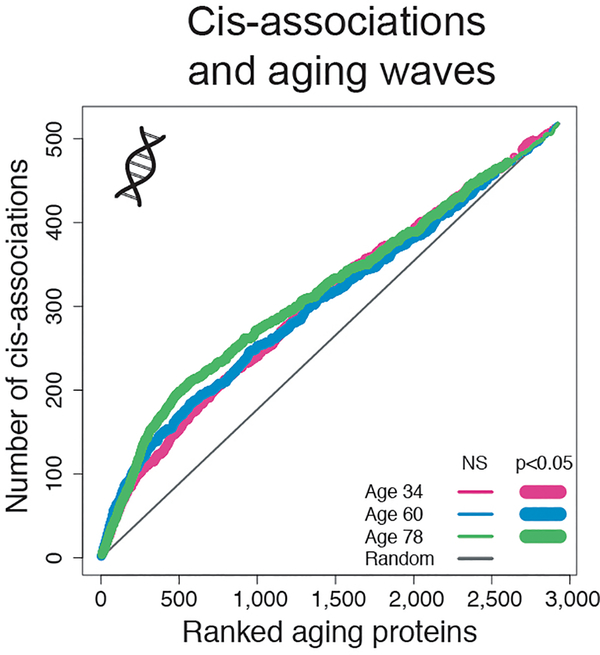
The plasma proteome is sensitive to the physiological state of an individual but is also genetically influenced27. To deconvolute complexity between genome, proteome, and physiology, we asked whether the top aging proteins change due to genetic polymorphisms or whether they are among the top predictors of disease, or phenotypic traits. More specifically, we sought to determine whether proteins that comprised the 3 waves of aging were uniquely linked to the genome or proteome of age-related diseases and traits (Fig. 4a). We used the ranked lists of the top proteins identified by DE-SWAN at each of the three crests (Fig. 3c and Supplementary Table 14) and summed the number of proteins linked to the genome and proteome of specific diseases and traits separately for each wave (i.e. the cumulative sum) (Fig. 4b–i and Extended Data 8). First, we mined the genomic atlas of the human plasma proteome27 (Fig. 4b) and discovered that the aging proteome is also genetically determined (Fig. 4c and Extended Data 8). However, the rank of proteins determined by trans-association appeared more random with aging (Fig. 4c), suggesting that other sources drive the aging plasma proteome. We then tested whether the waves of aging proteins were differentially linked with changes in cognitive and physical functions identified in Fig. 1h. Interestingly, the proteome associated with these traits overlapped with the proteome defining middle and old ages, when these functions decline the most (Fig. 4d and Fig. 4e). Finally, we used public datasets and summary statistics from SomaScan proteomic studies focused on age-related diseases, including Alzheimer’s Disease (AD)28, Down Syndrome (DS)29, and cardiovascular disease (CVD)30. A plasma proteomic study predicting body mass index (BMI)31 was used as a control since weight gain varies widely with age (Supplementary Figure 2). As expected, the proteome linked to BMI was not selectively enriched for proteins defining waves of aging (Fig. 4f). Conversely, CVD-associated proteins were strongly enriched in waves of proteins defining middle and old age compared to young age (Fig. 4g). This enrichment corresponded to an increased incidence of CVD after 55 years32. Finally, AD- and DS-associated proteins overlapped with the top proteins defining middle age and old age but not those defining young age (Fig. 4h–i). The fact that the proteome defining these two diseases also changed in old individuals of a separate disease-free cohort supports the notion of accelerated aging in DS and AD33,34. Altogether, these results show waves of proteomic aging are differentially linked to the genome and proteomic traits of various diseases.
Figure 4: Waves of aging proteins are differentially linked to the genome and proteome of disease and traits.
(a) Relevance of the aging waves. Schematic representation of analysis. The proteins changing at 34, 60 and 78y were ranked by p-value and were associated with the genome and the proteome of disease and traits.
(b) Association between the genome and the proteome. Network created using the pQTL associations identified by Sun et al. (2018).
(c) Enrichment for trans-association in the waves of aging proteins identified by DE-SWAN. Aging proteins at age 34, 60 and 78y were ranked based on p-value and the cumulative number of trans-associations was enumerated. One-sided permutation tests (1e+5 permutations) were used to assess significance.
Enrichment for proteins involved in cognitive and physical performance in the waves of aging proteins (d-e).
Enrichment for disease-associated proteins in the waves of aging proteins (f-i).
Discussion
Our analysis of the plasma proteome reveals complex, non-linear changes over the human lifespan. Although linear analysis provides information about the aging plasma proteome, modeling non-linear protein trajectories is necessary to fully appreciate these undulating changes.
It is well known that men and women age differently21, yet we were surprised that 2/3 of proteins changing with age also change with sex (895/1,379 proteins). This supports the National Institutes of Health (NIH) policy on the inclusion of women in clinical research and using sex as a biological variable in experiments. Nevertheless, a unique proteomic clock can be used to predict age in men and women, and deviations from this plasma proteomic clock are correlated with changes in clinical and functional parameters (Fig 1f–h). This panel of 373 proteins can be used to assess the relative health of an individual and to measure healthspan, analogous to epigenetic clocks based on DNA methylation patterns35. More large-scale plasma proteomic studies are required to establish the validity and utility of this clock and whether specific protein subsets are more appropriate to reflect particular clinical and functional parameters.
Blood is a sensitive marker of functional aging that also plays an active role in aging. Numerous studies have shown that soluble factors from young mouse blood reverse aspects of aging2–10,36. Here, we describe a 46-protein aging signature that is conserved in humans and mice, containing known aging proteins such GDF1537 and IGF1/INSR38 but also less investigated ones (Fig 1l). This conserved signature will allow deeper investigation of translational aging interventions in mice, such as heterochronic parabiosis, which partially reverses age-related changes of these proteins (Fig 1k).
By deep mining the aging plasma proteome, we identify undulating changes during the human lifespan. These changes are the result of clusters of proteins moving in distinct patterns, culminating in the emergence of 3 waves of aging. Unexpectedly, we find these clusters are often part of shared biological pathways, particularly cellular signaling (Fig. 2). In addition, we provide biological relevance for the 3 main waves of aging proteins (Fig. 3), which are characterized by key biological pathways with little overlap. In comparison, linear modeling fails to identify changes occurring late in the 8th decade of life (Fig. 3g). We conclude that linear modeling of aging based on omics data does not capture the complexity of biological aging across the lifespan. Thus, DE-SWAN will be invaluable for analyzing longitudinal datasets with linear and non-linear, quantifiable changes, and for integrating non-linear changes in the analysis of omics datasets.
Sources of variation of the plasma proteome can be diverse and under genetic control27,39. Intriguingly, we observed that the relative importance of trans-associations decreased with aging (Fig. 4c), which led us to investigate sources of variance with a focus on disease-associated proteomes. Proteins comprising the middle and old age waves differentially overlapped with proteins associated with cognitive and physical impairments. These proteins also discriminated patients from age-matched controls in Alzheimer’s disease, Down’s syndrome, and CVD (Fig. 4g–i), suggesting that the characteristic plasma proteins of aging are amplified in these age-related diseases. Using an disease-free cohort, we provide evidence of accelerated aging for these two diseases33,34. Further investigation of these proteins is warranted to determine whether these associations indicate aging biomarkers and/or causal mechanisms of disease. Nonetheless, these results suggest that variance within the aging plasma proteome slowly transitions from hard coding factors (i.e. genomic) to soft coding factors (e.g. diseases, environmental factors, and resulting changes in cognitive and physiological functions).
The undulating nature of the aging plasma proteome and its interactions with diseases ought to be considered when developing proteomic signatures for diagnostic purposes. Indeed, disease proteomes overlap significantly with the waves of aging proteins (Supplementary Table 15). Taking into account heterogeneous and complex changes to the plasma proteome during life will likely improve the sensitivity and specificity of prognostics and diagnostics tests. Moreover, these results are pertinent when considering the use of blood or blood products to treat aging and age-related diseases40. Specifically, identifying plasma proteins that promote or antagonize aging at different stages of life could lead to more targeted therapeutics and/or preventative therapies. Such reliable tests and treatments are urgently needed for numerous diseases and, in the future, we hope to describe plasma proteome changes that predict subjects transitioning to disease. Of particular interest are studies of Alzheimer’s disease for which blood-based biomarkers are unavailable and clinical symptoms are believed to occur up to two decades after disease onset.
Methods
Plasma proteomics measurements
The SomaScan platform was used to quantify relative levels of protein involved in several processes such as intercellular signaling, extracellular proteolysis, and metabolism. This platform was established to identify biomarker signatures of diseases and conditions, including cardiovascular risk30, cancer41 and neurodegenerative diseases28. The SomaScan platform is based on modified single-stranded DNA aptamers (SOMAmer® reagents) binding to specific protein targets. Assay details have been previously described19. Different versions of the SomaScan assay were used in the LonGenity, INTERVAL and the 4 independent human cohorts. These versions contained 5,284, 4,034, and 1,305 aptamers, respectively.
Out of the 4,034 aptamers measured in the INTERVAL cohort, 3,283 were contained in the publicly available dataset (https://ega-archive.org/studies/EGAS00001002555). Our study focuses on 2,925 aptamers with identical SeqId and SeqIdVersion in both INTERVAL and LonGenity cohorts (Supplementary Table 1). Out of the 2,925 aptamers, 888 were measured in the 4 independent cohorts and in mice (Supplementary Table 2).
Human cohorts characteristics
INTERVAL cohort
Participants in the INTERVAL randomized controlled trial (ISRCTN24760606) were recruited with the active collaboration of the National Health Service Blood and Transplant England (www.nhsbt.nhs.uk), which has supported field work and other elements of the trial. DNA extraction and genotyping was co-funded by the National Institute for Health Research (NIHR), the NIHR BioResource (http://bioresource.nihr.ac.uk/) and the NIHR Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust. The INTERVAL study was funded by NHSBT (11–01-GEN). The academic coordinating center for INTERVAL was supported by core funding from: NIHR Blood and Transplant Research Unit in Donor Health and Genomics (NIHR BTRU-2014–10024), UK Medical Research Council (MR/L003120/1), British Heart Foundation (RG/13/13/30194) and the NIHR Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust. Proteomic assays were funded by the academic coordinating center for INTERVAL and MRL, Merck & Co., Inc. A complete list of the investigators and contributors to the INTERVAL trial has been previously reported15. The academic coordinating center would like to thank blood donor center staff and blood donors for participating in the INTERVAL trial. For more information, see the Life Sciences Reporting Summary.
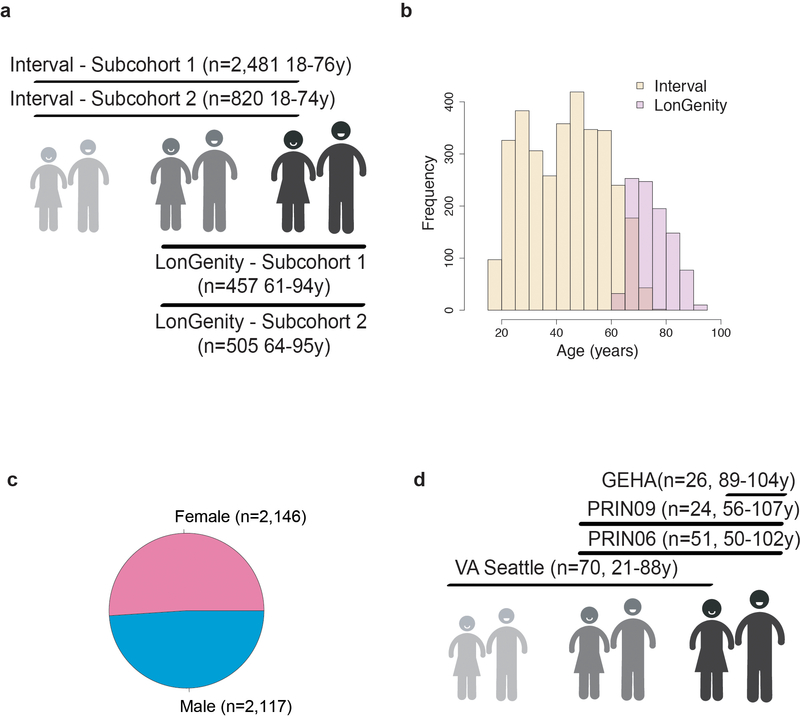
Proteomics measurements from 3,301 human plasma samples (1,685 males and 1,616 females) from 2 different subcohorts were used for this study. Age ranged from 18 to 76 years with a median age of 45 (1st Quartile=31, 3rd Quartile=55). Sample selection, processing and preparation were detailed previously27.
LonGenity cohort
LonGenity is an ongoing longitudinal study initiated in 2008, designed to identify biological factors that contribute to healthy aging16. The LonGenity study enrolls older adults of Ashkenazi Jewish descent, age 65–94 years at baseline. Approximately 50% of the cohort consists of offspring of parents with exceptional longevity (OPEL), defined by having at least one parent that survived to 95 years of age. The other half of the cohort includes offspring of parents with usual survival (OPUS), who do not have a parental history of exceptional longevity. Proteomics measurements from 1,030 human plasma samples (457 males and 573 females) collected at baseline in LonGenity participants were used for this study. Age ranged from 61 to 95 years with a median age of 74 (1st Quartile=69, 3rd Quartile=80). LonGenity subjects are thoroughly characterized demographically and phenotypically at annual visits that include collection of medical history and physical and neurocognitive assessments. Sixty-eight subjects without clinical and functional data were excluded from the analysis. The LonGenity study was approved by the Institutional Review Board (IRB) at the Albert Einstein College of Medicine.
Additional 4 independent cohorts
One hundred seventy-one human plasma samples (84 males and 87 females) were obtained from 4 different cohorts from the US and Europe (VASeattle, PRIN06, PRIN09, GEHA). Sample selection, processing and preparation of the VASeattle cohort were detailed previously42. Subjects from the PRIN06, PRIN09 and GEHA43 cohorts were enrolled by multiple Italian study centers. Subjects were mainly Caucasians. Age ranged from 21 to 107 years with a median of 70 years (1st Quartile=58, 3rd Quartile=89). Written informed consent was obtained for each subject. The IRB has determined that our research does not meet the definition of human subject research per Stanford’s Human Research Protection Program policy and no IRB approval was required for this study.
For these cohorts, all samples were stored at −80 °C and 150μl aliquots of plasma were sent on dry ice to SomaLogic Inc. (Boulder, Colorado, US). Plasma samples were analyzed in three different batches, 24 samples in 2015, 70 in 2016 and 77 in 2017. In addition to these 171 plasma samples, 12 additional aliquots from 4 of these samples were measured in the different batches to estimate intra- and inter-assay variability (Supplementary Table 3). Data for 1,305 SOMAmer probes were obtained. No sample or probe data were excluded. HybNorm.plateScale.medNorm files provided by SomaLogic Inc. were bridged to data from the 1st batch of samples using calibrators.
Normalization of INTERVAL and LonGenity datasets
Relative Fluorescent Units (RFU) of each plasma protein were log10-transformed. We normalized the levels of each protein within each subcohort based on the average of the subjects in the 6070 years range. Supplementary Figure 3 shows representative normalization examples. Note that this normalization is needed when fitting aging trajectories (Fig. 2) but does not affect the results when “subcohort” is included as covariate in the modeling.
The data from the 4 independent cohorts were log10-transformed and bridged together using SomaLogic procedure based on calibrators. However, the number of samples in the 60–70 years range was too small to reliably bridge this data to the INTERVAL and LonGenity cohorts.
Linear changes in the aging plasma proteome
To determine the effect of age and sex at the protein level, we used the following linear model:
Type II sum of squares (SS) were calculated using the Anova function of the R car package44. This SS type tests for each main effect after the other main effects. Q-values were estimated using Benjamini–Hochberg approach45. It should be noted that the age range differs between cohorts. If the adjustment for cohort effect decreases the number of false positives, it could also alter the true positive rate. In the 4 independent cohorts, the “Subcohort” covariate also accounts for batch effect since samples from different cohorts were measured in different batches (except for PRIN06 and GEHA that were measured together).
To determine the relative proportion of variance explained by age and sex, we calculated the partial Eta2 as follows:
Validation of the aging proteins
To provide confidence in the reproducibility of the protein assays, we compared our findings with the associations with age reported by Tanaka et al.22. To this end, we merged our results with those from Tanaka et al. using “SomaId”. Of note, Tanaka et al. used the same version of the SomaScan platform that we used for the 4 independent cohorts (1,305 proteins).
Sliding Enrichment Pathway Analysis (SEPA)
To determine the biological meaning of group of plasma proteins, we ranked the top 100 proteins based on the product of -log10(p-values) and beta age (or beta sex) and queried three of the most comprehensive biological annotation and pathway databases: Gene ontology - GO46, Kyoto Encyclopedia of Genes and Genomes – KEGG47 and Reactome48. Using these databases, we tested enrichment for pathways in the top 10 to top 100 proteins in increments of 1 protein. The 2,925 proteins measured in this study cover 90% of the human GOs, Reactome and KEGG terms containing more than 8 genes (Supplementary Figure 4).
To analyze each incremental list of proteins, we used the R topGO package49 for GO analysis and the R clusterProfiler package50 for KEGG and Reactome analyses. As input of SEPA, we used Gene Symbols provided by SomaLogic Inc. (Supplementary Table 1). The 2,925 proteins measured by SomaScan served as the background set of proteins against which to test for over-representation. Since several individual proteins (33 out of 2,925) were mapped to multiple Gene Symbols, we kept only the 1st Gene Symbol provided by Somalogic to prevent false positive enrichment. For KEGG and Reactome analysis, clusterProfiler requires EntrezID as input. Therefore, we mapped Gene Symbols to EntrezID using the org.Hs.eg.db package51. Again, to avoid false positive enrichment, only the 1st EntrezID was used when Gene Symbols were mapped to multiple EntrezID. Q-values were estimated using Benjamini–Hochberg approach45 for the different databases taken separately. For GO analysis, q-values were calculated for the three GOs classes (molecular function, cellular component, biological process) independently. To identify the most biologically meaningful terms and pathways, we reported only those with 20–500 proteins measured by the SomaScan assay. In addition, we focused on pathways consistently highly significant (q<0.05 for at least 20 different incremental list of proteins) and kept the top ten pathways per condition (e.g. for each wave of aging proteins). Ranking was performed based on the minimum fdr across the incremental lists of proteins. SEPA can be viewed as an extension of the GSEA approach52, with more control for true and false positives.
Validation of the aging signature in mice
Male and virgin female C57BL/6JN mice were shipped from the National Institute on Aging colony at Charles River (housed at 67–73 °F) to the Veterinary Medical Unit (VMU; housed at 68–76 °F)) at the VA Palo Alto (VA). At both locations, mice were housed on a 12-h light/dark cycle and provided food and water ad libitum. The diet at Charles River was NIH-31, and Teklad 2918 at the VA VMU. Littermates were not recorded or tracked. Mice 18-months-old and younger were housed at the VA VMU for no longer than 2 weeks before euthanasia, and mice older than 18-months were housed at the VA VMU until they reached the experimental age. After anaesthetization with 2.5% v/v Avertin, blood was drawn via cardiac puncture. All animal care and procedures were carried out in accordance with institutional guidelines approved by the VA Palo Alto Committee on Animal Research.
Heterochronic parabiosis was conducted as previously described3,10,53 with 3- and 18-month-old mice. Briefly, incisions in the flank were made through the skin and peritoneal cavity of both mice, and adjacent peritoneal cavities were sutured together. Adjacent knee and elbow joints were then sutured together to facilitate coordinated locomotion. Skin was then stapled together using surgical autoclips (9-mm, Clay Adams), and mice were placed under heat lamps to recover from anesthesia. Each individual mouse was injected subcutaneously with Baytril antibiotic (5 μg per g) and buprenorphine (0.05–0.1 μg ml−1) for pain management, and 0.9% (w/v) NaCl for hydration. Mice were monitored and administrated drugs and saline over the next week as previously described.
EDTA-plasma was isolated by centrifugation at 1,000g for 10 min at 4 °C before aliquoting and storing at −80 °C. A total of 110 plasma samples were analyzed and aliquots of 150μl of plasma were sent on dry ice to SomaLogic Inc. (Boulder, Colorado, US). Samples were sent in two different batches, 29 samples in 2016 and 81 in 2018. Data for 1,305 SOMAmer probes were obtained and no sample or probe data were excluded. RFUs of each plasma protein were log10-transformed.
The SomaScan assay has been developed and validated for human fluids but successfully used in mouse research6,25. To understand how similar mouse and human sequences are, we downloaded all homologies between mouse and human along with sequence identifiers for each species (HOM_MouseHumanSequence.rpt) from MGI (http://www.informatics.jax.org/) as plain text files. Then, the protein reference sequences for both organisms were extracted from UniProt (https://www.uniprot.org/). On these matched sequence pairs, for each protein we computed a global pairwise sequence alignment. The alignments have been calculated by using the R “Biostrings” library54. The average identity was 0.85, supporting the use of the SomaScan assay with mouse plasma.
To determine the effect of age and sex at the protein level, we used the 81 samples from 1 month to 30 months. To this end, we fitted the following linear model:
Type II sum of squares (SS) were calculated and Q-values were estimated using Benjamini– Hochberg approach.
To characterize the effects of young and old blood on the aging plasma proteome, normed scaled Principal Component Analysis (PCA) was performed using the R ade455 package
Prediction of human biological age using the plasma proteome
To determine whether the plasma proteome could predict biological age, we used glmnet56 and fitted a LASSO model (alpha=1, 100 lambda tested, “lamda.min” as the shrinkage variable estimated after 10-fold CV). Input variables consisted in Z-scaled log10 RFUs and sex information. Two-thirds (n=2,817) of the INTERVAL and LonGenity samples were used for training the model and the remaining 1,446 samples were used as a validation. In addition, the 171 samples from the 4 independent cohorts were used to further assess the robustness of the predictive model.
To estimate whether a subset of the aging clock can provide similar predictive results, we used a two-step approach as we described previously57. One hundred models (100 lambda) including 0 to 373 proteins were created in step 1 and we estimated the accuracy of each of these models on the discovery and validation datasets, separately. Broken stick regression was used to determine the best compromise between number of variables and prediction accuracy.
Associations between Δage and clinical and functions in old
We used the subjects from the LonGenity cohort to identify associations between deviations from the proteomic clock (Δage=predicted age-chronological age) and 334 clinical and functional variables (Supplementary Table 8). To this end, we tested the following linear model:
For binary outcomes, logistic regression was used. This analysis was separately performed in the discovery (n=2,817) and validation cohorts (n=1,446). Type II sum of squares were calculated using the Anova function of the R car package44.
Clustering of protein trajectories
To estimate protein trajectories during aging, plasma proteins levels were z-scored and LOESS (locally estimated scatterplot smoothing) regression was fitted for each plasma factor. To group proteins with similar trajectories, pairwise differences between LOESS estimates were calculated based on the Euclidian distance and hierarchical clustering was performed using the complete method. To understand the biological functions of each cluster, we queried Reactome, KEGG and GO databases, as described above.
Differential Expression - Sliding Window Analysis (DE-SWAN)
To identify and quantify linear and non-linear changes of the plasma proteome during aging, we developed a Differential Expression - Sliding Window ANalysis (DE-SWAN) approach. Considering, a vector l of k unique ages, we iteratively used lk as the center of a 20-year window and compared protein levels of subjects in parcels below and above lk i.e.[lk − 10y; lk[ vs ]lk; lk + 10y]. To test for differential expression, we used the following linear model:
Age being binarized according to the parcels. For each lk, q-values were estimated using Benjamini–Hochberg correction. Type II sum of squares were calculated using the Anova function of the R car package44.
To assess the robustness and relevance of DE-SWAN results, we tested multiple parcel widths (5, 10, 15 and 20 years). In addition, we used multiple q-values thresholds and compared these results with those obtained by chance. To this end, we randomly permutated the phenotypes of the subjects and applied DE-SWAN to this new dataset. To keep the data structure, age and sex were permuted together. In addition, we analyzed the INTERVAL and LonGenity cohorts separately (Supplementary Figure 1). Finally, we tested the same linear model when adjusting for subcohort. This led to a loss of statistical power when the age range of the INTERVAL and LonGenity cohorts were overlapping but the three waves of aging proteins remained and the ranks of the top proteins were nearly identical (Supplementary Figure 1). We used the model adjusted for subcohort when trying to understand the waves of aging proteins (Figure 3d–g and Figure 4). The significance levels of the intersections between aging plasma protein signatures identified by linear modeling and DE-SWAN at different ages were determined using the R SuperExactTest package58.
Relationships between the aging waves and the genome and the proteome of disease and traits
To quantify the overlap between proteins changing with age at different stages of life and the genome and the proteome of diseases and traits, we ranked DE-SWAN results based on p-values and created a k-ranked list of aging proteins, Lk. To reflect the degree to which the genome/proteome are linked with the waves of aging plasma proteins, we walked down Lk and counted the number of proteins associated with the genome or specific proteome. When different versions of the SomaScan platform were used, we walked down Lk until reaching the top 100 proteins measured in both studies.
To identify specific genetic variants associated with the aging plasma proteome, we mined the summary statistics generated by Sun et al.27, who found 1,927 associations with 1,104 plasma proteins. Qgraph59 R package was used to create a network between the genome and the 2,925 proteins analyzed in this study.
To determine whether the aging proteome is associated with the proteome of clinical and functional variables, we used the subjects from the LonGenity cohort and tested the following linear model for the top variables identified in Supplementary Table 8:
Type II sum of squares were calculated using the Anova function of the R car package44.
To determine whether the aging proteome is associated with disease proteomes, we integrated data and results from previous proteomic studies using the SomaScan platform. We re-analyzed one Alzheimer’s Disease - AD dataset publicly available by AddNeuroMed28 and used summary statistics from published studies focused cardiovascular disease - CVD30, Down Syndrom - DS29 and body mass index - BMI31.
AddNeuroMed is a European multi-center study in which the AD proteome was quantified in plasma samples from 681 controls, Mild Cognitive Impairment (MCI), and AD subjects using a previous version of the SomaScan assay. Files used were downloaded from the Synapse portal in March 2016 (syn5367752) and included measurements of 1016 plasma proteins from 931 samples. We limited our analysis to the 645 samples available at visit 1 (191 control, 165 MCI and 289 AD). Raw data were log10-transformed. Four samples (2 control and 2 AD) were considered as outliers based on visual inspection of the results of a Principal Component Analysis (PCA) and filtered out.
To identify plasma proteins associated with AD, we used linear models with diagnosis, age, sex and center as covariates:
Type II sum of squares were calculated using the Anova function of the R car package44.
To determine whether aging plasma proteins are involved in other disease signatures, we identified three studies using the SomaScan platform in large human cohorts providing detailed summary statistics. Carayol et al. mined the plasma proteome to obtain new insights into the molecular mechanisms of obesity. Out of 1,129 proteins measured, they identified 192 plasma proteins significantly associated with BMI (p<0.05 after Bonferroni correction). Summary statistics we used were obtained from their “Supplementary Data 1”31. Ganz et al. derived and validated a 9 protein risk-score to predict risk of cardiovascular outcomes30. In addition to these 9 proteins, 191 other proteins were significantly associated with cardiovascular risk (p<0.05 after Bonferroni correction). Summary statistics for these 200 proteins are available in their eTable 4 and the 1,130 proteins measured of this study are listed in their eTable 1. Finally, Sullivan et al. used an extended version of the SomaScan platform to study DS and identified a large number of dysregulated proteins29. We used the results for the Discovery cohort (sheet A of the “Supplementary File 1”) in which, 258 proteins out of 3,586 proteins are reported to be associated with DS (p<0.05 after Bonferroni correction).
Since detailed protein information was not available for all studies, we used either gene symbols or unitprotID to merge disease proteomes characterized in published studies with the aging proteome identified in this study. When multiple p-values were reported for the same gene symbol (or a combination of gene symbols), only the most significant p-value was retained.
Code availability
An R package for DE-SWAN is available in github: lehallib.github.io/DEswan/
Data availability
We created a searchable web interface to mine the human INTERVAL and LonGenity datasets (https://twc-stanford.shinyapps.io/aging_plasma_proteome/)
The Human independent cohorts and mouse protein data are available in Supplementary Tables 16 and 17. The INTERVAL data is available through the European Genome-Phenome Archive (https://ega-archive.org/studies/EGAS00001002555).
Extended Data
Extended Data Fig. 1. Sample demographics.
Age (a, b), cohort (a, b) and sex distributions (c) of the 4,263 subjects from the INTERVAL and LonGenity cohorts. (d) Age and cohort distributions of the 171 subjects from the 4 independent cohorts.
Extended Data Fig. 2. Comparing age and sex effects in independent cohorts.
(a) Age and sex effects in the INTERVAL and LonGenity studies (n=4,263) were compared to age and sex effects in 4 independent cohorts analyzed together (n=171) and to age effect from Tanaka et al. (n=240, 2018)
The aging plasma proteome was measured with the SomaScan assay in these cohorts and 888 proteins were measured in all studies
(b) Scatter plot representing the signed -log10(q value) of the sex effect in the INTERVAL/LonGenity cohorts (x axis, n=4,263) vs the 4 independent cohorts (y-axis, n=171).
Similar analysis for the age effect in the 4 independent cohorts (c, n=171) and in Tanaka et al study (d, n=240)
Extended Data Fig. 3. Deeper investigation of the aging proteomic clock.
a) Prediction of age in the 4 independent cohorts (n=171) using the proteomic clock. Only 141 proteins out of the 373 constituting the clock were measured in these samples. (b) Prediction of age in the discovery cohort (n=2,817) using the 373 plasma markers.
(c) Feature reduction of the aging model in the Discovery and Validation cohorts to estimate whether a subset of the aging signature can provide similar results to the 373 aging proteins. Dashed lines represent a broken stick model and indicate the best compromise between number of variables and prediction accuracy.
(d) Heatmap representing the associations between delta age and 334 clinical and functional variables. For quantitative traits, linear models adjusted for delta age, age and sex were used and significance was tested using F-test. For binary outcomes, binomial generalized linear models adjusted for delta age, age and sex were used and significance was tested using likelihood ratio chi-square test. As in (c) the analysis was performed for the top 2 to top 373 variables predicting age. The non-uniformity in the heatmaps suggests that specific subsets of proteins may best predict certain clinical and functional parameters
Extended Data Fig. 4. Proteins and proteome undulations in independent human cohorts and in mouse.
(a) Trajectories of 5 selected proteins based on the INTERVAL and LonGenity cohorts (n=4,263, left) and 4 independent human cohorts (n=171, right). Trajectories were estimated using LOESS regression.
Undulation of the 1,305 plasma proteins measured in 4 independent cohorts (b, n=171) and in mouse (c, n=81). Plasma proteins levels were z-scored and LOESS regression was fitted for each plasma factor.
Extended Data Fig. 5. Cluster trajectories in independent cohorts.
Protein trajectories for the 8 clusters identified in the INTERVAL and LonGenity cohorts (left column). Thicker lines represent the average trajectory for each cluster. Cluster trajectories for the subset of proteins measured in the 4 independent cohorts (middle column). Corresponding cluster trajectories in 4 independent cohorts (right column).
Extended Data Fig. 6. Pathways in clusters.
Pathway enrichment was tested using GO, Reactome and KEGG databases (n=4,263). Enrichment was tested using Fisher’s exact test (GO) and hypergeometric test (Reactome and KEGG). The top 4 pathways for each cluster are shown. Pathway IDs and number of plasma proteins associated are represented in the table.
Extended Data Fig. 7. DE-SWAN age effect for multiple q-values cutoffs, windows size and after phenotypes permutations.
Different Q-value cutoffs are represented in (a). Similar analysis with different after phenotype permutations (b) and different windows size in (c). The 3 local peaks identified at age 34, 60 and 78 are indicated by colored vertical lines.
Extended Data Fig. 8. Cis-associations and aging waves.
Enrichment for cis-association in the waves of aging proteins identified by DE-SWAN. Aging proteins were ranked based on p-values at age 34, 60 and 78 and the cumulative number of cis-associations was counted. One-sided permutation tests (1e+5 permutations) were used to assess significance.
Supplementary Material
Acknowledgements
We thank the members of the Wyss-Coray laboratory for feedback and support. We thank clinical staff for human blood-plasma collection/coordination. We thank A. Butterworth for his help to get access to the INTERVAL proteomics data.
The AddNeuroMed data are from a public-private partnership supported by EFPIA companies and European Union of the Sixth Framework program priority FP6-2004-LIFESCIHEALTH-5. Clinical leads responsible for data collection are I. Kłoszewska (Lodz), S. Lovestone (London), P. Mecocci (Perugia), H. Soininen (Kuopio), M. Tsolaki (Thessaloniki), and B. Vellas (Toulouse) and imaging leads A. Simmons (London), L. O. Wahlund (Stockholm) and C. Spenger (Zurich) and bioinformatics leads are R. Dobson (London) and S. Newhouse (London).
This work was supported by a National Institutes of Health National Institute on Aging (NIA) F32 1F32AG055255 01A1 (D.G.), Hungarian Brain Research Program Grant, No. 2017-1.2.1-NKP-2017-00002 (T.N), the Fulbright Foreign Student Program (T.N.), the Cure Alzheimer’s Fund (T.W.-C.), Nan Fung Life Sciences (T.W.-C.), the NOMIS Foundation (T.W.-C.), the Stanford Brain Rejuvenation Project (an initiative of the Stanford Wu Tsai Neurosciences Institute), the Paul F. Glenn Center for Aging Research (T.W.-C.), NIA R01 AG045034; DP1 AG053015 (T.W.-C.) and the NIA funded Stanford Alzheimer’s Disease Research Center P50AG047366, NIA K23AG051148 (S.M.), R01AG061155 (S.M.), American Federation for Aging Research (S.M.), R01AG044829 (J.V. and N.B), NIA R01AG057909 (N.B.), the Nathan Shock Center of Excellence for the Basic Biology of Aging P30AG038072 (N. B.), the Glenn Center for the Biology of Human Aging (N.B.).
Footnotes
Competing Interests
The authors declare no competing financial interests.
References
- 1.Harman D The aging process: major risk factor for disease and death. Proc Natl Acad Sci U S A 88, 5360–5363 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baht GS, et al. Exposure to a youthful circulaton rejuvenates bone repair through modulation of beta-catenin. Nat Commun 6, 7131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conboy IM, et al. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 433, 760–764 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Huang Q, et al. A Young Blood Environment Decreases Aging of Senile Mice Kidneys. J Gerontol A Biol Sci Med Sci 73, 421–428 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Katsimpardi L, et al. Vascular and neurogenic rejuvenation of the aging mouse brain by young systemic factors. Science 344, 630–634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loffredo FS, et al. Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell 153, 828–839 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salpeter SJ, et al. Systemic regulation of the age-related decline of pancreatic beta-cell replication. Diabetes 62, 2843–2848 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha M, et al. Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science 344, 649–652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villeda SA, et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature 477, 90–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villeda SA, et al. Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat Med 20, 659–663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valdes AM, Glass D & Spector TD Omics technologies and the study of human ageing. Nat Rev Genet 14, 601–607 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Stegeman R & Weake VM Transcriptional Signatures of Aging. J Mol Biol 429, 24272437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aramillo Irizar P, et al. Transcriptomic alterations during ageing reflect the shift from cancer to degenerative diseases in the elderly. Nat Commun 9, 327 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castellano JM, et al. Human umbilical cord plasma proteins revitalize hippocampal function in aged mice. Nature 544, 488–492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Angelantonio E, et al. Efficiency and safety of varying the frequency of whole blood donation (INTERVAL): a randomised trial of 45 000 donors. Lancet 390, 2360–2371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gubbi S, et al. Effect of Exceptional Parental Longevity and Lifestyle Factors on Prevalence of Cardiovascular Disease in Offspring. Am J Cardiol 120, 2170–2175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J & Rossi J Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov 16, 440 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Emilsson V, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 361, 769–773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gold L, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PloS one 5, e15004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Austad SN & Fischer KE Sex Differences in Lifespan. Cell metabolism 23, 10221033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostan R, et al. Gender, aging and longevity in humans: an update of an intriguing/neglected scenario paving the way to a gender-specific medicine. Clin Sci (Lond) 130, 1711–1725 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka T, et al. Plasma proteomic signature of age in healthy humans. Aging Cell 17, e12799 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen AA Aging across the tree of life: The importance of a comparative perspective for the use of animal models in aging. Biochimica et biophysica acta. Molecular basis of disease 1864, 2680–2689 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Somalogic. Non-human samples in the SOMAscan™ assay. Technical Note. (2015).
- 25.Guiraud S, et al. Identification of serum protein biomarkers for utrophin based DMD therapy. Scientific reports 7, 43697–43697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang RN, et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis 1, 87–105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun BB, et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sattlecker M, et al. Alzheimer’s disease biomarker discovery using SOMAscan multiplexed protein technology. Alzheimers Dement 10, 724–734 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Sullivan KD, et al. Trisomy 21 causes changes in the circulating proteome indicative of chronic autoinflammation. Sci Rep 7, 14818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganz P, et al. Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients With Stable Coronary Heart Disease. JAMA 315, 2532–2541 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Carayol J, et al. Protein quantitative trait locus study in obesity during weight-loss identifies a leptin regulator. Nat Commun 8, 2084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Go AS, et al. Heart disease and stroke statistics−−2013 update: a report from the American Heart Association. Circulation 127, e6–e245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franceschi C, Garagnani P, Parini P, Giuliani C & Santoro A Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat Rev Endocrinol 14, 576–590 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Franceschi C, et al. The Continuum of Aging and Age-Related Diseases: Common Mechanisms but Different Rates. Front Med (Lausanne) 5, 61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horvath S & Raj K DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet 19, 371–384 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Castellano JM, Kirby ED & Wyss-Coray T Blood-Borne Revitalization of the Aged Brain. JAMA neurology 72, 1191–1194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiklund FE, et al. Macrophage inhibitory cytokine-1 (MIC-1/GDF15): a new marker of all-cause mortality. Aging Cell 9, 1057–1064 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen E & Dillin A The insulin paradox: aging, proteotoxicity and neurodegeneration. Nat Rev Neurosci 9, 759–767 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suhre K, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun 8, 14357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sha SJ, et al. Safety, Tolerability, and Feasibility of Young Plasma Infusion in the Plasma for Alzheimer Symptom Amelioration Study: A Randomized Clinical Trial. JAMA neurology (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 41.Mehan MR, et al. Protein signature of lung cancer tissues. PloS one 7, e35157 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Britschgi M, et al. Modeling of pathological traits in Alzheimer’s disease based on systemic extracellular signaling proteome. Mol Cell Proteomics 10, M111 008862 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franceschi C, et al. Genetics of healthy aging in Europe: the EU-integrated project GEHA (GEnetics of Healthy Aging). Ann N Y Acad Sci 1100, 21–45 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Fox J & Weisberg S An R Companion to Applied Regression, (SAGE Publications, 2011). [Google Scholar]
- 45.Benjamini Y & Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300 (1995). [Google Scholar]
- 46.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanehisa M, Furumichi M, Tanabe M, Sato Y & Morishima K KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45, D353–D361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Croft D, et al. The Reactome pathway knowledgebase. Nucleic Acids Res 42, D472–477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexa A & Rahnenfuhrer J topGO: Enrichment Analysis for Gene Ontology (2016).
- 50.Yu G, Wang LG, Han Y & He QY clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlson M org.Hs.eg.db: Genome wide annotation for Human. (2017).
- 52.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castellano JM, et al. In vivo assessment of behavioral recovery and circulatory exchange in the peritoneal parabiosis model. Sci Rep 6, 29015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pagès H, Aboyoun P, Gentleman R & DebRoy S Biostrings: Efficient manipulation of biological strings. (2019).
- 55.Dray S & Dufour AB The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software 22, 1–20 (2007). [Google Scholar]
- 56.Friedman J, Hastie T & Tibshirani R Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 33, 1–22 (2010). [PMC free article] [PubMed] [Google Scholar]
- 57.Lehallier B, et al. Combined Plasma and Cerebrospinal Fluid Signature for the Prediction of Midterm Progression From Mild Cognitive Impairment to Alzheimer Disease. JAMA neurology 73, 203–212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang M, Zhao Y & Zhang B Efficient Test and Visualization of Multi-Set Intersections. Sci Rep 5, 16923 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Epskamp S, Cramer A, Waldorp L, Schmittmann V & Borsboom D qgraph: Network Visualizations of Relationships in Psychometric Data. Journal of Statistical Software, Articles 48, 1–18 (2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We created a searchable web interface to mine the human INTERVAL and LonGenity datasets (https://twc-stanford.shinyapps.io/aging_plasma_proteome/)
The Human independent cohorts and mouse protein data are available in Supplementary Tables 16 and 17. The INTERVAL data is available through the European Genome-Phenome Archive (https://ega-archive.org/studies/EGAS00001002555).