Abstract
Aim of the study
Micro-ribonucleic acids (miRNA) are small single stranded RNA molecules. They act as key regulators of several cellular processes such as proliferation, apoptosis, tumor differentiation, invasion and metastasis. Hepatocellular carcinoma (HCC) represents the most common primary liver cancer. miRNA-224 is an oncomiR that is highly upregulated in HCC tissues. The aim of the present study was to measure the relative expression of circulating miRNA-224 in the serum of patients with HCV-related liver cirrhosis and HCC and to assess its usefulness in the diagnosis of HCC.
Material and methods
Forty-eight patients were classified into two groups: 24 HCV-related HCC patients (HCC group), and 24 HCV-related liver cirrhosis patients (LC group). A third group included 24 healthy volunteers (control group). Clinical examination, imaging studies and routine laboratory investigations, including serum α-fetoprotein (AFP), were done. Quantification of serum miRNA-224 expression was performed using real time reverse transcription polymerase chain reaction (RT-PCR).
Results
The relative expression of serum miRNA-224 was significantly higher in HCC patients compared to LC patients and healthy control subjects. Its level correlated positively with the serum concentration of AFP and with Barcelona Clinic Liver Cancer (BCLC) stage of HCC. By combining miRNA-224 relative expression with AFP, their diagnostic sensitivity, specificity and accuracy increased significantly (95.0%, 92.1% and 93.2%, respectively) compared with either of the two markers alone in discriminating HCC from liver cirrhosis.
Conclusions
Serum miRNA-224 relative expression may aid in the diagnosis of HCC. Better diagnostic performance is obtained if miRNA-224 is combined with other tumor markers such as AFP.
Keywords: hepatocellular carcinoma, epigenetics, α-fetoprotein, miRNA-224
Introduction
According to a national Egyptian health survey in 2015, Egypt possesses one of the highest recorded prevalences of hepatitis C virus (HCV) in the world. Among the 15 to 59-year age group, the prevalence of positive HCV antibodies was 10% and that of positive HCV RNA was 7%, where 92.5% of patients are infected with genotype 4 [1, 2]. HCV is one of the main risk factors for developing hepatocellular carcinoma (HCC). HCC represents the most common primary liver cancer, the fifth most common cancer around the world and the third most frequent cause of cancer-related death, with about 600,000 cases discovered every year, of which more than 80% occur in developing countries [3]. Despite years of extensive research, HCC is still poorly understood, and very little progress has been made to improve its diagnosis and prognosis [4]. Identifying new clinical biomarkers for early detection of HCC would therefore contribute to increasing the survival rate of patients with this lethal disease.
Micro-ribonucleic acids (miRNAs) are small non-coding single stranded RNA molecules with a length of about 18-25 nucleotides. They have the ability of controlling the expression of their target messenger RNA (mRNA) by binding to its 3’ untranslated regions (3-UTRs), leading to alteration of its stability and translational ability [5]. They are known as key regulators of several cellular processes such as proliferation, apoptosis, tumor differentiation, invasion and metastasis. They have been detected in various human diseases including cancer, where they may be upregulated or downregulated in the neoplastic cells in comparison to their normal counterparts [6]. Therefore, they are now considered to be candidate non-invasive molecular biomarkers for cancer diagnosis. Recently, several studies have succeeded to detect miRNAs in the plasma/serum of HCC patients in a remarkably stable form. This step has opened a new window of opportunity for the development of non-invasive diagnostic tests of HCC without the need to obtain tissue samples from patients with severe liver diseases [7-9].
Micro-ribonucleic acid 224 (miRNA-224) is an oncomiR which plays a role in tumor development and progression by binding to 3’UTR regions of multiple genes that are involved in cell apoptosis, proliferation, migration, invasion and autophagy [10-13]. Previous studies have found that miRNA-224 had significantly higher expression in cervical cancer, cancer colon, pancreatic ductal adenocarcinoma, breast cancer and HCC [11, 14-17]. The aim of this study was to assess the expression of circulating miRNA-224 in serum of patients with HCV-related cirrhosis and hepatocellular carcinoma and to compare these results to normal control subjects in order to explore its usefulness as a non-invasive biomarker for identification of HCC.
Material and methods
Study population
The study was performed on 48 consecutive patients presenting to the Hepatology Unit of the Medical Research Institute Hospital, Alexandria University, Egypt enrolled during the period from December 2017 to April 2018. They were classified into two groups: 24 patients with hepatocellular carcinoma (HCC group) complicating HCV-related liver cirrhosis, and 24 patients with HCV-related liver cirrhosis (LC group) of matching age and sex. All patients had a negative history of receiving antiviral therapy, and all HCC patients had a negative history of interventional modalities for management of HCC (such as radiofrequency ablation, transarterial chemoembolization or cryotherapy). Serum samples were also collected from 24 healthy volunteers (control group) of matching age and sex. Liver cirrhosis was diagnosed based on clinical, laboratory and ultrasound findings, and the severity of liver disease was assessed using the Child-Turcotte-Pugh (CTP) score and class [18]. HCC cases were diagnosed according to the guidelines of the American Association for the Study of Liver disease (AASLD) published in 2011, which relied on detecting a hepatic focal lesion by ultrasound, further confirmed by a contrast-enhanced triphasic computed tomography (CT) scan study or magnetic resonance imaging (MRI) [19]. The Barcelona Clinic Liver Cancer (BCLC) system was used for staging of HCC cases [20]. Written informed consent forms were collected from all subjects involved in the study and the study protocol was approved by the local Ethics Committee of the Medical Research Institute in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans. Exclusion criteria included HBV infection or any other identifiable cause of chronic hepatitis other than HCV, any associated malignancies other than HCC and prior treatment for HCV.
Laboratory investigations
Routine laboratory investigations were done for all subjects, including complete blood count (CBC), international normalized ratio (INR), serum alanine transaminase (ALT), aspartate transaminase (AST), albumin, total bilirubin (T-Bil), alkaline phosphatase (ALP), γ-glutamyl transpeptidase (GGT), blood urea nitrogen (BUN) and creatinine. Routine chemistry tests were done on an Olympus AU 400 autoanalyzer (Beckman Coulter International SA, Nyon, Switzerland). Serum levels of a-fetoprotein (AFP) were measured by solid phase two sequential chemiluminescent immunometric assay using an Immulite 1000 system analyzer (Siemens Healthineers GmbH, Erlangen, Germany), and values up to 20 ng/ml were considered normal.
RNA extraction
Total RNA including small RNAs was extracted from 200 µl of serum using a QIAGEN miRNeasy mini kit (Cat. no. 217004) according to specified instructions, then total RNA including small RNAs was further diluted with 50 µl of RNase-free water. Total RNA concentration and purity were measured with the NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA), and samples were stored at –80°C.
RT-PCR quantification
Reverse transcription reaction was performed on 100 ng total RNA using a miRNA Select Hi/Lo-Rox kit according to the manufacturer’s instructions (BIOLINE). PCR amplification was performed after the miRNA was reverse transcribed into complementary DNA (cDNA), and the reaction setup of quantitative PCR (qPCR) was prepared as follows: 10 µl 2X SansiSMART PCR master mix, 2 µl miRNA-224 primer (EPIK miRNA Select Hi/low – ROX Kit), and 5 µl cDNA were mixed, then nuclease-free water was added to reach a final volume of 20 µl. The housekeeping miRNA-39 was used as an internal reference. The cycling conditions were: 1 cycle at 95°C for 10 minutes and 40°C for 5 minutes, then 40 cycles at 95°C for 10 seconds and 60°C for 30 seconds.
Raw Ct values were collected using the software supplied with the real time PCR instrument (Applied Biosystems 7000 Sequence Detector). The difference between the Ct values (ΔCt) of the target gene and housekeeping gene for each sample was calculated and the calibrated ΔCt value (ΔΔCt) for each sample was determined (ΔΔCt = ΔCt of target gene – ΔCt of control samples). The relative gene copy number was calculated by the expression 2-ΔΔCt [21].
Statistical methods
Data were analyzed using IBM SPSS advanced statistics version 20 (SPPS IBM Corp., Chicago, IL). Numerical data were expressed as mean ± standard deviation (SD) for parametric data or median (minimum-maximum) for nonparametric data as appropriate. Qualitative data were expressed as frequency and percentage. The chi-square test was used to examine the relation between qualitative variables. For non-normally distributed quantitative data, comparison between two groups was performed using the Mann-Whitney test (non-parametric t test). Comparison between three groups was done using the Kruskal-Wallis test (non-parametric ANOVA) then the post-hoc test was used for pair-wise comparison based on Kruskal-Wallis distribution. Correlation between the variables was analyzed using Pearson’s correlation coefficient. To evaluate the diagnostic value of serum miRNA-224 in HCC, receiver operating characteristic (ROC) curve analysis was performed with prediction of the best possible cutoff values in which sensitivity was plotted on the Y axis and 1-specificity on the X axis. In all tests, p value was considered significant if less than 0.05.
Results
Demographic data and clinical findings of the studied subjects are presented in Table 1. There was no statistically significant difference between the three groups regarding age and sex. There was also no significant difference between HCC and LC groups as regards CTP score and class (p > 0.05). BCLC staging of HCC patients revealed that more than half of them were at the end-stage of the disease (62.5%), while nearly one third of them (33.4%) were at early and very early stages. Triphasic CT evaluation of HCC patients showed that the majority of them had 2-3 or > 3 focal lesions on presentation (62.5%). The tumor involved both lobes of the liver in 54.2% of patients, with malignant portal vein thrombosis (macrovascular invasion) detected in 33.3%, lymph node involvement in 20.8%, and extrahepatic spread in only 4.2% of patients. The mean size of the largest focal lesion was 6.9 ±2.7 cm (Table 2).
Table 1.
Demographic and clinical data of the studied groups
| Parameter | HCC (n = 24) | LC (n = 24) | Control (n = 24) | p* |
|---|---|---|---|---|
| Age (years) | 56.4 ±7.9 | 55.5 ±6.5 | 54.4 ±5.3 | 0.122 |
| Male sex, n (%) | 15 (62.5) | 14 (58.33) | 15 (62.5) | 0.104 |
| Child score | 10.9 ±1.9 | 10.0 ±2.7 | – | 0.092 |
| Child class, n (%) | ||||
| A | 0 (0) | 0 (0) | – | 0.211 |
| B | 9 (37.5) | 11 (45.8) | – | |
| C | 15 (62.5) | 13 (54.2) | – | |
| BCLC stage of HCC, n (%) | ||||
| Very early | 4 (16.7) | – | – | – |
| Early | 4 (16.7) | – | – | – |
| Intermediate | 1 (4.2) | – | – | – |
| Advanced | 15 (62.5) | – | – | – |
HCC – hepatocellular carcinoma, LC – liver cirrhosis, BCLC – Barcelona Clinic Liver Cancer, n – number
statistically significant at p < 0.05
Table 2.
Radiologic tumor-related characteristics of hepatocellular carcinoma (HCC) group (n = 24)
| Parameter | |
|---|---|
| Number of focal lesions, n (%) | |
| Single | 9 (37.5) |
| Two/three | 7 (29.2) |
| More than three | 8 (33.3) |
| Size of largest focal lesion (cm) | 6.9 ±2.7 |
| Liver lobes involved, n (%) | |
| One lobe | 11 (45.8) |
| Both lobes | 13 (54.2) |
| Portal vein thrombosis, n (%) | 8 (33.3) |
| Lymph node metastasis, n (%) | 5 (20.8) |
| Extrahepatic spread, n (%) | 1 (4.2) |
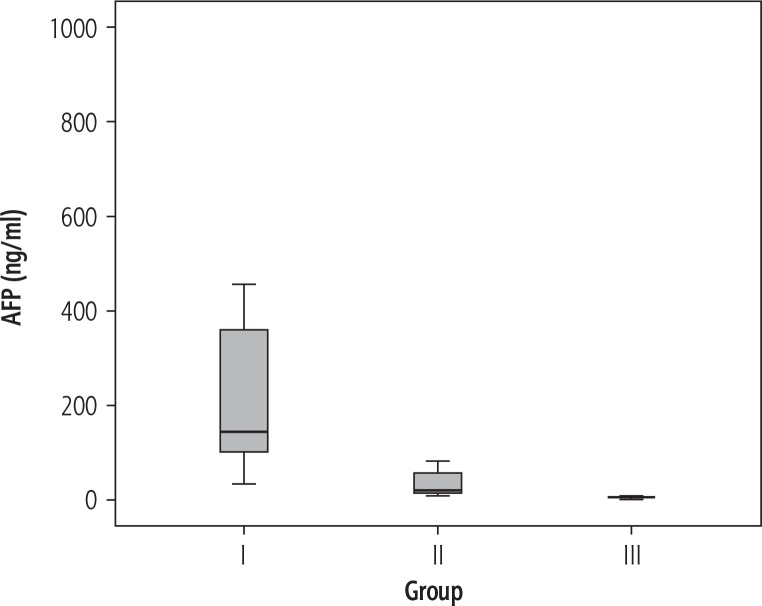
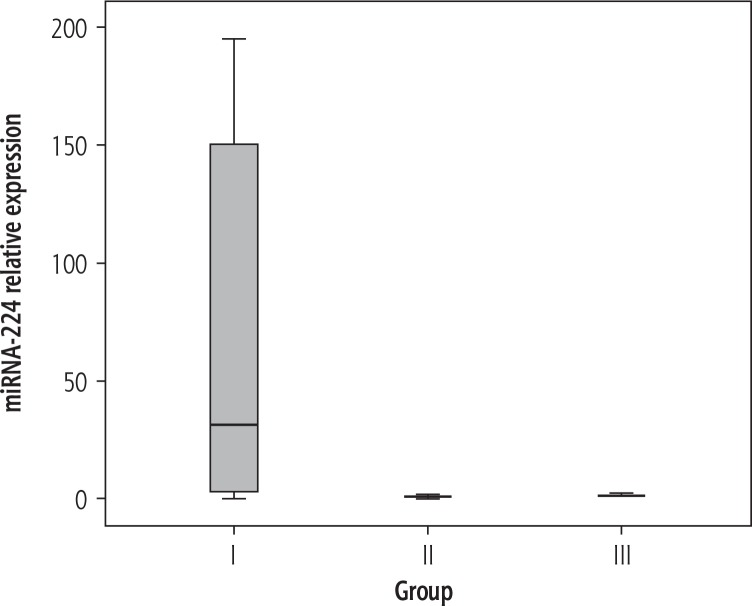
Table 3 shows the results of routine laboratory investigations as well as the relative expression of miRNA-224 of all groups. The serum levels of ALT, AST and BUN proved to be significantly higher in HCC compared to LC patients, while serum albumin, Hb concentration and platelet count were significantly lower (p < 0.05). Serum concentrations of AFP (Fig. 1) as well as the relative expression of miRNA-224 (Fig. 2) were significantly higher in HCC compared to the LC group and the control group (p < 0.05), while no significant difference in miRNA-224 expression was found between LC patients and control subjects (p = 0.296).
Table 3.
Laboratory data and miRNA-224 relative expression in the studied groups
| Parameter | HCC (n = 24) | LC (n = 24) | Ctrl (n = 24) | p1 HCC/LC | p2 HCC/Ctrl | p3 LC/Ctrl |
|---|---|---|---|---|---|---|
| ALT (U/l) | 96.4 ±159.1 | 33.7 ±22.9 | 17.0 ±2.6 | 0.031* | 0.009* | 0.001* |
| AST (U/l) | 185.3 ±340.9 | 61.3 ±45.7 | 16.6 ±3.9 | 0.042* | 0.010* | 0.000* |
| Albumin (g/dl) | 2.3 ±0.4 | 2.6 ±0.5 | 4.4 ±0.2 | 0.027* | 0.000* | 0.000* |
| T-Bil (mg/dl) | 7.5 ±7.7 | 5.7 ±8.7 | 0.6 ±0.1 | 0.228 | 0.000* | 0.003* |
| ALP (U/l) | 126.3 ±79.1 | 98.5 ±28.3 | 101.8 ±10.6 | 0.056 | 0.070 | 0.298 |
| GGT (U/l) | 49.0 ±33.1 | 41.5 ±20.8 | 17.4 ±4.2 | 0.178 | 0.000* | 0.000* |
| BUN (mg/dl) | 64.9 ±51.1 | 44.1 ±30.6 | 11.9 ±1.8 | 0.047* | 0.000* | 0.000* |
| Creatinine (mg/dl) | 1.3 ±0.6 | 1.1 ±0.2 | 0.8 ±0.1 | 0.062 | 0.000* | 0.000* |
| INR | 1.6 ±0.3 | 1.4 ±0.6 | 1.0 ±0.0 | 0.192 | 0.000* | 0.000* |
| Hb (g/dl) | 10.1 ±1.9 | 9.1 ±2.0 | 13.4 ±1.0 | 0.034* | 0.000* | 0.000* |
| WBC count (× 103/µl) | 6.9 ±3.0 | 6.7 ±4.1 | 6.3 ±1.4 | 0.433 | 0.177 | 0.302 |
| Platelets count (× 103/µl) | 97.7 ±52.2 | 58.5 ±127.2 | 226.5 ±69.8 | 0.018* | 0.00* | 0.013* |
| AFP (ng/ml) | 253.7 ±223.5 | 39.04 ±30.5 | 5.75 ±2.87 | 0.004* | 0.001* | 0.045* |
| miRNA-224 | 64.7 ±73.1 | 1.3 ±1.3 | 1.1 ±0.4 | 0.001* | 0.001* | 0.297 |
HCC – hepatocellular carcinoma, LC – liver cirrhosis, Ctrl – normal control group, n – number, ALT – alanine aminotransferase, AST – aspartate aminotransferase, T-Bil – total bilirubin, ALP – alkaline phosphatase, GGT – g-glutamyl transpeptidase, BUN – blood urea nitrogen, INR – international normalized ratio, Hb – hemoglobin, WBC – white blood cell, AFP – α-fetoprotein, miRNA-224 – micro-ribonucleic acid 224 (presented as relative expression), p1 – comparison between HCC and LC, p2 – comparison between HCC and normal control group, p3 – comparison between LC and normal control group
statistically significant at p < 0.05
Fig. 1.
Box plot of α-fetoprotein (AFP) in the different studied groups (group I – HCC, group II – liver cirrhosis, group III – normal controls)
Fig. 2.
Box plot of miRNA-224 relative expression in the different studied groups (group I – HCC, group II – liver cirrhosis, group III – normal controls)
A significant positive correlation was found between miRNA-224 expression and liver profile parameters (ALT, AST, ALP, and INR) as well as renal function tests (BUN and creatinine), while it correlated negatively with serum albumin and platelet count. Also, higher levels of miRNA-224 expression were associated with more severe liver disease (in terms of CTP class) and with higher BCLC stages of HCC. However, the association of miRNA-224 expression with the different radiologic characteristics of HCC was variable. It was positively associated only with extrahepatic spread of HCC (one patient). Other tumor characteristics revealed either a negative association (i.e. number of focal lesions, liver lobes involved and lymph node metastases) or completely no relation (i.e. size of largest focal lesion and malignant portal vein thrombosis) with miRNA-224 expression (Table 4).
Table 4.
Relation between miRNA-224 relative expression and the different laboratory and clinical parameters
| Parameter | r (p-value) | Parameter | Mean ±SD of miRNA-224 | p-value |
|---|---|---|---|---|
| ALT | 0.342 (0.003)* | Child class | 0.003* | |
| AST | 0.278 (0.018)* | B | 12.48 ±31.80 | |
| Albumin | –0.353 (0.002)* | C | 47.64 ±71.34 | |
| T-Bil | 0.123 (0.304) | BCLC stage | 0.013* | |
| ALP | 0.357 (0.002)* | Very early | 20.17 ±23.15 | |
| GGT | 0.062 (0.605) | Early | 5.69 ±6.73 | |
| BUN | 0.538 (0.00)* | Intermediate | 137.19 ±0.00 | |
| Creatinine | 0.256 (0.03)* | Advanced | 87.47 ±78.43 | |
| INR | 0.308 (0.008)* | Number of FLs | 0.003* | |
| Hb | –0.183 (0.123)* | Single | 100.88 ±63.79 | |
| WBCs | –0.083 (0.490)* | Two/three | 21.09 ±51.22 | |
| Platelets | –0.357 (0.002)* | More than three |
62.14 ±83.93 | |
| AFP involved |
0.26 (0.027)* | Liver lobes | 0.003* | |
| One lobe | 118.21 ±62.04 | |||
| Size of largest FL | –0.003 (0.987) | Both lobes | 42.66 ±66.78 | |
| PV thrombosis | 0.825 | |||
| Positive | 69.65 ±68.38 | |||
| Negative | 62.22 ±77.36 | |||
| LN metastasis | 0.001* | |||
| Positive | 18.77 ±16.10 | |||
| Negative | 76.78 ±77.60 | |||
| Extrahepatic spread |
0.013* | |||
| Positive | 137.19 ±0.00 | |||
| Negative | 61.54 ±73.01 |
ALT – alanine aminotransferase, AST – aspartate aminotransferase, T-Bil – total bilirubin, ALP – alkaline phosphatase, GGT – g-glutamyl-transpeptidase, BUN – blood urea nitrogen, PT – prothrombin time, INR – international normalized ratio, Hb – hemoglobin, WBCs – white blood cells, AFP – α-fetoprotein, miRNA-224 – micro-ribonucleic acid 224, FL – focal lesion, BCLC – Barcelona Clinic Liver Cancer, n – number, PV – portal vein, LN – lymph node, r – Pearson correlation coefficient
statistically significant at p < 0.05
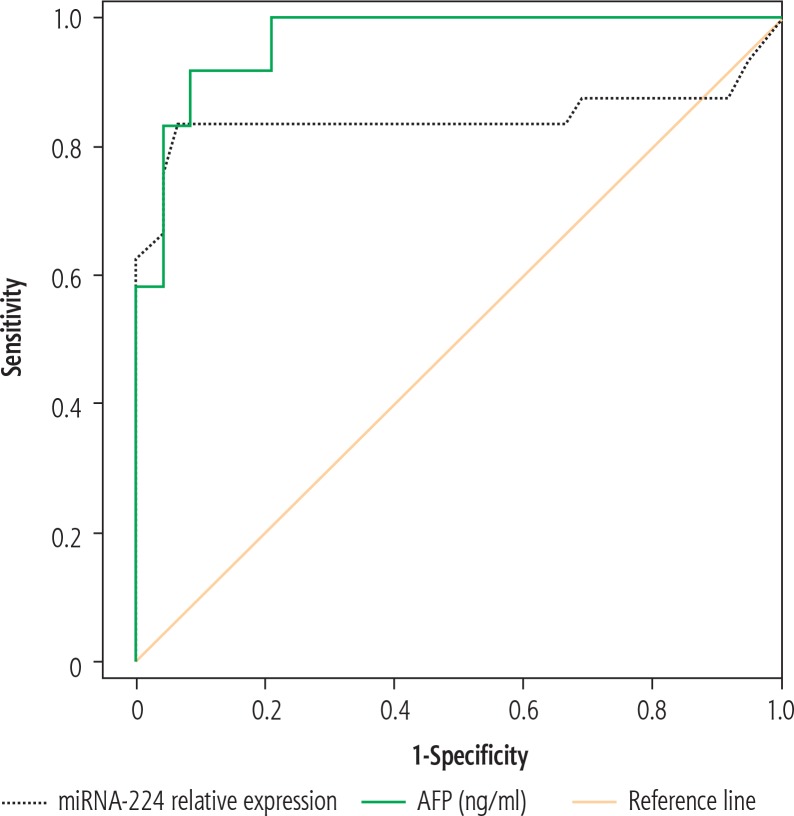
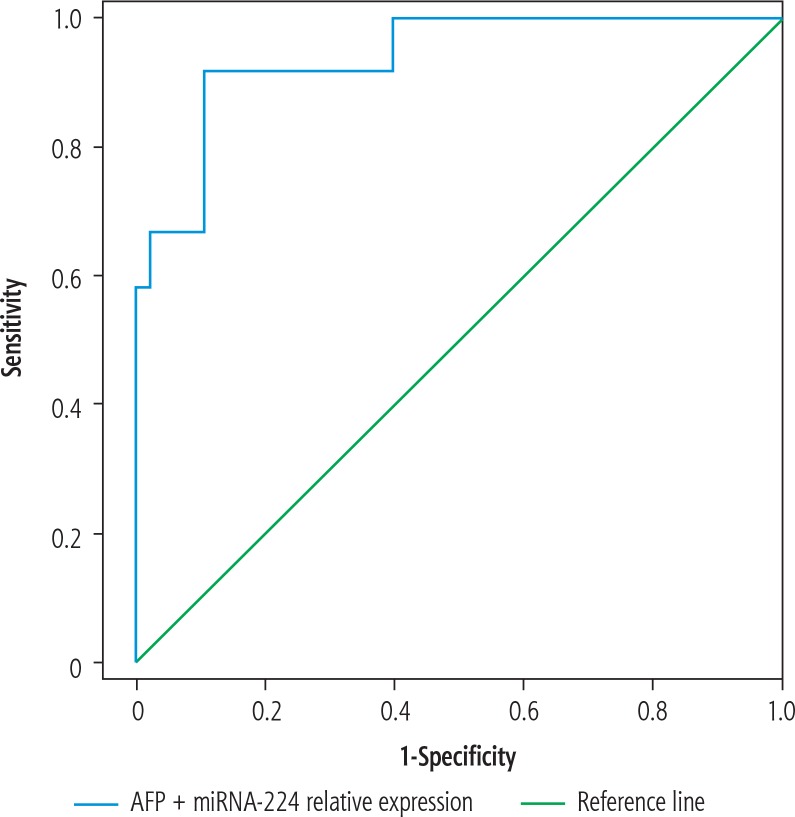
Nevertheless, a significant positive correlation existed between miRNA-224 expression and serum AFP level (p = 0.027), as shown in Table 4. ROC curve analysis also showed high sensitivity and specificity of both markers in distinguishing HCC patients from LC patients, being slightly higher for AFP than miRNA-224 (Table 5, Fig. 3). Combined together, both markers showed a sensitivity of 95.0% and a specificity of 92.1% in predicting HCC from LC patients (Table 5, Fig. 4).
Table 5.
Diagnostic performance of miRNA-224 relative expression and α-fetoprotein in predicting hepatocellular carcinoma (HCC) in liver cirrhosis patients
| Cut off | AUC | Sensitivity | Specificity | Accuracy | Asymptotic Sig. | Asymptotic 95% confidence interval | ||
|---|---|---|---|---|---|---|---|---|
| Lower bound | Upper bound | |||||||
| miRNA-224 | 2.00 | 0.844 | 86.0 | 81.0 | 83.0 | 0.001* | 0.710 | 0.977 |
| AFP (ng/ml) | 20.0 | 0.965 | 94.0 | 90.0 | 92.0 | 0.0001* | 0.930 | 0.99 |
| Combined miRNA-224 and AFP | 0.939 | 95.0 | 92.1 | 93.2 | 0.0001* | 0.885 | 0.994 | |
AFP – α-fetoprotein, miRNA-224 – micro-ribonucleic acid 224, AUC – area under the curve
Fig. 3.
Receiver-operator characteristic (ROC) curves comparing miRNA-224 relative expression and α-fetoprotein (AFP) in predicting hepatocellular carcinoma (HCC)
Fig. 4.
Receiver-operator characteristic (ROC) curve demonstrating combined miRNA-224 and α-fetoprotein (AFP) in predicting hepatocellular carcinoma (HCC)
Discussion
Hepatocellular carcinoma is a complex disease which is difficult to diagnose by a single biomarker. Even serum AFP, the only acceptable tumor marker of HCC, cannot be used alone for diagnosis, but rather must be combined with positive radiologic findings of the disease [19]. Moreover, the false negative results of AFP alone may be as high as 40% for patients with early stage HCC [22]. The discovery of microRNAs has opened new horizons of studying novel non-invasive molecular biomarkers for cancer diagnosis [23]. The aim of the present work was to measure the relative expression of circulating miRNA-224 in the serum of patients with HCV-related liver cirrhosis and HCC and to assess its usefulness in the diagnosis of HCC.
In the present study, serum miRNA-224 relative expression was found to be significantly upregulated in HCC patients compared to liver cirrhosis patients and healthy controls. These results were in agreement with Gui et al. [24] and Mamdouh et al. [25], who found that the circulating miRNA-224 was upregulated in the serum of HCC patients compared to control samples, and its expression was significantly higher in the serum of patients with high grade tumors.
The expression of miRNA-224 in HCC tissues and its role in the progression of the disease have been investigated by several researchers, who declared miRNA-224 as being one of the most commonly overexpressed miRNAs affecting crucial cellular pathways in HCC pathogenesis [4, 11]. One study demonstrated that this molecule acted as a master regulator of cell cycle progression and its overexpression accelerated cell growth [26]. Wang et al. reported that it inhibited tumor cell apoptosis by targeting apoptosis inhibitor 5 (API-5) and promoting cell growth [27]. Others stated that miR-224 functioned as an oncomiR in HCC through activating the AKT signaling pathway and promoting malignant hepatocyte proliferation and migration [10, 28]. Zhang et al. explained this malignant cell activation by targeting CDC42, CDH1, PAK2, BCL-2, and MAPK1 genes [11], while Scisciani et al. linked the miR-224 upregulation with the activation of lipopolysaccharide, lymphotoxin α and tumor necrosis factor α inflammatory pathways, which promoted cell migration/invasion in HCC [29].
The relation between tissue expression and circulating miRNA-224 in HCC was also investigated by a few researchers. Okajima et al. [9] and Lin et al. [30] revealed that miRNA-224 expression was significantly higher in HCC tissues and HCC cell lines than in normal hepatic tissues, and that the plasma miRNA-224 levels were also significantly higher in HCC compared with non-HCC patients, being significantly correlated with paired miR-224 levels of HCC tissues.
In the present work, serum miRNA-224 was positively correlated with several clinical aspects of the disease, including BCLC stage of HCC, as well as laboratory liver profile parameters (ALT, AST, ALP, INR) and – most importantly – serum concentration of AFP. These outcomes were very similar to the results of Zhuang et al., who found that serum miR-224 levels were significantly higher in the BCLC stage C patients compared to stage B and showed a significant relation with parameters of liver damage and serum AFP. They explained this by the link of miRNA-224 to liver inflammatory pathways. They also observed a positive relation of miRNA-224 with malignant portal vein thrombosis, but not with extra-liver metastasis [31]. In comparison, our results showed variable relation patterns of serum miRNA-224 with the different tumor characteristics. It was positively associated only with extrahepatic spread of HCC, while other characteristics were either negatively associated or had no relation to miRNA-224 expression. These inconclusive findings might be attributed to the relatively small number of HCC patients included in our work, and to the lack of facilities necessary for conducting a more detailed analysis of a possible relation between serum miRNA-224 and other tumor characteristics such as total tumor size, which is considered a limitation of our work.
We also evaluated the diagnostic performance of serum miRNA-224 expression in distinguishing HCC from liver cirrhosis patients by analysis of the receiver operating characteristic (ROC) curve, which showed 86% sensitivity and 81% specificity. These results were a bit lower than the conventional marker AFP which showed higher values (94% and 90%, respectively). However, by combining miR-224 expression with AFP, the diagnostic sensitivity and specificity increased significantly (95% and 92.1%, respectively) compared with either of the two markers alone in discriminating HCC from liver cirrhosis. These results were in agreement with the study of Lin et al. [30], who found that combining serum miRNA-224 with AFP had a better performance in discriminating HCC from liver cirrhosis and chronic hepatitis B, and the study of Amr et al. [32], who stated that miRNA-224 might be useful as a potential biomarker in predicting early HCC stages. Nevertheless, our study had a few other limitations. Despite the fact that about 92.5% of Egyptian patients are infected with HCV genotype 4 [2] and that genotyping is therefore not routinely done in Egypt, the current study should have conducted a more detailed analysis of the possible relation between serum miRNA-224 and several viral characteristics such as genotypes and HCV viral load.
Conclusions
The results of this study suggest the possible utility of serum miRNA-224 relative expression for the diagnosis of HCC in the future, although it does not yet seem to be ready as a diagnostic tool at the present time. Alone, it barely equals AFP, but in combination with the latter, some discriminative value is added. Further research involving a larger number of patients is recommended, which should allow testing a more elaborate panel of miRNAs and discovering more possible relations of these novel HCC markers to tumor and viral characteristics.
Disclosure
The authors report no conflict of interest.
References
- 1.Kandeel A, Genedy M, El-Refai S, et al. The prevalence of hepatitis C virus infection in Egypt 2015: implications for future policy on prevention and treatment. Liver Int. 2017;37:45–53. doi: 10.1111/liv.13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Omran D, Alboraie M, Zayed RA, et al. Towards hepatitis C virus elimination: Egyptian experience, achievements and limitations. World J Gastroenterol. 2018;24:4330–4340. doi: 10.3748/wjg.v24.i38.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Abd NE, Fawzy NA, El-Sheikh SM, et al. Circulating miRNA-122, miRNA-199a, and miRNA-16 as biomarkers for early detection of hepatocellular carcinoma in Egyptian patients with chronic hepatitis C virus infection. Mol Diagn Ther. 2015;19:213–220. doi: 10.1007/s40291-015-0148-1. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Lee CG. Role of miR-224 in hepatocellular carcinoma: a tool for possible therapeutic intervention? Epigenomics. 2011;3:235–243. doi: 10.2217/epi.11.5. [DOI] [PubMed] [Google Scholar]
- 5.Su H, Yang JR, Xu T, et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 6.Mourad L, El-Ahwany E, Zoheiry M, et al. Expression analysis of liver-specific circulating microRNAs in HCV-induced hepatocellular carcinoma in Egyptian patients. Cancer Biol Ther. 2018;19:400–406. doi: 10.1080/15384047.2018.1423922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ichikawa D, Komatsu S, Konishi H, et al. Circulating microRNA in digestive tract cancers. Gastroenterology. 2012;142:1074–1078.e1. doi: 10.1053/j.gastro.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Diaz G, Melis M, Tice A, et al. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int J Cancer. 2013;133:816–824. doi: 10.1002/ijc.28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okajima W, Komatsu S, Ichikawa D, et al. Circulating micro-RNA profiles in plasma: identification of miR-224 as a novel diagnostic biomarker in hepatocellular carcinoma independent of hepatic function. Oncotarget. 2016;7:53820–53836. doi: 10.18632/oncotarget.10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma D, Tao X, Gao F, et al. miR-224 functions as an onco-miRNA in hepatocellular carcinoma cells by activating AKT signaling. Oncol Lett. 2012;4:483–488. doi: 10.3892/ol.2012.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Takahashi S, Tasaka A, et al. Involvement of micro-RNA-224 in cell proliferation, migration, invasion, and anti-apoptosis in hepatocellular carcinoma. J Gastroenterol Hepatol. 2013;28:565–575. doi: 10.1111/j.1440-1746.2012.07271.x. [DOI] [PubMed] [Google Scholar]
- 12.Li Q, Ding C, Chen C, et al. miR-224 promotion of cell migration and invasion by targeting Homeobox D 10 gene in human hepatocellular carcinoma. J Gastroenterol Hepatol. 2014;29:835–842. doi: 10.1111/jgh.12429. [DOI] [PubMed] [Google Scholar]
- 13.Lan SH, Wu SY, Zuchini R, et al. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology. 2014;59:505–517. doi: 10.1002/hep.26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen SN, Wang LF, Jia YF, et al. Upregulation of microRNA-224 is associated with aggressive progression and poor prognosis in human cervical cancer. Diagn Pathol. 2013;8:69. doi: 10.1186/1746-1596-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mees ST, Mardin WA, Sielker S, et al. Involvement of CD40 targeting miR-224 and miR-486 on the progression of pancreatic ductal adenocarcinomas. Ann Surg Oncol. 2009;16:2339–2350. doi: 10.1245/s10434-009-0531-4. [DOI] [PubMed] [Google Scholar]
- 16.Zhang GJ, Zhou H, Xiao HX, et al. Up-regulation of miR-224 promotes cancer cell proliferation and invasion and predicts relapse of colorectal cancer. Cancer Cell Int. 2013;13:104. doi: 10.1186/1475-2867-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L, Dai T, Lin X, et al. MicroRNA-224 targets RKIP to control cell invasion and expression of metastasis genes in human breast cancer cells. Biochemv Biophys Res Commun. 2012;425:127–133. doi: 10.1016/j.bbrc.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 18.Cholongitas E, Papatheodoridis GV, Vangeli M, et al. Systematic review: The model for end-stage liver disease – should it replace Child-Pugh’s classification for assessing prognosis in cirrhosis? Aliment Pharmacol Ther. 2005;22:1079–1089. doi: 10.1111/j.1365-2036.2005.02691.x. [DOI] [PubMed] [Google Scholar]
- 19.Bruix J, Sherman M. American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forner A, Reig ME, de Lope CR, et al. Current strategy for staging and treatment: the BCLC update and future prospects. Semin Liver Dis. 2010;30:61–74. doi: 10.1055/s-0030-1247133. [DOI] [PubMed] [Google Scholar]
- 21.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C (T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 22.Tsuchiya N, Sawada Y, Endo I, et al. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2015;21:10573–10583. doi: 10.3748/wjg.v21.i37.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hauptman N, Glavac D. MicroRNAs and long non-coding RNAs: prospects in diagnostics and therapy of cancer. Radiol Oncol. 2013;47:311–318. doi: 10.2478/raon-2013-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gui J, Tian Y, Wen X, et al. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond) 2011;120:183–193. doi: 10.1042/CS20100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mamdouh S, Khorshed F, Aboushousha T, et al. Evaluation of Mir-224, Mir-215 and Mir-143 as serum biomarkers for HCV associated hepatocellular carcinoma. Asian Pac J Cancer Prev. 2017;18:3167–3171. doi: 10.22034/APJCP.2017.18.11.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An F, Olaru AV, Mezey E, et al. MicroRNA-224 induces G1/S checkpoint release in liver cancer. J Clin Med. 2015;4:1713–1728. doi: 10.3390/jcm4091713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Lee AT, Ma JZ, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205–13215. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- 28.Ladeiro Y, Couchy G, Balabaud C, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 29.Scisciani C, Vossio S, Guerrieri F, et al. Transcriptional regulation of miR-224 upregulated in human HCCs by NFκB inflammatory pathways. J Hepatol. 2012;56:855–861. doi: 10.1016/j.jhep.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 30.Lin L, Lu B, Yu J, et al. Serum miR-224 as a biomarker for detection of hepatocellular carcinoma at early stage. Clin Res Hepatol Gastroenterol. 2016;40:397–404. doi: 10.1016/j.clinre.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Zhuang LP, Meng ZQ. Serum miR-224 reflects stage of hepatocellular carcinoma and predicts survival. Biomed Res Int. 2015;2015:731781. doi: 10.1155/2015/731781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amr KS, Elmawgoud Atia HA, Elazeem Elbnhawy RA, et al. Early diagnostic evaluation of miR-122 and miR-224 as biomarkers for hepatocellular carcinoma. Genes Dis. 2017;4:215–221. doi: 10.1016/j.gendis.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]