Abstract
Eukaryotic cells are compartmentalized to form organelles, whose functions rely on proper phospholipid and protein transport. Here we determined the crystal structure of human VAT-1, a cytosolic soluble protein that was suggested to transfer phosphatidylserine, at 2.2 Å resolution. We found that VAT-1 transferred not only phosphatidylserine but also other acidic phospholipids between membranes in vitro. Structure-based mutational analyses showed the presence of a possible lipid-binding cavity at the interface between the two subdomains, and two tyrosine residues in the flexible loops facilitated phospholipid transfer, likely by functioning as a gate to this lipid-binding cavity. We also found that a basic and hydrophobic loop with two tryptophan residues protruded from the molecule and facilitated binding to the acidic-lipid membranes, thereby achieving efficient phospholipid transfer.
Keywords: endoplasmic reticulum (ER), mitochondria, lipid transport, lipid-protein interaction, X-ray crystallography
Introduction
Eukaryotic cells are highly compartmentalized to form membrane-bound organelles. Normal functions of each organelle rely on its specific set of resident proteins and optimized composition of membrane phospholipids. Phospholipids are made mainly in the endoplasmic reticulum (ER)3 membrane and partly in the mitochondrial inner membrane (IM) and other organelle membranes like Golgi membranes and then distributed specifically to all the cellular membranes. Thus, a cell needs to meet the demand of hydrophobic phospholipid molecules to be transferred between cellular membranes across the aqueous compartments and hence provides efficient and specific lipid trafficking systems (1–3). Generally, two distinct mechanisms can operate for phospholipid transfer between membranes: one by soluble lipid-transfer proteins and the other by membrane contact sites (4).
Biosynthesis of phosphatidylethanolamine (PE), a major phospholipid of mitochondrial membranes, occurs in spatially separated organelles, the ER and mitochondria. Phosphatidylserine (PS), a precursor of PE, is mainly generated in the ER and then transferred from the ER membrane to the mitochondrial IM via the mitochondrial outer membrane (OM) (5). PS decarboxylase, Psd1 in yeast, at the outer surface of the mitochondrial IM catalyzes decarboxylation of PS to generate PE (6). Similarly, cardiolipin (CL), a mitochondrial signature phospholipid, is generated in the mitochondrial IM from phosphatidic acid (PA), which is mainly made in the ER membrane (7), via several intermediate phospholipids. Production of mitochondrial PE and CL thus relies on efficient transport of PS and PA, respectively, from the ER membrane to the mitochondrial IM via the mitochondrial OM. PS and PA transfer between the mitochondrial OM and IM is mediated by soluble lipid-transfer protein complexes: the Ups2–Mdm35 and Ups1–Mdm35 complexes in yeast, respectively, (the SLMO2–TRIAP1 and PRELID1–TRIAP1 complexes in human) (8–11). PS in the OM at the contact sites between the OM and IM was also proposed to be converted to PE directly by the IM-localized Psd1 acting in trans (12).
Phospholipid transfer from the ER to mitochondria may involve direct physical contacts between the membranes of two organelles (13). In yeast, the best-characterized interorganelle contact site is formed by the ER–mitochondrial encounter structure (ERMES), a protein complex consisting of the integral ER membrane protein Mmm1, integral mitochondrial OM proteins Mdm10 and Mdm34, and the peripheral mitochondrial OM protein Mdm12 (14). Mdm12 and Mmm1 can bind to phospholipid (15, 16), and the Mdm12–Mmm1 complex can transfer lipids efficiently between lipid liposomes in vitro (16), and lacks of one of the ERMES subunits or mutations in Mmm1- or Mdm12-impaired PS transfer from the isolated ER membranes to mitochondria in vitro (16, 17). In mammalian cells, although accumulated evidence suggests the presence of ER–mitochondrial contact sites, identification of the components constituting those contact sites is still going on, including PDZD8, for example, as a putative mammalian ortholog of yeast Mmm1 (18).
VAT-1 (synaptic vesicle membrane protein VAT-1 homolog), a conserved cytosolic soluble protein in vertebrates (19), was shown to be involved in PS transfer from the ER to mitochondria using Xenopus egg components (20). Sequence analysis showed that VAT-1 belongs to the alcohol dehydrogenase family and possesses a conserved nicotinamide cofactor-binding motif (21). Overexpression of VAT-1 in HeLa cells induces mitochondrial fragmentation, whereas knockdown of VAT-1 induces extensive growth of mitochondrial network structures (22), suggesting the function of VAT-1 related to mitochondria. In addition to the proposed role in the PS transfer, VAT-1 was also found to be an acidic phospholipid-binding protein (23, 24), and cell fractionation of rat liver cell extracts showed that VAT-1 was localized mainly to the cytosol but partially to both the ER and mitochondria (22). However, how VAT-1 binds to membranes and transports PS from the ER to the mitochondrial OM remains largely unclear.
In the present study, we performed structural and functional analyses of human VAT-1. We determined the crystal structure of human VAT-1 at 2.2 Å resolution, which showed structural similarity to no other lipid-transfer proteins with known structures. The determined structure and structure-based mutational analyses suggested molecular mechanisms of VAT-1 binding to membranes and transfer of acidic phospholipids between membranes.
Results and discussion
Phospholipid transfer activity of VAT-1
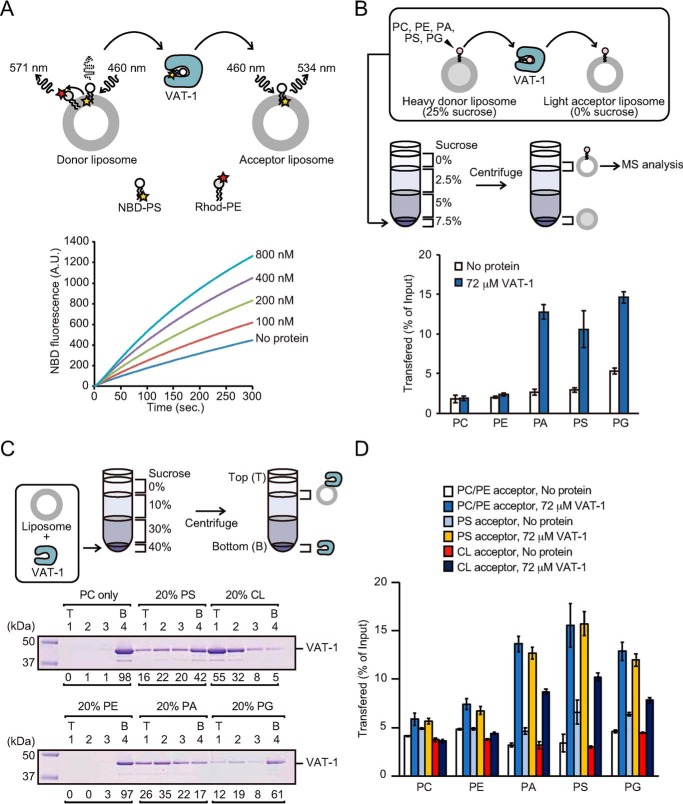
A previous study using Xenopus egg extracts showed that the recombinant VAT-1 protein stimulated PS transport from the ER into mitochondria in vitro (20). To test whether VAT-1 has a PS transfer activity between lipid membranes, we performed in vitro fluorescence-based PS transfer assays using purified human VAT-1 and lipid liposomes. Donor liposomes containing nitobenzoxadiazole-labeled PS (NBD-PS) and rhodamine-labeled PE (Rhod-PE), which quenched NBD fluorescence, were incubated with acceptor liposomes without fluorescent lipids in the presence of different concentrations of purified VAT-1, and transfer of NBD-PS to the acceptor liposomes was monitored as an increase in the NBD fluorescence. VAT-1 efficiently increased NBD fluorescence in a time- and dose-dependent manner, indicating that VAT-1 transports NBD-PS between liposomes (Fig. 1A). The NBD-PS transfer rates were obtained from the time course of the NBD fluorescence with and without VAT-1 (Fig. S2). The NBD-PS transfer rate is 0.061 and 0.148 nmol [NBD-PS]/min in the absence and presence of VAT-1, respectively (Fig. S2). Therefore, the NBD-PS transfer rate per VAT-1 molecule is 0.044 NBD-PS molecule/min/VAT-1 molecule. A previous NBD-phospholipid transfer assay using the Mmm1–Mdm12 complex of the ERMES complex, showed the NBD-phospholipids transfer rate per Mmm1–Mdm12 complex is 1.0–3.0 NBD-phospholipid molecule/min/Mmm1–Mdm12 molecule (16). The phospholipid transfer rate of VAT-1 is thus 23–68 times slower than that of Mmm1–Mdm12 complex.
Figure 1.
Phospholipid transfer and membrane-binding activities of VAT-1. A, PS transfer activity of VAT-1 was measured at 25 °C by the fluorescent-based PS transfer assay between liposomes shown in above schematic diagram. B, phospholipids transfer activities of VAT-1 were measured by the MS-based phospholipid transfer assay. VAT-1 was incubated with heavy donor liposomes containing PC, PE, PA, PS, and PG and light acceptor liposomes. Liposomes were separated by sucrose density-gradient ultracentrifugation. The amounts of each indicated phospholipids in acceptor liposomes relative to those in input donor liposomes were quantified by MS. The values are means ± S.D. of three independent experiments. C, VAT-1 was incubated with liposomes containing PC alone (PC only), PC/PS = 80/20 (20% PS), PC/CL = 80/20 (20% CL), PC/PE = 80/20, PC/PA = 80/20 (20% PA), or PC/PG = 80/20 (20% PG), and binding was analyzed by the liposome flotation assay. T, top; B, bottom. The amounts of VAT-1 in each fraction are shown below the gel (total VAT-1 amounts were set to 100). D, phospholipids transfer activities of VAT-1 were measured as in B. VAT-1 was incubated with heavy donor liposomes (0.25 mm; DOPC/DOPE/POPC/POPE/DOPA/DOPS/DOPG/18:1 NBD-PE = 55:19.5:5:5:5:5:5:0.5) and light acceptor liposomes (1 mm; DOPC/DOPE/18:1 Liss-Rhod-PE = 55:44.95:0.05) (1 mm; DOPC/DOPE/POPS/18:1 Liss-Rhod-PE = 55:34.95:10:0.05) or (1 mm; DOPC/DOPE/18:1 CL/18:1 Liss-Rhod-PE = 55:34.95:10:0.05), and the amounts of phospholipids in accepter liposomes were analyzed. The values are means ± S.D. of three independent experiments.
We then assessed phospholipid headgroup specificity of the VAT-1–mediated lipid transfer by the MS-based phospholipid transfer assay, which was free from the influence of the attached fluorescent NBD group. Heavy donor liposomes containing phospholipids with defined acyl chains and light acceptor liposomes lacking these phospholipids were incubated with VAT-1, and phospholipids in the acceptor liposomes were analyzed by MS analysis after their isolation from the donor liposomes by density-gradient centrifugation (Fig. 1B and Fig. S1). It is to be noted that phospholipid with a polar NBD-labeled acyl chain is less stable in the lipid bilayer than phospholipid without NBD (25), so that a transfer rate of NBD-free PS tended to be much slower than that of NBD-PS. Interestingly, negatively charged PA and phosphatidylglycerol (PG) as well as PS were transferred to acceptor liposomes, whereas neutral phosphatidylcholine (PC) and PE were not. Therefore, VAT-1 can transport negatively charged, not neutral, phospholipids between liposomes.
VAT-1 is a soluble protein but could partially associate with the ER and mitochondrial membranes (22). To assess the membrane association of VAT-1, we performed flotation experiments using liposomes composed of PC (80%) and various phospholipids (20%) (Fig. 1C). VAT-1 bound to liposomes containing negatively charged CL most efficiently and bound to those with acidic PA, PS, or PG, but not with neutral PC or PE, to some extent (Fig. 1C). However, the MS-based phospholipid transfer assay showed that VAT-1–mediated acidic lipid transfer was hardly affected by the presence of negatively charged phospholipids in the acceptor liposomes (Fig. 1D). Therefore, acidic phospholipids, especially CL, promote stable binding of VAT-1 to lipid membranes, yet such stable VAT-1 binding to membranes is not essential for efficient acidic phospholipid transfer by VAT-1.
Structure of VAT-1
To gain more insight into the mechanism of lipid transfer by VAT-1, we decided to determine the high-resolution structure of VAT-1ΔN40, a VAT-1 variant lacking the N-terminal 40 residues, which were not conserved among VAT-1 from different organisms (Fig. S3) and predicted to be disordered (26). VAT-1ΔN40 transferred NBD-PS between liposomes as efficiently as full-length VAT-1 (Fig. S4A). The crystal structure of VAT-1ΔN40 was determined by the molecular replacement method using the crystal structure of synaptic vesicle membrane protein VAT-1 homolog-like protein (VAT-1L; PDB code 4A27), which has a very low NBD-PS transfer activity compared with VAT-1 (Fig. S4B), as a search model and subsequently refined to 2.2 Å resolution (Table 1 and Fig. 2A).
Table 1.
Data collection, phasing and refinement statistics
One crystal was used for the collection of VAT-1 data for structure refinement. The statistic values for the highest resolution shell are shown in parentheses. Rpim is the precision-indicating merging R factor.
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 67.16, 131.72, 174.03 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Data set | |
| Wavelength (Å) | 1.00000 |
| Resolution range (Å) | 50.0–2.20 |
| Outer shell (Å) | 2.25–2.20 |
| Rmerge | 0.080 (1.042) |
| Rpim | 0.034 (0.441) |
| I/σI | 11.2 (1.6) |
| Completeness (%) | 99.7 (94.6) |
| Redundancy | 3.4 (3.3) |
| CC½ | 0.998 (0.648) |
| Refinement | |
| Resolution (Å) | 50–2.20 |
| No. reflections | 78589 |
| R/Rfree | 0.222/0.254 |
| No. atoms | |
| Protein | 9985 |
| Nitrate ion | 32 |
| Water | 158 |
| B-factors (Å2) | |
| Protein | 59.1 |
| Nitrate ion | 87.7 |
| Water | 56.8 |
| Root-mean-square deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.3 |
| PDB accession code | 6K9Y |
Figure 2.
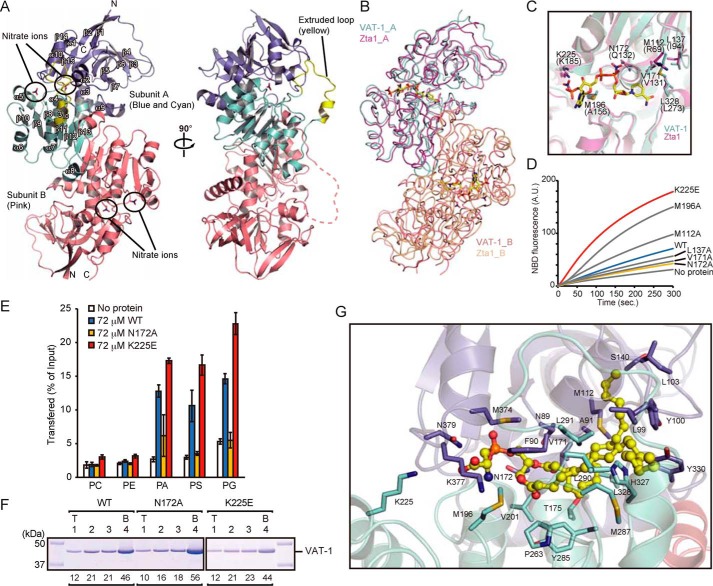
Crystal structure of VAT-1 and phospholipid-binding site. A, a ribbon diagram of the structure of VAT-1. Domain I, domain II, and the extruded loop region in subunit A are colored in blue, cyan, and yellow, respectively, and subunit B in pink. Nitrate ions are shown in stick form. The α-helices, β-strands, and a 310 helix are labeled. B, superposition of VAT-1 and Zta1. Subunit A and B of VAT-1 are colored in cyan and pink, respectively, and subunit A and B of Zta1 in magenta and light brown, respectively. Nitrate ions and NADPH molecules are shown in stick form with carbon in yellow. C, superposition of VAT-1 (cyan) and Zta1 (magenta) for the magnified nitrate-ion binding site of VAT-1. Nitrate ions and NADPH are shown in stick form with carbon in yellow. VAT-1 residues responsible for nitrate-ion binding are indicated. Zta1 residues are indicated in parentheses. D, PS transfer activities of VAT-1 mutants (1 μm) were measured at 25 °C as in Fig. 1A. E, phospholipids transfer activities of VAT-1 mutants were measured as in Fig. 1B. The values are means ± S.D. of three independent experiments. F, VAT-1 mutants were incubated with liposomes containing PC/PS = 80/20 (20% PS), and binding was analyzed as in Fig. 1C. T, top; B, bottom. The amounts of VAT-1 in each fraction are shown below the gel (total VAT-1 amounts were set to 100). G, a docking model of the VAT-1–DOPS complex. The DOPS molecule is shown in stick and ball representation with carbon in yellow. The side chains of Lys-225 and residues within 4 Å of the DOPS molecule are shown in stick form.
The crystallographic asymmetric unit contained four VAT-1ΔN40 molecules. Two molecules each assembled to form similar homodimers. Consistently, when we analyzed the oligomerization of VAT-1 and VAT-1ΔN40 by gel filtration, the apparent molecular weight of VAT-1ΔN40 corresponded to that of a dimer (Fig. S4C). Apparent molecular weight of full-length VAT-1 was much larger, yet this could be due to the presence of the N-terminal disordered 40-residue segment. The determined structure suggested that VAT-1ΔN40 was comprised of two structural units, domain I (residues 46–172 and 329–387) and domain II (residues 173–328) (Fig. 2A). Domain I formed a twisted β-sheet consisting of six β-strands (β3–β6 and β14–β15) flanked by five α-helices (α1–α3, α9, and α10) (Fig. S3). Domain II exhibited two repeated Rossmann folds (27) consisting of β8-α4-β9-α5-β10 and β11-α7-β12-α8-β13 (Fig. S3). Residual electron densities were observed in the internal cavity formed at the interface between domains I and II. The size of the electron densities is too large to be fitted to water molecules, and the shapes of the electron densities are different from those of tris or ethylene glycol in the cryo-protectant solution. On the other hand, nitrate ions derived from the crystallization solution were fitted rather well to the electron densities. Thus, we tentatively assigned nitrate ions to these electron densities (Fig. 2A).
Putative phospholipid-binding site
A query to the Dali server (28) revealed that NAD(P)H dependent quinone oxidoreductase and alcohol dehydrogenase had structural similarities with VAT-1ΔN40, but among them, Zta1, a yeast NADPH-dependent quinone oxidoreductase, complexed with NADPH (PDB code 3QWB) (29) exhibited high structural similarity (Fig. 2B). Interestingly, nitrate ions in the structure of VAT-1ΔN40 were positioned at the β-phosphate and 2′-phosphate groups of the bound NADPH molecule in the structure of Zta1 (Fig. 2C). However, gel-filtration analyses did not detect association of NADPH to VAT-1 (Fig. S4D), and NADPH reportedly gave little effect on in vitro PS transport activity of VAT-1 using Xenopus egg extracts (20). Replacement of Ala-156 in Zta1 with bulky Met-196 in VAT-1 may prevent NADPH from binding to VAT-1. In the Zta1 structure, side chains of Gln-132 and Lys-185 are hydrogen-bonded to the β-phosphate and 2′-phosphate oxygen atoms of the NADPH adenosine moiety, respectively, and a quinone molecule is suggested to bind to the nearby hydrophobic region (29). Likewise, in the VAT-1ΔN40 structure, side chains of Asn-172 and Lys-225 are within the distance that allows the formation of hydrogen bonds with two nitrate oxygen atoms, and hydrophobic residues (Met-112, Leu-137, Val-171, and Leu-328) are clustered in the proximity of the nitrate ions. We thus hypothesize that a phospholipid molecule can bind to VAT-1 with its phosphate group occupying the position of the nitrate ions and its acyl-chain binding to the hydrophobic region.
To test the validation of the role of the possible lipid-binding cavity of VAT-1, we made mutations N172A or K225E in the putative phosphate-binding site and M196A, M112A, L137A, or V171A in the putative hydrophobic acyl-chain binding site and tested their lipid transfer activities (Fig. 2C). Fluorescence-based NBD-PS transfer assays showed that although the N172A mutation decreased NBD-PS transfer activity, the K225E mutation even increased the activity (Fig. 2D). MS-based lipid transfer assays further showed that the N172A mutation suppressed the transfer of only acidic phospholipids (PA, PS, and PG), not neutral phospholipid (PC and PE) but that the K225E mutation enhanced the transfer of both neutral and acidic phospholipids (Fig. 2E). Therefore, although Asn-172 may bind to the negatively charged head group of acidic phospholipids, Lys-225 may not be directly involved in the headgroup binding. The liposome floatation assays showed that the N172A or K225E mutation did not affect the VAT-1 binding to PS-containing liposomes (Fig. 2F), suggesting that Asn-172 or Lys-225 is not involved in membrane binding. The mutations in the hydrophobic region gave divergent results; although mutations at hydrophobic residues 137 and 171 reduced the NBD-PS transfer, those at hydrophobic residues 112 and 196 enhanced the NBD-PS transfer (Fig. 2D). Perhaps hydrophobicity and steric hindrance of the hydrophobic region may counteract with each other to result in different consequences. CD spectra showed that L137A, V171A, and N172A mutations that decreased the PS transfer activity did not affect the overall structure of VAT-1 (Fig. S5A).
We generated a docking model of the VAT-1–DOPS complex using the AutoDock Vina suite (30). DOPS was docked into the putative phospholipid-binding cavity of the crystal structure of VAT-1 (Fig. 2G and Fig. S6A). As expected, the phosphoserine moiety of DOPS was positioned at the nitrate ion–binding site composed of Asn-172, Met-196, Met-374, Lys-377, and Asn-379, and acyl-chain tails were positioned at the neighboring hydrophobic region composed of mainly hydrophobic 19 residues (Fig. 2G). The docking model thus supports the PS binding to the hypothesized phospholipid-binding cavity.
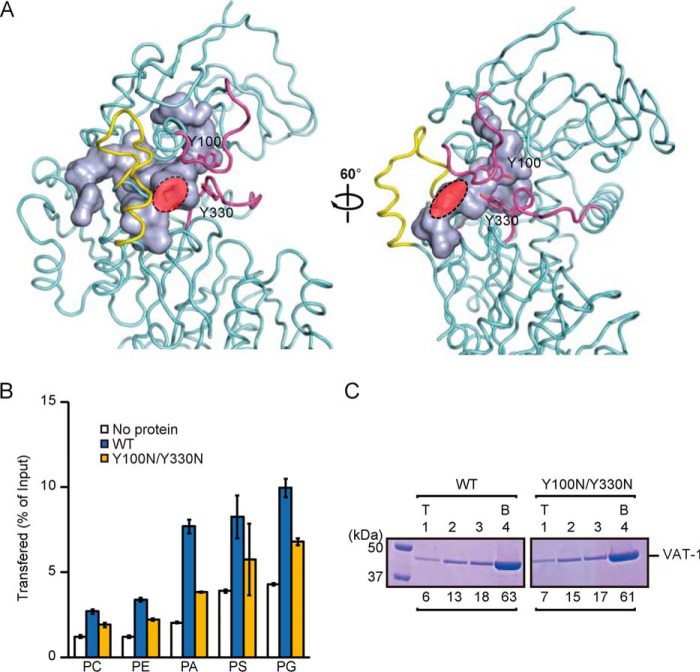
In the crystal structure of VAT-1, we found a twin-loop structure (segments 98–107 and 326–335) near the putative phospholipid-binding site, which showed relatively high crystallographic B-factor (Fig. S6B), indicating its flexible nature. The two tyrosine residues (Tyr-100 and Tyr-330) in the two loops interact with each other to form a “closed” conformation with a narrow opening (red region in Fig. 3A) for the internal hydrophobic cavity as a phospholipid-binding site (light blue in Fig. 3A). These Tyr-containing loops could function as a flexible gate for the phospholipid-binding site, like a flexible lid of other phospholipid-transfer proteins (31), to conceal the bound phospholipid from the aqueous solvent (Fig. 3A). To further assess the role of two tyrosine residues in the phospholipid transfer, we replaced these two tyrosine residues with a hydrophilic residue Asn (Y100N/Y330N) and performed the phospholipid transfer assay and liposome flotation assay using 20% PS-containing liposomes. The Y100N/Y330N mutation decreased the acidic phospholipids transfer activity but did not affect the binding to PS-containing liposomes (Fig. 3, B and C), suggesting that these two tyrosine residues act as a gate for the internal phospholipid-binding site. CD spectra confirmed that the Y100N/Y330N mutation did not affect the overall structure of VAT-1 (Fig. S5B).
Figure 3.
The twin-loop structure with two Tyr facilitates phospholipid transfer. A, the internal cavity and main-chain folding of VAT-1. The cavity is visualized by a light blue molecular surface. The twin-loop structure (residues 99–102 and 327–332) and the extruded loop (residues 291–306) are colored in magenta and yellow, respectively. Tyr-100 and Tyr-330 are shown in stick form. The opening of the cavity is indicated in red. B, phospholipid transfer activities of WT VAT-1 and the Y100N/Y330N mutant were measured as in Fig. 1B. The values are means ± S.D. of three independent experiments. C, WT VAT-1 and the Y100N/Y330N mutant were incubated with liposomes containing PC/PS = 80/20 (20% PS), and binding was analyzed as in Fig. 1C. T, top; B, bottom. The amounts of VAT-1 in each fraction are shown below the gel (total VAT-1 amounts were set to 100).
The extruded loop is responsible for phospholipid transfer and membrane binding
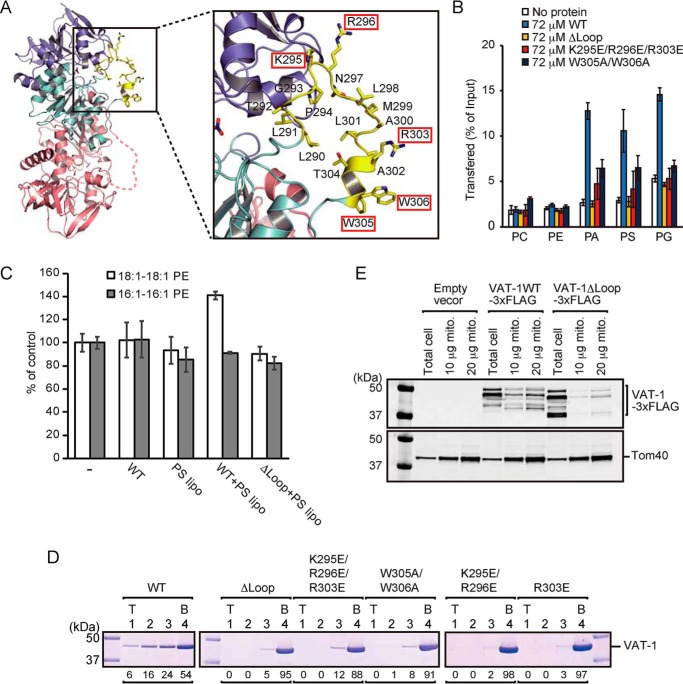
In the crystal structure, we noted that VAT-1 has, in addition to the above-described twin-loop structure, an extruded flexible loop (residues 290–306), whereas Zta1 does not have such a loop. This extruded loop of VAT-1 is rich in hydrophobic residues including two neighboring Trp residues (Trp-305 and Trp-306) and has three positively charged residues (Lys-295, Arg-296, and Arg-303) (Fig. 4A). Because VAT-1 is able to transfer acidic phospholipids, this extruded loop could play a role in acidic phospholipid transfer by VAT-1. To test this possibility, we made three types of VAT-1 mutants—the ΔLoop mutant lacking the loop; the K295E/R296E/R303E mutant, in which the positively charged residues in the loop are replaced with Glu; and the W305A/W306A mutant lacking twin Trp in the loop—and subjected them to MS-based phospholipid transfer assays between liposomes. Evidently, those VAT-1 mutants exhibited markedly reduced transfer activities for PA, PS, and PG, and the acidic lipid transfer by the ΔLoop mutant was even completely abolished (Fig. 4B). CD spectra confirmed that those mutations do not affect the overall structure of VAT-1 (Fig. S5C). Therefore, the extruded loop of VAT-1 is essential for the acidic phospholipid transfer activity between liposomes.
Figure 4.
The extruded flexible loop is responsible for the phospholipids transfer and the membrane binding. A, magnified view of the region around the extruded flexible loop, shown in stick form, of VAT-1. Lys-295, Arg-296, Arg-303, Trp-305, and Trp-306 are indicated by red boxes. B, phospholipid transfer activities of VAT-1 mutants were measured by the MS-based phospholipid transfer assay as in Fig. 1B. The values are means ± S.D. of three independent experiments. C, amounts of 16:1–16:1 PE, DOPE in isolated mitochondria incubated with DOPS-containing liposome (PS lipo), and purified WT VAT-1 (WT) or VAT-1ΔLoop (ΔLoop) relative to those in isolated mitochondria incubated without both liposomes and VAT-1 were measured by MS. The values are means ± S.D. of three independent experiments. D, VAT-1 mutants were incubated with liposomes containing PC/PS = 80/20 (20% PS), and binding was analyzed as in Fig. 1C. T, top; B, bottom. The amounts of VAT-1 in each fraction are shown below the gel (total VAT-1 amounts were set to 100). E, mitochondria were isolated from yeast cells overexpressing WT VAT-1 or VAT-1ΔLoop with C-terminal 3× FLAG tag. Total cell lysate and the indicated amounts of mitochondrial proteins were analyzed by SDS-PAGE followed by immunoblotting.
We then monitored lipid transfer from liposomes containing DOPS (18:1–18:1 PS) to isolated yeast mitochondria in the presence of VAT-1. After transfer from liposomes to mitochondria, DOPS will be converted to DOPE (18:1–18:1 PE) by PS decarboxylase Psd1 in the mitochondrial IM, which is monitored by MS analyses (Fig. 4C and Fig. S7). The amount of DOPE, but not 16:1–16:1 PE, increased only when mitochondria were incubated with DOPS-containing liposome and with WT VAT-1, not the VAT-1 ΔLoop mutant. The extruded loop of VAT-1 containing positively charged residues and two tryptophan residues is thus essential for acidic lipid transfer between liposomes or from liposomes to mitochondria like the case between liposomes.
How about membrane binding of these VAT-1 mutants? Liposome flotation assays using 20% PS-containing liposomes showed that the ΔLoop, K295E/R296E/R303E, and W305A/W306A mutants all failed to bind to PS containing liposomes (Fig. 4D). Although Lys-295 and Lys-296 are separated from Arg-303 by a distance of ∼16 Å (Fig. 4A), the VAT-1 mutants with the K295E/R296E mutation or R303E mutation alone failed to bind to PS-containing liposomes either (Fig. 4D). When we isolated mitochondria from yeast cells overexpressing VAT-1 with or without the extruded loop, the amount of VAT-1 recovered with mitochondria was significantly reduced by the loop deletion, although the flexible N terminus segment of VAT-1 appeared to be partially degraded (Fig. 4E). These results indicate that the extruded loop of VAT-1 is responsible for binding to mitochondria and acidic phospholipid membranes as well.
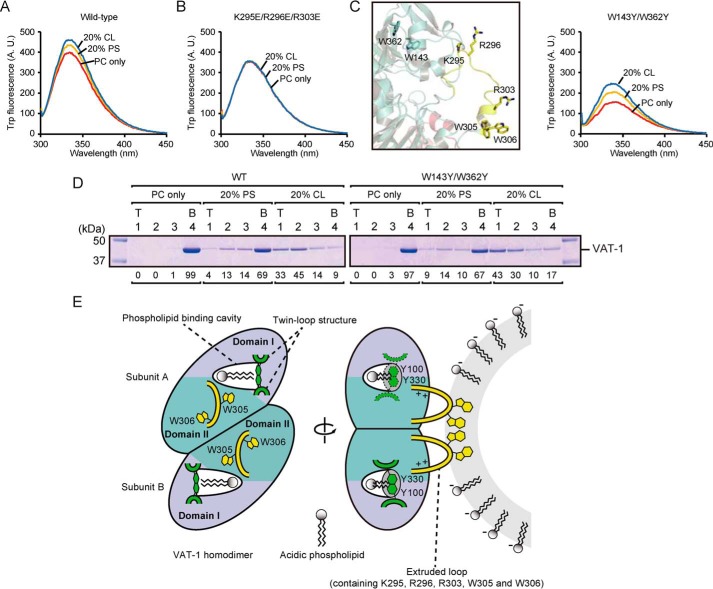
Twin Trp in the loop is inserted into the acidic membrane upon VAT-1 binding
We further focused on the roles of the two tryptophan residues (Trp-305 and Trp-306) in the extruded loop of VAT-1 in the membrane binding. Because Trp fluorescence is enhanced in the hydrophobic environment, we monitored the Trp fluorescence of VAT-1 upon binding to liposomes (Fig. 5A). When incubated with PC-only liposomes or PS- or CL-containing liposomes, intensities of Trp fluorescence of VAT-1 are in the order of CL-containing liposomes > PS-containing liposomes > PC-only liposomes (Fig. 5A), which corresponds to the order of liposome binding (Fig. 1C). On the other hand, the Trp fluorescence intensity of the K295E/R296E/R303E mutant, which lacks membrane-binding activity, did not change when incubated with PS- or CL-containing liposomes (Fig. 5B). In addition to Trp-305 and Trp-306 in the loop, VAT-1 has two more Trp residues (Trp-143 and Trp-362) in the interior of the molecule. We thus replaced Trp-143 and Trp-362 with Tyr residues (the W143Y/W362Y mutation) and measured Trp fluorescence of the W143Y/W362Y mutant (Fig. 5C). We could observe a more significant increase in the Trp fluorescence than WT VAT-1 upon binding to liposomes in the order of CL-containing liposomes > PS-containing liposomes > PC-only liposomes, which is consistent with the order of liposome binding (Fig. 5D). Therefore, Trp-305 and Trp-306 in the loop are directly involved in acidic lipid membrane binding.
Figure 5.
Role of two tryptophan residues in the extruded flexible loop upon binding to the acidic membrane. A and B, 25 μm WT VAT-1 (A) and K295E/R296E/R304E (B) was incubated with 1.5 mm liposomes containing PC alone (PC only), PC/PS = 80/20 (20% PS), or PC/CL = 80/20 (20% CL) at 25 °C for 10 min. Then tryptophan fluorescence spectra were recorded at 25 °C. λex = 295 nm, λem = 300–450 nm. C, left panel, a magnified view around Trp-143 and Trp-362, shown in stick form, in the extruded loop of VAT-1. Right panel, tryptophan fluorescence spectra of the W143Y/W362Y mutant with liposomes as in A and B. D, WT VAT-1 and the W143Y/W362Y mutant were incubated with liposomes containing PC alone (PC only), PC/PS = 80/20 (20% PS), or PC/CL = 80/20 (20% CL), and binding was analyzed as in Fig. 1C. T, top; B, bottom. The amounts of VAT-1 in each fraction are shown below the gel (total VAT-1 amounts were set to 100). E, left panel, a putative phospholipid-binding model of VAT-1. Two loops containing Tyr-100 and Tyr-330, shown in green, could function as a gate to the phospholipid binding cavity. Right panel, a membrane-binding model of VAT-1. The extruded flexible loops are shown in yellow. To extract acidic phospholipids from the membrane, VAT-1 recognizes acidic phospholipids in the membrane through basic residues in the loop, and then two tryptophan residues in the loop are inserted into the membrane.
Conclusion
We determined here the crystal structure of human VAT-1ΔN40. The determined structure, the substrate-docking model, and mutational analyses indicate several unique features of VAT-1 as a soluble phospholipid-transfer protein. First, substrate access to the possible acidic phospholipid-binding cavity of VAT-1 (Fig. 2, D, E, and G) is likely regulated by the two neighboring tyrosine residues (Tyr-100 and Tyr-330) in the twin-loop structure, which could perform a gating function (Fig. 5E). Second, the VAT-1 structure shows the presence of the extruded loop containing hydrophobic residues, two tryptophan residues (Trp-305 and Trp-306), and several positively charged residues, which is distinct from the above-mentioned gating twin-loop structure. This extruded loop appeared important for acidic phospholipid transfer as well as binding to membranes and mitochondria both in vitro and in vivo (Fig. 4). Binding of VAT-1 to the acidic phospholipid membrane through its extended loop is essential for acidic lipid transfer between the membranes. Because VAT-1ΔN40 and probably full-length VAT-1 form a dimer (Fig. S4C), the extruded flexible loop of each protomer is oriented toward the same direction in the VAT-1 dimer, so that the two protomers may cooperate to increase membrane-binding activity in the dimer (Fig. 5E).
In yeast, a counterpart of VAT-1 or a soluble PS transfer cytosolic protein is missing, but PS is transferred from the ER to mitochondria directly via the ER–mitochondrial physical contact site, ERMES (16, 17). In mammals, single well-defined ER–mitochondrial contact has not been established. PS transfer between the mammalian ER and mitochondria is thus most likely mediated by VAT-1 and perhaps other soluble PS transfer proteins, in addition to possible ER–mitochondria contact sites.
In summary, VAT-1 does not belong to any known superfamily of lipid-transfer proteins but was shown here to have a lipid transfer function. This suggests that there could be many more soluble proteins in the cytosol that function as lipid-transfer proteins but are hard to identify from the simple sequence analyses.
Experimental procedures
Construction of expression plasmids
To construct Escherichia coli expression plasmids for full-length VAT-1 (393 residues) or VAT-1ΔN40 (residues 41–393), the genes were amplified by PCR from the MegaMan human transcriptome cDNA Library (Agilent Technologies), digested with NdeI and XhoI, and cloned into the NdeI/XhoI site of pGBHPS (32). To construct E. coli expression plasmids for yeast Zta1, the ZTA1 gene was amplified by PCR from Saccharomyces cerevisiae W303-1A genome, digested with NdeI and XhoI, and cloned into the NdeI/XhoI site of pGBHPS. The pRS424/GAL1–VAT-1–3× FLAG plasmid was constructed as follows: the VAT-1 ORF was amplified by PCR, digested with BamHI and XhoI, and inserted into the BamHI/XhoI site between the GAL1 promoter and CYC1 terminator in the 2μ–TRP1 plasmid, pRS424, to yield pRS424/GAL1–VAT-1. A DNA fragment encoding the 3× FLAG sequence was inserted into pRS424/GAL1–VAT-1, just upstream of the stop codon of the VAT-1 gene, by inverse PCR, to yield pRS424/GAL1–VAT-1–3× FLAG. The expression plasmid for VAT-1ΔLoop (lacking residues 290–306) was constructed by inverse PCR. Mutations for the described amino acid substitutions were introduced by PCR-based site-directed mutagenesis.
Protein expression and purification
The B1 immunoglobulin domain of streptococcal protein G and hexahistidine (GB1-His6) fused N-terminally target proteins were expressed in E. coli strain C43 (DE3) (Lucigen) cultured in LB medium supplemented with 100 μg/ml ampicillin at 37 °C. When the A600 reached ∼0.8, isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.1 mm and cultured for 20 h at 20 °C to induce protein expression. The cells were then disrupted by sonication, and GB1-His6 tagged proteins were affinity-purified by a nickel–nitrilotriacetic acid Superflow column (Qiagen). The GB1-His6 tag was cleaved off with PreScission protease (GE Healthcare) and removed by a nickel–nitrilotriacetic acid Superflow column. For full-length VAT-1 and Zta1, the proteins were further purified by a HiLoad 26/60 Superdex 200 PG column (GE Healthcare) with elution buffer of 20 mm Tris-HCl, pH 7.5, and 150 mm NaCl. For VAT-1ΔN40, the protein was purified by a Resource Q anion-exchange column (GE Healthcare) with a linear NaCl gradient of 0–1000 mm in 20 mm Tris-HCl, pH 8.0, and then purified further by a HiLoad 26/60 Superdex 200 PG column (GE Healthcare) with elution buffer of 20 mm Tris-HCl, pH 7.5, 150 mm NaCl.
Liposomes
1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC, 850375C), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, 850457C), DOPE (850725C), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE, 850757C), 1,2-dioleoyl-sn-glycero-3-phosphate (DOPA, sodium salt, 840875C), DOPS (sodium salt, 840035C), 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DOPG, sodium salt, 840475C), 18:1 CL (1′,3′-bis[1,2-dioleoyl-sn-glycero-3-phospho-]-sn-glycerol, 710355C), 18:1–12:0 NBD-PS (1-oleoyl-2-{12-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphoserine, 810195C), 18:1 NBD-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2–1,3-benzoxadiazol-4-yl) ammonium salt, 810145C), and 18:1 Liss-Rhod-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl), ammonium salt, 810150C) were obtained from Avanti Polar Lipids, Inc. Liposomes were prepared as follows; lipids in chloroform stock solutions were mixed at a desired molar ratio, and the solvent was evaporated. Then the lipid film was hydrated in appropriate buffer at room temperature for 30 min, and the lipid suspension was extruded 20 times through a polycarbonate 0.1-μm filter using a mini extruder (Avanti Polar Lipids, Inc.).
Phospholipid transfer assay
NBD-PS transfer activities of VAT-1 and its mutants were measured by the fluorescent dequenching assay as described previously with several modifications (10). Donor liposomes (12.5 μm; DOPC/DOPE/18:1 Liss-Rhod-PE/18:1–12:0 NBD-PS = 50:40:2:8) were incubated with acceptor liposomes (50 μm; DOPC/DOPE/18:1 CL = 50:40:10) in the presence or absence of recombinant proteins in 2 ml of assay buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 2 mm EDTA) at 25 °C. NBD fluorescence was monitored by a FP-8500 fluorometer (JASCO). The MS-based phospholipid transfer assay was performed as described previously with several modifications (10). VAT-1 or its mutants was incubated with donor liposomes (0.25 mm; DOPC/DOPE/POPC/POPE/DOPA/DOPS/DOPG/18:1 NBD-PE = 55:19.5:5:5:5:5:5:0.5) containing 25% sucrose and acceptor liposomes (1 mm; DOPC/DOPE/POPS/18:1 Liss-Rhod-PE = 55:34.95:10:0.05) in 600 μl of assay buffer (20 mm Tris-HCl, pH 7.5, and 150 mm NaCl) at 16 °C. After 16 h of incubation at 16 °C, the samples were treated with 100 μg/ml proteinase K for 1 h at 37 °C. Then the samples were mixed with 200 μl of assay buffer containing 30% sucrose and incubated for 10 min on ice. The samples were placed on an ultracentrifuge tube, overlaid with 1200, 800, and 200 μl of assay buffer containing 5, 2.5, and 0% sucrose, respectively, and subjected to ultracentrifugation at 217,000 × g for 2.5 h. 1,000 μl fractions were collected from the top. Separation of donor and acceptor liposomes was assessed by fluorescence of NBD-PE and Liss-Rhod-PE (NBD-PE, 535 nm, excitation at 485 nm; Liss-Rhod-PE, 595 nm, excitation at 535 nm; Fig. S8). 200 μl of the top fraction was mixed with 800 μl of chloroform/methanol (2:1, v/v) and vortexed for 15 min. 200 μl of 0.1 m HCl and 0.1 m KCl was added to the sample and vortexed for 10 min. The organic phase was separated by centrifugation at 800 × g for 2 min, collected, and dried under N2 gas. The resulting lipid film was dissolved in 1 ml of 2-propanol containing 5 mm ammonium formate and 0.1% formate. Lipid samples were injected into a Turbo V electrospray ion source by using a syringe pump with a flow rate of 10 μl/min and analyzed by direct infusion MS using a 3200 QTrap system (SCIEX). POPC, POPE, DOPA, DOPS, and DOPG were analyzed by a multiple reaction monitoring (MRM) method. MRM analyses were performed in the negative-ion mode. For quantification, the singly charged precursors (POPC, m/z 804.6; POPE, m/z 716.6; DOPA, m/z 699.5; DOPS, m/z 786.6; DOPG, m/z 773.6) and the fragments corresponding to the 18:1 fatty acids (m/z 281.1) were selected as MRM transitions.
Liposome floatation assay
Liposome flotation assay was performed as described previously with several modifications (11). 2 mm liposomes (DOPC only, DOPC/DOPS = 80:20, DOPC/18:1 CL = 80:20, DOPC/DOPE = 80:20, DOPC/DOPA = 80/20, or DOPC/DOPG = 80/20) were incubated with 5 μm VAT-1 or its mutants in 100 μl of flotation buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl) at 25 °C for 10 min. 200 μl of ice-cold flotation buffer with 60% sucrose was added to the sample after incubation. The resulting liposome/protein mixture was put into a 5-ml ultracentrifuge tube and overlaid with 2 ml of flotation buffer with 30% sucrose, 2 ml of 10% sucrose and 0.5 ml of flotation buffer without sucrose. After centrifugation at 200,000 × g for 1.5 h, 1200-μl fractions were collected from the top. Proteins of each fraction were concentrated by TCA precipitation and analyzed by SDS-PAGE and Coomassie Brilliant Blue staining.
Crystallization and X-ray crystallography
A crystallization trial was performed using the sitting-drop vapor-diffusion method at 20 °C. Drops (0.5 μl) of ∼40 mg/ml VAT-1ΔN40 in 20 mm Tris-HCl, pH 7.5, and 150 mm NaCl were mixed with reservoir solution (0.2 m ammonium nitrate, 18–20% PEG 3350), and equilibrated against 70 μl of the same reservoir solution by vapor diffusion. The crystal was soaked in the reservoir solution supplemented with 15% ethylene glycol, flash-cooled, and kept in a stream of nitrogen gas at 100 K during data collection. X-ray diffraction data were collected with an EIGER 9M detector at SPring-8 Beamline BL32XU, using the helical data collection method with a 5 × 15-μm (width × height) microbeam. The diffraction data were processed using XDS (33). The structure of VAT-1ΔN40 was determined by the molecular replacement method with PHASER (34) in CCP4 suite (35), for which the crystal structure of synaptic vesicle membrane protein VAT-1 homolog-like protein (PDB code 4A27) was used as a search model. Further model building was performed manually with COOT (36), and crystallographic refinement was performed with CNS (37). Ramachandran-plot analysis with RAMPAGE (38) in the CCP4 suite showed that, among 1314 modeled residues, 1265 (96.3%) and 49 (3.7%) residues were in the favored and allowed regions, respectively, whereas no residues were in the outlier region.
CD measurements
CD spectra of proteins (5 μm) in 20 mm Tris-HCl, pH 8.0, containing 150 mm NaCl were recorded at room temperature on a J-820 spectropolarimeter (Jasco), using a cell with a path length of 0.2 cm.
In silico docking
AutoDock Vina (30) was used as a docking tool to generate a putative VAT-1–DOPS complex. Input files were prepared by using the Autodock Tools. The VAT-1 structure was set as rigid during docking, and the grid box with dimensions of 20 × 38 × 32 Å was placed into the phospholipid-binding site. All Autodock Vina parameters were kept as default.
Yeast strain and media
The yeast strain used in this study is W303-1A (39). Standard protocols were used for yeast manipulation (40). The cells were grown in YPLac (1% yeast extract, 2% polypeptone, and 3% lactic acid, pH 5.6) or SCGal (0.67% yeast nitrogen base without amino acids, 0.5% casamino acid, and 2% galactose) medium with appropriate supplements.
Isolation of mitochondria
Isolation of crude mitochondria was performed as described previously with several modifications (17). 5 ml of overnight yeast preculture was inoculated into 1 liter of YPGal or SCGal medium and cultivated at 30 °C for 21 h. Yeast cells were harvested and incubated in alkaline buffer (0.1 m Tris-HCl, pH 9.5, 10 mm DTT) for 15 min at 30 °C. After washing with spheroplast buffer (20 mm Tris-HCl, pH 7.5, 1.2 m sorbitol), the cells were treated with zymolyase 20T (5 mg for 1 g of yeast wet weight) in spheroplast buffer for 30 min at 30 °C. The resulting spheroplasts were resuspended in ice-cold breaking buffer (20 mm Tris-HCl, pH 7.5, 0.6 m mannitol, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride) and homogenized 20 times with a Dounce tissue grinder. After removing unbroken cells and cell debris including nuclear membranes by centrifugation at 2,000 × g for 5 min, crude mitochondria fractions were collected by centrifugation at 12,000 × g for 10 min. The crude mitochondria fractions were washed with SE buffer (250 mm sucrose, 10 mm MOPS-KOH pH 7.2, 1 mm EDTA), resuspended in SE buffer and frozen in liquid nitrogen. Protein concentrations of crude mitochondria were calculated by A280 measurements (A280 = 0.21 was assumed to be 10 mg/ml proteins).
PS transfer assay with mitochondria
1.35 mg/ml isolated mitochondria were incubated with 72 μm WT VAT-1 or VAT-1ΔLoop and DOPS-containing liposomes (0.25 mm; POPC/POPE/DOPS = 50:30:20) in 100 μl of SE buffer at 16 °C. After 16 h of incubation at 16 °C, the mitochondria were mixed with 900 μl of chloroform/methanol (2:1, v/v) and vortexed for 15 min. 200 μl of 0.1 m HCl and 0.1 m KCl was then added to the sample and vortexed for 10 min. The organic phase was separated and subjected to an MS-based phospholipid transfer assay as described above. DOPE and 16:1–16:1 PE were analyzed by a MRM method. For quantification, the singly charged precursors (DOPE, m/z 742.5; 16:1–16:1 PE, m/z 686.5) and the fragments corresponding to the 18:1 fatty acids (m/z 281.1) for DOPE or the 16:1 fatty acids (m/z 253.1) for 16:1–16:1 PE were selected as MRM transitions.
Immunoblotting
The anti-Tom40 antibodies were used in 1:2000 dilution for immunoblotting. An anti-FLAG antibody purchased from Sigma was used in 1:2000 dilutions. Proteins were visualized with fluorophore-conjugated secondary antibodies, goat anti-rabbit or mouse IgG (H + L) secondary antibody, and Cy5 conjugate (Life Technologies), and the signals were analyzed with a Typhoon FLA 9500 (GE Healthcare).
Trp fluorescence measurements
25 μm VAT-1 or its mutants was incubated with 1.5 mm liposomes containing PC alone, PC/PS = 80/20, or PC/CL = 80/20 in 2 ml of 20 mm Tris-HCl, pH 7.5, and 150 mm NaCl at 25 °C. After 10 min of incubation at 25 °C, Trp fluorescence spectra were recorded using an FP-8500 fluorometer (JASCO) at 25 °C with the excitation wavelength of 295 nm and emission wavelength of 300–450 nm.
Author contributions
Y. W. and T. E. conceptualization; Y. W. data curation; Y. W. formal analysis; Y. W., S. W., and T. E. funding acquisition; Y. W. investigation; Y. W. methodology; Y. W. and T. E. writing-original draft; Y. W. and T. E. writing-review and editing; Y. T., S. W., and T. E. supervision; C. K. resources; T. E. validation; T. E. project administration.
Supplementary Material
Acknowledgments
We thank the members of the Endo laboratory for discussions and critical comments on the manuscript. The synchrotron radiation experiments were performed at Beamline BL32XU at SPring-8, Japan, with the approval of the Japan Synchrotron Radiation Research Institute (Proposals 2017A2598 and 2017A2729).
This work was supported by Japan Society of the Promotion of Science KAKENHI Grants 16K21473 (to Y. W.), 15H05595 and 17H06414 (to Y. T.), and 15H05705 and 22227003 (to T. E.); a Japan Science and Technology Agency (JST) CREST Grant JPMJCR12M (to T. E.); a grant from the Takeda Science Foundation (to T. E.); and funds from the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8.
The atomic coordinates and structure factors (code 6K9Y) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ER
- endoplasmic reticulum
- PS
- phosphatidylserine
- PE
- phosphatidylethanolamine
- CL
- cardiolipin
- PA
- phosphatidic acid
- ERMES
- ER–mitochondrial encounter structure
- PG
- phosphatidylglycerol
- PC
- phosphatidylcholine
- PDB
- Protein Data Bank
- IM
- inner membrane
- OM
- outer membrane
- NBD-PS
- nitobenzoxadiazole-labeled PS
- Rhod-PE
- rhodamine-labeled PE
- DOPS
- 1,2-dioleoyl-sn-glycero-3-phospho-l-serine
- DOPE
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPE
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- DOPA
- 1,2-dioleoyl-sn-glycero-3-phosphate
- DOPG
- 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- MRM
- multiple reaction monitoring.
References
- 1. Henry S. A., Kohlwein S. D., and Carman G. M. (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349 10.1534/genetics.111.130286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vance J. E. (2015) Phospholipid synthesis and transport in mammalian cells. Traffic 16, 1–18 10.1111/tra.12230 [DOI] [PubMed] [Google Scholar]
- 3. Tatsuta T., and Langer T. (2017) Intramitochondrial phospholipid trafficking. Biochim. Biophys. Acta 1862, 81–89 10.1016/j.bbalip.2016.08.006 [DOI] [PubMed] [Google Scholar]
- 4. Tamura Y., Kawano S., and Endo T. (2019) Organelle contact zones as sites for lipid transfer. J. Biochem. 165, 115–123 10.1093/jb/mvy088 [DOI] [PubMed] [Google Scholar]
- 5. Vance J. E., and Tasseva G. (2013) Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim. Biophys. Acta 1831, 543–554 10.1016/j.bbalip.2012.08.016 [DOI] [PubMed] [Google Scholar]
- 6. Horvath S. E., Böttinger L., Vögtle F. N., Wiedemann N., Meisinger C., Becker T., and Daum G. (2012) Processing and topology of the yeast mitochondrial phosphatidylserine decarboxylase 1. J. Biol. Chem. 287, 36744–36755 10.1074/jbc.M112.398107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tamura Y., and Endo T. (2017) Role of intra- and inter-mitochondrial membrane contact sites in yeast phospholipid biogenesis. Adv. Exp. Med. Biol. 997, 121–133 10.1007/978-981-10-4567-7_9 [DOI] [PubMed] [Google Scholar]
- 8. Connerth M., Tatsuta T., Haag M., Klecker T., Westermann B., and Langer T. (2012) Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science 338, 815–818 10.1126/science.1225625 [DOI] [PubMed] [Google Scholar]
- 9. Potting C., Tatsuta T., König T., Haag M., Wai T., Aaltonen M. J., and Langer T. (2013) TRIAP1/PRELI complexes prevent apoptosis by mediating intramitochondrial transport of phosphatidic acid. Cell Metab. 18, 287–295 10.1016/j.cmet.2013.07.008 [DOI] [PubMed] [Google Scholar]
- 10. Miyata N., Watanabe Y., Tamura Y., Endo T., and Kuge O. (2016) Phosphatidylserine transport by Ups2–Mdm35 in respiration-active mitochondria. J. Cell Biol. 214, 77–88 10.1083/jcb.201601082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Watanabe Y., Tamura Y., Kawano S., and Endo T. (2015) Structural and mechanistic insights into phospholipid transfer by Ups1–Mdm35 in mitochondria. Nat. Commun. 6, 7922 10.1038/ncomms8922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aaltonen M. J., Friedman J. R., Osman C., Salin B., di Rago J. P., Nunnari J., Langer T., and Tatsuta T. (2016) MICOS and phospholipid transfer by Ups2–Mdm35 organize membrane lipid synthesis in mitochondria. J. Cell Biol. 213, 525–534 10.1083/jcb.201602007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lev S. (2012) Nonvesicular lipid transfer from the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 4, a013300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., and Walter P. (2009) An ER–mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jeong H., Park J., Jun Y., and Lee C. (2017) Crystal structures of Mmm1 and Mdm12–Mmm1 reveal mechanistic insight into phospholipid trafficking at ER–mitochondria contact sites. Proc. Natl. Acad. Sci. U.S.A. 114, E9502–E9511 10.1073/pnas.1715592114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kawano S., Tamura Y., Kojima R., Bala S., Asai E., Michel A. H., Kornmann B., Riezman I., Riezman H., Sakae Y., Okamoto Y., and Endo T. (2018) Structure–function insights into direct lipid transfer between membranes by Mmm1–Mdm12 of ERMES. J. Cell Biol. 217, 959–974 10.1083/jcb.201704119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kojima R., Endo T., and Tamura Y. (2016) A phospholipid transfer function of ER–mitochondria encounter structure revealed in vitro. Sci. Rep. 6, 30777 10.1038/srep30777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hirabayashi Y., Kwon S. K., Paek H., Pernice W. M., Paul M. A., Lee J., Erfani P., Raczkowski A., Petrey D. S., Pon L. A., and Polleux F. (2017) ER–mitochondria tethering by PDZD8 regulates Ca2+ dynamics in mammalian neurons. Science 358, 623–630 10.1126/science.aan6009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Linial M., Miller K., and Scheller R. H. (1989) VAT-1: an abundant membrane protein from Torpedo cholinergic synaptic vesicles. Neuron 2, 1265–1273 10.1016/0896-6273(89)90311-5 [DOI] [PubMed] [Google Scholar]
- 20. Junker M., and Rapoport T. A. (2015) Involvement of VAT-1 in phosphatidylserine transfer from the endoplasmic reticulum to mitochondria. Traffic 16, 1306–1317 10.1111/tra.12336 [DOI] [PubMed] [Google Scholar]
- 21. Linial M., and Levius O. (1993) VAT-1 from Torpedo is a membranous homologue of zeta crystallin. FEBS Lett. 315, 91–94 10.1016/0014-5793(93)81140-U [DOI] [PubMed] [Google Scholar]
- 22. Eura Y., Ishihara N., Oka T., and Mihara K. (2006) Identification of a novel protein that regulates mitochondrial fusion by modulating mitofusin (Mfn) protein function. J. Cell Sci. 119, 4913–4925 10.1242/jcs.03253 [DOI] [PubMed] [Google Scholar]
- 23. Tsujita K., Itoh T., Kondo A., Oyama M., Kozuka-Hata H., Irino Y., Hasegawa J., and Takenawa T. (2010) Proteome of acidic phospholipid-binding proteins: spatial and temporal regulation of Coronin 1A by phosphoinositides. J. Biol. Chem. 285, 6781–6789 10.1074/jbc.M109.057018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Faugaret D., Chouinard F. C., Harbour D., El azreq M. A., Bourgoin S. G. (2011) An essential role for phospholipase D in the recruitment of vesicle amine transport protein-1 to membranes in human neutrophils. Biochem. Pharmacol. 81, 144–156 10.1016/j.bcp.2010.09.014 [DOI] [PubMed] [Google Scholar]
- 25. Chattopadhyay A., and London E. (1987) Parallax method for direct measurement of membrane penetration depth utilizing fluorescence quenching by spin-labeled phospholipids. Biochemistry 26, 39–45 10.1021/bi00375a006 [DOI] [PubMed] [Google Scholar]
- 26. Jones D. T., and Cozzetto D. (2015) DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31, 857–863 10.1093/bioinformatics/btu744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rao S. T., and Rossmann M. G. (1973) Comparison of super-secondary structures in proteins. J. Mol. Biol. 76, 241–256 10.1016/0022-2836(73)90388-4 [DOI] [PubMed] [Google Scholar]
- 28. Holm L., and Laakso L. M. (2016) Dali server update. Nucleic Acids Res. 44, W351–W355 10.1093/nar/gkw357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo P. C., Ma X. X., Bao Z. Z., Ma J. D., Chen Y., and Zhou C. Z. (2011) Structural insights into the cofactor-assisted substrate recognition of yeast quinone oxidoreductase Zta1. J. Struct. Biol. 176, 112–118 10.1016/j.jsb.2011.07.010 [DOI] [PubMed] [Google Scholar]
- 30. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wong L. H., opič A., Levine T. P. (2017) Advances on the transfer of lipids by lipid transfer proteins. Trends Biochem. Sci. 42, 516–530 10.1016/j.tibs.2017.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kobashigawa Y., Kumeta H., Ogura K., and Inagaki F. (2009) Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method. J. Biomol. NMR 43, 145–150 10.1007/s10858-008-9296-5 [DOI] [PubMed] [Google Scholar]
- 33. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., and Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 10.1107/s0907444998003254 [DOI] [PubMed] [Google Scholar]
- 38. Lovell S. C., Davis I. W., Arendall W. B. 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., and Richardson D. C. (2003) Structure validation by Cα geometry: φ,ψ and Cβ deviation. Proteins 50, 437–450 10.1002/prot.10286 [DOI] [PubMed] [Google Scholar]
- 39. Thomas B. J., and Rothstein R. (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56, 619–630 10.1016/0092-8674(89)90584-9 [DOI] [PubMed] [Google Scholar]
- 40. Adams A., Gottschling D.E., Kaiser C., and Stearns T. (1998) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.