Abstract
In response to microbial invasion, the animal immune system generates hypochlorous acid (HOCl) that kills microorganisms in the oxidative burst. HOCl toxicity is amplified in the phagosome through import of the copper cation (Cu2+). In Escherichia coli and Salmonella, the transcriptional regulator RclR senses HOCl stress and induces expression of the RclA, -B, and -C proteins involved in bacterial defenses against oxidative stress. However, the structures and biochemical roles of the Rcl proteins remain to be elucidated. In this study, we first examined the role of the flavoprotein disulfide reductase (FDR) RclA in the survival of Salmonella in macrophage phagosomes, finding that RclA promotes Salmonella survival in macrophage vacuoles containing sublethal HOCl levels. To clarify the molecular mechanism, we determined the crystal structure of RclA from E. coli at 2.9 Å resolution. This analysis revealed that the structure of homodimeric RclA is similar to those of typical FDRs, exhibiting two conserved cysteine residues near the flavin ring of the cofactor flavin adenine dinucleotide (FAD). Of note, we observed that Cu2+ accelerated RclA-mediated oxidation of NADH, leading to a lowering of oxygen levels in vitro. Compared with the RclA WT enzyme, substitution of the conserved cysteine residues lowered the specificity to Cu2+ or substantially increased the production of superoxide anion in the absence of Cu2+. We conclude that RclA-mediated lowering of oxygen levels could contribute to the inhibition of oxidative bursts in phagosomes. Our study sheds light on the molecular basis for how bacteria can survive HOCl stress in macrophages.
Keywords: flavoprotein, copper, crystal structure, nicotinamide adenine dinucleotide (NADH), disulfide, oxidative stress, macrophage, flavoprotein disulfide reductase, hypochlorous acid, innate immune response, oxygen, RclA
Introduction
Hypochlorous acid (HOCl)2 is a powerful antimicrobial agent in the innate immune systems of higher eukaryotes (1–3). To kill invading bacteria, macrophages and neutrophils in the immune system employ myeloperoxidase to generate HOCl from H2O2, which is produced by the oxidative burst from molecular oxygen by NADPH oxidase (4). HOCl directly oxidizes amino acids containing amines and sulfurs and can cleave DNA once it is converted into a hydroxyl radical in the presence of iron or copper ions via Fenton(-like) reaction (5–8).
HypT, RclR, and NemR are HOCl-responsive transcriptional factors in bacteria that increase bacterial survival under HOCl stress (9–11). HypT, belonging to the LysR-type transcriptional regulators, employs a HOCl-sensitive methionine residue to repress genes related to the import of iron to the cytosol in Escherichia coli and Salmonella enterica serovar Typhimurium. HypT plays a critical role in the survival of Salmonella in macrophages (12). NemR of E. coli is a redox-regulated transcriptional repressor that utilizes the oxidation status of HOCl-sensitive cysteine residues. NemR expresses two enzymes, GloA and NemA, to detoxify methylglyoxal and reactive electrophiles (11).
RclR is a member of the AraC family of transcriptional regulators and senses HOCl with highly-conserved cysteine residues (9, 13). RclA, RclB, and RclC proteins were identified as RclR target genes in both E. coli and Salmonella. In E. coli, RclR, RclA, RclB, and RclC contribute to the HOCl resistance of the bacterium. Deletion of any one of these genes decreases the survival of E. coli under HOCl stress (9). RclA putatively belongs to the flavoprotein disulfide reductase (FDR) family. RclB is a small uncharacterized periplasmic protein that has the signal sequence for secretion. RclC is a multipass inner membrane protein with homology to quinone-binding proteins. However, little is known about the biochemical functions of RclA, RclB, and RclC.
A recent study suggested that the FDR RclA is widely conserved among bacteria that colonize epithelial surfaces, and it contributes to host colonization in environments with HOCl-mediated oxidative stress (14). FDRs usually form homodimers and use noncovalently-bound flavin adenine dinucleotide (FAD) and a nonflavin redox center, catalyzing NAD(P)H-dependent reduction of a variety of substrates, such as sulfur-containing peptides, metal ions, H2O2, and O2 (15). Like other FAD-dependent reductases, the reducing power of the bound FAD is supplied from the cytosolic NAD(P)H pool.
FDRs commonly have four domains (FAD-binding domain, NAD(P)H binding domain, central domain, and the interface domain). However, they can be divided into three subgroups depending on the redox disulfide motif and the presence of the additional nonflavin redox centers at the extra N- or C-terminal motif (15). Very recently, Derke et al. (14) reported that RclA from E. coli exhibits Cu2+ reductase activity, reducing Cu2+ to Cu+ to cope with HOCl stress. Here, we determined the crystal structures of RclA from E. coli, containing FAD at its active site. Based on the structures and the subsequent biochemical analyses, we investigated the functions of RclA in terms of HOCl resistance in bacteria.
Results
RclA contributes to the survival of Salmonella in macrophages
RclA is commonly found in Salmonella and E. coli (Fig. S1). Because HOCl is produced in macrophage phagosomes as a primary bactericidal agent, the function of RclA in resistance to HOCl suggests that RclA plays a role in the survival of the bacteria. Because Salmonella can proliferate in macrophages, we used S. enterica serovar Typhimurium to investigate the role of RclA in its proliferation in macrophages.
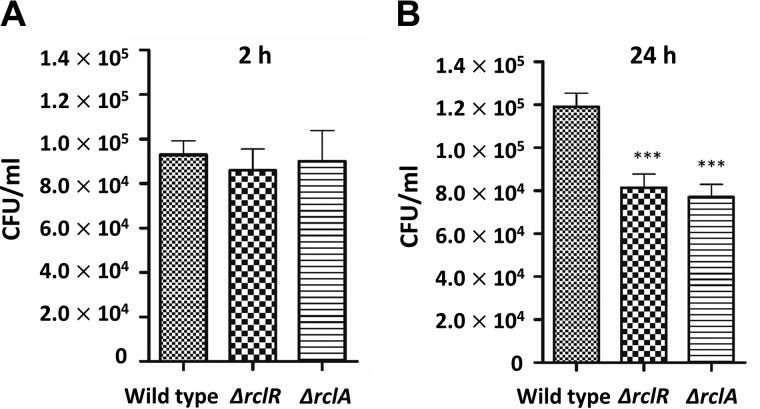
To confirm the role of RclA in the pathogenesis of Salmonella, we generated Salmonella mutant strains harboring the deletion of rclA or rclR. The WT and the RclR- or RclA-deletion mutant strains were precultured in LB broth and then incubated with macrophages. Deletion of rclA or rclR resulted in a 30% decrease in survival of Salmonella in macrophage after a 24-h incubation, indicating that RclA at least partly contributes to the survival of Salmonella in macrophages (Fig. 1). Taken together, our data indicate that RclA is involved in the pathogenesis of Salmonella by promoting survival in macrophage vacuoles containing sublethal doses of HOCl.
Figure 1.
Roles of RclR and RclA in macrophage survival. RAW264.7 macrophage cells were infected with Salmonella typhimurium SL1344 (WT) and its isogenic strains rclR and rclA. At 2 h (A) and 24 h (B) post-infection, the macrophage cells were lysed, and the number of intracellular bacteria was determined by plating on LB agar. The results are expressed as the mean ± S.D. of four independent experiments. Asterisks indicate a statistically significant difference between WT and mutant strains (***, p < 0.001).
Structural determination and overall structure
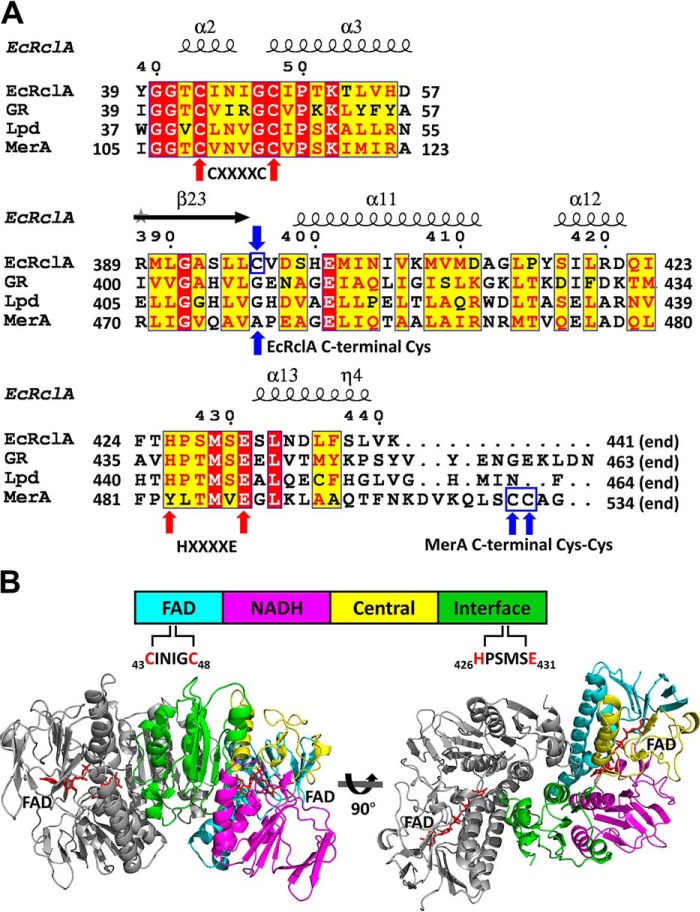
Sequence comparison suggested that RclA belongs to the group I FDRs due to the presence of two essential motifs: CXXXXC for the redox-active disulfide as a nonflavin redox center, and the essential HXXXXE that interacts with and promotes the disulfide formation of the CXXXXC motif with its histidine residue (Fig. 2A) (15). Like typical group I FDRs, RclA consists of the four domains and does not have the additional nonflavin redox center (Fig. 2B).
Figure 2.
Sequence analyses and the crystal structure of EcRclA. A, sequence alignment of FDR proteins with the secondary structure elements. GR, GSH-disulfide reductase from Bartonella henselae (group I FDR; GenBankTM accession no. WP_011180567); Lpd, dihydrolipoyl dehydrogenase from Mycobacterium multispecies (group I FDR; GenBankTM accession no. WP_003402301.1); MerA, mercuric reductase from Pseudomonas aeruginosa (group II FDR; GenBankTM accession no. WP_023980594.1). GR, Lpd, and MerA were chosen based on the highest score of DALI server structural analyses. The sequence alignment was performed using the T-Coffee server (35) and the ESPript server (36). For clarity, the regions containing the signature motifs are displayed. The full sequence alignment is presented in Fig. S2. The cysteine residues in the CXXXXC motifs and histidine and glutamate residues in the HXXXXE motif are indicated by red arrows. The C-terminal Cys in EcRclA and the C-terminal Cys–Cys motif in MerA are indicated by blue arrows and blue square. B, domain analysis and dimeric assembly of EcRclA. The schematic domain structure is shown at the top. The four domains are displayed in a bar containing a CXXXXC motif in the FAD-binding domain and the HXXXXE motif in the interface domain. The dimer is depicted by the ribbon representations at the bottom. One subunit is in the same color code of the top panel, and the other subunit is in gray. The bound FAD molecules are shown in the red stick representations.
To gain mechanistic insights on RclA from its 3D structures, we expressed the full-length RclA proteins (441 amino acids) of E. coli and Salmonella and successfully determined the crystal structure of RclA from E. coli (EcRclA) at 2.9-Å resolution (Fig. 2B and Table 1). The asymmetric unit contained four protomers consisting of two homodimers (Fig. S3). Each protomer contained a tightly-bound FAD molecule in its active site (Fig. 2B). Secondary structure elements of Fig. 2A were indicated in Fig. S4.
Table 1.
Data collection and refinement statistics
| RclA wildtype | RclA wildtype (Cu2+) | RclA C43S (Cu2+) | |
|---|---|---|---|
| Data collection | |||
| Beam line | PAL 5C | PAL 5C | PAL 5C |
| Wavelength (Å) | 0.97960 | 1.00003 | 1.00003 |
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 72.4, 189.4, 95.4 | 72.6, 188.4, 95.0 | 72.8, 191.5, 95.2 |
| α, β, γ (°) | 90, 107.3, 90 | 90, 107.6, 90 | 90, 107.4, 90 |
| Resolution (Å) | 50.0–2.90 (2.95–2.90) | 50.0–2.80 (2.85–2.80) | 50.0–3.00 (3.05–3.00) |
| Rpim | 0.044 (0.144) | 0.042 (0.198) | 0.045 (0.163) |
| Rmerge | 0.117 (0.281) | 0.065 (0.374) | 0.087 (0.235) |
| I/σ | 12.3 (4.21) | 17.4 (4.4) | 10.7 (2.7) |
| Completeness (%) | 98.5 (95.2) | 99.4 (97.6) | 92.8 (82.1) |
| Redundancy | 5.0 (3.2) | 3.6 (3.3) | 3.7 (2.3) |
| Refinement | |||
| Resolution (Å) | 40.37–2.9 | 47.24–2.8 | 46.61–3.0 |
| No. of reflections | 54,069 | 81985 | 45,241 |
| Rwork/Rfree | 0.2346/0.2806 | 0.2226/0.2707 | 0.2081/0.2565 |
| No. of total atoms | 13,746 | 13,709 | 13,845 |
| Wilson B-factor (Å) | 43.62 | 33.08 | 51.40 |
| R.M.S.D. | |||
| Bond lengths (Å) | 0.002 | 0.003 | 0.002 |
| Bond angles (°) | 0.511 | 0.520 | 0.516 |
| Ramachandran plot | |||
| Favored (%) | 94.5 | 95.41 | 95.67 |
| Allowed (%) | 5.5 | 4.59 | 4.33 |
| Outliers (%) | 0.0 | 0.0 | 0.0 |
| PDB code | 6KGY | 6KYY | 6KOD |
Structural comparison to group I FDRs
Two identical active sites are at the subunit interfaces of the homodimer, which consists of chain A and chain B protomers (Fig. 3A). The overall arrangement of the CXXXXC motif in chain A and the HXXXXE motif in chain B protomer in the active site of RclA is similar to that of group I FDR family proteins (Fig. 2A and Fig. S5). The two cysteines (Cys-43 and Cys-48) in the CXXXXC motif from chain A are in the reduced form, and the distance between the sulfur atoms of the two cysteine residues is ∼4.4 Å in the crystal structure. The two cysteine residues are near the isoalloxazine ring of FAD from chain A, and Cys-48 is closer to FAD than to Cys-43 (the distance between the Sγ of Cys-48 and the C4a atom of flavin is 3.5 Å). The HXXXXE motif from chain B is involved in the interaction with the CXXXXC motif from chain A protomer and the bound FAD from chain A (Fig. 3A). His-426 and Glu-431 in the HXXXXE motif from chain B protomer are paired with a distance of 2.5 Å, and His-426 makes a polar interaction with Cys-43 from chain A protomer with a distance of 3.0 Å (Fig. 3A). Two lysine residues in chain A protomer (Lys-13 and Lys-16), which are unique to RclA proteins, were found near Cys-43 from chain A protomer at the active site. Because group I FDR proteins reduce substrates via the disulfide formation of the CXXXXC motif together with the HXXXXE motif, our findings suggest that RclA shares the enzymatic mechanism of group I FDR proteins.
Figure 3.
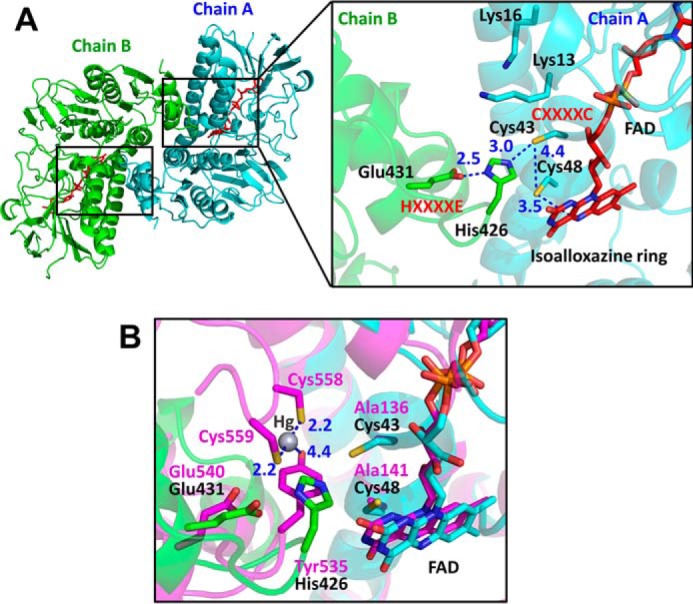
Structure of the active site of EcRclA. A, active site of RclA. Left, overall structure of RclA in the top view. Two identical active sites are marked as two small boxes. The box on the right is enlarged from one of the boxes from the overall structure on the left. Each subunit is depicted by the ribbon representation in cyan (chain A) or green (chain B). The bound FAD molecules are shown in the red stick representations. Each residue is labeled and indicated in the stick representations with the ribbon diagram in the background. The critical interactions between the two cysteines of CXXXXC from chain A, His-426 and Glu-431 of HXXXXE from chain B, and Cys-48 and the C4a atom of FAD are shown by blue dotted lines. Distances are in Å (blue). B, active-site comparison with MerA. The box is enlarged from Fig. S6. RclA (green and cyan) is aligned with MerA (magenta, PDB code 4K7Z) (37). Each residue aligned in the active site is labeled in a different color (black for RclA and magenta for MerA). His-426 in RclA is substituted with Tyr-535 in MerA. Mercury was bound to the two C-terminal cysteines of MerA, Cys5–58, and Cys-559. The interactions between the mercury, cysteine, and histidine residues are depicted by dashed lines. The two cysteine residues in the CXXXXC motif of MerA were substituted with alanine to determine the Hg2+-bound structure (37). Distances are in Å (blue).
Structural comparison to the group II FDR MerA
We searched for the closest protein to RclA in terms of structure using the DALI server (16). Mercuric reductase (MerA) (PDB code 4K7Z), a group II FDR, was ranked as the top entry (Z-score: 43.6). Like EcRclA reduces Cu2+ to Cu+, MerA converts Hg2+ to Hg0 (15). MerA contains the CXXXXC motif, similar to group I FDR proteins, and tyrosine replaces the histidine residue in the HXXXXE motif (Fig. 2A). The structural superposition of EcRclA onto MerA revealed a high structural similarity between the matched regions (sequence identity: 30%; RMSD: 1.522 between 320 Cα atoms; residues 5–433 of EcRclA, calculated with PyMOL (17)), except for the Cys–Cys motif for metal ion binding (Fig. S6 and Fig. 3B). The C-terminal end loop containing the Cys–Cys motif was folded into the active site near the CXXXXC motif and the tyrosine residue in the MerA structure (Figs. 2A and 3B). The Cys–Cys motif and the tyrosine residue interact with a mercuric ion at the active site (18).
Cu2+ and Hg2+ accelerate the oxidation of NADH in RclA
To elucidate the mechanism of RclA, we performed further biochemical analyses. We observed the basal level of the NADH oxidase activity of RclA in the absence of any additional material (control in Fig. 4A and 0 mm Cu2+ in Fig. 4B), which was found in many FDR enzymes (19–21). Previous studies of other FDRs suggested that the basal activity of RclA resulted from oxidation by the molecular oxygen in the buffer (19, 21–23).
Figure 4.
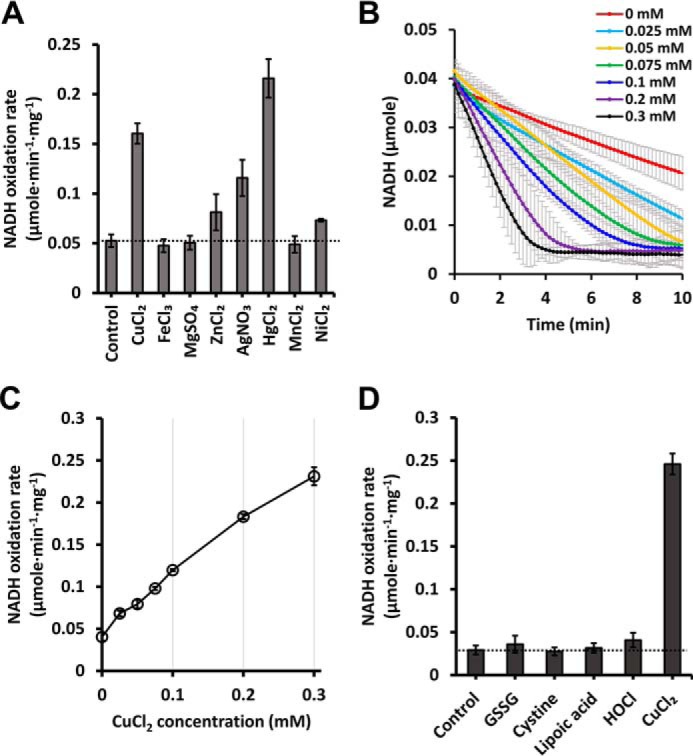
NADH oxidation of EcRclA in the presence of various substrates. A, NADH oxidation rates depend on various transition metal ions. NADH oxidation rates by RclA were measured over time spectrophotometrically (n = 3, mean ± S.D.). The reaction was initiated by injecting 200 μm NADH into a buffer containing 2 μm RclA with or without 100 μm of metal ions. A control experiment was performed without metal ion. The horizontal line indicates the reference NADH oxidation rate of control. B, NADH oxidation profiles with different concentrations of Cu2+. The reaction was carried out in a buffer containing 200 μm NADH and indicated concentrations of CuCl2 (from 0 to 0.3 mm). The amounts of the NADH oxidation were measured spectrophotometrically (n = 3, mean ± S.D.). C, initial NADH oxidation rates calculated from the NADH oxidation profile in Fig. 4B. The reaction was assayed with 200 μm NADH and marked concentrations of Cu2+. The NADH oxidation rates were measured over time spectrophotometrically (n = 3, mean ± S.D.). D, NADH oxidation rates with various substances. The reaction was initiated by injecting 200 μm NADH into a buffer containing 2 μm RclA and 200 μm of various materials. A control experiment was performed without material. NADH oxidation rates were measured over time spectrophotometrically (n = 3, mean ± S.D.)
Next, we tested whether RclA exhibits the MerA-like activity in which NADH oxidase activity is increased by adding metal ion (24, 25). We measured the NADH oxidase activity of RclA in the presence of various metal ions, such as Hg2+, Cu2+, Fe3+, Mn2+, Mg2+, and Zn2+ (Fig. 4A). Notably, Hg2+ and Cu2+ increased oxidation of NADH by ∼3-fold compared with the basal activity without the metal ions, and Ag+ resulted in an ∼2-fold increase. However, Fe2+, Mg2+, Zn2+, and Mn2+ did not affect the NADH oxidase activity. Similar results were observed in a preprint (14). We further determined that Cu2+ accelerates the NADH oxidation rate in a dose-dependent manner until reaching 0.3 mm CuCl2 (Fig. 4, B and C). The data suggest that at higher concentrations of CuCl2, the dose-dependent effect would continue.
To identify other substrates, we tested the NADH oxidation activity with oxidized GSH (GSSG), lipoic acid, cystine, and HOCl, which could be putative substrates of RclA. No increase was observed in the NADH oxidation rate by RclA with these substances (Fig. 4D). We thus focused on Cu2+ in the following analyses, because Hg2+ and Ag+ are not relevant in host immune responses, and Cu2+ is employed in phagosomes of host innate immune systems.
O2 is required for NADH oxidation in the presence of Cu2+
Although Derke et al. (14) reported the reduction of Cu2+ with the reducing power of NADH, the redox stoichiometry was not fully accounted for. Only 25–50% of the reducing power from NADH was used in the Cu2+ reduction to produce Cu+ (Fig. 4B in Ref. 14). Given that molecular oxygen dissolved in the reaction buffer is the oxidant candidate, we investigated molecular oxygen as the second oxidant in the RclA-mediated reaction.
Because a significant amount of O2 is dissolved in ambient conditions, we investigated the NADH oxidase activity of RclA under oxygen-depleted conditions by thoroughly purging the buffer with nitrogen gas (Fig. 5A). The NADH oxidation rate of EcRclA was compared in the presence and absence of Cu2+. The depletion of dissolved oxygen almost completely shut down the NADH oxidation by RclA regardless of the presence of Cu2+. Stoichiometric analyses of this reaction accounted for 50% of the NADH oxidation in the presence of Cu2+. Thus, our findings suggest that the presence of Cu2+ and molecular oxygen are interlinked in the oxidation of NADH by RclA, and O2 is required for Cu2+-mediated NADH oxidation by RclA.
Figure 5.
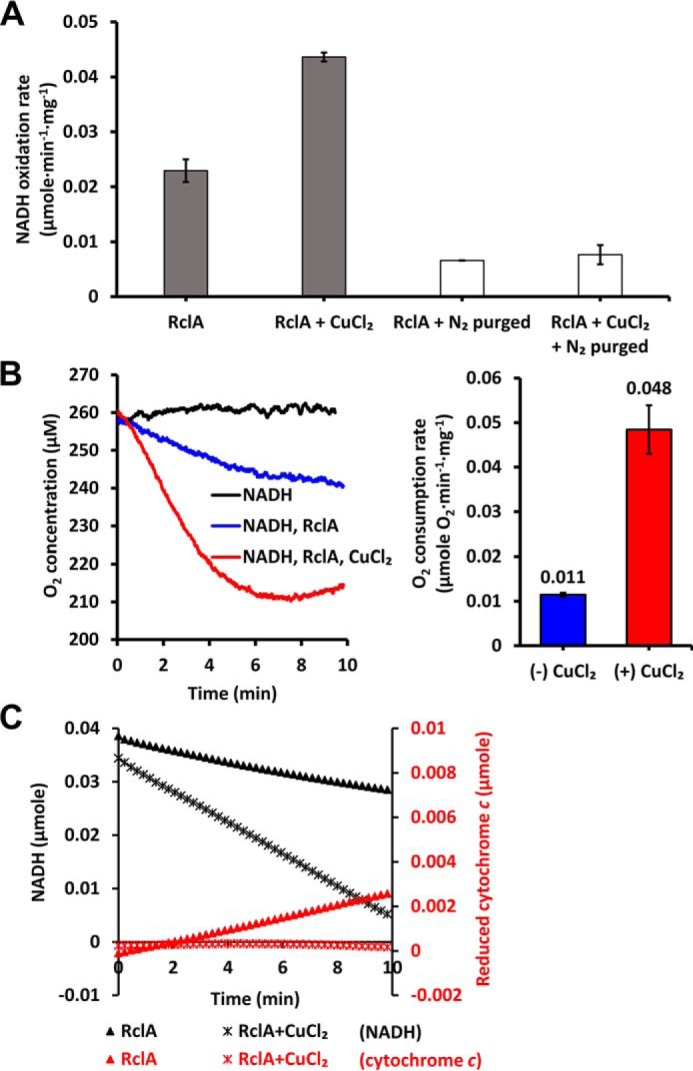
O2 depletion and consumption during NADH oxidation by EcRclA. A, NADH oxidation by RclA in O2-depleted conditions. The oxidation rates were measured with 200 μm NADH and 2 μm EcRclA WT, with or without 100 μm CuCl2 over time spectrophotometrically (n = 3, mean ± S.D.). O2 was depleted by N2 purging. B, O2 consumption rates during the NADH oxidation reaction by RclA. Left, O2 consumption profiles during the NADH oxidation reaction. The reaction was initiated by the addition of 200 μm NADH into the preincubated reaction solution containing 200 μm Cu2+ and 2 μm RclA. The O2 concentrations in the buffer were measured over time with an oxygen sensor. Data are the averages of three independent experiments. Right, O2 consumption rates calculated from the results of the left (n = 3, mean ± S.D.). C, superoxide anion production during NADH oxidation by the WT EcRclA protein. Production of superoxide anion was measured based on the amount of reduced cytochrome c at 550 nm (red), and NADH oxidation was measured at 340 nm (black) over time spectrophotometrically. The reaction was performed in the presence or absence of 100 μm Cu2+ in a buffer containing 200 μm NADH and 2 μm EcRclA WT. Data are the averages of three independent experiments.
Cu2+ promotes O2 consumption without producing superoxide anion
To determine whether O2 is the additional oxidant during the RclA reaction, we measured the amount of oxygen dissolved in the reaction buffer with an oxygen probe during the oxidation of NADH by RclA (Fig. 5B). The specific O2 consumption rate in the presence of 200 μm Cu2+ was about four times faster than that in the absence of Cu2+ (Fig. 5B), as observed in the NADH oxidation rates in the presence and absence of Cu2+ (Fig. 4C). These results indicate that Cu2+ catalyzes the reduction of oxygen during the oxidation of NADH. About 50 μm oxygen was consumed by the time 200 μm NADH was entirely used in the reaction (Figs. 4B and 5B), indicating that four NADH molecules were required for one oxygen molecule in this reaction of RclA.
Many FDR enzymes produce superoxide anion as the basal or side activity by a reaction between the bound FADH2 and molecular oxygen (22). We thus measured the amount of superoxide anion produced in the RclA-mediated reaction without Cu2+ using cytochrome c, which is rapidly reduced by superoxide anion with an increase in light absorbance at 550 nm. Like other FDR enzymes, EcRclA without Cu2+ produced superoxide anion (Fig. 5C). We performed similar experiments in the presence of Cu2+. Production of superoxide anion was not observed in the reaction, although more oxygen was consumed that in the absence of Cu2+ (Fig. 5C). These findings indicate that Cu2+ promotes the reaction between oxygen and FADH2 at the active site of RclA while suppressing the production of superoxide anion.
Roles of the two cysteine residues in the CXXXXC motif
To investigate the roles of the two conserved cysteine residues (Cys-43 and Cys-48) in the CXXXXC motif and the additional cysteine residue (Cys-396) at the C terminus, we produced EcRclA C43S, C48S, and C396S, and we then tested their activities in the presence or absence of Cu2+. The mutation at Cys-396 did not affect the tested RclA properties (data not shown), which is consistent with the structural analyses. The cysteine residue was buried and surrounded by two methionine residues in the hydrophobic region in the crystal structure (Fig. S7). Because the cysteine residue is far from the active site, it is unlikely that Cys-396 is involved in the catalysis.
However, analyses of the point mutation at Cys-43 or Cys-48 in the CXXXXC motif yielded interesting results (Fig. 6A). When Cys-48 was changed to a serine, the NADH oxidase activity was highly increased regardless of the presence of Cu2+, compared with those of WT enzyme (Fig. 6, A and B, and Fig. S8). These observations indicate that the Cu2+-dependent NADH oxidase activity of RclA is controlled by Cys-48. In terms of production of superoxide anion, C48S showed the increased basal superoxide production rate in the absence of Cu2+, indicating that Cys-48 may contribute to the suppression of the basal superoxide production by RclA (Fig. 6B).
Figure 6.

Activity of EcRclA mutants C43S and C48S compared with WT. A, initial NADH oxidation rate of RclA wildtype (WT) and mutants. The reaction was performed with 200 μm NADH and 2 μm EcRclA, with or without 100 μm Cu2+. Data are expressed as the mean ± S.D. of three independent experiments (μmol of NADH·min−1·mg−1 RclA). B, superoxide anion production during NADH oxidation by RclA WT and mutants in the absence of Cu2+. The reaction buffer contained 200 μm NADH, 2 μm EcRclA, and 50 μm cytochrome c. Superoxide anion production was measured using the amount of reduced cytochrome c at 550 nm (red); NADH oxidation was measured at 340 nm (black) over time spectrophotometrically at the same time points. Data are the averages of three independent experiments. C, NADH oxidation rate comparison between RclA C43S and WT with various metal ions. The reaction was performed with 200 μm NADH, 2 μm RclA, and 100 μm of metal ions. A control assay was performed without metal ion. Data are expressed as the mean ± S.D. of three independent experiments (μmol of NADH·min−1·mg−1 RclA).
The RclA C43S showed a dramatic increase in NADH oxidase activity in the presence of Cu2+ compared with the WT (Fig. 6A). However, the mutation at Cys-43 increased the NADH oxidase activity in the presence of other metal ions as well. Ag+ accelerated the oxidation rate of NADH as much as Cu2+, and Zn2+ and Hg2+ also further increased the NADH oxidase activity compared with that of WT enzyme (Fig. 6C). Thus our findings suggest that Cys-43 is involved in the metal specificity and further in the binding of Cu2+ at the active site.
Multiple Cu2+-binding sites
To analyze how Cu2+ affects the activity of RclA at the molecular level, we determined crystal structures of EcRclA in complex with Cu2+ in the presence of 5 mm CuCl2. To prevent the formation of the disulfide bond in the CXXXXC motif by Cu2+, we also determined the complex structure of the EcRclA C43S mutant (see below). We found that RclA is a histidine-rich protein containing 16 histidine residues (15 surface-exposed histidine residues and His-426 at the active site). Strong Cu2+ densities were observed near all of the histidine residues except for His-426 in both complex structures, indicating that Cu2+ is bound to almost all of the surface-exposed histidine residues of RclA (Fig. 7A). However, no apparent structural changes were observed in the complex structures despite the multiple Cu2+ binding at the histidine residues (Fig. S9). It is noteworthy that surface-exposed histidine residues are abundant in RclA. Because almost all of the histidine residues of RclA can bind to Cu2+ (Fig. 7A), RclA could lower the amount of free Cu2+ available to participate in the Fenton reaction with HOCl to produce highly-toxic hydroxyl radicals.
Figure 7.
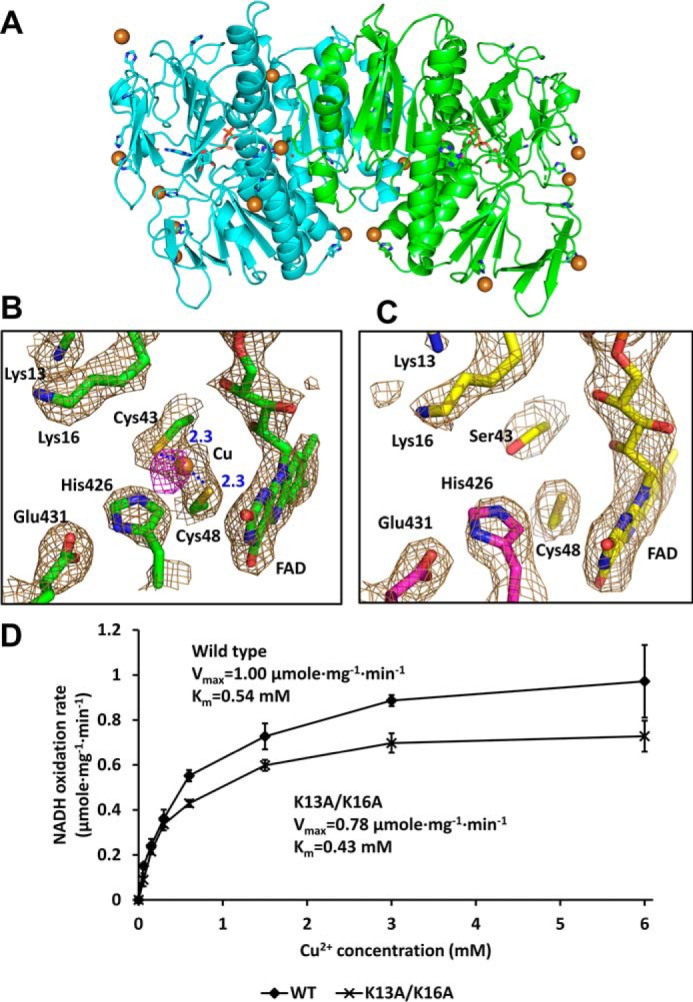
Extra electron density maps at the active site in Cu2+-soaked structures. A, overall structure of the RclA WT in complex with Cu2+. Each subunit is colored differently (cyan and green). Cu2+ is shown in the sphere (brown). Histidine residues and the bound FAD are shown in the stick representation. B, binding of Cu2+ at the active site of the WT RclA protein. Each subunit is colored differently (cyan and green). Cu2+ is shown in the sphere (brown). The electron density map is contoured at 1.5 σ (sand). Anomalous difference map of the bound Cu2+ is contoured at 3.7 σ (magenta). Distances are in Å (blue dash). C, active site of RclA C43S. Each subunit is colored differently (yellow and magenta). The electron density map is contoured at 1.5 σ (sand). D, biochemical analysis of Lys-13 and Lys-16. Initial velocities of the NADH oxidation by RclA WT and RclA K13A/K16A were measured for Km and Vmax. The reactions were performed with 2 μm RclA in a buffer containing 200 μm NADH and 0, 0.6, 0.15, 0.3, 0.6, 1.5, 3, or 6 mm Cu2+. NADH oxidation rates were measured over time spectrophotometrically (n = 3, mean ± S.D.). The Vmax or Km value was calculated by fitting the data with a Michaelis-Menten equation using an enzyme kinetics tool of the SigmaPlot Version 14.0 (Systat Software, San Jose, CA).
Cu2+ binding at the active site
Strong electron density maps near the cysteine residues at the active site also appeared in Cu2+-soaked crystals of the WT RclA protein. The maps were suggested as heavy-atom sites by a strong signal in the difference anomalous map. Thus, it is very likely that the strong electron density maps represent the Cu2+ binding (Fig. 7B). We noted that the Cu2+ density maps were between the sulfur atoms of Cys-43 and Cys-48 in the CXXXXC motif. Moreover, the densities were not found in the C43S mutant structure, indicating the importance of Cys-43 for the binding of Cu2+ (Fig. 7C). Combined with the biochemical results with the C43S mutant RclA, the binding between the two cysteine residues is essential in the selectivity to Cu2+ over the other metal ions.
Regarding Cys-48, the Cys-48 residue is located between the Cu2+ and the FAD molecule. This structure suggests that Cys-48 prevents the direct contact between Cu2+ and FAD, and it allows the electron flows from FAD to Cu2+ via Cys-48. This structural arrangement of Cu2+–Cys-48–FAD is well-supported by our biochemical results that Cys-48 contributed to the prevention of basal production of superoxide anion from oxygen.
Two lysine residues (Lys-13 and Lys-16), which are unique to RclA proteins, were located at the entrance to the active site. To examine the role of the lysine residues, we produced the mutant protein K13A/K16A and compared the Km and Vmax values for Cu2+ with those of the WT protein. The mutant protein showed both Km and Vmax values decreased by ∼1.3-fold with a similar Vmax/Km value (Fig. 7D). These biochemical results show that the two lysine residues provide the positively-charged environment to limit the prolonged staying of metal ions, promoting the turnover of the reaction cycles. Alternatively or concomitantly, this positively-charged environment would attract or capture negatively-charged reaction intermediates for the subsequent reactions.
Discussion
RclA from E. coli was reported to increase resistance to HOCl treatment (9). This study using Salmonella strains extends the role of RclA in bacterial survival in macrophage phagosomes. To understand the roles of RclA at the molecular level, we elucidated high-resolution structures of RclA from E. coli exhibiting the typical dimeric features of FDR family proteins. The structure of EcRclA showed intermingled properties of groups I and II FDRs. Although the overall structure of EcRclA was the most similar to the group II FDR MerA, the key motifs in the active site were more closely related to group I FDRs. While this manuscript was in preparation, Derke et al. (14) reported that EcRclA is a Cu2+ reductase that converts Cu2+ into Cu+ using the reducing power of NADH to cope with HOCl stress in Drosophila. Consistently, we found that Cu2+ boosted the NADH oxidation activity in a dose-dependent manner, and Cu2+ was bound to the active site. Furthermore, we discovered the O2 consumption activity of RclA in the presence of Cu2+, with a limited production of superoxide anion.
We further found that O2 reduction is required for the Cu2+ reduction activity of EcRclA and vice versa, indicating that the NADH oxidation is coupled with the reduction of O2 dissolved in the buffer. Furthermore, neither the reduced portions of Cu2+ nor O2 was able to account fully for the released electrons from NADH. These findings indicate that the reduction of Cu2+ and O2 should be simultaneously considered a reaction cycle. Because O2 is the starting substance to produce HOCl via oxidative burst in immune cells, the reduced level of O2 in the phagosomes would lower the level of HOCl. To decrease the level of HOCl would be an important role of RclA for bacterial survival in immune cells.
Despite the structural and chemical similarities to MerA mediating a two-electron transfer to a metal ion, a previous study (14) presented evidence for a one-electron transfer to Cu2+ producing Cu+. However, it seems odd that Cu+ is more active than Cu2+ in Fenton-like reactions to produce hydroxyl radicals from HOCl. We believe that more sophisticated reaction networks with other proteins would be involved.
FDRs are involved in many different chemical reactions using the bound FAD and various electron acceptors, such as disulfide, O2, or metal ions. In the group I FDR GSH reductase, electrons from NADPH are transferred to the disulfide of the oxidized GSH using the CXXXXC motif as the key intermediate. However, a small portion of electrons flows into oxygen to generate superoxide anion or H2O2 as a side reaction of FDRs (21). Furthermore, the reduction of GSH is strongly inhibited by Cu2+ (26), indicating that Cu2+ is bound at the active site, and thus it alters the electron flow in the reaction mechanism as a potential regulator. In the cases of MerA and RclA, the electrons flow into Hg2+ and Cu2+, respectively, as the final acceptor, which is the primary reaction of these enzymes. Thus, our findings suggest that binding of metal ions at the active site alters the pathway of electrons from FDRs toward the bound metal ions.
Our structural and mutational studies provide hints on the reaction mechanism of RclA (Fig. 8A). This study observed the Cu2+ binding in the presumably-oxidized FAD-bound structure, presenting the FAD–Cys-48–Cu2+–Cys-43 network as the critical electron flow route. The reaction would be triggered when the electrons are provided to FAD by NADH in the presence of O2. The electrons released from FADH2 are first transferred to Cys-48 and then transferred to the bound Cu2+ with the Cys-43. This Cys-48–mediated electron transfer to Cu2+ is consistent with the result that the Cu2+ dependence in NADH oxidation disappeared with increased levels of superoxide anion production in the C48S mutant. We thus propose that Cys-48 prevents the direct single electron transfer from FADH2 to O2 by absorbing electrons from FADH2, leading to suppression of superoxide anion production, and transfers the received electrons to Cu2+.
Figure 8.
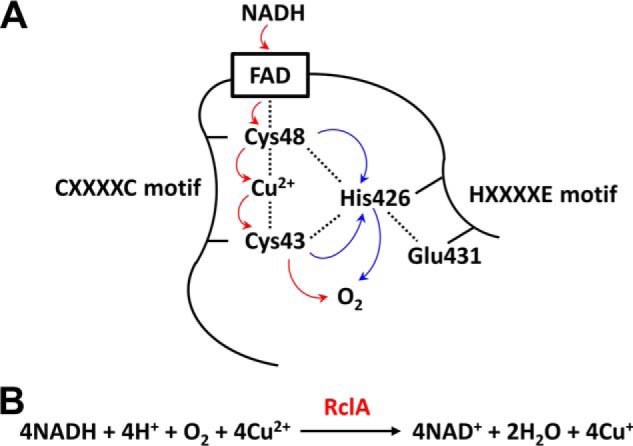
Proposed reaction order of O2 and Cu2+ reduction by EcRclA. A, red arrows represent the possible electron flow. Blue arrows indicate the possible proton flow. The bound FAD in EcRclA is presented in a black box. Interactions between two amino acid residues or between an amino acid residue and Cu2+ ion are shown as dashed lines. B, proposed reaction mediated by RclA.
The Cu2+-bound structure, together with the results of the C43S mutation, demonstrated that Cys-43 is directly involved in the binding of Cu2+. This C-terminal cysteine residue (Cys-43) in the CXXXXC motif would affect the specificity of various metal ions, as the C43S mutant showed different NADH oxidation rates with metal ions compared with the WT. Because the sulfur atom of Cys-43 and the imidazole ring of His-426 are within a distance to make a hydrogen bond, the His-426–Glu-431 pair in the HXXXXE motif would deprotonate Cys-43, promoting binding with the Cu2+. Furthermore, the His–Glu pair could also transfer protons to proton or electron acceptors during the catalysis. To reduce oxygen, the electron transfer should accompany the proton transfer. Thus, this proton transfer mediated by this HXXXXE motif would also be important for the catalysis. Because of the complexity of the reactions, we believe that multiple rounds might be required to complete the reaction. Combined with the previous result (14), we propose an overall reaction mechanism, which meets the stoichiometry (Fig. 8B). However, further studies are required to elucidate the complete reactions.
In conclusion, we used a high-resolution structure of RclA to investigate its biochemical roles and functions in the HOCl resistance of bacteria. Although further study is required to elucidate the molecular mechanism, our study provides a molecular basis for how bacteria can cope with our immune system, which may help in the creation of strategies to control pathogenic bacteria.
Experimental procedures
Plasmid construction and protein production
To overexpress the full-length (amino acids 1–441) RclA WT, the gene encoding the protein was amplified and cloned into the pProEX-HTa expression vector (Invitrogen). The resulting plasmid encodes the hexahistidine tag and the TEV protease cleavage site at the N terminus of the protein. For each RclA mutant protein (C43S, C48S, and K13A/K16A), site-directed mutagenesis was performed. The plasmids encoding RclA WT protein were transformed into the E. coli C43 (DE3) strain, and each plasmid encoding the RclA mutant was transformed into E. coli BL21 (DE3). The transformed cells were cultured in 1.5 liters of Luria Broth medium containing 100 μg/ml ampicillin at 37 °C until an OD600 of 0.7 was measured, and the protein expression was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 30 °C for 6 h. After harvesting, cells were resuspended in lysis buffer containing 20 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 2 mm 2-mercaptoethanol. The suspensions were disrupted using a continuous-type French press (Constant Systems Ltd., United Kingdom) at 23,000 p.s.i. After centrifugation at 19,000 × g for 30 min at 4 °C, the supernatant was loaded onto a nickel-nitrilotriacetic acid column (GE Healthcare). After washing the column with ∼300 ml of lysis buffer supplemented with 20 mm imidazole, the RclA WT protein was eluted with lysis buffer supplemented with 250 mm imidazole. The eluate was treated with TEV protease to cleave the hexahistidine tag. Cleaved proteins were diluted 4-fold with 20 mm Tris-HCl (pH 8.0) buffer containing 2 mm 2-mercaptoethanol and loaded onto a HiTrap Q column (GE Healthcare). A linear gradient of increasing NaCl concentrations was applied to the column. The fractions containing the protein were pooled, concentrated, and subjected to size-exclusion chromatography using a HiLoad 26/600 Superdex 200 pg column (GE Healthcare) equilibrated with 20 mm Tris-HCl (pH 8.0) buffer containing 150 mm NaCl and 2-mercaptoethanol. The purified protein was concentrated to 13 mg/ml and stored at −80 °C until used for crystallization. Protein concentrations were determined based on FAD content using a previously determined ϵ456 of 11.3 mm−1 cm−1.
Crystallization of RclA
RclA proteins (WT, C43S) were crystallized at 16 °C using the hanging-drop vapor–diffusion method after mixing 1 μl of protein solution and 1 μl of well solution. The RclA WT protein was first crystallized in a well solution containing 0.2 m ammonium sulfate, 0.1 m BisTris-HCl (pH 5.8), 25% (w/v) PEG 3350, and 2 mm TCEP. Subsequently, the crystals were disrupted for microseeding and diluted with the well solution containing 0.2 m ammonium sulfate, 0.1 m BisTris-HCl (pH 5.8), 13% (w/v) PEG 3350, and 2 mm TCEP. Microseeding was performed by mixing 1 μl of protein and 1 μl of microseeds diluted with the well solution. The RclA WT crystals were briefly incubated in the well solution supplemented with 20% (v/v) glycerol as a cryoprotectant, flash-frozen in liquid ethane at −88.5 °C, and subsequently transferred into liquid nitrogen to collect the dataset. For the Cu2+-soaked RclA WT structure, the crystals were incubated in a reservoir solution supplemented with 5 mm CuCl2 for 15 h. The crystals were incubated in the well solution supplemented with 30% (v/v) glycerol and 5 mm CuCl2 for 6 min, and flash-frozen in liquid nitrogen to collect the dataset. RclA C43S was crystallized in a solution containing 0.05 mm ammonium sulfate, 0.05 mm BisTris-HCl (pH 5.8), and 30% pentaerythritol ethoxylate (15/4_EO/OH). Microseeding was performed using disrupted RclA C43S crystals and was diluted with 0.05 mm ammonium sulfate, 0.05 mm BisTris-HCl (pH 5.8), 25% pentaerythritol ethoxylate (15/4_EO/OH). For Cu2+-soaked RclA C43S structure, the RclA C43S crystals were incubated in the well solution supplemented with 5 mm CuCl2 for 15 h. After incubation of the crystals in reservoir solution supplemented with 30% (v/v) glycerol and 5 mm CuCl2 for 30 s, crystals were flash-frozen in a nitrogen stream at −173 °C.
Data collection and structural determination
All diffraction datasets were collected at the Pohang Accelerator Laboratory Beamline 5C (27) and were processed using the HKL-2000 package (28, 29). The structure of the RclA WT was determined using the molecular replacement method with MOLREP in the CCP4 package (29), using a model built by the Galaxy web server (30, 31) as a search model. The initial model of the RclA WT was used to solve other structures of RclA (RclA WT-Cu2+ and RclA C43S-Cu2+) by the molecular replacement method. All of the structures were built using COOT and refined using PHENIX refine software (32, 33). The detailed statistics are shown in Table 1.
Enzyme kinetic assays
All assays were performed using UV-visible spectrophotometer, GeneQuant 1300 (GE Healthcare) or Multiscan Go (Thermo Fisher Scientific). The RclA activity was assayed by adding 2 μm protein (as measured based on flavin content) in 10 mm MES buffer (pH 7.0) containing 100 mm NaCl, 0.2 mm NADH, and 0.2 mm putative substrate. Initial rates of NADH oxidation by RclA with various metal ions were monitored by the loss of absorbance at 340 nm due to NADH oxidation (Δϵ340 = 6.2 mm−1 cm−1); the loss of absorbance was monitored for 10 min, and the oxidation profile was expressed in a time-dependent manner as the mean ± S.D. of three independent experiments. Control assays lacking the substrate or enzyme were routinely included as references. NADH oxidation by RclA mutants C43S and C48S were recorded as described previously for the RclA WT assay.
For the examination of enzyme dependence on Cu2+ concentration, NADH oxidation was measured at 340 nm with various concentrations of CuCl2. Before the assay, the RclA WT protein was desalted with 10 mm MES buffer (pH 7.0) supplemented with 100 mm NaCl to remove any remaining 2-mercaptoethanol from the RclA size-exclusion chromatography buffer. The activity assays were initiated by adding 2 μm desalted RclA WT protein to 10 mm MES buffer (pH 7.0) containing 100 mm NaCl, 0.2 mm NADH, and 0, 0.025, 0.05, 0.075, 0.1, 0.2, or 0.3 mm CuCl2. Initial rates were monitored by the loss of absorbance at 340 nm due to NADH oxidation (Δϵ340 = 6.2 mm−1 cm−1) and expressed as the mean ± S.D. of four independent experiments.
For the determination of Km and Vmax values of RclA WT and RclA K13A/K16A, NADH oxidation was measured at 340 nm with various concentrations of CuCl2. Before the assay, each RclA protein was desalted with 10 mm MES buffer (pH 7.0) supplemented with 100 mm NaCl. The activity assays were initiated by adding 2 μm desalted RclA WT or RclA K13A/K16A protein to 10 mm MES buffer (pH 7.0) containing 100 mm NaCl, 0.2 mm NADH, and 0, 0.6, 0.15, 0.3, 0.6, 1.5, 3, or 6 mm CuCl2. Initial rates were monitored by the loss of absorbance at 340 nm due to NADH oxidation (Δϵ340 = 6.2 mm−1 cm−1) and expressed as the mean ± S.D. of three independent experiments. The data were fit with a Michaelis-Menten equation, and the Km and Vmax values were determined using an enzyme kinetics tool of the SigmaPlot Version 14.0 (Systat Software, San Jose, CA).
NADH oxidation assay under low O2 conditions
The dissolved oxygen (DO) was removed by nitrogen purging. The buffer with 10 mm MES (pH 7.0), 100 mm NaCl, 0.2 mm NADH, and with or without 0.1 mm CuCl2 was contained in a 1-ml cuvette and sealed with parafilm. The solution was purged with N2 for 5 min. After nitrogen purging, 2 μm RclA was added. Initial rates of NADH oxidation by RclA were monitored by the loss of absorbance at 340 nm due to NADH oxidation (Δϵ340 = 6.2 mm−1 cm−1), monitored using a GeneQuant 1300 UV-visible spectrophotometer (GE Healthcare), and expressed in time-dependent manner as the mean ± S.D. of three independent experiments. A control assay in which the solution was not purged with N2 was included for comparison.
Detection of DO in water
DO concentrations were measured using the Gravity Analog Dissolved Oxygen Sensor (SKU:SEN0237; DFRobot) with an Arduino Nano board and the Arduino IDE program (34). The programming code was obtained from the DFRobot website. The reaction volume was 5 ml, containing 10 mm MES (pH 7.0) buffer with 100 mm NaCl, 0.2 mm NADH, and 1 μm RclA, with or without 0.2 mm CuCl2. The solution was stirred gently with a 1-cm magnetic stir bar to distribute DO evenly throughout the entire solution. After preincubation for 5 min with stirring for equilibrium, the RclA protein was added and the DO concentration was recorded for 10 min. The specific oxidation rate was measured within the initial 1 min and expressed as the mean ± S.D. of three independent experiments.
Measurement of O2˙̄ formation by cytochrome c assay
Superoxide anion (O2˙̄) formation was determined by monitoring the reduction of cytochrome c, which was isolated from the equine heart (Sigma). The reaction was performed with 200 μl of buffer containing 10 mm MES buffer (pH 7.0), 100 mm NaCl, 200 μm NADH, 100 μm CuCl2, 2 μm RclA, and 50 μm cytochrome c. The increase in the absorbance at 550 nm due to cytochrome c reduction was recorded at 37 °C for 10 min using an UV-visible spectrophotometer (Multiscan Go, Thermo Fisher Scientific). The amount of O2˙̄ released was calculated using an extinction coefficient of 21 mm−1 cm−1 and expressed as the mean ± S.D. of three independent experiments.
Intracellular survival assay
RAW264.7 macrophage-like cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, penicillin (50 units/ml), and streptomycin (50 units/ml). The cells were seeded at 2 × 105 cells/well in a 24-well tissue culture plate and incubated for 24 h at 37 °C with 5% CO2. A monolayer of cells was infected with bacteria at a multiplicity of infection of 10. After 30 min of incubation at 37 °C under 5% CO2, nonengulfed bacteria were removed by washing with PBS and incubated for 1 h with medium containing 100 μg/ml gentamycin to kill extracellular bacteria. Subsequently, the medium was exchanged with fresh medium containing 10 μg/ml gentamycin, and cells were cultured for the indicated times (2 or 24 h). For counting bacterial cells replicated within RAW264.7, the macrophage cells were lysed with 1% Triton X-100.
Author contributions
Y. B., J. A., I. J., S. R., and N.-C. H. conceptualization; Y. B. and J. K. resources; Y. B. and J. K. data curation; Y. B. and I. J. software; Y. B., J. K., J. A., S. H., and S. R. validation; Y. B., J. K., J. A., I. J., S. H., and N.-C. H. investigation; Y. B., J. K., and visualization; Y. B., J. K., J. A., and I. J. methodology; Y. B. and N.-C. H. writing-original draft; Y. B., S. H., and N.-C. H. writing-review and editing; J. K. and Y. B. formal analysis; N.-C. H. project administration; S. R., and N.-C. H. supervision; N.-C. H. funding acquisition.
Supplementary Material
Acknowledgment
We thank the Pohang Accelerator Laboratory (Pohang, Republic of Korea) for use of its Beamline 5C equipment.
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) Grant 710012-03-1-HD120, the ARC Program (to N. C. H.) funded by the Ministry of Agriculture, Food, and Rural Affairs, National Research Foundation of Korea Grant NRF-2017R1A2B2003992 (to N. C. H.), and by Korean Government (MSIT) Grant NRF-2019M3E5D6063871. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S9 and supporting Refs. 1 and 2.
The atomic coordinates and structure factors (codes 6KGY, 6KYY, and 6KOD) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- HOCl
- hypochlorous acid
- FDR
- flavoprotein disulfide reductase
- TCEP
- tris(2-carboxyethyl)phosphine
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- DO
- dissolved oxygen
- MerA
- mercuric reductase
- PDB
- Protein Data Bank
- RMSD
- root mean square deviation
- TEV
- tobacco etch virus.
References
- 1. Weiss S. J., and LoBuglio A. F. (1982) Phagocyte-generated oxygen metabolites and cellular injury. Lab. Invest. 47, 5–18 [PubMed] [Google Scholar]
- 2. Ha E. M., Oh C. T., Bae Y. S., and Lee W. J. (2005) A direct role for dual oxidase in Drosophila gut immunity. Science 310, 847–850 10.1126/science.1117311 [DOI] [PubMed] [Google Scholar]
- 3. Winterbourn C. C., and Kettle A. J. (2013) Redox reactions and microbial killing in the neutrophil phagosome. Antioxid. Redox Signal. 18, 642–660 10.1089/ars.2012.4827 [DOI] [PubMed] [Google Scholar]
- 4. Hampton M. B., Kettle A. J., and Winterbourn C. C. (1998) Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood 92, 3007–3017 10.1182/blood.V92.9.3007 [DOI] [PubMed] [Google Scholar]
- 5. Hawkins C. L., Pattison D. I., and Davies M. J. (2003) Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids 25, 259–274 10.1007/s00726-003-0016-x [DOI] [PubMed] [Google Scholar]
- 6. Davies M. J. (2005) The oxidative environment and protein damage. Biochim. Biophys. Acta 1703, 93–109 10.1016/j.bbapap.2004.08.007 [DOI] [PubMed] [Google Scholar]
- 7. Gray M. J., Wholey W. Y., and Jakob U. (2013) Bacterial responses to reactive chlorine species. Annu. Rev. Microbiol. 67, 141–160 10.1146/annurev-micro-102912-142520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Folkes L. K., Candeias L. P., and Wardman P. (1995) Kinetics and mechanisms of hypochlorous acid reactions. Arch. Biochem. Biophys. 323, 120–126 10.1006/abbi.1995.0017 [DOI] [PubMed] [Google Scholar]
- 9. Parker B. W., Schwessinger E. A., Jakob U., and Gray M. J. (2013) The RclR protein is a reactive chlorine-specific transcription factor in Escherichia coli. J. Biol. Chem. 288, 32574–32584 10.1074/jbc.M113.503516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gebendorfer K. M., Drazic A., Le Y., Gundlach J., Bepperling A., Kastenmüller A., Ganzinger K. A., Braun N., Franzmann T. M., and Winter J. (2012) Identification of a hypochlorite-specific transcription factor from Escherichia coli. J. Biol. Chem. 287, 6892–6903 10.1074/jbc.M111.287219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gray M. J., Wholey W.-Y., Parker B. W., Kim M., and Jakob U. (2013) NemR is a bleach-sensing transcription factor. J. Biol. Chem. 288, 13789–13798 10.1074/jbc.M113.454421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jo I., Kim D., No T., Hong S., Ahn J., Ryu S., and Ha N.-C. (2019) Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator. Proc. Natl. Acad. Sci. U.S.A. 116, 3740–3745 10.1073/pnas.1811509116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin R. G., and Rosner J. L. (2001) The AraC transcriptional activators. Curr. Opin. Microbiol. 4, 132–137 10.1016/S1369-5274(00)00178-8 [DOI] [PubMed] [Google Scholar]
- 14. Derke R. M., Barron A. J., Chaple I. F., Lapi S. E., Broderick N. A., and Gray M. J. (2019) RclA is a thermostable copper(II) reductase required for reactive chlorine resistance and host colonization in Escherichia coli. bioRxiv, 690669 10.1101/690669 [DOI]
- 15. Argyrou A., and Blanchard J. S. (2004) Flavoprotein disulfide reductases: advances in chemistry and function. Prog. Nucleic Acid Res. Mol. Biol. 78, 89–142 10.1016/S0079-6603(04)78003-4 [DOI] [PubMed] [Google Scholar]
- 16. Holm L. (2019) Benchmarking fold detection by DaliLite v. 5. Bioinformatics 35, 5326–5327 10.1093/bioinformatics/btz536 [DOI] [PubMed] [Google Scholar]
- 17. Schrodinger LLC. (2010) The PyMOL Molecular Graphics System, PyMOL version 1.7, Schrodinger, LLC, New York [Google Scholar]
- 18. Rennex D., Pickett M., and Bradley M. (1994) In vivo and in vitro effects of mutagenesis of active site tyrosine residues of mercuric reductase. FEBS Lett. 355, 220–222 10.1016/0014-5793(94)01180-X [DOI] [PubMed] [Google Scholar]
- 19. Angiulli G., Lantella A., Forte E., Angelucci F., Colotti G., Ilari A., and Malatesta F. (2015) Leishmania infantum trypanothione reductase is a promiscuous enzyme carrying an NADPH: O2 oxidoreductase activity shared by glutathione reductase. Biochim. Biophys. Acta 1850, 1891–1897 10.1016/j.bbagen.2015.05.022 [DOI] [PubMed] [Google Scholar]
- 20. Geueke B., Riebel B., and Hummel W. (2003) NADH oxidase from Lactobacillus brevis: a new catalyst for the regeneration of NAD. Enzyme Microbial Technology 32, 205–211 10.1016/S0141-0229(02)00290-9 [DOI] [Google Scholar]
- 21. Imlay J. A. (2013) The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454 10.1038/nrmicro3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Massey V., Strickland S., Mayhew S. G., Howell L. G., Engel P. C., Matthews R. G., Schuman M., and Sullivan P. A. (1969) The production of superoxide anion radicals in the reaction of reduced flavins and flavoproteins with molecular oxygen. Biochem. Biophys. Res. Commun. 36, 891–897 10.1016/0006-291X(69)90287-3 [DOI] [PubMed] [Google Scholar]
- 23. Liochev S. I., and Fridovich I. (1992) Superoxide generated by glutathione reductase initiates a vanadate-dependent free radical chain oxidation of NADH. Arch. Biochem. Biophys. 294, 403–406 10.1016/0003-9861(92)90703-Y [DOI] [PubMed] [Google Scholar]
- 24. Ledwidge R., Patel B., Dong A., Fiedler D., Falkowski M., Zelikova J., Summers A. O., Pai E. F., and Miller S. M. (2005) NmerA, the metal binding domain of mercuric ion reductase, removes Hg2+ from proteins, delivers it to the catalytic core, and protects cells under glutathione-depleted conditions. Biochemistry 44, 11402–11416 10.1021/bi050519d [DOI] [PubMed] [Google Scholar]
- 25. Ray S., Gachhui R., Pahan K., Chaudhury J., and Mandal A. (1989) Detoxification of mercury and organomercurials by nitrogen-fixing soil bacteria. J. Biosci. 14, 173–182 10.1007/BF02703169 [DOI] [Google Scholar]
- 26. Murakami K., Tsubouchi R., Fukayama M., and Yoshino M. (2014) Copper-dependent inhibition and oxidative inactivation with affinity cleavage of yeast glutathione reductase. Biometals 27, 551–558 10.1007/s10534-014-9731-x [DOI] [PubMed] [Google Scholar]
- 27. Park S.-Y., Ha S.-C., and Kim Y.-G. (2017) The protein crystallography beamlines at the pohang light source II. Biodesign 5, 30–34 [Google Scholar]
- 28. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 29. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shin W.-H., Lee G. R., Heo L., Lee H., and Seok C. (2014) Prediction of protein structure and interaction by GALAXY protein modeling programs. Bio. Design 2, 1–11 [Google Scholar]
- 31. Ko J., Park H., Heo L., and Seok C. (2012) GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 40, W294–W297 10.1093/nar/gks493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 33. Afonine P. V., Mustyakimov M., Grosse-Kunstleve R. W., Moriarty N. W., Langan P., and Adams P. D. (2010) Joint X-ray and neutron refinement with phenix. refine. Acta Crystallogr. D Biol. Crystallogr. 66, 1153–1163 10.1107/S0907444910026582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deleted in proof.
- 35. Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J.-M., Taly J.-F., and Notredame C. (2011) T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17 10.1093/nar/gkr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lian P., Guo H.-B., Riccardi D., Dong A., Parks J. M., Xu Q., Pai E. F., Miller S. M., Wei D.-Q., Smith J. C., and Guo H. (2014) X-ray structure of a Hg2+ complex of mercuric reductase (MerA) and quantum mechanical/molecular mechanical study of Hg2+ transfer between the C-terminal and buried catalytic site cysteine pairs. Biochemistry 53, 7211–7222 10.1021/bi500608u [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.