Abstract
Upon activation with pathogen-associated molecular patterns, metabolism of macrophages and dendritic cells is shifted from oxidative phosphorylation to aerobic glycolysis, which is considered important for proinflammatory cytokine production. Fragments of bacterial peptidoglycan (muramyl peptides) activate innate immune cells through nucleotide-binding oligomerization domain (NOD) 1 and/or NOD2 receptors. Here, we show that NOD1 and NOD2 agonists induce early glycolytic reprogramming of human monocyte-derived macrophages (MDM), which is similar to that induced by the Toll-like receptor 4 (TLR4) agonist lipopolysaccharide. This glycolytic reprogramming depends on Akt kinases, independent of mTOR complex 1 and is efficiently inhibited by 2-deoxy-d-glucose (2-DG) or by glucose starvation. 2-DG inhibits proinflammatory cytokine production by MDM and monocyte-derived dendritic cells activated by NOD1 or TLR4 agonists, except for tumor necrosis factor production by MDM, which is inhibited initially, but augmented 4 h after addition of agonists and later. However, 2-DG exerts these effects by inducing unfolded protein response rather than by inhibiting glycolysis. By contrast, glucose starvation does not cause unfolded protein response and, in normoxic conditions, only marginally affects proinflammatory cytokine production triggered through NOD1 or TLR4. In hypoxia mimicked by treating MDM with oligomycin (a mitochondrial ATP synthase inhibitor), both 2-DG and glucose starvation strongly suppress tumor necrosis factor and interleukin-6 production and compromise cell viability. In summary, the requirement of glycolytic reprogramming for proinflammatory cytokine production in normoxia is not obvious, and effects of 2-DG on cytokine responses should be interpreted cautiously. In hypoxia, however, glycolysis becomes critical for cytokine production and cell survival.
Keywords: macrophage, dendritic cell, glucose metabolism, Nod-like receptor (NLR), cytokine, unfolded protein response (UPR), 2-deoxy-d-glucose, metabolic reprogramming, NOD1, NOD2, glycolysis, muramyl peptides
Introduction
Metabolism of innate immune cells is profoundly reprogrammed upon their activation, so as to meet demands from diverse processes ongoing in activated cells, such as synthesis of cytokines and effector molecules, phagocytosis, and migration (1, 2). Specific characteristics of metabolic reprogramming depend on the activating stimulus. For example, resting macrophages use oxidative phosphorylation (OXPHOS)2 as their main energy source. Pro-inflammatory stimuli such as lipopolysaccharide (LPS) and interferon-γ, which trigger M1-type macrophage activation, induce a number of metabolic alterations including the switch to aerobic glycolysis, disruption of the Krebs cycle, and suppression of OXPHOS (1, 3, 4). Similar changes are observed in dendritic cells activated by LPS (5–7). M2-type macrophage activation induced by interleukin (IL)-4 is characterized by a transient increase in glycolysis (8) as well as augmentation of glutaminolysis and fatty acid oxidation (3, 9). It is proposed that the type of metabolism not only supports functional demands of innate immune cells, but also determines specific characteristics of their activation and differentiation (1, 3, 10). It follows that immune responses could be manipulated by targeting metabolism of innate immune cells, which would aid in treatment of acute and chronic inflammatory diseases (11, 12).
Aerobic glycolysis, also known as Warburg effect, is a type of metabolism whereby glycolysis serves as the main source of ATP, despite favorable conditions for OXPHOS (sufficient oxygen supply) (13). Increased glycolytic flux generates metabolites that serve as starting points for protein, nucleic acid, and lipid biosynthesis (13, 14). Glycolytic reprogramming of innate immune cells probably involves multiple mechanisms. One of them is more rapid and relies on the translocation of pre-existing hexokinase II (HK-II) onto the outer mitochondrial membrane (5). Another, slower mechanism is based on transcriptional up-regulation of glycolytic enzymes, mediated by a signaling pathway involving Akt kinase, mTOR complex 1 (mTORC1), and hypoxia-inducible factor (HIF)-1α (15). Aerobic glycolysis is viewed as a metabolic basis of inflammation (1).
Mechanisms of pro-inflammatory metabolic reprogramming have mostly been studied using LPS treatment in vitro or in vivo (3, 5, 6, 16, 17). However, LPS is a very strong stimulus that might not be representative of all the multitude of pathogen-associated molecular patterns recognized by diverse pattern recognition receptors (PRRs). In particular, little is known about metabolic reprogramming induced by muramyl peptides, i.e. fragments of bacterial peptidoglycan recognized by two closely related cytosolic receptors, nucleotide-binding oligomerization domain (NOD) 1 and NOD2 (18, 19). Importantly, NOD1 and NOD2 agonists are promising immunostimulants and adjuvants, some of which are used in the clinics (20).
Here, we have analyzed metabolic reprogramming induced by NOD1 and NOD2 agonists in human monocyte-derived macrophages (MDM) and monocyte-derived dendritic cells (MDDC) in vitro and in mouse peritoneal macrophages (PM) in vivo. We used N-acetylmuramyl-l-alanyl-d-isoglutamyl-meso-diaminopimelic acid (M-triDAP), which is an NOD1 agonist with a minor activity toward NOD2 (21, 22); two specific NOD2 agonists (N-acetylmuramyl-l-alanyl-d-isoglutamine, also known as muramyl dipeptide (MDP) and N-acetylglucosaminyl-N-acetylmuramyl-l-alanyl-d-isoglutamine, also known as glucosaminyl muramyl dipeptide (GMDP)) (21, 23); and LPS as a control. We show that NOD1/NOD2 and TLR4 agonists induce early glycolytic reprogramming of MDM. We also show that glycolytic reprogramming is not required for pro-inflammatory cytokine production by MDM and MDDC, at least within the time frame of our experiments (24 h).
Results
Expression of TNF, IL6, and IL1B by human MDM stimulated by M-triDAP, MDP, or LPS
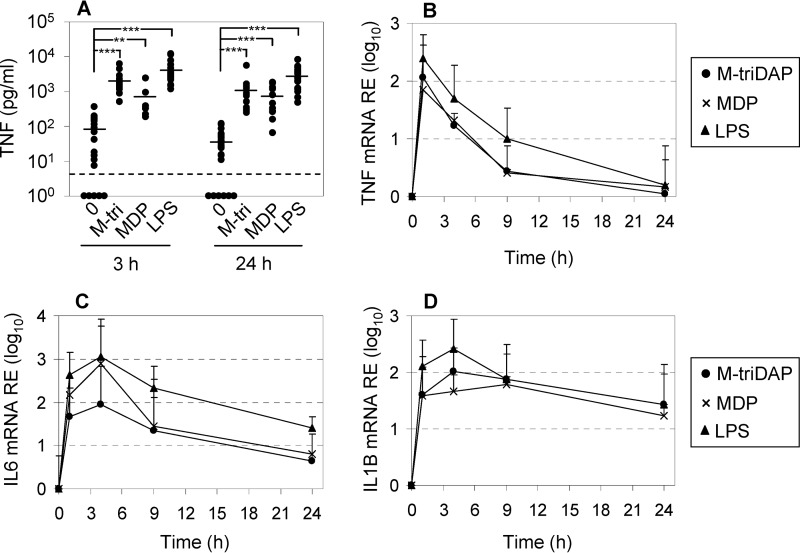
We generated MDM by culturing human donor monocytes with granulocyte-macrophage colony-stimulating factor (GM-CSF), and characterized cytokine responses of MDM to agonists of NOD1 (M-triDAP), NOD2 (MDP), and TLR4 (LPS). All three agonists induced tumor necrosis factor (TNF) secretion (Fig. 1A), with MDP being somewhat less potent than M-triDAP and LPS. All three agonists induced TNF, IL6, and IL1B mRNA expression (Fig. 1, B–D).
Figure 1.
Pro-inflammatory cytokine expression by human MDM. A, levels of TNF in MDM supernatants after 3 and 24 h of culture with medium alone (0), M-triDAP (10 μg/ml), MDP (1 μg/ml), or LPS (100 ng/ml). There were 18 donors, except for MDP (10 donors). Horizontal bars within each group are mean values; dashed line, lower limit of detection by ELISA. **, p < 0.01; ***, p < 0.001 by paired t test. B–D, kinetics of TNF, IL6, and IL1B mRNA expression by MDM stimulated as in A. Mean ± S.D. of log-transformed data, 9 donors.
NOD and TLR4 agonists trigger rapid glycolytic reprogramming of human MDM but differentially induce ACOD-1 expression
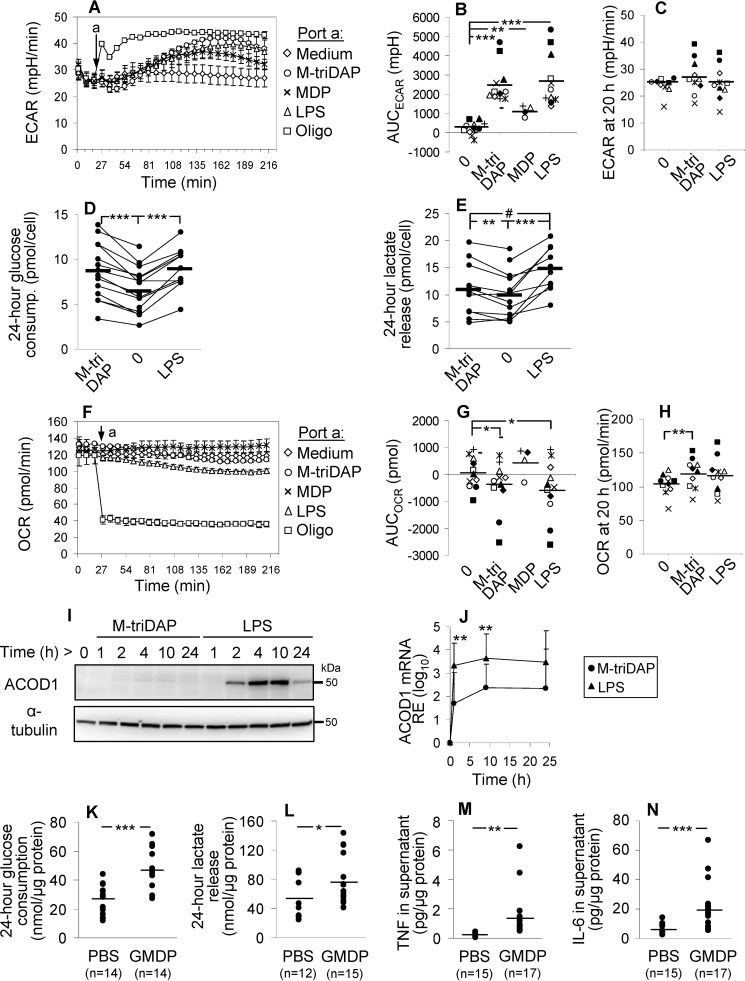
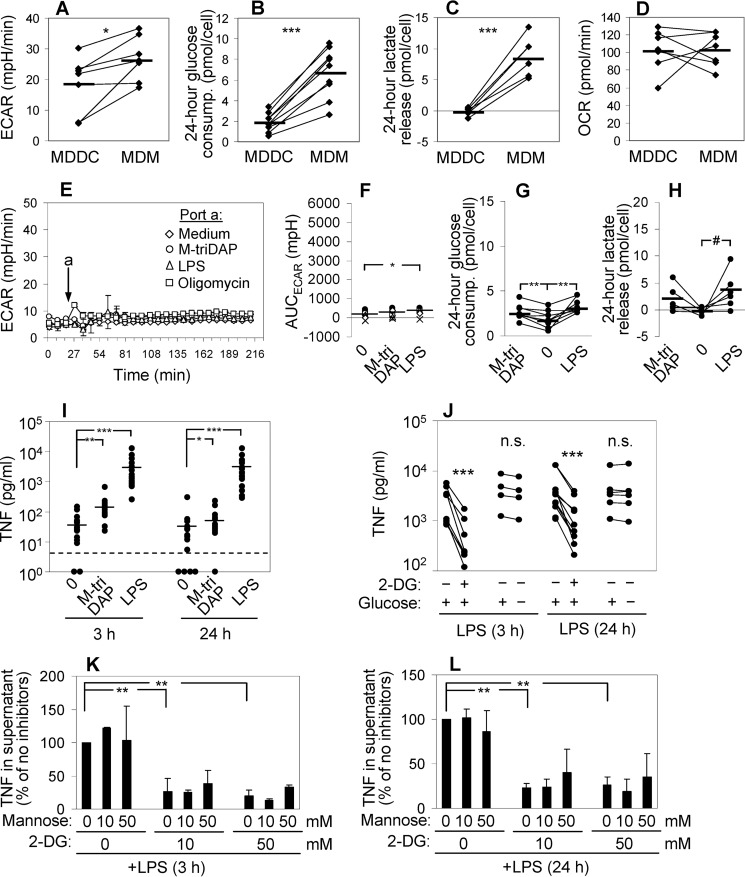
To assess glycolysis, we measured extracellular medium acidification rate (ECAR) before and after addition of M-triDAP, MDP, or LPS to MDM cultures. Within 1 h, all agonists induced elevation of ECAR (LPS ≈ M-triDAP > MDP) (Fig. 2, A and B). The response peaked at 2–2.5 h and gradually declined thereafter (Fig. 2A). To estimate glycolytic reserve, we treated MDM with oligomycin, a mitochondrial ATP synthase inhibitor. Oligomycin induced a more rapid elevation of ECAR, reaching a plateau at 30–40 min (Fig. 2A). Notably, at the peak of M-triDAP– or LPS-induced responses, MDM deployed most of their glycolytic reserve (Fig. 2A). At 20 h after addition of agonists, no differences in ECAR between unstimulated and stimulated MDM were found (Fig. 2C). Similar changes of ECAR upon M-triDAP or LPS treatment were observed in MDM generated with macrophage colony-stimulating factor (M-CSF) instead of GM-CSF (Fig. S1A).
Figure 2.
Metabolic reprogramming of macrophages activated by NOD1, NOD2 and TLR4 receptor agonists. A and F, kinetics of ECAR (A) and OCR (F) in human MDM after injection (arrow) of medium, M-triDAP (10 μg/ml), MDP (1 μg/ml), LPS (100 ng/ml), or oligomycin (2 μm). 1 representative experiment out of 12; mean ± S.D. values of quadruplicates. B and G, AUC for ECAR (B) and OCR (G) responses shown in A and F, respectively. Twelve donors, except for MDP (4 donors), each shown by a unique symbol. C and H, ECAR (C) and OCR (H) after a 20-h culture of MDM with medium alone, M-triDAP, or LPS (n = 10). D and E, 24-h glucose consumption (D) and lactate release (E) by MDM treated with medium, M-triDAP, or LPS (10–16 donors per data point; lines connect measurements from individual experiments). In B–E, G, and H, horizontal lines denote group means; #, p = 0.07; *, p < 0.05; **, p < 0.01; ***, p < 0.001 by paired t test. I, expression of ACOD1 protein by MDM stimulated with M-triDAP or LPS (Western blotting), 1 representative experiment out of 3. J, kinetics of ACOD1 mRNA expression upon treatment with M-triDAP or LPS (mean ± S.D., 6 donors). **, p < 0.01 for comparisons between M-triDAP- and LPS-induced expression at given time points. K–N, glucose metabolism and cytokine production by PM from C57BL/6 mice that had received PBS or GMDP (100 μg) subcutaneously. PM were sampled 2 h after injection and cultured for 24 h to assess glucose consumption (K), lactate (L), TNF (M), and IL-6 (N) release per μg cellular protein; horizontal lines denote group means; *, p < 0.05; ***, p < 0.001 by t test.
Additionally, M-triDAP and LPS treatment caused, on average, a 30–40% increase of glucose consumption by MDM (Fig. 2D; Fig. S1B). Interestingly, despite similar effects of M-triDAP and LPS on glucose consumption and ECAR, M-triDAP induced somewhat lower release of lactate as compared with LPS (Fig. 2E; Fig. S1C). Presumably, lactate released early on M-triDAP treatment (as evidenced by kinetics of ECAR) is partly re-utilized by MDM for further metabolism (24, 25).
Oxygen consumption rate (OCR) slowly decreased during first hours of stimulation with M-triDAP or LPS but not MDP, whereas after addition of oligomycin, OCR dropped abruptly by about two thirds (Fig. 2, F and G). When oligomycin was added 3 h after addition of agonists, differences in OCR between unstimulated and stimulated cells were abolished, indicating that the small decrease of OCR induced by M-triDAP or LPS was because of decrease of OXPHOS (Fig. S1D). Additionally, mitochondrial stress test indicated that MDM, irrespectively of preceding NOD1 or TLR4 agonist treatment, possess almost no spare respiratory capacity and use most of the oxygen consumed to sustain mitochondrial respiration (Fig. S1D).
After 20-hour stimulation, OCR in M-triDAP–treated but not LPS-treated MDM was slightly increased compared with untreated cells (Fig. 2H). Mitochondrial stress test did not reveal clear-cut differences in oxygen usage between stimulated and unstimulated cells (Fig. S1E).
A hallmark of metabolic reprogramming triggered by LPS is the induction of aconitate decarboxylase 1 (ACOD1) expression (3, 26). ACOD1 diverts carbon atom flow away from the Krebs cycle by metabolizing cis-aconitate, a Krebs cycle intermediate, into itaconate, an antimicrobial and immunomodulatory agent (17, 26–28). In unstimulated MDM, ACOD1 protein was undetectable (Fig. 2I). LPS strongly induced ACOD1 mRNA and protein expression, whereas M-triDAP induced much lower levels of ACOD1 mRNA and only trace amounts of ACOD1 protein (Fig. 2, I and J). In summary, both TLR4 and NOD receptor agonists induce rapid up-regulation of glycolysis in MDM, whereas ACOD1 is differentially induced and oxygen consumption is marginally affected.
A NOD2 agonist up-regulates glycolysis in mouse PM
To analyze metabolic reprogramming induced by a NOD receptor agonist in vivo, we injected C57BL/6 mice with 100 μg GMDP (a NOD2 agonist) (23), isolated PM 2 h after injection, and measured cytokine production and parameters of glycolysis ex vivo. GMDP increased both glucose consumption and lactate release (Fig. 2, K and L), which was paralleled by elevated TNF and IL-6 production (Fig. 2, M and N). Interestingly, there was a small but significant elevation of Ly6G+F4/80−CD11b+ cells (the phenotype of granulocytes) in GMDP-injected group compared with PBS-injected group (4.9 ± 4.4% versus 0.1 ± 0.1% of adherent cells; p < 0.001). However, no correlations were observed between percentages of Ly6G+ cells and parameters of PM metabolism and cytokine production. Thus, a NOD agonist injected in vivo enhances glucose metabolism in PM in a fashion similar to that observed in human MDM in vitro.
The role of Akt, mTORC1, and PFKFB3 in glycolytic reprogramming of MDM
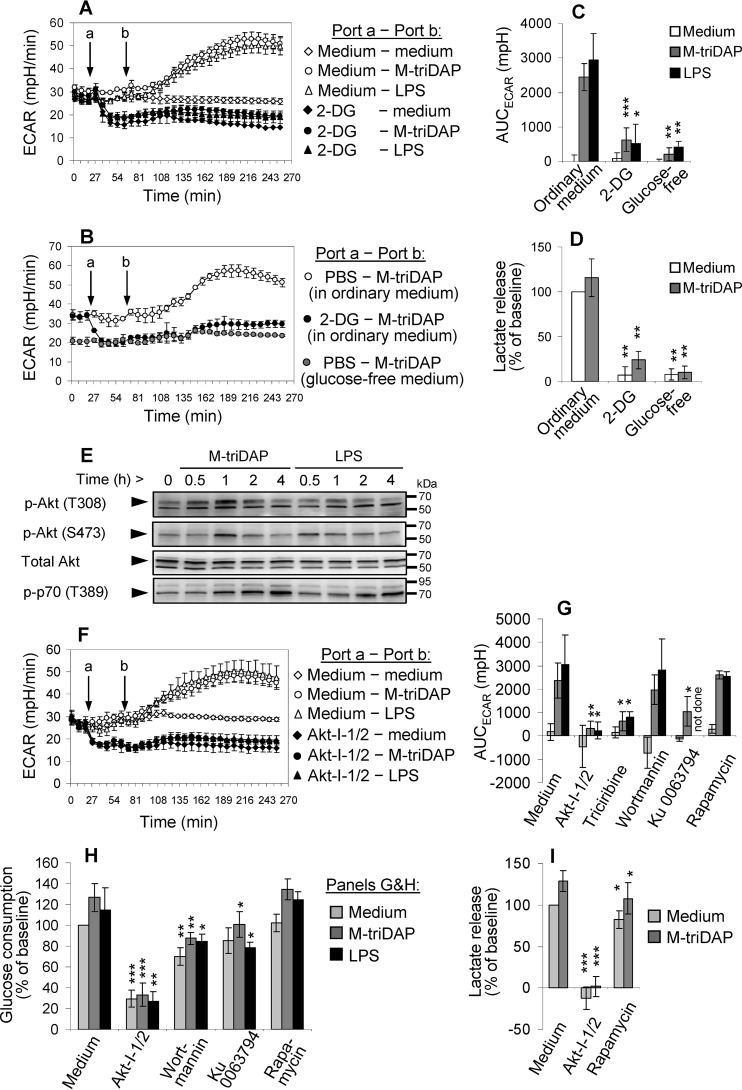
To verify that the elevation of ECAR induced by PRR agonists in human MDM is because of elevated glycolysis, we used either 2-DG, a competitive inhibitor of glycolysis, or glucose-free medium. As expected, 2-DG reduced basal ECAR (Fig. 3A), suppressed M-triDAP– or LPS-induced elevation of ECAR (Fig. 3, A and C), and strongly reduced lactate release (Fig. 3D). Glucose starvation had the same effect on glycolysis as 2-DG (Fig. 3, B–D).
Figure 3.
Mechanisms of glycolytic reprogramming induced by M-triDAP or LPS in MDM. A, ECAR responses induced by M-triDAP (10 μg/ml) or LPS (100 ng/ml) in ordinary glucose-replete medium without or with pre-injection of 2-DG (50 mm). 1 representative experiment out of 5. B, comparison of ECAR responses induced by M-triDAP in ordinary medium without or with 2-DG or in glucose-free medium (1 experiment out of 3). C, AUC for ECAR curves shown in A and C (mean ± S.D. of 5 donors for 2-DG and 3 donors for glucose-free medium). D, 24-hour lactate release in the presence of 2-DG or in glucose-free medium (8 and 6 donors, respectively). E, kinetics of Akt and p70 phosphorylation in MDM upon M-triDAP or LPS treatment (1 representative experiment out of 3). Arrowheads indicate specific bands for Akt/p-Akt (60 kDa) and p-p70 (70 kDa). F, kinetics of ECAR after injection of Akt-I-1/2 (10 μm) and subsequent injection of M-triDAP or LPS (1 experiment out of 4). G–I, effects of inhibitors of Akt and upstream and downstream kinases on AUCECAR (G), 24-h glucose consumption (H), and 24-h lactate release (I) by MDM. MDM were pre-treated with Akt-I-1/2 (10 μm), triciribine (20 μm), wortmannin (100 nm), Ku 0063794 (1 μm), or rapamycin (10 nm), after which stimulated with medium alone, M-triDAP (10 μg/ml), or LPS (100 ng/ml). Mean ± S.D., 4 to 8 experiments per data point. In plots C, D, G–I, asterisks denote comparisons to cells treated with the same PRR agonist in the absence of inhibitors (*, p < 0.05; **, p < 0.01; ***, p < 0.001 by paired t test).
As suggested by ECAR measurements, the elevation of glycolysis in MDM after addition of PRR agonists is rather fast and, therefore, might occur without de novo gene expression. In LPS-activated mouse DCs, a similarly fast elevation of ECAR was explained by redistribution of HK-II from cytosol to the outer mitochondrial membrane (5), where HK-II becomes fully active (29). To bind mitochondrial membrane, HK-II needs to be phosphorylated on threonine-473, which is accomplished by Akt kinase (5, 30). We observed a transient elevation of activated Akt (phosphorylated on threonine-308 and serine-473) at 30–60 min after addition of M-triDAP or LPS (Fig. 3E), i.e. near the time points when ECAR began to rise (see above). A specific Akt inhibitor (Akt-I-1/2) (31) suppressed basal parameters of glycolysis and cancelled their elevation upon M-triDAP or LPS treatment (Fig. 3, F–I); notably, effects of Akt-I-1/2 on glycolysis were very similar to those of 2-DG. Another Akt inhibitor, triciribine, also diminished the M-triDAP– or LPS-induced rise of ECAR (Fig. 3G; Fig. S2A). Inhibitors of kinases upstream of Akt, such as PI-3K (wortmannin) and mTORC2 (Ku 0063794), reduced Akt phosphorylation at Thr-308 and/or Ser-473 (Fig. S2B) and down-modulated parameters of glycolysis in basal and stimulated conditions (Fig. 3, G and H), although to a lesser extent than Akt-I-1/2. All these inhibitors also reduced phosphorylation of PRAS40 (Fig. S2, B and C), an inhibitory component of mTORC1 complex and a direct target of Akt (32).
A slower mechanism of glycolytic reprogramming, which also involves Akt, is based on up-regulation of glycolytic enzyme expression through activation of the Akt–mTORC1–HIF-1α signaling pathway (15). Both M-triDAP and LPS triggered mTORC1 activation in MDM, as indicated by increased phosphorylation of p70 kinase (S6 kinase), a target of mTORC1 (Fig. 3E). However, this pathway was probably not involved in up-regulation of glycolysis in MDM in our experiments. First, kinetics of p70 phosphorylation was much slower than that of Akt phosphorylation or ECAR. Second, Akt-I-1/2 did not interfere with phosphorylation of p70, unlike the mTORC1/2 inhibitor Ku 0063794 (Fig. S2B) or the specific mTORC1 inhibitor rapamycin (data not shown), suggesting Akt-independent mTORC1 activation (33–35). Interestingly, wortmannin abrogated p70 phosphorylation in unstimulated MDM, probably because of the existence of the PI-3K–PDK1–p70 pathway that bypasses Akt and mTORC1 (36–38); however, this effect of wortmannin was much weaker in M-triDAP– or LPS-stimulated MDM (Fig. S2B), indicating that kinases other than PDK1 or Akt, possibly MAP kinases or protein kinase C (34, 35, 39), were responsible for p70 phosphorylation in stimulated MDM. Third, rapamycin only marginally affected parameters of glycolysis (Fig. 3, G–I). Fourth, although HIF1A mRNA was induced at 4 h in M-triDAP– and LPS-stimulated MDM (Fig. S2B), this was not accompanied by accumulation of HIF-1α protein (Fig. S3A) because experiments were carried out in normoxic conditions that prevent HIF-1α stabilization. Notably, addition of CoCl2 to mimic hypoxia caused a substantial elevation of HIF-1α protein, which now corresponded with the rise of HIF1A mRNA expression at 4 h (Fig. S3B).
In agreement with these findings, we did not observe any significant changes of expression of genes encoding glucose transporters, glycolytic enzymes, and LDH in M-triDAP– or LPS-stimulated MDM, except for a minor elevation of GLUT1 mRNA at 9 h of LPS treatment (data not shown) and elevation of PFKFB3 mRNA at 1 h of M-triDAP treatment and at 1 and 9 h of LPS treatment (Fig. S3C).
PFKFB3 (the inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase) has been shown to play a role in glycolytic reprogramming of human and murine macrophages (40, 41), but not of murine DCs (5). The product of this enzyme, fructose-2,6-bisphosphate, enhances glycolysis through allosteric activation of phosphofructokinase-1. The rapid up-regulation of PFKFB3 (Fig. S3C) was consistent with the activation of the faster p38–MAPKAPK-2–SRF signaling pathway (42) rather than the slower mTORC1–HIF-1α pathway, and was inhibited by a p38 inhibitor (VX-745; Fig. S3D). However, neither VX-745 nor two highly specific PFKFB3 inhibitors (AZ PFKFB3 67 and PFK-15 at concentrations up to 100 times higher than IC50) (43, 44) affected basal and stimulated ECAR (Fig. S3E). Furthermore, AZ PFKFB3 67 did not affect 24-h glucose consumption by MDM, whereas VX-745 and PFK-15 actually increased both glucose consumption and lactate release (Fig. S3, F and G).
Collectively, these data point at a key role of Akt in the glycolytic reprogramming of MDM but do not support a significant role for mTORC1, HIF-1α or PFKFB3.
The influence of 2-DG, Akt-I-1/2, and glucose starvation on pro-inflammatory cytokine expression by MDM
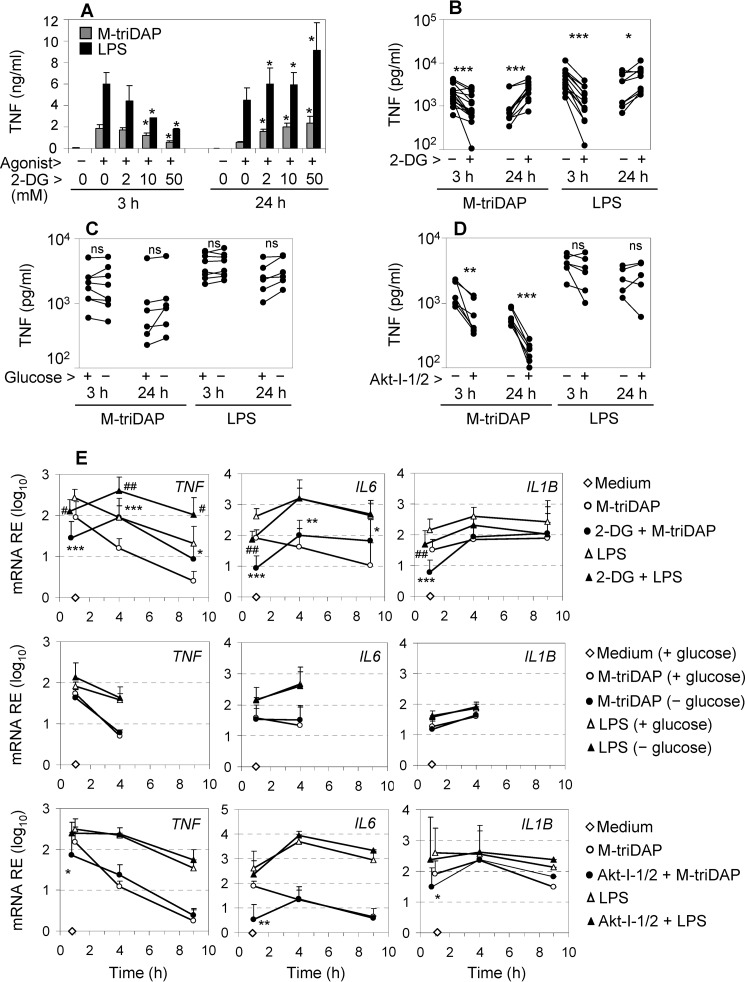
We then analyzed how inhibition of glycolysis by 2-DG, Akt-I-1/2, or glucose starvation affects cytokine responses induced by NOD1 and TLR4 agonists in MDM. 2-DG is widely used as an inhibitor of glycolysis, and several studies have suggested that 2-DG down-regulates LPS-induced pro-inflammatory cytokine expression and production (5, 45, 46). Unexpectedly, 2-DG had a dual effect on TNF production. At an early time point (3 h), 2-DG dose-dependently reduced levels of TNF in supernatants of M-triDAP– or LPS-stimulated MDM (Fig. 4, A and B). However, upon 24-h stimulation, levels of TNF were dose-dependently increased by 2-DG (Fig. 4, A and B), suggesting that 2-DG enhanced TNF synthesis between 3 and 24 h after PRR stimulation. 2-DG also inhibited production of IL-6 at either early or late time points (Fig. S4A) as well as production of pro-IL-1β (Fig. S4, C and D). Strikingly, the alterations of cytokine production were largely absent when glycolysis was inhibited by glucose starvation instead of 2-DG (Fig. 4C; Fig. S4, B–D), even though both approaches inhibited glycolysis with similar efficiency (Fig. 3, A–D). Finally, Akt-I-1/2 and triciribine suppressed M-triDAP–induced TNF production at both early and late time points, but did not affect LPS-induced TNF production (Fig. 4D; Fig. S4E).
Figure 4.
Effects of 2-DG, glucose-free medium, and Akt-I-1/2 on cytokine expression by M-triDAP- or LPS-stimulated MDM. A, MDM were pre-treated for 30 min with the indicated concentrations of 2-DG in ordinary medium and stimulated for 3 or 24 h with M-triDAP or LPS, after which TNF levels in supernatants were measured. Mean ± S.D., n = 3. *, p < 0.05 compared with cells treated with the same agonist in the absence of 2-DG (paired t test). B–D, MDM were pre-cultured for 30 min with 50 mm 2-DG in ordinary medium (B), in glucose-free medium (C), or with Akt-I-1/2 (10 μm), then stimulated with M-triDAP or LPS; TNF levels in supernatants were measured at 3 and 24 h (5 to 16 donors per data point). *, p < 0.05; **, p < 0.01; ***, p < 0.001 by paired t test. n.s., nonsignificant. E, effects of 2-DG (50 mm; upper row), glucose-free medium (middle row), and Akt-I-1/2 (10 μm; lower row) on TNF, IL6, and IL1B mRNA expression by MDM stimulated with M-triDAP or LPS. Mean ± S.D. of 4 to 10 donors per data point. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with MDM stimulated with M-triDAP without an inhibitor; #, p < 0.05; ##, p < 0.01 compared with MDM stimulated with LPS without an inhibitor at the same time point.
These alterations of cytokine expression were reproduced at the level of mRNA expression (Fig. 4E). Thus, 2-DG down-regulated TNF mRNA in MDM at an early time point (1 h), but up-regulated it at later time points (4 and 9 h) of stimulation with M-triDAP or LPS (Fig. 4E, upper left graph). 2-DG had a similar dual effect on M-triDAP–induced IL-6 mRNA expression, whereas LPS-induced IL-6 mRNA was suppressed by 2-DG at 1 h and not enhanced at later time points (Fig. 4E, upper middle graph). 2-DG down-modulated M-triDAP– or LPS-induced IL1B mRNA at 1 h but not at later time points (Fig. 4E, upper right).
Lack of glucose in the medium did not affect TNF, IL6, or IL1B mRNA expression triggered by M-triDAP or LPS (Fig. 4E, middle row). Finally, Akt-I-1/2 down-modulated M-triDAP–induced TNF, IL6, and IL1B mRNA expression at 1 h of stimulation, but not at later time points, and also did not affect LPS-induced cytokine mRNA expression (Fig. 4E, lower row), which indicated specific involvement of Akt in cytokine expression downstream of NOD1.
2-DG modulates pro-inflammatory cytokine expression by inducing endoplasmic reticulum stress
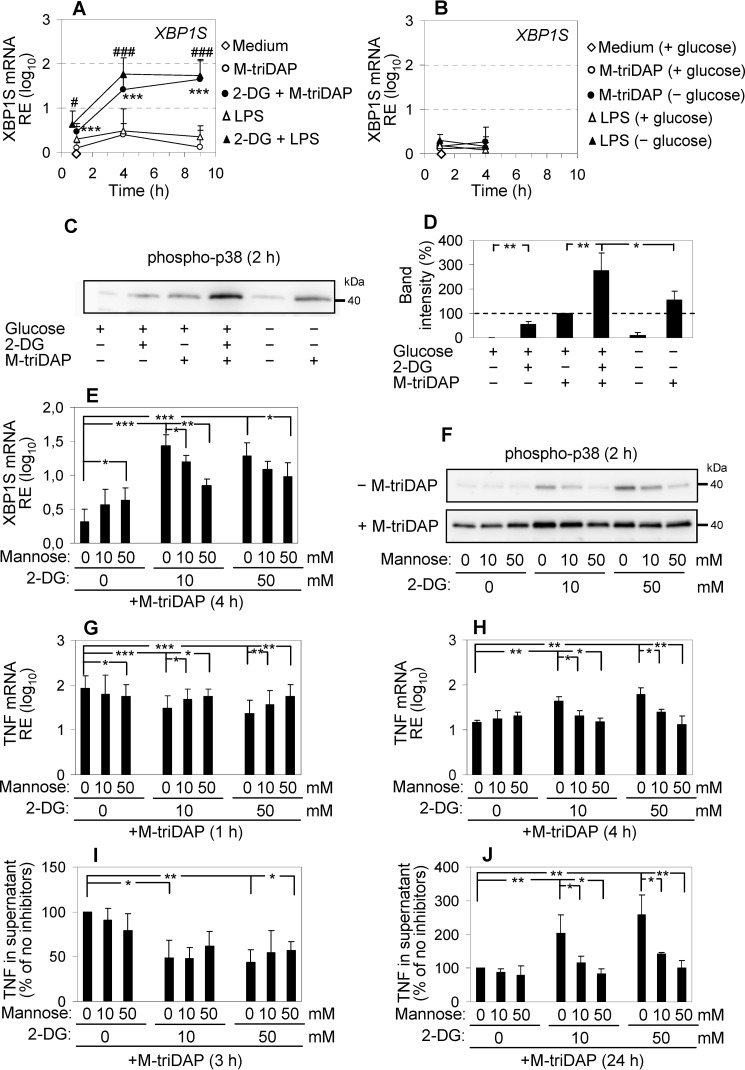
Akt kinases are involved in many aspects of macrophage activation and differentiation (47); therefore, effects of Akt inhibitors on cytokine expression are difficult to relate specifically to inhibition of glycolysis. However, the discrepancy between the effects of 2-DG and glucose starvation required an explanation. Chemically, 2-deoxy-d-glucose is identical to 2-deoxy-d-mannose and can therefore antagonize not only d-glucose but also d-mannose (48). By the latter activity, 2-DG can interfere with N-linked protein glycosylation in the endoplasmic reticulum (ER), which requires mannose. This, in turn, can disturb protein folding in ER, resulting in unfolded protein response (UPR) (48). UPR is a complex set of cellular responses which, on the one hand, aim to relieve overload of ER by incorrectly folded proteins (this includes degradation of most mRNA species and suppression of protein synthesis), and on the other hand, activate inflammatory pathways to inform the immune system about the presence of stressed cells (49). A hallmark of UPR is atypical splicing of XBP1 mRNA, i.e. the conversion of unspliced XBP1 mRNA (XBP1U) to its translationally competent spliced counterpart (XBP1S) (50). 2-DG alone and especially in combination with PRR agonists caused a fast (within 1 h) elevation of XBP1S mRNA, whereas PRR agonists alone did not induce XBP1 splicing (Fig. 5A; Fig. S4F). XBP1U mRNA as well as other genes involved in UPR, such as HSPA5/BiP and EIF2AK3/PERK, were up-regulated by 2-DG ± PRR agonists with a slower kinetics (Fig. S4F). When MDM were activated in glucose-free medium instead of 2-DG, XBP1 splicing was not induced (Fig. 5B). XBP1 protein can function as a positive regulator of pro-inflammatory cytokine expression (51). 2-DG also induced p38 MAPK phosphorylation and augmented M-triDAP–induced p38 phosphorylation, another pro-inflammatory aspect of UPR (Fig. 5, C and D). In glucose-free conditions, p38 phosphorylation was not significantly increased as compared with glucose-replete conditions (Fig. 5, C and D).
Figure 5.
2-DG modulates cytokine production by MDM by inducing UPR. A, levels of XBP1S mRNA in MDM stimulated with M-triDAP or LPS in ordinary medium without or with 2-DG (50 mm). Mean ± S.D., n = 8. ***, p < 0.001 compared with cells treated with M-triDAP alone; #, p < 0.05; ###, p < 0.001 compared with cells treated with LPS alone at the same time point. B, levels of XBP1S mRNA upon M-triDAP or LPS stimulation in ordinary or glucose-free medium. Mean ± S.D., n = 4. C and D, cells were pre-treated with 2-DG (50 mm) or glucose-free medium for 30 min, then cultured with or without M-triDAP for 2 h, after which p38 phosphorylation was assessed (C, a representative experiment, D, densitometry; mean ± S.D. of 5 donors). E–J, MDM were pre-treated for 30 min with 2-DG and d-mannose at indicated concentrations in ordinary medium, stimulated with M-triDAP for indicated time periods, after which levels of XPB1S mRNA (E), phospho-p38 (F), TNF mRNA (G and H) and TNF in supernatant (I and J) were assessed. Three experiments per data point, p values by paired t test. In I and J, results were normalized to cells treated with M-triDAP without 2-DG or mannose.
In cancer cells, the UPR induced by 2-DG is known to be alleviated by d-mannose (48). Indeed, d-mannose dose-dependently diminished the up-regulation of XBP1S mRNA (Fig. 5E) and phosphorylation of p38 induced by 2-DG in MDM (Fig. 5F). Interestingly, d-mannose at a high concentration by itself induced a minor up-regulation of XBP1S (Fig. 5E). Importantly, d-mannose cancelled or diminished most of the observed alterations of cytokine expression caused by 2-DG, including early down-regulation of TNF, IL6, and IL1B mRNA expression, early down-regulation of TNF secretion and pro-IL-1β expression, as well as late up-regulation of TNF mRNA expression and TNF secretion (Fig. 5, G–J; Fig. S5, A–G). However, it should be noted that the early 2-DG–induced suppression of TNF, IL-6, and IL-1β protein production was restored by d-mannose less efficiently than corresponding mRNA expression. In addition, the late augmenting effect of 2-DG on TNF expression was cancelled by p38 inhibition (Fig. S5H).
The observed effects of d-mannose indicated that 2-DG influenced cytokine expression through induction of UPR rather than inhibition of glycolysis. To verify that d-mannose did not cancel the effect of 2-DG on glycolysis, we measured ECAR of M-triDAP–activated MDM in the presence of different concentrations of d-mannose and/or 2-DG. d-mannose supported glycolysis (albeit less efficiently than d-glucose), but did not overcome the inhibitory effect of 2-DG on glycolysis (Fig. S6A).
2-DG has also been suggested to activate inflammasome and trigger the release of IL-1β (52), which could auto- or paracrinely up-regulate TNF expression at late stages of activation (3 h and later). However, we did not detect any significant IL-1β release by MDM in the presence of 2-DG during the first 3 h of stimulation (Fig. S6B), and IL-1RA did not interfere with the enhancing effect of 2-DG on M-triDAP–induced production of TNF (Fig. S6C).
Effects of glycolysis inhibitors on cytokine production by MDDC
To analyze the links between glycolysis and cytokine production in additional cell types, we studied glycolytic reprogramming of human MDDC obtained from the same donors as MDM. In basal conditions, MDDC showed lower level of glycolysis than MDM (Fig. 6, A–C), but comparable OXPHOS (Fig. 6D). MDDC only slightly increased their ECAR upon M-triDAP or LPS treatment, (Fig. 6, E and F) along with a very modest increase of glucose consumption (Fig. 6G) and inconsistent elevation of lactate release (Fig. 6H). Oligomycin-induced only a short spike of ECAR with a rapid return to the baseline (Fig. 6E), indicating low glycolytic reserve in MDDC.
Figure 6.
Metabolic reprogramming and cytokine production by human MDDC activated with M-triDAP or LPS. A–D, comparisons of glucose and energy metabolism in paired cultures of unstimulated MDDC and MDM: basal ECAR (A), 24-hour glucose consumption (B), 24-hour lactate release (C), basal OCR (D). 6 to 9 individual donors per data point. E, kinetics of ECAR in MDDC after injection of medium, M-triDAP (10 μg/ml), LPS (100 ng/ml), or oligomycin (2 μm). Same donor as in Fig. 2A. F–H, areas under ECAR curves (F), 24-hour glucose consumption (G), and lactate release (H) by MDDC treated with medium alone, M-triDAP, or LPS. MDDC were generated from the same donors as in Fig. 2, B, D, and E; 6 to 9 donors per data point. #, p = 0.07; *, p < 0.05; **, p < 0.01; ***, p < 0.001 by paired t test. In F–H, asterisks relate to comparisons with medium-treated MDDC. I, levels of TNF in MDDC supernatants after 3 and 24 h of culture with medium alone, M-triDAP (10 μg/ml), or LPS (100 ng/ml). 18 donors (same as in Fig. 1A). Horizontal bars within each group are mean values; dashed line, lower limit of detection by ELISA. J, effects of 2-DG (50 mm) or glucose-free medium on LPS-induced TNF secretion by MDDC (3 to 10 donors per data point). K and L, MDDC were pre-treated with indicated concentrations of 2-DG and d-mannose, then stimulated with LPS for 3 or 24 h. Levels of TNF in supernatants are expressed in relation to levels in the absence of 2-DG and mannose.
We demonstrated earlier that NOD1 and NOD2 agonists induce very low cytokine production by MDDC, whereas LPS triggers comparable secretion of TNF and IL-6 by MDDC and MDM (53). This was confirmed in the present study (compare Fig. 6I and Fig. 1A illustrating TNF production by MDDC and MDM, respectively, generated from the same individuals). Thus, MDDC and MDM are able to produce similar amounts of TNF in response to LPS, despite profound differences in glycolysis. As in MDM, glucose starvation did not significantly affect LPS-induced production of TNF, IL-6, and IL-12 by MDDC (Fig. 6J; Fig. S7, A and B). By contrast, 2-DG strongly suppressed LPS-induced TNF, IL-6, and IL-12 production by MDDC as well as compromised MDDC viability, and these effects of 2-DG were partially or fully reversed by d-mannose (Fig. 6, J–L; Fig. S7, C–E). 2-DG up-regulated UPR markers in MDDC to the same extent as in MDM (Fig. S7F).
2-DG and glucose starvation have similar effects in hypoxic MDM
The experiments described so far were performed in normoxic conditions. They showed that lack of glycolysis per se has relatively minor impact on cytokine production. However, glycolysis becomes a vital energy source in hypoxic conditions, which exist in inflammatory lesions. To mimic hypoxia, we treated MDM with oligomycin. Combinations of oligomycin with 2-DG or with glucose starvation completely suppressed M-triDAP– and LPS-induced TNF and IL-6 production by MDM (Fig. S8, A and B). At 3 h, when cytokine production was measured, most cells were still viable (Fig. S8C). At 24 h, oligomycin, 2-DG, or glucose starvation each had relatively minor impact on cell viability; however, combinations of oligomycin with glycolysis inhibitors caused massive cell death (Fig. S8, C and D).
Discussion
Metabolic reprogramming of innate immune cells is considered one of the driving forces of inflammation (1). Manipulation of metabolic pathways or metabolism-controlling signaling pathways is being experimentally tested as a means to treat or prevent life-threatening inflammatory conditions such as sepsis and lethal endotoxemia, with varying degrees of success (54–60). Much of the mechanistic data about pro-inflammatory metabolic reprogramming have been generated using LPS-stimulated macrophages or dendritic cells (1, 3, 5, 6, 46, 61). We have characterized, for the first time, metabolic reprogramming induced by agonists of NOD1 and NOD2 receptors in macrophages and dendritic cells.
We found that effects of a NOD1 agonist (M-triDAP) and a TLR4 agonist (LPS) on glycolysis in human MDM are largely similar. Both agonists trigger an early and robust augmentation of glycolysis along with a minor reduction of oxygen consumption. The effects of both agonists on glycolysis depend on Akt, but are independent of mTORC1 or de novo expression of glycolytic enzymes. It can be concluded from this and other studies (62) that early glycolytic reprogramming is probably a stereotypic response of macrophages to PRR stimulation. A striking metabolic difference between M-triDAP– and LPS-activated MDM is the lack of ACOD1 protein up-regulation upon M-triDAP treatment, which is in sharp contrast to the strong ACOD1 induction by LPS (Refs. 3, 26; Fig. 2, I and J). ACOD1 is the only enzyme capable of generating itaconate (27), a metabolite that potently inhibits growth of bacteria expressing citrate lyase (26). It will be important to compare bactericidal activity of M-triDAP– and LPS-activated macrophages against such microbes, and also to analyze ACOD1 induction by a wider range of microbial stimuli.
MDDC showed a markedly reduced glycolytic reserve compared with MDM and virtually no elevation of ECAR upon M-triDAP or LPS treatment. This is in some contrast to results by Everts et al. (5), who observed a significant increase of ECAR in LPS-stimulated mouse bone-marrow–derived DC (BMDC). In fact, ECAR responses of BMDC in the work by Everts et al. (5) resembled responses of MDM in our study. Therefore, murine data might not be directly applicable to human cells.
The links between glycolysis and pro-inflammatory cytokine production have been extensively investigated. Some glycolytic enzymes have been shown to function as direct transcriptional and posttranscriptional co-regulators of cytokine mRNA expression. For example, activated dimeric PKM2 can translocate to the nucleus and enhance LPS-induced IL1B and IL6 mRNA transcription (54, 63, 64), whereas GAPDH in its inactive state (under low-glucose conditions) can bind TNF mRNA and negatively regulate TNF translation (65). More generally, glycolysis is thought to supply ATP and carbon backbone for anabolic processes ongoing in activated immune cells, including cytokine synthesis. Many studies have shown that blockade of glycolysis is associated with down-regulated pro-inflammatory cytokine production by macrophages and dendritic cells (5, 15, 45, 46, 54, 66). However, most researchers inhibited glycolysis using 2-DG. Descriptions of the inhibitory effects of 2-DG on cytokine production are in fact contradictory. For example, Na et al. (45) reported that in murine macrophages, 2-DG reduces LPS-triggered TNF, IL6, and IL1B mRNA expression at early (1–4 h) stages of activation. According to Everts et al. (5), 2-DG does not influence pro-inflammatory cytokine mRNA expression induced by a 3-h treatment of murine BMDC with LPS, but does inhibit cytokine translation. Also, although most studies report negative effects of 2-DG on cytokine production (5, 45, 46, 54, 66), some studies show an enhancing effect (67) or no effect of 2-DG at all (68). As shown here, effects of 2-DG can actually depend on type of cell and cytokine measured as well as on sampling time.
In the present study, we found that cells with very different basal and induced levels of glycolysis (MDM and MDDC) are able to produce roughly similar amounts of TNF upon LPS treatment. Second, glycolysis in MDM was efficiently inhibited by three different approaches (glucose-free medium, 2-DG, and an Akt inhibitor), yet each of these conditions differently modulated TNF, IL6, and IL1B expression induced by PRR agonists, indicating that these modulatory effects (if present) were not because of inhibition of glycolysis. For example, Akt-I-1/2 suppressed glycolytic reprogramming induced in MDM by either M-triDAP or LPS; however, only M-triDAP–induced and not LPS-induced cytokine responses were inhibited. (To our knowledge, this is the first report about the role of Akt in cytokine production downstream of NOD1.) Next, we found that 2-DG applied in normoxic conditions modulates pro-inflammatory cytokine production by inducing a UPR rather than by inhibiting glycolysis. The ability of 2-DG to cause UPR through interference with N-linked protein glycosylation is well-known in molecular oncology and has been tested as a means to enhance cytotoxicity of traditional anti-cancer agents (69). We found that 2-DG augmented the expression of several UPR markers in MDM and MDDC, whereas d-mannose, by antagonizing the ability of 2-DG to induce UPR, cancelled or alleviated the effects of 2-DG on cytokine expression and production by MDDC and MDM as well as on MDDC viability. When glycolysis was inhibited by glucose starvation instead of 2-DG, no or very mild up-regulation of UPR markers occurred, and no or very mild changes of cytokine production by MDM and MDDC were observed. It should be noted that glucose starvation can also induce a mild ER stress, because glucose too is required for N-linked protein glycosylation (48).
The bidirectional effects of 2-DG on the expression of pro-inflammatory cytokines by MDM can be ascribed to different aspects of UPR. Thus, a group of processes within UPR aims to relieve the overload of ER by incorrectly folded proteins. This includes degradation of many mRNA species by IRE-1α (an ER stress sensor) and suppression of protein translation through phosphorylation of translation initiation factor EIF-2α by PERK (another ER stress sensor) (49, 70). These processes probably underlie the early inhibitory effect of 2-DG on cytokine mRNA expression and translation observed within the first 1–3 h after addition of 2-DG and PRR agonists. The late enhancing effect of 2-DG on TNF expression observed in MDM at 4 h and later can be explained by the pro-inflammatory aspects of UPR. Ongoing UPR is known to sensitize cells to PRR agonists (71). In particular, XBP1 protein translated from the spliced XBP1S mRNA can function as a positive regulator of cytokine mRNA transcription (51, 72). Similarly, p38 MAPK activated in 2-DG–treated cells may increase cytokine mRNA stability and translation (73, 74). As a proof of this, a p38 inhibitor (VX-745) abrogated the late enhancing effect of 2-DG on M-triDAP–induced TNF expression and secretion.
As already noted in “Results,” the early 2-DG–mediated suppression of cytokine protein production was restored by d-mannose less efficiently than corresponding mRNA expression (compare e.g. Fig. 5, G and I, for TNF mRNA and TNF protein, respectively). This indicates that 2-DG might modulate cytokine translation through mechanisms independent of competition with mannose. This effect on translation could not be explained by 2-DG–mediated inhibition of mTOR, because two specific mTOR inhibitors, rapamycin and Ku 0063794, did not inhibit TNF production by M-triDAP– or LPS-stimulated MDM3. 2-DG might affect cytokine translation, for example, by promoting of GAPDH binding to cytokine mRNA, as already described (65).
Taken together, these data indicate that effects of 2-DG on cytokine production in normoxic conditions should be interpreted with caution. At least within the time frame of our experiments (24 h), cells can obviously compensate for the lack of glycolysis, probably by deploying other metabolic pathways such as fatty acid oxidation or glutaminolysis, which feed into Krebs cycle and OXPHOS; and if no UPR is induced in parallel, cytokine production triggered through PRRs such as NOD1 and TLR4 remains largely unaffected. However, when OXPHOS is inhibited in hypoxic or hypoxia-like conditions, glycolysis becomes vital both for cytokine production and cell survival; hence 2-DG or glucose starvation combined with OXPHOS inhibition have similarly deleterious effects on MDM functions.
Experimental procedures
Reagents
M-triDAP and MDP were purchased from Invivogen (San Diego, CA) and reconstituted in endotoxin-free water supplied by the manufacturer. GMDP was kindly provided by E. A. Makarov (RAM Ltd., Moscow, Russia). LPS from Escherichia coli O111:B4 was from Merck Millipore (Billerica, MA). Recombinant human (rh) GM-CSF, M-CSF, and IL-4 were from Miltenyi Biotec (Bergisch Gladbach, Germany), rhIL-1RA from R&D Systems (Minneapolis, MN). d-mannose and cobalt (II) chloride were from Sigma. Enzyme inhibitors used in the study are listed in Table S1.
Culture and stimulation of MDDC and MDM
Studies utilizing human cells were approved by the local Ethical Committee of the Institute of Immunology (protocol No. 10/2017) and abided by the Declaration of Helsinki principles. To generate human immature MDDC, blood monocytes from anonymous donors were cultured in RPMI 1640 (Thermo Fisher Scientific) supplemented with 2 mm l-glutamine (Thermo Fisher), 10% fetal calf serum (PAA, Pasching, Austria), 80 ng/ml rhGM-CSF and 50 ng/ml rhIL-4 for 6 days, as described previously (53, 75). MDM were generated similarly, except that rhIL-4 was omitted (76). Whenever MDDC were examined, an MDM culture was set up from the same donor in parallel. When specifically noted, MDM were generated by culturing monocytes with 50 ng/ml rhM-CSF instead of rhGM-CSF). By flow cytometry, the phenotype of MDM was CD11c+CD14+CD206+, the phenotype of MDDC was CD11c+CD14−CD206−. The purity of cells with regard to these phenotypes was >90%.
On day 6, cells were replated in 96-well or 24-well plates without cytokines and stimulated with M-triDAP (10 μg/ml), MDP (1 μg/ml), or LPS (0.1 μg/ml) or left without stimulation. Working concentrations of the PRR agonists had been determined earlier (53). Enzyme inhibitors were added 30 min prior to PRR agonists. Glucose-free culture medium was prepared using glucose-free RPMI 1640 (Thermo Fisher), 10% fetal calf serum, and 2 mm l-glutamine and exchanged for ordinary culture medium 30 min prior to PRR stimulation. Glucose concentration in the glucose-free culture medium was <0.2 mm, as compared with 10.78 ± 0.41 mm in the ordinary medium. Supernatants and cells were harvested at indicated time points after addition of PRR agonists.
ELISA
Levels of human TNF in cell culture supernatants were determined by sandwich ELISA using reagent kits from Cytokine (St. Petersburg, Russia). Levels of human IL-1β, IL6, and IL-12p70 as well as mouse TNF and IL-6 were determined by kits from Thermo Fisher Scientific/eBioscience.
Determination of glucose and lactate
MDM and MDDC were seeded in 24-well plates at 250,000 cells per 500 μl (5*10−4 liter) complete culture medium without cytokines, and cultured with or without agonists for 24 h, after which supernatants were collected. Wells containing complete culture medium without cells served as a reference. Levels of glucose in the supernatants (mmol/liter) were measured using Synchron CX5 Pro (Beckman Coulter, Brea, CA). Levels of lactate were determined colorimetrically using a reagent kit from BioVision (Milpitas, CA). Glucose consumption and lactate release per cell were calculated as Cmedium − Csupernatant * 5*10−4/250,000.
Real-time measurements of cell metabolism
ECAR and OCR were measured using Seahorse XFe96 Analyzer (Agilent Technologies, Santa Clara, CA). ECAR and OCR characterize glycolysis and mitochondrial respiration, respectively. Cells were plated in complete culture medium in poly-d-lysine–coated Seahorse XF96 microplates (Agilent) at 16,000 cells/well and cultured overnight. Next morning, medium was exchanged to XF base medium supplemented with 2 mm l-glutamine, 10% fetal calf serum, and 10 mm d-glucose (Sigma), unless otherwise indicated. ECAR and OCR were measured every 9 min. After 3 basal measurements, stimuli (M-triDAP, LPS, oligomycin, or medium) were injected and a further 22 measurements were done. Enzyme inhibitors were injected after the basal measurements, followed by three to six additional measurements before the injection of stimuli. In some experiments, cells were pre-treated for 24 h with M-triDAP or LPS, after which ECAR and OCR were measured. Results were normalized by relative cellular protein content. Two types of parameters were calculated: (i) basal ECAR and OCR (a mean of the two measurements preceding addition of stimuli or inhibitors) and (ii) areas under time-response curves (AUC) after addition of stimuli, taking the last measurement preceding the addition of stimuli as the baseline. Mitochondrial stress test was performed as described (7) by measuring OCR upon sequential injections of 1 μm oligomycin, 2 μm carbonyl cyanide-4-(trifluoromethoxy)-phenylhydrazone (FCCP, Sigma), and 500 nm each of antimycin A and rotenone (both Sigma).
RT-PCR
MDM and MDDC were harvested after 1, 4, 9, or 24 h of stimulation with M-triDAP or LPS. Total RNA was extracted using TRI Reagent (Sigma). 0.5 μg total RNA was reverse transcribed using RevertAid Reverse Transcription Kit (Fermentas, Vilnius, Lithuania). Amplifications were done in a 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA) using a reagent mix containing SYBR Green (Evrogen, Moscow, Russia). Primer pairs spanning at least one intron or an exon-intron junction were designed using Primer-BLAST (Table S2). Cycling conditions were as follows: 95 °C (5 min), then 40 cycles of 95 °C (15 s) and 60 °C (45 s, detection step). Melting curves were analyzed after each amplification to confirm specificity of signal. Relative mRNA expression was calculated by the 2−ΔΔCt method using unstimulated MDM from the given donor as the reference sample and GAPDH expression for normalization. GAPDH was chosen because its expression was least variable across cell types and stimulation conditions (in six independent kinetic experiments, each employing paired MDM/MDDC cultures stimulated with M-triDAP and LPS and harvested at four time points, intra-experimental coefficient of variation of threshold cycle (Ct) for GAPDH was 2.18 ± 0.65% (mean ± S.D.); for comparison, same parameter for ACTB equaled 3.67 ± 0.6%). Relative mRNA expression of GAPDH itself was calculated by the 2−ΔCt formula without normalization. Differences in gene expression between groups or points were considered biologically significant if they met two criteria: (i) ratio of group or point means >2 or <0.5 and (ii) statistical significance with p < 0.05.
Western blotting
The procedure was as described in Ref. 53. Briefly, cells were washed with PBS and lysed in an ice-cold buffer containing 150 mm NaCl, 50 mm Tris, pH 8.0, 1% Triton X-100, and a mixture of protease and phosphatase inhibitors (MSSafe, Sigma). Proteins were resolved by SDS-PAGE using TGX Any kDa ready-made gels (Bio-Rad, Hercules, CA) and blotted onto polyvinylidene difluoride membranes in Bjerrum Schafer-Nielsen buffer. Membranes were blocked with 5% BSA or 5% nonfat dry milk (as recommended for specific Abs) and probed with primary antibodies overnight at 4 °C. Rabbit polyclonal Abs against human HIF-1α, ACOD1/IRG1, Akt, phospho-Akt (pT308), phospho-Akt (pS473), phospho-p70-S6K (pT389), phospho-PRAS40 (pT246), phospho-p38 (pT180/pY182) were purchased from Cell Signaling Technology (Danvers, MA). Goat polyclonal Ab against human IL-1β was from R&D Systems (Minneapolis, MN). α-Tubulin was detected with the DM1A mouse mAb (Merck Millipore) and used as a loading control. Primary Abs were detected by peroxidase-labeled secondary Abs (Jackson ImmunoResearch, West Grove, PA). The staining was developed using Immobilon Western substrate (Merck Millipore) and detected by an AI600 imager (Amersham Biosciences). Densitometry was done using ImageJ freeware (RRID:SCR_003070).
Animal experiments
Experiments were approved by the local Ethical Committee of the Institute of Immunology (protocol No. 1/18). C57BL/6 male mice weighing 18–20 g were injected subcutaneously with 100 μg GMDP in 200 μl endotoxin-free PBS. Control mice received an equal volume of PBS. 2 h later, samples of peritoneal lavage fluid were obtained. PM were isolated as described (77), with minor modifications. Peritoneal cells were washed with RPMI, adjusted to 2*106/ml in RPMI supplemented with penicillin-streptomycin, 2 mm l-glutamine, and 10% fetal calf serum, seeded in 24-well plates at 106 cells/well, and allowed to adhere for 2 h. Wells were washed three times with warm PBS and filled with 500 μl of the above medium. By flow cytometry, F4/80+CD11b+ macrophages constituted >85% of adherent cells. PM monolayers were incubated for 24 h, after which levels of glucose, lactate, TNF, and IL-6 in the supernatants were measured as described above and expressed as the amount of analyte released or consumed per 1 μg cellular protein.
Statistics
Data were analyzed by GraphPad Instat (GraphPad Software, San Diego, CA). Paired measurements were compared by paired t test.
Author contributions
N. E. M., A. S. B., Y. A. D., P. V. M., V. V. M., M. V. M., A. M. N., and M. V. P. formal analysis; N. E. M., A. S. B., Y. A. D., P. V. M., L. S. B., V. V. M., M. V. M., V. S. S., A. M. N., G. Z. C., and M. V. P. investigation; N. E. M., A. S. B., Y. A. D., P. V. M., L. S. B., V. V. M., M. V. M., V. S. S., A. M. N., G. Z. C., and M. V. P. methodology; N. E. M., A. S. B., B. V. P., and M. V. P. writing-review and editing; B. V. P. and M. V. P. conceptualization; M. V. P. supervision; M. V. P. funding acquisition; M. V. P. writing-original draft.
Supplementary Material
Acknowledgments
We are grateful to Prof. Boris Zhivotovsky and Vladimir Gogvadze (Moscow State University) for methodological help and valuable discussions, and to Dr. Irina Surina and Elena Drozdova (Institute of Immunology, Moscow) for help with determination of glucose.
This work was supported by Russian Science Foundation Grant 16-15-10314 (to N. E. M., A. S. B., Y. A. D., L. S. B., V. V. M., M. V. M., V. S. S., A. M. N., B. V. P., and M. V. P.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8 and Tables S1 and S2.
N. E. Murugina et al., unpublished observations.
- OXPHOS
- oxidative phosphorylation
- AUC
- area under curve
- BMDC
- bone-marrow–derived DC
- ECAR
- extracellular medium acidification rate
- ER
- endoplasmic reticulum
- GM-CSF
- granulocyte-macrophage colony-stimulating factor
- GMDP
- N-acetylglucosaminyl-N-acetylmuramyl-l-alanyl-d-isoglutamine (glucosaminyl muramyl dipeptide)
- HK
- hexokinase II
- LPS
- lipopolysaccharide
- M-triDAP
- N-acetylmuramyl-l-alanyl-d-isoglutamyl-meso-diaminopimelic acid
- MDDC
- monocyte-derived dendritic cells
- MDM
- monocyte-derived macrophages
- MDP
- N-acetylmuramyl-l-alanyl-d-isoglutamine (muramyl dipeptide)
- OCR
- oxygen consumption rate
- PM
- peritoneal macrophages
- PRR
- pattern recognition receptor
- rh
- recombinant human
- TNF
- tumor necrosis factor
- UPR
- unfolded protein response.
References
- 1. O'Neill L. A., and Pearce E. J. (2016) Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213, 15–23 10.1084/jem.20151570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buck M. D., O'Sullivan D., and Pearce E. L. (2015) T cell metabolism drives immunity. J. Exp. Med. 212, 1345–1360 10.1084/jem.20151159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jha A. K., Huang S. C., Sergushichev A., Lampropoulou V., Ivanova Y., Loginicheva E., Chmielewski K., Stewart K. M., Ashall J., Everts B., Pearce E. J., Driggers E. M., and Artyomov M. N. (2015) Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 10.1016/j.immuni.2015.02.005 [DOI] [PubMed] [Google Scholar]
- 4. Mills E. L., Kelly B., Logan A., Costa A. S. H., Varma M., Bryant C. E., Tourlomousis P., Däbritz J. H. M., Gottlieb E., Latorre I., Corr S. C., McManus G., Ryan D., Jacobs H. T., Szibor M., et al. (2016) Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167, 457–470.e13 10.1016/j.cell.2016.08.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Everts B., Amiel E., Huang S. C., Smith A. M., Chang C. H., Lam W. Y., Redmann V., Freitas T. C., Blagih J., van der Windt G. J., Artyomov M. N., Jones R. G., Pearce E. L., and Pearce E. J. (2014) TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKϵ supports the anabolic demands of dendritic cell activation. Nat. Immunol. 15, 323–332 10.1038/ni.2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krawczyk C. M., Holowka T., Sun J., Blagih J., Amiel E., DeBerardinis R. J., Cross J. R., Jung E., Thompson C. B., Jones R. G., and Pearce E. J. (2010) Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 115, 4742–4749 10.1182/blood-2009-10-249540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Everts B., Amiel E., van der Windt G. J., Freitas T. C., Chott R., Yarasheski K. E., Pearce E. L., and Pearce E. J. (2012) Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood 120, 1422–1431 10.1182/blood-2012-03-419747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang S. C., Smith A. M., Everts B., Colonna M., Pearce E. L., Schilling J. D., and Pearce E. J. (2016) Metabolic reprogramming mediated by the mTORC2-IRF4 signaling axis is essential for macrophage alternative activation. Immunity 45, 817–830 10.1016/j.immuni.2016.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang S. C., Everts B., Ivanova Y., O'Sullivan D., Nascimento M., Smith A. M., Beatty W., Love-Gregory L., Lam W. Y., O'Neill C. M., Yan C., Du H., Abumrad N. A., Urban J. F. Jr., Artyomov M. N., Pearce E. L., and Pearce E. J. (2014) Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 15, 846–855 10.1038/ni.2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van den Bossche J., O'Neill L. A., and Menon D. (2017) Macrophage immunometabolism: Where are we (going)? Trends Immunol. 38, 395–406 10.1016/j.it.2017.03.001 [DOI] [PubMed] [Google Scholar]
- 11. Stienstra R., Netea-Maier R. T., Riksen N. P., Joosten L. A. B., and Netea M. G. (2017) Specific and complex reprogramming of cellular metabolism in myeloid cells during innate immune responses. Cell Metab. 26, 142–156 10.1016/j.cmet.2017.06.001 [DOI] [PubMed] [Google Scholar]
- 12. Ip W. K. E., Hoshi N., Shouval D. S., Snapper S., and Medzhitov R. (2017) Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 356, 513–519 10.1126/science.aal3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diskin C., and Pålsson-McDermott E. M. (2018) Metabolic modulation in macrophage effector function. Front. Immunol. 9, 270 10.3389/fimmu.2018.00270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feingold K. R., Shigenaga J. K., Kazemi M. R., McDonald C. M., Patzek S. M., Cross A. S., Moser A., and Grunfeld C. (2012) Mechanisms of triglyceride accumulation in activated macrophages. J. Leukoc. Biol. 92, 829–839 10.1189/jlb.1111537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheng S. C., Quintin J., Cramer R. A., Shepardson K. M., Saeed S., Kumar V., Giamarellos-Bourboulis E. J., Martens J. H., Rao N. A., Aghajanirefah A., Manjeri G. R., Li Y., Ifrim D. C., Arts R. J., van der Veer B. M., et al. (2014) mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 10.1126/science.1250684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meiser J., Krämer L., Sapcariu S. C., Battello N., Ghelfi J., D'Herouel A. F., Skupin A., and Hiller K. (2016) Pro-inflammatory macrophages sustain pyruvate oxidation through pyruvate dehydrogenase for the synthesis of itaconate and to enable cytokine expression. J. Biol. Chem. 291, 3932–3946 10.1074/jbc.M115.676817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lampropoulou V., Sergushichev A., Bambouskova M., Nair S., Vincent E. E., Loginicheva E., Cervantes-Barragan L., Ma X., Huang S. C., Griss T., Weinheimer C. J., Khader S., Randolph G. J., Pearce E. J., Jones R. G., Diwan A., Diamond M. S., and Artyomov M. N. (2016) Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 24, 158–166 10.1016/j.cmet.2016.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Girardin S. E., Boneca I. G., Carneiro L. A., Antignac A., Jéhanno M., Viala J., Tedin K., Taha M. K., Labigne A., Zähringer U., Coyle A. J., DiStefano P. S., Bertin J., Sansonetti P. J., and Philpott D. J. (2003) Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300, 1584–1587 10.1126/science.1084677 [DOI] [PubMed] [Google Scholar]
- 19. Chamaillard M., Hashimoto M., Horie Y., Masumoto J., Qiu S., Saab L., Ogura Y., Kawasaki A., Fukase K., Kusumoto S., Valvano M. A., Foster S. J., Mak T. W., Nuñez G., and Inohara N. (2003) An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 4, 702–707 10.1038/ni945 [DOI] [PubMed] [Google Scholar]
- 20. Kager L., Pötschger U., and Bielack S. (2010) Review of mifamurtide in the treatment of patients with osteosarcoma. Ther. Clin. Risk Manag. 6, 279–286 10.2147/tcrm.s5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Girardin S. E., Travassos L. H., Hervé M., Blanot D., Boneca I. G., Philpott D. J., Sansonetti P. J., and Mengin-Lecreulx D. (2003) Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J. Biol. Chem. 278, 41702–41708 10.1074/jbc.M307198200 [DOI] [PubMed] [Google Scholar]
- 22. Dagil Y. A., Arbatsky N. P., Alkhazova B. I., L'vov V. L., Mazurov D. V., and Pashenkov M. V. (2016) The dual NOD1/NOD2 agonism of muropeptides containing a meso-diaminopimelic acid residue. PLoS One 11, e0160784 10.1371/journal.pone.0160784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meshcheryakova E., Makarov E., Philpott D., Andronova T., and Ivanov V. (2007) Evidence for correlation between the intensities of adjuvant effects and NOD2 activation by monomeric, dimeric and lipophylic derivatives of N-acetylglucosaminyl-N-acetylmuramyl peptides. Vaccine 25, 4515–4520 10.1016/j.vaccine.2007.04.006 [DOI] [PubMed] [Google Scholar]
- 24. Rogatzki M. J., Ferguson B. S., Goodwin M. L., and Gladden L. B. (2015) Lactate is always the end product of glycolysis. Front Neurosci. 9, 22 10.3389/fnins.2015.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hahn E. L., Halestrap A. P., and Gamelli R. L. (2000) Expression of the lactate transporter MCT1 in macrophages. Shock 13, 253–260 10.1097/00024382-200004000-00001 [DOI] [PubMed] [Google Scholar]
- 26. Michelucci A., Cordes T., Ghelfi J., Pailot A., Reiling N., Goldmann O., Binz T., Wegner A., Tallam A., Rausell A., Buttini M., Linster C. L., Medina E., Balling R., and Hiller K. (2013) Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. U.S.A. 110, 7820–7825 10.1073/pnas.1218599110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cordes T., Wallace M., Michelucci A., Divakaruni A. S., Sapcariu S. C., Sousa C., Koseki H., Cabrales P., Murphy A. N., Hiller K., and Metallo C. M. (2016) Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J. Biol. Chem. 291, 14274–14284 10.1074/jbc.M115.685792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mills E. L., Ryan D. G., Prag H. A., Dikovskaya D., Menon D., Zaslona Z., Jedrychowski M. P., Costa A. S. H., Higgins M., Hams E., Szpyt J., Runtsch M. C., King M. S., McGouran J. F., Fischer R., et al. (2018) Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117 10.1038/nature25986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. John S., Weiss J. N., and Ribalet B. (2011) Subcellular localization of hexokinases I and II directs the metabolic fate of glucose. PLoS One 6, e17674 10.1371/journal.pone.0017674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miyamoto S., Murphy A. N., and Brown J. H. (2008) Akt mediates mitochondrial protection in cardiomyocytes through phosphorylation of mitochondrial hexokinase-II. Cell Death Differ. 15, 521–529 10.1038/sj.cdd.4402285 [DOI] [PubMed] [Google Scholar]
- 31. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., and Cohen P. (2007) The selectivity of protein kinase inhibitors: A further update. Biochem. J. 408, 297–315 10.1042/BJ20070797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sancak Y., Thoreen C. C., Peterson T. R., Lindquist R. A., Kang S. A., Spooner E., Carr S. A., and Sabatini D. M. (2007) PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 25, 903–915 10.1016/j.molcel.2007.03.003 [DOI] [PubMed] [Google Scholar]
- 33. Wlodarski P., Kasprzycka M., Liu X., Marzec M., Robertson E. S., Slupianek A., and Wasik M. A. (2005) Activation of mammalian target of rapamycin in transformed B lymphocytes is nutrient dependent but independent of Akt, mitogen-activated protein kinase/extracellular signal-regulated kinase, insulin growth factor-I, and serum. Cancer Res. 65, 7800–7808 10.1158/0008-5472.CAN-04-4180 [DOI] [PubMed] [Google Scholar]
- 34. Tee A. R., Anjum R., and Blenis J. (2003) Inactivation of the tuberous sclerosis complex-1 and -2 gene products occurs by phosphoinositide 3-kinase/Akt-dependent and -independent phosphorylation of tuberin. J. Biol. Chem. 278, 37288–37296 10.1074/jbc.M303257200 [DOI] [PubMed] [Google Scholar]
- 35. Wang X., and Proud C. G. (1997) p70 S6 kinase is activated by sodium arsenite in adult rat cardiomyocytes: Roles for phosphatidylinositol 3-kinase and p38 MAP kinase. Biochem. Biophys. Res. Commun. 238, 207–212 10.1006/bbrc.1997.7273 [DOI] [PubMed] [Google Scholar]
- 36. Balendran A., Currie R., Armstrong C. G., Avruch J., and Alessi D. R. (1999) Evidence that 3-phosphoinositide-dependent protein kinase-1 mediates phosphorylation of p70 S6 kinase in vivo at Thr-412 as well as Thr-252. J. Biol. Chem. 274, 37400–37406 10.1074/jbc.274.52.37400 [DOI] [PubMed] [Google Scholar]
- 37. Romanelli A., Dreisbach V. C., and Blenis J. (2002) Characterization of phosphatidylinositol 3-kinase-dependent phosphorylation of the hydrophobic motif site Thr389 in p70 S6 kinase 1. J. Biol. Chem. 277, 40281–40289 10.1074/jbc.M205168200 [DOI] [PubMed] [Google Scholar]
- 38. Mora A., Komander D., van Aalten D. M., and Alessi D. R. (2004) PDK1, the master regulator of AGC kinase signal transduction. Semin. Cell Dev. Biol. 15, 161–170 10.1016/j.semcdb.2003.12.022 [DOI] [PubMed] [Google Scholar]
- 39. Tokuda H., Hatakeyama D., Shibata T., Akamatsu S., Oiso Y., and Kozawa O. (2003) p38 MAP kinase regulates BMP-4-stimulated VEGF synthesis via p70 S6 kinase in osteoblasts. Am. J. Physiol. Endocrinol. Metab. 284, E1202–E1209 10.1152/ajpendo.00300.2002 [DOI] [PubMed] [Google Scholar]
- 40. Tawakol A., Singh P., Mojena M., Pimentel-Santillana M., Emami H., MacNabb M., Rudd J. H., Narula J., Enriquez J. A., Través P. G., Fernández-Velasco M., Bartrons R., Martín-Sanz P., Fayad Z. A., Tejedor A., and Boscá L. (2015) HIF-1α and PFKFB3 mediate a tight relationship between proinflammatory activation and anerobic metabolism in atherosclerotic macrophages. Arterioscler. Thromb. Vasc. Biol. 35, 1463–1471 10.1161/ATVBAHA.115.305551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jiang H., Shi H., Sun M., Wang Y., Meng Q., Guo P., Cao Y., Chen J., Gao X., Li E., and Liu J. (2016) PFKFB3-driven macrophage glycolytic metabolism is a crucial component of innate antiviral defense. J. Immunol. 197, 2880–2890 10.4049/jimmunol.1600474 [DOI] [PubMed] [Google Scholar]
- 42. Novellasdemunt L., Bultot L., Manzano A., Ventura F., Rosa J. L., Vertommen D., Rider M. H., Navarro-Sabate À., and Bartrons R. (2013) PFKFB3 activation in cancer cells by the p38/MK2 pathway in response to stress stimuli. Biochem. J. 452, 531–543 10.1042/BJ20121886 [DOI] [PubMed] [Google Scholar]
- 43. Boyd S., Brookfield J. L., Critchlow S. E., Cumming I. A., Curtis N. J., Debreczeni J., Degorce S. L., Donald C., Evans N. J., Groombridge S., Hopcroft P., Jones N. P., Kettle J. G., Lamont S., Lewis H. J., et al. (2015) Structure-based design of potent and selective inhibitors of the metabolic kinase PFKFB3. J. Med. Chem. 58, 3611–3625 10.1021/acs.jmedchem.5b00352 [DOI] [PubMed] [Google Scholar]
- 44. Clem B. F., O'Neal J., Tapolsky G., Clem A. L., Imbert-Fernandez Y., Kerr D. A. 2nd, Klarer A. C., Redman R., Miller D. M., Trent J. O., Telang S., and Chesney J. (2013) Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol. Cancer Ther. 12, 1461–1470 10.1158/1535-7163.MCT-13-0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Na Y. R., Gu G. J., Jung D., Kim Y. W., Na J., Woo J. S., Cho J. Y., Youn H., and Seok S. H. (2016) GM-CSF induces inflammatory macrophages by regulating glycolysis and lipid metabolism. J. Immunol. 197, 4101–4109 10.4049/jimmunol.1600745 [DOI] [PubMed] [Google Scholar]
- 46. Tannahill G. M., Curtis A. M., Adamik J., Palsson-McDermott E. M., McGettrick A. F., Goel G., Frezza C., Bernard N. J., Kelly B., Foley N. H., Zheng L., Gardet A., Tong Z., Jany S. S., Corr S. C., et al. (2013) Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 10.1038/nature11986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vergadi E., Ieronymaki E., Lyroni K., Vaporidi K., and Tsatsanis C. (2017) Akt signaling pathway in macrophage activation and M1/M2 polarization. J. Immunol. 198, 1006–1014 10.4049/jimmunol.1601515 [DOI] [PubMed] [Google Scholar]
- 48. Xi H., Kurtoglu M., and Lampidis T. J. (2014) The wonders of 2-deoxy-d-glucose. IUBMB Life. 66, 110–121 10.1002/iub.1251 [DOI] [PubMed] [Google Scholar]
- 49. Grootjans J., Kaser A., Kaufman R. J., and Blumberg R. S. (2016) The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 16, 469–484 10.1038/nri.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yoshida H., Matsui T., Yamamoto A., Okada T., and Mori K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 107, 881–891 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- 51. Martinon F., Chen X., Lee A. H., and Glimcher L. H. (2010) TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat. Immunol. 11, 411–418 10.1038/ni.1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wolf A. J., Reyes C. N., Liang W., Becker C., Shimada K., Wheeler M. L., Cho H. C., Popescu N. I., Coggeshall K. M., Arditi M., and Underhill D. M. (2016) Hexokinase is an innate immune receptor for the detection of bacterial peptidoglycan. Cell 166, 624–636 10.1016/j.cell.2016.05.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pashenkov M. V., Balyasova L. S., Dagil Y. A., and Pinegin B. V. (2017) The role of the p38-MNK-eIF4E signaling axis in TNF production downstream of the NOD1 receptor. J. Immunol. 198, 1638–1648 10.4049/jimmunol.1600467 [DOI] [PubMed] [Google Scholar]
- 54. Hu K., Yang Y., Lin L., Ai Q., Dai J., Fan K., Ge P., Jiang R., Wan J., and Zhang L. (2018) Caloric restriction mimetic 2-deoxyglucose alleviated inflammatory lung injury via suppressing nuclear pyruvate kinase M2-signal transducer and activator of transcription 3 pathway. Front. Immunol. 9, 426 10.3389/fimmu.2018.00426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Idrovo J. P., Yang W. L., Jacob A., Corbo L., Nicastro J., Coppa G. F., and Wang P. (2016) Inhibition of lipogenesis reduces inflammation and organ injury in sepsis. J. Surg. Res. 200, 242–249 10.1016/j.jss.2015.06.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tsoyi K., Jang H. J., Nizamutdinova I. T., Kim Y. M., Lee Y. S., Kim H. J., Seo H. G., Lee J. H., and Chang K. C. (2011) Metformin inhibits HMGB1 release in LPS-treated RAW 264.7 cells and increases survival rate of endotoxaemic mice. Br. J. Pharmacol. 162, 1498–1508 10.1111/j.1476-5381.2010.01126.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tang G., Yang H., Chen J., Shi M., Ge L., Ge X., and Zhu G. (2017) Metformin ameliorates sepsis-induced brain injury by inhibiting apoptosis, oxidative stress and neuroinflammation via the PI3K/Akt signaling pathway. Oncotarget 8, 97977–97989 10.18632/oncotarget.20105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu Z., Bone N., Jiang S., Park D. W., Tadie J. M., Deshane J., Rodriguez C. A., Pittet J. F., Abraham E., and Zmijewski J. W. (2015) AMP-activated protein kinase and glycogen synthase kinase 3β modulate the severity of sepsis-induced lung injury. Mol. Med. 21, 937–950 10.2119/molmed.2015.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee P. S., Wilhelmson A. S., Hubner A. P., Reynolds S. B., Gallacchi D. A., Chiou T. T., and Kwiatkowski D. J. (2010) mTORC1-S6K activation by endotoxin contributes to cytokine up-regulation and early lethality in animals. PLoS One 5, e14399 10.1371/journal.pone.0014399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yen Y. T., Yang H. R., Lo H. C., Hsieh Y. C., Tsai S. C., Hong C. W., and Hsieh C. H. (2013) Enhancing autophagy with activated protein C and rapamycin protects against sepsis-induced acute lung injury. Surgery 153, 689–698 10.1016/j.surg.2012.11.021 [DOI] [PubMed] [Google Scholar]
- 61. Kelly B., and O'Neill L. A. (2015) Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 25, 771–784 10.1038/cr.2015.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tan Y., and Kagan J. C. (2019) Innate immune signaling organelles display natural and programmable signaling flexibility. Cell 177, 384–398.e11 10.1016/j.cell.2019.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Palsson-McDermott E. M., Curtis A. M., Goel G., Lauterbach M. A., Sheedy F. J., Gleeson L. E., van den Bosch M. W., Quinn S. R., Domingo-Fernandez R., Johnston D. G., Jiang J. K., Israelsen W. J., Keane J., Thomas C., Clish C., Vander Heiden M., Xavier R. J., and O'Neill L. A. (2015) Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the Warburg effect in LPS-activated macrophages. Cell Metab. 21, 65–80 10.1016/j.cmet.2014.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shirai T., Nazarewicz R. R., Wallis B. B., Yanes R. E., Watanabe R., Hilhorst M., Tian L., Harrison D. G., Giacomini J. C., Assimes T. L., Goronzy J. J., and Weyand C. M. (2016) The glycolytic enzyme PKM2 bridges metabolic and inflammatory dysfunction in coronary artery disease. J. Exp. Med. 213, 337–354 10.1084/jem.20150900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Millet P., Vachharajani V., McPhail L., Yoza B., and McCall C. E. (2016) GAPDH binding to TNF-α mRNA contributes to posttranscriptional repression in monocytes: A novel mechanism of communication between inflammation and metabolism. J. Immunol. 196, 2541–2551 10.4049/jimmunol.1501345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dietl K., Renner K., Dettmer K., Timischl B., Eberhart K., Dorn C., Hellerbrand C., Kastenberger M., Kunz-Schughart L. A., Oefner P. J., Andreesen R., Gottfried E., and Kreutz M. P. (2010) Lactic acid and acidification inhibit TNF secretion and glycolysis of human monocytes. J. Immunol. 184, 1200–1209 10.4049/jimmunol.0902584 [DOI] [PubMed] [Google Scholar]
- 67. Miller E. S., Koebel D. A., and Sonnenfeld G. (1993) The metabolic stressor 2-deoxy-d-glucose (2-DG) enhances LPS-stimulated cytokine production in mice. Brain Behav. Immun. 7, 317–325 10.1006/brbi.1993.1031 [DOI] [PubMed] [Google Scholar]
- 68. Wang A., Huen S. C., Luan H. H., Yu S., Zhang C., Gallezot J. D., Booth C. J., and Medzhitov R. (2016) Opposing effects of fasting metabolism on tissue tolerance in bacterial and viral inflammation. Cell 166, 1512–1525.e12 10.1016/j.cell.2016.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Maximchik P., Abdrakhmanov A., Inozemtseva E., Tyurin-Kuzmin P. A., Zhivotovsky B., and Gogvadze V. (2018) 2-deoxy-d-glucose has distinct and cell line-specific effects on the survival of different cancer cells upon antitumor drug treatment. FEBS J. 285, 4590–4601 10.1111/febs.14687 [DOI] [PubMed] [Google Scholar]
- 70. Hollien J., and Weissman J. S. (2006) Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313, 104–107 10.1126/science.1129631 [DOI] [PubMed] [Google Scholar]
- 71. Smith J. A. (2018) Regulation of cytokine production by the unfolded protein response; implications for infection and autoimmunity. Front. Immunol. 9, 422 10.3389/fimmu.2018.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kim S., Joe Y., Kim H. J., Kim Y. S., Jeong S. O., Pae H. O., Ryter S. W., Surh Y. J., and Chung H. T. (2015) Endoplasmic reticulum stress-induced IRE1α activation mediates cross-talk of GSK-3β and XBP-1 to regulate inflammatory cytokine production. J. Immunol. 194, 4498–4506 10.4049/jimmunol.1401399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fortin C. F., Mayer T. Z., Cloutier A., and McDonald P. P. (2013) Translational control of human neutrophil responses by MNK1. J. Leukoc. Biol. 94, 693–703 10.1189/jlb.0113012 [DOI] [PubMed] [Google Scholar]
- 74. Hitti E., Iakovleva T., Brook M., Deppenmeier S., Gruber A. D., Radzioch D., Clark A. R., Blackshear P. J., Kotlyarov A., and Gaestel M. (2006) Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol. Cell. Biol. 26, 2399–2407 10.1128/MCB.26.6.2399-2407.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sallusto F., and Lanzavecchia A. (1994) Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor α. J. Exp. Med. 179, 1109–1118 10.1084/jem.179.4.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pashenkov M. V., Popilyuk S. F., Alkhazova B. I., L'vov V. L., Murugin V. V., Fedenko E. S., Khaitov R. M., and Pinegin B. V. (2010) Muropeptides trigger distinct activation profiles in macrophages and dendritic cells. Int. Immunopharmacol. 10, 875–882 10.1016/j.intimp.2010.04.025 [DOI] [PubMed] [Google Scholar]
- 77. Zhang X., Goncalves R., and Mosser D. M. (2008) The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 83, 14.1.1–14.1.14 10.1002/0471142735.im1401s83 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.