Abstract
Exomic studies have demonstrated that innate immune genes exhibit an even higher degree of variation than the majority of other gene families. However, the phenotypic implications of this genetic variation are not well understood, with effects ranging from hypomorphic to silent to hyperfunctioning. In this work, we study the functional consequences of this variation by investigating polymorphisms in gasdermin D, the key pyroptotic effector protein. We find that, although SNPs affecting potential posttranslational modifications did not affect gasdermin D function or pyroptosis, polymorphisms disrupting sites predicted to be structurally important dramatically alter gasdermin D function. The manner in which these polymorphisms alter function varies from conserving normal pyroptotic function to inhibiting caspase cleavage to disrupting oligomerization and pore formation. Further, downstream of inflammasome activation, polymorphisms that cause loss of gasdermin D function convert inflammatory pyroptotic cell death into immunologically silent apoptotic cell death. These findings suggest that human genetic variation can alter mechanisms of cell death in inflammation.
Keywords: cell death, inflammasome, genetics, inflammation, genetic polymorphism, SNP
Introduction
Whole-genome sequencing studies have shown, surprisingly, a high amount of genetic diversity within the human population. Studies have even documented nucleotide variation at one in eight exonic residues, challenging the recently held dogma of a canonical genome sequence (1, 2). However, translating this wealth of genetic data into new therapeutic interventions has been constrained by a limited understanding of how genetic diversity alters biochemical interactions and translates into phenotypic variation.
The innate immune system utilizes inflammatory cell death, which is central to restricting pathogens and clearing damage. Pyroptotic cell death relies on activation of the inflammasome, a macromolecular cytosolic complex that senses perturbations within the cell via a receptor, such as NLRP1, NLRP3, AIM2, or NLRC4, and coordinates cleavage of caspase-1 (3–6). Activation of the inflammasome is classically a two-step process, with signal 1 acting as a priming event through transcriptional up-regulation of expression of NLRP3, pro-IL-1β, and pro-IL-18, followed by signal 2, which stimulates rapid assembly of the inflammasome complex in the cytosol (7). Activated caspase-1 cleaves the 52-kDa GSDMD into an inert C-terminal p20 fragment and a pore-forming N-terminal p30 fragment that oligomerizes and forms large pores in the membranes of the cell (8–11). Insertion of the GSDMD pore results in membrane failure and allows release of mature IL-1β, IL-18, and eicosanoids, culminating in a potent inflammatory response (12, 13).
During exome sequencing of 60,706 individuals, we found that cell death related pore-forming proteins demonstrate a high degree of variation across the population (2). As GSDMD is the critical executioner of pyroptotic cell death, here we ask whether these variants created biochemical changes resulting in phenotypic variation in pyroptotic function across the population (5, 6). Specifically, some of these SNPs disrupt sites of putative phosphorylation and ubiquitination or are found at sites that are structurally important for pyroptotic function. Using cell-based assays to test specific aspects of GSDMD-mediated pyroptotic function, we demonstrate that genetic diversity in GSDMD alters biochemical function to create phenotypic variation in pyroptotic function, resulting in differential inflammatory cell death responses.
Results
Gsdmd is highly polymorphic
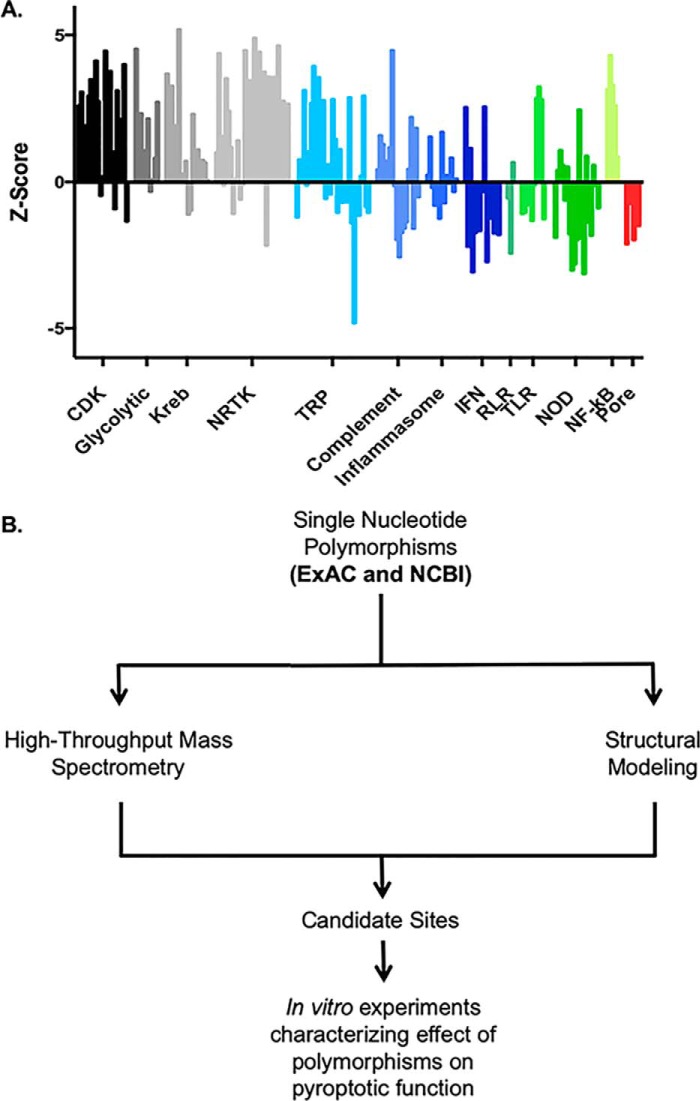
Exomic sequencing demonstrates a high degree of variability within the human genome. For each gene, the degree of genetic variation found within the human population can be measured by the Z score, a metric of the number of human variants found normalized to the length of the gene (Fig. 1A) (2). Genes critical for cellular function, such as those in the glycolytic pathway and Krebs cycle, demonstrate low variability and a high Z score. Conversely, many genes involved in the immune response including interferons, have a high degree of variability and a low Z score. Interestingly, many of the pore-forming immune proteins, including gasdermin family members and mixed lineage kinase domain-like pseudokinase, demonstrate a high degree of variability, potentially representing diversity in the immune response across the population. To study the potential impact of SNPs on pyroptotic cell death, GSDMD variants from two databases of human SNPs (curated by ExAC and NCBI) were screened for SNPs that disrupted sites of putative posttranslational modification (SNP-PTM)3 or SNPs that were potentially structurally disruptive (structural SNPs). Screening SNP sites through high-throughput MS databases identified four SNPs that disrupted sites of putative posttranslational modification (Fig. S1). 11 SNPs that were considered potentially structurally disruptive to GSDMD pyroptotic function were identified (5, 8, 11, 13–16). Interestingly, no residues predicted to enhance pyroptotic activity were found in the human population, suggesting that activating mutations in Gsdmd may be strongly selected against. Given these findings, we chose to investigate the phenotypic implications of GSDMD variation in the human population within these two distinct groups: SNP-PTM sites and structural SNP sites (Fig. 1B).
Figure 1.
Genomic variability and putative posttranslational modifications of GSDMD. A, comparison of genetic constraint between classes of proteins. The genetic constraint of a gene can be represented as a Z score, calculated as a function of the number of variants in a protein found in the exome normalized to the length of the protein. Higher Z scores indicate high constraint and low variability, whereas negative Z scores indicate low constraint and high variability. Each bar represents a single gene. Innate immune-sensing systems show a much higher tolerance of variation relative to kinase and metabolic enzymes. Among cell death–associated pore-forming proteins, the gasdermin family of proteins is comparable with MLKL, the necroptotic pore forming protein. CDK, cyclin-dependent kinase; NRTK, nonreceptor tyrosine kinase; TRP, transient receptor potential; RLR, RIG-I–like receptor; TLR, Toll-like receptor; NOD, nucleotide-binding oligomerization domain–like (2). B, screening of single nucleotides found in the human population for sites of putative posttranslational modification or structural significance with further workup of candidate sites.
SNPs disrupting putative phosphorylation and ubiquitination sites do not affect pyroptotic function
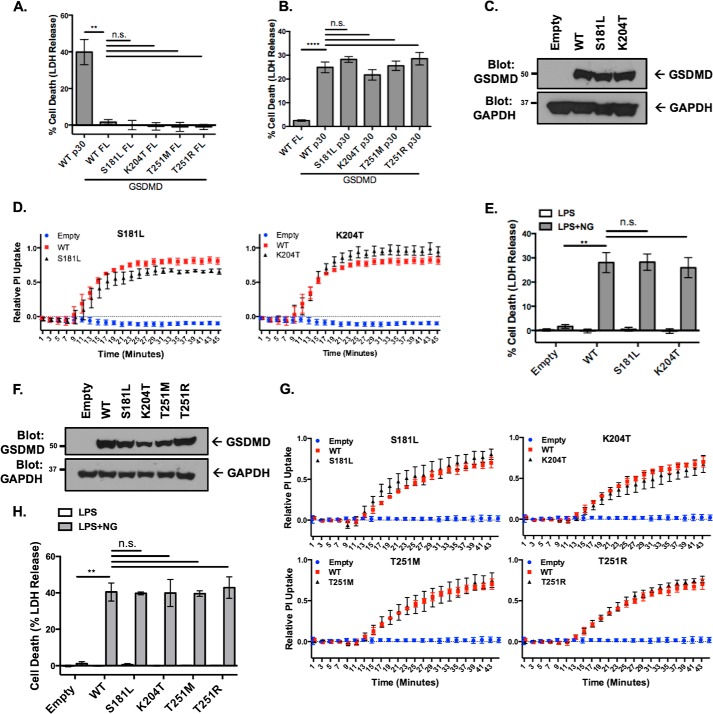
First, phenotypic alterations in genetic polymorphisms disrupting putative PTM sites were evaluated. Expression of full-length SNP-PTM variants in HEK293T cells demonstrated no cell death, indicating that autoinhibition is independent of posttranslational modification at these sites (Fig. 2A) (8, 17). Expression of SNP-PTM variant p30-GSDMD fragments in HEK293T cells demonstrated equivalent amounts of cell death as WT p30-GSDMD, indicating that p30 effector function was unaffected by modification at SNP-PTM sites (Fig. 2B). Two of the SNP-PTM variants are conserved in mice as S182L and K205T and were stably reconstituted in GSDMD−/− iBMDM cells (Fig. 2C) (16). LPS priming followed by nigericin stimulation resulted in S181L and K204T variants recapitulating WT pyroptotic pore formation, as measured by PI uptake, and cell death, as measured by LDH release (Fig. 2, D and E).
Figure 2.
GSDMD SNPs at PTM sites conserve normal pyroptotic function. A, cell death as measured by LDH release in response to expression of full-length (FL) SNP-PTM sites in HEK293T cells. Data represent three independent experiments, each with technical triplicates. **, p < 0.01; n.s., no significance. B, cell death resulting from expression of p30-GSDMD SNP-PTM sites in HEK293T cells. Data represent three independent experiments, each with technical duplicates. ****, p < 0.0001. C, Western blot of the stable reconstitution of SNP-PTM sites in Gsdmd−/− iBMDM cells. D, cell death measured by LDH release of reconstituted iBMDMs. E, PI uptake of SNP-PTM reconstituted iBMDMs in response to LPS with 10 μm compared with the same empty and WT GSDMD controls. F, Western blot of stable reconstitution of SNP-PTM sites in GSDMD−/− THP-1 cells. G, cell death measured by LDH release of reconstituted THP-1 cells. H, PI uptake of SNP-PTM reconstituted THP-1 cells in response to LPS with 10 μm compared with the same empty and WT GSDMD controls. In A, B, D, E, G, and H, data represent three or more independent experiments.
Polymorphisms disrupting sites of putative PTMs were investigated further in human monocytes. SNP-PTMs were stably reconstituted in GSDMD−/− THP-1 cells (Fig. 2F). SNP-PTMs recapitulated the WT phenotype in pore formation, as measured by PI uptake, and cell death, as measured by LDH release (Fig. 2, G and H). Thus, testing of the SNP-PTM variants demonstrated conservation of pyroptotic function, demonstrating that GSDMD tolerates variation at SNP-PTM sites and that pyroptotic cell death is independent of modification at these sites in human and murine cells of monocytic lineage.
Gsdmd structural SNPs cause phenotypic variation in pyroptotic cell death
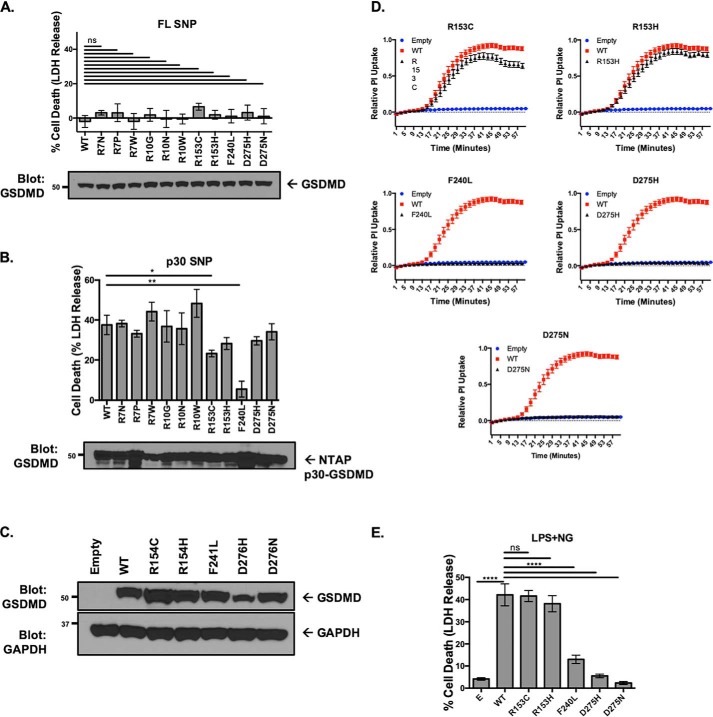
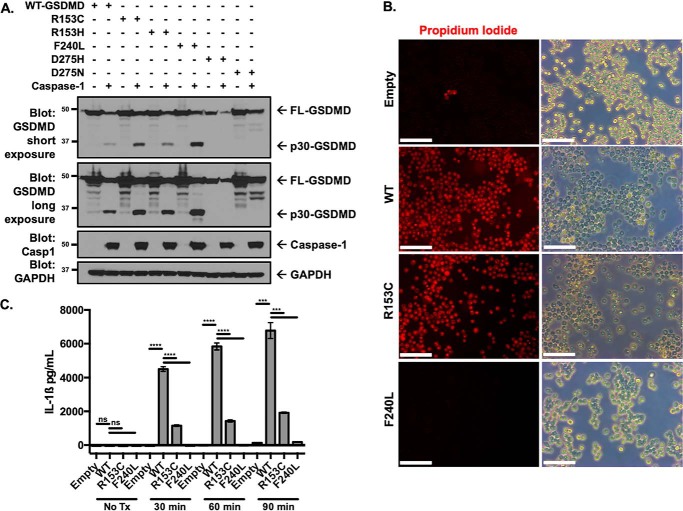
Second, phenotypic alterations in genetic polymorphisms disrupting key structural sites were evaluated. No spontaneous pyroptotic activity occurred with expression of the FL GSDMD structural SNPs, suggesting that none of these polymorphisms induced spontaneous activation (Fig. 3A). To determine loss of function, the p30-GSDMD fragment containing the structural SNPs was transfected into HEK293T cells. These experiments showed a significant decrease in the ability of R153C and F240L p30-GSDMD to induce cell death (Fig. 3B). Five SNPs (R153C, R153H, F240L, D275H, and D275N) were conserved in murine GSDMD. These SNPs were reconstituted in GSDMD−/− iBMDM cells with expression levels equivalent to reconstitution with WT GSDMD (Fig. 3C). Activation of the inflammasome with LPS and nigericin demonstrated almost complete suppression of pyroptotic pore formation and cell death with GSDMD SNPs F240L, D275H, and D275N after 1 h, whereas a minor decrease in pyroptotic pore formation was noted with R153C (Fig. 3, D and E).
Figure 3.
Genetic diversity in GSDMD leads to phenotypic variation in pyroptotic cell death. A, expression of FL structural SNPs in HEK293T cells. n.s., no significance. B, expression of p30 structural SNPs in HEK293T cells. *, p < 0.05; **, p < 0.01. C, stable reconstitution of SNPs into Gsdmd−/− iBMDM cells. D, pore formation as measured by PI uptake in reconstituted iBMDMs after priming with LPS and activation with nigericin. All SNPs were from the same experimental sets and compared against the same empty and WT controls. E, cell death as measured by LDH release in reconstituted iBMDM cells primed with LPS and activated with nigericin. ****, p < 0.0001. In A, B, D, and E, data represent three or more independent experiments.
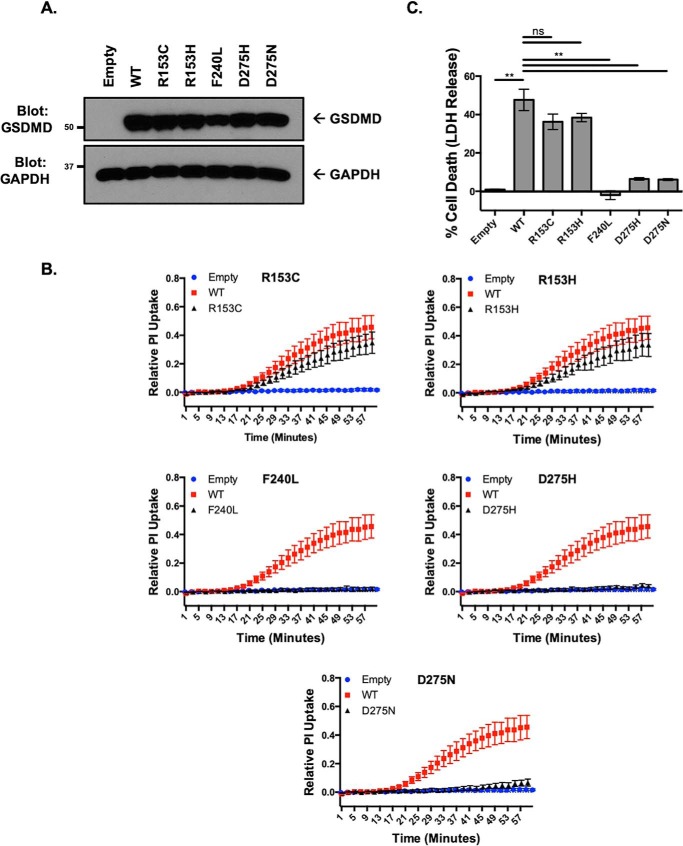
To determine whether these structural SNPs altered pyroptotic function in human cells of monocytic lineage, structural SNPs were stably reconstituted in GSDMD−/− THP-1 cells (Fig. 4A). Similar to the findings with murine iBMDMs, a minor but nonsignificant decrease was seen in cell death, as measured by LDH release with the R153C and R153H polymorphisms, which corresponded to a small decrease in kinetic assays of PI uptake in response to activation of inflammation with LPS and nigericin (Fig. 4, B and C). Total or near total loss of cell death, as measured by LDH release, and pore formation, as measured by PI uptake, were seen with F240L, D275H, and D275N polymorphisms following inflammasome activation (Fig. 4, B and C).
Figure 4.
Genetic diversity in GSDMD leads to phenotypic variation in pyroptotic cell death in human monocytes. A, stable reconstitution of structural SNPs into GSDMD−/− THP-1 cells. B, pore formation as measured by PI uptake in reconstituted THP-1 cells after priming with LPS and activation with nigericin. All SNPs were from the same experimental sets and compared against the same empty and WT controls. C, cell death as measured by LDH release in reconstituted THP-1 cells after priming with LPS and stimulation with nigericin. In B and C, data represent three or more independent experiments. **, p < 0.01; n.s., no significance.
To determine whether the lack of pyroptotic pore formation and cell death was due to inhibition of GSDMD cleavage with these polymorphisms, caspase-1 cleavage of GSDMD was assessed. Caspase-1 cleavage assays with the structural SNPs demonstrated normal cleavage of full-length WT GSDMD, R153C, R153H, and F240L but complete absence of cleavage with D275H and D275N, consistent with these SNPs disrupting the caspase cleavage site (Fig. 5A). This further suggested that the R153C and F240L polymorphisms disrupt GSDMD function downstream of cleavage. To further characterize this, the pyroptotic ability of R153C and F240L reconstituted cell lines was visualized using epifluorescence microscopy. WT and R153C cells demonstrated high levels of PI uptake as well as the membrane dysfunction characteristic of pyroptotic cell death following stimulation with LPS and nigericin, whereas the F240L structural SNP demonstrated loss of pyroptotic cell death (Fig. 5B). The ability of IL-1β to be released from cells expressing R153C and F240L polymorphisms was also impaired, further demonstrating the functional deficit resulting from these polymorphisms (Fig. 5C). As previous work has suggested a monomer-to-dimer step in the process of GSDMD pore formation, the absence of dimerization with the F240L polymorphism suggests that this SNP prevents monomer-to-dimer transition, which is likely a necessary step for higher-order oligomerization (18).
Figure 5.
Polymorphisms in GSDMD differ mechanistically by inhibiting caspase cleavage or altering p30 effector function. A, assessment of cleavage of GSDMD structural SNPs by caspase-1 in HEK293T cells. Full-length Gsdmd SNPs were coexpressed with caspase-1, with cleavage assessed by Western blotting. B, epifluorescence and phase-contrast microscopy of reconstituted Gsdmd−/− iBMDM cell lines following priming with LPS and 45-min stimulation with nigericin. Scale bars = 100 μm. C, IL-1β release in reconstituted cell lines following LPS priming and 10 μm nigericin treatment. n.s., no significance; ***, p < 0.001; ****, p < 0.0001. Data represent three or more independent experiments.
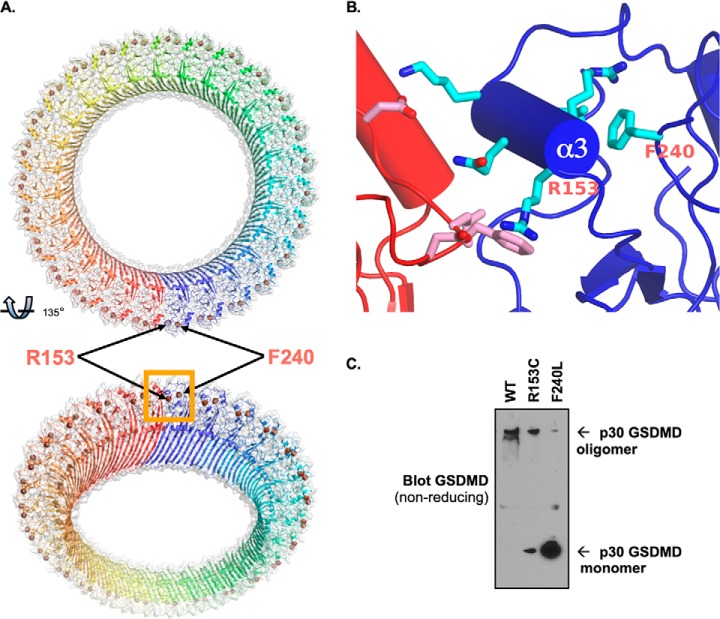
Structural modeling of the GSDMD pore revealed that the two residues that interfere with pyroptotic pore formation and cell death, R153C and F240L, lie at the interface between adjacent p30-GSDMD units at an interface that likely is critical for oligomerization (Fig. 6, A and B) (19, 20). Arg-153 is one part of a critical α-helix that is supported by Phe-240 and allows salt bridges to form between adjacent p30 segments. This was validated experimentally, as R153C demonstrated decreased oligomerization with F240L, demonstrating nearly complete loss of oligomer formation, consistent with the previous findings of pore formation and cell death (Fig. 6C).
Figure 6.
GSDMD polymorphisms alter oligomerization of the pyroptotic pore. A and B, structural modeling of the GSDMD pore, with structural SNPs Arg-153 and Phe-240 highlighted at the p30 oligomerization interface. C, the F240L polymorphism results in loss of p30-GSDMD oligomerization. WT and F240L p30-GSDMD were expressed in 293T cells. They were harvested under nonreducing conditions, and oligomerization was assessed by Western blotting.
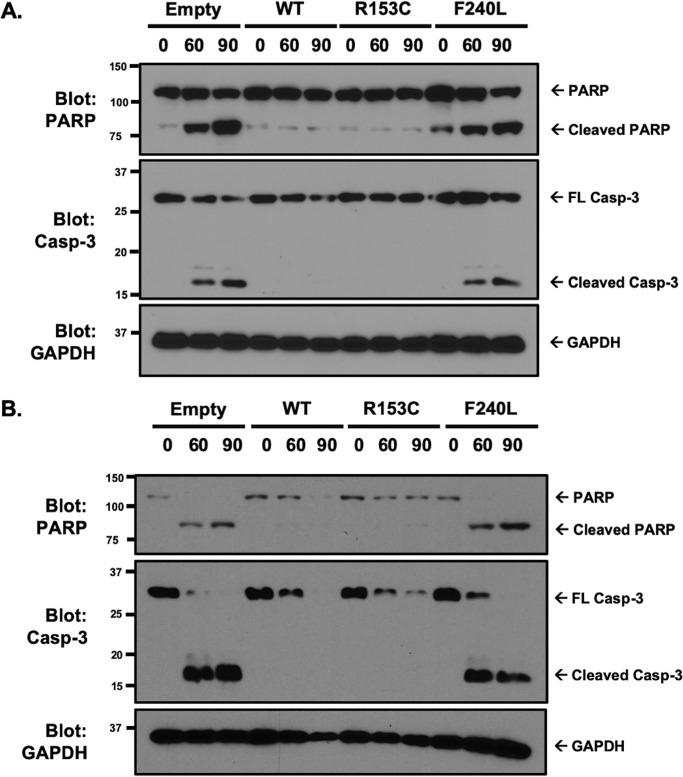
Loss of GSDMD, or classic pyroptotic cell death, switches the cell to caspase-3–dependent cell death (21, 22). Therefore, we asked whether caspase-3 was activated in structural SNPs that demonstrated loss of pyroptotic cell death because of loss of GSDMD function. To determine whether inhibition of pyroptotic cell death resulted in activation of apoptosis, cleavage of PARP and caspase-3 were assessed. Caspase-3 and PARP were not cleaved when WT or R153C GSDMD were expressed. However, PARP and caspase-3 were cleaved in GSDMD−/− and F240L cells in reconstituted GSDMD−/− iBMDMs and THP-1 cells (Fig. 7, A and B). Together, these findings suggest that human polymorphisms in GSDMD at structurally significant sites lead to loss of pyroptotic pore formation and cytokine release through loss of oligomerization and that, in some cases, these polymorphisms convert highly inflammatory pyroptotic cell death to the more immunologically silent apoptotic form of cell death.
Figure 7.
Structural SNPs convert pyroptotic cell death to apoptotic cell death. A and B, apoptotic cell death was investigated in reconstituted iBMDM (A) and THP-1 (B) cell lines by evaluating caspase-3 activation and PARP cleavage. Cells were primed with LPS for 4 h, followed by activation of the inflammasome with 10 μm nigericin. Cleavage of caspase-3 and PARP was assessed by Western blotting. Representative of three independent experiments.
Discussion
Genetic variation leading to phenotypic variation in pyroptotic function
The rapid expansion of available genetic data and our ability to sequence large numbers of patients has led to an inevitable bottleneck. Sequencing occurs at a far more rapid rate than functional characterization. By implementing a system to test the pyroptotic function of various polymorphisms, we found that, although GSDMD activity is independent of genetic regulation at sites of putative posttranslational modification, SNPs found within structurally important regions result in phenotypic variation in pyroptotic function. These polymorphisms in Gsdmd drive alterations in pyroptosis through a variety of mechanisms, including loss of caspase cleavage and complete disruption of p30 oligomerization and formation of the pyroptotic pore. Although the SNPs at Asp-275 alter the caspase 1/11 cleavage site, rendering it uncleavable, the R153C and F240L polymorphisms do not interfere with caspase cleavage but disrupt an interface between adjacent p30-GSDMD units that is important for oligomerization and formation of mature pores. A decrease in the pyroptotic pore results in loss of inflammatory cytokine release and diverts the cell to the less inflammatory apoptotic form of cell death.
Together, these findings demonstrate that genetic polymorphisms alter biochemical function and can dramatically alter inflammatory death responses downstream of inflammasome activation. Oligomerization of p30-GSDMD fragments, affinity for lipid membranes, and maturation into mature pores that substrates have to pass through are all facets of this process they may disrupt. As genetic data continue to accumulate at an exponential rate, new variants in GSDMD will be identified, necessitating delineation of their phenotypic significance. The role of caspases as regulators of inflammatory cell death and mediators of cellular secretion also continue to be defined (23, 24). As the field of inflammatory cell death and pyroptosis continues to be the focus of intense study, our understanding of the intricacies of GSDMD genetic variation and the relationship to human health and disease will continue to evolve.
Experimental procedures
Cells, plasmids, reagents, antibodies, and Western blotting
Expression assays were conducted using calcium phosphate or Lipofectamine transfection of HEK293T cells. Nlrp3−/− immortalized bone marrow–derived macrophages (iBMDMs) stably expressing NLRP3 and cerulean-ASC were a gift from Eicke Latz (University of Bonn). Gsdmd−/− iBMDM cells were generated using CRISPR-Cas9 as described previously (16, 25) using a LentiCRISPRv2 plasmid (Addgene) with the puromycin resistance marker exchanged for a hygromycin resistance marker (26). GSDMD−/− THP-1 cells were generated using CRISPR-Cas9 as described previously, starting with WT THP-1 cells (ATCC) (27). Cells were cloned out and individually tested for GSDMD expression by Western blotting using antibodies against murine GSDMDC1 (Santa Cruz, SC-393656) or human GSDMD (Santa Cruz, SC-81868; Sigma, HPA044487).
Murine or human GSDMD was Gibson-cloned into a reconstitution vector utilizing LentiCRISPRv2 as a template as described previously (16, 25, 28). Reconstituted GSDMD was made CRISPR-Cas9–resistant by silent mutation of the protospacer-adjacent motif NGG sequence using site-directed mutagenesis. GSDMD SNP-PTM variants and structural SNP mutants were generated using site-directed mutagenesis of the GSDMD lentiviral reconstitution vectors. Variants were confirmed by Sanger sequencing. GSDMD expression in the reconstituted iBMDM and THP-1 cell lines was probed using Western blotting with the respective antibodies. Stable reconstitution of cell lines was accomplished using a lentiviral expression construct with a neomycin resistance marker (28). CRISPR-Cas9 knockout or GSDMD reconstitution was performed using calcium phosphate transfection in HEK293T cells using the CRISPR-Cas9 or GSDMD reconstitution plasmid with PsPAX (Addgene) and PMD.2 (Addgene) at a 4:3:1.2 ratio. Viral supernatant was harvested from HEK293T cells spun at 1200 × g for 5 min, passaged through a 0.45-μm filter, combined with Polybrene infection/transfection reagent (Millipore), and used to transduce iBMDM cells. Reconstituted cell lines were selected using 1.0 mg/ml (iBMDMs) or 0.5 mg/ml (THP-1 cells) G418 (InvivoGen) for a minimum of 10 days.
Except where otherwise noted, cells were lysed using Triton lysis buffer with 1 mmol PMSF (Acros Organics), protease inhibitor mixture (Sigma), and calyculin (LC Laboratories). Cells used for probing p30-GSDMD were lysed in 8 m urea with 5% SDS or radioimmune precipitation assay buffer with 1% SDS combined with 1 mmol PMSF, protease inhibitor mixture, and calyculin and then passaged five times through a 25-gauge needle. Cells used in the oligomerization assay were lysed in NP-40 buffer in the absence of reducing agents, passaged five times through a 25-gauge needle, and run on a 4%–20% gradient gel (Bio-Rad).
Primary antibodies used were anti-GAPDH (Proteintech), anti-murine GSDMD (Santa Cruz and Abcam), anti-caspase-1 (Adipogen), anti-PARP (Cell Signaling Technology), anti-caspase-3 (Cell Signaling Technology), and anti-GFP (Cell Signaling Technology) and used according to the manufacturer's instructions. HRP-conjugated anti-mouse and anti-rabbit secondary antibodies (Cell Signaling Technology) were used according to the manufacturer's instructions.
Bioinformatics
SNPs were identified from the Exome Aggregation Consortium database originally published by Lek et al. (2) (RRID:SCR_014964). These polymorphisms were compared against the SNP database curated by the National Center for Biotechnology Information. Sites on GSDMD identified by high-throughput MS as sites of ubiquitination or phosphorylation modification were identified using PhosphoSitePlus (RRID:SCR_001837). For clarity, human polymorphisms conserved in mice are designated using their designated human positions.
Propidium iodide and cytotoxicity assays
Cells were plated out at 300,000 cells/well for the propidium iodide uptake assay and 100,000 cells/well for the cytotoxicity assay in 24-well plates. 4 h prior to the start of either assay, cells were stimulated with 1.0 μg/ml LPS (InvivoGen). At the start of each experiment, the medium was removed, and cells were washed once with PBS and cultured in Live Cell Imaging Solution (Invitrogen) or an imaging buffer containing 120 mm NaCl, 4 mm KCl, 1.5 mm CaCl2, 1 mm magnesium chloride, 25 mm HEPES, 5 mm glucose, and 0.1% BSA (pH 7.4). 10 μm nigericin (Sigma) was used to activate the NLRP3 inflammasome.
For the propidium iodide uptake assay, 1.0 μg/ml of propidium iodide (Molecular Probes) was added to the imaging buffer, and the background fluorescence was measured on a Spectramax i3x multimode microplate reader (Molecular Devices) with an excitation/emission measurement of 533/617. After obtaining background fluorescence readings, cells were stimulated with 10 μm nigericin (Sigma), and the PI fluorescence of each well measured at intervals of 1 min. Maximum PI fluorescence was measured after lysis using 1% Triton X-100 in each well. PI uptake was calculated relative to each well using the following formula: relative PI uptake = (fluorescence at time x − background fluorescence)/(maximum fluorescence − background fluorescence).
For the cytotoxicity assay, background LDH release was obtained in untreated cells, whereas maximum LDH release was obtained following lysis of the cells. After 1 h, LDH release was obtained using the Pierce LDH release assay (Invitrogen) according to the manufacture's instructions and read on the Spectramax i3x multimode microplate reader. Relative cell death was calculated according to the manufacturer's instructions based on the background and maximum LDH release controls for each cell type.
Imaging
Phase-contrast and epifluorescence images were taken on a Leica DMIL LED microscope with a ×20 objective. Cells were imaged without fixing in live cell imaging solution (Invitrogen) in medium containing 1.0 μg/ml propidium iodide.
Enzyme-linked immunosorbent assay
Release of IL-1β was determined by sandwich ELISA (BioLegend) according the manufacture's instructions. iBMDMs were plated at 200,000 cells/well in a 24-well plate. Cells were primed for 4 h with 1 μg/ml LPS, followed by treatment with 10 μm nigericin (Sigma). At specific time points following nigericin treatment, supernatants were collected, and cellular debris was cleared by centrifuging for 1 min at 13,000 rpm.
Structural modeling
A model of the human GSDMD pore formed by its N-terminal domain was created using the crystal structure of the N-terminal domain in the autoinhibited form (PDB code 6N9O) and the cryo-EM structure of mGSDMA3 in the pore form (6CB8) with the MMM server (RRID:SCR_018015) (19, 20). Fig. 6, A and B, was produced using PyMOL (RRID:SCR_000305).
Author contributions
J. K. R. and D. W. A. conceptualization; J. K. R., T. S. X., and D. W. A. data curation; J. K. R., T. S. X., and D. W. A. formal analysis; J. K. R., T. S. X., and D. W. A. investigation; J. K. R. and D. W. A. methodology; J. K. R. writing-original draft; T. S. X. software; D. W. A. supervision; D. W. A. funding acquisition; D. W. A. project administration; D. W. A. writing-review and editing.
Supplementary Material
Acknowledgments
We thank the Abbott, Dubyak, Ramakrishnan, and Xiao laboratories for insightful discussions. Input from George Dubyak (Case Western Reserve University), Xiaoxia Li (Cleveland Clinic), Bowen Zhou (Case Western Reserve University), Hannah Kondolf (Case Western Reserve University), and Steven Chirieleison (Case Western Reserve University, Stanford) and technical assistance from Sylvia Kertesy (Case Western Reserve University) greatly improved the manuscript.
This work was supported by NIDDK, National Institutes of Health Grant P01DK091222 (to D. W. A.); NIGMS, National Institutes of Health Grant R01GM086550 (to D. W. A.); and National Institutes of Health Grants R21AR069908 and R01GM127609 (to T. S. X.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Fig. S1.
- PTM
- posttranslational modification
- LDH
- lactate dehydrogenase
- FL
- full-length
- iBMDM
- immortalized bone marrow–derived macrophage
- LPS
- lipopolysaccharide
- PI
- propidium iodide.
References
- 1. 1000 Genomes Project Consortium, Auton A., Brooks L. D., Durbin R. M., Garrison E. P., Kang H. M., Korbel J. O., Marchini J. L., McCarthy S., McVean G. A., and Abecasis G. R. (2015) A global reference for human genetic variation. Nature 526, 68–74 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., Fennell T., O'Donnell-Luria A. H., Ware J. S., Hill A. J., Cummings B. B., Tukiainen T., Birnbaum D. P., Kosmicki J. A., Duncan L. E., Estrada K., et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lamkanfi M., and Dixit V. M. (2014) Mechanisms and functions of inflammasomes. Cell 157, 1013–1022 10.1016/j.cell.2014.04.007 [DOI] [PubMed] [Google Scholar]
- 4. Latz E., Xiao T. S., and Stutz A. (2013) Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 13, 397–411 10.1038/nri3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., and Shao F. (2015) Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- 6. Kayagaki N., Stowe I. B., Lee B. L., O'Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q. T., Liu P. S., Lill J. R., Li H., Wu J., Kummerfeld S., et al. (2015) Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- 7. He Y., Hara H., and Núñez G. (2016) Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem. Sci. 41, 1012–1021 10.1016/j.tibs.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ding J., Wang K., Liu W., She Y., Sun Q., Shi J., Sun H., Wang D.-C., and Shao F. (2016) Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116 10.1038/nature18590 [DOI] [PubMed] [Google Scholar]
- 9. Aglietti R. A., Estevez A., Gupta A., Ramirez M. G., Liu P. S., Kayagaki N., Ciferri C., Dixit V. M., and Dueber E. C. (2016) GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl. Acad. Sci. U.S.A. 113, 7858–7863 10.1073/pnas.1607769113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sborgi L., Rühl S., Mulvihill E., Pipercevic J., Heilig R., Stahlberg H., Farady C. J., Müller D. J., Broz P., and Hiller S. (2016) GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J. 35, 1766–1778 10.15252/embj.201694696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu X., Zhang Z., Ruan J., Pan Y., Magupalli V. G., Wu H., and Lieberman J. (2016) Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 10.1038/nature18629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. von Moltke J., Trinidad N. J., Moayeri M., Kintzer A. F., Wang S. B., van Rooijen N., Brown C. R., Krantz B. A., Leppla S. H., Gronert K., and Vance R. E. (2012) Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature 490, 107–111 10.1038/nature11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He W. T., Wan H., Hu L., Chen P., Wang X., Huang Z., Yang Z.-H., Zhong C.-Q., and Han J. (2015) Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 25, 1285–1298 10.1038/cr.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lei X., Zhang Z., Xiao X., Qi J., He B., and Wang J. (2017) Enterovirus 71 inhibits pyroptosis through cleavage of gasdermin D. J. Virol. 91, e01069–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen X., He W. T., Hu L., Li J., Fang Y., Wang X., Xu X., Wang Z., Huang K., and Han J. (2016) Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Res. 26, 1007–1020 10.1038/cr.2016.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rathkey J. K., Benson B. L., Chirieleison S. M., Yang J., Xiao T. S., Dubyak G. R., Huang A. Y., and Abbott D. W. (2017) Live-cell visualization of gasdermin D-driven pyroptotic cell death. J. Biol. Chem. 292, 14649–14658 10.1074/jbc.M117.797217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi P., Tang A., Xian L., Hou S., Zou D., Lv Y., Huang Z., Wang Q., Song A., Lin Z., and Gao X. (2015) Loss of conserved Gsdma3 self-regulation causes autophagy and cell death. Biochem. J. 468, 325–336 10.1042/BJ20150204 [DOI] [PubMed] [Google Scholar]
- 18. Rathkey J. K., Zhao J., Liu Z., Chen Y., Yang J., Kondolf H. C., Benson B. L., Chirieleison S. M., Huang A. Y., Dubyak G. R., Xiao T. S., Li X., and Abbott D. W. (2018) Chemical disruption of the pyroptotic pore-forming protein gasdermin D inhibits inflammatory cell death and sepsis. Sci. Immunol. 3, eaat2738 10.1126/sciimmunol.aat2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Z., Wang C., Yang J., Zhou B., Yang R., Ramachandran R., Abbott D. W., and Xiao T. S. (2019) Crystal structures of the full-length murine and human gasdermin D reveal mechanisms of autoinhibition, lipid binding, and oligomerization. Immunity 51, 43–49.e4 10.1016/j.immuni.2019.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruan J., Xia S., Liu X., Lieberman J., and Wu H. (2018) Cryo-EM structure of the gasdermin A3 membrane pore. Nature 557, 62–67 10.1038/s41586-018-0058-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Taabazuing C. Y., Okondo M. C., and Bachovchin D. A. (2017) Pyroptosis and apoptosis pathways engage in bidirectional crosstalk in monocytes and macrophages. Cell Chem. Biol. 24, 507–514.e4 10.1016/j.chembiol.2017.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schneider K. S., Groß C. J., Dreier R. F., Saller B. S., Mishra R., Gorka O., Heilig R., Meunier E., Dick M. S., iković T., Sodenkamp J., Médard G., Naumann R., Ruland J., Kuster B., et al. (2017) The inflammasome drives GSDMD-independent secondary pyroptosis and IL-1 release in the absence of caspase-1 protease activity. Cell Rep. 21, 3846–3859 10.1016/j.celrep.2017.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keller M., Rüegg A., Werner S., and Beer H. D. (2008) Active caspase-1 is a regulator of unconventional protein secretion. Cell 132, 818–831 10.1016/j.cell.2007.12.040 [DOI] [PubMed] [Google Scholar]
- 24. Newton K., Wickliffe K. E., Maltzman A., Dugger D. L., Reja R., Roose-girma M., Modrusan Z., Sagolla M. S., Webster J. D., and Dixit V. M. (2019) Activity of caspase-8 determines plasticity between cell death pathways. Nature 575, 679–682 10.1038/s41586-019-1752-8 [DOI] [PubMed] [Google Scholar]
- 25. Russo H. M., Rathkey J., Boyd-Tressler A., Katsnelson M. A., Abbott D. W., and Dubyak G. R. (2016) Active caspase-1 induces plasma membrane pores that precede pyroptotic lysis and are blocked by lanthanides. J. Immunol. 197, 1353–1367 10.4049/jimmunol.1600699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chirieleison S. M., Marsh R. A., Kumar P., Rathkey J. K., Dubyak G. R., and Abbott D. W. (2017) Nucleotide-binding oligomerization domain (NOD) signaling defects and cell death susceptibility cannot be uncoupled in X-linked inhibitor of apoptosis (XIAP)-driven inflammatory disease. J. Biol. Chem. 292, 9666–9679 10.1074/jbc.M117.781500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang J., Liu Z., Wang C., Yang R., Rathkey J. K., Pinkard O. W., Shi W., Chen Y., Dubyak G. R., Abbott D. W., and Xiao T. S. (2018) Mechanism of gasdermin D recognition by inflammatory caspases and their inhibition by a gasdermin D-derived peptide inhibitor. Proc. Natl. Acad. Sci. 115, 6792–6797 10.1073/pnas.1800562115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chirieleison S. M., Kertesy S. B., and Abbott D. W. (2016) Synthetic biology reveals the uniqueness of the RIP kinase domain. J. Immunol. 196, 4291–4297 10.4049/jimmunol.1502631 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.