Abstract
Bacteria account for 1000-fold more biomass than humans. They vary widely in shape and size. The morphological diversity of bacteria is due largely to the different peptidoglycan-based cell wall structures that encase bacterial cells. Although the basic structure of peptidoglycan is highly conserved, consisting of long glycan strands that are cross-linked by short peptide chains, the mature cell wall is chemically diverse. Peptidoglycan hydrolases and cell wall–tailoring enzymes that regulate glycan strand length, the degree of cross-linking, and the addition of other modifications to peptidoglycan are central in determining the final architecture of the bacterial cell wall. Historically, it has been difficult to biochemically characterize these enzymes that act on peptidoglycan because suitable peptidoglycan substrates were inaccessible. In this review, we discuss fundamental aspects of bacterial cell wall synthesis, describe the regulation and diverse biochemical and functional activities of peptidoglycan hydrolases, and highlight recently developed methods to make and label defined peptidoglycan substrates. We also review how access to these substrates has now enabled biochemical studies that deepen our understanding of how bacterial cell wall enzymes cooperate to build a mature cell wall. Such improved understanding is critical to the development of new antibiotics that disrupt cell wall biogenesis, a process essential to the survival of bacteria.
Keywords: peptidoglycan, Lipid II, hydrolase, teichoic acid, acetyltransferase, antibiotics, cell wall biochemistry, LytR-CpsA-Psr ligases, peptidoglycan hydrolase regulators, peptidoglycan hydrolase, O-acetyltransferase, antibiotic development
Introduction
At least a million different species of bacteria inhabit the earth, and they are incredibly diverse with respect to size, shape, life cycle, and ecological niche (1, 2). Bacteria can be as small as 0.2 μm in diameter or larger than 300 μm across (3). They come in many shapes, including spheres, rods, spirals, crescents, and even stars (4–6). Some bacteria can adopt alternate shapes in response to changes in the environment or during different phases of growth. Although many factors contribute to establishing bacterial size and shape, the peptidoglycan cell wall is central (7).
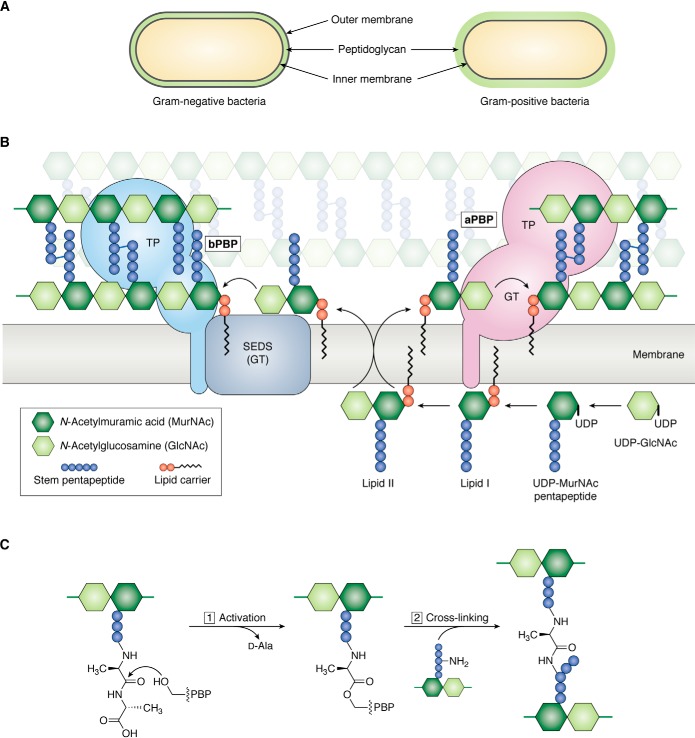
Peptidoglycan is a polymer composed of linear carbohydrate chains held together by peptide cross-links (8). This polymer, which is present in nearly all bacteria, forms a single, enormous macromolecule that surrounds the cell with the glycan chains oriented roughly parallel to the cytoplasmic membrane (9, 10). Gram-negative bacteria have a thin peptidoglycan layer between the inner and outer membrane, whereas Gram-positive bacteria have a thick peptidoglycan cell wall containing many layers of glycan chains (Fig. 1A) (11). Whether thin or thick, the peptidoglycan cell wall is essential not only for bacterial morphology but also for bacterial survival. Indeed, the role of peptidoglycan in bacterial survival has made peptidoglycan biosynthesis a target for important classes of antibiotics, including the β-lactams (e.g. the penicillins, cephalosporins, and carbapenems) and the glycopeptides (e.g. vancomycin) (12). Despite the importance of the cell wall both as a therapeutic target and as the key determinant of bacterial morphology, we still know relatively little about how it is assembled and remodeled during bacterial growth, division, and development (13–15). Moreover, much of what we do know about peptidoglycan assembly and remodeling is based on mutant phenotypes because the development of biochemical tools to study these processes has lagged behind the development of genetic and cell biological tools. Interpreting the biological functions of cell wall enzymes based on mutant phenotypes alone is problematic because these enzymes can have distinct biochemical and functional activities even if the mutants produce similar terminal phenotypes (e.g. cell growth or division defects). With recent advances in obtaining defined substrates for cell wall biochemistry, we are moving toward a more complete understanding of peptidoglycan biogenesis (16). This knowledge will facilitate the development of new antibiotics that specifically target and cripple cell wall assembly to ultimately kill the bacteria.
Figure 1.
Overview of peptidoglycan assembly pathway. A, Gram-negative bacteria have an inner (IM) and outer (OM) cell membrane; a thin layer of peptidoglycan is sandwiched between the cell membranes. Gram-positive bacteria only have an inner cell membrane that is surrounded by a thick layer of peptidoglycan. B, Lipid II, the undecaprenyl pyrophosphate lipid-linked precursor for peptidoglycan synthesis, is assembled on the inner leaflet of the inner cell membrane. Once fully assembled, Lipid II is flipped to the outer leaflet of the inner cell membrane by a flippase. Peptidoglycan GTs polymerize Lipid II into glycan strands, which are cross-linked by transpeptidases (TP) into the existing matrix. There are two families of glycosyltransferases: the GT module of aPBPs and SEDS proteins. Class A PBPs are bifunctional, meaning they have polymerization and cross-linking activities. SEDS proteins cooperate with a partner bPBP, which only has transpeptidase activity. C, the active-site serine of PBP transpeptidases attacks the terminal d-Ala–d-Ala amide bond in a stem peptide (donor), forming an acyl-enzyme covalent intermediate and kicking out the terminal d-Ala. Resolution of the covalent intermediate occurs upon reaction with a nucleophilic amine from an incoming stem peptide (acceptor), producing a new peptide bond that links two glycan strands.
Peptidoglycan is built from a lipid-linked disaccharide-peptide precursor called Lipid II, which is synthesized inside the cell in two stages (Fig. 1B) (17, 18). The first stage takes place in the cytoplasm, where soluble enzymes convert GlcNAc to N-acetylmuramic acid (MurNAc)2-pentapeptide (19). The pentapeptide varies between different bacterial species. The most common structures are l-Ala-d-isoGlu-mesoDAP-d-Ala-d-Ala in Gram-negative organisms and l-Ala-d-isoGlu-l-Lys-d-Ala-d-Ala in Gram-positive organisms. In the next stage, MurNAc-pentapeptide is coupled to a C55-undecaprenyl phosphate carrier lipid that is anchored in the cytoplasmic membrane. This pyrophosphate exchange reaction produces Lipid I, which is converted to Lipid II by the transfer of GlcNAc from UDP-GlcNAc to the C4 hydroxyl of Lipid I. In some species, the stem pentapeptide of Lipid II is additionally tailored. In many Gram-positive organisms, isoGlu at the second position is converted to isoGln by amidation of the α-carboxylate, and a short branch is attached to the l-Lys side chain (12). For example, Staphylococcus aureus has a pentaglycine branch, Streptococcus pneumoniae has an l-Ser-l-Ala or l-Ala-l-Ala branch, Enterococcus faecalis has an l-Ala-l-Ala branch, and Enterococcus faecium has a d-Asp branch. Once fully assembled, Lipid II is exported by the flippase MurJ across the cell membrane for incorporation into the cell wall (20, 21).
Cell wall assembly begins with polymerization of Lipid II by glycosyltransferases (GTs). The GTs add new Lipid II units to the reducing end of a growing glycan chain, releasing the carrier lipid in the process to be recycled back inside the cell (Fig. 1B) (22, 23). The polymer chains are then cross-linked by transpeptidases to the existing peptidoglycan matrix. Cross-linking occurs in two steps, starting with formation of a covalent intermediate via transpeptidase attack of the terminal amide bond in a donor stem peptide (Fig. 1C). A new peptide cross-link is formed when the intermediate reacts with the nucleophilic amine of an adjacent acceptor stem peptide. The transpeptidases are the targets of penicillin and other β-lactam antibiotics, which is why they are referred to as penicillin-binding proteins (PBPs) (24). There are two distinct families of glycosyltransferases that make the peptidoglycan polymers. The best-characterized glycosyltransferases are exemplified by the N-terminal domains of class A penicillin-binding proteins (aPBPs) (25). For over 60 years, it was believed that these glycosyltransferase domains were the only machinery that could polymerize Lipid II. This changed in 2016 when RodA, an essential integral membrane component of the cell elongation machinery, was shown to have Lipid II polymerization activity (26). In 2019, FtsW, a related protein essential for cell division, was also found to polymerize Lipid II (27). These SEDS (shape, elongation, division, and sporulation) proteins are structurally and mechanistically distinct from the aPBPs, and they function in complex with monofunctional bPBP transpeptidases (27–29). It is remarkable that it took this long to identify an enzymatic function for such a highly conserved family of proteins. A major impediment to progress was the lack of sufficient substrates for reconstitution.
The peptidoglycan synthases have been the subject of intense research, but they alone cannot assemble a complete cell wall. Cell wall maturation requires installing tailoring modifications, controlling the degree of cross-linking, trimming glycan strands to their final lengths post-synthesis, and growth-phase remodeling (12, 30). Peptidoglycan hydrolases3 play key roles in cell wall maturation (31–33). Many hydrolases that cleave different bonds in peptidoglycan have been identified and can be readily discovered by mining microbial genomes for characteristic hydrolase domains. However, we are only beginning to understand the many ways that these hydrolases act in cell wall assembly and remodeling. We have also begun to appreciate that understanding hydrolases requires identifying regulatory mechanisms that control their activities. Here we will use selected examples to highlight the activities, biological roles, and regulation of hydrolases. We will show how the ability to obtain defined substrates for biochemical study of hydrolases and other peptidoglycan-modifying enzymes has deepened our understanding of their biological functions.
Peptidoglycan hydrolases have diverse biochemical activities
For virtually every linkage in peptidoglycan, there is a hydrolase with the capacity to cleave it (31). The first peptidoglycan hydrolase was discovered in 1922 by Alexander Fleming, who isolated a bacteriolytic agent that he called “lysozyme” from the nasal mucus of a patient with rhinitis (34). Lysozyme belongs to a class of hydrolases known as the N-acetylmuramidases, which cleave the β-1,4-glycosidic linkage between MurNAc and GlcNAc. New classes of hydrolases have since been discovered.
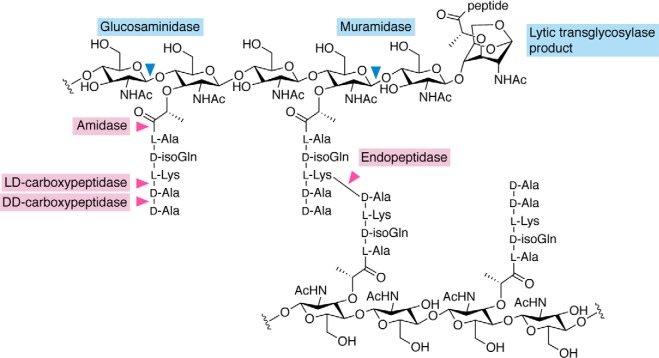
Peptidoglycan hydrolases can be broadly classified as glycosidases or peptidases based on their chemical cleavage specificity (Fig. 2) (31, 35). In addition to N-acetylmuramidases, there are two other types of glycosidases. N-Acetylglucosaminidases cleave the β-1,4-glycosidic bond between GlcNAc and MurNAc. Lytic transglycosylases nonhydrolytically cleave the β-1,4-glycosidic bond between MurNAc and GlcNAc with the concomitant conversion of MurNAc into anhMurNAc (1,6-anhydro-N-acetylmuramic acid) via an intramolecular reaction. Peptidases, on the other hand, attack linkages within the stem peptide of peptidoglycan. N-Acetylmuramoyl-l-alanine amidases release stem peptides from the glycan backbone by hydrolyzing the amide bond between l-Ala at the first position and the lactyl group of MurNAc. dd-Carboxypeptidases are exolytic peptidases that convert stem pentapeptides into tetrapeptides by cleaving the terminal d-Ala–d-Ala bond. There are also ld-carboxypeptidases that further convert stem tetrapeptides into tripeptides by cleaving the l-d bond between mesoDAP or l-Lys at position 3 and d-Ala at position 4. The other amide linkages within the stem peptide, and within interpeptide bridges of cross-linked peptidoglycan, are processed by endopeptidases.
Figure 2.
Cleavage sites of peptidoglycan hydrolases. Enzymes that hydrolyze peptidoglycan are broadly classified as glycosidases and peptidases, depending on where they cleave. Glycosidases (glucosaminidases, muramidases, and lytic transglycosylases) cleave within the glycan backbone. Peptidases (amidases, endopeptidases, ld-carboxypeptidases, and dd-carboxypeptidases) cleave peptide cross-links and within the stem peptides. The carets represent sites of hydrolysis.
Functional diversity of peptidoglycan hydrolases
The biochemical diversity of peptidoglycan hydrolases translates to even greater functional diversity. Bacterial predators such as bacteriophages produce peptidoglycan hydrolases to pierce the cell wall of their hosts during host cell infection (36–38). Bdellovibrio bacteriovorus is another bacterial predator that weaponizes peptidoglycan-modifying enzymes to manipulate the host cell niche (39). To compete with other bacteria, Pseudomonas aeruginosa uses a type VI secretion system to deliver the peptidoglycan hydrolase effectors Tse1 and Tse3 into the periplasmic compartment of an enemy cell, where they chew the cell from within (40). As a first line of defense, we produce lysozyme in our mucus membranes to kill bacterial invaders (34). However, hydrolases are not simply lytic enzymes that destroy the cell wall. Their activities are harnessed to support cell growth, division, and differentiation, enabling bacteria to propagate and adapt to changing environmental conditions (Fig. 3).
Figure 3.
Functions of peptidoglycan hydrolases. Bacterial predators weaponize hydrolases to degrade the peptidoglycan cell wall of their hosts, leading to host cell lysis. But hydrolases are more than just lysins. Bacteria harness endogenous hydrolases to support fundamental cellular processes. Peptidoglycan hydrolases play important roles in bacterial cell growth, differentiation, and the separation of daughter cells that have divided. They also tailor the peptidoglycan cell wall, controlling the length of glycan strands and the degree of cross-linking. Bacteria that recycle components of the cell wall use hydrolases to break the peptidoglycan matrix into smaller pieces that are transported back into the cell.
Many hydrolases function to effect daughter cell separation. When bacteria divide, they form a partition called a septum that is shared between daughter cells (41). Each daughter cell contains a membrane with a shared layer of peptidoglycan that must be split for separation to occur. This splitting is catalyzed by hydrolases. Bacterial mutants lacking splitting hydrolases fail to properly divide and form chains of unseparated cells. For example, an Escherichia coli triple deletion mutant lacking the amidases AmiA/B/C displays a filamentation phenotype in which cells that have divided remain stuck together (42).
Hydrolases are also needed for cell growth and development prior to cell division. As cells grow in size, hydrolases ensure that the peptidoglycan polymer surrounding the bacterium can stretch to accommodate cell expansion. These hydrolases break peptidoglycan cross-links in the existing matrix to make space for insertion of new peptidoglycan (43). For example, in Bacillus subtilis, CwlO and LytE are endopeptidases that break peptidoglycan cross-links along the lateral cell wall to support cell elongation. Deletion of either cwlO or lytE produces shorter cells, and a double deletion mutant is nonviable (44).
Bacteria also deploy hydrolases to chemically modify the structure of peptidoglycan to control its physical properties. Peptidoglycan in different organisms varies considerably in the length of glycan strands and the degree of cross-linking; the identity of the sugar at the ends of glycan strands can also vary. These physical properties affect cell wall rigidity, which in turn can have significant functional consequences. A rigid or stretchy cell wall can be the difference between life or death for a bacterium that is subjected to unfavorable osmotic environments (45). In E. coli, the lytic transglycosylase MltG has been proposed to control strand length by cleaving within a glycan strand. In so doing, MltG produces an anhMurNAc sugar at the reducing end of one strand and a GlcNAc sugar at the nonreducing end of the other strand (46). In S. aureus, the N-acetylglucosaminidase SagB has also been proposed to control strand length by cleaving within a glycan strand. But in this case, SagB produces a GlcNAc sugar at the reducing end of one strand and a MurNAc sugar at the nonreducing end of the other strand (47). The nature of the termini of glycan strands may dictate whether the strands can be further processed by downstream cell wall-tailoring enzymes (48). Of the S. aureus glucosaminidases, SagB is believed to play the predominant role in controlling peptidoglycan strand length, which is a key determinant of cell wall stiffness (49). Another property that determines cell wall stiffness is the degree of peptidoglycan cross-linking (45). Carboxypeptidases that trim stem pentapeptides into tetrapeptides, which can only act as acceptor substrates in dd-transpeptidase reactions, are believed to control the extent of peptidoglycan cross-linking. Other types of hydrolases that act early in cell wall assembly to trim stem peptides may also regulate peptidoglycan cross-linking (31).
Peptidoglycan recycling is another important function of hydrolases. As bacteria grow, they shed components of the cell wall. These turnover products are recycled for reincorporation into the cell wall (50). Finally, peptidoglycan hydrolases mediate transitions between cell states. In sporulating species, hydrolases promote mother cell lysis and spore germination (51, 52). In actinobacteria such as Mycobacterium tuberculosis, muralytic enzymes are involved in resuscitating cells from a dormant state (53, 54).
Peptidoglycan hydrolases therefore play essential and diverse functions across the life cycle of bacteria. What determines the function of any given hydrolase is a combination of intrinsic properties, including whether it recognizes uncross-linked peptidoglycan, cross-linked peptidoglycan, or peptidoglycan that is chemically modified in some other way and its regulation. When and where a hydrolase acts is crucial to its biological function, and hydrolases must be carefully controlled to avoid cell lysis. In the following section, we will summarize some regulatory mechanisms for cell wall hydrolases.
Regulation of peptidoglycan hydrolases
Several mechanisms exist to prevent hydrolases from compromising the structural integrity of the cell. One level of regulation is provided by transcriptional control over the expression of cell wall genes. In B. subtilis, the lytE and cwlO endopeptidase genes are regulated by the WalKR signal transduction pathway that governs cell wall homeostasis (55). In response to an unknown signal generated during cell wall metabolism, lytE and cwlO transcription is regulated to keep pace with growth (56). Protein localization provides another level of control over hydrolases. The activity of cell wall enzymes can be confined to specific sites in the cell. In E. coli, the amidases AmiB and AmiC are recruited to the site of division at midcell, whereas AmiA remains dispersed throughout the periplasm (57). Some hydrolases have tags that direct them to specific compartments of the cell. In S. aureus, the endopeptidase LytN contains a YSIRK signal that exports it to the cross-wall (septum) and a cell wall–binding domain that targets it to peptidoglycan so that it can split daughter cells (58). Proteolysis of a hydrolase can also contribute to its regulation. The steady-state levels of MepS, an E. coli endopeptidase that cleaves cross-links to enable cell wall expansion and consequently cell growth, depend on Prc protease-mediated degradation (59). Bacteria can also chemically modify their cell wall as a mechanism for regulating hydrolases that only act on certain substrates. For example, the peptidoglycan in many organisms is O-acetylated at the C6 hydroxyl group of MurNAc. This modification protects against cleavage by lytic enzymes, such as lysozyme (60), but is also used by some organisms to control the activity of endogenous hydrolases (61). The activity of lytic transglycosylases is especially sensitive to the acetylation state of peptidoglycan due to the nature of the reaction, which involves an intramolecular nucleophilic attack at C1 of MurNAc by the C6 hydroxyl group. Consequently, O-acetylation of the C6 hydroxyl group inhibits lytic transglycosylase activity because an unmodified group is required for the reaction. In Lactobacillus plantarum, high levels of MurNAc acetylation induce autolysis by a putative amidase. The peptidoglycan of L. plantarum is also O-acetylated at GlcNAc residues; this modification prevents cleavage by the glucosaminidase and major autolysin Acm2 (62). The mechanisms by which O-acetylation controls the activities of these endogenous hydrolases are unknown. Finally, it has recently become apparent that many cell wall enzymes are regulated by direct interaction with other proteins. Starting in the late 2000s, regulator proteins that directly interact with hydrolases to inhibit or activate their activities were discovered in rapid succession.
The first protein proposed to be a direct inhibitor of a cell wall hydrolase was IseA in B. subtilis. In 2008, Sekiguchi and co-workers (63) found that IseA inhibits the cell wall lytic activity of the endopeptidase LytF in vitro. Overexpression of IseA in cells phenocopied the cell-chaining defect of a ΔlytF mutant, consistent with a role for IseA in inhibiting LytF. A solution NMR structure of IseA suggested that an acidic loop in IseA reaches into and blocks the positively charged active site pocket of LytF (64). IseA is proposed to inhibit LytF in the transition from vegetative growth to stationary phase when less LytF activity is needed as cell growth slows. A direct inhibitor of a glycosidase has also been reported. This inhibitor, Ivy, is produced by bacteria that do not O-acetylate their peptidoglycan to block the activity of lytic transglycosylases (65).
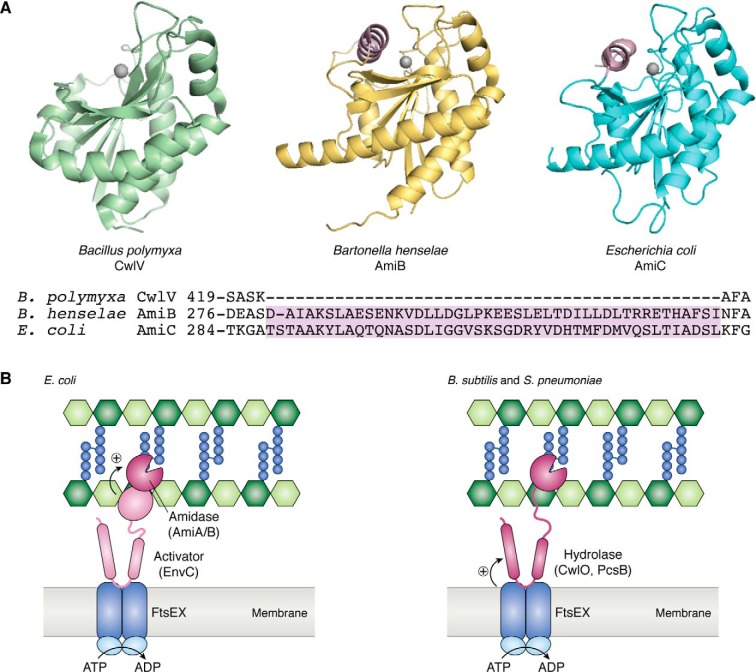
Compared with direct inhibitors, there are more examples of direct activators of peptidoglycan hydrolases. The best-characterized activators are EnvC and NlpD, which control the cell separation amidases AmiA/B/C (66–68). EnvC and NlpD resemble endopeptidases that cleave peptidoglycan cross-links, but the residues required for catalytic activity are missing, which suggested that they have a different function. Genetic studies provided clues to their function. An E. coli ΔenvC ΔnlpD mutant phenocopies a ΔamiABC triple deletion mutant, forming chains of unseparated cells (66). This result implied that EnvC and NlpD act in the same pathways as the amidases. Subsequent work showed that EnvC specifically activates AmiA and AmiB, whereas NlpD activates AmiC (67). Hydrolysis of a crude bacterial cell wall preparation took several hours when the amidases alone were added. However, in the presence of the appropriate activator protein, hydrolysis was rapid. Structural studies later hinted at a mechanism of activation. Crystal structures of AmiB and AmiC showed that their active sites are occluded by an α-helix (Fig. 4A) (69, 70). Amidases such as Bacillus polymyxa CwlV that are active on their own lack this α-helix (69). Moreover, removal of the α-helix sequence from AmiC rendered AmiC highly active even in the absence of its cognate activator NlpD. These data suggested that the amidases switch from an inactive to active conformation when the occluding α-helix is displaced through an interaction with the activator. How EnvC and NlpD physically interact with the amidases to lead to this proposed displacement of the α-helix is unclear (71).
Figure 4.
Direct regulation of peptidoglycan hydrolases. A (top), CwlV (PDB code 1JWQ) is an amidase that is active on its own. AmiB (PDB code 3NE8) and AmiC (PDB code 4BIN) are amidases that are directly activated by a regulator protein (69, 70). A helical domain, shown here in pink, sterically blocks the active site of AmiB and AmiC; displacement of this occluding helix activates the amidases. Bottom, a partial sequence alignment showing that the regulatory helix is present in AmiB and AmiC, but not CwlV. B, the E. coli amidases AmiA and AmiB are activated by direct interaction with EnvC, which itself is thought to be activated by FtsEX. In B. subtilis and S. pneumoniae, FtsEX is believed to regulate the hydrolases CwlO and PcsB, respectively. The ABC transporter FtsEX is presumed to harness ATP hydrolysis to adopt a conformation that is capable of activating the hydrolases.
Until recently, the only other factor proposed to directly activate hydrolases was the widely conserved FtsEX transmembrane complex. FtsEX is an ABC transporter in which FtsE forms the ATP-binding component and FtsX forms the transmembrane channel (72). Rather than acting as a transporter, FtsEX is thought to harness ATP hydrolysis to drive a conformational change in the transmembrane domain that somehow induces hydrolase activity. Two examples of hydrolases regulated by FtsEX are the S. pneumoniae hydrolase PcsB and the elongation-specific endopeptidase CwlO in B. subtilis (Fig. 4B) (73–75). Depletion of pneumococcal FtsEX phenocopies the cell division defects of a PcsB-depletion mutant (73). For B. subtilis, deletion of ftsEX produces shorter cells similar to deletion of cwlO (74, 75). Co-immunoprecipitation and bacterial two-hybrid assays further supported a direct interaction between FtsEX and the hydrolases. Recent structural and biochemical studies have proposed the following model for how the FtsEX complex regulates these hydrolases (76–78). In addition to a catalytic domain, both PcsB and CwlO have a coiled-coil domain, which appears to interact with two extracellular loops of FtsEX. The coiled-coil domain forms a V-shaped cleft that can lock around the catalytic domain like molecular tweezers. Interaction with FtsEX somehow releases the catalytic domain from the cleft to activate the hydrolase. This proposed mechanism for regulation by FtsEX has not been directly demonstrated through reconstitution.
Since the discovery of EnvC, NlpD, and FtsEX, new direct regulators of peptidoglycan hydrolases have been found in other organisms (79, 80). All but one are homologs of EnvC, NlpD, and FtsEX. The exception is a recently discovered S. aureus polytopic membrane protein called ActH, which activates an amidase, LytH, that controls S. aureus cell size (81). The LytH-ActH complex will be described below as the first example of how defined substrates can be used to characterize peptidoglycan hydrolase activity and to discover a new regulator protein. Given the diversity of hydrolases, we expect that other types of direct regulators with distinct structures and mechanisms remain to be identified.
Previous methods to characterize peptidoglycan hydrolases
Peptidoglycan hydrolases have traditionally been characterized using assays that involve bacterial sacculi, which are obtained by boiling bacteria in SDS and deproteinizing. Sacculi are highly heterogeneous and heavily cross-linked. A common assay to detect hydrolase activity is the zymogram (82, 83). Here, purified hydrolases are resolved on SDS-polyacrylamide gels containing sacculi in the gel matrix. Following separation, the gel is incubated in renaturation buffer to promote protein refolding and, as needed, subsequently stained with methylene blue to detect intact sacculi. Methylene blue staining is required to detect Gram-negative, but not Gram-positive, sacculi. Clear bands in the stained gel are interpreted as zones of hydrolase activity leading to cleaved sacculi. Zymography was for a long time the standard technique to assay hydrolase activity despite several drawbacks. One drawback is that charge-based exclusion of methylene blue leads to false positive results (84). As one example, E. coli EnvC was initially reported to have hydrolase activity by zymogram, but subsequent studies found that the signal was an artifact (67, 85). Another drawback is that zymograms cannot detect hydrolases that require an activator. The zymogram assay is also poorly suited for hydrolases that cannot spontaneously refold.
An alternative method to monitor peptidoglycan hydrolysis makes use of the dye Remazol Brilliant Blue (RBB), which under basic conditions forms a vinylsulfone species that can react with peptidoglycan (86). Isolated sacculi labeled with RBB are treated with a purified hydrolase, and reactions are terminated and centrifuged to pellet the undigested sacculi. The amount of dye released into the supernatant reports on hydrolytic activity. One drawback is that hydrolases that cannot act on cross-linked substrate would appear inactive in this dye release assay. Moreover, the RBB labeling is heterogeneous and, like other tailoring modifications, may interfere with the activities of some hydrolases. This assay also provides no information on the chemical cleavage specificity and product species. More recently, sacculi hydrolysis has been combined with LC-MS analysis to characterize the muropeptide cleavage products (87). But even with LC-MS analysis, the substrate preference of a hydrolase can only be indirectly deduced based on detectable changes in muropeptide species. The deficiencies in these assays underscore the need to obtain defined peptidoglycan substrates.
In vitro assembly of peptidoglycan enables characterization of cell wall hydrolases and tailoring enzymes
To study peptidoglycan hydrolases or tailoring enzymes ideally requires defined substrates and methods to detect reaction. This has been a challenge for several reasons. First, the peptidoglycan precursor, Lipid II, historically has been very challenging to obtain in practical quantities for biochemistry (88). Second, methods to build peptidoglycan substrates from Lipid II and label them for detection were lacking.
Starting in the late 1990s, several methods to obtain Lipid II were developed (89–94). The first methods involved chemical synthesis, but enzymatic methods were subsequently developed. These methods only provided access to Lipid II for research groups with the appropriate expertise, and even then, considerable effort was required. Moreover, each synthetic route was designed to make a particular Lipid II molecule. Painstaking modifications were required to acquire other variants. Because Lipid II structures vary and cross-linked peptidoglycan can only be made from variants having the correct stem peptide, the lack of a ready, general solution to obtain substrates hampered progress. This obstacle to progress has now been largely overcome.
The first finding that led to a breakthrough was that Lipid II can be accumulated in cells. Steady-state levels of the cell wall precursor Lipid II are normally low, but levels of undecaprenyl-phosphate, the carrier lipid on which Lipid II is built, are estimated at 1–3 × 105 molecules/cell (17, 18, 95). Therefore, cells have the capacity to make substantially more Lipid II. Antibiotics that block Lipid II utilization (e.g. moenomycin or vancomycin) were shown to cause a rapid (10–20 min), substantial increase in Lipid II pools (96, 97). The next challenge was to identify a strategy to isolate Lipid II. Because it has a C55 lipid chain, Lipid II is difficult to separate from other cellular lipids. However, it was found that Lipid II can be isolated at reasonable purity from large-scale bacterial cultures using a two-step extraction (Fig. 5A) (98). Lipid II accumulation and extraction have been successfully adapted to isolate Lipid II variants from diverse species, including S. aureus, B. subtilis, E. coli, S. pneumoniae, E. faecalis, and Mycobacterium smegmatis (Fig. 5B) (98–101). Lipid II has also been isolated from mutant strains of some of these species, expanding the repertoire of Lipid II structural variants. In S. aureus, the Fem proteins sequentially install five glycines to the side chain of l-Lys in the stem peptide: FemX adds the first glycine, FemA adds the second and third glycines, and FemB adds the final two glycines. Lipid II-monoglycine and Lipid II-triglycine have been prepared from S. aureus ΔfemAB and ΔfemB mutants, respectively (99). Similarly, an S. pneumoniae ΔmurMN mutant provides access to Lipid II with unmodified lysine at the third position (100). With sufficient quantities of different Lipid II in hand, previously inaccessible peptidoglycan substrates can now be easily obtained.
Figure 5.
Making and labeling peptidoglycan substrates to characterize cell wall enzymes. A, accumulation and extraction of Lipid II. Bacterial cultures are treated with an antibiotic that inhibits peptidoglycan synthesis to accumulate Lipid II in cells. The cells are spun down and resuspended in chloroform/methanol for the first extraction, which produces three layers. Lipid II is enriched in a thick interface fraction, whereas the majority of cellular phospholipids partition to the organic layer. A second extraction results in partitioning of Lipid II into the organic phase and UDP-MurNAc-pentapeptide, which is also present in the interface layer, into the aqueous phase. B, the two-step extraction allows isolation of Lipid II variants from the indicated species. C, uncross-linked glycan strands are synthesized from Lipid II using the monofunctional glycosyltransferase SgtB, which recognizes Lipid II variants with different stem peptides. To make cross-linked peptidoglycan, an appropriate bifunctional aPBP that recognizes the stem peptide is used. The aPBP polymerizes Lipid II and cross-links glycan strands. D, a chemical probe can be incorporated within the glycan backbone or stem peptide of peptidoglycan substrates to visualize and assess reaction of the substrates. i, one strategy to label the stem peptide of Lipid II or glycan strands makes use of the transpeptidase PBPX, which exchanges the terminal d-Ala in the stem peptide for a d-amino acid bearing a detectable tag. ii, another stem peptide labeling strategy selectively couples an amine-reactive probe to the side-chain primary amine at position 3 of the stem peptide. iii, a strategy to label the sugar backbone uses GalT to attach a [14C]galactose radiolabel at the nonreducing end of glycan strands. E, methods to digest peptidoglycan products for structural characterization by LC-MS. i, mutanolysin digestion and NaBH4 reduction of cross-linked peptidoglycan produce muropeptide species. ii, ColM treatment of uncross-linked glycan strands produces delipidated glycan diphosphate products.
Methods have been developed to build both uncross-linked and cross-linked peptidoglycan from Lipid II and to label the substrates for detection. To make uncross-linked glycan strands, Lipid II is polymerized with a monofunctional glycosyltransferase, such as S. aureus SgtB (Fig. 5C) (102). As this enzyme does not recognize the stem peptide, it can be used to polymerize diverse Lipid II variants. Glycan strand length can be controlled to some extent using glycosyltransferase mutants with processivity defects. Cross-linked peptidoglycan is made using an appropriate PBP that recognizes the stem peptide (98, 99, 103). Peptidoglycan substrates synthesized in vitro can be detected by incorporating a chemical probe within the glycan backbone or stem peptide (Fig. 5D). One approach for stem peptide labeling uses PBPX, a functionally unique transpeptidase from E. faecalis that does not cross-link peptidoglycan, to catalyze d-amino acid exchange (100). This reaction leads to an exchange of the terminal d-Ala on stem peptides for d-amino acids bearing a detectable tag. Alternatively, stem peptides can be labeled at the side-chain primary amine of the third amino acid with an amine-reactive probe (104, 105). These two strategies work for labeling Lipid II prior to polymerization or the stem peptides of preassembled glycan strands. Making labeled cross-linked peptidoglycan is also relatively straightforward. By incubating a d-amino acid probe with Lipid II and an appropriate PBP that both cross-links peptidoglycan and performs d-amino acid exchange, the PBP incorporates the probe as it synthesizes peptidoglycan (98). These labeling methods make it possible to detect reaction of peptidoglycan substrates in downstream enzymatic assays. In cases where stem peptide labeling interferes with enzymes that distinguish different stem peptides, a probe can instead be incorporated within the glycan backbone. One simple strategy uses the bovine β-1,4-galactosyltransferase GalT to attach a [14C]galactose radiolabel onto the C4 hydroxyl of GlcNAc at the nonreducing end of glycan strands (106).
In addition to labeling the substrates for detection, analytical methods allow for structural characterization of the peptidoglycan products (Fig. 5E). High-resolution MS is typically used for characterization because it can detect low amounts of material. For analysis of cross-linked peptidoglycan, material is first digested with mutanolysin, an N-acetylmuramidase that cleaves between MurNAc and GlcNAc units (98). Because cleavage results in anomeric mixtures of muropeptides with different retention times, the mutanolysin digests are reduced with sodium borohydride prior to LC-MS analysis. For uncross-linked peptidoglycan, degradation methods that remove the lipid from the reducing end of the polymers are required for good ionization. One method uses acid hydrolysis to remove the lipid but can produce mixtures of glycan mono- and diphosphates (98). An enzymatic method makes use of the protein toxin colicin M (ColM) from E. coli, which was originally shown to cleave Lipid II and yield a 1-pyrophospho-MurNAc-(peptide)-GlcNAc product (107). It has recently been shown that ColM can also cleave the lipid tail from uncross-linked glycan strands, generating glycan diphosphate products (23).
The ability to make labeled, defined peptidoglycan substrates now enables a systematic characterization of hydrolase activity (108). In the next section, we will highlight recent studies that have used defined substrates to characterize peptidoglycan hydrolases and cell wall–tailoring enzymes.
Connecting cell wall biochemistry with biological function
Some cell wall hydrolases act on cross-linked peptidoglycan, whereas others can only cleave uncross-linked substrates. Still others may cleave only substrates having particular stem peptides and tailoring modifications. Proving the biochemical activities of these enzymes is needed to uncover their functions in cells. The trend toward building substrates in vitro for biochemical reconstitution has started and already yielded new insights into cell wall biogenesis.
As one example, Lee et al. (48) reconstituted the activities of all known lytic transglycosylases from P. aeruginosa using defined peptidoglycan substrates. They showed that the lytic transglycosylases differ in substrate recognition. Some required an anhMurNAc terminus at the +2 position (i.e. two sugars toward the reducing end) from the cleavage site. One lytic transglycosylase, RlpA, preferred cleaving glycan strands lacking stem peptides (109). RlpA must therefore work in tandem with amidases in cells. Only some of these reconstituted lytic transglycosylases have been functionally characterized in cells. Understanding which substrates are recognized will help elucidate the biological functions of the remaining glycosidases.
A second example relates to a recent study that used synthetic peptidoglycan fragments to define the binding mechanism of the endopeptidase lysostaphin (110). Produced by Staphylococcus simulans biovar staphylolyticus, lysostaphin shows highly specific and potent lytic activity against other staphylococci (111). Lysostaphin has an SH3b cell wall–binding domain that recognizes the pentaglycine cross-bridges present exclusively in staphylococcal peptidoglycan, positioning the enzyme to cleave the cross-bridges. Within the SH3b domain resides a shallow groove that appears to sterically select for pentaglycine (112). Just this year, Gonzalez-Delgado et al. (110) discovered that the SH3b domain drives not only the specificity, but also high-affinity binding, of lysostaphin toward staphylococcal peptidoglycan. A co-crystal structure of the SH3b domain with a tetrapeptide-pentaglycine ligand revealed two separate binding sites that are on opposite sides of each SH3b monomer. Surprisingly, the ligand was found to simultaneously bind two SH3b monomers: the pentaglycine branch sits within one pocket of the first SH3b monomer, and the tetrapeptide stem protrudes into the opposite pocket on a second SH3b monomer. The authors proposed that this two-site binding mechanism enables lysostaphin to cluster on staphylococcal peptidoglycan, leading to the observed potency in lytic activity. Therefore, juxtaposition of substrate-binding sites serves as another way to regulate peptidoglycan hydrolase activity.
Another example is provided by the recently discovered S. aureus amidase-activator complex, LytH-ActH (Fig. 6) (81). LytH is a membrane-anchored amidase that was shown to remove stem peptides exclusively from uncross-linked glycan strands. This substrate preference suggested that LytH acts early in cell division because amidases that effect cell separation would need to cleave cross-linked peptidoglycan (113, 114). Accordingly, deletion of lytH led to defects in placement of nascent division sites as opposed to the final cell separation defects observed for mutants of amidases that separate daughter cells after cell division is complete. LytH trims stem peptides to control the density of peptidoglycan assembly sites at a given subcellular location. In doing so, LytH spatially regulates peptidoglycan synthesis to ensure cell growth is coordinated with cell division. Importantly, LytH was active only in the presence of a polytopic membrane protein partner, ActH, that was identified by co-immunoprecipitation. ActH is structurally distinct from EnvC, NlpD, and FtsEX and resembles a rhomboid serine protease, a family of proteins that is widespread in bacteria but whose biological roles have remained largely mysterious (115, 116). As such, ActH represents a new class of regulators that directly activate a partner cell wall hydrolase. Defined peptidoglycan substrates were instrumental in characterizing the LytH-ActH complex, as the complex only cleaves uncross-linked peptidoglycan. This work exemplifies how establishing the substrate preference of an enzyme is crucial in determining its biological function. How ActH activates LytH remains to be established.
Figure 6.
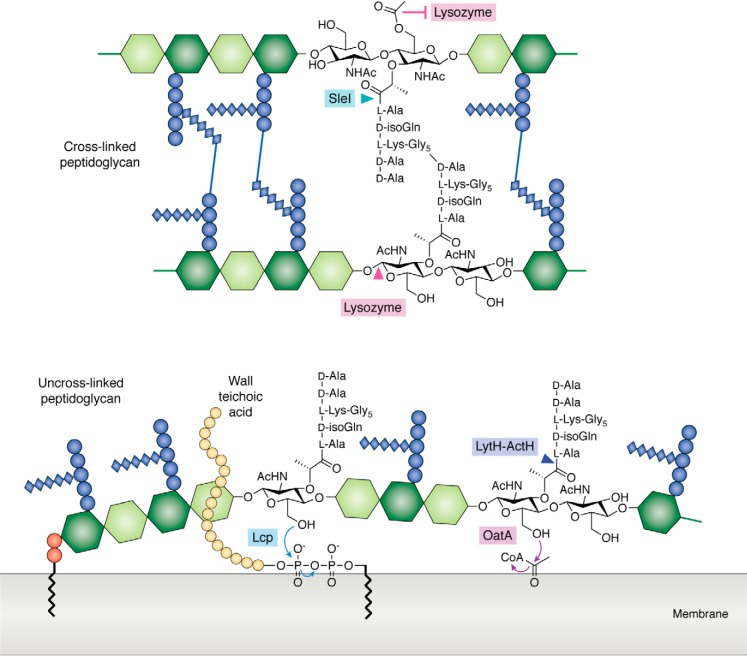
Peptidoglycan hydrolases and tailoring enzymes have distinct substrate preferences. In S. aureus, the LCP wall teichoic acid ligases and the LytH-ActH amidase-activator complex are membrane-anchored proteins that only act on uncross-linked peptidoglycan substrates. The S. aureus O-acetyltransferase OatA is also a membrane-anchored protein and may act preferentially on uncross-linked peptidoglycan. By contrast, S. aureus Sle1 is an amidase that acts on cross-linked peptidoglycan. Another example of substrate selectivity is provided by lysozyme, which cleaves unacetylated, but not O-acetylated, backbones. Substrate selectivity can offer insights into the functions of these enzymes, as those enzymes that act at an earlier stage of peptidoglycan synthesis may show a preference for nascent peptidoglycan substrates.
Access to defined substrates has advanced studies of other cell wall enzymes besides hydrolases that act on peptidoglycan (Fig. 6). In some species, the peptidoglycan is decorated with additional chemical modifications that are important for bacterial physiology (12). O-Acetylation of the MurNAc C6 hydroxyl and N-deacetylation of the GlcNAc C2 acetyl are two examples (117, 118). Both of these modifications protect pathogens that colonize a host from host-derived lysozyme. Other examples of tailoring modifications are the covalent attachment of glycopolymers, including wall teichoic acid (WTA), teichuronic acid, and capsular polysaccharide (CPS), to the glycan backbone of peptidoglycan (30, 119). These polymers are important for pathogenesis, contributing to immune evasion and host colonization. Other functions of these polymers include scaffolding cell wall proteins, conferring cell morphology, and protecting cells against antibiotics. A detailed characterization of the kinetics and substrate preferences of these cell wall–tailoring enzymes will help establish their biological roles. Once enzymatic activity has been confirmed in vitro, homologs can also be identified and rapidly characterized in other organisms.
Recent work has shown that peptidoglycan-tailoring enzymes exhibit clear substrate preferences, which influence when they act during cell wall assembly. Differences in substrate preferences between homologs may also reveal distinct biological roles. One class of enzymes for which substrate preferences have been studied are the LCP (LytR-CpsA-Psr) proteins that couple both WTA and CPS glycopolymers to peptidoglycan. For a long time, the identity of the coupling enzymes was not known. Genetic studies in 2011 and 2014 suggested that the LCP proteins could be the WTA and CPS ligases, but reconstitution to demonstrate ligase activity was not done because appropriate substrates were unavailable (120, 121). Recently, these activities were confirmed biochemically, when it was also found that the LCP proteins have strict substrate preferences. S. aureus LcpB, a prototypical LCP enzyme, only attaches WTA glycopolymers to uncross-linked peptidoglycan oligomers that are at least 4 sugars long (122). Neither Lipid II nor cross-linked peptidoglycan were substrates (123). Similarly, evidence suggests that S. aureus LcpC transfers CPS at an early stage of peptidoglycan synthesis as well (124). In this case, LcpC appears to couple CPS to Lipid II, although the products were not characterized. Structural studies have revealed a possible mechanism for substrate selection by the LCP proteins based on steric exclusion (123). The LCP protein has an extended narrow groove that likely excludes cross-linked substrates. Similar to LCP proteins, O-acetylation enzymes that install an acetyl group on the C6 hydroxyl of MurNAc also discriminate between different peptidoglycan substrates. The Gram-negative O-acetyltransferase PatB can only acetylate substrates that are at least 3 sugars in length (125, 126). The S. aureus O-acetyltransferase OatA prefers to acetylate uncross-linked glycan strands containing intact stem pentapeptides as opposed to those with trimmed peptides (127). Whether OatA can transfer acetyl groups to cross-linked peptidoglycan was not tested.
These mechanistic studies of LCP proteins and O-acetyltransferases have important biological implications for ordering the steps in cell wall assembly. In the final stages of cell wall assembly, several biosynthetic pathways converge to produce the mature cell wall (30). However, the sequence of cell wall assembly has been unclear. After Lipid II is transported across the inner cell membrane, it is polymerized into glycan strands that are cross-linked into the peptidoglycan matrix. In most cases, it is not known when tailoring modifications are made to peptidoglycan during its assembly. In principle, modifications may be added to Lipid II, uncross-linked polymers, and/or cross-linked polymers. One way to address the order of cell wall assembly is through in vitro reconstitution using a range of peptidoglycan intermediates. This type of logic has been commonly used in the field of natural product biosynthesis, where elucidating the order of assembly often relies on testing the ability of a biosynthetic enzyme to convert a pathway intermediate to a product that can be used in the subsequent step. In S. aureus, WTA glycopolymers can only be transferred to nascent (uncross-linked) peptidoglycan strands, and it has been shown that these WTA-modified polymers can then be cross-linked (123). Indeed, it has even been proposed that WTAs regulate the localization of the PBP4 transpeptidase to control peptidoglycan cross-linking (128). The clear preference of WTA ligases for modifying nascent peptidoglycan is consistent with their membrane-anchored location. S. aureus OatA is also anchored in the membrane and may also display a preference for uncross-linked peptidoglycan, although detailed studies of its substrate preferences that could help elucidate when it acts have not been done (127). The studies described here exemplify how using defined substrates that mimic peptidoglycan intermediates can provide temporal information on cell wall assembly. Moreover, access to peptidoglycan species containing post-synthetic modifications expands the ensemble of substrates available to assess cell wall hydrolase activity.
Conclusions and outlook
Nearly all bacteria are encased in layers of peptidoglycan, which is a central determinant of cell morphology (4–7). The basic structure of peptidoglycan is a simple carbohydrate backbone with peptide cross-links that are relatively conserved across bacterial species (8). Yet there is a vast diversity in the structure of the mature cell wall, owing to tailoring modifications and the trimming of peptidoglycan by hydrolases (12, 30). Peptidoglycan hydrolases are essential for cell wall biogenesis (31–33). However, due to their destructive potential, these hydrolases must be regulated to prevent cell wall damage. Elegant studies from the past decade established that bacteria encode regulator proteins that directly interact with hydrolases to control their activity. Yet only a few types of direct regulators have been identified despite the diversity of hydrolases. In part, the search for new regulators has been hindered by a lack of suitable peptidoglycan substrates to evaluate hydrolase activity. This problem has now been solved with new approaches to rapidly access large quantities of Lipid II variants from different bacterial species. Using Lipid II as a building block, uncross-linked and cross-linked peptidoglycan can be assembled in vitro and labeled for detection. Studies using these defined substrates have uncovered a new class of direct regulators of peptidoglycan hydrolases and provided mechanistic insights into other aspects of cell wall biogenesis.
Looking forward, there are many open questions that these new tools can help answer. A robust cell wall depends on the ordered convergence of multiple biosynthetic pathways. Many of the enzymes involved in cell wall assembly have been identified or can be computationally predicted, but we have yet to fully understand their biological functions. Determining when, where, and on what these enzymes act in cells is necessary to elucidate their functions. In the past decade, new microscopy techniques to track cell wall proteins have been developed that will provide valuable spatiotemporal information (129). The tools we outlined here to biochemically characterize cell wall enzymes will complement future imaging studies by reporting on substrate specificity. Finally, the ability to obtain defined substrates will also be useful for developing sensitive, high-throughput assays to monitor cell wall enzyme kinetics and screen for inhibitors. For peptidoglycan hydrolases, there are a few reported activity assays, but all use crude cell wall substrates. These assays can now be revisited with the tools we have described in hand, facilitating the identification of new antibiotics that disrupt the activity of enzymes essential to the assembly of a complete cell wall.
This work was supported by National Institutes of Health Grants R01 GM076710 and R01 AI148752 (to S. W.), T32GM007753 (to J. E. P.), and a National Science Foundation graduate research fellowship under Grant DGE1144152 (to T. D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
In this review, the term hydrolase refers to lytic enzymes that are hydrolases or lyases (e.g. lytic transglycosylases).
- MurNAc
- N-acetylmuramic acid
- GT
- glycosyltransferase
- PBP
- penicillin-binding protein
- aPBP and bPBP
- class A and B PBP, respectively
- RBB
- Remazol Brilliant Blue
- WTA
- wall teichoic acid
- CPS
- capsular polysaccharide
- SEDS
- shape, elongation, division, and sporulation
- ColM
- colicin M
- PDB
- Protein Data Bank
- anhMurNAc
- 1,6-anhydro-N-acetylmuramic acid.
References
- 1. Schloss P. D., Girard R. A., Martin T., Edwards J., and Thrash J. C. (2016) Status of the archaeal and bacterial census: an update. MBio 7, e00201–16 10.1128/mBio.00201-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bar-On Y. M., Phillips R., and Milo R. (2018) The biomass distribution on Earth. Proc. Natl. Acad. Sci. U.S.A. 115, 6506–6511 10.1073/pnas.1711842115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schulz H. N., and Jorgensen B. B. (2001) Big bacteria. Annu. Rev. Microbiol. 55, 105–137 10.1146/annurev.micro.55.1.105 [DOI] [PubMed] [Google Scholar]
- 4. Young K. D. (2006) The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70, 660–703 10.1128/MMBR.00001-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Young K. D. (2007) Bacterial morphology: why have different shapes? Curr. Opin. Microbiol. 10, 596–600 10.1016/j.mib.2007.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang D. C., Blair K. M., and Salama N. R. (2016) Staying in shape: the impact of cell shape on bacterial survival in diverse environments. Microbiol. Mol. Biol. Rev. 80, 187–203 10.1128/MMBR.00031-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cabeen M. T., and Jacobs-Wagner C. (2005) Bacterial cell shape. Nat. Rev. Microbiol. 3, 601–610 10.1038/nrmicro1205 [DOI] [PubMed] [Google Scholar]
- 8. Vollmer W., Blanot D., and de Pedro M. A. (2008) Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 10.1111/j.1574-6976.2007.00094.x [DOI] [PubMed] [Google Scholar]
- 9. Gan L., Chen S., and Jensen G. J. (2008) Molecular organization of Gram-negative peptidoglycan. Proc. Natl. Acad. Sci. U.S.A. 105, 18953–18957 10.1073/pnas.0808035105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim S. J., Chang J., and Singh M. (2015) Peptidoglycan architecture of Gram-positive bacteria by solid-state NMR. Biochim. Biophys. Acta 1848, 350–362 10.1016/j.bbamem.2014.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Silhavy T. J., Kahne D., and Walker S. (2010) The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 10.1101/cshperspect.a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rajagopal M., and Walker S. (2017) Envelope structures of Gram-positive bacteria. Curr. Top. Microbiol. Immunol. 404, 1–44 10.1007/82_2015_5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Typas A., Banzhaf M., Gross C. A., and Vollmer W. (2011) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136 10.1038/nrmicro2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Egan A. J. F., Cleverley R. M., Peters K., Lewis R. J., and Vollmer W. (2017) Regulation of bacterial cell wall growth. FEBS J. 284, 851–867 10.1111/febs.13959 [DOI] [PubMed] [Google Scholar]
- 15. Zhao H., Patel V., Helmann J. D., and Dörr T. (2017) Don't let sleeping dogmas lie: new views of peptidoglycan synthesis and its regulation. Mol. Microbiol. 106, 847–860 10.1111/mmi.13853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Taguchi A., Kahne D., and Walker S. (2019) Chemical tools to characterize peptidoglycan synthases. Curr. Opin. Chem. Biol. 53, 44–50 10.1016/j.cbpa.2019.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Heijenoort J. (2007) Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol. Mol. Biol. Rev. 71, 620–635 10.1128/MMBR.00016-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bouhss A., Trunkfield A. E., Bugg T. D. H., and Mengin-Lecreulx D. (2008) The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol. Rev. 32, 208–233 10.1111/j.1574-6976.2007.00089.x [DOI] [PubMed] [Google Scholar]
- 19. Barreteau H., Kovač A., Boniface A., Sova M., Gobec S., and Blanot D. (2008) Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 168–207 10.1111/j.1574-6976.2008.00104.x [DOI] [PubMed] [Google Scholar]
- 20. Ruiz N. (2008) Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 105, 15553–15557 10.1073/pnas.0808352105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sham L.-T., Butler E. K., Lebar M. D., Kahne D., Bernhardt T. G., and Ruiz N. (2014) MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345, 220–222 10.1126/science.1254522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perlstein D. L., Zhang Y., Wang T.-S., Kahne D. E., and Walker S. (2007) The direction of glycan chain elongation by peptidoglycan glycosyltransferases. J. Am. Chem. Soc. 129, 12674–12675 10.1021/ja075965y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Welsh M. A., Schaefer K., Taguchi A., Kahne D., and Walker S. (2019) Direction of chain growth and substrate preferences of shape, elongation, division, and sporulation-family peptidoglycan glycosyltransferases. J. Am. Chem. Soc. 141, 12994–12997 10.1021/jacs.9b06358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sauvage E., Kerff F., Terrak M., Ayala J. A., and Charlier P. (2008) The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 234–258 10.1111/j.1574-6976.2008.00105.x [DOI] [PubMed] [Google Scholar]
- 25. Goffin C., and Ghuysen J. M. (1998) Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62, 1079–1093 10.1128/MMBR.62.4.1079-1093.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meeske A. J., Riley E. P., Robins W. P., Uehara T., Mekalanos J. J., Kahne D., Walker S., Kruse A. C., Bernhardt T. G., and Rudner D. Z. (2016) SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 10.1038/nature19331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taguchi A., Welsh M. A., Marmont L. S., Lee W., Sjodt M., Kruse A. C., Kahne D., Bernhardt T. G., and Walker S. (2019) FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol. 4, 587–594 10.1038/s41564-018-0345-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rohs P. D. A., Buss J., Sim S. I., Squyres G. R., Srisuknimit V., Smith M., Cho H., Sjodt M., Kruse A. C., Garner E. C., Walker S., Kahne D. E., and Bernhardt T. G. (2018) A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLoS Genet. 14, e1007726 10.1371/journal.pgen.1007726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reichmann N. T., Tavares A. C., Saraiva B. M., Jousselin A., Reed P., Pereira A. R., Monteiro J. M., Sobral R. G., VanNieuwenhze M. S., Fernandes F., and Pinho M. G. (2019) SEDS-bPBP pairs direct lateral and septal peptidoglycan synthesis in Staphylococcus aureus. Nat. Microbiol. 4, 1368–1377 10.1038/s41564-019-0437-2 [DOI] [PubMed] [Google Scholar]
- 30. Hanson B. R., and Neely M. N. (2012) Coordinate regulation of Gram-positive cell surface components. Curr. Opin. Microbiol. 15, 204–210 10.1016/j.mib.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vollmer W., Joris B., Charlier P., and Foster S. (2008) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286 10.1111/j.1574-6976.2007.00099.x [DOI] [PubMed] [Google Scholar]
- 32. Uehara T., and Bernhardt T. G. (2011) More than just lysins: peptidoglycan hydrolases tailor the cell wall. Curr. Opin. Microbiol. 14, 698–703 10.1016/j.mib.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wyckoff T. J., Taylor J. A., and Salama N. R. (2012) Beyond growth: novel functions for bacterial cell wall hydrolases. Trends Microbiol. 20, 540–547 10.1016/j.tim.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fleming A. (1922) On a remarkable bacteriolytic element found in tissues and secretions. Proc. R. Soc. B Biol. Sci. 93, 306–317 10.1098/rspb.1922.0023 [DOI] [Google Scholar]
- 35. Vermassen A., Leroy S., Talon R., Provot C., Popowska M., and Desvaux M. (2019) Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front. Microbiol. 10, 331 10.3389/fmicb.2019.00331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krause R. M. (1957) Studies on bacteriophages of hemolytic streptococci. I. Factors influencing the interaction of phage and susceptible host cell. J. Exp. Med. 106, 365–384 10.1084/jem.106.3.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nelson D., Schuch R., Chahales P., Zhu S., and Fischetti V. A. (2006) PlyC: a multimeric bacteriophage lysin. Proc. Natl. Acad. Sci. U.S.A. 103, 10765–10770 10.1073/pnas.0604521103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gilmer D. B., Schmitz J. E., Euler C. W., and Fischetti V. A. (2013) Novel bacteriophage lysin with broad lytic activity protects against mixed infection by Streptococcus pyogenes and methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 57, 2743–2750 10.1128/AAC.02526-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lerner T. R., Lovering A. L., Bui N. K., Uchida K., Aizawa S.-I., Vollmer W., and Sockett R. E. (2012) Specialized peptidoglycan hydrolases sculpt the intra-bacterial niche of predatory Bdellovibrio and increase population fitness. PLoS Pathog. 8, e1002524 10.1371/journal.ppat.1002524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Russell A. B., Hood R. D., Bui N. K., LeRoux M., Vollmer W., and Mougous J. D. (2011) Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 10.1038/nature10244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adams D. W., and Errington J. (2009) Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7, 642–653 10.1038/nrmicro2198 [DOI] [PubMed] [Google Scholar]
- 42. Heidrich C., Templin M. F., Ursinus A., Merdanovic M., Berger J., Schwarz H., de Pedro M. A., and Höltje J. V. (2001) Involvement of N-acetylmuramyl-l-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41, 167–178 10.1046/j.1365-2958.2001.02499.x [DOI] [PubMed] [Google Scholar]
- 43. Vollmer W. (2012) Bacterial growth does require peptidoglycan hydrolases. Mol. Microbiol. 86, 1031–1035 10.1111/mmi.12059 [DOI] [PubMed] [Google Scholar]
- 44. Hashimoto M., Ooiwa S., and Sekiguchi J. (2012) Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of d,l-endopeptidase activity at the lateral cell wall. J. Bacteriol. 194, 796–803 10.1128/JB.05569-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Auer G. K., and Weibel D. B. (2017) Bacterial cell mechanics. Biochemistry 56, 3710–3724 10.1021/acs.biochem.7b00346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yunck R., Cho H., and Bernhardt T. G. (2016) Identification of MltG as a potential terminase for peptidoglycan polymerization in bacteria. Mol. Microbiol. 99, 700–718 10.1111/mmi.13258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chan Y. G. Y., Frankel M. B., Missiakas D., and Schneewind O. (2016) SagB glucosaminidase is a determinant of Staphylococcus aureus glycan chain length, antibiotic susceptibility, and protein secretion. J. Bacteriol. 198, 1123–1136 10.1128/JB.00983-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee M., Hesek D., Dik D. A., Fishovitz J., Lastochkin E., Boggess B., Fisher J. F., and Mobashery S. (2017) From genome to proteome to elucidation of reactions for all eleven known lytic transglycosylases from Pseudomonas aeruginosa. Angew. Chem. Int. Ed. Engl. 56, 2735–2739 10.1002/anie.201611279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wheeler R., Turner R. D., Bailey R. G., Salamaga B., Mesnage S., Mohamad S. A. S., Hayhurst E. J., Horsburgh M., Hobbs J. K., and Foster S. J. (2015) Bacterial cell enlargement requires control of cell wall stiffness mediated by peptidoglycan hydrolases. MBio 6, e00660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Johnson J. W., Fisher J. F., and Mobashery S. (2013) Bacterial cell-wall recycling. Ann. N.Y. Acad. Sci. 1277, 54–75 10.1111/j.1749-6632.2012.06813.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Smith T. J., and Foster S. J. (1995) Characterization of the involvement of two compensatory autolysins in mother cell lysis during sporulation of Bacillus subtilis 168. J. Bacteriol. 177, 3855–3862 10.1128/JB.177.13.3855-3862.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Popham D. L., Gilmore M. E., and Setlow P. (1999) Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties. J. Bacteriol. 181, 126–132 10.1128/JB.181.1.126-132.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mukamolova G. V., Kaprelyants A. S., Young D. I., Young M., and Kell D. B. (1998) A bacterial cytokine. Proc. Natl. Acad. Sci. U.S.A. 95, 8916–8921 10.1073/pnas.95.15.8916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mukamolova G. V., Murzin A. G., Salina E. G., Demina G. R., Kell D. B., Kaprelyants A. S., and Young M. (2006) Muralytic activity of Micrococcus luteus Rpf and its relationship to physiological activity in promoting bacterial growth and resuscitation. Mol. Microbiol. 59, 84–98 10.1111/j.1365-2958.2005.04930.x [DOI] [PubMed] [Google Scholar]
- 55. Bisicchia P., Noone D., Lioliou E., Howell A., Quigley S., Jensen T., Jarmer H., and Devine K. M. (2007) The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol. Microbiol. 65, 180–200 10.1111/j.1365-2958.2007.05782.x [DOI] [PubMed] [Google Scholar]
- 56. Dobihal G. S., Brunet Y. R., Flores-Kim J., and Rudner D. Z. (2019) Homeostatic control of cell wall hydrolysis by the WalRK two-component signaling pathway in Bacillus subtilis. Elife 8, e52088 10.7554/eLife.52088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bernhardt T. G., and de Boer P. A. J. (2003) The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48, 1171–1182 10.1046/j.1365-2958.2003.03511.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Frankel M. B., Hendrickx A. P. A., Missiakas D. M., and Schneewind O. (2011) LytN, a murein hydrolase in the cross-wall compartment of Staphylococcus aureus, is involved in proper bacterial growth and envelope assembly. J. Biol. Chem. 286, 32593–32605 10.1074/jbc.M111.258863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Singh S. K., Parveen S., SaiSree L., and Reddy M. (2015) Regulated proteolysis of a cross-link-specific peptidoglycan hydrolase contributes to bacterial morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 112, 10956–10961 10.1073/pnas.1507760112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bera A., Herbert S., Jakob A., Vollmer W., and Götz F. (2005) Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55, 778–787 10.1111/j.1365-2958.2004.04446.x [DOI] [PubMed] [Google Scholar]
- 61. Moynihan P. J., and Clarke A. J. (2011) O-Acetylated peptidoglycan: controlling the activity of bacterial autolysins and lytic enzymes of innate immune systems. Int. J. Biochem. Cell Biol. 43, 1655–1659 10.1016/j.biocel.2011.08.007 [DOI] [PubMed] [Google Scholar]
- 62. Bernard E., Rolain T., Courtin P., Guillot A., Langella P., Hols P., and Chapot-Chartier M.-P. (2011) Characterization of O-acetylation of N-acetylglucosamine: a novel structural variation of bacterial peptidoglycan. J. Biol. Chem. 286, 23950–23958 10.1074/jbc.M111.241414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yamamoto H., Hashimoto M., Higashitsuji Y., Harada H., Hariyama N., Takahashi L., Iwashita T., Ooiwa S., and Sekiguchi J. (2008) Post-translational control of vegetative cell separation enzymes through a direct interaction with specific inhibitor IseA in Bacillus subtilis. Mol. Microbiol. 70, 168–182 10.1111/j.1365-2958.2008.06398.x [DOI] [PubMed] [Google Scholar]
- 64. Arai R., Fukui S., Kobayashi N., and Sekiguchi J. (2012) Solution structure of IseA, an inhibitor protein of dl-endopeptidases from Bacillus subtilis, reveals a novel fold with a characteristic inhibitory loop. J. Biol. Chem. 287, 44736–44748 10.1074/jbc.M112.414763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Clarke C. A., Scheurwater E. M., and Clarke A. J. (2010) The vertebrate lysozyme inhibitor Ivy functions to inhibit the activity of lytic transglycosylase. J. Biol. Chem. 285, 14843–14847 10.1074/jbc.C110.120931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Uehara T., Dinh T., and Bernhardt T. G. (2009) LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J. Bacteriol. 191, 5094–5107 10.1128/JB.00505-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Uehara T., Parzych K. R., Dinh T., and Bernhardt T. G. (2010) Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 29, 1412–1422 10.1038/emboj.2010.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yang D. C., Peters N. T., Parzych K. R., Uehara T., Markovski M., and Bernhardt T. G. (2011) An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc. Natl. Acad. Sci. U.S.A. 108, E1052–E1060 10.1073/pnas.1107780108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yang D. C., Tan K., Joachimiak A., and Bernhardt T. G. (2012) A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division. Mol. Microbiol. 85, 768–781 10.1111/j.1365-2958.2012.08138.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rocaboy M., Herman R., Sauvage E., Remaut H., Moonens K., Terrak M., Charlier P., and Kerff F. (2013) The crystal structure of the cell division amidase AmiC reveals the fold of the AMIN domain, a new peptidoglycan binding domain. Mol. Microbiol. 90, 267–277 10.1111/mmi.12361 [DOI] [PubMed] [Google Scholar]
- 71. Peters N. T., Morlot C., Yang D. C., Uehara T., Vernet T., and Bernhardt T. G. (2013) Structure-function analysis of the LytM domain of EnvC, an activator of cell wall remodelling at the Escherichia coli division site. Mol. Microbiol. 89, 690–701 10.1111/mmi.12304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Crow A., Greene N. P., Kaplan E., and Koronakis V. (2017) Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily. Proc. Natl. Acad. Sci. U.S.A. 114, 12572–12577 10.1073/pnas.1712153114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sham L.-T., Barendt S. M., Kopecky K. E., and Winkler M. E. (2011) Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc. Natl. Acad. Sci. U.S.A. 108, E1061–E1069 10.1073/pnas.1108323108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Meisner J., Montero Llopis P., Sham L.-T., Garner E., Bernhardt T. G., and Rudner D. Z. (2013) FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol. Microbiol. 89, 1069–1083 10.1111/mmi.12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Domínguez-Cuevas P., Porcelli I., Daniel R. A., and Errington J. (2013) Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis. Mol. Microbiol. 89, 1084–1098 10.1111/mmi.12335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sham L.-T., Jensen K. R., Bruce K. E., and Winkler M. E. (2013) Involvement of FtsE ATPase and FtsX extracellular loops 1 and 2 in FtsEX-PcsB complex function in cell division of Streptococcus pneumoniae D39. MBio 4, e00431–13 10.1128/mBio.00431-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bartual S. G., Straume D., Stamsås G. A., Muñoz I. G., Alfonso C., Martínez-Ripoll M., Håvarstein L. S., and Hermoso J. A. (2014) Structural basis of PcsB-mediated cell separation in Streptococcus pneumoniae. Nat. Commun. 5, 3842 10.1038/ncomms4842 [DOI] [PubMed] [Google Scholar]
- 78. Rued B. E., Alcorlo M., Edmonds K. A., Martínez-Caballero S., Straume D., Fu Y., Bruce K. E., Wu H., Håvarstein L. S., Hermoso J. A., Winkler M. E., and Giedroc D. P. (2019) Structure of the large extracellular loop of FtsX and its interaction with the essential peptidoglycan hydrolase PcsB in Streptococcus pneumoniae. MBio 10, e02622–18 10.1128/mBio.02622-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mavrici D., Marakalala M. J., Holton J. M., Prigozhin D. M., Gee C. L., Zhang Y. J., Rubin E. J., and Alber T. (2014) Mycobacterium tuberculosis FtsX extracellular domain activates the peptidoglycan hydrolase, RipC. Proc. Natl. Acad. Sci. U.S.A. 111, 8037–8042 10.1073/pnas.1321812111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Meier E. L., Daitch A. K., Yao Q., Bhargava A., Jensen G. J., and Goley E. D. (2017) FtsEX-mediated regulation of the final stages of cell division reveals morphogenetic plasticity in Caulobacter crescentus. PLoS Genet. 13, e1006999 10.1371/journal.pgen.1006999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Do T., Schaefer K., Santiago A. G., Coe K. A., Fernandes P. B., Kahne D., Pinho M. G., and Walker S. (2020) Staphylococcus aureus cell growth and division are regulated by an amidase that trims peptides from uncrosslinked peptidoglycan. Nat. Microbiol. 10.1038/s41564-019-0632-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Leclerc D., and Asselin A. (1989) Detection of bacterial cell wall hydrolases after denaturing polyacrylamide gel electrophoresis. Can. J. Microbiol. 35, 749–753 10.1139/m89-125 [DOI] [PubMed] [Google Scholar]
- 83. Bernadsky G., Beveridge T. J., and Clarke A. J. (1994) Analysis of the sodium dodecyl sulfate-stable peptidoglycan autolysins of select Gram-negative pathogens by using renaturing polyacrylamide gel electrophoresis. J. Bacteriol. 176, 5225–5232 10.1128/JB.176.17.5225-5232.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Escobar C. A., and Cross T. A. (2018) False positives in using the zymogram assay for identification of peptidoglycan hydrolases. Anal. Biochem. 543, 162–166 10.1016/j.ab.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bernhardt T. G., and de Boer P. A. J. (2004) Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol. Microbiol. 52, 1255–1269 10.1111/j.1365-2958.2004.04063.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhou R., Chen S., and Recsei P. (1988) A dye release assay for determination of lysostaphin activity. Anal. Biochem. 171, 141–144 10.1016/0003-2697(88)90134-0 [DOI] [PubMed] [Google Scholar]
- 87. Desmarais S. M., De Pedro M. A., Cava F., and Huang K. C. (2013) Peptidoglycan at its peaks: how chromatographic analyses can reveal bacterial cell wall structure and assembly. Mol. Microbiol. 89, 1–13 10.1111/mmi.12266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lazar K., and Walker S. (2002) Substrate analogues to study cell-wall biosynthesis and its inhibition. Curr. Opin. Chem. Biol. 6, 786–793 10.1016/S1367-5931(02)00355-1 [DOI] [PubMed] [Google Scholar]
- 89. Ye X.-Y., Lo M.-C., Brunner L., Walker D., Kahne D., and Walker S. (2001) Better substrates for bacterial transglycosylases. J. Am. Chem. Soc. 123, 3155–3156 10.1021/ja010028q [DOI] [PubMed] [Google Scholar]
- 90. Schwartz B., Markwalder J. A., and Wang Y. (2001) Lipid II: total synthesis of the bacterial cell wall precursor and utilization as a substrate for glycosyltransfer and transpeptidation by penicillin binding protein (PBP) 1b of Escherichia coli. J. Am. Chem. Soc. 123, 11638–11643 10.1021/ja0166848 [DOI] [PubMed] [Google Scholar]
- 91. VanNieuwenhze M. S., Mauldin S. C., Zia-Ebrahimi M., Winger B. E., Hornback W. J., Saha S. L., Aikins J. A., and Blaszczak L. C. (2002) The first total synthesis of lipid II: the final monomeric intermediate in bacterial cell wall biosynthesis. J. Am. Chem. Soc. 124, 3656–3660 10.1021/ja017386d [DOI] [PubMed] [Google Scholar]
- 92. Breukink E., van Heusden H. E., Vollmerhaus P. J., Swiezewska E., Brunner L., Walker S., Heck A. J. R., and de Kruijff B. (2003) Lipid II is an intrinsic component of the pore induced by nisin in bacterial membranes. J. Biol. Chem. 278, 19898–19903 10.1074/jbc.M301463200 [DOI] [PubMed] [Google Scholar]
- 93. Huang L.-Y., Huang S.-H., Chang Y.-C., Cheng W.-C., Cheng T.-J. R., and Wong C.-H. (2014) Enzymatic synthesis of lipid II and analogues. Angew. Chem. Int. Ed. Engl. 53, 8060–8065 10.1002/anie.201402313 [DOI] [PubMed] [Google Scholar]
- 94. Lloyd A. J., Gilbey A. M., Blewett A. M., De Pascale G., El Zoeiby A., Levesque R. C., Catherwood A. C., Tomasz A., Bugg T. D. H., Roper D. I., and Dowson C. G. (2008) Characterization of tRNA-dependent peptide bond formation by MurM in the synthesis of Streptococcus pneumoniae peptidoglycan. J. Biol. Chem. 283, 6402–6417 10.1074/jbc.M708105200 [DOI] [PubMed] [Google Scholar]
- 95. Barreteau H., Magnet S., El Ghachi M., Touzé T., Arthur M., Mengin-Lecreulx D., and Blanot D. (2009) Quantitative high-performance liquid chromatography analysis of the pool levels of undecaprenyl phosphate and its derivatives in bacterial membranes. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877, 213–220 10.1016/j.jchromb.2008.12.010 [DOI] [PubMed] [Google Scholar]
- 96. Qiao Y., Lebar M. D., Schirner K., Schaefer K., Tsukamoto H., Kahne D., and Walker S. (2014) Detection of lipid-linked peptidoglycan precursors by exploiting an unexpected transpeptidase reaction. J. Am. Chem. Soc. 136, 14678–14681 10.1021/ja508147s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lee W., Schaefer K., Qiao Y., Srisuknimit V., Steinmetz H., Müller R., Kahne D., and Walker S. (2016) The mechanism of action of lysobactin. J. Am. Chem. Soc. 138, 100–103 10.1021/jacs.5b11807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Qiao Y., Srisuknimit V., Rubino F., Schaefer K., Ruiz N., Walker S., and Kahne D. (2017) Lipid II overproduction allows direct assay of transpeptidase inhibition by β-lactams. Nat. Chem. Biol. 13, 793–798 10.1038/nchembio.2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Srisuknimit V., Qiao Y., Schaefer K., Kahne D., and Walker S. (2017) Peptidoglycan cross-linking preferences of Staphylococcus aureus penicillin-binding proteins have implications for treating MRSA infections. J. Am. Chem. Soc. 139, 9791–9794 10.1021/jacs.7b04881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Welsh M. A., Taguchi A., Schaefer K., Van Tyne D., Lebreton F., Gilmore M. S., Kahne D., and Walker S. (2017) Identification of a functionally unique family of penicillin-binding proteins. J. Am. Chem. Soc. 139, 17727–17730 10.1021/jacs.7b10170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. García-Heredia A., Pohane A. A., Melzer E. S., Carr C. R., Fiolek T. J., Rundell S. R., Lim H. C., Wagner J. C., Morita Y. S., Swarts B. M., and Siegrist M. S. (2018) Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria. Elife 7, e37243 10.7554/eLife.37243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Rebets Y., Lupoli T., Qiao Y., Schirner K., Villet R., Hooper D., Kahne D., and Walker S. (2014) Moenomycin resistance mutations in Staphylococcus aureus reduce peptidoglycan chain length and cause aberrant cell division. ACS Chem. Biol. 9, 459–467 10.1021/cb4006744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lebar M. D., Lupoli T. J., Tsukamoto H., May J. M., Walker S., and Kahne D. (2013) Forming cross-linked peptidoglycan from synthetic Gram-negative Lipid II. J. Am. Chem. Soc. 135, 4632–4635 10.1021/ja312510m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Men H., Park P., Ge M., and Walker S. (1998) Substrate synthesis and activity assay for MurG. J. Am. Chem. Soc. 120, 2484–2485 10.1021/ja974221p [DOI] [Google Scholar]
- 105. Scherer K., Wiedemann I., Ciobanasu C., Sahl H. G., and Kubitscheck U. (2013) Aggregates of nisin with various bactoprenol-containing cell wall precursors differ in size and membrane permeation capacity. Biochim. Biophys. Acta 1828, 2628–2636 10.1016/j.bbamem.2013.07.014 [DOI] [PubMed] [Google Scholar]
- 106. Wang T.-S. A., Lupoli T. J., Sumida Y., Tsukamoto H., Wu Y., Rebets Y., Kahne D. E., and Walker S. (2011) Primer preactivation of peptidoglycan polymerases. J. Am. Chem. Soc. 133, 8528–8530 10.1021/ja2028712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. El Ghachi M., Bouhss A., Barreteau H., Touzé T., Auger G., Blanot D., and Mengin-Lecreulx D. (2006) Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J. Biol. Chem. 281, 22761–22772 10.1074/jbc.M602834200 [DOI] [PubMed] [Google Scholar]
- 108. Lupoli T. J., Taniguchi T., Wang T.-S., Perlstein D. L., Walker S., and Kahne D. E. (2009) Studying a cell division amidase using defined peptidoglycan substrates. J. Am. Chem. Soc. 131, 18230–18231 10.1021/ja908916z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Jorgenson M. A., Chen Y., Yahashiri A., Popham D. L., and Weiss D. S. (2014) The bacterial septal ring protein RlpA is a lytic transglycosylase that contributes to rod shape and daughter cell separation in Pseudomonas aeruginosa. Mol. Microbiol. 93, 113–128 10.1111/mmi.12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Gonzalez-Delgado L. S., Walters-Morgan H., Salamaga B., Robertson A. J., Hounslow A. M., Jagielska E., Sabała I., Williamson M. P., Lovering A. L., and Mesnage S. (2020) Two-site recognition of Staphylococcus aureus peptidoglycan by lysostaphin SH3b. Nat. Chem. Biol. 16, 24–30 10.1038/s41589-019-0393-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Schindler C. A., and Schuhardt V. T. (1964) Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc. Natl. Acad. Sci. U.S.A. 51, 414–421 10.1073/pnas.51.3.414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Mitkowski P., Jagielska E., Nowak E., Bujnicki J. M., Stefaniak F., Niedziałek D., Bochtler M., and Sabała I. (2019) Structural bases of peptidoglycan recognition by lysostaphin SH3b domain. Sci. Rep. 9, 5965 10.1038/s41598-019-42435-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Oshida T., Sugai M., Komatsuzawa H., Hong Y. M., Suginaka H., and Tomasz A. (1995) A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-β-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. U.S.A. 92, 285–289 10.1073/pnas.92.1.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kajimura J., Fujiwara T., Yamada S., Suzawa Y., Nishida T., Oyamada Y., Hayashi I., Yamagishi J.-I., Komatsuzawa H., and Sugai M. (2005) Identification and molecular characterization of an N-acetylmuramyl-l-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol. Microbiol. 58, 1087–1101 10.1111/j.1365-2958.2005.04881.x [DOI] [PubMed] [Google Scholar]
- 115. Rather P. (2013) Role of rhomboid proteases in bacteria. Biochim. Biophys. Acta 1828, 2849–2854 10.1016/j.bbamem.2013.03.012 [DOI] [PubMed] [Google Scholar]
- 116. Began J., Cordier B., Březinová J., Delisle J., Hexnerová R., Srb P., Rampírová P., Kožíšek M., Baudet M., Couté Y., Galinier A., Veverka V., Doan T., and Strisovsky K. (2020) Rhomboid intramembrane protease YqgP licenses bacterial membrane protein quality control as adaptor of FtsH AAA protease. EMBO J. 10.15252/embj.2019102935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Davis K. M., and Weiser J. N. (2011) Modifications to the peptidoglycan backbone help bacteria to establish infection. Infect. Immun. 79, 562–570 10.1128/IAI.00651-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Sychantha D., Brott A. S., Jones C. S., and Clarke A. J. (2018) Mechanistic pathways for peptidoglycan O-acetylation and de-O-acetylation. Front. Microbiol. 9, 2332 10.3389/fmicb.2018.02332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Brown S., Santa Maria J. P. Jr., and Walker S. (2013) Wall teichoic acids of Gram-positive bacteria. Annu. Rev. Microbiol. 67, 313–336 10.1146/annurev-micro-092412-155620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kawai Y., Marles-Wright J., Cleverley R. M., Emmins R., Ishikawa S., Kuwano M., Heinz N., Bui N. K., Hoyland C. N., Ogasawara N., Lewis R. J., Vollmer W., Daniel R. A., and Errington J. (2011) A widespread family of bacterial cell wall assembly proteins. EMBO J. 30, 4931–4941 10.1038/emboj.2011.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Chan Y. G.-Y., Kim H. K., Schneewind O., and Missiakas D. (2014) The capsular polysaccharide of Staphylococcus aureus is attached to peptidoglycan by the LytR-CpsA-Psr (LCP) family of enzymes. J. Biol. Chem. 289, 15680–15690 10.1074/jbc.M114.567669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Schaefer K., Matano L. M., Qiao Y., Kahne D., and Walker S. (2017) In vitro reconstitution demonstrates the cell wall ligase activity of LCP proteins. Nat. Chem. Biol. 13, 396–401 10.1038/nchembio.2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Schaefer K., Owens T. W., Kahne D., and Walker S. (2018) Substrate preferences establish the order of cell wall assembly in Staphylococcus aureus. J. Am. Chem. Soc. 140, 2442–2445 10.1021/jacs.7b13551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Rausch M., Deisinger J. P., Ulm H., Müller A., Li W., Hardt P., Wang X., Li X., Sylvester M., Engeser M., Vollmer W., Müller C. E., Sahl H. G., Lee J. C., and Schneider T. (2019) Coordination of capsule assembly and cell wall biosynthesis in Staphylococcus aureus. Nat. Commun. 10, 1404 10.1038/s41467-019-09356-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Moynihan P. J., and Clarke A. J. (2014) Substrate specificity and kinetic characterization of peptidoglycan O-acetyltransferase B from Neisseria gonorrhoeae. J. Biol. Chem. 289, 16748–16760 10.1074/jbc.M114.567388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wang Y., Lazor K. M., DeMeester K. E., Liang H., Heiss T. K., and Grimes C. L. (2017) Postsynthetic modification of bacterial peptidoglycan using bioorthogonal N-acetylcysteamine analogs and peptidoglycan O-acetyltransferase B. J. Am. Chem. Soc. 139, 13596–13599 10.1021/jacs.7b06820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Sychantha D., Jones C. S., Little D. J., Moynihan P. J., Robinson H., Galley N. F., Roper D. I., Dowson C. G., Howell P. L., and Clarke A. J. (2017) In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA). PLoS Pathog. 13, e1006667 10.1371/journal.ppat.1006667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Atilano M. L., Pereira P. M., Yates J., Reed P., Veiga H., Pinho M. G., and Filipe S. R. (2010) Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 107, 18991–18996 10.1073/pnas.1004304107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Radkov A. D., Hsu Y.-P., Booher G., and VanNieuwenhze M. S. (2018) Imaging bacterial cell wall biosynthesis. Annu. Rev. Biochem. 87, 991–1014 10.1146/annurev-biochem-062917-012921 [DOI] [PMC free article] [PubMed] [Google Scholar]