Abstract
Glucocorticoids are potent endogenous anti-inflammatory molecules, and their cognate receptor, glucocorticoid receptor (GR), is expressed in nearly all immune cells. Macrophages are heterogeneous immune cells having a central role in both tissue homeostasis and inflammation and also play a role in the pathogenesis of some inflammatory diseases. Paradoxically, glucocorticoids have only a limited efficacy in controlling the resolution of these macrophage-related diseases. Here, we report that the transcriptomes of monocyte-like THP-1 cells and macrophage-like THP-1 cells (THP1-MΦ) have largely conserved gene expression patterns. In contrast, the differentiation to THP1-MΦ significantly altered the sensitivity of gene transcription to glucocorticoids. Among glucocorticoid-regulated genes, we identified the exopeptidase dipeptidyl peptidase-4 (DPP4) as a critical glucocorticoid-responsive gene in THP1-MΦ. We found that GR directly induces DPP4 gene expression by binding to two glucocorticoid-responsive elements (GREs) within the DPP4 promoter. Additionally, we show that glucocorticoid-induced DPP4 expression is blocked by the GR antagonist RU-486 and by GR siRNA transfection and that DPP4 enzyme activity is reduced by DPP4 inhibitors. Of note, glucocorticoids highly stimulated macrophage mobility; unexpectedly, DPP4 mediated the glucocorticoid-induced macrophage migration, and siRNA-mediated knockdowns of GR and DPP4 blocked dexamethasone-induced THP1-MΦ migration. Moreover, glucocorticoid-induced DPP4 activation was also observed in proinflammatory M1-polarized murine macrophages, as well as peritoneal macrophages, and was associated with increased macrophage migration. Our results indicate that glucocorticoids directly up-regulate DPP4 expression and thereby induce migration in macrophages, potentially explaining why glucocorticoid therapy is less effective in controlling macrophage-dominated inflammatory disorders.
Keywords: glucocorticoid receptor, glucocorticoid, migration, macrophage, gene expression, immunology, inflammation, chromatin remodeling, nuclear receptor, dipeptidyl peptidase-4 (DPP4), DPP4 inhibitors, linagliptin, sitagliptin
Introduction
Glucocorticoids exert a wide array of systemic and tissue-specific effects, by signaling through the cognate glucocorticoid receptor (GR;2 NR3C1) in numerous tissues and cell types to systematically influence development, homeostasis, metabolism, and inflammation (1). One of the most important effects of both endogenous and exogenous glucocorticoids is immunomodulation, exerted mainly by suppressing transcription of pro-inflammatory genes and/or induction of anti-inflammatory genes (2). Synthetic glucocorticoids are commonly prescribed anti-inflammatory and immunomodulatory agents. Their therapeutic activity is substantial in a wide spectrum of diseases, including acute and chronic inflammation, autoimmune disorders (3), organ transplantation (4), and hematological cancers (5).
Contrary to well-known anti-inflammatory effects of glucocorticoids, there is emerging evidence of pro-inflammatory effects during inflammation (6–8). For example, glucocorticoid signaling in macrophages has been reported to up-regulate the expression of NLRP3 inflammasome component and to enhance the ATP-dependent secretion of cytokines such as TNFα and interleukin-6 (9). These findings suggest that glucocorticoids likely play a dual role regulating the innate and adaptive immune response differentially. These effects may depend on the type of inflammatory stimulus (10) and/or the timing of treatment (11), thus modulating the balance of the cellular state toward a net pro-inflammatory or anti-inflammatory state (7). These macrophage-intrinsic properties may explain why glucocorticoids are less effective in macrophage-mediated diseases (12), such as chronic obstructive pulmonary disease (11), ulcerative colitis (13), systemic lupus erythematosus (14), and rheumatoid arthritis (15).
Macrophages are involved in all phases of the inflammatory response, including alarm, mobilization, and resolution phases, and are able to drive either the propagation or resolution of inflammation (12, 16). The ontogeny of macrophages is still not fully understood; however, it is accepted that they can be grouped as tissue-resident macrophages (established independently of hematopoiesis) or infiltrating macrophages derived from circulating monocytes that are established following an inflammatory response (17–19). Thus, macrophages may contribute to the pathophysiology of several diseases, including inflammatory disorders (20, 21), cancer (22), and also states of low-grade inflammation such as obesity (23, 24).
The migratory capacity of macrophages has been studied in tumor-associated macrophages, because they have a critical role in different stages of tumor progression (25). In other inflammatory pathologies, such as multiple sclerosis, differences in activation and polarization of macrophages promote their migratory properties toward chemoattractants (26). This migration is associated with cytoskeleton rearrangements and also has been proposed to depend on the levels and type of integrin expression (26). For example, the expression of the chemokine receptor CXCR4 on mouse and human mature macrophages has been associated with migration toward lymph nodes during murine peritonitis resolution (27). Finally, in human macrophages (differentiated from CD14+ monocytes), CXCR4 expression and cell motility (showing a longer distance traveled) was induced upon dexamethasone stimulation. However, little attention has been directed to the identification of genes essential for macrophage movement (28).
Dipeptidyl peptidase-4 (DPP4), encoding for a membrane glycoprotein with exopeptidase activity, was recently described as being involved in the inflammatory macrophage profile associated with type 2 diabetes, obesity, and atherosclerosis (29, 30). This protein is multifunctional, with both enzymatic and nonenzymatic activities. The extracellular domain of DPP4 presents the catalytic site, primarily associated with inactivation of incretin hormones, such as glucagon-like peptides 1 and 2, and the gastric inhibitory polypeptide, as well as catalysis of chemokines (CCL2/MIP-1a, CXCL12/SDF-1, and CCL5/Rantes, among others) (31). DPP4 has also binding sites for adenosine deaminase and fibronectin, and their binding is associated with pro-inflammatory responses (32).
DPP4 inhibitors, such as linagliptin and sitagliptin (33–36), are useful for the control of blood glucose in type 2 diabetes patients (33) who do not respond well to metformin (34) and sulfonylureas (35) and patients suffering from the diabetogenic effects of glucocorticoids (36, 37).
In this study, using THP-1 cells, we show that the monocyte-to-macrophage differentiation was associated with both higher GR levels and greater sensitivity to glucocorticoids on macrophage-like THP-1 cells (THP1-MΦ) compared with monocyte-like THP-1 cells. These changes resulted in modifications of the glucocorticoid-dependent macrophage transcriptome. During the monocyte-to-THP1-MΦ differentiation, cells undergo chromatin remodeling, thus enhancing GR accessibility to glucocorticoid-response elements (GREs) within the DPP4 promoter. Furthermore, we show that DPP4 is a novel glucocorticoid-responsive gene specifically in human and mouse macrophages, but not regulated in monocytes. An in vitro migration assay using THP1-MΦ and M1 polarized bone marrow–derived macrophages (BMDMs) reveals that glucocorticoids regulate the macrophage movement via a DPP4-dependent process.
Results
Transcriptome analysis of monocyte-like THP-1 cells and macrophage-like THP-1 cells reveals a high conservation of gene expression between both cell types
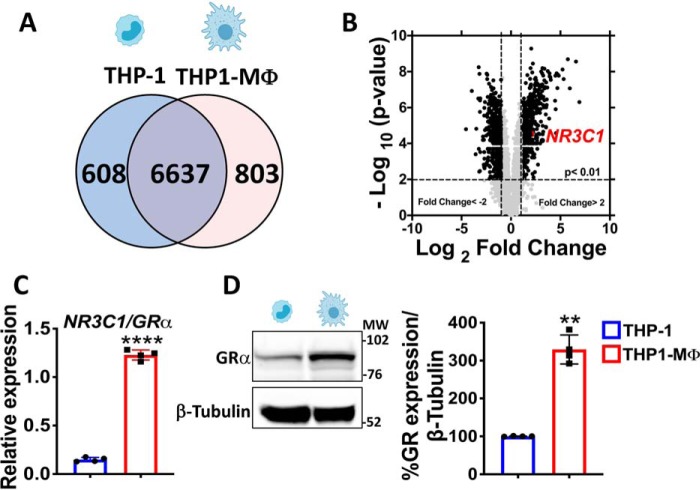
We investigated whether the state of cell differentiation among monocytes and macrophages modifies the gene expression, with profiles of monocyte-like THP-1 cells (THP-1) and macrophage-like THP-1 cells (THP1-MΦ) analyzed by genome-wide microarray (Fig. S1). Regulated genes were evaluated through a Venn diagram (Fig. 1A), which showed that a large number of genes are commonly expressed in THP-1 and THP1-MΦ (6,637 genes, corresponding to 82.4% of the total expressed genes). In contrast, 608 genes were only found in THP-1, whereas 803 were restricted to THP1-MΦ (Fig. 1A). Provocatively, a volcano plot comparing genes expressed in THP1-MΦ with monocyte-like THP-1 cells revealed that the gene NR3C1/GR was highly expressed in THP1-MΦ (Fig. 1B). These data were validated by qRT-PCR and Western blotting showing a 6-fold increase in the GR mRNA and a 3-fold increase in GR protein in THP1-MΦ compared with undifferentiated monocyte-like THP-1 cells (Fig. 1 (C and D), respectively). Interestingly, this phenomenon also was observed in primary murine BMDMs using M-CSF, where we found an increase in GR mRNA in macrophages post-differentiation from bone marrow monocytes (BMM) (Fig. S2).
Figure 1.
The transcriptome of macrophage-like THP-1 cells shows a higher GR expression compared with monocyte-like THP-1 cells. A, Venn diagram summarizing microarray data analysis shows the number of genes commonly expressed by THP-1 cells and MΦ-THP-1 and uniquely expressed in both cell types. B, volcano plot showing significantly differentially expressed genes in MΦ-THP-1 versus THP-1. In MΦ-THP-1 GR gene expression (also known as NR3C1/GRα) is 4-fold higher than THP-1. Shown is validation by qRT-PCR (C) and Western blotting (D) of GR expression that is higher in MΦ-THP-1 in comparison to THP-1. Data are mean ± S.D. (error bars) and are representative of four independent experiments. *, p < 0.05; two tailed unpaired Student's t test.
Monocyte-to-macrophage differentiation enhances their responsiveness to glucocorticoids in macrophages
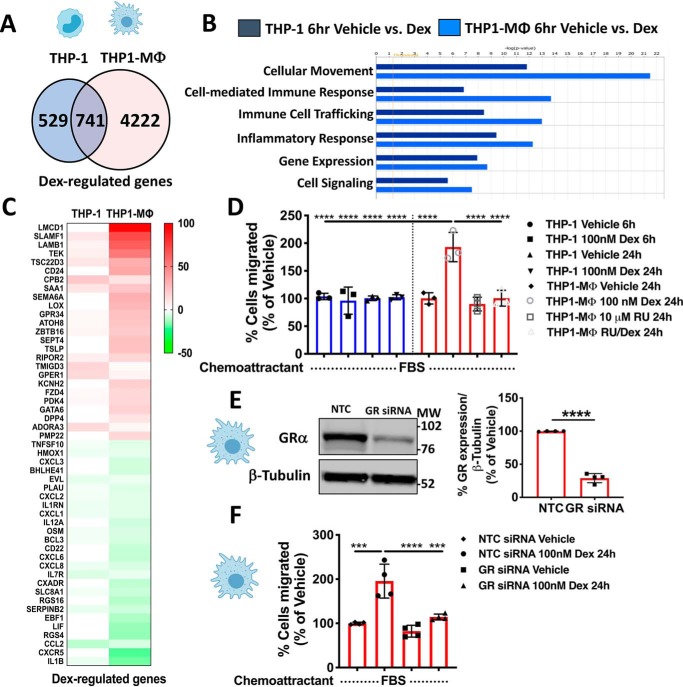
Due to the different expression levels of GR in monocyte-like THP-1 cells and THP1-MΦ, we investigated whether the state of cellular differentiation alters the sensitivity to glucocorticoids. For these experiments, monocyte-like THP-1 cells and THP1-MΦ were treated with dexamethasone (Dex) for 6 h, and total isolated mRNA was subsequently analyzed by a genome-wide microarray. Principal component analysis demonstrated considerable separation between treatment (control versus Dex) in both cell types. However, Dex-treated monocyte-like THP-1 cells and THP1-MΦ were dramatically separated, indicating differential glucocorticoid-regulated transcriptomes in these cells (Fig. S3A). To identify unique and common genes regulated by Dex among the cell types, the gene list was sorted by Venn diagram, revealing only 741 genes commonly regulated by Dex in monocyte-like THP-1 cells and THP1-MΦ (Fig. 2A). Moreover, the number of genes uniquely regulated by Dex in THP1-MΦ (4,222 genes) was 7-fold higher with respect to monocyte-like THP-1 cells (529 genes) (Fig. 2A). The most significant genes commonly and uniquely regulated by Dex in monocyte-like THP-1 cells and THP1-MΦ were further plotted on a volcano plot and validated by qRT-PCR (Fig. S3, B–G). To elucidate the significance of these findings in both cell types, gene sets were analyzed by Ingenuity Pathway Analysis® (IPA) software. Provocatively, cellular movement was overrepresented in THP1-MΦ compared with monocyte-like THP-1 cells (Fig. 2B). Moreover, cell-mediated immune response, immune cell trafficking, and inflammatory response pathways were enhanced in macrophages compared with monocytes (Fig. 2B and Fig. S4), suggesting that THP1-MΦ are more responsive than monocyte-like THP-1 cells to signaling by glucocorticoids.
Figure 2.
Glucocorticoids regulate the cell migration of macrophage-like THP-1 cells. A, a Venn diagram, summarizing microarray data analysis, shows the number of genes commonly regulated by Dex in THP-1 cells and in MΦ-THP-1 and uniquely regulated by Dex in both cell types. B, top 5 biological functions that are differentially regulated in THP-1 and MΦ-THP-1. C, heat map representing the top 25 up-regulated and the top 25 down-regulated genes by glucocorticoids in THP-1 and MΦ-THP-1 associated with the cellular movement pathway. D, in vitro transwell assay using 10% of FBS as chemoattractant was used to evaluate spontaneous cell migration of THP-1 and MΦ-THP-1 treated with vehicle, 100 nm Dex, 10 μm RU-486, or RU-486 with Dex. The graph shows that Dex treatment induces migration only in MΦ-THP-1, and this phenomenon is reversed by using the GR antagonist RU486. E, THP1-MΦ were transfected with NTC or GR siRNAs. 24 h after transfection, GR protein knockdown was evaluated by Western blotting (75% reduction). On the left, a representative immunoblot of GR and β-tubulin expression is shown. Right, densitometry analysis of GR normalized to β-tubulin. F, in vitro migration assay of THP1-MΦ transfected with NTC or GR siRNAs that have been treated for 24 h with or without 100 nm Dex. The histograms show that GR knockdown abolishes Dex-induced macrophage migration. Cell migration was calculated as percentage relative to vehicle-treated groups. Data are mean ± S.D. (error bars) and are representative of three independent experiments. ***, p < 0.001; ****, p < 0.0001; two-tailed unpaired Student's t test (E) and one-way ANOVA statistical test with Tukey's multiple-comparison test (D and F).
Glucocorticoids enhance macrophage-like THP-1 cell migration
Pathway analysis of the microarray data revealed that cell movement was the top biological function following Dex treatment in both monocyte-like THP-1 cells and THP1-MΦ, although cell movement annotation was more dramatically regulated in Dex-treated THP1-MΦ. The number of genes involved in cell movement and differentially regulated according to IPA was 1,102, 44.3% of which were induced, whereas the remaining 63.7% were inhibited in THP1-MΦ. Heat map analysis of the top 25 genes revealed that the magnitude of Dex-induced gene expression was greater in THP1-MΦ (Fig. 2C). Based on these findings, we evaluated how GR could impact THP1-MΦ migratory properties in a cell migration assay based on the Boyden Chamber principle. Spontaneous cell migration assay with cells seeded on the insert and serum as the chemoattractant at the bottom revealed that Dex treatment did not impact the monocyte-like THP-1 cell migratory properties at 6 and 24 h (Fig. 2D). However, Dex treatment of THP1-MΦ increased their migratory properties after 24 h (Fig. 2D). This effect was blocked by 1-h pretreatment with the GR antagonist, RU486 (Fig. 2D). This finding was corroborated by knocking down GR expression via siRNA. THP1-MΦ transfected with siRNA GR had ∼75% of GR silenced (Fig. 2E) and a significant reduction in Dex-induced cell migration compared with cells transfected with nontargeting control (NTC) siRNA (Fig. 2F). These results suggest that glucocorticoids regulate the migration of macrophage-like THP-1 cells.
The DPP4 is a new glucocorticoid-responsive gene regulated in macrophage-like THP-1 cells
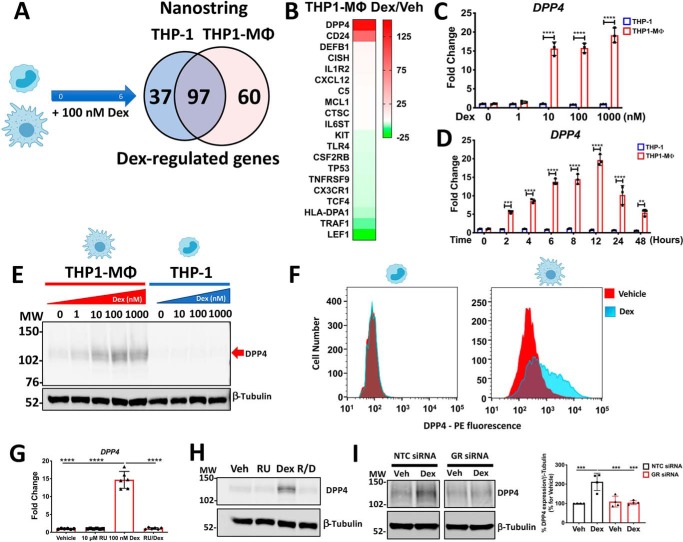
Data from our transcriptome analysis suggested that the pathways of cell migration and inflammation were highly regulated by glucocorticoids; thus, to substantiate this finding, we turned to the use of nanostring analysis. Among the 594 genes present in the code set Human Immunology, 194 genes were regulated by Dex, specifically 134 in monocyte-like THP-1 cells and 157 in THP1-MΦ (Fig. 3A). The expression pattern of genes related to the immune response sorted through a Venn diagram showed that 97 genes were commonly regulated by Dex in THP-1 and THP1-MΦ (corresponding to 50% of the total regulated genes) (Fig. 3A). Additionally, the number of genes regulated exclusively by Dex in THP-1 and THP1-MΦ was 37 and 60, respectively, indicating that almost twice as many genes were differentially regulated by Dex in THP1-MΦ (Fig. 3A). Among these genes, DPP4 was the most up-regulated by glucocorticoids in THP1-MΦ and also was part of the 100 genes mostly induced by microarray (Figs. 2A and 3B). Interestingly, using a small cohort of RNA samples (n = 9) from human monocyte–derived macrophages (38) that were treated with 100 nm dexamethasone for 6 h, we found that glucocorticoids significantly induced the mRNA expression of DPP4 in 6 of 9 samples analyzed (Fig. S5). Furthermore, DPP4 was not regulated by glucocorticoids in monocyte-like THP-1 cells (Fig. 3B). Glucocorticoid-dependent regulation of DPP4 was confirmed by qRT-PCR, through a dose-response and time-course analysis indicating that high levels of DPP4 mRNA were exclusively up-regulated in THP1-MΦ by 10, 100, and 1000 nm Dex (Fig. 3C) and maximally induced 12 h after Dex treatment (Fig. 3D). Additionally, Dex-induced DPP4 up-regulation was observed at the protein level by Western blotting and flow cytometry, observing a double immunoreactive band of ∼100 and 114 kDa (also induced by 10, 100, and 1,000 nm Dex) (Fig. 3E) and higher fluorescent intensity induced by Dex exclusively in THP1-MΦ (Fig. 3F). Finally, the functionality of GR in DPP4 induction was evaluated through pharmacological and genetic inhibition, using a pretreatment with GR antagonist RU-486 and transfection with GR siRNA, respectively. DPP4 induction by Dex in THP1-MΦ was blocked in the presence of RU486, both at the transcript level (6 h) (Fig. 3G) and protein level (24 h) (Fig. 3H), and inhibited by GR silencing, at the protein level at 24 h (50% down-regulation with respect to NTC (Fig. 3I).
Figure 3.
Glucocorticoids up-regulate DPP4 mRNA and protein levels in macrophage-like THP-1 cells by GR activation. A, a Venn diagram, summarizing NanoString Immunology code set data analysis, shows the number of genes commonly regulated by Dex in THP-1 cells and MΦ-THP-1 and uniquely regulated by Dex in both cell types. B, heat map of the top 10 up- and down-regulated genes by Dex in THP1-MΦ. C–F, glucocorticoid-induced DPP4 levels are uniquely expressed in THP1-MΦ. Shown are quantitative RT-PCR analyses of DPP4 mRNA levels in THP-1 and THP1-MΦ in Dex dose-response (C) and Dex time course (D) experiments. Shown are representative Western blotting (E) and flow cytometry (F) analysis of DPP4 protein from THP-1 and THP1-MΦ treated with 100 nm Dex for 24 h. The immunoreactive band is indicated by a red arrow. G–I, evaluation of GR participation in DPP4 up-regulation. Pharmacological inhibition of GR with RU-486 and GR knockdown by siRNA abolish Dex-induced expression of DPP4 at both mRNA (G) and protein (H and I) levels. Data are mean ± S.D. (error bars) and are representative of n = 3 to 6 independent experiments. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; two-way ANOVA test with Tukey's multiple-comparison test (C and D) and one-way ANOVA test with Tukey's multiple-comparison test (G and I).
Glucocorticoids regulate DPP4 expression by binding to GREs in regulatory regions of the DPP4 gene
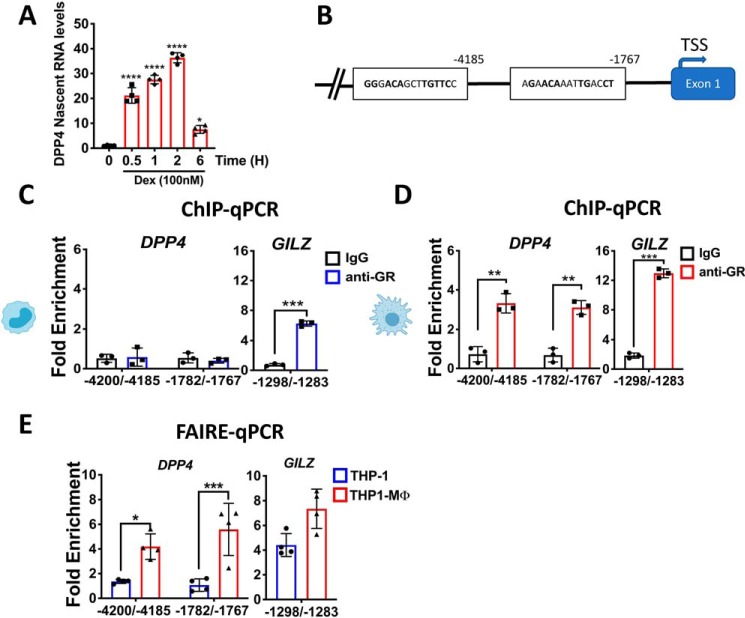
Through the determination of nascent RNA levels, we observed that glucocorticoids directly regulated the induction of DPP4, with an increase within just 30 min of treatment (Fig. 4A), thus indicating that DPP4 is a direct transcriptional target of the GR. In silico analysis of the human DPP4 gene revealed the presence of numerous putative GREs located in the regulatory region between 1.5 and 6.5 kb upstream of the transcription start site, each one with a score higher than 80% in relation to the consensus GRE sequence (Fig. 4B). Therefore, monocyte-like THP-1 cells and THP1-MΦ treated with 100 nm Dex or vehicle for 2 h were evaluated by ChIP coupled to real-time PCR (ChIP-qPCR), using anti-GR antibody and IgG isotype as negative control. In THP1-MΦ treated with Dex, an enrichment of GR was observed in GRE −4,200/−4,185 (3.6-fold) and GRE −1,782/−1,767 (3.2-fold) of the regulatory region of DPP4, suggesting a direct transcriptional control of the DPP4 gene by GR (Fig. 4D). In Dex-induced monocyte-like THP-1 cells, this effect was not observed (Fig. 4C). As a control to evaluate whether there is a differential specificity of GR in the binding to the DPP4 promoter, we analyzed the recruitment of liganded GR to the GRE located in the promoter region of the glucocorticoid target gene GILZ. We observed an enrichment of GR to the GRE of GILZ in both monocyte-like THP-1 cells and THP1-MΦ following Dex treatment (6- and 13-fold, respectively, Fig. 4 (C and D)). These data suggest that chromatin structure reorganization occurs during the monocyte-to-macrophage transition, thus allowing GR binding and serving as the mechanism through which DPP4 is differentially regulated in monocyte-like THP-1 cells and THP1-MΦ. To address this question, we evaluated chromatin accessibility using a formaldehyde-assisted isolation of regulatory elements (FAIRE) assay in the same regions of the DPP4 gene where GR occupied sites according to ChiP-qPCR in macrophages. Upon differentiation and Dex treatment, we observed an increase in DNA accessibility in both regions of the DPP4 gene flanking GRE −4,200/−4,185 and GRE −1,782/−1,767, respectively, in THP1-MΦ, whereas there was limited accessibility of them in monocyte-like THP-1 cells (Fig. 4E). As control, DNA accessibility was also observed in the site flanking GRE of the GILZ promoter, without differences between THP1-MΦ and monocyte-like THP-1 cells. The increase in FAIRE enrichment seen at the two GREs of DPP4 gene suggests that the chromatin remodeling during the monocyte-to-macrophage transition is required for GR to initiate Dex-induced DPP4 gene transcription.
Figure 4.
Glucocorticoids regulate DPP4 expression by binding to GREs in the DPP4 promoter. A, quantitative real-time PCR analysis of DPP4 nascent mRNA from macrophage-like THP-1 cells (THP1-MΦ) that have been treated with 100 nm Dex for the indicated times. Dex treatment stimulates the transcription of a significant amount of DPP4 nascent mRNA within 30 min following Dex exposure. B, schematic representation of a fragment of human DPP4 gene that highlights two identified GREs located at positions −4,200/−4,185 and −1,782/−1,767 from the transcription start site (TSS). Monocyte-like THP-1 cells (C) and THP1-MΦ (D) were treated with or without 100 nm Dex for 2 h to evaluate by ChIP-qPCR the amount of GR bound to DPP4 GREs exclusively in THP1-MΦ. GR-ChIP samples were analyzed by quantitative PCR, and the Ct values of each experimental group were analyzed in triplicate, compared with their respective inputs, and normalized to the IgG isotype control. GR recruitment to DPP4 GREs is expressed as -fold enrichment of Dex- versus vehicle-treated cells. C and D, ChIP-qPCR of GR-bound GRE located in the promoter region of GILZ gene was used as positive control. E, chromatin remodeling was measured by FAIRE-qPCR to evaluate GR accessibility to the GREs in the DPP4 gene and GILZ gene (as control) in response to the monocyte-to-macrophage differentiation process. For each GRE, chromatin accessibility was determined and compared with the respective DNA input. Data are mean ± S.D. (error bars) and are representative of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. Shown are a one-way ANOVA test with Tukey's multiple-comparison test (A) and a two-way ANOVA test with Sidak's multiple-comparison test (C–E).
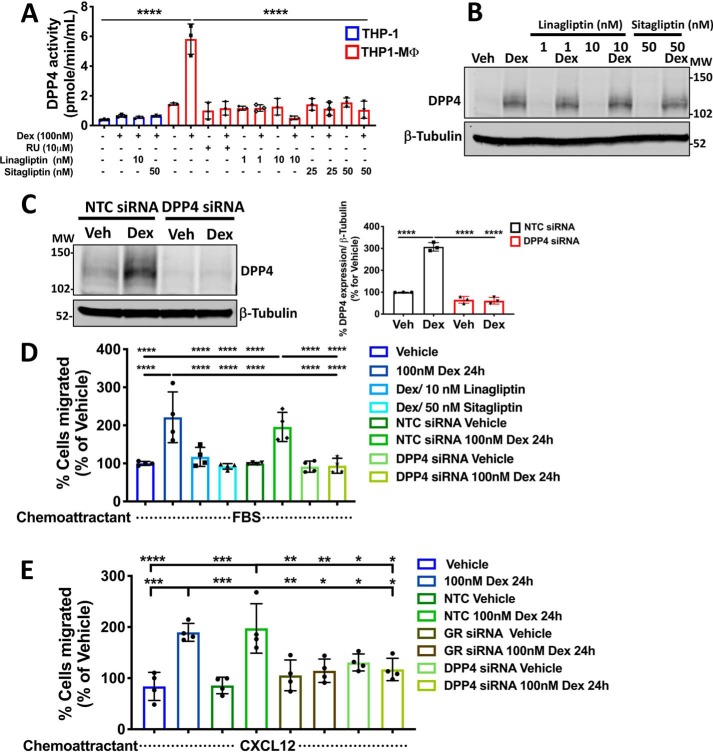
Glucocorticoids enhance DPP4 enzymatic activity in THP1-MΦ, and this effect is blocked by DPP4 inhibitors sitagliptin and linagliptin
The effects mediated by DPP4 in different cell types have been associated with both enzymatic and nonenzymatic functions. Based on this classification (and having shown that liganded GR up-regulates mRNA and protein DPP4 levels), we evaluated whether glucocorticoids could also modulate the DPP4 enzymatic activity. For these studies, monocyte-like THP-1 cells and THP1-MΦ were stimulated with Dex for 24 h or were pretreated with RU-486 or co-stimulated after 3 h with two concentrations of specific DPP4 inhibitors, sitagliptin and linagliptin. Cell lysates of each experimental condition were evaluated by fluorometric assay, showing no DPP4 activity from monocyte-like THP-1 cells under evaluated conditions (Fig. 5A). Conversely, in THP1-MΦ, we observed that Dex increased DPP4 enzymatic activity, and this effect is mediated by GR because it was blocked by RU486 pretreatment (Fig. 5A). In addition, using sitagliptin (25 and 50 nm) and linagliptin (1 and 10 nm), we found that both inhibitors completely block DPP4 enzymatic activity induced by Dex (Fig. 5A). Notably, treatment with both DPP4 inhibitors directly blocked DPP4 enzymatic activity without affecting Dex-induced DPP4 protein expression (Fig. 5B).
Figure 5.
The cell migration induced by dexamethasone in macrophage-like THP-1 cells is mediated through DPP4 activity. A, the enzymatic activity of DPP4 measured by fluorometric assay and induced by Dex was completely blocked by the DPP4 inhibitors sitagliptin (25 and 50 nm) and linagliptin (1 and 10 nm) in MΦ-THP-1. B, Western blotting for DPP4 showing that both DPP4 inhibitors do not affect the Dex-induced DPP4 protein expression. C, THP1-MΦ were transfected with NTC or DPP4 siRNAs. 24 h after transfection, cells were treated with or without Dex for 24 h, and DPP4 protein knockdown was evaluated by Western blotting (>70% reduction). On the left, a representative immunoblot of DPP4 and β-tubulin expression is shown. On the right, densitometry analysis of DPP4 normalized to β-tubulin is shown. D, Dex-induced THP1-MΦ migration is mediated by DPP4 expression. An in vitro migration assay shows that both pharmacological inhibition of DPP4 enzymatic activity, by linagliptin and sitagliptin, and silencing of DPP4 expression block the spontaneous migration of THP1-MΦ induced by Dex treatment. E, CXCL12-induced THP1-MΦ migration was blocked by GR and DPP4 knockdowns of cells following Dex treatment. Data are mean ± S.D. (error bars) and are representative of 3–4 independent experiments. ***, p < 0.001; ****, p < 0.0001; one-way ANOVA statistical test with Tukey's multiple-comparison test.
DPP4 promotes macrophage-like THP-1 cell migration
The multifunctionality of DPP4, enzymatic activity to regulate chemokine actions or nonenzymatic via interaction with other proteins, could also regulate, directly or indirectly, macrophage mobility. To investigate this idea, we silenced DPP4 in THP1-MΦ using siRNA or DPP4 inhibitors, evaluating cell migration behind glucocorticoid effects. Expression of DPP4 induced by Dex was decreased by 80% at the protein level in THP1-MΦ transfected with siRNA DPP4 with respect to NTC (Fig. 5C). Using this strategy to evaluate GR participation in cell migration, we compared the migratory potential of THP1-MΦ primed with Dex in the presence of DPP4 inhibitors. Cells were seeded in the insert, and medium with serum was added in the lower chamber as chemoattractant. Interestingly, the Dex-mediated increase in the migratory potential was inhibited in the presence of either 10 nm linagliptin or 50 nm sitagliptin, suggesting that this effect depended on DPP4 catalytic function (Fig. 5D). Finally, NTC and DPP4 siRNA macrophages, primed or unprimed with Dex for 24 h, were used to evaluate DPP4 participation in cell migration. An increase in the migratory potential induced by Dex in the NTC was suppressed by DPP4 knockdown (Fig. 5D), suggesting that DPP4 activity is necessary for enhancing macrophage migration. Interestingly, CXCL12 was able to promote the migration of cells treated with Dex for 24 h, and this effect was inhibited by both GR and DPP4 knockdown (Fig. 5E).
The induction of the Dpp4 gene by glucocorticoids is an exclusive mechanism in pro-inflammatory M1 macrophages
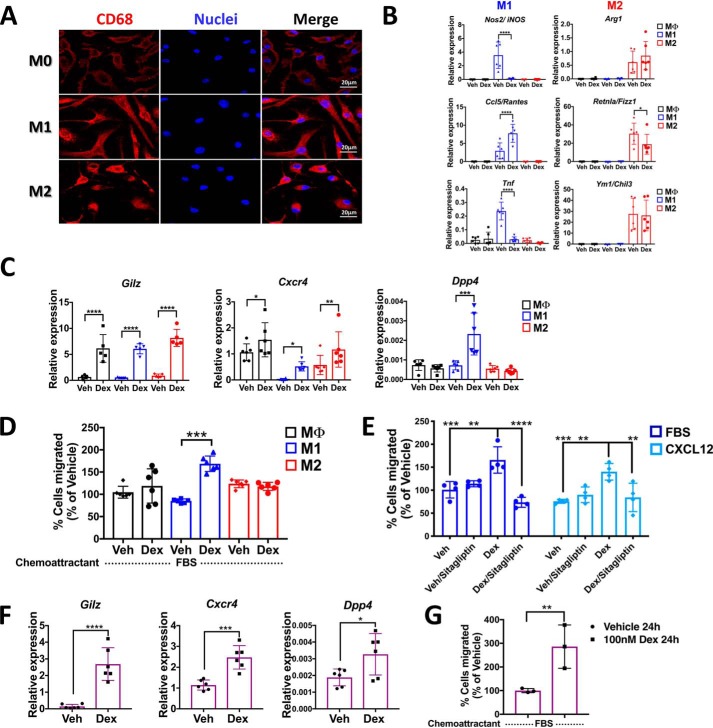
Our findings in THP1-MΦ indicate that glucocorticoids promote cell migration in part by inducing DPP4 expression. To examine whether this is a conserved mechanism, we assessed the effects of Dex treatment on mouse BMM and BMDMs. After inducing macrophage differentiation with M-CSF (M0, unstimulated macrophages), cell cultures were activated by treatment with lipopolysaccharide/interferon-γ (M1) or with interleukin-4 (M2). The phenotype of unpolarized (M0) macrophages as well as macrophages polarized to M1 and M2 was evaluated by confocal microscopy, showing the polarization-dependent morphological differences and the “classical” marker CD68 (Fig. 6A). Moreover, qRT-PCR analysis revealed that M1 macrophages exhibited the characteristic up-regulation of Nos2/iNOS, Ccl5/Rantes, and Tnf mRNA, whereas M2 macrophages exhibited increased expression of Arg1, Retnla, and Chil3 (Fig. 6B). To assess the impact of macrophage activation on glucocorticoid responsiveness, BMM, M0, M1, and M2 macrophage cultures were treated with 100 nm Dex for 6 h. Quantitative RT-PCR revealed that expression of M1-associated genes Nos2 and Tnf were suppressed 6 h after Dex treatment, whereas expression of M2-associated genes was not affected (Fig. 6B). Interestingly, whereas Dex treatment induced equivalent expression of the classic glucocorticoid target genes Gilz in each group and the induction of the chemokine receptor Cxcr4 in all groups of the macrophage populations, Dpp4 was exclusively induced only in M1 macrophages (Fig. 6C and Figs. S6 and S7). Finally, we examined the effects of glucocorticoid treatment on BMDM macrophage migration. For this purpose, M0, M1, and M2 macrophages were treated for 24 h with Dex, and their spontaneous migration was assessed. Consistent with our finding in THP1-MΦ, we found that Dex treatment enhanced spontaneous migration of M1 macrophages but did not affect the migration of M0 or M2 macrophages (Fig. 6D). Interestingly, M1 macrophages treated with Dex for 24 h and then pretreated or not for 1 h with 50 nm sitagliptin were used to evaluate the CXCL12-induced migration. We observe that the migration of M1 macrophages could also be induced by CXCL12, and this effect was blocked by pretreatment with sitagliptin (Fig. 6E). Finally, we evaluated glucocorticoid-induced effects in peritoneal macrophages that naturally show an ambulatory or motile phenotype. Together with Gilz induction, we observed Dpp4 induction upon Dex treatment (Fig. 6F) and higher cell migration induced by glucocorticoids (Fig. 6G), suggesting that glucocorticoid-induced migration in macrophages is mediated by DPP4.
Figure 6.
Glucocorticoids up-regulate Dpp4 expression in mouse BMDMs polarized to classically activated profile M1 and peritoneal macrophages promoting migration. BMM isolated by negative selection and differentiated into BMDM with 100 μg/ml M-CSF were evaluated by immunofluorescence microscopy. A, immunofluorescence staining of in vitro cultured mouse M0, M1, and M2 macrophages labeled with the macrophage marker CD68. Scale bar, 20 μm. B, gene expression profiles by qRT-PCR of M1 (Nos2, Ccl5, and Tnf) and M2 (Arg1, Retnla, and Ym1) markers at 6 h with 100 nm Dex. C, mRNA levels of Gilz, Cxcr4, and Dpp4 regulated by glucocorticoids in M0, M1, and M2 macrophages. D, in vitro migration assay, using 10% FBS as chemoattractant, of M0, M1, and M2 macrophages treated with or without dexamethasone. After 24 h, M1 macrophages show a significant increase in spontaneous migration upon Dex treatment. E, the cell migration of primary M1 macrophages induced by CXCL12 is blocked by pharmacological inhibition of DPP4 following dexamethasone treatment. F, mRNA levels of Gilz, Cxcr4, and Dpp4 regulated by glucocorticoids in mouse PM. G, in vitro migration assay, using 10% FBS as chemoattractant, of PM treated with or without dexamethasone shows an increase in the migratory potential induced by glucocorticoids. Data are mean ± S.D. (error bars) and are representative of 3–5 independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ordinary one-way ANOVA statistical test with Tukey's multiple-comparison test and unpaired two-tailed t test.
Discussion
The glucocorticoid receptor is expressed in almost all immune cells and mediates the actions of both endogenous or exogenous glucocorticoids, acting as potent regulators of inflammation (39). Interestingly, glucocorticoids have complex and different pleiotropic effects on monocytes and macrophages, but their contribution toward systemic anti-inflammatory effects is not yet fully understood. Here, we evaluated whether the process of monocyte-to macrophage differentiation modified glucocorticoid responsiveness. Transcriptome analysis of monocyte-like THP-1 and macrophage-like THP-1 cells revealed a higher GR expression in macrophage-like THP-1 compared with monocyte-like THP-1 cells. In addition, we report the identification of the pro-diabetic and pro-inflammatory exopeptidase DPP4 as a new glucocorticoid-responsive gene exclusively regulated in macrophages. Provocatively, DPP4 promotes the migration of macrophages that is induced by glucocorticoids.
Glucocorticoids suppress inflammation through the induction of potent anti-inflammatory effects and are frequently used to treat chronic inflammatory diseases involving lymphocytes, although they are less effective in suppressing macrophage-mediated diseases (11, 13, 14). Within macrophages, glucocorticoid action depends on the context and the timing, and they also have the capacity to mediate pro-inflammatory activities, such as enhancing leukocyte trafficking and pro-inflammatory cytokine production mainly during the first steps of the immune response (9).
One theory explaining this dual effect on immune cells gene regulation is that glucocorticoids initiate opposing forces simultaneously inducing pro- and anti-inflammatory pathways as a pro-resolutive strategy to quickly recover the cellular and tissue homeostasis (7, 40). The effect of GR activation is highly gene-, cell-, and stimulus-specific, as is evident from our transcriptome data. For example, the inhibition of CSF1 (colony-stimulating factor 1) and its receptor CSF1R could be one possible reason why glucocorticoids alone would not induce the differentiation of monocytes into macrophages.
The process of differentiation of monocytes to macrophages involves major structural and biochemical changes in the cell. However, transcriptome analysis between monocyte-like THP-1 cells and THP1-MΦ demonstrated a high number of genes commonly regulated between both cell types. These associations observed could be due to different gene expression levels, rather than de novo transcription of uniquely expressed genes, as is the case for GR. Moreover, higher GR levels observed in macrophages could be related to greater sensitivity toward glucocorticoids after differentiation of monocytes.
Genes involved in cell movement, trafficking, and chemotaxis were overrepresented among up- and down-glucocorticoid-regulated genes in THP1-MΦ, with DPP4 induced and differentially regulated by Dex exclusively in THP1-MΦ and primary murine polarized M1 macrophages. DPP4, also known as CD26, was originally described as a marker of T cell differentiation and activation (41). This study provides the first evidence that DPP4 expression is directly regulated by glucocorticoids, making it a promising candidate for glucocorticoid effects in human pro-inflammatory macrophages. Additionally, the presence of a highly conserved GRE motif in the position −4,200/−4,185 in humans and mice indicates possible shared mechanisms between species.
The importance of DPP4 for the medical community lies in the approval of the use of its inhibitors for treatment of type 2 diabetes, as monotherapy, or in combination with other oral anti-diabetes drugs (42) and the benefits in decreased risk of major cardiovascular events (43, 44). DPP4 inhibitors have anti-inflammatory effects, playing a critical role in obesity-induced inflammation and insulin resistance limiting macrophage infiltration in chronic inflammatory mouse models and regulating M1/M2 balance by mediating the reversion of one to the other (24). Previously, Zhong et al. showed that DPP4 expression was increased during monocyte differentiation into dendritic cells/macrophages and that nonenzymatic DPP4 function was associated with inflammation during obesity (32).
The exopeptidase DPP4 involved in the regulation of the immune system cleaves dipeptides from the N-terminal region of peptides and proteins (with a residue of Ala or Pro in the penultimate position) as well as various chemokines (45). The loss of two amino acids resulting from DPP4 enzymatic action can cause 1) increased or reduced biological peptide/protein activity, 2) increased specificity toward the receptor, 3) ligand inactivation, or 4) generation of receptor antagonists (46). Therefore, as chemokines direct leukocyte migration under homeostasis and inflammation, DPP4 proteolytic processing could have relevant consequences for correct functioning of the immune response. Our findings in macrophage-like THP-1 cells and murine polarized M1 macrophages indicate that glucocorticoids enhance spontaneous and CXCL12-induced migration in part by inducing DPP4 expression.
Noncatalytic DPP4 functions have also been related to adhesion and migration processes and interaction with extracellular matrix proteins (fibronectin and collagen). Moreover, DPP4 inhibitors ameliorate atherosclerosis by preventing monocyte recruitment and chemotaxis via modulation of RAC-1 (21). Additionally, DPP4 in T cells interacts with the chemokine receptor CXCR4 (41), selectively binding the chemokine CXCL12 (45). This binding could promote internalization of the complex DPP4/CXCR4 in the membrane, regulating local and temporal CXCL12 activity (41, 47). Regarding the nonenzymatic activity of DPP4 regulating the macrophage mobility, Hiromura et al. (48) have shown that DPP4 inhibitors affect DPP4 and caveolin-1 (CAV-1) interaction, resulting in the suppression of inflammation in mouse and human macrophages. In addition to this, it is well-known that CAV-1 activation of the GTP-binding protein RAC-1 plays a role in cell migration (49, 50). Thus, DPP4 inhibitors could block the interaction of the DPP4/CXCR4 axis, the activation of CXCR4 by CXCL12, and finally the consequential activation of CAV-1/RAC-1 in the promotion of macrophage mobility. Interestingly, we observed that CXCR4 and CXCL12 genes were enriched in the cell movement and migration pathways by IPA. Our data show that glucocorticoids also increased the expression of these genes in macrophage-like THP-1 cells and primary mouse macrophages and that they could be another downstream regulator of migratory capacity mediated by GR activation.
Finally, enzymatic DPP4 activity induced by Dex was completely blocked by specific DPP4 inhibitors (sitagliptin and linagliptin), suggesting the possibility that synthetic glucocorticoids would present a low efficacy in the resolution of macrophage-induced inflammation. Alternatively, these data also may indicate that glucocorticoids, through DPP4 induction, potentiate the retention and egress of macrophages from inflamed tissues, perhaps contributing to their anti-inflammatory properties of glucocorticoids.
Materials and methods
Reagents
Dex and RU486 were purchased from Steraloids, Inc (Newport, RI). Heat-inactivated fetal calf serum and charcoal-stripped heat-inactivated FBS were purchased from Gemini Bio-Products (West Sacramento, CA). RPMI medium, penicillin/streptomycin, HEPES (pH 7.0), and β-mercaptoethanol were purchased from Invitrogen (Thermo Fisher Scientific). Phorbol 12-myristate 13-acetate (PMA) was purchased from Sigma–Aldrich. Recombinant human and mouse CXCL12 were purchased from Biolegend (San Diego, CA). Human anti-GR and anti-DPP-4 antibodies were purchased from Cell Signaling Technology (Danvers, MA). The DPP-4 inhibitors sitagliptin and linagliptin were purchased from Selleckchem (Houston, TX). Dharmafect, NTC, siRNA GR, and siRNA DPP4 (ON-TARGETplus siRNA) were purchased from Horizon/Dharmacon (Lafayette, CO). TaqMan® RT-PCR primer probes were purchased from Applied Biosystems (Foster City, CA).
Mouse colony maintenance
All studies were performed with approval by the NIEHS, National Institutes of Health, animal care and use committee. The mice used for these studies were C57BL/6J purchased from the Jackson Laboratories (Bar Harbor, ME). Mice were maintained in a pathogen-free facility with 12-h day/night cycles. Standard mouse chow and water were provided ad libitum.
Cell culture
The human monocytic cell line THP-1 (ATCC®-TIB-202, American Type Culture Collection, Manassas, VA) together with their derived macrophages were maintained in RPMI medium supplemented with 10% heat-inactivated fetal bovine serum, 100 units of penicillin/streptomycin, 25 μm HEPES (pH 7.0), and 50 μm β-mercaptoethanol (complete medium) at a ratio of 2.5 × 105 cells/ml under conditions of humidity at 5% CO2 and 37 °C. Monocyte-like THP-1 cells were differentiated into THP1-MΦ. Briefly, monocyte-like THP-1 cells (2 × 106 cells/well) were activated with 0.5 μm PMA in serum-free medium supplemented with 25 μm HEPES for 3 h. Subsequently, adherent cells were washed with PBS and cultured first for 24 h with recovery medium without PMA (complete RPMI) and then for another 24 h in RPMI supplemented with 10% charcoal-stripped serum before being treated with dexamethasone. The success of the differentiation protocol was evaluated using phase-contrast microscopy and flow cytometry using a fluorophore-conjugated panel of antibodies against markers of monocyte and macrophage lineages (CD15s-BV510, CD11b-PE-Cy7, and CD11c-BV421) and Cell Tracker and 7-aminoactinomycin D for viability (BD Biosciences) (Fig. S1). Where indicated, cells were pretreated with 1 or 10 μm RU-486 for 1 h prior to the addition of Dex or pretreated with Dex for 3 h prior the addition of DPP4 inhibitor (linagliptin (1 and 10 nm) or sitagliptin (25 and 50 nm)).
Generation of knockout macrophage-like THP-1 cells by siRNA
For siRNA experiments, 3-day macrophage-like THP-1 cells completely differentiated were seeded in 6-well plates at a density of 1 × 106 cells/ml and then transfected with 25 nm NTC or with a 25 nm concentration of a mixture of four siRNAs provided as a single reagent of siRNA against GR or against DPP4 (ON-TARGETplus siRNA, Dharmacon) and DharmaFECT-1 in a mixture of Opti-MEM and medium without antibiotic. The transfection reaction was maintained at 37 °C in 5% CO2 for 24 h. The transfected cells were recovered in complete medium for another 24 h and maintained in charcoal-stripped medium for an additional 24 h prior to adding 100 nm Dex for 6 h for RNA or 24 h for protein analysis. The efficiency of the transfection was evaluated by Western blotting, and cells with GR or DPP4 silencing were used for functional analysis.
Analysis of gene expression using micrroarray, NanoString, and qRT-PCR
Cultures of 1 × 106/ml of monocytes THP-1 and THP1-MΦ were stimulated for 6 h with vehicle or 100 nm Dex for analysis of gene expression by microarray and NanoString (n = 3 biological replicates/condition), from 2 to 48 h with 1, 10, 100, and 1000 nm Dex for analysis of time course and dose response and from 0.5 to 6 h with 100 nm Dex for analysis of DPP4 nascent RNA levels (forward, 5′-GCTTCCCTCTAATTGGACTTGA-3′; probe, 5′-TTGCAGACACCGTGGAAGGTTCTT-3′; reverse, 5′-ACGGTGATGATGGTGACAAG-3′) by qRT-PCR. The data from microarray (GSE135130) and NanoString (GSE135165) were deposited in a GEO database (GSE).
Microarray analysis
Following Dex stimulation, cells were collected and lysed for total RNA extraction using a Qiagen RNeasy minikit (Qiagen, Hilden, Germany). Gene expression analysis by microarray was carried out using Agilent whole human genome 4 × 44 multiplex format oligonucleotide arrays (014850) (Agilent Technologies, Santa Clara, CA) following the Agilent one-color microarray-based gene expression analysis protocol. Starting with 500 ng of total RNA, complementary RNA labeled with the Cy3 probe was synthesized according to the manufacturer's protocol. For each sample, 1.65 μg of Cy3-labeled complementary RNA was fragmented and hybridized for 17 h in a rotating hybridization oven. The oligonucleotide arrays were washed and then scanned with an Agilent scanner. Data were obtained using the Agilent Feature Extraction software (version 12), performing the error modeled, adjusting for additive and multiplicative noise. The resulting data were processed using the OmicSoft Array Studio software (version 7.0) and visualized by principal component analysis. To identify the differentially expressed probes and to determine statistical differences between the means of the groups, an analysis of variance (ANOVA) was used. In addition, we used a multiple-test correction ANOVA and Benjamini–Hochberg with a value of p < 0.05 to reduce the number of false positives.
NanoString analysis
The analysis of gene expression using the NanoString® platform (NanoString, Seattle, WA) was carried out using the Human Immunology code set (NS_Immunology_C2328), which measures 547 endogenous RNAs and 14 housekeeping genes. 50 ng of each total RNA sample was used according to the manufacturer's instructions. RNA expression was quantified in an nCounter Digital Analyzer, and raw counts were generated and normalized with nSolver software (version 3.0). The data were normalized using the manufacturer's positive and negative control probes as well as two housekeeping genes (HPRT1 and PPIA). All samples passed the initial quality assurance/quality control of nSolver, and the replicates were well-correlated (R > 0.98). The raw and normalized compiled data (log2 of counts) were reanalyzed in Partek for statistical analysis (finding 159 probes with an average expression of less than 4 counts that were excluded), with 388 probes finally subjected to ANOVA in the treatment groups with p value corrected by false discovery rate post-hoc Benjamini–Hochberg for each comparison group.
qRT-PCR analysis
The analysis of the gene expression in dose response and time course using monocyte-like THP-1 cells and THP1-MΦ, human monocyte–derived macrophages (38), murine bone marrow monocytes, bone marrow-derived macrophages, and peritoneal macrophages by qRT-PCR was carried out using 50 ng of total RNA and the One-Step RT-PCR kit (Bio-Rad) together with sets of predesigned and validated TaqMan primer/probes for each analyzed transcript (Applied Biosystems). NR3C1 (Hs00230813_m1), GILZ/TSC22D3 (Hs00608272_m1), DPP4 (Hs00897391_m1), FKBP5 (Hs01561006_m1), PER1 (Hs00242988_m1), AREG (Hs00950669_m1), NLRP3 (Hs00918082_m1), HSD11B1 (Hs01547870_m1), TNF (Hs99999043_m1), CCL2 (Hs00234140_m1), HIST1H4C (Hs00543883_s1), CCL20 (Hs00355476_m1), CYP19A1 (Hs00903411_m1), CD86 (Hs01567026_m1), NOX1 (Hs00246589_m1), HSD11B2 (Hs00388669_m1), and PPIB (Hs00168719_m1) human genes and Nr3c1 (Mm00433832_m1), Gilz/Tsc22d3 (Mm00726417_s1), Dpp4 (Mm00494549_m1), Cxcr4 (Mm01996749_s1), Nos2 (Mm00440502_m1), Ccl5 (Mm01302427_m1), Tnf (Mm00443258_m1), Arg1 (Mm00475988_m1), Retnla/Fizz1 (Mm00445109_m1), Chil3/Ym1 (Mm00657889_mH), and Ppib (Mm00478295_m1) for mouse genes. The samples were run in duplicate in the real-time thermocycler model CFX96 from Bio-Rad. The Ct values from each transcript were normalized to the housekeeping gene PPIB and expressed relative to the level of the transcript in the unstimulated condition. As a positive control of the effect of Dex, the glucocorticoid-responsive genes FKBP5 and GILZ were used. Additionally, the activity of GR and levels of each transcript regulated by Dex were evaluated in the presence or absence of RU-486.
Analysis of canonical pathways using IPA
The lists of significantly regulated genes were annotated using IPA. Enrichment or overlap of canonical pathways and the top biological functions were determined by IPA, using Fisher's test (p < 0.05). Gene networks involved in the inflammatory response, cell movement, and chemotaxis were constructed using the Pathdesigner tool of IPA.
Protein analysis by Western blotting and flow cytometry
Total proteins were extracted in radioimmune precipitation buffer (25 mm Tris-HCl (pH 7.6), 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 0.1% SDS) supplemented with an inhibitor mixture of proteases (Roche, Rotkreuz, Switzerland). Equal amounts of protein were loaded and separated in precast Novex 10% Tris-glycine minigels (Thermo Fisher Scientific) and transferred to nitrocellulose membranes under a semidry rapid transfer system (Bio-Rad) and blocked with blocking buffer (LI-COR, Lincoln, NE) for 60 min at room temperature. Subsequently, the membranes were incubated overnight at 4 °C with primary antibodies anti-GR (1:1,000 dilution) and anti-DPP4 (1:1,000 dilution) in 5% skimmed powdered milk in TBS-T and 5% BSA in TBS-T, respectively. Blots were washed and incubated with goat anti-rabbit IRDye680-conjugated secondary antibody (LI-COR) for 1 h at room temperature and visualized with a LI-COR Odyssey Imaging scanner system. The obtained immunoreactivity was normalized to β-actin and/or β-tubulin proteins as a loading control and was expressed relative to the protein level of the unstimulated condition. To determine the expression at the protein level of activation markers in monocyte-like THP-1 cells and THP1-MΦ by flow cytometry, the cells were stimulated for 24 h with 100 nm Dex, fixed in paraformaldehyde for 10 min at room temperature, and permeabilized according to the surface or intracellular staining evaluated. The immunostaining process was performed using a panel of antibodies (BD Biosciences) conjugated against CD15s-BV510, CD11b-PE-Cy7, and CD26/DPP4-PE and their respective isotypes according to the manufacturer's specifications, prior to blocking Fc using a commercial blocker. The samples were evaluated in triplicate in the LSR II cytometer (BD Biosciences) and analyzed through the software FACSDiva version 6.1.3.
Determination of the enzymatic activity of DPP4 by fluorometric assay
Monocyte-like THP-1 cells and THP1-MΦ were treated with the GR antagonist RU486 before the stimulation of Dex for 24 h or with Dex during the first 3 h before adding two concentrations of the specific inhibitors of DPP4, sitagliptin and linagliptin. The cells were collected and lysed with lysis solution according to the manufacturer's instructions using the commercial kit DPP4 Activity Assay (Sigma–Aldrich). The results were plotted as pmol/ml/min (microunits/ml), where 1 unit of DPP4 is the amount of enzyme that hydrolyzes the DPP4 substrate to produce 1.0 μmol of AMC/min at 37 °C.
In silico analysis of GREs in the human DPP4 gene
Analysis in silico using the JASPAR software database revealed the presence of putative GREs in the promoter region. These GREs were mapped and analyzed by multiple alignments against the consensus sequence using the STAMP software, demonstrating a likelihood of GR binding in those regions of the DNA. According to this, primer probes flanking each of the GREs found in the promoter region of DPP4 were used for ChIP-qPCR and FAIRE analysis.
ChIP-qPCR and FAIRE analysis
Monocyte-like THP-1 cells and THP1-MΦ seeded at a density of 1 × 106 cells/ml stimulated with or without 100 nm Dex for 2 h were collected and evaluated by ChIP using the EZ-Magna ChIPTM A/G chromatin immunoprecipitation kit with immunomagnetic beads (EMD Millipore). For this, centrifuged and pelleted cells were cross-linked using 1% formaldehyde for 10 min at room temperature followed by quenching of the reaction with 1× glycine for 5 min and then lysed and homogenized with a Dounce homogenizer for isolation of the nuclear fraction in a solution containing cOmpleteTM protease inhibitor mixture (Sigma–Aldrich). The nuclear fraction isolated was sonicated in a Bioructor® with a controlled-temperature high-pressure cooling system (Diagenode, Sparta, NJ). A fraction of the fragmented chromatin was used to evaluate the quality of chromatin through agarose gel electrophoresis. DNA fragments sized between 0.2 and 0.5 kb were immunoprecipitated using 3 μg of anti-GR mAb (Cell Signaling) or the same concentration of anti-IgG as isotype control (EMD-Millipore). Subsequently, the immunocomplexes were isolated using magnetic beads of protein A/G-agarose; washed with solutions of low and high concentration of salts, LiCl solution, and TE buffer; and treated with RNase, proteinase K, and temperature to dissociate them for recovery and elution of the DNA. Aliquots of each DNA sample recovered were purified using columns, analyzed by quantitative PCR, using primers-probes flanking the two GREs: GRE −4,200/−4,185 (forward, 5′-CCTAGTGGAGCTGTGAGAAGA-3′; probe, 5′-TCCAGTTACACGGAACAAGCTGTCC-3′; reverse, 5′-CAGGCTGGCGTTGAGTATATG-3′) and GRE −1,782/−1,767 (forward, 5′-GCACAGGGTGTGAAGATATTTG-3′; probe, 5′-TGCCCTCCAGAGAACAAATTGACCT-3′; reverse, 5′-GAGGCTGGCTGACATCTAC-3′). The Ct values of each of the samples analyzed in triplicate were compared with respect to the initial input and normalized to the IgG isotype values and expressed as the -fold enrichment of the stimulated condition compared with the control. Additionally, GRE located in the promoter of the GILZ gene was used as a positive control. FAIRE analysis was performed according to Simon et al. (51) using the same set of primers-probes previously analyzed for ChIP-qPCR.
Experimental setup for mouse peritoneal macrophages and bone marrow–derived macrophages
Peritoneal macrophages (PM) and BMDMs were isolated from 8–12-week-old C57BL/6 mice by flushing the peritoneal cavity with 5 ml of ice-cold complete medium and flushing the femur and tibia with complete medium, respectively. The BMM were purified by negative selection using the EasySepTM mouse monocyte isolation kit (Stemcell Tech, Vancouver, Canada) and resuspended in complete medium supplemented with 100 ng/ml M-CSF (Miltenyi Biotec, Bergisch Gladbach, Germany). Cells at a density of 5.0 × 105 cells/well were incubated for 6 days at 37 °C and 5% CO2 with medium change every 3 days. The BMDM phenotype was analyzed by phase-contrast microscopy and confocal immunofluorescence using anti-CD68 antibody (Biolegend, San Diego, CA); by flow cytometry of the surface markers Ly6C-PerCP/Cy5.5, CD11b-FITC, F4/80-APC and DPP4-PE, as well as the M1 and M2 markers CD80-BV421 and CD206-PE/Cy7, respectively (Biolegend); and by the gene expression profile using qRT-PCR. For polarization to M1 and M2, BMDMs unpolarized (M0) were stimulated for 24 h with 10 ng/ml murine recombinant interferon-γ (Miltenyi Biotec) and 50 ng/ml lipopolysaccharide (Sigma) and with 10 ng/ml murine recombinant interleukin-4 (Miltenyi Biotec) to the M1 and M2, respectively, in 10% charcoal-stripped serum. For the experimental setup, M0, M1, and M2 macrophages were stimulated with 100 nm Dex for 6 h for the gene expression profile and until 24 h for protein analysis.
Analysis of migratory capacity of monocyte-like THP-1 cells, THP-1–derived macrophages, peritoneal macrophages, and BMDMs unpolarized and polarized to M1 and M2
Monocyte-like THP-1 cells, 6-day THP1-MO untransfected (mock) and transfected (NTC, GR siRNA and DPP4 siRNA), were treated with or without 100 nm Dex for 24 hours and then evaluated for their migratory properties. For some experiments, cells were preincubated with 10 μm RU-486 for 1 hour prior to the addition of Dex. PM and BMDM (unpolarized and polarized to M1 and M2) were treated in the same manner as described above Immediately after the stimulation, the supernatant was collected, and the cells were washed and detached with fresh and warmed 10 mm PBS-EDTA, collected in serum-free medium (without chemoattractant molecules) or in the presence of DPP4 inhibitors, counted, and reseeded at a density of 4 × 105 cells/ml in the insert of a QCMTM Chemotaxis 5-μm 24-well migration assay (with a 5-μm pore size for monocyte/macrophage movement) (EMDMillipore, Burlington, MA), and medium with 10% FBS or 100 ng/ml CXCL12/SDF-1a (human or mouse) as a chemoattractant in the lower chamber After 6 and 24 h, the migratory cells that adhered to the lower surface of the insert in the chamber were detached, lysed, and quantitated by the incorporation of a fluorescent probe CyQUANT® GR dye in a plate reader, according to the manufacturer's instructions. Each migration assay was repeated three times. The percentage of cells migrated was calculated in relation to unstimulated or untransfected conditions.
Immunofluorescence staining
M0, M1, and M2 macrophages were grown in glass-bottom culture dishes (MatTek Corp.). Then cells were washed with warm PBS, fixed with warm 4% paraformaldehyde for 20 min at room temperature, and permeabilized in PBS containing 2% BSA and 0.1% Triton X-100 for 30 min at room temperature. Cells were then blocked for 1 h with PBS containing 5% goat serum and 0.1% Triton X-100 at room temperature prior to incubation of the specimens at 4 °C overnight with anti-CD68 (Biolegend, San Diego, CA) antibody. The following morning, samples were washed with 1× PBS containing 0.1% Tween and incubated with the secondary antibody goat anti-rat AF594 for 1 h at room temperature. Samples were then washed, air-dried, and mounted with ProLong gold antifade mountant with 4′,6-diamidino-2-phenylindole (Thermo Scientific). A Zeiss laser-scanning confocal microscope (LSM 880; Carl Zeiss) was used to analyze CD68 expression.
Statistical analysis
GraphPad Prism version 7.0 was used to analyze the data. To determine the statistical significance of the results, the two-tailed unpaired Student's t test and one- or two-way ANOVA statistical test were performed with the ad hoc post-test according to the distribution of the data. Those comparisons whose value was p < 0.05 were considered statistically significant. In all of the experiments, the samples were analyzed in duplicate, and each experiment was performed at least three times independently.
Author contributions
D. D.-J., M. A. H., and J. A. C. conceptualization; D. D.-J. and J. A. C. data curation; D. D.-J. formal analysis; D. D.-J., M. G. P., J. T. B., and J. A. C. investigation; D. D.-J., M. G. P., J. T. B., and J. A. C. methodology; D. D.-J. and J. A. C. writing-original draft; D. D.-J., M. G. P., J. T. B., M. A. H., and J. A. C. writing-review and editing; M. A. H. and J. A. C. supervision; J. A. C. resources; J. A. C. funding acquisition.
Supplementary Material
Acknowledgments
We thank Carl Bortner and Maria Sifre of the Flow Cytometry core (NIEHS, National Institutes of Health (NIH)) for assistance with flow cytometry analysis and also Kevin Gerrish, Rickie Fanin, and Liwen Liu of the Molecular Genomics core (NIEHS, NIH) for assistance with microarray data. The figures were created with BioRender.
This work was supported by Intramural Research Program of the NIEHS, National Institutes of Health, Grant 1ZIAES090057. This work was also supported by CONICYT-PFCHA/ Doctorado Nacional/ 2015/ 21150264 and a supplemental predoctoral fellowship of the NIH Research Program (Grant 37432) (to D. D. J.), a postdoctoral research associate (PRAT) fellowship from NIGMS, NIH (Grant 1Fi2GM123974) (to J. T. B.), and FONDECYT Grant 1170648 (to M. A. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S7.
- GR
- glucocorticoid receptor
- THP-1 cell
- monocyte-like THP-1 cell
- THP1-MΦ
- macrophage-like THP-1 cell(s)
- IPA
- Ingenuity Pathway Analysis®
- TNF
- tumor necrosis factor
- DPP4
- dipeptidyl peptidase-4
- GRE
- glucocorticoid-responsive element
- BMDM
- bone marrow–derived macrophage
- qPCR
- quantitative PCR
- qRT-PCR
- quantitative RT-PCR
- M-CSF
- macrophage colony-stimulating factor
- BMM
- bone marrow monocytes
- Dex
- dexamethasone
- NTC
- nontargeting control
- FAIRE
- formaldehyde-assisted isolation of regulatory elements
- CAV-1
- caveolin-1
- PMA
- phorbol 12-myristate 13-acetate
- ANOVA
- analysis of variance
- PM
- peritoneal macrophages
- FBS
- fetal bovine serum.
References
- 1. Cohen D. M., and Steger D. J. (2017) Nuclear receptor function through genomics: lessons from the glucocorticoid receptor. Trends Endocrinol. Metab. 28, 531–540 10.1016/j.tem.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Necela B. M., and Cidlowski J. A. (2004) A single amino acid change in the first zinc finger of the DNA binding domain of the glucocorticoid receptor regulates differential promoter selectivity. J. Biol. Chem. 279, 39279–39288 10.1074/jbc.M405489200 [DOI] [PubMed] [Google Scholar]
- 3. Koenen M., Culemann S., Vettorazzi S., Caratti G., Frappart L., Baum W., Krönke G., Baschant U., and Tuckermann J. P. (2018) Glucocorticoid receptor in stromal cells is essential for glucocorticoid-mediated suppression of inflammation in arthritis. Ann. Rheum. Dis. 77, 1610–1618 10.1136/annrheumdis-2017-212762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guo B., Huang X., Cooper S., and Broxmeyer H. E. (2017) Glucocorticoid hormone-induced chromatin remodeling enhances human hematopoietic stem cell homing and engraftment. Nat. Med. 23, 424–428 10.1038/nm.4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Lange P., Segeren C. M., Koper J. W., Wiemer E., Sonneveld P., Brinkmann A. O., White A., Brogan I. J., de Jong F. H., and Lamberts S. W. (2001) Expression in hematological malignancies of a glucocorticoid receptor splice variant that augments glucocorticoid receptor-mediated effects in transfected cells. Cancer Res. 61, 3937–3941 [PubMed] [Google Scholar]
- 6. Lannan E. A., Galliher-Beckley A. J., Scoltock A. B., and Cidlowski J. A. (2012) Proinflammatory actions of glucocorticoids: glucocorticoids and TNFα coregulate gene expression in vitro and in vivo. Endocrinology 153, 3701–3712 10.1210/en.2012-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Desmet S. J., and De Bosscher K. (2017) Glucocorticoid receptors: finding the middle ground. J. Clin. Invest. 127, 1136–1145 10.1172/JCI88886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hermoso M. A., Matsuguchi T., Smoak K., and Cidlowski J. A. (2004) Glucocorticoids and tumor necrosis factor α cooperatively regulate toll-like receptor 2 gene expression. Mol. Cell Biol. 24, 4743–4756 10.1128/MCB.24.11.4743-4756.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Busillo J. M., Azzam K. M., and Cidlowski J. A. (2011) Glucocorticoids sensitize the innate immune system through regulation of the NLRP3 inflammasome. J. Biol. Chem. 286, 38703–38713 10.1074/jbc.M111.275370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frank M. G., Miguel Z. D., Watkins L. R., and Maier S. F. (2010) Prior exposure to glucocorticoids sensitizes the neuroinflammatory and peripheral inflammatory responses to E. coli lipopolysaccharide. Brain Behav. Immun. 24, 19–30 10.1016/j.bbi.2009.07.008 [DOI] [PubMed] [Google Scholar]
- 11. van de Garde M. D., Martinez F. O., Melgert B. N., Hylkema M. N., Jonkers R. E., and Hamann J. (2014) Chronic exposure to glucocorticoids shapes gene expression and modulates innate and adaptive activation pathways in macrophages with distinct changes in leukocyte attraction. J. Immunol. 192, 1196–1208 10.4049/jimmunol.1302138 [DOI] [PubMed] [Google Scholar]
- 12. Murray P. J., and Wynn T. A. (2011) Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 11, 723–737 10.1038/nri3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ishiguro Y., Ohkawara T., Sakuraba H., Yamagata K., Hiraga H., Yamaguchi S., Fukuda S., Munakata A., Nakane A., and Nishihira J. (2006) Macrophage migration inhibitory factor has a proinflammatory activity via the p38 pathway in glucocorticoid-resistant ulcerative colitis. Clin. Immunol. 120, 335–341 10.1016/j.clim.2006.05.010 [DOI] [PubMed] [Google Scholar]
- 14. Apostolopoulos D., and Morand E. F. (2017) It hasn't gone away: the problem of glucocorticoid use in lupus remains. Rheumatology 56, i114–i122 10.1093/rheumatology/kew406 [DOI] [PubMed] [Google Scholar]
- 15. Ayoub S., Hickey M. J., and Morand E. F. (2008) Mechanisms of disease: macrophage migration inhibitory factor in SLE, RA and atherosclerosis. Nat. Clin. Pract. Rheumatol. 4, 98–105 10.1038/ncprheum0701 [DOI] [PubMed] [Google Scholar]
- 16. Martinez F. O., Gordon S., Locati M., and Mantovani A. (2006) Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 177, 7303–7311 10.4049/jimmunol.177.10.7303 [DOI] [PubMed] [Google Scholar]
- 17. Ginhoux F., Schultze J. L., Murray P. J., Ochando J., and Biswas S. K. (2016) New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat. Immunol. 17, 34–40 10.1038/ni.3324 [DOI] [PubMed] [Google Scholar]
- 18. Bain C. C., Bravo-Blas A., Scott C. L., Perdiguero E. G., Geissmann F., Henri S., Malissen B., Osborne L. C., Artis D., and Mowat A. M. (2014) Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 15, 929–937 10.1038/ni.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jakubzick C., Gautier E. L., Gibbings S. L., Sojka D. K., Schlitzer A., Johnson T. E., Ivanov S., Duan Q., Bala S., Condon T., van Rooijen N., Grainger J. R., Belkaid Y., Ma'ayan A., Riches D. W., et al. (2013) Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity 39, 599–610 10.1016/j.immuni.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liao X., Shen Y., Zhang R., Sugi K., Vasudevan N. T., Alaiti M. A., Sweet D. R., Zhou L., Qing Y., Gerson S. L., Fu C., Wynshaw-Boris A., Hu R., Schwartz M. A., Fujioka H., et al. (2018) Distinct roles of resident and nonresident macrophages in nonischemic cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 115, E4661–E4669 10.1073/pnas.1720065115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shah Z., Kampfrath T., Deiuliis J. A., Zhong J., Pineda C., Ying Z., Xu X., Lu B., Moffatt-Bruce S., Durairaj R., Sun Q., Mihai G., Maiseyeu A., and Rajagopalan S. (2011) Long-term dipeptidyl-peptidase 4 inhibition reduces atherosclerosis and inflammation via effects on monocyte recruitment and chemotaxis. Circulation 124, 2338–2349 10.1161/CIRCULATIONAHA.111.041418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gordon S. R., Maute R. L., Dulken B. W., Hutter G., George B. M., McCracken M. N., Gupta R., Tsai J. M., Sinha R., Corey D., Ring A. M., Connolly A. J., and Weissman I. L. (2017) PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 545, 495–499 10.1038/nature22396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cucak H., Grunnet L. G., and Rosendahl A. (2014) Accumulation of M1-like macrophages in type 2 diabetic islets is followed by a systemic shift in macrophage polarization. J. Leukoc. Biol. 95, 149–160 10.1189/jlb.0213075 [DOI] [PubMed] [Google Scholar]
- 24. Zhuge F., Ni Y., Nagashimada M., Nagata N., Xu L., Mukaida N., Kaneko S., and Ota T. (2016) DPP-4 inhibition by linagliptin attenuates obesity-related inflammation and insulin resistance by regulating M1/M2 macrophage polarization. Diabetes 65, 2966–2979 10.2337/db16-0317 [DOI] [PubMed] [Google Scholar]
- 25. Bingle L., Brown N. J., and Lewis C. E. (2002) The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J. Pathol. 196, 254–265 10.1002/path.1027 [DOI] [PubMed] [Google Scholar]
- 26. Vogel D. Y., Heijnen P. D., Breur M., de Vries H. E., Tool A. T., Amor S., and Dijkstra C. D. (2014) Macrophages migrate in an activation-dependent manner to chemokines involved in neuroinflammation. J. Neuroinflammation 11, 23 10.1186/1742-2094-11-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Angsana J., Chen J., Liu L., Haller C. A., and Chaikof E. L. (2016) Efferocytosis as a regulator of macrophage chemokine receptor expression and polarization. Eur. J. Immunol. 46, 1592–1599 10.1002/eji.201546262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heideveld E., Hampton-O'Neil L. A., Cross S. J., van Alphen F. P. J., van den Biggelaar M., Toye A. M., and van den Akker E. (2018) Glucocorticoids induce differentiation of monocytes towards macrophages that share functional and phenotypical aspects with erythroblastic island macrophages. Haematologica 103, 395–405 10.3324/haematol.2017.179341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhong J., Maiseyeu A., Davis S. N., and Rajagopalan S. (2015) DPP4 in cardiometabolic disease: recent insights from the laboratory and clinical trials of DPP4 inhibition. Circ. Res. 116, 1491–1504 10.1161/CIRCRESAHA.116.305665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Röhrborn D., Wronkowitz N., and Eckel J. (2015) DPP4 in diabetes. Front. Immunol. 6, 386 10.3389/fimmu.2015.00386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Röhrborn D., Brückner J., Sell H., and Eckel J. (2016) Reduced DPP4 activity improves insulin signaling in primary human adipocytes. Biochem. Biophys. Res. Commun. 471, 348–354 10.1016/j.bbrc.2016.02.019 [DOI] [PubMed] [Google Scholar]
- 32. Zhong J., Rao X., Deiuliis J., Braunstein Z., Narula V., Hazey J., Mikami D., Needleman B., Satoskar A. R., and Rajagopalan S. (2013) A potential role for dendritic cell/macrophage-expressing DPP4 in obesity-induced visceral inflammation. Diabetes 62, 149–157 10.2337/db12-0230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goldstein B. J., Feinglos M. N., Lunceford J. K., Johnson J., Williams-Herman D. E., and Sitagliptin 036 Study Group (2007) Effect of initial combination therapy with sitagliptin, a dipeptidyl peptidase-4 inhibitor, and metformin on glycemic control in patients with type 2 diabetes. Diabetes Care 30, 1979–1987 10.2337/dc07-0627 [DOI] [PubMed] [Google Scholar]
- 34. Takahashi H., Nishimura R., Tsujino D., and Utsunomiya K. (2019) Which is better, high-dose metformin monotherapy or low-dose metformin/linagliptin combination therapy, in improving glycemic variability in type 2 diabetes patients with insufficient glycemic control despite low-dose metformin monotherapy? A randomized, cross-over, continuous glucose monitoring-based pilot study. J. Diabetes Investig. 10, 714–722 10.1111/jdi.12922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vilsbøll T., Ekholm E., Johnsson E., Dronamraju N., Jabbour S., and Lind M. (2019) Dapagliflozin plus saxagliptin add-on therapy compared with insulin in patients with type 2 diabetes poorly controlled by metformin with or without sulfonylurea therapy: a randomized clinical trial. Diabetes Care 42, 1464–1472 10.2337/dc18-1988 [DOI] [PubMed] [Google Scholar]
- 36. van Genugten R. E., van Raalte D. H., Muskiet M. H., Heymans M. W., Pouwels P. J., Ouwens D. M., Mari A., and Diamant M. (2014) Does dipeptidyl peptidase-4 inhibition prevent the diabetogenic effects of glucocorticoids in men with the metabolic syndrome? A randomized controlled trial. Eur. J. Endocrinol. 170, 429–439 10.1530/EJE-13-0610 [DOI] [PubMed] [Google Scholar]
- 37. Yata Y., Hosojima M., Kabasawa H., Ishikawa T., Kaseda R., Iino N., Suzuki Y., Saito A., and Narita I. (2017) The assessment of the efficacy of dipeptidyl peptidase-4 inhibitors in patients with glucocorticoid-induced diabetes by continuous glucose monitoring. Intern. Med. 56, 2555–2562 10.2169/internalmedicine.8296-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jewell C. M., Katen K. S., Barber L. M., Cannon C., Garantziotis S., and Cidlowski J. A. (2016) Healthy glucocorticoid receptor N363S carriers dysregulate gene expression associated with metabolic syndrome. Am. J. Physiol. Endocrinol. Metab. 311, E741–E748 10.1152/ajpendo.00105.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Busillo J. M., and Cidlowski J. A. (2013) The five Rs of glucocorticoid action during inflammation: ready, reinforce, repress, resolve, and restore. Trends Endocrinol. Metab. 24, 109–119 10.1016/j.tem.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cain D. W., and Cidlowski J. A. (2017) Immune regulation by glucocorticoids. Nat. Rev. Immunol. 17, 233–247 10.1038/nri.2017.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Herrera C., Morimoto C., Blanco J., Mallol J., Arenzana F., Lluis C., and Franco R. (2001) Comodulation of CXCR4 and CD26 in human lymphocytes. J. Biol. Chem. 276, 19532–19539 10.1074/jbc.M004586200 [DOI] [PubMed] [Google Scholar]
- 42. Crowley M. J., Williams J. W. Jr, Kosinski A. S., D'Alessio D. A., and Buse J. B. (2017) Metformin use may moderate the effect of DPP-4 inhibitors on cardiovascular outcomes. Diabetes Care 40, 1787–1789 10.2337/dc17-1528 [DOI] [PubMed] [Google Scholar]
- 43. Yen F. S., Chiang J. H., Pan C. W., Lin B. J., Wei J. C., and Hsu C. C. (2018) Cardiovascular outcomes of dipeptidyl peptidase-4 inhibitors in patients with type 2 diabetes on insulin therapy. Diabetes Res. Clin. Pract. 140, 279–287 10.1016/j.diabres.2018.04.012 [DOI] [PubMed] [Google Scholar]
- 44. Ou S. M., Chen H. T., Kuo S. C., Chen T. J., Shih C. J., and Chen Y. T. (2017) Dipeptidyl peptidase-4 inhibitors and cardiovascular risks in patients with pre-existing heart failure. Heart 103, 414–420 10.1136/heartjnl-2016-309687 [DOI] [PubMed] [Google Scholar]
- 45. Zhong J., and Rajagopalan S. (2015) Dipeptidyl peptidase-4 regulation of SDF-1/CXCR4 axis: implications for cardiovascular disease. Front. Immunol. 6, 477 10.3389/fimmu.2015.00477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blauenfeldt T., Petrone L., Del Nonno F., Baiocchini A., Falasca L., Chiacchio T., Bondet V., Vanini V., Palmieri F., Galluccio G., Casrouge A., Eugen-Olsen J., Albert M. L., Goletti D., Duffy D., and Ruhwald M. (2018) Interplay of DDP4 and IP-10 as a potential mechanism for cell recruitment to tuberculosis lesions. Front. Immunol. 9, 1456 10.3389/fimmu.2018.01456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Broxmeyer H. E., Capitano M., Campbell T. B., Hangoc G., and Cooper S. (2016) Modulation of hematopoietic chemokine effects in vitro and in vivo by DPP-4/CD26. Stem Cells Dev. 25, 575–585 10.1089/scd.2016.0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hiromura M., Nohtomi K., Mori Y., Kataoka H., Sugano M., Ohnuma K., Kuwata H., and Hirano T. (2018) Caveolin-1, a binding protein of CD26, is essential for the anti-inflammatory effects of dipeptidyl peptidase-4 inhibitors on human and mouse macrophages. Biochem. Biophys. Res. Commun. 495, 223–229 10.1016/j.bbrc.2017.11.016 [DOI] [PubMed] [Google Scholar]
- 49. Díaz J., Mendoza P., Ortiz R., Díaz N., Leyton L., Stupack D., Quest A. F., and Torres V. A. (2014) Rab5 is required in metastatic cancer cells for Caveolin-1-enhanced Rac1 activation, migration and invasion. J. Cell Sci. 127, 2401–2406 10.1242/jcs.141689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Grande-García A., Echarri A., de Rooij J., Alderson N. B., Waterman-Storer C. M., Valdivielso J. M., and del Pozo M. A. (2007) Caveolin-1 regulates cell polarization and directional migration through Src kinase and Rho GTPases. J. Cell Biol. 177, 683–694 10.1083/jcb.200701006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Simon J. M., Giresi P. G., Davis I. J., and Lieb J. D. (2013) A detailed protocol for formaldehyde-assisted isolation of regulatory elements (FAIRE). Curr. Protoc. Mol. Biol., Chapter 21, Unit 21.26 10.1002/0471142727.mb2126s102 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.