Pyrazinamide (PZA) is a cornerstone antimicrobial drug used exclusively for the treatment of tuberculosis (TB). Due to its ability to shorten drug therapy by 3 months and reduce disease relapse rates, PZA is considered an irreplaceable component of standard first-line short-course therapy for drug-susceptible TB and second-line treatment regimens for multidrug-resistant TB. Despite over 60 years of research on PZA and its crucial role in current and future TB treatment regimens, the mode of action of this unique drug remains unclear.
KEYWORDS: pyrazinamide, tuberculosis, drug resistance, drug susceptibility, mode of action, coenzyme A, antimicrobial activity, drug resistance mechanisms
SUMMARY
Pyrazinamide (PZA) is a cornerstone antimicrobial drug used exclusively for the treatment of tuberculosis (TB). Due to its ability to shorten drug therapy by 3 months and reduce disease relapse rates, PZA is considered an irreplaceable component of standard first-line short-course therapy for drug-susceptible TB and second-line treatment regimens for multidrug-resistant TB. Despite over 60 years of research on PZA and its crucial role in current and future TB treatment regimens, the mode of action of this unique drug remains unclear. Defining the mode of action for PZA will open new avenues for rational design of novel therapeutic approaches for the treatment of TB. In this review, we discuss the four prevailing models for PZA action, recent developments in modulation of PZA susceptibility and resistance, and outlooks for future research and drug development.
INTRODUCTION
In a 1954 urgent call to scientific action (1), Floyd M. Feldmann, medical director at the National Tuberculosis Association in New York, argued the case for accelerated research into fundamental aspects of tuberculosis (TB) therapy. Feldmann described knowledge gaps concerning the newly discovered sterilizing drug pyrazinamide (PZA). In highlighting areas of need for rapid progress, he posed some basic questions such as “Does [PZA] work in other animal species [besides mice]?” “What is the optimum drug dosage?” and “How [does PZA] work?” More than 65 years later, we know that Feldmann’s seemingly simple questions have rather complicated and currently incomplete answers.
Regarding mechanism of action, we know that PZA is a prodrug that is hydrolyzed to pyrazinoic acid (POA) in the mycobacterial cytoplasm by the Mycobacterium tuberculosis pyrazinamidase/nicotinamidase (PZase) (2, 3). This amidase, encoded by pncA (4), is involved in the salvage pathway for synthesis of the essential cofactor NAD. Since the NAD salvage pathway is nonessential for virulence of M. tuberculosis (5, 6), pncA loss-of-function mutations represent the most prevalent mechanism for PZA resistance in clinical isolates (4, 7–13). Mutations within pncA, including single-nucleotide polymorphisms (SNPs), multinucleotide polymorphisms, and indels, have been mapped along the entire span of the 561-bp open reading frame in PZA-resistant clinical isolates (14–20). Mutations in pncA have been shown to confer resistance largely through the loss of PncA enzymatic activity and/or protein abundance (21, 22). While loss-of-function mutations in pncA represent the major mechanism of PZA resistance in M. tuberculosis clinical isolates, up to 30% of PZA-resistant isolates show PZase activity and possess a wild-type pncA gene (15, 23, 24). The latter class of PZA-resistant strains indicates the existence of additional resistance mechanisms that remain to be defined.
Regarding in vivo efficacy, it is now well known that PZA is a sterilizing drug that is exquisitely selective against M. tuberculosis in multiple animal species, including mice (25, 26), rabbits (27, 28), nonhuman primates (29), humans (30, 31), and guinea pigs (32–34). Through extensive clinical trials, PZA has been found to shorten the required duration of TB therapy by 3 months (35–37) and shows activity against both replicating and slow-growing and nongrowing populations of M. tuberculosis bacilli (38–40). Despite the important role of PZA in TB therapy, a significant proportion of those receiving PZA treatment might not achieve the necessary concentrations required for therapeutic benefit due to differences in drug metabolism between individuals (41–43). Furthermore, patient adherence or early cessation of PZA treatment is influenced by its large dosing regimen (25 mg/kg of body weight daily) and adverse side effects, such as liver inflammation, gastrointestinal distress, and joint pain (44–46). Thus, development of more tolerable or more potent PZA or POA analogs may be necessary to treat some populations. Regardless of these unknowns, due to its unparalleled sterilizing activity in the majority of individuals, PZA has become an irreplaceable component of the first-line standard short-course therapy for drug-susceptible TB (47–50) and second-line treatment regimens for multidrug-resistant TB (MDR-TB) (51–53). Further, PZA is anticipated to be a component of future TB therapies (54, 55) involving novel drugs such as bedaquiline (TMC207) (56), the bicyclic nitromidazole pretomanid (PA-824) (57), and moxifloxacin (56).
Despite the indispensable role of PZA in modern TB drug therapy, the mechanistic basis for its action remains unresolved. Feldmann’s question “How [does PZA] work?” both fascinates and torments geneticists, microbiologists, and biochemists alike. Advances in mycobacterial genetics, transcriptomics, metabolomics, antibiotic resistance surveillance, and whole-genome sequencing have enabled researchers to identify multiple PZA-linked metabolic pathways that potentially converge on a single cellular process (58). We discuss the merits and drawbacks of four proposed models for the mechanism of PZA action, recent developments in modulation of PZA susceptibility and resistance, and outlooks for future research and drug development.
MODEL 1: PYRAZINOIC ACID FUNCTIONS AS A PROTONOPHORE
The discovery and implementation of PZA as a TB drug are a fascinating story and are covered in depth by Zhang and Mitchison (59) and Murray (60). In brief, PZA was discovered in a screen for antitubercular structural analogs of nicotinamide (vitamin B3) (61) following the unexpected observation that this vitamin had antitubercular activity in mice (62) and in humans (63, 64). Early experimental studies of the antitubercular action of PZA were largely restricted to mice infected with M. tuberculosis (25, 26, 61, 65) because PZA showed no inhibitory activity against the bacilli in standard mycobacterial culture medium (66). Reductionist bacterial culture-based approaches involving PZA were not possible until it was found that exposure of M. tuberculosis to mildly acidic conditions could induce PZA susceptibility (67). Dependence on an acidic environment to promote susceptibility to PZA was proposed as the major discrepancy between in vitro and in vivo environmental conditions. Consistent with this prediction, during initial infection, M. tuberculosis is engulfed by alveolar macrophages, in which the bacilli replicate within immature phagosomes with a pH of ∼6.2 (68–70). Upon interferon gamma-mediated activation, phagosomal acidification ensues (pH 4.5 to 5.0) (70, 71), rendering this niche well within the pH requirements for induction of PZA susceptibility of M. tuberculosis. Consistent with acidic pH as a driver for PZA susceptibility of M. tuberculosis in vivo, mice that produce tubercle lesions with alkaline pH respond poorly to treatment with PZA (72, 73).
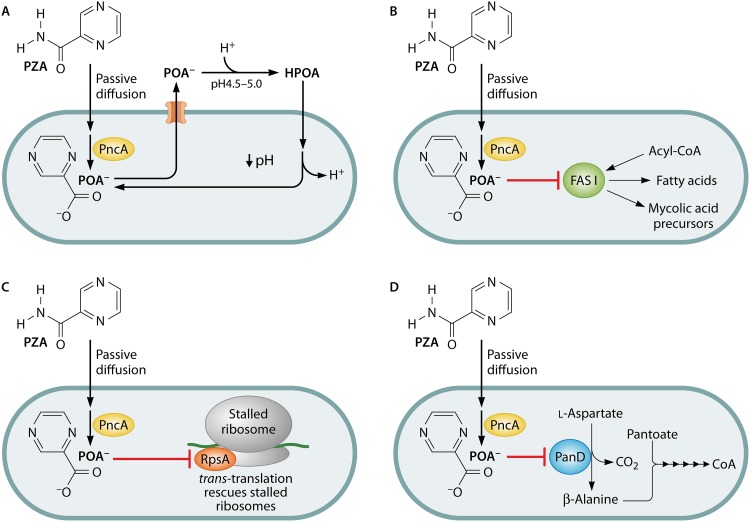
Acidic pH-driven susceptibility was the initial inspiration for the first mechanistic model for PZA action proposed by Zhang and colleagues (74–76) and has been extensively reviewed (77–79) (Fig. 1A). Under this model, PZA enters M. tuberculosis by passive diffusion across the cellular envelope to the cytoplasm (pH 7.2), where it is converted to the weak acid POA (pKa of 2.9) by PZase (76). POA anion is then exported from the bacillus through an unidentified weak efflux mechanism. In an acidic environment (e.g., activated phagosome or acidified culture medium), a small fraction of POA becomes protonated to form HPOA, which can permeate back across the bacterial envelope into the cytoplasm. By the Henderson-Hasselbalch equation {pH = pKa + log10 ([A−]/[HA])}, the theoretical amounts of protonated POA would be 0.1% at pH 5.8 and 0.008% at pH 7.0 (74). Once in the cytoplasm, HPOA dissociates to H+ and POA, and this cycle continues, resulting in cytoplasmic accumulation of protons, collapse of the cellular membrane potential, and acidification of the cytoplasm.
FIG 1.
Proposed modes of antitubercular action of pyrazinamide. Pyrazinamide enters the cell by diffusion and is activated by the cytoplasmic pyrazinamidase/nicotinamidase PncA. Pyrazinoic acid has been proposed to act as a protonophore leading to the acidification of the bacterial cytoplasm (A), an inhibitor of fatty acid synthase I (B), an inhibitor of trans-translation (C), and/or an inhibitor of coenzyme A biosynthesis (D).
Consistent with this model for POA as a protonophore, it has been shown that PZA treatment is associated with disruption of intrabacterial pH (pHIB) from 7.2 to below 6.5 within 48 h of treatment at pH 4.5 (80). Further, compounds that interfere with oxidative phosphorylation, such as the membrane potential uncoupling ionophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP; pKa, 4.8) and the FoF1-type ATP synthase inhibitor N,N′-dicyclohexylcarbodiimide (DCCD), have been reported to be synergistic with PZA when used in combination against M. tuberculosis in culture (74). Moreover, treatment of Mycobacterium bovis BCG with POA resulted in a progressive depletion in bacterial levels of ATP (81). While the protonophore model is widely cited within the literature, the studies described above did not directly address whether these physiologically relevant effects were the direct result of the proposed proton shuttling mechanism or a downstream effect of a yet-to-be-defined activity of POA. Consistent with the latter, collective evidence demonstrates that acidic pH is not strictly required for PZA action. Indeed, under near neutral culture conditions, PZA susceptibility can be promoted by overexpression of pncA (82, 83), inhibition of efflux pumps to prevent POA export from the bacilli (74, 84, 85), exposure of bacilli to conditions such as alkaline pH (67), nutrient limitation (83, 86), decreased temperature (87), and hypoxia (88–90), and replacement of PZA with POA (29). The nonessentiality of acidic pH for PZA and POA susceptibility of M. tuberculosis challenges the protonophore model as the principal basis for action of this drug.
Peterson et al. (83) recently compared the activity of bona fide ionophores with those of PZA and POA for the ability to disrupt membrane potential and pHIB under conditions that mediate susceptibility. pHIB homeostasis was assessed in an M. tuberculosis strain harboring a plasmid encoding a pH-sensitive ratiometric green fluorescent protein (pH-GFP) (91). This strain was treated with PZA, POA, CCCP (80, 92, 93), or monensin (93–95) under standard conditions used for PZA susceptibility testing (pH 5.8). While both CCCP and monensin led to rapid and dose-dependent intrabacterial acidification, there was no significant change in pHIB associated with POA and PZA treatment within the first 3 h (83). Since POA has a low pKa relative to those of CCCP (pKa, 4.8) and monensin (pKa, 6.6), it is not surprising that POA does not have robust protonophore activity under the experimental conditions that were used. Furthermore, PZA treatment of M. tuberculosis overexpressing pncA at neutral pH did not lead to a measurable decrease in pHIB despite full inhibition of bacterial growth (83). Membrane potential was also measured using a membrane-permeable fluorescent dye (DiOC2) in M. tuberculosis cells treated with CCCP or with POA at concentrations up to 10-fold over the MIC (83). As expected, CCCP treatment resulted in a dramatic loss of membrane potential. However, even at concentrations 10-fold above the MIC, POA failed to alter the membrane potential of M. tuberculosis in medium at pH 5.8 over the period that was evaluated (83).
The dispensability of acidic pH for PZA and POA action, lack of cytoplasmic acidification following exposure to inhibitory concentrations of PZA and POA, and lack of impact on membrane potential in treated and control cultures significantly undermine the protonophore model as the principal basis for POA action. It is likely that acidic conditions within the phagosomal compartment of activated macrophages provide the initial salvo for host-mediated potentiation of PZA action, but POA-dependent disruption of membrane potential and pHIB seem unlikely to be the driving forces behind the sterilizing activity of this drug.
MODEL 2: PYRAZINOIC ACID INHIBITS MYCOBACTERIAL FATTY ACID SYNTHASE I
In a second model for the mechanism of PZA action, it was proposed that POA selectively inhibits mycobacterial fatty acid synthase I (FAS-I) (96), a large multifunctional enzyme required for synthesis of C16 to C26 fatty acids (97–99) (Fig. 1B). Like its eukaryotic homolog, this enzyme contains all domains necessary for synthesis of fatty acids using acetyl coenzyme A (acetyl-CoA) as the primer unit and malonyl-CoA as two-carbon extender units (100, 101). In each round of extension, two molecules of NADPH are oxidized via the enoyl and beta-ketoacyl reductase activities of FAS-I (102). In mycobacterial species, FAS-I products can either be utilized for synthesis of cytoplasmic membrane lipids or be transferred to the fatty acid synthase II complex for synthesis of a diverse array of mycolic acids (101).
The FAS-I inhibition model for PZA action emerged from a study in which genomic DNA libraries from M. bovis BCG and Mycobacterium avium were expressed from multicopy cosmids in Mycobacterium smegmatis to screen for loci that conferred resistance to a structural analog of PZA, 5-chloropyrazinamide (5-Cl-PZA) (96). Subcloning analysis demonstrated that 5-Cl-PZA resistance was associated with fas-I overexpression (96). Likewise, overexpression of the M. tuberculosis fas-I gene in M. smegmatis also conferred resistance to 5-Cl-PZA (96). Since M. smegmatis is intrinsically resistant to PZA and POA, and overexpression of FAS-I was found to be toxic for M. tuberculosis, it was not possible to determine whether FAS-I overexpression could confer resistance to PZA or POA in mycobacteria (96). Through the use of [14C]acetate labeling studies, it was found that susceptible mycobacteria treated with PZA (82, 96, 103), 5-Cl-PZA (82, 96, 99, 103), and other PZA derivatives (103) showed a significant reduction in C16 to C26 fatty acid biosynthesis. Despite these findings, a direct association between PZA action and FAS-I inhibition was questioned by Boshoff et al. (104). Although 5-Cl-PZA was confirmed as a potent and irreversible FAS-I inhibitor, POA did not inhibit purified mycobacterial FAS-I at physiologically relevant concentrations (104). In a subsequent study, inhibition of recombinant M. tuberculosis FAS-I was confirmed, yet more than 9 mM POA was required to achieve 50% inhibition, in contrast to just 15 μM for 5-Cl-PZA (105). Since an intrabacterial concentration of 0.5 mM POA is sufficient for M. tuberculosis growth arrest (74), it seems unlikely that direct inhibition of FAS-I via POA is sufficient to explain mode of action. Enzymology and ligand interaction studies involving saturation transfer difference nuclear magnetic resonance (NMR) showed that PZA, 5-Cl-PZA, and other PZA analogs are competitive inhibitors of NADPH binding to purified mycobacterial FAS-I (105–107). In contrast, while POA was found to interact with FAS-I, it did not compete with NADPH for binding (107), indicating that association of 5-Cl-PZA and POA with FAS-I is mechanistically distinct. Together, these observations suggest that POA does not directly inhibit FAS-I, and inhibition of fatty acid synthesis by POA may be due to a linked metabolic disruption or inhibition of FAS-I by an as-yet-unidentified mycobacterial metabolite of POA.
MODEL 3: PYRAZINOIC ACID BINDS TO RpsA AND INHIBITS TRANS-TRANSLATION
In a third model for the mechanism of PZA action, it was suggested that POA selectively disrupts the process of trans-translation (108) (Fig. 1C). trans-Translation, discovered by Keiler et al. (109), is a ribosome salvage pathway used by nearly all bacterial species to free ribosomes that cannot disengage from the 3′ end of an mRNA lacking an in-frame stop codon (non-stop mRNA) (110). Without ribosome rescue, ribosomes can become sequestered by non-stop mRNAs, ultimately resulting in cell death due to arrest of protein synthesis (110). In the trans-translation pathway, SmpB and elongation factor Tu recruit tmRNA, a specialized RNA that has both tRNA and mRNA properties, to stalled ribosomes that lack an mRNA codon at the A site (111–113). Once recruited, alanine-charged tmRNA acts as a codon-independent tRNA and becomes linked to the nascent peptide through transpeptidation (109). Cotranslational switching then results in release of the non-stop mRNA with replacement by a loop of the tmRNA which encodes a degradation tag (109, 113). Following translation of this tag, the nascent peptide is released and targeted for proteolysis, and the ribosome disengages from tmRNA and is free to initiate translation of other available mRNA (109). This pathway is essential for viability of M. tuberculosis and many other bacterial pathogens and represents an outstanding novel target for drug discovery (114–117). Indeed, structurally related families of oxadiazole and tetrazole-based compounds have recently been identified that inhibit trans-translation in a large number of bacterial species, including Gram-negative, Gram-positive, and mycobacterial species (114–117).
The model for inhibition of trans-translation by POA emerged from a study focused on a presumed interaction between POA and the M. tuberculosis 30S ribosomal subunit protein S1 (108). In an attempt to identify interaction partners and putative targets of POA, Shi et al. (108) performed affinity chromatography studies in which the POA derivative 5-hydroxyl-2-pyrazinecarboxylic acid was covalently linked to a Sepharose column and used as a binding matrix for proteins from a whole-cell lysate of M. tuberculosis strain H37Ra. Nonspecific stripping of all proteins that had bound to the column using 25% ethylene glycol resulted in isolation of multiple proteins, of which RpsA (30S ribosomal protein subunit S1), Rv2783, Rv2731, and Rv3169 were identified by mass spectrometry (108). Consistent with a role for RpsA in PZA action, the authors stated that overexpression of rpsA conferred 5-fold resistance to PZA (108). In addition, the clinical isolate M. tuberculosis strain DHMH444, which shows 2-fold resistance to PZA and carries a wild-type pncA allele (12, 118), was found to harbor deletion of an alanine codon at position 438 (ΔA438) within the C-terminal region of the rpsA product (108). Isothermal titration calorimetry (ITC), an approach that can be used to determine ligand binding affinities through monitoring changes in free energy, was employed to evaluate a possible interaction between POA and purified recombinant RpsA (108). Titration of a saturated solution of POA (∼70 mM) into a solution of 10 μM wild-type M. tuberculosis RpsA showed a robust exothermic signal (108). When 100 μM POA was titrated into solutions of 10 μM M. smegmatis RpsA and M. tuberculosis RpsAΔA438, no signal was observed (108). While use of starkly different concentrations of POA in these assays makes it impossible to interpret these findings, it was concluded that POA bound wild-type M. tuberculosis RpsA with high affinity and failed to interact with M. smegmatis RpsA and M. tuberculosis RpsAΔA438 (108).
To evaluate whether POA could disrupt trans-translation, cell-free in vitro translation assays were conducted in reaction mixtures containing ribosomes isolated from M. tuberculosis, M. smegmatis, or Escherichia coli and supplemented with a charged tRNA mixture, M. tuberculosis SmpB, and unprocessed pre-tmRNA (108). Translation was assessed by detecting incorporation of [35S]methionine into dihydrofolate reductase (DHFR) expressed from an mRNA containing an in-frame stop codon (wild-type DHFR), or a similar message with the DHFR coding sequence followed by 8 rare AGG codons, 18 additional downstream codons, and an in-frame stop codon (DHFR 8×AGG) designed to induce translational stalling (108, 119). It is important to note that for rare codon-mediated translational stalling to trigger trans-translation, the culprit mRNA must be cleaved by an RNase in order to permit interaction between the ribosome and aminoacyl-tmRNA/SmpB complex (111, 119). In the assays reported by Shi et al. (108), if trans-translation were to ensue from stalling on the DHFR 8×AGG message, the resulting peptide would be extended by 13 amino acids corresponding to the tmRNA degradation tag. Since DHFR 8×AGG produced by standard translation would be extended by 26 amino acids, it would be critical to characterize the C-terminal residues of the resulting peptide. As expected, POA treatment had no impact on translation of wild-type DHFR by M. tuberculosis ribosomes or of DHFR 8×AGG with ribosomes from M. smegmatis and E. coli (108). In contrast, translation of DHFR 8×AGG by M. tuberculosis ribosomes was fully inhibited by the addition of POA at concentrations of 200 μM and greater (108). Unfortunately, the authors did not determine whether the shifted DHFR contained the tmRNA degradation tag or simply the 26 additional amino acids introduced by standard translation of DHFR 8×AGG (108). It is curious that these data differ from those in an earlier version of the manuscript that was deposited in the NCBI database (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3502614/), where signal for translation of DHFR 8×AGG is visible in the presence of as much as 800 μM POA. Regardless, it is of fundamental importance that since translation precedes trans-translation, inhibition of trans-translation would have resulted in synthesis of a nontagged DHFR. Thus, the reported results showing full inhibition of signal for protein synthesis are consistent with inhibition of translation, not inhibition of trans-translation.
Inspired by the findings of Shi et al. (108), several groups evaluated whether targeted sequencing of rpsA could be used to predict PZA resistance in M. tuberculosis clinical isolates bearing a wild-type pncA locus. Mutations within rpsA were identified in a limited number of strains, although no clear association with PZA resistance has been established (23, 108, 120–126). Alexander et al. (120) speculated that the RpsA C-terminal region is tolerant to amino acid substitutions and may be innocuous with respect to PZA action. In response to a comment posted by Simons and colleagues (121), Alexander et al. (127) cautioned that research attempting to attribute drug resistance to novel mutations must be tempered with experiments demonstrating linkage between phenotype and genotype. Indeed, two independent groups reconstructed the rpsA ΔA438 allele in M. tuberculosis, and both showed a <2-fold change in PZA susceptibility (128). Importantly, mutations in rpsA, including Δ438, have been identified in PZA-susceptible clinical isolates of M. tuberculosis (129, 130). Moreover, 10-fold overexpression of rpsA had no measurable impact on PZA susceptibility (128). Lack of an association between rpsA and PZA resistance is consistent with previous reports by Spiers et al. and Klemens et al., which demonstrated that M. tuberculosis strain DHMH444 is fully susceptible to POA in vitro (118) and to PZA in a murine model of infection (131), respectively. Collectively, these observations demonstrate that rpsA is not associated with PZA or POA susceptibility, and the low-level PZA resistance of M. tuberculosis strain DHMH444 is most likely due to its documented reduced level of PncA activity (118).
To reevaluate the possible interaction between POA and M. tuberculosis RpsA, Dillon et al. (128) repeated ITC ligand interaction studies described by Shi et al. (108). Studies with E. coli (132) and Pseudomonas (133) have shown that RpsA binds to single-stranded RNA and is important for translation initiation. When RpsA was titrated with poly(C) RNA, a robust bimodal interaction signal was observed, indicative of two high-affinity single-stranded RNA binding sites of RpsA. However, when 100 μM RpsA (pH 7.4) was titrated with a saturated solution of POA (pH 7.4), no change in free energy was detected, indicating that these solutes do not show a measurable interaction. In contrast, when the pH of the saturated solution of POA (pH 2.3) was not adjusted to match the diluent buffer and was titrated into near neutral (pH 7.4) phosphate buffer, a robust exothermic signal was observed (128), similar to that reported by Shi et al. (108). This signal was abolished when the pH of the saturated POA solution was adjusted to that of the diluent buffer (128). Thus, it is most probable that the signal reported by Shi et al. was a result of pH-dependent proton dissociation and not reflective of interaction between RpsA and POA.
Despite the ability of RpsA to interact with tmRNA (134), RpsA has been shown to be entirely dispensable for trans-translation in species in which its role has been evaluated, such as E. coli (135) and Thermus thermophilus (136, 137). To further examine the connection between POA action and mycobacterial trans-translation, Dillon et al. conducted cell-free in vitro trans-translation assays in reaction mixtures containing M. tuberculosis ribosomes supplemented with a charged tRNA mixture, appropriately processed and charged tmRNA, and M. tuberculosis SmpB (128). Rather than relying on translational stalling and mRNA cleavage to trigger trans-translation, a non-stop DHFR transcript was used (128). In these assays, trans-translational tagging of the non-stop DHFR was confirmed and tagging could be inhibited by an antisense oligonucleotide directed against tmRNA (115). Importantly, no inhibitory effect on trans-translation was observed with as much as 1 mM POA (128). Thus, the antitubercular activity of PZA is independent of trans-translation and RpsA.
MODEL 4: PYRAZINOIC ACID BLOCKS COENZYME A SYNTHESIS THROUGH INHIBITION OF l-ASPARTATE DECARBOXYLASE
In order to discover novel mechanisms for PZA resistance in M. tuberculosis, Zhang et al. isolated and characterized a large number of spontaneous PZA-resistant isolates (138). Of 174 strains that were analyzed, 169 had mutations in pncA, while 5 carried wild-type pncA and were subjected to full-genome resequencing. These pncA wild-type strains were found to harbor missense mutations within the panD (Rv3601c) gene, which encodes l-aspartate decarboxylase (138), a rate-limiting step in the CoA biosynthetic pathway (139, 140). In this pathway, β-alanine and l-pantoate are ligated by PanC (141) to form pantothenate, which is processed in five additional steps to afford CoA (Fig. 1D) (142). Further work revealed that PZA resistance phenotypes could be recapitulated in H37Ra overexpression of wild-type or mutant panD as well as panD from E. coli and M. smegmatis (143). Consistent with a role for POA in disruption of CoA biosynthesis, multiple recent studies have demonstrated that supplementation of culture medium with pathway intermediates, such as β-alanine, pantothenate, and pantetheine, can potently antagonize PZA- and POA-mediated growth inhibition of M. tuberculosis (143–145). In addition, Dillon et al. (144) demonstrated that other β-alanine-containing metabolites as well as the β-alanine structural analogs 3-aminopropanol and propanoic acid could antagonize PZA action. However, the β-alanine precursor, aspartate, and cosubstrate in pantothenate synthesis, pantoate, were not sufficient to induce an antagonistic effect (144). These data suggest that POA interacts in some way with the CoA biosynthetic pathway.
As CoA is an essential acyl carrier for hundreds of reactions in central metabolism (142), CoA depletion would provide an enticing explanation for the action of POA that unifies previous observations, such as the previously discussed impairments of energy metabolism and fatty acid synthesis. Notably, several groups have shown stress conditions that decrease cellular CoA pools, such as nutrient starvation and hypoxia, augment PZA susceptibility in M. tuberculosis (88–90). In fact, PZA treatment in anaerobic cultures of M. tuberculosis resulted in a 2-log reduction of bacterial CFU after 5 days (88). Recent work by Gopal et al. (145) has shown that wild-type M. bovis BCG displayed a significant decrease of cellular CoA after 12 and 24 h of POA treatment. Importantly, mutations in panD abrogated POA-mediated CoA depletion, resulting in CoA levels that were comparable to those of the no-drug control (145). Further, this study demonstrated that CoA depletion is specific for POA, as structural analogs, nicotinic acid and benzoic acid, did not significantly alter cellular CoA levels. In a separate study, Gopal et al. (146) conducted a metabolomic analysis on M. bovis BCG treated with POA to assess the effect of POA on intermediates of the CoA biosynthetic pathway. POA treatment resulted in a >10-fold reduction in β-alanine levels and depletion of numerous downstream intermediates in this pathway within 4 h. Additionally, depletion of CoA by POA resulted in the concomitant increase in the concentrations of medium-chain, dicarboxylate, and long-chain fatty acids within 24 h of treatment. Accumulation of fatty acids may contribute to bacterial cell death through impairment of oxidative phosphorylation and eventual collapse of membrane potential (147). Consistent with these findings, Rosen et al. showed that loss-of-function mutations in fadD2, an acyl-CoA ligase responsible for the detoxification of fatty acids, leads to hypersusceptibility of POA (148).
Based on genetic analysis of panD and the ability of POA to broadly disrupt CoA homeostasis, it is possible that POA interferes with activity of M. tuberculosis l-aspartate decarboxylase (Fig. 1D). In M. tuberculosis, PanD functions as a tetramer and shares sequence and structural similarity with other members of the PanD family (139). Yet M. tuberculosis PanD has a 13-amino-acid C-terminal extension which serves as the central contact point for tetramer formation (139). It is interesting that the majority of spontaneous panD mutations identified by Zhang et al. occurred within the portion corresponding to the last 13 amino acids of the C terminus (138). Similar panD missense mutations corresponding to the C terminus were subsequently described by Gopal et al. (145). In support of l-aspartate decarboxylase as a molecular target of POA, Gopal et al. (146) demonstrated interaction between PanD and POA (KD [equilibrium dissociation constant] = 6.1 μM ± 0.88 μM). Mutations within the N terminus and C terminus of PanD abrogated this interaction, which suggests that PanD-related PZA resistance is likely due to a loss of binding. Collectively, these studies support a model in which POA binds to PanD and inhibits synthesis of β-alanine, which ultimately leads to CoA insufficiency and broadly impaired central metabolism. However, it is important to note that an M. tuberculosis pantothenate auxotrophic strain (mc27000) containing a panD deletion remained susceptible to PZA when cultured in medium containing a subantagonistic concentration of panthetheine (144). Similar to the case with the parental strain, PZA susceptibility of M. tuberculosis mc27000 could be antagonized by exogenous pantothenate. However, unlike for the parental strain, PZA susceptibility of strain mc27000 was not antagonized by supplementation with β-alanine. These data demonstrate that if PanD is indeed a target of POA, additional targets likely exist within the CoA biosynthetic pathway. Future studies are necessary to further clarify the mechanism behind disruption of CoA biosynthesis and PZA activity.
Despite the isolation of POA-resistant panD missense mutants using laboratory strains of M. tuberculosis, analogous mutations have yet to be described for clinical isolates (123, 149). However, it is worth noting that the naturally PZA-resistant Mycobacterium canetti harbors a PanD M117T amino acid substitution (138). Importantly, recent work described by Gopal and colleagues (150) showed enrichment of POA-resistant M. tuberculosis strains from infected BALB/c mice that had been treated for 8 weeks with POA. Approximately 80% of M. tuberculosis POA-resistant isolates derived from infected mice contained mutations in panD, with the majority of these mutations corresponding to the C terminus. These recovered panD mutant strains were not reevaluated for PZA resistance in vivo. Yet infectivity of a previously characterized M. tuberculosis POA-resistant panD mutant (POAR 1) (145) was assessed using a low-dose aerosol infection in BALB/c mice. After 6 weeks of infection, this strain was found to have in vivo growth comparable to that of a matched wild-type control, suggesting that panD mutant strains remain infective (150). Furthermore, a recent study conducted by Ramirez-Busby et al. (125) analyzed 224 extensively drug-resistant (XDR) M. tuberculosis clinical isolates that showed PZA resistance, of which one pncA wild-type isolate contained a heterogeneous mutation (-G291) in panD. While the collective data demonstrate an incontrovertible association between POA action and CoA metabolism, the clinical relevance of panD to PZA resistance demands further analysis.
OTHER GENES ASSOCIATED WITH PZA RESISTANCE
Several other genes associated with PZA and POA resistance have recently been reported. Two independent laboratories have demonstrated a connection between mutations in clpC1 and POA resistance (150–153). ClpC1 (154, 155) is a class II AAA+ ATPase that provides chaperone activity for the essential cytoplasmic Clp protease (156–158). It is unclear whether clpC1-related POA resistance is due to a direct or indirect mechanism and how this relates to previous findings involving the CoA biosynthetic pathway.
Other research groups have expanded the list of potential M. tuberculosis targets responsible for PZA resistance. Njire et al. (159) have associated an Asp67Asn substitution in Rv2783 with PZA resistance. Rv2783 is a bifunctional enzyme that catalyzes the metabolism of RNA, single-stranded DNA, and ppGpp and was identified in a POA affinity chromatography assay by Shi et al. (108). Additional studies have associated PZA and POA resistance with mutations in numerous genes of unknown function (153). The roles of the corresponding functions of these various genes in resistance to PZA have yet to be elucidated but indicate that susceptibility and resistance of M. tuberculosis to PZA are quite complex.
FUTURE DIRECTIONS
Despite the identification of M. tuberculosis POA-resistant isolates in vitro, the in vivo relevance of the corresponding mutations to PZA resistance remains unclear. Correlation between in vitro findings and clinical efficacy are not yet straightforward and will require additional studies to resolve. As a first step, resistant strains identified in vitro should undergo extensive confirmation in animal models of TB infection in order to bridge the gap between in vitro and in vivo findings. Further, studies involving animals with defined impairments in cell-mediated immunity can help to elucidate the relevance of specific host responses that are critical for PZA efficacy. These animal experiments should utilize PZA and POA concentrations similar to those used in TB patients in order to represent standard treatment.
In addition to detailed characterization of novel PZA resistance mechanisms, future research should focus on other compounds that synergize with PZA. Niu and colleagues (160) screened a clinical drug library containing 1,524 substances for compounds that showed synergy with PZA. One hundred thirty hits were found to enhance PZA activity against stationary-phase cultures of M. tuberculosis strain H37Ra. Eighty-three of these hits were compounds that have FDA approval for other medical indications and should be evaluated for their potential in repurposing for enhancing PZA action. The identification and study of synergistic compounds will provide insight on the mode of action of PZA and could lead to shorter, more effective treatment regimens.
Recent elegant studies of PZA pharmacokinetics in TB patients (161) and animal models (28, 29, 162–164) have highlighted the importance of drug distribution and penetration into various lesion types as well as the reliance of the intracellular environment for PZA activity. In addition to measuring pH and tissue penetration, future studies should seek to characterize PZA metabolites in the caseum throughout the TB disease spectrum. Recently, Marakalala and colleagues have characterized the host proteomes of multiple lesion types and regions (caseous granuloma, caseous granuloma caseum, cavitary granuloma, cavitary granuloma caseum, and solid granuloma) (165). Interestingly, this study showed that greater differences occurred within regions of the same granuloma than among different lesion types. The centers of the granuloma were found to contain multiple proinflammatory signals, antimicrobial peptides, reactive oxygen species, and proinflammatory eicosanoids. In contrast, the tissue surrounding the caseum displayed an anti-inflammatory profile. Mapping of various granuloma landscapes should be expanded to include the characterization of resident M. tuberculosis subpopulations by single-cell analysis. Proteomic evaluation of heterologous granulomas and specific regions and M. tuberculosis subpopulations paired with targeted PZA pharmacokinetic data will provide researchers with a robust model of drug efficacy, potentiation by the host, and responsive or nonsusceptible bacterial cells. This model may be utilized to design antibiotic adjuvants and adjunctive therapeutics to enhance the host response and circumvent PZA resistance (166).
CONCLUDING REMARKS
While questions regarding PZA action that were posed by Feldman over 60 years ago have not been fully resolved, significant steps have been undertaken to understand this crucial drug. Recent advances have cleared some of the prevailing dogma that has surrounded PZA and indicated a correlation between metabolic activity and the drug’s activity. Future studies will expand upon these findings through examining the activity of PZA in the context of its associated host microenvironment. We are fortunate to be in a period of scientific research with unprecedented productivity bolstered by advances in genomics, high-throughput drug screens, and pharmacokinetics, all of which will be crucial to finally solve Feldmann’s 1954 questions concerning the basis for PZA activity against M. tuberculosis.
ACKNOWLEDGMENTS
This study was supported by NIH grant R01 AI123146 to A.D.B. E.A.L. was supported by a postdoctoral fellowship from the Ford Foundation of the National Academies of Science, Engineering, and Mathematics. N.A.D. was supported by NIH institutional training grant T32 HL07741.
All authors declare that they have no conflicts of interest.
REFERENCES
- 1.Feldmann FM. 1955. Can we accelerate tuberculosis research? Am Rev Tuberc 71:140–143. doi: 10.1164/artpd.1955.71.1.140. [DOI] [PubMed] [Google Scholar]
- 2.McClatchy JK, Tsang AY, Cernich MS. 1981. Use of pyrazinamidase activity on Mycobacterium tuberculosis as a rapid method for determination of pyrazinamide susceptibility. Antimicrob Agents Chemother 20:556–557. doi: 10.1128/aac.20.4.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler WR, Kilburn JO. 1983. Susceptibility of Mycobacterium tuberculosis to pyrazinamide and its relationship to pyrazinamidase activity. Antimicrob Agents Chemother 24:600–601. doi: 10.1128/aac.24.4.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scorpio A, Zhang Y. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med 2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 5.Boshoff HIM, Xu X, Tahlan K, Dowd CS, Pethe K, Camacho LR, Park T-H, Yun C-S, Schnappinger D, Ehrt S, Williams KJ, Barry CE. 2008. Biosynthesis and recycling of nicotinamide cofactors in Mycobacterium tuberculosis: an esssential role for NAD in nonreplicating bacilli. J Biol Chem 283:19329–19341. doi: 10.1074/jbc.M800694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vilchèze C, Weinrick B, Wong K-W, Chen B, Jacobs WR Jr. 2010. NAD(+) auxotrophy is bacteriocidal for the tubercle bacilli. Mol Microbiol 76:365–377. doi: 10.1111/j.1365-2958.2010.07099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allana S, Shashkina E, Mathema B, Bablishvili N, Tukvadze N, Shah NS, Kempker RR, Blumberg HM, Moodley P, Mlisana K, Brust JC, Gandhi NR. 2017. pncA gene mutations associated with pyrazinamide resistance in drug-resistant tuberculosis, South Africa and Georgia. Emerg Infect Dis 23:491–495. doi: 10.3201/eid2303.161034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng X, Ning Z, Drobniewski F, Yang J, Li Q, Zhang Z, Hu Y. 2017. pncA mutations are associated with slower sputum conversion during standard treatment of multidrug-resistant tuberculosis. Int J Antimicrob Agents 49:183–188. doi: 10.1016/j.ijantimicag.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Yoon JH, Nam JS, Kim KJ, Ro YT. 2014. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates from Korea and analysis of the correlation between the mutations and pyrazinamidase activity. World J Microbiol Biotechnol 30:2821–2828. doi: 10.1007/s11274-014-1706-0. [DOI] [PubMed] [Google Scholar]
- 10.Huang TS, Lee SS, Tu HZ, Huang WK, Chen YS, Huang CK, Wann SR, Lin HH, Liu YC. 2003. Correlation between pyrazinamide activity and pncA mutations in Mycobacterium tuberculosis isolates in Taiwan. Antimicrob Agents Chemother 47:3672–3673. doi: 10.1128/aac.47.11.3672-3673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huy NQ, Lucie C, Hoa TTT, Hung NV, Lan NTN, Son NT, Nhung NV, Anh DD, Anne-Laure B, Van Anh NT. 2017. Molecular analysis of pyrazinamide resistance in Mycobacterium tuberculosis in Vietnam highlights the high rate of pyrazinamide resistance-associated mutations in clinical isolates. Emerg Microbes Infect 6:e86. doi: 10.1038/emi.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, Zhang Y. 1997. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 41:540–543. doi: 10.1128/AAC.41.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Portugal I, Barreiro L, Moniz-Pereira J, Brum L. 2004. pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates in Portugal. Antimicrob Agents Chemother 48:2736–2738. doi: 10.1128/AAC.48.7.2736-2738.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitfield MG, Soeters HM, Warren RM, York T, Sampson SL, Streicher EM, van Helden PD, van Rie A. 2015. A global perspective on pyrazinamide resistance: systematic review and meta-analysis. PLoS One 10:e0133869. doi: 10.1371/journal.pone.0133869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoffels K, Mathys V, Fauville-Dufaux M, Wintjens R, Bifani P. 2012. Systematic analysis of pyrazinamide-resistant spontaneous mutants and clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother 56:5186–5193. doi: 10.1128/AAC.05385-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miotto P, Cabibbe AM, Feuerriegel S, Casali N, Drobniewski F, Rodionova Y, Bakonyte D, Stakenas P, Pimkina E, Augustynowicz-Kopec E, Degano M, Ambrosi A, Hoffner S, Mansjo M, Werngren J, Rusch-Gerdes S, Niemann S, Cirillo DM. 2014. Mycobacterium tuberculosis pyrazinamide resistance determinants: a multicenter study. mBio 5:e01819-14. doi: 10.1128/mBio.01819-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramirez-Busby SM, Valafar F. 2015. Systematic review of mutations in pyrazinamidase associated with pyrazinamide resistance in Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother 59:5267–5277. doi: 10.1128/AAC.00204-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sengstake S, Bergval IL, Schuitema AR, de Beer JL, Phelan J, de Zwaan R, Clark TG, van Soolingen D, Anthony RM. 2017. Pyrazinamide resistance-conferring mutations in pncA and the transmission of multidrug resistant TB in Georgia. BMC Infect Dis 17:491. doi: 10.1186/s12879-017-2594-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aung WW, Ei PW, Nyunt WW, Htwe MM, Win SM, Aye KT, Mon AS, Aung ST, Chang CL, Lee JS. 2018. Pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis clinical isolates in Myanmar. Antimicrob Agents Chemother 62:e01984-17. doi: 10.1128/AAC.01984-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sreevatsan S, Pan X, Zhang Y, Kreiswirth BN, Musser JM. 1997. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob Agents Chemother 41:636–640. doi: 10.1128/AAC.41.3.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yadon AN, Maharaj K, Adamson JH, Lai YP, Sacchettini JC, Ioerger TR, Rubin EJ, Pym AS. 2017. A comprehensive characterization of PncA polymorphisms that confer resistance to pyrazinamide. Nat Commun 8:588. doi: 10.1038/s41467-017-00721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aggarwal M, Singh A, Grover S, Pandey B, Kumari A, Grover A. 2018. Role of pncA gene mutations W68R and W68G in pyrazinamide resistance. J Cell Biochem 119:2567–2578. doi: 10.1002/jcb.26420. [DOI] [PubMed] [Google Scholar]
- 23.Bhuju S, Fonseca LDS, Marsico AG, de Oliveira Vieira GB, Sobral LF, Stehr M, Singh M, Saad MHF. 2013. Mycobacterium tuberculosis isolates from Rio de Janeiro reveal unusually low correlation between pyrazinamide resistance and mutations in the pncA gene. Infect Genet Evol 19:1–6. doi: 10.1016/j.meegid.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Wu X, Lu W, Shao Y, Song H, Li G, Li Y, Zhu L, Chen C. 2019. pncA gene mutations in reporting pyrazinamide resistance among the MDR-TB suspects. Infect Gen Evol 72:147–150. doi: 10.1016/j.meegid.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 25.Malone L, Schurr A, Lindh H, Mc KD, Kiser JS, Williams JH. 1952. The effect of pyrazinamide (aldinamide) on experimental tuberculosis in mice. Am Rev Tuberc 65:511–518. [PubMed] [Google Scholar]
- 26.McCune RM Jr, McDermott W, Tompsett R. 1956. The fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. II. The conversion of tuberculous infection to the latent state by the administration of pyrazinamide and a companion drug. J Exp Med 104:763–802. doi: 10.1084/jem.104.5.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kjellsson MC, Via LE, Goh A, Weiner D, Low KM, Kern S, Pillai G, Barry CE III, Dartois V. 2012. Pharmacokinetic evaluation of the penetration of antituberculosis agents in rabbit pulmonary lesions. Antimicrob Agents Chemother 56:446–457. doi: 10.1128/AAC.05208-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanc L, Sarathy JP, Alvarez Cabrera N, O’Brien P, Dias-Freedman I, Mina M, Sacchettini J, Savic RM, Gengenbacher M, Podell BK, Prideaux B, Ioerger T, Dick T, Dartois V. 2018. Impact of immunopathology on the antituberculous activity of pyrazinamide. J Exp Med 215:1975–1986. doi: 10.1084/jem.20180518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Via LE, Savic R, Weiner DM, Zimmerman MD, Prideaux B, Irwin SM, Lyon E, O’Brien P, Gopal P, Eum S, Lee M, Lanoix J-P, Dutta NK, Shim T, Cho JS, Kim W, Karakousis PC, Lenaerts A, Nuermberger E, Barry CE III, Dartois V. 2015. Host-mediated bioactivation of pyrazinamide: implications for efficacy, resistance, and therapeutic alternatives. ACS Infect Dis 1:203–214. doi: 10.1021/id500028m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeager RL, Munroe WG, Dessau FI. 1952. Pyrazinamide (aldinamide) in the treatment of pulmonary tuberculosis. Am Rev Tuberc 65:523–546. [PubMed] [Google Scholar]
- 31.Calix AA, White K. 1956. The role of pyrazinamide in the chemotherapy of chronic pulmonary tuberculosis; a clinical evaluation of 39 cases treated with rotation therapy. J Med Assoc State Ala 26:81–86. [PubMed] [Google Scholar]
- 32.Dessau FI, Yeager RL, Burger FJ, Williams JH. 1952. Pyrazinamide (aldinamide) in experimental tuberculosis of the guinea pig. Am Rev Tuberc 65:519–522. [PubMed] [Google Scholar]
- 33.Dessau FI, Yeager RL, Burger F. 1953. Further studies with aldinamide in experimental tuberculosis of the guinea pig. Tuberculol Thorac Dis 14:149–154. [PubMed] [Google Scholar]
- 34.Ahmad Z, Fraig MM, Bisson GP, Nuermberger EL, Grosset JH, Karakousis PC. 2011. Dose-dependent activity of pyrazinamide in animal models of intracellular and extracellular tuberculosis infections. Antimicrob Agents Chemother 55:1527–1532. doi: 10.1128/AAC.01524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geiter LJ, O’Brien RJ, Combs DL, Snider DE Jr. 1987. United States Public Health Service Tuberculosis Therapy Trial 21: preliminary results of an evaluation of a combination tablet of isoniazid, rifampin and pyrazinamide. Tubercle 68:41–46. doi: 10.1016/S0041-3879(87)80021-1. [DOI] [PubMed] [Google Scholar]
- 36.Aquinas M. 1982. Short-course therapy for tuberculosis. Drugs 24:118–132. doi: 10.2165/00003495-198224020-00002. [DOI] [PubMed] [Google Scholar]
- 37.Fox W. 1981. Whither short-course chemotherapy? Br J Dis Chest 75:331–357. doi: 10.1016/0007-0971(81)90022-x. [DOI] [PubMed] [Google Scholar]
- 38.Vocat A, Hartkoorn RC, Lechartier B, Zhang M, Dhar N, Cole ST, Sala C. 2015. Bioluminescence for assessing drug potency against nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother 59:4012–4019. doi: 10.1128/AAC.00528-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grosset J, Almeida D, Converse PJ, Tyagi S, Li SY, Ammerman NC, Pym AS, Wallengren K, Hafner R, Lalloo U, Swindells S, Bishai WR. 2012. Modeling early bactericidal activity in murine tuberculosis provides insights into the activity of isoniazid and pyrazinamide. Proc Natl Acad Sci U S A 109:15001–15005. doi: 10.1073/pnas.1203636109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heifets L, Lindholm-Levy P. 1992. Pyrazinamide sterilizing activity in vitro against semidormant Mycobacterium tuberculosis bacterial populations. Am Rev Respir Dis 145:1223–1225. doi: 10.1164/ajrccm/145.5.1223. [DOI] [PubMed] [Google Scholar]
- 41.Swaminathan S, Pasipanodya JG, Ramachandran G, Hemanth Kumar AK, Srivastava S, Deshpande D, Nuermberger E, Gumbo T. 2016. Drug concentration thresholds predictive of therapy failure and death in children with tuberculosis: bread crumb trails in random forests. Clin Infect Dis 63:S63–S74. doi: 10.1093/cid/ciw471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srivastava S, Pasipanodya JG, Meek C, Leff R, Gumbo T. 2011. Multidrug-resistant tuberculosis not due to noncompliance but to between-patient pharmacokinetic variability. J Infect Dis 204:1951–1959. doi: 10.1093/infdis/jir658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiruy H, Rogers Z, Mbowane C, Adamson J, Ngotho L, Karim F, Gumbo T, Bishai W, Jeena P. 2015. Subtherapeutic concentrations of first-line anti-TB drugs in South African children treated according to current guidelines: the PHATISA study. J Antimicrob Chemother 70:1115–1123. doi: 10.1093/jac/dku478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anonymous. 2006. Pyrazinamide, Drugs and Lactation Database (LactMed). National Library of Medicine, Bethesda, MD. [Google Scholar]
- 45.Chang KC, Leung CC, Yew WW, Lau TY, Tam CM. 2008. Hepatotoxicity of pyrazinamide: cohort and case-control analyses. Am J Respir Crit Care Med 177:1391–1396. doi: 10.1164/rccm.200802-355OC. [DOI] [PubMed] [Google Scholar]
- 46.Centers for Disease Control and Prevention. 2001. Fatal and severe hepatitis associated with rifampin and pyrazinamide for the treatment of latent tuberculosis infection—New York and Georgia, 2000. MMWR Morb Mortal Wkly Rep 50:289–291. [PubMed] [Google Scholar]
- 47.Fox W, Mitchison DA. 1975. Short-course chemotherapy for pulmonary tuberculosis. Am Rev Respir Dis 111:845–848. doi: 10.1164/arrd.1975.111.6.845. [DOI] [PubMed] [Google Scholar]
- 48.Anonymous. 1973. Short-course treatment in pulmonary tuberculosis. East Afr Med J 50:672–680. [PubMed] [Google Scholar]
- 49.Anonymous. 1974. Controlled clinical trial of four short-course (6-month) regimens of chemotherapy for treatment of pulmonary tuberculosis. Third report. East African-British Medical Research Councils. Lancet ii:237–240. [PubMed] [Google Scholar]
- 50.Lecoeur HF, Truffot-Pernot C, Grosset JH. 1989. Experimental short-course preventive therapy of tuberculosis with rifampin and pyrazinamide. Am Rev Respir Dis 140:1189–1193. doi: 10.1164/ajrccm/140.5.1189. [DOI] [PubMed] [Google Scholar]
- 51.Piubello A, Harouna SH, Souleymane MB, Boukary I, Morou S, Daouda M, Hanki Y, Van Deun A. 2014. High cure rate with standardised short-course multidrug-resistant tuberculosis treatment in Niger: no relapses. Int J Tuber Lung Dis 18:1188–1194. doi: 10.5588/ijtld.13.0075. [DOI] [PubMed] [Google Scholar]
- 52.Petty TL, Mitchell RS. 1962. Successful treatment of advanced isonizid- and streptomycin-resistant pulmonary tuberculosis with ethionamide, pyrazinamide, and isoniazid. Am Rev Respir Dis 86:503–512. doi: 10.1164/arrd.1962.86.4.503. [DOI] [PubMed] [Google Scholar]
- 53.Franke MF, Becerra MC, Tierney DB, Rich ML, Bonilla C, Bayona J, McLaughlin MM, Mitnick CD. 2015. Counting pyrazinamide in regimens for multidrug-resistant tuberculosis. Ann Am Thorac Soc 12:674–679. doi: 10.1513/AnnalsATS.201411-538OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dawson R, Diacon A. 2013. PA-824, moxifloxacin and pyrazinamide combination therapy for tuberculosis. Expert Opin Invest Drugs 22:927–932. doi: 10.1517/13543784.2013.801958. [DOI] [PubMed] [Google Scholar]
- 55.Li SY, Tasneen R, Tyagi S, Soni H, Converse PJ, Mdluli K, Nuermberger EL. 2017. Bactericidal and sterilizing activity of a novel regimen with bedaquiline, pretomanid, moxifloxacin, and pyrazinamide in a murine model of tuberculosis. Antimicrob Agents Chemother 61:e00913-17. doi: 10.1128/AAC.00913-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diacon AH, Dawson R, von Groote-Bidlingmaier F, Symons G, Venter A, Donald PR, van Niekerk C, Everitt D, Hutchings J, Burger DA, Schall R, Mendel CM. 2015. Bactericidal activity of pyrazinamide and clofazimine alone and in combinations with pretomanid and bedaquiline. Am J Respir Crit Care Med 191:943–953. doi: 10.1164/rccm.201410-1801OC. [DOI] [PubMed] [Google Scholar]
- 57.Diacon AH, Dawson R, von Groote-Bidlingmaier F, Symons G, Venter A, Donald PR, van Niekerk C, Everitt D, Winter H, Becker P, Mendel CM, Spigelman MK. 2012. 14-day bactericidal activity of PA-824, bedaquiline, pyrazinamide, and moxifloxacin combinations: a randomised trial. Lancet 380:986–993. doi: 10.1016/S0140-6736(12)61080-0. [DOI] [PubMed] [Google Scholar]
- 58.Anthony RM, den Hertog AL, van Soolingen D. 2018. ‘Happy the man, who, studying nature’s laws, Thro’ known effects can trace the secret cause.’ Do we have enough pieces to solve the pyrazinamide puzzle? J Antimicrob Chemother 73:1750–1754. doi: 10.1093/jac/dky060. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Mitchison D. 2003. The curious characteristics of pyrazinamide: a review. Int J Tuber Lung Dis 7:6–21. [PubMed] [Google Scholar]
- 60.Murray MF. 2003. Nicotinamide: an oral antimicrobial agent with activity against both Mycobacterium tuberculosis and human immunodeficiency virus. Clin Infect Dis 36:453–460. doi: 10.1086/367544. [DOI] [PubMed] [Google Scholar]
- 61.Kushner S, Dalalian H, Sanjurjo JL, Bach FL, Safir SR, Smith VK, Williams JH. 1952. Experimental chemotherapy of tuberculosis. II. The synthesis of pyrazinamides and related compounds. J Am Chem Soc 74:3617–3621. doi: 10.1021/ja01134a045. [DOI] [Google Scholar]
- 62.McKenzie BS, Malone L, Kushner S, Oleson JJ, SubbaRow Y. 1948. The effect of nicotinic acid amide on experimental tuberculosis of white mice. J Lab Clin Med 33:1249–1253. [PubMed] [Google Scholar]
- 63.Haunt E. 1945. Note sur l’action de tres fortes doses d’amide nicotinique dans les lesion bacillaires. Gazette Hopitau 118:259–260. [Google Scholar]
- 64.Chorine V. 1945. Action de l’amide nicotinique sur les bacilles du genre mycobacterium. C R Hebd Seances Acad Sci 220:150–151. [Google Scholar]
- 65.Solotorovsky M, Gregory FJ, Ironson EJ, Bugie EJ, O’Neill RC, Pfister R III. 1952. Pyrazinoic acid amide; an agent active against experimental murine tuberculosis. Proc Soc Exp Biol Med 79:563–565. doi: 10.3181/00379727-79-19447. [DOI] [PubMed] [Google Scholar]
- 66.Tarshis MS, Weed WA Jr. 1953. Lack of significant in vitro sensitivity of Mycobacterium tuberculosis to pyrazinamide on three different solid media. Am Rev Tuberc 67:391–395. doi: 10.1164/art.1953.67.3.391. [DOI] [PubMed] [Google Scholar]
- 67.McDermott W, Tompsett R. 1954. Activation of pyrazinamide and nicotinamide in acidic environments in vitro. Am Rev Tuberc 70:748–754. doi: 10.1164/art.1954.70.4.748. [DOI] [PubMed] [Google Scholar]
- 68.Russell DG. 2007. Who puts the tubercle in tuberculosis? Nat Rev Microbiol 5:39–47. doi: 10.1038/nrmicro1538. [DOI] [PubMed] [Google Scholar]
- 69.Elkington PT, Friedland JS. 2015. Permutations of time and place in tuberculosis. Lancet Infect Dis 15:1357–1360. doi: 10.1016/S1473-3099(15)00135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.MacMicking JD, Taylor GA, McKinney JD. 2003. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science 302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 71.Sprick MG. 1956. Phagocytosis of M. tuberculosis and M. smegmatis stained with indicator dyes. Am Rev Tuberc 74:552–565. doi: 10.1164/artpd.1956.74.4.552. [DOI] [PubMed] [Google Scholar]
- 72.Lanoix JP, Ioerger T, Ormond A, Kaya F, Sacchettini J, Dartois V, Nuermberger E. 2016. Selective inactivity of pyrazinamide against tuberculosis in C3HeB/FeJ mice is best explained by neutral pH of caseum. Antimicrob Agents Chemother 60:735–743. doi: 10.1128/AAC.01370-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lanoix JP, Lenaerts AJ, Nuermberger EL. 2015. Heterogeneous disease progression and treatment response in a C3HeB/FeJ mouse model of tuberculosis. Dis Model Mech 8:603–610. doi: 10.1242/dmm.019513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, Scorpio A, Nikaido H, Sun Z. 1999. Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Bacteriol 181:2044–2049. doi: 10.1128/JB.181.7.2044-2049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z. 2003. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J Antimicrob Chemother 52:790–795. doi: 10.1093/jac/dkg446. [DOI] [PubMed] [Google Scholar]
- 76.Zhang Y, Shi W, Zhang W, Mitchison D. 2014. Mechanisms of pyrazinamide action and resistance. Microbiol Spectr 2:MGM2-0023-2013. doi: 10.1128/microbiolspec.MGM2-0023-2013. [DOI] [PubMed] [Google Scholar]
- 77.Singh P, Mishra AK, Malonia SK, Chauhan DS, Sharma VD, Venkatesan K, Katoch VM. 2006. The paradox of pyrazinamide: an update on the molecular mechanisms of pyrazinamide resistance in mycobacteria. J Commun Dis 38:288–298. [PubMed] [Google Scholar]
- 78.Njire M, Tan Y, Mugweru J, Wang C, Guo J, Yew W, Tan S, Zhang T. 2016. Pyrazinamide resistance in Mycobacterium tuberculosis: review and update. Adv Med Sci 61:63–71. doi: 10.1016/j.advms.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 79.Stehr M, Elamin AA, Singh M. 2015. Pyrazinamide: the importance of uncovering the mechanisms of action in mycobacteria. Expert Rev Anti Infect Ther 13:593–603. doi: 10.1586/14787210.2015.1021784. [DOI] [PubMed] [Google Scholar]
- 80.Darby CM, Ingólfsson HI, Jiang X, Shen C, Sun M, Zhao N, Burns K, Liu G, Ehrt S, Warren JD, Andersen OS, Anderson OS, Brickner SJ, Nathan C. 2013. Whole cell screen for inhibitors of pH homeostasis in Mycobacterium tuberculosis. PLoS One 8:e68942. doi: 10.1371/journal.pone.0068942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu P, Haagsma AC, Pham H, Maaskant JJ, Mol S, Lill H, Bald D. 2011. Pyrazinoic acid decreases the proton motive force, respiratory ATP synthesis activity, and cellular ATP levels. Antimicrob Agents Chemother 55:5354–5357. doi: 10.1128/AAC.00507-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baughn AD, Deng J, Vilcheze C, Riestra A, Welch JT, Jacobs WR Jr, Zimhony O. 2010. Mutually exclusive genotypes for pyrazinamide and 5-chloropyrazinamide resistance reveal a potential resistance-proofing strategy. Antimicrob Agents Chemother 54:5323–5328. doi: 10.1128/AAC.00529-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peterson ND, Rosen BC, Dillon NA, Baughn AD. 2015. Uncoupling environmental pH and intrabacterial acidification from pyrazinamide susceptibility in Mycobacterium tuberculosis. Antimicrob Agents Chemother 59:7320–7326. doi: 10.1128/AAC.00967-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y, Permar S, Sun Z. 2002. Conditions that may affect the results of susceptibility testing of Mycobacterium tuberculosis to pyrazinamide. J Med Microbiol 51:42–49. doi: 10.1099/0022-1317-51-1-42. [DOI] [PubMed] [Google Scholar]
- 85.Zhang Y, Zhang J, Cui P, Zhang Y, Zhang W. 2017. Identification of novel efflux proteins Rv0191, Rv3756c, Rv3008, and Rv1667c involved in pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 61:e00940-17. doi: 10.1128/AAC.00940-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang Q, Chen ZF, Li YY, Zhang Y, Ren Y, Fu Z, Xu SQ. 2007. Nutrient-starved incubation conditions enhance pyrazinamide activity against Mycobacterium tuberculosis. Chemotherapy 53:338–343. doi: 10.1159/000107723. [DOI] [PubMed] [Google Scholar]
- 87.den Hertog AL, Menting S, Pfeltz R, Warns M, Siddiqi SH, Anthony RM. 2016. Pyrazinamide is active against Mycobacterium tuberculosis cultures at neutral pH and low temperature. Antimicrob Agents Chemother 60:4956–4960. doi: 10.1128/AAC.00654-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wade MM, Zhang Y. 2004. Anaerobic incubation conditions enhance pyrazinamide activity against Mycobacterium tuberculosis. J Med Microbiol 53:769–773. doi: 10.1099/jmm.0.45639-0. [DOI] [PubMed] [Google Scholar]
- 89.Hu Y, Coates AR, Mitchison DA. 2006. Sterilising action of pyrazinamide in models of dormant and rifampicin-tolerant Mycobacterium tuberculosis. Int J Tuber Lung Dis 10:317–322. [PubMed] [Google Scholar]
- 90.Piccaro G, Giannoni F, Filippini P, Mustazzolu A, Fattorini L. 2013. Activities of drug combinations against Mycobacterium tuberculosis grown in aerobic and hypoxic acidic conditions. Antimicrob Agents Chemother 57:1428–1433. doi: 10.1128/AAC.02154-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vandal OH, Pierini LM, Schnappinger D, Nathan CF, Ehrt S. 2008. A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat Med 14:849–854. doi: 10.1038/nm.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kasianowicz J, Benz R, McLaughlin S. 1984. The kinetic mechanism by which CCCP (carbonyl cyanide m-chlorophenylhydrazone) transports protons across membranes. J Membr Biol 82:179–190. doi: 10.1007/bf01868942. [DOI] [PubMed] [Google Scholar]
- 93.Rao M, Streur TL, Aldwell FE, Cook GM. 2001. Intracellular pH regulation by Mycobacterium smegmatis and Mycobacterium bovis BCG. Microbiology 147:1017–1024. doi: 10.1099/00221287-147-4-1017. [DOI] [PubMed] [Google Scholar]
- 94.Huczyński A, Janczak J, Lowicki D, Brzezinski B. 2012. Monensin A acid complexes as a model of electrogenic transport of sodium cation. Biochim Biophys Acta 1818:2108–2119. doi: 10.1016/j.bbamem.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 95.Greenstein RJ, Su L, Whitlock RH, Brown ST. 2009. Monensin causes dose dependent inhibition of Mycobacterium avium subspecies paratuberculosis in radiometric culture. Gut Pathog 1:4. doi: 10.1186/1757-4749-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zimhony O, Cox JS, Welch JT, Vilcheze C, Jacobs WR Jr. 2000. Pyrazinamide inhibits the eukaryotic-like fatty acid synthetase I (FASI) of Mycobacterium tuberculosis. Nat Med 6:1043–1047. doi: 10.1038/79558. [DOI] [PubMed] [Google Scholar]
- 97.Brindley DN, Matsumura S, Bloch K. 1969. Mycobacterium phlei fatty acid synthetase—a bacterial multienzyme complex. Nature 224:666–669. doi: 10.1038/224666a0. [DOI] [Google Scholar]
- 98.Kikuchi S, Rainwater DL, Kolattukudy PE. 1992. Purification and characterization of an unusually large fatty acid synthase from Mycobacterium tuberculosis var. bovis BCG. Arch Biochem Biophys 295:318–326. doi: 10.1016/0003-9861(92)90524-z. [DOI] [PubMed] [Google Scholar]
- 99.Ciccarelli L, Connell SR, Enderle M, Mills DJ, Vonck J, Grininger M. 2013. Structure and conformational variability of the Mycobacterium tuberculosis fatty acid synthase multienzyme complex. Structure 21:1251–1257. doi: 10.1016/j.str.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 100.Schweizer E, Hofmann J. 2004. Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol Mol Biol Rev 68:501–517. doi: 10.1128/MMBR.68.3.501-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pawelczyk J, Kremer L. 2014. The molecular genetics of mycolic acid biosynthesis. Microbiol Spectr 2:MGM2-0003-2013. doi: 10.1128/microbiolspec.MGM2-0003-2013. [DOI] [PubMed] [Google Scholar]
- 102.Wakil SJ. 1989. Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry 28:4523–4530. doi: 10.1021/bi00437a001. [DOI] [PubMed] [Google Scholar]
- 103.Zimhony O, Vilcheze C, Arai M, Welch JT, Jacobs WR Jr. 2007. Pyrazinoic acid and its n-propyl ester inhibit fatty acid synthase type I in replicating tubercle bacilli. Antimicrob Agents Chemother 51:752–754. doi: 10.1128/AAC.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boshoff HI, Mizrahi V, Barry CE III. 2002. Effects of pyrazinamide on fatty acid synthesis by whole mycobacterial cells and purified fatty acid synthase I. J Bacteriol 184:2167–2172. doi: 10.1128/jb.184.8.2167-2172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ngo SC, Zimhony O, Chung WJ, Sayahi H, Jacobs WR Jr, Welch JT. 2007. Inhibition of isolated Mycobacterium tuberculosis fatty acid synthase I by pyrazinamide analogs. Antimicrob Agents Chemother 51:2430–2435. doi: 10.1128/AAC.01458-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sayahi H, Pugliese KM, Zimhony O, Jacobs WR Jr, Shekhtman A, Welch JT. 2012. Analogs of the antituberculous agent pyrazinamide are competitive inhibitors of NADPH binding to M. tuberculosis fatty acid synthase I. Chem Biodivers 9:2582–2596. doi: 10.1002/cbdv.201200291. [DOI] [PubMed] [Google Scholar]
- 107.Sayahi H, Zimhony O, Jacobs WR, Shekhtman A, Welch JT. 2011. Pyrazinamide, but not pyrazinoic acid, is a competitive inhibitor of NADPH binding to Mycobacterium tuberculosis fatty acid synthase I. Bioorg Med Chem Lett 21:4804–4807. doi: 10.1016/j.bmcl.2011.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE III, Wang H, Zhang W, Zhang Y. 2011. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 333:1630–1632. doi: 10.1126/science.1208813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Keiler KC, Waller PRH, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 110.Keiler KC, Feaga HA. 2014. Resolving nonstop translation complexes is a matter of life or death. J Bacteriol 196:2123–2130. doi: 10.1128/JB.01490-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Neubauer C, Gillet R, Kelley AC, Ramakrishnan V. 2012. Decoding in the absence of a codon by tmRNA and SmpB in the ribosome. Science 335:1366–1369. doi: 10.1126/science.1217039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Valle M, Gillet R, Kaur S, Henne A, Ramakrishnan V, Frank J. 2003. Visualizing tmRNA entry into a stalled ribosome. Science 300:127–130. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- 113.Ramrath DJ, Yamamoto H, Rother K, Wittek D, Pech M, Mielke T, Loerke J, Scheerer P, Ivanov P, Teraoka Y, Shpanchenko O, Nierhaus KH, Spahn CM. 2012. The complex of tmRNA-SmpB and EF-G on translocating ribosomes. Nature 485:526–529. doi: 10.1038/nature11006. [DOI] [PubMed] [Google Scholar]
- 114.Alumasa JN, Manzanillo PS, Peterson ND, Lundrigan T, Baughn AD, Cox JS, Keiler KC. 2017. Ribosome rescue inhibitors kill actively growing and nonreplicating persister Mycobacterium tuberculosis cells. ACS Infect Dis 3:634–644. doi: 10.1021/acsinfecdis.7b00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ramadoss NS, Alumasa JN, Cheng L, Wang Y, Li S, Chambers BS, Chang H, Chatterjee AK, Brinker A, Engels IH, Keiler KC. 2013. Small molecule inhibitors of trans-translation have broad-spectrum antibiotic activity. Proc Natl Acad Sci U S A 110:10282–10287. doi: 10.1073/pnas.1302816110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Goralski TDP, Dewan KK, Alumasa JN, Avanzato V, Place DE, Markley RL, Katkere B, Rabadi SM, Bakshi CS, Keiler KC, Kirimanjeswara GS. 2016. Inhibitors of ribosome rescue arrest growth of Francisella tularensis at all stages of intracellular replication. Antimicrob Agents Chemother 60:3276–3282. doi: 10.1128/AAC.03089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Alumasa JN, Goralski TDP, Keiler KC. 2017. Tetrazole-based trans-translation inhibitors kill Bacillus anthracis spores to protect host cells. Antimicrob Agents Chemother 61:e01199-17. doi: 10.1128/AAC.01199-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Speirs RJ, Welch JT, Cynamon MH. 1995. Activity of n-propyl pyrazinoate against pyrazinamide-resistant Mycobacterium tuberculosis: investigations into mechanism of action of and mechanism of resistance to pyrazinamide. Antimicrob Agents Chemother 39:1269–1271. doi: 10.1128/aac.39.6.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li X, Hirano R, Tagami H, Aiba H. 2006. Protein tagging at rare codons is caused by tmRNA action at the 3′ end of nonstop mRNA generated in response to ribosome stalling. RNA 12:248–255. doi: 10.1261/rna.2212606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Alexander DC, Ma JH, Guthrie JL, Blair J, Chedore P, Jamieson FB. 2012. Gene sequencing for routine verification of pyrazinamide resistance in Mycobacterium tuberculosis: a role for pncA but not rpsA. J Clin Microbiol 50:3726–3728. doi: 10.1128/JCM.00620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Simons SO, Mulder A, van Ingen J, Boeree MJ, van Soolingen D. 2013. Role of rpsA gene sequencing in diagnosis of pyrazinamide resistance. J Clin Microbiol 51:382. doi: 10.1128/JCM.02739-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tan Y, Hu Z, Zhang T, Cai X, Kuang H, Liu Y, Chen J, Yang F, Zhang K, Tan S, Zhao Y. 2014. Role of pncA and rpsA gene sequencing in detection of pyrazinamide resistance in Mycobacterium tuberculosis isolates from southern China. J Clin Microbiol 52:291–297. doi: 10.1128/JCM.01903-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gu Y, Yu X, Jiang G, Wang X, Ma Y, Li Y, Huang H. 2016. Pyrazinamide resistance among multidrug-resistant tuberculosis clinical isolates in a national referral center of China and its correlations with pncA, rpsA, and panD gene mutations. Diagn Microbiol Infect Dis 84:207–211. doi: 10.1016/j.diagmicrobio.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 124.Akhmetova A, Kozhamkulov U, Bismilda V, Chingissova L, Abildaev T, Dymova M, Filipenko M, Ramanculov E. 2015. Mutations in the pncA and rpsA genes among 77 Mycobacterium tuberculosis isolates in Kazakhstan. Int J Tuber Lung Dis 19:179–184. doi: 10.5588/ijtld.14.0305. [DOI] [PubMed] [Google Scholar]
- 125.Ramirez-Busby SM, Rodwell TC, Fink L, Catanzaro D, Jackson RL, Pettigrove M, Catanzaro A, Valafar F. 2017. A multinational analysis of mutations and heterogeneity in PZase, rpsA, and panD associated with pyrazinamide resistance in M/XDR Mycobacterium tuberculosis. Sci Rep 7:3790. doi: 10.1038/s41598-017-03452-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pang Y, Zhu D, Zheng H, Shen J, Hu Y, Liu J, Zhao Y. 2017. Prevalence and molecular characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates from Southern China. BMC Infect Dis 17:711. doi: 10.1186/s12879-017-2761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Alexander DC, Ma JH, Guthrie JL, Blair J, Chedore P, Jamieson FB. 2013. Reply to “Role of rpsA gene sequencing in diagnosis of pyrazinamide resistance.” J Clin Microbiol 51:383. doi: 10.1128/JCM.02760-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dillon NA, Peterson ND, Feaga HA, Keiler KC, Baughn AD. 2017. Anti-tubercular activity of pyrazinamide is independent of trans-translation and RpsA. Sci Rep 7:6135. doi: 10.1038/s41598-017-06415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Iwamoto T, Murase Y, Yoshida S, Aono A, Kuroda M, Sekizuka T, Yamashita A, Kato K, Takii T, Arikawa K, Kato S, Mitarai S. 2019. Overcoming the pitfalls of automatic interpretation of whole genome sequencing data by online tools for the prediction of pyrazinamide resistance in Mycobacterium tuberculosis. PLoS One 14:e0212798. doi: 10.1371/journal.pone.0212798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xia Q, Zhao LL, Li F, Fan YM, Chen YY, Wu BB, Liu ZW, Pan AZ, Zhu M. 2015. Phenotypic and genotypic characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates in Zhejiang, China. Antimicrob Agents Chemother 59:1690–1695. doi: 10.1128/AAC.04541-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Klemens SP, Sharpe CA, Cynamon MH. 1996. Activity of pyrazinamide in a murine model against Mycobacterium tuberculosis isolates with various levels of in vitro susceptibility. Antimicrob Agents Chemother 40:14–16. doi: 10.1128/AAC.40.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Qu X, Lancaster L, Noller HF, Bustamante C, Tinoco I Jr. 2012. Ribosomal protein S1 unwinds double-stranded RNA in multiple steps. Proc Natl Acad Sci U S A 109:14458–14463. doi: 10.1073/pnas.1208950109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tchufistova LS, Komarova AV, Boni IV. 2003. A key role for the mRNA leader structure in translational control of ribosomal protein S1 synthesis in gamma-proteobacteria. Nucleic Acids Res 31:6996–7002. doi: 10.1093/nar/gkg883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wower IK, Zwieb CW, Guven SA, Wower J. 2000. Binding and cross‐linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome. EMBO J 19:6612–6621. doi: 10.1093/emboj/19.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.McGinness KE, Sauer RT. 2004. Ribosomal protein S1 binds mRNA and tmRNA similarly but plays distinct roles in translation of these molecules. Proc Natl Acad Sci U S A 101:13454–13459. doi: 10.1073/pnas.0405521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Takada K, Takemoto C, Kawazoe M, Shirouzu M, Yokoyama S, Muto A, Himeno H. 2007. Thermus thermophilus tmRNA and trans-translation. Nucleic Acids Symp Ser (Oxf) 51:369–370. doi: 10.1093/nass/nrm185. [DOI] [PubMed] [Google Scholar]
- 137.Qi H, Shimizu Y, Ueda T. 2007. Ribosomal protein S1 is not essential for the trans-translation machinery. J Mol Biol 368:845–852. doi: 10.1016/j.jmb.2007.02.068. [DOI] [PubMed] [Google Scholar]
- 138.Zhang S, Chen J, Shi W, Liu W, Zhang W, Zhang Y. 2013. Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg Microbes Infect 2:e34. doi: 10.1038/emi.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chopra S, Pai H, Ranganathan A. 2002. Expression, purification, and biochemical characterization of Mycobacterium tuberculosis aspartate decarboxylase, PanD. Protein Expr Purif 25:533–540. doi: 10.1016/s1046-5928(02)00039-6. [DOI] [PubMed] [Google Scholar]
- 140.Jackowski S, Rock CO. 1981. Regulation of coenzyme A biosynthesis. J Bacteriol 148:926–932. doi: 10.1128/JB.148.3.926-932.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Webb ME, Smith AG, Abell C. 2004. Biosynthesis of pantothenate. Nat Prod Rep 21:695–721. doi: 10.1039/b316419p. [DOI] [PubMed] [Google Scholar]
- 142.Leonardi R, Zhang YM, Rock CO, Jackowski S. 2005. Coenzyme A: back in action. Prog Lipid Res 44:125–153. doi: 10.1016/j.plipres.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 143.Shi W, Chen J, Feng J, Cui P, Zhang S, Weng X, Zhang W, Zhang Y. 2014. Aspartate decarboxylase (PanD) as a new target of pyrazinamide in Mycobacterium tuberculosis. Emerg Microbes Infect 3:e58. doi: 10.1038/emi.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dillon NA, Peterson ND, Rosen BC, Baughn AD. 2014. Pantothenate and pantetheine antagonize the antitubercular activity of pyrazinamide. Antimicrob Agents Chemother 58:7258–7263. doi: 10.1128/AAC.04028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gopal P, Yee M, Sarathy J, Low JL, Sarathy JP, Kaya F, Dartois V, Gengenbacher M, Dick T. 2016. Pyrazinamide resistance is caused by two distinct mechanisms: prevention of coenzyme A depletion and loss of virulence factor synthesis. ACS Infect Dis 2:616–626. doi: 10.1021/acsinfecdis.6b00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gopal P, Nartey W, Ragunathan P, Sarathy J, Kaya F, Yee M, Setzer C, Manimekalai MSS, Dartois V, Gruber G, Dick T. 2017. Pyrazinoic acid inhibits mycobacterial coenzyme A biosynthesis by binding to aspartate decarboxylase PanD. ACS Infect Dis 3:807–819. doi: 10.1021/acsinfecdis.7b00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Desbois AP, Smith VJ. 2010. Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl Microbiol Biotechnol 85:1629–1642. doi: 10.1007/s00253-009-2355-3. [DOI] [PubMed] [Google Scholar]
- 148.Rosen BC, Dillon NA, Peterson ND, Minato Y, Baughn AD. 2017. Long-chain fatty acyl coenzyme A ligase FadD2 mediates intrinsic pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 61:e02130-16. doi: 10.1128/AAC.02130-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Maslov DA, Zaĭchikova MV, Chernousova LN, Shur KV, Bekker OB, Smirnova TG, Larionova EE, Andreevskaya SN, Zhang Y, Danilenko VN. 2015. Resistance to pyrazinamide in Russian Mycobacterium tuberculosis isolates: pncA sequencing versus Bactec MGIT 960. Tuberculosis (Edinb) 95:608–612. doi: 10.1016/j.tube.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 150.Gopal P, Tasneen R, Yee M, Lanoix JP, Sarathy JP, Rasic G, Li L, Dartois V, Nuermberger E, Dick T. 2017. In vivo-selected pyrazinoic acid-resistant M. tuberculosis strains harbor missense mutations in the aspartate decarboxylase PanD and the unfoldase ClpC1. ACS Infect Dis 3:492–501. doi: 10.1021/acsinfecdis.7b00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yee M, Gopal P, Dick T. 2017. Missense mutations in the unfoldase ClpC1 of the caseinolytic protease complex are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 61:e02342-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang S, Chen J, Shi W, Cui P, Zhang J, Cho S, Zhang W, Zhang Y. 2017. Mutation in clpC1 encoding an ATP-dependent ATPase involved in protein degradation is associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg Microbes Infect 6:e8. doi: 10.1038/emi.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shi W, Chen J, Zhang S, Zhang W, Zhang Y, Shi W, Chen J, Zhang S, Zhang W, Zhang Y. 2018. Identification of novel mutations in LprG (rv1411c), rv0521, rv3630, rv0010c, ppsC, and cyp128 associated with pyrazinoic acid/pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 62:e00430-18. doi: 10.1128/AAC.00430-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kar NP, Sikriwal D, Rath P, Choudhary RK, Batra JK. 2008. Mycobacterium tuberculosis ClpC1: characterization and role of the N-terminal domain in its function. FEBS J 275:6149–6158. doi: 10.1111/j.1742-4658.2008.06738.x. [DOI] [PubMed] [Google Scholar]
- 155.Li M, Kandror O, Akopian T, Dharkar P, Wlodawer A, Maurizi MR, Goldberg AL. 2016. Structure and functional properties of the active form of the proteolytic complex, ClpP1P2, from Mycobacterium tuberculosis. J Biol Chem 291:7465–7476. doi: 10.1074/jbc.M115.700344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schmitz KR, Sauer RT. 2014. Substrate delivery by the AAA+ ClpX and ClpC1 unfoldases activates the mycobacterial ClpP1P2 peptidase. Mol Microbiol 93:617–628. doi: 10.1111/mmi.12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Akopian T, Kandror O, Raju RM, UnniKrishnan M, Rubin EJ, Goldberg AL. 2012. The active ClpP protease from M. tuberculosis is a complex composed of a heptameric ClpP1 and a ClpP2 ring. EMBO J 31:1529–1541. doi: 10.1038/emboj.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Raju RM, Jedrychowski MP, Wei J-R, Pinkham JT, Park AS, O’Brien K, Rehren G, Schnappinger D, Gygi SP, Rubin EJ. 2014. Post-translational regulation via Clp protease is critical for survival of Mycobacterium tuberculosis. PLoS Pathog 10:e1003994. doi: 10.1371/journal.ppat.1003994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Njire M, Wang N, Wang B, Tan Y, Cai X, Liu Y, Mugweru J, Guo J, Hameed HMA, Tan S, Liu J, Yew WW, Nuermberger E, Lamichhane G, Liu J, Zhang T. 2017. Pyrazinoic acid inhibits a bifunctional enzyme in Mycobacterium tuberculosis. Antimicrob Agents Chemother 61:e00070-17. doi: 10.1128/AAC.00070-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Niu H, Ma C, Cui P, Shi W, Zhang S, Feng J, Sullivan D, Zhu B, Zhang W, Zhang Y. 2017. Identification of drug candidates that enhance pyrazinamide activity from a clinical compound library. Emerg Microbes Infect 6:e27. doi: 10.1038/emi.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Prideaux B, Via LE, Zimmerman MD, Eum S, Sarathy J, O’Brien P, Chen C, Kaya F, Weiner DM, Chen P-Y, Song T, Lee M, Shim TS, Cho JS, Kim W, Cho SN, Olivier KN, Barry CE, Dartois V. 2015. The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat Med 21:1223–1227. doi: 10.1038/nm.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Driver ER, Ryan GJ, Hoff DR, Irwin SM, Basaraba RJ, Kramnik I, Lenaerts AJ. 2012. Evaluation of a mouse model of necrotic granuloma formation using C3HeB/FeJ mice for testing of drugs against Mycobacterium tuberculosis. Antimicrob Agents Chemother 56:3181–3195. doi: 10.1128/AAC.00217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Irwin SM, Driver E, Lyon E, Schrupp C, Ryan G, Gonzalez-Juarrero M, Basaraba RJ, Nuermberger EL, Lenaerts AJ. 2015. Presence of multiple lesion types with vastly different microenvironments in C3HeB/FeJ mice following aerosol infection with Mycobacterium tuberculosis. Dis Model Mech 8:591–602. doi: 10.1242/dmm.019570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Irwin SM, Prideaux B, Lyon ER, Zimmerman MD, Brooks EJ, Schrupp CA, Chen C, Reichlen MJ, Asay BC, Voskuil MI, Nuermberger EL, Andries K, Lyons MA, Dartois V, Lenaerts AJ. 2016. Bedaquiline and pyrazinamide treatment responses are affected by pulmonary lesion heterogeneity in Mycobacterium tuberculosis infected C3HeB/FeJ mice. ACS Infect Dis 2:251–267. doi: 10.1021/acsinfecdis.5b00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Marakalala MJ, Raju RM, Sharma K, Zhang YJ, Eugenin EA, Prideaux B, Daudelin IB, Chen PY, Booty MG, Kim JH, Eum SY, Via LE, Behar SM, Barry CE III, Mann M, Dartois V, Rubin EJ. 2016. Inflammatory signaling in human tuberculosis granulomas is spatially organized. Nat Med 22:531–538. doi: 10.1038/nm.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wright GD. 2016. Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol 24:862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]