Viruses and mobile genetic elements are molecular parasites or symbionts that coevolve with nearly all forms of cellular life. The route of virus replication and protein expression is determined by the viral genome type. Comparison of these routes led to the classification of viruses into seven “Baltimore classes” (BCs) that define the major features of virus reproduction. However, recent phylogenomic studies identified multiple evolutionary connections among viruses within each of the BCs as well as between different classes.
KEYWORDS: ICTV, evolution, megataxonomy, phylogenomics, phylogeny, realm, virosphere, virus classification, virus nomenclature, virus taxonomy
SUMMARY
Viruses and mobile genetic elements are molecular parasites or symbionts that coevolve with nearly all forms of cellular life. The route of virus replication and protein expression is determined by the viral genome type. Comparison of these routes led to the classification of viruses into seven “Baltimore classes” (BCs) that define the major features of virus reproduction. However, recent phylogenomic studies identified multiple evolutionary connections among viruses within each of the BCs as well as between different classes. Due to the modular organization of virus genomes, these relationships defy simple representation as lines of descent but rather form complex networks. Phylogenetic analyses of virus hallmark genes combined with analyses of gene-sharing networks show that replication modules of five BCs (three classes of RNA viruses and two classes of reverse-transcribing viruses) evolved from a common ancestor that encoded an RNA-directed RNA polymerase or a reverse transcriptase. Bona fide viruses evolved from this ancestor on multiple, independent occasions via the recruitment of distinct cellular proteins as capsid subunits and other structural components of virions. The single-stranded DNA (ssDNA) viruses are a polyphyletic class, with different groups evolving by recombination between rolling-circle-replicating plasmids, which contributed the replication protein, and positive-sense RNA viruses, which contributed the capsid protein. The double-stranded DNA (dsDNA) viruses are distributed among several large monophyletic groups and arose via the combination of distinct structural modules with equally diverse replication modules. Phylogenomic analyses reveal the finer structure of evolutionary connections among RNA viruses and reverse-transcribing viruses, ssDNA viruses, and large subsets of dsDNA viruses. Taken together, these analyses allow us to outline the global organization of the virus world. Here, we describe the key aspects of this organization and propose a comprehensive hierarchical taxonomy of viruses.
The eternal mystery of the world is its comprehensibility … The fact that it is comprehensible is a miracle.
—Albert Einstein, 1936
INTRODUCTION
All cellular life-forms, with the exception of some intracellular bacterial parasites, host distinct repertoires of viruses and other mobile genetic elements (MGEs). Viruses appear to be the dominant biological entities on our planet, with the total count of virus particles in aquatic environments alone at any given point in time reaching the staggering value of 1031, a number that is at least an order of magnitude greater than the corresponding count of cells (1–5). Accordingly, lytic infections of cellular organisms, primarily bacteria, by viruses play a central role in the biological matter turnover in the biosphere (1, 5–10). The genetic diversity of viruses is harder to assess, but, beyond doubt, the gene pool of viruses is, in the least, comparable to that of hosts. The estimates of the number of distinct prokaryotes on earth differ widely, in the range of 107 to 1012 (11–14), and accordingly, estimation of the number of distinct viruses infecting prokaryotes at 108 to 1013 is reasonable. Even assuming the lowest number in this range and even without attempting to count viruses of eukaryotes, these estimates represent vast diversity. Furthermore, the genomes of most viruses accumulate mutations much faster than genomes of cellular organisms due to both the typically low fidelity of the virus replication machinery that stems, in part, from the absence of proofreading activity in many viruses and the strong selection pressure on virus populations (15–20). Some viruses are able to rapidly explore sequence space via recombination, whereas those with segmented or multipartite genomes can complement the accumulation of mutations through replication with reassortment of genome segments (21, 22). Thus, viruses encompass an enormous pool of rapidly evolving genes that appears to continuously contribute to the emergence of new genes in cellular life-forms through the exchange of genetic material between cells and viruses. However, despite the rapid short-term evolution of viruses, the key genes responsible for virion formation and virus genome replication are conserved over the long term due to selective constraints (23, 24).
Theoretical arguments and mathematical models of replicator evolution indicate that genetic parasites inescapably emerge even in the simplest molecular replicator systems and persist through their subsequent evolution (25–30). Together with the ubiquity and enormous diversity of viruses in the extant biosphere, these findings lead to the conclusion that viruses and other MGEs played major roles in the evolution of life ever since its earliest stages (8, 31–33). In a sense, all evolution of life is a history of coevolution between MGEs and their cellular hosts. This coevolution involves both the perennial arms race that leads to the evolution of the extremely diverse and elaborate defense systems of the hosts and various forms of cooperation, in particular exaptation of MGE genetic material for diverse and novel roles in hosts and the converse hijacking of defense system components by MGEs (34, 35, 235). The interactions between MGEs and their cellular hosts span the entire range from mutualism, e.g., between bacteria or archaea and numerous plasmids that carry genes essential for their hosts, via various forms of symbiotic relationships, to full antagonism as in the case of lytic viruses (36–38). Considering the enormous abundance and diversity of viruses and other MGEs, and the ubiquitous interactions between MGEs and cellular hosts, a thorough investigation of the evolutionary relationships among viruses and MGEs is essential to advance our understanding of the evolution of life (8, 32, 39–41).
In this article, we summarize the emerging understanding of the evolutionary relationships among diverse groups of viruses and argue that a coherent account of the global organization of the virus world is now within reach. Prior to this work, only a few attempts at virus megataxonomy were undertaken. For instance, in 1943, Ruska advocated for a virus classification based on certain morphological characteristics of virions (42). In 1948, Holmes outlined an all-virus-encompassing order with various subranks on the basis of virus-host tropism (43). In 1961, Cooper suggested a system on the basis of virus genome type and the absence or presence of a virion lipid envelope (44). Others subsequently modified this taxonomy by adding virion pH sensitivity and virion shape as classification criteria (45–48). In the most consistent virus taxonomy effort of the pregenomic era, from 1962 to 1966, Lwoff, Horne, and Tournier developed an alternative system that also focused on the type of nucleic acid that is encapsidated into virions and virion morphology (“LHT system”) (49–51).
The advances of virus genomics and metagenomics create both the challenge and the opportunity to develop a new, genome-based virus megataxonomy (52). Recently, the International Committee on Taxonomy of Viruses (ICTV) formally approved the possibility to classify viruses known only from genomic sequence (53) and to create taxa above the rank of order (54), thus opening the door for a comprehensive taxonomic scheme. Here, we describe the emerging global view of the organization of the virus world and propose a complete, even though preliminary, outline of the forthcoming virus megataxonomy.
THE BALTIMORE CLASSES OF VIRUSES, VIRUS HALLMARK GENES, AND MAJOR EVOLUTIONARY TRENDS IN THE VIRUS WORLD
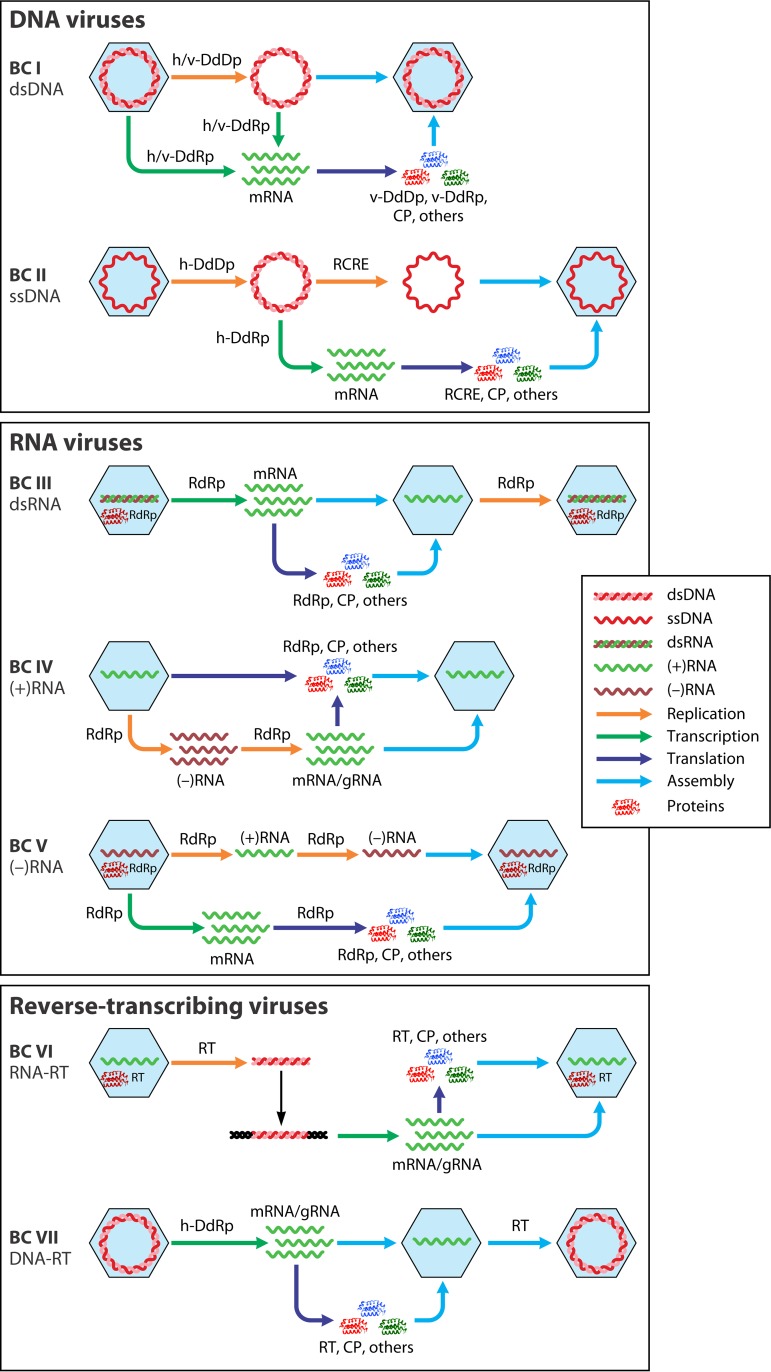
In sharp contrast with cellular organisms, viruses and MGEs use effectively all possible genome replication and expression strategies (Fig. 1). The only general exclusion principle that governs the information flow in cellular life-forms and MGEs is the central dogma of molecular biology according to which information is transferred from nucleic acids to proteins but never in the reverse direction (55, 56). In a seminal 1971 article, Baltimore classified all then-known viruses into six distinct classes that became known as Baltimore classes (BCs) (a seventh class was introduced later), on the basis of the structure of the virion's nucleic acid (traditionally called the virus genome) (57–59):
Double-stranded DNA (dsDNA) viruses, with the same replication-expression strategy as in cellular life forms
Single-stranded DNA (ssDNA) viruses that replicate mostly via a rolling-circle mechanism
dsRNA viruses
Positive-sense RNA [(+)RNA] viruses that have ssRNA genomes with the same polarity as the virus mRNA(s)
Negative-sense RNA [(−)RNA] viruses that have ssRNA genomes complementary to the virus mRNA(s)
RNA reverse-transcribing viruses that have (+)RNA genomes that replicate via DNA intermediates synthesized by reverse transcription of the genome
DNA reverse-transcribing viruses replicating via reverse transcription but incorporating into virions a dsDNA or an RNA-DNA form of the virus genome.
FIG 1.
The seven Baltimore classes (BCs): information flow. For each BC, the processes of replication, transcription, translation, and virion assembly are shown by color-coded arrows (see the inset). Host enzymes that are involved in virus genome replication or transcription are prefixed with “h-,” and in cases when, in a given BC, one of these processes can be mediated by either a host- or a virus-encoded enzyme, the latter is prefixed with “v-.” Otherwise, virus-encoded enzymes are not prefixed. CP, capsid protein; DdDp, DNA-directed DNA polymerase; DdRp, DNA-directed RNA polymerase; gRNA, genomic RNA; RdRp, RNA-directed RNA polymerase; RT, reverse transcriptase; RCRE, rolling-circle replication (initiation) endonuclease.
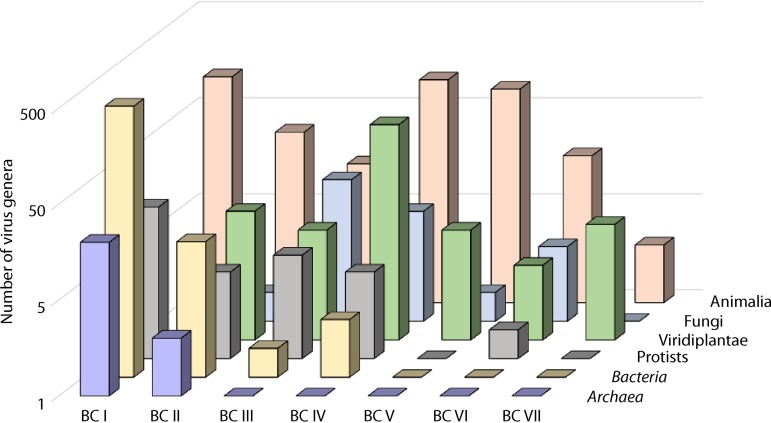
Ever since the publication of Baltimore’s paradigmatic article (57), the BCs have been informally used as the top classification rank for viruses, and a proposal has been made to formalize this status (54). Recently, all RNA viruses (BCs III, IV, and V) have been formally unified under the realm taxon, which is equivalent to the domain rank used for cellular organisms (60). From a formal information transfer perspective, each of the BCs can be viewed as equivalent in stature to all cellular life-forms (59). In other words, all cellular life-forms use a strategy of genome replication and expression that corresponds to a single BC, namely, BC I. Viruses of the different BCs widely differ in abundance, diversity, and distribution across the diversity of cellular hosts. The most abundant and diverse viruses (at least to our current knowledge) are BC I viruses, which are most common in prokaryotic hosts, and BC IV viruses, which dominate the viromes of eukaryotes (61). However, none of the BCs can be considered “exotic,” because all BCs include viruses and virus-related MGEs that are abundant in at least some hosts (Fig. 2).
FIG 2.
Distribution of the seven BCs of viruses in the major divisions of prokaryotes and eukaryotes. The virus genera are from the ICTV report (https://talk.ictvonline.org/ictv-reports/ictv_online_report/).
Although pragmatic, the classification of viruses into the BCs does not necessarily accurately reflect evolutionary relationships. The proposition that the BCs are monophyletic is arguably attractive and at face value plausible. However, evidence supports monophyly for some of the BCs but refutes it for others. Generally, the evolution of viruses and MGEs is studied with methods of molecular evolutionary analysis that are also used for cellular organisms (62, 63). However, the organizations of the genetic spaces dramatically differ between viruses and their cellular hosts. Even among cellular life-forms, the genomes display remarkable plasticity and evolve in highly dynamic regimes. Especially in prokaryotes, the dominant evolutionary process is horizontal gene transfer (HGT), which results in massive gene gain and loss (64–67). Notwithstanding these turbulent evolutionary dynamics, the genomes of all cellular organisms encompass about a hundred universal genes that encode, almost exclusively, protein and RNA components of the translation system (68). The presence of this universal gene core is strong evidence for the origin of all cellular life from a last universal cellular ancestor (LUCA) (68–70). Furthermore, phylogenetic analysis of these universal genes reveals a central vertical trend in the evolution of cellular life-forms and provides a scaffold for reconstructions of evolution by gene gain, gene loss, and HGT (71).
In the genetic space of viruses and MGEs, no genes are universal or even conserved in the majority of viruses (72). Viruses have several distinct points of origin, so there has never been a last common ancestor of all viruses. Moreover, even when a gene is represented in many diverse groups of viruses, it cannot be automatically assumed that it originated in their last common ancestor. The cause of the uncertainty is that viruses acquire numerous genes from their cellular hosts, and independent capture of homologous genes by diverse viruses appears to be fairly common (73, 74). Furthermore, rapid virus genome evolution often leads to extreme divergence of homologous sequences in distinct viruses, so the successful detection of such homologs using even the most powerful methods of sequence analysis is not guaranteed.
Notwithstanding the highly dynamic character of virus evolution and the ensuing methodological problems, large-scale reconstruction of virus evolution is not a hopeless pursuit. Despite the lack of a common ancestor of all viruses, evolutionary analyses of large virus groups have proven productive. The prime approach in these studies is phylogenetic analysis of virus hallmark genes (VHGs), i.e., genes that are broadly conserved among diverse groups of viruses (72).
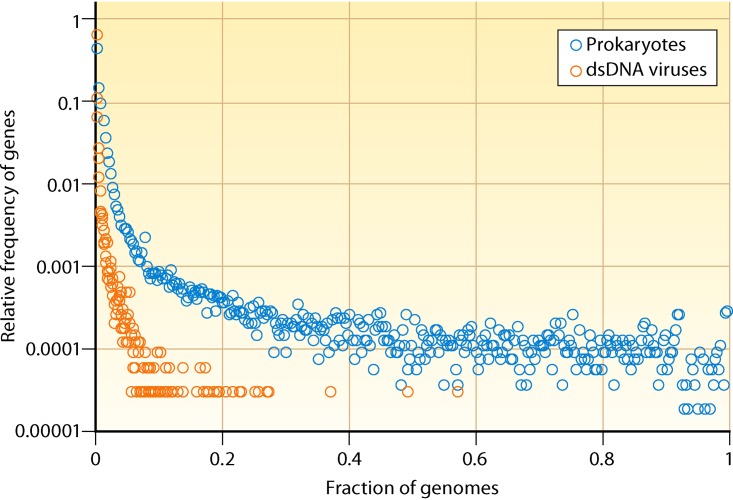
For cellular life-forms, in particular archaea and bacteria, gene frequency plots show a characteristic, asymmetrical U shape that includes a small uptick on the right that corresponds to (nearly) universal genes, a comparatively flat middle part consisting of moderately conserved genes, and a large ascending part on the left that includes rare and unique genes (Fig. 3). This characteristic distribution of gene frequencies is observed at all evolutionary depths among prokaryotes and can be accurately described by a model of evolution that incorporates a particular distribution of gene replacement rates (i.e., deleterious effects of gene loss) (75–77). The gene frequency distribution for virus genomes of dsDNA viruses, the only BC for which the number of genes is sufficiently large to analyze this type of distribution, is qualitatively similar but quantitatively different, with a notably greater fraction of viral genes contained within the poorly conserved/unique cloud and a minuscule core of conserved genes (78, 79) (Fig. 3). The genes comprising this core, together with the (nearly) ubiquitous genes in genomes of viruses of the other BCs, can be defined as VHGs.
FIG 3.
Comparison of gene frequency distributions for prokaryotes and dsDNA viruses (Baltimore class I). The horizontal axis shows the fraction of genomes in which a given family of orthologous genes is represented. The vertical axis (logarithmic scale) shows the relative frequency of orthologous gene families that are represented in the given number of genomes. Thus, common genes are on the right of the plot, and rare genes are on the left.
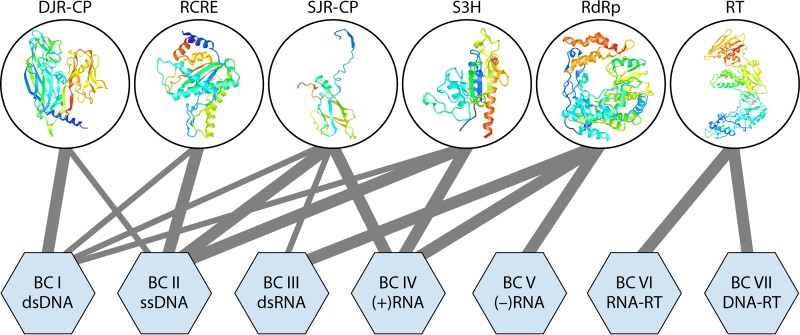
Not surprisingly, VHGs encode proteins involved in central functions of virus replication and morphogenesis (72). Most VHGs are restricted to one BC, but several are present in enormously diverse viruses and span two or even three BCs (Fig. 4). These “super-VHGs” encode the following:
RNA-directed RNA polymerases (RdRps) that form an apparently monophyletic group of palm domain-containing polymerases and unite the three BCs (III, IV, and V) of RNA viruses
RNA-directed DNA polymerases, or reverse transcriptases (RTs), that unify the two BCs (VI and VII) of reverse-transcribing viruses, along with the related MGEs, and belong to the same branch of the palm domain polymerases as the RdRps
Superfamily 3 helicases (S3Hs) that are encoded almost exclusively by MGEs, including (+)RNA (BC IV), most of the ssDNA (BC II), and diverse groups of dsDNA (BC I) viruses
Single-jelly-roll capsid proteins (SJR-CPs), which are the most common form of CPs among (+)RNA (BC IV) and ssDNA (BC II) viruses
Double-jelly-roll capsid proteins (DJR-CPs), widespread among dsDNA viruses and also found in some ssDNA viruses
Rolling-circle replication initiation endonucleases (RCREs) that are encoded by the great majority of ssDNA viruses but are also present in some dsDNA viruses.
FIG 4.
Representation of the 6 “superviral hallmark genes” in virus genomes of the seven Baltimore classes. The “superviral hallmark proteins” are shown by ribbon diagrams of the representative protein structures. The lines connect the proteins with the viruses of BCs in which they are present. The thickness of each connecting line roughly reflects the abundance of a given “superhallmark” gene in a given BC. DJR-CP, double-jelly-roll capsid protein; RCRE, rolling-circle replication (initiation) endonuclease; RdRp, RNA-directed RNA polymerase; RT, reverse transcriptase; S3H, superfamily 3 helicase; SJR-CP, single-jelly-roll capsid protein.
The origins of super-VHGs appear to be widely different. In particular, RdRps, RTs, and RCREs most likely represent the heritage of the primordial, precellular replicator pool as indicated by the absence of orthologs of these proteins in cellular life-forms (except for some that, obviously, have been captured from MGEs) (72, 80). The S3Hs and SJR-CPs seem to have been acquired from cellular hosts at the earliest stages of virus evolution, whereas the DJR-CPs probably evolved from SJR-CPs in a common ancestor of an ancient virus lineage (80, 81).
The evolution of the rest of the virus genomes can be reconstructed using VHG phylogenies as scaffolds (74, 82). A complementary approach is the dissection of bipartite gene-genome networks of viruses into distinct modules (79, 83, 84). The (super-)VHGs are hubs of such networks. Arguably, a comprehensive account of virus evolution can be achieved only through the combination of phylogenetic and network approaches.
During the last few years, the application of these approaches to different large groups of viruses, often initially defined as BCs, has resulted in fairly detailed and well-supported evolutionary classifications for most of these groups (61, 82, 85). Thus, we unexpectedly find ourselves in the position of mapping the major regions of the virus world with reasonable accuracy, albeit with many blank/unresolved spots on the map.
In the sections that follow, we summarize the emerging understanding of the evolutionary connections among viruses within and across BCs. The coherent assemblages of related viruses identified through a combination of VHG phylogenies and networks of gene sharing could become the taxa of multiple ranks in a new, internally consistent, hierarchical system of virus taxonomy. In this forthcoming megataxonomy, novel top-rank virus taxa will displace the BCs from their current status as informal highest-rank taxonomic units. Nevertheless, the BCs will remain an essential framework for the study of virus biology, much like the concepts of prokaryotes and eukaryotes, which do not reflect the phylogeny of cellular life-forms and which are not taxa of any rank but nevertheless are key staples of comparative cell biology.
Note that, here, we use “-virids” and “-virins” (as outlined in reference 86) as specific suffixes to denote the collective members of a virus family and subfamily, respectively, and to differentiate them from the collective members of a virus genus (“-viruses”).
EVOLUTIONARY LINKS AMONG VIRUSES WITHIN AND ACROSS THE BALTIMORE CLASSES
RNA Viruses
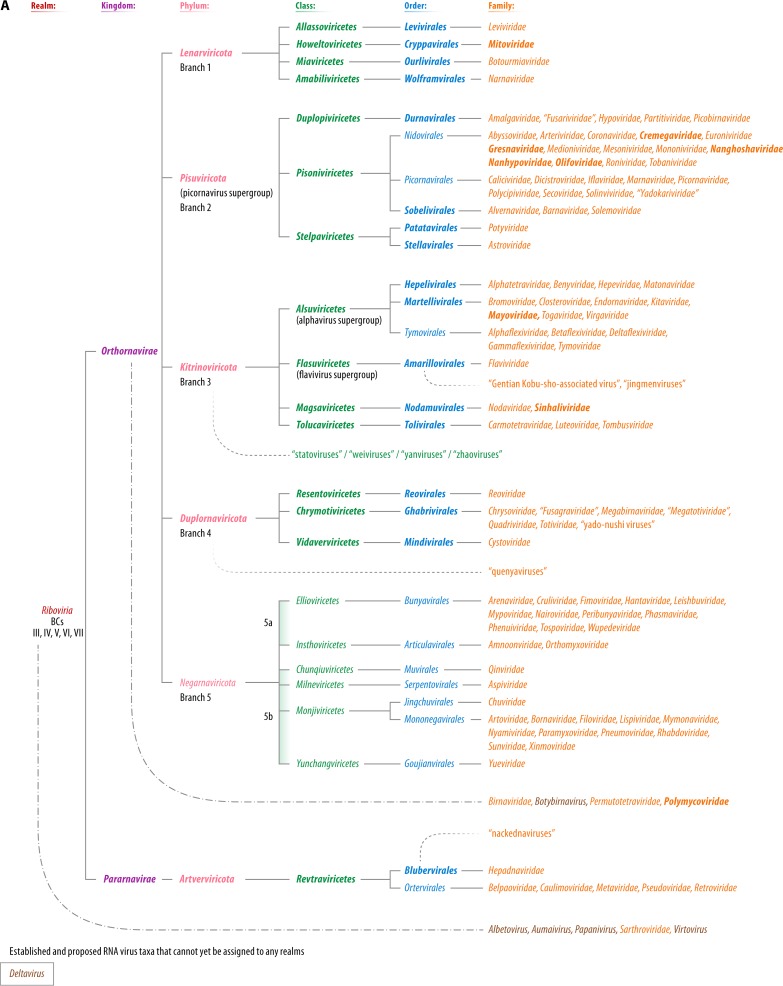
Viruses in BCs III, IV, and V share a single VHG, the RdRp (87, 88). The RdRp sequences are highly divergent among the three BCs, and the very demonstration of their homology has been far from a trivial task (89, 90). Nevertheless, comparative analyses of thousands of sequences of diverse RdRps and RTs in BCs III, IV, and V and of the structures of these polymerases demonstrate the monophyly of these enzymes, all of which contain the so-called palm domain (80, 82, 91). Accurate phylogenetic analyses of protein families that consist of protein sequences as highly divergent as those of RdRps and RTs require the utmost caution (92, 93). Yet, notwithstanding all the caveats, the RdRp phylogenetic tree, which was rooted using the highly conserved RTs of group II introns as the outgroup, has a well-defined structure (82, 94) (Fig. 5). Although the placement of the root on the RT branch is forced, the tree topology appears to be fully compatible with this assumption because the monophyly of the RdRps is strongly supported.
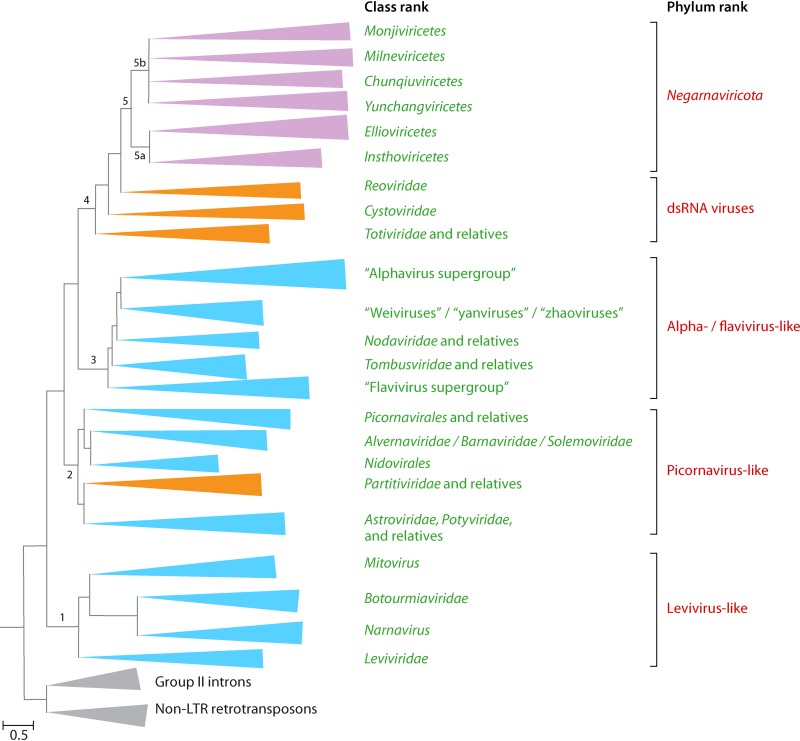
FIG 5.
Schematic representation of the phylogenetic tree of the RNA-directed RNA polymerases (RdRps) of RNA viruses (realm Riboviria). The five major branches discussed in the text are labeled. Only selected clades including the best-characterized virus groups are shown within each major branch. Each branch represents collapsed sequences of the respective set of RdRps. The reverse transcriptase sequences from group II introns and non-LTR retrotransposons were used as the outgroup to root the tree. The three BCs included in the tree are color-coded (orange, BC III; blue, BC IV; purple, BC V). (Adapted from reference 82.)
The RdRps form five major branches (82). Two branches consist entirely of (+)RNA viruses (branches 1 and 3), one branch combines (+)RNA and dsRNA viruses (branch 2), another one consists of dsRNA viruses (branch 4), and the last branch includes (−)RNA viruses (branch 5) (Fig. 5). The monophyly of each of the five branches is strongly supported, but the evolutionary relationships among the five branches are less certain. The RdRp is the hallmark of RNA viruses that brings them all together, whereas other genes are specific to each of the five branches and their major subbranches. Multiple gene exchanges between RNA viruses of different branches appear to have occurred, complicating evolutionary reconstructions.
Branch 1.
Branch 1 includes RdRps of viruses assigned to the Leviviridae and is the only branch of (+)RNA viruses infecting prokaryotes, specifically a limited diversity of bacteria of the phylum Proteobacteria. Levivirids remain a compact group of viruses despite the recent expansion of “levi-like” viruses discovered through metagenomics sequence analysis (95, 236).
In addition to levivirids, branch 1 includes the RdRps of their apparent descendants infecting eukaryotes (Fig. 5). The RdRp tree topology suggests that levivirids first gave rise to “minimal” RNA replicons, known as mitoviruses, which encode only RdRps (96). Mitoviruses reproduce in the mitochondria of some protists, fungi, and plants, as befits descendants of prokaryotic viruses, and could have been acquired by protoeukaryotes together with the mitochondrial endosymbiont (97). Mitoviruses are the apparent ancestors of another group of capsidless RNA replicons, narnaviruses, which escaped to the eukaryotic cytosol. Subsequently, mitoviruses and narnaviruses were apparently lost from most eukaryotic lineages.
Descendants of narnaviruses reacquired a gene encoding a distinct capsid protein, namely, SJR-CP, and some also captured genes encoding a helicase and a movement protein (98). These derivatives of the levivirid-related naked replicons, members of the Botourmiaviridae, were initially thought to infect only plants but subsequently were identified in fungi and invertebrates as well (99, 100). Thus, the history of branch 1 seems to have involved a loss of the levivirid structural module (the unique major and minor CP genes), yielding capsidless replicons, some of which subsequently reacquired an unrelated CP (SJR-CP) from plant viruses. The natural history of branch 1 is the simplest among the RNA viruses and prompts the creation of a high-rank taxon for these viruses. Furthermore, the taxonomy within this forthcoming taxon will have to be revised because the family Botourmiaviridae is located between the genera Mitovirus and Narnavirus of the current family Narnaviridae.
Branch 2.
Branch 2 of the RdRp tree (Fig. 5) includes diverse (+)RNA viruses and two groups of dsRNA viruses that are sometimes collectively referred to as the “picornavirus supergroup” (88, 101–103), named after the Picornavirales, an expansive, well-characterized order of protist, animal, and plant viruses that includes many human pathogens (104). Viruses in this branch show an enormous diversity of genome lengths, gene repertoires, and genome organizations and form virions with staggeringly different structures. Branch 2 includes viruses with the largest known RNA genomes (e.g., abyssovirids and planarian secretory cell nidovirus, both classified as Nidovirales) (105, 106), along with viruses with some of the smallest RNA genomes, such as sobemoviruses (Sobemoviridae) (107). Notwithstanding this enormous diversity and despite the fact that no genes except for those encoding RdRp are shared by all viruses in branch 2, the VHGs encoding SJR-CPs and trypsin-like proteases appear to derive from a common virus ancestor of this branch (82).
The evolutionary scenario for viruses in branch 2 includes the parallel acquisition of helicases of three superfamilies and a number of additional genes, especially by viruses of the Nidovirales. Perhaps the most prominent feature of branch 2 is the interleaving of two BCs. A strongly supported clade of dsRNA viruses within branch 2 consists of the members of the Partitiviridae and Picobirnaviridae (part of BC III) embedded among (+)RNA viruses (BC IV). Partitivirids and picobirnavirids have small, bipartite genomes, of which one segment encodes the RdRp, whereas the other segment encodes the CP that is apparently homologous to the CPs of other dsRNA viruses such as the Reoviridae (108, 109) (see below). This clade also includes capsidless RNA replicons that encode only the RdRp (100, 110, 111). These replicons reproduce inside mitochondria or chloroplasts of algae and use the mitochondrial genetic code, resembling mitoviruses of branch 1 with respect to genome content and lifestyle. Thus, the dsRNA virus clade in branch 2 most likely evolved via a scenario similar to that outlined above for the botourmiavirids in branch 1, namely, the combination of an RdRp-only capsidless replicon inherited from prokaryotes and a eukaryotic dsRNA virus CP gene.
Picobirnavirid genes are preceded by evolutionarily conserved Shine-Dalgarno sequences, suggesting the possibility that the actual hosts of these viruses discovered in association with animals are bacteria (112, 113).
An additional, striking case of mixing different BCs within the same virus genome is presented by viruses of the Amalgaviridae, a family of unsegmented dsRNA viruses infecting plants and protists that encode an RdRp confidently affined with those of partitivirids, but their putative CP is derived from nucleocapsid proteins of the (−)RNA viruses of the bunyaviral Phenuiviridae (BC V) (114).
From a taxonomic standpoint, the dissection of branch 2 shows the inevitability of the creation of taxa that cross the boundaries between the BCs. This branch alone spans enormous phylogenetic depth, necessitating the creation of a multirank taxonomy.
Branch 3.
Branch 3 of the RdRp tree consists of viruses that are about as diverse, in terms of gene composition and genome architecture, as those comprising branch 2 but currently includes only (+)RNA viruses infecting eukaryotes (Fig. 5). Branch 3 contains two clades consisting of thoroughly characterized plant and animal viruses, some of which possess long and highly complex genomes (by RNA virus standards). These clades are informally assigned to the “alphavirus supergroup” and “flavivirus supergroup” (88, 102, 115) (Fig. 5). The “alphavirus supergroup” includes an extremely broad range of plant viruses and a narrower variety of animal viruses, all of which share a conserved array of VHGs that encode capping enzymes, superfamily 1 helicases, and RdRps (61, 102). The conservation of this block of VHGs strongly supports the monophyly of the supergroup.
The “flavivirus supergroup” is smaller than the “alphavirus supergroup” and, until recently, included only animal-infecting viruses of the Flaviviridae. However, metaviromic studies have greatly expanded this supergroup by adding numerous plant and animal viruses with diverse genome organizations, including multipartite (+)RNA flavivirus-related agents tentatively called “jingmenviruses” and even a putative dsRNA flavivirus-related agent associated with gentian Kobu-sho syndrome (100, 116–120). Similar to the “alphavirus supergroup,” the majority of the “flavivirus supergroup” viruses encode capping enzymes and helicases. However, these proteins (domains) belong to protein families that are distinct from those that include the counterpart enzymes in the “alphavirus supergroup” and clearly have been acquired independently (82).
In addition to the two supergroups, branch 3 includes a number of lineages that all represent viruses with typically short genomes that encode RdRps, CPs, and a few additional proteins. Two strongly supported, well-characterized clades among these short-genome viruses are formed by members of the Nodaviridae and Tombusviridae and their relatives, but many more groups will have to be added for viruses discovered by metavirome sequencing, such as “statoviruses,” “weiviruses,” “yanviruses,” and “zhaoviruses” (100, 116, 121, 122). Again, a multirank, hierarchical taxonomic structure is necessary to adequately represent this branch.
Branch 4.
Branch 4 of the RdRp tree thus far includes only dsRNA viruses (Fig. 5), but a novel group of (+)RNA viruses (“quenyaviruses”) that might belong at the root of this branch was recently described (123). In a pattern mimicking the genomic diversity in branches 2 and 3, dsRNA viruses in branch 4 span wide ranges of genome lengths and complexities. This branch contains the “minimal” genomes of viruses of the family Totiviridae that encode only RdRp and CP, along with the highly complex reovirid genomes that consist of up to 12 dsRNA segments and encompass numerous genes. The key observation that supports the monophyly of dsRNA viruses in branch 4 is the conservation of the CP structure and the capsid architecture, in which 60 homo- or heterodimers of the CP are organized on an unusual, pseudo-T=2 lattice (109, 124). The simpler totivirids and the complex reovirids and their immediate relatives comprise the two main clades within branch 4. Notably, the reovirid-like clade includes cystovirids, the only known group of dsRNA viruses infecting prokaryotes (125). Thus, the same trend of apparent evolution of eukaryotic viruses from prokaryotic ancestors as that identified in branch 1 and suggested for branch 2 is recapitulated among branch 4 dsRNA viruses, and a multirank, hierarchical taxonomic structure can be readily applied.
Branch 5.
Branch 5 (established phylum Negarnaviricota [52, 60]) consists of all known (−)RNA viruses with the exception of deltaviruses and RdRp-less satellite viruses (albetoviruses, aumaiviruses, papaniviruses, sarthrovirids, and virtoviruses). Perhaps, unexpectedly, branch 5 is lodged within branch 4, where it forms the sister group of the reovirid-like clade. The monophyly of the (−)RNA virus RdRps is unambiguously supported by all used phylogenetic methods, but the position of the (−)RNA branch amid the dsRNA viruses is less confidently supported (82). Nevertheless, the placement of (−)RNA viruses within the dsRNA branch appears to be reaffirmed by comparison of the structures of the RdRps of (−)RNA and dsRNA viruses (126, 127). Furthermore, this tree topology is compatible with the apparent logical course of virus evolution: from the (+)RNA viruses that are ubiquitous in eukaryotes, to the dsRNA viruses that are less widely spread, to the (−)RNA viruses that, despite their diversity, appear to have a limited host range (Fig. 2) and are absent from prokaryotes. In contrast to (+)RNA viruses, dsRNA viruses and (−)RNA viruses share a signature of the reproduction cycle: among viruses of both of these BCs, the replication and transcription machineries are packaged into virions, and this packaging is essential for the replication of the respective RNA forms.
Branch 5 splits into two strongly supported clades (52, 60, 82), one of which consists mostly of viruses with segmented genomes that use the mRNA “cap-snatching” mechanism (subbranch 5a, subphylum Polyploviricotina, classes Ellioviricetes [Bunyavirales] and Insthoviricetes [Orthomyxoviridae, Tilapinevirus]), whereas the other clade includes viruses with primarily unsegmented genomes that encode their own capping enzymes (subbranch 5b, subphylum Haploviricotina, classes Monjiviricetes [Chuviridae, Mononegavirales] and Milneviricetes [Ophiovirus]) (Fig. 5).
The position of the phylum Negarnaviricota within the dsRNA virus branch creates a taxonomic conundrum because dsRNA viruses of branch 4 are paraphyletic with respect to branch 5. Conceivably, branch 4 could be accommodated taxonomically by at least three phyla.
Classified or officially proposed groups of RNA viruses remaining unassigned.
Several groups of currently classified RNA viruses or RNA viruses that have officially been proposed to be classified remain unassignable to any of the five major branches due to the extreme divergence of their RdRps. These groups include the (+)RNA Permutotetraviridae and the dsRNA Birnaviridae, Botybirnavirus, and proposed Polymycoviridae (formerly known as “tetramycoviruses” [128]). Monophyly of two families of viruses with circular domain permutation within their RdRps, Permutotetraviridae and Birnaviridae, has been suggested (129, 130), but a robust placement of these unusual RdRps in the phylogenetic tree requires further investigation. Also unclassified remains the only (+)RNA virus that has been sequenced from a hyperthermal environment and is suspected to infect the crenarchaeote Saccharolobus solfataricus (131). The forthcoming expansion of the RNA virus genome collection, in particular resulting from massive metagenomic analyses, is expected to place the currently unassigned groups into existing or new groups.
Reverse-Transcribing Viruses
Reverse-transcribing viruses (BCs VI and VII) are widespread in protists, fungi, plants, and animals but, at least so far, are not known to infect prokaryotes. However, prokaryotes and most eukaryotes host several types of capsidless, nonviral retroelements. In prokaryotes and many eukaryotic organelles, these retroelements include group II introns and retrons (132, 133), whereas eukaryotes host several types of non-long terminal repeat (LTR) retrotransposons (short and long interspersed nuclear elements [SINEs and LINEs, respectively]) (134, 135). In close analogy to RNA viruses, reverse-transcribing viruses (some of which are traditionally called LTR retrotransposons) and the LINEs among non-LTR retrotransposons share a single gene coding for the key genome replication enzyme, the RT. Thus, similar to the RNA virus RdRp, RT is the only phylogenetic marker that can be analyzed to deduce the evolution of reverse-transcribing viruses and retrotransposons (91, 132).
The RT phylogenetic tree includes five major branches, four of which consist of nonviral retroelements (132, 136). The fifth strongly supported branch harbors all currently known reverse-transcribing viruses. Thus, the RT phylogeny establishes the monophyly of reverse-transcribing viruses that appear to share an ancestry from a retrotransposon, conceivably at an early stage in the evolution of eukaryotes.
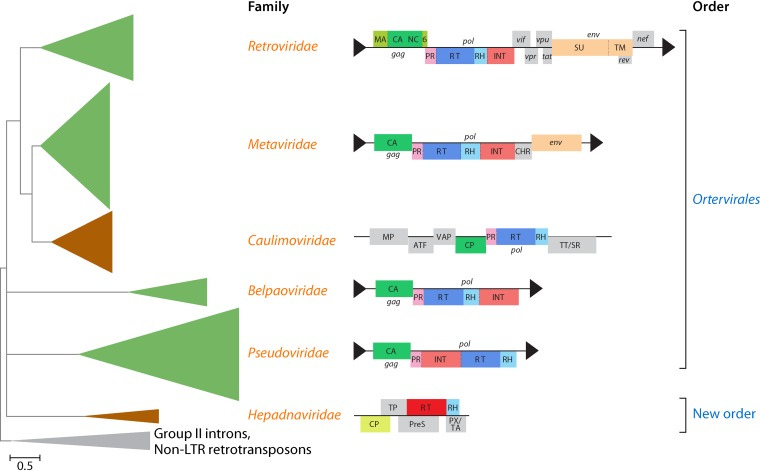
The virus branch of the RT tree consists of four clades, three of which represent, respectively, viruses of the Hepadnaviridae (together with “nackednaviruses,” a recently discovered group of nonenveloped hepadna-like viruses [137]), Belpaoviridae, and Pseudoviridae, whereas the fourth clade unites the viruses of the Caulimoviridae, Metaviridae, and Retroviridae (Fig. 6) (136, 137). In addition to the RT, all these families of viruses (and LTR retrotransposons), except for hepadnavirids/“nackednaviruses,” also share the genes encoding two core virion components, namely, the CP and the nucleocapsid protein. The latter appears to have been lost in the spumaretrovirin subclade of the retrovirids (138). Furthermore, belpaovirids, caulimovirids, metavirids, pseudovirids, and retrovirids share a distinct replication-priming mechanism, by which host tRNA molecules are recruited as primers for reverse transcription (136). In contrast, hepadnavirids and, by inference, “nackednaviruses” use a protein-priming mechanism involving the terminal protein domain of the RT (137). Therefore, the five families of reverse-transcribing viruses are most likely monophyletic (justifying their current taxonomic classification into the Ortervirales [136]), to the exclusion of Hepadnaviridae and “nackednaviruses” that comprise the earliest-branching clade of reverse-transcribing viruses.
FIG 6.
Schematic phylogenetic subtree of the reverse transcriptases (RTs) of reverse-transcribing viruses (Caulimoviridae, Hepadnaviridae, and Retroviridae) and long terminal repeat (LTR) retrotransposons (Belpaoviridae, Metaviridae, and Pseudoviridae). The tree is rooted using sequences from nonviral retroelements (prokaryotic group II introns and non-LTR retrotransposons). Genome organizations of selected representatives of reverse-transcribing viruses and LTR retrotransposons are shown near the corresponding clades. LTRs are shown as black triangles. 6, 6-kDa protein; ATF, aphid transmission factor; CA/CP, capsid protein; CHR, chromodomain (contained only in the integrases of certain clades of metavirids from plants, fungi, and several vertebrates); gag, group-specific antigen; env, envelope gene; INT, integrase; MA, matrix protein; MP, movement protein; NC, nucleocapsid; nef, tat, rev, vif, vpr, and vpu, genes expressing regulatory proteins via spliced mRNAs; P, polymerase; pol, polymerase gene; PR, protease; PreS, pre-surface protein (envelope); PX/TA, protein X/transcription activator; RH, RNase H; SU, surface glycoprotein; TM, transmembrane glycoprotein; TP, terminal protein domain; TT/SR, translation trans-activator/suppressor of RNA interference; VAP, virion-associated protein. The two BCs included in the tree are color-coded (green, BC VI; brown, BC VII). (Adapted from reference 136.)
Notably, both caulimovirids and hepadnavirids/“nackednaviruses” (BC VII; often collectively called “pararetroviruses”) package dsDNA genomes into their virions but are far apart in the RT tree (Fig. 6). Thus, BC VII is most likely polyphyletic, mimicking the case of dsRNA viruses. Given the monophyly of all viruses in the RT tree (Fig. 6), joining the Ortervirales and Hepadnaviridae/“nackednaviruses” together appears logical and could form another order in a single, higher-rank taxon.
Single-Stranded DNA Viruses
Most ssDNA viruses possess small, circular genomes that replicate via the rolling-circle replication (RCR) mechanism and infect a wide variety of prokaryotic and eukaryotic hosts (139). Several recent studies have revealed the high diversity and abundance of ssDNA viruses in various habitats, showing that these viruses comprise a major component of the earth’s virome (140–146). The ssDNA viruses are currently classified into four families of viruses infecting bacteria and archaea (Inoviridae, Microviridae, Pleolipoviridae, and Spiraviridae) and nine families of viruses infecting eukaryotes (Anelloviridae, Bacilladnaviridae, Bidnaviridae, Circoviridae, Geminiviridae, Genomoviridae, Nanoviridae, Parvoviridae, and Smacoviridae). The ssDNA viruses with circular genomes that encode a replication initiator protein (Rep) of the His-hydrophobic-His (HUH) endonuclease superfamily and are suspected or shown to infect eukaryotes, i.e., those classified as Bacilladnaviridae, Circoviridae, Geminiviridae, Genomoviridae, Nanoviridae, and Smacoviridae, have been collectively referred to as circular Rep-encoding single-stranded DNA (CRESS-DNA) viruses (139, 147). Recent metagenomics studies reveal a far greater diversity of CRESS-DNA viruses than previously observed, with several family-rank clades (139, 148, 149).
Unlike most of the ssDNA viruses, parvovirids and bidnavirids possess linear ssDNA genomes (150, 151). Whereas parvovirid genomes are replicated by the rolling-hairpin mechanism (152), a variation of RCR, bidnavirids encode protein-primed family B DNA polymerases (151). Notably, different pleolipovirids have either ssDNA or dsDNA genomes (153, 154), once again demonstrating evolutionary mixing of different BCs, in this case within the same family.
Thus, BC II (ssDNA viruses) consists primarily of “minimal” viruses with genomes that often encode a single protein involved in replication and a structural protein (and, in some groups, one or two accessory proteins). Except for anellovirids, bidnavirids, spiravirids, and certain inovirids, all ssDNA virus genomes encode the HUH endonuclease domain that cleaves the genomic ssDNA at specific sites and initiates RCR (or rolling-hairpin replication) (155, 156). The eukaryotic CRESS-DNA viruses also encode S3Hs that typically are fused to HUH endonucleases to form two-domain Reps. In contrast, prokaryotic ssDNA viruses encode single-domain HUH endonucleases (85, 157, 158). With the exception of inovirids, pleolipovirids, spiravirids, and the recently discovered Flavobacterium-infecting, lipid-containing phage (FLiP) (proposed Finnlakeviridae) (159), the capsids of ssDNA viruses are made of SJR-CPs.
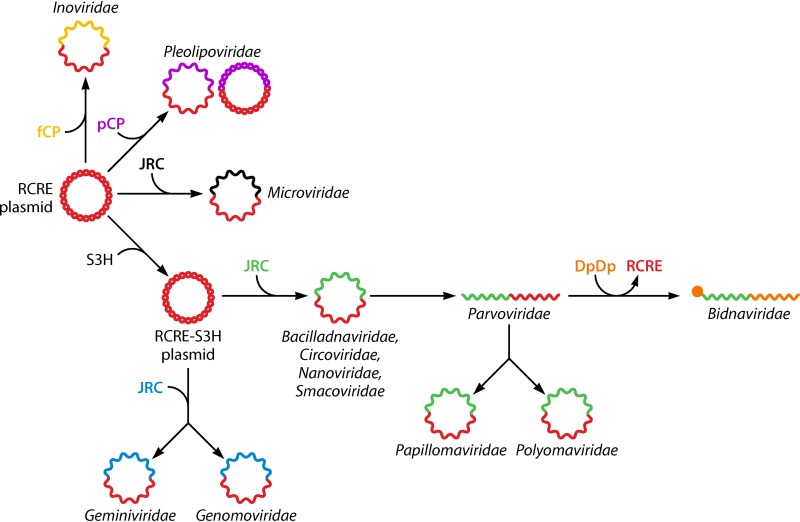
Sequence comparisons and phylogenetic analyses of HUH endonucleases, S3Hs, and CPs of ssDNA viruses suggest multiple origins of these viruses via a chimeric scenario (Fig. 7) (85, 158, 160–163). The Reps of CRESS-DNA viruses are most closely related to the homologous Reps of small bacterial RCR plasmids that also encode HUH endonucleases and S3Hs. Phylogenetic analyses of the Reps show that the replication apparatus of the CRESS-DNA viruses evolved from the plasmid replication machinery on at least three independent occasions (Fig. 7) (85). Similarly, the HUH endonucleases of CRESS-DNA viruses infecting prokaryotes appear to have evolved from plasmid endonucleases that are not fused to S3Hs.
FIG 7.
Multiple, chimeric origins of ssDNA viruses (and two groups of dsDNA viruses [Papillomaviridae and Polyomaviridae]). The genomes are shown with two colors each, to emphasize the distinct origins of the genes encoding proteins involved in replication (Reps) and those encoding capsid proteins (CPs). JRC, jelly roll capsid protein (nonorthologous JRC genes are shown with different colors); fCP, filamentous capsid protein; pCP, polymorphic capsid protein; DdDp, DNA-directed DNA polymerase; RCRE, rolling-circle replication (initiation) endonuclease; S3H, superfamily 3 helicase. (Adapted from reference 85.)
The CPs of eukaryotic CRESS-DNA viruses seem to come from a completely different source: these proteins are most closely related to the CPs of diverse plant and animal (+)RNA viruses (85, 164, 165). Thus, eukaryotic CRESS-DNA viruses apparently evolved by recombination between a bacterial plasmid and a cDNA copy of a (+)RNA virus on multiple, independent occasions. The realization of this scenario in the evolution of multiple groups of CRESS-DNA viruses is one of the most remarkable cases of widespread convergent evolution among viruses and other MGEs. In addition, this scenario implies that the diversification of the eukaryotic (+)RNA viruses preceded the emergence of CRESS-DNA viruses. The provenance of the divergent microvirid SJR-CPs remains uncertain (81).
Phylogenetic analysis of the Reps reveals two large branches of eukaryotic CRESS-DNA viruses, each including several families/clades, that could form a higher-ranked taxon, and all CRESS-DNA viruses can be unified at an even higher rank (85). Eukaryotic CRESS-DNA viruses are the likely ancestors of parvovirids, with their linear ssDNA genomes. Remarkably, members of the Papillomaviridae and Polyomaviridae, viruses with small, circular dsDNA genomes, seem to have evolved from ssDNA viruses, most likely parvovirids, in a major transition in which the HUH domain lost its endonuclease activity but retained the sequence-specific DNA-binding activity (166). Concomitantly, the replication mechanism of these viruses switched from the RCR to the “theta” bidirectional mechanism that operates in plasmid and bacterial chromosome replication. The CPs of papillomavirids and polyomavirids are highly divergent, so it remains unclear whether they evolved from parvovirid CPs or were independently recruited (81).
Parvovirids, papillomavirids, and polyomavirids could join the assemblage of CRESS-DNA viruses at the top taxonomic rank. Although rare, this would not be the only case of the same taxon including viruses with different nucleic acid forms encapsidated into virions. For example, as pointed out above, the Ortervirales include reverse-transcribing viruses that encapsidate RNA and DNA genomes, whereas members of the Pleolipoviridae possess either ssDNA or dsDNA genomes. In terms of the evolutionary status of the BCs, ssDNA viruses present a unique conundrum, as these viruses constitute a polyphyletic assemblage with multiple, diverse origins of different VHGs.
Double-Stranded DNA Viruses
The dsDNA viruses make up the most diverse and expansive of the BCs (BC I). These viruses dominate the prokaryotic virome. dsDNA viruses are common among eukaryotes as well despite their low abundance and diversity compared to RNA viruses (61) (Fig. 2). Not a single gene is conserved in all dsDNA viruses, which makes the construction of a phylogenetic tree for the entire BC I impossible, in principle. However, gene-sharing networks, combined with phylogenies of the genes that are conserved in large groups of viruses, provide a working approach for dsDNA virus classification (79, 83, 84). The diversity of unclassified dsDNA viruses is enormous and seems to include completely uncharted parts of the virus world (167–169). Thus, we can cover this BC class here in only a coarse-grain outline.
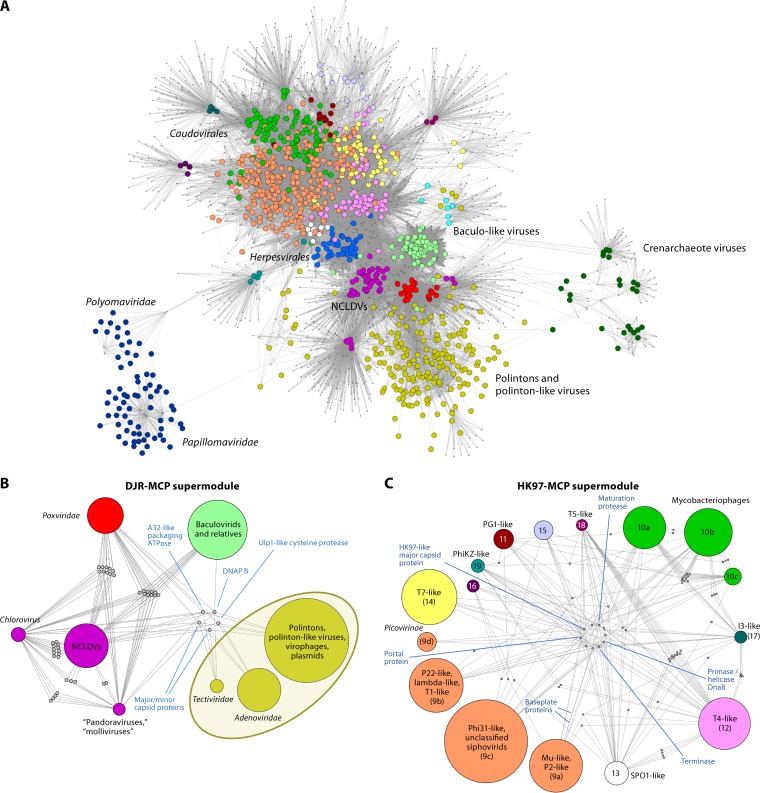
The gene-sharing network of the dsDNA viruses splits into two expansive supermodules and three smaller modules (Fig. 8). The supermodules are held together by sets of VHGs that encode structural proteins and enzymes involved in virion morphogenesis, and each supermodule appears to be monophyletic, at least with respect to these core genes.
FIG 8.
Bipartite gene-genome network of dsDNA viruses. (A) Complete network. The larger circles show nodes corresponding to genomes, and dots show nodes corresponding to core gene families. An edge connecting a circle and a dot indicates that a genome contains a representative of a core gene family. Virus genomes that belong to different modules of the network are shown in different colors. (B and C) Internal organizations of the DJR-MCP (B) and HK97-MCP (C) supermodules. The individual modules within each supermodule are shown by circles. The positions of some major groups of viruses are indicated. In panel C, the numbers correspond to the module numbering described previously (79); the unnamed modules are small groups of tailed bacteriophages. DNAP, DNA polymerase. (Adapted from reference 79.)
DJR-MCP supermodule: dsDNA viruses with vertical jelly roll capsid proteins.
The double-jelly-roll major capsid protein (DJR-MCP) supermodule consists of numerous groups of viruses with icosahedral virions that infect bacteria, archaea, and eukaryotes (170, 171) (Fig. 9). In addition to the eponymous DJR-MCPs, most of these viruses encode minor SJR-CPs (e.g., penton proteins) and FtsK-HerA superfamily genome-packaging ATPases (172–175). Apart from this morphogenetic gene triad, the majority of the viruses in the DJR-MCP supermodule encode DNA-directed DNA polymerases (DdDps) and often S3Hs (61, 176) or replication initiation proteins (corticovirids) (177).
FIG 9.
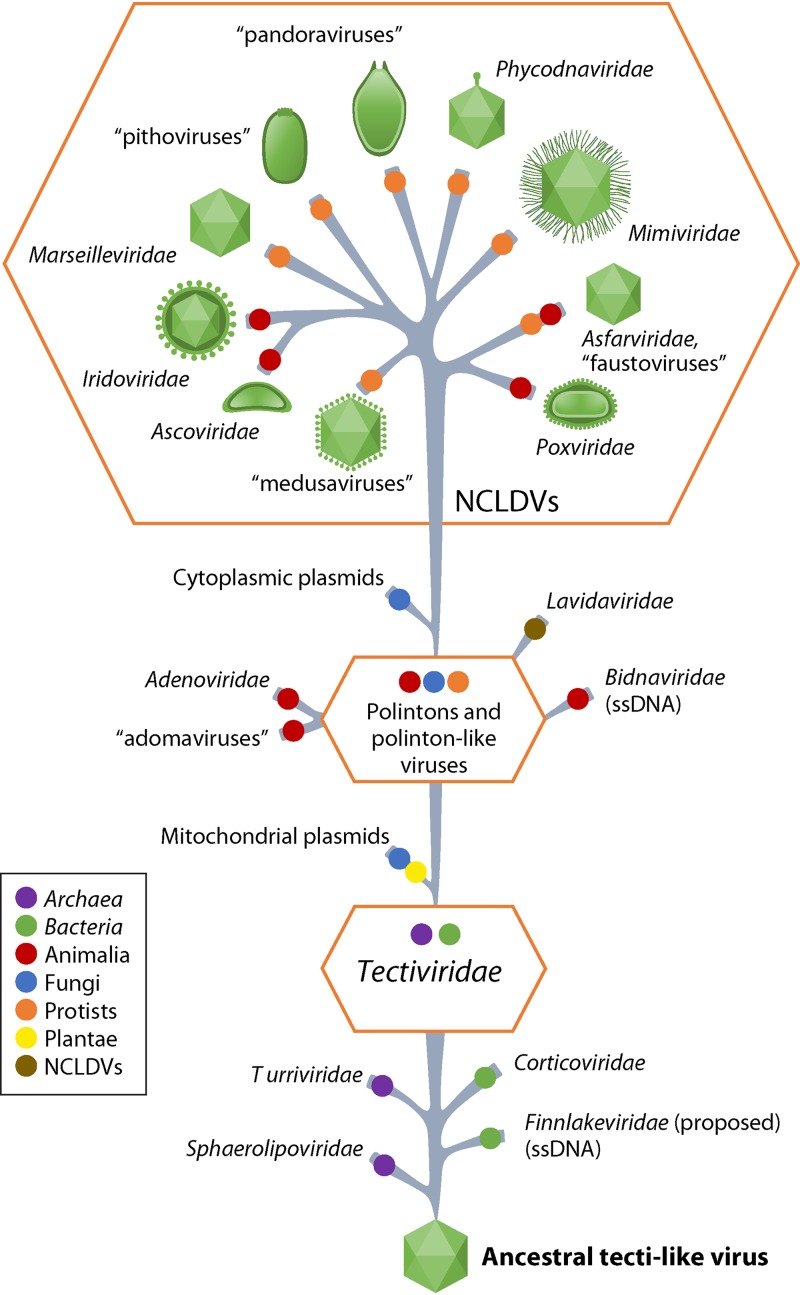
Proposed evolutionary scenario for DJR-MCP supermodule viruses. The host ranges of viruses are shown by colored circles (see the color-code on the left). For nucleocytoplasmic large DNA viruses (NCLDVs), schematic virion shapes are shown. (Adapted from reference 176.)
The DJR-MCP supermodule includes tail-less bacteriophages that are currently classified as Corticoviridae and Tectiviridae as well as archaeal viruses of the Turriviridae. However, metagenomic searches for putative viruses encoding DJR-MCPs revealed an enormous diversity of uncharacterized members of this supermodule that apparently infect prokaryotes (one of these new groups has been proposed as “Autolykiviridae”) (167, 178). These recent findings suggest that DJR-MCP viruses are a major, possibly even dominant, although not-well-characterized part of the prokaryotic virome.
Archaeal and bacterial viruses of the Sphaerolipoviridae (179) encode two vertical single-jelly-roll MCPs (along with minor SJR-CPs) (180, 181) that form a capsid lattice structurally resembling the DJR-MCP-based capsid lattice of, for example, Pseudomonas phage PRD1 (Tectiviridae: Alphatectivirus), rather than those of (+)RNA and ssDNA viruses composed of horizontal SJR-CPs (182, 183). Therefore, it appears likely that sphaerolipovirids retain the ancestral state from which the vertical DJR-MCPs evolved via gene fusion (81).
Eukaryotes host an even greater diversity of DJR-MCP supermodule viruses (Fig. 9). Polintons, self-synthesizing transposon-like elements that are integrated into diverse eukaryotic genomes but are predicted to form virions and diverse polinton-like viruses and virophages, appear to descend directly from tectivirids or their immediate relatives (176). Adenovirids are a derived offshoot of the same line of descent. Similar to polintons, besides the major and minor CPs, adenovirids encode a distinct protease that is most likely derived from a host deubiquitinylating enzyme and is involved in virion morphogenesis (176). Notably, tectivirid-like and “polinton-like” viruses additionally gave rise to two groups of capsidless elements, mitochondrial and cytoplasmic linear plasmids, respectively, that are found in some protists, fungi, and plants (176).
Nucleocytoplasmic large DNA viruses (NCLDVs) comprise an expansive assemblage of several families of eukaryotic viruses, including the Phycodnaviridae, extended Ascoviridae, Asfarviridae, Iridoviridae, Marseilleviridae, Mimiviridae, and Poxviridae, and currently unclassified giant viruses such as “cedratviruses,” “faustoviruses,” “medusaviruses,” “Mininucleoviridae,” “molliviruses,” “orpheoviruses,” “pacmanviruses,” “pandoraviruses,” and “pithoviruses” (74, 184–191, 237). All these viruses have long or “giant” genomes (up to 2.5 Mb) and corresponding large virions. Most of these viruses retain the DJR structural module, the SJR minor CP, and the genome-packaging ATPase and protease, although some viruses, such as “pandoraviruses,” have secondarily lost the DJR-MCP and form irregularly shaped virions instead of virions based on typical icosahedral capsids (192–194). Other NCLDV groups, such as “molliviruses,” “pithoviruses,” and poxvirids, retain the DJR-MCP but do not incorporate it into virions that, again, assume unusual shapes (74, 195–198). Comparative genomic reconstructions of NCLDV evolution have mapped ∼40 genes to the common ancestor of this virus group that, in all likelihood, existed prior to the divergence of the major eukaryote divisions (199). Phylogenetic analysis of the highly conserved core genes of the NCLDVs yields a well-resolved tree in which most of the unassigned groups belong to strongly supported branches together with known viruses (74). Based on these analyses, “cedratviruses,” “orpheoviruses,” and “pithoviruses” form a clade that joins the larger assemblage within the NCLDV group that also includes the Ascoviridae, Iridoviridae, and Marseilleviridae. “Molliviruses” and “pandoraviruses” also are related groups and form a distinct clade with coccolithoviruses and phaeoviruses currently included in the Phycodnaviridae. Only “medusaviruses,” which appear to have acquired an unusually rich repertoire of eukaryotic genes (191), remain an unassigned group. These observations provide a comprehensive hierarchical taxonomy of the NCLDVs that will include a number of new taxa of different ranks and requires splitting of some existing taxa, such as the apparently polyphyletic family Phycodnaviridae (see below).
Notwithstanding all the genomic embellishments as well as unusual gene losses, the NCLDVs appear to descend from “polinton-like viruses” (74, 176) (Fig. 9). The giant viruses, despite their “cell-like” genome lengths and the diverse functional repertoire of genes, appear to have evolved from smaller viruses independently, in different branches of the NCLDV clade (200–202).
Recently, a novel group of viruses, “yaraviruses,” has been reported. The type virus “yaravirus” has a 45-kb genome, i.e., a genome intermediate in length between NCLDVs and polinton-like viruses. The “yaravirus” genome shares several distinctive genes (e.g., those encoding DJR-MCP, packaging ATPase, and primase-S3H) with NCLDVs whereas others are apparently lacking. The provenance of this unusual virus remains to be established through detailed genome analysis (238).
Finally, an ssDNA virus, FLiP, encoding a DJR-MCP with close structural similarity to those of dsDNA viruses such as Pseudoalteromonas phage PM2, has been discovered, revealing a connection between dsDNA and ssDNA viruses (159, 183).
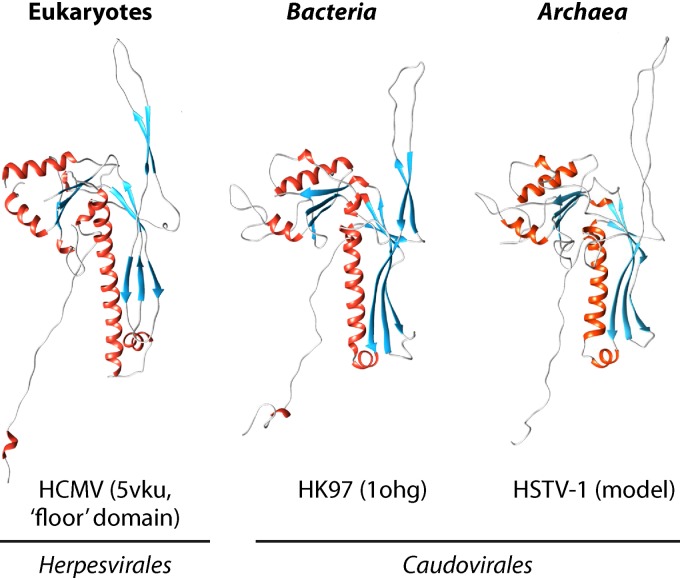
HK97-MCP supermodule.
The HK97-MCP supermodule, named after Escherichia coli phage HK97 (Caudovirales: Siphoviridae: Hendrixvirus), for which the capsid structure was among the first resolved in this virus assemblage (203), unites tailed viruses infecting bacteria and archaea and a single, albeit large, group of animal viruses that are currently classified as Herpesvirales (Fig. 8C). The icosahedral capsids of these viruses are made of proteins that are unrelated to DJR-MCPs (203–206) (Fig. 10). In addition to the HK97-MCP, viruses in this supermodule possess conserved portal proteins and two subunits (large and small) of the terminase (207, 208). The large terminase subunits are a distinct variety of packaging ATPases that share only the general structural design of the P-loop fold with the packaging ATPases of the DJR-MCP supermodule viruses and are not directly related to the latter (209).
FIG 10.
Comparison of the structures of the major capsid proteins of bacterial, archaeal, and eukaryotic viruses of the HK97-MCP supermodule. For HCMV and HK97, the Protein Data Bank (PDB) accession numbers are indicated in parentheses (5VKU and 1OHG). HCMV, human cytomegalovirus; HK97, Escherichia coli phage HK97; HSTV-1, Haloarcula sinaiiensis tailed virus 1.
Despite the conservation of the structural and morphogenetic proteins, the HK97-MCP supermodule viruses display enormous ranges of genome complexity and gene content. Some of the viruses in this supermodule encode nearly complete replication systems, whereas the genomes of other viruses effectively lack genes involved in replication and completely rely on the host replication machineries (210). The prokaryotic HK97-MCP viruses currently comprise the vast order Caudovirales, which includes five established families (Ackermannviridae, Herelleviridae, Myoviridae, Podoviridae, and Siphoviridae) and five proposed families (Autographiviridae, Chaseviridae, Demerecviridae, Drexlerviridae, and Zobellviridae).
This taxonomy is currently under reorganization and, in the near future, can be expected to expand into many new families, to adequately reflect the extreme heterogeneity of these viruses with respect to gene composition and phylogenetic relationships (211, 212). Bipartite network and comparative genomics analyses identified multiple modules consisting of various tailed bacterial and archaeal viruses that potentially could form new families (Fig. 8C) (79, 83, 213). However, even this picture appears to be dramatically incomplete as new groups of HK97-MCP viruses continue to be discovered by metagenomics and single-cell genomics. Recent examples include the putative family of cross-assembly (crAss)-like viruses (168, 214, 215), “magroviruses” that infect mesophilic euryarchaeotes (216), and several groups of Lak-like megaphages (169, 217). All these virus groups are only distantly related to other HK97-MCP viruses and can be fully expected to require new taxa of different ranks.
Baculovirid/nudivirid module.
A distinct, comparatively small, stand-alone module consists of several families of arthropod viruses, including the Baculoviridae, Hytrosaviridae, Nimaviridae, Polydnaviridae (genus Bracovirus), and Nudiviridae (218–220). These viruses encompass several genes that are distantly related to core genes of the NCLDVs and thus could be highly derived members of the DJR-MCP supermodule, despite the absence of the DJR-MCP and formation of odd-shaped virions (61, 79).
Crenarchaeote virus module.
A large module that is nearly disconnected from the rest of the gene-sharing network of dsDNA viruses consists of viruses that infect hyperthermophilic crenarchaeotes. These viruses adopt various unique virion morphologies, with CPs that are unrelated to structural proteins of other viruses or to each other (83, 221–224) (Fig. 8A). Most of the crenarchaeote viruses also lack genes for recognizable homologs of proteins that comprise the replication machineries of other viruses (210). Nevertheless, these viruses are linked in the network through shared genes for transcription factors, uncharacterized ATPases, and glycosyltransferases (83). However, these genes are not orthologous among crenarchaeote viruses from different families and, in all likelihood, have been acquired horizontally from either other viruses or the respective hosts. Given the apparent independent origins of most crenarchaeote virus groups, several highest-rank taxa would be needed to adequately reflect their places in the virus world.
Papillomavirid/polyomavirid module.
A stand-alone module of dsDNA viruses includes papillomavirids and polyomavirids that apparently evolved from ssDNA viruses (see the section above) and thus are disconnected from the rest of the dsDNA viruses. Remarkably, a recently discovered unusual group of viruses, “adomaviruses,” bridges papillomavirids/polyomavirids with the typical dsDNA viruses of the DJR-MCP supermodule. “Adomaviruses” combine a gene encoding a polyomavirid-like replication protein with the genes encoding the morphogenetic protease and at least one other virion protein with homologs in the dsDNA viruses of the Adenoviridae (225–227).
Taken together, the analyses summarized above indicate that, similarly to the ssDNA viruses, the dsDNA viruses (BC I) are polyphyletic, that is, apparently evolved from unrelated components on multiple independent occasions. The monophyletic groups among the dsDNA viruses are the two major supermodules, several groups of crenarchaeote viruses, and the papillomavirids/polyomavirids.
PROPOSED MEGATAXONOMY OF VIRUSES
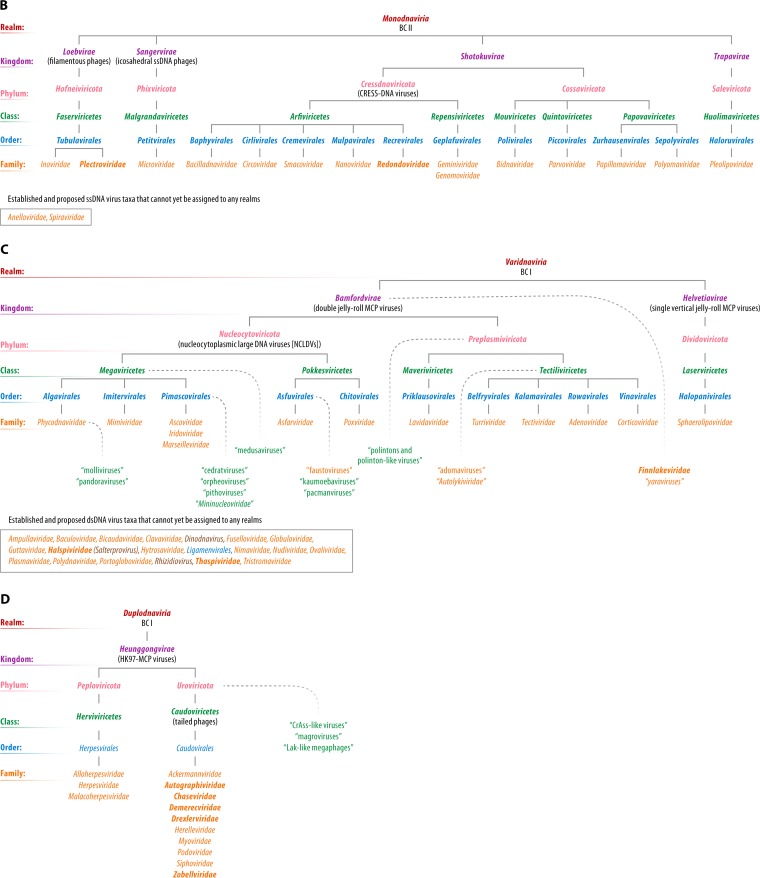
The ICTV currently approves eight principal/primary taxon ranks (in descending order, realm, kingdom, phylum, class, order, family, genus, and species) and seven secondary ranks (subrealm, subkingdom, subphylum, subclass, suborder, subfamily, and subgenus). As of 2019, only a single realm, Riboviria, has been established (228). This realm currently consists of all (+)RNA, dsRNA, and (−)RNA viruses that comprise the only formally established virus phylum (Negarnaviricota). This phylum, in turn, includes the only established subphyla and classes of viruses (52, 60). The rest of the current virus taxonomy begins at the rank of disconnected orders or families. The emerging view of the global organization of the virus world outlined above naturally translates into a hierarchical scheme of an expanded virus taxonomy. In this section, we explicitly present such a hierarchy, suggesting the need for numerous taxa above the rank of family or order. Figure 11 outlines a megataxonomy from realm to family rank, which we officially proposed via four proposals to the ICTV in 2019 (229–232). The hierarchy presented in Fig. 11, which fills all principal/primary available ranks, was approved by the ICTV Executive Committee in December 2019 but must be considered tentative until formally ratified by the entire ICTV in the spring of 2020.
FIG 11.
Proposed megataxonomy of the virus world. Officially proposed taxon names are printed in boldface type; names of taxa that have already been accepted by the ICTV are rendered in normal font. The names of the virus groups that have not been officially proposed or formally accepted are shown in quotation marks. Taxon and virus group positions in the hierarchy, when uncertain, are indicated by fading curved dotted lines. The etymology of newly proposed taxon names associated with this article is outlined in File S1 in the supplemental material. Official taxa or officially proposed taxa that can be assigned only to top ranks at this time are indicated by curved dashed lines in the absence of quotation marks. All taxa/virus groups are to be considered sensu lato; i.e., they may include numerous “-like” viruses not yet named and/or classified. Proposed taxon names should be seen as provisional. Definitive names will be approved and ratified by the ICTV and will be found at the ICTV website (https://talk.ictvonline.org/).
RNA Viruses and Reverse-Transcribing Viruses
RdRps and RTs are the only proteins that are universally conserved among both RNA viruses and reverse-transcribing viruses and are the key replication enzymes of viruses of BCs III, IV, and V (RdRps) and BCs VI and VII (RTs). The monophyly of these proteins calls for the unification of these viruses in a single, highest-rank taxon, the established realm Riboviria (228), with two naturally defined kingdoms for RdRp- and RT-encoding viruses, respectively (Fig. 11A). In practice, this implies the inclusion of the reverse-transcribing viruses into the recently established realm Riboviria, which currently consists of only BCs III, IV, and V. The proposed taxonomic structure within each kingdom is based on the topologies of the phylogenetic tree of RdRps and RTs (Fig. 5 and 6, respectively) and the gene composition of viruses within the major branches.
The megataxonomy of RNA viruses and reverse-transcribing viruses entails at least two conceptual problems. First, capsidless elements related to viruses are treated inconsistently. Endornavirids, hypovirids, mitoviruses, narnaviruses, and umbraviruses are all included in the taxonomic scheme of RNA viruses, whereas non-LTR retrotransposons are currently not classified with the RT viruses. This nonuniform approach can be justified by the fact that the capsidless RNA replicons clearly are derivatives of bona fide RNA viruses, whereas non-LTR retroelements are outside the RT virus tree and are the apparent ancestors of reverse-transcribing viruses. Furthermore, at present, belpaovirids, metavirids, and pseudovirids are mostly referred to as “LTR retrotransposons.” However, in these cases, the arguments for classifying them as virus families appear compelling.
The second problem is the possible paraphyly of dsRNA viruses with respect to (−)RNA viruses. The origin of (−)RNA viruses from within dsRNA viruses requires further validation but, if confirmed, will make “reovirid-like” dsRNA viruses a paraphyletic taxon. In principle, this inconsistency can be resolved through the creation of three phyla of dsRNA viruses.
ssDNA Viruses
Most ssDNA viruses encode homologous replication proteins, HUH endonucleases, and many groups additionally encode S3Hs that are fused to these endonucleases. Numerous ssDNA viruses also encode homologous capsid proteins, SJR-MCPs. However, phylogenomic analyses show that ssDNA viruses do not share a single origin, as these viruses evolved by the recombination of plasmids with cDNA copies of RNA viruses on multiple, independent occasions (Fig. 7). Strikingly, even among viruses within a single, rapidly expanding, recognized family, Inoviridae, multiple replacements of genes encoding the HUH endonuclease with homologous but also with unrelated plasmid genes seem to have occurred (146).
The natural history of ssDNA viruses, despite its complexity, is becoming clear through recent phylogenomic analyses and challenges the very concepts of monophyly and polyphyly (85). Indeed, both the replication and structural genes of different groups of ssDNA viruses originate from plasmids and RNA viruses, respectively, but stem from different families of these elements. An additional complication is that, apart from the ssDNA viruses, HUH endonucleases are encoded by a number of dsDNA viruses. Thus, ssDNA viruses are neither monophyletic nor polyphyletic in the traditional sense, a type of relationship that does not seem to have analogs among other viruses or cellular life-forms. This conundrum questions the legitimacy of the unification of ssDNA viruses into a single taxonomic hierarchy. Nonetheless, not unlike RNA viruses or reverse-transcribing viruses, the majority of ssDNA viruses, along with papilloma- and polyomavirids with small dsDNA genomes, share the key replication enzyme, HUH endonuclease, that is conducive to phylogenetic analysis. Thus, we rely on the HUH endonuclease phylogeny and clustering to propose a megataxonomy that also includes the small dsDNA viruses, papillomavirids and polyomavirids, which apparently derive from ssDNA viruses (Fig. 11B). The extent of divergence between different groups of ssDNA viruses suggests elevating several of the currently recognized families (Inoviridae, Microviridae, and Parvoviridae) to higher taxonomic ranks, those of order or even class.
There are several groups of ssDNA viruses that do not encode Reps of the HUH endonuclease superfamily, including annelovirids, bidnavirids, a substantial fraction of inovirids, and the recently discovered bacteriophage FLiP. The origin of annelovirids remains mysterious, whereas the evolutionary trajectories of bidnavirids, inovirids, and FLiP appear to be traceable. Bidnavirids have apparently evolved from parvovirids via the replacement of the HUH endonuclease domain, but not the S3H domain, of the Rep with a protein-primed DNA polymerase from polintons (151). The evolution of inovirids featured multiple Rep replacements with nonorthologous and nonhomologous genes from plasmids and transposons, but the morphogenetic module firmly unifies all filamentous phages (146). The ancestral state of the FLiP Rep (Rep_trans superfamily) remains uncertain because FLiP is currently the sole representative of the proposed Finnlakeviridae. However, this virus is structurally related to DJR-MCP viruses and can thus be legitimately included in the corresponding realm.
dsDNA Viruses
dsDNA viruses account for the bulk of the diversity in the virus world and accordingly comprise the most complex network of relationships among all viruses. Furthermore, at least two major divisions of dsDNA viruses, the DJR-MCP and the HK97-MCP supermodules, appear to have genuinely independent origins, despite sharing some homologous genes. Accordingly, these two major groups of dsDNA viruses merit classification as separate, highest-rank taxa, i.e., realms (Fig. 11C and D). In some parts of the dsDNA domain of the virus world, the currently available information is insufficient for us to develop more than only a general outline of the taxonomic hierarchies. Furthermore, among the dsDNA viruses of hyperthermophilic crenarchaeotes, the few shared genes, such as those encoding DNA-binding proteins and glycosylases, in all likelihood spread via HGT (221, 222). The lack of apparent common ancestry among most crenarchaeote viruses would justify their placement into several separate realms. However, due to sparse sampling of these virus groups, it appears premature to establish highest-rank megataxonomies for these viruses at this point.
The Evolutionary and Taxonomic Status of the Baltimore Classes
The phylogenomic analysis of all known groups of viruses discussed above places the BCs into an evolutionary framework. The BCs fare quite differently as potential high-rank evolutionary units. Only BC V, (−)RNA viruses, survived unscathed as a monophyletic taxon. In contrast, the dsRNA viruses are a polyphyletic group, with at least two major branches derived from different parts of the (+)RNA virus tree. Thus, (+)RNA viruses, although formally monophyletic, comprise a paraphyletic taxon with respect to dsRNA viruses and (−)RNA viruses. Among the reverse-transcribing viruses, the RNA-containing viruses are similarly paraphyletic relative to one group of DNA-containing viruses (caulimovirids), whereas the second group of DNA-containing viruses (hepadnavirids/“nackednaviruses”) forms a distinct taxon.
Apart from this splitting and blending of the BCs, the global organization of the virus world that we attempt to reconstruct here (Fig. 11 and 12). features the unification of all RNA viruses with reverse-transcribing viruses into a single, monophyletic group that joins five BCs, a construct that was not in any form implied by Baltimore’s system.
FIG 12.
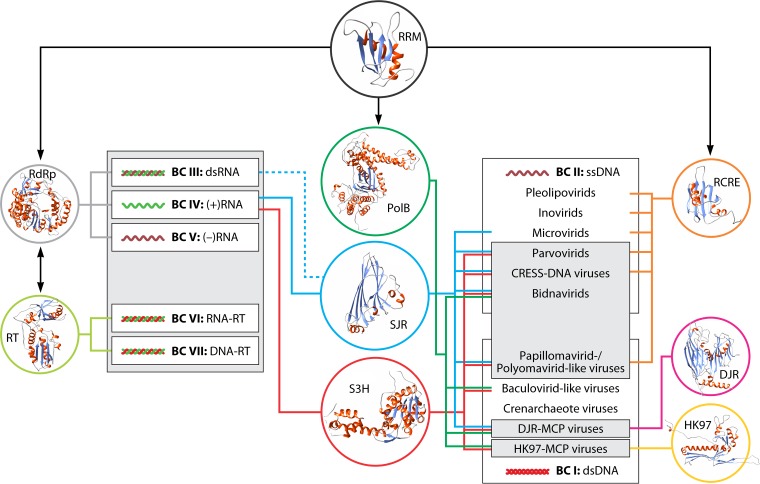
Virus realms, Baltimore classes, and the global network of the virus world. Gray shapes denote the four virus realms, and white rectangles denote the seven Baltimore classes. The colored circles contain ribbon diagram representations of the structures of hallmark virus proteins and the RNA recognition motif (RRM) domain. Circles are connected to groups of viruses, in which a given viral hallmark protein or domain is represented, and to BCs by lines of the corresponding color. Abbreviations: DJR, double jelly roll; PolB, family B DNA-directed DNA polymerase; RCRE, rolling-circle replication (initiation) endonuclease; RdRp, RNA-directed RNA polymerase; RT, reverse transcriptase; S3H, superfamily 3 helicase; SJR, single jelly roll.
The ssDNA viruses present a special case of chimeric origins that involve parallel recruitment of key genes from common ancestral pools and do not fit the traditional concepts of either monophyly or polyphyly. Thus, the creation of a highest-rank taxon encompassing all ssDNA viruses is, to a degree, a matter of convenience. Somewhat paradoxically, apart from this major conundrum, the small dsDNA viruses (papillomavirids and polyomavirids) are clearly derived from ssDNA viruses and accordingly should be included in the taxonomic hierarchy of the latter, once again mixing two BCs (Fig. 11 and 12). Inversely, the FLiP (ssDNA virus) genome should be classified with dsDNA viruses of the DJR-MCP supermodule.
The dsDNA viruses are the most obvious case of polyphyly of a BC, where at least two independent realms do not share an origin, along with the several groups of crenarchaeote viruses that might qualify as independent realms and papillomavirids and polyomavirids that are derived from ssDNA viruses and are classified accordingly.
The overall conclusion is clear: the BCs are a useful, coherent classification of viruses by genome type that correlates with the evolutionary relationships revealed by phylogenomic analysis. However, this correlation is obviously imperfect. In general, the BCs are not monophyletic and thus cannot be adopted as the top-rank virus taxa.
The Ultimate Network Architecture of the Virus World
The final question that we must address here is whether evolutionary connections across the entire virus world exist that would make the megataxon “Viruses” a legitimate concept. As discussed above, some major groups of viruses, for example, the two realms of dsDNA viruses, do not share an ancestry. Yet the matter is not unambiguous. Indeed, three simple protein domains that are present in multiple (super-)VHGs, namely, (i) the RNA recognition motif (RRM) that forms the cores of all virus polymerases; (ii) the jelly roll, the core domain of SJR- and DJR-MCPs; and (iii) S3H, the replicative helicase of diverse viruses, link the majority of the viruses in all BCs and in three of the four proposed realms (Fig. 12) (80). Thus, underneath the vastly different evolutionary histories and the polyphyly of viruses, fundamental structural and evolutionary unity likely harkens back to the primordial pool of genetic elements. In particular, primordial RNA replicons, including reverse-transcribing replicons akin to extant group II introns, were the likely ancestors of the realm Riboviria (80).
CONCLUSIONS
The principal and, in our view, striking conclusion of this work is that the global organization of the virus world is becoming transparent. Evidently, there are numerous, sometimes vast, white spots on the map of the virosphere. These gaps are being gradually filled thanks to increasingly sensitive analyses of genomic and metagenomic data coupled with comparisons of virus protein structures. A comprehensive, internally consistent, and stable hierarchical taxonomy of viruses seems to be within the reach of the current generation of virologists. We find it unexpected and remarkable that, notwithstanding the enormous diversity of viruses that is rapidly expanding through the advances of metaviromics, the contours of such a hierarchical virus taxonomy appear to be solidifying. At the top of the megataxonomy are the four effectively independent realms that, however, are connected at an even higher rank of unification through the super-VHG domains. Within each of the virus realms (Riboviria, “Monodnaviria,” “Varidnaviria,” and “Duplodnaviria”), the readily traceable hierarchy of evolutionary relationships calls for the construction of a comprehensive hierarchical taxonomy. Here, we outlined this entire hierarchy and presented a sketch of a megataxonomy (Fig. 11), which we have recently proposed to the ICTV in a series of four formal taxonomic proposals (229–232) that were approved by the ICTV Executive Committee in December 2019. We emphasize that this megataxonomy is an open system, in which the emergence of new high-rank taxa, including realms, can be anticipated. The BCs, which, for decades, have been informally construed as the highest-rank virus taxa, do not accurately represent the evolution of viruses and thus lack taxonomic relevance. Nevertheless, the BCs remain a fundamental and useful classification of viruses by genome type and replication-expression strategies that reflects important aspects of virus biology and evolution.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jiro Wada (Integrated Research Facility at Fort Detrick) for help with figure preparation.
E.V.K., Y.I.W., and N.Y. are supported by the Intramural Research Program of the U.S. National Institutes of Health (National Library of Medicine). M.K. was supported by l’Agence Nationale de la Recherche (project ENVIRA, no. ANR-17-CE15-0005-01). J.H.K. is supported through Battelle Memorial Institute’s prime contract with the U.S. National Institute of Allergy and Infectious Diseases (NIAID) under contract no. HHSN272200700016I.
The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Department of Health and Human Services or of the institutions and companies affiliated with the authors. In no event shall any of these entities have any responsibility or liability for any use, misuse, inability to use, or reliance upon the information contained herein. The U.S. Department of Health and Human Services does not endorse any products or commercial services mentioned in this publication.
Biographies

Eugene V. Koonin received his B.Sc. and Ph.D. from Moscow State University, Russia. He was a Postdoctoral Fellow at the Institute of Poliomyelitis of the Russian Academy of Medical Sciences and a Laboratory Chief at the Institute of Microbiology of the Russian Academy of Sciences. He is currently the Leader of the Evolutionary Genomics Group and an NIH Distinguished Investigator at the National Center for Biotechnology Information (NCBI) at the National Institutes of Health (NIH) in Bethesda, MD. He is a Fellow of the American Academy of Microbiology and the American Academy of Arts and Sciences, a Member of the National Academy of Sciences of the United States, and a Foreign Associate of the European Molecular Biology Organization; received honorary doctorates from L’Université d’Aix-Marseille in France and Wageningen University in the Netherlands; and is a recipient of the National Library of Medicine Board of Regents Award, the NIH Director’s Award, and the Benjamin Franklin Award. He serves as Editor in Chief for Biology Direct and on the editorial boards of BMC Biology, Nucleic Acids Research, and RNA Biology and is a member of the ICTV Negarnaviricota Study Group. He has published over 800 articles.

Valerian V. Dolja graduated from Moscow State University, Russia, in 1974 and received his Ph.D. and D.Sc. in 1980 and 1987, respectively. He moved to the United States in 1991 as a Visiting Scientist at Texas A&M University. In 1994, he joined the faculty of the Department of Botany and Plant Pathology at Oregon State University, Corvallis, OR, and was promoted to Full Professor in 2001. His laboratory staff worked in the areas of plant cell biology (myosin transport) and plant RNA virology (virus-host interactions, functional genomics, and gene expression vectors). The current focus of his research is on virus evolution (comparative genomics and virus origins). Dr. Dolja serves on the editorial boards of Biology Direct, Frontiers in Plant Science, the Journal of Virology, and Virology, and he is an elected Fellow of the American Association for the Advancement of Science and the American Academy of Microbiology. He has published more than 140 articles.

Mart Krupovic is the Head of the Archaeal Virology Unit in the Department of Microbiology at the Institut Pasteur of Paris, France. He received his Ph.D. in 2010 in general microbiology from the University of Helsinki, Finland. His current research focuses on the diversity, origin, and evolution of viruses as well as molecular mechanisms of virus-host interactions in archaea. He has published over 150 journal articles and serves as an editor or on the editorial boards of Biology Direct, Research in Microbiology, Scientific Reports, Virology, and Virus Evolution. He is also an Elected Member of the Executive Committee of the International Committee on Taxonomy of Viruses (ICTV), chairs the ICTV Bidnaviridae and Plasmaviridae Study Groups, and is a member of the ICTV Amalgaviridae and Negarnaviricota Study Groups.

Arvind Varsani is a molecular virologist at Arizona State University. He received his Ph.D. (Molecular and Cell Biology) from the University of Cape Town (South Africa) in 2003. He works across ecosystems, from plants to animals, and from the tropics to the Antarctic, with a strong focus on viral evolution and dynamics and viral metagenomics. His research includes a combination of traditional virology, microscopy (including transmission electron microscopy), and molecular and cellular biology techniques in conjunction with modern techniques, including high-throughput sequencing, synthetic biology, and bioinformatics. He has published over 240 journal articles and serves as Associate Editor for Virus Evolution and editor for Microorganisms and Phytopathology. He also served on the advisory board for Virology. He is also an Elected Member of the Executive Committee of the International Committee on Taxonomy of Viruses (ICTV).

Yuri I. Wolf graduated from Moscow State University, Russia, in 1989 with a B.Sc. in virology and received a Ph.D. in genetics from the Institute of Cytology and Genetics in Novosibirsk in 1996. Since 1997, he has worked at the National Center for Biotechnology Information (NCBI) of the National Institutes of Health (NIH) in Bethesda, MD. He is currently a Lead Scientist in Eugene V. Koonin’s group and a member of the ICTV Negarnaviricota Study Group. Dr. Wolf has published over 200 articles.

Natalya Yutin graduated from Kharkov State University, Ukraine, with an M.Sc. in biophysics and received a Ph.D. in molecular microbiology from Technion, Haifa, Israel. In 2007, she joined the National Center for Biotechnology Information (NCBI), National Institutes of Health (NIH), in Bethesda, MD, where she currently works as a Staff Scientist in Eugene V. Koonin’s group. She has published over 50 articles.

F. Murilo Zerbini received a B.Sc. in agronomy in 1988 from the Federal University of Viçosa, Brazil, and a Ph.D. in plant pathology from the University of California, Davis, in 1996. He has been a Full Professor in the Department of Plant Pathology of the Federal University of Viçosa since 2014. His laboratory studies the evolution of begomoviruses (whitefly-transmitted geminivirids) and begomovirus-host interactions. Dr. Zerbini has published more than 120 papers and advised 27 doctorate students. He was editor in chief of Tropical Plant Pathology (2012 to 2017) and Associate Editor of the Archives of Virology (2006 to 2019), the Journal of General Virology (2008 to 2013), and currently is an Associate Editor of Virology (2013 to the present). He is the Plant Virus Subcommittee Chair of the Executive Committee of the International Committee on Taxonomy of Viruses (ICTV) and is a member of the ICTV Geminiviridae and Potyviridae Study Groups.

Jens H. Kuhn received two Ph.D.’s and an M.D. from Freie Universität, Berlin, Germany. He is currently a Research Leader at Battelle Memorial Institute, Columbus, OH, tasked as the Virology Lead (Contractor) at NIH/NIAID/DCR’s Biosafety Level 4 Integrated Research Facility at Fort Detrick in Frederick, MD. Dr. Kuhn authored Filoviruses: a Compendium of 40 Years of Epidemiological, Clinical, and Laboratory Studies (2008); coauthored The Soviet Biological Weapons Program—a History (2012); and published close to 210 articles. He is the Animal dsRNA and ssRNA Viruses Subcommittee Chair of the Executive Committee of the International Committee on Taxonomy of Viruses (ICTV); chairs the ICTV Bunyavirales, Filoviridae, and Negarnaviricota Study Groups; and is a member of the ICTV Arenaviridae, Artoviridae, Bornaviridae, Monjiviricetes, Nairoviridae, and Nyamiviridae Study Groups.
APPENDIX
GLOSSARY
- megataxonomy
Classification of viruses into taxa at or above the order rank to reflect the known macroevolutionary relationships of large virus groups plus nomenclature for these megataxa. Derived from similar terms used in cellular taxonomies (e.g., see references 233 and 234).
- monophyletic
Belonging to a group of organisms that consists of all the descendants from a common ancestor.
- paraphyletic
Consisting of all the descendants of the group’s last common ancestor excluding one or more monophyletic subgroups; the group is then considered paraphyletic with respect to the excluded subgroups.
- polyphyletic
Not being connected to a common ancestor.
- superviral hallmark genes (super-VHGs)
VHGs that are conserved across different Baltimore classes of viruses and define realms .
- viral hallmark genes (VHGs)
genes or parts of genes that are conserved in diverse groups of viruses; here used to define subrealm megataxa.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Chow C-E, Suttle CA. 2015. Biogeography of viruses in the sea. Annu Rev Virol 2:41–66. doi: 10.1146/annurev-virology-031413-085540. [DOI] [PubMed] [Google Scholar]
- 2.Edwards RA, Rohwer F. 2005. Viral metagenomics. Nat Rev Microbiol 3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 3.Rohwer F. 2003. Global phage diversity. Cell 113:141. doi: 10.1016/s0092-8674(03)00276-9. [DOI] [PubMed] [Google Scholar]
- 4.Rosario K, Breitbart M. 2011. Exploring the viral world through metagenomics. Curr Opin Virol 1:289–297. doi: 10.1016/j.coviro.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Suttle CA. 2005. Viruses in the sea. Nature 437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 6.Cobián Güemes AG, Youle M, Cantú VA, Felts B, Nulton J, Rohwer F. 2016. Viruses as winners in the game of life. Annu Rev Virol 3:197–214. doi: 10.1146/annurev-virology-100114-054952. [DOI] [PubMed] [Google Scholar]
- 7.Danovaro R, Dell’Anno A, Corinaldesi C, Rastelli E, Cavicchioli R, Krupovic M, Noble RT, Nunoura T, Prangishvili D. 2016. Virus-mediated archaeal hecatomb in the deep seafloor. Sci Adv 2:e1600492. doi: 10.1126/sciadv.1600492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koonin EV, Dolja VV. 2013. A virocentric perspective on the evolution of life. Curr Opin Virol 3:546–557. doi: 10.1016/j.coviro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rohwer F, Thurber RV. 2009. Viruses manipulate the marine environment. Nature 459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 10.Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol 5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 11.Amann R, Rosselló-Móra R. 2016. After all, only millions? mBio 7:e00999-16. doi: 10.1128/mBio.00999-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curtis TP, Sloan WT, Scannell JW. 2002. Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci U S A 99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locey KJ, Lennon JT. 2016. Scaling laws predict global microbial diversity. Proc Natl Acad Sci U S A 113:5970–5975. doi: 10.1073/pnas.1521291113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinatzer BA, Tian L, Heath LS. 2017. A proposal for a portal to make earth’s microbial diversity easily accessible and searchable. Antonie Van Leeuwenhoek 110:1271–1279. doi: 10.1007/s10482-017-0849-z. [DOI] [PubMed] [Google Scholar]
- 15.Drake JW, Charlesworth B, Charlesworth D, Crow JF. 1998. Rates of spontaneous mutation. Genetics 148:1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanjuán R, Domingo-Calap P. 2016. Mechanisms of viral mutation. Cell Mol Life Sci 73:4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R. 2010. Viral mutation rates. J Virol 84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. 1982. Rapid evolution of RNA genomes. Science 215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 19.Geoghegan JL, Holmes EC. 2018. Evolutionary virology at 40. Genetics 210:1151–1162. doi: 10.1534/genetics.118.301556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domingo E, Perales C. 2019. Viral quasispecies. PLoS Genet 15:e1008271. doi: 10.1371/journal.pgen.1008271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald SM, Nelson MI, Turner PE, Patton JT. 2016. Reassortment in segmented RNA viruses: mechanisms and outcomes. Nat Rev Microbiol 14:448–460. doi: 10.1038/nrmicro.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varsani A, Lefeuvre P, Roumagnac P, Martin D. 2018. Notes on recombination and reassortment in multipartite/segmented viruses. Curr Opin Virol 33:156–166. doi: 10.1016/j.coviro.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 23.Koonin EV, Gorbalenya AE. 1989. Evolution of RNA genomes: does the high mutation rate necessitate high rate of evolution of viral proteins? J Mol Evol 28:524–527. doi: 10.1007/bf02602932. [DOI] [PubMed] [Google Scholar]
- 24.Holmes EC. 2011. What does virus evolution tell us about virus origins? J Virol 85:5247–5251. doi: 10.1128/JVI.02203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berezovskaya F, Karev GP, Katsnelson MI, Wolf YI, Koonin EV. 2018. Stable coevolutionary regimes for genetic parasites and their hosts: you must differ to coevolve. Biol Direct 13:27. doi: 10.1186/s13062-018-0230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iranzo J, Puigbò P, Lobkovsky AE, Wolf YI, Koonin EV. 2016. Inevitability of genetic parasites. Genome Biol Evol 8:2856–2869. doi: 10.1093/gbe/evw193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koonin EV, Wolf YI, Katsnelson MI. 2017. Inevitability of the emergence and persistence of genetic parasites caused by evolutionary instability of parasite-free states. Biol Direct 12:31. doi: 10.1186/s13062-017-0202-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szathmáry E, Maynard Smith J. 1997. From replicators to reproducers: the first major transitions leading to life. J Theor Biol 187:555–571. doi: 10.1006/jtbi.1996.0389. [DOI] [PubMed] [Google Scholar]
- 29.Takeuchi N, Hogeweg P. 2007. The role of complex formation and deleterious mutations for the stability of RNA-like replicator systems. J Mol Evol 65:668–686. doi: 10.1007/s00239-007-9044-6. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi N, Hogeweg P. 2012. Evolutionary dynamics of RNA-like replicator systems: a bioinformatic approach to the origin of life. Phys Life Rev 9:219–263. doi: 10.1016/j.plrev.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forterre P, Prangishvili D. 2009. The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann N Y Acad Sci 1178:65–77. doi: 10.1111/j.1749-6632.2009.04993.x. [DOI] [PubMed] [Google Scholar]
- 32.Forterre P, Prangishvili D. 2013. The major role of viruses in cellular evolution: facts and hypotheses. Curr Opin Virol 3:558–565. doi: 10.1016/j.coviro.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Koonin EV, Wolf YI. 2012. Evolution of microbes and viruses: a paradigm shift in evolutionary biology? Front Cell Infect Microbiol 2:119. doi: 10.3389/fcimb.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koonin EV, Krupovic M. 2018. The depths of virus exaptation. Curr Opin Virol 31:1–8. doi: 10.1016/j.coviro.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 35.Roossinck MJ, Bazán ER. 2017. Symbiosis: viruses as intimate partners. Annu Rev Virol 4:123–139. doi: 10.1146/annurev-virology-110615-042323. [DOI] [PubMed] [Google Scholar]
- 36.Jalasvuori M. 2012. Vehicles, replicators, and intercellular movement of genetic information: evolutionary dissection of a bacterial cell. Int J Evol Biol 2012:874153. doi: 10.1155/2012/874153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jalasvuori M, Koonin EV. 2015. Classification of prokaryotic genetic replicators: between selfishness and altruism. Ann N Y Acad Sci 1341:96–105. doi: 10.1111/nyas.12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koonin EV, Starokadomskyy P. 2016. Are viruses alive? The replicator paradigm sheds decisive light on an old but misguided question. Stud Hist Philos Biol Biomed Sci 59:125–134. doi: 10.1016/j.shpsc.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.López-García P. 2012. The place of viruses in biology in light of the metabolism- versus-replication-first debate. Hist Philos Life Sci 34:391–406. [PubMed] [Google Scholar]
- 40.Moreira D, López-García P. 2009. Ten reasons to exclude viruses from the tree of life. Nat Rev Microbiol 7:306–311. doi: 10.1038/nrmicro2108. [DOI] [PubMed] [Google Scholar]
- 41.Koonin EV. 2016. Viruses and mobile elements as drivers of evolutionary transitions. Philos Trans R Soc Lond B Biol Sci 371:20150442. doi: 10.1098/rstb.2015.0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruska H. 1943. Versuch zu einer Ordnung der Virusarten. Arch Gesamte Virusforsch 2:480–498. doi: 10.1007/BF01244584. [DOI] [Google Scholar]
- 43.Holmes FO. 1948. Order Virales. The filterable viruses, p 1125–1286 (supplement no. 2). In Breed RS, Murray EGD, Hitchens AP (ed), Bergey’s manual of determinative bacteriology, 6th ed Williams & Wilkins Company, Baltimore, MD. [Google Scholar]
- 44.Cooper PD. 1961. A chemical basis for the classification of animal viruses. Nature 190:302–305. doi: 10.1038/190302a0. [DOI] [PubMed] [Google Scholar]
- 45.Wildy P. 1962. Classifying viruses at higher levels: symmetry and structure of virus particles as criteria. Symp Soc Gen Microbiol XII:145–163. [Google Scholar]
- 46.Tauraso NM, Shelokov A. 1967. Arboviruses—a problem in classification. Arch Gesamte Virusforsch 22:273–279. doi: 10.1007/bf01240522. [DOI] [PubMed] [Google Scholar]
- 47.Horne RW, Wildy P. 1961. Symmetry in virus architecture. Virology 15:348–373. doi: 10.1016/0042-6822(61)90366-x. [DOI] [PubMed] [Google Scholar]
- 48.Hamparian VV, Hilleman MR, Ketler A. 1963. Contributions to characterization and classification of animal viruses. Proc Soc Exp Biol Med 112:1040–1050. doi: 10.3181/00379727-112-28247. [DOI] [PubMed] [Google Scholar]
- 49.Lwoff A. 1967. Principles of classification and nomenclature of viruses. Nature 215:13–14. doi: 10.1038/215013a0. [DOI] [PubMed] [Google Scholar]
- 50.Lwoff A, Horne RW, Tournier P. 1962. Un système des virus. C R Hebd Seances Acad Sci 254:4225–4227. [PubMed] [Google Scholar]
- 51.Lwoff A, Tournier P. 1966. The classification of viruses. Annu Rev Microbiol 20:45–74. doi: 10.1146/annurev.mi.20.100166.000401. [DOI] [PubMed] [Google Scholar]
- 52.Kuhn JH, Wolf YI, Krupovic M, Zhang Y-Z, Maes P, Dolja VV, Koonin EV. 2019. Classify viruses—the gain is worth the pain. Nature 566:318–320. doi: 10.1038/d41586-019-00599-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simmonds P, Adams MJ, Benkő M, Breitbart M, Brister JR, Carstens EB, Davison AJ, Delwart E, Gorbalenya AE, Harrach B, Hull R, King AMQ, Koonin EV, Krupovic M, Kuhn JH, Lefkowitz EJ, Nibert ML, Orton R, Roossinck MJ, Sabanadzovic S, Sullivan MB, Suttle CA, Tesh RB, van der Vlugt RA, Varsani A, Zerbini FM. 2017. Consensus statement: virus taxonomy in the age of metagenomics. Nat Rev Microbiol 15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- 54.Gorbalenya AE. 2018. Increasing the number of available ranks in virus taxonomy from five to ten and adopting the Baltimore classes as taxa at the basal rank. Arch Virol 163:2933–2936. doi: 10.1007/s00705-018-3915-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crick F. 1970. Central dogma of molecular biology. Nature 227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 56.Koonin EV. 2015. Why the central dogma: on the nature of the great biological exclusion principle. Biol Direct 10:52. doi: 10.1186/s13062-015-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baltimore D. 1971. Expression of animal virus genomes. Bacteriol Rev 35:235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Condit R. 2013. Principles of virology, p 21–51. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 59.Agol VI. 1974. Towards the system of viruses. Biosystems 6:113–132. doi: 10.1016/0303-2647(74)90003-3. [DOI] [PubMed] [Google Scholar]
- 60.Siddell SG, Walker PJ, Lefkowitz EJ, Mushegian AR, Adams MJ, Dutilh BE, Gorbalenya AE, Harrach B, Harrison RL, Junglen S, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Nibert M, Rubino L, Sabanadzovic S, Sanfaçon H, Simmonds P, Varsani A, Zerbini FM, Davison AJ. 2019. Additional changes to taxonomy ratified in a special vote by the International Committee on Taxonomy of Viruses (October 2018). Arch Virol 164:943–946. doi: 10.1007/s00705-018-04136-2. [DOI] [PubMed] [Google Scholar]
- 61.Koonin EV, Dolja VV, Krupovic M. 2015. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479–480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Felsenstein J. 2004. Inferring phylogenies. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 63.Li W-H. 1997. Molecular evolution. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 64.Doolittle WF. 1999. Phylogenetic classification and the universal tree. Science 284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 65.Doolittle WF. 1999. Lateral genomics. Trends Cell Biol 9:M5–M8. doi: 10.1016/S0962-8924(99)01664-5. [DOI] [PubMed] [Google Scholar]
- 66.Koonin EV, Makarova KS, Aravind L. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol 55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Puigbò P, Lobkovsky AE, Kristensen DM, Wolf YI, Koonin EV. 2014. Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes. BMC Biol 12:66. doi: 10.1186/s12915-014-0066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koonin EV. 2003. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol 1:127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- 69.Glansdorff N, Xu Y, Labedan B. 2008. The last universal common ancestor: emergence, constitution and genetic legacy of an elusive forerunner. Biol Direct 3:29. doi: 10.1186/1745-6150-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weiss MC, Preiner M, Xavier JC, Zimorski V, Martin WF. 2018. The last universal common ancestor between ancient Earth chemistry and the onset of genetics. PLoS Genet 14:e1007518. doi: 10.1371/journal.pgen.1007518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Puigbò P, Wolf YI, Koonin EV. 2009. Search for a ‘tree of life’ in the thicket of the phylogenetic forest. J Biol 8:59. doi: 10.1186/jbiol159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koonin EV, Senkevich TG, Dolja VV. 2006. The ancient virus world and evolution of cells. Biol Direct 1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yutin N, Koonin EV. 2012. Hidden evolutionary complexity of nucleo-cytoplasmic large DNA viruses of eukaryotes. Virol J 9:161. doi: 10.1186/1743-422X-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koonin EV, Yutin N. 2019. Evolution of the large nucleocytoplasmic DNA viruses of eukaryotes and convergent origins of viral gigantism. Adv Virus Res 103:167–202. doi: 10.1016/bs.aivir.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 75.Iranzo J, Cuesta JA, Manrubia S, Katsnelson MI, Koonin EV. 2017. Disentangling the effects of selection and loss bias on gene dynamics. Proc Natl Acad Sci U S A 114:E5616–E5624. doi: 10.1073/pnas.1704925114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koonin EV, Wolf YI. 2008. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lobkovsky AE, Wolf YI, Koonin EV. 2013. Gene frequency distributions reject a neutral model of genome evolution. Genome Biol Evol 5:233–242. doi: 10.1093/gbe/evt002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kristensen DM, Waller AS, Yamada T, Bork P, Mushegian AR, Koonin EV. 2013. Orthologous gene clusters and taxon signature genes for viruses of prokaryotes. J Bacteriol 195:941–950. doi: 10.1128/JB.01801-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iranzo J, Krupovic M, Koonin EV. 2016. The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. mBio 7:e00978-16. doi: 10.1128/mBio.00978-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krupovic M, Dolja VV, Koonin EV. 2019. Origin of viruses: primordial replicators recruiting capsids from hosts. Nat Rev Microbiol 17:449–458. doi: 10.1038/s41579-019-0205-6. [DOI] [PubMed] [Google Scholar]
- 81.Krupovic M, Koonin EV. 2017. Multiple origins of viral capsid proteins from cellular ancestors. Proc Natl Acad Sci U S A 114:E2401–E2410. doi: 10.1073/pnas.1621061114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wolf YI, Kazlauskas D, Iranzo J, Lucía-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV. 2018. Origins and evolution of the global RNA virome. mBio 9:e02329-18. doi: 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iranzo J, Koonin EV, Prangishvili D, Krupovic M. 2016. Bipartite network analysis of the archaeal virosphere: evolutionary connections between viruses and capsidless mobile elements. J Virol 90:11043–11055. doi: 10.1128/JVI.01622-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iranzo J, Krupovic M, Koonin EV. 2017. A network perspective on the virus world. Commun Integr Biol 10:e1296614. doi: 10.1080/19420889.2017.1296614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kazlauskas D, Varsani A, Koonin EV, Krupovic M. 2019. Multiple origins of prokaryotic and eukaryotic single-stranded DNA viruses from bacterial and archaeal plasmids. Nat Commun 10:3425. doi: 10.1038/s41467-019-11433-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vetten HJ, Haenni A-L. 2006. Taxon-specific suffixes for vernacular names. Arch Virol 151:1249–1250. doi: 10.1007/s00705-006-0743-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kamer G, Argos P. 1984. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res 12:7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koonin EV. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol 72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 89.Koonin EV, Gorbalenya AE, Chumakov KM. 1989. Tentative identification of RNA-dependent RNA polymerases of dsRNA viruses and their relationship to positive strand RNA viral polymerases. FEBS Lett 252:42–46. doi: 10.1016/0014-5793(89)80886-5. [DOI] [PubMed] [Google Scholar]
- 90.Poch O, Sauvaget I, Delarue M, Tordo N. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J 8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xiong Y, Eickbush TH. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zanotto PM, Gibbs MJ, Gould EA, Holmes EC. 1996. A reevaluation of the higher taxonomy of viruses based on RNA polymerases. J Virol 70:6083–6096. doi: 10.1128/JVI.70.9.6083-6096.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holmes EC, Duchêne S. 2019. Can sequence phylogenies safely infer the origin of the global virome? mBio 10:e00289-19. doi: 10.1128/mBio.00289-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wolf YI, Kazlauskas D, Iranzo J, Lucia-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV. 2019. Reply to Holmes and Duchêne, “Can sequence phylogenies safely infer the origin of the global virome?”: deep phylogenetic analysis of RNA viruses is highly challenging but not meaningless. mBio 10:e00542-19. doi: 10.1128/mBio.00542-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krishnamurthy SR, Janowski AB, Zhao G, Barouch D, Wang D. 2016. Hyperexpansion of RNA bacteriophage diversity. PLoS Biol 14:e1002409. doi: 10.1371/journal.pbio.1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hillman BI, Cai G. 2013. The family Narnaviridae: simplest of RNA viruses. Adv Virus Res 86:149–176. doi: 10.1016/B978-0-12-394315-6.00006-4. [DOI] [PubMed] [Google Scholar]
- 97.Embley TM, Martin W. 2006. Eukaryotic evolution, changes and challenges. Nature 440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 98.Rastgou M, Habibi MK, Izadpanah K, Masenga V, Milne RG, Wolf YI, Koonin EV, Turina M. 2009. Molecular characterization of the plant virus genus Ourmiavirus and evidence of inter-kingdom reassortment of viral genome segments as its possible route of origin. J Gen Virol 90:2525–2535. doi: 10.1099/vir.0.013086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Donaire L, Rozas J, Ayllón MA. 2016. Molecular characterization of Botrytis ourmia-like virus, a mycovirus close to the plant pathogenic genus Ourmiavirus. Virology 489:158–164. doi: 10.1016/j.virol.2015.11.027. [DOI] [PubMed] [Google Scholar]
- 100.Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, Qin XC, Li J, Cao JP, Eden JS, Buchmann J, Wang W, Xu J, Holmes EC, Zhang YZ. 2016. Redefining the invertebrate RNA virosphere. Nature 540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 101.Goldbach R, Wellink J. 1988. Evolution of plus-strand RNA viruses. Intervirology 29:260–267. doi: 10.1159/000150054. [DOI] [PubMed] [Google Scholar]
- 102.Koonin EV, Dolja VV. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol 28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 103.Koonin EV, Wolf YI, Nagasaki K, Dolja VV. 2008. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat Rev Microbiol 6:925–939. doi: 10.1038/nrmicro2030. [DOI] [PubMed] [Google Scholar]
- 104.Le Gall O, Christian P, Fauquet CM, King AMQ, Knowles NJ, Nakashima N, Stanway G, Gorbalenya AE. 2008. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T=3 virion architecture. Arch Virol 153:715–727. doi: 10.1007/s00705-008-0041-x. [DOI] [PubMed] [Google Scholar]
- 105.Bukhari K, Mulley G, Gulyaeva AA, Zhao L, Shu G, Jiang J, Neuman BW. 2018. Description and initial characterization of metatranscriptomic nidovirus-like genomes from the proposed new family Abyssoviridae, and from a sister group to the Coronavirinae, the proposed genus Alphaletovirus. Virology 524:160–171. doi: 10.1016/j.virol.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Saberi A, Gulyaeva AA, Brubacher JL, Newmark PA, Gorbalenya AE. 2018. A planarian nidovirus expands the limits of RNA genome size. PLoS Pathog 14:e1007314. doi: 10.1371/journal.ppat.1007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sõmera M, Sarmiento C, Truve E. 2015. Overview on sobemoviruses and a proposal for the creation of the family Sobemoviridae. Viruses 7:3076–3115. doi: 10.3390/v7062761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.El Omari K, Sutton G, Ravantti JJ, Zhang H, Walter TS, Grimes JM, Bamford DH, Stuart DI, Mancini EJ. 2013. Plate tectonics of virus shell assembly and reorganization in phage Φ8, a distant relative of mammalian reoviruses. Structure 21:1384–1395. doi: 10.1016/j.str.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Luque D, Gómez-Blanco J, Garriga D, Brilot AF, González JM, Havens WM, Carrascosa JL, Trus BL, Verdaguer N, Ghabrial SA, Castón JR. 2014. Cryo-EM near-atomic structure of a dsRNA fungal virus shows ancient structural motifs preserved in the dsRNA viral lineage. Proc Natl Acad Sci U S A 111:7641–7646. doi: 10.1073/pnas.1404330111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Koga R, Fukuhara T, Nitta T. 1998. Molecular characterization of a single mitochondria-associated double-stranded RNA in the green alga Bryopsis. Plant Mol Biol 36:717–724. doi: 10.1023/a:1005907310553. [DOI] [PubMed] [Google Scholar]
- 111.Koga R, Horiuchi H, Fukuhara T. 2003. Double-stranded RNA replicons associated with chloroplasts of a green alga, Bryopsis cinicola. Plant Mol Biol 51:991–999. doi: 10.1023/a:1023003412859. [DOI] [PubMed] [Google Scholar]
- 112.Boros A, Polgár B, Pankovics P, Fenyvesi H, Engelmann P, Phan TG, Delwart E, Reuter G. 2018. Multiple divergent picobirnaviruses with functional prokaryotic Shine-Dalgarno ribosome binding sites present in cloacal sample of a diarrheic chicken. Virology 525:62–72. doi: 10.1016/j.virol.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 113.Krishnamurthy SR, Wang D. 2018. Extensive conservation of prokaryotic ribosomal binding sites in known and novel picobirnaviruses. Virology 516:108–114. doi: 10.1016/j.virol.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 114.Krupovic M, Dolja VV, Koonin EV. 2015. Plant viruses of the Amalgaviridae family evolved via recombination between viruses with double-stranded and negative-strand RNA genomes. Biol Direct 10:12. doi: 10.1186/s13062-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goldbach R. 1987. Genome similarities between plant and animal RNA viruses. Microbiol Sci 4:197–202. [PubMed] [Google Scholar]
- 116.Shi M, Zhang Y-Z, Holmes EC. 2018. Meta-transcriptomics and the evolutionary biology of RNA viruses. Virus Res 243:83–90. doi: 10.1016/j.virusres.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Qin X-C, Shi M, Tian J-H, Lin X-D, Gao D-Y, He J-R, Wang J-B, Li C-X, Kang Y-J, Yu B, Zhou D-J, Xu J, Plyusnin A, Holmes EC, Zhang Y-Z. 2014. A tick-borne segmented RNA virus contains genome segments derived from unsegmented viral ancestors. Proc Natl Acad Sci U S A 111:6744–6749. doi: 10.1073/pnas.1324194111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang Z-D, Wang B, Wei F, Han S-Z, Zhang L, Yang Z-T, Yan Y, Lv X-L, Li L, Wang S-C, Song M-X, Zhang H-J, Huang S-J, Chen J, Huang F-Q, Li S, Liu H-H, Hong J, Jin Y-L, Wang W, Zhou J-Y, Liu Q. 2019. A new segmented virus associated with human febrile illness in China. N Engl J Med 380:2116–2125. doi: 10.1056/NEJMoa1805068. [DOI] [PubMed] [Google Scholar]
- 119.Shi M, Lin X-D, Vasilakis N, Tian J-H, Li C-X, Chen L-J, Eastwood G, Diao X-N, Chen M-H, Chen X, Qin X-C, Widen SG, Wood TG, Tesh RB, Xu J, Holmes EC, Zhang Y-Z. 2016. Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the Flaviviridae and related viruses. J Virol 90:659–669. doi: 10.1128/JVI.02036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kobayashi K, Atsumi G, Iwadate Y, Tomita R, Chiba K, Akasaka S, Nishihara M, Takahashi H, Yamaoka N, Nishiguchi M, Sekine K-T. 2013. Gentian Kobu-sho-associated virus: a tentative, novel double-stranded RNA virus that is relevant to gentian Kobu-sho syndrome. J Gen Plant Pathol 79:56–63. doi: 10.1007/s10327-012-0423-5. [DOI] [Google Scholar]
- 121.Shi M, Lin X-D, Chen X, Tian J-H, Chen L-J, Li K, Wang W, Eden J-S, Shen J-J, Liu L, Holmes EC, Zhang Y-Z. 2018. The evolutionary history of vertebrate RNA viruses. Nature 556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 122.Janowski AB, Krishnamurthy SR, Lim ES, Zhao G, Brenchley JM, Barouch DH, Thakwalakwa C, Manary MJ, Holtz LR, Wang D. 2017. Statoviruses, a novel taxon of RNA viruses present in the gastrointestinal tracts of diverse mammals. Virology 504:36–44. doi: 10.1016/j.virol.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Obbard DJ, Shi M, Roberts KE, Longdon B, Dennis AB. 2020. A new lineage of segmented RNA viruses infecting animals. Virus Evol 6:vez061. doi: 10.1093/ve/vez061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mata CP, Luque D, Gómez-Blanco J, Rodríguez JM, González JM, Suzuki N, Ghabrial SA, Carrascosa JL, Trus BL, Castón JR. 2017. Acquisition of functions on the outer capsid surface during evolution of double-stranded RNA fungal viruses. PLoS Pathog 13:e1006755. doi: 10.1371/journal.ppat.1006755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mäntynen S, Sundberg L-R, Poranen MM. 2018. Recognition of six additional cystoviruses: Pseudomonas virus phi6 is no longer the sole species of the family Cystoviridae. Arch Virol 163:1117–1124. doi: 10.1007/s00705-017-3679-4. [DOI] [PubMed] [Google Scholar]
- 126.Pflug A, Guilligay D, Reich S, Cusack S. 2014. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 127.Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SPJ. 2015. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kotta-Loizou I, Coutts RHA. 2017. Studies on the virome of the entomopathogenic fungus Beauveria bassiana reveal novel dsRNA elements and mild hypervirulence. PLoS Pathog 13:e1006183. doi: 10.1371/journal.ppat.1006183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gorbalenya AE, Pringle FM, Zeddam J-L, Luke BT, Cameron CE, Kalmakoff J, Hanzlik TN, Gordon KHJ, Ward VK. 2002. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J Mol Biol 324:47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gibrat J-F, Mariadassou M, Boudinot P, Delmas B. 2013. Analyses of the radiation of birnaviruses from diverse host phyla and of their evolutionary affinities with other double-stranded RNA and positive strand RNA viruses using robust structure-based multiple sequence alignments and advanced phylogenetic methods. BMC Evol Biol 13:154. doi: 10.1186/1471-2148-13-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bolduc B, Shaughnessy DP, Wolf YI, Koonin EV, Roberto FF, Young M. 2012. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J Virol 86:5562–5573. doi: 10.1128/JVI.07196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gladyshev EA, Arkhipova IR. 2011. A widespread class of reverse transcriptase-related cellular genes. Proc Natl Acad Sci U S A 108:20311–20316. doi: 10.1073/pnas.1100266108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zimmerly S, Wu L. 2015. An unexplored diversity of reverse transcriptases in bacteria. Microbiol Spectr 3:MDNA3-0058-2014. doi: 10.1128/microbiolspec.MDNA3-0058-2014. [DOI] [PubMed] [Google Scholar]
- 134.Gilbert C, Feschotte C. 2018. Horizontal acquisition of transposable elements and viral sequences: patterns and consequences. Curr Opin Genet Dev 49:15–24. doi: 10.1016/j.gde.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Platt RN II, Vandewege MW, Ray DA. 2018. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res 26:25–43. doi: 10.1007/s10577-017-9570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Krupovic M, Blomberg J, Coffin JM, Dasgupta I, Fan H, Geering AD, Gifford R, Harrach B, Hull R, Johnson W, Kreuze JF, Lindemann D, Llorens C, Lockhart B, Mayer J, Muller E, Olszewski NE, Pappu HR, Pooggin MM, Richert-Pöggeler KR, Sabanadzovic S, Sanfaçon H, Schoelz JE, Seal S, Stavolone L, Stoye JP, Teycheney P-Y, Tristem M, Koonin EV, Kuhn JH. 2018. Ortervirales: new virus order unifying five families of reverse-transcribing viruses. J Virol 92:e00515-18. doi: 10.1128/JVI.00515-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lauber C, Seitz S, Mattei S, Suh A, Beck J, Herstein J, Börold J, Salzburger W, Kaderali L, Briggs JAG, Bartenschlager R. 2017. Deciphering the origin and evolution of hepatitis B viruses by means of a family of non-enveloped fish viruses. Cell Host Microbe 22:387–399. doi: 10.1016/j.chom.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Krupovic M, Koonin EV. 2017. Homologous capsid proteins testify to the common ancestry of retroviruses, caulimoviruses, pseudoviruses, and metaviruses. J Virol 91:e00210-17. doi: 10.1128/JVI.00210-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhao L, Rosario K, Breitbart M, Duffy S. 2019. Eukaryotic circular Rep-encoding single-stranded DNA (CRESS DNA) viruses: ubiquitous viruses with small genomes and a diverse host range. Adv Virus Res 103:71–133. doi: 10.1016/bs.aivir.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 140.Labonté JM, Suttle CA. 2013. Previously unknown and highly divergent ssDNA viruses populate the oceans. ISME J 7:2169–2177. doi: 10.1038/ismej.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Malathi VG, Renuka Devi P. 2019. ssDNA viruses: key players in global virome. Virusdisease 30:3–12. doi: 10.1007/s13337-019-00519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rosario K, Mettel KA, Benner BE, Johnson R, Scott C, Yusseff-Vanegas SZ, Baker CCM, Cassill DL, Storer C, Varsani A, Breitbart M. 2018. Virus discovery in all three major lineages of terrestrial arthropods highlights the diversity of single-stranded DNA viruses associated with invertebrates. PeerJ 6:e5761. doi: 10.7717/peerj.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Roux S, Krupovic M, Poulet A, Debroas D, Enault F. 2012. Evolution and diversity of the Microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PLoS One 7:e40418. doi: 10.1371/journal.pone.0040418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Roux S, Solonenko NE, Dang VT, Poulos BT, Schwenck SM, Goldsmith DB, Coleman ML, Breitbart M, Sullivan MB. 2016. Towards quantitative viromics for both double-stranded and single-stranded DNA viruses. PeerJ 4:e2777. doi: 10.7717/peerj.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Creasy A, Rosario K, Leigh BA, Dishaw LJ, Breitbart M. 2018. Unprecedented diversity of ssDNA phages from the family Microviridae detected within the gut of a protochordate model organism (Ciona robusta). Viruses 10:404. doi: 10.3390/v10080404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Roux S, Krupovic M, Daly RA, Borges AL, Nayfach S, Schulz F, Sharrar A, Matheus Carnevali PB, Cheng J-F, Ivanova NN, Bondy-Denomy J, Wrighton KC, Woyke T, Visel A, Kyrpides NC, Eloe-Fadrosh EA. 2019. Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth’s biomes. Nat Microbiol 4:1895–1906. doi: 10.1038/s41564-019-0510-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rosario K, Dayaram A, Marinov M, Ware J, Kraberger S, Stainton D, Breitbart M, Varsani A. 2012. Diverse circular ssDNA viruses discovered in dragonflies (Odonata: Epiprocta). J Gen Virol 93:2668–2681. doi: 10.1099/vir.0.045948-0. [DOI] [PubMed] [Google Scholar]
- 148.Tisza MJ, Pastrana DV, Welch NL, Stewart B, Peretti A, Starrett GJ, Pang Y-YS, Varsani A, Krishnamurthy SR, Pesavento PA, McDermott DH, Murphy PM, Whited JL, Miller B, Brenchley J, Rosshart SP, Rehermann B, Doorbar J, Ta’ala BA, Pletnikova O, Troncoso JC, Resnick SM, Bolduc B, Sullivan MB, Varsani A, Segall AM, Buck CB. 2020. Discovery of several thousand highly diverse circular DNA viruses. eLife 9:e51971. doi: 10.7554/eLife.51971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kazlauskas D, Varsani A, Krupovic M. 2018. Pervasive chimerism in the replication-associated proteins of uncultured single-stranded DNA viruses. Viruses 10:187. doi: 10.3390/v10040187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cotmore SF, Tattersall P. 2014. Parvoviruses: small does not mean simple. Annu Rev Virol 1:517–537. doi: 10.1146/annurev-virology-031413-085444. [DOI] [PubMed] [Google Scholar]
- 151.Krupovic M, Koonin EV. 2014. Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Sci Rep 4:5347. doi: 10.1038/srep05347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tattersall P, Ward DC. 1976. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature 263:106–109. doi: 10.1038/263106a0. [DOI] [PubMed] [Google Scholar]
- 153.Atanasova NS, Senčilo A, Pietilä MK, Roine E, Oksanen HM, Bamford DH. 2015. Comparison of lipid-containing bacterial and archaeal viruses. Adv Virus Res 92:1–61. doi: 10.1016/bs.aivir.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 154.Pietilä MK, Roine E, Sencilo A, Bamford DH, Oksanen HM. 2016. Pleolipoviridae, a newly proposed family comprising archaeal pleomorphic viruses with single-stranded or double-stranded DNA genomes. Arch Virol 161:249–256. doi: 10.1007/s00705-015-2613-x. [DOI] [PubMed] [Google Scholar]
- 155.Chandler M, de la Cruz F, Dyda F, Hickman AB, Moncalian G, Ton-Hoang B. 2013. Breaking and joining single-stranded DNA: the HUH endonuclease superfamily. Nat Rev Microbiol 11:525–538. doi: 10.1038/nrmicro3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ilyina TV, Koonin EV. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res 20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gorbalenya AE, Koonin EV, Wolf YI. 1990. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett 262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- 158.Krupovic M. 2012. Recombination between RNA viruses and plasmids might have played a central role in the origin and evolution of small DNA viruses. Bioessays 34:867–870. doi: 10.1002/bies.201200083. [DOI] [PubMed] [Google Scholar]
- 159.Laanto E, Mäntynen S, De Colibus L, Marjakangas J, Gillum A, Stuart DI, Ravantti JJ, Huiskonen JT, Sundberg L-R. 2017. Virus found in a boreal lake links ssDNA and dsDNA viruses. Proc Natl Acad Sci U S A 114:8378–8383. doi: 10.1073/pnas.1703834114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Koonin EV, Ilyina TV. 1992. Geminivirus replication proteins are related to prokaryotic plasmid rolling circle DNA replication initiator proteins. J Gen Virol 73:2763–2766. doi: 10.1099/0022-1317-73-10-2763. [DOI] [PubMed] [Google Scholar]
- 161.Krupovic M. 2013. Networks of evolutionary interactions underlying the polyphyletic origin of ssDNA viruses. Curr Opin Virol 3:578–586. doi: 10.1016/j.coviro.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 162.Krupovic M, Ravantti JJ, Bamford DH. 2009. Geminiviruses: a tale of a plasmid becoming a virus. BMC Evol Biol 9:112. doi: 10.1186/1471-2148-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Roux S, Enault F, Bronner G, Vaulot D, Forterre P, Krupovic M. 2013. Chimeric viruses blur the borders between the major groups of eukaryotic single-stranded DNA viruses. Nat Commun 4:2700. doi: 10.1038/ncomms3700. [DOI] [PubMed] [Google Scholar]
- 164.Diemer GS, Stedman KM. 2012. A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Biol Direct 7:13. doi: 10.1186/1745-6150-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Krupovic M, Zhi N, Li J, Hu G, Koonin EV, Wong S, Shevchenko S, Zhao K, Young NS. 2015. Multiple layers of chimerism in a single-stranded DNA virus discovered by deep sequencing. Genome Biol Evol 7:993–1001. doi: 10.1093/gbe/evv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Iyer LM, Koonin EV, Leipe DD, Aravind L. 2005. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res 33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yutin N, Bäckström D, Ettema TJG, Krupovic M, Koonin EV. 2018. Vast diversity of prokaryotic virus genomes encoding double jelly-roll major capsid proteins uncovered by genomic and metagenomic sequence analysis. Virol J 15:67. doi: 10.1186/s12985-018-0974-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yutin N, Makarova KS, Gussow AB, Krupovic M, Segall A, Edwards RA, Koonin EV. 2018. Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat Microbiol 3:38–46. doi: 10.1038/s41564-017-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Al-Shayeb B, Sachdeva R, Chen L-X, Ward F, Munk P, Devoto A, Castelle CJ, Olm MR, Bouma-Gregson K, Amano Y, He C, Méheust R, Brooks B, Thomas A, Lavy A, Matheus-Carnevali P, Sun C, Goltsman DSA, Borton MA, Nelson TC, Kantor R, Jaffe AL, Keren R, Farag IF, Lei S, Finstad K, Amundson R, Anantharaman K, Zhou J, Probst AJ, Power ME, Tringe SG, Li W-J, Wrighton K, Harrison S, Morowitz M, Relman DA, Doudna JA, Lehours A-C, Warren L, Cate JHD, Santini JM, Banfield JF. 2019. Clades of huge phage from across Earth’s ecosystems. bioRxiv doi: 10.1101/572362. [DOI] [PMC free article] [PubMed]
- 170.Abrescia NGA, Cockburn JJ, Grimes JM, Sutton GC, Diprose JM, Butcher SJ, Fuller SD, San Martín C, Burnett RM, Stuart DI, Bamford DH, Bamford JK. 2004. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 432:68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- 171.Abrescia NGA, Bamford DH, Grimes JM, Stuart DI. 2012. Structure unifies the viral universe. Annu Rev Biochem 81:795–822. doi: 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]
- 172.Burroughs AM, Iyer LM, Aravind L. 2007. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn 3:48–65. doi: 10.1159/000107603. [DOI] [PubMed] [Google Scholar]
- 173.Fang Q, Zhu D, Agarkova I, Adhikari J, Klose T, Liu Y, Chen Z, Sun Y, Gross ML, Van Etten JL, Zhang X, Rossmann MG. 2019. Near-atomic structure of a giant virus. Nat Commun 10:388. doi: 10.1038/s41467-019-08319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Veesler D, Ng T-S, Sendamarai AK, Eilers BJ, Lawrence CM, Lok S-M, Young MJ, Johnson JE, Fu C-Y. 2013. Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography. Proc Natl Acad Sci U S A 110:5504–5509. doi: 10.1073/pnas.1300601110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Iyer LM, Makarova KS, Koonin EV, Aravind L. 2004. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res 32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Krupovic M, Koonin EV. 2015. Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution. Nat Rev Microbiol 13:105–115. doi: 10.1038/nrmicro3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Männistö RH, Kivelä HM, Paulin L, Bamford DH, Bamford JK. 1999. The complete genome sequence of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262:355–363. doi: 10.1006/viro.1999.9837. [DOI] [PubMed] [Google Scholar]
- 178.Kauffman KM, Hussain FA, Yang J, Arevalo P, Brown JM, Chang WK, VanInsberghe D, Elsherbini J, Sharma RS, Cutler MB, Kelly L, Polz MF. 2018. A major lineage of non-tailed dsDNA viruses as unrecognized killers of marine bacteria. Nature 554:118–122. doi: 10.1038/nature25474. [DOI] [PubMed] [Google Scholar]
- 179.Pawlowski A, Rissanen I, Bamford JKH, Krupovic M, Jalasvuori M. 2014. Gammasphaerolipovirus, a newly proposed bacteriophage genus, unifies viruses of halophilic archaea and thermophilic bacteria within the novel family Sphaerolipoviridae. Arch Virol 159:1541–1554. doi: 10.1007/s00705-013-1970-6. [DOI] [PubMed] [Google Scholar]
- 180.de Colibus L, Roine E, Walter TS, Ilca SL, Wang X, Wang N, Roseman AM, Bamford D, Huiskonen JT, Stuart DI. 2019. Assembly of complex viruses exemplified by a halophilic euryarchaeal virus. Nat Commun 10:1456. doi: 10.1038/s41467-019-09451-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Santos-Pérez I, Charro D, Gil-Carton D, Azkargorta M, Elortza F, Bamford DH, Oksanen HM, Abrescia NGA. 2019. Structural basis for assembly of vertical single β-barrel viruses. Nat Commun 10:1184. doi: 10.1038/s41467-019-08927-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Demina TA, Pietilä MK, Svirskaitė J, Ravantti JJ, Atanasova NS, Bamford DH, Oksanen HM. 2016. Archaeal Haloarcula californiae icosahedral virus 1 highlights conserved elements in icosahedral membrane-containing DNA viruses from extreme environments. mBio 7:e00699-16. doi: 10.1128/mBio.00699-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Oksanen HM, Abrescia NGA. 2019. Membrane-containing icosahedral bacteriophage PRD1: the dawn of viral lineages. Adv Exp Med Biol 1215:85–109. doi: 10.1007/978-3-030-14741-9_5. [DOI] [PubMed] [Google Scholar]
- 184.Iyer LM, Aravind L, Koonin EV. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol 75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Iyer LM, Balaji S, Koonin EV, Aravind L. 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res 117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 186.Koonin EV, Yutin N. 2010. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology 53:284–292. doi: 10.1159/000312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Andreani J, Aherfi S, Bou Khalil JY, Di Pinto F, Bitam I, Raoult D, Colson P, La Scola B. 2016. Cedratvirus, a double-cork structured giant virus, is a distant relative of pithoviruses. Viruses 8:300. doi: 10.3390/v8110300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rodrigues RAL, Andreani J, Andrade ACDSP, Machado TB, Abdi S, Levasseur A, Abrahão JS, La Scola B. 2018. Morphologic and genomic analyses of new isolates reveal a second lineage of cedratviruses. J Virol 92:e00372-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Andreani J, Khalil JYB, Sevvana M, Benamar S, Di Pinto F, Bitam I, Colson P, Klose T, Rossmann MG, Raoult D, La Scola B. 2017. Pacmanvirus, a new giant icosahedral virus at the crossroads between Asfarviridae and faustoviruses. J Virol 91:e00212-17. doi: 10.1128/JVI.00212-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Andreani J, Khalil JYB, Baptiste E, Hasni I, Michelle C, Raoult D, Levasseur A, La Scola B. 2017. Orpheovirus IHUMI-LCC2: a new virus among the giant viruses. Front Microbiol 8:2643. doi: 10.3389/fmicb.2017.02643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yoshikawa G, Blanc-Mathieu R, Song C, Kayama Y, Mochizuki T, Murata K, Ogata H, Takemura M. 2019. Medusavirus, a novel large DNA virus discovered from hot spring water. J Virol 93:e02130-18. doi: 10.1128/JVI.02130-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Philippe N, Legendre M, Doutre G, Couté Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie J-M, Abergel C. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 193.Yutin N, Koonin EV. 2013. Pandoraviruses are highly derived phycodnaviruses. Biol Direct 8:25. doi: 10.1186/1745-6150-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Legendre M, Fabre E, Poirot O, Jeudy S, Lartigue A, Alempic J-M, Beucher L, Philippe N, Bertaux L, Christo-Foroux E, Labadie K, Couté Y, Abergel C, Claverie J-M. 2018. Diversity and evolution of the emerging Pandoraviridae family. Nat Commun 9:2285. doi: 10.1038/s41467-018-04698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Legendre M, Lartigue A, Bertaux L, Jeudy S, Bartoli J, Lescot M, Alempic J-M, Ramus C, Bruley C, Labadie K, Shmakova L, Rivkina E, Couté Y, Abergel C, Claverie J-M. 2015. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc Natl Acad Sci U S A 112:E5327–E5335. doi: 10.1073/pnas.1510795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Legendre M, Bartoli J, Shmakova L, Jeudy S, Labadie K, Adrait A, Lescot M, Poirot O, Bertaux L, Bruley C, Couté Y, Rivkina E, Abergel C, Claverie J-M. 2014. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci U S A 111:4274–4279. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hyun J-K, Accurso C, Hijnen M, Schult P, Pettikiriarachchi A, Mitra AK, Coulibaly F. 2011. Membrane remodeling by the double-barrel scaffolding protein of poxvirus. PLoS Pathog 7:e1002239. doi: 10.1371/journal.ppat.1002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bahar MW, Graham SC, Stuart DI, Grimes JM. 2011. Insights into the evolution of a complex virus from the crystal structure of vaccinia virus D13. Structure 19:1011–1020. doi: 10.1016/j.str.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yutin N, Wolf YI, Raoult D, Koonin EV. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J 6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yutin N, Wolf YI, Koonin EV. 2014. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology 466–467:38–52. doi: 10.1016/j.virol.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Koonin EV, Yutin N. 2018. Multiple evolutionary origins of giant viruses [version 1; peer review: 4 approved]. F1000Res 7(F1000 Faculty Rev):1840. doi: 10.12688/f1000research.16248.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bäckström D, Yutin N, Jørgensen SL, Dharamshi J, Homa F, Zaremba-Niedwiedzka K, Spang A, Wolf YI, Koonin EV, Ettema TJG. 2019. Virus genomes from deep sea sediments expand the ocean megavirome and support independent origins of viral gigantism. mBio 10:e02497-18. doi: 10.1128/mBio.02497-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. 2000. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 204.Bayfield OW, Klimuk E, Winkler DC, Hesketh EL, Chechik M, Cheng N, Dykeman EC, Minakhin L, Ranson NA, Severinov K, Steven AC, Antson AA. 2019. Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids. Proc Natl Acad Sci U S A 116:3556–3561. doi: 10.1073/pnas.1813204116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. 2005. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci U S A 102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Dai X, Zhou ZH. 2018. Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science 360:eaao7298. doi: 10.1126/science.aao7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Feiss M, Rao VB. 2012. The bacteriophage DNA packaging machine. Adv Exp Med Biol 726:489–509. doi: 10.1007/978-1-4614-0980-9_22. [DOI] [PubMed] [Google Scholar]
- 208.Rao VB, Feiss M. 2008. The bacteriophage DNA packaging motor. Annu Rev Genet 42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 209.Iyer LM, Leipe DD, Koonin EV, Aravind L. 2004. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol 146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 210.Kazlauskas D, Krupovic M, Venclovas Č. 2016. The logic of DNA replication in double-stranded DNA viruses: insights from global analysis of viral genomes. Nucleic Acids Res 44:4551–4564. doi: 10.1093/nar/gkw322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Barylski J, Enault F, Dutilh BE, Schuller MBP, Edwards RA, Gillis A, Klumpp J, Knezevic P, Krupovic M, Kuhn JH, Lavigne R, Oksanen HM, Sullivan MB, Jang HB, Simmonds P, Aiewsakun P, Wittmann J, Tolstoy I, Brister JR, Kropinski AM, Adriaenssens EM. 2020. Analysis of spounaviruses as a case study for the overdue reclassification of tailed phages. Syst Biol 69:110–123. doi: 10.1093/sysbio/syz036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Low SJ, Džunková M, Chaumeil P-A, Parks DH, Hugenholtz P. 2019. Evaluation of a concatenated protein phylogeny for classification of tailed double-stranded DNA viruses belonging to the order Caudovirales. Nat Microbiol 4:1306–1315. doi: 10.1038/s41564-019-0448-z. [DOI] [PubMed] [Google Scholar]
- 213.Aiewsakun P, Adriaenssens EM, Lavigne R, Kropinski AM, Simmonds P. 2018. Evaluation of the genomic diversity of viruses infecting bacteria, archaea and eukaryotes using a common bioinformatic platform: steps towards a unified taxonomy. J Gen Virol 99:1331–1343. doi: 10.1099/jgv.0.001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GGZ, Boling L, Barr JJ, Speth DR, Seguritan V, Aziz RK, Felts B, Dinsdale EA, Mokili JL, Edwards RA. 2014. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun 5:4498. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Shkoporov AN, Khokhlova EV, Fitzgerald CB, Stockdale SR, Draper LA, Ross RP, Hill C. 2018. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat Commun 9:4781. doi: 10.1038/s41467-018-07225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Philosof A, Yutin N, Flores-Uribe J, Sharon I, Koonin EV, Béjà O. 2017. Novel abundant oceanic viruses of uncultured marine group II euryarchaeota. Curr Biol 27:1362–1368. doi: 10.1016/j.cub.2017.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Devoto AE, Santini JM, Olm MR, Anantharaman K, Munk P, Tung J, Archie EA, Turnbaugh PJ, Seed KD, Blekhman R, Aarestrup FM, Thomas BC, Banfield JF. 2019. Megaphages infect Prevotella and variants are widespread in gut microbiomes. Nat Microbiol 4:693–700. doi: 10.1038/s41564-018-0338-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang Y, Bininda-Emonds ORP, Jehle JA. 2012. Nudivirus genomics and phylogeny, p 33–52. In Garcia ML, Romanowski V (ed), Viral genomes—molecular structure, diversity, gene expression mechanisms and host-virus interactions. IntechOpen Ltd, London, United Kingdom. doi: 10.5772/27793. [DOI] [Google Scholar]
- 219.Wang Y, Jehle JA. 2009. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: new insights on an old topic. J Invertebr Pathol 101:187–193. doi: 10.1016/j.jip.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 220.Wang H-C, Hirono I, Maningas MBB, Somboonwiwat K, Stentiford G, ICTV Report Consortium . 2019. ICTV virus taxonomy profile: Nimaviridae. J Gen Virol 100:1053–1054. doi: 10.1099/jgv.0.001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Krupovic M, Cvirkaite-Krupovic V, Iranzo J, Prangishvili D, Koonin EV. 2018. Viruses of archaea: structural, functional, environmental and evolutionary genomics. Virus Res 244:181–193. doi: 10.1016/j.virusres.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Prangishvili D, Bamford DH, Forterre P, Iranzo J, Koonin EV, Krupovic M. 2017. The enigmatic archaeal virosphere. Nat Rev Microbiol 15:724–739. doi: 10.1038/nrmicro.2017.125. [DOI] [PubMed] [Google Scholar]
- 223.Prangishvili D, Garrett RA, Koonin EV. 2006. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res 117:52–67. doi: 10.1016/j.virusres.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 224.Hartman R, Munson-McGee J, Young MJ, Lawrence CM. 2019. Survey of high-resolution archaeal virus structures. Curr Opin Virol 36:74–83. doi: 10.1016/j.coviro.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 225.Dill JA, Camus AC, Leary JH, Ng TFF. 2018. Microscopic and molecular evidence of the first elasmobranch adomavirus, the cause of skin disease in a giant guitarfish, Rhynchobatus djiddensis. mBio 9:e00185-18. doi: 10.1128/mBio.00185-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Welch NL, Yutin N, Dill JA, Camus AC, Pang Y-YS, Schiller JT, An P, Cantalupo PG, Pipas JM, Delwart E, Koda S, Subramaniam K, Waltzek TB, Bian C, Shi Q, Ruan Z, Koonin EV, Buck CB, Ng TFF. 2018. Adomaviruses: an emerging virus family provides insights into DNA virus evolution. bioRxiv doi: 10.1101/341131. [DOI]
- 227.Welch NL, Tisza MJ, Belford A, Pastrana DV, Pang Y-YS, Schiller JT, An P, Cantalupo PG, Pipas JM, Koda S, Subramaniam K, Waltzek TB, Bian C, Shi Q, Ruan Z, Ng TFF, Starrett GJ, Buck CB. 2019. Identification of “missing link” families of small DNA tumor viruses. bioRxiv doi: 10.1101/697771. [DOI]
- 228.Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Dempsey DM, Dutilh BE, Harrach B, Harrison RL, Hendrickson RC, Junglen S, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Nibert M, Rubino L, Sabanadzovic S, Simmonds P, Varsani A, Zerbini FM, Davison AJ. 2019. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2019). Arch Virol 164:2417–2429. doi: 10.1007/s00705-019-04306-w. [DOI] [PubMed] [Google Scholar]
- 229.Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini M, Kuhn JH. 2019. Create a megataxonomic framework, filling all principal taxonomic ranks, for realm Riboviria. ICTV TaxoProp 2019.006G.A.v1.Riboviria. https://talk.ictvonline.org/files/proposals/taxonomy_proposals_general1/m/gen04/9348.
- 230.Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini M, Kuhn JH. 2019. Create a megataxonomic framework, filling all principal taxonomic ranks, for ssDNA viruses. ICTV TaxoProp 2019.005G.A.v1.Monodnaviria. https://talk.ictvonline.org/files/proposals/taxonomy_proposals_general1/m/gen04/9346.
- 231.Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini M, Kuhn JH. 2019. Create a megataxonomic framework, filling all principal taxonomic ranks, for DNA viruses encoding vertical jelly roll-type major capsid proteins. ICTV TaxoProp 2019.003G.A.v1.Varidnaviria. https://talk.ictvonline.org/files/proposals/taxonomy_proposals_general1/m/gen04/9342.
- 232.Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini M, Kuhn JH. 2019. Create a megataxonomic framework, filling all principal/primary taxonomic ranks, for dsDNA viruses encoding HK97-type major capsid proteins. ICTV TaxoProp 2019.004G.A.v1.Duplodnaviria. https://talk.ictvonline.org/files/proposals/taxonomy_proposals_general1/m/gen04/9344.
- 233.Cavalier-Smith T. 2002. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol 52:7–76. doi: 10.1099/00207713-52-1-7. [DOI] [PubMed] [Google Scholar]
- 234.Сердюк ИН, Галзитская ОВ. 2007. Неструктурированные области в элонгационных факторах EF1А трех надцарств живого мира. Мол Биол (Моск) 41:1042–1055. [Serdyûk IN, Galzitskayaâ OV. 2007. Unstructured areas in the EF1A elongation factors of the three kingdoms of the living world. Mol Biol (Mosk) 41: 1042–1055.] [PubMed] [Google Scholar]
- 235.Koonin EV, Makarova KS, Wolf YI, Krupovic M. 2020. Evolutionary entanglement of mobile genetic elements and host defence systems: guns for hire. Nat Rev Genet 21:119–131. doi: 10.1038/s41576-019-0172-9. [DOI] [PubMed] [Google Scholar]
- 236.Callanan J, Stockdale SR, Shkoporov A, Draper LA, Ross RP, Hill C. 2020. Expansion of known ssRNA phage genomes: from tens to over a thousand. Sci Adv 6:eaay5981. doi: 10.1126/sciadv.aay5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Subramaniam K, Behringer DC, Bojko J, Yutin N, Clark AS, Bateman KS, van Aerle R, Bass D, Kerr RC, Koonin EV, Stentiford GD, Waltzek TB. 2020. A new family of DNA viruses causing disease in crustaceans from diverse aquatic biomes. mBio 11:e02938-19. doi: 10.1128/mBio.02938-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Boratto PVM, Oliveira GP, Machado TB, Andrade ACSP, Baudoin J-P, Klose T, Schulz F, Azza S, Decloquement P, Chabrière E, Colson P, Levasseur A, La Scola B, Abrahão JS. 2020. A mysterious 80 nm amoeba virus with a near-complete “ORFan genome” challenges the classification of DNA viruses. bioRxiv doi: 10.1101/2020.01.28.923185. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.