Kang et al. show that the hijacking of normally transiently induced pathways of myeloid regeneration by low Notch and high Wnt activity in hematopoietic stem cells is a common feature shared by myeloid leukemia that could be therapeutically targeted to control aberrant myeloid cell production.
Abstract
Targeting commonly altered mechanisms in leukemia can provide additional treatment options. Here, we show that an inducible pathway of myeloid regeneration involving the remodeling of the multipotent progenitor (MPP) compartment downstream of hematopoietic stem cells (HSCs) is commonly hijacked in myeloid malignancies. We establish that differential regulation of Notch and Wnt signaling transiently triggers myeloid regeneration from HSCs in response to stress, and that constitutive low Notch and high Wnt activity in leukemic stem cells (LSCs) maintains this pathway activated in malignancies. We also identify compensatory crosstalk mechanisms between Notch and Wnt signaling that prevent damaging HSC function, MPP production, and blood output in conditions of high Notch and low Wnt activity. Finally, we demonstrate that restoring Notch and Wnt deregulated activity in LSCs attenuates disease progression. Our results uncover a mechanism that controls myeloid regeneration and early lineage decisions in HSCs and could be targeted in LSCs to normalize leukemic myeloid cell production.
Graphical Abstract
Introduction
Myeloid leukemias are blood cancers that affect the production of myeloid lineage cells, with disease entities classified as chronic or acute based on their progression characteristics (Arber et al., 2016). Chronic diseases are indolent malignancies that include myeloproliferative neoplasms (MPNs) such as chronic myelogenous leukemia (CML), which are defined by excessive production of myeloid cells, and myelodysplastic syndrome (MDS) characterized by insufficient production of healthy mature cells. MPN or MDS patients can progress to acute myeloid leukemia (AML), or AML can directly occur de novo, and is a fast-growing malignancy caused by accumulation of immature myeloblasts (Döhner et al., 2015). Tremendous efforts have focused on developing therapies for myeloid leukemia by targeting recurrent driver mutations with tyrosine kinase inhibitors in MPNs (Tefferi and Pardanani, 2015) or unique disease features with differentiating agents in AML (Ma et al., 2017). Targeted therapies have revolutionized leukemia treatment, although they are not curative in most cases, as the leukemic stem cell (LSC) population driving disease development and often recurrence is usually not eradicated (Holyoake and Vetrie, 2017). However, their success in controlling disease development and progression has shown the clinical importance of normalizing blood production in leukemic contexts. Therefore, a better understanding of the mechanisms of myeloid cell expansion, a shared feature of myeloid leukemia, could help develop new treatment approaches to be used in combination with current targeted therapies.
Myeloid cell production, or myelopoiesis, is a complex and highly inducible process regulated at many levels along the hierarchy of early hematopoietic stem and progenitor cells (HSPCs; Pietras et al., 2015; Hérault et al., 2017). At steady state, the blood composition reflects the differential production by rare self-renewing hematopoietic stem cells (HSCs) of a small number of myeloid-biased multipotent progenitors (MPPs; MPP2 and MPP3) and a large amount of lymphoid-biased MPP (MPP4), which both generate granulocyte macrophage progenitors (GMPs) and give rise to myeloid cells. During blood regeneration, HSCs are induced to overproduce MPP2/MPP3, and MPP4 is redirected toward an almost exclusive myeloid output (Pietras et al., 2015). An important consequence of the activation of this myeloid regeneration axis is the formation of GMP clusters in the bone marrow (BM), which drives the local overproduction of granulocytes (Hérault et al., 2017). Altogether, the remodeling of the MPP compartment and the induction of GMP clusters represent pathways of myeloid regeneration that are transiently triggered during stress and appear to be continuously activated in myeloid diseases (Hérault et al., 2017). However, the molecular pathways regulating the differential production of lineage-biased MPPs by HSCs during steady state, during regeneration, and in myeloid leukemia are currently unknown.
Developmental pathways such as Notch (Bigas and Espinosa, 2012) and Wnt (Clevers, 2006) are essential in controlling the fate and differentiation potential of many stem cell populations across organisms. Both Notch and Wnt have been extensively studied for their role in adult HSC function and blood production, but often with confusing or conflicting results (Lampreia et al., 2017; Lento et al., 2013). A seminal study has reconciled some of these findings by showing that different dosages of canonical Wnt signaling have different effects on HSC engraftment and self-renewal activity (Luis et al., 2011). Crosstalk has also been reported between Notch and Wnt (Duncan et al., 2005), which add further complexity to the understanding of the specific role played by each pathway in driving HSC fate decision and lineage commitment. Despite this, increased Wnt activity has consistently been associated with increased myeloid cell production (Luis et al., 2011) and activation of GMP cluster formation (Hérault et al., 2017), and decreased Notch activity with myeloid expansion in leukemic conditions (Lobry et al., 2014). In fact, decreased Notch and increased Wnt activities are well-documented events in the development of myeloid leukemia in both mouse models and human patients. Mice with reduced Notch activity due to either defective Notch ligand/receptor binding (Zhou et al., 2008; Yao et al., 2011), impaired Notch intracellular domain (NICD) cleavage (Klinakis et al., 2011), or altered expression of Notch signaling transcription factor (Wang et al., 2014) consistently develop MPN phenotypes. Along the same line, HSCs from mice lacking the junB transcription factor, which all succumb to a CML-like disease, display a striking lack of responsiveness to Notch stimulation (Santaguida et al., 2009), and HSCs directly isolated from an inducible BCR/ABL-driven CML mouse model show molecular features of low Notch activity (Reynaud et al., 2011). Reduced Notch signaling has also been reported in AML patients, with blast cells having low expression of NICD protein (Kannan et al., 2013). Concerning the Wnt pathway, β-catenin is one of the most critical genes for LSC activity in both CML (Zhao et al., 2007; Heidel et al., 2012) and AML (Wang et al., 2010; Guezguez et al., 2016) mouse models, with high levels of nuclear β-catenin found in GMPs from blast crisis and imatinib-resistant CML patient samples (Jamieson et al., 2004). High Wnt activity is also implicated in AML, with hypermethylation of Wnt antagonist genes such as SFRPs and DKKs observed in patient BM samples (Valencia et al., 2009). Despite this extensive knowledge, the underlying cellular processes and the early stem and progenitor cell populations that are affected by these changes in Wnt and Notch signaling activities and drive myeloid cell expansion are still poorly understood.
Here, we show that Notch and Wnt are essential to control the differential production of lineage-biased MPPs by HSCs, with high Notch and low Wnt activity favoring the production of lymphoid-biased MPP4 at steady state. During regeneration, we find that a transient decrease in Notch and increase in Wnt activity in HSCs drive MPP3 expansion and contribute to a transitory increase in myeloid cell production. In disease conditions, we demonstrate that constitutively low Notch and high Wnt activity in LSCs enforces the constant activation of this emergency pathway, hence fueling leukemic myeloid cell production. Moreover, we identify compensatory crosstalk mechanisms between Notch and Wnt signaling that normalize blood production in certain contexts and directly show that restoring deregulated Notch and Wnt activity in LSCs attenuates disease progression. Our results elucidate the function of Notch and Wnt signaling pathways in triggering the early stage of myeloid regeneration and their implication in myeloid malignancies.
Results
Constitutive activation of myeloid regeneration pathways in myeloid malignancies
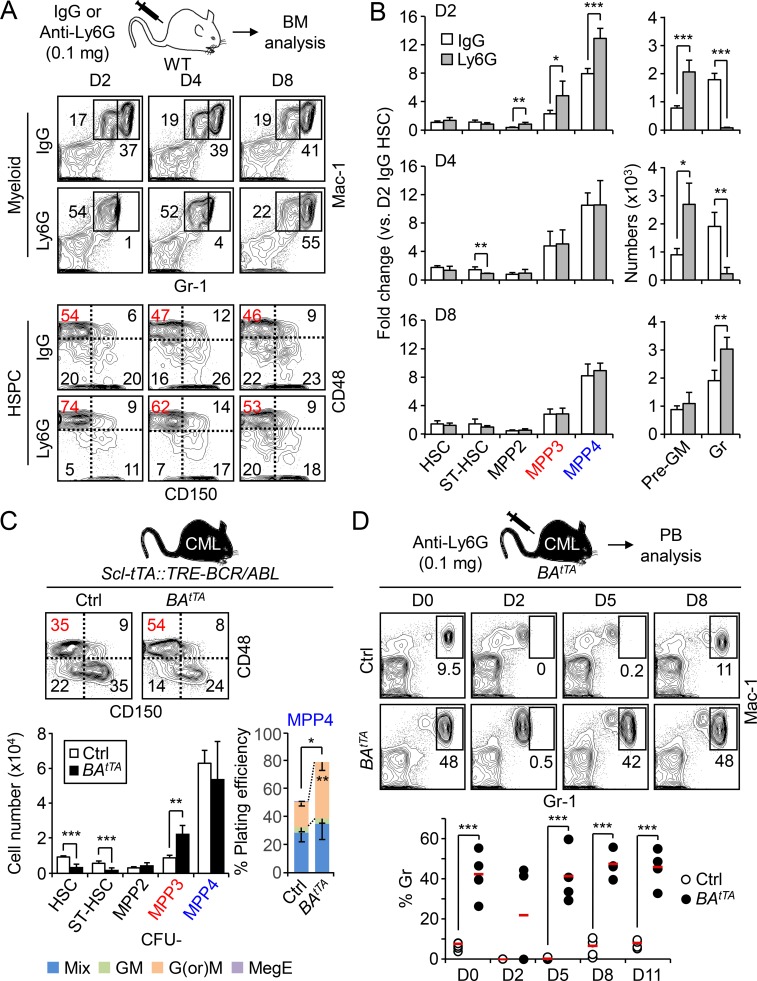
We previously reported the activation of a myeloid regeneration pathway characterized by expansion of myeloid-biased MPP2 and MPP3 and altered differentiation or myeloid reprograming of lymphoid-biased MPP4 to rebuild the blood system upon HSC transplantation (Tplx; Pietras et al., 2015). To determine whether this mechanism is also activated upon other myeloid demands, we used an anti-Ly6G antibody to selectively deplete granulocytes (Daley et al., 2008). WT C57Bl/6 mice were injected once with 0.1 mg of control IgG or anti-Ly6G antibodies and followed over time for changes in HSPC and mature BM populations (Figs. 1, A and B; and Fig. S1 A). At day 2 following Ly6G injection, Mac-1+/Gr-1+ granulocytes were completely depleted, while the Mac-1+/Gr-1low population containing granulocyte precursors (pre-GMs) was already expanded. Similarly, all MPP subsets were expanded in day 2 Ly6G-treated mice, including MPP2 (Lin−/c-Kit+/Sca-1+/Flk2−/CD48+/CD150+), MPP3 (Lin−/c-Kit+/Sca-1+/Flk2−/CD48+/CD150−), and MPP4 (Lin−/c-Kit+/Sca-1+/Flk2+). In contrast, the number of HSCs (Lin−/c-Kit+/Sca-1+/Flk2−/CD48−/CD150+) remained unchanged during the entire kinetics, although the frequency of the metabolically activated CD34+ MPP1 subset was significantly increased at day 2 (Fig. S1 B). Granulocytes started to come back at day 4, when MPPs normalized to steady state levels, and were overproduced by day 8, when all immature stem and progenitor cells, including pre-GM, had restored homeostatic levels. We also showed that day 2 MPP4 produced more mature granulocyte and macrophage colonies in methylcellulose, which were resolved in day 8 MPP4, and confirmed the transient myeloid redirection of MPP4 in this regenerative context (Fig. S1 C). These results demonstrate that the myeloid regeneration pathway we defined in a Tplx setting also operates in native conditions to respond to more specific myeloid demands.
Figure 1.
Hijacking of myeloid regeneration pathways in leukemia. (A) Experimental scheme and representative FACS plots of myeloid cells and HSPCs in the BM of IgG- and Ly6G-injected mice at the indicated time points. The percentage of MPP3 within the Flk2− LSK gate is shown in red. D, day. (B) Quantification of HSPCs and myeloid cells in the BM of Ly6G- and IgG-injected mice (six and four mice per group, respectively, in two independent experiments). Results are expressed as fold change relative to D2 IgG HSCs (set to 1). Gr, granulocyte; ST-HSC, short-term HSC. (C) Analyses of age-matched control (Ctrl) and BAtTA mice with representative FACS plots and quantification of BM HSPCs (eight mice per group, left, in four independent experiments), and myeloid differentiation of MPP4 in methylcellulose (n = 3, right, in three independent experiments). Mix: all lineages; GM, granulocyte/macrophage; G(or)M, granulocyte (or) macrophage; MegE, megakaryocyte/erythrocyte. (D) Representative FACS plots and quantification of granulocyte regeneration in the PB of Ly6G-injected Ctrl and BAtTA mice at the indicated time points (six and four mice per group, respectively, in two independent experiments). Average values are indicated with a red bar. Results are expressed as mean ± SD; unpaired Student’s t test was used. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Figure S1.
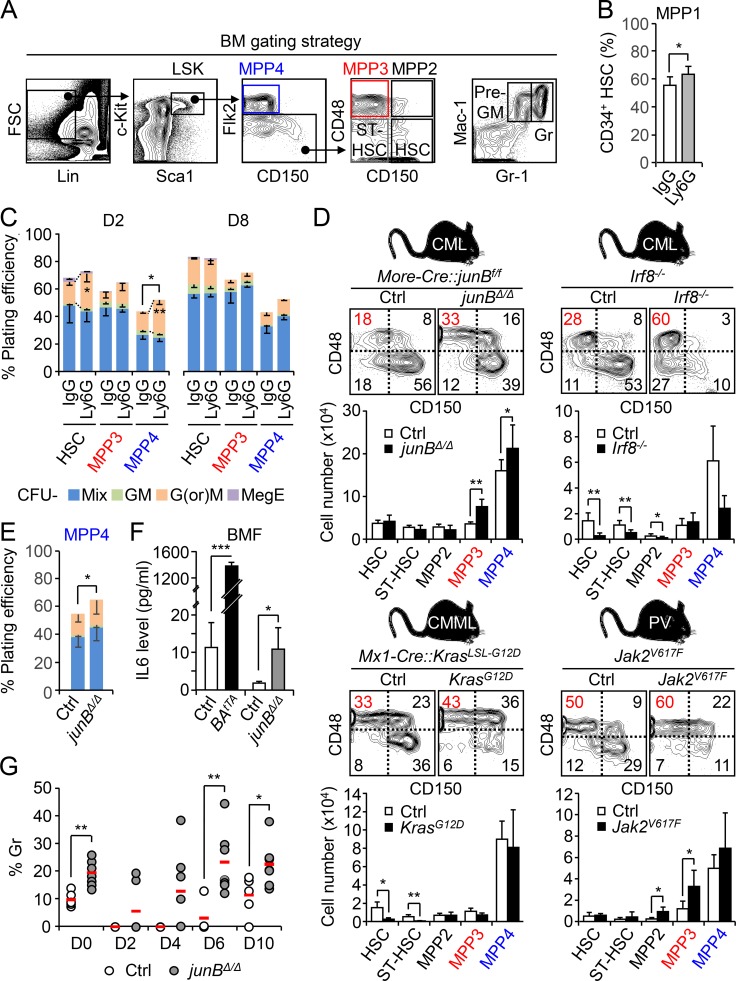
Constitutive activation of myeloid regeneration pathways in leukemic mouse models. (A) Gating strategy used for identifying and isolating HSC, short-term HSC (ST-HSC), MPP2, MPP3, MPP4, pre-GM, and granulocyte (Gr). (B) Percentage of activated CD34+ MPP1 within the HSC pool in D2 IgG- or Ly6G-injected mice (four mice per group, in two independent experiments). (C) Myeloid differentiation of HSCs, MPP3, and MPP4 isolated from D2 (n = 4) and D8 (n = 3) IgG- or Ly6G-injected WT mice in methylcellulose (in two independent experiments). GM, granulocyte/macrophage; G(or)M, granulocyte or macrophage; MegE, megakaryocyte/erythrocyte; Mix, all lineages. (D) Analyses of various independent cohorts of diseased leukemic mice and age-matched controls (Ctrls) with representative FACS plots and quantification of BM HSPCs (11 and 8 mice per group for Ctrl and junBΔ/Δ, respectively; seven mice per group for Ctrl and Irf8−/−; three and four mice per group for Ctrl and KrasG12D, respectively; five mice per group for Ctrl and Jak2V617F). CMML, chronic myelomonocytic leukemia; PV, polycythemia vera. The percentage of MPP3 within the Flk2− LSK gate is shown in red. (E) Myeloid differentiation of Ctrl and junBΔ/Δ MPP4 in methylcellulose (n = 5, in two independent experiments). (F) IL-6 level in BM fluid (BMF) of BAtTA and junBΔ/Δ mice and their respective age-matched Ctrl (n = 3, in one experiment). (G) Quantification of granulocyte regeneration in the PB of Ctrl and junBΔ/Δ mice at the indicated times after Ly6G injections (five and seven mice per group, respectively, in two independent experiments). Average values are indicated with a red bar. Results are expressed as mean ± SD; unpaired Student’s t test was used. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
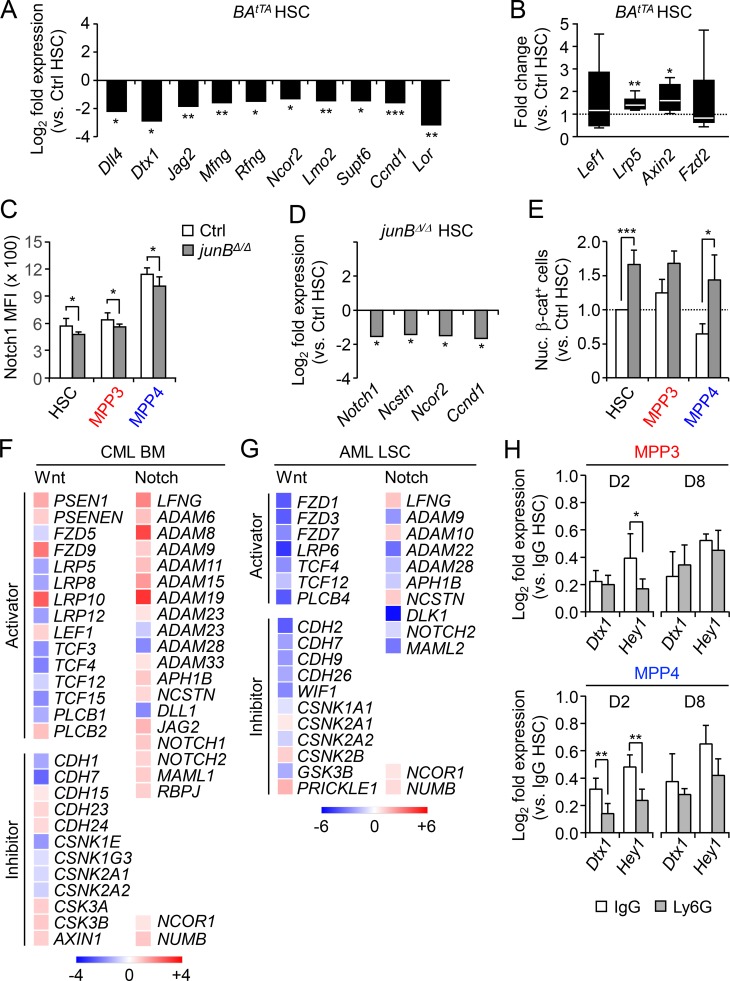
Next, we investigated the state of this emergency pathway in genetic mouse models of myeloid leukemia, including the inducible Scl-tTA::TRE-BCR/ABL (BAtTA) and constitutive More-Cre::junBf/f (junBΔ/Δ) and Irf8−/− models of CML (Reynaud et al., 2011; Santaguida et al., 2009; Holtschke et al., 1996), the inducible Mx1-Cre::KrasLSL-G12D (KrasG12D) model of chronic myelomonocytic leukemia (Braun et al., 2004), and the constitutive Jak2V617F knock-in model of polycythemia vera (Mullally et al., 2010; Fig. 1 C and Fig. S1 D). In every case, we observed increased MPP3 frequency that was either associated with a clear expansion of MPP3 numbers (i.e., BAtTA, junBΔ/Δ, and Jak2V617F mice) or the maintenance of this MPP subset in the context of loss of HSCs (i.e., Irf8−/− and KrasG12D mice). Both MPP2 and MPP4 were also affected, but not in a consistent manner as observed for MPP3 expansion. For the rest of these studies, we focused on the BAtTA and junBΔ/Δ leukemic models and first confirmed the altered myeloid differentiation of their MPP4 populations using methylcellulose assays (Fig. 1 C and Fig. S1 E). We also showed increased IL-6 production in the BM fluid of these leukemic animals, which likely drives enhanced myeloid differentiation from MPP4 (Fig. S1 F; Reynaud et al., 2011).
To demonstrate that activation of myeloid regeneration plays a role in the constitutive overproduction of leukemic myeloid cells, we followed granulocyte regeneration in the peripheral blood (PB) of Ly6G-treated BAtTA mice. As expected, diseased BAtTA mice had massively increased numbers of circulating granulocytes, which in some cases could not be completely depleted upon injection of 0.1 mg of anti-Ly6G antibody (two of four mice). However, Ly6G-depleted BAtTA mice showed a significantly faster granulocyte recovery compared with controls (Fig. 1 D). Similar results were obtained with Ly6G-treated junBΔ/Δ mice (Fig. S1 G) and directly supported the idea that constant activation of myeloid regeneration contributes to the overproduction of myeloid cells in disease conditions. Taken together, these results indicate that MPP3 expansion and myeloid reprogramming of MPP4 are common features of myeloid leukemia and reflect the hijacking of a normally transiently activated pathway of emergency myelopoiesis.
Leukemic MPPs share common features with regenerating MPPs
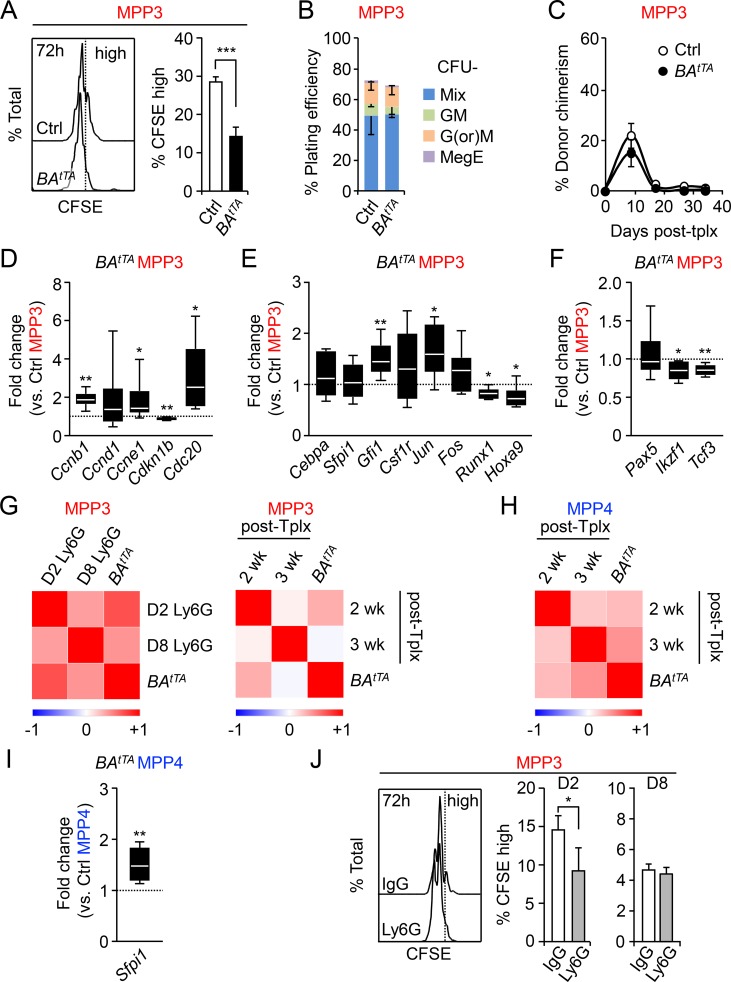
We previously described the aberrant features of leukemic BAtTA HSCs and MPP4 (Reynaud et al., 2011). Here, we performed detailed functional and molecular characterization of leukemic BAtTA MPP3. Similar to leukemic MPP4, BAtTA MPP3 showed accelerated cell division compared with control MPP3, as illustrated by the decreased frequency of CFSE-retaining (CFSE high) cells after 72 h culture (Fig. 2 A). However, BAtTA MPP3 did not exhibit major differences in overall plating efficiency and colony type in methylcellulose assays compared with control MPP3 (Fig. 2 B). Moreover, transplanted BAtTA MPP3 showed the same PB chimerism as control MPP3 in short-term lineage-tracing experiments in sublethally irradiated mice (Fig. 2 C), thus confirming similar myeloid output from diseased and normal MPP3 in vivo. Next, we conducted molecular analysis using a custom-made 96-gene Fluidigm multiplexing quantitative PCR (qPCR) platform and pools of 100 MPP3 isolated from individual diseased BAtTA and control mice (Table S1). Consistent with their active cell cycle status, we observed increased expression of cyclin B1 (Ccnb1), cyclin E1 (Ccne1), and the cell division cycle protein 20 (Cdc20), together with decreased expression of the cyclin dependent kinase inhibitor p27 (Cdk1nb) in BAtTA MPP3 (Fig. 2 D). We also observed enhanced myeloid poising in BAtTA MPP3 as illustrated by the increased expression of Gfi1 and c-Jun myeloid commitment genes, downregulation of Runx1 and Hoxa9 self-renewal genes, and further decreased expression of lymphoid commitment genes such as Ikzf1 and Tcf3 (Fig. 2, E and F). Interestingly, Pearson correlation analyses showed a high similarity in the molecular characteristics of BAtTA MPP3 and actively regenerating MPP3 obtained from either day 2 Ly6G-treated or 2-wk post-Tplx mice (Pietras et al., 2015), times that correspond in both cases to the main phase of myeloid lineage rebuilding (Fig. 2 G). In contrast, BAtTA MPP4 were more similar to regenerating MPP4 from 3-wk post-Tplx mice (Fig. 2 H), which still have a strong myeloid output but are starting to recover their lymphoid potential. In fact, BAtTA MPP4 showed significantly higher Sfpi1 (PU.1) expression compared with control MPP4 in support of its altered differentiation toward the myeloid lineage (Fig. 2 I). Regenerative MPP3 also showed significantly faster division rates with a lower CFSE high population at day 2, which was restored to normal levels at day 8 after Ly6G treatment (Fig. 2 J). Taken together, these results indicate that leukemic MPP3 share common features with regenerating MPP3, including faster division kinetics and enhanced myeloid commitment. However, despite their molecular poising, neither leukemic BAtTA MPP3 nor actively regenerating MPP3 from 2-wk post-Tplx mice (Pietras et al., 2015) had increased myeloid output, likely reflecting the fact that they both have accelerated rather than augmented myeloid cell production. This reinforces the idea that leukemia hijacks myeloid regeneration pathways that are initiated at the level of HSCs rather than directly in MPPs.
Figure 2.
Characterization of leukemic MPP3. (A) Representative FACS plots and percentage of remaining CFSE high cells after 72 h culture of control (Ctrl) and BAtTA MPP3 (n = 3, in two independent experiments). (B) Myeloid differentiation of Ctrl and BAtTA MPP3 (n = 3, in three independent experiments). GM, granulocyte/macrophage; G(or)M, granulocyte (or) macrophage; MegE, megakaryocyte/erythrocyte; Mix, all lineages. (C) Donor-derived chimerism in the PB of sublethally irradiated recipients after Tplx with 2,000 Ctrl or BAtTA MPP3 (four and five mice per group, respectively, in two independent experiments). (D–F) Fluidigm analysis of (D) cell cycle, (E) myeloid, and (F) lymphoid gene expression in Ctrl and BAtTA MPP3 (n = 6, four pools of 100 cells/mouse, in two independent experiments). Results are expressed as fold change relative to Ctrl MPP3 (set to 1). (G) Pearson correlation of Fluidigm gene expression dataset comparing BAtTA MPP3 with regenerating MPP3 isolated from either D2 and D8 Ly6G-injected mice (left) or 2 and 3 wk after HSC Tplx (right, Pietras et al., 2015). (H) Pearson correlation of Fluidigm gene expression dataset comparing BAtTA MPP4 with MPP4 at 2 and 3 wk after Tplx (Pietras et al., 2015). (I) Fluidigm analysis of Sfpi1 (PU.1) expression in Ctrl and BAtTA MPP4 (n = 6, four pools of 100 cells per mouse, in two independent experiments). Results are expressed as fold change relative to Ctrl MPP4 (set to 1). (J) Percentage of remaining CFSE high cells after 72 h culture of MPP3 isolated from D2 and D8 IgG- or Ly6G-injected mice (n = 4, in two independent experiments). Results are expressed as mean ± SD; unpaired Student’s t test was used. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Table S1. Fluidigm gene expression level in BAtTA MPP3.
| Gene | Fold change (versus control MPP3) | Gene | Fold change (versus control MPP3) |
|---|---|---|---|
| Axin2 | 2.89 ± 1.74a | Hhip | 8.10 ± 5.65b |
| Bax | 1.17 ± 0.40 | Hif1a | 1.27 ± 0.27b |
| Bbc3 | 0.74 ± 0.54 | Hmga2 | 0.80 ± 0.21 |
| Bcl2 | 1.15 ± 0.25 | Hoxa9 | 0.76 ± 0.22b |
| Bcl2l1 | 0.97 ± 0.24 | Id1 | 2.87 ± 1.04a |
| Birc2 | 0.93 ± 0.13 | Ikzf1 | 0.85 ± 0.12b |
| Bmi | 0.96 ± 0.17 | Il1b | 4.34 ± 2.44a |
| Cbx7 | 2.05 ± 1.65 | Il6 | 2.82 ± 0.87a |
| Ccl3 | 2.05 ± 1.65 | Il6ra | 0.69 ± 0.34 |
| Ccna2 | 1.04 ± 0.39 | Il7r | 3.91 ± 1.95a |
| Ccnb1 | 1.83 ± 0.42a | Irf8 | 0.58 ± 0.30 |
| Ccnd1 | 1.75 ± 1.82 | Jun | 1.65 ± 0.52b |
| Ccne1 | 1.86 ± 1.09b | Lef1 | 0.83 ± 0.68 |
| Cd34 | 0.74 ± 0.21b | Lrp5 | 0.95 ± 0.40 |
| Cd48 | 0.94 ± 0.39 | Mcl1 | 1.02 ± 0.48 |
| Cdc20 | 2.97 ± 1.81b | Meis1 | 0.77 ± 0.16b |
| Cdk2 | 1.05 ± 0.42 | Mfng | 0.91 ± 0.19 |
| Cdkn1a | 1.08 ± 0.16 | Mki67 | 0.81 ± 0.32 |
| Cdkn1b | 0.81 ± 0.08a | Mpl | 1.53 ± 1.02 |
| Cdkn1c | 0.66 ± 0.42 | Myc | 1.04 ± 0.30 |
| Cebpa | 1.18 ± 0.42 | Nfe2l2 | 1.37 ± 0.34b |
| Csf1r | 1.37 ± 0.72 | Nfkbia | 1.46 ± 0.51 |
| Csf2ra | 0.78 ± 0.28 | Notch1 | 1.08 ± 0.19 |
| Csf3r | 0.79 ± 0.22 | Pai1 | 10.64 ± 9.71b |
| Dnmt1 | 0.92 ± 0.26 | Pax5 | 1.06 ± 0.33 |
| Dnmt3a | 0.83 ± 0.21 | Pdk4 | 2.53 ± 1.20a |
| Ebf1 | 1.34 ± 0.74 | Pmaip1 | 4.02 ± 2.37b |
| Egr1 | 2.24 ± 1.37b | Ppargc1a | 0.54 ± 0.18a |
| Epor | 2.02 ± 1.60 | Prkdc | 0.74 ± 0.23 |
| Evi1 | 0.55 ± 0.20b | Ptch1 | 1.25 ± 0.51 |
| Ezh1 | 1.02 ± 0.07 | Ptch2 | 3.03 ± 3.66 |
| Ezh2 | 0.79 ± 0.25 | Rad51 | 0.79 ± 0.54 |
| Flt3 | 1.07 ± 0.41 | Rpa1 | 0.80 ± 0.34 |
| Fn1 | 0.70 ± 0.67 | Runx1 | 0.83 ± 0.11b |
| Fos | 1.27 ± 0.44 | Sfpi1 | 1.06 ± 0.35 |
| Foxo3 | 0.65 ± 0.20b | Slamf1 | 1.41 ± 1.06 |
| Fzd2 | 3.30 ± 2.28b | Smad7 | 1.25 ± 0.54 |
| Gata1 | 4.32 ± 2.83a | Tcf3 | 0.86 ± 0.07a |
| Gata2 | 2.23 ± 1.26b | Tnf | 2.14 ± 1.16 |
| Gfi1 | 1.50 ± 0.35a | Traf1 | 1.41 ± 0.82 |
| Gfi1b | 1.00 ± 0.57 | Vwf | 4.05 ± 2.38a |
| Gli1 | 5.17 ± 3.26 | Xiap | 0.85 ± 0.18 |
| Hes1 | 6.46 ± 7.29b | Xrcc5 | 0.99 ± 0.24 |
| Hes5 | 1.47 ± 0.75 | Xrcc6 | 1.15 ± 0.29 |
| Hey1 | 2.67 ± 1.43b | Zfpm1 | 0.60 ± 0.30b |
| Hey2 | 1.10 ± 0.88 |
Fluidigm analyses in BAtTA MPP3 (n = 6; four pools of 100 cells per mouse, in two independent experiments). Results are expressed as fold change relative to control MPP3 (set to 1) and are mean ± SD; unpaired Student’s t test was used.
P < 0.01.
P < 0.05.
Signaling pathways controlling MPP production from HSCs
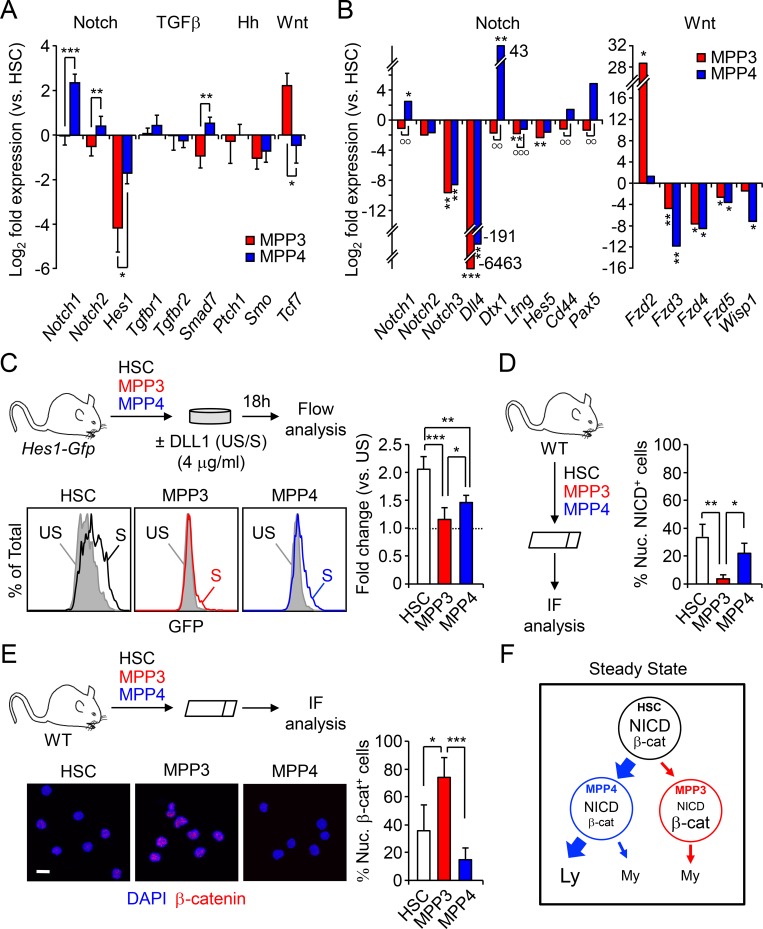
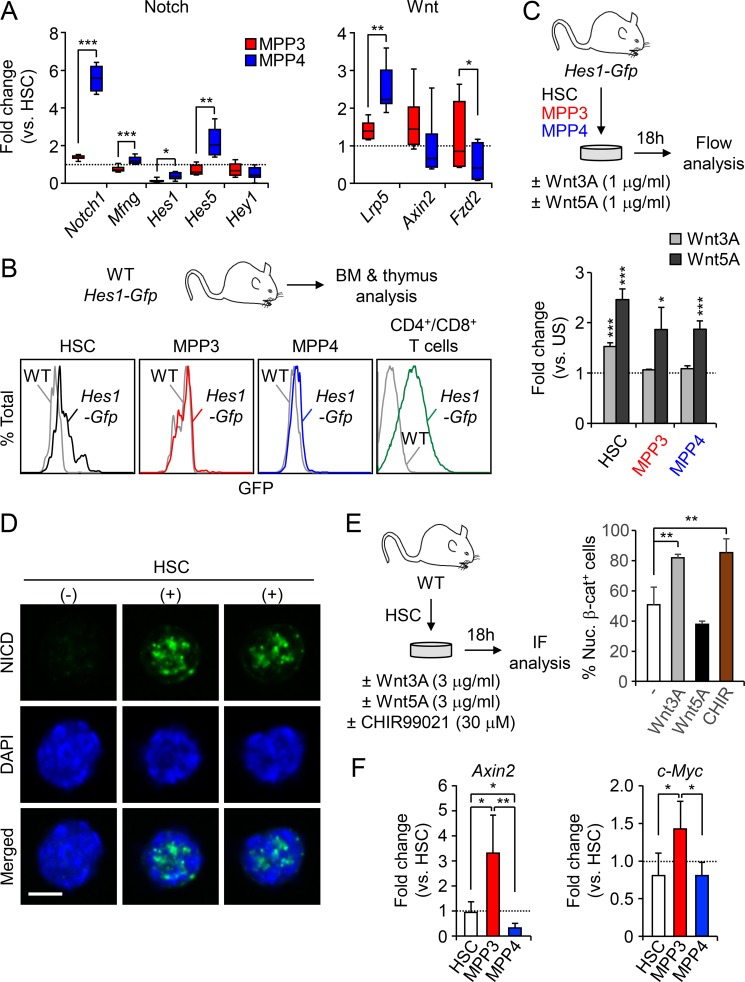
To start investigating the mechanisms by which HSCs control the differential production of MPP subsets, we compared by quantitative RT-PCR the expression levels of key genes reflecting activation of Notch, TGFβ, hedgehog (Hh), and Wnt signaling pathways in HSCs, MPP3, and MPP4 freshly isolated from WT mice (Fig. 3 A). Interestingly, Notch, Wnt, and, to some extent, TGFβ pathway genes showed opposite expression levels in MPP3 and MPP4, with low Notch and high Wnt expression in MPP3 and high Notch and low Wnt expression in MPP4. We then used the commercial Notch signaling pathway SABioscience PCR array and our custom made Fluidigm multiplexing qPCR panel to confirm these results (Fig. 3 B and Fig. S2 A). As expected, the Notch signaling gene Notch1, Notch ligand Dtx1, Notch receptor processing enzymes Lfng and Mfng, and Notch targets Cd44, Pax5, Hes1, and Hes5 were all expressed at significantly lower levels in MPP3 than in MPP4. Conversely, several Wnt receptors, in particular Fzd2, were highly expressed in MPP3 compared with MPP4. To test whether these differences in expression levels directly correlate with changes in signaling activity, we first used Hes1-Gfp reporter mice as a readout for Notch activity. As previously reported (Oh et al., 2013), HSCs displayed higher Notch activity than MPP3 and MPP4, with both showing almost no discernable activity, although it remained largely below the level of Notch activity observed in T cells (Fig. S2 B). However, Hes1 expression per se is notoriously promiscuous and can be induced by other signaling mechanisms including the Wnt pathway (Duncan et al., 2005), which we directly confirmed by incubating Hes1-Gfp HSCs, MPP3, and MPP4 with both canonical Wnt3a and noncanonical Wnt5a ligands (Fig. S2 C).
Figure 3.
Differential Notch and Wnt activity in lineage-biased MPP subsets. (A) Quantitative RT-PCR analysis of canonical Notch, TGFβ, Hedgehog (Hh), and Wnt pathway genes in HSPCs (n = 3, in one experiment). Results are expressed as log2 fold expression relative to HSCs (set to 0). (B) SABiosciences PCR array of Notch and Wnt pathway-related genes in HSPCs (n = 3, in two independent experiments). Results are expressed as log2 mean fold expression relative to HSCs (set to 0). (C) Schematic of the ex vivo stimulation of Hes1-Gfp HSPCs by plate-bound DLL1 ligand, representative FACS plots and quantification of Notch activity (n = 6 for HSCs, n = 5 for MPP3 and MPP4, in four independent experiments). Results are expressed as fold changes in GFP mean fluorescence intensity (MFI) relative to respective unstimulated population (set to 1). S, stimulated; US, unstimulated. (D) Schematic of the NICD IF analysis and percentage of nuclear NICD–positive (nuc. NICD+) cells in HSPCs (n = 3, in two independent experiments). (E) Schematic of the IF analysis with representative images and percentage of nuclear β-catenin–positive (nuc. β-cat+) cells in HSPCs (n = 5 for HSC and MPP4, n = 4 for MPP3, in five independent experiments). Scale bar, 10 µm. (F) Model depicting differential Notch and Wnt activity in HSPCs at steady state. Results are expressed as mean ± SD except in B; unpaired Student’s t test was used. *, P ≤ 0.05; **/°°, P ≤ 0.01; ***/°°°, P ≤ 0.001; *, versus HSC levels; °, in between MPPs.
Figure S2.
Measuring Notch and Wnt activity in HSPCs. (A) Fluidigm analysis of Notch and Wnt pathway genes in WT HSPCs (n = 6, four pools of 100 cells per mouse, in two independent experiments). Results are expressed as fold change relative to HSCs (set to 1). (B) Representative FACS plots of Notch activity in freshly isolated Hes1-Gfp HSPCs and T cells. Gray histograms show the baseline GFP level in WT cells. (C) Schematic of the ex vivo stimulation of Hes1-Gfp HSPCs by Wnt ligands and quantification of Notch activity (n = 4 for HSC and MPP4, n = 3 for MPP3, in two independent experiments). Results are expressed as fold change in GFP MFI relative to respective unstimulated (US) population (set to 1). (D) Representative IF images of nuclear NICD-positive (+) and -negative (−) HSCs. Scale bar, 5 µm. (E) Schematic of the ex vivo stimulation of WT HSCs by Wnt ligands and the GSK3 inhibitor CHIR99021 and quantification of the percentage of Nuc. β-cat+ cells (n = 3 for untreated and Wnt5A, n = 4 for Wnt3A and CHIR, in three independent experiments). (F) Quantitative RT-PCR analysis of Wnt target genes in WT HSPCs (n = 4, in one experiment). Results are expressed as fold change relative to HSC (set to 1). Results are expressed as mean ± SD; unpaired Student’s t test was used. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
To directly test the response to Notch stimulation, we next incubated Hes1-Gfp HSCs, MPP3, and MPP4 with plate-bound DLL1 in vitro for 18 h (Fig. 3 C). In these conditions, we found the highest level of Notch induction in HSCs followed by MPP4, while MPP3 still had negligible Notch activity. Moreover, we confirmed this gradient of Notch activity in freshly isolated HSCs, MPP3, and MPP4 using NICD immunofluorescence (IF) staining (Fig. 3 D and Fig. S2 D).
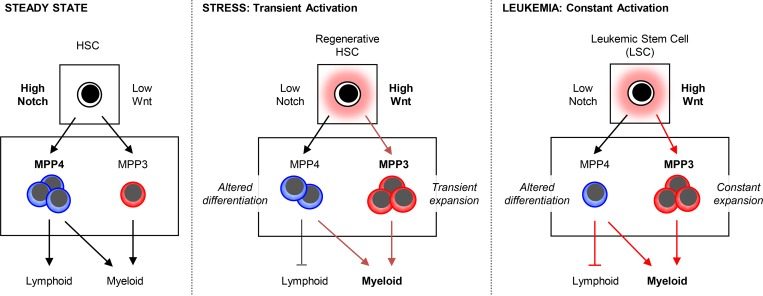
To assess Wnt activity, we used IF staining for nuclear β-catenin and first confirmed the specificity of this approach by incubating HSCs with either Wnt3a, Wnt5a, or the GSK3 inhibitor CHIR 99021 as a positive control (Fig. S2 E). Consistently, nuclear β-catenin levels were increased by ∼60% upon exposure to the canonical Wnt3a ligand, which was in the range of CHIR 99021 treatment, and remained unchanged upon exposure to the noncanonical Wnt5a ligand. We then measured Wnt activity in freshly isolated HSCs, MPP3, and MPP4 and found that MPP3 had a significantly higher constitutive level of nuclear β-catenin compared with HSCs and, even more, to MPP4 (Fig. 3 E). Furthermore, MPP3 displayed the highest level of expression of Wnt target genes, including Axin2 and c-Myc, compared with HSC and MPP4 (Fig. S2 F), hence confirming enhanced Wnt activity at the transcriptional level in MPP3. Altogether, these results indicate that at steady state, high Notch and low Wnt activities in HSCs correlate with large production of MPP4, which maintain a relatively similar, albeit attenuated, signaling activity with that of HSCs and small production of MPP3 with downregulated Notch activity and augmented Wnt activity compared with both HSCs and MPP4 (Fig. 3 F). Moreover, these results suggest that differential levels of Notch and Wnt signaling activity may dictate the ratio of MPP3 and MPP4 generated by HSCs.
Lowering Notch activity triggers the early stage of myeloid regeneration pathways
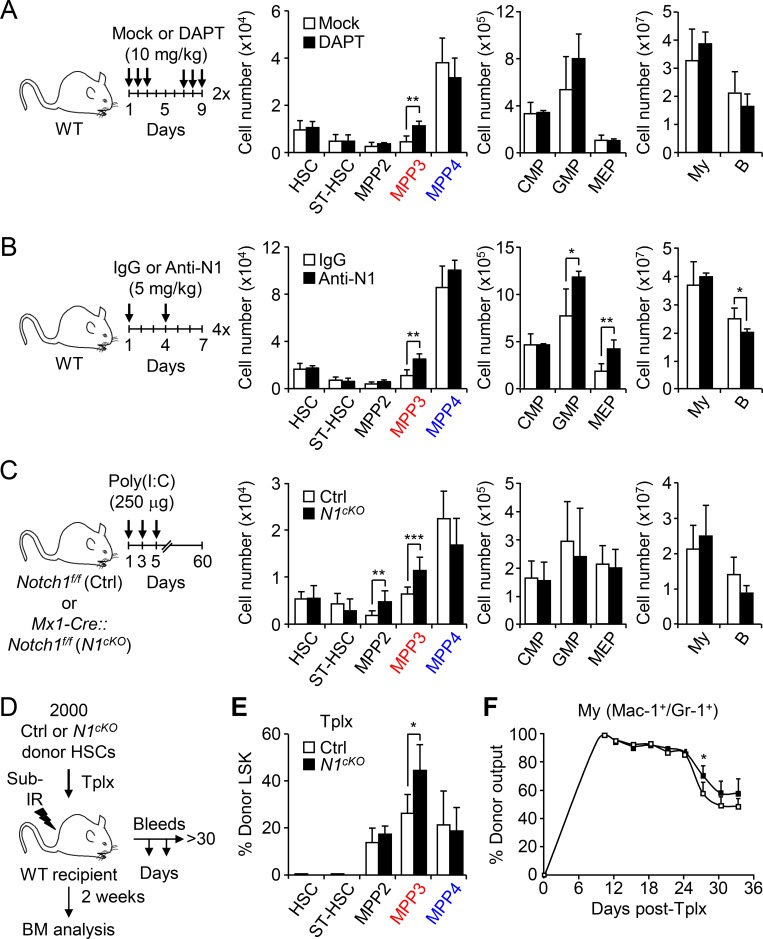
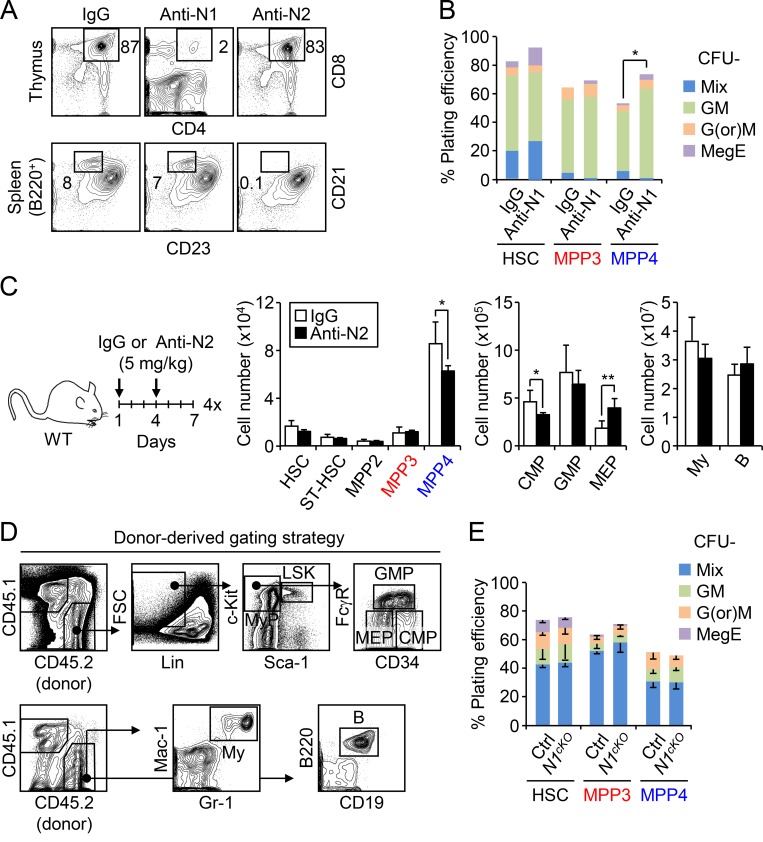
To directly test whether modulating Notch activity could change the production of MPPs by HSCs, we first treated WT mice with the γ-secretase inhibitor N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT), which blocks Notch signaling by preventing NICD cleavage. Remarkably, administration of DAPT specifically expands MPP3 numbers without affecting other BM populations (Fig. 4 A). We then used the more specific anti-Notch1 and anti-Notch2 blocking antibodies to treat WT mice and validated our treatment protocol by showing decreased CD4+/CD8+ thymic T cells and marginal zone CD21+/CD23− splenic B cells, respectively (Fig. S3 A; Wu et al., 2010). In these conditions, blockade of Notch signaling by anti-Notch 1 resulted in a specific increase in MPP3 numbers, which was accompanied by GMP expansion and altered myeloid differentiation of MPP4 as measured in methylcellulose (Fig. 4 B and Fig. S3 B). Activation of myeloid regeneration was specific to decreased signaling through the Notch1 receptor, as treatment with anti-Notch2 did not change MPP3 numbers (Fig. S3 C). We confirmed the involvement of decreased Notch1 signaling directly in hematopoietic cells using Mx1-Cre::Notch1f/f conditional knockout (N1cKO) mice, where again we observed a significantly expanded MPP3 compartment compared with age-matched controls (Fig. 4 C).
Figure 4.
Lowering Notch activity drives MPP3 expansion. (A and B) Treatment scheme and BM cellularity upon pharmacological inhibition of the Notch pathway in WT mice injected with either (A) the γ-secretase inhibitor DAPT or mock treatment (three and four mice per group, respectively, in one experiment) or (B) anti-Notch1 (Anti-N1) blocking antibody or control IgG (three mice per group, in one experiment). Arrows indicate the injection days, and “x” indicates the number of treatment cycles. B, B cell; CMP, common myeloid progenitor; MEP, megakaryocyte/erythrocyte progenitor; My, myeloid cell; ST-HSC, short-term HSC. (C) Treatment scheme and BM cellularity upon genetic inactivation of the Notch pathway in Notch1 conditional knockout (N1cKO) mice compared with age-matched control (Ctrl) mice (four and three mice per group, respectively, in two independent experiments). Arrows indicate the days of Poly(I:C) injections. (D–F) Cell-intrinsic effect of low Notch activity with (D) schematic of the short-term lineage tracking experiment with Tplx of Ctrl and N1cKO HSCs into sublethally irradiated (sub-IR) recipients, (E) frequency of donor-derived BM HSPCs at 2 wk after Tplx (five and four mice per group, respectively, in two independent experiments), and (F) frequency of donor-derived myeloid cells in PB over time (four and five mice per group, respectively, in two independent experiments). Results are expressed as mean ± SD; unpaired Student’s t test was used. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Figure S3.
Notch modulation and myeloid regeneration. (A) Representative FACS plots validating the effects of anti-Notch1 (Anti-N1) and anti-Notch2 (Anti-N2) blocking antibody treatment in WT mice. (B) Myeloid differentiation of HSPCs from IgG- and Anti-N1–treated mice in methylcellulose. Results are mean for one of two experiments performed in triplicate. GM, granulocyte/macrophage; G(or)M, granulocyte or macrophage; MegE, megakaryocyte/erythrocyte; Mix, all lineages. (C) Treatment scheme and BM cellularity in IgG- and Anti-N2–treated mice (three mice per group). B, B cell; CMP, common myeloid progenitor; MEP, megakaryocyte/erythrocyte progenitor; My, myeloid cell; ST-HSC, short-term HSC. (D) Gating strategy used for identifying donor-derived stem, progenitor, and mature cells in transplanted mice. (E) Myeloid differentiation of control (Ctrl) and N1cKO HSPCs in methylcellulose (n = 4, in one experiment). Results are expressed as mean ± SD except in B; unpaired Student’s t test was used. *, P ≤ 0.05; **, P ≤ 0.01.
To demonstrate the cell-autonomous effect of lowering Notch activity in HSCs in enhancing myeloid regeneration, we used the same short-term lineage-tracing approach that we previously employed to demonstrate increased myeloid output from regenerating HSCs (Pietras et al., 2015). We transplanted control or N1cKO HSCs into sublethally irradiated recipient mice and measured donor-derived lineage output over 1 mo after Tplx, and BM reconstitution at 2 wk after Tplx when myeloid regeneration pathways were fully activated (Fig. 4 D and Fig. S3 D). Strikingly, we observed an expanded MPP3 compartment and more sustained myeloid cell production from transplanted N1cKO HSCs (Fig. 4, E and F), thus directly confirming that lowering Notch activity in HSCs drives myeloid regeneration associated with increased MPP3 production. In contrast, we did not observe altered differentiation potential from N1cKO MPP4 when cultured in methylcellulose (Fig. S3 E), thereby indicating that it is likely a non–cell-autonomous effect occurring as an indirect consequence of lowering Notch activity in cells that are not targeted by Mx1-Cre deletion. In fact, Notch inhibition in BM endothelial cells has been shown to increase the production of proinflammatory cytokines in the niche (Wang et al., 2014), including the known MPP4 myeloid instructing factor IL-6.
Increasing Wnt activity also activates early myeloid regeneration pathways
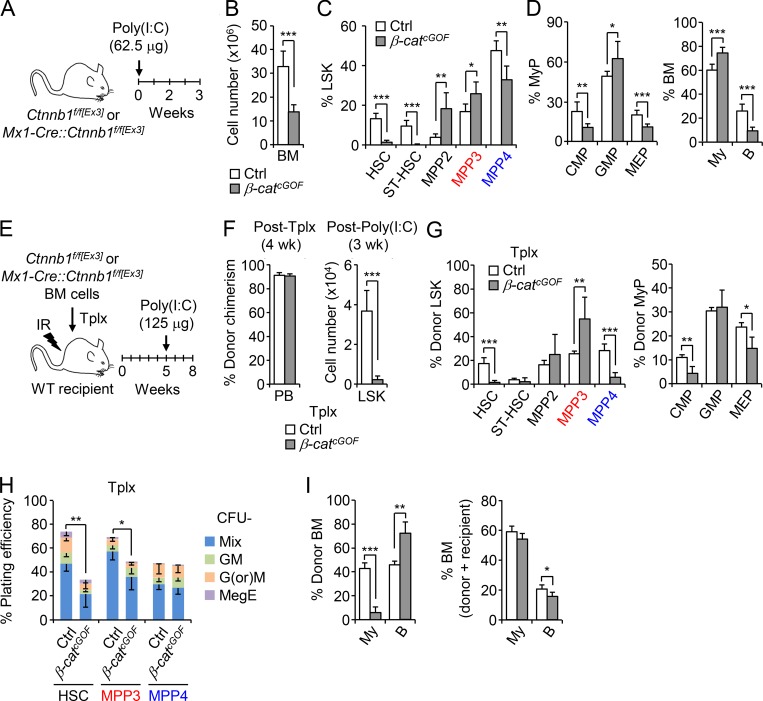
To investigate the implication of the Wnt pathway, we employed a conditional β-catenin gain-of-function (GOF) approach using Mx1-Cre::Ctnnb1f/f[Ex3] (β-catcGOF) mice, which express a nondegradable form of β-catenin. After a single injection of a low dose of Poly(I:C) (polyinosinic:polycytidylic acid), β-catcGOF mice quickly became sick, with massively decreased BM cellularity by 3 wk, and died shortly thereafter (Figs. 5, A and B). Despite this BM attrition, we observed a significant increase in MPP3 and mature myeloid cell frequency associated with a loss of the HSC compartment in β-catcGOF mice (Fig. 5, C and D). To confirm these results, we transplanted untreated Ctnnb1f/f[Ex3] and Mx1-Cre::Ctnnb1f/f[Ex3] BM cells into lethally irradiated WT recipients, waited 4 wk for full engraftment with ∼90% chimerism in both cases, then treated with Poly(I:C) and analyzed Tplx controls and β-catcGOF mice after 3 wk (Fig. 5, E–G). As expected from previous data showing impaired HSC self-renewal and differentiation potential in high Wnt genetic models (Luis et al., 2011), we observed a much-reduced Lin−/Sca-1+/c-Kit+ (LSK) HSPC compartment in Tplx β-catcGOF mice (Fig. 5 F). However, the frequency of MPP3 was specifically increased while the frequency of MPP4 was decreased, suggesting that high Wnt activity forces HSCs to differentiate into MPP3, likely at the expense of MPP4 production (Fig. 5 G). In addition, the ability of donor-derived MPP4 isolated from Tplx β-catcGOF mice to produce myeloid colonies in methylcellulose was maintained, while the differentiation potential of donor-derived HSCs and MPP3 was severely compromised (Fig. 5 H), directly supporting the idea of enhanced differentiation along the HSC/MPP3 axis in response to increased Wnt activity. Surprisingly, the frequency of donor-derived mature myeloid cells was reduced in Tplx β-catcGOF mice, likely due to a competitive disadvantage of high Wnt versus recipient-derived WT myeloid cells (Fig. 5 I). In support of this idea, the total frequency of BM myeloid cells was unchanged between Tplx control and β-catcGOF mice. Furthermore, while the frequency of donor-derived B cells was increased in Tplx β-catcGOF mice, the total frequency of BM B cells was significantly reduced, hence confirming decreased lymphoid cell output from Tplx β-catcGOF HSCs (Fig. 5 I). These results indicate that increasing Wnt activity forces HSCs to undergo myeloid differentiation, leading to MPP3 expansion but at the cost of HSC maintenance, MPP4 production, and optimal generation of myeloid cells. In fact, we already showed that while increased Wnt activity is also needed for GMP cluster formation, a subsequent decrease in Wnt activity is then required to drive myeloid expansion (Hérault et al., 2017), thus highlighting the importance of transient changes in signaling activity for proper myeloid regeneration. Taken together, these findings demonstrate that lowering Notch signaling and increasing Wnt activity are sufficient to trigger the early stage of myeloid regeneration from HSCs.
Figure 5.
Increasing Wnt activity also forces MPP3 expansion. (A–D) Consequences of genetic activation of the Wnt pathway in β-catenin conditional GOF (β-catcGOF) mice compared with age-matched control (Ctrl) mice (five and six mice per group, respectively, in four independent experiments) with (A) treatment scheme, (B) BM cellularity, (C) frequency of HSPCs, and (D) frequency of myeloid progenitors (MyPs) and mature cells. B, B cell; CMP, common myeloid progenitor; MEP, megakaryocyte/erythrocyte progenitor; My, myeloid cell; ST-HSC, short-term HSC. (E–I) Cell-intrinsic effect of high Wnt activity with (E) treatment scheme of genetic activation of the Wnt pathway in a Tplx model of β-catcGOF compared with Ctrl mice (four mice per group, in three independent experiments), (F) donor chimerism 4 wk after Tplx (left) and donor-derived LSK cellularity 3 wk after Poly(I:C) injections (right), (G) frequency of donor-derived HSPCs and MyPs, (H) myeloid differentiation of donor-derived HSPCs in methylcellulose (n = 3), and (I) frequency of donor-derived mature cells (left) and total percentage of BM mature cells (right). GM, granulocyte/macrophage; G(or)M, granulocyte (or) macrophage; MegE, megakaryocyte/erythrocyte; Mix, all lineages; IR, irradiation. Results are expressed as mean ± SD; unpaired Student’s t test was used. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Increasing Notch and decreasing Wnt activity instead induces compensatory crosstalk mechanisms
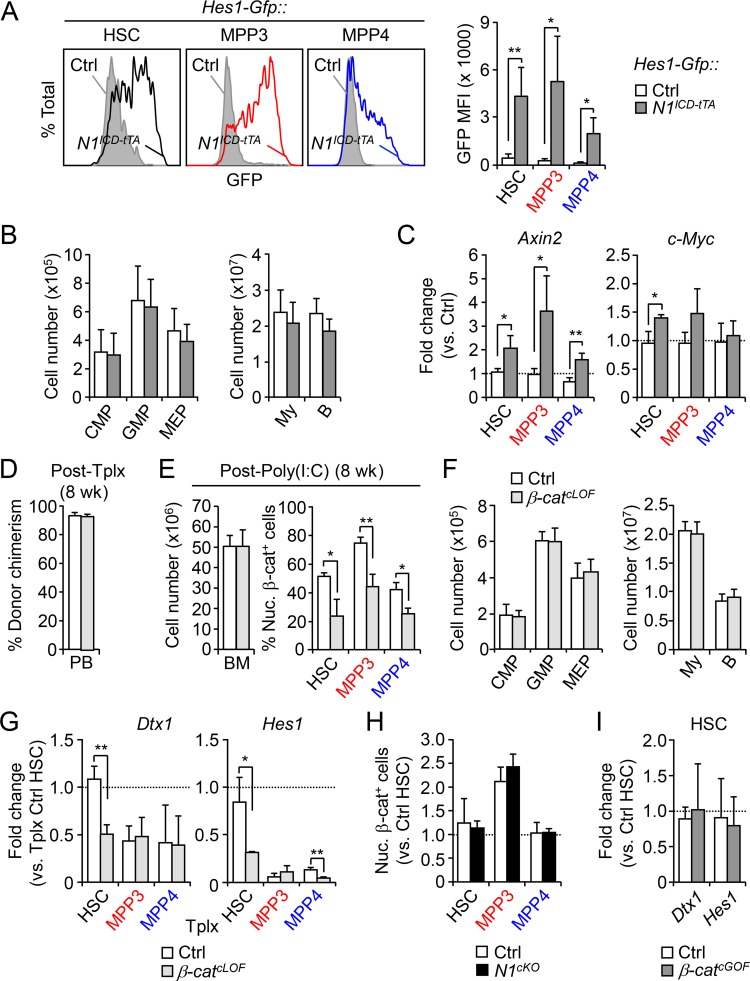
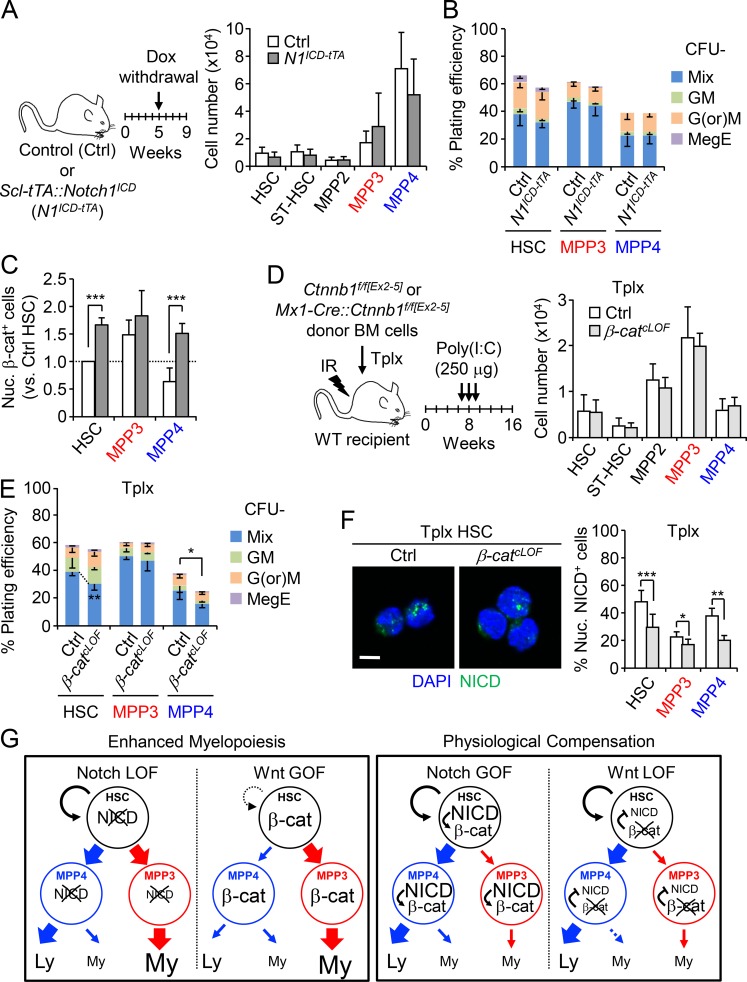
To further explore the connection between Notch and Wnt pathways, we next asked whether increasing Notch and decreasing Wnt activity could lead to opposite effects and decrease MPP3 production. We first expressed Notch1 intracellular domain (N1ICD) under the control of the HSC-specific Scl-tTA driver and confirmed increased Notch activity in HSCs, MPP3, and MPP4 using Scl-tTA::N1ICD (N1ICD-tTA) mice crossed to the Hes1-Gfp reporter line (Fig. S4 A). Surprisingly, we found that increasing Notch activity did not decrease MPP3 numbers or change any other BM populations in N1ICD-tTA mice (Fig. 6 A and Fig. S4 B). Of note, these mice also did not develop T cell acute lymphoblastic leukemia (T-ALL), as the Scl-tTA driver is not expressed in T cell populations requiring high Notch activity to drive T-ALL development (Bockamp et al., 2006; King et al., 2013). Methylcellulose assays also showed no difference in myeloid lineage output from N1ICD-tTA MPP4 compared with control MPP4 (Fig. 6 B). We then measured nuclear β-catenin levels and expression of the Wnt target genes Axin2 and c-Myc in HSCs, MPP3, and MPP4 isolated from N1ICD-tTA mice and found significantly higher Wnt activity in all N1ICD-tTA cells compared with control populations (Fig. 6 C and Fig. S4 C). These results suggest that a compensatory increase in Wnt activity could normalize the effects of elevated Notch activity, leading to unchanged MPP3 production and MPP4 myeloid differentiation potential in vivo.
Figure S4.
Cross-regulation of Notch and Wnt activity in HSPCs. (A) Representative FACS plots and quantification of Notch reporter activity (GFP MFI) in Ctrl::Hes1-Gfp and N1ICD-tTA::Hes1-Gfp HSPCs (four and three mice per group, respectively, in two independent experiments). Ctrl, control. (B and C) Consequences of genetic activation of the Notch pathway in conditional NICD (N1ICD-tTA) transgenic mice compared with age-matched Ctrl mice (five mice per group; in two independent experiments) with (B) BM myeloid progenitors and mature cell cellularity and (C) quantitative RT-PCR analysis of Wnt target genes in HSPCs (n = 4 for Ctrl and n = 3 for N1ICD-tTA, in one experiment). Results are expressed as fold change relative to respective Ctrl populations (set to 1). B, B cell; CMP, common myeloid progenitor; MEP, megakaryocyte/erythrocyte progenitor; My, myeloid cell. (D–G) Consequences of genetic inhibition of the Wnt pathway in a Tplx model of β-catenin conditional LOF (β-catcLOF) compared with Ctrl mice (five mice per group, in three independent experiments) with (D) donor chimerism at 8 wk after Tplx, (E) donor-derived BM cellularity (left) and quantification of Wnt activity based on nuclear β-catenin staining in HSPCs (right) at 8 wk after Poly(I:C) injections, (F) BM donor-derived myeloid progenitors and mature cell cellularity, and (G) quantitative RT-PCR analysis of Notch target genes in donor-derived HSPCs (n = 3 and 4, respectively). Results are expressed as fold change relative to HSC from Tplx Ctrl mice (set to 1). (H) Quantification of Wnt activity based on nuclear β-catenin staining in Ctrl and N1cKO HSPCs (n = 4, in three independent experiments). Results are expressed as fold changes relative to Ctrl HSCs (set to 1). (I) Quantitative RT-PCR analysis of Notch target genes in Ctrl and β-catcGOF HSCs (n = 3 and 4, respectively, in one experiment). Results are expressed as fold change relative to Ctrl HSC (set to 1). Results are expressed as mean ± SD; unpaired Student’s t test was used. *, P ≤ 0.05; **, P ≤ 0.01.
Figure 6.
Compensatory crosstalk between Notch and Wnt activity in controlling MPP production. (A–C) Consequences of genetic activation of the Notch pathway in conditional NICD (N1ICD-tTA) transgenic mice compared with age-matched Ctrl mice (five mice per group, in two independent experiments) with (A) treatment scheme and HSPC cellularity, (B) myeloid differentiation of Ctrl and N1ICD-tTA HSPCs in methylcellulose (n = 5, in three independent experiments), and (C) quantification of nuclear β-catenin positive (nuc. β-cat+) cells in Ctrl and N1ICD-tTA HSPCs (n = 3, in two independent experiments). Results are expressed as fold changes relative to Ctrl HSCs (set to 1). GM, granulocyte/macrophage; G(or)M, granulocyte (or) macrophage; MegE, megakaryocyte/erythrocyte; Mix, all lineages; ST-HSC, short-term HSC. (D–F) Consequences of genetic inhibition of the Wnt pathway in a Tplx model of β-catenin conditional LOF (β-catcLOF) mice compared with Ctrl mice (five mice per group; in three independent experiments) with (D) treatment scheme and donor-derived HSPC cellularity, (E) myeloid differentiation of donor-derived HSPCs from Tplx Ctrl and β-catcLOF in methylcellulose (n = 5, in two independent experiments), and (F) representative images and quantification of nuclear NICD–positive (nuc. NICD+) cells from Tplx Ctrl or β-catcLOF mice at 6 wk after Poly(I:C) injection (n = 8 for HSCs and n = 4 for MPP3 and MPP4, in two independent experiments). IR, irradiation. Scale bar, 5 µm. (G) Model depicting the two outcomes observed when manipulating Notch and Wnt signaling in HSPCs. GOF is indicated by bigger letters and LOF by cross out. Ly, lymphoid; My, myeloid. Results are expressed as mean ± SD; unpaired Student’s t test was used. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Next, we assessed the effects of decreasing Wnt activity using a conditional β-catenin loss-of-function (LOF) approach in Mx1-Cre::Ctnnb1f/f[Ex2–5] (β-catcLOF) mice, which deletes exons 2–5 of β-catenin. However, β-catcLOF mice died within 2 wk after Poly(I:C) injection, likely due to gastrointestinal issues (Fevr et al., 2007), which prevented their direct analyses. For this reason, we transplanted untreated control and Mx1-Cre::Ctnnb1f/f[Ex2–5] BM cells into lethally irradiated WT recipients, waited 8 wk for full engraftment, treated with Poly(I:C), and analyzed Tplx control and β-catcLOF mice after 2 mo (Fig. 6 D and Fig. S4, D–F). Surprisingly, lowering Wnt activity by ∼56% in HSPCs from Tplx β-catcLOF mice did not change MPP3 production or any other BM populations, although we observed reduced myeloid colony-forming capacity from MPP4 in methylcellulose assays (Fig. 6 E). Interestingly, NICD IF staining and quantitative RT-PCR analyses of Notch target genes revealed significantly decreased Notch activity in Tplx β-catcLOF HSCs and MPPs (Fig. 6 F and Fig. S4 G), suggesting that a compensatory reduction in Notch activity could help maintain unchanged MPP3 production in vivo.
Finally, we measured Wnt activity upon decreasing Notch activity in N1cKO HSCs and MPPs (Fig. S4 H) and Notch activity upon increasing Wnt activity in β-catcGOF HSCs (Fig. S4 I) but could not document any compensatory effects in these conditions of enhanced myelopoiesis. This is in sharp contrast to increasing Notch and decreasing Wnt activity, which would damage blood production and appears to be resisted much more robustly through these physiological compensation mechanisms. Taken together, these results demonstrate complex crosstalk between Notch and Wnt signaling in controlling the induction of the myeloid regeneration pathway from HSCs (Fig. 6 G). They show that both lowering Notch activity and increasing Wnt activity are needed to drive HSCs to overproduce MPP3. Moreover, they identify important compensatory crosstalk mechanisms that prevent damaging blood production and maintain the size of the MPP3 compartment and myeloid cell production in situations where Notch is overactivated or Wnt is inactivated.
Rewiring of Notch and Wnt signaling activity in leukemic and regenerating HSCs
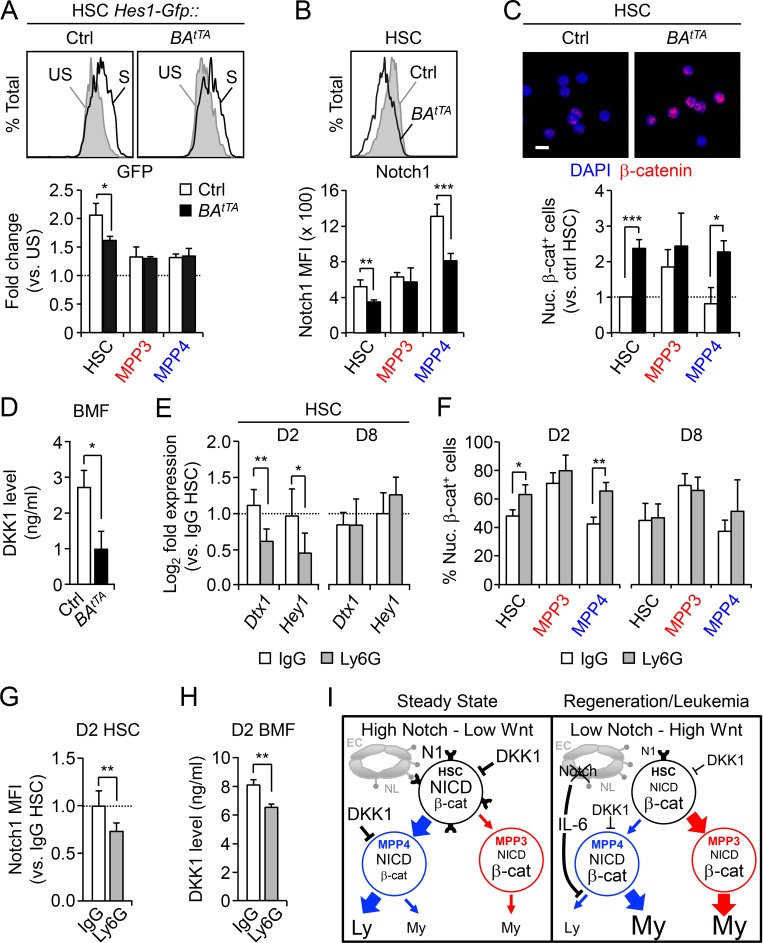
To determine whether such coordinated changes in Notch and Wnt signaling activity accompany the activation of myeloid regeneration pathways in LSCs, we focused on our inducible BAtTA CML model. To confirm decreased Notch activity (Reynaud et al., 2011), we crossed BAtTA mice with the Hes1-Gfp reporter line and stimulated freshly isolated cells with plate-bound DLL1 in vitro (Fig. 7 A). As expected, BAtTA::Hes1-Gfp HSCs were significantly less responsive to Notch stimulation than control Hes1-Gfp HSCs, while neither MPP3 nor MPP4 was significantly affected. We also found that BAtTA HSCs, like BAtTA MPP4, had significantly lower surface expression of the Notch1 receptor, which likely mediated their reduced Notch signaling activity (Fig. 7 B). Moreover, we observed a striking upregulation of nuclear β-catenin levels in both BAtTA HSCs and MPP4 (Fig. 7 C), associated with significantly reduced levels of the Wnt inhibitor DKK1 present in the BM fluid of BAtTA mice (Fig. 7 D), which probably directly contributed to their increased Wnt signaling activity. Molecular analyses of BAtTA HSCs further confirmed the downregulation of Notch pathway genes, especially for genes involved in Notch ligand-receptor binding processes, and upregulation of Wnt pathway genes including the ligand coreceptor Lrp5 and Wnt target gene Axin2 (Fig. S5, A and B). These deregulated features were largely shared by leukemic junBΔ/Δ HSCs, with decreased Notch1 surface expression and reduced Notch signaling activity in LSCs and increased Wnt activity in leukemic junBΔ/Δ HSCs and MPP4 (Fig. S5, C–E). Moreover, we observed similar molecular trends of decreased Notch activity and increased Wnt activity in both CML (GSE4170) and AML (GSE17054) patient samples (Fig. S5, F and G). Altogether, these data suggest that lower Notch activity and higher Wnt activity are common features of LSCs in both mice and humans and are likely the result of changes in the way transformed cells respond to regulators of these pathways expressed by their leukemic BM niche microenvironment.
Figure 7.
Lower Notch and higher Wnt activity in leukemic and regenerating HSCs. (A) Representative FACS plots and quantification of Notch reporter activity in Ctrl::Hes1-Gfp and BAtTA::Hes1-Gfp HSCs cultured for 18 h ± plate-bound DLL1 ligands (n = 3, in two independent experiments). Results are expressed as fold changes in GFP MFI relative to unstimulated conditions (set to 1). Ctrl, control; S, stimulated; US, unstimulated. (B) Representative FACS plots and quantification of Notch1 surface expression in Ctrl and BAtTA HSPCs (four and five mice per group, respectively, in two independent experiments). (C) Representative images and quantification of nuclear β-catenin–positive (nuc. β-cat+) cells in freshly isolated Ctrl and BAtTA HSPCs (n = 5 for HSC, n = 4 for MPP3, n = 3 for MPP4, in five independent experiments). Results are expressed as fold changes relative to Ctrl HSCs (set to 1). Scale bar, 10 µm. (D) DKK1 level in the BM fluid (BMF) of Ctrl and BAtTA mice (n = 4, in four independent experiments). (E) Quantitative RT-PCR analysis of Notch target genes in HSCs from D2 and D8 IgG- or Ly6G-injected mice (n = 5 for D2 IgG and Ly6G, n = 3 for D8 IgG, n = 5 for D8 Ly6G, in two independent experiments). Results are expressed as fold change relative to HSCs from IgG-injected mice (set to 1). (F) Percentage of nuclear β-catenin positive cells in HSCs from D2 and D8 IgG- or Ly6G-injected mice (n = 3, in three independent experiments). (G) Notch1 surface expression in HSCs from D2 IgG- or Ly6G-injected mice (four and six mice per group, respectively, in two independent experiments). Results are expressed as fold change relative to IgG HSCs (set to 1). (H) DKK1 level in BMF of D2 IgG- or Ly6G-injected mice (n = 3, in one experiment). (I) Model depicting the cell-intrinsic and cell-extrinsic regulations of Notch and Wnt pathways in HSPCs at steady state and during regeneration or leukemia development. EC, endothelial cell; Ly, lymphoid; My, myeloid; NL, Notch ligand; N1, Notch1 receptor. Results are expressed as mean ± SD; unpaired Student’s t test was used. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Figure S5.
Deregulated Notch and Wnt activity in leukemic HSCs. (A) SABiosciences PCR array of Notch pathway–related genes in control (Ctrl) and BAtTA HSCs (n = 6, in two independent experiments). Results are expressed as log2 mean fold expression relative to Ctrl HSCs (set to 0). (B) Fluidigm analysis of Wnt pathway genes in Ctrl and BAtTA HSCs (n = 6, four pools of 100 cells per mouse, in two independent experiments). Results are expressed as fold change relative to Ctrl HSCs (set to 1). (C) Notch1 surface expression in Ctrl and junBΔ/Δ HSPCs (five mice per group, in one experiment). (D) SABiosciences PCR array of Notch pathway–related genes in Ctrl and junBΔ/Δ HSCs (n = 3, in one experiment). Results are expressed as log2 mean fold expression relative to Ctrl HSCs (set to 0). (E) Percentage of Nuc. β-cat+ cells in Ctrl and junBΔ/Δ HSPCs (n = 3, in three independent experiments). Results are expressed as fold change relative to Ctrl HSCs (set to 1). (F and G) Heat map of significantly differentially expressed Wnt and Notch pathway–related genes in CML (GSE4170) and AML (GSE17054) patient samples (false discovery rate <0.05). Results are expressed as fold change of (F) chronic phase CML BM relative to healthy CD34+ BM cells and (G) isolated AML LSCs (Lin−/CD34+/CD38−/CD123+) relative to healthy BM HSCs (Lin−/CD34+/CD38−/CD90low). (H) Quantitative RT-PCR analysis of Notch target genes in MPP3 and MPP4 from D2 and D8 IgG- or Ly6G-injected mice (n = 4 for D2 IgG, n = 6 for D2 Ly6G, n = 3 for D8 IgG and Ly6G, in one experiment). Results are expressed as fold change relative to HSCs from IgG-injected mice (set to 1). Results are expressed as mean ± SD except in A and D; unpaired Student’s t test was used. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Next, we investigated Notch and Wnt signaling activity in regenerating Ly6G-treated WT HSCs. Similar to leukemic HSCs, we observed the downregulation of Notch target genes such as Dtx1 and Hey1 in day 2 HSPCs and increased nuclear β-catenin levels in both day 2 HSCs and MPP4 (Fig. 7, E and F; and Fig. S5 H). However, in contrast to the constant activation observed in disease context, these changes in signaling activity were transient and were fully restored to normal levels at day 8. In addition, day 2 HSCs had lower levels of Notch1 surface expression, likely contributing to their reduced Notch signaling activity, and the BM fluid of day 2 Ly6G-treated mice showed significantly reduced DKK1 levels, also likely contributing to the increased Wnt activity observed in regenerating day 2 HSCs and MPP4 (Fig. 7, G and H). Taken together, these results highlight the similarity between regeneration and leukemia development in activating myeloid regeneration, with the engagement of similar cell-intrinsic and cell-extrinsic regulatory mechanisms modulating Notch and Wnt activity in both acute and chronic contexts (Fig. 7 I).
Restoring Notch and Wnt deregulated activity attenuates disease progression
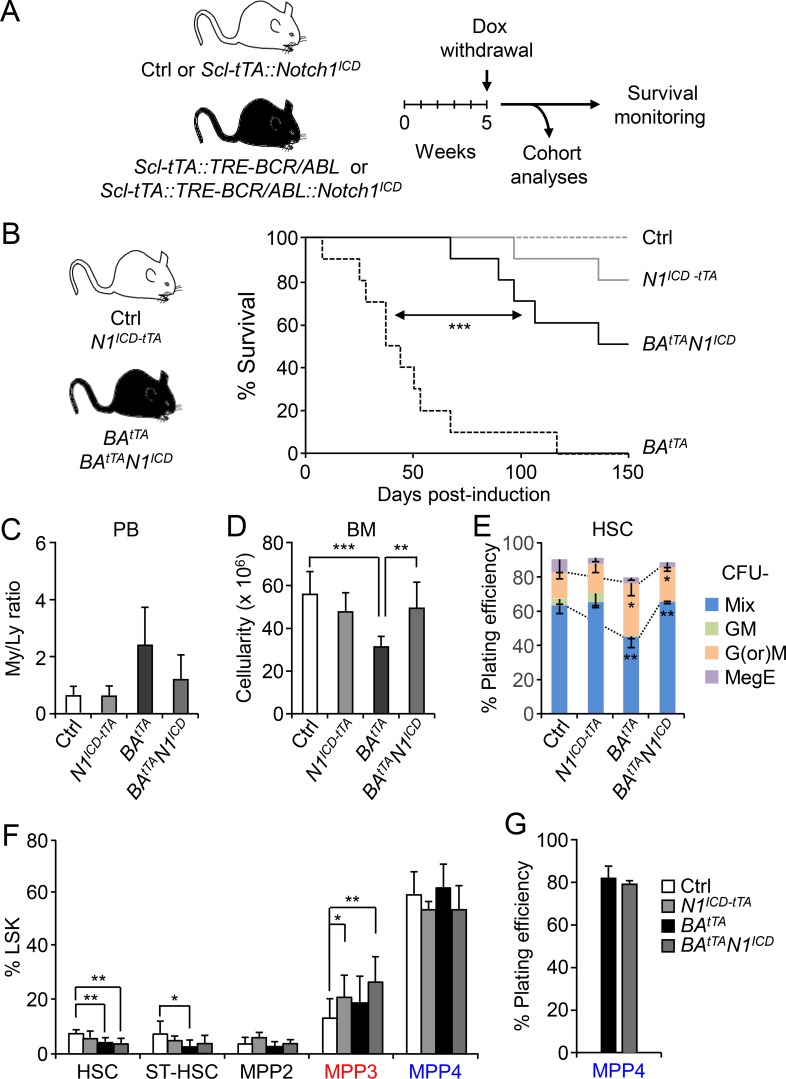
Finally, we investigated whether restoring the deregulated activity of the Notch and Wnt pathways in LSCs could have beneficial effects on disease progression. To increase Notch activity, we crossed N1ICD with BAtTA mice to generate BAtTAN1ICD and littermate BAtTA diseased mice. In parallel, we used N1ICD-tTA and littermate controls that were similarly induced by doxycycline (Dox) withdrawal 5 wk after birth and followed them for up to 150 d (Fig. 8 A). Remarkably, restoring Notch activity significantly delayed disease onset and provided a clear survival benefit, with 60% of BAtTAN1ICD mice (6 of 10) surviving at a time when all of the BAtTA mice (10 of 10) were dead (Fig. 8 B). To understand the mechanisms driving this rescue, we analyzed all the genotypes in each cohort when BAtTA animals reached a disease burden of ∼40%–50% granulocytes in their PB (average of 50 d after induction; n = 5 independent cohorts). Strikingly, increasing Notch activity in BAtTAN1ICD mice decreased the relative number of circulating myeloid cells and allowed recovery of BM cellularity (Fig. 8, C and D). The myeloid output of BAtTAN1ICD HSCs was also restored, with reduced mature granulocyte or macrophage colonies formed in the methylcellulose assay compared with BAtTA HSCs (Fig. 8 E). However, we did not observe a reduction in MPP3 frequency, which actually increased, or a change in MPP4 myeloid differentiation potential in BAtTAN1ICD mice compared with BAtTA mice (Fig. 8, F and G), which was likely due to the high Wnt activity already displayed by these populations and the engagement of compensatory crosstalk mechanisms designed to maintain MPP3 production. As such, these results demonstrate that increasing Notch activity in LSCs can directly attenuate the aberrant activation of the myeloid regeneration pathway and reduce myeloid cell production, without significantly reducing the size of the leukemic MPP3 compartment.
Figure 8.
Restoring Notch activity attenuates leukemia progression. (A) Schematic for the generation and analyses of BAtTAN1ICD mice and associated cohort littermates. Ctrl, control. (B) Survival curve upon genetic activation of the Notch pathway by crossing Ctrl and BAtTA mice with the conditional N1ICD-tTA transgenic mice (10 mice per group, in five independent experiments). Induction, Dox withdrawal. (C–E) Analyses of five independent cohorts showing (C) the ratio of myeloid/lymphoid (My/Ly) cells in PB (11 mice per group for Ctrl, 3 for N1ICD-tTA and BAtTAN1ICD, and 10 for BAtTA), (D) BM cellularity (11 mice per group for Ctrl, 5 for N1ICD-tTA and BAtTAN1ICD, and 9 for BAtTA), and (E) HSC myeloid differentiation in methylcellulose (n = 4 for Ctrl, n = 3 for all the other groups). GM, granulocyte/macrophage; G(or)M, granulocyte (or) macrophage; MegE, megakaryocyte/erythrocyte; Mix, all lineages. (F) Quantification of BM HSPCs in BAtTAN1ICD mice and cohort littermates (11 mice/group for Ctrl, 5 for N1ICD-tTA and BAtTAN1ICD, and 9 for BAtTA, in five independent experiments). ST-HSC, short-term HSC. (G) Myeloid differentiation of MPP4 from BAtTA and BAtTAN1ICD mice in methylcellulose (n = 3, in three independent experiments). Results are expressed as mean ± SD; unpaired Student’s t test was used. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
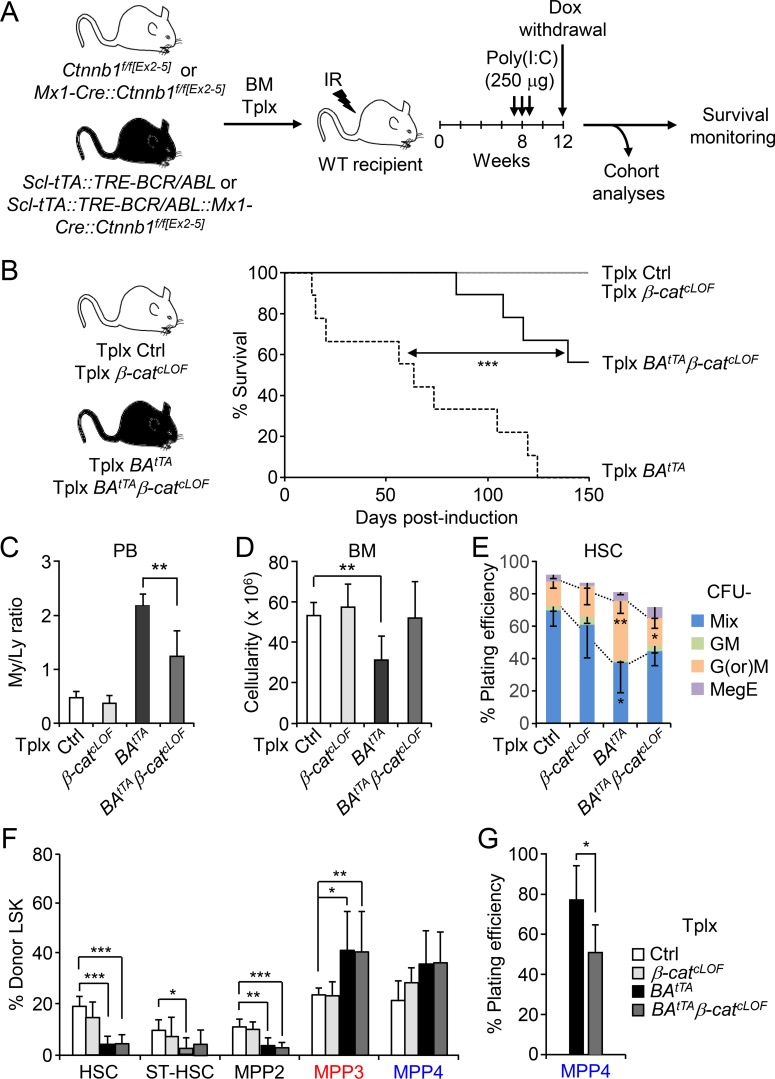
To decrease Wnt activity, we first crossed Mx1-Cre::Ctnnb1f/f[Ex2–5] mice with Scl-tTA::TRE-BCR/ABL mice to generate Scl-tTA::TRE-BCR/ABL::Mx1-Cre::Ctnnb1f/f[Ex2–5] mice. We then transplanted BM cells from these animals and littermate controls, Mx1-Cre::Ctnnb1f/f[Ex2–5] and Scl-tTA::TRE-BCR/ABL controls, into lethally irradiated recipients, waited 2 mo for full engraftment, injected Poly(I:C) to inactivate β-catenin, and a month later withdrew Dox to induce BCR/ABL expression and followed these mice for up to 150 d (Fig. 9 A). Similar to Notch restoration, decreasing Wnt activity also significantly delayed disease onset and provided a clear survival benefit, with ∼67% of Tplx BAtTAβ-catcLOF mice (six of nine) surviving at a time when all of the Tplx BAtTA mice (nine of nine) were dead (Fig. 9 B). Cohort analyses performed when Tplx BAtTA mice reached ∼40–50% disease burden (average of 60 d after induction; n = 4 independent cohorts) revealed significantly reduced levels of circulating myeloid cells in the PB and recovery of BM cellularity in Tplx BAtTAβ-catcLOF mice compared with Tplx BAtTA mice and partial restoration of the myeloid output of Tplx BAtTAβ-catcLOF HSCs in methylcellulose, with decreased production of mature granulocyte and macrophage colonies (Fig. 9, C–E). However, we also found no changes in MPP3 frequency in Tplx BAtTAβ-catcLOF mice, as predicted from the low Notch activity already displayed by these populations and the engagement of compensatory crosstalk mechanisms designed to maintain MPP3 production, but we observed reduced myeloid differentiation of Tplx BAtTAβ-catcLOF MPP4, again as expected for a loss of Wnt signaling activity in this population (Fig. 9, F and G). Taken together, these results show that increasing Notch activity or decreasing Wnt activity in LSCs has very similar effects in correcting myeloid cell production, delaying disease progression, and improving overall survival.
Figure 9.
Normalizing Wnt activity also mitigates leukemia progression. (A) Schematic for the generation and analyses of Tplx BAtTAβ-catcLOF and associated cohort littermates. IR, irradiation. (B) Survival curve upon genetic inhibition of the Wnt pathway by crossing control (Ctrl) and BAtTA mice with the β-catenin conditional LOF (β-catcLOF) mice and using a Tplx model (10 mice per group for β-catcLOF, 9 for all the other groups in two independent experiments). Induction, Dox withdrawal. (C–E) Analyses of four independent cohorts showing (C) the ratio of myeloid/lymphoid (My/Ly) cells in PB (five mice per group for Ctrl and BAtTAβ-catcLOF, four for β-catcLOF and BAtTA), (D) BM cellularity (five mice per group for Ctrl and BAtTAβ-catcLOF, four for β-catcLOF and BAtTA), and (E) HSC myeloid differentiation in methylcellulose (n = 4 for Ctrl; n = 3 for all the other groups). GM, granulocyte/macrophage; G(or)M, granulocyte (or) macrophage; MegE, megakaryocyte/erythrocyte; Mix, all lineages. (F) Quantification of BM HSPCs in Tplx BAtTAβ-catcLOF and cohort littermates (five mice per group for Tplx Ctrl and BAtTAβ-catcLOF, four for Tplx β-catcLOF and BAtTA, in four independent experiments). ST-HSC, short-term HSC. (G) Myeloid differentiation of MPP4 from Tplx BAtTA and BAtTAβ-catcLOF mice in methylcellulose (n = 4, in four independent experiments). Results are expressed as mean ± SD; unpaired Student’s t test was used. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Discussion
Here, we provide a novel understanding of the role of the Notch and Wnt pathways in controlling HSC fate decisions, particularly the differential production of lineage-biased MPPs and their function in regulating myeloid cell production. We show that lowering Notch and increasing Wnt activity in HSCs triggers the early stage of myeloid regeneration associated with increased production of MPP3. We also identify compensatory crosstalks between Notch and Wnt activities that prevents damaging MPP function and blood production when Notch is overactivated or Wnt inactivated. Moreover, we demonstrate that myeloid leukemia hijacks such emergency pathways, which are normally used transiently for myeloid regeneration, and repurpose both their cellular mechanisms and their molecular regulators for malignant outgrowth. Altogether, we identify new targets that could be used to limit myeloid cell production and aberrant LSC activity for the treatment of leukemia patients.
Despite a long-lasting and broad interest in addressing Notch and Wnt functions in HSC biology, the emerging picture has remained quite confusing (Lampreia et al., 2017; Lento et al., 2013). This is in large part because, with a few exceptions, most studies have investigated these two signaling mechanisms in isolation, focusing mainly on changes in HSC Tplx or “self-renewal” ability. In fact, HSC Tplx is a complex process that is affected by changes in different aspects of HSC biology, including cell cycle and metabolic activation, survival rates, homing, and engraftment ability (Orford and Scadden, 2008). Since both Notch and Wnt play important, sometime divergent, and often redundant roles in many of these individual aspects, which are not yet all fully decrypted, it is therefore not surprising that no clear trend has yet emerged regarding their role in a complex process such as HSC self-renewal. Here, we focus on the function of Notch and Wnt in controlling lineage commitment and differential production of lineage-biased MPPs downstream of HSCs. We show that relatively modest but consistent changes in their activity levels (i.e., decreased Notch and increased Wnt activity levels) trigger early myeloid regeneration pathways associated with the overproduction and likely accelerated differentiation of MPP3. In particular, lowering Notch activity increases MPP3 production without overtly altering HSC maintenance, while increasing Wnt activity also increases MPP3 production but at the direct expense of HSC maintenance and engraftment potential (Luis et al., 2011) and MPP4 production. In fact, increasing Wnt activity directly activates the cell cycle machinery and likely drives HSC functional exhaustion through deregulated proliferation (Orford and Scadden, 2008), while decreasing Notch activity essentially impacts quiescence-enforcing mechanisms, particularly in inflammatory contexts, which might allow HSCs to start cycling without losing their functionality (Pietras, 2017). This might explain why changes in Notch activity are often observed in LSCs, as they do not lead to compromised HSC fitness while still increasing myelopoiesis (Lobry et al., 2014).
We also found that lowering Notch activity through the Notch1 receptor, but not the Notch2 receptor, increases MPP3 production. This is in contrast to a previous study reporting a role for Notch2, but not Notch1, in inhibiting myeloid differentiation (Varnum-Finney et al., 2011). This discrepancy could be due to differences in the experimental system (in vitro versus in vivo) or the readout used in each study (MPP levels in the BM versus mature myeloid cells in the PB), differences in Notch1 deletion efficiency, which likely underlie many other conflicting results in the field, or the confounding effects of cytokine cocktails used in vitro as already described (Bigas et al., 1998). Interestingly, neither decreasing Notch activity nor increasing Wnt activity lead to compensatory effects on the other signaling pathways, clearly highlighting their unique role in HSC regulation. This is in contrast to increasing Notch activity, which results in increased Wnt activity, or lowering Wnt activity, which leads to a concomitant decrease in Notch activity. These physiological compensatory effects result in unchanged levels of MPP production and lineage output at steady state, and may also contribute to some of the confounding effects that were observed when manipulating these pathways, including the increased HSC engraftment found in various Notch GOF mice (Kunisato et al., 2003; Calvi et al., 2003; Varnum-Finney et al., 2003). They raise the intriguing possibility that HSC self-renewal may be directly regulated by the ratio of Notch versus Wnt activity and not just by the absolute level of Notch or Wnt signaling. This concept of a balance between Notch and Wnt levels determining the net outcome of HSC functionality might also help untangle some of the current controversy regarding the role of these two pathways in HSC biology.
Our results also demonstrate the important intrinsic role played by the Wnt pathway in tuning the lineage potential of MPP4 in both regenerative and leukemic settings, with increased Wnt activity blocking lymphoid commitment and decreased Wnt activity impairing myeloid commitment from this highly reprogrammable population. In contrast, Notch signaling appears dispensable for modulating MPP4 fate intrinsically, although it can act extrinsically to modulate MPP4 myeloid output by controlling the production of proinflammatory cytokines like IL-6 by BM niche cells (Wang et al., 2014). Strikingly, MPP3s are mostly refractory to changes in Notch and Wnt activity, with a rigid myeloid lineage identity and differentiation rates that can be accelerated by extrinsic signaling through pro-myeloid differentiation cytokines like IL-1 (Pietras et al., 2016). This more prominent role of Wnt in controlling the lineage output of downstream progenitors, compared with Notch, extends to GMPs, where it plays a key role in GMP cluster formation (Hérault et al., 2017). Taken together, our results identify a complex interplay between Notch and Wnt signaling and cell-intrinsic versus-extrinsic regulations in controlling the differential production of lineage-biased MPPs by HSCs and in shaping blood output at steady state and in regenerating contexts. They show that attempts to perturb myeloid pathways through increased Notch or decreased Wnt are resisted much more robustly than attempts at enhancing myelopoiesis through decreased Notch or increased Wnt, most likely via the physiological compensatory mechanisms that we describe here. It will be interesting to determine whether similar mechanisms and changes in MPP production also occur during ontogeny (Bigas et al., 2013) and aging (Geiger et al., 2014), two other stages of hematopoietic development where both Notch and Wnt have also been shown to play important functions. Moreover, given the crucial roles of Notch and Wnt pathways in stem cell biology, our findings may have broad implications for other stem cell populations and for malignancy development in a range of tissues.
Our findings further emphasize that malignancies do not create a new modality of cell production but hijack existing cellular and molecular mechanisms of regeneration, which they repurpose for disease output. We show that aberrant myeloid cell production in leukemia results from the chronic activation of normally transiently induced myeloid regeneration pathways, with the same activation mechanisms converging on decreased Notch and increased Wnt activity. Here, we found that reduced expression of the Notch1 receptor is a mechanism likely contributing to dampening Notch signaling in HSCs, acutely in regenerating HSCs, and in a constitutive manner in leukemic HSCs. However, similar outcomes could be elicited by other mechanisms, including by decreasing the expression of ligands for the Notch1 receptor in BM niche cells, especially osteoblastic lineage and endothelial cells (Schepers et al., 2015). We also found decreased levels of the Wnt inhibitor DKK1 in the BM niche as one possible mechanism contributing to higher Wnt activity in both regenerating and leukemic HSCs. Again, this is not an exclusive mechanism, since other ways of increasing Wnt activity could result in similar activations of myeloid regeneration pathways. In fact, it is likely that each disease entity will exhibit its own set of molecular alterations, converging on decreased Notch and increased Wnt activity in LSCs as already described (Abrahamsson et al., 2009; Majeti et al., 2009) and confirmed here in reanalyzed published patient datasets. It will now be important to confirm that such low Notch and high Wnt activity in human LSCs triggers the constitutive activation of similar myeloid regeneration pathways at the cellular level. This is an important, but not easy, task since the current understanding of lineage-biased MPPs in the human system is still very limited (Notta et al., 2016).
Finally, our results identify a common mechanism for myeloid leukemia development that could provide new treatment options to limit aberrant myeloid cell production. This is particularly relevant for patients with no identified driver mutations or who relapsed with resistance mutations, and to provide a complementary, likely safer, approach for myeloid tumor debulking and prolonging time to progression. Here, we show that correcting the deregulated signaling activity in CML LSCs by increasing Notch and decreasing Wnt activity limits myeloid cell production, delays disease progression, and improves survival, hence providing a proof of concept in mouse models that tuning down myeloid regeneration pathways can have direct benefits. This aligns well with previous studies showing that β-catenin inhibition can potentiate the effects of CML treatment by reversing tyrosine kinase inhibitor resistance (Zhou et al., 2017) or synergizes with FLT-3 inhibition in AML treatment (Jiang et al., 2018), hence reenforcing our idea that modulating the myeloid regeneration pathway by restoring Notch and Wnt activity could complement current therapies. We also demonstrate that the same signaling restoration has no effects on normal HSC function while dampening aberrant myeloid cell production from LSCs. In this context, it is interesting to note that the size of the leukemic MPP3 compartment is a poor predictor of treatment efficacy, likely as a side consequence of the engagement of physiological crosstalk mechanisms between these signaling pathways designed to dampen the consequence of elevated Notch and decreased Wnt activity. The next step in translating these findings for clinical applications will be to identify druggable targets that could appropriately increase Notch activity while limiting Wnt activity. This would provide an exciting new avenue for leukemia treatment and a specific anti-LSC therapy that would block their enhanced and overly biased differentiation potential.
Materials and methods
Experimental model and subject details
Mice
CD45.2 C57BL/6J, CD45.1 C57BL/6-BoyJ, B6.Cg-Tg(Mx1-cre)1Cgn/J mice were originally purchased from the Jackson Laboratory and bred in our animal facilities, either at University of California San Francisco (UCSF) or Columbia University Irving Medical Center (CUIMC). Ctnnb1flox/flox[Ex3] (Harada et al., 1999) and Ctnnb1flox/flox[Ex2–5] (Brault et al., 2001) mice were obtained from Dr. Diana Laird (UCSF, San Francisco, CA); TRE-Notch1ICD (Carlson et al., 2005) and Notch1tm2Rko/GridJ (Radtke et al., 1999) mice from Dr. Rong Wang (UCSF, San Francisco, CA); Hes1-Gfp reporter mice (Oh et al., 2013) from Dr. Ioannis Aifantis (New York University, New York, NY); and Irf8−/− mice (Holtschke et al., 1996) from Dr. Harrinder Singh (Cincinnati Children’s Hospital, Cincinnati, OH), and were bred in our animal facilities. Scl-tTA::TRE-BCR/ABL (Reynaud et al., 2011) and More-Cre::JunBflox/flox (Santaguida et al., 2009) mice were described previously. Scl-tTA::TRE-BCR/ABL mice were bred with TRE-Notch1ICD mice to produce Scl-tTA::TRE-BCR/ABL::TRE-Notch1ICD mice, and Mx1-CRE::Ctnnb1flox/flox[Ex2–5] mice with Scl-tTA::TRE-BCR/ABL to produce Scl-tTA::TRE-BCR/ABL::Mx1-CRE::Ctnnb1flox/flox[Ex2–5]mice. Six- to 12-wk-old mice were used as donors for cell isolation and in vivo experiments, and 8- to 12-wk-old congenic mice were used as recipients for Tplx experiments. Respective littermates or age-matched animals were used as controls. Bones from Mx1-CRE::KrasLSL-G12D mice (Braun et al., 2004) were provided by Dr. Benjamin Braun (UCSF, San Francisco, CA), and bones from Vav-CRE::Jak2+/V617F mice (Mullally et al., 2010) by Dr. Anne Mullally (Dana-Farber Cancer Institute, Boston, MA).
No specific randomization or blinding protocol was used, and both male and female animals were used indifferently in the study. For Mx1-cre-mediated deletion, 4- to 6-wk-old Mx1-Cre–positive and –negative mice were injected intraperitoneally three times every other day with 250 µg Poly(I:C) (27473201; GE Healthcare) in PBS, except for Ctnnb1flox/flox[Ex3] mice, which were only injected once with 62.5 µg Poly(I:C). Poly(I:C) injections were completed at least 4 wk before each experiment, and Poly(I:C)-injected Mx1-Cre–negative mice were used as controls. For BCR/ABL induction, mice were withdrawn from Dox containing water at 5 wk of age. All mice were maintained in mouse facilities at UCSF or CUIMC in accordance with Institutional Animal Care and Use Committee protocols approved at each institution.
Method details
In vivo assays
For granulocyte depletion, mice were injected once intraperitoneally with 0.1 mg of anti-Ly6G antibody or IgG control (UCSF monoclonal antibody core) in 200 µl PBS. For Notch inhibition, mice were treated with either the γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycine t-butyl ester (DAPT; D5942; Sigma-Aldrich) or specific anti-Notch1 or Notch2 blocking antibodies (Wu et al., 2010). DAPT was reconstituted in DMSO at 20 mg/ml and diluted with 10% ethanol in corn oil (Sigma-Aldrich) to be administered intraperitoneally at a dose of 10 mg/kg per day, using a 3 d ON/3 d OFF schedule for up to 21 d to reduce the gastrointestinal toxicity associated with γ-secretase inhibition. Mice similarly injected with vehicle (10 µl DMSO diluted with 10% ethanol in corn oil) were used as controls. Anti-Notch1 or Notch2 blocking antibodies were administered intraperitoneally at a dose of 5 mg/kg every 3 d for up to 28 d. Mice similarly injected with anti-ragweed mouse IgG2a isotype were used as controls. For Tplx experiments, recipient mice were either lethally irradiated (11 Gy, delivered in split doses 3 h apart) and injected retro-orbitally with 2 × 106 BM cells or sublethally irradiated (8.5 Gy, delivered in split doses 3 h apart) and injected retro-orbitally with 2,000 HSCs within the next 12 h following irradiation. A 137Cs source irradiator (J.L. Shepherd) or an x-ray irradiator (Faxitron) was used at UCSF and Columbia University, respectively. Transplanted mice were maintained on antibiotic-containing water for 4 wk and analyzed for donor-derived chimerism by bleeding. PB was obtained from the retro-orbital plexus and collected in tubes containing 4 ml of ammonium-chloride-potassium lysis buffer (150 mM NH4Cl and 10 mM KHCO3) supplemented with 10 mM EDTA for flow cytometry analyses.
Flow cytometry
Staining of hematopoietic cells was performed as described previously (Hérault et al., 2017). Briefly, BM cells were obtained by either crushing leg, arm, and pelvic bones together or flushing leg bones in HBSS containing 2% heat-inactivated FBS (Sigma-Aldrich), staining buffer that was then used for all subsequent incubation and washing steps. Single-cell suspensions of thymocytes and splenocytes were obtained upon mechanical dissociation. Erythrocytes were lysed with ammonium-chloride-potassium lysis buffer (150 mM NH4Cl, and 10 mM KHCO3), and contaminating crushed bone fragments were removed by centrifugation on a Ficoll gradient (Histopaque 1119; Sigma-Aldrich). Cellularity was determined by ViCELL-XR automated cell counter (Beckman-Coulter). For sorting, BM cells were labeled with CD117 (c-Kit) microbeads (Miltenyi Biotec) and enriched for c-Kit+ cells with an automated AutoMACS (Miltenyi Biotec). For immunophenotyping or sorting for immature cells, unfractionated or c-Kit–enriched BM cells were incubated with a cocktail of unconjugated rat anti-lineage (Lin) antibodies (CD3, CD5, CD8, B220 from BioLegend, and CD4, Ter-119, Gr-1, and Mac-1 from eBioscience) followed by goat anti–rat-PE-Cy5 (A10691; Invitrogen) and subsequent blocking with purified rat IgG (Sigma-Aldrich). Cells were then stained with Sca-1-PB (108120; BioLegend), c-Kit-APC-Cy7 (105826; BioLegend), CD48-A647 (103416; BioLegend), CD150-PE (115904; BioLegend), and Flk2-Bio (13–1351-85; eBioscience) followed by SA-Qdot605 (Q10101MP; Invitrogen), CD34-FITC (11–0341-85; eBioscience), and FcγR-PerCP-eFluor710 (46–0161-82; eBioscience). For mature cell analyses, BM cells were stained with Gr-1-eFluor450 (48–5931-82; eBioscience), Mac-1-PE-Cy7 (25–0112-82; eBioscience), B220-APC-eFluor780 (47–0452-82; eBioscience), and CD19-PerCP-Cy5.5 (551001; BD PharMingen). For immunophenotyping or sorting from Hes1-Gfp reporter mice, BM cells were stained with Lin/PE-Cy5, c-Kit-APC-Cy7, Sca-1-PB, Flk2-Bio/SA-Qdot605, CD48-A647, and CD150-PE. For thymus T cells, thymocytes were stained with CD4-Bio (13–0041-82; eBioscience)/SA-Qdot605 and CD8-PE (100708; BioLegend). For Notch1 surface staining, BM cells were stained with Lin/PE-Cy5, c-Kit-APC-Cy7, Sca-PE-Cy7 (108114; BioLegend), Flk2-PE (12–1351-82; eBioscience), CD48-PB (103418; BioLegend), CD150-APC (115910; BioLegend), and Notch1-Bio (130605; BioLegend) followed by SA-Qdot605. To follow granulocyte recovery after anti-Ly6G depletion, PB cells were stained with Gr-1-eFluor450, Mac-1-PE-Cy7, and Ter-119-PE-Cy5 (15–5921-83; eBioscience). For donor-derived chimerism analyses in transplanted mice, PB cells were stained with Gr-1-eFluor450, Mac-1-PE-Cy7, B220-APC-eFluor780, CD3-eFluor660 (50–0032-82; eBioscience), Ter-119-PE-Cy5, CD45.1-PE (12–0453-83; eBioscience), and CD45.2-FITC (11–0454-85; eBioscience). For donor-derived HSPC analyses, BM cells were stained with Lin/PE-Cy5, c-Kit-APC-Cy7, Sca-1-PB, Flk2-Bio/SA-Qdot605, CD48-A647, CD150-PE, CD45.1-PE, and CD45.2-FITC. For validation of anti-Notch1 and Notch2 blocking antibody effects, thymocytes were stained with CD4-FITC (553729; BD PharMingen) and CD8-PE and splenocytes with B220-APC-eFluor780 (47–0452-82; eBioscience), CD21-PE (12–0219-42; eBioscience), and CD23-PE-Cy7 (25–0232-82; eBioscience). Cells were finally resuspended in 2% FBS/HBSS containing 1 µg/ml propidium iodide to exclude dead cells. Cell sorting was performed on FACS Aria II (Becton Dickinson), and each population was double-sorted to maximize cell purity. All data were collected on FACS Aria II, LSR II, or Celesta (Becton Dickinson) and analyzed using FlowJo (Treestar). We also confirmed that changing Notch and Wnt activity did not change the expression of surface markers used to identify HSCs, MPP3, and MPP4.
Ex vivo assays
All cultures were performed at 37°C in a 5% CO2 water jacket incubator (Thermo Scientific). Cells were grown in IMDM containing 5% FBS (StemCell Technology), 50 U/ml penicillin, 50 µg/ml streptomycin, 2 mM l-glutamine, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, and 50 µM 2-mercaptoethanol, supplemented with stem cell factor (25 ng/ml), thrombopoietin (25 ng/ml), Flt3-L (25 ng/ml), IL-11 (25 ng/ml), IL-3 (10 ng/ml), GM-CSF (10 ng/ml), and erythropoietin (4 U/ml; all from PeproTech). Wnt3a (1324-WNP/CF; R&D Systems) and Wnt5a (645-WN/CF; R&D Systems) were added to the liquid culture at 1 µg/ml for testing Notch activity upon Wnt ligand stimulation using cells isolated from Hes1-Gfp reporter mice or at 3 µg/ml for measuring nuclear β-catenin levels and the Wnt inhibitor CHIR99021 (S1263; Selleckchem) at 30 μΜ. Cells were directly sorted into 96-well plates (3,000 cells per well) containing 200 µl of culture media with the indicated recombinant protein or drug and incubated for 18 h before analyses by flow cytometry or IF staining. For DLL1 (5026-DL-050; R&D Systems) treatment, the 96-well plate was first precoated with 4 µg/ml DLL1 using fibronectin (F1141; Sigma-Aldrich) for 1 h at 37°C and washed with PBS three times, and then cells were sorted into the plate (3,000 cells per well) containing 200 µl of culture media and incubated for 18 h before analyses by flow cytometry or IF staining. For methylcellulose colony assays, cells were plated into 35-mm dish (100 cells per dish) containing 1 ml methylcellulose (M3231; StemCell Technologies) supplemented with penicillin/streptomycin and the cytokines described above. For clonogenic methylcellulose colony assays, single cells were directly sorted into each well of a 96-well plate containing 100 µl of methylcellulose. Colonies were manually counted under a microscope after 8–9 d of culture. For CFSE dilution assay, sorted MPP3 were labeled with 2.5 µM CFSE (C1157; Molecular Probes) as described previously (Pietras et al., 2016) and cultured for 3 d before analyses.
IF staining
For β-catenin staining, cells (2,000–3,000 cells per slide) were pipetted onto poly-l-lysine–coated slides (P0425-72EA; Sigma-Aldrich), settled down for 15 min at room temperature, fixed with 4% paraformaldehyde for 10 min at room temperature, then washed three times with PBS and permeabilized and blocked for 1 h at room temperature with 0.1% Tween-20 in 10% FBS/PBS, which was then used as antibody incubation buffer for all the subsequent steps. Cells were then incubated overnight at 4°C with a rabbit anti-mouse β-catenin (9582S; Cell Signaling) primary antibody, washed three times with PBS, and incubated for 1 h at room temperature with a donkey anti-rabbit-A555 (A31572; Invitrogen) secondary antibody. After washing three times with PBS, slides were mounted with VectaShield (H-1200; Vector Laboratories) containing 1 µg/ml DAPI. For NICD staining, cells (2,000–3,000 cells per slide) were pipetted onto poly-l-lysine–coated slides, settled down for 15 min, fixed with ice-cold 95% ethanol supplemented with 5% acetic acid for 10 min at −20°C, and washed three times with PBS. Cells were then blocked with 3% BSA/PBS for 1 h at room temperature and incubated overnight at 4°C with a rabbit anti-cleaved Notch1 (Val1744; 4147; Cell Signaling) primary antibody in 3% BSA/PBS. Cells were washed three times with PBS and treated with 2% H2O2 for 5 min to quench endogenous peroxidase activity. After washing three times with PBS, cells were incubated with poly-HRP conjugated anti-rabbit IgG (B40943; Invitrogen) secondary antibody for 1 h at room temperature, washed three times with PBS, and incubated with Alexa Fluor 488 tyramide reagent (B40943; Invitrogen) for 10 min at room temperature according to the manufacturer’s instruction for signal amplification. Cells were washed three times with PBS and stained with 1 µg/ml DAPI for 10 min at room temperature and finally washed three times with PBS; the slides were mounted with VectaShield (H-1000; Vector Laboratories). Cells were imaged on a SP5 Leica upright confocal microscope (63× objective) for β-catenin images and a Nikon Ti Eclipse inverted confocal microscope for NICD images (60× objective), and images were processed using Fiji (https://fiji.sc). For quantification, at least 100 cells per condition were randomly captured, and nuclear β-catenin and NICD staining were scored by eye.
Gene expression analyses
Fluidigm gene expression analyses were done on the 96.96 Dynamic Array Integrated Fluidic Circuit and analyses were performed as previously described (Pietras et al., 2015). In brief, 100 cells were directly sorted per well of 96-well plates containing 5 µl CellsDirect lysis buffer (11753–100; Invitrogen), reverse-transcribed and preamplified for 18 cycles using SuperScript III Platinum Taq Mix (12574–026; Invitrogen) with a custom-made set of 96 proprietary target-specific primers (Fluidigm). The resulting cDNA was analyzed on a Biomark system (Fluidigm) using EvaGreen SYBR dye (172–5211; Bio-Rad). Data were collected with Biomark Data Collection Software (Fluidigm) and analyzed using Biomark qPCR software with a quality threshold of 0.65 and linear baseline correction. Melt curves and melting temperature values for each assay reaction were checked individually, and reactions with melt curves showing multiple peaks or poor quality were discarded, leaving 91 genes excluding housekeeping genes (Actb, Gapdh, Gusb, and Hprt) for further analyses. For gene expression quantification, data were exported as a Microsoft Excel .csv file and analyzed by the ΔΔCt method using Gusb for normalization. Box plots were generated using Prism (GraphPad). For similarity matrix, Pearson correlation coefficient was calculated using 27 overlapping genes with 2-wk and 3-wk post-Tplx data (Pietras et al., 2015) or 83 overlapping genes with D2 and D8 anti-Ly6G depletion data and visualized with Morpheus (Broad Institute). For quantitative RT-PCR analyses, cells (5,000–8,000 cells/condition) were directly sorted into 500 µl Trizol LS (10296–010; Life Technologies) or 350 µl RLT lysis buffer containing 1% β-mercaptoethanol (74034; Qiagen). RNA was isolated according to the manufacturer’s instructions, treated with DNase I (18068–015; Invitrogen), and reverse-transcribed using the SuperScript III kit and random hexamers (18080–051; Invitrogen). Runs were performed on a 7900HT Fast Real-Time PCR system or QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems) using SYBR Green reagents (KK4603 or KK4620; Kapa Biosystems) and cDNA equivalent of 200 cells per reaction. Values were normalized to Actb expression levels, and relative changes were calculated using the ΔΔCt method. For SABiosciences Notch PCR array, 5,000 cells were used to extract RNA using the Arcturus PicoPure RNA Isolation kit (KIT0204; Applied Biosystems) according to the manufacturer’s instructions. RNA concentration was measured using a bioanalyzer, and 1 ng of RNA was used to make cDNA as described above. All cDNA samples were preamplified for pathway specific genes, and PCR arrays were performed according to the manufacturer’s instructions (PAMM-059ZE-4; Qiagen). Runs were performed on a 7900HT Fast Real-Time PCR system, and data were analyzed using web-based SABiosciences RT2 Profiler PCR Array Data Analysis software. Values were normalized to Gusb expression.
Cytokine analyses
For collecting BM fluids, the four long bones (two femurs and two tibiae) of each mouse were flushed with the same 200 µl of 2% FBS/HBSS using a 0.3-ml insulin syringe with a 28G needle and spun at 500 ×g for 5 min to remove BM cells. Supernatants were further clarified by spinning down at 12,000 ×g for 10 min, and samples were stored at −80°C until use. For cytokine measurement, 50 µl of two times– and four times–diluted samples for IL6 (50–172-18; eBioscience), and three times–diluted samples for DKK1 (EMDKK1; Invitrogen) were analyzed with an ELISA kit according to the manufacturer’s instructions.
Reanalyses of leukemia patient samples
The comparison between chronic phase CML and normal CD34+ BM cells used the GSE4170 dataset and was performed using Limma (Bioconductor) with a significance cutoff of a Benjamini-Hochberg false discovery rate of ≤0.05. The comparison between AML LSCs (Lin−/CD34+/CD38−/CD123+) and normal HSC (Lin−/CD34+/CD38−/CD90low) used the GSE17054 dataset and was performed similarly. Results are expressed as fold change relative to controls and visualized as a heat map using Morpheus (Broad Institute).
Quantitation and statistical analysis
All experiments were repeated as indicated; n indicates the number of independent biological repeats. Data are expressed as mean ± SD. Mice for treatment and Tplx were randomized, samples were alternated whenever possible, and no blinding protocol was used. No statistical method was used to predetermine sample size. Pairwise statistical significance was evaluated by two-tailed Student’s t test. P values ≤0.05 were considered statistically significant.
Data and software availability
Source data for all the figures are provided with the paper. All other data are available from the corresponding author upon reasonable request.
Online supplemental material
Fig. S1 shows that early-stage myeloid regeneration pathways are activated in various leukemic mouse models. Fig. S2 shows the steady state levels of Notch and Wnt signaling activity in HSPC populations. Fig. S3 shows the effect of decreased Notch signaling on HSPC differentiation. Fig. S4 shows the compensatory crosstalk mechanism between Notch and Wnt signaling in maintaining MPP3 production. Fig. S5 shows low Notch and high Wnt activity in LSCs in mouse models and human patient samples. Table S1 shows 96 custom-made Fluidigm gene expression levels in leukemic BAtTA MPP3.
Acknowledgments
We thank D. Laird for β-catenin GOF mice; J. Rock (UCSF, San Francisco, CA) for β-catenin LOF mice; I. Aifantis for Hes1-Gfp mice; B. Braun for KrasG12D bones; A. Mullally for Jak2V617F bones; H. Singh for Irf8−/− mice; R. Wang (UCSF, San Francisco, CA) for TRE-Notch1ICD and Notch1tm2Rko/GridJ mice; C. Siebel (Genentech, South San Francisco, CA) for anti-Notch blocking antibodies; M. Lee (UCSF) and M. Kissner (Columbia University) for management of our flow cytometry core facilities; S.Y. Zhang, L. Smith, and E. Slobodenyuk for technical assistance; and all members of the Passegué Laboratory both at UCSF and CUIMC for critical insights and suggestions. We are particularly grateful to Dr. D. Reynaud (Cincinnati Children’s Hospital) for his work initiating these studies when he was in the Passegué Laboratory at UCSF. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, E. Passegué. Anti-Notch1 and Notch2 blocking antibodies were obtained from Genentech under an material transfer agreement.
Y-A. Kang was supported by an Leukemia & Lymphoma Society special fellowship and E.M. Pietras by National Institutes of Health grants F32 HL106989 and K01 DK09831. This work was supported by National Institutes of Health grants 2R01HL092471, R01HL111266, and 1R35HL135763 and an LLS Scholar Award to E.M. Pietras.
Author contributions: Conceptualization, Y-A. Kang and E. Passegué; methodology, Y-A. Kang; formal analysis, Y-A. Kang; investigation, Y-A. Kang and E.M. Pietras; writing – original draft, Y-A. Kang and E. Passegué; writing – review and editing, Y-A. Kang and E. Passegué; visualization, Y-A. Kang and E. Passegué; supervision, E. Passegué; funding acquisition, E. Passegué.
References
- Abrahamsson A.E., Geron I., Gotlib J., Dao K.H., Barroga C.F., Newton I.G., Giles F.J., Durocher J., Creusot R.S., Karimi M., et al. 2009. Glycogen synthase kinase 3beta missplicing contributes to leukemia stem cell generation. Proc. Natl. Acad. Sci. USA. 106:3925–3929. 10.1073/pnas.0900189106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., Le Beau M.M., Bloomfield C.D., Cazzola M., and Vardiman J.W.. 2016. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 127:2391–2405. 10.1182/blood-2016-03-643544 [DOI] [PubMed] [Google Scholar]
- Bigas A., and Espinosa L.. 2012. Hematopoietic stem cells: to be or Notch to be. Blood. 119:3226–3235. 10.1182/blood-2011-10-355826 [DOI] [PubMed] [Google Scholar]
- Bigas A., Martin D.I., and Milner L.A.. 1998. Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines. Mol. Cell. Biol. 18:2324–2333. 10.1128/MCB.18.4.2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigas A., Guiu J., and Gama-Norton L.. 2013. Notch and Wnt signaling in the emergence of hematopoietic stem cells. Blood Cells Mol. Dis. 51:264–270. 10.1016/j.bcmd.2013.07.005 [DOI] [PubMed] [Google Scholar]
- Bockamp E., Antunes C., Maringer M., Heck R., Presser K., Beilke S., Ohngemach S., Alt R., Cross M., Sprengel R., et al. 2006. Tetracycline-controlled transgenic targeting from the SCL locus directs conditional expression to erythrocytes, megakaryocytes, granulocytes, and c-kit-expressing lineage-negative hematopoietic cells. Blood. 108:1533–1541. 10.1182/blood-2005-12-012104 [DOI] [PubMed] [Google Scholar]
- Brault V., Moore R., Kutsch S., Ishibashi M., Rowitch D.H., McMahon A.P., Sommer L., Boussadia O., and Kemler R.. 2001. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 128:1253–1264. [DOI] [PubMed] [Google Scholar]
- Braun B.S., Tuveson D.A., Kong N., Le D.T., Kogan S.C., Rozmus J., Le Beau M.M., Jacks T.E., and Shannon K.M.. 2004. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc. Natl. Acad. Sci. USA. 101:597–602. 10.1073/pnas.0307203101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi L.M., Adams G.B., Weibrecht K.W., Weber J.M., Olson D.P., Knight M.C., Martin R.P., Schipani E., Divieti P., Bringhurst F.R., et al. 2003. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 425:841–846. 10.1038/nature02040 [DOI] [PubMed] [Google Scholar]
- Carlson T.R., Yan Y., Wu X., Lam M.T., Tang G.L., Beverly L.J., Messina L.M., Capobianco A.J., Werb Z., and Wang R.. 2005. Endothelial expression of constitutively active Notch4 elicits reversible arteriovenous malformations in adult mice. Proc. Natl. Acad. Sci. USA. 102:9884–9889. 10.1073/pnas.0504391102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. 2006. Wnt/beta-catenin signaling in development and disease. Cell. 127:469–480. 10.1016/j.cell.2006.10.018 [DOI] [PubMed] [Google Scholar]
- Daley J.M., Thomay A.A., Connolly M.D., Reichner J.S., and Albina J.E.. 2008. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J. Leukoc. Biol. 83:64–70. 10.1189/jlb.0407247 [DOI] [PubMed] [Google Scholar]
- Döhner H., Weisdorf D.J., and Bloomfield C.D.. 2015. Acute Myeloid Leukemia. N. Engl. J. Med. 373:1136–1152. 10.1056/NEJMra1406184 [DOI] [PubMed] [Google Scholar]
- Duncan A.W., Rattis F.M., DiMascio L.N., Congdon K.L., Pazianos G., Zhao C., Yoon K., Cook J.M., Willert K., Gaiano N., et al. 2005. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat. Immunol. 6:314–322. 10.1038/ni1164 [DOI] [PubMed] [Google Scholar]
- Fevr T., Robine S., Louvard D., and Huelsken J.. 2007. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol. Cell. Biol. 27:7551–7559. 10.1128/MCB.01034-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger H., Denkinger M., and Schirmbeck R.. 2014. Hematopoietic stem cell aging. Curr. Opin. Immunol. 29:86–92. 10.1016/j.coi.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Guezguez B., Almakadi M., Benoit Y.D., Shapovalova Z., Rahmig S., Fiebig-Comyn A., Casado F.L., Tanasijevic B., Bresolin S., Masetti R., et al. 2016. GSK3 deficiencies in hematopoietic stem cells initiate pre-neoplastic state that is predictive of clinical outcomes of human acute leukemia. Cancer Cell. 29:61–74. 10.1016/j.ccell.2015.11.012 [DOI] [PubMed] [Google Scholar]
- Harada N., Tamai Y., Ishikawa T., Sauer B., Takaku K., Oshima M., and Taketo M.M.. 1999. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 18:5931–5942. 10.1093/emboj/18.21.5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel F.H., Bullinger L., Feng Z., Wang Z., Neff T.A., Stein L., Kalaitzidis D., Lane S.W., and Armstrong S.A.. 2012. Genetic and pharmacologic inhibition of β-catenin targets imatinib-resistant leukemia stem cells in CML. Cell Stem Cell. 10:412–424. 10.1016/j.stem.2012.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérault A., Binnewies M., Leong S., Calero-Nieto F.J., Zhang S.Y., Kang Y.A., Wang X., Pietras E.M., Chu S.H., Barry-Holson K., et al. 2017. Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis. Nature. 544:53–58. 10.1038/nature21693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtschke T., Löhler J., Kanno Y., Fehr T., Giese N., Rosenbauer F., Lou J., Knobeloch K.P., Gabriele L., Waring J.F., et al. 1996. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 87:307–317. 10.1016/S0092-8674(00)81348-3 [DOI] [PubMed] [Google Scholar]
- Holyoake T.L., and Vetrie D.. 2017. The chronic myeloid leukemia stem cell: stemming the tide of persistence. Blood. 129:1595–1606. 10.1182/blood-2016-09-696013 [DOI] [PubMed] [Google Scholar]
- Jamieson C.H., Ailles L.E., Dylla S.J., Muijtjens M., Jones C., Zehnder J.L., Gotlib J., Li K., Manz M.G., Keating A., et al. 2004. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 351:657–667. 10.1056/NEJMoa040258 [DOI] [PubMed] [Google Scholar]
- Jiang X., Mak P.Y., Mu H., Tao W., Mak D.H., Kornblau S., Zhang Q., Ruvolo P., Burks J.K., Zhang W., et al. 2018. Disruption of Wnt/β-Catenin Exerts Antileukemia Activity and Synergizes with FLT3 Inhibition in FLT3-Mutant Acute Myeloid Leukemia. Clin. Cancer Res. 24:2417–2429. 10.1158/1078-0432.CCR-17-1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S., Sutphin R.M., Hall M.G., Golfman L.S., Fang W., Nolo R.M., Akers L.J., Hammitt R.A., McMurray J.S., Kornblau S.M., et al. 2013. Notch activation inhibits AML growth and survival: a potential therapeutic approach. J. Exp. Med. 210:321–337. 10.1084/jem.20121527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King B., Trimarchi T., Reavie L., Xu L., Mullenders J., Ntziachristos P., Aranda-Orgilles B., Perez-Garcia A., Shi J., Vakoc C., et al. 2013. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 153:1552–1566. 10.1016/j.cell.2013.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinakis A., Lobry C., Abdel-Wahab O., Oh P., Haeno H., Buonamici S., van De Walle I., Cathelin S., Trimarchi T., Araldi E., et al. 2011. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 473:230–233. 10.1038/nature09999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisato A., Chiba S., Nakagami-Yamaguchi E., Kumano K., Saito T., Masuda S., Yamaguchi T., Osawa M., Kageyama R., Nakauchi H., et al. 2003. HES-1 preserves purified hematopoietic stem cells ex vivo and accumulates side population cells in vivo. Blood. 101:1777–1783. 10.1182/blood-2002-07-2051 [DOI] [PubMed] [Google Scholar]
- Lampreia F.P., Carmelo J.G., and Anjos-Afonso F.. 2017. Notch Signaling in the Regulation of Hematopoietic Stem Cell. Curr. Stem Cell Rep. 3:202–209. 10.1007/s40778-017-0090-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lento W., Congdon K., Voermans C., Kritzik M., and Reya T.. 2013. Wnt signaling in normal and malignant hematopoiesis. Cold Spring Harb. Perspect. Biol. 5:a008011 10.1101/cshperspect.a008011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry C., Oh P., Mansour M.R., Look A.T., and Aifantis I.. 2014. Notch signaling: switching an oncogene to a tumor suppressor. Blood. 123:2451–2459. 10.1182/blood-2013-08-355818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis T.C., Naber B.A., Roozen P.P., Brugman M.H., de Haas E.F., Ghazvini M., Fibbe W.E., van Dongen J.J., Fodde R., and Staal F.J.. 2011. Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell. 9:345–356. 10.1016/j.stem.2011.07.017 [DOI] [PubMed] [Google Scholar]
- Ma P., Song W., and Hess J.L.. 2017. A new target for differentiation therapy in AML. Cell Res. 27:9–10. 10.1038/cr.2016.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti R., Becker M.W., Tian Q., Lee T.L., Yan X., Liu R., Chiang J.H., Hood L., Clarke M.F., and Weissman I.L.. 2009. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc. Natl. Acad. Sci. USA. 106:3396–3401. 10.1073/pnas.0900089106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullally A., Lane S.W., Ball B., Megerdichian C., Okabe R., Al-Shahrour F., Paktinat M., Haydu J.E., Housman E., Lord A.M., et al. 2010. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell. 17:584–596. 10.1016/j.ccr.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F., Zandi S., Takayama N., Dobson S., Gan O.I., Wilson G., Kaufmann K.B., McLeod J., Laurenti E., Dunant C.F., et al. 2016. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 351:aab2116 10.1126/science.aab2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh P., Lobry C., Gao J., Tikhonova A., Loizou E., Manet J., van Handel B., Ibrahim S., Greve J., Mikkola H., et al. 2013. In vivo mapping of notch pathway activity in normal and stress hematopoiesis. Cell Stem Cell. 13:190–204. 10.1016/j.stem.2013.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orford K.W., and Scadden D.T.. 2008. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 9:115–128. 10.1038/nrg2269 [DOI] [PubMed] [Google Scholar]
- Pietras E.M. 2017. Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood. 130:1693–1698. 10.1182/blood-2017-06-780882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras E.M., Reynaud D., Kang Y.A., Carlin D., Calero-Nieto F.J., Leavitt A.D., Stuart J.M., Göttgens B., and Passegué E.. 2015. Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell. 17:35–46. 10.1016/j.stem.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras E.M., Mirantes-Barbeito C., Fong S., Loeffler D., Kovtonyuk L.V., Zhang S., Lakshminarasimhan R., Chin C.P., Techner J.M., Will B., et al. 2016. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18:607–618. 10.1038/ncb3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H.R., and Aguet M.. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 10:547–558. 10.1016/s1074-7613(00)80054-0 [DOI] [PubMed] [Google Scholar]
- Reynaud D., Pietras E., Barry-Holson K., Mir A., Binnewies M., Jeanne M., Sala-Torra O., Radich J.P., and Passegué E.. 2011. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer Cell. 20:661–673. 10.1016/j.ccr.2011.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida M., Schepers K., King B., Sabnis A.J., Forsberg E.C., Attema J.L., Braun B.S., and Passegué E.. 2009. JunB protects against myeloid malignancies by limiting hematopoietic stem cell proliferation and differentiation without affecting self-renewal. Cancer Cell. 15:341–352. 10.1016/j.ccr.2009.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers K., Campbell T.B., and Passegué E.. 2015. Normal and leukemic stem cell niches: insights and therapeutic opportunities. Cell Stem Cell. 16:254–267. 10.1016/j.stem.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A., and Pardanani A.. 2015. Myeloproliferative Neoplasms: A Contemporary Review. JAMA Oncol. 1:97–105. 10.1001/jamaoncol.2015.89 [DOI] [PubMed] [Google Scholar]
- Valencia A., Román-Gómez J., Cervera J., Such E., Barragán E., Bolufer P., Moscardó F., Sanz G.F., and Sanz M.A.. 2009. Wnt signaling pathway is epigenetically regulated by methylation of Wnt antagonists in acute myeloid leukemia. Leukemia. 23:1658–1666. 10.1038/leu.2009.86 [DOI] [PubMed] [Google Scholar]
- Varnum-Finney B., Brashem-Stein C., and Bernstein I.D.. 2003. Combined effects of Notch signaling and cytokines induce a multiple log increase in precursors with lymphoid and myeloid reconstituting ability. Blood. 101:1784–1789. 10.1182/blood-2002-06-1862 [DOI] [PubMed] [Google Scholar]
- Varnum-Finney B., Halasz L.M., Sun M., Gridley T., Radtke F., and Bernstein I.D.. 2011. Notch2 governs the rate of generation of mouse long- and short-term repopulating stem cells. J. Clin. Invest. 121:1207–1216. 10.1172/JCI43868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Krivtsov A.V., Sinha A.U., North T.E., Goessling W., Feng Z., Zon L.I., and Armstrong S.A.. 2010. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 327:1650–1653. 10.1126/science.1186624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhang H., Rodriguez S., Cao L., Parish J., Mumaw C., Zollman A., Kamoka M.M., Mu J., Chen D.Z., et al. 2014. Notch-dependent repression of miR-155 in the bone marrow niche regulates hematopoiesis in an NF-κB-dependent manner. Cell Stem Cell. 15:51–65. 10.1016/j.stem.2014.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Cain-Hom C., Choy L., Hagenbeek T.J., de Leon G.P., Chen Y., Finkle D., Venook R., Wu X., Ridgway J., et al. 2010. Therapeutic antibody targeting of individual Notch receptors. Nature. 464:1052–1057. 10.1038/nature08878 [DOI] [PubMed] [Google Scholar]
- Yao D., Huang Y., Huang X., Wang W., Yan Q., Wei L., Xin W., Gerson S., Stanley P., Lowe J.B., et al. 2011. Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions. Blood. 117:5652–5662. 10.1182/blood-2010-12-326074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Blum J., Chen A., Kwon H.Y., Jung S.H., Cook J.M., Lagoo A., and Reya T.. 2007. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 12:528–541. 10.1016/j.ccr.2007.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Li L.W., Yan Q., Petryniak B., Man Y., Su C., Shim J., Chervin S., and Lowe J.B.. 2008. Notch-dependent control of myelopoiesis is regulated by fucosylation. Blood. 112:308–319. 10.1182/blood-2007-11-115204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Mak P.Y., Mu H., Mak D.H., Zeng Z., Cortes J., Liu Q., Andreeff M., and Carter B.Z.. 2017. Combined inhibition of β-catenin and Bcr-Abl synergistically targets tyrosine kinase inhibitor-resistant blast crisis chronic myeloid leukemia blasts and progenitors in vitro and in vivo. Leukemia. 31:2065–2074. 10.1038/leu.2017.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Source data for all the figures are provided with the paper. All other data are available from the corresponding author upon reasonable request.