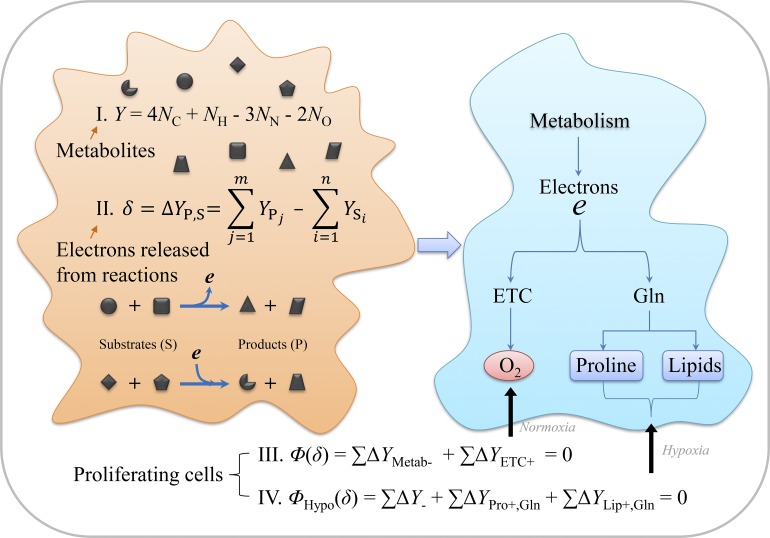
Liu et al. build up an electron balance model for central carbon metabolism in cancer cells and reveal that simultaneous inhibitions of proline biosynthesis and lipogenesis can block cancer cell growth under hypoxia and in vivo tumor progress.
Abstract
Cancer cells often proliferate under hypoxia and reprogram their metabolism. However, how to find targets to effectively block the hypoxia-associated metabolic pathways remains unclear. Here, we developed a tool to conveniently calculate electrons dissipated in metabolic transformations. Based on the law of conservation of electrons in chemical reactions, we further built up an electron balance model for central carbon metabolism, and it can accurately outline metabolic plasticity under hypoxia. Our model specifies that glutamine metabolism reprogrammed for biosynthesis of lipid and/or proline actually acts as the alternative electron bin to enable electron transfer in proliferating cells under hypoxia. Inhibition of both proline biosynthesis and lipogenesis can synergistically suppress cancer cell growth under hypoxia and in vivo tumor onset. Therefore, our model helps to reveal combinations of potential targets to inhibit tumor growth by blocking hypoxia-rewired metabolism and provides a useful tool for future studies on cancer metabolism.
Graphical Abstract
Introduction
Life is a self-organizing and self-adaptive system. Therefore, it is not surprising that proliferating cells, such as cancer cells, develop significant metabolic heterogeneity (Boroughs and DeBerardinis, 2015; Martinez-Outschoorn et al., 2017; Pavlova and Thompson, 2016) to cope with the changing microenvironment. Even though the specific pathways are blocked in some subsets of cells, alternatively functional counterparts are usually developed for compensation. This kind of plasticity seriously frustrates our efforts to control cell proliferation by targeting some specific metabolic pathways.
As a pool of integrated chemical transformations in cells, metabolism must comply with chemical laws. If we can find out the chemical law underlying metabolic reprogramming in proliferating cancer cells, we may can design a cocktail of broad-spectrum treatments for cancers regardless of the heterogeneity. Classically, chemical reactions involve the forming and breaking of chemical bonds between atoms. Since chemical bonds result from a redistribution of the outer electrons of atoms, chemical reactions may encompass electron transfer between each other or release electrons to the system. In mammalian cells, the transfer of electrons of compounds to oxygen occurs directly, semi-directly, or indirectly by using electron acceptors. Direct and semi-direct oxidations are catalyzed by dioxygenase and monooxygenase and are mainly involved in the catabolism of some compounds with aromatic groups or long aliphatic chains (Harayama et al., 1992). In contrast, most redox transformations are performed indirectly. The final transfer of indirect electrons to oxygen is coupled to mitochondrial ATP generation, which is much more efficient than the other energy-producing pathway, aerobic glycolysis with glucose being converted to lactate.
The electron acceptor flavin adenine dinucleotide (FAD)/flavin mononucleotide (FMN) tightly or covalently binds to its enzymes. After accepting electrons, reduced flavin adenine dinucleotide (FADH2)/reduced flavin mononucleotide (FMNH2) usually immediately transfers electrons to nicotinamide adenine dinucleotide (NAD+) and nicotinamide adenine dinucleotide phosphate (NADP+) or directly to the electron transport chain (ETC; Lienhart et al., 2013). Therefore, NAD+ and NADP+ are the major free electron acceptors that link up oxidative and reductive reactions. Electrons in reduced nicotinamide adenine dinucleotide (NADH) and/or reduced nicotinamide adenine dinucleotide phosphate (NADPH) are compartmentally generated in cells, but they can be easily translocated spatially by various biochemical shuttles or transhydrogenated between each other (Gaude et al., 2018; Jackson et al., 2015; Lewis et al., 2014). Based on the conservation of electrons in chemical reactions, the global electrons produced have to be equated to those dissipated in a cell, which is indispensable to support cell survival and growth.
As for mammalian cells, the established fact is that oxygen is used to accept the net electrons released from the whole cellular metabolism, which is mediated by the mitochondrial ETC. Therefore, the mitochondrion plays an essential role as the electron acceptor in enabling cell proliferation in addition to energy generation (Birsoy et al., 2015; Sullivan et al., 2015; Titov et al., 2016). However, when the ETC is disabled by hypoxia or pathological/pharmacological inhibition, how to enable electron transfer to support cell proliferation remains elusive. Here, we built up an electron balance model to reveal the chemical mechanism of metabolic reprogramming under hypoxia. Based on our model, we reveal that both proline biosynthesis and lipogenesis can act as the alternative electron acceptor and that blocking them synergistically suppresses tumor growth in vivo and in vitro.
Results
A tool to calculate electron production in a metabolic transformation
To comprehensively understand electron transfer in cells, we introduced a concept from the previous established degree of reduction in bioprocess engineering (Shuler and Kargi, 2001). The degree of reduction of any element in a compound is equal to the valence of this element; thus, the degrees of reduction of some elements are C = +4, H = +1, N = −3, and O = −2 in CO2, H2O, and NH3 (Shuler and Kargi, 2001). Here, we used Y as the symbol for the degree of reduction of a compound. Therefore, Y characterizes the potential ability of a typical biological metabolite, with the molecular formula of CCHHOONN, to release electrons upon its complete oxidation to CO2, H2O, and NH3 in cells. Hence,
| (1) |
in which NC, NH, NN, and NO are the number of carbon, hydrogen, oxygen, and nitrogen atoms in the molecule. Here, we listed Y values of intracellular metabolites involved in the metabolism of glucose, amino acids, nucleic acids, and lipids (Table 1 and Table S1). In aerobic organisms, the end metabolites, such as carbon dioxide, water, and urea/ammonia, do not release any electron because their Y is 0, whereas oxygen can only function as the electron acceptor due to its negative YO2 value (Table 1).
Table 1. The degree of reduction of metabolites.
| Metabolites | Formula | Y value | |
|---|---|---|---|
| Carbohydrate | Glucose | C6H12O6 | 24 |
| Fructose | C6H12O6 | 24 | |
| Glyceraldehyde | C3H6O3 | 12 | |
| Glyceric acid | C3H6O4 | 10 | |
| Pyruvate | C3H4O3 | 10 | |
| Lactate | C3H6O3 | 12 | |
| Oxaloacetate | C4H4O5 | 10 | |
| Malate | C4H6O5 | 12 | |
| Fumarate | C4H4O4 | 12 | |
| Succinate | C4H6O4 | 14 | |
| α-Ketoglutarate | C5H6O5 | 16 | |
| Citrate | C6H8O7 | 18 | |
| Nonessential amino acids | Glycine | C2H5NO2 | 6 |
| Serine | C3H7NO3 | 10 | |
| Aspartate | C4H7NO4 | 12 | |
| Asparagine | C4H8N2O3 | 12 | |
| Glutamine | C5H10N2O3 | 18 | |
| Glutamate | C5H9NO4 | 18 | |
| Alanine | C3H7NO2 | 12 | |
| Proline | C5H9NO2 | 22 | |
| Acetyl-CoA | Acetate | C2H4O2 | 8 |
| Lipid components | Glycerol | C3H8O3 | 14 |
| Palmitate | C16H32O2 | 92 | |
| Farnesola | C15H26O | 84 | |
| End metabolites | Carbon dioxide | CO2 | 0 |
| Water | H2O | 0 | |
| Urea/ammonia | CH4N2O/NH3 | 0 | |
| Oxygen | O2 | −4 |
Farnesol (farnesyl pyrophosphate) is the precursor of sterols.
Eq. 1 can be used to quantitatively calculate electrons required for metabolic conversions by Y change, ΔYP,S, between Y values of initial substrates and those of final products. A transformation is shown with
where substrates (S) and products (P) are the carbon-containing metabolites with the quantity of carbon atoms being made conserved. ΔYP,S can be expressed as
| (2) |
A positive Y change means the consumption of electrons, while a negative value indicates the release of electrons in this conversion. Notably, ΔY only depends on initiators and objects independently of detailed metabolic routes (a formal derivation detailed in Materials and methods). Phosphate group and co-enzyme are often used to facilitate metabolic transformations; however, their addition to metabolites does not involve the transfer of electrons. Thus, we only need to simply focus on the form of compounds without phosphate and co-enzyme when ΔY is calculated.
An electron balance model for proliferating cells
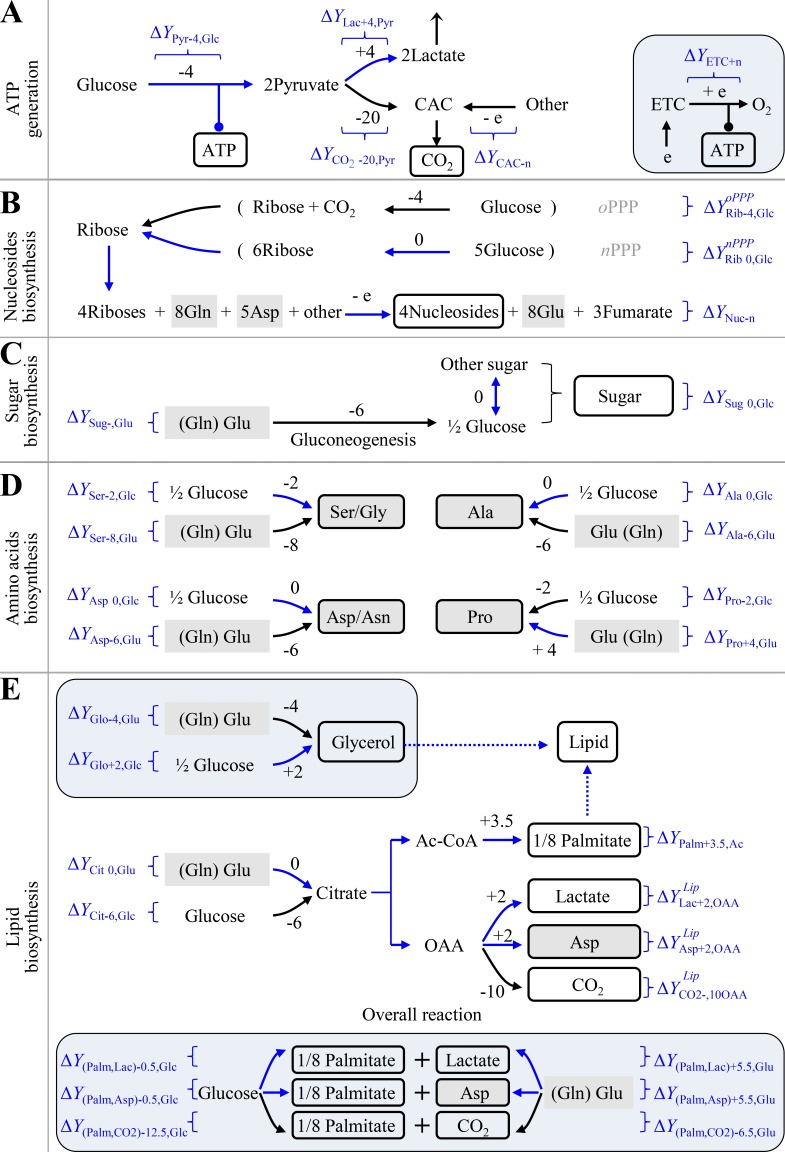
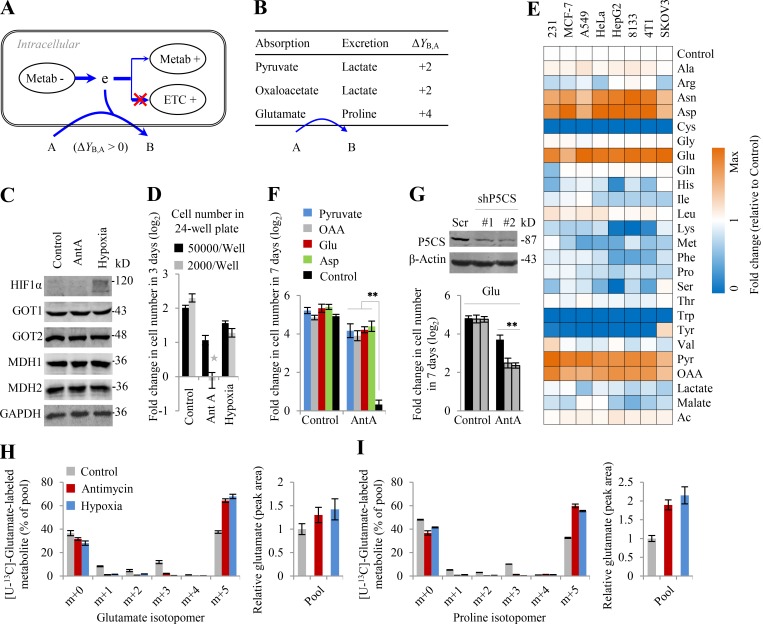
Proliferating cells must produce ATP and duplicate all the building bricks to make new cells. Mammalian and bacterial cells share similar chemical compositions (Alberts et al., 2008; Table S2). Macromolecules including proteins, nucleic acids, lipids, and polysaccharides account for 87%, and their precursors involving amino acids, nucleotides, fatty acids, sugars, and the related intermediates make up 9.3% of cell mass. The remaining chemicals are inorganic ions (3%) that are unrelated to electron transfer and other small molecules (0.7%) that could be negligible to the analysis of global intracellular electrons. Protein synthesis from amino acids, nucleic acid synthesis from nucleosides, polysaccharide synthesis from sugars, and lipid synthesis from fatty acids do not involve the transfer of electrons. Therefore, here we mainly focused on the synthesis of amino acids (nonessential amino acids), nucleosides, fatty acids, and sugars. The major carbon source in blood includes glucose and glutamine. We calculated the possible ΔY of these building bricks, including ATP generation, nucleotide biosynthesis, sugar biosynthesis, amino acid synthesis, and lipid biosynthesis for cell proliferation based on Y values of the metabolites in Table 1 and Table S1 (Fig. 1, A–E; Figs. S1 and S2; and Table S3).
Figure 1.
The dissipation of electrons (ΔY) in metabolic transformations. ΔY is calculated by Eq. 2 based on the Y values in Table 1 and Table S1. (A) ΔY in ATP generation. The inset is mitochondrial ATP generation. (B) ΔY in the biosynthesis of nucleosides. Ribose can be generated in oxidative PPP (oPPP) or nonoxidative PPP (nPPP). The exact ΔY values of ribose-initiated biosynthesis of nucleosides depend on the specified nucleoside. See Fig. S2 and Table S3 for details. (C) ΔY in the biosynthesis of sugar. (D) ΔY in the biosynthesis of nonessential amino acids. The carbons of amino acids could be from glucose or glutamine. (E) ΔY in the biosynthesis of lipids. Palmitate could be synthesized from glucose or glutamine. The insets are the biosynthesis of glycerol (glycerol-3-phosphate) and the overall reactions of palmitate from glucose or glutamine. In all panels, numbers are ΔY values in one reaction. “−” means the production of electrons, while “+” refers to the consumption of electrons. White box indicates the building bricks for proliferating cells. Gray box shows amino acids. Curved arrows indicate optional pathways. Brackets represents the coupled pathways. Blue routes show the metabolic pathways prevailing under hypoxia. Ac-CoA, acetyl-CoA; Cit, citrate; e, electron; Lac, lactate; OAA, oxaloacetate; Palm, palmitate; Pyr, pyruvate; Rib, ribose.
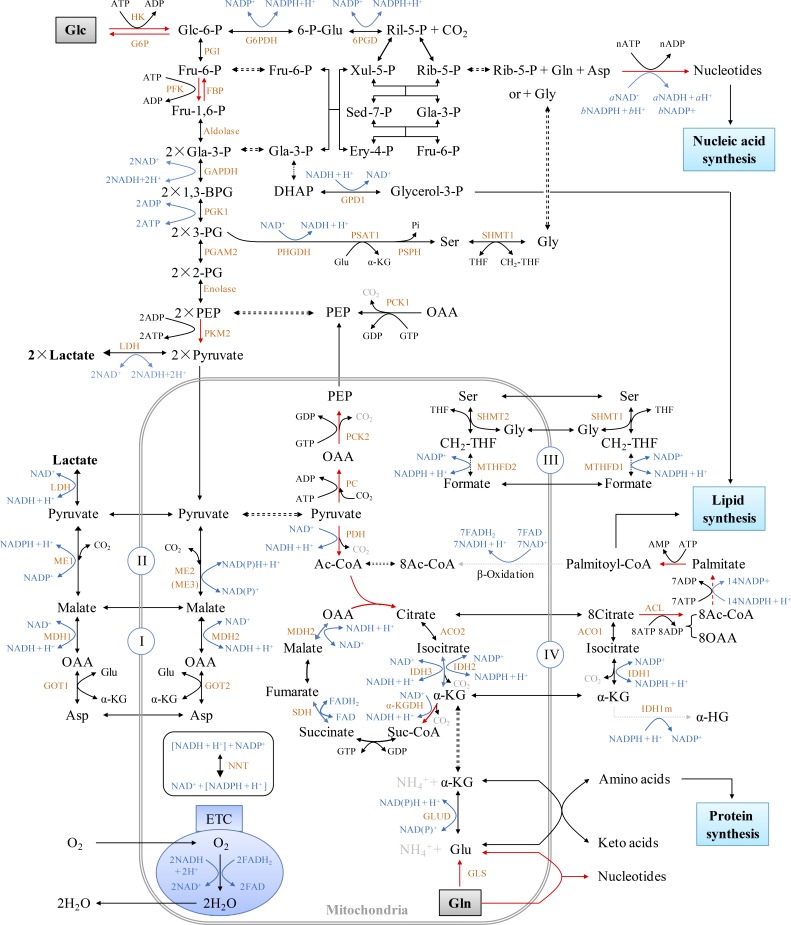
Figure S1.
Intracellular metabolic overview. The metabolic pathways were organized based on Berg et al. (2011). The red arrows show the irreversible direction of the reactions. The synthesis of each nucleotide is summarized in Fig. S2 and Table S3. 1,3-BPG, 1,3-bisphosphoglycerate; 2-PG, 2-phosphoglycerate; 3-PG, 3-phosphoglycerate; 6-p-Glu, 6-phosphogluconate; Ac-CoA, acetyl-CoA; Asp, aspartate; DHAP, dihydroxyacetone phosphate; Ery-4-P, erythrose-4-phosphate; Fru-1,6-P, fructose-1,6-bisphosphate; Fru-6-P, fructose-6-phosphate; Gla-3-P, glyceraldehydes-3-phosphate; Glc, glucose; Glc-6-P, glucose-6-phosphate; OAA, oxaloacetate; PEP, phosphoenolpyruvate; Rib-5-P, ribose-5-phosphate; Ril-5-P, ribulose-5-phosphate; Sed-7-P, sedoheptulose 7-phosphate; Suc-CoA, succinyl-Co enzyme A; THF, tetrahydrofolate; Xul-5-P, xylulose-5-phosphate; α-KG, α-ketoglutarate. The full names of enzymes used here are listed in Table S5.
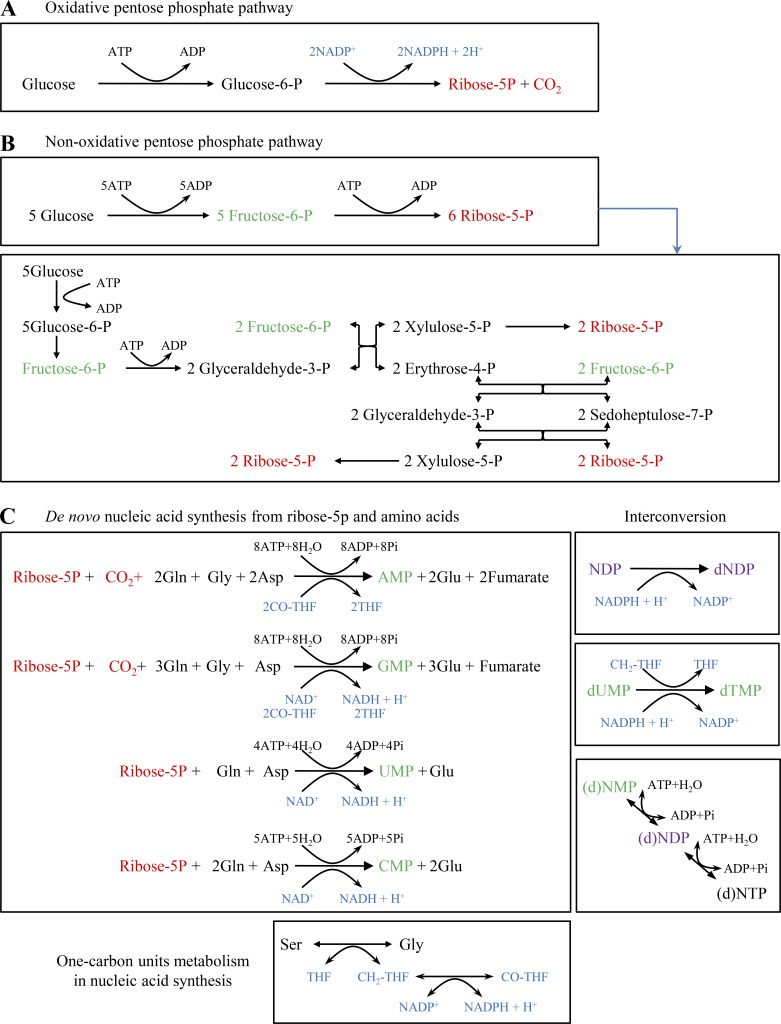
Figure S2.
De novo biosynthesis of nucleosides. (A) Oxidative PPP. See Fig. S1 for the detailed processes. (B) Non-oxidative PPP. The upper box is the overall conversion of glucose to ribose-5-phosphate, and the detailed reactions are shown in the lower box. (C) De novo biosyntheses of nucleotides from ribose-5-phosphate and amino acids. Ribonucleotide, ribonucleoside monophosphate (NMP), AMP, GMP, UMP, or CMP, was synthesized from ribose-5-phosphate, glutamine, aspartate, and/or glycine. NMP was converted to ribonucleoside diphosphate (NDP) that was further reduced to deoxyribonucleotide diphosphate (dNDP). dUMP was processed to dTMP. dNMP/NMP, dNDP/NDP, and dNTP/NTP could be converted into each other, and these reactions required ATP but did not involve electrons. The syntheses of some nucleotides required one-carbon units. All the reactions were summarized based on Berg et al. (2011). Asp, aspartate.
We can calculate the total electrons consumed by ATP generation (Fig. 1 A), nucleotide biosynthesis (Fig. 1 B), sugar biosynthesis (Fig. 1 C), amino acid synthesis (Fig. 1 D), and lipid biosynthesis (Fig. 1 E). Based on the law of electron conservation in chemical reactions, the total electrons produced in a cell [Φ(δ)] should be around 0. Hence,
| (3) |
∑ means the sum of electrons from all the intracellular reactions, and variables with slashes from different metabolic pathways are optional depending on the availability of nutrients and cellular states (Fig. 1). As a cell is a self-organizing system, Eq. 3 can be flexibly set up to 0 by using different metabolic pathways. When oxygen is sufficient, the ETC works efficiently. To support ATP generation coupled to the ETC, cells may use the metabolic pathways producing electrons.
Biosyntheses of proline and lipids are rewired to consume electrons under hypoxia
Under the condition of ETC dysfunction, such as hypoxia and ETC inhibition, the electron flow to oxygen is blocked (here let ∑ΔYETC+ = 0), thus
It could lead to accumulated electrons that unlikely rise infinitely. Therefore, to equate Φ(δ) with 0 while still enabling anabolic processes, proliferating cells have to rewire their metabolic reactions to balance electron transfer by reducing production and enhancing the dissipation of electrons (Fig. 2 A). In doing so, Φ(δ) is reset up to 0 by selecting higher ΔY values from alternative metabolic pathways, thus giving
| (4) |
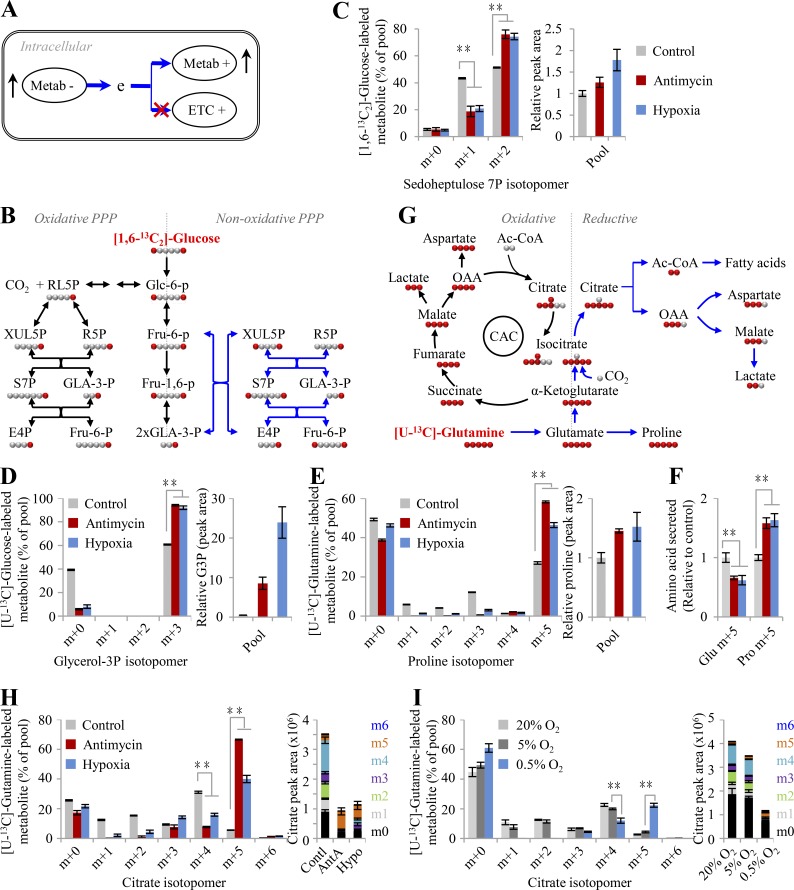
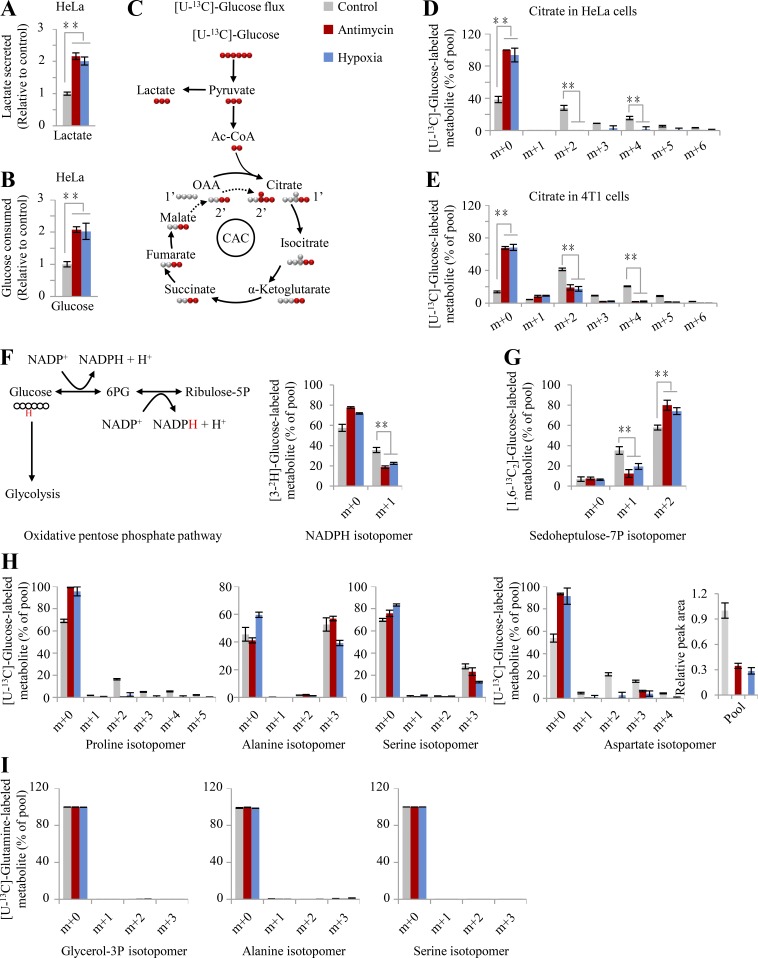
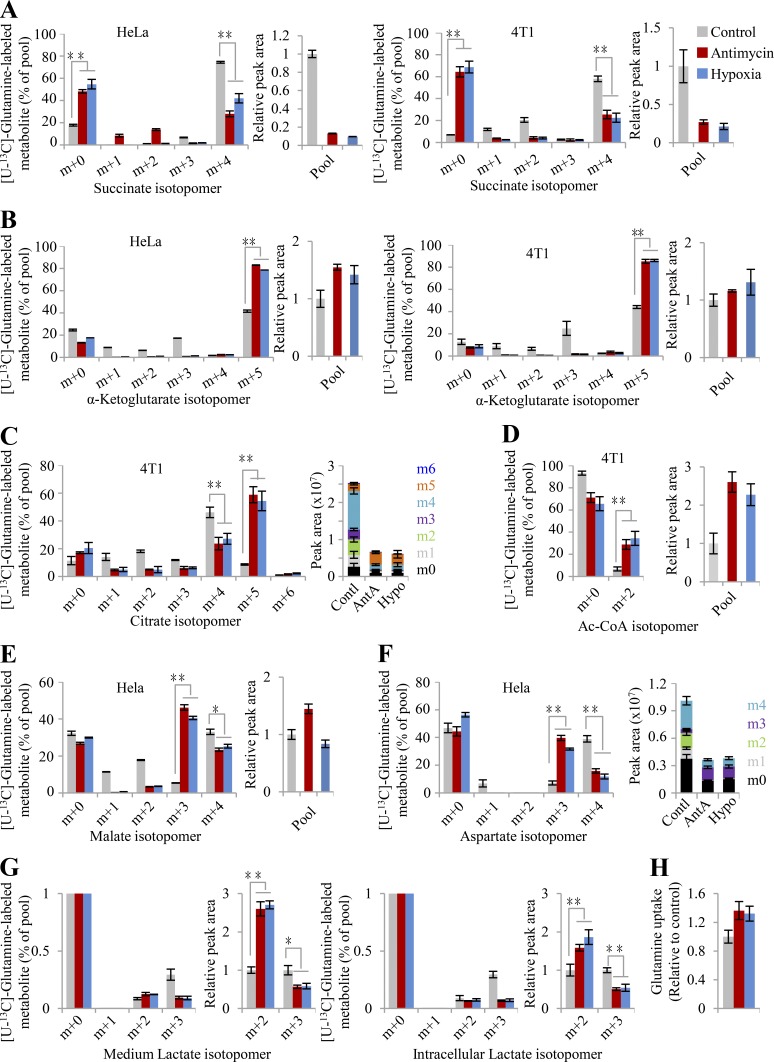
To test this model, herein we used the mitochondrial inhibitor antimycin A to block the ETC and cultured cells under hypoxia (0.5% oxygen) to reduce the electron acceptor. Based on Φ′(δ), glycolytic pyruvate shunts to lactate (∑ΔYLac+,Pyr) and avoids entering the citric acid cycle (CAC) to produce electrons (∑ΔYCO2−,Pyr), which results in increased lactate production. Upon anaerobic glycolysis, cells could generate enough ATP by increasing the metabolic flux of glucose to lactate. These typical hypoxic effects are widely observed and were also confirmed here (Fig. S3, A and B). This pathway does not release any electrons (ΔY = 0) because ΔYPyr−,Glc is negatively equal to ΔYLac+,Pyr (Fig. 1 A). Normally, the CAC produces a great many electrons for mitochondrial ATP generation by completely oxidizing its participants to carbon dioxides, but also provides on purpose some essential intermediates for biosynthesis. Therefore, upon ETC dysfunction, the CAC had to be shaped to a state of low activity required for cell proliferation, endowed with a ΔY′ CAC- in Eq. 4. Our results showed that both [U-13C]-glucose–derived citrate m+2 and [U-13C]-glutamine–derived succinate m+4 were significantly reduced in the condition of hypoxia and ETC inhibition (Fig. S3, C–E; and Fig. S4 A), indicating a decrease in the metabolic flux of both glucose and glutamine in the oxidative CAC.
Figure 2.
Glutamine-initiated proline biosynthesis and reductive metabolism upon ETC dysfunction. (A) A model to show that cells adapt to ETC dysfunction by maximizing Φ(δ). To do so, cells optimize their metabolic pathways by using electron-consuming and avoiding electron-producing metabolic pathways. (B) Schematic for the oxidative PPP and the nonoxidative PPP. Black arrows show the oxidative PPP and glycolytic pathway; blue arrows indicate the nonoxidative PPP. (C) Mass isotopomer analysis of sedoheptulose 7-phosphate in HeLa cells cultured with [1,6-13C2]-glucose for 8 h under the conditions of hypoxia or inhibition by antimycin A (1 µM). (D) Mass isotopomer analysis of glycerol 3-phosphate in HeLa cells cultured with [U-13C]-glucose for 8 h under the condition of hypoxia or inhibition by antimycin A (1 µM). The relative pool size is also shown. (E) Mass isotopomer analysis of proline in HeLa cells cultured with [U-13C]-glutamine for 8 h under the condition of hypoxia or inhibition by antimycin A (1 µM). The relative pool size is also shown. (F) Excretion of glutamate and proline from HeLa cells cultured with [U-13C]-glutamine for 8 h under the condition of hypoxia or inhibition by antimycin A (1 µM). (G) Schematic of glutamine oxidative or reductive metabolism. Black arrows show oxidative glutamine metabolism; blue arrows show reductive glutamine metabolism. (H) Mass isotopomer analysis of citrate in HeLa cells cultured with [U-13C]-glutamine for 8 h under the condition of hypoxia or inhibition by antimycin A (1 µM). The pool size is also shown. (I) Mass isotopomer analysis of citrate in HeLa cells cultured in the HPLM with [U-13C]-glutamine for 8 h under different concentrations of oxygen. The pool size is also shown. Data were from three independent cultures. Error bars represent mean ± SD. **, P < 0.01; Student’s t test. Ac-CoA, acetyl-CoA; AntA, antimycin A; Contl, control; e, electron; G3P, glycerol 3-phosphate; Hypo, hypoxia; OAA, oxaloacetate.
Figure S3.
Glucose–derived metabolites upon ETC dysfunction. (A and B) Glucose utilization and lactate secretion in HeLa cells cultured with [U-13C]-glucose under the conditions of hypoxia or inhibition by antimycin A (1 µM). (C) Schematic for glucose metabolism. (D and E) Mass isotopomer analysis of citrate in HeLa and 4T1 cells cultured with [U-13C]-glucose under the conditions of hypoxia or inhibition by antimycin A (1 µM). The decreased fraction of [U-13C]-glucose–derived citrate and increased fraction of unlabeled citrate by [U-13C]-glucose suggested the reduced metabolic flux of glucose to the CAC. (F) Mass isotopomer analysis of NADPH in HeLa cells cultured with [3-2H]-glucose under the condition of hypoxia or inhibition by antimycin A (1 µM). Left schematic indicates [3-2H]-glucose metabolism in the oxidative PPP (oPPP). Red H represents deuterium atom in/from [3-2H]-glucose. (G) Mass isotopomer analysis of sedoheptulose 7-phosphate in 4T1 cells cultured with [1,6-13C2]-glucose under hypoxia or inhibition by antimycin A (1 µM). (H) Mass isotopomer analysis of proline, alanine, serine, and aspartate in HeLa cells cultured with [U-13C]-glucose under the conditions of hypoxia or inhibition by antimycin A (1 µM). (I) Mass isotopomer analysis of glycerol 3-phosphate, alanine, and serine in HeLa cells cultured with [U-13C]-glutamine under the condition of hypoxia or inhibition by antimycin A (1 µM). Data were from three independent cultures. Error bars represent mean ± SD. **, P < 0.01; Student’s t test. Ac-CoA, acetyl-CoA; OAA, oxaloacetate.
Figure S4.
Glutamine-derived metabolites upon ETC dysfunction. (A–F) Mass isotopomer analysis of succinate, α-ketoglutarate, citrate, acetyl-CoA, malate, and aspartate in HeLa or 4T1 cells cultured with [U-13C]-glutamine under the condition of hypoxia or inhibition by antimycin A (1 µM). The relative pool size of the metabolite is shown on the right. (G) Mass isotopomer analysis of intracellular or secretory lactate in HeLa cells cultured with [U-13C]-glutamine under hypoxia or inhibition by antimycin A (1 µM). The relative pool size of the metabolite is shown on the right. (H) Secretion of lactate from HeLa cells cultured with [U-13C]-glutamine under the conditions of hypoxia or inhibition by antimycin A (1 µM). Data were from three independent cultures. Error bars represent mean ± SD. *, P < 0.05; **, P < 0.01; Student’s t test. Ac-CoA, acetyl-CoA; AntA, antimycin A; Contl, control; Hypo, hypoxia.
As shown in Eq. 4, Φ′(δ) prefers the nonoxidative pentose phosphate pathway (PPP; ∑ΔYnPPP Rib_0,Glc) to reduce the production of electrons in the condition of hypoxia and ECT inhibition (Fig. 1 B). Our results showed that the fraction of [3-2H]-glucose–derived [2H]-labeled NADPH m+1 via the oxidative PPP was observed to decline upon hypoxia and ETC inhibition (Fig. S3 F). The analysis of enrichment of [1,6-13C2]-glucose in sedoheptulose 7-phosphate, a specific intermediate of PPP, further demonstrated that sedoheptulose 7-phosphate m+2 in the nonoxidative PPP increased while sedoheptulose 7-phosphate m+1 via the oxidative PPP decreased in both HeLa and 4T1 cells under hypoxia or ETC inhibition (Fig. 2, B and C; and Fig. S3 G). In fact, even in the normal condition, the fraction of sedoheptulose 7-phosphate m+2 was more than that of m+1. These data suggest that the nonoxidative PPP, instead of the oxidative PPP, is preferentially used by cancer cells, in particular under hypoxia.
Sugar, serine, alanine, and glycerol are directly derived from glucose or glycolytic intermediates; thus, their biosynthesis initiated from glucose prevails in proliferating cells (Fig. 2 D and Fig. S3, H and I). In contrast, proline is readily synthesized from glutamate that is mainly produced from glutamine (Fig. 2 E and Fig. S3, H and I). Consistently, these metabolic preferences support Φ′(δ). In addition, hypoxia and ETC inhibition promoted glucose–derived glycerol 3-phosphate and glutamine–derived proline and meanwhile increased their cellular levels because of their positive ΔY values (ΔYGlo+2,Glc and ΔYPro+4,Glu > 0; Fig. 2, D and E), which contributes to Φ′(δ). Glutamate is the product of glutamine degradation and can be directly excreted if it is beyond the cellular requirement. Therefore, we measured the level of glutamate and proline in the medium and found that glutamine–derived glutamate was reduced while glutamine–derived proline was significantly enhanced in the medium under hypoxia or ETC inhibition (Fig. 2 F), further supporting the role of proline biosynthesis from glutamate.
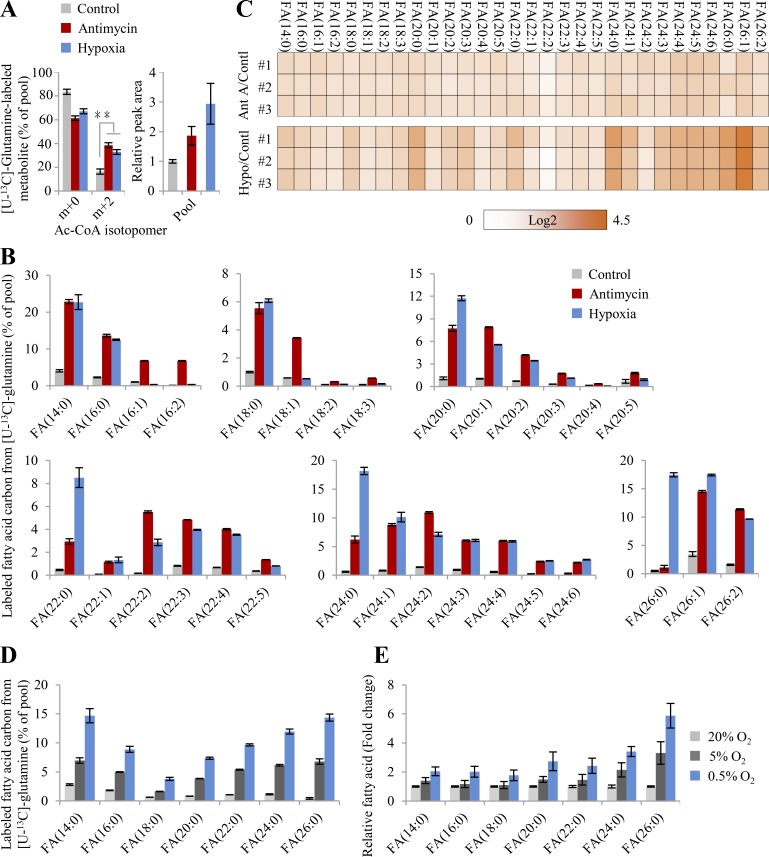
As suggested by Eq. 4, glutamine is rewired for the synthesis of fatty acid through the reductive pathway than CAC-mediated oxidation under ETC dysfunction. We next traced cellular metabolism using [U-13C]-glutamine tracing analysis (Fig. 2 G). Through the forward CAC, glutamine–derived α-ketoglutarate m+5 was predominantly oxidized to succinate m+4, malate m+4, oxaloacetate m+4 that could be converted into aspartate m+4, citrate m+4, and isocitrate m+4 (the oxidative pathway in Fig. 2 G). Through the backward CAC, glutamine–derived α-ketoglutarate m+5 was reduced by carboxylation to isocitrate m+5 and citrate m+5 that was further lysed to acetyl coenzyme A (acetyl-CoA) m+2 and oxaloacetate for the generation of aspartate m+3 and malate m+3 (the reductive pathway in Fig. 2 G). In fact, several recent reports already showed that hypoxia or ETC inhibition induced reductive glutamine metabolism (Metallo et al., 2011; Mullen et al., 2014; Mullen et al., 2011; Wang et al., 2019; Wise et al., 2011). Here, these observations were confirmed again using HeLa and 4T1 cells in the conditions of hypoxia and antimycin A inhibition (Fig. S4, A–F). Diagnostically, hypoxia and antimycin A significantly promoted the fraction and level of glutamine–derived citrate m+5 through the reductive pathway, while it decreased those of citrate m+4 in the oxidative pathway (Fig. 2 H and Fig. S4 C). Finally, glutamine-13C was significantly enriched in almost all of the measured fatty acids that were synthesized from their precursor, glutamine–derived acetyl-CoA, upon ETC dysfunction (Fig. 3, A and B). Moreover, the contents of these fatty acids were also enhanced under these conditions (Fig. 3 C).
Figure 3.
Glutamine-initiated lipogenesis upon ETC dysfunction. (A) Mass isotopomer analysis of acetyl-CoA in HeLa cells cultured with [U-13C]-glutamine for 8 h under the condition of hypoxia or inhibition by antimycin A (1 µM). (B) Labeled carbons in fatty acids in HeLa cells cultured with [U-13C]-glutamine for 48 h under the condition of hypoxia or inhibition by antimycin A (1 µM). (C) Relative cellular levels of fatty acids in HeLa parallel with (B). Fold changes (log2) relative to the control are colored. (D) Labeled carbons in fatty acids in HeLa cells cultured in the HPLM with [U-13C]-glutamine for 8 h under different concentrations of oxygen. (E) Relative cellular levels of fatty acids in HeLa parallel with (D). Data were from three independent cultures. Error bars represent mean ± SD. **, P < 0.01; Student’s t test. Ac-CoA, acetyl-CoA; AntA, antimycin A; Contl, control; FA, fatty acid; Hypo, hypoxia.
To facilitate fatty biosynthesis, glutamine–derived oxaloacetate, as the concomitant of acetyl-CoA (Fig. 2 G), has to be removed. Since the CAC is inactivated under ETC dysfunction, oxaloacetate could be converted to secretory lactate. Indeed, we observed that glutamine–derived lactate m+2 from malate m+3 in the reductive pathway increased while that of m+3 from malate m+4 in the oxidative pathway decreased inside cells or in the medium under ETC dysfunction (Fig. S4 G). However, glutamine–derived lactate was negligible, <0.5% of the pool (Fig. S4 G), suggesting an alternative pathway to scavenge oxaloacetate. Recently, we identified a specific metabolic pathway where glutamine–derived oxaloacetate was converted to aspartate using its amine group and then further integrated, with its amide nitrogen, into secretory dihydroorotate under hypoxia (Wang et al., 2019). Therefore, through such a pathway, cells can use glutamine as the carbon source only for acetyl-CoA and safely remove the remaining carbon and nitrogen.
Furthermore, simplifying Eq. 4 by removing all the variables with ΔY = 0 and rearranging it yielded
| (5) |
Electrons were minimally produced by the anabolic transformations, including CAC, serine synthesis, and nucleic acid synthesis, here designated as ∑ΔY−. The assimilation of glycerol 3-phoshphate is coupled to glutamate-initiated lipid synthesis (Fig. 1 E), with electron production being denoted as ∑ΔYLip+,Glu. Thus, Eq. 5 is simplified as Eq. 6:
| (6) |
Obviously, glutamine/glutamate–initiated syntheses of both lipid and proline essentially play a determinative role in metabolically dissipating electrons to support cell proliferation under the condition of hypoxia and ETC inhibition. Therefore, ETC dysfunction provoked the uptake of glutamine (Fig. S4 H), consistent with previous reports (Metallo et al., 2011; Wang et al., 2019). Furthermore, we applied human plasma–like medium (HPLM; Cantor et al., 2017; Che et al., 2020 to test cellular metabolism at physiological (5%), hypoxic (0.5%), and normal oxygen (20%) concentrations. Compared with normal oxygen concentration, 5% oxygen slightly increased the fraction and content of [U-13C]-glutamine–derived citrate m+5 (Fig. 2 I) but obviously induced the 13C-labled fatty acids (Fig. 3, D and E). By contrast, hypoxia significantly triggered glutamine reductive metabolism (Fig. 2 I) and glutamine-initiated fatty acid biosynthesis (Fig. 3, D and E) relative to 5% or 20% oxygen concentration in HeLa cells cultured in HPLM.
Potential electron consumption by secretory proline synthesized from glutamate
In addition to metabolic rewiring, cells may also survive ETC dysfunction by excreting metabolites that were reduced from the absorbed nutrients, with these metabolic transformations consuming electrons (ΔY > 0; Fig. 4 A). Based on Y of metabolites listed in Table 1, the conversions of glutamate to proline and pyruvate/oxaloacetate to lactate have positive ΔY values (Fig. 4 B). Therefore, the supplementation of glutamate, pyruvate, or oxaloacetate may facilitate cell survival under ETC dysfunction. Under some conditions, cell proliferation was seriously blocked by ETC inhibition. This was reported to result from decreased cellular aspartate by accumulated electrons (Birsoy et al., 2015; Sullivan et al., 2015). Our recent report demonstrated that accumulated electrons largely pushed aspartate into secretory dihydroorotate, which led to the reduced cellular aspartate (Wang et al., 2019). Herein, our 13C-tracing results again confirmed that the cellular level of aspartate significantly decreased, although it was actively synthesized from glutamine, but not glucose, through the reductive pathway upon ETC dysfunction, featured by the enhanced fraction and level of [U-13C]-glutamine–labeled aspartate m+3 under ETC dysfunction (Figs. S3 H and S4 F). Cells require a large amount of aspartate for biosynthesis and protein to initiate proliferation (Garcia-Bermudez et al., 2018; Sullivan et al., 2018) that could dissipate electrons by reprogrammed metabolic pathways according to Eq. 4. More directly, as a necessary component of the malate-aspartate shuttle that accounts for electron homeostasis between the cytosol and mitochondria, the scarcity of aspartate could globally impair electron consumption in cells, given that the involved enzymes, GOT1, GOT2, MDH1, and MDH2, were not significantly affected by hypoxia and ETC inhibition (Fig. 4 C). Recently, it was reported that the addition of aspartate, pyruvate, and oxaloacetate rescued cells from ETC inhibition (Birsoy et al., 2015; Sullivan et al., 2015; Titov et al., 2016). Here, we found that HeLa cells at a low cell density were somehow arrested upon ETC inhibition by antimycin A but not under hypoxia (Fig. 4 D). Therefore, we used this condition to further test the rescuing effects of glutamate, as well as aspartate, pyruvate, oxaloacetate, and some related metabolites including other amino acids, lactate, malate, and acetate, on proliferation of several cell lines, MDA-MB-231, MCF-7, A549, HeLa, HepG2, 8133, SKOV3, and 4T1 cells that were treated with antimycin A. We found that like aspartate, pyruvate, or oxaloacetate, supplementation of glutamate, not other metabolites tested, indeed rescued proliferation of cells treated with antimycin A (Fig. 4 E). These results were verified again in HeLa cells (Fig. 4 F). Moreover, knockdown of P5CS, the critical enzyme involved in the proline biosynthesis pathway, significantly suppressed the rescuing effect of glutamate on antimycin A inhibition (Fig. 4 G).
Figure 4.
Glutamate flux to proline rescues cell proliferation under ETC dysfunction. (A) A model to show that cells adapt to ETC dysfunction by metabolically excreting electrons. (B) Electron consumption of conversion of pyruvate, oxaloacetate, and glutamate to lactate and proline. (C) Western blot for HIF1α, GOT1/2, and MDH1/2. The whole lysis was collected for Western blot after HeLa cells were treated under hypoxia or with antimycin A (1 µM) for 8 h. 30 µg protein was loaded for each lane. (D) Effects of hypoxia and antimycin A on cell proliferation. Fold change in cell number (log2) of HeLa cells at different cell densities as indicated after a 3-d treatment with 1 µM of antimycin A or hypoxia. (E) Effects of exogenous nutrients on proliferation of cancer cells inhibited by antimycin A (1 µM). 2,000 cells per well in a 24-well plate were cultured in DMEM. 1 ml of treatment media containing 5 mM of nutrient as indicated and 1 µM of antimycin A were added at day 0. Final cell counts were measured 7 d after treatment, and fold changes in cell number relative to the control without additional nutrient were colored. (F) Supplementation of pyruvate, oxaloacetate, and glutamate rescues HeLa cells from proliferation inhibition by antimycin A. HeLa cells (2,000 cells per well in a 24-well plate) were seeded and treated in the same conditions described in D. Fold changes in cell number (log2) were calculated. (G) Effects of exogenous glutamate on proliferation of HeLa cells with or without P5CS knockdown inhibited by antimycin A (1 µM). The experimental condition was the same as in E. (H and I) Mass isotopomer analysis of glutamate and proline in HeLa cells cultured with 2 mM of [U-13C]-glutamate for 8 h under the condition of hypoxia or inhibition by antimycin A (1 µM). The pool size is also shown. Data were from three independent cultures. (D–G) Data were from triplicate experiments, and all experimental data were verified in at least two independent experiments, except in E. Error bars represent mean ± SD. *, P < 0.05; **, P < 0.01; Student’s t test. AntA, antimycin A; Asp, aspartate; e, electron; OAA, oxaloacetate.
Furthermore, we traced cellular glutamate and proline using [U-13C]-glutamate and observed that hypoxia and antimycin A promoted glutamate absorption and its flux to proline (Fig. 4, H and I). These results suggest that glutamate–derived proline biosynthesis, not proline itself, supports cell proliferation under hypoxia, considering that proline supplementation did not rescue cells from antimycin A inhibition. However, like aspartate (Wuu et al., 1988), the concentrations of pyruvate and oxaloacetate in human blood are very low (Krebs, 1950); thus, they unlikely exert in vivo a positive role in removing intracellular electrons in proliferating cells suffering from hypoxia. By contrast, blood glutamate is relatively rich; thus, glutamate flux to proline could be potentially employed by cells, which is actually incorporated into Eq. 6.
Electron accumulation drives glutamine-initiated proline biosynthesis and lipogenesis
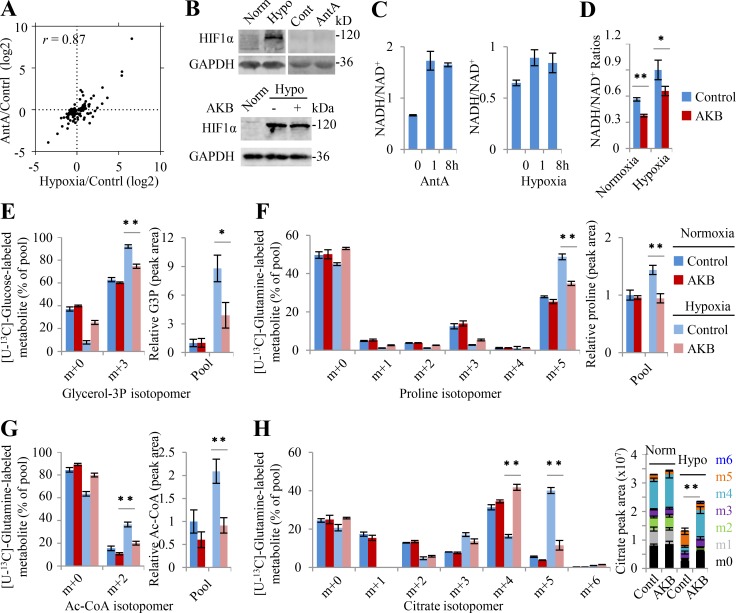
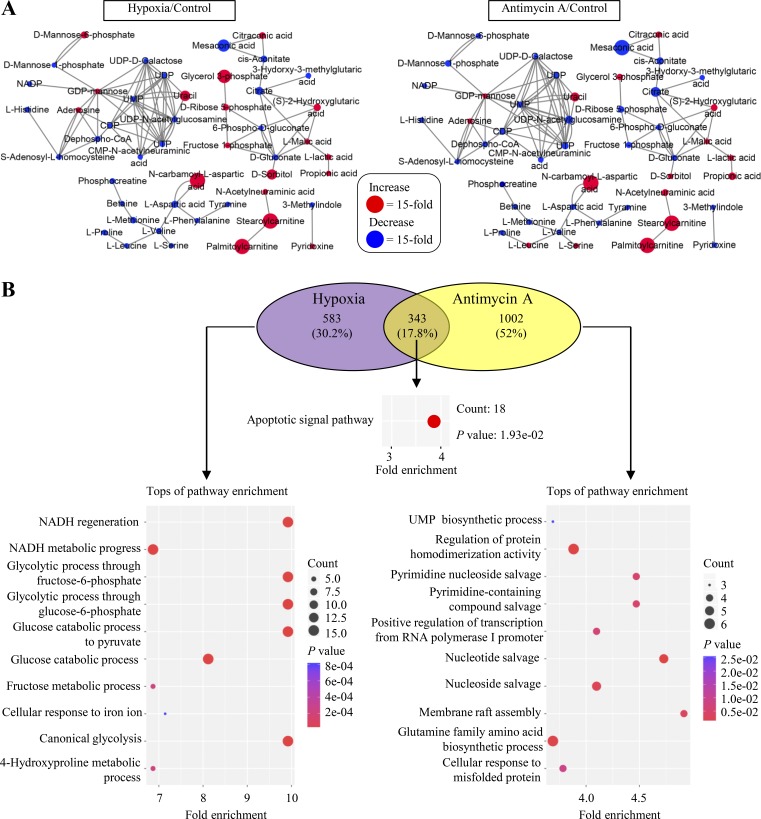
Collectively, hypoxia and ETC inhibition rewired metabolic pathways almost in the same fashion according to Φ′(δ) in Eq. 4, and they also induced similar metabolic profiling with correlation coefficient r = 0.87 (Figs. 5 A and S5 A). These results suggest that hypoxia and antimycin A may share the same mechanism underlying metabolic reprogramming. However, the gene ontology analysis on RNA sequencing data showed that no overlap gene was enriched in the metabolic pathways (Fig. S5 B), suggesting that the regulation of gene transcription could not be the direct cause of metabolic reprogramming under hypoxia and ETC inhibition. Indeed, unlike hypoxia, antimycin A did not increase the protein level of transcriptional factor HIF1α (Fig. 5 B). It was well characterized that antimycin A blocked the ETC to accumulate cellular NADH and thus enhance the ratio of NADH/NAD+ (Fig. 5 C). Here our data showed that hypoxia also increased the cellular NADH/NAD+ ratio (Fig. 5 C), hinting as a possibility that NADH accumulation plays a critical role.
Figure 5.
Electron accumulation drives glutamine-initiated proline biosynthesis and lipogenesis. (A) 105 metabolites in HeLa cells significantly changed by hypoxia or antimycin A. Cells were treated under hypoxia or with antimycin A (1 µM) for 8 h. Metabolites were measured by LC-MS (n = 6). r, the correlation coefficient for hypoxia- and antimycin A–induced metabolites. (B) Western blot for HIF1α. The whole lysis was collected for Western blot after HeLa cells were treated under hypoxia or with antimycin A (1 µM) for 8 h in the absence or presence of 1 mM α-ketobutyrate (AKB). 30 µg protein was loaded for each lane. (C) NAD+/NADH ratios in HeLa cells were measured. Cells were treated under hypoxia or with antimycin A (1 µM) for 1 h or 8 h. (D) The ratios of NADH/NAD+ and NADPH/NADP+ in HeLa cells cultured under normoxia or hypoxia for 8 h in the absence or presence of 1 mM AKB. (E) Mass isotopomer analysis of glycerol 3-phosphate (GP3) in HeLa cells cultured with [U-13C]-glucose under normoxia or hypoxia for 8 h in the absence or presence of 1 mM AKB. The relative pool size is also shown. (F–H) Mass isotopomer analysis of proline, acetyl-CoA, and citrate in HeLa cells cultured with [U-13C]-glutamine under normoxia or hypoxia for 8 h in the absence or presence of 1 mM AKB. The relative pool size is also shown. Data were from three independent cultures, except in C and D. Data in C and D were from triplicate experiments, and all experimental data were verified in at least two independent experiments. Error bars represent mean ± SD. *, P < 0.05; **, P < 0.01; Student’s t test. Ac-CoA, acetyl-CoA; AntA, antimycin A; Cont/Contl/Contrl, control; Hypo, hypoxia; Norm, normoxia.
Figure S5.
Hypoxia and antimycin A induce similar changes in metabolites but not in gene expressions. (A) Metamapp visualization of metabolic changes in HeLa cells under the condition of hypoxia or inhibition by antimycin A (1 µM). Red nodes represent metabolites with increased signal intensity in stem cells; blue nodes represent metabolites with decreased signal intensity in stem cells (P < 0.05). Node sizes scale with fold change. (B) Gene ontology biological process analysis on RNA-sequencing data of HeLa cells under the condition of hypoxia or inhibition by antimycin A (1 µM). Up-regulated genes were analyzed here, and no overlap gene was enriched in metabolic pathways.
To test whether NADH accumulation accounted for hypoxia-induced metabolic rewiring, α-ketobutyrate was used to attenuate the NADH/NAD+ ratio. α-Ketobutyrate is an analogue of pyruvate and can be reduced by lactate dehydrogenases (LDHs) to α-hydroxybutyrate, which is secreted out of cells (Sullivan et al., 2015). This process concomitantly converts NADH + H+ to NAD+, consuming electrons. As expected, α-ketobutyrate did not affect hypoxia-induced HIF1α but significantly reversed the increased NADH/NAD+ ratio under hypoxia (Fig. 5, B and D). As a result, α-ketobutyrate significantly restored the fractions and levels of glucose–derived glycerol 3-phosphate m+3 (Fig. 5 E), glutamine–derived proline m+5 (Fig. 5 F), acetyl-CoA m+2 (Fig. 5 G), and citrate m+5/m+4 (Fig. 5 H) under hypoxia. Taken together, these data strongly suggest that electron accumulation drives glutamine–initiated proline biosynthesis and lipogenesis under hypoxia.
Proline biosynthesis and lipogenesis compensate each other under hypoxia
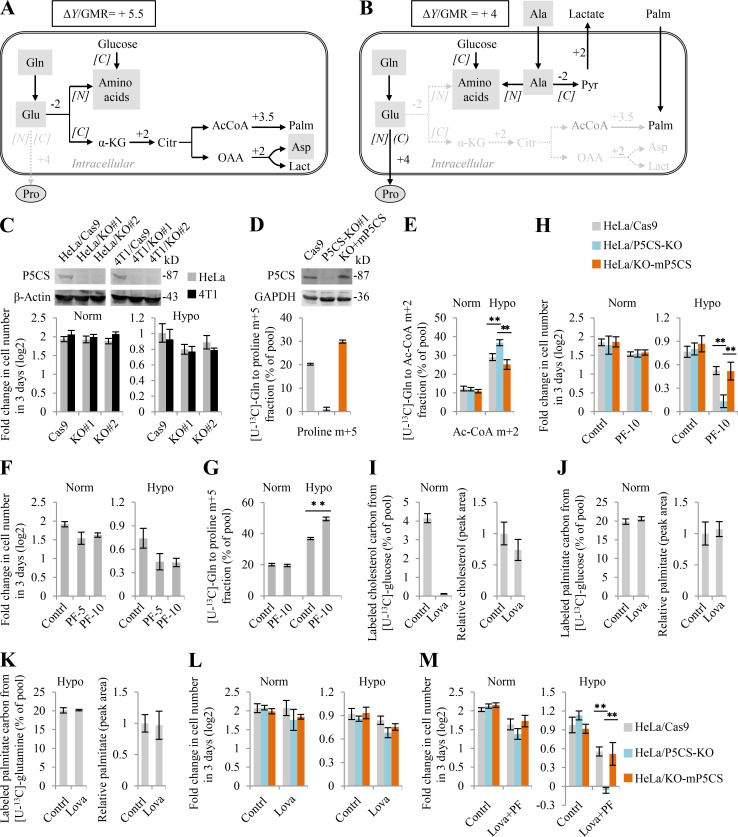
As revealed in Eq. 6, cancer cells can accommodate to hypoxia by initiating glutamine/glutamate–derived lipid and/or proline. The total available glutamate could be primarily diverted to synthesize lipids to consume more electrons under hypoxia (+5.5 in Fig. 6 A versus +4 in Fig. 6 B). Nevertheless, the conversion of glutamate to proline has a more efficient ΔY/ATP ratio of +4 compared with glutamate-initiated palmitate synthesis with a ΔY/ATP of +2.93 (Table S4). Therefore, cells may use extracellular lipid and shift glutamate from lipid synthesis to secretory proline synthesis in the hypoxic in vivo microenvironment, only if they can directly acquire amino acids or obtain a nitrogen source for amino acid synthesis, such as alanine, the second-most abundant amino acid in human blood (Fig. 6 B). The carbon atoms of alanine can be converted to excretory lactate, and this process does not produce electrons (ΔY = 0; Fig. 6 B).
Figure 6.
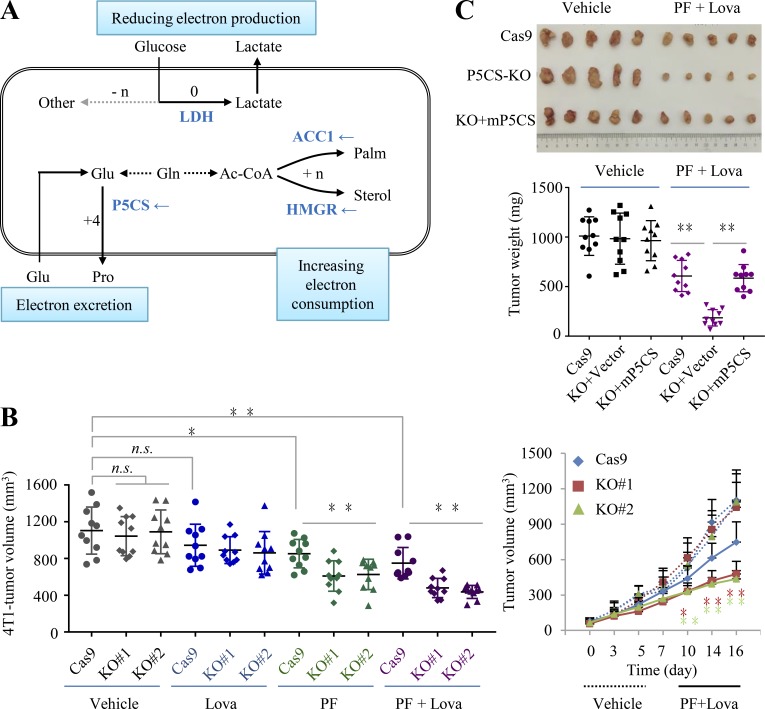
Glutamine–derived proline biosynthesis and lipogenesis compensate each other under hypoxia. ΔY/GMR refers to the overall ΔY value for the equivalent of each glutamate-associated metabolic reaction. (A) A model to show the glutamate-initiated palmitate biosynthesis. (B) A model to show glutamate–derived secretory proline. Cells use exogenous lipids/palmitates, and glutamate is used to synthesize proline that is secreted out of cells. Amino acids are absorbed or assumed to synthesize from the alanine amino group. (C) Proliferation rate of P5CS-expressing (HeLa/Cas9 and 4T1/Cas9) and P5CS-null (HeLa/KO#1, HeLa/KO#2, 4T1/KO#1, and 4T1/KO#2) cells. Gray boxes show amino acids; black arrows show the active pathways. (D and E) Mass isotopomer analysis of proline and acetyl-CoA in HeLa/wt, HeLa/P5CS-KO, and HeLa/KO-mP5CS cells cultured with [U-13C]-glutamine in the proline-contained medium under hypoxia or normoxia. (F) Proliferation rate of HeLa cells treated with or without ACC1 inhibitor PF-05175157 (PF; 5 µM or 10 µM). (G) Mass isotopomer analysis of proline in HeLa cells cultured with [U-13C]-glutamine in the proline-contained medium under hypoxia or normoxia. Cells were treated with or without 10 µM of PF-05175157. (H) Proliferation rate of HeLa/wt, HeLa/P5CS-KO, and HeLa/KO-mP5CS cells in the proline-contained medium treated with or without 10 µM of PF-05175157 under normoxia and hypoxia. (I) Labeled carbons in cholesterol in HeLa cells cultured with [U-13C]-glucose for 48 h under the normal condition in the presence or absence of lovastatin (1 µM). The relative pool size is also shown. (J) Labeled carbons in palmitate in HeLa cells cultured with [U-13C]-glucose for 48 h under the normal condition in the presence or absence of lovastatin (1 µM). The relative pool size is also shown. (K) Labeled carbons in palmitate in HeLa cells cultured with [U-13C]-glutamine for 48 h under hypoxia in the presence or absence of lovastatin (1 µM). The relative pool size is also shown. (L and M) Proliferation rate of HeLa/wt, HeLa/P5CS-KO, and HeLa/KO-mP5CS cells in the proline-contained medium treated with or without 1 µM of lovastatin and/or 10 µM of PF under normoxia and hypoxia. Data in C, F, H, L, and (M were from triplicate experiments, and all experimental data were verified in at least two independent experiments. Data in D, E, G, and I–K were from three independent cultures. Error bars represent mean ± SD. **, P < 0.01; Student’s t test. AcCoA/Ac-CoA, acetyl-CoA; Contrl, control; Hypo, hypoxia; Lact, lactate; Lova, lovastatin; Norm, normoxia; OAA, oxaloacetate; Palm, palmitate.
To further determine the effects of glutamine–initiated lipogenesis and –derived proline biosynthesis on cell proliferation under hypoxia, we tried to suppress the two pathways respectively or in combination. Since no inhibitor against proline synthesis is available, we knocked out P5CS in HeLa and 4T1 cells that proliferated normally in the proline-contained medium (Fig. 6 C), although glutamine–derived proline was completely blocked (Fig. 6 D). However, inhibition of proline synthesis by depleting P5CS marginally sensitized cells to hypoxia (Fig. 6 C). This should be attributable to the fact that cells usually have active de novo lipogenesis, the major electron-dissipated process. Moreover, we observed that P5CS KO even promoted glutamine–derived acetyl-CoA in P5CS-null HeLa cells under hypoxia (Fig. 6 E). Reexpression of P5CS in the P5CS-null cells restored proline biosynthesis and declined acetyl-CoA production from glutamine under hypoxia (Fig. 6, D and E). Furthermore, we used the inhibitor PF-05175157 to suppress acetyl-CoA carboxylase (ACC1), the rate-limiting enzyme for fatty acid synthesis (Griffith et al., 2014), and found that it inhibited cell proliferation but also promoted glutamine–derived proline biosynthesis under hypoxia (Fig. 6, F and G). Moreover, ACC1 inhibition in combination with P5CS KO significantly increased its killing ability in HeLa cells under hypoxia but not in the normal condition (Fig. 6 H). In addition to fatty acids, sterols can also assimilate acetyl-CoA by the rate-limiting enzyme HMGR (Fig. 7 A). However, only ∼4% [U-13C]-glucose–labeled cholesterol was measured under normoxia, and even no [U-13C]-glutamine–labeled cholesterol was detected under hypoxia (Fig. 6 I). By contrast, in the same samples, ∼20% of fatty acid carbons were derived from glucose or glutamine (Fig. 6, J and K). Lovastatin was a potent HMGR inhibitor (Tobert, 2003) and almost completely inhibited the biosynthesis of cholesterol in HeLa cells at 1 μM (Fig. 6 I), but it did not affect fatty biosynthesis (Fig. 6, J and K). Under such a concentration, lovastatin did not affect or marginally affected cell growth alone (Fig. 6 L) or in combination with PF-05175157 (Fig. 6 M) under normoxia and hypoxia. Taken together, these results show that glutamine-initiated lipogenesis and proline biosynthesis could compensate for each other upon their blockage under hypoxia.
Figure 7.
Blocking proline biosynthesis sensitizes in vivo tumor to lipogenesis inhibition. (A) Potential targets to block electron-consuming metabolic pathways. (B and C) Simultaneous inhibition of P5CS, ACC1, and HMGR synergistically suppressed tumor growth in vivo. P5CS is the key enzyme in the synthesis of proline, and it is knocked out in 4T1 and HeLa cells. PF-05175157 (PF) and lovastatin (Lova) are the inhibitors of ACC1 and HMGR. Here, tumors from vehicle-treated and 30 mg/kg Lova plus 100 mg/kg PF-treated nu/nu mice on day 16 for 4T1 tumors and day 28 for HeLa tumors are shown. Mice with 4T1–derived tumors were dosed with intraperitoneal injection (Per IP) once every other day, while those with HeLa–derived tumors were treated once a day with the indicated drugs. The ruler with centimeter scale was used as a reference. Error bars represent mean ± SD. *, P < 0.05; **, P < 0.01; Student’s t test (n = 10). n.s., nonsignificant. Ac-CoA, acetyl-CoA; Lova, lovastatin; Palm, palmitate.
Blocking P5CS sensitizes in vivo tumor growth to inhibition of lipogenesis
Considering the hypoxic microenvironment of in vivo cancers, we next tested whether the simultaneous blockage of both proline and lipid syntheses can disable tumor growth. Based on the above results in the cultured HeLa cells, consuming electrons seemed to be majorly afforded by fatty acid synthesis, but it could be context dependent, in particular in the in vivo conditions. Recently, cholesterol biosynthesis was reported to support the growth of liver tumor with depletion of fatty acid synthase, another critical enzyme involved in fatty acid synthesis, in an animal model (Che et al., 2020). Although the detailed mechanism remained unknown, now it was reasonable to be attributed to their compensation effects on electron consumption as described in the current study.
Statins, the inhibitors of HMGR, are not good cancer drugs, but they could suppress the potential compensation pathway for lipogenesis inhibition. Therefore, herein we used both PF-05175157 and lovastatin to block lipogenesis. In the xenograph model, we observed that P5CS KO did not affect tumor growth but did sensitize 4T1–derived tumors to PF-05175157 and/or lovastatin (Fig. 7 B), confirming that proline biosynthesis at least can partially compensate the blockage of lipogenesis. Furthermore, the combination of PF-05175157 with lovastatin significantly suppressed P5CS KO HeLa–derived tumor growth, and this effect was reversed by the reexpression of P5CS (Fig. 7 C). These results suggest that blocking proline biosynthesis can sensitize in vivo tumor growth to lipogenesis inhibition.
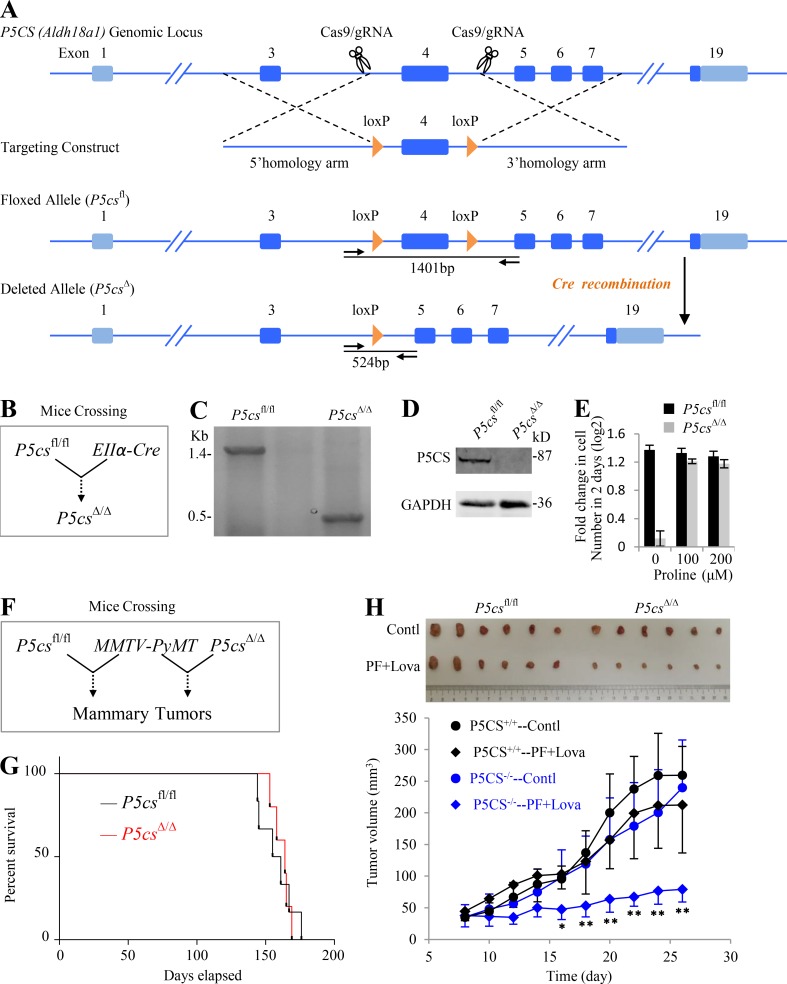
To comprehensively understand the physiological function of P5CS, we generated a mouse model that allows conditional deletion of P5CS exon 4 using CRISPR-mediated homologous recombination (Fig. 8 A). Mice with homozygous P5CS conditional alleles (P5CSfl/fl) were crossed with those (EIIα-Cre) expressing Cre recombinase driven by EIIα promoter, which can drive gene expression in most mouse tissues, including the germ cells (Lakso et al., 1996), to produce progeny containing the heterozygous P5CS conditional allele and EIIα-Cre (P5CS+/fl EIIα-Cre); these heterozygous mice were then crossed with each other to delete P5CS exon 4 (Fig. 8 B). The P5CSΔ/Δ genotype was verified using a PCR-based strategy and Western blot in MEFs (Fig. 8, C and D). Surprisingly, P5CSΔ/Δ progeny normally reproduced, were healthy, and had no overt phenotype, although P5CSΔ/Δ MEFs proliferated in vitro depending on proline supplementation (Fig. 8 E). These results suggest that the nutritional proline source was sufficient to support the development and growth of P5CSΔ/Δ mice.
Figure 8.
Inhibiting proline biosynthesis and lipogenesis synergistically suppressed in vivo tumor growth. (A) A schematic describes generation of a floxed (loxP flanked P5CS exon 4) mouse, P5CSfl/fl, using CRISPR/Cas9. (B) A schematic shows generation of a P5CS exon 4–deleted mouse, P5CSΔ/Δ, by crossing P5CSfl/fl to EIIα-Cre mice. (C) PCR genotyping of genomic DNA from P5CSfl/fl and P5CSΔ/Δ MEFs. Primers anneal flanks of the loxP sites as indicated by arrows in (A) and produce amplicons of 1,401 bp for the P5CSfl/fl allele and 524 bp for the P5CSΔ/Δ allele. (D) Western blot analysis of P5CS protein from P5CSfl/fl and P5CSΔ/Δ MEFs. (E) Proliferation rate of P5CSfl/fl and P5CSΔ/Δ MEFs in the absence or presence of proline supplementation as indicated. Experiments were done in triplicate in two independent experiments. Error bars represent mean ± SD. (F) A schematic shows generation of a model of breast cancer by crossing P5CSfl/fl and P5CSΔ/Δ mice, respectively, to MMTV-PyMT mice. (G) Kaplan-Meier survival curve comparing P5CSfl/fl (n = 6) and P5CSΔ/Δ (n = 5) mice expressing MMTV-PyMT. (H) Effects of lipogenesis inhibition on P5CSfl/fl and P5CSΔ/Δ tumor growth. Tumors were transplanted to nude mice and treated with vehicle or 30 mg/kg Lova plus 100 mg/kg PF once a day for 19 d. The ruler with centimeter scale was used as a reference. Error bars represent mean ± SD. *, P < 0.05; **, P < 0.01; Student’s t test (n = 6). Contl, control; Lova, lovastatin; PF, PF-05175157.
Next, to investigate the role of P5CS in tumor formation and progression, P5CSfl/fl and P5CSΔ/Δ mice were respectively crossed to an established model of breast cancer (mammary tumor virus [MMTV]–polyoma middle-T antigen [PyMT]; Guy et al., 1992; Fig. 8 F). MMTV-PyMT in mice can lead to formation of mammary tumors by ∼10–12 wk. Our results indicated that P5CSΔ/Δ MMTV-PyMT mice showed similar tumor-associated mortality compared with their P5CSfl/fl MMTV-PyMT counterparts (Fig. 8 G), again confirming that P5CS deletion alone did not affect tumor onset. We then transplanted P5CSΔ/Δ MMTV-PyMT and P5CSfl/fl MMTV-PyMT tumors to the groins of nude mice. These tumors expanded at a similar rate (Fig. 8 H). Surprisingly, P5CSfl/fl MMTV-PyMT tumors scarcely responded to the treatment of PF-05175157 and lovastatin. By contrast, P5CSΔ/Δ MMTV-PyMT tumors were dramatically suppressed by them (Fig. 8 H). Taken together, our data strongly suggest that inhibitions of the biosynthesis of proline and lipid can synergistically suppress in vivo tumor growth.
Discussion
Due to a fast growth rate and poor vasculature, tumor cells often suffer from an insufficient supply of nutrition and oxygen (Martinez-Outschoorn et al., 2017). To maintain uptake of nutrients, tumor cells usually enhance expression of their transporters (Flier et al., 1987; Wise et al., 2008; Wise and Thompson, 2010). However, oxygen passively diffuses into cells; thus, tumor cells are unable to acquire enough oxygen under hypoxia. Instead, they adapt to such harsh conditions by optimizing their metabolism, as revealed by our electron balance model.
Our model clearly shows that to enable electron transfer under hypoxia, all the strategies taken by cells mainly converge on three points, reducing electron production, increasing electron consumption, and increasing electron excretion (Fig. 7 A). It is important to point out that although our model can dictate the possibilities, it is unable to predict which metabolic route(s) could be used alone or in combination by a cell. In fact, metabolic reprogramming can be promoted by numerous signal transduction pathways with a high degree of heterogeneity in cancer cells (Boroughs and DeBerardinis, 2015; Martinez-Outschoorn et al., 2017; Pavlova and Thompson, 2016). Bypassing these intricate and interchangeable cascades, we can now mainly control electron transfer under hypoxia by inhibiting the synthesis of both proline and lipid through a cocktail of treatments. Moreover, these inhibitors are expected to be nontoxic to normal cells that can survive on blood lipid and proline supplies. In addition, blockage of LDH A/B can force glucose to be intracellularly catabolized and produce electrons, which should synergistically work with suppression of electron dissipation. Therefore, a combination of LDH, ACC1, HMGR, and P5CS could be a promising broad-spectrum treatment for solid tumors.
Hypoxia-induced metabolic reprogramming is highly associated with and most likely driven by the accumulation of electrons (Fig. 5), which is also supported by our recent finding that hypoxia-induced NADH accumulation rewires glutamine nitrogen metabolism (Wang et al., 2019). However, hypoxia up-regulates expression of genes enriched in NADH-associated metabolic processes (Fig. S5 B), suggesting that the hypoxia-associated signal pathways could help maintain cells in a reprogrammed metabolic state adapting to ETC dysfunction. Intriguingly, hypoxia, as a physiological condition, usually triggers the generation of ROS that results from insufficient electrons transferring to electron acceptors, such as oxygen (Bell et al., 2007). Therefore, it can actually potentiate the hypoxic situation. Nonetheless, the intracellular metabolic intermediate ROS may activate various signal pathways (Saikolappan et al., 2019) to rewire cellular metabolism according to the electron balance model. For example, under hypoxia, cells require an increased level of ROS to stabilize HIF1α protein (Bell et al., 2007). In return, HIF1α signal can stimulate glucose fermentation to lactate (Doherty and Cleveland, 2013), inactivate α-ketoglutarate dehydrogenase and pyruvate dehydrogenase (Kim et al., 2006; Sun and Denko, 2014), and increase the nonoxidative PPP (Shukla et al., 2017), which assists in reducing electron production. It also elevates fatty acid synthase expression (Furuta et al., 2008) and thus lipogenesis to consume electrons.
There are currently controversial opinions on the roles of NADPH produced in the oxidative PPP in cancers. Supporting data on oxidative PPP mainly focus on its antioxidant activity by producing NADPH (Patra and Hay, 2014). However, epidemiological investigations demonstrated that G6PD deficiency, the rate-limiting enzyme in the oxidative PPP, did not affect the incidence of cancers (Cocco et al., 2007; Ferraris et al., 1988). Recently, some reports showed that oncogenic signals, such as KRAS and HIF1α, can increase nonoxidative PPP activity (Shukla et al., 2017; Ying et al., 2012). Here, our results show that hypoxia and ETC inhibition can shift from oxidative PPP to nonoxidative PPP and emphasize that the nonoxidative PPP, compared with the oxidative PPP, reduces electron production to facilitate electron transfer in cancer cells under hypoxia. Complementary to several recent reports showing that proline synthesis may play a critical role in supporting in vivo growth of some cancers (Liu et al., 2012; Loayza-Puch et al., 2016), our results show that hypoxia stimulates glutamine–derived proline biosynthesis and its excretion. Moreover, our model reveals that the process of proline biosynthesis, rather than proline itself, is important for cancer cell growth under hypoxic conditions, especially in situations where lipogenesis is inhibited. As for the main components of lipids, fatty acid and cholesterol are synthesized from acetyl-CoA, and the processes consume electrons, with ΔY = +3.5/acetyl-CoA regarding palmitate biosynthesis. However, if cells make acetyl-CoA from glucose, electrons are released (ΔY = −4/acetyl-CoA) and even exceed those dissipated in lipid biosynthesis. When acetyl-CoA biosynthesis starts from glutamine via the reductive pathway, it expends electrons (ΔY = +2/acetyl-CoA) concomitantly with production of aspartate or lactate (Fig. 1 E). Therefore, glutamine-initiated de novo lipogenesis can really dissipate electrons. Alternatively, if acetate is used to generate acetyl-CoA without electrons involved (ΔY = 0/acetyl-CoA), it can also render de novo lipogenesis as the electron acceptor. It could explain why cancer cells addict to acetate under hypoxia (Kamphorst et al., 2014; Schug et al., 2016). In addition, our results show that hypoxia stimulates the biosynthesis of glucose–derived glycerol 3-phosphate (ΔY = +2/glycerol 3-phosphate), another component of lipid. Taken together, our model establishes the conditioned biosynthesis of proline and lipid as the electron bin to enable electron transfer under hypoxia. To ensure their biosynthesis, proline and lipid are possibly either excreted out or accumulated if beyond the requirement for biomass, as observed in the current study (Figs. 2 F and 3 C). However, we could not rule out the possibility that hypoxia meanwhile activates the absorption of lipids in cancer cells. Very recently, a report showed that hypoxia and ETC inhibitions can induce lipid accumulation in or secretion from cells, although it focused on the effect of polyunsaturated fatty acid biosynthesis on maintaining cellular NAD+ recycling (Kim et al., 2019). These observations also exactly support our hypothesis.
Lipid synthesis occurs in the cytosol and expends NADPH. Increasing evidence supports that the major intracellular NADPH source in cancer cells is not the oxidative PPP but some substitutive metabolic reactions (Boroughs and DeBerardinis, 2015), such as oxidation of malate to pyruvate (Jiang et al., 2013; Son et al., 2013), conversion of isocitrate to α-ketoglutarate (Metallo et al., 2011; Mullen et al., 2011), and serine/one carbon metabolism (Fan et al., 2014; Lewis et al., 2014; Ye et al., 2014). The continuous supplement of NADPH by these reactions relies on metabolic shuttles between the cytosol and mitochondria. Notably, these shuttles essentially convert mitochondrial NADPH/NADH to cytosolic NADPH; thus, they do not really produce net electrons but just redistribute them and/or change their forms. In addition, the malate-aspartate shuttle between the cytosol and mitochondria can maintain NADH homeostasis between the cytosol and mitochondria (LaNoue and Williamson, 1971), and NADH and NADPH can be easily converted to each other in mitochondria by some enzymes, such as NAD(P)+ transhydrogenase, IDH2/IDH3, glutamate dehydrogenase, and ME2/ME3 (Fig. S1; full list of abbreviations listed in Table S5). Therefore, with these metabolic shuttles and reversible conversions, intracellular electrons can be harmoniously used by electron-consuming reactions, such as lipogenesis and proline synthesis under hypoxia.
It should be noted that our current model only focused on central carbon metabolism in the cultured cells. It could be very complicated for the in vivo metabolic transformations. However, Eq. 2 can be conveniently used to analyze the additional metabolic pathways. To facilitate electron transfer, any metabolic transformation with ΔY > 0 could be of benefit for the survival of cancer cells under hypoxia or with deficient mitochondria. Therefore, treatments for tumors could be potentiated by disabling electron transfer. One may expect some other strategies for counteracting electron dissipation, blocking angiogenesis to reduce supply with oxygen as the electron acceptor, impairing the ETC with biguanides to exacerbate hypoxic effects, starving with serine/glycine and nucleotides to force cells to synthesize these metabolites (ΔY < 0), and so on. Generally, any means to neutralize Φ′(δ) established on Eq. 2 could be a potential treatment for cancers alone or in combination with inhibition biosynthesis of lipid and proline.
Overall, our model suggests a promising combination of targets to control tumor growth under hypoxia. Moreover, this treatment can be further intensified by blocking electron transfer established on our concept. Therefore, it is anticipated that the model of electron balance developed in the current study could help us to better understand cancer metabolism and its therapeutic application in the future.
Materials and methods
Derivation of Eq. 2
To conveniently follow the states of cellular metabolites involved in transformations, we only need to focus on carbon-containing metabolites because only the oxidation states of carbon atoms are changeable, with the electron acceptors or donors being excluded. Therefore, as for a transformation,
| (7) |
all the carbon-containing compounds are listed so that the quantity of carbon atoms is conserved. Electrons produced by a metabolic conversion can be quantitatively calculated by Y change, ΔYP,S, between Y values of the initial substrates and those of the final products.
| (2) |
To prove Eq. 2, we then make the quantities of nitrogen and oxygen atoms conserved by adding x moles of NH3 and y moles of H2O as the products and thus change the reaction in Eq. 7 to
| (8) |
In the reaction in Eq. 8, the quantities of carbon, nitrogen, and oxygen atoms are conserved. Notably, x and y could be negative, meaning that NH3 and H2O are actually substrates. Thus,
| (9) |
| (10) |
| (11) |
NC–S, NN–S, and NO–S are the numbers of carbon, nitrogen, and oxygen atoms in substrates, while NC-P, NN-P, and NO-P are those in the products. Based on Eq. 1, Y = 4NC + NH − 3NN − NO, for metabolites,
| (12) |
| (13) |
In Eq. 12 and Eq. 13, NH–P and NH–S are the numbers of hydrogen atoms in substrates and products.
As for the reaction in Eq. 8, where Y of NH3 and H2O is 0,
| (14) |
Hence,
| (2) |
By inserting Eq. 12 and Eq. 13 into Eq. 2, we obtained
| (15) |
Substituting Eqs. 9, 10, and 11 into Eq. 15 yields
| (16) |
In Eq. 16,
is actually the difference of the quantities of hydrogen atoms between substrates and products in the reaction in Eq. 8). Based on the law of conservation of mass, these hydrogen atoms (the electron carriers) must be integrated into electron acceptors such as NAD(P)+ and FMN/FAD if the difference is positive. Otherwise, they come from the electron donor, for example, NAD(P)H + H+ or FMNH2/FADH2. Therefore, ΔYP,S represents the number of electrons produced or consumed in the reactions in Eq. 7 and Eq. 8.
The conversion of substrates to products may have several metabolic routes with different energy requirements, but ΔY between substrates and products keeps constant based on the above analyses. For example, glucose can be oxidized to carbon dioxide via glycolysis and CAC,
or through the PPP,
These processes consume or generate energy, but they still produce the same number of electrons determined by Eq. 2.
The ΔY value is exactly the same as those calculated from the detailed metabolic conversions. 1 mol of glucose produces 10 mol of NADH + H+ and 2 mol of FADH2 in glycolysis and CAC, or 12 mol of NADPH + H+ in the oxidative PPP (Fig. S1). Taken all together, the constant ΔY reflects the chemical nature of biochemical transformations.
As for most redox reactions, NAD(P)+, FMN/FAD, or their reductive forms are used in electron transfer. However, in some cases, reactions may directly use other electron donors, such as tetrahydrofolic acid and ascorbic acid. The resultant oxidized electron donors, dihydrofolic acid and dehydroascorbic acid, need to be finally reduced back for the sake of redox homeostasis, usually using NADPH + H+ as the electron donor (Schnell et al., 2004; Wilson, 2002). Therefore, in essence, these reactions equivalently consume NADPH + H+. In some other cases, oxygen could be directly used as the electron acceptor in the reactions. Oxygen is expected to accept electrons from NADH + H+ through the mitochondrial electron transport chain. If oxygen accepts electrons from metabolites in reactions, the same equivalents of NADH + H+ are left in the cells, in particular under hypoxic conditions. Thus, when used as an electron acceptor, oxygen is essentially equal to NAD+. Taken together, the analyses on the transfer of global electrons in cells are not restricted to the forms of electron acceptor/donor.
General reagents
PF-05175157 was provided by ChemicalBook, and lovastatin was obtained from MedChem Express. [U-13C]-glucose, [1,6-13C]-glucose, [3-2H]-glucose, [1-2H]-glucose, [6,6-2H]-glucose, and [U-13C]-glutamine were purchased from Cambridge Isotope Laboratories. Antimycin A, amino acids, pyruvate, oxaloacetate, α-ketoglutarate, citrate, malate, acetate, dimethyl α-ketoglutarate, triethyl citrate, dimethyl malate, and all general chemicals were obtained from Sigma unless otherwise described.
Cell culture
HeLa, MDA-MB-231, MCF-7, A549, HepG2, 8133, SKOV3, and mouse 4T1 cells were obtained from the American Type Culture Collection. All cell lines tested negative for mycoplasma contamination. All the cells were maintained in DMEM or HPLM supplemented with 10% fetal bovine serum (BioInd) and 50 IU penicillin/streptomycin (Invitrogen) in a humidified atmosphere with 5% CO2 at 37°C. DMEM contained 25 mM glucose, 2 mM glutamine, and 1 mM pyruvate. HPLM was prepared according to previous reports (Cantor et al., 2017; Che et al., 2020), containing 5 mM glucose, 0.55 mM glutamine, and 0.05 mM pyruvate. Hypoxia studies were performed at 0.5% oxygen.
Cell proliferation assay
As for the cell proliferation assay in Fig. 4, D–F, 2 × 103 cells per well were seeded in 24-well plates in DMEM medium without pyruvate supplemented with 10% of dialyzed serum. After overnight incubation for cells to adhere, 3 wells were measured to determine initial count at time of treatment. Cells were washed twice in PBS and 1 ml of treatment media containing 5 mM of nutrient as indicated, and 1 µM of antimycin A was added. After the desired treatments, medium was aspirated from wells, and cells were fixed by adding 500 µl of 4% paraformaldehyde in PBS and incubated at room temperature for 30 min. Cells were stained with 500 µl of 1 µg/ml Hoechst 33345 after paraformaldehyde solution was aspirated. Plates covered with sealing aluminum foil were stored at 4°C before cell counting. Images were collected using an NIS-element AR Nikon ECLIPSE Ti microscope. Four areas (5 × 5 mm2) were scanned and taken to cover the whole well. Images were analyzed, and nuclei number per well was counted with the NIS-Element AR Operation System. The cell-counting method showed a linear range from 1,000 to 100,000 cells per well as determined by using a hemocytometer and the Cell Titer-Glo Luminescent Cell Viability Assay kit (Cat #7570; Promega).
As for the proliferation assay in other experiments, 1 × 105 cells per well was seeded in 12-well plates. Cell number was determined by using the Cell Titer-Glo Luminescent Cell Viability Assay kit.
Generation of P5CS-knockout cell lines
pCDH-Cas9-2A-GFP-BSD was used to express Cas9. Single-guide RNA was cloned into pLentiGuide-puro-Vector (Sanjana et al., 2014) linearized with BsmBI. Two target sequences, 5′-ACGAACATGTCTAATAGCTG-3′ and 5′-GCGCAGGCGCTGTTTGCCGA-3′, were used for mouse P5CS and 5′-ACAGGCAATTCCAGTCTTAG-3′ and 5′-GGGCAGCACAGATGCTGTAC-3′ for human P5CS. We first generated cell lines stably expressing Cas9-2A-GFP-BSD. Viral packaging was done according to a previously described protocol (Li et al., 2010). Briefly, pCDH-Cas9-2A-GFP-BSD was cotransfected with pCMV-dR8.91 and pCMV-VSV-G into 293T cells using calcium phosphate coprecipitation at 20:10:10 µg (for a 10-cm dish). The transfection medium containing calcium phosphate and plasmid mixture was replaced with fresh complete medium after incubation for 5 h. Media containing virus were collected 48 h after transfection passed through a 0.45-mm filter to eliminate cells. Target cells in 6-well tissue culture plates were infected with the virus-containing medium in the presence of polybrene (10 µg/ml) for 48 h. After infection, virus was removed and single cells were colonized. Sub-colonies with a low level of Cas9, 4T1/Cas9, and HeLa/Cas9 were used. Second, 4T1/Cas9 and HeLa/Cas9 were transiently transfected with sgP5CS plasmids using Lipofectamine-3000. Cells were single-cell sorted with a flow cytometer into the wells of a 96-well plate containing 200 ml of RPMI supplemented with 10% FBS. Two plates of single cells were collected for each sgP5CS sequence. Cells were grown for weeks, and the resultant colonies were trypsinized and expanded. Clones were validated for KO of P5CS via immunoblotting and sequencing.
Metabolite isotope tracing
Liquid chromatography–mass spectrometry (LC/MS) analyses were conducted on a TSQ Quantiva triple quadrupole mass spectrometer networked to a Dionex UltiMate 3000 ultra-performance liquid chromatography system (Thermo Fisher Scientific) at the Metabolomics Facility at Tsinghua University Branch of China National Center for Protein Sciences. Multiple reaction monitoring mode was developed using chemical standards. Experiments were performed in medium containing 10% dialyzed FBS (Gibco). Medium was prepared to contain 100% of either the glucose or glutamine pool labeled with 13C and unlabeled other pool. DMEM without glucose, glutamine, pyruvate, and phenol red was prepared from powder (Cat #D5030; Sigma) by adding 3.7 g NaHCO3 per liter and adjusting the pH to 7.4, then supplemented with 10 mM [U-13C]-glucose and 1 mM unlabeled glutamine or 10 mM unlabeled glucose and 1 mM [U-13C]-glutamine. HPLM was prepared to contain 0.55 mM [U-13C]-glutamine, 5 mM unlabeled glucose, and 0.05 mM unlabeled pyruvate.
As for polar metabolite analysis, cells were grown in 60-mm dishes until 80% confluent, then rinsed with PBS and cultured with 2 ml 13C-containing medium for 8 h under normoxia or hypoxia or in the presence of 1 µM antimycin A. Cells were extracted by freeze-thawing three times in 0.5 ml 80% methanol (prechilled to −80°C) after they were rinsed twice with 2 ml room temperature PBS, or 100 µM medium was extracted with 400 µM 100% methanol (prechilled to −80°C). Macromolecules and debris were removed by centrifugation at 14,000 g for 20 min at 4–8°C, and the metabolite-containing supernatants were dried under nitrogen gas. Dried samples were stored at −80°C and then resuspended in 50 µl water and prepared for LC/MS analyses. 1 µl of each sample was injected onto a Synergi Hydro-RP 100A 2.1 × 100–mm column (Phenomenex) for metabolite separation with column temperature at 35°C. Mobile phases A and B were 10 mM thiobarbituric acid in aqueous with pH 5.0 and 100% methanol, respectively. The chromatographic gradient was set for mobile phase B as follows: 0–3.5 min: 1% B; 3.5–22 min: from 1% to 70% B; 22–23 min: from 70% to 90% B; 23–25 min: 90% B; 25–30 min: 1% B. Data were acquired using a positive/negative switching method. Spray voltages of 3.5 kV and 2.5 kV were applied for positive and negative modes, respectively. Q1 and Q3 resolution was set at 0.7, and 1 s of cycle time was used in the method.
Two levels of identification were performed simultaneously using Tracefinder (Thermo Fisher Scientific). Metabolites were first potentially assigned according to endogenous mass spectrometry (MS) database by accurate masses. At the same time, those that could match with the spectra in the fragment database were confirmed in the tandem MS (MS/MS) level. Therefore, both MS/MS-confirmed and potentially assigned metabolites were displayed in the results. 10-ppm and 15-ppm mass tolerance was applied for precursor and fragment matching. Still, a 0.25-min retention time shift was allowed for quantitation.
As for saponified fatty acid and cholesterol analysis, 5 × 105 cells per well were seeded in 6-well plates overnight, then rinsed with PBS and cultured with 3 ml 13C-containing medium for 48 h under normoxia or hypoxia or in the presence of 1 µM antimycin A. Samples were prepared as previously described (Kamphorst et al., 2013). Media were aspirated, and cells were rinsed twice with 2 ml room temperature PBS and then extracted with 1 ml 50% methanol solution containing 0.1 M HCl (prechilled to −80°C). The resulting liquid and cell debris were scraped into a glass vial. Chloroform (0.5 ml) was added, the mixture was vortexed for 1 min and then centrifuged at 3,000 rpm for 15 min, and the chloroform layer was transferred to a glass vial. The extract was dried under N2, reconstituted into 1 ml 90% methanol solution containing 0.3 M KOH, incubated at 80°C for 1 h to saponify fatty acids, acidified with 0.1 ml of formic acid, extracted twice with 1 ml of hexane, and dried under N2. Dried samples were stored at −80°C and then resuspended in 150 µl dichloromethane:methanol and prepared for LC/MS analyses (Xu et al., 2018). Cortecs C18 column (2.1 × 100 mm; Waters) was applied for analysis. Mobile phase A was prepared by dissolving 0.77 g of ammonium acetate in 400 ml of HPLC-grade water, followed by adding 600 ml of HPLC-grade acetonitrile. Mobile phase B was prepared by mixing 100 ml of acetonitrile with 900 ml isopropanol. The gradient was as below: 0 min, 37% B; 1.5 min, 37% B; 4 min, 45% B; 5 min, 52% B; 8 min, 58% B; 11 min, 66% B; 14 min, 70% B; 18 min, 75% B; 20 min, 98% B; 22 min, 98% B; 22.1 min, 37% B; 25 min, 37% B. Some MS parameters were different from polar compound analysis as below: spray voltage, 3.2 kV for positive and 2.8 kV for negative; capillary temperature, 320°C; aux gas flow rate (arb), 10; and mass range (m/z), 240–2,000 for positive and 200–2,000 for negative. Multiple reaction monitoring data were analyzed using Tracefinder. Lipids were assigned by home-built lipid database in “screening” mode and quantified in “quan” mode. Lipids were identified based on matching precursor and characteristic fragment masses. 5-ppm and 10-ppm mass tolerance was used for precursor and fragment, respectively. Only the lipids with chromatographic area >5E6 were considered as confident identification. A 0.25-min retention time shift was allowed for quantitation.
Retention times and mass fragmentation signatures of all metabolites were validated using pure standards. Ion pairs with various 13C labels were derived based on chemical structures of precursors and fragments. The abundance of each mass isotopomer was then mathematically corrected to eliminate natural abundance isotopes and finally converted into a percentage of the total pool. The percentage of labeled fatty acid or cholesterol carbons of the total pool (P) was calculated as the following:
where n is the number of labeled carbons in an m-carbon fatty acid or cholesterol and Pn is the percentage of n-carbon–labeled fatty acid of the total pool.
Untargeted metabolomics
Analyses were performed using an ultra-high-performance liquid chromatography (1290 Infinity LC; Agilent Technologies) coupled to a quadrupole time-of-flight (TripleTOF 6600; AB Sciex) at Shanghai Applied Protein Technology. For hydrophilic interaction liquid chromatography separation, samples were analyzed using a 2.1 × 100–mm ACQUIY ultra-performance liquid chromatography BEH 1.7-µm column (Waters). In both ESI positive and negative modes, the mobile phase contained A = 25 mM ammonium acetate and 25 mM ammonium hydroxide in water and B = acetonitrile. The gradient was 85% B for 1 min and was linearly reduced to 65% in 11 min; it then was reduced to 40% in 0.1 min and kept for 4 min and then increased to 85% in 0.1 min, with a 5-min reequilibration period employed. The ESI source conditions were set as follows: Ion Source Gas1 (Gas1) as 60, Ion Source Gas2 (Gas2) as 60, curtain gas (CUR) as 30, source temperature: 600°C, and IonSpray Voltage Floating ±5,500 V. In MS-only acquisition, the instrument was set to acquire over the m/z range 60–1,000 daltons, and the accumulation time for the time-of-flight MS scan was set at 0.20 s/spectra. In auto MS/MS acquisition, the instrument was set to acquire over the m/z range 25–1,000 daltons, and the accumulation time for product ion scan was set at 0.05 s/spectra. The product ion scan is acquired using information-dependent acquisition with the high-sensitivity mode selected. The collision energy was fixed at 35 V with ±15 eV. Declustering potential was set as ±60 V. The raw MS data (wiff.scan files) were converted to MzXML files using ProteoWizard MSConvert and were processed using XCMS Online (https://xcmsonline.scripps.edu) for feature detection, retention time correction, and alignment. The metabolites were identified by accuracy mass (<25 ppm) and MS/MS data, which were matched with our standards database. In the extracted ion features, only the variables having >50% of the nonzero measurement values in at least one group were kept. For the multivariate statistical analysis, the MetaboAnalyst (http://www.metaboanalyst.ca) web-based system was used. After Pareto scaling, principal component analysis and partial least-squares-discriminant analysis were performed. Leave-one-out cross-validation and response permutation testing were used to evaluate the robustness of the model. The significantly different metabolites were determined based on the combination of a statistically significant threshold of variable influence on projection values obtained from the partial least-squares-discriminant analysis model and two-tailed Student’s t test (P value) on the raw data, and the metabolites with variable influence on projection values >1.0 and P values <0.1 were considered significant.
Lactate production, glucose uptake, and glutamine uptake assay
Measurement of lactate, glucose, and glutamine concentrations in cell culture medium was performed using the M-100 Automatic Biosensors Analyzer (Shenzhen Siemen Technology Co., Ltd) according to user instructions. Cells were grown in 12-well plates at 2 × 105 cells per well for 24 h and then were incubated with the treatment medium containing 10% dialyzed FBS, 10 mM glucose, 1 mM glutamine, and no pyruvate. Cell media were collected at 0 h and 24 h. Aliquots of 100 µl medium were used to measure the concentration of glucose, lactate, and glutamine. Aliquots of the medium without cells under the same conditions were used to measure the concentration of metabolites as the background control. The increased or reduced amount of metabolite in the medium, normalized for area under the curve, was the excretion or uptake of metabolite by a cell per hour. The values were either directly presented or normalized for comparison with the control.
Intracellular NADPH/NADP+ and NADH/NAD+ assay
NADH/NAD+ and NADPH/NADP+ were done using the NAD/NADH-Glo Assay (Cat #G9072; Promega) and NADP/NADPH-Glo Assays (Cat #G9082; Promega) according to the manual instructions with modifications. Briefly, 1.6 × 105 cells per well were seeded in 12-well plates for 24 h and then incubated with the treatment medium for the desired period. Cells were quickly washed once with PBS, extracted with ice-cold lysis buffer (1% dodecyltrimethylammonium bromide in 0.2 N NaOH diluted 1:1 with PBS), immediately frozen at −80°C, and thawed at room temperature. To measure NADH and NADPH, 150 µl of the samples was moved to wells of 48-well plates and incubated at 60°C for 15 min, where basic conditions selectively degrade NAD+ and NADP+. To measure NAD+ and NADP+, another 150 µl of the samples was moved to wells of 48-well plates containing 75 µl of 0.4 N HCl and incubated at 60°C for 15 min, and the acidic conditions selectively degraded NADH and NADPH. Following incubations, samples were allowed to equilibrate to room temperature and then were quenched by neutralizing with 150 µl of HCl/Trizma solution (0.25 M Tris in 0.2 N HCl; NADH and NADPH measurements) or 75 µl of 0.5 M Tris base (NAD+ and NADP+ measurements). 50 µl of the neutralized samples was moved to wells of 96-well white luminometer plates (Cat #3925; CoStar) and mixed with 50 µl of the newly prepared NAD/NADH or NADP/NADPH-Glo Detection Reagent. The mixtures were gently shaken for 30 min at room temperature, and the luminescence was measured using a Synergy H1 Hybrid Multi-Mode Reader (BioTek).
Western blot
After desired treatments as specified as indicated, cells were washed twice with PBS and lysed in buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM b-glycer-ophosphate, 1 mM sodium vanadate, 1 mg ml−1 leupeptin, and 1 mM phenylmethylsulfonyl fluoride). Equal amounts of protein (30 µg) were loaded onto 10% SDS-PAGE gels. Western detection was performed using a Li-Cor Odyssey image reader. The goat anti-rabbit IgG (Cat #P/N 926–68071, 1:10,000 dilution) secondary antibodies were obtained from Li-Cor. GAPDH (Cat #60004–1-lg, 1:5,000 dilution), HIF1α (Cat #20960-1-AP, 1:1,000 dilution), GOT1 (Cat #14886–1-AP, 1:1,000 dilution), GOT2 (Cat #14800–1-AP, 1:1,000 dilution), MDH1 (Cat #15904–1-AP, 1:5,000 dilution), MDH2 (Cat #15462–1-AP, 1:500 dilution), and P5CS (Cat #17719-1-AP, 1:500 dilution) antibodies were obtained from Proteintech.
Animal experiments
The animal protocol was approved by the Institutional Animal Care and Use Committee (IACUC) at Tianjin Medical University, in accordance with the principles and procedures outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The IACUC approval number is E2015093. Female nude mice (4–5 wk, 19–20 g) were purchased from the Experimental Animal Center of Nanjing Biomedical Research Institute at Nanjing University. Mice were administered with a standard and housed and maintained in a pathogen-free house in a 12:12 h light–dark cycle. Temperature and humidity were maintained at 24°C ± 2°C and 50% ± 5%, respectively.
As for 4T1/cas9, P5CS-KO#1, or P5CS-KO#2 cells, 2 × 106 cells were injected subcutaneously into the groins, and 35 mice were used for each cell line. After establishment of palpable tumors, tumor growth was measured every 3 d using digital calipers, and volumes were calculated using the formula 1/2 × L × W2. Tumor prevention studies began at 12 d after tumor cell injection. Mice were randomized into their treatment groups based on similar average tumor volumes. The average tumor volume at the initiation of treatment was 80 mm3. Seven treatment groups were designed: vehicle (n = 5), 15 mg/kg lovastatin (n = 5), 30 mg/kg lovastatin (n = 5), 50 mg/kg PF-05175157 (n = 5), 100 mg/kg PF-05175157 (n = 5), 15 mg/kg lovastatin plus 50 mg/kg PF-05175157 (n = 5), and 30 mg/kg lovastatin plus 100 mg/kg PF-05175157 (n = 5). As for HeLa/Cas9, HeLa/P5CS-KO, and HeLa/KO+mP5CS cells, 2 × 106 cells were injected subcutaneously into the groins, and 10 mice were used for each cell line and divided into two groups: vehicle (n = 5) and 30 mg/kg lovastatin plus 100 mg/kg PF-05175157 (n = 5). Mice with 4T1–derived tumors were dosed by intraperitoneal injection once every other day, while those with HeLa–derived tumors were treated once a day with the indicated drugs. Vehicle solution (0.9% NaCl, 1% Tween 80, and 30% PEG400), lovastatin, and PF-05175157 were dissolved in vehicle solution. All of the mice were killed at the end, and tumors were harvested and weighed.
Generation of transgenic mice
The animal protocol was approved by the IACUC at Tianjin Medical University in accordance with the principles and procedures outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The IACUC approval number is 2016082. The Cre-loxP technology was used to generate P5CS (Aldh18a1) KO mice. The genomic DNA of P5CS exon 4 was amplified and flanked by loxP (floxed) to constructed targeting vector by infusion cloning, carrying 5′ homologous arm (5.4 kb), floxp region (0.8 kb), and 3′ homologous arm (5.7 kb) as shown in the schema model (Fig. 8 A). The CRISPR-mediated homologous recombination was used for generation of floxed P5CS exon 4–knock-in mice. Two guide RNA sequences were, respectively, for cutting the flanks of floxed P5CS exon 4: 5′-TCCCCCAATGCTAGGATTGAAGG-3′ and 5′-TCCTTCAATCCTAGCATTGGGGG-3′ used for the 5′ homologous arm; and 5′-CGGCTCTAGCAATTAGGTTGGGG-3′ and 5′-CTGCCGTGGACCTAAGAGAACGG-3′ for the 3′ homologous arm. Cas9 mRNA, targeting constructed vector and single-guide RNAs, were comicroinjected into the zygote of the C57BL/6J mouse to generate chimeric mice progeny, P5CSfl/+ mice. The phenotypes of P5CS alleles were verified by DNA sequencing. The homozygote (P5CSfl/fl) mice were generated from self-crosses between heterozygote (P5CSfl/+) mice. To harvest P5CSΔ/Δ mice, P5CSfl/fl mice were crossed with EIIa-Cre transgenic mice, in which Cre recombinase was expressed under the control of the adenovirus EIIa promoter during the early embryo period (Lakso et al., 1996) to induce the deletion of P5CS exon 4 at the targeted allele in a wide range of tissues. To explore the role of P5CS in breast cancer, MMTV-PyMT mice were used as murine breast cancer models, which express the PyMT controlled by the MMTV (Guy et al., 1992). P5CSΔ/Δ MMTV-PyMT and P5CSfl/fl MMTV-PyMT female mice were generated by P5CSΔ/Δ and P5CSfl/fl mice mated to MMTV-PyMT mice, respectively, and intercrossed with themselves. The genotypes of the resulting progeny were identified by PCR analysis using specific primers to detect the efficiency of KO P5CS. The primers (P1 and P2) for floxed P5CS and deleted P5CS alleles were 5′-GCTACAACTCCCCTGTCCAA-3′ and 5′-TGGTGGCTCACAACCCTCT-3′. The 1,401-bp and 524-bp amplification fragments of PCR were indicated as P5CSΔ/Δ MMTV-PyMT and P5CSfl/fl MMTV-PyMT female mice. The animals were followed weekly for breast tumors to inoculate.
RNA-seq and data analysis
Construction of an RNA-seq library and RNA-seq was completed by KangChen Bio-tech Company. The sequencing library was determined by Agilent 2100 Bioanalyzer using the Aglient DNA 1000 chip kit, and sequencing was performed on an Illumina Hiseq 4000. Raw data were processed using Solexa Pipeline Version 1.8. The quality control of samples was accomplished using FASTQC V0.11.5. Clean reads were aligned to the human reference genome HG19 by HISAT42. StringTie V1.2.3 was used to calculate gene expression, and Ballgown was used to determine differential expression. Differential expressed genes with log2 (fold change) >1 and false discovery rate <0.001 were considered significant, and then Gene Ontology enrichment analysis was performed. RNA-seq data have been deposited into the Gene Expression Omnibus under accession no. GSE123571.
Statistics
Data are given as means ± SD. Statistical analyses were performed using unpaired two-tailed Student’s t test for comparison between two groups. Asterisks in the figures indicated statistical significances (*, P < 0.05; **, P < 0.01).
Online supplemental material
Fig. S1 shows the metabolic overview. Fig. S2 indicates de novo biosynthesis of nucleosides. Fig. S3 shows glucose–derived metabolites upon ETC dysfunction. Fig. S4 shows glutamine–derived metabolites upon ETC dysfunction. Fig. S5 demonstrates that hypoxia and antimycin A induced similar changes in metabolites but not in gene expressions. Table S1 lists the degree of reduction of metabolites. Table S2 shows the composition of a typical bacterium or mammalian cell. Table S3 calculates the change in the degree of reduction during nucleic acid synthesis. Table S4 shows ΔY/ATP for glutamate-initiated biosynthesis of palmitate or proline. Table S5 lists the full names of enzymes used in Fig. S1.
Supplementary Material
lists the degree of reduction of metabolites.
shows the composition of a typical bacterium or mammalian cell.
shows the change in the degree of reduction during nucleic acid synthesis.
shows ΔY/ATP for glutamate-initiated biosynthesis of proline or palmitate.
lists the full names of enzymes used in Fig. S1.
Acknowledgments
We thank Dr. Wei Du (University of Chicago), Dr. Xinhua Feng (Zhejiang University, China), and Dr. Qunying Lei (Fudan University, China) for carefully reading through the manuscript and valuable discussion. We thank Dr. Xiaohui Liu (Metabolomics Facility at Tsinghua University Branch of China National Center for Protein Sciences, China) for technical help and also thank Dr. Suling Liu (Fudan University, Shanghai, China) for providing materials.
This work is supported by Natural Science Foundation of China grants 81622037, 81672762, and 81372185, and Support Project of High-Level Teachers in Beijing Municipa Universities in the Period of the 13th Five–Year Plan grant CIT&TCD20190333.
The authors declare no competing financial interests.
Author contributions: B. Li conceived and designed the study, developed the model, and derived the equations; M. Liu, Y. Wang, C. Yang, and Y. Ruan performed experiments; Y. Wang, M. Liu, and B. Li analyzed the data; Y. Ruan, Q. Chu, C. Bai, and Y. Cui created some constructs and cell lines; C. Chen and C. Yang provided constructive advice; G. Ying provided constructive advice and thoughtful discussion, and the first letter of his last name, “Y”, was used as the symbol for the degree of reduction of a compound; B. Li wrote the paper.
References
- Alberts B., Wilson J.H., and Hunt T.. 2008. Molecular Biology of the Cell. Garland Science, New York. 1690 pp. [Google Scholar]
- Bell E.L., Klimova T.A., Eisenbart J., Moraes C.T., Murphy M.P., Budinger G.R., and Chandel N.S.. 2007. The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J. Cell Biol. 177:1029–1036. 10.1083/jcb.200609074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J.M., Tymoczko J.L., and Stryer L.. 2011. Biochemistry. Seventh edition Palgrave Macmillan, New York. [Google Scholar]
- Birsoy K., Wang T., Chen W.W., Freinkman E., Abu-Remaileh M., and Sabatini D.M.. 2015. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 162:540–551. 10.1016/j.cell.2015.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroughs L.K., and DeBerardinis R.J.. 2015. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 17:351–359. 10.1038/ncb3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor J.R., Abu-Remaileh M., Kanarek N., Freinkman E., Gao X., Louissaint A. Jr., Lewis C.A., and Sabatini D.M.. 2017. Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell. 169:258–272.e17. 10.1016/j.cell.2017.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che L., Chi W., Qiao Y., Zhang J., Song X., Liu Y., Li L., Jia J., Pilo M.G., Wang J., et al. 2020. Cholesterol biosynthesis supports the growth of hepatocarcinoma lesions depleted of fatty acid synthase in mice and humans. Gut. 69:177–186. 10.1136/gutjnl-2018-317581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco P., Ennas M.G., Melis M.A., Sollaino C., Collu S., Fadda D., Gabbas A., Massarelli G., Rais M., Todde P., and Angelucci E.. 2007. Glucose-6-phosphate dehydrogenase polymorphism and lymphoma risk. Tumori. 93:121–123. 10.1177/030089160709300201 [DOI] [PubMed] [Google Scholar]
- Doherty J.R., and Cleveland J.L.. 2013. Targeting lactate metabolism for cancer therapeutics. J. Clin. Invest. 123:3685–3692. 10.1172/JCI69741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Ye J., Kamphorst J.J., Shlomi T., Thompson C.B., and Rabinowitz J.D.. 2014. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 510:298–302. 10.1038/nature13236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraris A.M., Broccia G., Meloni T., Forteleoni G., and Gaetani G.F.. 1988. Glucose 6-phosphate dehydrogenase deficiency and incidence of hematologic malignancy. Am. J. Hum. Genet. 42:516–520. [PMC free article] [PubMed] [Google Scholar]
- Flier J.S., Mueckler M.M., Usher P., and Lodish H.F.. 1987. Elevated levels of glucose transport and transporter messenger RNA are induced by ras or src oncogenes. Science. 235:1492–1495. 10.1126/science.3103217 [DOI] [PubMed] [Google Scholar]
- Furuta E., Pai S.K., Zhan R., Bandyopadhyay S., Watabe M., Mo Y.Y., Hirota S., Hosobe S., Tsukada T., Miura K., et al. 2008. Fatty acid synthase gene is up-regulated by hypoxia via activation of Akt and sterol regulatory element binding protein-1. Cancer Res. 68:1003–1011. 10.1158/0008-5472.CAN-07-2489 [DOI] [PubMed] [Google Scholar]
- Garcia-Bermudez J., Baudrier L., La K., Zhu X.G., Fidelin J., Sviderskiy V.O., Papagiannakopoulos T., Molina H., Snuderl M., Lewis C.A., et al. 2018. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol. 20:775–781. 10.1038/s41556-018-0118-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude E., Schmidt C., Gammage P.A., Dugourd A., Blacker T., Chew S.P., Saez-Rodriguez J., O’Neill J.S., Szabadkai G., Minczuk M., and Frezza C.. 2018. NADH Shuttling Couples Cytosolic Reductive Carboxylation of Glutamine with Glycolysis in Cells with Mitochondrial Dysfunction. Mol. Cell. 69:581–593.e7. 10.1016/j.molcel.2018.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith D.A., Kung D.W., Esler W.P., Amor P.A., Bagley S.W., Beysen C., Carvajal-Gonzalez S., Doran S.D., Limberakis C., Mathiowetz A.M., et al. 2014. Decreasing the rate of metabolic ketone reduction in the discovery of a clinical acetyl-CoA carboxylase inhibitor for the treatment of diabetes. J. Med. Chem. 57:10512–10526. 10.1021/jm5016022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy C.T., Cardiff R.D., and Muller W.J.. 1992. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12:954–961. 10.1128/MCB.12.3.954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Kok M., and Neidle E.L.. 1992. Functional and evolutionary relationships among diverse oxygenases. Annu. Rev. Microbiol. 46:565–601. 10.1146/annurev.mi.46.100192.003025 [DOI] [PubMed] [Google Scholar]
- Jackson J.B., Leung J.H., Stout C.D., Schurig-Briccio L.A., and Gennis R.B.. 2015. Review and Hypothesis. New insights into the reaction mechanism of transhydrogenase: Swivelling the dIII component may gate the proton channel. FEBS Lett. 589:2027–2033. 10.1016/j.febslet.2015.06.027 [DOI] [PubMed] [Google Scholar]
- Jiang P., Du W., Mancuso A., Wellen K.E., and Yang X.. 2013. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 493:689–693. 10.1038/nature11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst J.J., Cross J.R., Fan J., de Stanchina E., Mathew R., White E.P., Thompson C.B., and Rabinowitz J.D.. 2013. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc. Natl. Acad. Sci. USA. 110:8882–8887. 10.1073/pnas.1307237110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst J.J., Chung M.K., Fan J., and Rabinowitz J.D.. 2014. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab. 2:23 10.1186/2049-3002-2-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.W., Tchernyshyov I., Semenza G.L., and Dang C.V.. 2006. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3:177–185. 10.1016/j.cmet.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Kim W., Deik A., Gonzalez C., Gonzalez M.E., Fu F., Ferrari M., Churchhouse C.L., Florez J.C., Jacobs S.B.R., Clish C.B., and Rhee E.P.. 2019. Polyunsaturated Fatty Acid Desaturation Is a Mechanism for Glycolytic NAD+ Recycling. Cell Metab. 29:856–870.e7. 10.1016/j.cmet.2018.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs H.A. 1950. Chemical composition of blood plasma and serum. Annu. Rev. Biochem. 19:409–430. 10.1146/annurev.bi.19.070150.002205 [DOI] [PubMed] [Google Scholar]
- Lakso M., Pichel J.G., Gorman J.R., Sauer B., Okamoto Y., Lee E., Alt F.W., and Westphal H.. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA. 93:5860–5865. 10.1073/pnas.93.12.5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaNoue K.F., and Williamson J.R.. 1971. Interrelationships between malate-aspartate shuttle and citric acid cycle in rat heart mitochondria. Metabolism. 20:119–140. 10.1016/0026-0495(71)90087-4 [DOI] [PubMed] [Google Scholar]
- Lewis C.A., Parker S.J., Fiske B.P., McCloskey D., Gui D.Y., Green C.R., Vokes N.I., Feist A.M., Vander Heiden M.G., and Metallo C.M.. 2014. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell. 55:253–263. 10.1016/j.molcel.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Gordon G.M., Du C.H., Xu J., and Du W.. 2010. Specific killing of Rb mutant cancer cells by inactivating TSC2. Cancer Cell. 17:469–480. 10.1016/j.ccr.2010.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienhart W.D., Gudipati V., and Macheroux P.. 2013. The human flavoproteome. Arch. Biochem. Biophys. 535:150–162. 10.1016/j.abb.2013.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Le A., Hancock C., Lane A.N., Dang C.V., Fan T.W., and Phang J.M.. 2012. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc. Natl. Acad. Sci. USA. 109:8983–8988. 10.1073/pnas.1203244109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza-Puch F., Rooijers K., Buil L.C., Zijlstra J., Oude Vrielink J.F., Lopes R., Ugalde A.P., van Breugel P., Hofland I., Wesseling J., et al. 2016. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature. 530:490–494. 10.1038/nature16982 [DOI] [PubMed] [Google Scholar]
- Martinez-Outschoorn U.E., Peiris-Pages M., Pestell R.G., Sotgia F., and Lisanti M.P.. 2017. Cancer metabolism: a therapeutic perspective. Nat. Rev. Clin. Oncol. 14:11–31. [DOI] [PubMed] [Google Scholar]
- Metallo C.M., Gameiro P.A., Bell E.L., Mattaini K.R., Yang J., Hiller K., Jewell C.M., Johnson Z.R., Irvine D.J., Guarente L., et al. 2011. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 481:380–384. 10.1038/nature10602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen A.R., Wheaton W.W., Jin E.S., Chen P.H., Sullivan L.B., Cheng T., Yang Y., Linehan W.M., Chandel N.S., and DeBerardinis R.J.. 2011. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 481:385–388. 10.1038/nature10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen A.R., Hu Z., Shi X., Jiang L., Boroughs L.K., Kovacs Z., Boriack R., Rakheja D., Sullivan L.B., Linehan W.M., et al. 2014. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Reports. 7:1679–1690. 10.1016/j.celrep.2014.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra K.C., and Hay N.. 2014. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 39:347–354. 10.1016/j.tibs.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova N.N., and Thompson C.B.. 2016. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 23:27–47. 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikolappan S., Kumar B., Shishodia G., Koul S., and Koul H.K.. 2019. Reactive oxygen species and cancer: A complex interaction. Cancer Lett. 452:132–143. 10.1016/j.canlet.2019.03.020 [DOI] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell J.R., Dyson H.J., and Wright P.E.. 2004. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu. Rev. Biophys. Biomol. Struct. 33:119–140. 10.1146/annurev.biophys.33.110502.133613 [DOI] [PubMed] [Google Scholar]
- Schug Z.T., Vande Voorde J., and Gottlieb E.. 2016. The metabolic fate of acetate in cancer. Nat. Rev. Cancer. 16:708–717. 10.1038/nrc.2016.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S.K., Purohit V., Mehla K., Gunda V., Chaika N.V., Vernucci E., King R.J., Abrego J., Goode G.D., Dasgupta A., et al. 2017. MUC1 and HIF-1alpha Signaling Crosstalk Induces Anabolic Glucose Metabolism to Impart Gemcitabine Resistance to Pancreatic Cancer. Cancer Cell. 32:71–87.e7. 10.1016/j.ccell.2017.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuler M.L., and Kargi F.. 2001. Bioprocess Engineering: Basic Concepts. Second edition. Prentice Hall, Upper Saddle River, NJ. 576 pp. [Google Scholar]
- Son J., Lyssiotis C.A., Ying H., Wang X., Hua S., Ligorio M., Perera R.M., Ferrone C.R., Mullarky E., Shyh-Chang N., et al. 2013. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 496:101–105. 10.1038/nature12040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan L.B., Gui D.Y., Hosios A.M., Bush L.N., Freinkman E., and Vander Heiden M.G.. 2015. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 162:552–563. 10.1016/j.cell.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan L.B., Luengo A., Danai L.V., Bush L.N., Diehl F.F., Hosios A.M., Lau A.N., Elmiligy S., Malstrom S., Lewis C.A., and Vander Heiden M.G.. 2018. Aspartate is an endogenous metabolic limitation for tumour growth. Nat. Cell Biol. 20:782–788. 10.1038/s41556-018-0125-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R.C., and Denko N.C.. 2014. Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab. 19:285–292. 10.1016/j.cmet.2013.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titov D.V., Cracan V., Goodman R.P., Peng J., Grabarek Z., and Mootha V.K.. 2016. Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science. 352:231–235. 10.1126/science.aad4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobert J.A. 2003. Lovastatin and beyond: the history of the HMG-CoA reductase inhibitors. Nat. Rev. Drug Discov. 2:517–526. 10.1038/nrd1112 [DOI] [PubMed] [Google Scholar]
- Wang Y., Bai C., Ruan Y., Liu M., Chu Q., Qiu L., Yang C., and Li B.. 2019. Coordinative metabolism of glutamine carbon and nitrogen in proliferating cancer cells under hypoxia. Nat. Commun. 10:201 10.1038/s41467-018-08033-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.X. 2002. The physiological role of dehydroascorbic acid. FEBS Lett. 527:5–9. 10.1016/S0014-5793(02)03167-8 [DOI] [PubMed] [Google Scholar]
- Wise D.R., and Thompson C.B.. 2010. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 35:427–433. 10.1016/j.tibs.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise D.R., DeBerardinis R.J., Mancuso A., Sayed N., Zhang X.Y., Pfeiffer H.K., Nissim I., Daikhin E., Yudkoff M., McMahon S.B., and Thompson C.B.. 2008. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA. 105:18782–18787. 10.1073/pnas.0810199105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise D.R., Ward P.S., Shay J.E., Cross J.R., Gruber J.J., Sachdeva U.M., Platt J.M., DeMatteo R.G., Simon M.C., and Thompson C.B.. 2011. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. USA. 108:19611–19616. 10.1073/pnas.1117773108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuu J.A., Wen L.Y., Chuang T.Y., and Chang G.G.. 1988. Amino acid concentrations in serum and aqueous humor from subjects with extreme myopia or senile cataract. Clin. Chem. 34:1610–1613. [PubMed] [Google Scholar]
- Xu L., Wang X., Jiao Y., and Liu X.. 2018. Assessment of potential false positives via orbitrap-based untargeted lipidomics from rat tissues. Talanta. 178:287–293. 10.1016/j.talanta.2017.09.046 [DOI] [PubMed] [Google Scholar]
- Ye J., Fan J., Venneti S., Wan Y.W., Pawel B.R., Zhang J., Finley L.W., Lu C., Lindsten T., Cross J.R., et al. 2014. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 4:1406–1417. 10.1158/2159-8290.CD-14-0250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H., Kimmelman A.C., Lyssiotis C.A., Hua S., Chu G.C., Fletcher-Sananikone E., Locasale J.W., Son J., Zhang H., Coloff J.L., et al. 2012. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 149:656–670. 10.1016/j.cell.2012.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the degree of reduction of metabolites.
shows the composition of a typical bacterium or mammalian cell.
shows the change in the degree of reduction during nucleic acid synthesis.
shows ΔY/ATP for glutamate-initiated biosynthesis of proline or palmitate.
lists the full names of enzymes used in Fig. S1.