Chen et al. identify patients with homozygous essential loss-of-function variants in human IL6ST that result in a complete loss of signaling of the whole family of GP130-dependent cytokines. This study identifies GP130 deficiency as the cause of an extended Stüve-Wiedemann syndrome.
Abstract
The gene IL6ST encodes GP130, the common signal transducer of the IL-6 cytokine family consisting of 10 cytokines. Previous studies have identified cytokine-selective IL6ST defects that preserve LIF signaling. We describe three unrelated families with at least five affected individuals who presented with lethal Stüve-Wiedemann–like syndrome characterized by skeletal dysplasia and neonatal lung dysfunction with additional features such as congenital thrombocytopenia, eczematoid dermatitis, renal abnormalities, and defective acute-phase response. We identified essential loss-of-function variants in IL6ST (a homozygous nonsense variant and a homozygous intronic splice variant with exon skipping). Functional tests showed absent cellular responses to GP130-dependent cytokines including IL-6, IL-11, IL-27, oncostatin M (OSM), and leukemia inhibitory factor (LIF). Genetic reconstitution of GP130 by lentiviral transduction in patient-derived cells reversed the signaling defect. This study identifies a new genetic syndrome caused by the complete lack of signaling of a whole family of GP130-dependent cytokines in humans and highlights the importance of the LIF signaling pathway in pre- and perinatal development.
Introduction
GP130 is a ubiquitously expressed cytokine receptor subunit and a key component of the IL-6 cytokine family receptors mediating signaling of 10 cytokines including IL-6, IL-11, IL-27, oncostatin M (OSM), cardiotrophin-1, cardiotrophin-like cytokine, neuropoietin, and leukemia inhibitory factor (LIF; Kang et al., 2019; Murakami et al., 2019). Collectively, these cytokines are important in inflammation, organogenesis, and cell survival. GP130 forms a hexameric receptor complex after binding the IL-6–activated IL-6R or IL-11–activated IL-11R and heterotrimeric GP130-LIF receptor (LIFR)-LIF or GP130-OSMR-OSM receptor complexes. Upon cytokine recognition, signal transduction via GP130 is mediated through the JAK-STAT pathway that principally results in STAT3 phosphorylation (pSTAT3) and activation (Kang et al., 2019; Murakami et al., 2019).
The importance of GP130-dependent signaling has been highlighted by gene deletion studies of Il6st or its cytokine-specific coreceptors (Akira, 2000). Homozygous deletion of Il6st is lethal in utero for mice, due to ventricular myocardial hypoplasia, disrupted placental architecture, and a loss of fetal liver hematopoiesis (Yoshida et al., 1996). This phenotype is similar to animals with targeted disruption of Stat3 (Takeda et al., 1997) or Lifr (Ware et al., 1995). Mice with conditional deletion of Il6st have elucidated the role of GP130 in other organs (Yoshida et al., 1996), including the lungs, central nervous system, immune system, and skeleton (Itoh et al., 2006). Studies of knock-in mouse mutants with impaired activation of either STAT or MAPK pathways by GP130 show that GP130 signaling ensures normal bone growth in a STAT-dependent manner (Sims et al., 2004).
We have previously shown that cytokine-selective defects in GP130 due to homozygous missense variants in IL6ST (c.1210A>T; p.Asn404Tyr and c.1493C>T; p.Pro498Leu) cause immunodeficiency with elevated IgE and craniosynostosis (Schwerd et al., 2017; Shahin et al., 2019). These variants cause selective defects in IL-6, IL-11, IL-27, and OSM signaling, while LIF signaling is preserved. Many features of the phenotype in those patients overlap with the autosomal dominant hyper-IgE recurrent infection syndrome (MIM #147060) that is associated with STAT3 mutations (Holland et al., 2007; Minegishi et al., 2007). It is thought that the skeletal phenotype is caused by defective IL-11 signaling, since patients with pathogenic IL11RA variants present with craniosynostosis and dental anomalies (MIM #614188; Kreiborg-Pakistani syndrome; Keupp et al., 2013; Nieminen et al., 2011). Based on the lethality in mice and the strong evolutionary selection against loss-of-function variants in human genomic databases, it has been assumed that complete absence of GP130 would be lethal in humans (Schwerd et al., 2017; Shahin et al., 2019).
Stüve-Wiedemann syndrome (STWS; MIM #601559) is a semilethal ultra-rare autosomal recessive condition with features of skeletal dysplasia (short bowed tubular bones, osteoporosis, and camptodactyly), respiratory distress, feeding difficulties, hyperthermic episodes, myotonia, and decreased sensitivity to pain. In most patients, STWS is caused by biallelic loss-of-function variants in the LIF receptor gene (LIFR; Dagoneau et al., 2004). Yet some STWS patients lack mutations in LIFR, suggesting genetic heterogeneity of this condition (Jung et al., 2010).
Here, we describe three unrelated families with seven affected individuals that presented with a lethal congenital disorder resembling an extended STWS (or STWS type II). In five patients with available DNA, the molecular diagnosis of homozygous loss-of-function variants in the IL6ST gene was established.
Results and discussion
Pathogenic IL6ST variants in patients with extended STWS
Family A is a consanguineous family from Afghanistan (Fig. 1 A; affected individuals are referred to as A-II-1 and A-II-2). The first fetus (A-II-1) had features of lethal skeletal dysplasia during routine ultrasound at gestational age (GA) 19+4 wk, and pregnancy was terminated (Table 1). Postmortem examination and skeletal radiographs confirmed severe bent bone dysplasia (Fig. 1, B and C). During the second pregnancy (A-II-2), ultrasound at GA 16+0 wk showed similar abnormalities as in the first fetus. The female baby was born via planned Caesarean section at GA 36+3 wk. She had a small thorax, short bowed extremities, and skeletal radiographical features consistent with STWS (Fig. 1, D and E), resembling the bone phenotype of A-II-1 (Fig. 1, B and C). Within a few hours of birth, thrombocytopenia was noted (A-II-2 platelet counts: day 1, 27 × 109 platelets/liter; day 2, 57 × 109/liter; normal range, 80–350 × 109/liter). She developed progressive respiratory insufficiency, heart failure, and irreversible pulmonary hypertension, from which she died 2 d after delivery. Peripheral blood investigations identified normal C-reactive protein (A-II-2 C-reactive protein: day 1, <0.60 mg/liter; day 2, <0.60 mg/liter; normal range, <3.0 mg/liter) and low fibrinogen (A-II-2 fibrinogen: day 1, 0.5 g/liter; day 2, 1.8 g/liter; normal range, 2.0–4.0 g/liter). Postmortem examination confirmed extensive skeletal deformities with uneven bone trabecular pattern in the long bones of extremities (Fig. 1 F; representative normal control for A-II-1 is shown in Fig. S1 A), growth retardation, and membranous lung disease.
Figure 1.
Clinical phenotypes and genetic findings in families A, B, and C. (A) Pedigree of family A. Patients affected by an extended STWS and homozygous for the pathogenic variant NM_002184.3; c.841C>T; p.Arg281* in IL6ST are shown in solid symbols (A-II-1, female fetus from terminated pregnancy; A-II-2, live-born female infant; A-II-3, fetus). (B–E) Radiographs of patient A-II-1 and A-II-2 with a skeletal manifestation with bent bone dysplasia. Radiograph of A-II-1 (B) shows severe bowing of the long bones, particularly striking in the lower extremities. Magnified views of the extremities of A-II-1 and A-II-2 (C, D, and E) demonstrate broadening of the metaphyses with a triangular zone of irregular trabecular pattern and cortical thickening of the concave side of the diaphyses. (F) Overview of H&E staining of femur sections from A-II-1, featuring uneven bone trabeculation pattern in the tubular bones. Scale bar indicates 5 mm. (G) Sanger sequencing of heterozygous c.841C>T variant in the parents (A-I-1 and A-I-2) and unaffected offspring (A-II-3) and homozygous variant in both A-II-1 and A-II-2. (H) Pedigree of family B. Patients with c.841C>T; p.Arg281* in IL6ST are shown in solid black symbols (B-II-3, male fetus; B-II-4, male fetus). (I–K) Fetal ultrasound pictures of patient B-II-3 and B-II-4 showing bent long bones: B-II-3 (I) shows obtuse bending of the femur, and B-II-4 (J and K) shows sharp bending of the tibia, fibula, radius, and ulna. (L) Sequencing of heterozygous c.841C>T variant in the parents (B-I-1 and B-I-2) and homozygous variant in B-II-4. (M) Pedigree of family C. Patient with homozygous intronic IL6ST pathogenic variant (c.1699+4A>G) in solid black symbols (C-II-3). C-II-1 is in solid gray because he was clinically affected but his DNA was not available for sequencing. (N and O) Radiographs of patients C-II-1 and C-II-3, demonstrating mild broadening of the metaphyses with mild undermineralization and diaphyseal cortical thickening. (P) Sequencing trace of the homozygous c.1699+4A>G variant in IL6ST of patient C-II-3. (Q) Sequences covering the end of exon 13 and the start of intron 13 of the IL6ST gene in humans, in comparison to other species from the UCSC genome browser. In A, H, and M, black symbols represent affected status with confirmed mutations; gray solid symbols indicate clinically affected patient whose DNA was not available for genetic analyses. Where genotype has been confirmed by sequencing, it is referred to as either WT/V or V/V under the symbol. WT, wild-type (black); V, variant (red).
Table 1. Clinical features of the patients in families A, B, and C.
| Family A | Family B | Family C | ||||
|---|---|---|---|---|---|---|
| Case | A-II-1 | A-II-2 | B-II-3 | B-II-4 | C-II-1 | C-II-3 |
| Ethnic origin | Afghanistan | Afghanistan | Saudi Arabia | Saudi Arabia | Netherlands | Netherlands |
| Consanguinity | Yes | Yes | Yes | Yes | No | No |
| IL6ST mutations (all homozygous) | c.841C>T; p.Arg281* | c.841C>T; p.Arg281* | c.841C>T;p.Arg281* | c.841C>T; p.Arg281* | c.1699+4A>G | c.1699+4A>G |
| Gender | Female | Female | Male | Male | Male | Female |
| Outcome | Termination of pregnancy | Death at day 2 | Early neonatal death (2–3 h) | Death in utero | Death at day 2 | Death at 6 yr |
| Major clinical phenotype | STWS | STWS | STWS | STWS | STWS | STWS |
| Clinical features | ||||||
| Bone manifestations | Limb bowing; severe deformities; growth defect | Limb bowing; growth defect | Bowed tubular bones | Short tubular bones | Bowing of legs; camptodactyly | Bowing of femora; restricted limb extension; camptodactyly; joint instability; severe scoliosis; short stature |
| Respiratory and dysautonomia symptoms | NA | Breathing difficulties; irreversible pulmonary hypertension; membranous disease in lungs; heart failure | NA | NA | Respiratory distress | Hyperthermia; hyporeflexia; motor delay; sucking and swallowing difficulties |
| Immune and hematologic phenotype | NA | No eczema; no eosinophilia; thrombocytopenia | NA | NA | NA | Eczema at age 1 yra |
| Other | Disseminated intravascular coagulation | Multicystic dysplastic right kidney; dilated cisterna magna; small chest | ||||
| Weight | NA | Birth (GA36+3): 1,734 g; z-score: −2.29 | NA | NA | Birth (GA 40+0): 2,700 g; z-score: −2.09 | Birth (GA 40+0): 2,995 g; z-score −1.13 |
| 2 8/12 yr: 8.9 kg; z-score: −2.33 | ||||||
| Height/length | NA | Birth (GA36+3): 36 cm; z-score: −4.77 | NA | NA | Birth 47 cm (GA 40); z-score: −2.39 | Birth (GA 40+0): 49 cm; z-score −1.23 |
| 2 8/12 yr: 78 cm; z-score: −3.91 | ||||||
BW, birth weight; GA given in weeks + days; NA, not available. Z-scores of weight and length/height were calculated using Voigt et al. (2006) for fetuses and newborns and World Health Organization child growth standards (https://www.who.int/childgrowth/standards/en/) for C-II-3 in childhood.
No clinical records available.
Figure S1.
Bone phenotype, MLPA analysis of the IL6ST gene, RNA-seq expression data of multiple cytokine receptor units in control amniocytes, the parental analysis of GP130 and pSTAT3 response, and PCR analysis. (A) H&E staining of femur bones from affected patient A-II-1 and an age-matched control. Several anatomic regions are labeled and color-coordinated, for guidance in the other panels. Scale bar indicates 1 mm. (B) Growth plate zones following safranin O/fast green (SOFG) staining show resting (left), proliferative (middle), and hypertrophic (right panel) zones. Scale bar indicates 50 µm. (C) Multinuclear osteoclasts in the primary spongiosa were visualized by immunostaining for Cathepsin K and counterstaining with hematoxylin. Scale bar indicates 20 µm. (D) Quantification of the number of nuclei in cathepsin K–positive cells, with only cells containing two or more nuclei counted. (E) Quantification of the size of cathepsin K–positive cells containing two or more nuclei. (C–E) Six visual fields analyzed with three images from each side of the bone; black bars indicate control; gray bars indicate patient A-II-1. Note that colored boxes surrounding images in B and C correspond to regions indicated in A. (F) Ratio for each probe (deletions would cause a ratio of <0.75 and duplications of >1.3). Exons 4, 8, 13, and 17 in IL6ST and genes upstream and downstream of IL6ST (ANKRD55 and IL31RA) were investigated. (G) RNA-seq expression of multiple cytokine receptor units in control amniocytes. In case of multiple RNA-Seq Probe Set IDs of each receptor, the one with highest expression was shown. Blue lines represent the median of n = 11 individual amniocyte samples. GP130-dependent cytokine receptors are shown in red. Each dot represents one individual sample, data taken from Kang et al. (2015). (H–J) GP130 surface expression in ex vivo CD3+CD4+, CD3+CD8+, and CD14+ cells gated from peripheral blood mononuclear cells of heterozygous parents compared with four healthy donors. Representative histograms of one control and the father (left) and geometric mean florescence intensity (gMFI; right) are shown. (K–M) Response of pSTAT3 in heterozygous parents after stimulation with IL-6 (100 ng/ml for 15 min) compared with four healthy controls (gray, unstimulated; blue and red, stimulated). Representative histograms are shown on the left. (H–M) Blue, controls; red, father (upper triangle) and mother (lower triangle). Dashed line indicates the lowest value from the healthy controls. (O–P) Agarose gels for RT-PCR and nested PCR using four primer pairs (pair 1, exon 11 and boundary of exons 14–15; pair 2, exons 12–16; pair 3, exons 12–14; pair 4 exons 11–16). 1, amplicon from cDNA from EBV-LCLs of patient C-II-3 with c.1699+4A>G variant; 2–3, control EBV-LCLs, 100-bp ladder on the side of each gel. (P) Nested PCR using amplicons from exons 12–14 with primer pair 3. Amplicon of expected 218-bp size in control samples versus amplicon of ∼70-bp in patient sample confirming skipping of exon 13. 1:100 and 1:500 indicate dilution ratio for the RT-PCR products.
DNA of patient A-II-2 was investigated by prenatal array-CGH and gene panel sequencing for osteogenesis imperfecta genes, but no pathogenic variants were detected. Similarly, no pathogenic variants were found by whole genome sequencing (WGS) analyzed using a panel of known skeletal dysplasia genes (Genomics England PanelApp; https://panelapp.genomicsengland.co.uk/panels/309/). Subsequent WGS analysis on the two patients and their parents revealed a homozygous nonsense variant in both affected offspring in IL6ST (NM_002184.3: c.841C>T; p.Arg281*), confirmed by Sanger sequencing (Fig. 1 G). The parents and a third fetus (A-II-3, ongoing pregnancy) were heterozygous carriers (Fig. 1 A). The nonsense variant is predicted to delete the C-terminal half of GP130. Irrespective of whether nonsense-mediated decay occurs or a stable version of the truncated protein is expressed, a lack of the membrane-binding region would prevent signaling. The c.841C>T variant has not been observed in our in-house database (n = 1,432), gnomAD, or 1000 Genomes. No other genetic variants in family A explained the phenotype (Table S1).
Comparing bone histology from patient A-II-1 with a control showed grossly normal organization of the growth plate, with swollen chondrocytes and decreased levels of proteoglycans (Fig. S1 B). Less bone matrix was observed in the primary spongiosa (bone volume/total volume was 22.7 ± 1.2% in control versus 12.4 ± 2.0% in A-II-1). This was associated with similar numbers of DMP-1+ late osteoblasts but a 3.5-fold increase in DMP-1+ osteocytes in this region (the number of DMP-1+ cells/mm2 was 1,390 ± 520 in the control versus 4,970 ± 820 in A-II-1). The size and the number of nuclei per cathepsin-K+ osteoclasts were greater in the primary spongiosa of A-II-1 compared with control (Fig. S1, C–E). All these changes suggest decreased activity of the growth plate. There was a substantial increase in diaphyseal bone volume (bone volume/total volume was 9.20 ± 2.36% in control versus 63.0 ± 5.2% in A-II-1), but no changes in the number of osteoblasts, osteocytes, or osteoclasts were detected between the patient and control samples.
We have subsequently identified a second consanguineous family (family B) from Saudi Arabia, with at least two fetuses affected by STWS-like skeletal dysplasia having the same homozygous mutation in c.841C>T; p.Arg281* in IL6ST as family A (Fig. 1 H). This family was previously included in a large-scale, first-tier clinical exome sequencing in a highly consanguineous population, where IL6ST was identified as a candidate gene but not further functionally characterized (Monies et al., 2019). The heterozygous parents had four pregnancies. Two fetuses (B-II-3 and B-II-4) were homozygous for c.841C>T; p.Arg281* (variants identified by exome and Sanger sequencing respectively; sequence of B-II-4 is shown in Fig. 1 L). B-II-3 was born spontaneously at term but died 2–3 h after delivery. He had short femora (less then fifth centile), bowed tibia (Fig. 1 I), multicystic dysplastic right kidney, dilated cisterna magna, and a small chest. Fetal ultrasound of B-II-4 showed normal nuchal translucency scan at 12 wk and short bowed bones (Fig. 1, J and K), and he died in utero. The same couple had two other pregnancies: the first fetus (B-II-1) died in utero with multiple anomalies and bowed bone deformities and was likely affected with the same condition, but no DNA sample was available. The second pregnancy was a miscarriage (B-II-2). Analysis of single nucleotide polymorphism data and runs of homozygosity regions between the two families, A and B, carrying the mutation c.841C>T; p.Arg281*, suggested that the two families were not related.
The third family (family C) is nonconsanguineous, of Dutch origin (Fig. 1 M). This family had two affected children (C-II-1 and C-II-3) and one healthy child (C-II-2). The first child was born at term with bowed femora and tibia (Fig. 1 N) as well as camptodactyly. He died 2 d after birth due to respiratory failure. The second affected child (C-II-3) was born at term and had mildly bowed femora (Fig. 1 O), camptodactyly, hyperhidrotic hands, tented upper lip, and high palate. The patient had infantile sucking, swallowing, and feeding difficulties but survived 6 yr. She had short stature, scoliosis, and motor delay and presented with multiple bouts of hyperthermia that on one occasion caused a febrile convulsion. C-II-3 had normal dentition but was troubled by eczematoid dermatitis from the age of 1 yr onward. At the age of 6 yr, she had scoliosis surgery and died 2 d later due to cardiorespiratory arrest. In family C, no mutations in LIFR were identified by Sanger sequencing (Jung et al., 2010). Subsequently, whole exome sequencing identified a homozygous intronic splice site variant within the IL6ST locus (NC_000005.10:g.55247753A>G; Hg19, c.1699+4A>G) in C-II-3. Among all homozygous or compound heterozygous variants (Table S1), the IL6ST variant was considered the top candidate. The main features of the patients are summarized in Table 1.
Sanger sequencing confirmed the homozygous c.1699+4A>G variant in DNA from EBV-transformed lymphoblastoid cell lines (EBV-LCLs) of C-II-3 (Fig. 1 P). The family has been lost to follow-up, and no DNA sequencing or clinical data are available from the parents, C-II-1, or unaffected C-II-2. To confirm that the apparently homozygous variant in the unrelated family is not caused by a compound heterozygosity with a deletion of the corresponding allele, we performed multiplex ligation-dependent probe amplification (MLPA) with four probes in the IL6ST gene (exons 4, 8, 13, and 17) and two probes in the two genes adjacent to IL6ST (ANKRD55 and IL31RA). No deletion was identified (Fig. S1 F).
The c.1699+4A>G variant is located at the splice donor site of intron 13 of IL6ST (Fig. 1 Q). Natural donor splice site variants at +4 are functionally relevant and highly conserved (Caminsky et al., 2014). The variant is predicted to prevent intron splicing (MaxEntScan tool drop in the score from 4.88 down to 0.52 caused by the variant). The transcript inferred pathogenicity score indicated a damaging splicing effect, with a score of 0.955 (cutoff is 0.4; highest is 1). This site is highly conserved among vertebrates (Fig. 1 Q) and in humans, as no variation was observed in 220,000 alleles of gnomAD, 1000 Genomes, or in-house databases.
Nonsense variant p.Arg281* abrogates GP130 protein expression and signaling
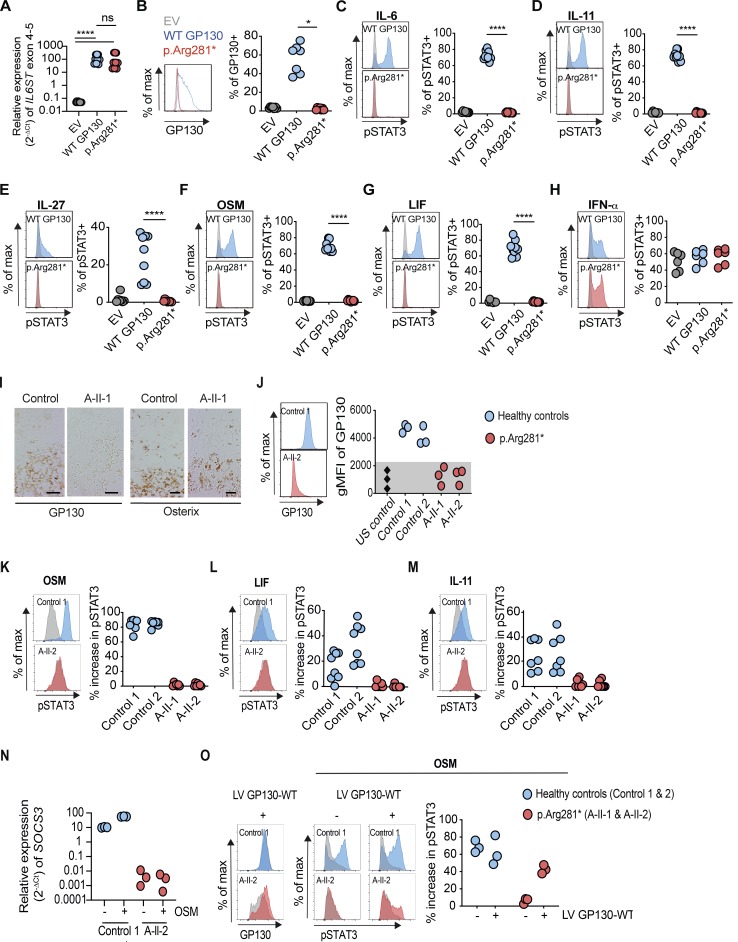
To investigate the in vitro effects of the p.Arg281* variant, we used a GP130-deficient HEK293 cell line in transfection experiments (Fig. 2). As the nonsense variant is predicted to delete the C-terminal half of GP130 (mutation site is in exon 8), we first measured the expression of IL6ST exons 4–5 by quantitative RT-PCR. Exons 4–5 of IL6ST were comparably detectable in GP130-deficient HEK293 cells transfected with plasmid encoding WT GP130 and p.Arg281*, confirming the presence of the truncated GP130 receptor mRNA (Fig. 2 A). Transfection with WT GP130 restored GP130 surface expression, whereas it remained absent in the p.Arg281* transfected cells (Fig. 2 B). Functionally, transfection with WT GP130 plasmid restored pSTAT3 in response to the GP130-dependent cytokines IL-6, IL-11, IL-27, OSM, and LIF, whereas cells transfected with the p.Arg281* variant remained unresponsive (Fig. 2, C–G). By contrast, the GP130-independent pSTAT3 response to IFN-α was unaffected in all conditions (Fig. 2 H).
Figure 2.
Homozygosity for IL6ST p.Arg281* variant completely abrogates GP130-dependent IL-6, IL-11, IL-27, OSM, and LIF signaling. (A) Gene expression (2−ΔCt) of IL6ST exons 4–5 from p.Arg281* transfected GP130-deficient HEK293 cells 24 h after transfection, relative to endogenous control gene RPLPO. Data represent nine technical repeats from three independent experiments. (B) GP130 surface expression in p.Arg281* transfected GP130-deficient HEK293 cells. Results are representative of seven independent experiments. (C–H) Response of pSTAT3 in p.Arg281* transfected GP130-deficient HEK293 after stimulations with IL-6, IL-11, IL-27, OSM, LIF, and IFN-α stimulation (all 100 ng/ml for 15 min). For assessment of IL-6 and IL-11 signaling, cells were cotransfected with plasmids encoding IL-6Rα and IL-11Rα, respectively. Successfully transfected cells were gated based on GFP expression. Representative histograms are shown on the left (gray, unstimulated; blue and red, stimulated). Quantification is based on six independent experiments per cytokine, with one to two replicates per experiment. (A–H) EV, empty vector; WT GP130, wild type. Statistical differences were determined by means of Mann–Whitney U test, ****, P ≤ 0.0001; *, P ≤ 0.05. (I) Immunostaining of the growth plate and primary spongiosa with absent GP130 expression and normal osterix at the border of the hypertrophic zone of A-II-1 compared with an age-matched control fetus. Scale bar indicates 50 µm. (J) GP130 expression in amniocytes from patient A-II-1 or A-II-2 compared with healthy controls by flow cytometry. Geometric mean fluorescence intensity (gMFI) values are shown from representative histograms (left), and dot plots summarize data from three experiments (right). US, unstained. (K–M) Response of pSTAT3 in A-II-1 or A-II-2 amniocytes after stimulations with OSM, LIF, or IL-11 (all 100 ng/ml for 15 min) compared with healthy controls (gray, unstimulated; blue and red, stimulated). Representative histograms (left) are shown with percentage increase of pSTAT3 in cytokine-stimulated cells determined by comparison with unstimulated cells (right). Dot plots summarize data from five experiments per cytokine (right), with one to two replicates per experiment. (N) Gene expression (2−ΔCt) of SOCS3 from amniocytes in the presence or absence of OSM (100 ng/ml) for 24 h, relative to endogenous control gene RPLPO. Data represent measurements from two experiments. (O) GP130 surface expression after lentiviral transduction with WT GP130. Representative histograms of control 1 and A-II-2 are shown on the left (gray, untransduced; blue and red, transduced). Response of pSTAT3 after OSM stimulation (100 ng/ml for 15 min) in control and patient amniocytes that were infected with or without GP130-expressing lentivirus (gray, unstimulated; blue and red, OSM). Three experiments were performed on cells from healthy controls and patients. LV, lentivirus.
Consistent with homozygous nonsense variant, expression of GP130 in femur bone sections of A-II-1 was absent, whereas staining for the osteoprogenitor marker osterix was comparable to controls (Fig. 2 I). Next, we measured the surface expression of GP130 in patient-derived amniocytes by flow cytometry. GP130 protein expression was absent in amniocytes from A-II-1 and A-II-2 but present in healthy controls (Fig. 2 J). Although OSM, LIF, and IL-11 stimulation resulted in variable intensities of pSTAT3 in control amniocytes, the patient-derived amniocytes consistently displayed a complete loss of signaling (Fig. 2, K–M). In agreement with the minimal IL6RA expression (Kang et al., 2015; Fig. S1 G), IL-6-induced pSTAT3 was undetectable in both control and patient amniocytes. We noticed higher pSTAT3 baseline levels in A-II-1 and A-II-2 amniocytes compared with controls (Fig. 2, K–M). This may be explained by a feedback mechanism similar to the negative regulation of GP130 signaling by SOCS3 (Babon et al., 2014; Milner et al., 2015). Consequently, we found a reduction in SOCS3 mRNA expression in unstimulated amniocytes from A-II-2 compared with controls (Fig. 2 N). To confirm the causal relationship between the IL6ST variant and the defective GP130-dependent cytokine response, we performed a rescue experiment via lentiviral transduction of WT GP130. Transduction of WT GP130 partially restored GP130 expression as well as GP130-dependent OSM signaling, as indicated by the increase in pSTAT3 in patient-derived amniocytes (Fig. 2 O).
To investigate the genotype-phenotype segregation in the healthy parents of family A, we measured GP130 expression and pSTAT3 in response to IL-6 stimulation from CD4+, CD8+ T cells, and CD14+ monocytes (Fig. S1, H–M). Both father and mother displayed a mild reduction of GP130 protein expression in primary T cells and monocytes compared with healthy control individuals (Fig. S2, H–J). Monocytes from both parents showed a mild reduction of pSTAT3 in response to IL-6 (Fig. S2, K–M).
Figure S2.
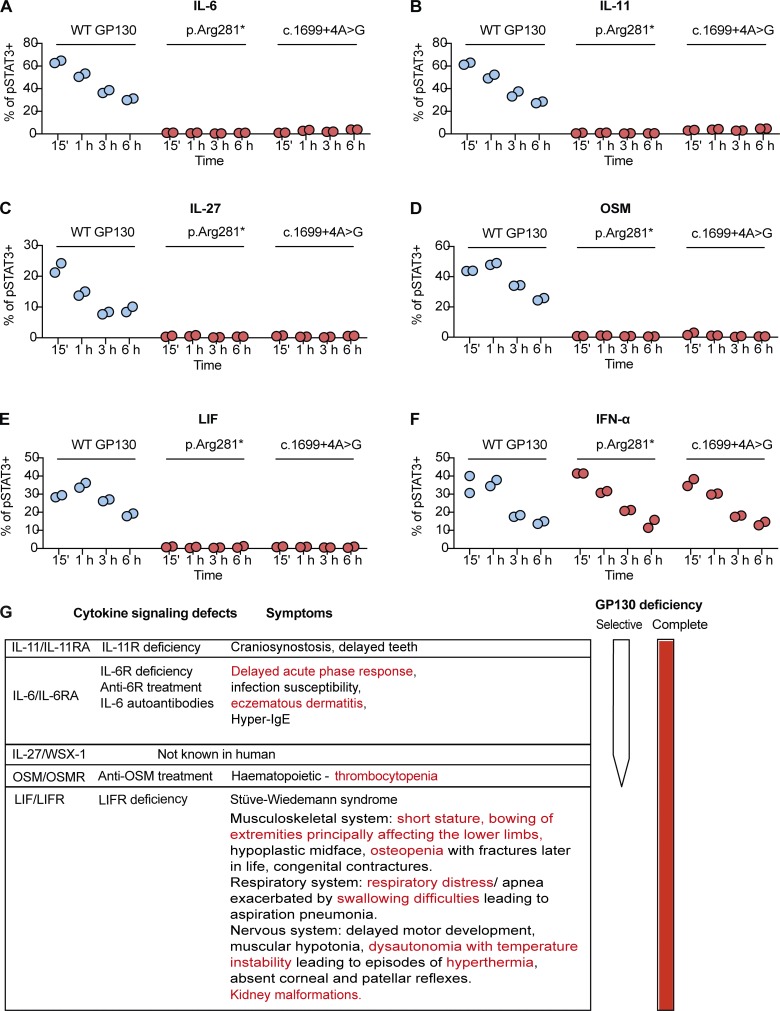
The complete loss of GP130-dependent IL-6, IL-11, IL-27, OSM, and LIF signaling in p.Arg281* and c.1699+4A>G variant is not due to delayed kinetics; and a schematic summary of GP130 deficiency, the relationship between loss of cytokine signaling and clinical symptoms. (A–F) Response of pSTAT3 in c.1699+4A>G transfected GP130-deficient HEK293 after stimulations with IL-6, IL-11, IL-27, OSM, LIF, and IFN-α stimulation (all 100 ng/ml) over a time course covering 15 min, 1 h, 3 h, and 6 h. For assessment of IL-6 and IL-11 signaling, cells were cotransfected with plasmids encoding IL-6Rα and IL-11Rα, respectively. Successfully transfected cells were gated based on GFP expression. Quantification is based on individual independent experiments per cytokine, with duplicates per experiment. (G) A schematic summary of GP130 deficiency and the relationship between loss of cytokine signaling and clinical symptoms. The symptoms described in red were noticed in our patients.
Intronic variant c.1699+4A>G GP130 leads to loss of function despite membrane expression
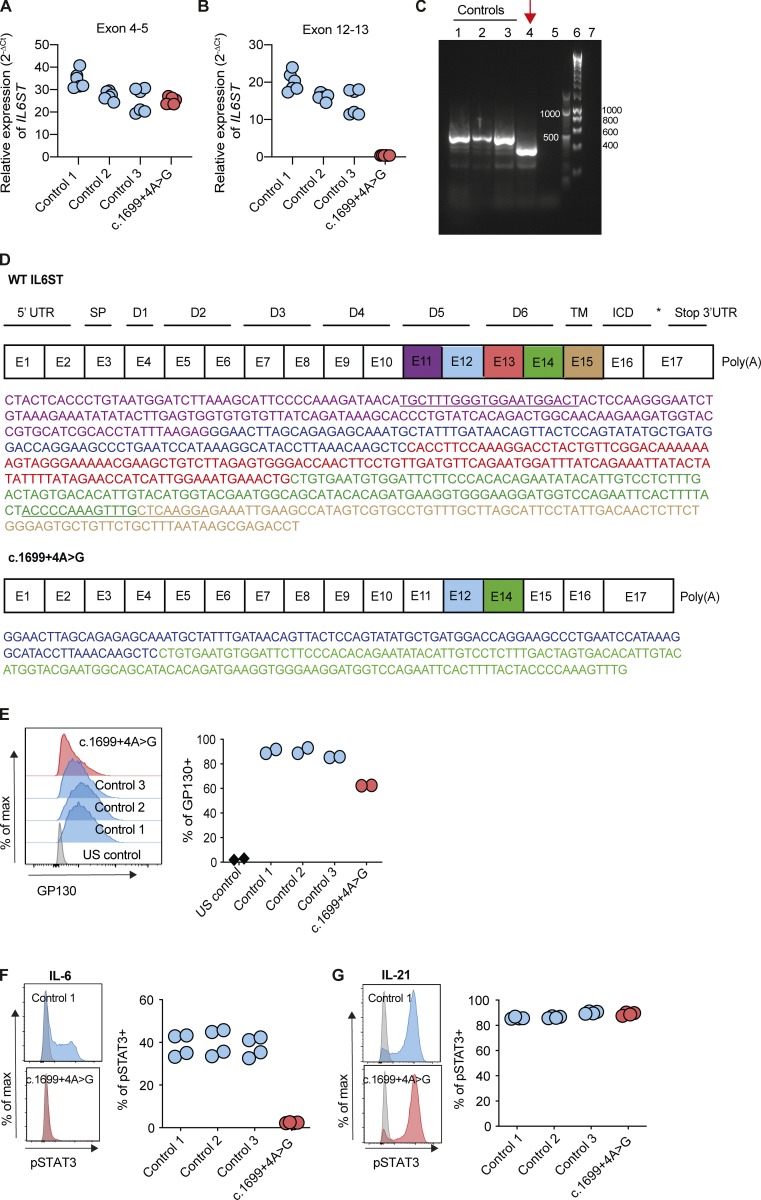
We next investigated the consequences of the intronic pathogenic c.1699+4A>G variant in family C. The variant is located at the splice donor site of intron 13 of IL6ST (Fig. 1 Q). To investigate whether exon 13 is expressed, we first measured mRNA expression of IL6ST exons 4–5 and exons 12–13, respectively. Exons 4–5 of IL6ST were detectable in EBV-LCLs of patient C-II-3 and three controls (Fig. 3 A), while expression of exons 12–13 of IL6ST was absent in C-II-3 (Fig. 3 B), suggesting skipping of exon 13. By fragment size analysis in agarose gel and Sanger sequencing, the PCR generated a longer product in healthy EBV-LCLs, compared with that from patient C-II-3 (Fig. 3 C, lane 4). cDNA sequencing of C-II-3 using primers on exons 11 and 14–15 (primer pair locations underlined in Fig. 3 D) showed absence of exon 13 sequence. A deletion of exon 13 is in-frame and is predicted to lead to a truncated protein retaining its membrane spanning and intracellular domains. Nested RT-PCR confirmed there was no full-length splice product from C-II-3, as indicated by lack of exon 13 amplification (Fig. S1, O and P; primer pair 3). Therefore, at the age when the cells from C-II-3 were obtained, the mutation was not leaky.
Figure 3.
The c.1699+4A>G intronic variant of IL6ST results in exon skipping and a complete loss of IL-6 signaling in EBV-LCLs. (A and B) Gene expression (2−ΔCt) of IL6ST exons 4–5 and exons 12–13 from EBV-LCLs of healthy controls and c.1699+4A>G patient, relative to endogenous control gene RPLPO. Data represent six technical repeats from two experiments. (C) Agarose gel with the amplicon from cDNA of patient C-II-3 c.1699+4A>G variant shows a smaller PCR product. Lane 1–3, control EBV-LCLs; lane 4, c.1699+4A>G variant; lane 5, negative control containing no cDNA; lane 6–7, 1-kb and 100-bp ladder, respectively. (D) A schematic figure of the exon-domain organization of the human IL6ST gene and the loss of exon 13 (red) in the c.1699+4A>G variant, as confirmed by Sanger sequencing. Purple, blue, red, green, and brown correspond to the sequence of exon 11, 12, 13, 14, and 15, respectively. The primer pair sequences used for the PCR products are underlined. E, exon; D, domain; UTR, untranslated region; SP, the hydrophobic signal peptide sequence; TM, transmembrane domain; ICD, intracellular cytoplasmic domain. (E) GP130 surface expression in patient-derived EBV-LCLs. Results are representative of two experiments. Gray, US, unstained; blue, healthy EBV-LCLs, control 1–3; red, c.1699+4A>G variant. (F and G) Response of pSTAT3 in c.1699+4A>G EBV-LCLs after stimulations with IL-6 and IL-21 (50 ng/ml for 15 min) compared with healthy controls. Representative histograms are shown on the left (gray, unstimulated; blue and red, stimulation) and quantification is based on two experiments per cytokine, with duplicates per experiment.
Consequently, we detected the presence of GP130 protein on the surface of EBV-LCLs of C-II-3, but with a 20–30% reduction compared with three healthy control EBV-LCLs (Fig. 3 E). The presence of GP130 raised the possibility of residual function. We stimulated EBV-LCLs from C-II-3 with IL-6 or IL-21 and recorded the presence of pSTAT3 in response to IL-21 but not IL-6 (Fig. 3, F and G).
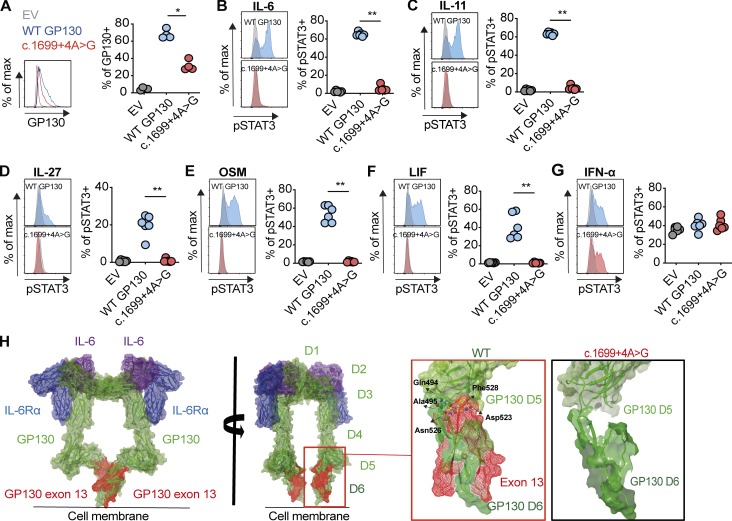
To confirm the functional consequence of c.1699+4A>G mutation on GP130-dependent cytokines in particular LIF signaling, we transfected GP130-deficient HEK293 cells with plasmids encoding WT GP130 and c.1699+4A>G (Fig. 4). GP130 surface expression was detectable but reduced in c.1699+4A>G transfected cells, as well as in EBV-LCLs from C-II-3 (Figs. 4 A and 3 E). Functionally, cells transfected with plasmids encoding c.1699+4A>G did not respond to GP130-dependent cytokines, including LIF (Fig. 4, B–F). Stimulation with IFN-α resulted in comparable pSTAT3 response regardless of the transfection conditions (Fig. 4 G). We conclude that the c.1699+4A>G variant leads to a truncated extracellular GP130 receptor that is expressed but nonfunctional.
Figure 4.
c.1699+4A>G intronic variant of IL6ST completely abrogates GP130-dependent IL-6, IL-11, IL-27, OSM, and LIF signaling and the structural remodeling. (A) GP130 surface expression in c.1699+4A>G transfected GP130-deficient HEK293 cells 24 h after transfection. Results are representative of four independent experiments. (B–G) Response of pSTAT3 in c.1699+4A>G transfected GP130-deficient HEK293 after stimulations with IL-6, IL-11, IL-27, OSM, LIF, and IFN-α stimulation (all 100 ng/ml for 15 min). For assessment of IL-6 and IL-11 signaling, cells were cotransfected with plasmids encoding IL-6Rα and IL-11Rα, respectively. Successfully transfected cells were gated based on GFP expression. Representative histograms are shown on the left (gray, unstimulated; blue and red, stimulated). Quantification is based on four independent experiments per cytokine, with one to two replicates per experiment. EV, empty vector; WT GP130, wildtype. Statistical differences were determined by means of Mann–Whitney U test, **, P ≤ 0.01, *, P ≤ 0.05. (H) In silico model of the hexameric structure of the extracellular region of GP130 (green ribbon) in complex with IL-6 (purple ribbon) and IL6Rα (blue ribbon). GP130 contains six ectodomains: an N-terminal Ig-like domain (D1), two cytokine binding domains (D2–D3), and three membrane-proximal fibronectin type III domains (D4–D6). Comparison of the D5/D6 interface (light and dark green ribbon) in WT and c.1699+4A>G variant predicts a major structural consequence to the inflexible junction of D5/D6 interface and the overall integrity of D6 in the absence of exon 13. In the deletion mutant, the entire contact region between D5 and D6 is significantly reduced, resulting in a weaker interdomain interface and perhaps preventing the formation of multimeric complexes of GP130. Key interdomain polar contacts identified in the crystal structures made by Asp523 and Asn526 to Ser439 and Gln494 in the WT protein are not present. Hydrophobic interactions between Phe528 and backbone of residues Asn526, Gly527, Phe528, and Tyr491 are also absent. Residues in D5 and D6 are highlighted as green and yellow carbon spheres, respectively. D6 is no longer a well-ordered domain, resulting in a “significantly impaired feet” of the tall GP130 receptor that perhaps does not bring the two cytoplasmic domains into position in close proximity to allow transphosphorylation of receptor-associated JAKs. In addition, “leg-leg” dimer contacts could be further reduced, resulting in more conformational flexibility, less rigidity of the hexamer structure upon IL-6 binding, and reduced interaction between the two legs. D, domain.
To examine the structural consequences of the c.1699+4A>G variant, we modeled the truncated protein using available x-ray crystal and EM images (Boulanger et al., 2003; Skiniotis et al., 2005, 2008; Xu et al., 2010). We mapped the peptide sequence coded by exon 13 (domain 6) onto the structure (Fig. 4 H). The structural analysis suggests that cytokine binding could occur in the mutant receptor (and the antibody epitopes are largely intact, explaining the detection on the surface) but predicts defective signal transduction via domain 5 and 6 interactions.
To confirm that any residual pSTAT3 response in both the p.Arg281* and c.1699+4A>G variants was not missed at our 15-min measurement point due to delayed kinetics, we stimulated transfected cells with indicated cytokines over a time course (Fig. S2, A–F). GP130-deficient HEK293 cells transfected with p.Arg281* and c.1699+4A>G variants consistently failed to respond to GP130-dependent cytokine stimulations at all time points tested, whereas pSTAT3 response over IFN-α stimulation decreased over time, mirroring the WT.
Many cytokine families have evolved highly conserved common receptor chains that define them, while cytokine-specific receptors show less evolutionary conservation. By nature of this combinatorial biology, mutations in the common chain have detrimental consequences, since they affect signaling of multiple cytokines. Genetic defects in IL2RG cause X-linked severe combined immunodeficiency due to defects in the entire IL-2 cytokine family (Leonard et al., 2019; Liao et al., 2011), and mutations in IL10RB result in infantile-onset inflammatory bowel disease, largely due to defective IL-10 signaling, while also associated with defective IL-22, IL-26, and IL-28 signaling (Ouyang and O’Garra, 2019).
Here we report a syndrome that is caused by the complete absence of signaling by the IL-6 cytokine family. We conclude the following. (a) Deleterious variants in IL6ST lead to complex phenotype with lethal cardiopulmonary defects, skeletal abnormalities, autonomous dysfunction and additional variable clinical features, an extended STWS phenotype (or STWS type II). (b) Patients with IL6ST loss-of-function variants can survive the intrauterine period and, in a few rare cases, the first few years of life. This is in contrast with embryonically lethal phenotype in gp130 knockout mice that may be explained by differences in pluripotent stem cell development (Onishi and Zandstra, 2015; Schnerch et al., 2010) and/or differences in placental support (Malassiné et al., 2003). (c) The absence of symptoms in the parents with heterozygous IL6ST and 20–44% reduction in GP130 surface expression suggest that one defective copy of IL6ST is not compensated for by the remaining WT allele, and that haploinsufficiency is not deleterious. (d) The similarities of the phenotype of our patients with IL6ST protein-truncating variants and those of classical STWS caused by biallelic LIFR mutations suggest that most of symptoms of our reported patients were due to defective LIF signaling (Nicola and Babon, 2015). Patient C-II-3 survived until 6 yr, in agreement with the fact that some patients with classical STWS survive during childhood, since their dysautonomia improves after the first year of life.
It is likely that different parts of defective cytokine signaling through GP130 explain the phenotypic components of the condition. In addition to the pathognomonic features of STWS caused by defective LIF signaling, this may explain the kidney abnormalities seen in B-II-3, since LIFR deficiency was similarly associated with kidney malformations in patients and in vivo mouse models (Kosfeld et al., 2017). Defective LIF signaling explains why six of seven patients with confirmed skeletal abnormalities were unable to survive the intrauterine or neonatal period. This confirms the phenotypic similarity of our patients with those with classic STWS due to LIFR deficiency and contrasts with previously reported GP130 missense variants (p.Asn404Tyr and p.Pro498Leu). These two variants have defective IL-6 and IL-11 signaling, reduced IL-27 and OSM signaling, and largely normal LIF signaling. We can only speculate as to the selectivity of these previously reported mutations in contrast with the phenotype of our patients. Both p.Asn404Tyr and p.Pro498Leu mutations are predicted to affect the secondary and tertiary structure of the domain 4 and 5 of the GP130 receptor. This flexibility could be more important for the hexameric complexes that form the IL-6 and IL-11 receptor complexes compared with the trimeric IL-27, OSM, and LIF receptor complexes. Defective IL-11 and OSM signaling may contribute to skeletal abnormalities (Sims, 2016). Defective IL-6 signaling may explain dermatitis and lack of acute-phase response, as indicated by the low fibrinogen and normal C-reactive protein despite a neonatal stress response. Our patients did not survive long enough to develop a phenotype similar to IL-6R defects (Spencer et al., 2019). Loss of OSM signaling may explain the severe congenital thrombocytopenia in A-II-2. Genetic ablation of either OSM (Sato et al., 2014) or OSM receptor (Tanaka et al., 2003) in mice is associated with thrombocytopenia, and a first-time-in-human study of an anti-OSM monoclonal antibody (GSK2330811) was associated with thrombocytopenia (Reid et al., 2018). Patient C-II-3 survived infancy and developed additional immunologic symptoms of eczematoid dermatitis reminiscent of defective IL-6/GP130/STAT3 signaling, as observed in previous GP130 homozygous patients with missense variants in IL6ST (Schwerd et al., 2017; Shahin et al., 2019).
In summary, our findings explain a complex human syndrome due to the functional defect of a whole class of IL-6 family cytokines. The phenotypic overlap between the conditions with global IL6ST deficiency, cytokine selective IL6ST defects, and those caused by isolated cytokine defects caused by genetic or therapeutic targeting of the IL-6, IL-11, OSM, and LIF signaling axis is summarized in Fig. S2 G.
Materials and methods
Patients and samples
The study was approved by the Ethics Committee of the Karolinska Institutet (2014/983-31/1); the patients of family A were clinically examined at Lund University, Sweden. The molecular analyses for family A were performed at the Departments of Clinical Genetics, Lund University, and Karolinska University Hospital on genomic DNA from amniocytes of A-II-1 and from blood of the parents and A-II-2. The patients from Saudi Arabia (family B) were recruited with informed consent under an institutional review board–approved research protocol (KFSRHC RAC #2121053). The patients from the Netherlands (family C) initially participated in a French study, protocol number IRB0000388. Control individuals were recruited via the Oxford GI illnesses biobank (REC 16/YH/0247). Control fetal samples were obtained from Biobank at the Department of Laboratory Medicine, Division of Pathology, Karolinska Institutet, Huddinge, Sweden. Control amniocytes were obtained commercially from Coriell Institute.
Array genomic hybridization
Genomic DNA was extracted according to standard protocols from peripheral blood from A-II-2 and parents and from amniocytes of A-II-1. CytoScan HD Array was used (Thermo Fisher Scientific) according to manufacturer’s instructions. In a normal setting (parents and A-II-2), the resolution is ≈12 kb for deletions and ≈36 kb for duplications. In a prenatal setting (A-II-1), we only report >1 Mb for deletions and >2 Mb for duplications.
WGS
For family A, clinical WGS was performed as previously described (Stranneheim et al., 2014). A negative clinical gene-screening panel, targeting 377 known skeletal disease genes, was performed at Science for Life laboratory, and data were analyzed at the Department of Clinical Genetics, Stockholm, Sweden, for A-II-2. Next, family members were added to the same analysis: first parents, and then the DNA from the affected fetus (A-II-1). WGS was performed using HiSeq X (Illumina) with a TruSeq DNA PCR-free library preparation at Clinical Genomics (SciLifeLab, Stockholm, Sweden) to yield minimum 30× coverage. The variants were ranked according to expected disease-causing potential, their presence in coding exons ±20 base pairs intronic sequence, and a minor allele frequency of <0.005 according to public databases (1000 Genomes, dbSNP, and ExAC/gnomAD) and our local in-house database. Family analysis of parents and the affected patients revealed a short list of very rare variants (Table S1).
Exome sequencing
Exome sequencing in family B and positional mapping were performed as previously described (Monies et al., 2019). Exome sequencing in family C was performed at the genomic platform of the IMAGINE Institute (Paris, France) with the SureSelect Human All Exon kit (Agilent Technologies). Agilent SureSelect Human All Exon (V4) libraries were prepared from 3 µg of genomic DNA according to instructions of the manufacturer. Pooled barcoded exome libraries were sequenced using a HiSeq2500 (Illumina) to generate paired-end reads. After demultiplexing, sequences were mapped to the human genome reference (NCBI build37/hg 19 version) with Burrows-Wheeler Aligner (Li and Durbin, 2010). The mean depth of coverage obtained for each sample was ≥80×, with 95% of the exome covered at least 15×. Variant calling was performed with the Genome Analysis Toolkit (GATK; McKenna et al., 2010), Sequence Alignment/Map tools (Li et al., 2009), and Picard Tools. Single nucleotide variants were called with GATK UnifiedGenotyper, whereas indel calls were made with the GATK IndelGenotyper_v2. All variants with a read coverage ≤2× and a Phred-scaled quality of ≤20 were filtered out. Variants were annotated and filtered using an in-house annotation software system (Polyweb, unpublished). The analyses focused on nonsynonymous variants, splice variants, and coding indels. Variant pathogenicity was evaluated using the prediction algorithms SIFT (sorting intolerant from tolerant; cutoff ≤0.05), PolyPhen2 (HumVar scores, cutoff ≥0.447), and MutationTaster (cutoff: qualitative prediction as pathogenic). The variant frequency in control datasets was assessed, including the ExAC database, dbSNP129, 1000 Genomes, ClinVar, HGMD, and in-house exome data. IL6ST variants were confirmed by Sanger sequencing, and segregation was verified.
The following primers were used. For family A: IL6ST forward primer, 5′-GTTGTGTTGAGCCAAGATCG-3′, and IL6ST reverse primer, 5′-AAAACCAGAACCAGTTCACATC-3′. For family B: IL6ST forward primer, 5′-CACCTGTAATCCCAGCCACT-3′ and IL6ST reverse primer, 5′-GCATCTGGCCTCCATGATAA-3′. For family C: IL6ST forward primer, 5′-TGTAAAACGACGGCCAGTGTGGGACCAACTTCCTGTTG-3′, and IL6ST reverse primer, 5′-CAGGAAACAGCTATGACCAACCATAAGGCAAATTCTCAGG-3′. The variants have been reported to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/): SCV000925665 for c.841C>T and SCV000924636 for c.1699+4A>G.
Online resources
The following genetic databases were used: gnomAD (https://gnomad.broadinstitute.org); 1000 Genomes (https://www.internationalgenome.org); and ClinVar.
Splice variant analysis
Splice variant analysis was performed using MaxEnt (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html) and the transcript-inferred pathogenicity score tool (http://trap-score.org/Search?query=5-55247753-T-C).
Cell culture
Healthy amniotic fluid–derived amniocytes were derived from Coriell Institute (GM00472 and GM00957) and patients A-II-1 and A-II-2 were obtained from the Department of Clinical Genetics, Lund University Hospital, Sweden. Amniocytes were maintained in AmnioMAX-II complete medium (Thermo Fisher Scientific) at a 1:2 passage ratio. EBV-LCLs from the patient carrying c.1699+4A>G variant and three healthy controls were generated from peripheral blood mononuclear cells with supernatant from the EBV-producing marmoset cell line B95-8 according to standard protocols. EBV-LCLs were maintained in RPMI 1640 (Sigma-Aldrich), whereas HEK293 and GP130-deficient HEK293 cell lines were cultured in DMEM (Sigma-Aldrich), both supplemented with 10% FCS.
GP130 surface expression
Staining of surface GP130 was performed using anti-GP130-biotin (AN-G30; eBioscience) followed by staining with streptavidin-PE CF594, and expression was determined by flow cytometry. Cells were stained with Fixable Viability Dye eFluor 780 (eBioscience), and dead cells were excluded.
Cytokine stimulation
Cells were stimulated with indicated concentrations of recombinant human IL-6, OSM, LIF, IFN-α, IL-11, IL-27, and IL-21 (Miltenyi Biotec, R&D Systems, and PeproTech).
GP130 reconstitution via lentivirus transduction
GP130 reconstitution in amniocytes was performed essentially as previously described (Schwerd et al., 2017). Sequence encoding WT IL6ST was cloned into the pcDNA3.1(+) vector without C- or N-terminal tags (GenScript). The WT IL6ST gene was custom cloned into pLENTI7.3V5-DEST (Geneart; Thermo Fisher Scientific). Lipofection on HEK293T cells was performed using plasmid encoding WT IL6ST and Lipofectamine 2000 (Thermo Fisher Scientific). Lentiviral supernatant was generated from HEK293T cells and filtered using a 0.45-µm filter before adding to amniocytes on 24-well plates. The transduction efficiency of amniocytes was >40% as confirmed by GFP fluorescence measurements by flow cytometry.
Plasmid expression experiments
GP130-deficient HEK293 cell line was generated using CRISPR/Cas9 as previously described (Schwerd et al., 2017). GP130-deficient HEK293 cells were transiently transfected with plasmids encoding empty vector, WT, or mutant p.Arg281* or c.1699+4A>G GP130 (mutagenesis created by GenScript; pcDNA3.1(+)) using Lipofectamine 2000 (Thermo Fisher Scientific). To fully access IL-6 and IL-11 signaling, cells were cotransfected with IL-6Rα and IL-11Rα expression vectors. Cotransfection with GFP-encoding plasmid allowed gating on successfully transfected cells. After 24 h, cells were serum starved for 2 h before stimulation with indicated cytokines at 100 ng/ml. Phosflow STAT3 assay was performed as described below.
Phosflow STAT3 signaling assay
Phosphorylation of STAT3 (pSTAT3) was determined by Phosflow. Analysis of primary lymphocytes and monocytes was performed using peripheral blood. Peripheral blood mononuclear cells were separated by Lymphoprep and density gradient. Cells were subsequently stained with anti-CD3-FITC (clone: UCHT1; BioLegend), anti-CD4-BV605 (clone: OKT4; BioLegend), anti-CD8-PE CF594 (clone:RPA-T8; BD Biosciences), anti-CD14-BV650 (clone: M5E2; BioLegend), and anti-CD19-BV711 (clone: SJ25C1; BD Biosciences) for 20 min on ice. 5 min through surface staining, cells were stimulated for 15 min with 100 ng/ml IL-6 before fixation. For the analysis of amniocytes, cells were plated in 96-well plates and serum starved overnight to reduce background. Next day, cells were washed three times with serum-free DMEM (Sigma-Aldrich) and further serum-starved for 2 h. Cells were subsequently stained with Fixable Viability Dye eFluor 780 (eBioscience) and stimulated with 100 ng/ml OSM, 100 ng/ml IL-11, and 100 ng/ml LIF for 15 min before detachment with trypsin-EDTA solution and fixation in prewarmed Cytofix buffer. Levels of pSTAT3 in EBV-LCLs were determined in serum-starved cells, followed by stimulation with 50 ng/ml IL-6 or IL-21 before fixation. After fixation, amniocytes, HEK293 cell lines and EBV-LCLs were washed and permeabilized with ice-cold Perm Buffer III (BD Biosciences) and stained with anti-STAT3 (pY705)-Alexa Fluor 647 (clone 4/P-STAT3) at room temperature. LSRII or LSR Fortessa was used to acquire samples, and data were analyzed using Flowjo software (v.10.4.0).
Quantitative RT-PCR
Total RNA was extracted from EBV-LCLs, amniocytes, or transfected GP130-deficient HEK293 cells using the RNeasy mini kit (Qiagen) accordingly to the manufacturer’s instructions, and cDNA was synthesized using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). Gene expression was determined by Taqman assays from Applied Biosystems: SOCS3 (Hs02330328_s1), IL6ST exons 4–5 (Hs01006736_g1), and exons 12–13 (Hs01006729_g1). Quantitative RT-PCR was performed in triplicate using a Bio-Rad machine. Changes in gene expression were normalized using an endogenous control RPLPO (Hs00420895_gH), and data were expressed in accordance with the 2−ΔCt method.
PCR and agarose gel
PCR was performed using a standard protocol for Phusion High Fidelity DNA Polymerase (New England BioLabs). The designed primer pair with a combination of IL6ST_cDNA_exon11_forward (5′-TGCTTTGGGTGGAATGGACT-3′) and IL6ST_cDNA_exon 14–15_reverse (5′-TCCTTGAGCAAACTTTGGGGT-3′) were used for PCR (Thermo Fisher Scientific). Thermocycling conditions were denaturation at 98°C for 3 min, 35 cycles of repeated 98°C for 10 s, 60°C for 30 s, and 72°C for 30 s, followed by final extension at 72°C for 10 min. The DNA fragments were subsequently run on a 1.5% agarose gel and purified using a QIAquick Gel Extraction Kit according to manufacturer’s instructions (Qiagen) before being Sanger sequenced (Source Bioscience).
Nested RT-PCR
cDNA derived from C-II-3 was used to perform nested PCR using a number of IL6ST primer pairs. Primer pair 1: exon 11_forward (5′-TGCTTTGGGTGGAATGGACT-3′) and exon 14–15_reverse (5′-TCCTTGAGCAAACTTTGGGGT-3′). Primer pair 2: exon 12_forward (5′-ATGCTGATGGACCAGGAAGC-3′) and exon 16_reverse (5′-GAGTGTGAGGTGACCACTGG-3′). Primer pair 3: exon 12_forward (5′-CAGGAAGCCCTGAATCCATA-3′) and exon 14_reverse (5′-TTCTGTGTGGGAAGAATCCA-3′). Primer pair 4: exon 11_forward (5′-TGCTTTGGGTGGAATGGACT-3′) and exon 16_reverse (5′-GAGTGTGAGGTGACCACTGG-3′). Thermocycling conditions for both rounds of nested RT-PCR were denaturation at 95°C for 5 min, 35 cycles of repeated 95°C for 45 s, 58°C for 30 s, and 72°C for 45 s, followed by final extension at 72°C for 10 min. The DNA fragments were subsequently run on a 1% agarose gel.
MLPA
DNA was isolated from EBV-LCLs using the DNeasy Blood & Tissue kit (Qiagen) according to the manufacturer’s instructions. The variants in IL6ST (exons 4, 8, 13, and 17) and genes adjacent to IL6ST (ANKRD55 and IL31RA) were amplified with PCR primers, and multiple ligation-dependent probe amplification was performed according to the manufacturer’s protocol (MRC Holland). The sequences of the probes are as follows: GABRA4_fam-pilot_A, 5′-GGGTTCCCTAAGGGTTGGACAGCCTGTTGTCATAACCATCG-3′; GABRA4_fam-pilot_B, [Phos] 5′-AGCAAACTGTCCAGGATGCGTCTAGATTGGATCTTGCTGGCAC-3′; IL6ST_ex4_A, 5′-GGGTTCCCTAAGGGTTGGAGTGAACTTCTAGATCCATGT-3′; IL6ST_ex4_B, [Phos] 5′-GGTTATATCAGTCCTGAATCTCCAGTCTAGATTGGATCTTGCTGGCAC-3′. ANKRD55_downst_A, 5′-GGGTTCCCTAAGGGTTGGAGACTTGGAATTGCAGGAAGCACTCG-3′; ANKRD55_downst_B, [Phos] 5′-GTTTGTCCGCAGAGGTTCTCCAGTCTAGATTGGATCTTGCTGGCAC-3′. IL6ST_ex8_A, 5′-GGGTTCCCTAAGGGTTGGACTGTCCAAGACCTTAAACCTTTTACA-3′; IL6ST_ex8_B, [Phos] 5′-GAATATGTGTTTAGGATTCGCTGTATGTCTAGATTGGATCTTGCTGGCAC-3′. IL6ST_ex17_3UTR_A, 5′-GGGTTCCCTAAGGGTTGGAGCTGCGACTGATGAAGGCATGCCTAAAAG-3′; IL6ST_ex17_3UTR_B, [Phos] 5′-TTACTTACCACAGACTGTACGGCAAGGCTCTAGATTGGATCTTGCTGGCAC-3′. IL31RA_upstr_A, 5′-GGGTTCCCTAAGGGTTGGAGCTAAACCTGAAGGAGTCTGATGACTCTGT-3′; IL31RA_upstr_B, [Phos] 5′-GAACACAGAAGACAGGATCTTAAAACCATGTCTAGATTGGATCTTGCTGGCAC-3′. Stern PCLN13q_A, 5′-GGGTTCCCTAAGGGTTGGAGACACAAGGGTGTAAAATGCACG-3′; Stern PCLN13q_B, [Phos] 5′-TTTCAGGGTGTGTTTGCATATGATTTAATCAATCAGTATGTCTAGATTGGATCTTGCTGGCAC-3′. IL6ST_ex13_A, 5′-GGGTTCCCTAAGGGTTGGACACCTTCCAAAGGACCTACTGTTCGGACAAAAAAAGTAG-3′; IL6ST_ex13_B, [Phos] 5′-GGAAAAACGAAGCTGTCTTAGAGTGGGACCAACTTCCTCTAGATTGGATCTTGCTGGCAC-3′. RB1ex23_A, 5′-GGGTTCCCTAAGGGTTGGAGTCACCAATACCTCACATTCCTCGAAGCCCTTACAAGTTTCCT-3′; RB1ex23_B, [Phos] 5′-AGTTCACCCTTACGGATTCCTGGAGGGAACATCTATATTTCACCTCTAGATTGGATCTTGCTGGCAC-3′. MRPL41_e1_132_A, 5′-GGGTTCCCTAAGGGTTGGAGACCCTGACAACCTGGAAAAGTACGGCTTCGAGCCCACACAGGAG-3′; MRPL41_e1_132_B, [Phos] 5′-GGAAAGCTCTTCCAGCTCTACCCCAGGAACTTCCTGCGCTAGCTGTCTAGATTGGATCTTGCTGGCAC-3′.
Histology
3-µm sections from paraffin-embedded tissue from epiphyses of lower femur were stained with H&E and examined by a pediatric histopathologist. Variations in the intensities of H&E staining can be attributed to protocols being conducted at different centers. Safranin O/Fast green staining was performed as previously described (Newton et al., 2015). The primary spongiosa was defined as being 50 µm from the growth plate.
Immunostaining
Slides were dewaxed at 54°C for 1 h and rehydrated from xylene through an ethanol gradient to distilled water. Antigen retrieval was performed by submerging slides in Target retrieval solution (DAKO) and heating to 2 psig (103.6°C) in a pressure cooker (All American 1925x). After rinsing in distilled water, a peroxidase quench was performed using 3% hydrogen peroxide (Merck) in methanol. The slides were rinsed in distilled water and blocked with 3% normal horse serum (Vector Laboratories) in PBST (PBS containing 0.1% Tween 20 [Merck]) for 1 h. Slides were incubated overnight with primary antibodies to IL6ST (Sigma-Aldrich; HPA010558, 1:50, gently rocking), osterix (Abcam; ab22552, 1:50, gently rocking), DMP-1 (Abcam; ab103203, 1:50, at 2–8°C), or Cathepsin K (a gift from Goran Andersson [Karolinska Institutet, Stockholm, Sweden], previously developed and validated; Kamiya et al., 1998; Ljusberg et al., 2005; 1:100, at 2–8°C). Slides were then rinsed with PBST and incubated with biotin-conjugated goat anti-rabbit antibodies (Jackson ImmunoResearch Laboratories; 1:400), with gentle rocking for 1 h. Rinsing was performed with PBST, and slides were incubated with ABC reagent (Vector Laboratories), with gentle rocking for 1 h. After PBST rinsing, slides were exposed to TSA solution (Perkin Elmer) and gently rocked for 15 min, and rinsed again with PBST before incubation with ABC reagents (Vector laboratories) with gentle rocking for a further 1 h. Development with DAB solution (DAKO) for test and control slides was performed for the same duration. After dehydration and mounting with Pertex (Histolab), sections were imaged using an Axioskop 2 Plus microscope with Axiovision software (Zeiss).
Computational structural modeling
A structural model of GP130 nonfunctional variant was built based on the crystal structure and EM images of the full ectodomain and Fibronectin III domains of GP130 (PDB accession nos. 3L5H, 3L5I, 3L5J, 3E0G, 1P9M, and 1PVH). Rigid body structural alignments were performed using the crystal structure of the IL6-IL6Rα-GP130 domain D1–D3 hexameric complex as a template and aligning the full ectodomain structure. Then the peptide sequence that corresponds to exon 13 was deleted. GP130 adopts a C-shaped conformation with an Ig-like domain (D1) at the top and three membrane-proximal FNIII domains (D4–D6) supporting the two membrane-distal ligand-binding regions (D2 and D3). Sequence alignment and structural modeling were performed using BioLuminate and Maestro software packages (2019-1 release, Schrödinger). Structures were prepared and minimized following Protein Preparation Wizard workflow in Maestro with the OPLS3e force field. All protein structures were modeled, analyzed, and rendered in Maestro (release 2019-1, v.11.9.011, MMshare Version 4.5.011, Platform Linux-x86_64). Coordinate files were obtained from PDB (accession no. 1P9M for IL-6/IL-6Rα/GP130; accession no. 3L5H for GP130 D1-D6).
Statistical analysis
Results were analyzed using Prism 8 (GraphPad Software). Significance of differences was determined by Mann–Whitney U test, and P values < 0.05 were considered significant. The bone histology values presented in the text are mean ± SEM (to three significant figures) of values collected from six discrete visual fields, analyzed for each patient.
Online supplemental material
Fig. S1 shows the bone phenotype, MLPA analysis of the IL6ST gene, RNA-seq expression data of multiple cytokine receptor units in control amniocytes, the parental analysis of GP130 and pSTAT3 response, and PCR analysis. Fig. S2 shows that the complete loss of GP130-dependent IL-6, IL-11, IL-27, OSM, and LIF signaling in p.Arg281* and c.1699+4A>G variant is not due to delayed kinetics and a schematic summary of GP130 deficiency, and the relationship between loss of cytokine signaling and clinical symptoms. Table S1 lists the rare sequence variants identified using whole genome variant analysis in family A (quadro) and whole exome variant analysis in family B and C (trios).
Supplementary Material
lists rare sequence variants identified using whole-genome variant analysis in family A (quadro) and whole exome variant analysis in families B and C (trios).
Acknowledgments
We thank the families and blood donors for participation in this study. Whole exome sequencing analysis for family B was done at the Imagine Institute (France). We acknowledge GeneMatcher Exchange for enabling the collaboration and Saskia van der Velde-Visser and Dr. Petr Jira for their support. We thank Anette Niklasson and Annica Westlund at Fetal Pathology laboratory at Karolinska University Hospital for help with sectioning the samples for histological analysis.
We acknowledge the contribution of the Oxford Gastrointestinal Biobank, which is supported by the National Institute for Health Research Oxford Biomedical Research Centre. This project was supported through the regional agreement on medical training and clinical research between Stockholm County Council and Karolinska Institutet (ALF20150143 and 20180131), by grants from the Swedish Research Council (G. Grigelioniene and A.S. Chagin), H.K.H. Kronprinsessan Lovisas Förening för Barnasjukvård/Stiftelsen Axel Tielmans minnesfond, Stiftelsen Sällskapet Barnavård (A. Hammarsjö and P.T. Newton), Stiftelsen Promobilia (G. Grigelioniene), and Stiftelsen Frimurare Barnhuset i Stockholm (G. Grigelioniene and P.T. Newton). Bone analysis was supported by a grant from the Russian Science Foundation (19-15-00241). Computational infrastructure and assistance in massively parallel sequencing were provided from Clinical Genomics, Science for Life Laboratory. H.H. Uhlig is supported by the Biomedical Research Centre Oxford.
Author contributions: Y.-H. Chen and G. Grigelioniene contributed equally and should be considered shared first authors. F.S. Alkuraya, A. Laurence, and V. Cormier-Daire contributed equally. G. Grigelioniene, Y.-H. Chen, J. Brodszki, J. Gullander, K. Jedrycha, M. Elving, D. Gisselsson, A. Hammarsjö, H.S. Alsaif, W.I.Y. Kurdi, F. Abdulwahab, F.S. Alkuraya, S. Bacrot, C. Huber, B. Hamel, B. Gürtl-Lackner, and V. Cormier-Daire performed genetic and clinical evaluations. Y.-H. Chen, A. Laurence, D. Aschenbrenner, P.T. Newton, D. Batkovskyte, and H.H. Uhlig performed functional analysis. G. Nishimura is responsible for interpretation of radiographical diagnosis for family 1. N Papadogiannakis., A.S. Chagin, and P.T. Newton performed and interpreted histology and immunohistochemistry analyses. V. Shanmugasundaram performed structural remodeling. L. Devey developed the hypothesis and discussed the experimental data. G. Grigelioniene, V. Cormier-Daire, and H.H. Uhlig directed research. G. Grigelioniene, Y.-H. Chen, J. Gullander, M. Elving, P.T. Newton, and H.H. Uhlig wrote the manuscript. All authors have revised and approved the manuscript.
References
- Akira S. 2000. Roles of STAT3 defined by tissue-specific gene targeting. Oncogene. 19:2607–2611. 10.1038/sj.onc.1203478 [DOI] [PubMed] [Google Scholar]
- Babon J.J., Varghese L.N., and Nicola N.A.. 2014. Inhibition of IL-6 family cytokines by SOCS3. Semin. Immunol. 26:13–19. 10.1016/j.smim.2013.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger M.J., Bankovich A.J., Kortemme T., Baker D., and Garcia K.C.. 2003. Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130. Mol. Cell. 12:577–589. 10.1016/S1097-2765(03)00365-4 [DOI] [PubMed] [Google Scholar]
- Caminsky N., Mucaki E.J., and Rogan P.K.. 2014. Interpretation of mRNA splicing mutations in genetic disease: review of the literature and guidelines for information-theoretical analysis. F1000 Res. 3:282 10.12688/f1000research.5654.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagoneau N., Scheffer D., Huber C., Al-Gazali L.I., Di Rocco M., Godard A., Martinovic J., Raas-Rothschild A., Sigaudy S., Unger S., et al. . 2004. Null leukemia inhibitory factor receptor (LIFR) mutations in Stuve-Wiedemann/Schwartz-Jampel type 2 syndrome. Am. J. Hum. Genet. 74:298–305. 10.1086/381715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland S.M., DeLeo F.R., Elloumi H.Z., Hsu A.P., Uzel G., Brodsky N., Freeman A.F., Demidowich A., Davis J., Turner M.L., et al. . 2007. STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 357:1608–1619. 10.1056/NEJMoa073687 [DOI] [PubMed] [Google Scholar]
- Itoh S., Udagawa N., Takahashi N., Yoshitake F., Narita H., Ebisu S., and Ishihara K.. 2006. A critical role for interleukin-6 family-mediated Stat3 activation in osteoblast differentiation and bone formation. Bone. 39:505–512. 10.1016/j.bone.2006.02.074 [DOI] [PubMed] [Google Scholar]
- Jung C., Dagoneau N., Baujat G., Le Merrer M., David A., Di Rocco M., Hamel B., Mégarbané A., Superti-Furga A., Unger S., et al. . 2010. Stüve-Wiedemann syndrome: long-term follow-up and genetic heterogeneity. Clin. Genet. 77:266–272. 10.1111/j.1399-0004.2009.01314.x [DOI] [PubMed] [Google Scholar]
- Kamiya T., Kobayashi Y., Kanaoka K., Nakashima T., Kato Y., Mizuno A., and Sakai H.. 1998. Fluorescence microscopic demonstration of cathepsin K activity as the major lysosomal cysteine proteinase in osteoclasts. J. Biochem. 123:752–759. 10.1093/oxfordjournals.jbchem.a022001 [DOI] [PubMed] [Google Scholar]
- Kang J.H., Park H.J., Jung Y.W., Shim S.H., Sung S.R., Park J.E., Cha D.H., and Ahn E.H.. 2015. Comparative Transcriptome Analysis of Cell-Free Fetal RNA from Amniotic Fluid and RNA from Amniocytes in Uncomplicated Pregnancies. PLoS One. 10:e0132955 10.1371/journal.pone.0132955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Tanaka T., Narazaki M., and Kishimoto T.. 2019. Targeting Interleukin-6 Signaling in Clinic. Immunity. 50:1007–1023. 10.1016/j.immuni.2019.03.026 [DOI] [PubMed] [Google Scholar]
- Keupp K., Li Y., Vargel I., Hoischen A., Richardson R., Neveling K., Alanay Y., Uz E., Elcioğlu N., Rachwalski M., et al. . 2013. Mutations in the interleukin receptor IL11RA cause autosomal recessive Crouzon-like craniosynostosis. Mol. Genet. Genomic Med. 1:223–237. 10.1002/mgg3.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosfeld A., Brand F., Weiss A.C., Kreuzer M., Goerk M., Martens H., Schubert S., Schäfer A.K., Riehmer V., Hennies I., et al. . 2017. Mutations in the leukemia inhibitory factor receptor (LIFR) gene and Lifr deficiency cause urinary tract malformations. Hum. Mol. Genet. 26:1716–1731. 10.1093/hmg/ddx086 [DOI] [PubMed] [Google Scholar]
- Leonard W.J., Lin J.X., and O’Shea J.J.. 2019. The γc Family of Cytokines: Basic Biology to Therapeutic Ramifications. Immunity. 50:832–850. 10.1016/j.immuni.2019.03.028 [DOI] [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 26:589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W., Lin J.X., and Leonard W.J.. 2011. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr. Opin. Immunol. 23:598–604. 10.1016/j.coi.2011.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljusberg J., Wang Y., Lång P., Norgård M., Dodds R., Hultenby K., Ek-Rylander B., and Andersson G.. 2005. Proteolytic excision of a repressive loop domain in tartrate-resistant acid phosphatase by cathepsin K in osteoclasts. J. Biol. Chem. 280:28370–28381. 10.1074/jbc.M502469200 [DOI] [PubMed] [Google Scholar]
- Malassiné A., Frendo J.L., and Evain-Brion D.. 2003. A comparison of placental development and endocrine functions between the human and mouse model. Hum. Reprod. Update. 9:531–539. 10.1093/humupd/dmg043 [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., and DePristo M.A.. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner J.D., Vogel T.P., Forbes L., Ma C.A., Stray-Pedersen A., Niemela J.E., Lyons J.J., Engelhardt K.R., Zhang Y., Topcagic N., et al. . 2015. Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood. 125:591–599. 10.1182/blood-2014-09-602763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y., Saito M., Tsuchiya S., Tsuge I., Takada H., Hara T., Kawamura N., Ariga T., Pasic S., Stojkovic O., et al. . 2007. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 448:1058–1062. 10.1038/nature06096 [DOI] [PubMed] [Google Scholar]
- Monies D., Abouelhoda M., Assoum M., Moghrabi N., Rafiullah R., Almontashiri N., Alowain M., Alzaidan H., Alsayed M., Subhani S., et al. . 2019. Lessons Learned from Large-Scale, First-Tier Clinical Exome Sequencing in a Highly Consanguineous Population. Am. J. Hum. Genet. 104:1182–1201. 10.1016/j.ajhg.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M., Kamimura D., and Hirano T.. 2019. Pleiotropy and Specificity: Insights from the Interleukin 6 Family of Cytokines. Immunity. 50:812–831. 10.1016/j.immuni.2019.03.027 [DOI] [PubMed] [Google Scholar]
- Newton P.T., Vuppalapati K.K., Bouderlique T., and Chagin A.S.. 2015. Pharmacological inhibition of lysosomes activates the MTORC1 signaling pathway in chondrocytes in an autophagy-independent manner. Autophagy. 11:1594–1607. 10.1080/15548627.2015.1068489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola N.A., and Babon J.J.. 2015. Leukemia inhibitory factor (LIF). Cytokine Growth Factor Rev. 26:533–544. 10.1016/j.cytogfr.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen P., Morgan N.V., Fenwick A.L., Parmanen S., Veistinen L., Mikkola M.L., van der Spek P.J., Giraud A., Judd L., Arte S., et al. . 2011. Inactivation of IL11 signaling causes craniosynostosis, delayed tooth eruption, and supernumerary teeth. Am. J. Hum. Genet. 89:67–81. 10.1016/j.ajhg.2011.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi K., and Zandstra P.W.. 2015. LIF signaling in stem cells and development. Development. 142:2230–2236. 10.1242/dev.117598 [DOI] [PubMed] [Google Scholar]
- Ouyang W., and O’Garra A.. 2019. IL-10 Family Cytokines IL-10 and IL-22: from Basic Science to Clinical Translation. Immunity. 50:871–891. 10.1016/j.immuni.2019.03.020 [DOI] [PubMed] [Google Scholar]
- Reid J., Zamuner S., Edwards K., Rumley S.A., Nevin K., Feeney M., Zecchin C., Fernando D., and Wisniacki N.. 2018. In vivo affinity and target engagement in skin and blood in a first-time-in-human study of an anti-oncostatin M monoclonal antibody. Br. J. Clin. Pharmacol. 84:2280–2291. 10.1111/bcp.13669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato F., Miyaoka Y., Miyajima A., and Tanaka M.. 2014. Oncostatin M maintains the hematopoietic microenvironment in the bone marrow by modulating adipogenesis and osteogenesis. PLoS One. 9:e116209 10.1371/journal.pone.0116209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnerch A., Cerdan C., and Bhatia M.. 2010. Distinguishing between mouse and human pluripotent stem cell regulation: the best laid plans of mice and men. Stem Cells. 28:419–430. [DOI] [PubMed] [Google Scholar]
- Schwerd T., Twigg S.R.F., Aschenbrenner D., Manrique S., Miller K.A., Taylor I.B., Capitani M., McGowan S.J., Sweeney E., Weber A., et al. . 2017. A biallelic mutation in IL6ST encoding the GP130 co-receptor causes immunodeficiency and craniosynostosis. J. Exp. Med. 214:2547–2562. 10.1084/jem.20161810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahin T., Aschenbrenner D., Cagdas D., Bal S.K., Conde C.D., Garncarz W., Medgyesi D., Schwerd T., Karaatmaca B., Cetinkaya P.G., et al. . 2019. Selective loss of function variants in IL6ST cause Hyper-IgE syndrome with distinct impairments of T-cell phenotype and function. Haematologica. 104:609–621. 10.3324/haematol.2018.194233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims N.A. 2016. Cell-specific paracrine actions of IL-6 family cytokines from bone, marrow and muscle that control bone formation and resorption. Int. J. Biochem. Cell Biol. 79:14–23. 10.1016/j.biocel.2016.08.003 [DOI] [PubMed] [Google Scholar]
- Sims N.A., Jenkins B.J., Quinn J.M., Nakamura A., Glatt M., Gillespie M.T., Ernst M., and Martin T.J.. 2004. Glycoprotein 130 regulates bone turnover and bone size by distinct downstream signaling pathways. J. Clin. Invest. 113:379–389. 10.1172/JCI19872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiniotis G., Boulanger M.J., Garcia K.C., and Walz T.. 2005. Signaling conformations of the tall cytokine receptor gp130 when in complex with IL-6 and IL-6 receptor. Nat. Struct. Mol. Biol. 12:545–551. 10.1038/nsmb941 [DOI] [PubMed] [Google Scholar]
- Skiniotis G., Lupardus P.J., Martick M., Walz T., and Garcia K.C.. 2008. Structural organization of a full-length gp130/LIF-R cytokine receptor transmembrane complex. Mol. Cell. 31:737–748. 10.1016/j.molcel.2008.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S., Köstel Bal S., Egner W., Lango Allen H., Raza S.I., Ma C.A., Gürel M., Zhang Y., Sun G., Sabroe R.A., et al. . 2019. Loss of the interleukin-6 receptor causes immunodeficiency, atopy, and abnormal inflammatory responses. J. Exp. Med. 216:1986–1998. 10.1084/jem.20190344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranneheim H., Engvall M., Naess K., Lesko N., Larsson P., Dahlberg M., Andeer R., Wredenberg A., Freyer C., Barbaro M., et al. . 2014. Rapid pulsed whole genome sequencing for comprehensive acute diagnostics of inborn errors of metabolism. BMC Genomics. 15:1090 10.1186/1471-2164-15-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Noguchi K., Shi W., Tanaka T., Matsumoto M., Yoshida N., Kishimoto T., and Akira S.. 1997. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA. 94:3801–3804. 10.1073/pnas.94.8.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Hirabayashi Y., Sekiguchi T., Inoue T., Katsuki M., and Miyajima A.. 2003. Targeted disruption of oncostatin M receptor results in altered hematopoiesis. Blood. 102:3154–3162. 10.1182/blood-2003-02-0367 [DOI] [PubMed] [Google Scholar]
- Voigt M., Fusch C., Olbertz D., Hartmann K., Rochow N., Renken C., and Schneider K.T.M.. 2006. Analyse des Neugeborenenkollektivs der Bundesrepublik Deutschland. Geburtshilfe Frauenheilkd. 66:956–970. 10.1055/s-2006-924458 [DOI] [Google Scholar]
- Ware C.B., Horowitz M.C., Renshaw B.R., Hunt J.S., Liggitt D., Koblar S.A., Gliniak B.C., McKenna H.J., Papayannopoulou T., Thoma B., et al. . 1995. Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects and results in perinatal death. Development. 121:1283–1299. [DOI] [PubMed] [Google Scholar]
- Xu Y., Kershaw N.J., Luo C.S., Soo P., Pocock M.J., Czabotar P.E., Hilton D.J., Nicola N.A., Garrett T.P., and Zhang J.G.. 2010. Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes. J. Biol. Chem. 285:21214–21218. 10.1074/jbc.C110.129502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Taga T., Saito M., Suematsu S., Kumanogoh A., Tanaka T., Fujiwara H., Hirata M., Yamagami T., Nakahata T., et al. . 1996. Targeted disruption of gp130, a common signal transducer for the interleukin 6 family of cytokines, leads to myocardial and hematological disorders. Proc. Natl. Acad. Sci. USA. 93:407–411. 10.1073/pnas.93.1.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists rare sequence variants identified using whole-genome variant analysis in family A (quadro) and whole exome variant analysis in families B and C (trios).