An unexpected BDCA-2+CD123+CD1a+ DC subset comprises a major DC population in acute human sterile skin inflammation. scRNA-seq shows these to be activated DCs able to express markers normally associated with plasmacytoid DCs, prompting a re-evaluation of previous studies.
Abstract
Plasmacytoid dendritic cells (pDCs) produce type I interferon (IFN-I) and are traditionally defined as being BDCA-2+CD123+. pDCs are not readily detectable in healthy human skin, but have been suggested to accumulate in wounds. Here, we describe a CD1a-bearing BDCA-2+CD123int DC subset that rapidly infiltrates human skin wounds and comprises a major DC population. Using single-cell RNA sequencing, we show that these cells are largely activated DCs acquiring features compatible with lymph node homing and antigen presentation, but unexpectedly express both BDCA-2 and CD123, potentially mimicking pDCs. Furthermore, a third BDCA-2–expressing population, Axl+Siglec-6+ DCs (ASDC), was also found to infiltrate human skin during wounding. These data demonstrate early skin infiltration of a previously unrecognized CD123intBDCA-2+CD1a+ DC subset during acute sterile inflammation, and prompt a re-evaluation of previously ascribed pDC involvement in skin disease.
Introduction
Skin barrier compromise can serve as a portal of entry for microorganisms to the underlying tissue, or results in the initiation of inflammatory skin disorders, such as atopic dermatitis. Sensing and restoring skin tissue integrity involves crosstalk between epithelial cells, stromal cells, and hematopoietic cells that are responsible for clearing apoptotic/necrotic cell debris and protecting the host against microbial invasion (Martin, 1997; Singer and Clark, 1999; Wilgus, 2008). Furthermore, through secretory mediators, such as cytokines and growth factors, immune cells also support re-epithelialization, angiogenesis, and scar formation during the course of the wound-healing process (Eming et al., 2014; Greaves et al., 2013). Dysregulation of immune responses contributes to delayed or improper wound repair and the prolongation or exacerbation of skin inflammatory diseases (Guo and Dipietro, 2010; Segre, 2006); hence, understanding the underlying mechanisms of skin wound sensing and repair is of therapeutic relevance.
Plasmacytoid dendritic cells (pDCs), a BDCA-2–expressing dendritic cell (DC) subset, are generally not found in healthy human skin (Wollenberg et al., 2002). However, they have been reported to infiltrate skin during viral infection (Donaghy et al., 2009; Gerlini et al., 2006), inflammatory diseases (Nestle et al., 2005; Wollenberg et al., 2002), or skin injuries (Gregorio et al., 2010). When skin is damaged, keratinocytes at the wound edge express increased levels of cathelicidins, contributing to the inhibition of pathogen growth (Dorschner et al., 2001). Cathelicidins can form complexes with DNAs and RNAs released from the dying cells, which then serve as TLR7 and TLR9 agonists to be captured by skin-infiltrated pDCs (Ganguly et al., 2009; Lande et al., 2007), leading to secretion of robust quantities of type I interferon (IFN-I; Gilliet et al., 2008; Reizis et al., 2011). In mouse models, IFN-I can activate T cells to produce several effector cytokines, such as IL-17A and IL-22, that help modulate the function of human keratinocytes to promote the restoration of skin barrier function and microbial defense (Avitabile et al., 2015; Boniface et al., 2005; Wolk et al., 2004). The impairment of IFN-I signaling contributes to delayed re-epithelization of skin wounds (Gregorio et al., 2010; Nestle et al., 2005).
While T cells recognizing peptides have been well studied, it is becoming increasingly clear that T cells can also recognize nonpeptide antigens, for example, lipids and lipopeptides presented by MHC-like molecules, including CD1a (Bourgeois et al., 2015; de Jong et al., 2010; Jarrett et al., 2016; Moody et al., 2004). CD1a is able to present endogenous skin lipid antigens, such as squalene, wax esters, lysophospholipids, and fatty acids (de Jong et al., 2014), which are enriched in the epidermis, as well as exogenous lipid ligands from pollen (Agea et al., 2005), plant sap (Kim et al., 2016), and bacteria (Peña-Cruz et al., 2003). CD1a is abundantly expressed by human Langerhans cells (LCs) and thymocytes and is inducible by subsets of human DCs and innate lymphoid cells. It plays a role in the regulation of T cell–mediated inflammatory responses in skin disease (Hardman et al., 2017; Jarrett et al., 2016; Subramaniam et al., 2016). Furthermore, CD1a blockade led to the alleviation of psoriatic and dermatitis skin inflammation in a transgenic murine model, suggesting therapeutic relevance (Kim et al., 2016).
Despite the circumstantial evidence supporting the existence of BDCA-2–expressing pDCs and contribution to human skin integrity, the transcriptomic analysis of this population remains lacking in humans. Here, we use human skin challenge systems to present the discovery of a CD1a-expressing BDCA-2+ subpopulation with conventional DC (cDC)–activating properties, yet absent broad pDC signature transcription profiles. These data prompt re-evaluation of the role previously ascribed to pDCs in the skin, and could represent a potential therapeutic target in promoting wound repair or alleviating inflammatory skin disease.
Results
CD1a-positive, BDCA-2–expressing DCs accumulated near skin wound sites
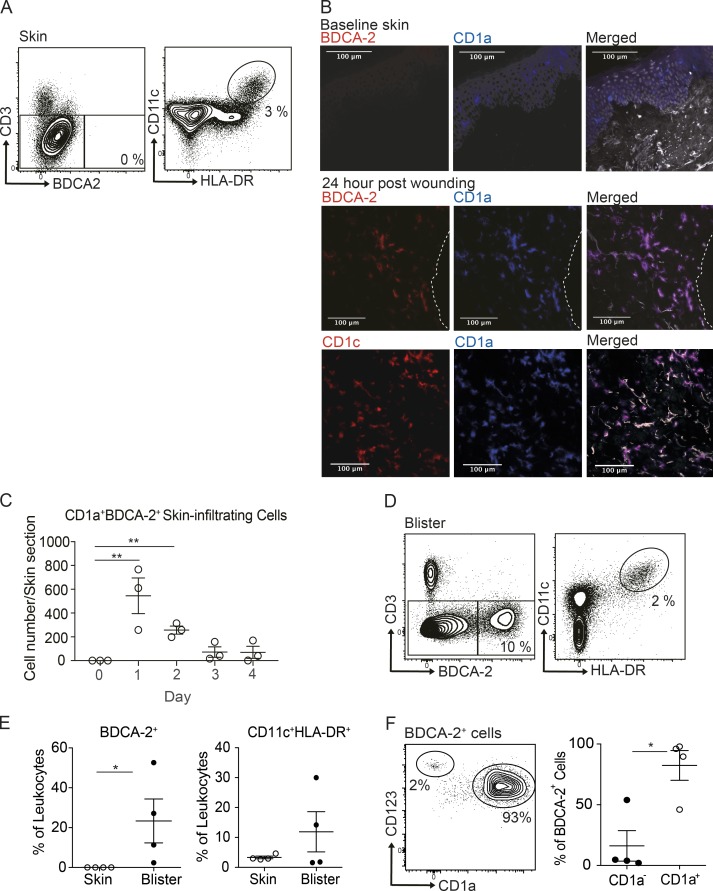
pDCs are normally characterized as CD123+BDCA-2+ hematopoietic cells and have been observed to accumulate in skin wounds, but are rarely observed in normal skin. Indeed, we confirmed that healthy human skin had undetectable levels of pDCs (Fig. 1 A). Meanwhile, broad populations of cDCs and monocytes, defined as CD11c+HLA-DR+, were present in healthy skin (Fig. 1 A). To investigate the distribution of DC populations infiltrating into skin wounds, baseline full-thickness punch biopsies were performed on the flank of healthy donors to create skin wounds. This was followed by larger biopsies to encompass the original wounds, taken 1–4 d later, and the frozen tissue sections were generated. In prewound skin, there were no BDCA-2+ cells, while CD1a was observed in the epidermis associated with LCs (Fig. 1 B). Unexpectedly, an accumulation of BDCA-2+CD1a+CD1c+ cells was detected in the dermis along the wound margin as early as 1 d after wounding and sustained up to 48 h (Fig. 1, B and C). The results provided architectural and longitudinal evidence that, in human skin wounds, BDCA-2+ DCs can unexpectedly be found expressing CD1a, which hitherto has been shown to be largely restricted to cDC subsets, LCs, innate lymphoid cells, and thymocytes.
Figure 1.
CD1a-positive BDCA-2+ DCs can be found in the skin suction blisters and near skin wound. (A) Mononuclear cells isolated from normal skin were analyzed for DC subsets by flow cytometry. One representative result is shown of three independent experiments; n = 4. (B and C) Immunohistology of skin biopsies collected at day 0 to day 4 after bilateral skin wounds on the flank from healthy volunteers. The results of day 1 after wounding are shown (B). DAPI (gray), CD1a+ (blue), BDCA-2+ or CD1c+ (red), and CD1a+BDCA-2+ cells (purple). Dashed white lines represent the edge of the wound. One representative result is shown of three independent analyses. Time-course frequency of CD1a+BDCA-2+ DC per biopsy is shown (C). Each dot represents a biopsy. (D and E) Mononuclear cells isolated from skin suction blisters were analyzed for monocyte and DC subsets by flow cytometry. Percentages of BDCA-2+ DC and CD11c+HLA-DR+ cDCs/monocytes in the skin biopsies or blisters. (F) Expression of CD123 and CD1a by BDCA-2+ cells. Percentages of CD1a+ DCs and CD1a− DCs in the blisters. One representative blister result is shown of four independent experiments; each dot represents a donor. One to two blisters per challenge condition were created per upper arm of each donor. Scale bars, 100 µm. Lines represent the mean + SEM. *, P < 0.05; **, P < 0.01; Student’s t test or one-way ANOVA.
Distinct subsets of BDCA-2–expressing DCs rapidly infiltrate skin wounds
Human skin suction blisters were used as a complementary in vivo human skin traumatic wound model (Jarrett et al., 2016; Salimi et al., 2013). Skin suction blisters represent a specific form of wounding associated with dermo-epidermal separation, and cells harvested from the blisters do not require processing or prolonged enzymatic treatments required by conventional skin biopsies; therefore, while being labor intensive, this technique does offer the ability to capture tissue cells directly ex vivo. In contrast to healthy skin, detectable levels of BDCA-2+ DCs and CD11c+HLA-DR+ cells were found in the skin blister fluid (Fig. 1, D and E). On further analysis of the skin-infiltrating BDCA-2+ DC population, two distinct subsets were revealed: CD123hiCD1a− and CD123intCD1a+ populations, of which the latter vastly predominated (Fig. 1 F). As an unexpected heterogeneity was observed within skin BDCA-2+ DCs, their gene expression profiles and the functional consequences were the primary target of subsequent single-cell studies.
Single-cell RNA profiling of skin-infiltrating BDCA-2+ DCs in sterile skin wounds
By using single-cell RNA sequencing (scRNA-seq), we were able to further investigate the nature of the BDCA-2+ cells isolated from the skin suction blisters, and to investigate their niche roles in sterile skin inflammation.
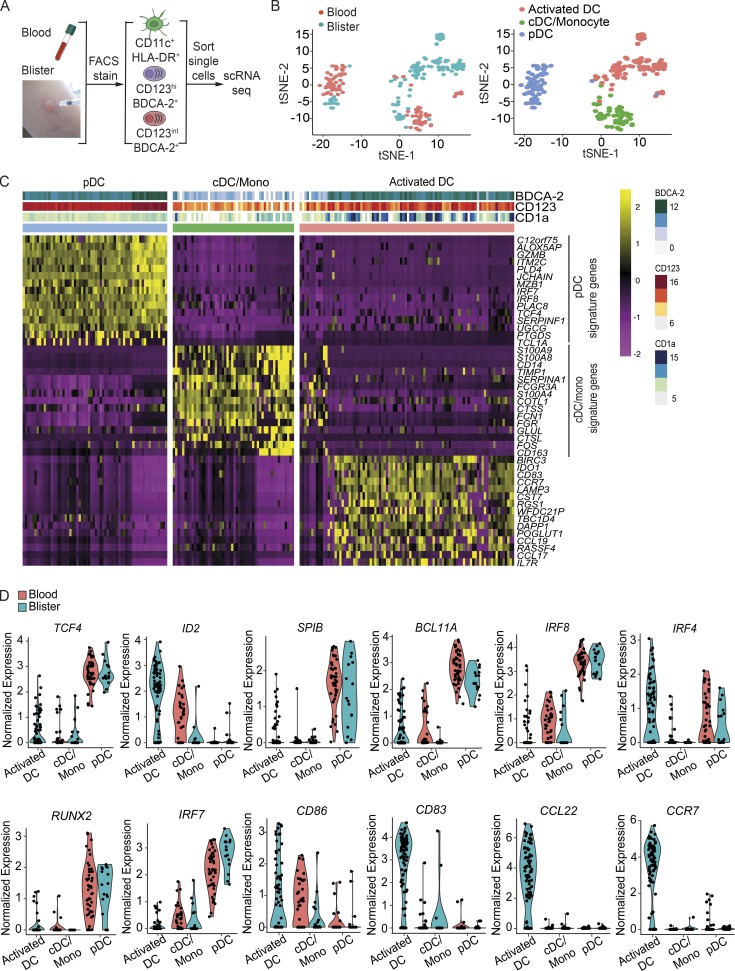
Blood- and skin blister–derived cDCs/monocytes, BDCA-2+CD123hi DCs, and BDCA-2+CD123int DCs were isolated by FACS based on surface marker expression, followed by scRNA-seq and RNA expression quantification (Fig. 2 A). Unbiased dimensionality reduction of the scRNA-seq data with t-distributed stochastic neighbor embedding (t-SNE) showed three distinct clusters (Fig. 2 B). We identified differentially expressed DC-related genes after comparing gene expression profiles between each subset, with the relevant top cDC/monocyte and pDC signature genes shown in Fig. 2 C. While BDCA-2+CD123hi DCs expressed pDC gene profiles, skin BDCA-2+CD123int DCs formed a distinct cluster that lacked cDC and pDC signature genes, but had an increased expression of genes associated with DC activation; hence, they are termed “activated DCs” hereafter. 10 blood cells were clustered with activated DCs; however, they expressed cDC/monocyte signature genes and showed low CD123 and BDCA-2 expression (Fig. 2 C), and were therefore not included in the subsequent analysis.
Figure 2.
Human saline blister-derived BDCA-2+ DC heterogeneity analysis by scRNA-seq. (A) Workflow of experimental strategy. 43 blood CD11c+HLA-DR+ cells, 47 blood BDCA-2+ cells, 19 skin saline blister CD11c+HLA-DR+ cells, and 97 skin saline blister BDCA-2+ cells were successfully profiled from a single donor. (B) Unbiased t-SNE analysis of all DC subsets. Each dot represents an individual cell. (C) Heatmap colored by normalized gene expression in individual cells of each cluster. Top margin color bars report the index fluorescent values of CD123, BDCA-2, and CD1a. (D) Violin plots demonstrate the expression distribution of indicated genes across DC subsets in blood and blisters. 10 blood cells within the activated DC cluster were removed because they expressed cDC/monocyte (mono) signature genes and showed low CD123 and BDCA-2 expression. Each dot represents an individual cell.
Transcription factor 4 (TCF4) is essential for pDC development and phenotype maintenance, and the alteration of the balance between TCF4 and its inhibitor ID2 expression leads to the up-regulation of cDC-related genes. In addition, losing TCF4 expression results in the reduction of several genes related to the development, survival, and activation of pDCs and their progenitors (Cisse et al., 2008; Ghosh et al., 2010; Robbins et al., 2008), including SPIB (Karrich et al., 2012; Schotte et al., 2004), BCL11A (Ippolito et al., 2014), and IRF8 (Esashi et al., 2008). Here, we noted that the TCF4 expression level was low in skin activated DCs, coinciding with reduced levels of SPIB, BCL11A, and IRF8, compared with skin BDCA-2+CD123hi DCs, while the ID2 level was high (Fig. 2 D). Meanwhile, IRF4, which is highly expressed in certain cDC subsets (Naik et al., 2015) and pDCs, remained at a high level in skin activated DCs (Fig. 2 D).
In terms of genes relating to DC functions, runt related transcription factor 2 (RUNX2) and its downstream signaling molecule IRF7, which are key regulators of IFN-I production (Chopin et al., 2016; Sawai et al., 2013), were both decreased, while costimulatory marker genes were elevated in skin activated DCs (Fig. 2 D). Notably, skin activated DCs expressed very high levels of CCL22, a chemoattractant for C-C chemokine receptor type 4 (CCR4)–expressing cells that facilitates migration into the skin (Elhussein Mohamed et al., 2016; Katou et al., 2001; Kusumoto et al., 2007; Layseca-Espinosa et al., 2013). CCR7 directs the lymph node homing of DCs and was one of the most significantly up-regulated transcripts in skin activated DCs (Fig. 2 D). Collectively, these data supported the idea that skin wound BDCA-2+CD123int DCs, expressing markers previously ascribed to pDCs, had “activated DC” functionality to present antigens, migrate to lymph nodes, and recruit T cells. Skin BDCA-2+CD123hi DCs, on the other hand, were functionally more closely related to their blood ancestry cells and were most likely to represent bona fide pDCs.
Transcriptomic analysis of skin BDCA-2+ DCs in skin inflammatory conditions
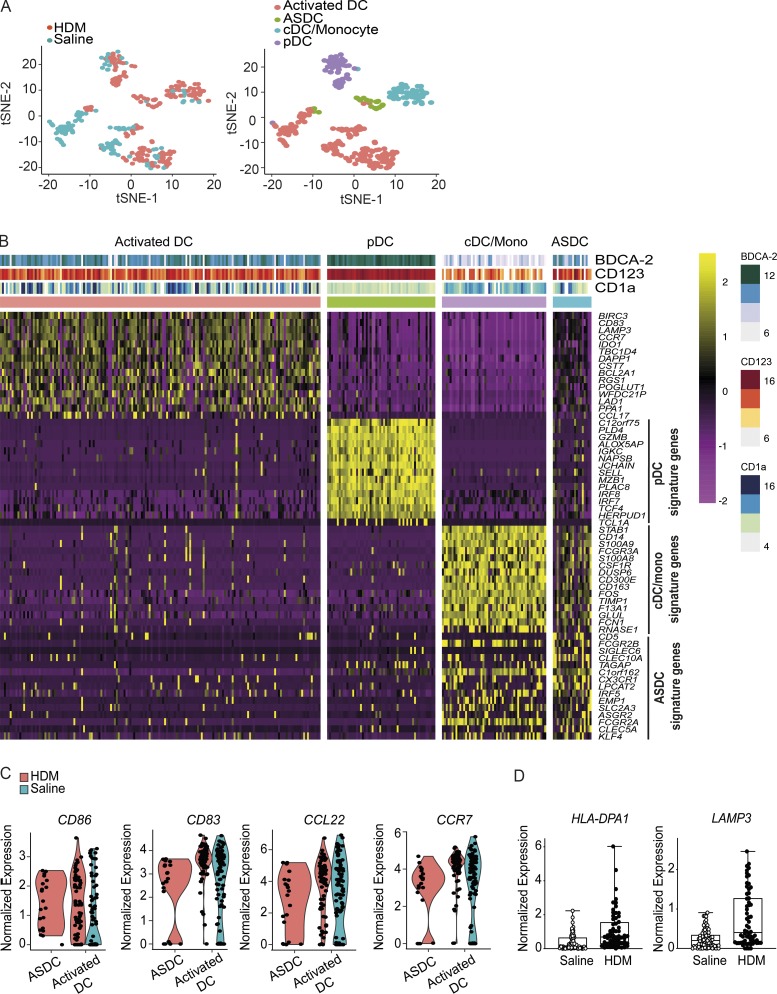
Skin barrier disruption can serve as an onset of inflammatory skin disorders. House dust mite (HDM) is a common environmental allergen, and cutaneous HDM exposure has been shown to associate with the pathology of atopic dermatitis (Norris et al., 1988). Skin suction blisters provided an accessible tool to investigate human sterile skin inflammation after HDM skin challenge. Comparing the scRNA-seq dataset of saline- and HDM-challenged skin, we observed four clusters comprising cDCs/monocytes, pDCs, and activated DCs, Axl+Siglec-6+ DCs (ASDCs; Fig. 3 A). ASDCs represent a noncanonical DC subset that lacks IFN-I production ability but can prime T cells (Villani et al., 2017). The ASDCs detected herein from skin blisters were highly activated, expressing CCL22 and CD83, and may be recruited into skin in response to inflammatory cues (Fig. 3, B and C). In addition, activated DCs derived from HDM-challenged skin blisters expressed a higher level of genes related to DC maturation, such as HLA-DPA1 and LAMP3, which appear during DC maturation (de Saint-Vis et al., 1998), suggesting that they were responsive toward environmental allergens (Fig. 3 D).
Figure 3.
Human HDM blister-derived BDCA-2+ DC heterogeneity analysis by scRNA-seq. (A) 180 cells were successfully profiled from HDM-challenged skin blisters from a single donor, and unbiased t-SNE analysis of all DC subsets isolated from saline and HDM blisters is shown. Each dot represents an individual cell. (B) Heatmap colored by normalized gene expression in individual cells of each cluster. Top margin color bars report the index fluorescent values of CD123, BDCA-2, and CD1a. Mono, monocyte. (C) Violin plots demonstrate the expression distribution of indicated genes across ASDCs and activated DCs. Each dot represents an individual cell. (D) HLA-DPA1 and LAMP3 expression of activated DCs from saline- and HDM-challenged blisters. Boxes show 25–75% percentiles; whiskers show minimum to maximum.
Phenotypic analysis of skin-infiltrating BDCA-2+ DCs
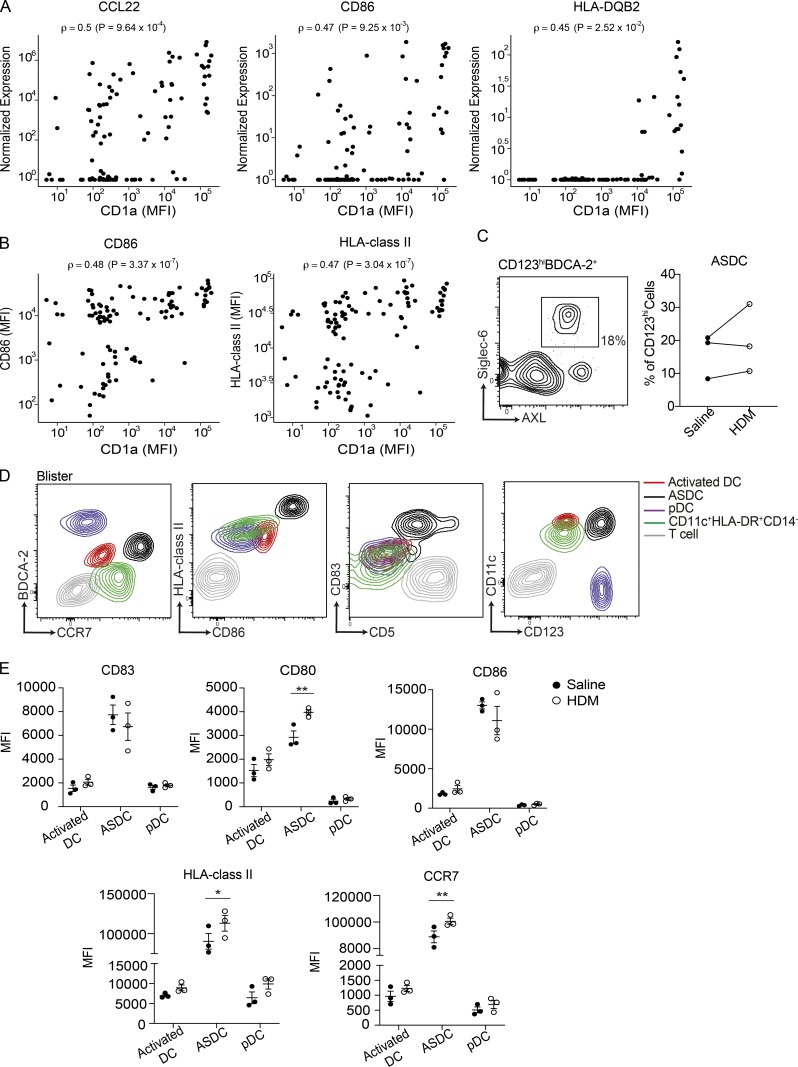
Notably, a wide spectrum of gene expression among the skin-infiltrating BDCA-2+CD123int population was shown in the transcriptomic analysis. Here, we observed that genes involved in antigen presentation and recruitment of T cells, such as CCL22, HLA class II, and CD86, were positively correlated with CD1a protein levels (Fig. 4 A). This was further validated with CD1a, CD86, and HLA class II surface protein expression on skin activated DCs (Fig. 4 B), suggesting a correlation between activation of skin BDCA-2+CD123int DCs and higher levels of CD1a expression. ASDCs were identified in both saline- and HDM-challenged blisters (Fig. 4 C); compared with other DC subsets and T cells, ASDCs expressed higher levels of activation markers (Fig. 4, D and E). In addition, ASDCs up-regulated CD80, HLA class II, and CCR7 after HDM stimulation (Fig. 4 E). In comparison to the BDCA-2+CD123hi population, the BDCA-2+CD123int population exhibited both pDC and cDC surface markers, along with a more active phenotype, and expressed molecules associated with antigen presentation (Fig. 4, D and E). The minor population of BDCA-2+CD123hi DCs were more compatible with canonical pDCs. These data suggested a potential acquisition of an activation phenotype in response to skin-derived environmental cues.
Figure 4.
Human skin activated DCs and ASDCs express genes and proteins relating to antigen presentation. (A) CD1a protein expression is significantly correlated with gene expression of CCL22 (ρ = 0.5, P < 0.05), CD86 (ρ = 0.47, P < 0.05), and HLA-DQB2 (ρ = 0.45, P < 0.05). Each dot represents an individual cell. Spearman correlation was performed. (B) Flow cytometry of CD1a, CD86, and HLA class II expression of BDCA-2+ DCs from skin suction blisters. Correlations between CD86 and CD1a (ρ = 0.48, P < 0.05) and HLA-DR and CD1a (ρ = 0.47, P < 0.05) are shown. Each dot represents an individual cell. Spearman correlation was performed. One representative result of three independent experiments is shown. (C–E) Mononuclear cells isolated from skin suction blisters were analyzed for DC subsets by flow cytometry. The percentage of ASDCs (C) and expression of BDCA-2, CCR7, HLA class II, CD86, CD83, CD123, and CD11c of activated DCs (red), ASDCs (black), bona fide pDCs (purple), cDCs (green), and T cells (gray) in the saline- and HDM-challenged blisters (D and E) are shown. One representative result of two independent experiments is shown; each dot represents a donor. One to two blisters per challenge condition were created per upper arm of each donor. MFI, mean fluorescence intensity. Lines represent the mean + SEM. *, P < 0.05; **, P < 0.01; Student’s t test or two-way ANOVA.
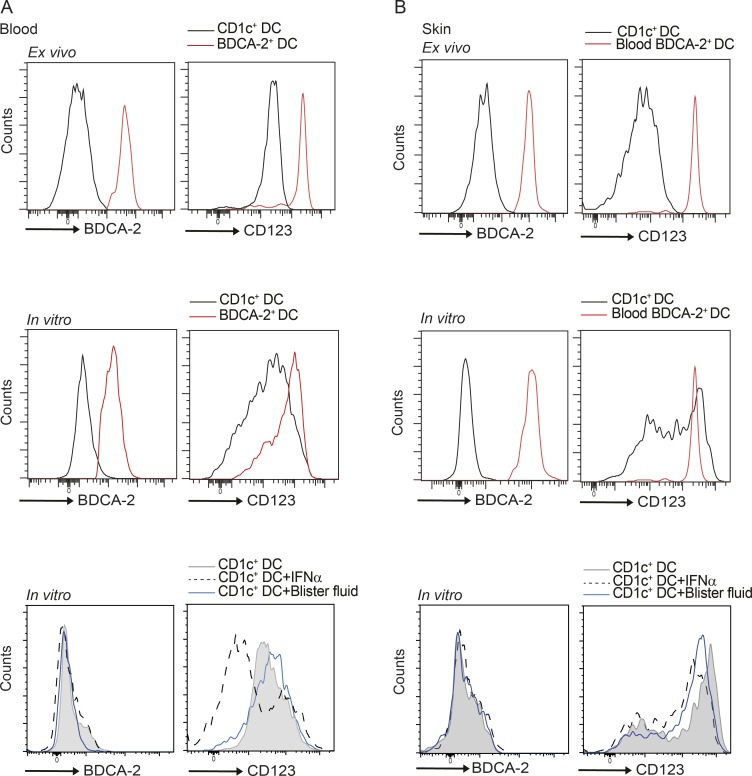
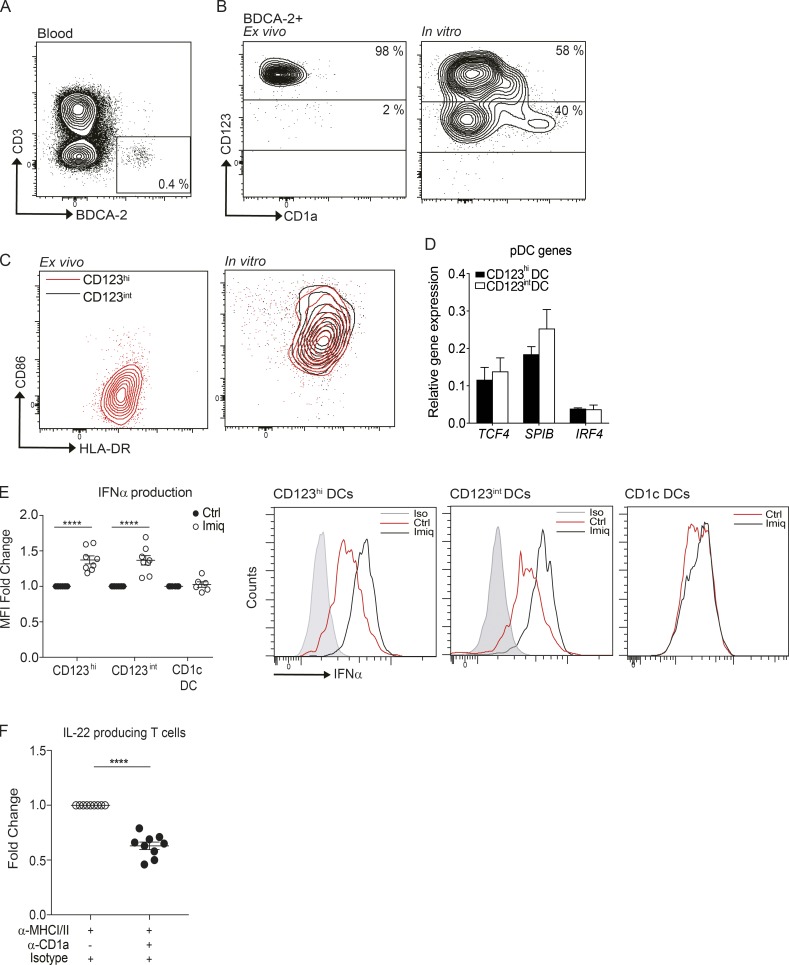
In view of the limited cell numbers obtained from skin blisters to undertake functional analyses, we first acquired DC subsets from blood or larger skin samples from plastic surgery procedures to analyze their functional phenotype in response to relevant stimuli. As there are no BDCA-2+ cells in healthy skin (Fig. 1 B, upper panels), and the BDCA-2+ cells in skin wounds were CD1c+ (Fig. 1 B, lower panels), we used CD1c+ cells as the skin source population. Blood and skin CD1c+ DCs were isolated and cultured in vitro with IFNα or skin blister fluid for 48 h, and up-regulated CD123 expression was observed; however, neither blood nor skin CD1c+ DCs expressed BDCA-2 after culture (Fig. S1, A and B). The freshly isolated BDCA-2+ DCs from the blood were predominantly CD123hi with low levels of CD1a on the surface, consistent with being a typical pDC population (Fig. S2, A and B). After 48-h culture, both CD123hiBDCA-2+ and CD123intBDCA-2+ live cell populations were identified with elevated expression of CD1a, particularly on the latter (Fig. S2, B and C). However, a similar transcription factor profile was exhibited by both BDCA-2+ subpopulations, contrasting with blister-derived activated DCs (Fig. S2 D). In vitro cultured blood DC subsets were subjected to imiquimod (TLR7 agonist) treatment, and both BDCA-2+ subsets produced similar levels of IFNα (Fig. S2 E; raw mean fluorescence intensity values of CD123hi [Ctrl, 5,218 + 538; Imiq, 7,115 + 1,112], CD123int [Ctrl, 5,516 + 400; Imiq, 7,090 + 885], and CD1c+ DC [Ctrl, 5,043 + 1,115; Imiq, 5,040 + 1,029]). To evaluate if the CD1a on in vitro cultured BDCA-2+ cells was functionally relevant, blood polyclonal T cells were incubated with in vitro cultured autologous BDCA-2+ DCs, and IL-22 cytokine production was detected, after blocking human MHC class I and class II signaling. Neutralizing CD1a signaling attenuated IL-22 secretion from the T cells (Fig. S2 F; raw spot numbers from 50,000 T cells of anti-MHC I/II [73.83 + 11.59] and anti-MHC I/II + anti-CD1a [48.89 + 9.49]), suggesting the dependence on CD1a for autoreactive T cell activation.
Figure S1.
Expression levels of BDCA-2 and CD123 on DCs in skin and blood. (A and B) BDCA-2 and CD123 expression was analyzed by flow cytometry on ex vivo and in vitro IL-3, IFNα, and blister fluid–cultured blood CD1c+ DCs and blood BDCA-2+ DCs (A) and ex vivo and in vitro IL-3, IFNα, and blister fluid–cultured skin CD1c+ DCs and blood BDCA-2+ DCs (B). One representative result of three independent experiments is shown; n = 3.
Figure S2.
Phenotypic analysis of in vitro–derived blood BDCA-2+ DCs. (A) PBMCs were analyzed for BDCA-2+ DCs by flow cytometry. (B) Expression of CD123 and CD1a on freshly isolated BDCA-2+ DCs (ex vivo) or blood BDCA-2+ DCs treated with IL-3 (50 ng/ml) for 48 h (in vitro). (C) CD86 and HLA-DR expression on in vitro BDCA-2+ DCs analyzed by flow cytometry. One representative result of 10 independent experiments is shown for B and C; n = 16. (D) TCF4, SPI-B, and IRF4 expression level of CD123hi and CD123int BDCA-2+ DCs after in vitro culture with IL-3. One representative result of three independent experiments is shown; n = 3. (E) Expression of IFNα production by each BDCA-2+ DC subset and CD1c+ DCs analyzed by flow cytometry. Fold-induction of IFNα by CD123hi and CD123int BDCA-2+ DCs and cDCs after in vitro imiquimod (Imiq) stimulation. One representative result of three independent experiments is shown; each dot represents a donor. Ctrl, control. (F) IL-22 production from blood T cells co-cultured overnight with CD1a-positive blood BDCA-2+ DCs. IL-22 was measured by ELISpot in the absence or presence of anti-CD1a antibody and anti-MHC I/II antibodies. Data represent the fold reduction of five independent experiments; each dot represents a donor. Lines represent the mean + SEM. MFI, mean fluorescence intensity. **, P < 0.01; ****, P < 0.0001; Student’s t test or one-way ANOVA.
In summary, these results show that neither blood BDCA-2+ DCs nor blood/skin CD1c+ DCs fully recapitulated the phenotype and function of the skin activated DCs under in vitro manipulation, suggesting that microenvironmental cues in the skin are indispensable for BDCA-2–expressing activated DCs to acquire broad functional capabilities to support the early critical events during sterile skin inflammation in humans.
pDC involvement in skin inflammation
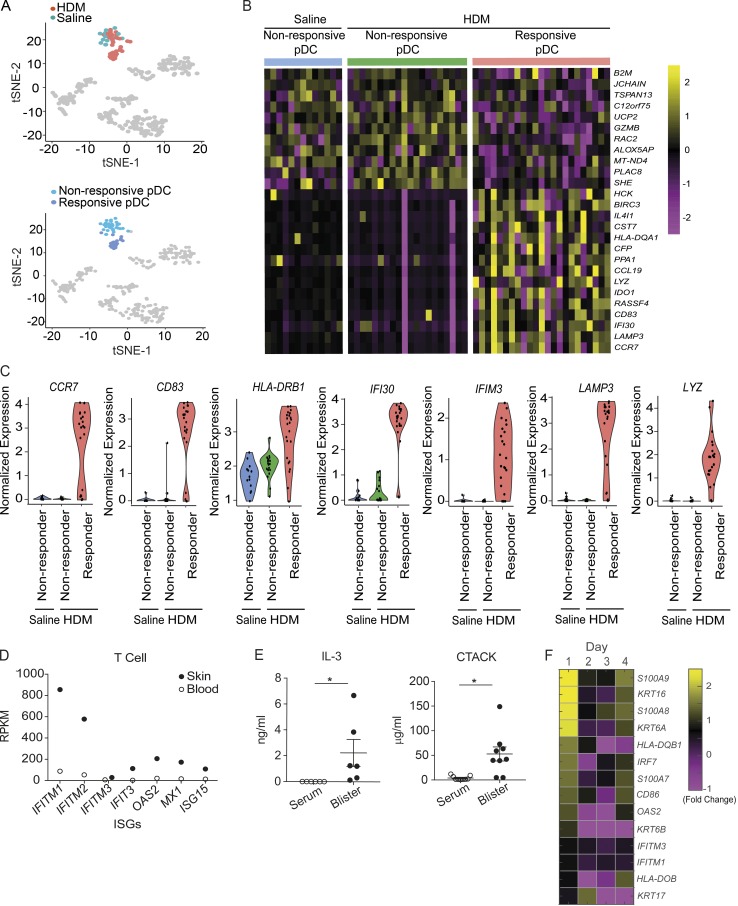
To further confirm the contribution of bona fide pDC in sterile acute skin inflammation, we analyzed the pDC cluster, which subclustered into two populations based on activation status (Fig. 5 A). The majority of pDCs isolated from the saline-challenged skin were less activated, while approximately half of the pDCs isolated from the HDM-challenged skin expressed genes related to DC activation and maturation, including chemokine receptor and costimulatory markers, and genes related to lysosomal trafficking, potentially supporting antigen processing (Fig. 5, B and C). Bulk RNA sequencing analysis was undertaken on T cells isolated directly ex vivo from the HDM-challenged skin blisters. This analysis showed a significant increase in signature IFN-stimulated genes, such as IFITM1, IFITM2, IFIT3, MX1, and OAS2 (Fig. 5 D), confirming that IFN-I was present at functionally relevant levels. Exposure of pDCs to IL-3 increased the expression of CCR10 and induced pDC recruitment to inflamed murine skin lesion sites, and IL-3 is also an essential cytokine involved in pDC survival and activation (Mathan et al., 2013; Sisirak et al., 2011). Therefore, concentrations of CCR10 ligand, cutaneous T cell–attracting chemokine (CTACK), and IL-3 were measured in the skin HDM-challenged blister fluid, compared with their levels in the serum. Both CTACK and IL-3 were significantly up-regulated in the interstitial blister fluid, potentially contributing to pDC recruitment to the skin (Fig. 5 E). Targeted RNA sequencing analysis was performed on the skin wound biopsies to determine wound-related gene profiles. Fold changes of gene expression from each day compared with baseline skin are shown (Fig. 5 F); we detected high induction of keratin proteins KRT6, KRT16, and KRT17 and antimicrobial peptides S100A7, S100A8, and S100A9 in early wounds, consistent with the reports of gene expression profiles of activated keratinocytes in skin wounds (Aragona et al., 2017; Freedberg et al., 2001). Moreover, we noted that several IFN-stimulated genes, such as OAS2, IFITM3, and IFITM1, and costimulatory marker genes, including HLA-DQB, HLA-DOB, and CD86, were up-regulated 1 d after wounding, suggesting the potential involvement of pDCs and other APCs in the early stage of the human skin wound-healing process. Overall, while our data showed that the majority of previously identified “pDCs” are likely to be activated DCs, there is nevertheless a small population of bona fide pDCs infiltrating into skin wounds, and these pDCs are functionally active.
Figure 5.
Skin-infiltrating pDCs and their relevant cytokines can be found in skin suction blisters. (A) t-SNE analysis of pDCs isolated from saline and HDM blisters is shown. Each dot represents an individual cell. (B) Heatmap colored by normalized gene expression in individual cells for pDC status. (C) Violin plots demonstrate the expression of indicated genes across pDCs from saline- and HDM-challenged blisters. Each dot represents an individual cell. (D) T cells were isolated from skin suction blisters or blood and subjected to RNA sequencing analysis. Reads of skin-infiltrated cells were compared with those of blood counterparts. RPKM, reads per kilobase of transcript per million mapped reads. (E) Concentration of CTACK and IL-3 in the blister fluid or serum was evaluated by multiplex bead array. Data are pooled from six to nine donors. Error bars show mean and SEM. (F) Targeted RNA sequencing was performed on skin biopsies collected at day 0 to day 4 after bilateral skin wounds on the flank from healthy volunteers. Heatmap colored by log fold-change values for genes on each day after wounding. Average result of an experiment using three skin biopsies at each day is shown.
Discussion
DC subsets are traditionally defined according to their surface markers, functional properties, and developmental ancestry. However, it is now accepted that the preexistent classification underestimated the heterogeneity of blood and tissue DC populations. Despite sharing some surface marker features and transcription factors, subsets are likely to be functionally specialized (Chu et al., 2012; Haniffa et al., 2012). Recent studies using scRNA-seq and high-dimensional phenotypic mapping have revealed newly discovered subtypes of blood DCs (Alcantara-Hernandez et al., 2017; Alculumbre et al., 2018; Villani et al., 2017). For example, pDCs have previously been mapped as a relatively homogeneous group, until recent reports noted the existence of phenotypically distinct human blood pDC subsets. Several studies on human and mouse pDC heterogeneity have suggested the existence of noncanonical populations that can feature an intermediate status between pDC and cDC (Alcantara-Hernandez et al., 2017; Bar-On et al., 2010; Reizis, 2019; Villani et al., 2017; Zhang et al., 2017); for example, ASDCs exhibit a spectrum between cDC-like and pDC-like cells (See et al., 2017; Villani et al., 2017). Alcantara-Hernandez et al. (2017) have observed ASDCs in the blood and lymphoid tissues, such as tonsil and spleen, but none in steady-state skin. Here, we identify a skin DC subset that is an early infiltrator in two models of human skin sterile inflammation, which was functionally distinct from traditionally defined pDCs, yet bearing some pDC surface features. The expression of CD123, BDCA-2, and CD1a by this activated DC subset in the skin prompts a re-evaluation of cells previously considered to be pDCs in skin pathology.
CD1a is expressed constitutively by LCs and is inducible by dermal DCs, which both exist in steady-state skin and are enriched in skin inflammatory conditions, consistent with a functional role for CD1a in skin inflammation (Angel et al., 2006; Haniffa et al., 2015; Katou et al., 2000; Klechevsky et al., 2008; Korenfeld et al., 2017; Wollenberg et al., 1996; Wollenberg et al., 2011). Studies have reported functional differences between human LCs and CD1a+ dermal APCs. CD1a+ dermal APCs localized more closely to lymphatic vessels with chemotactic abilities, and arrived earlier in cutaneous lymph nodes than LCs (Angel et al., 2006; Kissenpfennig et al., 2005; Klechevsky et al., 2008). These reports indicate the importance of having multiple cell types that can be rapidly recruited and acquire the necessary functionality to contribute to the inflammatory response. Furthermore, CD1a antigen presentation to T cells has been demonstrated to associate with the inflammation of atopic dermatitis, venom allergy, contact dermatitis, and psoriasis (Bourgeois et al., 2015; Cheung et al., 2016; Hardman et al., 2017; Jarrett et al., 2016; Kim et al., 2016). The robust infiltration, rapid turnover rate, and specialized features of skin CD1a-bearing BDCA-2+CD123int DCs defined herein, which are not found in healthy skin, suggest that they also contribute to the skin immune network and could potentially have a broad role in skin wound healing and disease pathology.
Tang-Huau et al. (2018) recently conducted scRNA-seq analyses on ascites macrophages, ascites DCs, and tonsil DCs and described an activated cDC2 subset in the tonsil expressing high levels of genes similar to those found in skin BDCA-2+CD123int DCs, including CCR7, CCL19, LAMP3, IDO1, CD83, and HLA class II. However, CD123 and BDCA-2 surface markers were not included during isolation of the DCs from the tonsil, and further analysis showed that genes, including RELB, IER2, and SPI1, were different between tonsil activated cDC2s and skin BDCA-2+CD123int DCs, suggesting that the environmental milieu may play a role in shaping the phenotypic and functional features of tissue DC subsets.
A recent report of therapeutic intervention of targeting BDCA-2 in individuals with systemic lupus erythematosus highlighted the involvement of BDCA-2–expressing cells in human skin inflammatory conditions (Furie et al., 2019). Upon binding, BIIB059, a humanized anti–BDCA-2 antibody, induced a rapid internalization of BDCA-2 from the pDC surface, leading to the attenuation of IFN-I secretion by pDCs. In this clinical trial, administration of BIIB059 decreased IFN-I responses and cellular infiltration in the skin, leading to clinical benefits. It is unclear if other BDCA-2–expressing DCs coexist with bona fide pDCs in the skin lesions of systemic lupus erythematosus patients. In addition, given the data presented herein, it is of interest whether anti–BDCA-2 antibody targets activated DCs and ASDCs as well as pDCs. It may be of clinical interest to evaluate the mechanisms and efficacy of the anti–BDCA-2 antibody in chronic skin inflammatory conditions where activated DCs and ASDCs play a role.
In conclusion, skin BDCA-2+CD123int DCs represent a specialized tissue-specific DC status. Our study identifies a major previously unrecognized activated DC population in humans that infiltrates early into skin wounds and contributes to acute sterile inflammation. In addition, the identification of ASDCs after skin wounding suggests a role in skin disease and prompts further investigation in this and other forms of skin inflammation. The overlapping surface phenotype of these two noncanonical DCs with pDCs necessitates a re-examination of disease models where modulation of previously ascribed “pDC” numbers and function have been suggested. Furthermore, the cells and pathways may provide a novel target for therapeutic intervention in conditions of aberrant tissue repair and skin disorders.
Materials and methods
Isolation of immune cells from the blood or skin of humans
Human blood samples were obtained from healthy donors under local ethics approval (14/SC/0106, National Research Ethics Service [NRES]). Peripheral blood mononuclear cells (PBMCs) were isolated using Lymphoprep (Stem Cell Technologies) gradient isolation. Skin samples were obtained from healthy donors undergoing plastic surgery. Samples were taken under good clinical practice guidance with ethical approval of the NRES Committee South Central. Subcutaneous fat was removed completely before the sample was dissected and incubated in collagenase P (Roche) overnight at 37°C. Cold 10 mM EDTA solution was added to the sample to stop the digestion. The digested tissue was passed through a 70-µm strainer and washed with EDTA solution, and mononuclear cells were harvested with Lymphoprep gradient isolation before further procedures.
Antibodies, flow cytometry, and cell isolation
Surface staining was performed at 4°C in FACS staining buffer (0.5% FBS in PBS). Intracellular staining was performed using Foxp3/Transcription Factor Staining Buffer Set (Invitrogen), following the manufacturer’s instructions. The following anti-human antibodies were used: Brilliant Violet 650-anti-CD3 (OCT3; BioLegend), PE/Cy7-anti-CD123 (6H6; BioLegend), APC-anti-CD1a (HI149; BD Biosciences), Alexa Fluor 700 anti-CD1c (L161; BioLegend), PerCP-anti-CD11b (DCIS1/18; BioLegend), Brilliant Violet 421-anti-CD11c (3.9; BioLegend), Brilliant Violet 650-anti-CD11c (3.9; BioLegend), Brilliant Violet 785-anti-HLA-DR (L243; BioLegend), APC/Fire 750-anti-HLA-DR (L243; BioLegend), PE anti-BDCA-2 (201A; BioLegend), Brilliant Violet 421 anti-BDCA-2 (201A; BioLegend), PE/Dazzle 594 anti-CD86 (IT2.2; BioLegend), Brilliant Violet 650-anti-CD1a (HI149; BD Biosciences), Brilliant Violet 605-anti-CD83 (HB15e; BioLegend), BUV805 anti-CD5 (UCHT2; BD Biosciences), BUV805-Anti-CD4 (SK3; BD Biosciences), BUV661-Anti-CD3 (UCHT1; BD Biosciences), Brilliant Violet 421-anti-CD80 (2D10; BioLegend), Alexa Fluor 488-anti-Axl (108724; Bio-Techne), Alexa Fluor 700-anti-Siglec-6 (767329; Bio-Techne), Brilliant Violet 785-anti-CCR7 (G043H7; BioLegend), APC/Cy7 anti-CD14 (63D3; BioLegend), Brilliant Violet 570-anti-CD14 (M5E2; BioLegend), live/dead aqua (Invitrogen), and live/dead violet (Invitrogen). Flow data were acquired using FACSDiva on LSRFortessa and further analyzed with FlowJo (Tree Star) software. Blister BDCA-2+ DCs were sorted with the BD FACSAria IIIu as BDCA-2+CD123hi and BDCA-2+CD123int cells. Blood BDCA-2+ DCs and T cells were isolated by magnetic-activated cell sorting with anti-BDCA-4 and anti-CD3 MicroBeads (Miltenyi Biotec), respectively, according to the manufacturer’s instructions. Blood CD1c+ DCs were isolated using BDCA-1+ DC isolation kit (Miltenyi Biotec) with BDCA-4+, CD19+, and CD14+ cells depleted from the blood before CD1c separation. For ensuring higher purity of both DC subsets, the positively labeled cells underwent a second column run.
Skin sample collection
Healthy donors were recruited, and bilateral 3-mm baseline full-thickness punch biopsies on the flank of the lower back were performed to create skin wounds. Larger biopsies (6 mm) to encompass the bilateral original wounds were taken 1–4 d later. The study was conducted under ethics approval [14/SC/0106 NRES].
Skin section and confocal microscopy analysis
The skin biopsy specimens were snap-frozen in optimal cutting temperature embedding compound and stored at −80°C. The tissues were cut into 4-µm sections and placed on slides to air-dry for 30 min before being stored at −80°C. Slides were rehydrated in PBS for 10 min before staining. The endogenous peroxidase activity of the sample was quenched by adding 0.3% hydrogen peroxide solution with a 5-min incubation at room temperature. The endogenous biotin was blocked with Avidin/Biotin Blocking Kit (Vector Laboratories Ltd), and 10% goat serum was used to reduce nonspecific binding of antibodies. Antibodies used for confocal microscopy include CD1a (1:100, OKT6; in-house production and conjugated with Alexa Fluor 647 dye) and BDCA2-Biotin (1:2, AC144; Miltenyi Biotec). Alexa Fluor 594 Tyramide SuperBoost kit (streptavidin; Thermo Fisher Scientific) was used to enhance the signals. Slides were incubated at 4°C with primary antibodies overnight. After washing, HRP-conjugated streptavidin was added to the sections and incubated at 4°C overnight. Excess streptavidin was washed away, the tissues were incubated with tyramide working solution for 8 min at room temperature, and the reaction was stopped with Reaction Stop Reagent. After staining, slides were mounted using antifade mounting medium with DAPI (Vector Laboratories Ltd), coverslips were applied, and slides were refrigerated in the dark until analyzed by confocal microscopy (Zeiss LSM 780 Confocal Microscope-Inverted Microscope; 25×/0.8 Imm Korr DIC M27; room temperature; Axiocam camera; Zen software), and Fiji was used for image processing.
Suction blister technique
Saline or HDM extract was delivered to the upper arm of the volunteers by intraepidermal skin prick test, and suction blister cups were applied to the sites with vacuum pressure of 200 mmHg to create blisters. Blisters were protected overnight, and the fluid was aspirated 24 h later to collect skin-infiltrated cells and tissue fluid. The concentration of indicated cytokines was measured by multiplex array (multiplex bead array), and the cells were stained with cell surface antibodies for flow cytometric analysis of DC subsets or for bulk or scRNA-seq analysis.
scRNA-seq
Human PBMCs and blister cells were stained with antibodies in PBS for 10 min, and each targeted cell of interest was sorted into each well of 96-well plates containing 2 µl of 0.2% Triton plus 1 U/µl RNaseOUT (Thermo Fisher Scientific). 182 blood cells and 273 skin cells were collected, and single-cell libraries were prepared using the Smart-Seq2 protocol (Picelli et al., 2014) with minor modifications. We performed reverse transcription using Smartscribe RT enzyme (50 U/cell; Takara). For cDNA amplification, we performed 25 PCR cycles. All the other parameters were as described in the original paper. Not all the wells were successfully profiled, and some of the wells were excluded from the data analysis after applying quality control (QC) filters.
scRNA-seq data analysis
Gene expression for individual cells was quantified using Salmon (Patro et al., 2017). Cells were kept if they had a mapping rate >40%, expressed >1,200 detected genes, and contained <20% of their reads associated to ERCC spike-ins. Genes were removed from the analysis if they were expressed (>1 count) in less than five cells. Lastly, the total read count was regressed from the expression values to normalize the data. t-SNE was performed in R (v3.4) using the Seurat package (v2.3.4; Satija et al., 2015). In the blood and skin data, clusters were called at a resolution of 1.8 using the first six principal components, generated using highly variable genes. In the saline and HDM data, clusters were called at a resolution of 2.4 using the first six principal components, generated using all genes. In both cases, clusters were merged if they presented similar markers, indicating a similar function. This is the case for subclusters of activated DCs and the responsive/nonresponsive pDCs (Fig. 5 A). Differentially expressed genes were obtained by using the FindMarkers function and considered differentially expressed if the corrected P value was <0.05. scRNA-seq data have been deposited in ArrayExpress under the accession no. E-MTAB-8498.
Bulk RNA sequencing
Suction blister cells and PBMCs were harvested, and T cells were sorted with the BD FACSAria IIIu and collected directly into TRIzol LS. The mRNA extraction, RNA sequencing, and data analysis were performed as previously described (Hardman et al., 2017). GEO data were deposited under accession identifier GSE139952.
Skin-targeted RNA sequencing
The skin biopsy specimens were subjected to targeted RNA sequencing analysis. Skin samples were collected directly into RNA Later reagent (Ambion) and frozen at −80°C until further use. Tissue disruption and homogenization was performed in RLT buffer (Qiagen) using TissueRuptor (Qiagen) on ice. RNA was extracted with Rneasy Fibrous Tissue Mini Kit (Qiagen) according to the manufacturer’s instructions. Next Generation Sequencing was performed with Illumina reagents (TruSeq Targeted RNA Index Kit and TruSeq Targeted RNA Custom Panel Kit) at the Deep Seq Facility (University of Nottingham, Nottingham, UK). Following QC analysis with the fastQC package, reads were aligned using STAR against the human genome assembly (UCSC GRCh38/hg38). Gene expression levels were quantified as read counts using the featureCounts function from the Subread package with default parameters (Ramos et al., 2011). The read counts were used for the identification of global differential gene expression between specified populations using the edgeR package. Values for reads per kilobase of transcript per million mapped reads were also generated using the edgeR package. GEO data were deposited under accession identifier GSE139834.
In vitro DC culture and T cell assay
Magnetic-activated cell sorting–purified BDCA-2+ DCs or CD1c+ DCs were seeded into 96-well U-bottom plates with 1–2.5 × 105 cells/well density in RPMI medium (supplied with 10% FCS, 2 mM L-glutamine, 100 IU/ml penicillin, 100 µg/ml streptomycin, 100 µM sodium pyruvate, nonessential amino acids, 50 µM β-mercaptoethanol, and 10 mM Hepes) and IL-3 (50 ng/ml; BioLegend) for 48 h. imiquimod (50 ng/ml; Invivogen) was used as stimulus in some conditions, and brefeldin A (5 µg/ml; BioLegend) was added for 12 h to block the cytokine transport process to enhance intracellular cytokine staining signals. Cells were then centrifuged at 1,500 rpm for 5 min at 4°C and used for flow cytometric analysis. For T cell co-culture assays, IL-22 ELISpot (Mabtech AB) was used to assess CD1a reactivity. The MultiScreen-IP sterile filter plates with hydrophobic poly(vinylidene fluoride) (Millipore) were coated with capture antibodies at 4°C overnight. In vitro cultured BDCA-2+ DCs (1.5 × 104) were co-cultured with T cells (5 × 104) per well with anti-HLA-A,B,C (10 µg/ml, W6/32; BioLegend) and HLA-DR (10 µg/ml, L243; BioLegend) antibodies and incubated at 37°C and 5% CO2 overnight. In some wells, BDCA-2+ DCs were pretreated with anti-CD1a blocking antibody (OKT-6, 10 µg/ml; produced in house) or IgG1 isotype control (10 µg/ml; BioLegend) 1 h before the addition of T cells. PMA (5 ng/ml) and ionomycin (5 ng/ml) stimulation or T cells alone were set up as positive controls or negative controls, respectively. After incubation, plates were washed and incubated with biotin-linked anti–IL-22 monoclonal antibody for 2 h. After washing, the plates were incubated for 2 h with streptavidin–alkaline phosphatase and were developed using the alkaline phosphatase conjugate substrate kit (Bio-Rad). The numbers of IL-22–producing T cells were counted using the automated ELISpot reader (ELISpot Reader Classic; Autimmun Diagnostika gmbh). The result graphs were calculated as the fold change of each condition to the anti-HLA/IgG1 isotype control treatment.
Quantitative real-time PCR
The mRNA of the cells was prepared using a TurboCapture kit (Qiagen). The cDNA was synthesized with M-MLV reverse transcription (Invitrogen) and then subjected to quantitative real-time PCR using TaqMan Probe (Life Technologies) to TCF4 (Hs00162613_m1), SPI-B (Hs00162150_m1), IRF4 (Hs00180031_m1), and GAPDH (Hs99999905_m1). Reactions were performed in a 7500 Fast Thermal Cycler (Applied Biosystems). Data were normalized to GAPDH.
Statistical analyses
All values are shown as means. Error bars represent SEMs. Paired one- or two-tailed t tests, one-way ANOVA, and Pearson correlation were performed using GraphPad Prism version 7.00 (GraphPad Software) to assess statistical significance: *, P < 0.05; **, P < 0.01; ***, P < 0.005; and ****, P < 0.0001.
Online supplemental material
Fig. S1 shows a characterization of BDCA-2 and CD123 expression on skin and blood DC subsets in in vitro assays. Fig. S2 presents phenotypic analysis of in vitro–derived blood BDCA-2+ DCs, including marker expression, gene profile, and functionality.
Acknowledgments
We thank the staff of the Medical Research Council Weatherall Institute of Molecular Medicine FACS facility, and Wolfson Imaging Centre, Wellcome Sanger Institute Sequencing pipelines, all of the research participants and research nurses, Melanie Westmoreland, and Teena Mackenzie.
We were supported by National Institute for Health Research Clinical Research Network, British Association of Dermatologists, British Skin Foundation, Misses Barrie Charitable Trust, Medical Research Council (U105178805; to A.N.J. McKenzie), Wellcome Trust (100963/Z/13/Z), National Institute for Health Research Oxford Biomedical Research Centre, UCB Pharma, Medical Research Council Human Immunology Unit core funding (to M. Salimi and V. Cerundolo), and Cancer Research UK program grant C399/A2291 (to V. Cerundolo). T. Gomes was supported by the European Union’s H2020 research and innovation program “ENLIGHT-TEN” under the Marie Sklodowska-Curie grant agreement 675395. This publication is part of the Human Cell Atlas (http://www.humancellatlas.org/publications). The views expressed are those of the authors and not necessarily those of the National Health Services, the National Institute for Health Research, or the Department of Health.
G. Ogg has served on advisory boards or holds consultancies or equity with Eli Lilly, Novartis, Janssen, Orbit Discovery, and UCB Pharma, and has undertaken clinical trials for Atopix, Regeneron/Sanofi, Roche, and AnaptysBio. A.N.J. McKenzie has grants from GSK and MedImmune/AZ. The authors declare no further competing financial interests.
Author contributions: Y.-L. Chen, T. Gomes, C.S. Hardman, F.A. Vieira Braga, D. Gutowska-Owsiak, M. Salimi, N. Gray, D.A. Duncan, G. Reynolds, D. Johnson, M. Salio, and J.L. Barlow performed the experiments and analyses and wrote the manuscript. V. Cerundolo, A.N.J. McKenzie, S. Teichmann, M. Haniffa, and G. Ogg designed and supervised the experiments and wrote the manuscript.
References
- Agea E., Russano A., Bistoni O., Mannucci R., Nicoletti I., Corazzi L., Postle A.D., De Libero G., Porcelli S.A., and Spinozzi F.. 2005. Human CD1-restricted T cell recognition of lipids from pollens. J. Exp. Med. 202:295–308. 10.1084/jem.20050773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcántara-Hernández M., Leylek R., Wagar L.E., Engleman E.G., Keler T., Marinkovich M.P., Davis M.M., Nolan G.P., and Idoyaga J.. 2017. High-dimensional phenotypic mapping of human dendritic cells reveals interindividual variation and tissue specialization. Immunity. 47:1037–1050.e6. 10.1016/j.immuni.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alculumbre S.G., Saint-André V., Di Domizio J., Vargas P., Sirven P., Bost P., Maurin M., Maiuri P., Wery M., Roman M.S., et al. . 2018. Diversification of human plasmacytoid predendritic cells in response to a single stimulus. Nat. Immunol. 19:63–75. 10.1038/s41590-017-0012-z [DOI] [PubMed] [Google Scholar]
- Angel C.E., George E., Brooks A.E., Ostrovsky L.L., Brown T.L., and Dunbar P.R.. 2006. Cutting edge: CD1a+ antigen-presenting cells in human dermis respond rapidly to CCR7 ligands. J. Immunol. 176:5730–5734. 10.4049/jimmunol.176.10.5730 [DOI] [PubMed] [Google Scholar]
- Aragona M., Dekoninck S., Rulands S., Lenglez S., Mascré G., Simons B.D., and Blanpain C.. 2017. Defining stem cell dynamics and migration during wound healing in mouse skin epidermis. Nat. Commun. 8:14684 10.1038/ncomms14684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitabile S., Odorisio T., Madonna S., Eyerich S., Guerra L., Eyerich K., Zambruno G., Cavani A., and Cianfarani F.. 2015. Interleukin-22 promotes wound repair in diabetes by improving keratinocyte pro-healing functions. J. Invest. Dermatol. 135:2862–2870. 10.1038/jid.2015.278 [DOI] [PubMed] [Google Scholar]
- Bar-On L., Birnberg T., Lewis K.L., Edelson B.T., Bruder D., Hildner K., Buer J., Murphy K.M., Reizis B., and Jung S.. 2010. CX3CR1+ CD8α+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. USA. 107:14745–14750. 10.1073/pnas.1001562107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniface K., Bernard F.X., Garcia M., Gurney A.L., Lecron J.C., and Morel F.. 2005. IL-22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J. Immunol. 174:3695–3702. 10.4049/jimmunol.174.6.3695 [DOI] [PubMed] [Google Scholar]
- Bourgeois E.A., Subramaniam S., Cheng T.Y., De Jong A., Layre E., Ly D., Salimi M., Legaspi A., Modlin R.L., Salio M., et al. . 2015. Bee venom processes human skin lipids for presentation by CD1a. J. Exp. Med. 212:149–163. 10.1084/jem.20141505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.L., Jarrett R., Subramaniam S., Salimi M., Gutowska-Owsiak D., Chen Y.L., Hardman C., Xue L., Cerundolo V., and Ogg G.. 2016. Psoriatic T cells recognize neolipid antigens generated by mast cell phospholipase delivered by exosomes and presented by CD1a. J. Exp. Med. 213:2399–2412. 10.1084/jem.20160258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin M., Preston S.P., Lun A.T.L., Tellier J., Smyth G.K., Pellegrini M., Belz G.T., Corcoran L.M., Visvader J.E., Wu L., and Nutt S.L.. 2016. RUNX2 mediates plasmacytoid dendritic cell egress from the bone marrow and controls viral immunity. Cell Reports. 15:866–878. 10.1016/j.celrep.2016.03.066 [DOI] [PubMed] [Google Scholar]
- Chu C.C., Ali N., Karagiannis P., Di Meglio P., Skowera A., Napolitano L., Barinaga G., Grys K., Sharif-Paghaleh E., Karagiannis S.N., et al. . 2012. Resident CD141 (BDCA3)+ dendritic cells in human skin produce IL-10 and induce regulatory T cells that suppress skin inflammation. J. Exp. Med. 209:935–945. 10.1084/jem.20112583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse B., Caton M.L., Lehner M., Maeda T., Scheu S., Locksley R., Holmberg D., Zweier C., den Hollander N.S., Kant S.G., et al. . 2008. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 135:37–48. 10.1016/j.cell.2008.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong A., Peña-Cruz V., Cheng T.Y., Clark R.A., Van Rhijn I., and Moody D.B.. 2010. CD1a-autoreactive T cells are a normal component of the human αβ T cell repertoire. Nat. Immunol. 11:1102–1109. 10.1038/ni.1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong A., Cheng T.Y., Huang S., Gras S., Birkinshaw R.W., Kasmar A.G., Van Rhijn I., Peña-Cruz V., Ruan D.T., Altman J.D., et al. . 2014. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nat. Immunol. 15:177–185. 10.1038/ni.2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint-Vis B., Vincent J., Vandenabeele S., Vanbervliet B., Pin J.J., Aït-Yahia S., Patel S., Mattei M.G., Banchereau J., Zurawski S., et al. . 1998. A novel lysosome-associated membrane glycoprotein, DC-LAMP, induced upon DC maturation, is transiently expressed in MHC class II compartment. Immunity. 9:325–336. 10.1016/S1074-7613(00)80615-9 [DOI] [PubMed] [Google Scholar]
- Donaghy H., Bosnjak L., Harman A.N., Marsden V., Tyring S.K., Meng T.C., and Cunningham A.L.. 2009. Role for plasmacytoid dendritic cells in the immune control of recurrent human herpes simplex virus infection. J. Virol. 83:1952–1961. 10.1128/JVI.01578-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschner R.A., Pestonjamasp V.K., Tamakuwala S., Ohtake T., Rudisill J., Nizet V., Agerberth B., Gudmundsson G.H., and Gallo R.L.. 2001. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J. Invest. Dermatol. 117:91–97. 10.1046/j.1523-1747.2001.01340.x [DOI] [PubMed] [Google Scholar]
- Elhussein Mohamed O.Y., Elazomi A., Mohamed M.S., and Abdalla F.B.. 2016. Local elevation of CCL22: a new trend in immunotherapy (skin model). Journal of Cellular Immunotherapy. 2:79–84. 10.1016/j.jocit.2015.12.001 [DOI] [Google Scholar]
- Eming S.A., Martin P., and Tomic-Canic M.. 2014. Wound repair and regeneration: mechanisms, signaling, and translation. Sci. Transl. Med. 6:265sr6 10.1126/scitranslmed.3009337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esashi E., Wang Y.H., Perng O., Qin X.F., Liu Y.J., and Watowich S.S.. 2008. The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity. 28:509–520. 10.1016/j.immuni.2008.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedberg I.M., Tomic-Canic M., Komine M., and Blumenberg M.. 2001. Keratins and the keratinocyte activation cycle. J. Invest. Dermatol. 116:633–640. 10.1046/j.1523-1747.2001.01327.x [DOI] [PubMed] [Google Scholar]
- Furie R., Werth V.P., Merola J.F., Stevenson L., Reynolds T.L., Naik H., Wang W., Christmann R., Gardet A., Pellerin A., et al. . 2019. Monoclonal antibody targeting BDCA2 ameliorates skin lesions in systemic lupus erythematosus. J. Clin. Invest. 129:1359–1371. 10.1172/JCI124466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly D., Chamilos G., Lande R., Gregorio J., Meller S., Facchinetti V., Homey B., Barrat F.J., Zal T., and Gilliet M.. 2009. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J. Exp. Med. 206:1983–1994. 10.1084/jem.20090480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlini G., Mariotti G., Bianchi B., and Pimpinelli N.. 2006. Massive recruitment of type I interferon producing plasmacytoid dendritic cells in varicella skin lesions. J. Invest. Dermatol. 126:507–509. 10.1038/sj.jid.5700052 [DOI] [PubMed] [Google Scholar]
- Ghosh H.S., Cisse B., Bunin A., Lewis K.L., and Reizis B.. 2010. Continuous expression of the transcription factor e2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity. 33:905–916. 10.1016/j.immuni.2010.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliet M., Cao W., and Liu Y.J.. 2008. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 8:594–606. 10.1038/nri2358 [DOI] [PubMed] [Google Scholar]
- Greaves N.S., Ashcroft K.J., Baguneid M., and Bayat A.. 2013. Current understanding of molecular and cellular mechanisms in fibroplasia and angiogenesis during acute wound healing. J. Dermatol. Sci. 72:206–217. 10.1016/j.jdermsci.2013.07.008 [DOI] [PubMed] [Google Scholar]
- Gregorio J., Meller S., Conrad C., Di Nardo A., Homey B., Lauerma A., Arai N., Gallo R.L., Digiovanni J., and Gilliet M.. 2010. Plasmacytoid dendritic cells sense skin injury and promote wound healing through type I interferons. J. Exp. Med. 207:2921–2930. 10.1084/jem.20101102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., and Dipietro L.A.. 2010. Factors affecting wound healing. J. Dent. Res. 89:219–229. 10.1177/0022034509359125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniffa M., Shin A., Bigley V., McGovern N., Teo P., See P., Wasan P.S., Wang X.N., Malinarich F., Malleret B., et al. . 2012. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 37:60–73. 10.1016/j.immuni.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniffa M., Gunawan M., and Jardine L.. 2015. Human skin dendritic cells in health and disease. J. Dermatol. Sci. 77:85–92. 10.1016/j.jdermsci.2014.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman C.S., Chen Y.L., Salimi M., Jarrett R., Johnson D., Järvinen V.J., Owens R.J., Repapi E., Cousins D.J., Barlow J.L., et al. . 2017. CD1a presentation of endogenous antigens by group 2 innate lymphoid cells. Sci. Immunol. 2:eaan5918 10.1126/sciimmunol.aan5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito G.C., Dekker J.D., Wang Y.H., Lee B.K., Shaffer A.L. III, Lin J., Wall J.K., Lee B.S., Staudt L.M., Liu Y.J., et al. . 2014. Dendritic cell fate is determined by BCL11A. Proc. Natl. Acad. Sci. USA. 111:E998–E1006. 10.1073/pnas.1319228111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett R., Salio M., Lloyd-Lavery A., Subramaniam S., Bourgeois E., Archer C., Cheung K.L., Hardman C., Chandler D., Salimi M., et al. . 2016. Filaggrin inhibits generation of CD1a neolipid antigens by house dust mite-derived phospholipase. Sci. Transl. Med. 8:325ra18 10.1126/scitranslmed.aad6833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrich J.J., Balzarolo M., Schmidlin H., Libouban M., Nagasawa M., Gentek R., Kamihira S., Maeda T., Amsen D., Wolkers M.C., and Blom B.. 2012. The transcription factor Spi-B regulates human plasmacytoid dendritic cell survival through direct induction of the antiapoptotic gene BCL2-A1. Blood. 119:5191–5200. 10.1182/blood-2011-07-370239 [DOI] [PubMed] [Google Scholar]
- Katou F., Ohtani H., Saaristo A., Nagura H., and Motegi K.. 2000. Immunological activation of dermal Langerhans cells in contact with lymphocytes in a model of human inflamed skin. Am. J. Pathol. 156:519–527. 10.1016/S0002-9440(10)64756-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou F., Ohtani H., Nakayama T., Ono K., Matsushima K., Saaristo A., Nagura H., Yoshie O., and Motegi K.. 2001. Macrophage-derived chemokine (MDC/CCL22) and CCR4 are involved in the formation of T lymphocyte-dendritic cell clusters in human inflamed skin and secondary lymphoid tissue. Am. J. Pathol. 158:1263–1270. 10.1016/S0002-9440(10)64077-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Hu Y., Yongqing T., Kim J., Hughes V.A., Le Nours J., Marquez E.A., Purcell A.W., Wan Q., Sugita M., et al. . 2016. CD1a on Langerhans cells controls inflammatory skin disease. Nat. Immunol. 17:1159–1166. 10.1038/ni.3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissenpfennig A., Henri S., Dubois B., Laplace-Builhé C., Perrin P., Romani N., Tripp C.H., Douillard P., Leserman L., Kaiserlian D., et al. . 2005. Dynamics and function of Langerhans cells in vivo: dermal dendritic cells colonize lymph node areas distinct from slower migrating Langerhans cells. Immunity. 22:643–654. 10.1016/j.immuni.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Klechevsky E., Morita R., Liu M., Cao Y., Coquery S., Thompson-Snipes L., Briere F., Chaussabel D., Zurawski G., Palucka A.K., et al. . 2008. Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity. 29:497–510. 10.1016/j.immuni.2008.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenfeld D., Gorvel L., Munk A., Man J., Schaffer A., Tung T., Mann C., and Klechevsky E.. 2017. A type of human skin dendritic cell marked by CD5 is associated with the development of inflammatory skin disease. JCI Insight. 2:96101 10.1172/jci.insight.96101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumoto M., Xu B., Shi M., Matsuyama T., Aoyama K., and Takeuchi T.. 2007. Expression of chemokine receptor CCR4 and its ligands (CCL17 and CCL22) in murine contact hypersensitivity. J. Interferon Cytokine Res. 27:901–910. 10.1089/jir.2006.0064 [DOI] [PubMed] [Google Scholar]
- Lande R., Gregorio J., Facchinetti V., Chatterjee B., Wang Y.H., Homey B., Cao W., Wang Y.H., Su B., Nestle F.O., et al. . 2007. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 449:564–569. 10.1038/nature06116 [DOI] [PubMed] [Google Scholar]
- Layseca-Espinosa E., Korniotis S., Montandon R., Gras C., Bouillié M., Gonzalez-Amaro R., Dy M., and Zavala F.. 2013. CCL22-producing CD8α- myeloid dendritic cells mediate regulatory T cell recruitment in response to G-CSF treatment. J. Immunol. 191:2266–2272. 10.4049/jimmunol.1202307 [DOI] [PubMed] [Google Scholar]
- Martin P. 1997. Wound healing--aiming for perfect skin regeneration. Science. 276:75–81. 10.1126/science.276.5309.75 [DOI] [PubMed] [Google Scholar]
- Mathan T.S., Figdor C.G., and Buschow S.I.. 2013. Human plasmacytoid dendritic cells: from molecules to intercellular communication network. Front. Immunol. 4:372 10.3389/fimmu.2013.00372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody D.B., Young D.C., Cheng T.Y., Rosat J.P., Roura-Mir C., O’Connor P.B., Zajonc D.M., Walz A., Miller M.J., Levery S.B., et al. . 2004. T cell activation by lipopeptide antigens. Science. 303:527–531. 10.1126/science.1089353 [DOI] [PubMed] [Google Scholar]
- Naik S., Bouladoux N., Linehan J.L., Han S.J., Harrison O.J., Wilhelm C., Conlan S., Himmelfarb S., Byrd A.L., Deming C., et al. . 2015. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 520:104–108. 10.1038/nature14052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestle F.O., Conrad C., Tun-Kyi A., Homey B., Gombert M., Boyman O., Burg G., Liu Y.J., and Gilliet M.. 2005. Plasmacytoid predendritic cells initiate psoriasis through interferon-α production. J. Exp. Med. 202:135–143. 10.1084/jem.20050500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris P.G., Schofield O., and Camp R.D.. 1988. A study of the role of house dust mite in atopic dermatitis. Br. J. Dermatol. 118:435–440. 10.1111/j.1365-2133.1988.tb02440.x [DOI] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., and Kingsford C.. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 14:417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Cruz V., Ito S., Dascher C.C., Brenner M.B., and Sugita M.. 2003. Epidermal Langerhans cells efficiently mediate CD1a-dependent presentation of microbial lipid antigens to T cells. J. Invest. Dermatol. 121:517–521. 10.1046/j.1523-1747.2003.12429.x [DOI] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., and Sandberg R.. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9:171–181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- Ramos E., Kautz L., Rodriguez R., Hansen M., Gabayan V., Ginzburg Y., Roth M.P., Nemeth E., and Ganz T.. 2011. Evidence for distinct pathways of hepcidin regulation by acute and chronic iron loading in mice. Hepatology. 53:1333–1341. 10.1002/hep.24178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizis B. 2019. Plasmacytoid dendritic cells: development, regulation, and function. Immunity. 50:37–50. 10.1016/j.immuni.2018.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizis B., Bunin A., Ghosh H.S., Lewis K.L., and Sisirak V.. 2011. Plasmacytoid dendritic cells: recent progress and open questions. Annu. Rev. Immunol. 29:163–183. 10.1146/annurev-immunol-031210-101345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins S.H., Walzer T., Dembélé D., Thibault C., Defays A., Bessou G., Xu H., Vivier E., Sellars M., Pierre P., et al. . 2008. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 9:R17 10.1186/gb-2008-9-1-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi M., Barlow J.L., Saunders S.P., Xue L., Gutowska-Owsiak D., Wang X., Huang L.C., Johnson D., Scanlon S.T., McKenzie A.N., et al. . 2013. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J. Exp. Med. 210:2939–2950. 10.1084/jem.20130351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., and Regev A.. 2015. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33:495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai C.M., Sisirak V., Ghosh H.S., Hou E.Z., Ceribelli M., Staudt L.M., and Reizis B.. 2013. Transcription factor Runx2 controls the development and migration of plasmacytoid dendritic cells. J. Exp. Med. 210:2151–2159. 10.1084/jem.20130443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotte R., Nagasawa M., Weijer K., Spits H., and Blom B.. 2004. The ETS transcription factor Spi-B is required for human plasmacytoid dendritic cell development. J. Exp. Med. 200:1503–1509. 10.1084/jem.20041231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- See P., Dutertre C.A., Chen J., Günther P., McGovern N., Irac S.E., Gunawan M., Beyer M., Händler K., Duan K., et al. . 2017. Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 356:eaag3009 10.1126/science.aag3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre J.A. 2006. Epidermal barrier formation and recovery in skin disorders. J. Clin. Invest. 116:1150–1158. 10.1172/JCI28521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A.J., and Clark R.A.. 1999. Cutaneous wound healing. N. Engl. J. Med. 341:738–746. 10.1056/NEJM199909023411006 [DOI] [PubMed] [Google Scholar]
- Sisirak V., Vey N., Vanbervliet B., Duhen T., Puisieux I., Homey B., Bowman E.P., Trinchieri G., Dubois B., Kaiserlian D., et al. . 2011. CCR6/CCR10-mediated plasmacytoid dendritic cell recruitment to inflamed epithelia after instruction in lymphoid tissues. Blood. 118:5130–5140. 10.1182/blood-2010-07-295626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S., Aslam A., Misbah S.A., Salio M., Cerundolo V., Moody D.B., and Ogg G.. 2016. Elevated and cross-responsive CD1a-reactive T cells in bee and wasp venom allergic individuals. Eur. J. Immunol. 46:242–252. 10.1002/eji.201545869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang-Huau T.L., Gueguen P., Goudot C., Durand M., Bohec M., Baulande S., Pasquier B., Amigorena S., and Segura E.. 2018. Human in vivo-generated monocyte-derived dendritic cells and macrophages cross-present antigens through a vacuolar pathway. Nat. Commun. 9:2570 10.1038/s41467-018-04985-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani A.C., Satija R., Reynolds G., Sarkizova S., Shekhar K., Fletcher J., Griesbeck M., Butler A., Zheng S., Lazo S., et al. . 2017. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 356:eaah4573 10.1126/science.aah4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilgus T.A. 2008. Immune cells in the healing skin wound: influential players at each stage of repair. Pharmacol. Res. 58:112–116. 10.1016/j.phrs.2008.07.009 [DOI] [PubMed] [Google Scholar]
- Wolk K., Kunz S., Witte E., Friedrich M., Asadullah K., and Sabat R.. 2004. IL-22 increases the innate immunity of tissues. Immunity. 21:241–254. 10.1016/j.immuni.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Wollenberg A., Kraft S., Hanau D., and Bieber T.. 1996. Immunomorphological and ultrastructural characterization of Langerhans cells and a novel, inflammatory dendritic epidermal cell (IDEC) population in lesional skin of atopic eczema. J. Invest. Dermatol. 106:446–453. 10.1111/1523-1747.ep12343596 [DOI] [PubMed] [Google Scholar]
- Wollenberg A., Wagner M., Günther S., Towarowski A., Tuma E., Moderer M., Rothenfusser S., Wetzel S., Endres S., and Hartmann G.. 2002. Plasmacytoid dendritic cells: a new cutaneous dendritic cell subset with distinct role in inflammatory skin diseases. J. Invest. Dermatol. 119:1096–1102. 10.1046/j.1523-1747.2002.19515.x [DOI] [PubMed] [Google Scholar]
- Wollenberg A., Räwer H.C., and Schauber J.. 2011. Innate immunity in atopic dermatitis. Clin. Rev. Allergy Immunol. 41:272–281. 10.1007/s12016-010-8227-x [DOI] [PubMed] [Google Scholar]
- Zhang H., Gregorio J.D., Iwahori T., Zhang X., Choi O., Tolentino L.L., Prestwood T., Carmi Y., and Engleman E.G.. 2017. A distinct subset of plasmacytoid dendritic cells induces activation and differentiation of B and T lymphocytes. Proc. Natl. Acad. Sci. USA. 114:1988–1993. 10.1073/pnas.1610630114 [DOI] [PMC free article] [PubMed] [Google Scholar]