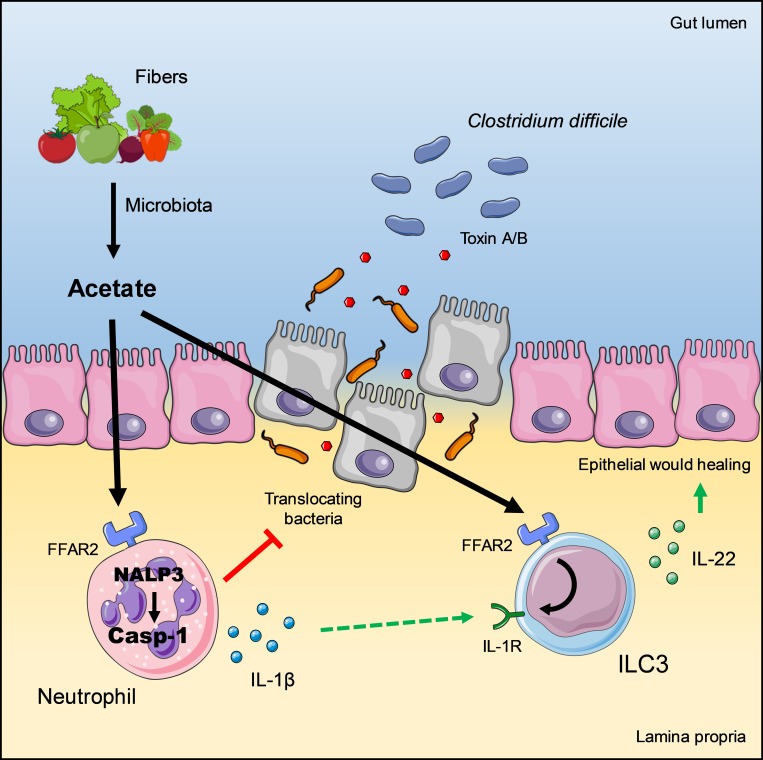
Microbiota-derived acetate coordinates innate immune responses during intestinal Clostridium difficile infection through its cognate receptor FFAR2. Acetate accelerates early neutrophil recruitment and increases ILC3 expression of the IL-1 receptor, boosting ILC3 production of IL-22 in response to neutrophil-derived IL-1β.
Abstract
Antibiotic-induced dysbiosis is a key predisposing factor for Clostridium difficile infections (CDIs), which cause intestinal disease ranging from mild diarrhea to pseudomembranous colitis. Here, we examined the impact of a microbiota-derived metabolite, short-chain fatty acid acetate, on an acute mouse model of CDI. We found that administration of acetate is remarkably beneficial in ameliorating disease. Mechanistically, we show that acetate enhances innate immune responses by acting on both neutrophils and ILC3s through its cognate receptor free fatty acid receptor 2 (FFAR2). In neutrophils, acetate-FFAR2 signaling accelerates their recruitment to the inflammatory sites, facilitates inflammasome activation, and promotes the release of IL-1β; in ILC3s, acetate-FFAR2 augments expression of the IL-1 receptor, which boosts IL-22 secretion in response to IL-1β. We conclude that microbiota-derived acetate promotes host innate responses to C. difficile through coordinate action on neutrophils and ILC3s.
Graphical Abstract
Introduction
The intestinal microbiota plays a crucial role in the maintenance of host homeostasis by shaping and supporting multiple physiological functions, such as nutrient absorption, metabolism, and the development of the immune system (Ding et al., 2019; Richard and Sokol, 2019; Sonnenburg and Sonnenburg, 2019; Sovran et al., 2019; Zmora et al., 2019). The microbiota also outcompetes intestinal colonization by microbial pathogens and thus provides an exogenous defense mechanism against infections (Rodriguez et al., 2015; Stecher and Hardt, 2008). Indeed, quantitative and/or qualitative changes in the composition of the microbiota, known as dysbiosis, are associated with expansion of pathobionts, damage to the intestinal epithelium, bacterial translocation into deeper organs, and ultimately development of numerous pathologies (Ferreira et al., 2014; Johanesen et al., 2015; Theriot et al., 2014).
Clostridium difficile is an anaerobic bacterium that forms spores, which can be found in the intestines of up to 17% of healthy adult individuals. C. difficile is resistant to several antimicrobial agents, including clindamycin, ampicillin, and third-generation cephalosporins, and hence acquires a selective advantage over other microorganisms during antibiotic therapy (Johanesen et al., 2015; Rea et al., 2011; Theriot et al., 2014). Thus, antibiotic-induced dysbiosis can lead to C. difficile infections (CDIs) that cause intestinal disease ranging from mild diarrhea to pseudomembranous colitis (Chen et al., 2008; Rea et al., 2011; Theriot et al., 2014). C. difficile exerts pathogenicity through spore germination and production of toxins, particularly toxins A (enterotoxin) and B (cytotoxin), which cause irreversible damage to the colonic epithelium (Bibbò et al., 2014; Voth and Ballard, 2005). Loss of epithelial integrity results in increased intestinal permeability and translocation of bacteria from the gut lumen into deeper tissues (Hasegawa et al., 2012; Naaber et al., 1998). Emphasizing the importance of healthy microbiota in preventing CDI, several studies have demonstrated that patients with recurrent CDI benefit more from transplantation of fecal microbiota derived from healthy donors than from conventional antimicrobial therapies (Lawley et al., 2012; van Nood et al., 2013).
Innate immune responses are essential for host resistance against CDI. Neutrophils provide an early line of defense: once recruited into the intestinal mucosa, neutrophils carry out several protective functions, such as producing reactive oxygen species (El-Zaatari et al., 2014) and secreting IL-1β (Hasegawa et al., 2012). Depletion of neutrophils in mice results in acute mortality following infection (Jarchum et al., 2012). Two subsets of innate lymphoid cells (ILCs) have also been shown to contribute to early host defense against C. difficile (Abt et al., 2015). ILC1s secrete IFNγ, which activates macrophage defense functions; ILC3s secrete IL-22, which induces the production of antimicrobial peptides in the gut (Sadighi Akha et al., 2015) and activates the complement pathway in the lung and liver to clear translocated bacteria (Hasegawa et al., 2014). The importance of ILCs in CDI is underscored by the observation that Rag1−/− mice, which lack T and B cells, can recover from acute infection as well as WT mice, whereas Rag1−/− × Il2rg−/− mice, which lack ILCs as well as adaptive lymphocytes, succumb to infection (Abt et al., 2015; Hasegawa et al., 2014).
Production of short-chain fatty acids (SCFAs) through the fermentation of dietary fibers is a major mechanism through which the microbiota promotes intestinal immunity (Corrêa-Oliveira et al., 2016). SCFAs include acetate, propionate, and butyrate; these molecules can act through disparate mechanisms, such as activating specific G protein–coupled receptors, inhibiting histone deacetylases, stimulating histone acetyltransferase, and stabilizing hypoxia inducible factor 1 (Donohoe and Bultman, 2012; Fellows et al., 2018; Kelly et al., 2015; Kim et al., 2013; Vinolo et al., 2011). Through these pathways, SCFAs influence multiple immune cell functions, including chemotaxis, energy production, gene expression, cell differentiation, and cell proliferation (Donohoe and Bultman, 2012; Fellows et al., 2018; Kelly et al., 2015; Kim et al., 2013; Tan et al., 2014; Vinolo et al., 2011). While the role of SCFAs has been studied in various proinflammatory and antiinflammatory intestinal immune responses (Donohoe and Bultman, 2012; Fujiwara et al., 2018; Furusawa et al., 2013; Kaiko et al., 2016; Kim et al., 2013; Maslowski et al., 2009), only the impact of butyrate was recently demonstrated during CDI (Fachi et al., 2019). The influence of other SCFAs on immune responses against C. difficile is presently unknown.
Here, we examined the impact of acetate on an acute mouse model of CDI. We found that administration of acetate is remarkably beneficial in ameliorating disease. Mechanistically, we show that acetate enhances innate immune responses by acting on both neutrophils and ILC3s through its cognate receptor free fatty acid receptor 2 (FFAR2). In neutrophils, acetate-FFAR2 signaling facilitates inflammasome activation and promotes the release of IL-1β; in ILC3s, acetate-FFAR2 augments expression of the IL-1 receptor (IL-1R), which boosts IL-22 secretion in response to IL-1β. We conclude that microbiota-derived acetate promotes host innate responses to C. difficile through coordinate action on neutrophils and ILC3s.
Results
A fiber-rich diet and oral administration of acetate protect mice against CDI
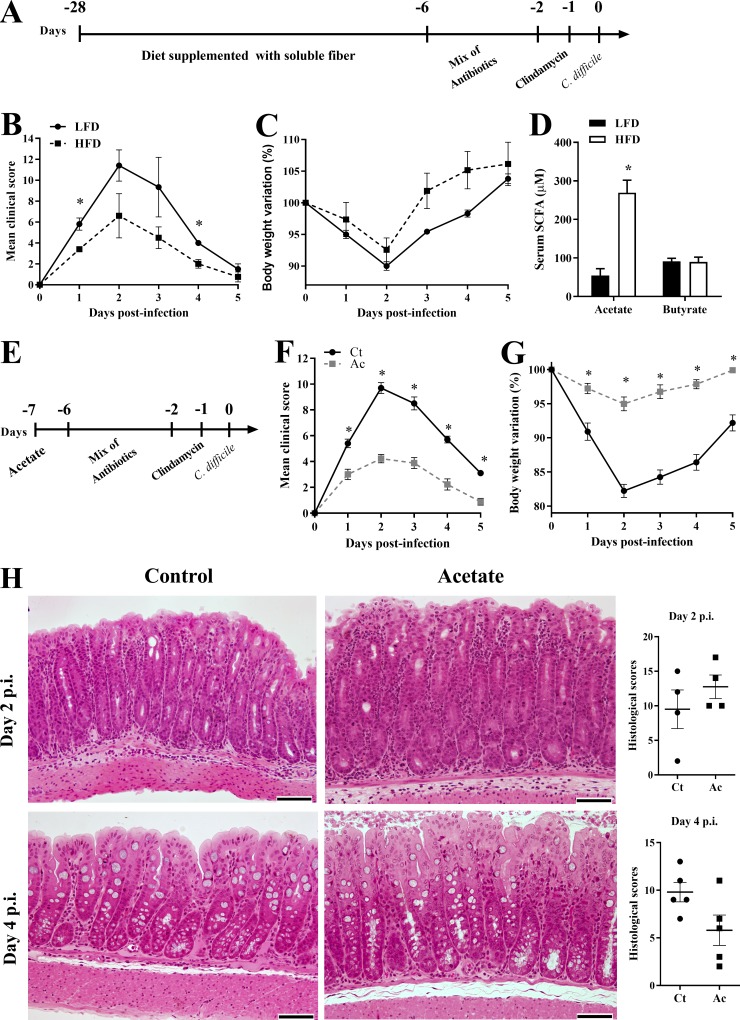
We tested the clinical course of CDI in WT mice fed a soluble fiber-rich diet (10% pectin) in comparison to mice fed control diet (AIN93M; Fig. 1 A). Mice fed the pectin-rich diet had improved clinical scores and lost less weight than mice fed the control diet (Fig. 1, B and C). The protective effect of the fiber-rich diet was associated with an increase in intestinal bacteria content (Fig. S1 A), as well as higher concentrations of acetate in serum (Fig. 1 D). Because acetate is a metabolite produced by intestinal bacteria through soluble fiber fermentation, this suggests that bacterial-derived acetate may be protective during CDI. Corroborating this hypothesis, addition of 150 mM acetate to the drinking water before and throughout the infection was sufficient to protect mice against CDI, resulting in improved clinical scores and less-severe weight loss (Fig. 1, E–G) without impacting intestinal bacterial content (Fig. S1 B). Histologically, we observed increased inflammatory infiltrate in acetate-treated mice 2 d postinfection (p.i.) as well as attenuated histopathology and mucosal hyperplasia 4 d p.i. (Fig. 1 H). Since acetate did not inhibit C. difficile growth (Fig. S1 C) or toxin production in vitro or in vivo (not depicted) and had rapid course of action by improving clinical scores and weight loss even when it was administered at the time of infection (Fig. S1, D–F), we conclude that acetate improves host resistance to C. difficile rather than directly interfering with bacterial replication.
Figure 1.
High-fiber diet (HFD) and acetate administration protect against CDI. (A) WT mice were fed a HFD or low-fiber diet (LFD) before and throughout the experiment. Mice were treated with a mix of antibiotics for 4 d and then received a single i.p. dose of clindamycin. 1 d later, mice were infected with 108 CFU of C. difficile (day 0). (B and C) Mice were monitored for clinical score (B) and weight change (C) until day 5 p.i. (D) Serum SCFA concentrations were measured on day 0 before infection (n = 5). (E) Mice received 150 mM acetate in the drinking water from 1 d before antibiotic treatment until the end of the infection. (F and G) Mice were monitored for clinical score (F) and weight change (G; n = 10). (H) Representative histological sections of colons stained with hematoxylin and eosin and blinded histopathological scoring of mice on day 2 and 4 p.i. that were either treated (Ac) or not (Ct) with acetate (n = 4). Scale bars = 200 µm. Results are representative of two independent experiments with four to five mice in each experimental group (A–C and H) or pooled results from two experiments with four to five mice in each experimental group (D–G). Results are presented as mean ± SEM. *, P < 0.05.
Figure S1.
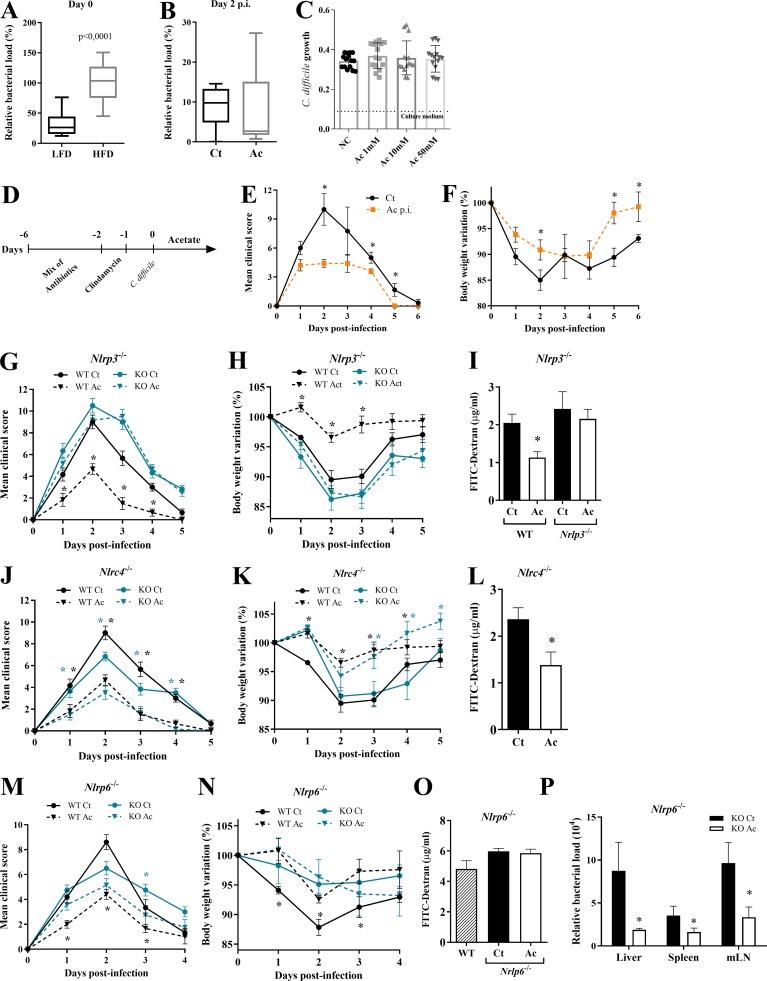
Acetate protects against CDI through NLRP3 activation. (A and B) Analysis by qPCR of relative bacterial load in the feces. Mice were fed with a HFD or LFD before and after antibiotic treatment and analyzed on day 0 before infection (A; n = 10); or treated (Ac) or not (Ct) with acetate in the drinking water throughout the experiment and analyzed on day 2 p.i (B; n = 6). The bar indicates median ± minimum to maximum. (C) In vitro growth of C. difficile measured by optical density at 600 nm in the presence of different concentrations of acetate (n = 15). The dashed line represents the optical density of culture medium without bacteria. (D–F) WT mice were treated with a mix of antibiotics for 4 d and then received a single i.p. dose of clindamycin. 1 d later (day 0), mice started to receive 150 mM acetate in the drinking water and were infected with 108 CFU of C. difficile. Mice were monitored for clinical symptoms (E) and weight changes (F) until day 6 p.i. (n = 5). (G–I) Clinical score (G), body weight variations (H), and intestinal permeability (I) of Nlrp3−/− and WT mice that were treated or not with acetate and infected with C. difficile (n = 5–6). (J–L) Clinical score (J), body weight variations (K), and intestinal permeability (L) of Nlrc4−/− mice treated or not with acetate and infected with C. difficile (n = 6). Nlrp3−/−, Nlrc4−/−, and WT mice were bred in the same animal facility, matched for sex and age, and infected at the same time to avoid batch effects. WT graphs included with Nlrp3−/− and Nlrc4−/− graphs are the same included with Casp1/11−/− graphs (see Fig. 3) and are presented in all figures for comparison. (M and N) Clinical score (M) and body weight variations (N) of Nlrp6−/− mice that were treated or not with acetate and infected with C. difficile (n = 5). WT graphs included with Nlrp6−/− graphs are the same included with Il22−/− data (see Fig. 5) and shown in both figures for comparison. (O) Analysis of intestinal permeability of Nlrp6−/− mice on day 2 p.i. (n = 5). (P) Relative bacterial load translocated to the peripheral organs on day 2 p.i. in Nlrp6−/− mice treated or not with acetate (n = 5). Results are representative of two independent experiments with four mice in each experimental group (B and D–F) or pooled results from two experiments with five mice in each group (A), three experiments with five replicates in each one (C), or one experiment with five to six mice in each group (G–P). Results are presented as mean ± SEM. *, P < 0.05.
Acetate enhances neutrophil accumulation and prevents bacterial translocation
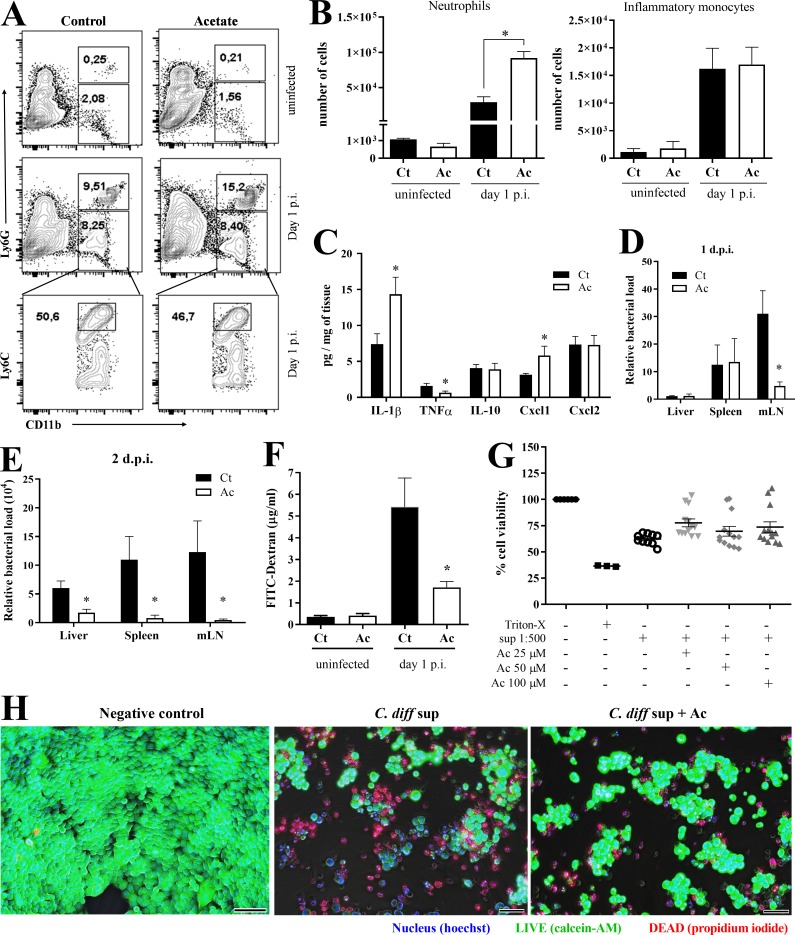
We next examined the impact of acetate on the host response to CDI. Neutrophils have been shown to be crucial early during the host response to C. difficile (Jarchum et al., 2012). In accordance with the histological observations, as early as 1 d p.i., mice treated with acetate had more neutrophils infiltrating the colon, but not inflammatory monocytes, than untreated controls (Figs. 1 H and 2, A and B). This initial effect of acetate treatment was associated with more colonic content of CXCL1, a neutrophil chemoattractant produced by intestinal epithelia and neutrophils themselves (Fig. 2 C). The colons of acetate-treated mice also contained more IL-1β (Fig. 2 C), a cytokine produced by intestinal epithelia and neutrophils that has a prominent role in impeding the translocation of gut bacteria and their systemic dissemination (Hasegawa et al., 2012). The content of proinflammatory TNFα was slightly reduced (Fig. 2 C). Consistent with the increase in neutrophil infiltration and IL-1β production, acetate-treated mice showed a detectable reduction of bacteria translocation into mesenteric lymph nodes (mLNs) on day 1 p.i., which became obvious in several organs on day 2 p.i. (Fig. 2, D and E). Further substantiating a role in protecting the intestinal mucosa from CDI-induced damage, acetate also curtailed systemic permeability of FITC-dextran after oral administration in infected mice (Fig. 2 F). In vitro, acetate did not prevent intestinal epithelial cell death by C. difficile toxins, as measured by propidium iodide and calcein-AM staining (Fig. 2, G and H). Together, these data suggest that acetate bolsters host resistance to C. difficile–mediated intestinal damage by enhancing early intestinal immune responses, such as neutrophil accumulation and production of IL-1β.
Figure 2.
Acetate increases colonic neutrophil recruitment and IL-1β content during CDI. (A and B) Representative plots (A) and absolute number of neutrophils and inflammatory monocytes in the colonic lamina propria (B) of naive and C. difficile–infected mice on day 1 p.i. (n = 4). (C) Quantification of cytokines in colon samples on day 1 p.i. Mice were treated (Ac) or not (Ct) with acetate in the drinking water. Results are normalized according to the tissue weight (n = 8). (D and E) Bacterial translocation into the mLNs, spleen, and liver assessed by qPCR on day 1 (D) or 2 (E) p.i. (n = 5–7). (F) Analysis of intestinal permeability using FITC-dextran. Mice received FITC-dextran by gavage on day 1 p.i. Serum samples were collected 4 h later (n = 4). (G and H) Effect of acetate on the intestinal epithelial cell line HCT-116 exposed to supernatant containing C. difficile toxins. (G) Percentage of live cells after 48 h of incubation with C. difficile supernatant ± acetate at 25, 50, and 100 µM (n = 10–13). Cells lysed with Triton X-100 were included as control. (H) Images of HCT-116 monolayer stained with Hoechst (blue, nucleus), calcein-AM (green, viable cells), and propidium iodide (red, dead cells) after 48 h of incubation with C. difficile supernatant ± acetate (C. diff sup ± Ac) at 25 µM. Scale bars = 100 µm. Results are representative of at least two independent experiments with three to four mice in each experimental group (A and E), pooled results from two independent experiments with two to four mice in each experimental group (B–D), and three experiments with three to five replicates in each group (F and G). Results are presented as mean ± SEM. *, P < 0.05.
The protective effect of acetate depends on inflammasome activation
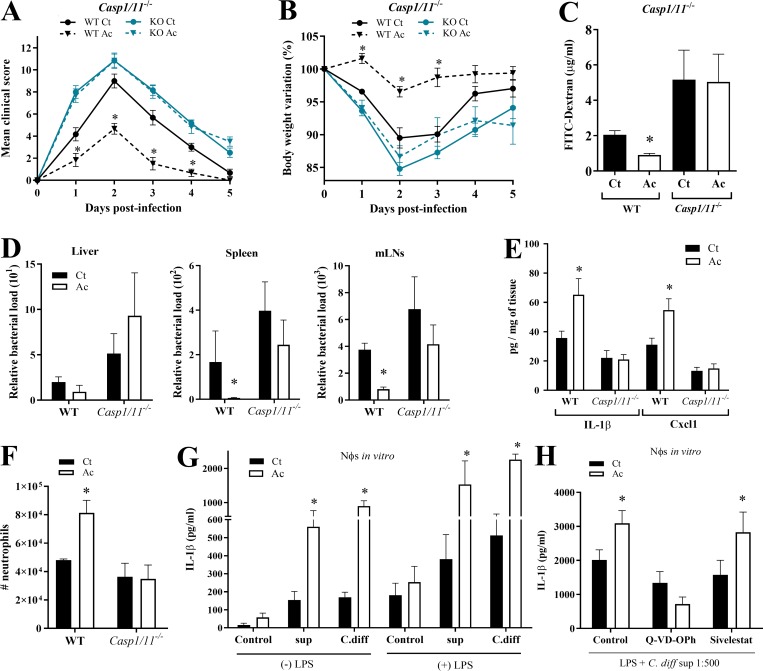
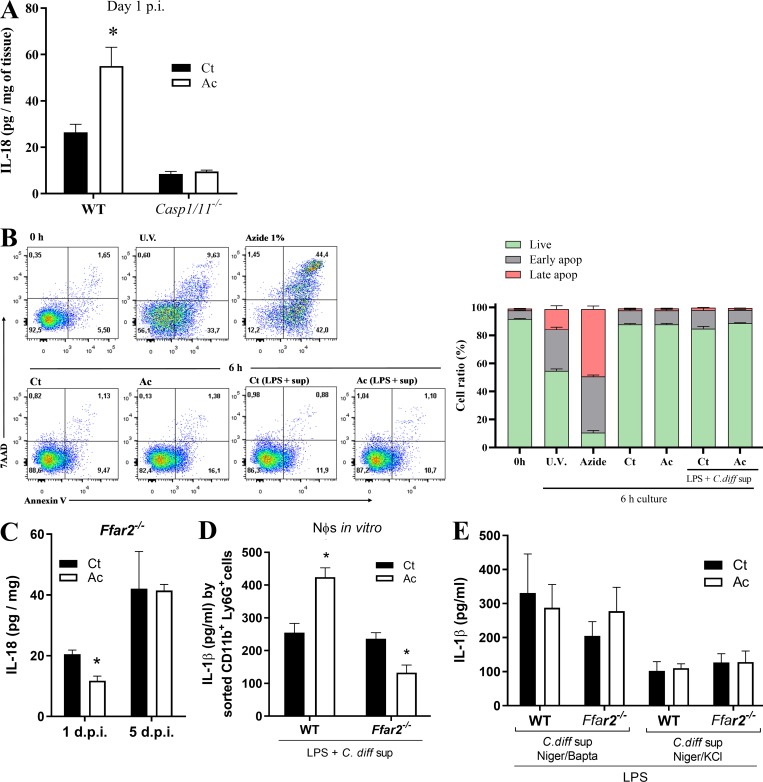
It has been shown that C. difficile toxins trigger IL-1β production through inflammasome activation. C. difficile toxins glucosylate RhoA-GTPase, activating the Pyrin inflammasome (Xu et al., 2014). In addition, they trigger the NLRP3 inflammasome through an as-yet-undefined mechanism, perhaps by inducing reactive oxygen species (Ng et al., 2010). To determine whether acetate enhances inflammasome activation, we examined the impact of acetate on CDI in mice lacking either the effector caspase-1 or one of various inflammasomes implicated in caspase-1 activation. The beneficial effect of acetate on clinical score, weight change, and intestinal permeability during CDI was abrogated in Casp1/11−/− mice (Fig. 3, A–C), as well as Nlrp3−/− mice, which lack one of the inflammasomes that sense C. difficile toxins (Fig. S1, G–I). In contrast, acetate maintained its protective effect in Nlrc4−/− mice and in large part, in Nlrp6−/− mice (Fig. S1, J–P). Acetate also failed to prevent translocation of bacteria into the liver, spleen, or mLNs of Casp1/11−/− mice (Fig. 3 D). Additionally, acetate did not enhance the content of IL-1β, CXCL1 (Fig. 3 E), or neutrophil infiltration (Fig. 3 F) in the colons of Casp1/11−/− mice. Since caspase-1 activation also triggers IL-18 production, we examined the impact of acetate on IL-18 during CDI. Increased amounts of IL-18 were detected in the colons of WT mice on day 1 p.i. (Fig. S2 A). However, acetate did not enhance colonic IL-18 content in Casp1/11−/− mice, providing further evidence that acetate acts by promoting inflammasome-regulated cytokine production during CDI.
Figure 3.
Acetate augments caspase-1 activation and IL-1β secretion in response to C. difficile toxins. (A–C) Clinical score (A), body weight changes (B), and intestinal permeability (C) in Casp1/11−/− and WT mice that were treated (Ac) or not (Ct) with acetate and infected with C. difficile (n = 5–6). Casp1/11−/− and WT were bred in the same animal facility, matched for sex and age, and infected at the same time to avoid batch effects. (D) Bacterial translocation into the peripheral organs in Casp1/11−/− and WT mice on day 1 p.i. (n = 6). (E) Quantification of IL-1β and CXCL1 in colon samples of Casp1/11−/− and WT mice on day 1 p.i. Results were normalized according to sample weight (n = 6). (F) Absolute numbers of CD11b+Ly6G+ neutrophils in the colonic lamina propria of Casp1/11−/− mice and WT mice on day 1 p.i. (n = 6). (G) Quantification of IL-1β production by neutrophils in vitro. Neutrophils were incubated with or without LPS ± acetate, followed by stimulation in vitro with supernatant containing whole C. difficile (C.diff), toxin-containing supernatant (sup), or medium as indicated (n = 8). (H) IL-1β production by neutrophils after incubation in vitro with LPS, C. difficile supernatant, and either caspase-1 inhibitor (Q-VD-OPh), elastase inhibitor (Sivelestat; n = 6), or DMSO (control). Results are representative of two independent experiments with three to six mice in each experimental group (A, B, E, and F), one experiment with six mice in each group (C and D), or pooled results from two independent experiments with three to four mice in each group (G and H). Results are presented as mean ± SEM. *, P < 0.05.
Figure S2.
Acetate augments IL-1β and IL-18 secretion in response to C. difficile toxins. (A) IL-18 content in the colon of WT and Casp1/11−/− mice that were treated (Ac) or not (Ct) with acetate (n = 5–6). Mice were examined on day 1 p.i. The results were normalized according to sample weight. (B) Neutrophil viability measured by 7-AAD and Annexin-V staining before and after 6-h in vitro incubation. Neutrophils were isolated from bone marrow and stimulated as follows: 0 h, staining after BM isolation; U.V., 1 min ultraviolet irradiation and then 6-h incubation; Azide 1%, 6-h incubation in 1% sodium azide medium; Ct, 6-h culture in RPMI and 10% bovine calf serum; Ac, 6-h incubation in medium with 25 mM acetate; Ct (LPS + sup), 2 h of incubation with LPS, followed by 4 h with C. difficile supernatant; Ac (LPS + sup), 2 h of incubation with LPS + acetate, followed by C. difficile supernatant (n = 6). (C) IL-18 content in the colon of Ffar2−/− mice that were treated or not with acetate and infected with C. difficile (n = 5–6). Mice were examined on day 1 and 5 p.i. (D) IL-1β production by neutrophils after incubation in vitro with LPS ± acetate followed by stimulation with C. difficile supernatant. Neutrophils were isolated from the bone marrow of WT and Ffar2−/− mice by FACS sorting CD11b+Ly6G+ cells (n = 12). (E) IL-1β production by neutrophils in vitro from bone marrow of WT or Ffar2−/− mice. Neutrophils were preincubated with LPS ± acetate and then stimulated with C. difficile supernatant together with nigericin and BAPTA-AM or together with nigericin and KCl (n = 6). Results are representative of at least two independent experiments with four to five mice in each experimental group (A) or pooled results from two independent experiments with three to five mice in each experimental group (B–E). Results are presented as mean ± SEM. *, P < 0.05.
Acetate augments inflammasome activation and IL-1β secretion in neutrophils
Given the major role of neutrophils in IL-1β secretion and in controlling bacterial translocation during CDI (Hasegawa et al., 2012; Fig. 2), we wanted to examine the effects of acetate on inflammasome activation by C. difficile in neutrophils. Stimulation of neutrophils purified from the bone marrow of WT mice with acetate markedly potentiated the production of IL-1β in response to C. difficile or its supernatant containing a defined amount of toxins (Fig. 3 G), without affecting neutrophil viability (Fig. S2 B). The enhancing effect of acetate on IL-1β secretion was evident whether or not neutrophils were primed with LPS (Fig. 3 G). Moreover, acetate effect was blocked by an inhibitor of caspase-1, whereas it was unaffected by an inhibitor of neutrophil elastase (Fig. 3 H). We conclude that acetate augments inflammasome-regulated IL-1β secretion by neutrophils in response to C. difficile toxins.
The protective effect of acetate during CDI is FFAR2 dependent
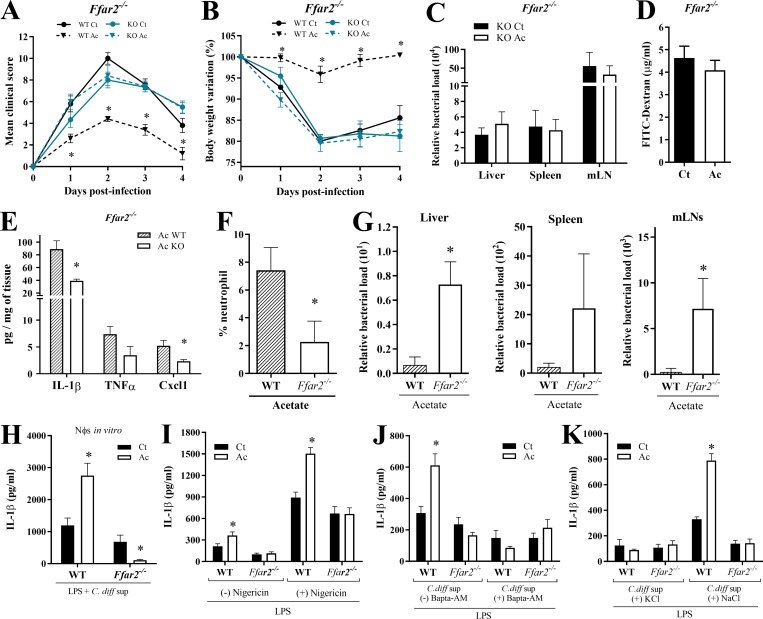
Among various mechanisms, acetate can modulate cell functions through activation of FFAR2 (also known as GPR43) in epithelial cells and neutrophils (Corrêa-Oliveira et al., 2016). Given that acetate generates faster outcomes through FFAR2 signaling than through other mechanisms requiring chromatin remodeling, and that acetate acts rapidly during CDI (Fig. S1, D–F), we sought to examine the impact of FFAR2 in acetate-mediated protection against CDI. We found that the beneficial effects of acetate on clinical score, weight change, systemic translocation of bacteria, and intestinal permeability during CDI were abrogated in Ffar2−/− mice (Fig. 4, A–D). Additionally, the increased colonic content of CXCL1, IL-1β, and IL-18; the enhanced infiltration of neutrophils in the colon; and the reduced systemic bacterial translocation mediated by acetate in WT mice were not evident in Ffar2−/− mice (Fig. 4, E–G; and Fig. S2 C). Together, these findings suggest that the effects of acetate on host resistance to CDI depend on FFAR2.
Figure 4.
Acetate acts through FFAR2. (A and B) Clinical score (A) and weight changes (B) of Ffar2−/− and WT mice infected with C. difficile and treated (Ac) or not (Ct) with acetate (n = 3–6). Ffar2−/− and WT littermates were infected at the same time. (C) Bacterial translocation into peripheral organs 2 d p.i. in Ffar2−/− treated or not with acetate (n = 3). (D) Intestinal permeability in Ffar2−/− mice treated or not with acetate (n = 4). (E–G) Cytokine content (E), neutrophil percentage (F), and bacterial load translocated into the peripheral organs (G) on day 1 p.i. in WT or Ffar2−/− mice treated with acetate (n = 5). (H) IL-1β production by WT and Ffar2−/− neutrophils after priming in vitro with LPS ± acetate followed by stimulation with C. difficile supernatant (C. diff sup; n = 8). (I) IL-1β production by WT and Ffar2−/− neutrophils after incubation in vitro with LPS ± acetate followed by stimulation with Nigericin or no stimulation (n = 8). (J and K) IL-1β production by neutrophils from WT and Ffar2−/− mice. Cells were preincubated with LPS ± acetate for 2 h as indicated, and then stimulated with C. difficile supernatant ± BAPTA-AM (J); C. difficile supernatant ± KCl (K); or C. difficile supernatant ± NaCl (L; n = 6). Results are representative of at least two independent experiments with three to five mice in each experimental group (A–G) or pooled results from two independent experiments with three to four mice in each group (H–K). Results are presented as mean ± SEM. *, P < 0.05.
FFAR2 enhances neutrophil secretion of IL-1β in an inflammasome-dependent fashion
We sought to determine whether acetate enhances neutrophil production of IL-1β through FFAR2. We prepared neutrophils by density gradient centrifugation from the bone marrow of WT and Ffar2−/− mice, primed them in vitro with LPS in the presence or absence of acetate, stimulated them with C. difficile toxins, and then measured IL-1β production. The ability of acetate to augment IL-1β production by neutrophils was entirely dependent on FFAR2 (Fig. 4 H). Similar results were obtained using neutrophils highly purified from bone marrow by cell sorting (Fig. S2 D). We also tested whether acetate could enhance IL-1β production by neutrophils that are stimulated with a known inflammasome activator, the potassium (K+) ionophore nigericin. Indeed, acetate enhanced the K+ efflux–induced IL-1β production, and this effect was dependent on FFAR2 (Fig. 4 I). Thus, acetate enhances inflammasome activation and IL-1β secretion in neutrophils through FFAR2.
We further corroborated that acetate-FFAR2 signaling enhances IL-1β production in neutrophils through inflammasome activation. Treatment of neutrophils with BAPTA-AM (1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid acetyloxymethyl ester), which inhibits NLRP3 by blocking Ca2+ influx, abrogated the ability of acetate to potentiate IL-1β production through FFAR2 signaling in response to LPS and C. difficile toxins or nigericin (Figs. 4 J and S2 E). A similar result was obtained by treating neutrophils with extracellular KCl, which inhibits NLRP3 by blocking K+ efflux (Figs. 4 K and S2 E). In contrast, incubation of neutrophils with NaCl, which boosts inflammasome activation by promoting K+ efflux, facilitated acetate enhancement of IL-1β production through FFAR2 (Fig. 4 K). Taken together, these results suggest that the ability of acetate–FFAR2 axis to potentiate the production of IL-1β in response to C. difficile toxins depends on inflammasome activation.
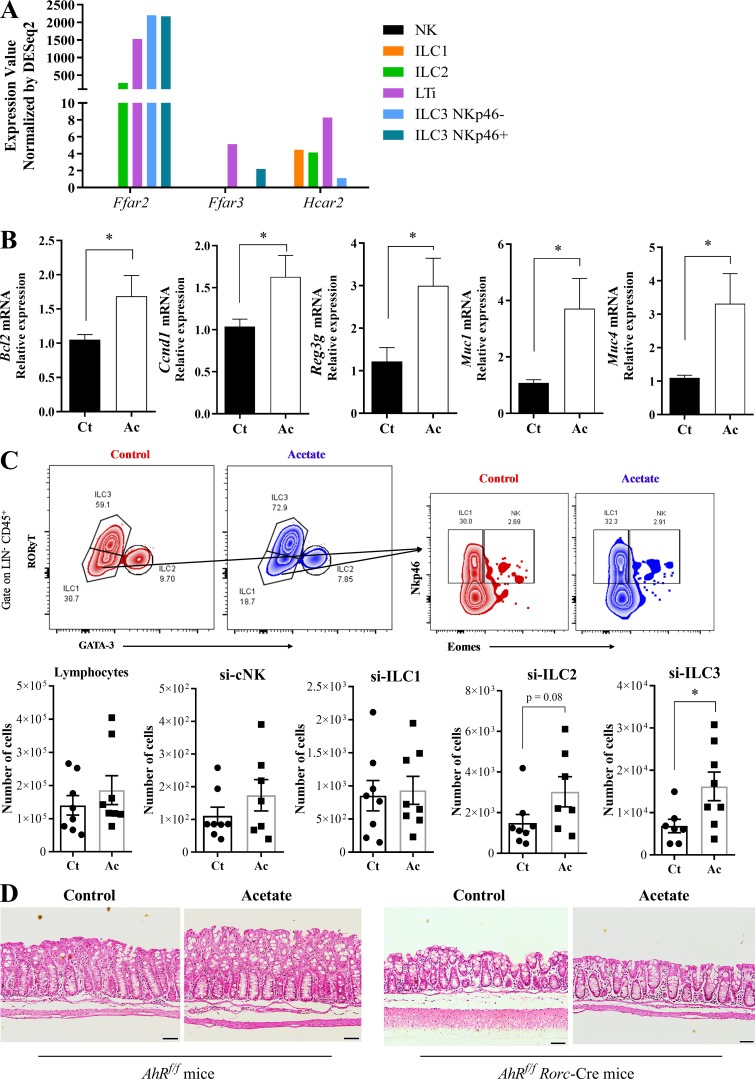
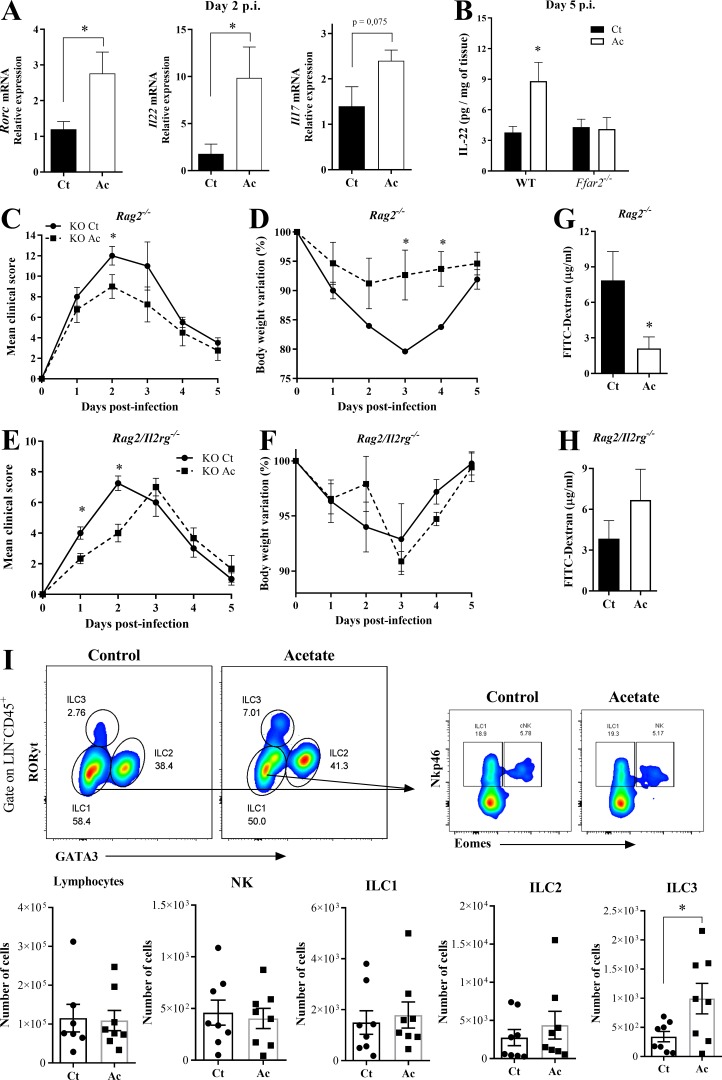
Acetate-mediated protection during CDI depends on IL-22 producing ILC3
It has been shown that ILC3s protect from CDI through IL-22 production (Abt et al., 2015; Hasegawa et al., 2014). Moreover, ILC3s express high amounts of FFAR2 (Fig. S3 A and http://www.immgen.org). Therefore, we asked whether acetate also impacts this protective mechanism. We found that acetate treatment increased the expression of Rorc and Il22 in the colon of WT mice 2 d p.i. (Fig. 5 A), as well as IL-22 target genes (Bcl2, Ccnd1, Reg3g, Muc1, and Muc4) 5 d p.i. (Fig. S3 B). Increased amounts of IL-22 protein were detected in the colons of WT mice but not Ffar2−/− mice on day 5 p.i. (Fig. 5 B), suggesting that acetate may promote IL-22 production through FFAR2. Although IL-22 was previously shown to be primarily produced by ILC3s during CDI (Abt et al., 2015), whether acetate enhances IL-22 secretion by ILC3s and/or T cells was not clear. To address this question, we examined the impact of acetate in Rag2−/− and Rag2/Il2rg−/− mice with CDI. The protective effect of acetate was evident in Rag2−/− mice, which lack T and B lymphocytes but retain ILCs, whereas it was undetectable in Rag2/Il2rg−/− mice, which lack both innate and adaptive lymphocytes (Fig. 5, C–F). Reduction of intestinal permeability by acetate was also maintained in Rag2−/− but not Rag2/Il2rg−/− mice (Fig. 5, G and H). Thus, acetate-mediated protection of infected mice depends on ILCs.
Figure S3.
Acetate increases ILC3s number and activity during CDI. (A) SCFA receptor expression in ILC subsets. RNA sequencing data available at http://www.immgen.org. (B) IL-22 target genes expression in the colon of WT mice on day 5 p.i. Mice were treated (Ac) or not (Ct) with acetate and infected with C. difficile (n = 7–8). (C) Percentages of total lymphocytes, natural killer (NK), ILC1, ILC2, and ILC3 subsets in the small intestine (si) from mice 5 d p.i. (n = 7–8). A representative flow cytometry plot depicting the strategy to identify the subsets is presented on the top. (D) Representative histological sections of colons stained with hematoxylin and eosin of ILC3-deficient mice on day 5 p.i. treated or not with acetate (n = 3). Scale bars = 200 µm. Results are representative of at least two independent experiments with three mice in each experimental group (D) or pooled results from two independent experiments with three to four mice in each experimental group (B and C). Results are presented as mean ± SEM. *, P < 0.05.
Figure 5.
The protective effect of acetate against CDI depends on ILC3s. (A) Rorc, Il22, and Il17 mRNA expression in the colon of WT mice on day 2 p.i. Mice were treated (Ac) or not (Ct) with acetate and infected with C. difficile (n = 5). (B) IL-22 content in the colon of WT and Ffar2−/− mice on day 5 p.i. Mice were treated or not with acetate and infected with C. difficile (n = 5–6). (C–F) Clinical score and weight changes of Rag2−/− mice (C and D) and Rag2/Il2rg−/− mice (E and F) that were treated or not with acetate and infected with C. difficile (n = 4). (G and H) Intestinal permeability of Rag2−/− (G) and Rag2/Il2rg−/− (H) mice treated or not with acetate on day 2 p.i. (n = 4). (I) Percentages of total lymphocytes, natural killer (NK) cells, ILC1s, ILC2s, and ILC3s in the colon of mice 5 d p.i. (n = 7–8). A representative flow cytometry plot depicting the strategy for identifying NK-ILC subsets is presented at the top. Results are pooled results from two independent experiments with three to six mice in each group (A, B, and I). Results are presented as mean ± SEM. *, P < 0.05.
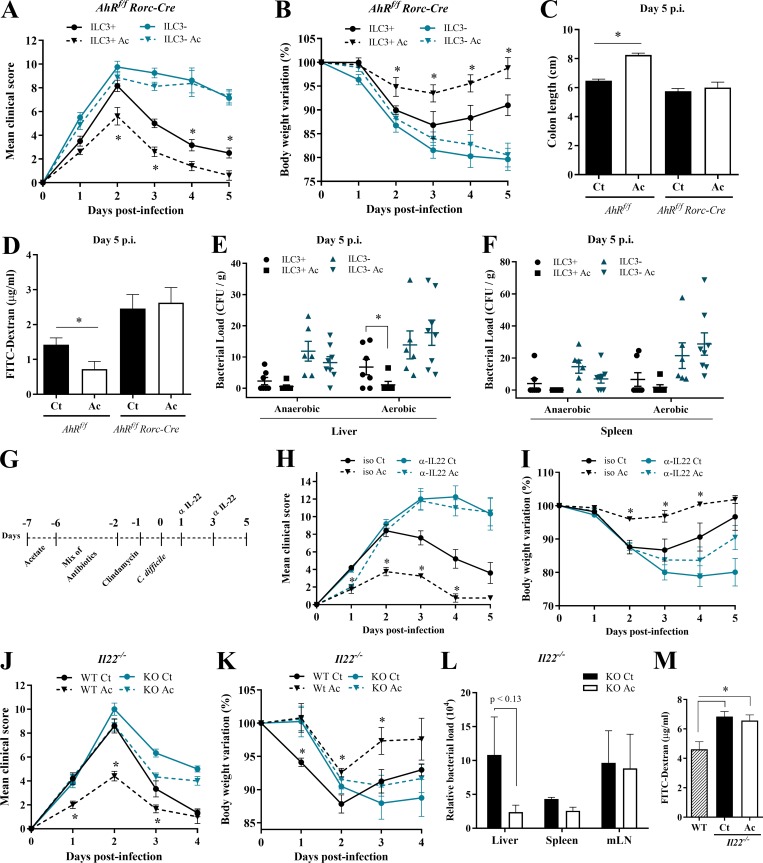
We next examined the impact of acetate on the abundance of ILC subsets on day 5 p.i. Acetate-treated mice showed a moderate increase in the number of ILC3s, but not of other ILCs, in the colon (Fig. 5 I) and small intestine (Fig. S3 C). To validate that acetate-mediated protection is dependent on ILC3s, we infected AhRf/f Rorc-Cre mice, which have a strong reduction of ILC3s (Song et al., 2015). We found that the beneficial effects of acetate on clinical score, weight change, colon length, intestinal permeability, and systemic translocation of bacteria during CDI were strongly reduced in the absence of ILC3s (Fig. 6, A–F). These mice also did not show clinical recovery and mucosal hyperplasia on day 5, in contrast to what normally occurs in WT mice (Fig. 6, A and B; and Fig. S3 D), indicating that ILC3s are important not only to mitigate the disease but also for a complete resolution.
Figure 6.
IL-22–producing ILC3s are essential for acetate-mediated protection during CDI. (A–D) Clinical score (A), body weight changes (B), colon length (C), and intestinal permeability (D) in ILC3-sufficient (AhRfl/fl) and -deficient (AhRfl/fl × Rorc-Cre) mice that were treated (Ac) or not (Ct) with acetate and infected with C. difficile (n = 5–8). AhRfl/fl × Rorc-Cre and AhRfl/fl littermates were infected at the same time. (E and F) Bacterial translocation into the liver (E) and spleen (F) in ILC3-sufficient and -deficient mice treated or not with acetate and infected with C. difficile. Samples were collected 5 d p.i., plated in blood agar, and incubated for 4 d at 37°C in aerobic or anaerobic conditions (n = 5–8). (G) Infected mice were treated with or without acetate and received an i.p. dose of anti–IL-22 neutralizing antibody or isotype (iso) control IgG2a on days 1 and 3 p.i. (H and I) Mice were monitored for clinical score (H) and weight change (I) until day 5 p.i. (n = 5). (J and K) Clinical score (J) and weight changes (K) of IL22−/− and WT mice infected with 108 CFU of C. difficile that were either treated or not with acetate (n = 5–6). IL22−/− and WT mice were bred in the same animal facility, matched for sex and age, and infected at the same time to avoid batch effects. (L) Bacterial translocation into peripheral organs on day 2 p.i. in IL22−/− mice treated or not with acetate (n = 5–6). (M) Intestinal permeability of IL22−/− mice treated or not with acetate and infected with C. difficile (n = 5–6). Infected WT mice not treated with acetate are shown as controls. Results are representative of at least two independent experiments with four to six mice in each experimental group (G–I) or pooled results from two independent experiments with three to four mice in each group (A–F). Results are presented as mean ± SEM. *, P < 0.05.
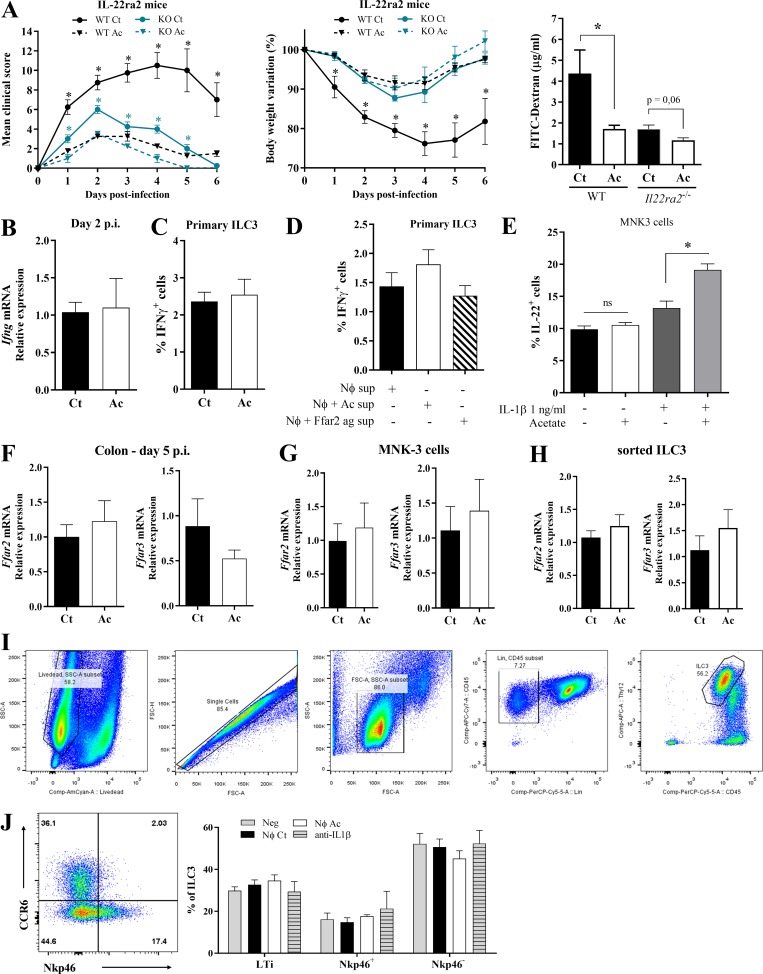
Since ILC3s produce IL-22, which promotes epithelial cell defense mechanisms and mucus production, we asked whether acetate-mediated protection against CDI depends on IL-22 production. To test this, acetate-treated and untreated mice received an i.p. dose of anti–IL-22 neutralizing antibody or isotype control on day 1 and 3 after CDI (Fig. 6 G). Neutralization of IL-22 entirely reduced the protective effect of acetate, raising the clinical score and body weight loss of acetate-treated mice to levels comparable to those seen in untreated mice that also received anti-IL-22 (Fig. 6, H and I). Similarly, beneficial effects of acetate on clinical score, body weight, bacterial translocation, and epithelial permeability during CDI were strongly reduced in Il22−/− mice (Fig. 6, J–M). Conversely, newly generated mice with genetic disruption of IL-22 binding protein (Il22ra2), which acts as an IL-22 decoy (Huber et al., 2012), were as resistant to CDI as acetate-treated WT mice. Acetate treatment did not further improve CDI resistance of Il22ra2−/− mice (Fig. S4 A). Together, these data highlight the importance of IL-22 production by ILC3s in the protective effect of acetate.
Figure S4.
Acetate-mediated protection during CDI involves IL-22 secretion by ILC3s. (A) Clinical score, body weight variations, and intestinal permeability of Il22ra2−/− and WT mice that were treated (Ac) or not (Ct) with acetate and infected with C. difficile (n = 5–6). (B) Ifng mRNA expression in the colon of WT mice on day 2 p.i. that were treated or not with acetate (n = 5). (C) IFNγ production by primary ILC3s ex vivo (n = 5). Cells were directly treated with C. difficile supernatant ± acetate. (D) IFNγ secretion by primary ILC3s incubated with neutrophil supernatants. Neutrophils were stimulated with LPS+ C. difficile supernatant (Nφ sup); LPS+ C. difficile supernatant and acetate (Nφ + Ac sup); and LPS+ C. difficile supernatant and FFAR2 agonist (Nφ + Ffar2 ag sup; n = 5). (E) IL-22–producing MNK3 cells after 3 h of treatment with different combinations of acetate and IL-1β, as indicated (n = 5). ns, not significant. (F–H) Ffar2 and Ffar3 mRNA expression in the colon of WT mice on day 5 p.i. (F; n = 7–8); MNK3 cells after incubation with acetate (G; n = 5–6); and FACS sorted-ILC3s from antibiotic-treated mice (H; day 0, before CDI). Mice were either treated or not with acetate in the drinking water (n = 4). (I) Gating strategy used to identify primary ILC3s from the small intestine of WT mice (Bando et al., 2018; Robinette et al., 2017). SSC, side scatter. (J) Percentage of CCR6+NKp46− (LTi), CCR6−NKp46−, CCR6−NKp46+ subsets within primary ILC3s treated ex vivo with neutrophil supernatants. A representative flow cytometry plot depicting the strategy to identify the subsets is presented on the left. Results are representative of at least two independent experiments with two to five mice in each experimental group (A–F) or are pooled from two independent experiments with three to five mice in each experimental group (F–I). Results are presented as mean ± SEM. *, P < 0.05.
Acetate did not increase IFNγ expression in the colon or IFNγ production by primary ILC3 ex vivo (Fig. S4, B–D), suggesting no obvious impact on ILC1s. We conclude that ILC3s production of IL-22 has a major role in acetate-induced protection against CDI.
Acetate-FFAR2 signaling augments IL-22 production by increasing ILC3 responsiveness to neutrophil-derived IL-1β
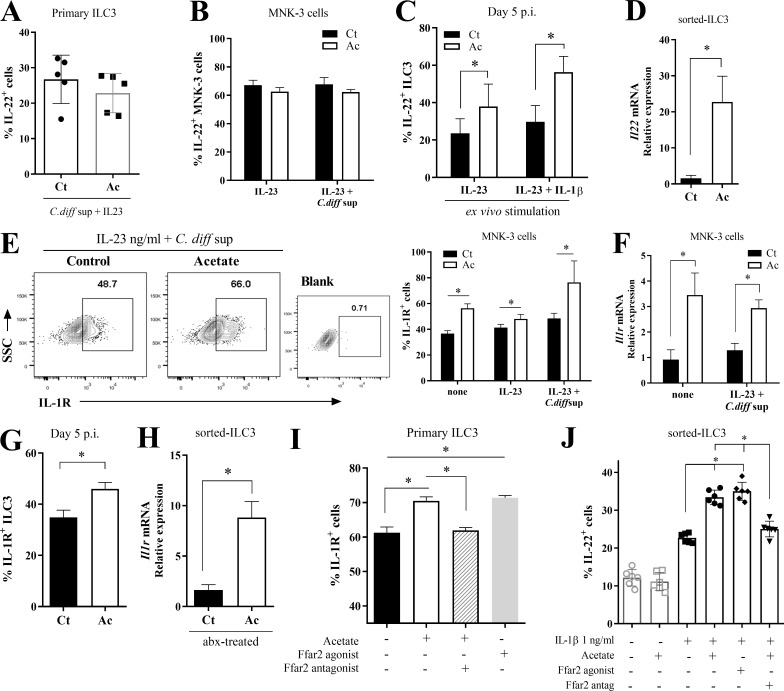
Since ILC3s express FFAR2, and acetate protection was dependent on these cells and FFAR2, we decided to test the direct effect of acetate on the production of IL-22. We incubated primary ILC3s in vitro with acetate, C. difficile supernatant, and IL-23 and measured the percentage of IL-22–producing cells by intracellular staining. Acetate had no impact on IL-22 production in these cultures (Fig. 7 A). Similar results were obtained using the ILC3 line MNK3 (Allan et al., 2015; Fig. 7 B). However, the percentage of IL-22–producing ILC3s was increased in acetate-treated mice 5 d p.i. (Fig. 7 C). Similarly, acetate treatment increased the expression of IL-22 mRNA in ILC3s FACS sorted from antibiotic-treated mice (Fig. 7 D). It is known that ILC3s secrete IL-22 in response to IL-1β and IL-23 (Takatori et al., 2009; Vivier et al., 2018). We sought to determine whether acetate stimulated ILC3s indirectly through enhanced expression of IL-23 receptor or IL-1R. We found that acetate increased the expression of IL-1R in (a) MNK3 cells (Fig. 7, E and F); (b) primary ILC3s isolated from C. difficile–infected mice 5 d p.i. (Fig. 7 G); (c) primary ILC3s isolated from antibiotic-treated mice (Fig. 7 H); and (d) primary ILC3s treated with acetate in vitro (Fig. 7 I). These results suggested that acetate may increase ILC3 responsiveness to IL-1β. To confirm that IL-1R expression is induced by acetate via FFAR2, we assessed the impact of synthetic FFAR2 agonist and antagonist on the stimulatory activity in primary ILC3s. FFAR2 agonist increased the percentages of IL-1R–expressing primary ILC3s, as did acetate, while FFAR2 antagonist inhibited such effect (Fig. 7 I). To corroborate that acetate increases the responsiveness of ILC3s to IL-1β, we examined IL-22 production of FACS sorted-ILC3s (Fig. 7 J) and MNK3 cells (Fig. S4 E) stimulated in vitro with different combinations of acetate and IL-1β. Acetate-FFAR2 signaling clearly enhanced the percentage of IL-22–producing cells induced by IL-1β (Fig. 7 J). Further, we also found that acetate does not change the expression of Ffar2 and Ffar3 in the colon of mice 5 d p.i. (Fig. S4 F), as well as both MNK3 (Fig. S4 G) and ILC3s FACS-sorted from antibiotic-treated mice (Fig. S4 H).
Figure 7.
Acetate enhances ILC3 production of IL-22 by increasing their responsiveness to neutrophil IL-1β. (A) Percentages of IL-22–producing primary ILC3s incubated ex vivo with C. difficile supernatant, IL-23, and acetate as indicated (n = 5). (B) Percentages of IL-22–producing MNK3 cells incubated with C. difficile supernatant and/or IL-23, ± acetate, as indicated (n = 5). (C) Percentages of IL-22–producing ILC3s from C. difficile–infected mice on day 5 p.i. (n = 8). Cells were stimulated with Golgi plug and IL-23 (10 ng/ml) ± IL-1β (1 ng/ml) for 3 h. (D) Il22 mRNA expression of ILC3s FACS sorted from antibiotic-treated mice (day 0, before CDI). Mice were either treated (Ac) or not (Ct) with acetate in the drinking water (n = 4). (E and F) Percentages of IL-1R–expressing MNK3 cells (E) and Il1r mRNA expression (F) after incubation with C. difficile supernatant and/or IL-23 ± acetate (n = 5–6). Representative plots are presented on the left. Blank, control antibody; SSC, side scatter. (G) Percentages of IL-1R–expressing ILC3s from C. difficile–infected mice on day 5 p.i. (n = 8). (H) Il1r mRNA expression by FACS-sorted ILC3s from antibiotic (abx)-treated mice. Mice were either treated or not with acetate (n = 4). (I) Percentages of IL-1R–expressing primary ILC3s after ex vivo treatment with acetate, Ffar2-agonist, and Ffar2-antagonist, as indicated (n = 5). (J) IL-22–producing cells within sorted ILC3s treated with different combinations of acetate, IL-1β, Ffar2-agonist, or Ffar2-antagonist, as indicated (n = 5). Results are representative of at least two independent experiments with three to four mice/replicates in each group (A and I–K) or pooled results from two independent experiments with two to five mice/replicates in each one (B–H). All mice used were littermates. Results are presented as mean ± SEM. *, P < 0.05.
Acetate promotes neutrophil-ILC3 cross-talk through IL-1β
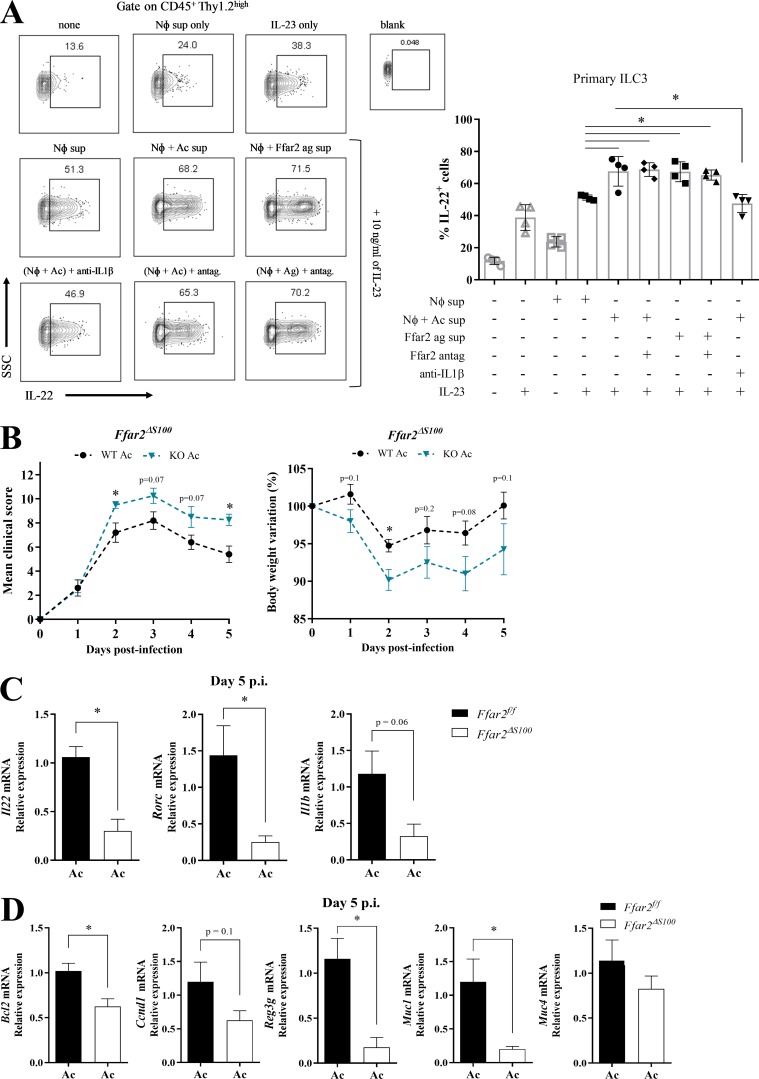
We sought to directly demonstrate that acetate activates primary ILC3s through IL-1β produced by neutrophils. We isolated ILC3s from the intestinal lamina propria of Rag1−/− mice, which lack T and B cells, and incubated them ex vivo with IL-23 together with culture supernatants from neutrophils that had been stimulated with LPS and C. difficile toxins, with or without acetate, or with synthetic FFAR2 agonist (Fig. 8 A). ILC3s stimulated with supernatant of neutrophils treated with acetate or FFAR2 agonist produced more IL-22 than did ILC3s cultured with supernatant of neutrophils treated with LPS and C. difficile toxins only. This effect could be blocked with a neutralizing anti-IL-1β antibody, but not by a FFAR2 antagonist (Fig. 8 A), demonstrating that ILC3 production of IL-22 is induced by neutrophil-derived IL-1β and not FFAR2 activation of ILC3s. In contrast to the stimulating effect on cytokine secretion, the supernatant of neutrophils had no impact on the abundance of total ILC3s or any of the CCR6+NKp46−, CCR6−NKp46−, or CCR6−NKp46+ subsets included in the ILC3 population purified from the intestine using a standard cell sorting protocol (Fig. S4, I and J).
Figure 8.
Acetate promotes neutrophil–ILC3 cross-talk through IL-1β. (A) Percentages of IL-22–producing primary ILC3s that were isolated from Rag1−/− mice and triggered in vitro with IL-23 and supernatant of neutrophils in the indicated combinations. Neutrophils were stimulated as follows: Nφ sup, LPS + C. difficile supernatant; Nφ + Ac sup, LPS + C. difficile supernatant + acetate; Nφ + Ffar2 ag sup: LPS + C. difficile supernatant + Ffar2-agonist. Anti–IL-1β blocking antibody or Ffar2-antagonist (antag.) was added, as indicated. Representative flow cytometry plots are presented on the left (n = 4). Top: Stimulations with IL-23 or neutrophil supernatant only. Middle: Stimulations combining IL-23 and various neutrophil supernatants. Bottom: Addition of anti–IL-1β or Ffar2 antagonist. SSC, side scatter. (B) Clinical score and body weight variation of Ffar2fl/fl and Ffar2ΔS100 (Ffar2fl/fl × S100-Cre) mice that were treated with acetate and infected with C. difficile (n = 4–5). Ffar2ΔS100 and Ffar2fl/fl littermates were infected at the same time. (C and D) Il22, Rorc, Il1b (C), and IL-22 target genes mRNA expression (D) in the colon of acetate-treated Ffar2fl/fl and Ffar2ΔS100 mice on day 5 p.i. (n = 4–5). Results are representative of at least two independent experiments with three to four mice in each (A) or pooled results from two independent experiments with two to three mice in each group (B–D). All mice used were littermates. Results are presented as mean ± SEM. *, P < 0.05.
Finally, examination of mice with a deletion of the Ffar2 gene in neutrophils (Ffar2ΔS100) provided in vivo demonstration that acetate-mediated protection against CDI depends on neutrophils. The beneficial effect of acetate treatment on clinical score and weight change in CDI was reduced in Ffar2ΔS100 mice compared with Ffar2f/f mice (Fig. 8 B). Notably, Ffar2ΔS100 mice exhibited no clinical recovery on day 5 in contrast to Ffar2f/f mice. Moreover, acetate-treated Ffar2ΔS100 mice showed decreased expression of Il22, Rorc, and Il1b in the colon compared with acetate-treated Ffar2f/f mice 5 d p.i. (Fig. 8 C), as well as reduced expression of IL-22 target genes (Bcl2, Ccnd1, Reg3g, and Muc1) in the colon (Fig. 8 D), demonstrating that ILC3-mediated protection during CDI is at least in part due to acetate-FFAR2 mediated activation of neutrophils. Altogether, these data corroborate that acetate protects from CDI by promoting neutrophil-ILC3 cross-talk through IL-1β.
Discussion
The presence of commensal gut microbiota is essential to suppress C. difficile expansion and pathogenicity (Carroll and Bartlett, 2011; Koenigsknecht and Young, 2013; Lewis and Pamer, 2017). Our study demonstrates that the protective impact of the microbiota depends at least in part on the production of acetate through fiber fermentation, which activates the FFAR2 signaling pathway in both neutrophils and ILC3s coordinating their protective functions. Acetate-FFAR2 signaling in neutrophils enhances inflammasome-mediated secretion of active IL-1β, which provides a first line of defense against C. difficile toxins. Acetate-FFAR2 signaling in ILC3s augments expression of IL-1R, and the elevated levels of IL-1R boost IL-1β–induced production of IL-22, which elicits antimicrobial and repair mechanisms in intestinal epithelial cells. Thus, microbiota-derived acetate promotes coordinate activation of neutrophils and ILC3s through FFAR2, which bolsters host innate inflammatory and repair responses to C. difficile.
By using newly generated Ffar2ΔS100 mice, we conclusively demonstrated that acetate provides protection against CDI by activating neutrophil FFAR2. Since FFAR2 acts as a chemotactic receptor (Schulenberg and Maslowsky, 2009; Vinolo et al., 2011), its engagement by acetate may be important to increase the initial recruitment of neutrophils after CDI and/or their survival at C. difficile–damaged sites. Moreover, we found that acetate-FFAR2 signaling augmented IL-1β production by neutrophils in response to C. difficile toxin–mediated activation of the inflammasome. Indeed, the impact of acetate on IL-1β was negligible in Ffar2−/− as well as Casp1/11−/− and Nlrp3−/− mice. How acetate-FFAR2 signaling enhances C. difficile toxin–induced inflammasome activation remains to be investigated. It is possible that acetate-FFAR2 signaling facilitates K+ efflux and/or Ca2+ influx that trigger NLRP3 activation. Consistent with this, engagement of FFAR2 on colonic epithelial cells by acetate was shown to stimulate NLRP3 through Ca2+ influx and cell hyperpolarization (Macia et al., 2015). Acetate-FFAR2 signaling may also activate other downstream mediators that interact with inflammasomes, such as microfilaments, tubulin, and protein kinase C μ–dependent integrin-linked kinase, as shown for other G protein–coupled receptors that impact inflammasome activation (Boro and Balaji, 2017). Although not tested in our study, acetate may also enhance pyrin inflammasome activation in response to C. difficile toxins. While inflammasome activation was mainly detected in neutrophils, we do not exclude that intestinal macrophages may contribute to caspase-1–mediated IL-1β production. We also noticed that acetate can enhance caspase-1–mediated production of IL-18. This effect may be relevant in other models of colitis, such as dextran sulfate sodium–induced colitis, in which IL-18 has a prominent role in promoting epithelial repair (Macia et al., 2015).
In addition to boosting the IL-1β response, acetate enhanced IL-22 response during CDI. IL-22 promotes antimicrobial and repair mechanisms in epithelial cells (Ouyang and Valdez, 2008; Pickert et al., 2009), as well as systemic immune responses via activation of the C3 complement pathway (Hasegawa et al., 2014). IL-22 induction contributed considerably to the overall protection mediated by acetate, which indeed was markedly reduced in Il22−/− mice, as well as WT mice treated with a neutralizing anti-IL-22 antibody. We confirmed a previous report that ILC3s are a major source of IL-22 during CDI (Abt et al., 2015) and demonstrated that mice specifically lacking ILC3s have a defect in control of the acute phase and in the resolution of CDI. We further examined how acetate induces IL-22 in ILC3s. Although ILC3s express FFAR2, we found no direct effect of acetate-FFAR2 signaling on IL-22 production. However, the acetate-FFAR axis enhanced ILC3 expression of IL-1R, thereby heightening ILC3 sensitivity to IL-1β, a known inducer of IL-22 that is produced by neutrophils during CDI and perhaps by other myeloid cells. Thus, acetate stimulates IL-22 through a coordinate increase in IL-1β bioavailability and ILC3 responsiveness to IL-1β. These results complement the recent observation that FFAR2 signaling promotes in situ proliferation of colonic ILC3 (Chun et al., 2019). It was shown that butyrate, but not acetate or propionate, suppresses ILC2 function and ameliorates ILC2-driven airway inflammation through histone deacetylase inhibition and H3 acetylation, independent of FFAR2 and FFAR3 (Thio et al., 2018). Whether acetate impacts epigenetic regulation of ILC3s, neutrophils, or other immune cells responding to C. difficile remains to be explored.
Currently, the first line treatment for C. difficile–infected patients includes vancomycin, fidaxomicin, and/or metronidazole (McDonald et al., 2018). However, patients suffer relapsing disease refractory to standard antibiotic treatment benefit from fecal transplant (McDonald et al., 2018). Our discovery of the impact of acetate treatment on CDI in mice may further contribute to innovate the treatment of C. difficile. Future studies will be necessary to determine whether acetate is similarly effective in humans. In summary, our data demonstrate a marked protective effect of microbial-derived acetate against CDI and show that acetate activates FFAR on both neutrophils and ILC3s, resulting in a coordinate increase of inflammasome-induced production of IL-1β in neutrophils and responsiveness to IL-1β in ILC3s. Overall, this study highlights the considerable impact and the complex mechanisms of action of intestinal microbiota metabolites in the control of dysbiosis and intestinal infections.
Materials and methods
Mice
8-wk-old C57BL/6, Rag2/Il2rg-deficient, Rag1−/−, and S100-Cre male mice were purchased from Jackson Laboratory or the Multidisciplinary Centre for Biological Investigation. Caspase1/11, Nlpr3, and Rag2-deficient mice were purchased from the Centre for Development of Experimental Models for Medicine and Biology of the Federal University of Sao Paulo. Nlrp6−/−, Nlrc4−/−, Il22−/−, and Ffar2−/− mice were provided by our collaborators from the Institute of Biological Science of the Federal University of Minas Gerais. AhRfl/fl RorcCre mice were previously reported (Song et al., 2015). Ffar2fl/fl mice were provided by our collaborator from the University of Illinois at Chicago, IL. All strains were maintained in a C57BL/6 background and were kept in regular filter-top cages with free access to sterile water and food. Animal procedures were approved by the Ethics Committee on Animal Use of the Institute of Biology (protocol numbers 3230-1, 3742-1, and 4886-1) and by Washington University Animal Studies Committee.
Generation of Il22ra2 (IL-22 binding protein) null LacZ and floxed mice
To generate Il22ra2tm1b (null LacZ) and Il22ra2tm1c (flox) mice, two Il22ra2tm2a(EUCOMM)Wtsi ES cell clones (ES line JM8A3.N1) were obtained from European Conditional Mouse Mutagenesis Consortium; one correctly targeted clone, confirmed by Southern blot analysis, was introduced into B6-albino (C57BL/6J-Tyrc-2J/J) eight-cell embryos by laser-assisted injection. Male chimeras were initially bred to B6-albino mice to assess germline transmission; those transmitting were bred to CMV-Cre transgenic mice (B6.C-Tg(CMV-Cre)1Cgn/J; >99% C57BL/6) to delete exon 3 of Il22ra2 and the neomycin-resistance cassette. To delete lacZ as well as the neomycin-resistance cassette and generate mice with loxP sites flanking exon 3 of Il22ra2, chimeras were bred to CAG-FLPe C57BL/6 mice. The CMV-Cre and FLPe transgenes were subsequently bred out of the lines.
Bacteria
Toxigenic C. difficile VPI 10463 strain was cultivated in brain-heart infusion (BHI) blood agar supplemented with hemin (5 µg/ml) and menadione (1 µg/ml) at 37°C in anaerobic atmosphere (AnaeroGen, Oxoid; Thermo Fisher Scientific) in jars.
Cells
Human colon carcinoma cells (HCT116) were cultivated in DMEM supplemented with 10% FBS, 2 mM l-glutamine, 100 U ml−1 penicillin, and 100 µg/ml streptomycin (Vitrocell Embriolife) until the 10th passage. The MNK3 cell line was previously described and provides an attractive in vitro system to study the function of ILC3/LTi cells (Allan et al., 2015). MNK3 cells were cultured in DMEM with 10% FBS, 2 mM GlutaMAX, 1 mM sodium pyruvate, 55 µM 2-mercaptoethanol, 50 µg/ml gentamicin, and 10 mM Hepes, from HyClone with 4% IL-7 and 2% IL-2.
Model of infection
Mice infections were performed as previously described (Chen et al., 2008). Mice were pretreated with antibiotic mixture (0.4 mg/ml kanamycin, 0.035 mg/ml gentamicin, 0.035 mg/ml colistin, 0.215 mg/ml metronidazole, and 0.045 mg/ml vancomycin; Sigma-Aldrich) added to drinking water for 4 d. Next, mice received clindamycin (10 mg/kg, i.p.; Sigma-Aldrich). After 1 d, mice were infected with 108 CFU of C. difficile by gavage. Mice were weighed and monitored daily during the entire protocol with a clinical severity score that varied from 0 (normal) to 15, as described (Li et al., 2012; Table S1).
Table S1. Clinical score.
| Category | Score | |||
|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |
| Activity | Normal | Alert/slow moving | Lethargic/shaky | Inactive unless prodded |
| Posture | Normal | Back slanted | Hunched | Hunched/nose down |
| Coat | Normal | Piloerection | Rough skin | Very ruffled/puff/ungroomed |
| Diarrhea | Normal | Soft stool/discolored (yellowish) | Wet stained tail/mucous ± blood | Liquid/no stool (ileus) |
| Eyes/nose | Normal | Squinted/half closed | Squinted/discharge | Closed/discharge |
Clinical score = sum of all parameter scores. Total possible score is 15.
Diets and acetate treatment
Animals received oral pretreatment with acetate at 150 mM or placebo in the drinking water, as reported in other studies (Vieira et al., 2017). Acetate treatment started 1 d before addition of antibiotics and continued throughout the protocol. Acetate solution was prepared using acetic acid (Sigma-Aldrich) at a concentration of 1.5 M in water (stock solution 10×, pH adjusted to 7.2–7.4, filtered at 0.22 µm). Mice had ad libitum access to water and food during the entire protocol. In dietary experiments, mice were maintained in diets with different amounts of fiber: a control diet, based on American Institute of Nutrition (AIN93) recommendations containing 5% cellulose; and a diet that in addition to 5% cellulose was supplemented with 10% of soluble fiber pectin from citrus (Vieira et al., 2017). Mice were prefed with the different diets for 21 d (Fig. 1 A).
Measurement of SCFAs
Blood samples were harvested from mice, and the serum was used for measurement of SCFA concentration. Samples were prepared as previously described (Fellows et al., 2018). Chromatographic analyses were performed using a GCMS-QP2010 Ultra mass spectrometer (Shimadzu; Thermo Fisher Scientific) and a 30 m × 0.25 mm fused-silica capillary Stabilwax column (Restek Corp.) coated with 0.25 µm polyethylene glycol. Samples (100 µl) were injected at 250°C using a 25:1 split ratio. High-grade pure helium was used as carrier gas at 1.0 ml/min constant flow. Mass conditions were as follows: ionization voltage, 70 eV; ion source temperature, 200°C; full scan mode, 35–500 m/z with 0.2-s scan velocity. The runtime was 11.95 min.
Determination of fecal bacterial load
Fecal samples (50 mg) were used for extraction of microbial genomic DNA using the PureLink Microbiome DNA Purification kit (Thermo Fisher Scientific). For bacterial load measurement, DNA was quantified by quantitative PCR (qPCR) using primers complementary to Eubacteria 16S rDNA conserved region (E338F sense 5′-ACTCCTACGGGAGGCAGCAGT-3′ and U1407R anti-sense 5′-ATTACCGCGGCGGCTGCTGGC-3′; Durand et al., 2010). A standard curve was constructed with serial dilutions of Escherichia coli genomic DNA. Results were normalized by percentage of the control.
C. difficile in vitro assay
For the in vitro assay with C. difficile and acetate, bacteria were cultured 3 d earlier on BHI blood agar supplemented with hemin (5 µg/ml) and menadione (1 µg/ml) in anaerobic chamber at 37°C. Initially, bacteria were resuspended and washed twice with sterile PBS. After that, the bacterial concentration was adjusted and maintained at 0.5 × 107 CFU/ml in a final volume of 5 ml BHI broth with or without acetate at different concentrations. Cultures were incubated for 72 h in anaerobiosis at 37°C and had the optical density of the medium read at 600 nm wavelength. To verify possible contaminations of the liquid culture, the medium was plated on BHI blood agar, and bacterial growth and gram staining of the colonies were observed.
Histological analysis
Mouse colons were harvested, opened longitudinally, and fixed in 4% formalin/0.1% glutaraldehyde. Tissues were processed into historesin, and 5-µm sections were prepared for staining with hematoxylin and eosin or Giemsa/Rosenfeld solution. Slides were analyzed using an Olympus microscope (model U-LH100HG). Samples were analyzed blindly using histological scores for each parameter (Table S2). Overall scores were the sums of each component and varied from 0 to 30.
Table S2. Histological score.
| Category | Finding | 0 (none) | 1 (mild) | 2 (moderate) | 3 (severe) |
|---|---|---|---|---|---|
| Mucosal epithelium | Ulceration | None | Mild surface | Moderate | Extensive full thickness |
| Crypts | Mitotic activity | Lower third | Mild middle third | Moderate middle third | Upper third |
| Mucus depletion | None | Mild | Moderate | Severe | |
| Lamina propria | Mononuclear infiltrate | None | Mild | Moderate | Severe |
| Granulocyte infiltrate | None | Mild | Moderate | Severe | |
| Vascularity | None | Mild | Moderate | Severe | |
| Fibrin deposition | None | Mucosal | Submucosal | Transmural | |
| Submucosal | Mononuclear infiltrate | None | Mild | Moderate | Severe |
| Granulocyte infiltrate | None | Mild | Moderate | Severe | |
| Edema | None | Mild | Moderate | Severe |
Histological score = sum of all parameter scores. Total possible score is 30.
Analysis of neutrophil population
The lamina propria of the colon was collected and processed for neutrophil analysis. First, the colon was extracted and washed twice with 1× HBSS supplemented with 5% FBS and 2 mM EDTA in a 50-ml Falcon tube under horizontal shaking in an orbital mixer at 250 rpm for 20 min at 37°C. Then, enzymatic digestion was performed in solution containing 1× HBSS supplemented with 2 mM EDTA and collagenase IV (40 U/ml) at 200 rpm for 15 min at 37°C. The obtained cells were then separated from the tissue using a 70-µm filter (Cell Strainer; BD Bioscience). Samples were counted in Neubauer’s chamber, and 106 cells were labeled with specific antibodies coupled to different fluorochromes and analyzed by flow cytometry. Anti-Ly6G FITC (Gr-1) clone 1A8-Ly6g, anti-Ly6C PE clone HK1.4, and anti-CD11b APC clone M1/70 anti-mouse antibodies (eBioscience, Thermo Fisher Scientific) were used for analysis of the neutrophil population on the lamina propria. Data were analyzed using FlowJo software.
Measurement of cytokines in tissues
Colon samples (100 mg) were homogenized in PBS containing protease inhibitors (Thermo Fisher Scientific). Samples were centrifuged for 10 min at 2,000 g, and supernatants used for measurement of IL-1β, TNF-α, CXCL1, CXCL2, IL-10, IL-22, and IL-18 using the Duo Set ELISA kit (R&D Systems).
Bacterial translocation
Spleen, liver, and mLNs were harvested on day 1 or 2 of infection. Bacterial 16S rDNA were extracted using the PureLink Microbiome DNA Purification kit (Thermo Fisher Scientific), and gene levels were quantified by qPCR using primers complementary to Eubacteria 16S rDNA conserved region (Table S3). The bacterial load was determined by a standard curve with serial dilutions of E. coli genomic DNA, and the CFU per gram of tissue was determined by dividing gene levels by sample weight.
Table S3. Primers used for qPCR.
| Primer | Sequence (5′ to 3′) |
|---|---|
| 16s of eubacteria | |
| Sense | ACTCCTACGGGAGGCAGCAGT |
| Antisense | ATTACCGCGGCTGCTGGC |
| β2-Microglobulin (B2m) | |
| Sense | CCCCACTGAGACTGATACATACG |
| Antisense | CGATCCCAGTAGACGGTCTTG |
| Il17 | |
| Sense | TCAGCGTGTCCAAACACTGAG |
| Antisense | GACTTTGAGGTTGACCTTCACAT |
| Il22 | |
| Sense | AGAATGTCAGAAGGCTGAAGG |
| Antisense | AGGAGCAGTTCTTCGTTTTCTAG |
| ROR-γt (Rorc) | |
| Sense | TCCACTACGGGGTTATCACCT |
| Antisense | AGTAGGCCACATTACACTGCT |
| Il1r | |
| Sense | GCACGCCCAGGAGAATATGA |
| Antisense | AGAGGACACTTGCGAATATCAA |
| Il1b | |
| Sense | GGCAGCTACCTGTGTCTTTCCC |
| Antisense | ATATGGGTCCGACAGCACGAG |
| Bcl2 | |
| Sense | TGAGTACCTGAACCGGCATCT |
| Antisense | GCATCCCAGCCTCCGTTAT |
| Cyclin D1 (Ccnd1) | |
| Sense | GCAAGCATGCACAGACCTT |
| Antisense | GTTGTGCGGTAGCAGGAGA |
| g3g | |
| Sense | TTCCTGTCCTCCATGATCAAAA |
| Antisense | CATCCACCTCTGTTGGGTTCA |
| Muc1 | |
| Sense | CCCTACCTACCACACTCACGGACG |
| Antisense | GTGGTCACCACAGCTGGGTTGGT |
| Muc4 | |
| Sense | GAGGGCTACTGTCACAATGGAGGC |
| Antisense | AGGGTTCCGAAGAGGATCCCGTAG |
| Ffar2 | |
| Sense | GGCTCCCTGCCAACCTGCTG |
| Antisense | GTGCACAGGGGCAGGCTGAG |
| Ffar3 | |
| Sense | GGGTTACACACAGAGGTGGC |
| Antisense | CATCACGTTGAGGGGGAGTC |
| IFNγ (Ifng) | |
| Sense | ATGAACGCACACACTGCATC |
| Antisense | CCATCCTTTTGCCAGTTCCTC |
Measurement of intestinal permeability with FITC-dextran
Mice received 200 µl FITC-dextran (70,000 D; Sigma-Aldrich) suspension (250 mg/kg) by gavage on day 2 of infection. After 4 h, mice were anesthetized, blood was collected by caudal puncture, and fluorescence readings were performed in a Multi-Mode Microplate Reader (Synergy HT) at 485/528 nm (excitation/emission). A standard curve was prepared with serial dilutions of FITC-dextran in PBS.
Epithelial cell culture and live/dead assay
Human colon carcinoma cells (HCT116) were cultivated in 1:500 bacterial supernatant and 10, 25, or 50 µM of acetate in 96-well plates (1.0 × 105 cells/well). After 48 h, cells were washed gently using Dulbecco’s PBS (DPBS), and medium was replaced by 100 liters Calcein-AM and propidium iodide (2 µM) in DPBS and incubated for 30 min at 37°C under 5% CO2. Images were obtained using the Cytation 5 Cell Imaging Multi-Mode Reader, and green fluorescence (485/530 nm, excitation/emission) of viable cells and red fluorescence (530/645 nm) of dead cells were quantified using Gen5 software (Biotek). As a positive control, cells were preincubated with 0.1% Triton X-100 for 15 min. To obtain C. difficile supernatants, the toxigenic strain VPI 10463 was cultured 24 h at 37°C in anaerobic conditions in BHI medium supplemented with hemin and menadione. The culture was centrifuged at 10,000 g for 5 min, and supernatant was used for treatment of HCT116 culture (1:500 ratio).
Quantitative gene expression
Total RNA was extracted from tissue using the PureLink RNA kit (Ambion). RNA was converted to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems), and qPCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) and primers indicated in Table S3. Quantification of gene expression was performed using the 2ΔΔ Ct method, with β2-microglobulin as a reference gene.
Neutrophil culture
Bone marrow neutrophils were isolated using a Percoll (GE Healthcare) gradient (55/65%). The purity of this preparation was >70%. FACS-sorted Ly6G+CD11b+ cells were also used in some analyses. Cells (4 × 106 cells/ml) were plated at 37°C in RPMI (Vitrocell) containing 10% FBS and without antibiotics. Initially, cells were incubated for 2 h with or without addition of LPS (0.1 µg/ml, E. coli 0111:B4) and 25 mM acetate. C. difficile (1:1) or its supernatant (1:500) was then added to the culture for 4 h. Subsequently, IL-1β cytokine released in the culture was quantified by ELISA. To elucidate the pathways of acetate on inflammasome activation, after incubation in vitro with LPS ± acetate, neutrophils were treated with a known inflammasome activator, the potassium (K+) ionophore nigericin (1 µM), or with BAPTA-AM (25 µM), which inhibits NLRP3 by blocking Ca2+ influx, ± C. difficile culture supernatant (1:500). Similarly, neutrophils were followed by stimulation with extracellular KCl (45 mM), which inhibits NLRP3 by blocking K+ efflux, or NaCl (45 mM), which boosts inflammasome activation by promoting K+ efflux. We also performed an experiment with either caspase-1 (10 µM Q-VD-Oph) or neutrophil elastase (10 µM Sivelesat sodium salt) inhibitors. To determine the cellular viability of neutrophils, BioLegend’s FITC Annexin V Apoptosis Detection Kit with 7-aminoactinomycin D (7-AAD) was used, and the supplier’s instructions were followed.
IL-22 in vivo neutralization
C. difficile–infected mice, treated or not with acetate in the drinking water, received an i.p. dose of 50 µl PBS with 150 g anti-mouse IL-22 neutralizing antibody (clone 8E11; Genentech) or an equivalent amount of isotype control IgG2a (BioXcell) on days 1 and 3 p.i. Mice were clinically evaluated until the fifth day of infection.
ILCs isolation, MNK3, and cytokine detection
Adult small intestine or colon samples were harvested from C57BL/6 male mice, and mesenteric adipose tissue, Peyer’s patches, and intraepithelial lymphocytes were first removed by dissection and two EDTA extraction washes. Intestinal lamina propria immune cells were isolated using Collagenase 4 (Sigma-Aldrich) digestion (40 min) and were enriched at the interface between a gradient of 40% and 70% of Percoll (GE Healthcare) in HBSS. For functional experiments, small intestine lamina propria cells were cultured in a 96-well plate in complete medium and stimulated ex vivo in the presence of Golgi Plug (BD) for 3 h at 37°C. Following incubation, Live+CD45+LIN– (CD3–CD5–CD19–B220–) CD45lowThy1.2high ILC3s were stained for surface molecules, fixed, and stained for intracellular IL-22 (clone Poly5164; BioLegend). For IL-1R expression, cells were stained with anti-mouse CD121a (IL-1R, clone JAMA-147; BioLegend). Data were analyzed using FlowJo software. The MNK3 cell line was previously described (Allan et al., 2015). MNK3 was stimulated and stained for intracellular IL-22 as described for primary ILC3s.
Statistical analysis
Analyses were performed using GraphPad software 5.0. All data are presented as means ± SEM, and n represents the number of samples per group. The exact value of n in each experiment is indicated in the corresponding figure legends. Differences were considered significant for P < 0.05. Results were first analyzed using D’Agostino/Shapiro–Wilk normality tests and compared by Student’s t test or Mann–Whitney U test, as appropriate. For more than two groups, differences were compared by one-way analysis of variance followed by Tukey’s post hoc test.
Online supplemental material
Fig. S1 shows the relative bacterial load in the feces on days 0 and 2 d p.i., in vitro growth of C. difficile after treatment with acetate, and clinical scores and body weight changes of mice treated with acetate after infection and of infected Nlrp3−/−, Nlrc4−/−, and Nlrp6−/− mice. Fig. S2 shows IL-18 content in the colon of WT and Casp1/11−/− and Ffar2−/− mice, neutrophils viability before and after in vitro incubation, and IL-1β production by neutrophils isolated from the bone marrow and stimulated in vitro. Fig. S3 shows expression of SCFA receptors in ILC populations, IL-22 target genes expression in the colon 5 d p.i., ILC subsets in the small intestine 5 d p.i., and representative histological sections of colons of ILC3-deficient mice 5 d p.i. Fig. S4 shows clinical scores, body weight changes, and intestinal permeability of IL-22 binding protein–deficient mice (Il22ra2−/−); effect of acetate on IFNγ production; in vitro treatment of MNK3 cells with acetate and IL-1β; effect of acetate on Ffar2 and Ffar3 expression; and ILC3 subsets. Table S1 describes the clinical score criteria used to evaluate infected mice. Table S2 lists all parameters used for histopathological analyses. Table S3 includes a list of primers used to analyze the gene expression by qPCR. Table S4 lists the reagents used in our study.
Table S4. Reagents.
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD11b, APC | eBioscience | Clone M1/70,17-0112-82 |
| Anti-mouse Ly6G, FITC | eBioscience | Clone 1A8,11-9668-80 |
| Anti-mouse Ly6C, PE | BioLegend | Clone HK1.4,128007 |
| Anti-mouse CD45 APC/Cy7 | BioLegend | Clone 30-F11,103115 |
| Anti-mouse CD45 PerCP/Cy5.5 | BioLegend | Clone 30-F11,103131 |
| Anti-mouse CD3 PerCP/Cy5.5 | BioLegend | Clone 17A2,100217 |
| Anti-mouse CD5 PerCP/Cy5.5 | BioLegend | Clone 53-7.3,100623 |
| Anti-mouse CD19 PerCP/Cy5.5 | BioLegend | Clone 6D5,115533 |
| Anti-mouse B220 PerCP/Cy5.5 | BioLegend | Clone RA3-6B2,103235 |
| Anti-mouse CD90.2 (Thy-1.2) FITC | BioLegend | Clone 30-H12,105305 |
| Anti-mouse CD90.2 (Thy-1.2) APC | BioLegend | Clone 53-2.1,140311 |
| Anti-mouse CD196 (CCR6) Brilliant Violet 421 | BioLegend | Clone 29-2L17,129817 |
| Anti-mouse CD335 (NKp46) PE | BioLegend | Clone 29A1.4,137603 |
| Anti-mouse CD335 (NKp46) PE-Cy7 | BioLegend | Clone 29A1.4,137617 |
| Anti-mouse Rorgt APC | Thermo Fisher Scientific | Clone B2D,17-6981-82 |
| Anti-mouse GATA3 Alexa Fluor 488 | eBioscience | Clone L50-823,560077 |
| Anti-mouse EOMES PE | Thermo Fisher Scientific | Clone Dan11mag,12-4875-82 |
| Anti-mouse IL22 PE | BioLegend | Clone Poly5164,516404 |
| Anti-mouse IL22 APC | BioLegend | Clone Poly5164,516409 |
| Anti-mouse IFNy APC-Cy7 | eBioscience | 50-5773-82 |
| Anti-mouse CD121a (IL-1 R, Type I/p80) biotinylated | BioLegend | JAMA-147,113503 |
| PE/Cy7 Streptavidin | BioLegend | 405206 |
| Brilliant Violet 421 Streptavidin | BioLegend | 405226 |
| IL-1 beta monoclonal anti-mouse | eBioscience | 16-7012-81 |
| IL-22 monoclonal anti-mouse neutralizing antibody | Genentech | Clone 8E11 |
| InVivo MAb mouse IgG2a isotype control | BioXcell | Clone C1.18.4,BE0085 |
| Bacterial strains | ||
| C. difficile VPI 10463 | Dr. Mário Júlio Ávila Campos (University of São Paulo, São Paulo, Brazil) | N/A |
| E. coli | Dr. Marcelo Lancelotti (University of Campinas, Campinas, Brazil) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Kanamycin | Sigma-Aldrich | K1876-5G |
| Gentamicin | Sigma-Aldrich | G1914-5G, G1397-10ML |
| Colistin | Sigma-Aldrich | C4461-100MG |
| Metronidazole | Sigma-Aldrich | M3761-5G |
| Vancomycin | Sigma-Aldrich | V2002-1G |
| Clindamycin | Sigma-Aldrich | C5269-50MG |
| Acetic acid | Sigma-Aldrich | A6283-1L |
| Pectin fiber diet | This paper | N/A |
| Control LFD | This paper | N/A |
| BHI agar | Sigma-Aldrich | 70138-500G |
| BHI broth | Sigma-Aldrich | 53286-500G |
| Yeast extract | Sigma-Aldrich | 92144-500G-F |
| Hemin | Sigma-Aldrich | H9039-1G |
| Menadione | Sigma-Aldrich | M5625-25G |
| Formaldehyde solution | Sigma-Aldrich | 252549-1L |
| Glutaraldehyde solution | Sigma-Aldrich | G7651-10ML |
| Histological historesin | Leica | 7592 |
| Giemsa stain, modified solution | Sigma-Aldrich | 32884-250ML |
| Hematoxylin solution, Harris modified | Sigma-Aldrich | HHS16 |
| Eosin stain solution 5% | Sigma-Aldrich | R03040-74 |
| Protease inhibitors | Thermo Fisher Scientific | 78430 |
| Power SYBR Green PCR Master Mix | Applied Biosystems | 4367659 |
| RPMI-1640 | Vitrocell Embriolife | N/A |
| DMEM | Thermo Fisher Scientific | 11966-025 |
| FBS | Corning | MT35016CV |
| l-Glutamine | Sigma-Aldrich | G3126 |
| Streptomycin sulfate | Sigma-Aldrich | S6501 |
| Penicillin | Sigma-Aldrich | 1502701 |
| GlutaMAX Supplement | Thermo Fisher Scientific | 35050061 |
| Sodium pyruvate | Sigma-Aldrich | P2256 |
| β-Mercaptoethanol | Sigma-Aldrich | M6250-10ML |
| Hepes | HyClone | SH30237.01 |
| Deoxyribonuclease I from bovine pancreas | Sigma-Aldrich | DN25 |
| 0.5 M EDTA, pH 8.0 | Invitrogen | AM9261 |
| dl-DTT | Sigma-Aldrich | D9779 |
| Collagenase IV | Sigma-Aldrich | C5138-500MG |
| Tris-HCl | Sigma-Aldrich | T5941-500G |
| SDS | Sigma-Aldrich | L3771-100G |
| Dulbecco’s PBS | Sigma-Aldrich | D4031-1L |
| Calcein-AM | Sigma-Aldrich | 17783-1MG |
| Propidium iodide | Sigma-Aldrich | P4170-10MG |
| Triton X-100 | Merck Millipore | 1086431000 |
| HBSS 10× | Sigma-Aldrich | H4641-500ML |
| Percoll density gradient medium | Ge Healthcare | 17089101 |
| LPS from E. coli 0111:B4 | Sigma-Aldrich | LPS25 |
| Nigericin sodium salt | Sigma-Aldrich | N7143-10MG |
| BAPTA-AM | Thermo Fisher Scientific | B1205 |
| Sodium chloride | Sigma-Aldrich | S9888-500G |
| Potassium chloride | Sigma-Aldrich | 746436-500G |
| Recombinant mouse IL-23 protein | R&D Systems | 1887-ML-010 |
| Recombinant mouse IL-1β/IL-1F2 protein | R&D Systems | 401-ML-010 |
| GPR43 (FFA2) agonist, Calbiochem | Millipore | 371725-10MG |
| GPR43 (FFA2) antagonist, GLPG 0974 | Tocris | 5621 |
| Critical commercial assays | ||
| PureLink Microbiome DNA Purification | Thermo Fisher Scientific | A29790 |
| Duo Set Kit: TNF-α | R&D Systems | DY008 |
| Duo Set Kit: IL-1β | R&D Systems | DY401 |
| Duo Set Kit: IL-10 | R&D Systems | DY417 |
| Duo Set Kit: MIP-2 (Cxcl-2) | R&D Systems | DY452 |
| Duo Set Kit: KC (Cxcl-1) | R&D Systems | DY453 |
| Duo Set Kit: IL-18 | R&D Systems | DY122-05 |
| Duo Set Kit: IL-22 | R&D Systems | DY582-05 |
| Fixation/Permeabilization Solution Kit with BD GolgiPlug | BD Biosciences | 555028 |
| PE Annexin V Apoptosis Detection Kit with 7-AAD | BioLegend | 640934 |
| Experimental models: cell lines | ||
| HCT-116 | Dr. Patrick Varga-Weisz (University of Essex, Colchester, UK) | N/A |
| MNK3 | This paper | N/A |
| Experimental models: organisms/strains | ||
| C57BL/6 male mice | CEMIB - UNICAMP | N/A |
| Rag2/Il2rg-deficient mice | Jackson Laboratory | Jax# 014593 |
| Rag2-deficient mice | Federal University of Sao Paulo, Brazil | N/A |
| Rag1-deficient mice | Washington University School of Medicine, St Louis, MO | N/A |
| Caspase1/11-deficient mice | Federal University of Sao Paulo, Brazil | N/A |
| Nlpr3-deficient mice | Federal University of Sao Paulo, Brazil | N/A |
| Nlrp6-deficient mice | Federal University of Minas Gerais, Brazil | N/A |
| Nlrc4-deficient mice | Federal University of Minas Gerais, Brazil | N/A |
| Il22-deficient mice | Federal University of Minas Gerais, Brazil | N/A |
| Ffar2-deficient mice | Federal University of Minas Gerais, Brazil | N/A |
| AhR floxed mice | Washington University School of Medicine, St Louis, MO | N/A |
| Ffar2 floxed mice | University of Illinois at Chicago, IL | N/A |
| S100-Cre mice | Jackson Laboratory | Jax# 021614 |
| Rorc-Cre mice | Jackson Laboratory | Jax# 022791 |
| Il22ra2-deficient mice | This paper | |
| Oligonucleotides | ||
| Eubacteria 16S rDNA E338F sense and U1407R antisense | Durand et al., 2010 | N/A |
| Primers listed in Table S3 | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism v5.0 | GraphPad Software | N/A |
| FlowJo LLC v10.1 | Becton Dickinson | N/A |
| FACSDiva | BD Biosciences | N/A |
| ImageJ | National Institutes of Health | N/A |
| Gen5 software | Biotek | N/A |
| Applied Biosystems 7500 Real-Time PCR System | Thermo Fisher Scientific | N/A |
| Other | ||
| AnaeroGen Oxoid | Thermo Fisher Scientific | AN0025A |
| 70- and 40-µm cell strainers | BD Biosciences | CLS431751-50EA |
| LIVE/DEAD fixable dead stain | Thermo Fisher Scientific | L34962 |
| FITC–dextran | Sigma-Aldrich | 46944-100MG-F |
| Hoechst 33258 solution | Thermo Fisher Scientific | 62249 |
N/A, not applicable.
Acknowledgments
We thank Drs. Jennifer K. Bando, Marina Cella, and Susan Gilfillan for critical comments. We also thank Marcelo Pires Amaral for helping with mouse experiments. We thank Tangsheng Yi (Department of Immunology Discovery, Genentech, South San Francisco, CA) and Genentech for the kind gift of anti–IL-22.
This study was supported by a research grant from Fundação de Amparo à Pesquisa do Estado de São Paulo (12/10653-9, 17/16280-3). The study was also financed by the Conselho Nacional de Desenvolvimento Científico e Tecnológico and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Finance Code 001. J.L. Fachi, P.B. Rodrigues, and L.P. Pral are recipient of fellowships from Fundação de Amparo à Pesquisa do Estado de São Paulo (2017/06577-9, 2019/14342-7, 2018/02208-1). M. Colonna was supported by Mucosal Immunology Studies Team, National Institute of Allergy and Infectious Diseases, National Institutes of Health (U01 AI095542). B.T. Layden is supported by the National Institutes of Health under award number R01DK104927-01A1; University of Chicago Diabetes Research and Training Center (P30DK020595); and Department of Veterans’ Affairs, Veterans Health Administration, Office of Research and Development, VA merit (grant no. 1I01BX003382-01-A1).
Author contributions: Conceptualization: J.L. Fachi, M. Colonna, M.A.R. Vinolo; Methodology: J.L. Fachi, C. Sécca, P.B. Rodrigues, F.C.P. Mato, J.S. Felipe, B. Di Luccia, L.P. Pral, M. Rungue, V.M. Rocha, F.T. Sato, U. Sampaio, M. Colonna, S.R. Consonni; Investigation: J.L. Fachi, K.R. Bortoluci, M. Colonna, M.A.R. Vinolo; Original draft: J.L. Fachi, M. Colonna, M.A.R. Vinolo, Review & editing: J.L. Fachi, M.T.P.S. Clerici, H.G. Rodrigues, N.O.S. Câmara, S.R. Consonni, A.T. Vieira, S.C. Oliveira, C.R. Mackay, K.R. Bortoluci, B.T. Layden, M. Colonna, M.A.R. Vinolo; Visualization: all authors; Funding acquisition: M. Colonna, M.A.R. Vinolo; Resources: A.T. Vieira, S.C. Oliveira, H.G. Rodrigues, N.O.S. Câmara, C.R. Mackay, B.T. Layden, K.R. Bortoluci, M. Colonna, M.A.R. Vinolo; Supervision: M. Colonna, M.A.R. Vinolo.
References
- Abt M.C., Lewis B.B., Caballero S., Xiong H., Carter R.A., Sušac B., Ling L., Leiner I., and Pamer E.G.. 2015. Innate Immune Defenses Mediated by Two ILC Subsets Are Critical for Protection against Acute Clostridium difficile Infection. Cell Host Microbe. 18:27–37. 10.1016/j.chom.2015.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan D.S., Kirkham C.L., Aguilar O.A., Qu L.C., Chen P., Fine J.H., Serra P., Awong G., Gommerman J.L., Zúñiga-Pflücker J.C., and Carlyle J.R.. 2015. An in vitro model of innate lymphoid cell function and differentiation. Mucosal Immunol. 8:340–351. 10.1038/mi.2014.71 [DOI] [PubMed] [Google Scholar]
- Bibbò S., Lopetuso L.R., Ianiro G., Di Rienzo T., Gasbarrini A., and Cammarota G.. 2014. Role of microbiota and innate immunity in recurrent Clostridium difficile infection. J. Immunol. Res. 2014:462740 10.1155/2014/462740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boro M., and Balaji K.N.. 2017. CXCL1 and CXCL2 Regulate NLRP3 Inflammasome Activation via G-Protein-Coupled Receptor CXCR2. J. Immunol. 199:1660–1671. 10.4049/jimmunol.1700129 [DOI] [PubMed] [Google Scholar]
- Carroll K.C., and Bartlett J.G.. 2011. Biology of Clostridium difficile: implications for epidemiology and diagnosis. Annu. Rev. Microbiol. 65:501–521. 10.1146/annurev-micro-090110-102824 [DOI] [PubMed] [Google Scholar]
- Chen X., Katchar K., Goldsmith J.D., Nanthakumar N., Cheknis A., Gerding D.N., and Kelly C.P.. 2008. A mouse model of Clostridium difficile-associated disease. Gastroenterology. 135:1984–1992. 10.1053/j.gastro.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Chun E., Lavoie S., Fonseca-Pereira D., Bae S., Michaud M., Hoveyda H.R., Fraser G.L., Gallini Comeau C.A., Glickman J.N., Fuller M.H., et al. 2019. Metabolite-Sensing Receptor Ffar2 Regulates Colonic Group 3 Innate Lymphoid Cells and Gut Immunity. Immunity. 51:871–884.e6. 10.1016/j.immuni.2019.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrêa-Oliveira R., Fachi J.L., Vieira A., Sato F.T., and Vinolo M.A.. 2016. Regulation of immune cell function by short-chain fatty acids. Clin. Transl. Immunology. 5:e73 10.1038/cti.2016.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R.X., Goh W.R., Wu R.N., Yue X.Q., Luo X., Khine W.W.T., Wu J.R., and Lee Y.K.. 2019. Revisit gut microbiota and its impact on human health and disease. Yao Wu Shi Pin Fen Xi. 27:623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe D.R., and Bultman S.J.. 2012. Metaboloepigenetics: interrelationships between energy metabolism and epigenetic control of gene expression. J. Cell. Physiol. 227:3169–3177. 10.1002/jcp.24054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand L., Zbinden M., Cueff-Gauchard V., Duperron S., Roussel E.G., Shillito B., and Cambon-Bonavita M.A.. 2010. Microbial diversity associated with the hydrothermal shrimp Rimicaris exoculata gut and occurrence of a resident microbial community. FEMS Microbiol. Ecol. 71:291–303. 10.1111/j.1574-6941.2009.00806.x [DOI] [PubMed] [Google Scholar]
- El-Zaatari M., Chang Y.M., Zhang M., Franz M., Shreiner A., McDermott A.J., van der Sluijs K.F., Lutter R., Grasberger H., Kamada N., et al. 2014. Tryptophan catabolism restricts IFN-γ-expressing neutrophils and Clostridium difficile immunopathology. J. Immunol. 193:807–816. 10.4049/jimmunol.1302913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachi J.L., Felipe J.S., Pral L.P., da Silva B.K., Corrêa R.O., de Andrade M.C.P., da Fonseca D.M., Basso P.J., Câmara N.O.S., de Sales E Souza É.L., et al. 2019. Butyrate Protects Mice from Clostridium difficile-Induced Colitis through an HIF-1-Dependent Mechanism. Cell Reports. 27:750–761.e7. 10.1016/j.celrep.2019.03.054 [DOI] [PubMed] [Google Scholar]
- Fellows R., Denizot J., Stellato C., Cuomo A., Jain P., Stoyanova E., Balázsi S., Hajnády Z., Liebert A., Kazakevych J., et al. 2018. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 9:105 10.1038/s41467-017-02651-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira C.M., Vieira A.T., Vinolo M.A., Oliveira F.A., Curi R., and Martins F.S.. 2014. The central role of the gut microbiota in chronic inflammatory diseases. J. Immunol. Res. 2014:689492 10.1155/2014/689492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara H., Docampo M.D., Riwes M., Peltier D., Toubai T., Henig I., Wu S.J., Kim S., Taylor A., Brabbs S., et al. 2018. Microbial metabolite sensor GPR43 controls severity of experimental GVHD. Nat. Commun. 9:3674 10.1038/s41467-018-06048-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., et al. 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 504:446–450. 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Kamada N., Jiao Y., Liu M.Z., Núñez G., and Inohara N.. 2012. Protective role of commensals against Clostridium difficile infection via an IL-1β-mediated positive-feedback loop. J. Immunol. 189:3085–3091. 10.4049/jimmunol.1200821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Yada S., Liu M.Z., Kamada N., Muñoz-Planillo R., Do N., Núñez G., and Inohara N.. 2014. Interleukin-22 regulates the complement system to promote resistance against pathobionts after pathogen-induced intestinal damage. Immunity. 41:620–632. 10.1016/j.immuni.2014.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S., Gagliani N., Zenewicz L.A., Huber F.J., Bosurgi L., Hu B., Hedl M., Zhang W., O’Connor W. Jr., Murphy A.J., et al. 2012. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 491:259–263. 10.1038/nature11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarchum I., Liu M., Shi C., Equinda M., and Pamer E.G.. 2012. Critical role for MyD88-mediated neutrophil recruitment during Clostridium difficile colitis. Infect. Immun. 80:2989–2996. 10.1128/IAI.00448-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanesen P.A., Mackin K.E., Hutton M.L., Awad M.M., Larcombe S., Amy J.M., and Lyras D.. 2015. Disruption of the Gut Microbiome: Clostridium difficile Infection and the Threat of Antibiotic Resistance. Genes (Basel). 6:1347–1360. 10.3390/genes6041347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiko G.E., Ryu S.H., Koues O.I., Collins P.L., Solnica-Krezel L., Pearce E.J., Pearce E.L., Oltz E.M., and Stappenbeck T.S.. 2016. The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell. 167:1137 10.1016/j.cell.2016.10.034 [DOI] [PubMed] [Google Scholar]
- Kelly C.J., Zheng L., Campbell E.L., Saeedi B., Scholz C.C., Bayless A.J., Wilson K.E., Glover L.E., Kominsky D.J., Magnuson A., et al. 2015. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe. 17:662–671. 10.1016/j.chom.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.H., Kang S.G., Park J.H., Yanagisawa M., and Kim C.H.. 2013. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 145:396–406.e1: 10. 10.1053/j.gastro.2013.04.056 [DOI] [PubMed] [Google Scholar]
- Koenigsknecht M.J., and Young V.B.. 2013. Faecal microbiota transplantation for the treatment of recurrent Clostridium difficile infection: current promise and future needs. Curr. Opin. Gastroenterol. 29:628–632. 10.1097/MOG.0b013e328365d326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley T.D., Clare S., Walker A.W., Stares M.D., Connor T.R., Raisen C., Goulding D., Rad R., Schreiber F., Brandt C., et al. 2012. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 8:e1002995 10.1371/journal.ppat.1002995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.B., and Pamer E.G.. 2017. Microbiota-Based Therapies for Clostridium difficile and Antibiotic-Resistant Enteric Infections. Annu. Rev. Microbiol. 71:157–178. 10.1146/annurev-micro-090816-093549 [DOI] [PubMed] [Google Scholar]
- Li Y., Figler R.A., Kolling G., Bracken T.C., Rieger J., Stevenson R.W., Linden J., Guerrant R.L., and Warren C.A.. 2012. Adenosine A2A receptor activation reduces recurrence and mortality from Clostridium difficile infection in mice following vancomycin treatment. BMC Infect. Dis. 12:342 10.1186/1471-2334-12-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia L., Tan J., Vieira A.T., Leach K., Stanley D., Luong S., Maruya M., Ian McKenzie C., Hijikata A., Wong C., et al. 2015. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 6:6734 10.1038/ncomms7734 [DOI] [PubMed] [Google Scholar]
- Maslowski K.M., Vieira A.T., Ng A., Kranich J., Sierro F., Yu D., Schilter H.C., Rolph M.S., Mackay F., Artis D., et al. 2009. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 461:1282–1286. 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J.A.K., Mullish B.H., Pechlivanis A., Liu Z., Brignardello J., Kao D., Holmes E., Li J.V., Clarke T.B., Thursz M.R., and Marchesi J.R.. 2018. Inhibiting Growth of Clostridioides difficile by Restoring Valerate, Produced by the Intestinal Microbiota. Gastroenterology. 155:1495–1507.e15. 10.1053/j.gastro.2018.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naaber P., Mikelsaar R.H., Salminen S., and Mikelsaar M.. 1998. Bacterial translocation, intestinal microflora and morphological changes of intestinal mucosa in experimental models of Clostridium difficile infection. J. Med. Microbiol. 47:591–598. 10.1099/00222615-47-7-591 [DOI] [PubMed] [Google Scholar]
- Ng J., Hirota S.A., Gross O., Li Y., Ulke-Lemee A., Potentier M.S., Schenck L.P., Vilaysane A., Seamone M.E., Feng H., et al. 2010. Clostridium difficile toxin-induced inflammation and intestinal injury are mediated by the inflammasome. Gastroenterology. 139:542–552: 552.e1–552.e3. 10.1053/j.gastro.2010.04.005 [DOI] [PubMed] [Google Scholar]
- Ouyang W., and Valdez P.. 2008. IL-22 in mucosal immunity. Mucosal Immunol. 1:335–338. 10.1038/mi.2008.26 [DOI] [PubMed] [Google Scholar]
- Pickert G., Neufert C., Leppkes M., Zheng Y., Wittkopf N., Warntjen M., Lehr H.A., Hirth S., Weigmann B., Wirtz S., et al. 2009. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 206:1465–1472. 10.1084/jem.20082683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea M.C., Dobson A., O’Sullivan O., Crispie F., Fouhy F., Cotter P.D., Shanahan F., Kiely B., Hill C., and Ross R.P.. 2011. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc. Natl. Acad. Sci. USA. 108(Suppl 1):4639–4644. 10.1073/pnas.1001224107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M.L., and Sokol H.. 2019. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 16:331–345. 10.1038/s41575-019-0121-2 [DOI] [PubMed] [Google Scholar]
- Rodriguez C., Taminiau B., Van Broeck J., Delmée M., and Daube G.. 2015. Clostridium difficile infection and intestinal microbiota interactions. Microb. Pathog. 89:201–209. 10.1016/j.micpath.2015.10.018 [DOI] [PubMed] [Google Scholar]
- Sadighi Akha A.A., McDermott A.J., Theriot C.M., Carlson P.E. Jr., Frank C.R., McDonald R.A., Falkowski N.R., Bergin I.L., Young V.B., and Huffnagle G.B.. 2015. Interleukin-22 and CD160 play additive roles in the host mucosal response to Clostridium difficile infection in mice. Immunology. 144:587–597. 10.1111/imm.12414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenberg J.E., and Maslowsky J.. 2009. Taking substance use and development seriously: Developmentally distal and proximal influences on adolescent drug use. Monogr. Soc. Res. Child Dev. 74:121–130. 10.1111/j.1540-5834.2009.00544.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Lee J.S., Gilfillan S., Robinette M.L., Newberry R.D., Stappenbeck T.S., Mack M., Cella M., and Colonna M.. 2015. Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. J. Exp. Med. 212:1869–1882. 10.1084/jem.20151403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg J.L., and Sonnenburg E.D.. 2019. Vulnerability of the industrialized microbiota. Science. 366:eaaw9255 10.1126/science.aaw9255 [DOI] [PubMed] [Google Scholar]
- Sovran B., Hugenholtz F., Elderman M., Van Beek A.A., Graversen K., Huijskes M., Boekschoten M.V., Savelkoul H.F.J., De Vos P., Dekker J., and Wells J.M.. 2019. Age-associated Impairment of the Mucus Barrier Function is Associated with Profound Changes in Microbiota and Immunity. Sci. Rep. 9:1437 10.1038/s41598-018-35228-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B., and Hardt W.D.. 2008. The role of microbiota in infectious disease. Trends Microbiol. 16:107–114. 10.1016/j.tim.2007.12.008 [DOI] [PubMed] [Google Scholar]
- Takatori H., Kanno Y., Watford W.T., Tato C.M., Weiss G., Ivanov I.I., Littman D.R., and O’Shea J.J.. 2009. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J. Exp. Med. 206:35–41. 10.1084/jem.20072713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., McKenzie C., Potamitis M., Thorburn A.N., Mackay C.R., and Macia L.. 2014. The role of short-chain fatty acids in health and disease. Adv. Immunol. 121:91–119. 10.1016/B978-0-12-800100-4.00003-9 [DOI] [PubMed] [Google Scholar]
- Theriot C.M., Koenigsknecht M.J., Carlson P.E. Jr., Hatton G.E., Nelson A.M., Li B., Huffnagle G.B., Z Li J., and Young V.B.. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 5:3114 10.1038/ncomms4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thio C.L., Chi P.Y., Lai A.C., and Chang Y.J.. 2018. Regulation of type 2 innate lymphoid cell-dependent airway hyperreactivity by butyrate. Allergy Clin. Immunol, 10.1016/j.jaci.2018.02.032 [DOI] [PubMed] [Google Scholar]
- van Nood E., Vrieze A., Nieuwdorp M., Fuentes S., Zoetendal E.G., de Vos W.M., Visser C.E., Kuijper E.J., Bartelsman J.F., Tijssen J.G., et al. 2013. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368:407–415. 10.1056/NEJMoa1205037 [DOI] [PubMed] [Google Scholar]
- Vieira A.T., Galvão I., Macia L.M., Sernaglia E.M., Vinolo M.A.R., Garcia C.C., Tavares L.P., Amaral F.A., Sousa L.P., Martins F.S., et al. 2017. Dietary fiber and the short-chain fatty acid acetate promote resolution of neutrophilic inflammation in a model of gout in mice. J. Leukoc. Biol. 101:275–284. 10.1189/jlb.3A1015-453RRR [DOI] [PubMed] [Google Scholar]
- Vinolo M.A., Ferguson G.J., Kulkarni S., Damoulakis G., Anderson K., Bohlooly-Y M., Stephens L., Hawkins P.T., and Curi R.. 2011. SCFAs induce mouse neutrophil chemotaxis through the GPR43 receptor. PLoS One. 6:e21205 10.1371/journal.pone.0021205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N.J., Mebius R.E., et al. 2018. Innate Lymphoid Cells: 10 Years On. Cell. 174:1054–1066. 10.1016/j.cell.2018.07.017 [DOI] [PubMed] [Google Scholar]
- Voth D.E., and Ballard J.D.. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18:247–263. 10.1128/CMR.18.2.247-263.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Yang J., Gao W., Li L., Li P., Zhang L., Gong Y.N., Peng X., Xi J.J., Chen S., et al. 2014. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 513:237–241. 10.1038/nature13449 [DOI] [PubMed] [Google Scholar]
- Zmora N., Suez J., and Elinav E.. 2019. You are what you eat: diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 16:35–56. 10.1038/s41575-018-0061-2 [DOI] [PubMed] [Google Scholar]