ncRNA-encoded polypeptide plays a big role in triple-negative breast cancer.
Abstract
A new concept is emerging in the non-coding RNA (ncRNA) field: an increasing number of ncRNAs in fact codify for short peptides that have biological activities. In this issue of JEM, Wang et al. (https://doi.org/10.1084/jem.20190950) report the identification of a long ncRNA (lncRNA)–encoded 60–amino acid polypeptide that they name ASRPS, as well as its ability to inhibit angiogenesis in the deadly triple-negative breast cancer (TNBC).
Non-coding RNAs (ncRNAs) have a novel and additional property: they can code for short peptides with regulatory ability. As for the name, ncRNAs were considered to be lacking in the ability to code for proteins, but to have important gene expression regulatory function in both healthy and cancer cells. In the early 2000s, the identification of transcripts in cancer cells that did not codify for proteins but did regulate protein expression took the cancer community by storm (Calin et al., 2002). For decades, it was supposed that targeting specific proteins at the center of cancer hallmarks could represent the big therapeutic advance able to significantly reduce cancer mortality (Wood et al., 2007). However, increasing evidence shows that altered expression levels of ncRNA play a relevant role in cancer cell biology, are associated with poor clinical outcome of cancer patients, and may represent valuable targets for novel and more effective cancer therapies.

Insights from Simone Anfossi and George A. Calin.
In this issue of JEM, Wang et al. report on an important study based on a screening specifically designed to identify differentially expressed long ncRNAs (lncRNAs) that have protein-coding potential in triple-negative breast cancer (TNBC), the breast cancer subtype with the poorest clinical outcome. They used eight different assays to identify and confirm the existence of the 60–amino acid-long peptide ASRPS as a transcript of the lncRNA LINC00908. The experimental approach included the following: screening of sequences encoding for open reading frames (ORFs) using ORFFinder; ORF concordance with the peaks of the ribosome profiling data from the GWIPS-viz database; the cloning of putative ORFs in pcDNA3.1 tagged with FLAG for Western blot identification; the use of a plasmid with the fusion of GFPmut ORF (mutated ATG start codon) at the C-terminus of ASRPS to determine if the in-frame starting codon of ASRPS was functional; the polysome profiling for the identification of the endogenous expression of ASRPS; in situ hybridization for both micropeptide and lncRNA to detect co-expression; and the production of a rabbit polyclonal antibody specific for ASRPS for identification by blot. Finally, to prove that ASRPS is not a by-product resulting from the processing of a longer protein, the authors used antisense oligos to specifically block the lncRNA ORF translation. This is a truly extensive research work for the identification of a micropeptide and strengthens the confidence of the reader on the newly identified short peptide.
Interestingly, low expression of ASRPS was associated with poor survival in two independent large cohorts of TNBC patients, and the LINC00908 inhibited TNBC tumor growth mainly through the micropeptide. By performing a large set of genomic and mechanistic studies, Wang et al. (2019) identified that the estrogen receptor alpha directly regulates the transcription of lncRNA and accordingly the micropeptide, which explains their specific low expression in TNBC, but not in non-TNBC samples. STAT3–IL-6–VEGF signaling is an essential cancer pathway that is downstream of ASRPS. Using overexpression and knock-down experiments, the authors proved that ASRPS regulates VEGF expression via direct binding to the STAT3 coiled-coil domain with consequent inhibition of auto-phosphorylation and repression of tumor angiogenesis. The lncRNA LINC00908 did not play any role in this regulatory pathway and had no effect on angiogenesis, as demonstrated by a mutation in the start codon ATG in the lncRNA (no ASRPS translation) and pull-down assay (no direct interaction between LINC00908 and ASRPS). MMTV-PyMT mouse mammary tumor model proved that transgenic expression of ASRPS inhibited breast cancer angiogenesis and the intratumoral injection of ASRPS significantly improved survival of mice in TNBC mouse xenograft model.
These results are important, as they introduce the paradigm of micropeptides codified by lncRNAs in TNBC, their expression regulation by the estrogen receptor pathway, and the production of an angiogenesis-regulating micropeptide. All these represent new findings that have led to an exciting therapeutic potential, such as the development of ASRPS-based anti-angiogenesis for TNBC therapy. Such advances are of interest due to the lack of specific delivery strategies and the pleiotropic effects of targeting ncRNAs through RNA mimics or RNA sponges. Furthermore, unlike microRNAs, micropeptides do not potentially bind and activate Toll-like receptors with consequent induction of the cytokine storm and disastrous effects especially in metastatic cancer patients with poor kidney and liver function. Given their very small size, the micropeptides can easily shuttle among cell compartments and be involved in cell-to-cell communication through extracellular vesicles. Therefore, the use of tumor suppressor micropeptides could be a safer approach to be further developed.
The research on ncRNAs is progressing rapidly, with an increasing number of new molecules identified. Indeed, advances in bioinformatics and deep sequencing technology have allowed the cloning and annotation of numerous short and long ncRNAs (microRNAs, PIWI-interacting RNAs, small nucleolar RNAs, transfer RNA–derived small RNAs, endogenous small interfering RNAs, enhancer ncRNAs, natural antisense transcripts, circular RNAs, long intergenic ncRNAs, transcribed ultraconserved regions, or primate-specific pyknon transcripts) that have exceeded the number of protein-coding transcripts (Hon et al., 2017; Mattick, 2018; Lee et al., 2009; Rigoutsos et al., 2017). This plethora of transcripts are codified from the non-exonic part of the genome that accounts for up to 97% of the human genome. An exponentially growing number of papers describes their functions in each cancer hallmark, along with their potential as biomarkers and therapeutic targets (Schmitt and Chang, 2016). Still, a fundamental question has to be answered: if proteins composed by the combinations of twenty different amino acids have notably higher numbers of versatile functions, such as the more diverse enzymatic and structural features, compared with RNAs, then why does almost all of the human genome codify for ncRNAs instead of proteins?
Answers to this question are not straightforward, as we are still at the beginning of understanding the functionality of the “dark matter” of the human genome. Yet, quite unexpected findings were revealed over the last years: several ncRNAs (including lncRNAs or microRNA precursors) codify for small peptides, also named micropeptides (defined as peptides with less than 100 amino acids; Makarewich and Olson, 2017). Most of them were discovered by chance and in non-cancer models some begin with the ATG start codon whereas others do not, and the length ranges may vary considerably (Table 1). These peptide sequences were described under various names, including small peptides, micropeptides, miPEP, and others, proving the need of nomenclature self-organization for this new field that stands at the junction between transcriptomics and proteomics. One possibility is to use the term “micropeptides” for the short peptides under 100 amino acids long codified by ncRNA loci, and “small peptides” for peptides under 100 amino acids codified by protein-coding loci. Many small peptides are known to have important functional effects, but they derive from the cleavage of longer amino acidic sequences from proteins codified by classic coding genes rather than coded by ncRNAs. This is the case of the 28–amino acid–long vasoactive intestinal peptide (VIP) cleaved from a much longer precursor molecule, prepro-VIP, codified by a locus on the chromosomal region 6q24, and exerting a plethora of functions including vasodilatory effects or contribution to associative learning (Krabbe et al., 2019).
Table 1. Examples of micropeptides codified by human lncRNAs.
| Name (abbreviation) | Number of amino acids | Function | lncRNA | Reference |
|---|---|---|---|---|
| Myoregulin (MRLN) | 46 | Regulates Ca2+ handling by inhibiting the pump activity of SERCA | LINC00948 | Anderson et al., 2015 |
| Small regulatory polypeptide of amino acid response (SPAR) | 90 | Regulates mTORC1 and muscle regeneration | LINC00961 | Matsumoto et al., 2017 |
| Non-annotated P-body dissociating polypeptide (NOBODY) | 68 | Interacts with mRNA-decapping proteins, which are the molecular components of the first enzymatic step in the 5′-to-3′ mRNA decay pathway | LINC01420 | D’Lima et al., 2017 |
| HOXB cluster antisense RNA 3 peptides (HOXB-AS3 peptide) | 53 | Suppresses colon cancer growth; competitively binds to the arginine residue in RGG motif of hnRNP A1 antagonizing its regulation of pyruvate kinase splicing; low levels correlate with poor prognosis | HOXB-AS3 | Huang et al., 2017 |
| Exon 2 of LINC-PINT endogenous circRNA (PINT87aa) | 87 | Suppresses glioblastoma cell proliferation by interaction with polymerase associated factor complex and inhibition of transcriptional elongation of multiple oncogenes | Circular form of LINC-PINT | Zhang et al., 2018 |
SECA, sarcoplasmic reticulum Ca2+-ATPase; hnRNP, heterogeneous nuclear ribonucleoprotein; circRNA, circular RNA.
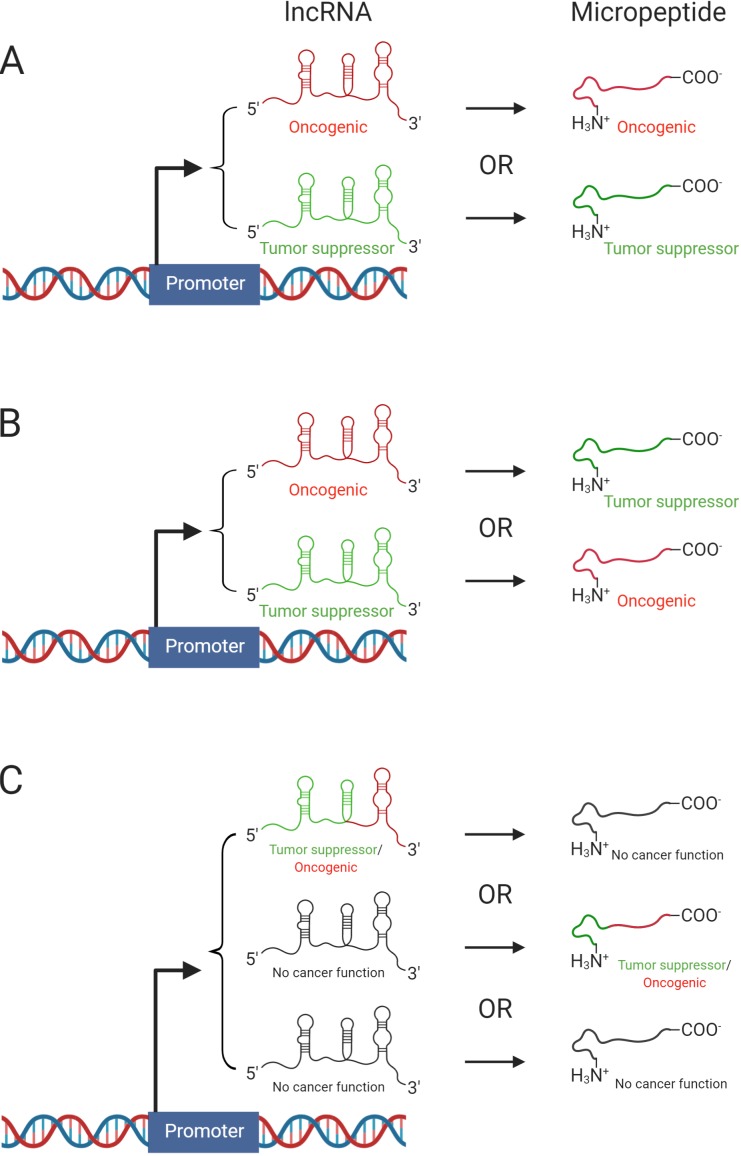
The “duo” cancer effects of the tandem lncRNA-encoding micropeptide and the micropeptide itself. Synergistic effects (A); antagonistic effects (B); and no functional overlap (C). Created with BioRender.com.
A lot remains to be learned. What is the proportion of micropeptides that do not have the classic ATG starting codon? The more such micropeptides exist in nature, the more difficult it will be to identify the sequences they are coded from using ORF-related approaches. How short could these micropeptides be to keep their functionality? Proteins as small as a few tens of amino acids were identified to be functional, so, by analogy, ncRNA-derived micropeptides of 20–30 amino acids in length could potentially be functional too. A better understanding of the relationship between the host lncRNAs and the codified micropeptides is needed. There are three potential situations: a synergistic effect with both the micropeptide and lncRNA working as oncogenes or as tumor suppressors; an antagonistic effect between the two types of transcripts, one tumor suppressor and the other oncogenic; or no functional overlap. The latter is the case for ASRPS and LINC00908, and this could be an important advantage for developing ASRPR restitution therapeutics. Such innovative therapeutic approaches can be expanded from cancer to any type of disease in which such micropeptides are identified, and this is another reason why the study of Wang et al. (2019) is relevant. This is an exciting time in cancer genomics and biology; after the era of protein dominance followed by the focus on non-coding transcripts, we can now contribute to the unexpected rise of the micropeptide era.
References
- Anderson D.M., et al. Cell. 2015 doi: 10.1016/j.cell.2015.01.009. [DOI] [Google Scholar]
- Calin G.A., et al. Proc. Natl. Acad. Sci. USA. 2002 doi: 10.1073/pnas.242606799. [DOI] [Google Scholar]
- D’Lima N.G., et al. Nat. Chem. Biol. 2017 doi: 10.1038/nchembio.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C.C., et al. Nature. 2017 doi: 10.1038/nature21374. [DOI] [Google Scholar]
- Huang J.Z., et al. Mol. Cell. 2017 doi: 10.1016/j.molcel.2017.09.015. [DOI] [Google Scholar]
- Krabbe S., et al. Nat. Neurosci. 2019 doi: 10.1038/s41593-019-0508-y. [DOI] [Google Scholar]
- Lee Y.S., et al. Genes Dev. 2009 doi: 10.1101/gad.1837609. [DOI] [Google Scholar]
- Makarewich C.A., and Olson E.N. Trends Cell Biol. 2017 doi: 10.1016/j.tcb.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A., et al. Cell Cycle. 2017 doi: 10.1080/15384101.2017.1304735. [DOI] [Google Scholar]
- Mattick J.S. Noncoding RNA. 2018 [Google Scholar]
- Rigoutsos I., et al. Genome Biol. 2017 doi: 10.1186/s13059-017-1224-0. [DOI] [Google Scholar]
- Schmitt A.M., and Chang H.Y. Cancer Cell. 2016 doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., et al. J. Exp. Med. 2019 doi: 10.1084/jem.20190950. [DOI] [Google Scholar]
- Wood L.D., et al. Science. 2007 doi: 10.1126/science.1145720. [DOI] [Google Scholar]
- Zhang M., et al. Nat. Commun. 2018 doi: 10.1038/s41467-018-06862-2. [DOI] [Google Scholar]