Abstract
Background & Aims:
Functions of intestinal stem cells (ISCs) are regulated by diet and metabolic pathways. Hepatocyte nuclear factor 4 (HNF4) are transcription factors that bind fatty acids. We investigated how HNF4 transcription factors regulate metabolism and their functions in ISCs in mice.
Methods:
We performed studies with Villin-CreERT2, Lgr5-EGFP-IRES-CreERT2, Hnf4af/f, and Hnf4gCrispr/Crispr mice. Villin-CreERT2; Lgr5-EGFP-IRES-CreERT2; Hnf4af/f; Hnf4gCrispr/Crispr mice are hereafter referred to Hnf4agDKO. Mice were given tamoxifen to induce Cre recombinase. Mice transgenic with only Cre alleles (Villin-CreERT2, Lgr5-EGFP-IRES-CreERT2, Hnf4a+/+, and Hnf4g+/+) or mice given vehicle were used as controls. Crypt and villus cells were isolated, incubated with fluorescently labeled fatty acid or glucose analog, and analyzed by confocal microscopy. Fatty acid oxidation activity and tricarboxylic acid (TCA) metabolites were measured in cells collected from the proximal half of the small intestine of Hnf4agDKO and control mice. We performed chromatin immunoprecipitation and gene expression profiling analyses to identify genes regulated by HNF4 factors. We established organoids from duodenal crypts, incubated them with labeled palmitate or acetate, and measured production of TCA metabolites or fatty acids. Acetate, a precursor of acetyl-CoA (a product of fatty acid beta-oxidation [FAO]), or dichloroacetate, a compound that promotes pyruvate oxidation and generation of mitochondrial acetyl-CoA, were used for metabolic intervention.
Results:
Crypt cells rapidly absorbed labeled fatty acid, and mRNA levels of Lgr5+ stem cell markers (Lgr5, Olfm4, Smoc2, Msi1, and Ascl2) were downregulated in organoids incubated with etomoxir, an inhibitor of FAO, indicating that FAO was required for renewal of ISCs. HNF4A and HNF4G were expressed in ISCs and throughout the intestinal epithelium. Single knockout of either HNF4A or HNF4G did not affect maintenance of ISCs, but double-knockout of HNF4A and HNF4G resulted in ISC loss; stem cells failed to renew. FAO supports ISC renewal, and HNF4 transcription factors directly activate FAO genes, including Acsl5 and Acsf2 (encode regulators of acyl-CoA synthesis), Slc27a2 (encodes a fatty acid transporter), Fabp2 (encodes fatty acid binding protein), and Hadh (encodes hydroxyacyl-CoA dehydrogenase). In the intestinal epithelium of Hnf4agDKO mice, expression levels of FAO genes, FAO activity, and metabolites of TCA were all significantly decreased, but fatty acid synthesis transcripts were increased, compared with control mice. The contribution of labeled palmitate or acetate to the TCA cycle was reduced in organoids derived from Hnf4agDKO mice, compared with control mice. Incubation of organoids derived from double-knockout mice with acetate or dichloroacetate restored stem cells.
Conclusions:
In mice, the transcription factors HNF4A and HNF4G regulate expression of genes required for fatty acid oxidation and are required for renewal of intestinal stem cells.
Keywords: regulatory networks, stemness, mitochondria, gene expression
Graphical Abstract
Lay Summary:
We identified proteins that regulate metabolic processes required for intestinal stem cell renewal and replacement of the intestinal epithelium as cells die.
Introduction
The intestinal epithelium has the capacity to self-renew and generate differentiated cells. Its turnover occurs every 3–5 days, fueled by an actively cycling stem cell population characterized by Lgr5 expression1. Intestinal stem cells (known as Lgr5+ crypt base columnar cells or ISCs) and their proliferating progeny are nestled into cup-like structures termed crypts, while luminal projections called villi consist entirely of differentiated cells. Lgr5+ ISCs function as the primary cell of origin of colon tumors2, are essential for intestinal regeneration following injury3, and function dynamically in response to different nutritional environments.
Nutritional regulation and metabolic control have profound roles in stem cell biology4–8. Studies have shown that ISCs respond to caloric restriction9, 10 and dietary nutrients including fatty acids11, glucose12, N-acetyl-D-glucosamine13 and amino acids14, 15. Work from our lab16, 17 and others18 have revealed that Lgr5+ cells are particularly dependent upon mitochondrial metabolism, and others have suggested that fatty acid oxidation could fuel oxidative phosphorylation in ISCs10. These studies demonstrate that cellular metabolism is a fundamental regulator of intestinal stem cell homeostasis. However, it remains unclear how the unique metabolic state of intestinal stem cells is established.
Since fatty acid β-oxidation (FAO) is crucial for stem cell maintenance in multiple tissues10, 19–22, there is a clear imperative to understand the regulatory mechanisms through which ISCs implement FAO for self-renewal. Hepatocyte nuclear factor 4 (HNF4) factors have a conserved ligand-binding domains that are constitutively bound to fatty acids23–26. HNF4A is an important regulator of metabolic homeostasis, and transcribes genes involved in hepatic lipid metabolism27–29. Genes associated with acyl-CoA metabolism were identified as putative HNF4A targets28, and other studies have also shown that HNF4A binds acyl-CoA-binding proteins and fatty acyl-CoA thioesters30, 31. In Drosophila, dHNF4 is responsive to dietary signals, functions as a sensor for free fatty acids, and is essential for lipid catabolism and β-oxidation for energy production32.
The intestinal stem cell has been implicated in intestinal disease and cancer. Understanding the regulatory mechanisms controlling intestinal stem cell physiology is of great importance to human health and regenerative medicine33–37. However, the mechanisms controlling ISC metabolism are poorly understood. Given the potential contribution of FAO in stem cell maintenance10, 19–22 and the importance of HNF4 in regulating lipid metabolic processes32, 38, 39, we investigated how HNF4 transcription factors impact ISC metabolism, and whether HNF4-dependent metabolic regulation is required for ISC maintenance.
Methods
Additional experimental details on cell isolation, histology and immunostaining, western blot, RNA extraction and qRT-PCR, organoid culture, counting and immunofluorescence, FAO activity assay, fatty acid extraction and saponification, LC-MS analysis, and bioinformatics can be found in supplemental materials.
Mice
The Villin-CreERT2 transgene40, Lgr5-EGFP-IRES-CreERT2 knock-in1, Hnf4αf/f38 and Hnf4ɣCrispr/Crispr41 alleles were integrated to generate the conditional compound-mutants and controls. Experimental mice (8–12 weeks old) were treated with tamoxifen (Sigma T5648) at 50mg/kg/day or vehicle by intraperitoneal injection. To avoid the non-specific phenotype caused by tamoxifen on Villin-CreERT2 models42, tamoxifen-induced Cre-only controls were also tested in this study. All mouse protocols and experiments were approved by the Rutgers Institutional Animal Care and Use Committee. All samples were collected between 12:00 and 14:00 to avoid circadian variability.
Fatty acid and glucose analog uptake
Crypt and villus cells were freshly isolated from mouse intestine as described above. Cells were incubated with different durations (0, 2, 5, 30 and 60 min, in dark, on ice) of fluorescent BODIPY™ FL C16 fatty acid (4,4-Difluoro-5,7-Dimethyl-4-Bora-3a,4a-Diaza-s-Indacene-3-Hexadecanoic Acid, also known as palmitic acid, 10 μM, Invitrogen D3821), BODIPY™ FL C12 fatty acid (4,4-Difluoro-5-(2-Thienyl)-4-Bora-3a,4a-Diaza-s-Indacene-3-Dodecanoic Acid, 10 μM, Invitrogen D3835) or 2-NBDG (2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose, 10 μM, Invitrogen N13195) on a rocker. There were no obvious differences of fatty acid/glucose uptake when we compared the incubation condition on ice and at room temperature. To increase viability of isolated crypt and villus cells, cells were incubated with BODIPY™ FL C16, FL C12, or 2-NBDG on ice in this study. Trypan blue staining was used to verify cell viability throughout the experimental timecourse. After incubation, live cells were washed with PBS twice, and then stained with DraQ5 (Cell signaling 4084L) at room temperature for 5 min. Live cells were mounted and viewed on a Zeiss LSM 510 Meta confocal microscope. Only intact intestinal crypt or villus units were used to measure uptake. We noted that fixation can quench the fluorescence of fatty acid with short-term incubation (2 min and 5 min) but works fine for the long-term incubation (30 min and 60 min). When fixation was used to maintain cell morphology for next day imaging, cells were fixed with 4% paraformaldehyde at room temperature for 20 min, washed with PBS for twice, and then stained with DAPI (Biotium 40043, 1:5000) at room temperature for 5 min. After nuclear staining, cells were washed with PBS, resuspended in fluorescent mounting medium, and mounted for subsequent imaging. All images comparing tissues were acquired at the same time and using identical imaging conditions. ImageJ was used to quantify the uptake of fluorescent fatty acid/glucose analog.
TCA metabolite extraction
Villus cells were scraped and collected from the proximal half of the small intestine of Hnf4αɣDKO mice (3 days post tamoxifen injection) and their littermate controls. Cells were resuspended in ice cold extraction buffer (40:40:20 methanol:acetonitrile:H2O with 0.5% V/V formic acid). 500 μl of extraction buffer was added per 50 mg tissue, incubated on ice for 5 min, and followed by adding 25 μl 15% (m/V) NH4HCO3. Samples were centrifuged at 15000 g for 10 min at 4°C to pellet cell debris and proteins. The supernatant was transferred into a phospholipid removal column (Phenomenex 8B-S133-TAK) for further cleanup, and then stored at −80°C for subsequent LC-MS analysis. Tissue weight was used to normalize the corrected ion counts of TCA metabolites.
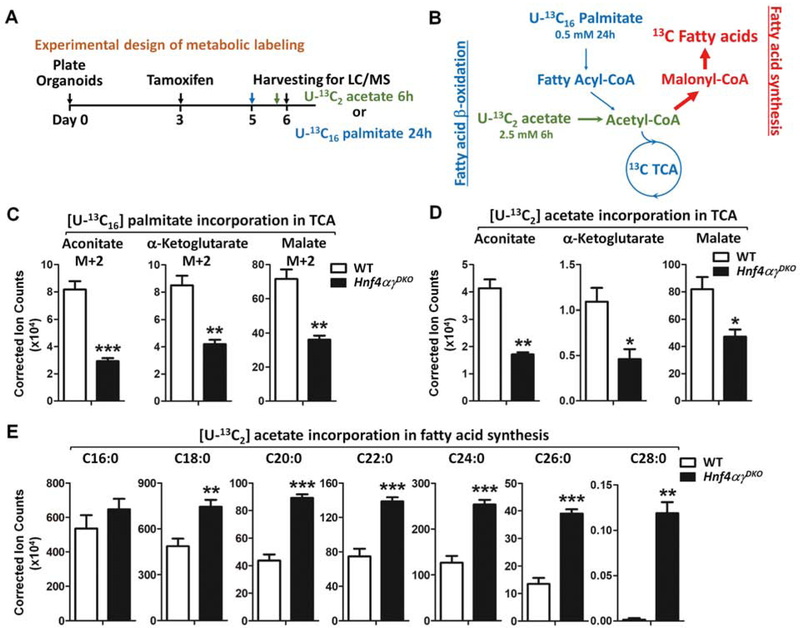
For U-13C16 palmitate organoid labeling experiments, tamoxifen (1 μM) or vehicle was added to culture medium for 12 hours on Day 3 after seeding. On Day 5, organoids were treated with 0.5 mM U-13C16 palmitate (Sigma 605573) and 1 mM carnitine (Sigma C0158) for 24 hours prior to harvesting. For U-13C2 acetate organoid labeling experiments, tamoxifen (1 μM) or vehicle was added to culture medium for 12 hours on Day 3 after seeding. On Day 6, organoids were treated with 2.5 mM U-13C2 acetate (1,2-13C2, Cambridge Isotope Laboratories, Inc. CLM-440–1) for 6 hours prior to harvesting. 250 μl of extraction buffer was used to extract metabolites from 10 mg of organoid pellets (10 μl pellet size ≈ 10 mg), and followed the same protocol as descried above. Protein concentration was determined by Pierce BCA Protein Assay Kit (Thermo), and was used to normalize the corrected ion counts of TCA metabolites.
Statistical analysis
The data are presented as mean ± SEM, and statistical comparisons were performed using one-way ANOVA followed by Dunnett’s post test with the GraphPad Prism version 7.02 or Student’s t-test at P < 0.001***, P < 0.01** or P < 0.05*. Bioinformatics-related statistical analysis was done with the embedded statistics in each package, including GSEA43, Cuffdiff44 and HOMER45. P < 0.05 or FDR < 0.05 (95% confidence interval) was considered statistically significant.
Results
Disruption of FAO impairs self-renewal of ISCs
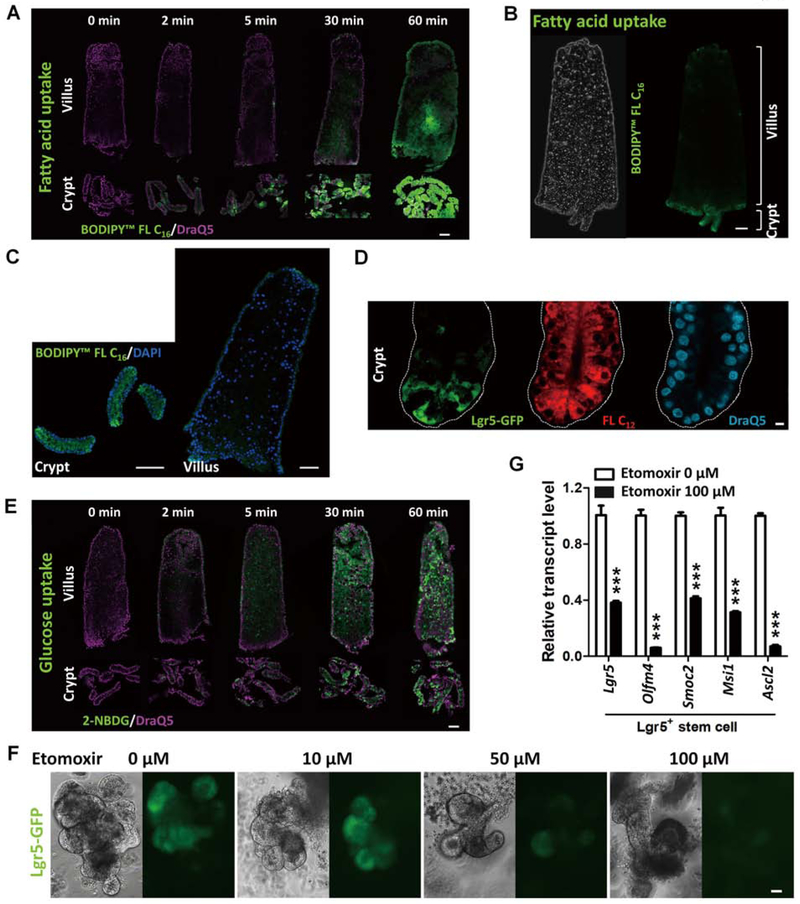
Studies have begun to delineate the impact of fatty acids on intestinal stem cell function10, 11, yet the capacity of crypt cells to import fatty acids is unclear. Palmitic acid accounts for 20–30% of total fatty acids in humans, and can be attained from the diet or synthesized endogenously46. To evaluate fatty acid uptake, we isolated crypt and villus epithelial cells from mouse intestine and incubated them with fluorescent palmitic acid. Crypt cells rapidly absorbed labeled palmitic acid within 2 minutes of incubation (Figures 1A–D), and fluorescent intensity increased over time (Figure 1A and S1A). By comparison, villus cells absorbed the labeled fatty acids more slowly, lacking fluorescent signal until 30–60 minutes (Figure 1A and S1B). By contrast, a fluorescent glucose analog was absorbed by the villus compartment within 5 minutes of incubation (Figure 1E and S1C). Reports of treatment of Lgr5+ stem cells with glycolysis inhibitors showed a mild effect in organoid reconstitution18, thus leaving open a possible role for fatty acids in ISC energy metabolism. To investigate fatty acid uptake of ISCs, crypts were isolated from Lgr5-EGFP-IRES-CreERT2 knock-in1 mice and incubated with BODIPY™ fatty acid. Red fluorescent labeled dodecanoic acid was indeed observed in Lgr5-GFP+ stem cells (Figures 1D), as well as in other crypt cells. To further investigate the functional requirement of FAO in ISCs, Etomoxir, a FAO inhibitor, was applied using the well-established intestinal organoid assay47. Lgr5-GFP+ stem cells were observed in the budding crypt domains of organoids, as expected. However, upon treatment with the FAO inhibitor, Lgr5-GFP expression diminished in a dose-dependent manner (Figure 1F). The transcript levels of Lgr5+ stem cell markers, including Lgr5, Olfm4, Smoc2, Msi1 and Ascl2, were also dramatically downregulated with FAO inhibition by Etomoxir (Figure 1G), suggesting that FAO is required for self-renewal of ISCs10.
Figure 1. FAO is required for intestinal stem cell renewal.
(A-E) Intestinal crypts uptake fatty acid more readily than villi (n=3 independent experiments). Crypt and villus epithelial cells were isolated from the proximal half of the small intestine (from the midpoint between gastroduodenal junction to ileal-cecal junction) and incubated with 10 μM BODIPY™ FL C16 (fluorescent palmitic acid), FL C12 (fluorescent dodecanoic acid), or 10 μM 2-NBDG (fluorescent glucose analog), respectively. Similar results are seen in duodenum, jejunum and ileum (Fig. S1B). (A) Time-course study of fluorescent fatty acid (BODIPY™ FL C16) uptake shows that intestinal crypt cells uptake more fatty acid than villus cells (scale bar, 50 μm). (B-C) Fatty acid (green fluorescence) accumulates more in the crypt cells than in villus cells (scale bars, 50 μm). (D) Red fluorescent fatty acids (10 μM BODIPY™ FL C12, 5 min) are observed in Lgr5-GFP stem cells in the crypts (scale bar, 5 μm). (E) Glucose uptake in crypt and villus cells (scale bar, 50 μm). (F) Fewer Lgr5-GFP cells are observed in organoids treated with Etomoxir (fatty acid oxidation inhibitor) for 72 hours (n=3 independent experiments). (G) qRT-PCR shows a significant decrease in the transcript levels of intestinal Lgr5+ stem cell markers in the organoids treated with 100 μM Etomoxir for 72 hours. Data are presented as mean ± SEM (n=3–4 independent organoid cultures per treatment, Student’s t-test, two-sided at P < 0.001***).
HNF4 factors directly activate FAO genes and support β-oxidation in the intestinal epithelium
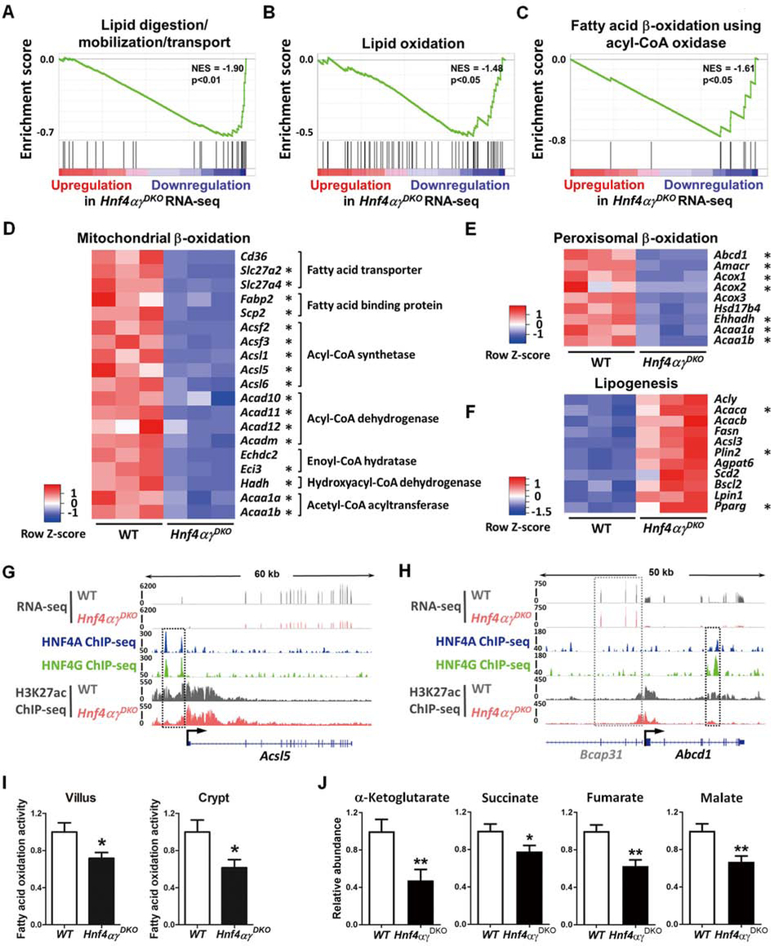
HNF4A has been implicated in fatty acid metabolism, with severe lipid deposits accumulating upon conditional loss of Hnf4α in the liver38. Compared to the liver, loss of function studies of HNF4A in the intestine show relatively minor phenotypes48–51. We recently demonstrated that the transcription factor HNF4A acts redundantly with an intestine-restricted HNF4 paralog, HNF4G, to activate enhancer chromatin and stabilize enterocyte cell identity41. Expression profiling of Hnf4αɣDKO (Hnf4αf/f; Villin-CreERT2; Hnf4ɣCrispr/Crispr) mice indicated that the top gene ontology categories of HNF4-dependent genes are associated with lipid metabolism41. GSEA indicates that genes associated with lipid digestion and transport (Figure 2A), as well as fatty acid oxidation (Figure 2B and 2C), are all significantly downregulated in Hnf4αɣDKO. FAO, also termed β-oxidation, refers to the sequential removal of 2-carbon units from acyl chains by oxidation at the β-carbon position. The TCA cycle utilizes acetyl-CoA derived from FAO. Together, these processes generate NADH and FADH2, which fuel oxidative phosphorylation to generate ATP. In mammals, FAO occurs in both mitochondria and peroxisomes, and transcript levels of genes that function in virtually every step in the process of FAO are compromised upon loss of HNF4 factors (Figure 2D and 2E). Conversely, transcripts associated with lipogenesis are elevated in Hnf4αɣDKO epithelium (Figure 2F). Notably, genes promoting FAO are more likely to have nearby HNF4-binding sites than lipogenesis genes based upon ChIP-seq analysis (MACS P ≤ 10−3, HNF4 direct targets are indicated by asterisks in Figure 2D–F). For example, acyl-CoA synthetases catalyze the initial reaction in fatty acid metabolism by forming a thioester bond between a fatty acid and coenzyme A, and HNF4A and HNF4G directly bind to the acyl-CoA synthetase genes Acsl5 and Acsf2 (see HNF4 ChIP-seq tracks in Figure 2G and S2A respectively). HNF4 paralogs can also bind to other β-oxidation genes (Figure S2A), such as Slc27a2 (fatty acid transporter), Fabp2 (fatty acid binding) and Hadh (hydroxyacyl-CoA dehydrogenase). The protein encoded by Abcd1, a member of the superfamily of ATP-binding cassette transporters, is involved in peroxisomal import of very long chain fatty acids. In peroxisomal FAO, the first reaction is catalyzed by an acyl-CoA oxidase (ACOX), which is regarded as the main enzymatic step controlling flux through the pathway. We find HNF4A and HNF4G directly bind to Abcd1, Acox1 and other important peroxisomal FAO genes including Ehhadh (see HNF4 ChIP-seq tracks in Figure 2H and S2A). These mitochondrial and peroxisomal β-oxidation genes are robustly expressed in WT, but are dramatically downregulated in Hnf4αɣDKO (FDR < 0.05, see RNA-seq tracks in Figure 2G, 2H, and S2). Consistent with direct activation by HNF4, accessible chromatin structure is lost at HNF4-bound regions of these β-oxidation gene loci in Hnf4αɣDKO (see MNase-H3K27ac ChIP-seq tracks in Figure 2G, 2H, and S2A).
Figure 2. HNF4 paralogs bind to and activate FAO genes, and FAO is compromised upon HNF4 loss.
(A-C) GSEA of RNA-seq (duodenal epithelium, 2–3 days post tamoxifen-induced knockout, n=3 biological replicates, Kolmogorov-Smirnov test) reveals significantly reduced expression of genes related to lipid digestion and fatty acid β-oxidation in Hnf4αɣDKO. Heatmaps of RNA-seq data show (D-E) decreased transcript levels of mitochondrial and peroxisomal β-oxidation genes but (F) increased transcript levels of lipogenesis genes in Hnf4αɣDKO (n=3 biological replicates, FDR < 0.05). Asterisks indicate genes that are directly bound by HNF4 within 50kb of annotated TSSs by ChIP-seq. (G) ChIP-seq (n=2 biological replicates) and RNA-seq (n=3 biological replicates) tracks show that HNF4 factors bind to and activate Acsl5 (acyl-CoA synthetase gene), and loss of HNF4 results in reduced active chromatin signal (H3K27ac ChIP-seq, n=2 biological replicates, see black dashed rectangles). (H) HNF4 factors can also bind to and activate Abcd1 (peroxisomal β-oxidation related transporter). The neighboring gene locus is not bound by HNF4 and shows no change in expression levels, serving as an internal control (Bcap31, see gray dashed rectangles). More examples are shown in Fig. S2. (I) Hnf4αɣDKO mice show decreased FAO activity in both villus and crypt cells. The FAO assay measures palmitoyl-CoA-induced oxidation, as detected by NADH generation. Data are presented as mean ± SEM (villus, n=9 biological replicates; crypt, n= 3–5 biological replicates; Student’s t-test, two-sided at P < 0.05*). (J) TCA metabolites are reduced upon HNF4 loss. Metabolites were extracted from villus cells scraped from proximal small intestine of Hnf4αɣDKO and their WT littermates after 3 consecutive days of tamoxifen or vehicle injection, respectively. Corrected ion counts were normalized by tissue weight, and further normalized relative to the average abundance of metabolite from WT littermates of the same experimental batch. Data are presented as mean ± SEM (LC-MS, n=10 WT and 9 mutants, Student’s t-test, two-sided at P < 0.01** and P < 0.05*).
These findings suggest that HNF4 factors directly activate genes that contribute to fatty acid catabolism (summary schematic of results in Figure S2B). We therefore measured palmitoyl-CoA-induced enzymatic activity in cell extracts of Hnf4αɣDKO intestinal epithelium versus controls. Consistent with reduced levels of FAO gene expression, Hnf4αɣDKO epithelium exhibits impaired FAO activity compared to controls (Figure 2I). We hypothesized that compromised FAO upon HNF4 loss therefore leads to a subsequent reduction in TCA metabolites. Indeed, TCA metabolites of villus cells were reduced in Hnf4αɣDKO, indicated by LC-MS measurements of α-ketoglutarate, succinate, fumarate and malate (Figure 2J). In addition to fatty acid oxidation, acetyl-CoA can be generated by glycolysis, followed by pyruvate oxidation in the mitochondria. However, neither glycolysis pathway transcripts nor genes contributing to pyruvate oxidation are compromised upon HNF4 loss (Figure S3). Taken together, HNF4 paralogs bind to and activate FAO genes, and loss of HNF4 leads to compromised transcript levels of FAO genes, impaired palmitoyl-CoA dependent enzymatic activity, and reduced levels of TCA metabolites.
Loss of HNF4 paralogs in the intestinal epithelium triggers Lgr5+ stem cell loss
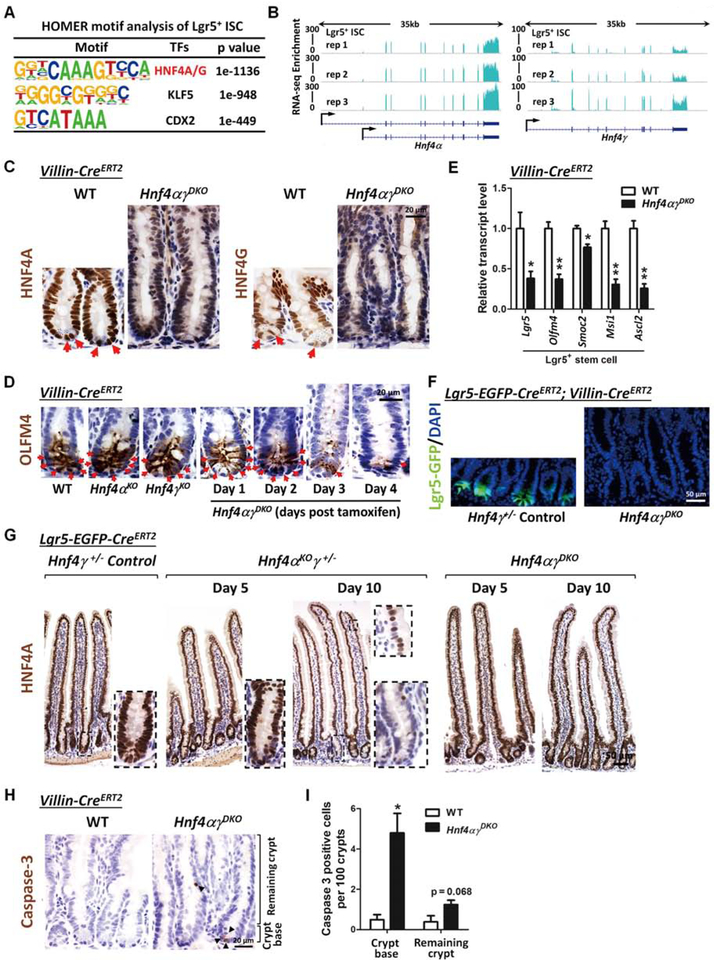
Since HNF4 factors appear to contribute strongly to β-oxidation, a process important for intestinal stem cell renewal10, we next explored whether HNF4 factors could be contributing to ISC function. We applied DNA-binding motif analysis to ATAC-seq data of isolated ISCs52 and found that the HNF4A/G motif is the top-scoring factor (Figure 3A), suggesting that HNF4 factors could function as broad regulators of gene expression in ISCs. RNA-seq data of ISCs52 demonstrate high Hnf4α and Hnf4ɣ expression in the Lgr5+ intestinal stem cells (Figure 3B). Immunoreactivity of HNF4A and HNF4G was observed throughout the intestinal epithelium, including in crypt base columnar stem cells (Figure 3C). On the Hnf4ɣ mutant background, induced deletion of Hnf4α throughout the intestinal epithelium using the Villin-CreERT2 driver (Figure S4A) triggers loss of stem cells within 4 days of tamoxifen treatment, as indicated by the absence of OLFM4-expressing cells at the base of crypts throughout the intestine (Figure 3D and S4B). However, single knockout of either HNF4A or HNF4G does not affect the maintenance of intestinal stem cells (Figure 3D), suggesting redundant functions of HNF4 paralogs in stem cell renewal. Reduced Lgr5+ stem cell transcripts, Lgr5, Olmf4, Smoc2, Msi1 and Ascl2, are observed in the crypts of Hnf4αɣDKO at day 2 post tamoxifen induced knockout (Figure 3E), and Hnf4αɣDKO mice had to be euthanized within 5 days due to compromised epithelial functions. To further monitor the Lgr5-expressing cell population, we generated mice in which we could both inactivate HNF4 throughout the epithelium and visualize the Lgr5-expressing stem cells with an Lgr5-GFP knock-in allele1: Lgr5-EGFP-IRES-CreERT2; Villin-CreERT2; Hnf4αf/f; Hnf4ɣCrispr/Crispr. Consistent with OLFM4 staining, there are no detectible Lgr5-GFP+ stem cells remaining in the Hnf4αɣDKO mice at day 4 after tamoxifen treatment (Figure 3F and S4C).
Figure 3. Loss of HNF4 paralogs in the intestinal epithelium triggers Lgr5+ stem cell loss.
(A) ATAC-seq of isolated intestinal stem cells reveals that HNF4A and HNF4G DNA-binding motifs (HOMER de novo) are most abundant at accessible chromatin regions of intestinal stem cells (GSE83394, n=2 biological replicates). (B) RNA-seq tracks of Hnf4α and Hnf4ɣ transcript levels in the intestinal stem cells (GSE83394, n=3 biological replicates) show robust expression of these paralogs. (C) Immunostaining of HNF4A and HNF4G in WT and Hnf4αɣDKO (representative of 3 biological replicates). (D) Immunostaining of OLFM4 (stem cell marker). Stem cells (red arrows) are lost in the crypts following Hnf4α and Hnf4ɣ double knockout. (E) qRT-PCR shows a significant decrease in the Lgr5+ stem cell markers in Hnf4αɣDKO. Crypts were isolated from duodenal epithelium of Hnf4αɣDKO and their WT littermates after 2 consecutive days of tamoxifen or vehicle injection, respectively. qPCR data are presented as mean ± SEM (n=3 biological replicates, Student’s t-test, two-sided at P < 0.01** and P < 0.05*). (F) Lgr5-GFP+ cells are observed in the crypt bottom of control mice, but are completely lost in Hnf4αɣDKO after 4 days of tamoxifen-induced knockout (representative of 3 biological replicates). (G) Immunostaining of HNF4A shows that Cre-mediated recombination of the Hnf4α locus is initiated via Lgr5-EGFP-CreERT2 ( a stem cell-specific Cre driver).However, most recombined HNF4A-negative cells are not preserved in the epithelium of the Hnf4αɣDKO mice (representative of 3 biological replicates), suggesting that Hnf4αɣDKO mutant stem cells are replaced by neighboring cells. (H) Immunostaining of cleaved Caspase-3 in WT and Hnf4αɣDKO. (I) Hnf4αɣDKO shows a significant increase of cleaved Caspase-3 postive cells in the crypt base of Hnf4αɣDKO compared to WT littermates. Data are presented as mean ± SEM (n=3–4 biological replicates, Student’s t-test, two-sided at P < 0.05*).
To test for a cell-autonomous role for HNF4 in stem cells, we inactivated HNF4 using the Lgr5-EGFP-CreERT2, a stem cell-specific Cre driver that is mosaically expressed in a subset of crypts1. HNF4A-negative cells are observed in the crypts and villi of Lgr5-EGFP-CreERT2; Hnf4αf/f; Hnf4ɣ+/− mice after 5 days and 10 days post tamoxifen treatment, indicating efficient knockout of HNF4A in the stem cells and their progeny. However, most HNF4 recombination is not preserved in Lgr5-EGFP-CreERT2; Hnf4αɣDKO mice, implying that the mutant stem cells are replaced by neighboring non-recombined healthy cells. These findings suggest that HNF4 factors are required in the ISCs in a cell-autonomous fashion (Figure 3G and S4D). We also find increased apoptosis in Hnf4αɣDKO (Figure 3H), particularly in the crypt base (Figure 3I), corresponding to loss of stem cell survival without HNF4.
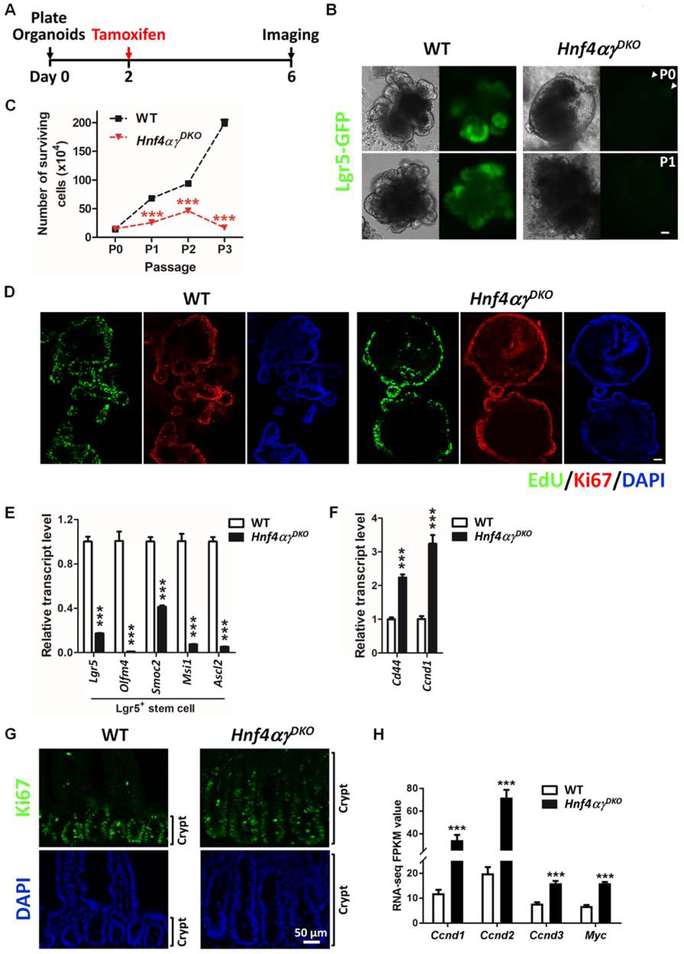
We utilized the organoid model to investigate the role of HNF4 in Lgr5+ stem cells. Organoids derived from Hnf4αɣDKO crypts after 2 days of tamoxifen injection do not establish viable cultures (Figure S4E), so we derived organoids from uninjected Lgr5-EGFP-CreERT2; Villin-CreERT2; Hnf4αf/f; Hnf4ɣCrispr/Crispr crypts and treated with tamoxifen in vitro. Unlike wild type organoids, which grow indefinitely with branched crypt domains containing Lgr5+ stem cells, Hnf4αɣDKO organoids initially form spherical organoids, but die out after passaging (Figure 4A and 4B). Live imaging of Lgr5-GFP in Hnf4αɣDKO organoids reveals diminished Lgr5-GFP+ cells compared to controls (Figure 4A and S4F), suggesting HNF4 factors are also required to maintain Lgr5 stem cells in organoid cultures. We hypothesized that the absence of HNF4 could skew the balance of a stem cell’s decision to renew versus enter the proliferating cell population towards lineage commitment. Knockout of HNF4 resulted in spherical organoids homogeneously composed of proliferative cells, as revealed by EdU and Ki67 staining (Figure 4C). Consistent with this phenotype, expression of Lgr5 stem cell markers is downregulated (Figure 4D), whereas the expression of proliferative cell markers is upregulated (Figure 4E). As with the in vitro data, in vivo data suggest that loss of HNF4 factors triggers a burst in cell proliferation, crypt expansion, and an increased population of Ki67+ cells by day 4 after tamoxifen treatment (Figure 4F), coincident with the timing of stem cell loss. This finding is corroborated by elevated transcript levels of proliferative cell markers observed in Hnf4αɣDKO mice (Figure 4G). Taken together, both in vitro (Figure 4A–E) and in vivo (Figure 4F and 4G) data suggest that in the absence of HNF4 paralogs, stem cells fail to renew and instead contribute to a population of proliferating cells, presumably representing the transit-amplifying population.
Figure 4. Transit amplifying-like proliferating cells predominate in the absence of Lgr5+ cells upon HNF4 loss.
(A) WT organoids exhibit branched structures and express robust Lgr5-GFP, whereas Hnf4αɣDKO organoids form spherical structures and exhibit little Lgr5-GFP fluorescence (see arrows). 1 μM tamoxifen or vehicle control was added to culture medium of primary organoids on Day 2 after seeding. After passage, Hnf4αɣDKO organoids failed to survive (n=6 independent experiments). Scale bar, 50 μm. Tamoxifen-induced Cre-only controls were also tested and shown in Fig. S4F. (B) WT and Hnf4αɣDKO organoids were passaged three times, and the number of surviving cells of organoids (n=4 independent organoid cultures) was counted at each passage, from primary (P0) passage to the third passage (P3). While HNF4 double-mutant organoids grow initially as proliferative spheres, they cannot be passaged indefinetly, such as their control counterparts. (C) Co-staining of EdU and Ki67 (proliferative markers). Organoids were treated with 10 μM EdU 6 hours prior to fixation. Scale bar, 50 μm. qRT-PCR shows a significant decrease of Lgr5+ stem cell markers (D) but a significant increase of proliferative cell markers (E) in Hnf4αɣDKO organoids. 1 μM tamoxifen or vehicle control was added to culture medium of primary organoids on Day 2 after seeding. All the primary organoids were harvested at Day 6 after seeding. Data are presented as mean ± SEM (n=3 independent organoid cultures, Student’s t-test, two-sided at P < 0.001***). (F) Crypt elongation is observed in Hnf4αɣDKO at day 4 post tamoxifen-induced knockout as shown in Ki67 immunofluorescent staining (representative of 4 biological replicates). (G) RNA-seq data reveal an elevation of proliferative cell markers in Hnf4αɣDKO compared to their littermate controls (n=3 biological replicates). Statistical tests were embedded in Cuffdiff at FDR < 0.001***.
ISCs lacking HNF4 paralogs are rescued by metabolic intervention
The importance of FAO in stem cell maintenance has been reported in several tissues10, 19–22, and corroborated in this study (Figure 1). Since we found that HNF4 factors bind to and activate FAO genes (Figure 2 and Figure S2), we examined whether the contribution of U-13C16 palmitate to TCA cycle metabolites was altered in Hnf4αɣDKO organoids (schematic see Figure 5A and 5B). Indeed, contribution of labeled palmitate to the TCA cycle was substantially reduced in Hnf4αɣDKO organoids (Figure 5C and Figure S5A), consistent with diminished FAO activity in HNF4 mutants. To determine the fate of acetate (a precursor of acetyl-CoA, a major product pf FAO) in Hnf4αɣDKO mutants, we cultured primary organoids with U-13C2 acetate for 6 hours, and measured the incorporation of 13C in TCA metabolites as well as in saponified lipid (schematic see Figure 5A and 5B). 13C-labeled TCA metabolites were indeed reduced in Hnf4αɣDKO mutants compared to the WT organoids (Figure 5Dand Figure S5B). In contrast, there was a stark increase in the 13C incorporation in fatty acid synthesis in Hnf4αɣDKO mutants (Figure 5E). These data suggest that HNF4 loss redirects acetyl-CoA away from the TCA cycle and towards fatty acid synthesis.
Figure 5. Loss of HNF4 paralogs leads to compromised TCA cycle contribution and elevated fatty acid synthesis.
(A-B) Schematics of 13C metabolic labeling experiments. (C) 24 hours of labeling with U-13C16 palmitate (0.5 mM) indicates reduced contribution of β-oxidation to the TCA cycle metabolites in Hnf4αɣDKO organoids (n=4 independent organoid cultures, LC-MS, see expanded panels in Fig. S5A). (D-E) 6 hours of U-13C2 acetate (2.5 mM) labeling shows (D) reduced 13C labeled TCA metabolites and (E) elevated 13C labeled fatty acids upon HNF4 loss (n=4 independent organoid cultures). Data are presented as mean ± SEM (Student’s t-test, two-sided at P < 0.001***, P < 0.01** and P < 0.05*).
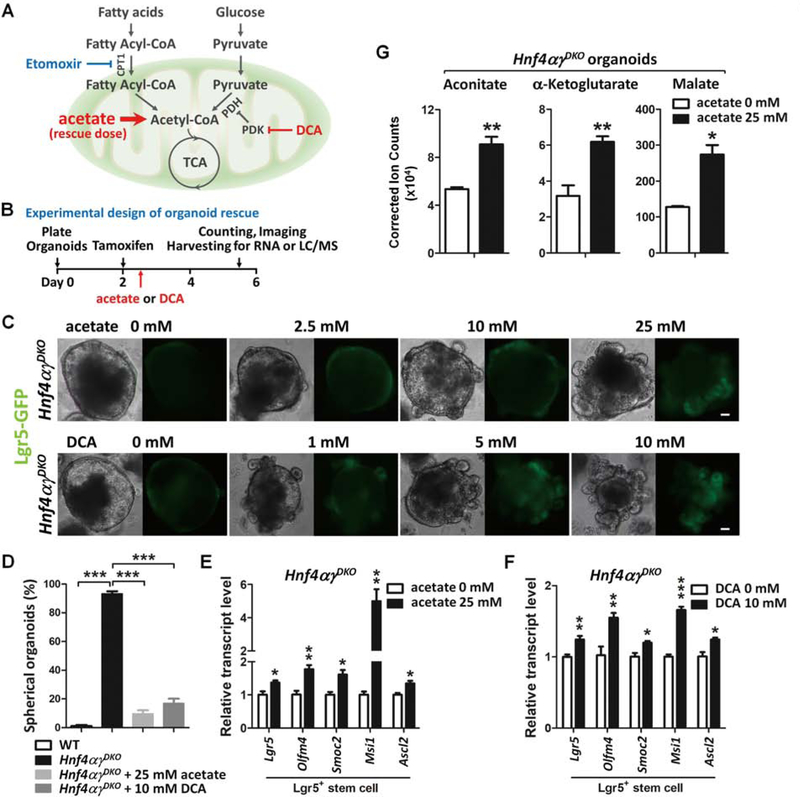
To test whether impaired FAO in the Hnf4αɣDKO was contributing to stem cell loss, we tested whether mutant organoids were sensitized to FAO inhibition by Etomoxir (schematic see Figure 6A). Hnf4αɣDKO organoids take on a spherical morphology consisting of proliferative cells and a lack of Lgr5+-cells (Figure 4A and Figure 4C). FAO inhibition accelerates the formation of spherical organoids in Hnf4αɣDKO within 24 hours in the presence of Etomoxir (Figure S5C and S5D). If a FAO defect was responsible for stem cell loss in Hnf4αɣDKO organoids, we wondered whether supplement of acetate could rescue the phenotype (schematic see Figure 6A and 6B). Lower doses of acetate treatment (2.5 mM) could not rescue the spherical organoid structure but did induce visible Lgr5-GFP fluorescence compared to the untreated group. Higher doses of acetate, approaching levels observed in the colonic lumen53, rescued both the spherical morphology and expression of the Lgr5-GFP reporter in Hnf4αɣDKO organoids (Figure 6C, top panel). More than 80% of Hnf4αɣDKO organoids exhibited stem cell-deficient, spherical structures upon Hnf4 loss, but acetate rescued the organoid morphology, with only 10% of Hnf4αɣDKO organoids showing spherical structures after 25 mM acetate treatment (Figure 6D), forestalling the collapse of the Hnf4αɣDKO organoids for several days. Corroborating these findings, the transcript levels of Lgr5 stem cell markers are also rescued upon acetate treatment in Hnf4αɣDKO (Figure 6E).
Figure 6. Acetate supplementation restores TCA cycle metabolites and ISC renewal.
(A) Schematic of agonists/antagonists used in the fatty acid/pyruvate oxidation-TCA pathways. CPT1: Carnitine palmitoyltransferase I; PDH: Pyruvate dehydrogenase; PDK: Pyruvate dehydrogenase kinase. (B) Schematics of acetate/DCA rescue experiments. (C-F) In Hnf4αɣDKO, intestinal stem cell maintenance is prolonged with addition of acetate (a precursor of acetyl-CoA) or dichloroacetate (DCA, a promoter of pyruvate oxidation and generation of acetyl-CoA). (C) Representative morphology (n=4 independent experiments) and (D) percentage of spherical organoids arising from organoid culture of Hnf4αɣDKO with or without acetate or DCA treatment (n=4 independent organoid cultures, one-way ANOVA followed by Dunnett’s post test at P < 0.001***). qRT-PCR shows rescued Lgr5+ stem cell markers in the Hnf4αɣDKO organoids upon (E) 25 mM acetate treatment (n=3 independent organoid cultures) or (F) 10 mM DCA treatment (n=4 independent organoid cultures). (G) TCA metabolite levels are restored in 25 mM acetate treated Hnf4αɣDKO organoids (n=4 independent organoid cultures). Scale bars, 50 μm.
Disruption of FAO in HNF4 mutants, and subsequent rescue of stem cell renewal by acetate suggested that a deficiency in acetyl-CoA could be limiting stem cell renewal. Pyruvate oxidation is an alternative source of acetyl-CoA for the TCA cycle. As pyruvate oxidation genes were not compromised in Hnf4αɣDKO (Figure S3), we tested whether dichloroacetate (DCA), a compound promoting pyruvate oxidation and generation of mitochondrial acetyl-CoA (schematic see Figure 6A and 6B) could also rescue the Hnf4αɣDKO mutant phenotype. Indeed, treatment with DCA also rescued stem cell loss in Hnf4αɣDKO (Figure 6C, bottom panel, 6D and 6F). Interestingly, the rescue dose of acetate (25 mM) or DCA (10 mM) that prolongs stem cell maintenance in Hnf4αγDKO mutant organoids impairs the growth of WT organoids (Figure S5E–G).
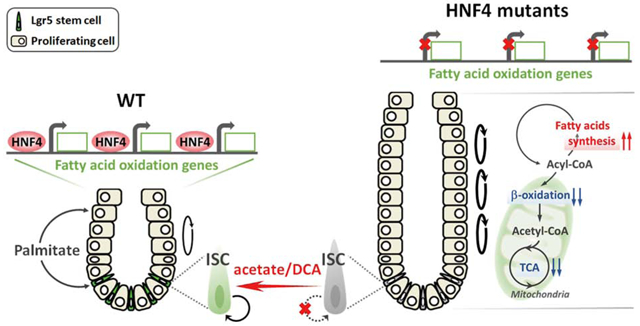
Exogenous acetate supplementation has been shown to rescue cellular functions dependent upon FAO by substituting for acetyl-CoA54–56. Given that FAO and TCA metabolites are reduced in Hnf4αɣDKO mutants (Figure 2, 5C, 5D and Figure S2, S5A, S5B), we wondered whether elevated levels of acetate could rescue stem cell function in Hnf4αɣDKO organoids by supporting the TCA cycle. Indeed, acetate supplementation restored TCA metabolite levels in Hnf4αɣDKO (Figure 6G). In summary, we demonstrate that FAO promotes stem cell renewal. HNF4 factors bind to and activate FAO genes, and upon loss of HNF4 factors, fatty acid synthesis is elevated at the expense of β-oxidation. Acetate supplementation suppresses stem cell loss in the Hnf4αɣDKO, suggesting that HNF4 factors may be required for stem cell renewal via their role in promoting β-oxidation.
Discussion
Disruption of FAO results in hematopoietic stem cell (HSC) exhaustion and lineage commitment of HSC daughter cells22. Quiescent neural stem and progenitor cells (NSPCs) require high levels of FAO, and FAO inhibition induces NSPCs exit from quiescence to proliferation57. Together with our work and recent findings, there is an emerging theme that links FAO to stem cell maintenance. Conditions leading to elevated fatty acid levels and mitochondrial metabolism correspond to enhanced ISC renewal: Fasting augments ISC numbers by shifting metabolism toward more FAO, whereas chronic disruption of Cpt1a, the rate-limiting enzyme in FAO, decreases ISC numbers and function10. Meanwhile, a high-fat diet elevates Lgr5+ cell numbers, and culturing with fatty acids bolsters self-renewal of intestinal organoids11. Lgr5+ ISCs exhibit high mitochondrial activity, and inhibition of mitochondrial activity in Lgr5+ ISCs leads to compromised stem cell function16–18. Collectively, these studies imply that the high mitochondrial content of ISCs12, 18 is likely coordinated with a requirement for fatty acid oxidation. Our studies now provide a regulatory link through which intestinal transcription factors promote expression of genes that provide the metabolic infrastructure to carry out FAO.
Glucose and fatty acids are two major fuels that drive ATP production for energy requirement. Fatty acid oxidation and pyruvate oxidation may have opposing effects on renewal of intestinal Lgr5+ stem cells; inhibition of pyruvate oxidation enhances stem cell maintenance12, whereas activation of fatty acid oxidation promotes stem cell renewal10, 11. Our findings suggest FAO is required for ISC renewal under normal conditions (Figure 1). Limiting mitochondrial pyruvate metabolism via genetic ablation of the mitochondrial pyruvate carrier has been shown to enhance ISC proliferation12. However, DCA treatment, which promotes pyruvate oxidation, restores stem cell renewal in the Hnf4αɣDKO. We suspect that DCA treatment has this effect under conditions of dramatic FAO deficiency, as we observe in Hnf4αɣDKO cells (Figure 2 and Figure S2). The Hnf4αɣDKO stem cell can partially restore acetyl-CoA that is limited by loss of FAO via elevated pyruvate oxidation upon DCA treatment, which could thus help restore ISC renewal.
In our previous study41, we observed an expansion of the crypt domain in Hnf4αɣDKO mice, which may be attributed to both an increased contribution of ISCs to the progenitor domain (this study) and a failure of crypt cells to differentiate into enterocytes41. Our current study finds that HNF4 transcription factors are essential for intestinal stem cell renewal, and thus provides a link between the fundamental transcriptional regulatory networks governing intestinal identity41, 58, 59, with a metabolic program essential for intestinal stem cell function. The critical role for HNF4 factors in intestinal stem cell renewal was not previously appreciated in single HNF4 mutants due to redundancy between Hnf4α and Hnf4ɣ paralogs. These unappreciated HNF4 functions broaden our understanding of metabolic regulation in intestinal stem cell homeostasis, and may provide a regulatory conduit for ISCs during conditions in which elevated free fatty acids are present (such as during fasting or in the presence of high fat diet).
Supplementary Material
What you need to know:
BACKGROUND AND CONTEXT:
Nutritional and metabolic changes can alter intestinal stem cell numbers and their ability to renew. Hepatocyte nuclear factor 4 (HNF4) proteins are transcription factors that regulate hundreds of genes in the intestinal epithelium. HNF4 factors are expressed in intestinal stem cells, but their function in intestinal stem cells is unclear.
NEW FINDINGS:
In mice, the transcription factors HNF4A and HNF4G directly activated genes that regulate fatty acid oxidation and are required for renewal of intestinal stem cells.
LIMITATIONS:
The inability of intestinal stem cells in mice with double knockout of HNF4A and HNF4G to survive long term made it difficult to collect and study these cells.
IMPACT:
We identified HNF4 transcription factors regulate intestinal stem cell renewal and metabolic pathways required for intestinal stem cell functions.
Acknowledgments
We thank Noriko Goldsmith and Josh Thackray for helpful imaging support.
Funding
This research was funded by a grant from the NIH (R01CA190558, M.P.V.). M.P.V is also supported by the Intestinal Stem Cell Consortium from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health under grant number U01 DK103141. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. E.M.B. was kindly supported by a Rutgers Newark Chancellor’s SEED grant. Support was also received from the Sequencing Facility and Metabolomics Shared Resource of the Rutgers Cancer Institute of New Jersey (P30CA072720) and imaging core facility of Human Genetics Institute of New Jersey. L.C. was supported by New Jersey Commission on Cancer Research grant (DFHS18PPC051). R.P.V., N.H.T., A.P. and S.L. were supported by MacMillan Summer Undergraduate Research Fellowships.
Abbreviations used in this paper:
- ACOX
acyl-CoA oxidase
- ATAC-seq
assay for transposase accessible chromatin sequencing
- ChIP
chromatin immunoprecipitation
- CPT1
carnitine palmitoyltransferase I
- DAB
3,3’-diaminobenzidine
- DCA
dichloroacetate
- FAO
fatty acid β-oxidation
- FDR
false discovery rate
- FPKM
fragments per kilobase of transcript per million mapped reads
- GSEA
gene set enrichment analysis
- Hadh
hydroxyacyl-CoA dehydrogenase
- HNF4A
hepatocyte nuclear factor 4 alpha
- HNF4G
hepatocyte nuclear factor 4 gamma
- HSC
hematopoietic stem cell
- IGV
integrative genomics viewer
- IHC
immunohistochemistry
- ISC
intestinal stem cell
- LC-MS
liquid chromatography-mass spectrometry
- Lgr5
leucine rich repeat containing G protein-coupled receptor 5
- MACS
model-based analysis of ChIP-Seq
- MNase
micrococcal nuclease
- NSPCs
neural stem and progenitor cells
- PDH
pyruvate dehydrogenase
- PDK
pyruvate dehydrogenase kinase
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- TCA
tricarboxylic acid
- TSSs
transcription start sites
- UHPLC
ultra-high-performance liquid chromatography
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors declare no competing interests.
Data availability
The accession number of the Hnf4 mutants versus WT transcriptome, HNF4 ChIP-seq and H3K27ac MNase-ChIP-seq of mouse intestinal epithelial cells have been deposited in GEO (GSE112946). GSE83394 was used to analyze chromatin accessibility and RNA-seq data of Lgr5+ intestinal stem cells.
References
- 1.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007;449:1003–7. [DOI] [PubMed] [Google Scholar]
- 2.Barker N, Ridgway RA, van Es JH, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 2009;457:608–11. [DOI] [PubMed] [Google Scholar]
- 3.Metcalfe C, Kljavin NM, Ybarra R, et al. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell 2014;14:149–59. [DOI] [PubMed] [Google Scholar]
- 4.Mihaylova MM, Sabatini DM, Yilmaz OH. Dietary and metabolic control of stem cell function in physiology and cancer. Cell Stem Cell 2014;14:292–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei P, Dove KK, Bensard C, et al. The Force Is Strong with This One: Metabolism (Over)powers Stem Cell Fate. Trends Cell Biol 2018;28:551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shyh-Chang N, Ng HH. The metabolic programming of stem cells. Genes Dev 2017;31:336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shim J, Gururaja-Rao S, Banerjee U. Nutritional regulation of stem and progenitor cells in Drosophila. Development 2013;140:4647–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alonso S, Yilmaz OH. Nutritional Regulation of Intestinal Stem Cells. Annu Rev Nutr 2018;38:273–301. [DOI] [PubMed] [Google Scholar]
- 9.Yilmaz OH, Katajisto P, Lamming DW, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 2012;486:490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mihaylova MM, Cheng CW, Cao AQ, et al. Fasting Activates Fatty Acid Oxidation to Enhance Intestinal Stem Cell Function during Homeostasis and Aging. Cell Stem Cell 2018;22:769–778 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beyaz S, Mana MD, Roper J, et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 2016;531:53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schell JC, Wisidagama DR, Bensard C, et al. Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nat Cell Biol 2017;19:1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattila J, Kokki K, Hietakangas V, et al. Stem Cell Intrinsic Hexosamine Metabolism Regulates Intestinal Adaptation to Nutrient Content. Dev Cell 2018;47:112–121 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng H, Gerencser AA, Jasper H. Signal integration by Ca(2+) regulates intestinal stem-cell activity. Nature 2015;528:212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obata F, Tsuda-Sakurai K, Yamazaki T, et al. Nutritional Control of Stem Cell Division through S-Adenosylmethionine in Drosophila Intestine. Dev Cell 2018;44:741–751 e3. [DOI] [PubMed] [Google Scholar]
- 16.Perekatt AO, Valdez MJ, Davila M, et al. YY1 is indispensable for Lgr5+ intestinal stem cell renewal. Proc Natl Acad Sci U S A 2014;111:7695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivillibhuthur M, Warder BN, Toke NH, et al. TFAM is required for maturation of the fetal and adult intestinal epithelium. Dev Biol 2018;439:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez-Colman MJ, Schewe M, Meerlo M, et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 2017;543:424–427. [DOI] [PubMed] [Google Scholar]
- 19.Xie Z, Jones A, Deeney JT, et al. Inborn Errors of Long-Chain Fatty Acid beta-Oxidation Link Neural Stem Cell Self-Renewal to Autism. Cell Rep 2016;14:991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryall JG, Dell’Orso S, Derfoul A, et al. The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell 2015;16:171–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoll EA, Makin R, Sweet IR, et al. Neural Stem Cells in the Adult Subventricular Zone Oxidize Fatty Acids to Produce Energy and Support Neurogenic Activity. Stem Cells 2015;33:2306–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito K, Carracedo A, Weiss D, et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med 2012;18:1350–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandra V, Huang P, Potluri N, et al. Multidomain integration in the structure of the HNF-4alpha nuclear receptor complex. Nature 2013;495:394–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhe-Paganon S, Duda K, Iwamoto M, et al. Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem 2002;277:37973–6. [DOI] [PubMed] [Google Scholar]
- 25.Duda K, Chi YI, Shoelson SE. Structural basis for HNF-4alpha activation by ligand and coactivator binding. J Biol Chem 2004;279:23311–6. [DOI] [PubMed] [Google Scholar]
- 26.Wisely GB, Miller AB, Davis RG, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure 2002;10:1225–34. [DOI] [PubMed] [Google Scholar]
- 27.Bolotin E, Liao H, Ta TC, et al. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology 2010;51:642–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang B, Mane-Padros D, Bolotin E, et al. Identification of a binding motif specific to HNF4 by comparative analysis of multiple nuclear receptors. Nucleic Acids Res 2012;40:5343–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watt AJ, Garrison WD, Duncan SA. HNF4: a central regulator of hepatocyte differentiation and function. Hepatology 2003;37:1249–53. [DOI] [PubMed] [Google Scholar]
- 30.Hertz R, Magenheim J, Berman I, et al. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4alpha. Nature 1998;392:512–6. [DOI] [PubMed] [Google Scholar]
- 31.Petrescu AD, Payne HR, Boedecker A, et al. Physical and functional interaction of Acyl-CoA-binding protein with hepatocyte nuclear factor-4 alpha. J Biol Chem 2003;278:51813–24. [DOI] [PubMed] [Google Scholar]
- 32.Palanker L, Tennessen JM, Lam G, et al. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metab 2009;9:228–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barker N Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol 2014;15:19–33. [DOI] [PubMed] [Google Scholar]
- 34.Bitar KN, Zakhem E. Bioengineering the gut: future prospects of regenerative medicine. Nat Rev Gastroenterol Hepatol 2016;13:543–56. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura T, Sato T. Advancing Intestinal Organoid Technology Toward Regenerative Medicine. Cell Mol Gastroenterol Hepatol 2018;5:51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vermeulen L, Snippert HJ. Stem cell dynamics in homeostasis and cancer of the intestine. Nat Rev Cancer 2014;14:468–80. [DOI] [PubMed] [Google Scholar]
- 37.Yousefi M, Li L, Lengner CJ. Hierarchy and Plasticity in the Intestinal Stem Cell Compartment. Trends Cell Biol 2017;27:753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayhurst GP, Lee YH, Lambert G, et al. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 2001;21:1393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Odom DT, Zizlsperger N, Gordon DB, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science 2004;303:1378–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.el Marjou F, Janssen KP, Chang BH, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 2004;39:186–93. [DOI] [PubMed] [Google Scholar]
- 41.Chen L, Toke NH, Luo S, et al. A reinforcing HNF4-SMAD4 feed-forward module stabilizes enterocyte identity. Nat Genet 2019;51:777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bohin N, Carlson EA, Samuelson LC. Genome Toxicity and Impaired Stem Cell Function after Conditional Activation of CreER(T2) in the Intestine. Stem Cell Reports 2018;11:1337–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7:562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carta G, Murru E, Banni S, et al. Palmitic Acid: Physiological Role, Metabolism and Nutritional Implications. Front Physiol 2017;8:902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009;459:262–5. [DOI] [PubMed] [Google Scholar]
- 48.Babeu JP, Darsigny M, Lussier CR, et al. Hepatocyte nuclear factor 4alpha contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am J Physiol Gastrointest Liver Physiol 2009;297:G124–134. [DOI] [PubMed] [Google Scholar]
- 49.Cattin AL, Le Beyec J, Barreau F, et al. Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol Cell Biol 2009;29:6294–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.San Roman AK, Aronson BE, Krasinski SD, et al. Transcription factors GATA4 and HNF4A control distinct aspects of intestinal homeostasis in conjunction with transcription factor CDX2. J Biol Chem 2015;290:1850–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verzi MP, Shin H, San Roman AK, et al. Intestinal master transcription factor CDX2 controls chromatin access for partner transcription factor binding. Mol Cell Biol 2013;33:281–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jadhav U, Saxena M, O’Neill NK, et al. Dynamic Reorganization of Chromatin Accessibility Signatures during Dedifferentiation of Secretory Precursors into Lgr5+ Intestinal Stem Cells. Cell Stem Cell 2017;21:65–77 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.den Besten G, van Eunen K, Groen AK, et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 2013;54:2325–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schoors S, Bruning U, Missiaen R, et al. Fatty acid carbon is essential for dNTP synthesis in endothelial cells. Nature 2015;520:192–U113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong BW, Wang X, Zecchin A, et al. The role of fatty acid beta-oxidation in lymphangiogenesis. Nature 2017;542:49–54. [DOI] [PubMed] [Google Scholar]
- 56.Xiong J, Kawagishi H, Yan Y, et al. A Metabolic Basis for Endothelial-to-Mesenchymal Transition. Mol Cell 2018;69:689–698 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knobloch M, Pilz GA, Ghesquiere B, et al. A Fatty Acid Oxidation-Dependent Metabolic Shift Regulates Adult Neural Stem Cell Activity. Cell Rep 2017;20:2144–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindeboom RG, van Voorthuijsen L, Oost KC, et al. Integrative multi-omics analysis of intestinal organoid differentiation. Mol Syst Biol 2018;14:e8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davison JM, Lickwar CR, Song L, et al. Microbiota regulate intestinal epithelial gene expression by suppressing the transcription factor Hepatocyte nuclear factor 4 alpha. Genome Res 2017;27:1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.