Abstract
Background
Expression levels of SYK, a critical signaling tyrosine kinase in basophils, are uniquely low relative to all other circulating leukocytes and levels are highly variable in the population.
Hypothesis
Transcriptional regulation of SYK through unique silencing of the SYK gene determines its basophil-specific expression patterns.
Methods
Basophils (CD34B) were derived from cultures of CD34+ progenitor cells by two methods (G1 or G3). Peripheral blood basophils (PBB, relative SYK protein level = 1), B-cell (SYK = 8), CD34B G1 (SYK = 11) and G3 (SYK = 5) were examined by ATACseq methods and the transcriptomes of 6 cell types, PBB, eosinophils (PBE, SYK = 11), dendritic cells (PDC, SYK = 30), CD34+ progenitors (SYK = 11), CD34B G1 and G3 were analyzed for patterns that matched patterns of SYK expression in these cells, with a focus on transcription factors.
Results
ATACseq showed that PBB have multiple open regions in the SYK gene suggesting a non-silenced state with: 1 region unique to PBB (low SYK expression), one region unique to both PBB (low SYK expression) and G1/G3 CD34B (high and moderate SYK expression, respectively) and 5 regions unique to B-cells (high SYK expression). SYK expression across the 6 cell types explored showed a unique pattern that was matched to the expression patterns of 3 transcription factors, KLF5, ZNF608, and c-MAF.
Conclusions
Two new potential regulatory pathways for SYK expression were identified. One appears independent of transcriptional regulation and one appears to be dependent on transcriptional control in the SYK gene.
Keywords: Human, Basophil, Allergy, Development, Signal Transduction
Capsule Summary
Expression of the early signaling kinase, SYK, that allows IgE-mediated secretion from basophils and mast cells, is uniquely low and heterogeneous in humans. Two new potential pathways regulating SYK expression were identified.
Introduction
IgE-mediated secretion from human basophils (and mast cells) requires the activity of SYK, a tyrosine kinase in the SYK/ZAP-70 2-member family 1–3. SYK expression in human basophils is uniquely low despite the absolute requirement for SYK expression in generating an IgE-mediated response 4. With one exception – the circulating αβ T-cell, which uses ZAP-70 -- the basophil shows the lowest expression of SYK of all the common circulating leukocytes. Indeed, the level of SYK lies at the cusp of functionality for FceRI-mediated activation of these cells 5, 6. For example, there are subjects whose basophils don’t express enough SYK to drive an IgE-mediated reaction despite there being an excess of FceRI (so-called non-releasers) 5, 7–10. SYK expression therefore correlates well with the maximum secretion possible through FceRI. There are only 2 known mechanisms of regulation of SYK expression in human basophils. The first is a post-translational process of down-regulation following the crosslinking of cell surface FceRI through a process of ubiquitination and proteasomal degradation 11. While this is a strong mechanism, as a mechanistic genesis explanation for the highly variable expression of SYK in basophils among the general population, it is unsatisfactory. For example, recent studies have not been able to detect auto-antibodies that could drive SYK down-regulation in most individuals (while possibly explaining low SYK in basophils from patients with chronic spontaneous urticaria) 12. A second mechanism, up-regulation by exposure to high concentrations of IL-3, is weak 5, 13. Changes in SYK protein expression induced by IL-3 are in the range of 30%, not the 30-fold range found in the general population or the 100-fold range when comparing peripheral blood basophils to other leukocytes.
All leukocytes are derived from a CD34+ progenitor cell that responds to factors that include the cytokine environment. A simple in vitro culture of CD34+ cells with IL-3 alone will drive the production of a highly heterogeneous mixture of cells with the overall characteristics of mature basophils 4. But the phenotype is not precisely that of mature circulating basophils. For example, SYK protein expression in the culture-derived basophils (CD34B) is unchanged from the CD34+ progenitor cell, levels that are 11 fold higher that the mature peripheral blood basophil (PBB). With the exception of dendritic cells and monocytes, most other leukocytes express SYK at levels similar to CD34+ progenitors, suggesting that there is no program during differentiation that modifies the basic mechanics of SYK transcription or translation during maturation. The monocyte and dendritic cell express approximately 3 times the SYK of the CD34+ progenitor suggesting there is a program to up-regulate expression of this kinase. Therefore, in contrast, basophils appear to undergo a program of down-regulation during maturation.
The mechanism of SYK down-regulation during maturation is not clear. As noted above, one strong mechanism of SYK regulation is related to FceRI aggregation. But previous studies have also determined that SYK protein expression was reasonably well correlated with steady state SYK mRNA levels amongst subjects’ basophils or amongst leukocyte cell types 4. Preliminary studies suggested that mRNA stability is not different between CD34B and PBB 4,14 (and unpublished studies) nor was there any evidence for a miRNA that may regulate SYK mRNA 4, 14. Together, the various observations suggest that SYK protein expression may be under transcriptional control of the SYK gene, control that is unique to the human basophil.
The studies in this report provide a first look at how SYK may be regulated outside of the IgE-dependent processes. During preliminary studies of CD34+ cells maturing into basophils, we discovered that these cells can be induced to down-regulate SYK during maturation by an IgE-independent mechanism. This report characterizes this discovery and explores some of its implications. In addition, this study posed the hypothesis that the SYK gene is silenced during maturation, either completely with no active open/accessible chromatin, or through a unique pattern of “open” chromatin.
Methods
Materials, buffers and methods that have been used extensively in prior publications are presented in detail in the online supplement.
Generation of CD34-derived basophil from CD34+− progenitors
Two sources of CD34+ cells were used in these studies. For most of the studies, residual leukocytes from plateletpheresis were enriched by a combination of Percoll gradient separation (53%/62% Percoll) and positive selection. The upper layer of the gradient separation was found to enriched in CD34+ cells (personal communication from Dr. John Schroeder). Magnetic positive selection of CD34+ cells (Miltenyi) followed the manufacturers guidelines with slight modifications. Generally, no more than 1×109 total cells were incubated for 30 minutes at 4˚C with 200 microliters of kit reagent before passage over StemCell columns. In early pilot studies, the CD34+ purity after column separation was determined by flow cytometry using a Miltenyi antibody designed to work with the purification reagents. Typically, post-column purities were 42±13% and yields ranged from 0.2 to 3.0 million CD34+ cells (average of 1.0±0.7×106). For some preliminary studies, CD34+ cells were obtained from the lab of Dr. Shelley Heimfeld (Fred Hutchinson Cancer Research Center, Seattle, WA, USA) and IL-3 cultures started without further handling.
The CD34+ progenitors were re-suspended at density of 0.1 – 0.2×106 cells/ml in StemPro serum-free medium, which contained the provided nutrient supplement, 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 5 ng/ml rhIL-3, and different concentrations of Flt3L and SCF (stem cell factor). Cells were cultured at 37°C with 5% CO2. Cell counts and viability were checked on day 14–21 by erythrocin B staining. The cells generated are heterogeneous in size, granularity, and nuclear morphology. Although not presented in the figures, a rough set of categories (large vs. small, strong alcian blue positivity vs. weak, etc) were enumerated to monitor qualitative differences. By-and-large, there were only minor differences in these characteristics for the culture conditions tested. In many cases, a low G centrifugation (40G for 1 minute in a 70 μl volume) was used to remove a significant fraction of dead cells from the harvested cultures prior to use in subsequent assays. Previous studies of these cells established that dead cells do not contribute mRNA, therefore cell lysis procedures for mRNA were based on the live cell counts. However, protein remains present in dead cells, so lysis for protein measurements was based on the total cell count. See also Tables E1 and E2 for details about the culture conditions.
ATAC-seq Analysis
The samples for ATAC-seq were prepared as described in the literature [ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide]. Briefly, 2×105 cells were washed with cold PBS and re-suspended in 50 μl of cold lysis buffer. The cell pellets were harvested by centrifugation for 10 min at 500 × g (4°C), and placed on ice. The transposition reaction mix was prepared according to the manufacturer’s guidelines with adjustments to optimize the fragmentation of the DNA for deep sequencing. Each cell type studied was examined with a range of cell number/reagent ratios to optimize the outcome. Generally, the reaction mixture was 25 μl TD (2 × reaction buffer from Nextera kit), 1 μl TDE1 (Nextera Tn5 Transposase from Nextera kit) and 24 μl nuclease-free H2O. The nuclei pellet was re-suspended in the transposition reaction mix, and incubated at 37°C for 30 min. Immediately following transposition, DNA was purified using a Qiagen MinElute PCR Purification Kit, and transposed DNA was eluted in 10 μl elution buffer (Buffer EB from the MinElute kit consisting of 10 mM Tris · Cl, pH 8). The transposed DNA fragments were amplified using PCR, and the amplified library was purified using Qiagen MinElute PCR Purification Kit, eluted in 20 μl elution buffer, and separated by gel electrophoresis. The gel was excised in the 200–1000 bp region and DNA eluted and the DNA suspended in 20 ul TE buffer. Sequencing was performed by Illumina Nextseq Sequencing using the high-output 75-cycle reagent kit for single 75bp reads. Sequencing included 150–400 million reads, depending on the number of samples (with their unique bar coding) combined for a particular run. Data analysis workflow included Bowtie2 (2.2.5), Samtools (0.1.19) and peak calling with MACS2 using the hg19 human genome build for reference, and HOMER used for peak annotation. Images of the ‘.bedgra’ files produced from the workflow were generated with the UCSC genome browser.
Data Preparation
All microarray and ATACseq data relevant to the paper are located in three repositories, The primary repository is the Gene Expression Omnibus (accession # GSE138610). Additional data, including an in-house designed application for exploring the ultimate raw data set, are available via ‘http://www.basophil.net’ and ‘http://162.129.217.250/basophilMicroarrays’. The website includes a variety of files one of which provides a detailed description of the analysis methodology for the microarray results.
Results
IgE-independent down-regulation of SYK in CD34+ derived basophils
Previous studies used only IL-3 during the culture of CD34+ progenitors to generate basophils whose characteristics have partially described 4. These cells were phenotypically similar to basophils but SYK levels were similar to the CD34+ progenitors (and other leukocytes), not PBB. Other studies have shown that during the production of eosinophils from CD34+ progenitors, that high concentrations of Flt3L and SCF during the first 3 days would enhance recovery of mature eosinophils 15. Evidence suggests that basophil and eosinophil have a similar lineage 16–18, so we examined whether this same methodology could be used to enhance recovery of mature basophils.
For the purposes of presenting these results, several labels were adopted to describe the different methods of culture that were explored. The traditional method of culture (5 ng/ml of IL-3 only for 2–3 weeks) was labeled Group 1 (G1). A second method used Flt3L, SCF and IL-3 during the first 3 days of culture and 5 ng/ml IL-3 alone thereafter for 2–3 weeks was labeled group 2 (G2). A third group (G3) incorporated Flt3L, SCF and IL-3 throughout the culture period. The cultured cells were evaluated for overall cell viability, alcian blue positive (ABP) cell/live cell ratio, ABP cell numbers, surface markers, histamine content, and SYK levels. Figure E1 in the online repository shows the various culture schemes tested.
These studies developed iteratively towards a method where the resulting preparations contained a greater number of ABP appearing morphologically similar to PBB, with similar cell surface expression of FceRI to G1 cells, but expression levels of SYK protein that were reduced considerably compared to G1 method. The online supplement presents in greater detail the progression of these studies. Following the three experimental series presented in the online supplement, the final approach incorporated a low concentration of Flt3L and SCF throughout the culture.
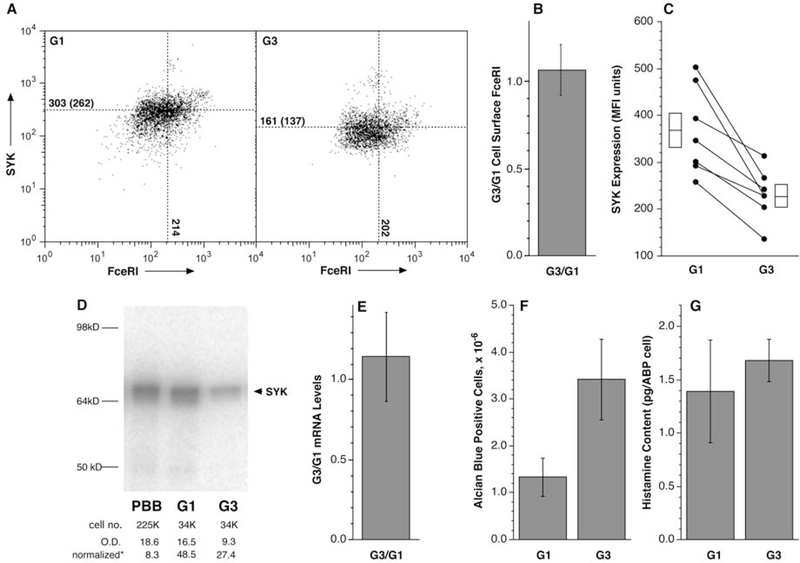
Figure 1 summarizes the results from the experimental series that were used to generate Figure E4 and a subsequent distinct experimental series only comparing G1 and G3 preparations. As shown in Figures 1, E2–E4, these two culture models generate ABP-cells that have basophil characteristics but are distinguished by a down-regulation of SYK expression in the G3 group (Figure 1A & C). In addition, as noted above, the cultures with Flt3L and SCF consistently generated more ABP cells (Figure 1F) but with similar histamine content per cell (Figure 1G) and FceRI expression (Figure 1B). Furthermore, based on morphology (size and kidney-shaped condensed nucleus) there were 10-fold greater numbers of PBB-like cells in G3 cultures versus G1 cultures (data not shown). Western blots (Figure 1D) comparing SYK expression in the G1 and G3 groups supported the conclusions of flow cytometry where SYK expression was lower in the G3 group. With respect to other cell markers of mature basophils (e.g., CD203c, CD63, CD32b), the online supplement includes other tests of similarity between the G1 and G3 groups that suggest that the two culture methods generate cells similar in phenotype.
Figure 1:
Primary characteristics of G1 and G3 cultures. Using the methodologies described in the text and similar to Figure E4, G1 and G3 CD34B were generated and analyzed for various metrics of the basophil phenotype. Panel A: an example of SYK and FceRI expression for the G1 and G3 cultures as determined by flow cytometry (the gating for these plots was the exclusion of “debris” or dead cells using forward and side scatter followed by gating for CD123+ cells which represented ≈80% of the live cells). Panel B: relative FceRI expression for the G1 & G3 comparison. Panel C: compilation of SYK expression studies as assessed by flow cytometry. Panel D: example Western blots comparing CD34B G1 and G3 cell preparations compared to a PBB standard. The number of cells represented in the lysate run in the gel is noted as cell number (cell no.). Below this line is the optical density (O.D.) for the bands and the normalized OD per 100,000 cells. As found previously, SYK expression in the CD34B was significantly greater than PBB (≈6-fold in this instance). Panel E: rato of SYK mRNA expression for G1/G3 cells (see online repository Table E4 for details). Panel F: the total number of alcian-blue positive (ABP) cells. Panel G: histamine content per ABP cell counted.
The G1 and G3 cultures represented a contrasting model of basophil development with a particular focus on SYK expression. These two culture conditions were explored for characteristics that might provide insights on the regulation of SYK expression.
ATACseq Differences
The difference in SYK expression when comparing CD34B and PBB at both the protein and mRNA level 4 and the similarity in the decay rate of the mRNA between PBB and CD34B (unpublished experiments and reference 4) suggested that there is a transcriptional rate difference for SYK mRNA between these two cell types. Polymorphonuclear cells like the peripheral blood basophil are characterized by their apparent inability to engage in further replication. In addition, differentiation can silence non-relevant genes. These observations raise the possibility that low SYK expression results from rendering the SYK gene inaccessible to transcription, perhaps late in basophil differentiation (since there is some mRNA with an unaltered decay rate). As a step in characterizing the SYK gene, PBB were examined with ATACseq methods. To provide context, two cells, B-cells and CD34B-G1, that express similar levels of SYK protein to each other but 10-times greater than PBB were also examined. The B-cell was chosen because it represented a non-polymorphonuclear leukocyte, unlike the basophil. Finally, CD34B-G3 cells (which were developed in parallel from the same progenitors used for the G1 results) were examined for differences that may explain the transition from G1 to G3 under the influence of low levels Flt3L and SCF to down-regulate SYK protein.
ATACseq generates a wealth of information about the cell’s DNA state. These results represent the first look at the human basophil’s “open/accessible” and “closed” DNA status. A first pass through the results showed that there were many accessible genes in PBB that were consistent with the mRNA being present and many “closed” genes for mRNA (or protein) that were not present in basophils but present in B cells or CD34B cells. For example, for PBB, IL-4, HDC (histidine decarboxylase), MS4A2 (beta subunit of FceRI), and FCERIA showed peak-calls in PBB (and CD34B) that were not present in B cells. Conversely, CD79A, BLNK, and EBF1 were present in the B-cell profiles and not PBB.
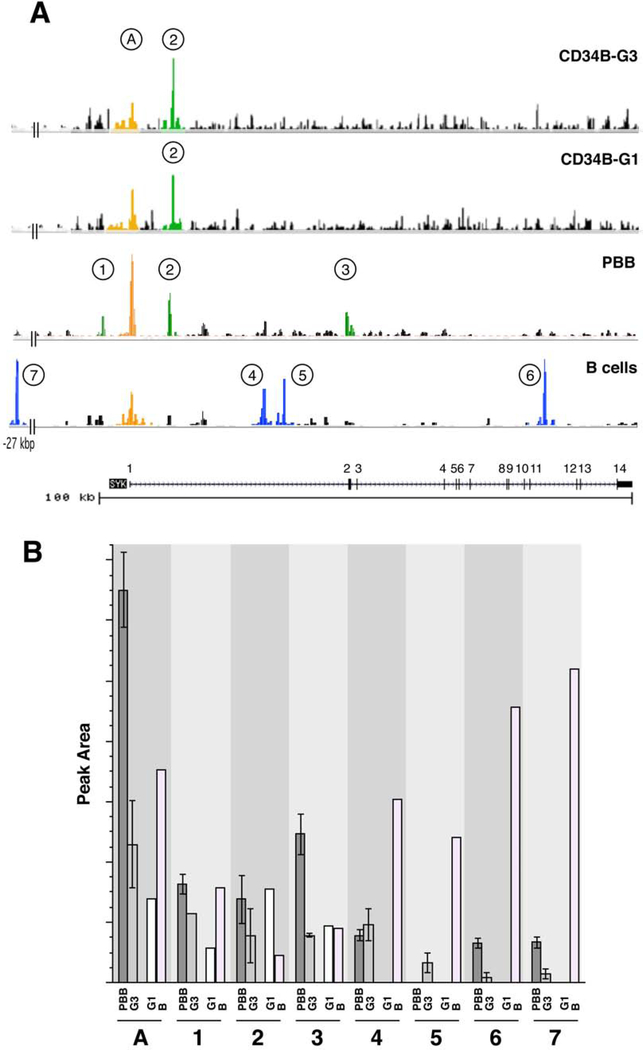
Figure 2, panel A shows the SYK gene region. The same region is shown for B cells and CD34B G1 and G3 cells. There are shared active regions in these profiles and several that are not shared; regions of interest are labeled 1–7 and ‘A’. The region labeled ‘A’ includes the transcription start site that was identified in the FANTOM5 project (http://fantom.gsc.riken.jp). Although there are no obvious start sites (TATA or CAAT boxes) in the region immediately upstream of exon 1, the FANTOM5 project included a TSS analysis of the human SYK gene in mast cells and basophils and 99% of the TSSs for this gene did lie 27 bp inside of the identified exon 1. Figure E5 shows peak ‘A’ expanded and the sequence information as it relates to the 5’ end of basophil SYK mRNA. Note that the identified exon 1 contains the transcription start site but encodes none of the sequence found in the mRNA (in data not presented, this mRNA 5’UTR was roughly mapped using sequential PCR primers). The 5’UTR begins downstream of the Genebank-identified exon 1, stops at 141 bp 3’ of the TSS and continues in exon2 (see Figure E6).
Figure 2:
ATACseq information for the SYK gene. Panel A: Four cell types, B cells, peripheral blood basophils (PBB), CD34B culture derived basophils using the G1 and G3 protocols, were analyzed by ATACseq and the data for the SYK gene (shown at the bottom of the figure, both exons (black regions) and introns (line with arrows indicating the direction of transcription), with numbered exons) collated using the UCSC browser. Eight peaks (identified by MACS2/HOMER annotation of the peak calls) are labeled: ‘A’ is the peak associated with the generally identified TSS for SYK and ‘1’ through ‘7’ peaks 5’ and throughout the gene. The bottom-most trace is the hypersensitivity profile identified in Genebank. Panel B: Summary of the ATAC results for the 8 identified peaks in the SYK gene. The ATACseq results were averaged for preparations that were of high quality; PBB (n=3), CB34B-G3 (n=2), CD34B-G1 (n=1), and B-cells (n=1). See online repository for methodological details.
The bottom of Figure 2, panel A shows the Genebank assignments of SYK exons and introns; there are 14 expected exons covering approximately 100 kbps and the known hypersensitivity regions are also shown in the tracing in Figure 3A. The peak calls colored blue (numbered 4–7) are most evident in the B cells. The peak calls colored green (numbered 1–3) are most evident in basophils and the orange region is at the common exon 1 region. Peak-call 1 may be shared between B cells and basophils. It is notable that CD34B cells are similar to PBB; in particular the peak labeled ‘2’ is found in both CD34B and PBB but not B cells. Peak 3 is weak but present in PBB but not apparent in CD34B. An expanded version of the peak 2 and peak 3 regions is shown in Figure E6. It is also notable that basophils and B cells show differences in activity in intron 1 or at exon 2 (peaks 2, 3 (basophil unique), 4 and 5 (B cell unique)). B cells also show a peak at −27 kbp upstream of exon1 (peak 7) that isn’t apparent in basophils as well as a peak in intron 11 (peak 6). Activity in basophils, B cells, and CD34B cells centers around the TSS and includes approximately 1200 bp, but only 500–600 bp upstream of the TSS. The average results for these peaks is shown in figure 2, panel B.
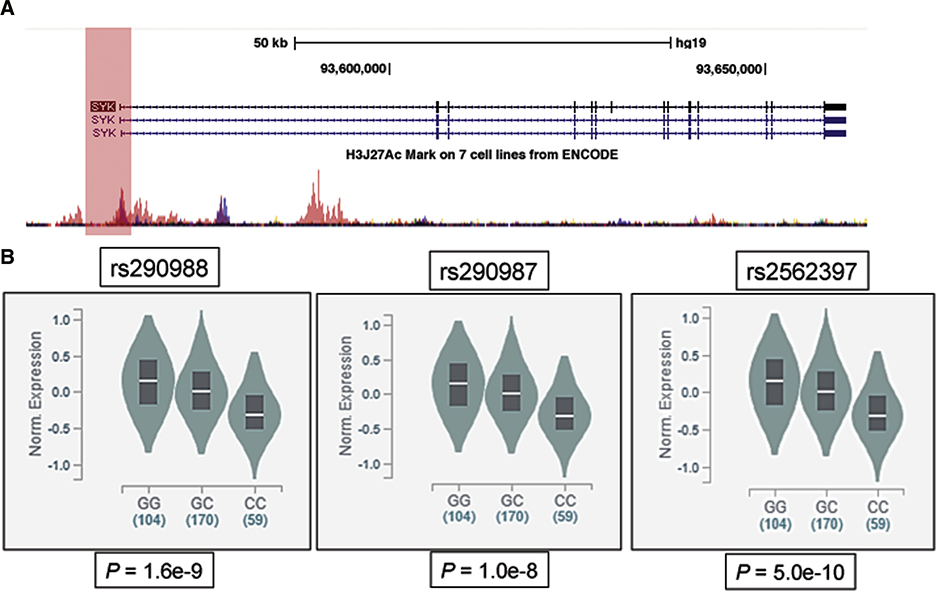
Figure 3:
ENCODE regulation tracks on the SYK gene region (chromosome 9: 90,795,787–90,898,549) and eQTL analysis for SYK promoter single nucleotide variants. Panel A illustrates the SYK gene structure, position of 3 variants (rs290988 (−1.1kb), rs290987 (−771bp) and rs2562397 (−76bp), in complete linkage disequilibrium (LD), (positions relative to transcription start site, TSS) within 1.1kb upstream of SYK (depicted by the User Track), and regulatory regions with ENCODE regulation tracks, including UCSC Gene (SYK), layered H3K27Ac and DNase Clusters. Panel B demonstrates significant SNP-gene eQTL association in tissues for these SYK promoter variants (with the CC homozygote showing lower SYK expression).
Regulatory regions for SYK were evaluated using the ENCODE regulatory tracks19. Interestingly, 3 single nucleotide variants (rs290988, rs290987 and rs2562397, in complete linkage disequilibrium (LD), located at −1084, −771 and −46 bp of the TSS) within 1.1kb upstream of SYK overlaps with many regulatory features, including enhancer histone H3K27Acmarks (which are often found near active regulatory elements) and DNAseI hypersensitivity clusters (Figure 3, panel A). The ATACseq results for basophils showed a strong peak spanning the likely transcriptional start site within exon1 ±600 bp (labeled ‘A’ in Figure 2).
By analyzing global RNA expression within individual tissues or cells and treating the expression levels of genes as quantitative traits, variations in gene expression that are highly correlated with genetic variation can be identified as expression quantitative trait loci, or eQTLs. The publicly available database (GTEx), with newly released RNA sequencing data from 1,641 samples across 43 human tissues from 175 individuals, was searched for SYK eQTL-related SNPs. Significant SNP-gene eQTL association was observed in samples from multiple tissue types including skin (P < 10−8) and EBV-transformed lymphocytes (P < 10−3) for three SYK promoter variants (rs290988, rs290987 and rs2562397) that are in complete LD (Figure 3, panel B). Homozygous carriers had significantly lower gene expression compared to the wild-type. These findings provided additional evidence suggesting SYK promoter variants could be potential modifiers of SYK expression. Indeed, one of the three SNPs (rs2562397) is located within the first GCbox motif (GGGCGG) that centered on the peak around exon1 (Figure E5) (see next section).
Transcriptomes of CD34, CD34B (G1 and G3), PBB, PBE and PDC
As a next step, the differences in the transcriptomes of various cell types that differentially express SYK protein were of interest. Specifically, this experimental series represented a search for transcription factors whose expression was consistent with the pattern of SYK expression in 6 cell types. The transcriptome for PBB has been published 13. This report will add information about CD34B cells and to enrich the comparison, add information about the peripheral blood eosinophil (PBE) and plasmacytoid dendritic cell (PDC). The utility of the PBE is that during development from CD34+progenitors, basophils and eosinophils are thought to develop along similar pathways16–18 and in these artificial culture conditions, eosinophil-like cells are observed to be generated transiently (unpublished reports). In addition, PBE express SYK at levels 10 times the levels in PBB and equal to the CD34+ progenitor 13. The implication is that the genetic program that determines SYK expression in the CD34B cells may be more similar to PBE than PBB (see results in the online repository and Figure E7). PDC’s express 30 times more SYK than PBB 13 and provide a useful perspective on regulation of this gene when it is not simply maintained at the level of the CD34+ progenitor but up-regulated during maturation to the cell obtained in circulation. The final comparisons included the changes that occur between CD34+, day 0 vs. day 21, CD34B and the differences between the G1 and G3 culture conditions that lead to lower SYK levels in G3. For some of the issues analyzed, the focus was on the presence of transcription factors and whether a pattern of mRNA expression for a particular transcription factor fit with the pattern of SYK expression across these various cell types.
There were several approaches to the general data analysis of the transcriptomes of these cells. Four issues related to general issues of transcriptomics are presented in the online supplement (see also Tables E5, E6 and E7). The fifth comparison relates to using all 6 transcriptomes to explore patterns consistent with SYK expression.
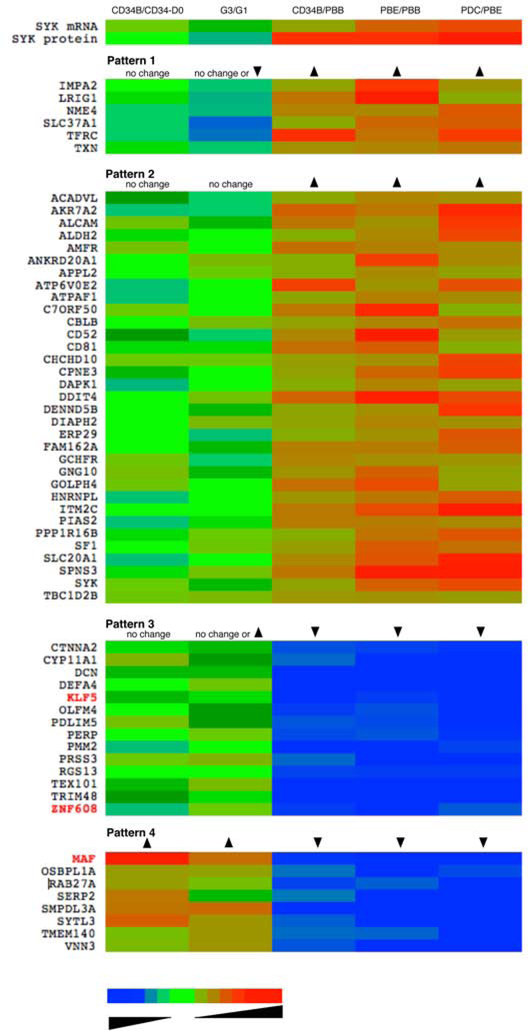
The entire data set was used to discover a pattern in specific genes that could be used to guide future studies and to suggest possible mechanisms of regulation, transcription factors that could be matched to the accessible regions of the SYK gene shown in Figure 2. Figure 4 summarizes the results of this analysis as a heatmap and Table E3 enumerates the specific data. The analysis began by defining a list of 2-fold changes when comparing mRNA expression in PBE vs. PBB. In order to capture a broad selection of differences, 2-fold would be considered a very liberal threshold since the Bonferroni, Benjamini-Hochberg false discovery rate (FDR), and paired t-test thresholds were 31-, 20- and 2.67-fold respectively. This list was then used to generate ratio data for 4 additional comparisons. At the top of the heatmap is the pattern for SYK mRNA levels. Although not part of this study (except for the G3/G1 comparison), the comparative levels for SYK protein are also noted (derived from 4, 14). The remainder of the heatmap shows genes that follow this pattern, either directly or in an inverted manner. There were 5 Boolean logic gates in the algorithm to generate the four transcript patterns noted in the figure. For example, one logic gate would be a test of whether the ratio of CD34B/CD34-D0 for a gene was unchanged (in these studies, high purity CD34 progenitors (D0=day0) were obtained from the Fred Hutchinson Cancer Research Center and used to generate the G1 culture transcriptomes). The logic gate for the G3/G1 difference was allowed to either reflect no change (defined as a fold-change between 0.5 and 1.5) or a decrease/increase (>2-fold). The logic gates for CD34B/PBB, PBE/PBB and PDC/PBB were set for an increase or decrease. The heatmap and table (Table E3) shows the logic gates and then list the transcripts that followed the pattern. The reasoning for this specific analysis is that 1) there may be changes in other elements that either follow the pattern of SYK and therefore be part of a similar regulatory network, 2) they are controlling elements in SYK regulation, or 3) they may be inhibitory elements that directly or indirectly influence SYK expression. There were 61 transcripts that fit these patterns of which 3 were known transcription factors; KLF5, ZNF608 and MAF (to be discussed below). Interestingly, JUN, FOS, AND MAF are transcription factors that regulate the IL-4 gene, which is only weakly expressed in the CD34B cells (see discussion).
Figure 4:
Pattern matching SYK expression with transcript behavior across different cell types, displayed as a heat-map (see the online repository for a tabulated presentation of the same data, Table E3. Using microarray results for 5 types of cells [peripheral blood basophils (PBB), peripheral blood eosinophils (PBE), peripheral blood plasmacytoid dendritic cells (PDC), CD34-derived basophils (CD34B) developed with the G1 or G3 protocols] transcripts were analyzed for following 4 patterns of expression. The short gene name is shown to the left of the heat-map and columns in the heat-map are colored for the ratio of the presence of the transcripts for the comparison shown at the top of the column. The arrows in the patterns indicate whether the ratio for a particular comparison is greater than 1.0, less than 1.0 or relatively unchanged. The colors represent the log-fold-change of the transcript for the particular comparison, ranging from 0.01 to 100, blue to red with shaded green representing changes near 1.0-fold (where unchanged refers to ratios between 0.75- and 1.33-fold).
The microarray results also provided a first look at whether there were differences in SYK mRNA between CD34B-G1 and G3 cells. To confirm the absence of a difference, qPCR was used on the 2 paired preparations used for microarray and one additional G1/G3 sample pair. Figure 1, panel E summarizes the results and Table E4 provides greater detail on these results, showing that there was no difference in SYK mRNA between the two types of cells. See Table E8 for a broader context of the differences between these two cell types.
Discussion
As noted in the introduction, SYK expression in peripheral blood basophils is uniquely low relative to nearly all other leukocytes despite being a required early tyrosine kinase linking FceRI aggregation to secretion. A model of basophil development (CD34B) that involved culture of CD34+ progenitors cells with IL-3 alone develops characteristics roughly consistent with PBB. However, it is clear that these cultures only generate basophil-like cells that are morphologically identical to PBB at a low frequency. And in the context of SYK, these cells clearly are not phenotypically like PBB. In our efforts to induce more productive CD34B cultures, we discovered that the addition of low concentrations of Flt3L and SCF would induce 2.5-fold increases in alcian blue positive cells and nearly 10-fold greater numbers of cells with condensed bi-lobed nuclei and cell size consistent with circulating basophils. In addition, this change was accompanied by decreases in SYK expression. This second model of basophil maturation allowed a side-by-side comparison to explore potential regulatory pathways. When coupled with comparisons to eosinophils, plasmacytoid dendritic cells, PBB or B cells, it becomes apparent that there are multiple potential controlling elements in the regulation of SYK expression.
Therefore, these studies created a new model of basophil generation from CD34+ progenitors. While our interest in the Flt3/SCF-induced changes in SYK expression motivates our interest in this model, it is apparent that these cytokines lead to cultures more enriched in cells resembling circulating basophils. Profiling the transcriptome shows that the G1 culture model produced a cell that resembles the basophil more than an eosinophil. Two-thirds of the transcripts that were used to test the similarity between CD34B and PBB move further towards the basophil profile in the G2/G3 culture model. However, the difference between G3 and G1 is not always in the direction of the basophil, with one-third of the transcripts moving away from the phenotype of the PBB, although not necessarily towards the eosinophil.
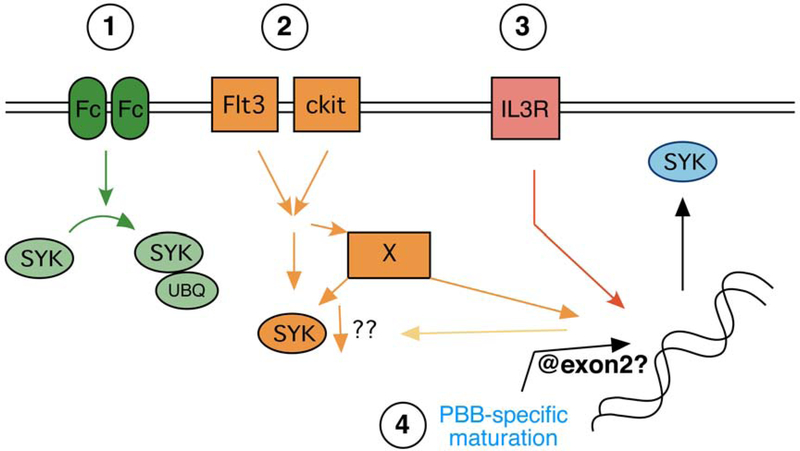
Figure 5 summarizes the pathways that this study and prior studies now suggest result in regulation of SYK expression. At the far left of the cartoon is the well-established FceRI-mediated ubiquinylation of SYK as a post-translational process of down-regulation. This mechanism of SYK down-regulation is well studied and aggregation is found to have a significant effect; the median reduction following an optimal level of FceRI aggregation will result in 70% losses of SYK 20 but in some preparations reductions of 90% are observed in vitro. Studies of this specific process led to a recent study that explored the possibility that auto-antibodies could be a driver of the high variability of SYK protein expression in the general population 12. With some caveats, this recent study concluded that there was no evidence for such a process in most subjects. However, when it was possible to demonstrate the presence of auto-antibodies, as found in 5–10% of patients with chronic spontaneous urticaria (CSU), it was evident that such a mechanism could be a possible cause of significant SYK reduction. These auto-antibodies could also reduce SYK expression in basophils maturing in CD34B cultures. But, in general, this is not a satisfactory explanation for the low levels of SYK expression in PBB compared to other circulating leukocytes nor a reasonable explanation for the high variability in SYK expression among non-CSU subjects.
Figure 5:
Synopsis of the possible mechanisms of regulating SYK expression in basophils. There are four proposed mechanisms based on available information for regulation of SYK expression.
The current studies frame the regulatory pathways in a general sense. Previous studies showed that mRNA for SYK showed similar rates of disappearance in PBB and CD34B following the addition of actinomyin D 4, 14 (and unpublished results) suggesting that the increased steady-state level of mRNA in CD34B relative to PBB was not a result of differences in mRNA stability. The levels of SYK mRNA show a reasonable correlation with protein expression (R2 = 0.29)4, also suggesting a dependence on transcription (with no evidence for mRNA stability differences). But the current studies do reveal that there is at least one mechanism, exposure to Flt3L and/or SCF, that is operating independently of a mechanism that regulates SYK mRNA levels. Further work will be needed to illuminate the precise mechanisms. The Flt3/ckit receptors together contribute to a down-regulation of SYK but not through a down-regulation of SYK mRNA. These two receptors have been shown, in some cellular contexts, to interact with c-CBL and therefore may mediate a loss of SYK by mechanisms similar to FceRI (although this requires further study). The G1/G3 results also re-raise the possibility of a miRNA that regulates SYK translation without degradation of the mRNA. It is also possible that Flt3L and SCF act indirectly to alter SYK generation (represented by the ‘X’ factor in the cartoon or the arrow leading to change indirectly through the nucleus). IL-3 is known to induce an increase in SYK mRNA (with more modest effects on protein synthesis 5, 13), suggesting another regulatory pathway that acts on transcription. In other words, IL-3 increases SYK mRNA with only minor effects on SYK protein while Flt3L/SCF result in no change in SYK mRNA with marked effects on SYK protein. Finally, the ATACseq results suggest some regulation at the level of DNA organization (see discussion below).
If there is transcriptional regulation of the SYK, which has been suggested in previous studies, the pattern matching shown in Figure 4 and Table E3 suggests 3 transcription factors that may participate in this process. A bioinformatics-based screening identified several SYK promoter transcription factors (TFs) and cis-elements in the regions identified in the ATACseq. Even with some stringency, there are, not surprisingly, many potential transcription factors that could regulate this gene. Some of the possibilities are notable for their connections to other pieces of information. Five merit some discussion and are further explored in the online repository.
The tabulated patterns for transcription factors suggest a connection with c-JUN whose expression is high in CD34B and PBE and low in basophils. Indeed, basophils don’t express c-JUN protein. Perhaps linked with c-JUN are two transcription factors whose patterns of expression are tightly matched with the expression pattern of SYK in the cells studied (see Figure 4). For the pattern displayed by c-MAF, the prediction is that c-MAF is a repressor of transcription. It is not uncommon to find an inverted relationship for c-MAF and c-JUN in the regulation of a gene. This is an interesting possibility because c-MAF is also a transcription factor important in IL-4 secretion 21, a cytokine that is a hallmark of basophil secretion 22. It may be relevant that T cells also use c-MAF for IL-4 gene transcription and T cells also suppress SYK expression, as noted above. Also, CD34B express IL-4 poorly relative to PBB, a result consistent with the poor expression of c-MAF in PBE and CD34B. But, as seen in Figure 4 and Table E3, c-MAF mRNA is increased in G3 cells relative to G1 cells while mRNA for SYK was unchanged. There are canonical c-MARE or c-MAF cis-elements within the range of ATACseq peaks ‘A’, 1 and 2. See Figures E5 and E6 for transcription factor cis-element motifs relevant to c-MAF.
The transcription factor that most closely associates with the pattern of SYK expression is KLF5. There is little known about this transcription factor except that its target cis-element is a GC box. There are two such DNA sequences near the TSS for SYK (see figure E5). The pattern for KLF5 is consistent with a repressive element. Similarly, ZNF608 shows an inverse pattern consistent with repression of SYK expression (see Figures E5 and E6). It is interesting that of all the open regions identified in basophils and B-cell, only the regions in basophils show canonical sequences known to bind c-MAF, KLF5 or ZNF608 (in the latter case, the motifs for ZNF608 can only be guessed from other zinc finger transcription factors 23, 24 because there has been insufficient study of ZNF608).
A potentially interesting peak is labeled ‘3’ in Figure 2. It should be noted that the MACS2 algorithm as employed does not generate a peak-call for a peak of this size but it is present in all 3 basophils tracks and not in B-cells or CD34B. As noted above, this region is located around the start of exon 2 of the SYK gene. Splicing of most proximal stretch (but most distal of the TSS) of the 5’UTR of mature SYK mRNA occurs in this region so it may represent a region that provides an opportunity for regulation. For example, there is a canonical c-MARE (c-MAF response element) located in this region (see Figure E6). Additional study would be needed to draw stronger conclusions regarding its presence or linkage to SYK expression and why it is more evident in mature PBB and not CD34B or B-cells (given that SYK transcription would require splicing in this region). The very strong peaks associated only with B-cells are interesting although given the similar expression of SYK in B cells and CD34B cells, it is not clear how these regions are influencing B-cells to produce SYK levels higher than PBB, but levels that are only similar to CD34B.
These studies also provide a more complete perspective on the CD34B model of basophil development. It has been know for some time that the cultures were imperfect in producing a cell with all the characteristics of the mature peripheral blood basophil. These results expand on the prior impressions with transcriptome and ATACseq information; both indicate that there are many similarities to PBB but the model remains imperfect. However, the studies also add a new tool, the G3 cultures, to explore regulation of SYK expression. These studies identify a potentially unique region of regulation in the SYK gene in PBB and expose three potential transcription factors, c-MAF, KLF5, and ZNF608, whose expression may modulate the expression of SYK and may be linked to basophil differentiation.
Supplementary Material
Key Messages.
The tyrosine kinase SYK, a critical early signal transduction element in IgE-mediated secretion, is regulated by two pathways.
Flt3L and SCF down-regulate SYK expression during basophil development from CD34+ stem cell progenitors.
From ATACseq results, the SYK gene in peripheral blood basophils is accessible for transcription, despite the low level of SYK expression in these cells, with two regions in the gene that are uniquely present when compared to B-cells where SYK is expressed at 10-times higher levels.
Transcription factors, KLF5, ZNF608 and c-MAF were identified as potential repressor elements in the low expression of SYK in peripheral blood basophils.
Acknowledgements
The authors would like to thank Valerie Alexander for her excellent technical assistance and Dr. John Schroder for supplying samples of 8-week CD34-derived human mast cells for comparisons and plasmacytoid dendritic cells for mRNA. This research was supported by NIH grant AI100952.
Funding: These studies were funded by NIH AI100952.
Abbreviations
- SYK
spleen tyrosine kinase
- SCF
Stem Cell Factor
- Flt3L
Flt3 ligand
- PBB
peripheral blood basophil
- PBE
peripheral blood eosinophil
- PDC
plasmacytoid dendritic cell
- TSS
transcription start site
- UTR
untranslated region
- eQTL
expression quantitative trait loci
Footnotes
COI: All authors have no conflicts of interest relevant to this manuscript to report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vilarino N, MacGlashan D Jr. Transient transfection of human peripheral blood basophils. J Immunol Methods 2005; 296:11–8. [DOI] [PubMed] [Google Scholar]
- 2.MacGlashan DW Jr., Undem BJ. Inducing an Anergic State in Mast Cells and Basophils without Secretion. J. Allergy Clin. Immunol 2008; 121:1500–6. [DOI] [PubMed] [Google Scholar]
- 3.Havard S, Scola AM, Kay LJ, Ishmael SS, Macglashan DW Jr., Peachell PT. Characterization of syk expression in human lung mast cells: relationship with function. Clin Exp Allergy 2011; 41:378–88. [DOI] [PubMed] [Google Scholar]
- 4.Ishmael SS, MacGlashan DW Jr. Syk expression in peripheral blood leukocytes, CD34+ progenitors, and CD34-derived basophils. J Leukoc Biol 2010; 87:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacGlashan DW Jr. Relationship Between Syk and SHIP Expression and Secretion from Human Basophils in the General Population. J. Allergy Clin. Immunol 2007; 119:626–33. [DOI] [PubMed] [Google Scholar]
- 6.Ishmael S, MacGlashan D Jr. Early signal protein expression profiles in basophils: a population study. J Leukoc Biol 2009; 86:313–25. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen KL, Gillis S, MacGlashan DW Jr. A comparative study of releasing and nonreleasing human basophils: nonreleasing basophils lack an early component of the signal transduction pathway that follows IgE cross-linking. J Allergy Clin Immunol 1990; 85:1020–9. [DOI] [PubMed] [Google Scholar]
- 8.Kepley CL, Youssef L, Andrews RP, Wilson BS, Oliver JM. Syk deficiency in nonreleaser basophils. J Allergy Clin Immunol 1999; 104:279–84. [DOI] [PubMed] [Google Scholar]
- 9.Lavens-Phillips SE, MacGlashan DW Jr. The tyrosine kinases, p53/56lyn and p72syk are differentially expressed at the protein level but not at the mRNA level in non-releasing human basophils. Amer. J. Resp. Cell Mol. Biol 2000; 23:566–71. [DOI] [PubMed] [Google Scholar]
- 10.MacGlashan DW Jr., Lavens-Phillips S Characteristics of the free cytosolic calcium timelag following IgE-mediated stimulation of human basophils: Significiance for the non-releasing basophil phenotype. J. Leuk. Biol 2001; 69:224–32. [PubMed] [Google Scholar]
- 11.MacGlashan D, Miura K. Loss of syk kinase during IgE-mediated stimulation of human basophils. J Allergy Clin Immunol 2004; 114:1317–24. [DOI] [PubMed] [Google Scholar]
- 12.MacGlashan D Autoantibodies to IgE and FcepsilonRI and the natural variability of spleen tyrosine kinase expression in basophils. J Allergy Clin Immunol 2019; 143:1100–7 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacGlashan D Jr. Expression profiling of human basophils: modulation by cytokines and secretagogues. PLoS One 2015; 10:e0126435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacGlashan D Jr. Stability of Syk protein and mRNA in human peripheral blood basophils. J Leukoc Biol 2016; 100:535–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bedi R, Du J, Sharma AK, Gomes I, Ackerman SJ. Human C/EBP-epsilon activator and repressor isoforms differentially reprogram myeloid lineage commitment and differentiation. Blood 2009; 113:317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denburg JA, Tanno Y, Bienenstock J. Growth and differentiation of human basophils, eosinophils, and mast cells In: Mast cell differentiation and heterogeneity. New York: Raven Press; 1986. p. 71–83. [Google Scholar]
- 17.Otsuka H, Dolovich J, Befus AD, Telizyn S, Bienenstock J, Denburg JA. Basophilic cell progenitors, nasal metachromatic cells, and peripheral blood basophils in ragweed-allergic patients. J Allergy Clin Immunol 1986; 78:365–71. [DOI] [PubMed] [Google Scholar]
- 18.Denburg JA, Dolovich J, Harnish D. Basophil mast cell and eosinophil growth and differentiation factors in human allergic disease. Clin Exp Allergy 1989; 19:249–54. [DOI] [PubMed] [Google Scholar]
- 19.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012; 22:1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacGlashan DW Jr., Ishmael S, Macdonald SM, Langdon JM, Arm JP, Sloane DE. Induced Loss of Syk in Human Basophils by Non-IgE-Dependent Stimuli. J Immunol 2008; 180:4208–17. [DOI] [PubMed] [Google Scholar]
- 21.Lai CY, Lin SY, Wu CK, Yeh LT, Sytwu HK, Miaw SC. Tyrosine phosphorylation of c-Maf enhances the expression of IL-4 gene. J Immunol 2012; 189:1545–50. [DOI] [PubMed] [Google Scholar]
- 22.MacGlashan D Jr., White JM, Huang SK, Ono SJ, Schroeder JT, Lichtenstein LM. Secretion of IL-4 from human basophils. The relationship between IL-4 mRNA and protein in resting and stimulated basophils. J Immunol 1994; 152:3006–16. [PubMed] [Google Scholar]
- 23.Yang S, Chen J, Guo Y, Lin H, Zhang Z, Feng G, et al. Identification of prognostic biomarkers for response to radiotherapy by DNA microarray in nasopharyngeal carcinoma patients. Int J Oncol 2012; 40:1590–600. [DOI] [PubMed] [Google Scholar]
- 24.Vandevenne M, Jacques DA, Artuz C, Nguyen CD, Kwan AH, Segal DJ, et al. New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217. J Biol Chem 2013; 288:10616–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.