Abstract
Lafora disease (LD) is a rare, fatal form of progressive myoclonus epilepsy. The molecular basis of this devastating disease is still poorly understood and no treatment is available yet, which leads to the death of the patients around 10 years from the onset of the first symptoms. The hallmark of LD is the accumulation of insoluble glycogen-like inclusions in the brain and peripheral tissues, as a consequence of altered glycogen homeostasis. In addition, other determinants in the pathophysiology of LD have been suggested, such as proteostasis impairment, with the reduction in autophagy, and oxidative stress, among others. In order to gain a general view of the genes involved in the pathophysiology of LD, in this work, we have performed RNA-Seq transcriptome analyses of whole-brain tissue from two independent mouse models of the disease, namely Epm2a−/− and Epm2b−/− mice, at different times of age. Our results provide strong evidence for three major facts: first, in both models of LD, we found a common set of upregulated genes, most of them encoding mediators of inflammatory response; second, there was a progression with the age in the appearance of these inflammatory markers, starting at three months of age; and third, reactive glia was responsible for the expression of these inflammatory genes. These results clearly indicate that neuroinflammation is one of the most important traits to be considered in order to fully understand the pathophysiology of LD, and define reactive glia as novel therapeutic targets in the disease.
Keywords: Lafora disease, progressive myoclonus epilepsy, transcriptome, neuroinflammation, reactive astrocytes, activated microglia
INTRODUCTION
Lafora progressive myoclonus epilepsy (Lafora disease, LD, OMIM254780, ORPHA#501) is a rare, fatal autosomal recessive neurological disorder characterized by epilepsy, neurodegeneration and the accumulation of insoluble aberrant forms of glycogen called Lafora bodies (LBs) in brain and other peripheral tissues [[1], [2]]. LD occurs during childhood or early adolescence and it is characterized by generalized tonic-clonic seizures, myoclonus, absences, and visual seizures. Patients present a rapid progression of dementia with amplification of seizures, leading to death after a decade from the onset of the first symptoms [[1], [2]]. LD is caused by mutations in two genes: EPM2A, encoding the glucan phosphatase laforin [[3], [4]], and EPM2B, encoding the E3-ubiquitin ligase malin [5]. Laforin and malin form a functional complex [[6], [7], [8]] which is involved in the regulation of glycogen synthesis, the homeostasis of glucose transporters, the maintenance of proteostasis and the response to oxidative stress, among others physiological pathways [[1], [2], [9]]. The formation of this functional complex would explain why patients with mutations in laforin or malin are neurologically and histologically indistinguishable [10].
In spite of the efforts made by different groups during the last decade, the molecular basis of the pathophysiology of LD is still poorly understood and no treatment is available yet for this devastating disease. In order to gain knowledge on the pathophysiology of LD, several animal models of the disease have been used (e.g. dogs, mice). Among them, the murine models of LD have been extensively used: Epm2a−/− mice lack exon 4 from the Epm2a gene [11] and Epm2b−/− mice lack the single exon present in the Epm2b gene [[12], [13], [14]]. Both mouse models present similar pathophysiological phenotypes, i.e., they show similar behavioral impairments [15], they are more sensitive to the effects of the pro-epileptic drug pentylenetetrazole (PTZ) [16], they accumulate LBs in the brain and other peripheral tissues [[12], [13], [14]], they show altered autophagy [[14], [17], [18]] and they show reactive astrocytes and microglia [[13], [19], [20], [21]]. In these LD mouse models, the disease progresses with age, being the pathological phenotypes more severe as the animals get older [[11], [15], [20]].
Since LD is quite a complex disease, we decided to use an unbiased “omic” approach in order to obtain a general view of gene expression alterations and to identify possible markers that could elucidate the mechanisms underlying this disorder. With this aim, we analyzed the genes that were differentially expressed in the brain of Epm2a−/− and Epm2b−/− mice in comparison to control animals by using RNA-Seq technology, at two different ages: at 3 months of age (at the beginning of the development of the pathophysiological phenotypes) and at 16 months of age, when a florid pathophysiology is presented. It is assumed that RNA-Seq technique is superior to microarrays since it provides wide coverage of the differentially expressed genes and allows also the detection of more genes with greater fold-change, avoiding the problem of signal oversaturation [[22], [23]]. Our results indicate that Epm2a−/− and Epm2b−/− mouse brains overexpress a common set of genes mostly related to inflammation and that the expression of these genes increases with the age of the animals. We also provide evidence that reactive glia is responsible for the expression of these genes. These results are consistent with the presence of pro-inflammatory markers (i.e., IL-1β, TNFα and IL-6) in the brain of Epm2a−/− and Epm2b−/− mice [20] and suggest that neuroinflammation should be considered as one of the most important traits in Lafora disease.
MATERIAL AND METHODS
Ethic statement, animal care, mice and husbandry
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Consejo Superior de Investigaciones Cientificas (CSIC, Spain) and approved by the Conselleria de Agricultura, Medio Ambiente, Cambio Climático y Desarrollo Rural from the Generalitat Valenciana. All mouse procedures were approved by the animal committee of the Instituto de Biomedicina de Valencia-CSIC [Permit Number: (IBV-16), 2015/VSC/PEA/00029]. All efforts were made to minimize animal suffering. Homozygous Epm2a−/− and Epm2b−/− in a pure C57BL/6JRccHsd background and the corresponding controls of 3, 5, 7, 12 and 16 months of age were used in this study. The presence of polyglucosan inclusions (the hallmark of LD) in the brain of the Epm2a−/− and Epm2b−/− mouse lines was confirmed by PAS staining (Supplementary Fig. S1). Mice were maintained in the IBV-CSIC facility on a 12/12 light/dark cycle under constant temperature (23°C) with food and water provided ad libitum. Male mice were sacrificed by cervical dislocation and whole brains rinsed twice with cold PBS and frozen immediately in liquid nitrogen. When planning the experiments the principles outlined in the ARRIVE guidelines and the Basel declaration including the 3R concept have been considered.
Whole RNA extraction from mouse brain
Frozen brains were pulverized in a liquid nitrogen-cooled stainless steel mortar and pestle. Then, 1 ml of TRIzol™ Reagent (Thermo Fisher Scientific, Madrid, Spain) was added per 100 mg of pulverized brain and homogenated by pipetting up/down five times with 1 ml tip to break tissue into small pieces. The homogenates were passed ten times through a 21 gauge needle in 2 ml syringe and incubated 5 min at room temperature (RT) on a plate shaker to allow complete lysis. Samples were centrifuged for 10 min at 12,000×g and 4°C. Supernatants were transferred to a new tube. Next, 0.2 ml of chloroform per 1 ml of TRIzol™ Reagent was added to each tube, mixed by inversion ten times and incubated for 3 min at RT. Samples were centrifuged at 12,000×g for 15 min at 4°C and the top aqueous phase (approx. 0.6 ml) was transferred to a new tube. Finally, RNAs were precipitated by adding 1 volume of isopropanol to the aqueous phase, mixed by inversion and incubated for 10 min at RT. Samples were centrifuged for 15 min at 12,000×g and 4°C. RNA pellets were washed twice with ethanol 75%, air-dried and resuspended in 75 μL RNase-free water. RNA purity was assessed using a NanoDropONE spectrophotometer (ThermoFisher, Madrid, Spain) and quality was evaluated using RNA 6000 Nano kit and Agilent 2100 Bioanalyzer System (Agilent, Madrid, Spain). All samples had an RNA Quality Number higher than 9.0, except one sample corresponding to Epm2a−/− mouse of 16 months of age that was excluded from the downstream analyses.
RNA-Seq analysis
The RNA-Seq experiment was conducted by the Multigenic Analysis Unit from the UCIM-INCLIVA (University of Valencia, Valencia Spain). 1 μg of total RNA was used for library preparation. cDNA libraries were prepared using the TruSeq Stranded mRNA LP (48 Spl) (Illumina, San Diego, CA), according to the manufacturer’s instructions. The concentration of cDNA libraries was measured using Qubit 3.0 and the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, USA). The length distribution of library fragments was determined using the Agilent 2100 Bioanalyzer using the Agilent DNA 1000 Kit (Agilent, USA). Libraries were then sequenced on the Illumina® NextSeq 550 to generate 75-bp single reads using the Illumina® NSQ 500 High Output KT v2 (75 CYS) (Illumina, San Diego, CA), according to the manufacturer’s protocol.
RNA-Seq data analysis
Data analysis was performed with the support of EpiDisease S.L. (a spin-off from the Center for Biomedical Network Research, CIBER-SCIII, Spain). All reads were aligned using the mouse reference genome (GRCm38 version) from Ensembl. Subsequently, the intersection between the aligned position of reads and the gene coordinates was performed, i.e. counting the number of reads overlapping a gene. These steps were performed using the Subread/Rsubread packages (http://subread.sourceforge.net/) which comprise a suite of high-performance software programs for processing next-generation sequencing data. Genes with very low counts across all libraries provide little evidence for differential expression. These genes were filtered out prior to further analysis. In our analysis, a gene should have more than 1 count-per-million (CPM) in at least 3 samples (the size of the smallest group) to be considered; otherwise, the expression of the gene was discarded. Subsequently, trimmed mean of M-values (TMM) normalization [24] was performed to eliminate composition biases between libraries. The specific dispersions per gene were estimated with a negative binomial distribution [[25], [26]]. The differential expression analysis was executed using a quasi-likelihood F-test [27]. Raw p-values were corrected for multiple testing using the Benjamini-Hochberg method and the FDR (false discovery rate) calculated accordingly [28]. Before carrying out the differential expression analysis, data were explored by generating a multi-dimensional scaling (MDS) plot. This visualizes the differences between the expression profiles of different samples in two dimensions. The number of unique and common differentially expressed genes in Epm2a−/− and Epm2b−/− samples compared to the control group was represented in Venn diagrams. Besides, samples were hierarchically clustered by their gene expression pattern similarity and represented in a heatmap. Volcano plots were used to represent the proportion of differentially expressed genes obtained from the Epm2a−/− vs control and Epm2b−/− vs control comparisons. The 30 most upregulated genes in both comparisons (FC > 2 and FDR < 0.01) were labeled.
Functional gene clustering and network analysis
Gene enrichment analysis was applied to classify differentially expressed genes according to different GO Biological Processes. Pathways that were enriched in the list of differentially expressed genes were identified using the DAVID tools v6.7 (Database for Annotation, Visualization and Integrated Discovery; https://david.ncifcrf.gov/tools.jsp) [29].
Gene expression validation by RT-qPCR analyses
Nine genes were chosen and tested to confirm their expression levels using SYBR green-based RT-qPCR. For each reaction, a total of 60 ng of total RNA from each brain sample was reverse transcribed and PCR amplified by qPCR in a total volume of 12 μl using PowerUp SYBR Green Master Mix (Applied Biosystems, Madrid, Spain), Protector RNase Inhibitor (Roche, Barcelona, Spain), MultiScribe Reverse Transcriptase (Applied Biosystems, Madrid, Spain) and specific primers, according to the manufacturer’s protocol. The gene-specific primers were designed using Primer3Plus software version 2.4.2 [30], validated by BLAST (Basic Local Alignment Search Tool) analysis on NCBI and synthesized by Sigma-Aldrich (Madrid, Spain). The primer sequences are listed in Supplementary Table S1. SYBR green-based one-step RT-qPCR was performed under the following conditions: 48° C for 30 min and 95° C for 10 min, followed by 40 cycles of 95° C for 15 s, 60° C for 1 min, and 60° C to 95° C in increments of 0.5° C for 30 s to generate melting curves, on a StepOnePlus Real-Time PCR System (Applied Biosystems, Madrid, Spain). The data were processed using StepOnePlus software version 2.3 and expression values were calculated using the comparative Ct method. Each qPCR reaction was performed on eight biological and three technical replicates each in samples from 16 months old animals. The β-Actin (Actb) gene was used as the endogenous reference control to normalize target gene expression. The comparative analyses for every gene among the biological groups were assessed by a one-way nested analysis of variance (ANOVA), followed by a post-hoc Tukey’s comparison test using GraphPad Prism 5 software. Values were expressed as means and horizontal bars indicate standard error of the means (SEM).
Western blot analyses
Mouse brain homogenates were lysed in RIPA buffer [50 mM Tris-HCl, pH 8; 150 mM NaCl; 0.5% sodium deoxycholate; 0.1% SDS; 1% Nonidet P40; 1mM PMSF; 100 μM leupeptin and complete protease inhibitor cocktail (Roche, Barcelona, Spain)] for 30 min at 4°C with occasional vortexing. The homogenates were passed ten times through a 25 gauge needle in 2 ml syringe and centrifuged at 13,000xg for 15 min at 4°C. Supernatants were collected and a total of 25 μg protein was subjected to SDS-PAGE and transferred onto a PVDF membrane. Membranes were blocked with 5% (w/v) nonfat milk in Tris-buffered saline (TBS-T: 50 mM Tris-HCl, 150 mM NaCl, pH 7.4) with 0.1% Tween-20 for 1 h at room temperature and incubated overnight at 4°C with the corresponding primary antibodies: anti-Lcn2 (Abcam, ab63929), anti-Ccl5 (Abcam, ab 189841), anti-Cxcl10 (Preprotech, 500-P129) and anti-Actin (Sigma, A2066). Then, membranes were probed with suitable secondary antibodies for 1 h at room temperature. Signals were obtained by chemiluminescence using an image reader Fuji-LAS-4000 (GE Healthcare, Barcelona, Spain) and Lumi-Light Western Blotting Substrate (Roche Applied Science, Barcelona, Spain). The results were analyzed using the software Image Studio version 5.2 (LI-COR Biosciences, Germany). Experiments were performed in at least three individuals from each genotype. Protein levels related to the levels of actin were referred to the levels found in control samples. Since in the case of the anti-Ccl5 and anti-Cxcl10 antibodies they recognize different forms of the chemokines, probably depending on their oxidative conditions, in the densitometry analyses we quantified the region of the gels containing all the forms. Results are shown as means +/− standard error of the mean (SEM). Differences between paired samples were analyzed by two-tailed Student’s t-tests using Graph Pad Prism version 5.0 statistical software (La Jolla, CA, USA). P values have been considered as *P<0.05 and ***P< 0.001.
Immunohistofluorescence (IHF) analyses
Sixteen-months-old mice were sacrificed by cervical dislocation. Brains were recovered and the right hemisphere was fixed in 4% paraformaldehyde in phosphate buffer saline (PBS) for 24 hours at 4°C. After three washes with PBS, the samples were dehydrated and embedded in paraffin and sectioned at 4 μm using a microtome HM-340E (Microm, Madrid, Spain). Sections were deparaffined, rehydrated and microwave antigen retrieval was performed for 10 min in 10 mM citrate buffer pH 6.0. Sections were immersed in blocking buffer (1% BSA, 10% FBS, 0.2% Triton X100, in PBS) and incubated O/N at 4°C with primary antibody diluted in blocking buffer: anti-Lcn2 (1/25, Novus Biologicals NBP1-05183), anti-Cxcl10 (1/25, Abcam ab8098), anti-Gfap (1/300, Synaptic Sytems 173 004), anti-Gfap (1/200, Sigma G3893) and anti-Iba1 (1/50, Waco 019-19741). After three washes of 10 min in PBS, sections were incubated for one hour at room temperature with the appropriate secondary antibody diluted at 1/500 in blocking buffer without Triton X100, washed once with PBS, incubated with DAPI (Sigma, Madrid, Spain), washed twice with PBS and mounted in AquaPolymount (Polysciences Inc., USA). Images were acquired in a Confocal Spectral Leica TCS SP8 microscope (Leica, Wetzlar, Germany).
RESULTS
Identification of genes differentially expressed in the brain of 16 months-old Epm2a−/− and Epm2b−/− mice compared to control animals
In order to identify the genes that were differentially expressed in the brain of Epm2a−/− and Epm2b−/− mice compared to control animals we performed RNA-Seq analyses from whole brain tissue homogenates of 16 months-old animals. Analyses were initially designed for 4 samples of each biological group but one Epm2a−/− sample was discarded because of its poor quality (see Material and Methods). The principal component analysis (PCA) showed that samples with the same genotype clustered together and were separated from the other groups (Supplementary Fig. S1). Differential expression analysis between Epm2a−/−, Epm2b−/− and control groups identified only 28 and 19 genes down-regulated with more than 0.5 fold change (FC) in the Epm2a−/− and Epm2b−/− mouse brains, respectively. As expected, the most down-regulated gene in the Epm2a−/− mice was Epm2a and the one in Epm2b−/− mice was Epm2b, thus validating the quality of the RNA-Seq analyses. The rest of down-regulated genes corresponded mainly to either in silico predicted genes or those with unknown function (Supplementary Table S2).
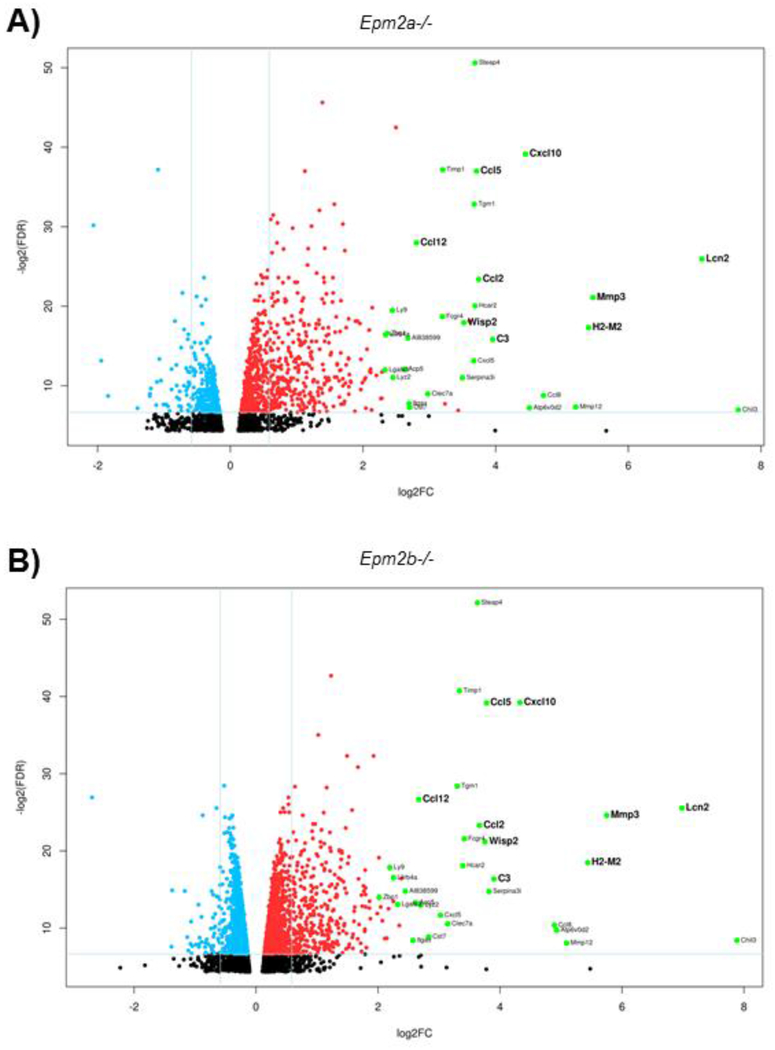
More importantly, the analyses identified 456 genes in the Epm2a−/− samples and 442 genes in the Epm2b−/− samples that were upregulated more than 1.5 FC as compared to the control samples (Supplementary Table S3 and S4, respectively), with a false discovery rate (FDR) lower than 0.05. Among these, 334 genes were similarly upregulated in both Epm2a−/− and Epm2b−/− mice, 122 genes were specifically upregulated in the Epm2a−/− mice whereas 108 genes were specifically upregulated in the Epm2b−/− mice. A volcano plot of the differentially expressed genes with an FC higher than 1.5 or lower than 0.5 and an FDR lower than 0.01 is shown in Fig. 1.
Figure 1. Differentially upregulated genes outnumber the downregulated ones.
Volcano plots of differentially expressed genes in Epm2a−/− vs control (A) and Epm2b−/− vs control (B) animals of 16 months of age. Horizontal lines indicate the relative expression fold change (log2FC) compared to controls. The vertical line represents the FDR value (−log2FDR). The red and blue points are significantly upregulated and downregulated genes, respectively, in Epm2a−/− vs control and Epm2b−/− vs control samples (FDR <0.01, FC ≥1.5 or ≤0.5). The most upregulated genes in Epm2a−/− and Epm2b−/− vs control are labeled in green and the name of the gene written on their right side. Genes selected for validation by RT-qPCR are shown in bold. Black points belong to genes whose FDR values were higher than 0.01 and FC values were lower than 1.5. Values corresponding to Epm2a (FC 0.04, FDR 1.76E-08) and Epm2b (FC 0.00, FDR 4.50E-21) genes have been omitted from the corresponding plots to allow a better comparative representation between the differentially expressed genes in Epm2a−/− and Epm2b−/− samples.
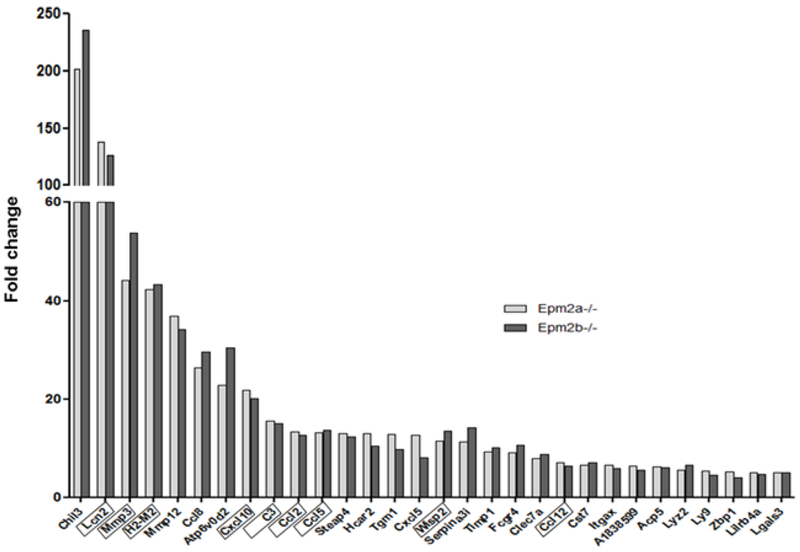
Using a more selective criteria of up-regulation (>2.0 FC increase in both groups, or when up-regulation occurred with a >2.0 FC increase in at least one of the groups), we identified 229 genes that were upregulated in both Epm2a−/− and Epm2b−/− samples and, surprisingly, each of them exhibited a similar FC in both groups (Supplementary Table S5). Fig. 2 and Table I describe the 30 common genes with the highest up-regulation fold in both Epm2a−/− and Epm2b−/− groups having an FDR lower than 0.01. These genes are also indicated in the volcano plots (Fig. 1). The fact that under these conditions of stringency (FC higher than 2.0 in at least one sample and an FDR lower than 0.01) the coincidences reached around 80% of the upregulated genes in both Epm2a−/− and Epm2b−/− samples with respect to controls is compatible with the fact that both Epm2a−/− and Epm2b−/− mice show similar pathological phenotypes (see Introduction). These results were also in agreement with the similar clinical presentation of patients with mutations in either EPM2A or EPM2B genes (see Introduction).
Figure 2. Genes commonly upregulated in both LD models show similar levels of expression.
Representation of the 30 genes with the most upregulated fold change in both Epm2a−/− and Epm2b−/− mice of 16 months of age with an FDR <0.01 are arranged according to their fold change. The genes used for validation analyses by qRT-PCR are boxed.
Table I:
30 genes with the most upregulation fold in both Epm2a−/− and Epm2b−/− samples of 16 months old animals and with an FDR <0.01 in both samples. The gene name, false discovery rate (FDR), fold change (FC), description and MGI Id (mouse genome informatics) are indicated. Genes are arranged according to FC. In grey, genes validated by RT-qPCR (see text for details).
| Epm2a−/− | Epm2b−/− | |||||
|---|---|---|---|---|---|---|
| Gene | FDR | FC | FC | FDR | Description [MGI Id] | |
| Chil3 | 0.0079 | 201.73 | 236.11 | 0.0029 | chitinase-like 3 [Id:1330860] | |
| Lcn2 | 1.57E-08 | 138.06 | 126.29 | 2.05E-08 | lipocalin 2 [Id:96757] | |
| Mmp3 | 4.46E-07 | 44.19 | 53.78 | 3.91E-08 | matrix metallopeptidase 3 [Id:97010] | |
| H2-M2 | 6.15E-06 | 42.23 | 43.35 | 2.71E-06 | histocompatibility 2. M region locus 2 [Id:95914] | |
| Mmp12 | 0.0062 | 36.94 | 34.09 | 0.0037 | matrix metallopeptidase 12 [Id:97005] | |
| Ccl8 | 0.0022 | 26.36 | 29.66 | 0.0007 | chemokine (C-C motif) ligand 8 [Id:101878] | |
| Atp6v0d2 | 0.0068 | 22.77 | 30.48 | 0.0011 | ATPase. H+ transporting. lysosomal V0 subunit D2 [Id:1924415] | |
| Cxcl10 | 1.65E-12 | 21.84 | 20.06 | 1.57E-12 | chemokine (C-X-C motif) ligand 10 [Id:1352450] | |
| C3 | 1.73E-05 | 15.51 | 14.94 | 1.18E-05 | complement component 3 [Id:88227] | |
| Ccl2 | 9.12E-08 | 13.40 | 12.68 | 9.69E-08 | chemokine (C-C motif) ligand 2 [Id:98259] | |
| Ccl5 | 7.37E-12 | 13.10 | 13.71 | 1.61E-12 | chemokine (C-C motif) ligand 5 [Id:98262] | |
| Steap4 | 5.84E-16 | 12.91 | 12.37 | 2.03E-16 | STEAP family member 4 [Id:1923560] | |
| Hcar2 | 9.29E-07 | 12.91 | 10.49 | 3.62E-06 | hydroxycarboxylic acid receptor 2 [Id:1933383] | |
| Tgm1 | 1.32E-10 | 12.80 | 9.85 | 2.86E-09 | transglutaminase 1. K polypeptide [Id:98730] | |
| Cxcl5 | 0.0001 | 12.73 | 8.13 | 0.0003 | chemokine (C-X-C motif) ligand 5 [Id:1096868] | |
| Wisp2 | 4.03E-06 | 11.47 | 13.49 | 4.24E-07 | WNT1 inducible signaling pathway protein 2 [Id:1328326] | |
| Serpina3i | 0.0004 | 11.30 | 14.12 | 3.66E-05 | serine (or cysteine) peptidase inhibitor. clade A. member 3I [Id:2182841] | |
| Timp1 | 6.47E-12 | 9.19 | 10.09 | 5.48E-13 | tissue inhibitor of metalloproteinase 1 [Id:98752] | |
| Fcgr4 | 2.37E-06 | 9.15 | 10.65 | 3.22E-07 | Fc receptor. IgG. low affinity IV [Id:2179523] | |
| Clec7a | 0.0020 | 7.87 | 8.82 | 0.0006 | C-type lectin domain family 7. member a [Id:1861431] | |
| Ccl12 | 3.82E-09 | 6.99 | 6.36 | 9.35E-09 | chemokine (C-C motif) ligand 12 [Id:108224] | |
| Cst7 | 0.0064 | 6.50 | 7.11 | 0.0021 | cystatin F (leukocystatin) [Id:1298217] | |
| Itgax | 0.0046 | 6.47 | 5.95 | 0.0029 | integrin alpha X [Id:96609] | |
| AI838599 | 1.54E-05 | 6.40 | 5.46 | 3.56E-05 | expressed sequence AI838599 [Id:3510989] | |
| Acp5 | 0.0002 | 6.17 | 6.12 | 0.0001 | acid phosphatase 5. tartrate resistant [Id:87883] | |
| Lyz2 | 0.0004 | 5.47 | 6.51 | 0.0001 | lysozyme 2 [Id:96897] | |
| Ly9 | 1.35E-06 | 5.44 | 4.57 | 4.22E-06 | lymphocyte antigen 9 [Id:96885] | |
| Zbp1 | 1.02E-05 | 5.14 | 4.05 | 6.18E-05 | Z-DNA binding protein 1 [Id:1927449] | |
| Lilrb4a | 1.13E-05 | 5.07 | 4.76 | 1.06E-05 | leukocyte immunoglobulin-like receptor. subfamily B. member 4A [Id:102701] | |
| Lgals3 | 0.0002 | 5.03 | 5.00 | 0.0001 | lectin. galactose binding. soluble 3 [Id:96778] |
Under these stringent conditions, 26 genes were specifically upregulated only in Epm2a−/− mice and 26 genes were specifically upregulated in only Epm2b−/− mice (Supplementary Table S6).
Most upregulated genes in Epm2a−/− and Epm2b−/− samples from 16 months-old animals are related to inflammatory processes
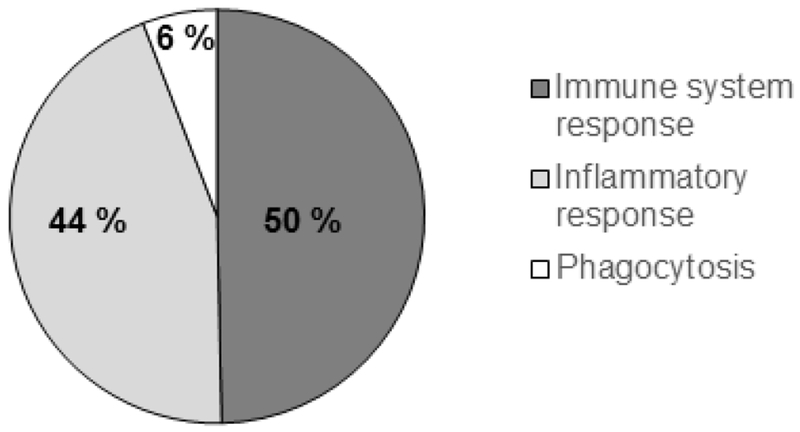
We explored the biological function of the 229 common upregulated genes in the Epm2a−/− and Epm2b−/− samples, using the DAVID tools v6.7 (see Material and Methods), in order to find gene families of specific biological functions. Ten of the genes encoded hypothetical proteins or proteins with unknown function and were discarded from further analysis. The rest (219 genes) were classified according to different GO (Gene Ontology) Biological Processes. In Fig. 3 and Table II we describe the first 20 most representative GO Biological Processes in both Epm2a−/− and Epm2b−/− samples. These included categories related to immune system response (with entries like “immune system process”, “innate immune response”, “immune response”, and “antigen processing and presentation of exogenous peptide antigen via MHC class II” and “antigen processing and presentation”), inflammatory response (with entries like “inflammatory response”, “cellular response to interferon gamma”, “response to interferon gamma”, “cellular response to interferon beta”, “neutrophil chemotaxis”, “chemotaxis”, “chemokine-mediated signaling pathway”, “response to virus”, “defense response to virus”, “defense response to Gram(+) bacterium”, “positive regulation of inflammatory response”, “positive regulation of angiogenesis” and “positive regulation of ERK1 and ERK2 cascade”) and phagocytosis (with entries like “positive regulation of phagocytosis” and “phagocytosis”). In Supplementary Table S7 we describe the upregulated genes according to functional categories. We would like to point out the presence of genes encoding cytokines and their receptors, chemokines and their receptors, complement proteins, Toll-like receptors, interferon activated genes, inflammasome mediators and histocompatibility complex proteins, among other inflammation-related genes (Table II and Supplementary Table S7). The genes upregulated more than 2.0 fold only in Epm2a−/− samples or only in Epm2b−/− samples encoded mainly additional pro-inflammatory markers (Supplementary Table S6).
Figure 3. Most of the shared upregulated genes in LD models are related to the immune and inflammatory response.
Representation of the 20 most representative GO (Gene Ontology) biological processes covered by the shared upregulated genes identified in Epm2a−/− and Epm2b−/− animals of 16 months of age respect to control.
Table II:
Classification of the shared upregulated genes in Epm2a−/− and Epm2b−/− samples of 16 months old animals in comparison to controls according to GO Biological processes using the DAVID tools v6.7 (see Methods). Only the first 20 most representative GO Biological Processes are indicated.
| GOTERM Biological Process | Number related genes | P-value | Gene names |
|---|---|---|---|
| Immune system process | 50 | 1.2E-38 | Oas2, Oasl1, Oasl2, Btk, Clec4a2, Cd300lf, Cd300c2, Cd74, Cd84, Cd86, Fcgr1, Mx1, Nlrc5, Naip2, Naip5, Naip6, Zbp1, Adgre1, Bst2, Casp4, C1qc, C1qa, C1qb, C1s1, C3, Gbp5, Hck, H2-D1, H2-K1, H2-Q7, H2-Aa, H2-Ab1, H2-Eb1, Ifi30, Irf5, Irf7, Ift1, Il1rl2, Lgals3, Lilrb4a, Lcn2, Ly9, Myo1g, Psmb8, Rsad2, Themis2, Tlr1, Tlr2, Tlr7, Tnfaip8l2. |
| Innate immune response | 43 | 1.8E-29 | Oas1a, Oas2, Oasl1, Oasl2, Btk, Clec4a, Clec7a, Cd84, Eif4, Fcgr1, Mx1, Nlrc5, Naip2, Naip5, Naip6, Tyrobp, Zbp1, Ang, Bst2, Casp4, C1qc, C1qa, C1qb, C1s1, C3, C4b, Cyba, Cybb, Hck, Irf5, Irf7, Ifit1, Il1rl2, Lgals3, Lcn2, Ly9, Ptx3, Rsad2, Tlr1, Tlr2, Tlr7, Trem2, Tnfaip8l2. |
| Inflammatory response | 36 | 4.1E-24 | Clec7a, Fas, Naip2, Naip5, Naip6, Aif1, Cnr2, Casp4, Ccl12, Ccl2, Ccl5, Ccl6, Ccl8, Cxcl10, Cxcl5, Cxcr6, Chil1, Chil3, C3, C3ar1, C4b, Cyba, Cybb, Gbp5, Hck, Il1a, Il1b, Il1rl2, Ncf1, Slc11a1, S1pr3, Themis2, Tlr1, Tlr2, Tlr7, Tnfrsf9. |
| Immune response | 31 | 9.1E-22 | Oas2, Oasl1, Oasl2, Cd74, Fyb, Fas, Fcgr2b, Was, Ctss, Ccl12, Ccl2, Ccl5, Ccl6, Ccl8, Cxcl10, Cxcl5, H2-D1, H2-K1, H2-Oa, H2-Aa, H2-Ab1, H2-Eb1, Irf8, Il1a, Il1b, Tlr1, Tlr2, Tlr7, Tnfsf8, Tnfrsf9, Vav1. |
| Cellular response to interferon gamma | 14 | 3.6E-13 | Aif1, Ccl12, Ccl2, Ccl5, Ccl6, Ccl8, Gbp2, Gbp3, Gbp5, Gbp6, Gbp10, Gbp8, H2-Q7, H2-Ab1. |
| Response to virus | 14 | 6.1E-12 | Oas1a, Oas2, Oasl1, Oasl2, Mx1, Batf3, Bst2, Ccl5, cxcl10, Hspb1, Ifi27l2a, Ifit1, Lcn2, Rsad2. |
| Defense response to virus | 17 | 3.9E-11 | Oas1a, Oas2, Oasl1, Oasl2, Cd86, Isg15, Mx1, Nlrc5, Zbp1, Bst2, Cxcl10, Itgax, Irf5, Ifit1, Ifit3b, Rsad2, Tlr7. |
| Neutrophil chemotaxis | 11 | 3.5E-9 | Fcgr3, Ccl12, Ccl2, Ccl5, Ccl6, Ccl8, Csf3r, Itgb2, Il1b, Lgals3, Vav1. |
| Antigen processing and presentation of exogenous peptide antigen via MHC classII | 7 | 4.2E-9 | Cd74, Fcgr2b, H2-Oa, H2-Aa, H2-Ab1, Ifi30. |
| Defense response to Gram(+) bacterium | 12 | 5.2E-9 | Acp5, Ang, Gbp2, Gbp3, Gbp6, Gbp10, Hck, Lyz2, Myolf, Ncfl, Tlr2, Tnfsf8. |
| Chemotaxis | 13 | 5,9E-9 | Rac2, Ccl12, Ccl2, Ccl5, Ccl6, Ccl8, Cxcl10, Cxcl16, Cxcl5, Cxcr6, Cmklr1, C3ar1, Dock2. |
| Positive regulation of phagocytosis | 9 | 4.3E-8 | Fcgr1, Fcgr3, Fcgr2b, C3, Cyba, Dock2, Il1b, Ptx3, Slc11a1. |
| Antigen processing and presentation | 9 | 1.1E-7 | Cd74, Ctss, H2-D1, H2-K1, H2-Oa, H2-Aa, H2-Ab1, H2-Eb1, Psmb8. |
| Response to interferon gamma | 7 | 3.9E-7 | Cd86, Bst2, Ccl5, Cxcl16, H2-Aa, H2-Eb1, Slc11a1. |
| Positive regulation of inflammatory response | 9 | 3.8E-7 | Adora3, Ctss, Ccl12, Ccl2, Ccl5, Ccl6, Ccl8, Il1rl1, Tlr2. |
| Positive regulation of angiogenesis | 11 | 7.9E-7 | Ctsh, Ccl5, Chil1, C3, C3ar1, Cybb, Hspb1, Itgb2, Il1a, Il1b, Lgals3. |
| Positive regulation of ERK1 and ERK2 cascade | 13 | 1.0E-6 | Cd74, Ccl12, Ccl2, Ccl5, Ccl6, Ccl8, Chil1, C3, Gpnmb, Il1a, Il1b, Tlr2, Trem2. |
| Phagocytosis | 8 | 1.4E-6 | Hck, Itgal, Itgb2, Irf8, Myo1g, Pld4, Slc11a1, Vav1 |
| Chemokine-mediated signaling pathway | 8 | 2.1E-6 | Gpr35, ccl12, Ccl2, Ccl5, Ccl6, Ccl8, Cxcl10, Cxcl5. |
| Cellular response to interferon beta | 7 | 4.9E-6 | Gbp2, Gbp3, Gbp6, Ifi204, Ifi209, Ifi47, ifit1. |
In summary, considering the general genetic landscape depicted by these transcriptomic results, inflammation emerges as a clear hallmark that contributes to the pathophysiology of Lafora disease.
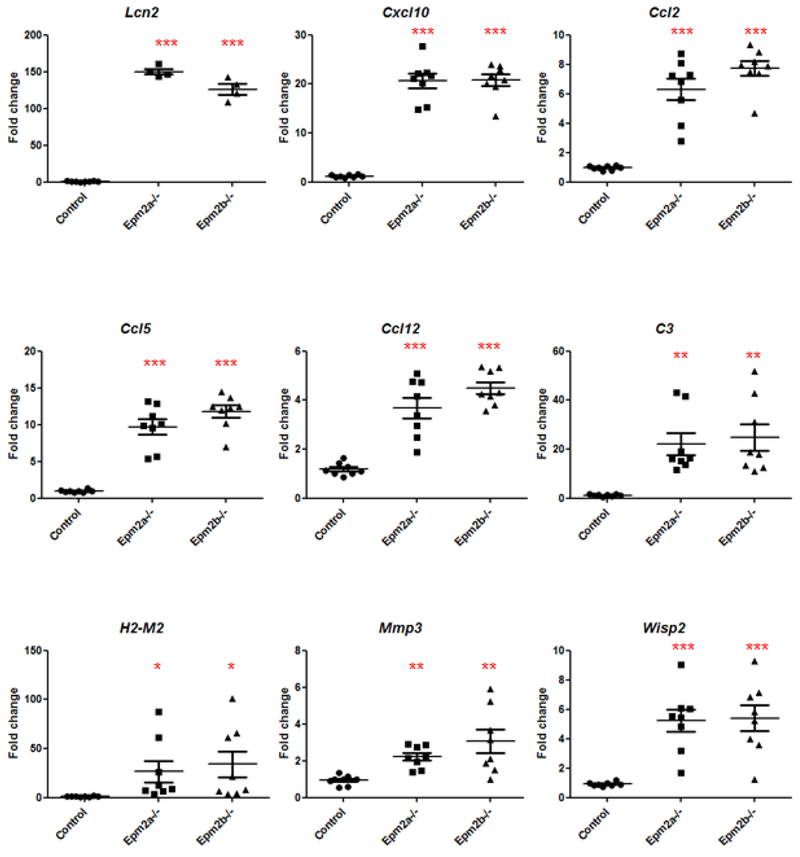
Validation of upregulated genes in Epm2a−/− and Epm2b−/− samples
In order to validate the results obtained by RNA-Seq, we measured the relative expression levels of nine selected genes (Lcn2, Cxcl10, Ccl2, Ccl5, Ccl12, C3, H2-M2, Mmp3 and Wisp2) by RT-qPCR. This selection was based on their higher FC and lower FDR value (Table I), as well as in their belonging to a higher number of GO Biologically Processes classified by DAVID bioinformatics tool (Table II) in both Epm2a−/− and Epm2b−/− samples (their position is highlighted in bold in Fig. 1 and boxed in Fig. 2). We also included Wisp2 in the selected group in order to analyze alternative signaling pathways different from inflammation (i.e., WNT-inducible signaling pathway). To perform these analyses, we used RNA samples from the RNA-Seq experiments and also new ones isolated from independent mice. As shown in Fig. 4, all selected genes showed a statistically higher relative expression in both Epm2a−/− and Epm2b−/− samples as compared to controls, corroborating the previous RNA-Seq results.
Figure 4. qRT-PCR validation panel of some overexpressed genes.
Relative expression levels of nine selected genes (Lcn2, Cxcl10, Ccl2, Ccl5, Ccl12, C3, H2-M2, Mmp3 and Wisp2) from brains of control, Epm2a−/− and Epm2b−/− mice of 16 months of age was analyzed by RT-qPCR in eight biological samples (4 samples used to perform RNA-Seq analyses and 4 new independent samples) and three technical replicates. The expression levels of each gene were normalized to those of the actin beta gene (Actβ). The expression values are represented as circles (control), squares (Epm2a−/−) and triangles (Epm2b−/−) and expressed as a fold change (FC) respect to a control sample. Horizontal bars indicate mean values and standard error of the mean (SEM). Statistical analyses were performed by One-way ANOVA with a post-hoc Tukey’s comparison test for control vs Epm2a−/− and control vs Epm2b−/− using GraphPad Prism 5 software. P-value *<0.05, **<0.01 and ***<0.001.
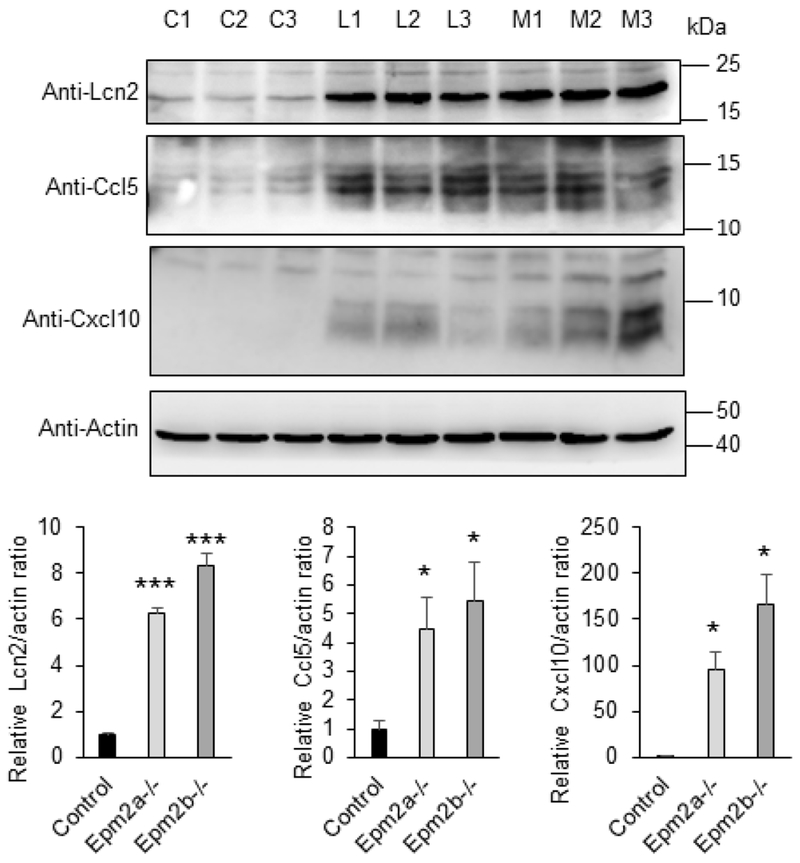
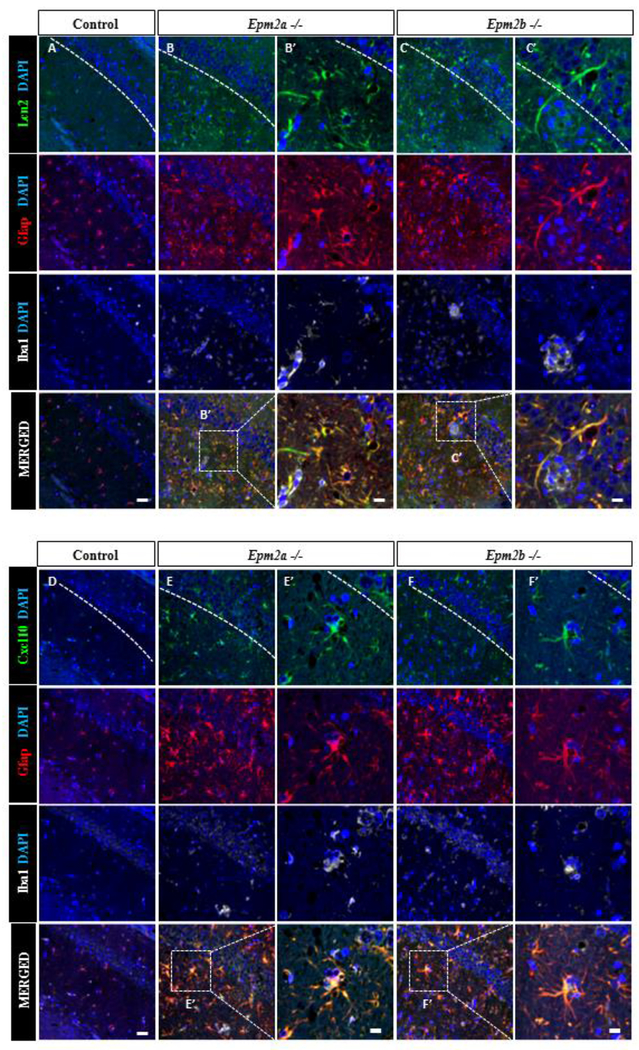
Additionally, we analyzed by Western blotting the protein levels of some genes with high fold change expression. We observed increased levels of Lipocalin 2 (Lcn2), Ccl5 and Cxcl10 in brain extracts from Epm2a−/− and Epm2b−/− mice of 16 months of age in comparison to control animals (Fig. 5). To figure out which cells were responsible for the production of some of these inflammatory mediators, brain sections of 16 months old mice were assessed by immunofluorescence. As shown in Fig. 6, reactive astrocytes that were positive for Gfap (glial fibrillary acidic protein, a marker of activated astrocytes) were also positive for the expression of Lipocalin 2 and Cxcl10, indicating the participation of these cells in the inflammatory response. All these results validated the RNA-Seq experiments and further corroborated that in the brain of the two LD mouse models of 16 months of age, clear neuroinflammation was present.
Figure 5: Lcn2, Ccl5 and Cxcl10 proteins are more abundant in LD-brain mutants.
Protein levels of Lipocalin2 (Lcn2), Ccl5 and Cxcl10 in brain extracts from 16 months-old control (C), Epm2a−/− (L) and Epm2b−/− mice (M) were analyzed by Western blot using the indicated antibodies and densitometric quantification of the corresponding blots were carried out as described in Material and Methods. Three independent samples from each genotype were analyzed. Protein levels related to the levels of actin were referred to the levels found in control samples. Results are expressed as means +/− standard error of the mean (SEM). Differences between paired samples were analyzed by two-tailed Student’s t-tests using Graph Pad Prism version 5.0 statistical software (La Jolla, CA, USA). P values have been considered as *P<0.05 and ***P< 0.001. No significant differences were observed between Epm2a−/− and Epm2b−/− brain extracts.
Figure 6: LD-reactive astrocytes express Lcn2 and Cxcl10 chemokines.
Confocal images of CA1-hippocampus of control, Epm2a −/− and Epm2b −/− mice of 16 months of age. Immunofluorescence against Gfap, Iba1 and chemokines Lcn2 (A-C) or Cxcl10 (D-F) is shown. Samples from LD mice have higher number Lcn2+ cells (B-C) or Cxcl10+cells (E-F) compared to control animals (A) or (D) respectively. Lcn2 expression co-localizes with the astrocyte marker Gfap but not with Iba1 microglia marker. The white dashed line highlights hippocampus CA1 region. Scale bar (shown in A and D): 25μm. Squared dashed line indicates the area that is magnified in the corresponding B’, C’, E’ and F’ pictures; scale bar: 75 μm.
Progression of the expression of different genes with age
Given the landscape depicted by the aforementioned transcriptomic and protein expression data, and since Lafora disease is a type of progressive myoclonus epilepsy that aggravates with age, we wondered whether the expression of inflammation-related genes could also take place at earlier stages. For this reason, we decided to repeat the RNA-Seq analyses in samples from 3 months-old Epm2a−/− and Epm2b−/− animals since at this age they are at the beginning of the development of the pathophysiological phenotypes. In this way, we sought to characterize initial changes in the expression of genes related to the beginning of the disease. We analyzed four independent samples from each genotype. However, differential expression analyses between Epm2a−/− and control groups identified only 2 genes that were upregulated, with an FDR lower than 0.05 and an FC higher than 1.2, and 7 genes that were downregulated, with a FDR lower than 0.05 and a FC lower than 0.8, one of them being Epm2a, validating the analysis (Supplementary Table 8). Differential expression analyses between Epm2b−/− and control groups identified 40 upregulated genes, with an FDR lower than 0.05 and an FC higher than 1.2, and 108 downregulated genes, with a FDR lower than 0.05 and a FC lower than 0.8, one of them being Epm2b, validating the analysis (Supplementary Table 8). Among all these genes, only 2 were upregulated and 2 were downregulated in both Epm2a−/− and Epm2b−/− samples respect to controls (Table III). One of the two upregulated genes was Gfap (encoding glial fibrillary acidic protein), being these results in agreement with previous results from our group in which, by immunohistochemistry analyses, we detected higher levels of this protein in the brain from 3 months-old Epm2a−/− and Epm2b−/− mice in comparison to controls [20]. The other common upregulated gene was Spon1, which encoded spondin-1, a cell adhesion protein involved in axon guidance. However, the differential expression obtained for Spon1 and the two downregulated genes Per2 and Tnfrsf25 was not observed in the RNAseq dataset at 16 months of age.
Table III:
Shared genes differentially expressed in both Epm2a−/− and Epm2b−/− samples of 3 months old animals with an FDR <0.05 in both samples. The gene name, false discovery rate (FDR), fold change (FC), description and MGI Id (mouse genome informatics) are indicated. Genes are arranged according to FC.
| Epm2a−/− | Epm2b−/− | |||||
|---|---|---|---|---|---|---|
| Gene | FDR | FC | FC | FDR | Description [MGI Id] | |
| Spon1 | 0.0086 | 1.26 | 1.23 | 0.0068 | spondin 1 [Id:2385287] | |
| Gfap | 0.0434 | 1.23 | 1.31 | 0.0012 | glial fibrillary acidic protein [Id:95697] | |
| Per2 | 0.0018 | 0.70 | 0.79 | 0.0180 | period circadian clock 2 [Id:1195265] | |
| Tnfrsf25 | 0.0315 | 0.51 | 0.51 | 0.0068 | tumor necrosis factor receptor superfamily, member 25 [Id:1934667] |
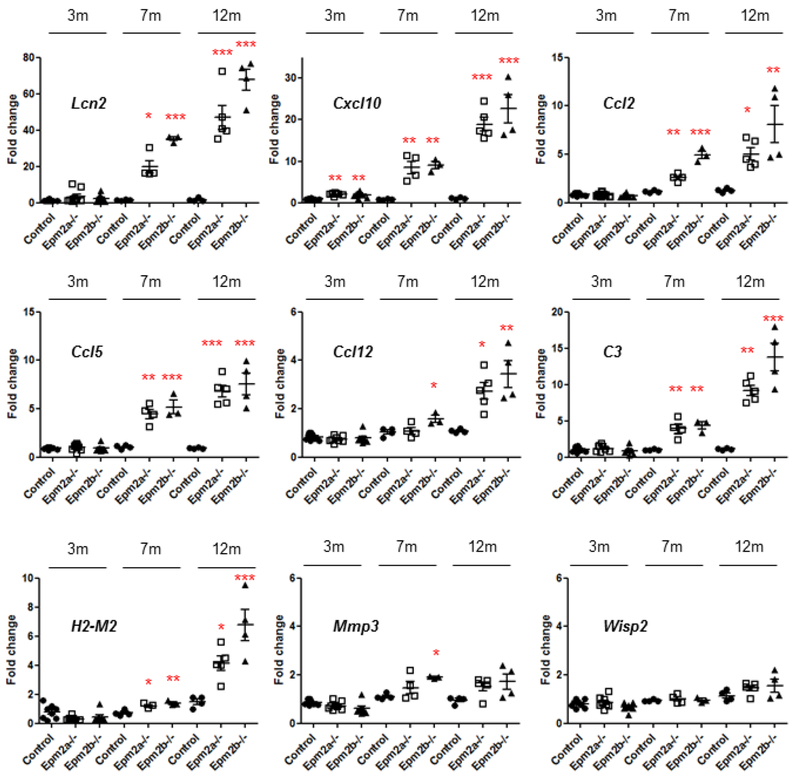
Although not present in the RNA-seq data at three months of age, we measured by RT-qPCR the expression of the nine genes (Lcn2, Cxcl10, Ccl2, Ccl5, Ccl12, C3, H2-M2, Mmp3 and Wisp2) used in the validation experiments described above in the samples from 3 months-old Epm2a−/− and Epm2b−/− mice. As shown in Fig. 7, only the expression of the Cxcl10 gene was significantly higher (around 2.5 FC) in Epm2a−/− and Epm2b−/− samples as compared to controls. All these results indicate that at three months of age there are minor changes in the differential expression of genes between the two LD mice and controls, in agreement with the absence of a clear pathophysiological phenotype at this age.
Figure 7. The expression of the selected upregulated genes starts at early stages.
Relative expression levels of nine selected genes (Lcn2, Cxcl10, Ccl2, Ccl5, Ccl12, C3, H2-M2, Mmp2 and Wisp2) from brains of control, Epm2a−/− and Epm2b−/− mice of 3, 7 and 12 months of age were analyzed by RT-qPCR in four biological samples and three technical replicates. The expression levels of each gene were normalized to those of the actin beta gene (Actp). The expression values are represented as circles (control), empty squares (Epm2a−/−) and triangles (Epm2b−/−) and are expressed as a fold change (FC) respect to a control sample. Horizontal bars indicate mean values and standard error of the mean (SEM). Statistical analysis was performed by One-way ANOVA with a post-hoc Tukey’s comparison test for control vs Epm2a−/− and control vs Epm2b−/− was performed using GraphPad Prism 5 software. P-value *<0.05, **<0.01 and ***<0.001.
We then repeated the RT-qPCR experiments in samples from mice of 7 and 12 months of age and found a clear progression of the differential expression of Lcn2, Cxcl10, Ccl2, Ccl5, Ccl12, C3 and H2-M2, and a tendency to increase in the case of Mmp3 and Wisp2 (Fig. 7). We also performed the RT-qPCR experiments in samples from Epm2b−/− mice of 5 months of age and only the expression of Cxcl10 was significantly increased (around 5.8 FC; n:7, p:0.022).
These results suggested a progressive increase in the inflammatory response which correlates with age, providing a possible explanation of the worsening of the pathophysiological symptoms.
DISCUSSION
This work was aimed to identify possible markers that could elucidate the mechanisms underlying LD. Whole-brain tissue from two different Lafora disease mouse models, Epm2a−/− and Epm2b−/−, were analyzed by RNA-Seq technology. The analyses were initially performed in 16 months-old mice to magnify the possible alterations of the LD phenotype. In 2005, Ganesh et al., [31] described a transcriptional profiling in six months old Epm2a−/− mice, derived from 129SvJ isogenic background, using GenChip analysis (Affymetrix Murine Genome U74A and U74B GeneChips with 24,935 probe sets). They described that only 46 probe sets were increased and 23 probe sets were decreased in the brain from Epm2a−/− mice. The authors indicated that the differentially expressed probe sets were involved in gene expression or modification of proteins, post-translational modification of proteins and transcriptional regulation. As far as inflammation is concerned, they only described 2 probe sets. However, in our dataset we could not find any of the 69 genes described by these authors. Perhaps differences in the background of the mice (129SvJ in their case, C57BL6 in ours), the age of the animals (6 months in their study, 16 and 3 months in ours) or the technology used (GeneChips in their study vs. RNA-seq in ours) could account for the different results.
Our RNA-Seq results indicate that in both models of LD, there is a common set of upregulated genes, most of them encoding mediators of inflammatory response, thus suggesting that the observed changes in expression of these shared genes could reflect the similar pathophysiological phenotypes present in both mice (see Introduction). In addition, they also indicate a progression with age, since the first changes in these inflammatory markers starts at three months of age and reach maximum differences at 16 months of age. RNA-Seq results were confirmed by RT-qPCR showing in nine selected genes a statistically significant increase in their relative expression levels at 16 months of age in Epm2a−/− and Epm2b−/− samples respect to controls. Higher protein levels of Lcn2, Ccl5 and Cxcl10 were also found, and noteworthy, we also observe co-localization between reactive astrocytes, Lcn2 and Cxcl10.
We have compared our results with other datasets obtained in different transcriptomic analyses aimed to understand changes in gene expression related to inflammation, finding important similarities in the set of upregulated genes we describe herein. For example, after acute treatment with LPS (lipopolysaccharide, to induce brain inflammation) or in mice models of neurodegeneration, there was a clear upregulation of Apoe, Axl, Ccl2, Ccl5, Clec7a, Csfl, Csf3, Cxcl10, Cybb, Il-1b, Itgax, Lcn2, Lgals3/Galectin3, Mmp3, Spp1, Steap4, Timp1, Tlr2, and Zbp1, a group of genes consistently upregulated in activated microglia which could mediate the regulation of immune-, phagosome-, lysosome-, oxidative phosphorylation- and antigen presentation-signaling pathways [[32], [33], [34]]. In addition, Zamanian et al, [35] indicate that under conditions that trigger astrocyte reactivity there was an upregulation of Lcn2, Steap4, Timpl, Cxcl10 and Ccl2, among other genes. These last authors suggested that the upregulation of these genes would be the result of an inflammatory response. Interestingly, our dataset also indicates an upregulation of all these genes in both LD mouse models (Table I and Supplementary Table S5), supporting the idea that reactive astrocytes and activated microglia play a major role in the upregulation of genes which could account, at least in part, for the pathophysiology of LD. Moreover, these results are also in agreement with previous indications, pointing out to a clear increase in the levels of reactive astrocytes (Gfap+ cells) and activated microglia (Iba1+ cells) in LD mouse models, which became more intense as the animals aged [[13], [19], [20], [21]].
An additional proof of the possible involvement of reactive astrocytes and microglia in the pathogenesis of LD was found when we analyzed our dataset of upregulated genes in Epm2a−/− and Epm2b−/− samples using the RNA-Seq transcriptome and splicing database of glia, neurons and vascular cells in mice (http://web.stanford.edu/group/barres_lab/brain_error_3.html) [36]. We found that among the 229 commonly upregulated (>2.0 fold) genes in Epm2a−/− and Epm2b−/− samples, more than 60% of them encoded microglial specific proteins, and 26% additional genes showed a shared expression in microglia and other cell types (astrocytes and endothelial cells).
Interestingly, a recent report analyzed the upregulation of genes in response to kainic acid, a pro-convulsant drug [37]. Among the set of genes upregulated under these conditions, we found some that were also upregulated in LD mice samples (i.e, Il1rl1, Tnfsf8, Tnfaip2, Il1rl2, Osmr, Csf2rb2, Ccl8, Cxcl10, Ccl2, Ccl5, Cxcr6, C3, C4b, C1qa, C1qc, C1qb, Tlr1, Irf7, Irf5, H2-M2, H2-Q4, H2-Q6, H2-K1, H2-D1, Cd300lf, Cd22, Lyz2, Adora3, Tmem106a and Trem2). The authors also suggested a clear correlation between the upregulation of these genes and the appearance of seizures, probably as a consequence of inflammation and activation of microglia. Since in our LD samples, most of these genes were upregulated, we suggest that inflammation, astrocyte reactivity and activation of microglia may play a major role triggering seizures in LD. This is in agreement with a direct relationship that exists between neuroinflammation and epilepsy in humans. Immunohistological and biochemical studies of samples obtained from patients with different types of drug-resistant epilepsies have demonstrated the presence of neuroinflammation, which appears even before the onset of epilepsy (reviewed in [38]).
The presence of neuroinflammatory markers in LD mouse models is comparable to the ones present in a mouse model (Cstb−/−) of Unverricht-Lundborg disease (ULD), another form of progressive myoclonus epilepsy (EPM1) due to mutations in the CSTB gene, which encodes cystatin B, an inhibitor of lysosomal cathepsin B. In Cstb−/− mice there is an activation of microglia which aggravates with age and an upregulation of genes related to immune response, what led to the hypothesis that glial-derived pro-inflammatory chemokines and cytokines contributed to recurrent excitation and seizures [[39], [40], [41], [42]]. Therefore, two different forms of progressive myoclonus epilepsy (EPM1-ULD and EPM2-LD) present similar reactive glial-derived neuroinflammatory markers, that could contribute to disease.
Since Lafora disease is a type of progressive myoclonus epilepsy that aggravates with age, we repeated the RNA-Seq analyses in animals of three months of age, since we sought to characterize the initial changes in the expression of genes related to the beginning of the disease. However, at this age only minor changes were observed in the differential expression of genes: we could only detect increased levels of Spon1 and Gfap in both Epm2a−/− and Epm2b−/− mice (Table III). In the case of Gfap, its expression was also commonly upregulated in the samples from LD mice of 16 months of age (around 2.5 FC), and it encodes a protein related to the presence of reactive astrocytes. In fact, in previous work we detected higher levels of this protein in the brain of Epm2a−/− and Epm2b−/− mice by immunohistochemistry, at three months of age [20]. These results seem to suggest that at three months of age astrocytes have begun to be activated, although to a minor extent. We also studied the expression of the same set of validated genes described above, in samples from LD mice of 3, 5, 7 and 12 months of age and found that only the expression of Cxcl10 gene started at 3 months of age (2.5 FC), suggesting that the expression of this gene could be considered as one of the early determinants in the pathophysiology of LD.
We would like to point out that LD mice of three months of age already contain large amounts of polyglucosan inclusions in the brain [43]. Therefore, in our opinion, the dysregulation of the synthesis of polyglucosans seems to be independent of the changes observed in the transcriptome at this age. Taking all these results together, we suggest that an early unknown insult (e.g. the formation of polyglucosan inclusions?) would activate astrocytes/microglia promoting the expression of at least Gfap and the Cxcl10 chemokine. This would trigger a subsequent inflammatory response that would aggravate with age.
In summary, we present strong evidence of an upregulation of the expression of common genes in two independent mouse models of LD at 16 months of age. Most upregulated genes encoded pro-inflammatory mediators, suggesting that inflammation may play a crucial role in the pathophysiology of LD. Our results also suggest that the use of anti-inflammatory agents that target specific epileptogenic pathways that are altered in LD could be beneficial in a disease that has no treatment yet. This hypothesis is supported by different reports in the literature which indicate that the use of anti-inflammatory drugs have clinical effects on different forms of epilepsy [[38], [44], [45]]. Furthermore, we propose an early-stage window for application of this type of treatments, based in the parallelism found between gene expression alterations, increase in neuroinflammatory markers, and disease progression with age found in LD.
Supplementary Material
ACKNOWLEDGMENTS
We want to thank Miguel Heredia (CIBERER) for his help in obtaining the brain samples and Ursula Estada (Multigenic Analysis Unit from the UCIM-INCLIVA; University of Valencia, Valencia Spain) for his help in constructing libraries and performing RNA-Seq experiments. This work was supported by grants from the Spanish Ministry of Economy and Competitiveness SAF2014-54604-C3-1-R and SAF2017-83151-R, a grant from Fundación Ramón Areces (CIVP18A3935) and a grant from the National Institute of Health (NIH-NINDS) P01NS097197, which established the Lafora Epilepsy Cure Initiative (LECI), to PS. We also acknowledge a grant from the Spanish Ministry of Economy and Competitiveness SAF2014-54604-C3-2-R to EK and a grant from Generalitat Valenciana Prometeo2018/135 to PS and FVP.
ABBREVIATIONS
- FC
fold change
- FDR
false discovery rate
- LD
Lafora disease
- RT-qPCR
real-time quantitative polymerase chain reaction
Footnotes
COMPETING INTERESTS
None of the authors has any conflict of interest to disclose.
REFERENCES
- 1.Turnbull J, Tiberia E, Striano P, Genton P, Carpenter S, Ackerley CA, Minassian BA (2016) Lafora disease. Epileptic Disord 18 (S2):38–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Gimeno MA, Knecht E, Sanz P (2018) Lafora Disease: A Ubiquitination-Related Pathology. Cells 7 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minassian BA, Lee JR, Herbrick JA, Huizenga J, Soder S, Mungall AJ, Dunham I, Gardner R, Fong CY, Carpenter S, Jardim L, Satishchandra P, Andermann E, Snead OC 3rd, Lopes-Cendes I, Tsui LC, Delgado-Escueta AV, Rouleau GA, Scherer SW (1998) Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat Genet 20 (2): 171–174. [DOI] [PubMed] [Google Scholar]
- 4.Serratosa JM, Gomez-Garre P, Gallardo ME, Anta B, de Bernabe DB, Lindhout D, Augustijn PB, Tassinari CA, Malafosse RM, Topcu M, Grid D, Dravet C, Berkovic SF, de Cordoba SR (1999) A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2). Hum Mol Genet 8 (2):345–352. [DOI] [PubMed] [Google Scholar]
- 5.Chan EM, Young EJ, Ianzano L, Munteanu I, Zhao X, Christopoulos CC, Avanzini G, Elia M, Ackerley CA, Jovic NJ, Bohlega S, Andermann E, Rouleau GA, Delgado-Escueta AV, Minassian BA, Scherer SW (2003) Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat Genet 35 (2):125–127. [DOI] [PubMed] [Google Scholar]
- 6.Gentry MS, Worby CA, Dixon JE (2005) Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc Natl Acad Sci U S A 102 (24):8501–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohi H, Ianzano L, Zhao XC, Chan EM, Turnbull J, Scherer SW, Ackerley CA, Minassian BA (2005) Novel glycogen synthase kinase 3 and ubiquitination pathways in progressive myoclonus epilepsy. Hum Mol Genet 14 (18):2727–2736. [DOI] [PubMed] [Google Scholar]
- 8.Solaz-Fuster MC, Gimeno-Alcaniz JV, Ros S, Fernandez-Sanchez ME, Garcia-Fojeda B, Criado Garcia O, Vilchez D, Dominguez J, Garcia-Rocha M, Sanchez-Piris M, Aguado C, Knecht E, Serratosa J, Guinovart JJ, Sanz P, Rodriguez de Cordoba S (2008) Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway. Hum Mol Genet 17 (5):667–678. [DOI] [PubMed] [Google Scholar]
- 9.Roma-Mateo C, Aguado C, Garcia-Gimenez JL, Knecht E, Sanz P, Pallardo FV (2015) Oxidative stress, a new hallmark in the pathophysiology of Lafora progressive myoclonus epilepsy. Free Radic Biol Med 88 (Pt A):30–41. [DOI] [PubMed] [Google Scholar]
- 10.Gómez-Abad C, Gomez-Garre P, Gutierrez-Delicado E, Saygi S, Michelucci R, Tassinari CA, Rodriguez de Cordoba S, Serratosa JM (2005) Lafora disease due to EPM2B mutations. A clinical and genetic study. Neurology 64:982–986. [DOI] [PubMed] [Google Scholar]
- 11.Ganesh S, Delgado-Escueta AV, Sakamoto T, Avila MR, Machado-Salas J, Hoshii Y, Akagi T, Gomi H, Suzuki T, Amano K, Agarwala KL, Hasegawa Y, Bai DS, Ishihara T, Hashikawa T, Itohara S, Cornford EM, Niki H, Yamakawa K (2002) Targeted disruption of the Epm2a gene causes formation of Lafora inclusion bodies, neurodegeneration, ataxia, myoclonus epilepsy and impaired behavioral response in mice. Hum Mol Genet 11 (11):1251–1262. [DOI] [PubMed] [Google Scholar]
- 12.DePaoli-Roach AA, Tagliabracci VS, Segvich DM, Meyer CM, Irimia JM, Roach PJ (2010) Genetic depletion of the malin E3 ubiquitin ligase in mice leads to lafora bodies and the accumulation of insoluble laforin. J Biol Chem 285 (33):25372–25381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valles-Ortega J, Duran J, Garcia-Rocha M, Bosch C, Saez I, Pujadas L, Serafin A, Canas X, Soriano E, Delgado-Garcia JM, Gruart A, Guinovart JJ (2011) Neurodegeneration and functional impairments associated with glycogen synthase accumulation in a mouse model of Lafora disease. EMBO molecular medicine 3 (11):667–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Criado O, Aguado C, Gayarre J, Duran-Trio L, Garcia-Cabrero AM, Vernia S, San Millan B, Heredia M, Roma-Mateo C, Mouron S, Juana-Lopez L, Dominguez M, Navarro C, Serratosa JM, Sanchez M, Sanz P, Bovolenta P, Knecht E, Rodriguez de Cordoba S (2012) Lafora bodies and neurological defects in malin-deficient mice correlate with impaired autophagy. Hum Mol Genet 21 (7):1521–1533. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Cabrero AM, Marinas A, Guerrero R, de Cordoba SR, Serratosa JM, Sanchez MP (2012) Laforin and malin deletions in mice produce similar neurologic impairments. J Neuropathol Exp Neurol 71 (5):413–421. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Cabrero AM, Sanchez-Elexpuru G, Serratosa JM, Sanchez MP (2014) Enhanced sensitivity of laforin- and malin-deficient mice to the convulsant agent pentylenetetrazole. Front Neurosci 8:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguado C, Sarkar S, Korolchuk VI, Criado O, Vernia S, Boya P, Sanz P, de Cordoba SR, Knecht E, Rubinsztein DC (2010) Laforin, the most common protein mutated in Lafora disease, regulates autophagy. Hum Mol Genet 19 (14):2867–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puri R, Ganesh S (2010) Laforin in autophagy: a possible link between carbohydrate and protein in Lafora disease? Autophagy 6 (8):1229–1231. [DOI] [PubMed] [Google Scholar]
- 19.Turnbull J, DePaoli-Roach AA, Zhao X, Cortez MA, Pencea N, Tiberia E, Piliguian M, Roach PJ, Wang P, Ackerley CA, Minassian BA (2011) PTG depletion removes Lafora bodies and rescues the fatal epilepsy of Lafora disease. PLoS genetics 7 (4):e1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Gonzalez I, Viana R, Sanz P, Ferrer I (2017) Inflammation in Lafora Disease: Evolution with Disease Progression in Laforin and Malin Knock-out Mouse Models. Mol Neurobiol 54 (5):3119–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rai A, Mishra R, Ganesh S (2017) Suppression of leptin signaling reduces polyglucosan inclusions and seizure susceptibility in a mouse model for Lafora disease. Hum Mol Genet 26 (24):4778–4785. [DOI] [PubMed] [Google Scholar]
- 22.Bottomly D, Walter NA, Hunter JE, Darakjian P, Kawane S, Buck KJ, Searles RP, Mooney M, McWeeney SK, Hitzemann R (2011) Evaluating gene expression in C57BL/6J and DBA/2J mouse striatum using RNA-Seq and microarrays. PLoS One 6 (3):e17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, Gong B, Bushel PR, Thierry-Mieg J, Thierry-Mieg D, Xu J, Fang H, Hong H, Shen J, Su Z, Meehan J, Li X, Yang L, Li H, Labaj PP, Kreil DP, Megherbi D, Gaj S, Caiment F, van Delft J, Kleinjans J, Scherer A, Devanarayan V, Wang J, Yang Y, Qian HR, Lancashire LJ, Bessarabova M, Nikolsky Y, Furlanello C, Chierici M, Albanese D, Jurman G, Riccadonna S, Filosi M, Visintainer R, Zhang KK, Li J, Hsieh JH, Svoboda DL, Fuscoe JC, Deng Y, Shi L, Paules RS, Auerbach SS, Tong W (2014) The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotechnol 32 (9):926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11 (3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson MD, Smyth GK (2007) Moderated statistical tests for assessing differences in tag abundance. Bioinformatics (Oxford, England) 23 (21):2881–2887. [DOI] [PubMed] [Google Scholar]
- 26.Robinson MD, Smyth GK (2008) Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9 (2):321–332. [DOI] [PubMed] [Google Scholar]
- 27.Lun AT, Chen Y, Smyth GK (2016) It’s DE-licious: A Recipe for Differential Expression Analyses of RNA-seq Experiments Using Quasi-Likelihood Methods in edgeR. Methods Mol Biol 1418:391–416. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini YaH Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B 57:289–300. [Google Scholar]
- 29.Huang da W, Sherman BT, Stephens R, Baseler MW, Lane HC, Lempicki RA (2008) DAVID gene ID conversion tool. Bioinformation 2 (10):428–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3--new capabilities and interfaces. Nucleic Acids Res 40 (15):e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganesh S, Tsurutani N, Amano K, Mittal S, Uchikawa C, Delgado-Escueta AV, Yamakawa K (2005) Transcriptional profiling of a mouse model for Lafora disease reveals dysregulation of genes involved in the expression and modification of proteins. Neurosci Lett 387 (2):62–67. [DOI] [PubMed] [Google Scholar]
- 32.Holtman IR, Raj DD, Miller JA, Schaafsma W, Yin Z, Brouwer N, Wes PD, Moller T, Orre M, Kamphuis W, Hol EM, Boddeke EW, Eggen BJ (2015) Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: a co-expression meta-analysis. Acta Neuropathol Commun 3:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang SS, Ren Y, Liu CC, Kurti A, Baker KE, Bu G, Asmann Y, Fryer JD (2018) Lipocalin-2 protects the brain during inflammatory conditions. Mol Psychiatry 23 (2):344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srinivasan K, Friedman BA, Larson JL, Lauffer BE, Goldstein LD, Appling LL, Borneo J, Poon C, Ho T, Cai F, Steiner P, van der Brug MP, Modrusan Z, Kaminker JS, Hansen DV (2016) Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat Commun 7:11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, Barres BA (2012) Genomic analysis of reactive astrogliosis. J Neurosci 32 (18):6391–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34 (36):11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bosco DB, Zheng J, Xu Z, Peng J, Eyo UB, Tang K, Yan C, Huang J, Feng L, Wu G, Richardson JR, Wang H, Wu LJ (2018) RNAseq analysis of hippocampal microglia after kainic acid-induced seizures. Mol Brain 11 (1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vezzani A, Balosso S, Ravizza T (2019) Neuroinflammatory pathways as treatment targets and biomarkers in epilepsy. Nat Rev Neurol 15 (8):459–472. [DOI] [PubMed] [Google Scholar]
- 39.Tegelberg S, Kopra O, Joensuu T, Cooper JD, Lehesjoki AE (2012) Early microglial activation precedes neuronal loss in the brain of the Cstb−/− mouse model of progressive myoclonus epilepsy, EPM1. J Neuropathol Exp Neurol 71 (1):40–53. [DOI] [PubMed] [Google Scholar]
- 40.Joensuu T, Tegelberg S, Reinmaa E, Segerstrale M, Hakala P, Pehkonen H, Korpi ER, Tyynela J, Taira T, Hovatta I, Kopra O, Lehesjoki AE (2014) Gene expression alterations in the cerebellum and granule neurons of Cstb(−/−) mouse are associated with early synaptic changes and inflammation. PLoS One 9 (2):e89321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okuneva O, Korber I, Li Z, Tian L, Joensuu T, Kopra O, Lehesjoki AE (2015) Abnormal microglial activation in the Cstb(−/−) mouse, a model for progressive myoclonus epilepsy, EPM1. Glia 63 (3):400–411. [DOI] [PubMed] [Google Scholar]
- 42.Okuneva O, Li Z, Korber I, Tegelberg S, Joensuu T, Tian L, Lehesjoki AE (2016) Brain inflammation is accompanied by peripheral inflammation in Cstb (−/−) mice, a model for progressive myoclonus epilepsy. J Neuroinflammation 13 (1):298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubio-Villena C, Viana R, Bonet J, Garcia-Gimeno MA, Casado M, Heredia M, Sanz P (2018) Astrocytes: new players in progressive myoclonus epilepsy of Lafora type. Hum Mol Genet 27 (7):1290–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matin N, Tabatabaie O, Falsaperla R, Lubrano R, Pavone P, Mahmood F, Gullotta M, Serra A, Di Mauro P, Cocuzza S, Vitaliti G (2015) Epilepsy and innate immune system: A possible immunogenic predisposition and related therapeutic implications. Hum Vaccin Immunother 11 (8):2021–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aronica E, Bauer S, Bozzi Y, Caleo M, Dingledine R, Gorter JA, Henshall DC, Kaufer D, Koh S, Loscher W, Louboutin JP, Mishto M, Norwood BA, Palma E, Poulter MO, Terrone G, Vezzani A, Kaminski RM (2017) Neuroinflammatory targets and treatments for epilepsy validated in experimental models. Epilepsia 58 Suppl 3:27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.