Abstract
Chitin deacetylase (CDA) can hydrolyse the acetamido group of chitin polymers to produce chitosans, which are used in various fields including the biomedical and pharmaceutical industries, food production, agriculture, and water treatment. CDA represents a more environmentally-friendly and easier to control alternative to the chemical methods currently utilised to produce chitosans from chitin; however, the majority of identified CDAs display activity toward low-molecular-weight oligomers and are essentially inactive toward polymeric chitin or chitosans. Therefore, it is important to identify novel CDAs with activity toward polymeric chitin and chitosans. In this study, we isolated the bacterium Rhodococcus equi F6 from a soil sample and showed that it expresses a novel CDA (ReCDA), whose activity toward 4-nitroacetanilide reached 19.20 U/mL/h during fermentation and was able to deacetylate polymeric chitin, colloidal chitin, glycol-chitin, and chitosan. Whole genome sequencing revealed that ReCDA is unique to the R. equi F6 genome, while phylogenetic analysis indicated that ReCDA is evolutionarily distant from other CDAs. In conclusion, ReCDA isolated from the R. equi F6 strain expands the known repertoire of CDAs and could be used to deacetylate polymeric chitosans and chitin in industrial applications.
Subject terms: Carbohydrates, Biopolymers, Applied microbiology, Bacteria
Introduction
Chitin is the second most abundant biopolymer after cellulose and is mainly obtained as a waste product of the seafood industry at a relatively low cost1. The chitin derivative, chitosan, is a linear polysaccharide comprised of β-(1→4)-linked glucosamine and N-acetyl glucosamine units which are randomly arranged within the chitosan polysaccharide chain2. Due to its ability to dissolve in dilute acids, chitosan is more useful than its crystalline precursor chitin in various industrial applications3, including the biomedical and pharmaceutical industries, food production, agriculture, and water treatment4.
Chitosan occurs naturally and is mainly found in the cell walls of certain fungi, the exoskeletons of certain insects (such as the abdominal wall of termite queens), and in some yeasts5,6. Although chitosan can be extracted from fungal sources7, the method is commercially inapplicable as it provides too low a yield at too high a cost. Therefore, chitosan is still obtained by treating marine-derived chitin with thermo-alkaline8, a method that is inexpensive and results in high yields but is environmentally unsafe and difficult to control, producing a heterogeneous range of products depending on the degree of deacetylation9. The enzymatic production of chitosan using microbial chitin deacetylase (CDA; EC 3.5.1.41), which can hydrolyse the acetamido group in chitin polymers to produce chitosans10, is an environmentally-friendly process that is easy to control and results in a high yield of homogeneous end products11.
CDAs have been identified in marine and soil bacteria, several fungi, a few insects, and at least one virus12,13. Fungal CDAs exist mostly as N-glycosylated (20–70%) glycoproteins, while bacterial CDAs are mainly chitin oligosaccharide deacetylases (CODs) active toward low molecular weight CODs1; however, some bacterial CDAs also show broad substrate specificity14. Only a few microbial strains are known to produce CDAs, and they require specialised fermentation conditions for CDA production15. Since, CDAs have not yet been industrialised, the identification of a bacterial CDA with high activity toward polymeric chitin and chitosans would provide a straightforward approach for enzymatically converting chitin into chitosan.
In this study, we collected soil from different environmental sources and screened its CDA activity, identifying a strain producing high levels of CDA which was characterised by whole genome sequencing and designated as Rhodococcus equi F6. We then examined the CDA-producing capacity of R. equi F6 during fermentation and evaluated the activity of crude CDA isolated from R. equi F6 (ReCDA) toward polymeric chitin and chitosans.
Results and Discussion
Novel chitin-deacetylase-producing bacterial strain identified from soil
More than 100 soil samples were collected from different regions of China, including Chengdu, Shenyang, Xi’an, and numerous cities in Shandong province. The samples were selected randomly to obtain a wide range of strains. The primary screen determined the deacetylation activity of microorganisms present in each soil sample using a plate-based enzymatic assay with 4-nitroacetanilide as the colour indicator. After 2–3 days of incubation, colonies with a yellow circle indicating the occurrence of deacetylation were selected for secondary screening, during which their enzymatic activity was determined following fermentation in LB medium. The F6 strain exhibited the greatest CDA activity of the 16 positive isolates (Supplementary Table S1); therefore, this strain was selected for further taxonomic, physiological, and biochemical analyses.
Phylogenetic classification and characteristics of the novel CDA-producing strain F6
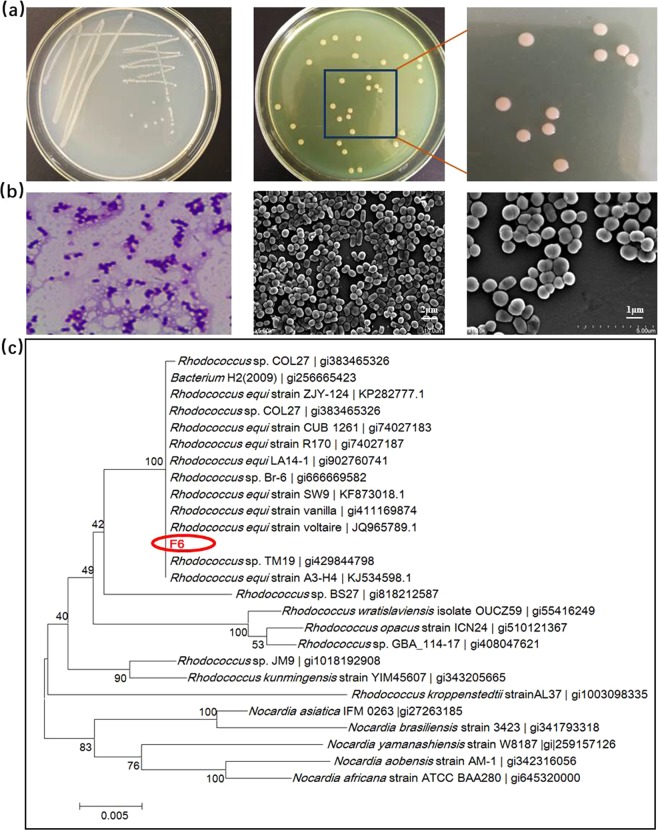
When incubated for 2–3 days at 37 °C, colonies of the F6 strain appeared light pink, round, smooth, opaque, glistening, and mucoid (Fig. 1a). In addition, we found that the F6 strain was gram-positive (Fig. 1b), non-motile, aerobic, and did not produce spores. Scanning electron microscopy (SEM) images revealed that the F6 strain was coccoid in shape, with a diameter of 0.5–1.0 μm (Fig. 1b). Biolog-based identification assays indicated that the F6 strain utilised 15 different carbon sources, while the strain also grew in the presence of acetic acid, which is the by-product of CDA-catalysed deacetylation. The phenotypic and chemotaxonomic characteristics of the F6 strain are summarised in Table 1.
Figure 1.
Morphological and phylogenetic characteristics of the F6 strain. (a) Colony morphology of the F6 strain. (b) Gram staining and SEM image showing the coccoid shape of the F6 strain. (c) Phylogenetic analysis of the F6 strain based on BLAST results.
Table 1.
Phenotypic and chemotaxonomic characteristics of R. equi F6.
| Characteristic | Reaction | Characteristic | Reaction |
|---|---|---|---|
| Colonies: | Light pink, opaque, smooth | Cells: | Distinct cocci |
| pH: | 4.0–9.0 | Temperature: | 25–40 °C |
| Utilisation as sole carbon and energy source: | |||
| D-Ribose | + | Xylitol | − |
| D-Fructose | − | Acetic Acid | + |
| D-Glucose | + | α-Hydroxybutyric Acid | + |
| Sucrose | − | β-Hydroxybutyric Acid | + |
| D-Mannitol | − | α-Ketovaleric Acid | + |
| Dextrin | + | D-Cellobiose | − |
| D-Arabinose | − | D-Sorbitol | − |
| D-Xylose | + | L-Malic Acid | + |
| D-Galactose | − | Pyruvic Acid Methyl Ester | + |
| L-Rhamnose | − | Succinic Acid Mono-Methyl Ester | + |
| Lactamide | + | Propionic Acid | + |
| D-Lactic Acid Methyl Ester | + | Pyruvic Acid | + |
| L-Lactic Acid | + | Tween 40 | + |
| D-Turanose | − | Tween 80 | + |
| Arabitol | − | Lactose | − |
| myo-Inositol | − | Maltose | − |
| Inulin | − | Melezitose | − |
| N-Acetylglucosamine | − | Glycerol | + |
+, Positive; −, negative.
Based on the results of the physiological, biochemical, and 16S rDNA sequence analyses (Fig. 1c), the F6 strain was classified as belonging to the genus Rhodococcus. Further alignment of the 16S rDNA of the F6 strain with other 16S rDNA sequences in the GenBank database revealed that the strain had 100% identity with several Rhodococcus equi strains. Therefore, the F6 strain was designated as Rhodococcus equi F6 (R. equi F6) and deposited into the China General Microbiological Culture Collection as Rhodococcus equi under the accession number 14861. However, as shown in Supplementary Table S2, the biochemical and physiological characteristics of R. equi F6 were not always consistent with those of previously described Rhodococcus strains16–21, suggesting that R. equi F6 may be a novel strain.
Rhodococcus species are known to biodegrade compounds that are not easily degraded by other organisms22–24, making them promising biocatalysts for industrial applications. In addition, members of the Rhodococcus genus can degrade natural hydrophobic compounds and xenobiotics, thus have been used in environmental, pharmaceutical, and chemical fields in addition to energy25,26. To the best of our knowledge, this is the first study to report CDA production by a novel R. equi strain.
CDA production capacity of R. equi F6
Since the CDA produced by R. equi F6 (ReCDA) was localised intracellularly, we investigated the kinetics of ReCDA production by sampling the fermentation broth every few hours and determining enzymatic activity. The kinetics of R. equi F6 cell growth and CDA production in the main culture are shown in Fig. 2. CDA activity reached 87.2 U/mL during the first 5 h and then increased to the maximum level of 157.6 U/mL after 12 h. Moreover, CDA production by R. equi F6 exceeded that previously reported in other microorganisms27–29, which could lower the cost of recycling fermenters and bulk production. Biolog-based analysis indicated that the F6 strain did not have complex fermentation requirements for CDA production, making it suitable for industrial applications, while further mutation breeding of R. equi F6 could produce a strain with even higher CDA yields.
Figure 2.
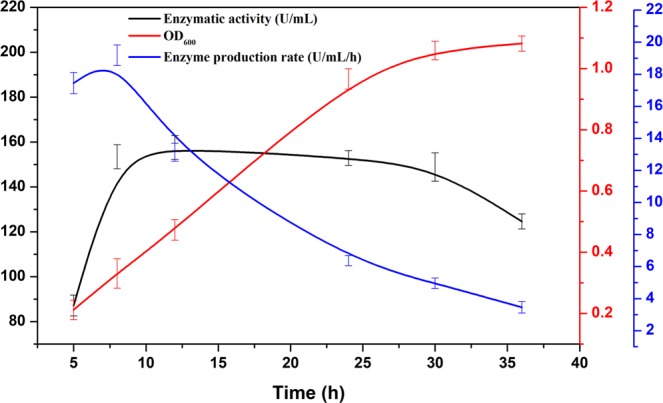
Cell growth and CDA production curves for R. equi F6.
Deacetylation of polymeric chitosans and chitin by crude ReCDA
We measured acetic acid production by HPLC analysis to assess whether crude ReCDA could hydrolyse different polymeric substrates. While crude ReCDA was able to hydrolyse colloidal chitin, glycol-chitin, and chitosan, it had no effect on water-insoluble chitin (Supplementary Fig. S1a). ReCDA hydrolysis was further characterised by MALDI-TOF MS, finding that the average molecular weight of glycol-chitin decreased significantly after 6 h, resulting in a mixture of compounds with different degrees of deacetylation (DD) (Supplementary Fig. S1b).
The majority of known bacterial CDAs are active only toward low-molecular-mass oligomers1; for instance Vibrionaceae CDA can only deacetylate substrates with degrees of polymerisation (DP) of 2–630, while the activity of Shewanella CDA decreases from DP2 to DP4, with no activity at DP531. ReCDA displays several advantages over these other bacterial CDAs since it can hydrolyse polymeric substrates and has high activity toward other substrates, as shown in Table 2. Moreover, ReCDA retained its enzymatic activity at high temperatures, low pH, and in presence of most divalent cations (Supplementary Fig. S2). Although we used a crude ReCDA preparation instead of the pure enzyme in this study, it is worth noting that crude enzymes are often used directly in industry32; thus, ReCDA could be used to prepare chitosan in combination with partial chemical treatment or dissolving with pretreated chitin, which will be reported in the near future. Consequently, we will purify and functionally characterise ReCDA in our future study.
Table 2.
Relative activity of crude ReCDA toward different substrates.
| Substrate | Peak area for acetate (mAU*s) | Relative activity (%)a |
|---|---|---|
| 4-Nitroacetanilide | 71.36 ± 1.09 | 100.00 |
| N-Acetyl-DL-methionine | 139.72 ± 2.85 | 204.12 ± 1.49 |
| Chitosan (DAs, 85%) | 92.99 ± 1.05 | 132.94 ± 0.93 |
| N-acetylglucosamine | 91.40 ± 1.42 | 130.52 ± 0.90 |
| Colloidal chitin | 78.73 ± 2.78 | 111.20 ± 2.40 |
| Glycol-chitin | 64.23 ± 2.01 | 89.12 ± 1.72 |
| N-Acetyl-DL-tryptophan | 63.44 ± 1.65 | 88.36 ± 1.57 |
| N-Acetyl-L-leucine | 46.74 ± 1.58 | 62.32 ± 1.91 |
| N -Acetyl-L-cysteine | 44.70 ± 1.97 | 59.36 ± 2.03 |
| 3-Acetylindole | 34.98 ± 1.46 | 44.19 ± 1.92 |
| Beta-D-Ribofuranose 1-acetate 2,3,5-tribenzoate | 24.54 ± 1.26 | 28.56 ± 2.06 |
| Chitooligosaccharides (2–6) | 21.38 ± 1.51 | 23.85 ± 1.97 |
| Powdered chitin | 0.00 | 0.00 |
aRelative activity was determined by HPLC analysis and activity with 4-nitroacetanilide as the substrate was used as the standard.
Whole genome sequencing, assembly, and annotation of R. equi F6
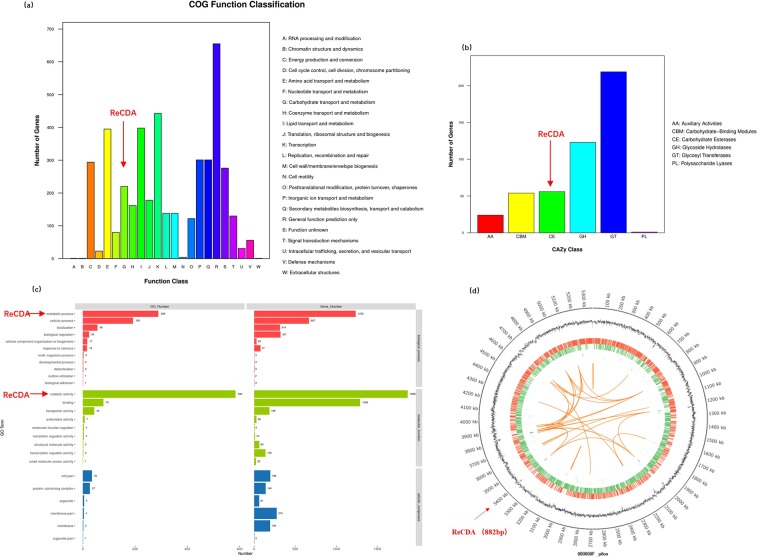
To identify genes involved in chitin deacetylation, we performed whole genome sequencing on R. equi F6, with de-novo genome assembly revealing that the R. equi F6 genome is 5,354,717 bp in length, has a GC content of 68.62%, and contains 4,996 coding sequences and 126 total ncRNAs. CAZy annotation was successfully performed on 477 protein encoding genes (PEGs), 4,348 COG genes, 1,404 GO entries, and 3,679 Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways. Functional PEG annotation revealed a total of 23 classifications, with most predicted to be involved in general functions such as transcription, lipid transport, metabolism, amino acid transport, and metabolism. A total of 220 genes were involved in carbohydrate transport and metabolism (including ReCDA), representing 5.06% of the total number of genes (Fig. 3a), while the 477 genes identified using CAZy represented 10.97% of the total number of genes, indicating that R. equi F6 can metabolise carbohydrates33. Next, we performed PEG analysis with Blastp to identify CAZymes encoded by ReCDA (Fig. 3b) and annotated ReCDA using CE 4. A histogram of target gene distribution using GO terms is shown in Fig. 3c. ReCDA was annotated for biological processes (carbohydrate metabolic processes) and molecular functions (catalytic and hydrolase activity toward carbon-nitrogen, but not peptide, bonds), and its genetic sequence was annotated uniquely in the whole genome database, suggesting that it is the only chitin deacetylase in R. equi F6. The ReCDA gene (Supplementary Data 5) was 882 bp in length and located at 3,400 kb in the genomic map (Fig. 3d).
Figure 3.
Whole genome analysis of R. equi F6. (a) COG functional gene classification. Abscissa represents COG functional classification and ordinate represents the number of genes annotated within each classification. (b) CAZy family distribution map. Abscissa represents CAZy family classification and ordinate represents the number of genes. (c) Histogram showing the distribution of Gene Ontology (GO) terms. Abscissa represents the gene number and ordinate represents GO terms. Different colours are used to distinguish biological processes, cellular components, and molecular functions. (d) Genomic map. From outside to inside: first circle shows genome location information; second circle shows GC content; third circle (red) shows genes encoded on positive strand; fourth circle (green) shows genes encoded on negative strand; fifth circle (blue) shows ncRNAs on positive strand; sixth circle (purple) shows ncRNAs on negative strand; and seventh circle (orange) shows long genome segment repetitive sequences.
To further explore whether R. equi F6 could be used to degrade chitin, we analysed its whole genome for the presence of enzymes involved in chitin deacetylation and hydrolysis. A total of 51 genes were annotated as deacetylases or chitinases (Supplementary Table S3), accounting for 10.69% of the 477 genes annotated in the CAZy family. In addition to the single ReCDA gene, four genes encoding N-acetylglucosamine deacetylases and 16 encoding diacetylchitobiose deacetylases were annotated within the 23 deacetylases, indicating that R. equi F6 is a potent chitin decomposer. Notably, only five bacterial CDAs have been reported to date13,14,31,34.
Phylogenetic analysis for ReCDA
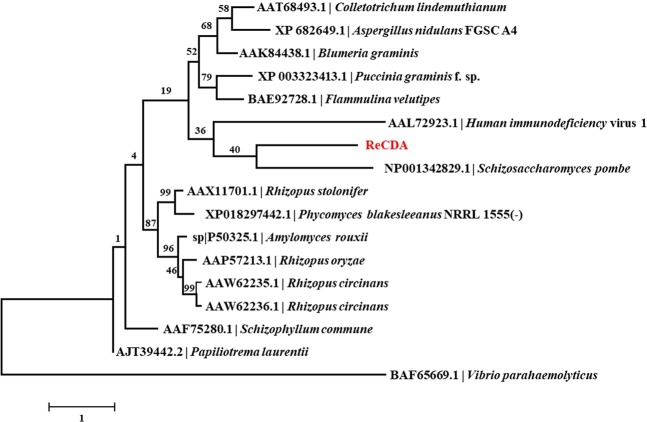
Finally, we used phylogenetic analysis to compare ReCDA with known CDAs from other microorganisms. ReCDA was located in the same node as the CDA from fission yeast Schizosaccharomyces pombe (NCBI Reference Sequence: NP_001342829.1; Fig. 4), with multiple-sequence alignments indicating that ReCDA has 87% query coverage and 21.77% identity with S. pombe CDA. The identity of ReCDA with other known CDAs was below 32%, while the majority of the bootstrap values were below 60%, indicating that the node was not well supported. Taken together, these low degrees of homology indicate that ReCDA is a novel enzyme.
Figure 4.
Molecular phylogenetic analysis of CDAs from R. equi F6 and several other known bacteria. Evolutionary history was inferred using the maximum likelihood method based on a JTT matrix-based model52. The amino acid sequences used in this tree were reported in37–47. Clustal-X2 (Version 1.83) was used for multiple-sequence alignment.
Methods
Isolation of CDA-producing organisms
Soil samples were collected from different ecological areas in north, central, and south China, including Chengdu, Shenyang, Xi’an, and numerous cities in Shandong province. The samples were screened for microbial strains with high CDA activity by diluting each soil sample (5 g) in sterilised water (50 mL) and incubating the sample at 37 °C for 30 min at 200 rpm. Each supernatant was then serially diluted and sprayed onto agar plates containing colloidal chitin as the carbon source and 4-nitroacetanilide (200 mg/L), which can easily penetrate bacterial cell walls and indicate deacetylation by changing colour. The plates were incubated at 37 °C for 2–3 days and desirable strains were screened based on the relative sizes of the yellow circles in the primary screen and using an enzymatic activity assay in the secondary screen.
Culture conditions and phenotypic analysis of the strains
To allow fermentation, the strains were cultured in LB medium for 24 or 36 h at 37 °C and 200 rpm, after which their OD600 value and crude enzyme activity were measured, with each experiment performed in triplicate. The growth characteristics of each strain were determined using a Biolog system consisting of a microplate with 95 different carbon sources and a computer-driven automatic plate reader, wherein the four azole redox dye is reduced when a carbon source is consumed by the organism, causing a colour change from colourless to purple35.
Scanning electron microscopy
SEM was performed as described previously36 with the following modifications. Briefly, the F6 strain was grown in LB medium at 37 °C for 24 h, centrifuged at 8000 r/min for 5 min, and the bacterial pellets washed five times with phosphate-buffered saline (PBS). The samples were fixed overnight using 2.5% (v/v) glutaraldehyde, washed thrice with PBS, and then covered with a gold layer prior to observation using an SU1510 FE-SEM (Hitachi, Japan).
Phylogenetic analysis
The 16S rDNA of the identified candidate strain was amplified using universal primers (27 F and 1492 R) under the following colony PCR conditions: 94 °C for 5 min; 30 cycles of 94 °C for 30 s, 52 °C for 30 s, and 72 °C for 90 s; followed by 72 °C for 10 min. The purified PCR products were sequenced and their sequencing data subjected to phylogenetic analysis. The CDA amino acid sequences isolated from Rhodococcus equi F6 (ReCDA) were compared to those of known CDAs from other microorganisms37–47 by phylogenetic analysis, with a phylogenetic tree constructed using MEGA (version 7.0)48.
Enzyme assays
The bacterial cells were washed, diluted with 0.2 M phosphate buffer (pH 7.0), and homogenised by grinding in a pestle and mortar in presence of liquid nitrogen. The enzymatic activity assay was performed as described previously28. Briefly, the reaction mixture was incubated at 37 °C for 1 h, with one unit of CDA activity defined as the amount of enzyme needed to catalyse the release of 4-nitroaniline per hour from 4-nitroacetanilide28.
Analysis of polymeric chitosan and chitin deacetylation by crude ReCDA
To determine the ability of ReCDA to hydrolyse the acetamido group, we used a series of polymeric substrates, including chitin powder, colloidal chitin (prepared using chitin powder), chitosan polymers with 80 and 90% DD, and glycol-chitin (Wako, Japan). Crude ReCDA (5 mL) was stirred into an excess of substrate solution and the mixture was allowed to react for 12 h at 37 °C and pH 4.0 with agitation. Samples were then boiled for 5 min and the relative activity of the crude ReCDA toward different substrates was assessed by HPLC to measure the production of acetic acid49,50. Glycol-chitin and the product of glycol-chitin hydrolysis were also analysed by MALDI-TOF MS using an Ultraflex II TOF/TOF MALDI-TOF mass spectrometer (Bruker Daltonics, Germany) alongside an N2 laser with a 337-nm wavelength at a frequency of 50 Hz and positive ion-reflector mode at an accelerating voltage of 20 kV. All analytes were spotted on a 384-spot stainless steel plate and 2,5-dihydroxybenzoic acid (DHB) was used as the matrix, with 0.6 μL of matrix solution applied to each spot29.
Whole genome sequencing, assembly, and annotation
The whole genome sequencing of R. equi F6 was performed by Suzhou Genewiz Biotechnology Co. Ltd. PCR products from R. equi F6 were cleaned up and validated using an Agilent 2100 Bioanalyser (Agilent Technologies, Palo Alto, CA, USA) and quantified using a Qubit 3.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). Libraries with different indices were multiplexed and loaded using an Illumina HiSeq instrument (Illumina, San Diego, CA, USA), according to the manufacturer’s instructions. Sequencing was carried out using a 2 × 150 paired-end (PE) configuration, while image analysis and base calling were conducted using HiSeq Control Software (HCS)+OLB+GAPipeline-1.6 (Illumina) on a HiSeq instrument (Illumina). The library was also sequenced on a PacBio RSII/Sequel SMRT instrument51. Coding genes were annotated using BLAST in the National Centre for Biotechnology Information (NCBI) NR database. Gene function was annotated using the GO database, while pathways were annotated using the KEGG database. Proteins were classified phylogenetically using the COG/KOG Clusters of Orthologous Groups (COG/KOG) database.
Nucleotide sequence accession number
The nucleotide sequence of R. equi F6 was deposited in the NCBI SRA database under the accession number PRJNA526377.
Supplementary information
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Grant No. 21706194), Tianjin Municipal Science and Technology Commission (17PTGCCX00190 and 17PTSYJC00080), and the Innovative Research Team of Tianjin Municipal Education Commission (TD13-5013). We would like to thank Editage (www.editage.cn) for English language editing.
Author contributions
Q.Y.M. and M.W. designed this study. Q.Y.M., X.Z.G., and Y.B.S. conceived and planned the experiments. Q.Y.M., X.Z.G., and X.Y.B. performed the experiments. Q.Y.M., X.Z.G., Y.B.S., and M.L.X. analysed the data. Q.Y.M., X.Z.G., and L.N.T. wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Qinyuan Ma and Xiuzhen Gao.
Contributor Information
Yanbing Shen, Email: shenyb@tust.edu.cn.
Min Wang, Email: minw@tust.edu.cn.
Supplementary information
is available for this paper at 10.1038/s41598-020-61349-9.
References
- 1.Grifoll-Romero L, Pascual S, Aragunde H, Biarnés X, Planas A. Chitin deacetylases: Structures, specificities, and biotech applications. Polymers. 2018;10:352–381. doi: 10.3390/polym10040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis LL, Bartnicki-Garcia S. Chitosan synthesis by the tandem action of chitin synthetase and chitin deacetylase from Mucor rouxii. Biochemistry. 1984;23:1065–1073. doi: 10.1021/bi00301a005. [DOI] [Google Scholar]
- 3.Philibert T, Lee BH, Fabien N. Current status and new perspectives on chitin and chitosan as functional biopolymers. Appl. Biochem. Biotechnol. 2017;181:1314–1337. doi: 10.1007/s12010-016-2286-2. [DOI] [PubMed] [Google Scholar]
- 4.Rinaudo M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006;31:603–632. doi: 10.1016/j.progpolymsci.2006.06.001. [DOI] [Google Scholar]
- 5.Pillai, S. K. & Ray, S. S. Chitosan-based Nanocomposites in Natural Polymers: Volume 2: Nanocomposites. 33–68 (Royal Society of Chemistry (2012).
- 6.Dhillon GS, Kaur S, Brar SK, Verma M. Green synthesis approach: extraction of chitosan from fungus mycelia. Crit. Rev. Biotechnol. 2013;33:379–403. doi: 10.3109/07388551.2012.717217. [DOI] [PubMed] [Google Scholar]
- 7.Ghormade V, Pathan EK, Deshpande MV. Can fungi compete with marine sources for chitosan production? Int. J. Biol. Macromol. 2017;104:1415–1421. doi: 10.1016/j.ijbiomac.2017.01.112. [DOI] [PubMed] [Google Scholar]
- 8.Hamed I, Özogul F, Regenstein JM. Industrial applications of crustacean by-products (chitin, chitosan, and chitooligosaccharides): A review. Trends Food Sci. Technol. 2016;48:40–50. doi: 10.1016/j.tifs.2015.11.007. [DOI] [Google Scholar]
- 9.Tsigos I, Martinou A, Kafetzopoulos D, Bouriotis V. Chitin deacetylases: new, versatile tools in biotechnology. Trends Biotechnol. 2000;18:305–312. doi: 10.1016/S0167-7799(00)01462-1. [DOI] [PubMed] [Google Scholar]
- 10.Aranda-Martinez A, et al. Expression and specificity of a chitin deacetylase from the nematophagous fungus Pochonia chlamydosporia potentially involved in pathogenicity. Sci. Rep. 2018;8:2170–2181. doi: 10.1038/s41598-018-19902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aktuganov GE, Melent’ev AI. Specific features of chitosan depolymerization by chitinases, chitosanases, and nonspecific enzymes in the production of bioactive chitooligosaccharides (Review) Appl. Biochem. Microbiol. 2017;53:611–627. doi: 10.1134/S0003683817060023. [DOI] [Google Scholar]
- 12.Schmitz C, et al. Conversion of Chitin to Defined Chitosan Oligomers: Current Status and Future Prospects. Mar. Drugs. 2019;17:452. doi: 10.3390/md17080452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.John M, Röhrig H, Schmidt J, Wieneke U, Schell J. Rhizobium NodB protein involved in nodulation signal synthesis is a chitooligosaccharide deacetylase. Proc. Natl. Acad. Sci. 1993;90:625–629. doi: 10.1073/pnas.90.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuveng TR, et al. Structure and function of a CE4 deacetylase isolated from a marine environment. Plos One. 2017;12:e0187544. doi: 10.1371/journal.pone.0187544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suresh PV, Sakhare PZ, Sachindra NM, Halami PM. Extracellular chitin deacetylase production in solid state fermentation by native soil isolates of Penicillium monoverticillium and Fusarium oxysporum. J. Food Sci. Technol. 2014;51:1594–1599. doi: 10.1007/s13197-012-0676-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuyama H, Yumoto I, Kudo T, Shida O. Rhodococcus tukisamuensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2003;53:1333–1337. doi: 10.1099/ijs.0.02523-0. [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi M, Hatano K, Sedlácek I, Pácová Z. Rhodococcus jostii sp. nov., isolated from a medieval grave. Int. J. Syst. Evol. Microbiol. 2002;52:409–413. doi: 10.1099/00207713-52-2-409. [DOI] [PubMed] [Google Scholar]
- 18.Yoon JH, et al. Rhodococcus koreensis sp. nov., a 2,4-dinitrophenol-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2000;50:1193–1201. doi: 10.1099/00207713-50-3-1193. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Zhang Y, Xiao C, Liu Z, Goodfellow M. Rhodococcus maanshanensis sp. nov., a novel actinomycete from soil. Int. J. Syst. Evol. Microbiol. 2002;52:2121–2126. doi: 10.1099/00207713-52-6-2121. [DOI] [PubMed] [Google Scholar]
- 20.Yoon JH, et al. Rhodococcus pyridinivorans sp. nov., a pyridine-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2000;50:2173–2180. doi: 10.1099/00207713-50-6-2173. [DOI] [PubMed] [Google Scholar]
- 21.Klatte S, Kroppenstedt RM, Rainey FA. Rhodococcus opacus sp.nov., An Unusual Nutritionally Versatile Rhodococcus-species. Syst. Appl. Microbiol. 1994;17:355–360. doi: 10.1016/S0723-2020(11)80051-2. [DOI] [Google Scholar]
- 22.McLeod MP, et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. 2006;103:15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MartÍnková L, Křen V. Nitrile- and amide-converting microbial enzymes: Stereo-, regio- and chemoselectivity. Biocatal. Biotransformation. 2002;20:73–93. doi: 10.1080/10242420290018069. [DOI] [Google Scholar]
- 24.Singh R, Sharma R, Tewari NG, Rawat DS. Nitrilase and its application as a “green” catalyst. Chemi. Biodivers. 2006;3:1279–1287. doi: 10.1002/cbdv.200690131. [DOI] [PubMed] [Google Scholar]
- 25.Van der Geize R, Dijkhuizen L. Harnessing the catabolic diversity of Rhodococci for environmental and biotechnological applications. Curr. Opin. Microbiol. 2004;7:255–261. doi: 10.1016/j.mib.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Larkin MJ, Kulakov LA, Allen CCR. Biodegradation and Rhodococcus-masters of catabolic versatility. Curr. Opin. Biotechnol. 2005;16:282–290. doi: 10.1016/j.copbio.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Pareek N, Vivekanand V, Dwivedi P, Singh RP. Penicillium oxalicum SAEM-51: a mutagenised strain for enhanced production of chitin deacetylase for bioconversion to chitosan. N. Biotechnol. 2011;28:118–124. doi: 10.1016/j.nbt.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 28.Sun Y, Zhang J, Wu S, Wang S. Statistical optimization for production of chitin deacetylase from Rhodococcus erythropolis HG05. Carbohydr. Polym. 2014;102:649–652. doi: 10.1016/j.carbpol.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Ramos-Puebla A, et al. Addition of abscisic acid increases the production of chitin deacetylase by Colletotrichum gloeosporioides in submerged culture. Process Biochem. 2016;51:959–966. doi: 10.1016/j.procbio.2016.05.003. [DOI] [Google Scholar]
- 30.Li X, Wang L-X, Wang X, Roseman S. The chitin catabolic cascade in the marine bacterium Vibrio Cholerae: Characterization of a unique chitin oligosaccharide deacetylase. Glycobiology. 2007;17:1377–1387. doi: 10.1093/glycob/cwm096. [DOI] [PubMed] [Google Scholar]
- 31.Hirano T, Uehara R, Shiraishi H, Hakamata W, Nishio T. Chitin oligosaccharide deacetylase from Shewanella woodyi ATCC51908. J. Appl. Glycosci. 2015;62:153–157. doi: 10.5458/jag.jag.JAG-2015_014. [DOI] [Google Scholar]
- 32.Sashiwa H, et al. Production of N-acetyl-d-glucosamine from α-chitin by crude enzymes from Aeromonas hydrophila H-2330. Carbohydr. Res. 2002;337:761–763. doi: 10.1016/S0008-6215(02)00034-4. [DOI] [PubMed] [Google Scholar]
- 33.Yadav S, Dubey SK. Cellulose degradation potential of Paenibacillus lautus strain BHU3 and its whole genome sequence. Bioresour. Technol. 2018;262:124–131. doi: 10.1016/j.biortech.2018.04.067. [DOI] [PubMed] [Google Scholar]
- 34.Ohishi K, et al. Purification and properties of two deacetylases produced by Vibrio alginolyticus H-8. Biosci. Biotechnol. Biochem. 1997;61:1113–1117. doi: 10.1271/bbb.61.1113. [DOI] [Google Scholar]
- 35.Wragg P, Randall L, Whatmore AM. Comparison of Biolog GEN III MicroStation semi-automated bacterial identification system with matrix-assisted laser desorption ionization-time of flight mass spectrometry and 16S ribosomal RNA gene sequencing for the identification of bacteria of veterinary interest. J. Microbiol. Methods. 2014;105:16–21. doi: 10.1016/j.mimet.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Ong KS, et al. Burkholderia paludis sp. nov., an antibiotic-siderophore producing novel Burkholderia cepacia complex species, isolated from Malaysian tropical peat swamp soil. Front. Microbiol. 2016;7:2046. doi: 10.3389/fmicb.2016.02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuo Y, Tanaka K, Matsuda H, Kawamukai M. cda1+, encoding chitin deacetylase is required for proper spore formation in Schizosaccharomyces pombe. FEBS Lett. 2005;579:2737–2743. doi: 10.1016/j.febslet.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Gauthier C, Clerisse F, Dommes J, Jaspar-Versali M-F. Characterization and cloning of chitin deacetylases from Rhizopus circinans. Protein Expr. Purif. 2008;59:127–137. doi: 10.1016/j.pep.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, et al. Cloning of a Heat-Stable Chitin Deacetylase Gene from Aspergillus nidulans and its Functional Expression in Escherichia coli. Appl. Biochem. Biotechnol. 2009;162:843–854. doi: 10.1007/s12010-009-8772-z. [DOI] [PubMed] [Google Scholar]
- 40.Chakraborty W, et al. Expression of a chitin deacetylase gene, up-regulated in Cryptococcus laurentii strain RY1, under nitrogen limitation. J. Basic Microbiol. 2016;56:576–579. doi: 10.1002/jobm.201500596. [DOI] [PubMed] [Google Scholar]
- 41.Shrestha B, Blondeau K, Stevens WF, Hegarat FL. Expression of chitin deacetylase from Colletotrichum lindemuthianum in Pichia pastoris: purification and characterization. Protein Expr. Purif. 2004;38:196–204. doi: 10.1016/j.pep.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 42.Sarkar S, Gupta S, Chakraborty W, Senapati S, Gachhui R. Homology modeling, molecular docking and molecular dynamics studies of the catalytic domain of chitin deacetylase from Cryptococcus laurentii strain RY1. Int. J. Biol. Macromol. 2017;104:1682–1691. doi: 10.1016/j.ijbiomac.2017.03.057. [DOI] [PubMed] [Google Scholar]
- 43.Yamada M, et al. Isolation and characterization of a gene coding for chitin deacetylase specifically expressed during fruiting body development in the basidiomycete Flammulina velutipes and its expression in the yeast Pichia pastoris. FEMS Microbiol. Lett. 2008;289:130–137. doi: 10.1111/j.1574-6968.2008.01361.x. [DOI] [PubMed] [Google Scholar]
- 44.Tokuyasu K, Kaneko S, Hayashi K, Mori Y. Production of a recombinant chitin deacetylase in the culture medium of Escherichia coli cells. FEBS Lett. 1999;458:23–26. doi: 10.1016/S0014-5793(99)01113-8. [DOI] [PubMed] [Google Scholar]
- 45.Jeraj N, Kunič B, Lenasi H, Breskvar K. Purification and molecular characterization of chitin deacetylase from Rhizopus nigricans. Enzyme Microb. Technol. 2006;39:1294–1299. doi: 10.1016/j.enzmictec.2006.03.017. [DOI] [Google Scholar]
- 46.Naqvi S, et al. A Recombinant Fungal Chitin Deacetylase Produces Fully Defined Chitosan Oligomers with Novel Patterns of Acetylation. Appl. Environ. Microbiol. 2016;82:6645. doi: 10.1128/AEM.01961-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Z, et al. Structure and function of a broad-specificity chitin deacetylase from Aspergillus nidulans FGSC A4. Sci. Rep. 2017;7:1746. doi: 10.1038/s41598-017-02043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naqvi S, et al. A recombinant fungal chitin deacetylase produces fully defined chitosan oligomers with novel patterns of acetylation. Appl. Environ. Microbiol. 2016;82:6645–6655. doi: 10.1128/AEM.01961-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amorim RVS, Ledingham WM, Fukushima K, Campos-Takaki GM. Screening of chitin deacetylase from Mucoralean strains (Zygomycetes) and its relationship to cell growth rate. J. Ind. Microbiol. Biotechnol. 2005;32:19–23. doi: 10.1007/s10295-004-0197-7. [DOI] [PubMed] [Google Scholar]
- 51.McCarthy A. Third Generation DNA Sequencing: Pacific Biosciences’ Single Molecule Real Time Technology. Chem. Biol. 2010;17:675–676. doi: 10.1016/j.chembiol.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 52.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.