Abstract
Wildfires can encourage the establishment of invasive plants by releasing potent germination stimulants, such as karrikins. Seed germination of Brassica tournefortii, a noxious weed of Mediterranean climates, is strongly stimulated by KAR1, the archetypal karrikin produced from burning vegetation. In contrast, the closely-related yet non-fire-associated ephemeral Arabidopsis thaliana is unusual because it responds preferentially to KAR2. The α/β-hydrolase KARRIKIN INSENSITIVE 2 (KAI2) is the putative karrikin receptor identified in Arabidopsis. Here we show that B. tournefortii expresses three KAI2 homologues, and the most highly-expressed homologue is sufficient to confer enhanced responses to KAR1 relative to KAR2 when expressed in Arabidopsis. We identify two amino acid residues near the KAI2 active site that explain the ligand selectivity, and show that this combination has arisen independently multiple times within dicots. Our results suggest that duplication and diversification of KAI2 proteins could confer differential responses to chemical cues produced by environmental disturbance, including fire.
Subject terms: Small molecules, Plant genetics, Plant hormones, Plant signalling
Karrikins are germination stimulants perceived by KAI2 in Arabidopsis. Here the authors show that Brassica tournefortii, a close relative to Arabidopsis, has multiple copies of KAI2 with amino acid substitutions that confer responsiveness to the specific karrikin compounds found in wildfire smoke.
Introduction
Environmental disturbance promotes the establishment of invasive species, posing a potent threat to global biodiversity. Changing wildfire regimes, such as increasing frequency of fires, is one of the most-relevant disturbance factors contributing to elevated invasion threat1. Wildfires create germination opportunities in part by releasing seed germination stimulants, such as karrikins, from burning vegetation2,3. Karrikins comprise a family of butenolides with six known members. In samples of smoke water generated by burning grass straw, KAR1 is the most abundant karrikin, whereas KAR2 is six times less abundant4, although these proportions may differ depending on source material, preparation method, age and storage conditions5. These two analogues differ only by the presence of a methyl group on the butenolide ring in KAR1, which is absent in KAR2 (Supplementary Fig. 1a). Invasive plant species that are responsive to karrikins could utilise natural and human-induced fires to facilitate their establishment6,7.
Karrikins can overcome seed dormancy and promote seed germination in a number of smoke-responsive species, as well as others that are not associated with fire regimes8–10. Furthermore, karrikins also influence light-dependent seedling development. Karrikins enhance the sensitivity of Arabidopsis thaliana seedlings to light by inhibiting hypocotyl elongation, stimulating cotyledon expansion and promoting chlorophyll accumulation in a dose-dependent manner11. Karrikins have been reported to enhance seedling survival and biomass in species such as tomato, rice and maize12,13. Such growth-promoting effects of karrikins, especially at critical early stages of the life cycle, have the potential to further encourage the establishment of invasive species after fire events.
Brassica tournefortii (Brassicaceae; Sahara mustard) is native to northern Africa and the Middle East, but is an invasive weed that blights many ecosystems with a Mediterranean climate and chaparral-type vegetation that are prone to wildfires in North America, Australia and South Africa. B. tournefortii seeds can persist in the soil for many seasons, undergoing wet–dry cycling that can influence dormancy and contribute to boom-bust cycles that outcompete native species9,14. B. tournefortii plants may radically alter fire frequency and intensity by influencing fuel profiles15,16, further exacerbating the impact of fire on susceptible native ecosystems. In addition, seeds of B. tournefortii are particularly responsive to smoke-derived karrikins, and show a positive germination response to KAR1 in the nanomolar range10. Accordingly, B. tournefortii is particularly well positioned to invade areas disturbed by fire events17,18.
The putative karrikin receptor KARRIKIN INSENSITIVE 2 (KAI2) was identified in Arabidopsis, a weedy ephemeral that originated in Eurasia but is now widely distributed throughout the northern hemisphere19–21. Arabidopsis is not known to colonise fire-prone habitats, but nevertheless seeds germinate in response to karrikins in the micromolar range22. Unlike most smoke-responsive species that respond more readily to KAR18,23, Arabidopsis responds preferentially to KAR222. KAI2 is an evolutionarily ancient α/β-hydrolase and a paralogue of DWARF14 (D14), the receptor for strigolactones24,25. Karrikins and strigolactones are chemically similar by virtue of a butenolide moiety that is necessary for bioactivity26,27. KAI2 and D14 have dual functions as both enzyme and receptor, but the functional significance of the enzymatic activity remains contested28–32. Furthermore, the basis for ligand specificity by these two highly congruent proteins remains essentially unknown.
Orthologues of KAI2 are ubiquitous in land plants, and are normally present as a single gene copy within an ancient and highly conserved ‘eu-KAI2' clade33. There is growing evidence that, beyond its ability to mediate karrikin responses, KAI2 has a core ancestral role in perceiving an endogenous KAI2 ligand ('KL') that regulates seed germination, seedling development, leaf shape and cuticle development34–36. Since its divergence from the Arabidopsis lineage, the tribe Brassiceae, which includes the genus Brassica, underwent a whole genome triplication event 24–29 million years ago37–39. This process might have allowed additional KAI2 copies to gain altered ligand specificity, potentially enhancing perception of environmental signals such as karrikins from smoke. Here, we report that two out of three KAI2 homologues expressed in B. tournefortii show distinct preferences for different karrikins. We take advantage of the relatively recent genome triplication event in the Brassiceae to identify two amino acids that are sufficient to explain these karrikin preferences and confirm this by mutagenesis. Beyond demonstrating the potential ecological significance of diversity among KAI2 homologues, our findings also reveal active site regions critical for ligand selectivity among KAI2 receptor-enzymes that are found in all land plants.
Results
B. tournefortii is most sensitive to KAR1
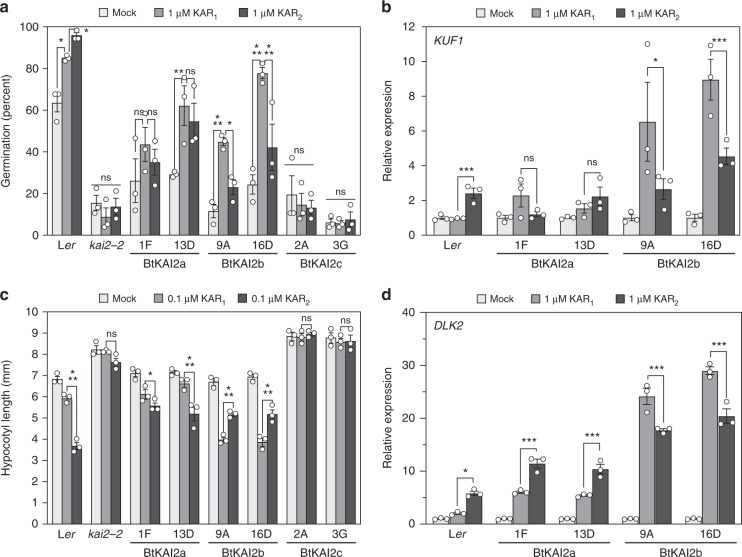
To characterise the karrikin response of B. tournefortii, we performed multiple physiological and molecular assays comparing KAR1 activity with that of KAR2. First, germination of B. tournefortii seeds was consistently more responsive to KAR1 than KAR2 at 10 nM, 100 nM, and 1 µM (Fig. 1a and Supplementary Fig. 1b–d). Second, homologues of two karrikin-responsive transcripts, DWARF14-LIKE2 (BtDLK2) and SALT TOLERANCE HOMOLOG7 (BtSTH7), identified on the basis of close sequence homology to their Arabidopsis counterparts11,20, were significantly more highly expressed when treated with 1 µM KAR1 than with 1 µM KAR2 (Fig. 1b). These observed differences in seed response are not owing to differential karrikin uptake, as both KAR1 and KAR2 were taken up from solution at similar rates by B. tournefortii seeds during imbibition, as was also true for Arabidopsis seeds (Supplementary Fig. 1e–f). Besides promoting germination of primary-dormant seeds, karrikins also inhibited hypocotyl elongation in B. tournefortii seedlings, as is the case in Arabidopsis; again, KAR1 showed a stronger effect than KAR2 (Fig. 1c, d). Levels of BtDLK2 transcripts in seedlings were also more responsive to KAR1 than KAR2 at a given concentration (Fig. 1e). Therefore, we conclude that B. tournefortii is more sensitive to KAR1 than to KAR2, a ligand preference that is a feature of seed germination in many karrikin-responsive species from ecosystems prone to fires8,23.
Fig. 1. Brassica tournefortii is highly sensitive to KAR1, the major karrikin analogue isolated from plant-derived smoke.
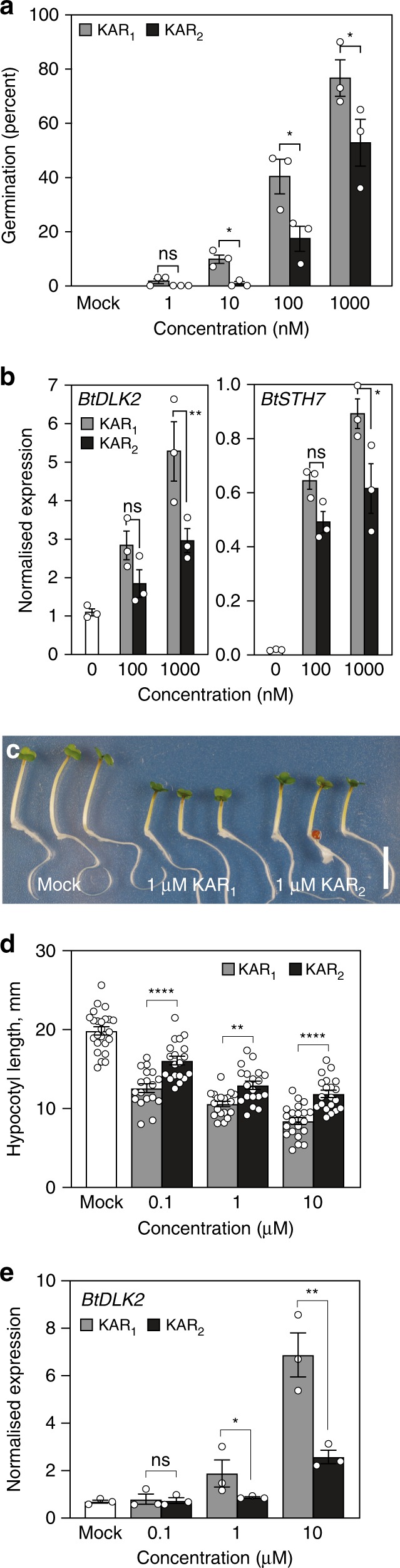
a Germination responses of B. tournefortii seed to KAR1 and KAR2. Data are cumulative germination after 11 days (mean ± SE; n = 3 biological replicates per treatment, ≥35 seeds per replicate). b Levels of karrikin-responsive transcripts BtDLK2 and BtSTH7 in B. tournefortii seed. Seeds were imbibed for 24 h in the dark supplemented with KAR1 and KAR2. Transcripts were normalised to BtCACS reference transcripts. Data are means ± SE, n = 3 biological replicates, ≥50 seeds per replicate. c, d Hypocotyl elongation responses of B. tournefortii seedlings grown for 4 days under continuous red light on water-saturated glass fibre filter paper containing 0.1% acetone (mock), KAR1 or KAR2. Data are means ± 95% CI of n = 18–24 seedlings. Scale bar: 10 mm. e Levels of BtDLK2 transcripts in B. tournefortii seedlings grown under the same conditions as for d. Data are means ± SE, n = 3 biological replicates, ≥20 seedlings per replicate. In all panels, asterisks denote significance levels (ANOVA) between indicated conditions: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
BtKAI2a and BtKAI2b are functional in Arabidopsis
To establish whether there are multiple KAI2 homologues present in B. tournefortii, we examined transcriptomes from seeds and seedlings. Three putative KAI2 homologues were identified (BtKAI2a, BtKAI2b and BtKAI2c; Fig. 2a; Supplementary Figs. 2, 3). BtKAI2a grouped with AtKAI2 and those of other Brassicaceae within a single clade, whereas BtKAI2b and BtKAI2c grouped within a clade unique to Brassica. This taxonomic distribution implies that BtKAI2a is most similar to AtKAI2, and that BtKAI2b and BtKAI2c are paralogues that arose via genome triplication during the evolution of Brassica. All three BtKAI2 transcripts were expressed in B. tournefortii seeds, but only BtKAI2a and BtKAI2b could be detected in seedlings (Fig. 2b). In seeds and seedlings, BtKAI2b transcripts were the most abundant of the three, but there were no consistent effects on any of the transcripts upon treatment with 1 µM KAR1. We also identified two BtD14 homologues, at least one of which is functionally orthologous to AtD14 (Supplementary Figs. 2, 4).
Fig. 2. Two differentially expressed B. tournefortii KAI2 homologues are functional in Arabidopsis.
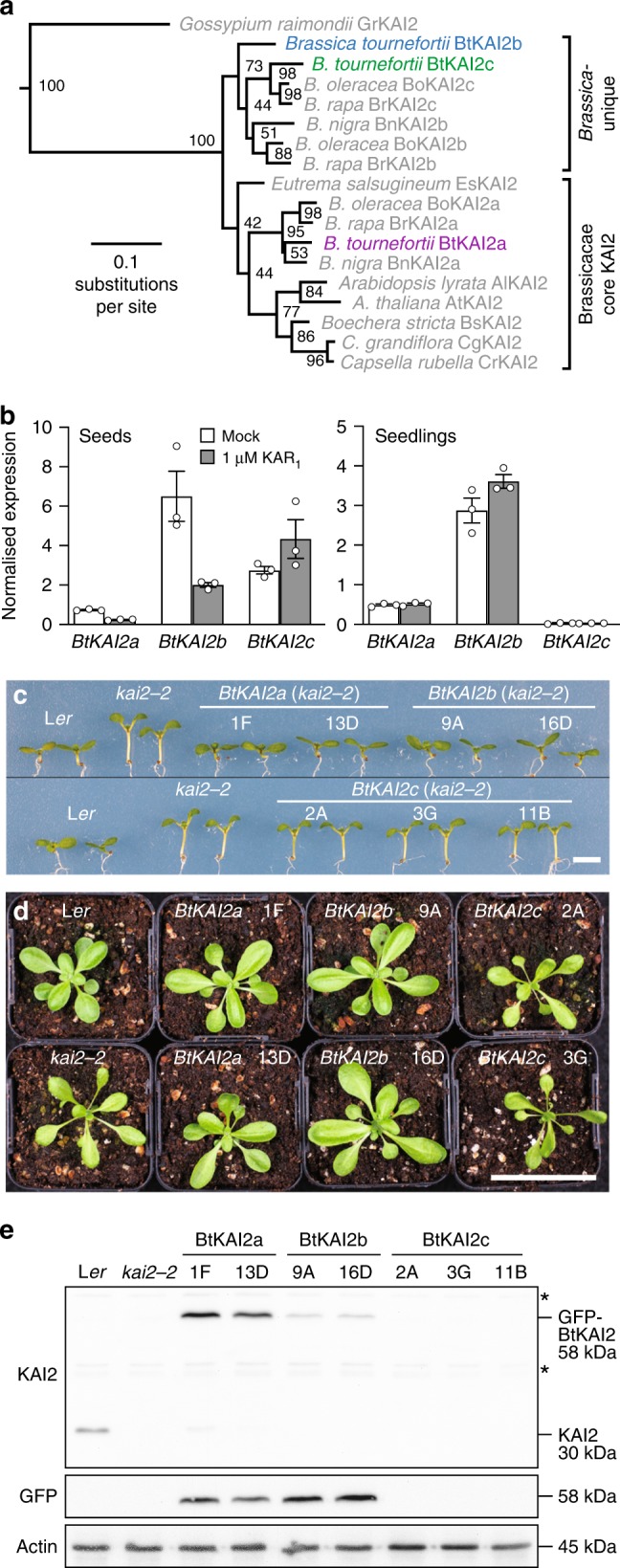
a Maximum likelihood phylogeny of KAI2 homologues in the Brassicaceae, based on nucleotide data. Node values represent bootstrap support from 100 replicates. A eu-KAI2 sequence from Gossypium raimondii (Malvaceae) serves as an outgroup. Tree shown is a subset of a larger phylogeny in Supplementary Fig. 2. b Transcript levels of the three BtKAI2 homologues in B. tournefortii seeds imbibed for 24 h (left) and 4-day-old seedlings (right) treated with 0.1% acetone (mock) or with 1 µM KAR1 for 24 h. c, d Seedling and rosette phenotypes of two independent transgenic lines of Arabidopsis homozygous for KAI2pro:GFP-BtKAI2 transgenes. Seedlings were 7 days old and rosettes 22 days old. Scale bars: 5 mm c; 50 mm d. e Immunoblots of soluble proteins challenged with antibodies against KAI2 (upper panel), GFP (middle panel) or actin as a loading control (lower panel). Anti-KAI2 detects both the native AtKAI2 protein (30 kDa) and the GFP-BtKAI2 fusion proteins (58 kDa) but detects BtKAI2b relatively poorly. Non-specific bands are marked with asterisks. Protein was isolated from pools of ∼50 7-day-old seedlings. Source data are provided as a Source Data file.
We performed transgenic complementation of the Arabidopsis kai2-2 null mutant by expressing each of the three isoforms as a GFP fusion protein driven by the Arabidopsis KAI2 promoter and 5′-UTR (KAI2pro:GFP-BtKAI2) to accurately reflect the native expression profile of KAI2 in Arabidopsis. Such fusions of GFP with KAI2 proteins have been used previously to analyse KAI2 activity19,40,41. Both BtKAI2a and BtKAI2b complemented the seedling and leaf phenotypes of kai2-2, whereas BtKAI2c did not (Fig. 2c, d). GFP-BtKAI2a and GFP-BtKAI2b accumulated at consistent levels when detected using both an anti-KAI2 antibody and an anti-GFP antibody (which negated the effects of sequence differences in the anti-KAI2 epitope region; Fig. 2e and Supplementary Fig. 3). However, we could not detect GFP-BtKAI2c protein in three independent transgenic lines with either antibody. We verified the GFP-BtKAI2c transcript sequences expressed in the transgenic plant material by RT-PCR and Sanger sequencing (Supplementary Fig. 5). As the GFP-BtKAI2c mRNA is processed faithfully in Arabidopsis, the apparent absence of protein is most likely a result of posttranslational events. We then tested whether BtKAI2c was generally poorly expressed in plant cells by transient overexpression in tobacco leaves using Agrobacterium-mediated infiltration and tobacco mosaic virus-based plant expression vectors42. Even with the extremely high levels of expression supported by this system, levels of myc-tagged BtKAI2c protein were substantially lower than equivalently tagged BtKAI2a and BtKAI2b (Supplementary Fig. 5). This result implies that BtKAI2c protein may be inherently unstable and/or prone to degradation in plant cells.
BtKAI2c carries a destabilising mutation at position 98
Numerous mutations identified by forward genetics destabilise AtKAI2 and render the protein undetectable in plant extracts43. Ten residues in the BtKAI2c sequence are unique to BtKAI2c within the genus Brassica (Supplementary Fig. 3). We predicted the likely effect of each of these residues upon protein function, relative to the Brassica consensus residues at the same positions, using the PROVEAN tool44. Among the 10 sites, R98 and E129 exceeded the cutoff value of − 2.5 and were thus predicted to be highly deleterious to protein function, whereas D137 narrowly missed this threshold (Supplementary Table 2). Therefore, based on this predictive analysis, the native BtKAI2c protein carries at least two and possibly three substitutions that have the potential to render the protein non-functional.
R98 is potentially highly deleterious because it is situated in the active site adjacent to the catalytic serine, whereas E129 and D137 are located on the flexible hinge that links the lid and core domains (Supplementary Fig. 6a). In AtKAI2, the G133E mutation is highly destabilising20,41, suggesting that non-conserved substitutions in the hinge region have the potential to be deleterious as well. To test the effect of these residues on BtKAI2c stability, we constructed three variants that reverted the native BtKAI2c sequence to consensus amino acids within Brassica at position 98 and/or in the hinge region: BtKAI2cR98V, a quadruple mutant BtKAI2cE129D;D130V;E133Q;D137E (BtKAI2cQUAD), and a quintuple mutant that combined all five substitutions (BtKAI2cQUINT). Should these amino acids be crucial to protein stability, we expected to restore BtKAI2c expression.
First, we tried expressing the native and mutated BtKAI2c variants in tobacco leaves using Agrobacterium-mediated infiltration. The vectors were based on those used to generate the GFP-BtKAI2c Arabidopsis transgenics, allowing us to compare expression of the same GFP fusion proteins. As was the case for stably transformed Arabidopsis, we found that native GFP-BtKAI2c protein was undetectable, as was the BtKAI2cQUAD variant (Supplementary Fig. 6b). However, BtKAI2cR98V and BtKAI2cQUINT were clearly detected. We also examined the effect of the equivalent V96R mutation on GFP-AtKAI2, and found that this mutation completely abolished expression compared with the native GFP-AtKAI2 control (Supplementary Fig. 6b). These results suggest that the R98 residue causes the loss of protein expression in BtKAI2c, and that arginine at this position is not tolerated by KAI2 proteins in general.
Early in our investigation, we had tried without success to express and purify BtKAI2c from E. coli for biochemical characterisation. Having restored BtKAI2c expression in plants with the R98V mutation, we next tried to express these proteins in bacteria. We found that SUMO-BtKAI2c R98V was expressed at ~10-fold higher levels than native SUMO-BtKAI2c in crude lysates, suggesting that the R98V mutation enhanced protein folding and/or stability in bacterial cells. Although it was detectable in crude lysates, SUMO-BtKAI2c was consistently intransigent to recovery following affinity chromatography; in contrast, the R98V and quintuple mutant versions were stably recovered at high levels and purity (Supplementary Fig. 6c). From these results, we conclude that native BtKAI2c is non-functional owing to protein instability induced by a highly non-conservative substitution at the active site.
BtKAI2a and BtKAI2b show differential ligand specificity
We further characterised the ligand specificity of BtKAI2 homologues by performing physiological and molecular assays with the stable transgenic Arabidopsis lines. Primary-dormant Arabidopsis seeds homozygous for the transgenes were tested for germination response (Fig. 3a and Supplementary Fig. 7). None of the transgenic lines completely restored germination levels to those of wild type, perhaps reflecting the need for additional, seed-specific regulatory elements not included in our transgenes, or the importance of genomic context for proper KAI2 expression in seeds. Nevertheless, germination responses to karrikins were evident in the transgenic seeds: germination of BtKAI2b seeds was more responsive to KAR1 than KAR2 at 1 μM, whereas no significant difference was observed for BtKAI2a seeds. As expected, BtKAI2c seeds were indistinguishable from the kai2 control and insensitive to karrikins. We also found that levels of KAR-UP F-BOX 1 (KUF1) transcripts, which are responsive to karrikins in Arabidopsis seeds11, were only slightly induced by karrikins in BtKAI2a transgenic seeds, but were strongly enhanced by KAR1 in BtKAI2b seeds (Fig. 3b).
Fig. 3. Functional divergence between BtKAI2 homologues.
a Germination responses of primary-dormant Arabidopsis seed homozygous for KAI2pro:GFP-BtKAI2 transgenes in the kai2-2 background. Germination values were determined 120 h after sowing. Extended germination curves are shown in Supplementary Fig. 7. Data are means ± SE, n = 3 independent seed batches, 75 seeds per batch. b Levels of KUF1 transcripts in KAI2pro:GFP-BtKAI2 seeds treated with 1 µM KAR1 or KAR2 for 96 h (see Methods). Expression was normalised to CACS reference transcripts and scaled to the value for mock-treated seed within each genotype. Data are means ± SE of n = 3 biological replicates. c Hypocotyl elongation responses of KAI2pro:GFP-BtKAI2 seedlings treated with KAR1 or KAR2. Data are means ± SE of n = 3 biological replicates, 12–18 seedlings per replicate. d Levels of DLK2 transcripts in 8-day-old KAI2pro:GFP-BtKAI2 seedlings treated with KAR1 or KAR2 for 8 h. Expression was normalised to CACS reference transcripts and scaled to the value for mock-treated seedlings within each genotype. Data are means ± SE of n = 3 biological replicates. Pairwise significant differences: *P < 0.05, **P < 0.01, ***P < 0.001; ns, P >0.05 (ANOVA). Source data are provided as a Source Data file.
In seedlings, both BtKAI2a and BtKAI2b, but not BtKAI2c, could complement the kai2 hypocotyl elongation phenotype (Fig. 3c). Responses to karrikins in terms of hypocotyl elongation (Fig. 3c) and levels of DLK2 transcripts (Fig. 3d) broadly agreed with the germination response with respect to karrikin preferences, with BtKAI2b transgenics showing a clear preference for KAR1 over KAR2. However, BtKAI2a seedlings showed a preference for KAR2 that was not evident in seeds. From these experiments, we conclude that BtKAI2a is similar to AtKAI2 in terms of ligand specificity, whereas BtKAI2b has a clear preference for KAR1 over KAR2. As BtKAI2b is more highly expressed than BtKAI2a in B. tournefortii seeds and seedlings (Fig. 2b), we conclude that the ligand specificity of BtKAI2b substantially contributes to the enhanced KAR1-responsiveness of this species at these stages of the life cycle.
Positions 98 and 191 account for ligand specificity
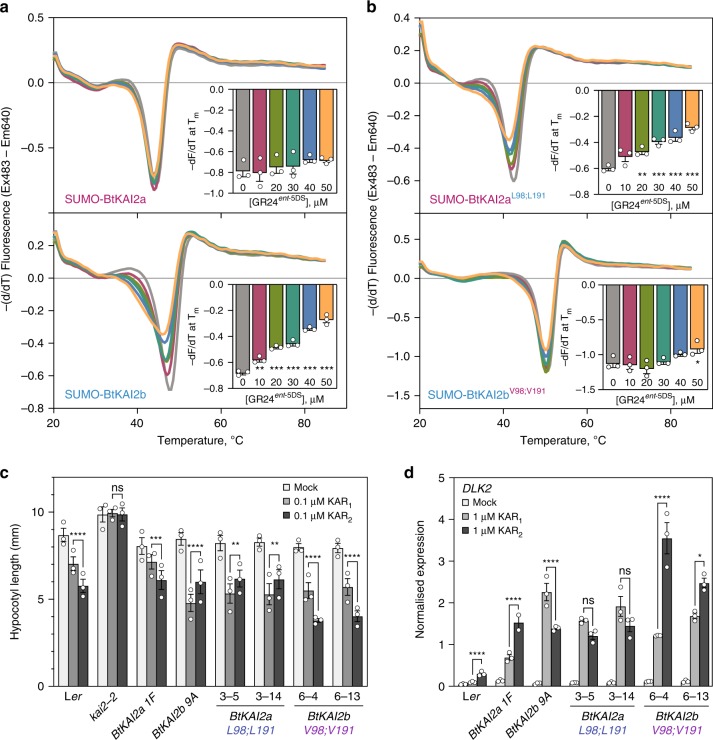
To investigate interactions between BtKAI2 homologues and ligands, we performed differential scanning fluorimetry (DSF) assays on purified recombinant proteins (Supplementary Fig. 8). DSF has been used extensively for inferring the interaction of strigolactone-like compounds with D14- and KAI2-related proteins29,41,45–48. Racemic GR24 is a widely used synthetic strigolactone analogue that consists of two enantiomers (Supplementary Fig. 1a), of which GR24ent-5DS is bioactive via AtKAI240,49. Catalytically inactive D14 and KAI2 variants do not respond to GR24 in DSF, suggesting that the shift in thermal response results from ligand hydrolysis and a corresponding conformational change in the receptor29,41,46. In DSF assays, AtKAI2 shows a specific response to >100 µM GR24ent-5DS but, for unclear reasons, no response to karrikins41. Likewise, we found that both BtKAI2a and BtKAI2b were also unresponsive to karrikins in DSF (Supplementary Fig. 9a). Therefore, we used DSF assays with GR24ent-5DS as a surrogate substrate as a means to analyse differences in ligand interactions between BtKAI2a and BtKAI2b. We used N-terminal 6xHIS-SUMO as a solubility tag to aid expression in E. coli and enhance stability. We found that the presence of the tag did not compromise the thermal destabilisation response of BtKAI2b to GR24ent-5DS (Supplementary Fig. 9b), and therefore, we used intact 6xHIS-SUMO fusion proteins (prefixed with ‘SUMO' throughout) for all other experiments.
We found that SUMO-AtKAI2 and SUMO-BtKAI2a showed similar responses to >100 µM GR24ent-5DS, but the response of SUMO-BtKAI2b was clear at >25 µM (Supplementary Fig. 9c). We then used a lower range of GR24ent-5DS concentrations (0–50 µM) to determine the threshold for response, which we defined as a statistically significant reduction in the maximal rate of change in fluorescence at the melting temperature of the protein (Tm). Although SUMO-BtKAI2a showed only a weak and non-significant response at 40 and 50 µM GR24ent-5DS, SUMO-BtKAI2b responded significantly at 10 µM and above (Fig. 4a). These results suggest that BtKAI2b is more sensitive than BtKAI2a to GR24ent-5DS, and that BtKAI2a is most like AtKAI2 in this respect.
Fig. 4. Two residues account for ligand specificity between BtKAI2a and BtKAI2b.
a DSF curves of SUMO-BtKAI2a and SUMO-BtKAI2b fusion proteins treated with 0–50 µM GR24ent-5DS, a KAI2-bioactive ligand. Each curve is the average of three sets of reactions, each comprising four technical replicates. Insets plot the minimum value of −(dF/dT) at the melting point of the protein as determined in the absence of ligand (means ± SE, n = 3). Significant differences from untreated control: *P < 0.05, **P < 0.01, ***P < 0.001 (ANOVA). b DSF curves of SUMO-BtKAI2aL98;L191 and SUMO-BtKAI2bV98;V191 fusion proteins treated with 0–50 µM GR24ent-5DS. c Hypocotyl elongation responses of Arabidopsis expressing GFP-BtKAI2aL98;L191 and GFP-BtKAI2bV98;V191 fusion proteins and treated with KAR1 or KAR2. Data are a summary of three experimental replicates performed on separate occasions, each comprising 25–40 seedlings per genotype/treatment combination. Data for each replicate are shown in Supplementary Fig. 11. Error bars are SE, n = 3 experimental replicates; each dot corresponds to the mean value derived from each replicate. Asterisks denote significant differences: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (linear mixed model with experimental replicate as a random effect; specific pairwise comparisons using Tukey’s HSD correction). d Levels of DLK2 transcripts in 8-day-old seedlings of the same transgenic lines as above treated with 1 µM KAR1, 1 µM KAR2, or 0.1% acetone (mock) and harvested 8 h later. Expression was normalised to CACS reference transcripts. Data are means ± SE of n = 3 pools of ~50 seedlings treated in parallel. Asterisks denote significant differences as above (two-way ANOVA; specific pairwise comparisons using Tukey’s HSD correction). Source data are provided as a Source Data file.
BtKAI2a and BtKAI2b differ in primary amino-acid sequence at just 14 positions (Supplementary Fig. 3). We postulated that differences in ligand specificity might be determined by amino acids in the vicinity of the ligand-binding pocket. Protein structural homology models revealed only two residues that differ in this region: V98 and V191 in BtKAI2a, and L98 and L191 in BtKAI2b; the corresponding residues in AtKAI2 and AtD14 are valines (Supplementary Fig. 8). Residue 98 is immediately adjacent to the catalytic serine at the base of the pocket. Residue 191 is located internally on αT4 of the lid domain which, in AtD14, is associated with a major rearrangement of protein structure upon ligand binding that reduces the size of the pocket31. Homology modelling suggests only subtle differences in the size and shape of the primary ligand binding pocket of BtKAI2a and BtKAI2b (Supplementary Fig. 8). To determine whether these residues are pertinent to ligand specificity, we replaced the two valine residues of BtKAI2a with leucine residues, generating the variant BtKAI2aL98;L191, and vice versa for BtKAI2b, generating the variant BtKAI2bV98;V191 (Supplementary Fig. 8). In DSF assays, we found that exchanging the two residues was sufficient to switch the original responses, such that SUMO-BtKAI2aL98;L191 responded sensitively to GR24ent-5DS, but SUMO-BtKAI2bV98;V191 did not (Fig. 4b). We also assessed the function of the stable SUMO-BtKAI2cR98V and SUMO-BtKAI2cQUINT variants by DSF against GR24ent-5DS ligand, and found that they were only weakly destabilised by this ligand, when compared with the highly sensitive BtKAI2b (Supplementary Fig. 6e). Overall, these results indicate that BtKAI2a and BtKAI2b have distinct response profiles, albeit to a synthetic ligand, and that residues 98 and 191 contribute to this difference.
We reasoned that the most robust and informative means to assess the effect of residues 98 and 191 upon the response to karrikins was to test their function directly in planta. We expressed BtKAI2aL98;L191 and BtKAI2bV98;V191 as GFP fusion proteins in the Arabidopsis kai2-2 null mutant background driven by the Arabidopsis KAI2 promoter (KAI2pro:GFP-BtKAI2aL98;L191 and KAI2pro:GFP-BtKAI2bV98;V191), and selected two independent homozygous transgenic lines for each, on the basis of protein expression level and segregation ratio (Supplementary Fig. 10). Using two different assays, we found that substitutions between BtKAI2a and BtKAI2b at positions 98 and 191 also reversed karrikin preference in Arabidopsis seedlings both in terms of hypocotyl elongation and DLK2 transcript levels (Fig. 4c, d; Supplementary Fig. 11). Most prominently, the clear preference of BtKAI2b for KAR1 was unambiguously reversed to a preference for KAR2 in BtKAI2bV98;V191 transgenics, effectively recapitulating the response of the native BtKAI2a protein. We also examined the responses of seeds in these transgenic lines, and found that BtKAI2aL98;L191 seeds germinated with a clear preference for KAR1, whereas BtKAI2bV98;V191 seeds showed no clear preference for either karrikin (Supplementary Fig. 12). Notably, this germination response pattern is the opposite to that observed for seeds expressing the native BtKAI2 proteins, with BtKAI2a exhibiting no preference for either karrikin and BtKAI2b a preference for KAR1 (Fig. 3a, Supplementary Fig. 7). Taken together, these results demonstrate that the residues at positions 98 and 191 largely determine differences in karrikin specificity between BtKAI2a and BtKAI2b, at least in the context of Arabidopsis seedlings.
Co-dependency between positions 96 and 189 among angiosperms
We wished to gain further insight into the functional importance of diversity at positions 96 and 189 of KAI2 proteins (based on AtKAI2 notation, equivalent to positions 98 and 191 in BtKAI2a and BtKAI2b). We analysed 476 distinct KAI2 homologues from 441 angiosperm species (spanning eudicots, monocots and magnoliids, but excluding B. tournefortii) using data collected from the 1000 Plant Transcriptomes project50. We found that at positions 96 and 189, valine was by far the most common amino acid (93.1% and 93.5% of sequences, respectively; Fig. 5a and Supplementary Data 1). L96 was observed in 6.1% of sequences, whereas L189 was even rarer (2.1%). The L96;V189 combination was observed in 14 sequences (2.9%), whereas V96;L189 was observed in only one homologue from Ternstroemia gymanthera (Pentaphylacaceae; 0.2%). Notably, this species also expressed a second KAI2 homologue with the canonical V96;V189 combination. If the frequencies of valine and leucine at positions 96 and 189 were independent, the L96;L189 combination would be expected to occur less than once among 476 sequences (0.13%). However, the coincidence of L96 and L189 was observed in nine sequences (1.9%), representing a significant 15-fold enrichment for the L96;L189 pair over that expected by chance alone (χ2 = 115.6; P < 0.0001).
Fig. 5. Diversity at positions 96 and 189 in KAI2 homologues across angiosperms.
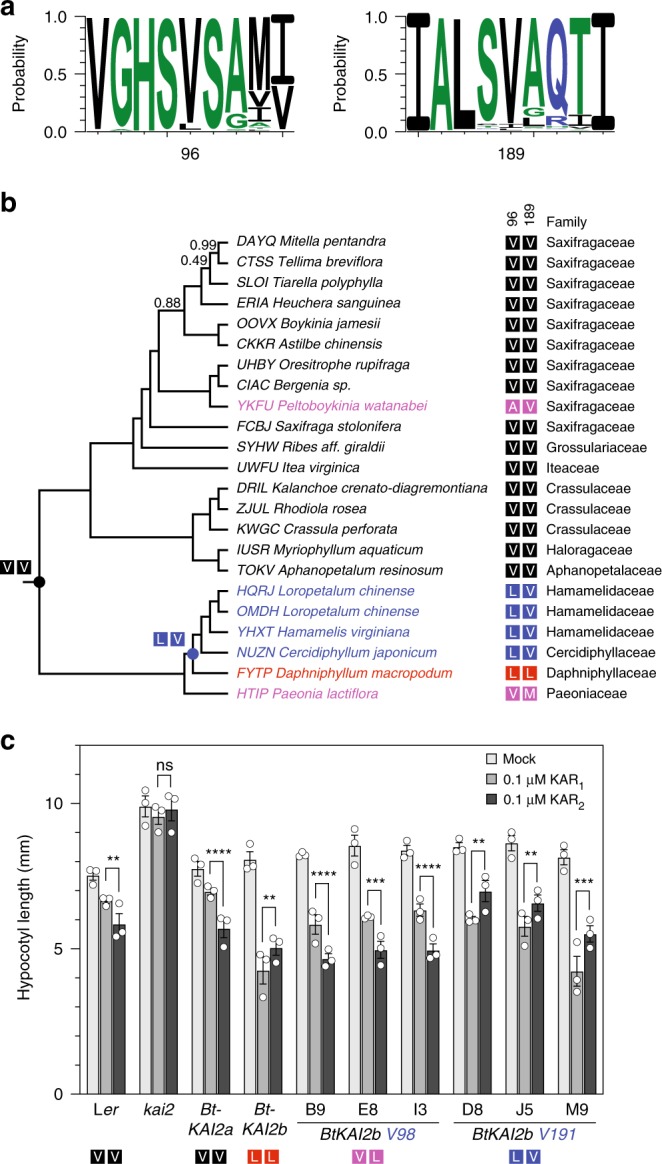
a Sequence logos generated from 476 aligned KAI2 homologues sampled from eudicots, monocots and magnoliids and centred on position 96 (left) and 189 (right). b Species tree of the Saxifragales based upon 410 single-copy nuclear genes estimated using the ASTRAL gene tree method50. Branch support values are shown only where <1. The four-letter taxon prefix corresponds to the 1000 Plant Transcriptomes project Sample ID. Taxa are highlighted by colour according to identity of amino acids of KAI2 orthologues at positions 96 and 189, respectively: black, VV; blue, LV; red, LL; magenta, other. Postulated ancestral identities at specific nodes are indicated with filled circles. c Hypocotyl elongation responses to karrikins in three independent transgenic lines homozygous for GFP-BtKAI2aL98;V191 and three lines for GFP-BtKAI2bV98;L191. Data shown are a summary of three experimental replicates performed on separate occasions, each comprising ~20 seedlings per genotype/treatment combination. Data from each replicate are presented in Supplementary Fig. 13. Error bars are SE, n = 3 experimental replicates; each dot corresponds to the mean value derived from each replicate. Asterisks denote significant differences: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (linear mixed model with experimental replicate as a random effect; specific pairwise comparisons using Tukey’s HSD correction). Source data are provided as a Source Data file.
Among the nine newly-identified KAI2L96;L189 sequences, eight were restricted to eudicots, distributed evenly between both the superrosids (Brassicales, Fabales and Saxifragales; four species) and superasterids (Caryophyllales; four species). The ninth sequence, co-expressed with another transcript encoding KAI2V96;V189, was identified from the basal dicot Hakea drupacea (syn. Hakea suaveolens; Proteaceae). Strikingly, this species is native to southwestern Australia, but is recognised as an invasive weed in South Africa that relies upon fire to liberate seeds from the canopy51. The L96;L189 combination therefore seems to have arisen independently on multiple occasions. Furthermore, within the order Saxifragales, Daphniphyllum macropodum (KAI2L96;L189) belongs to a clade of species that all express KAI2L96;V189 homologues (Fig. 5b). Although this clade is only represented by five sequences and four species, this distribution pattern suggests that a V96L mutation occurred first in the common ancestor of the Hamamelidaceae, Cercidiphyllaceae and Daphyniphyllaceae, followed by V189L in the lineage leading to D. macropodum. Overall, this analysis suggests that positions 96 and 189, while not in direct contact with each other in the protein tertiary structure (Supplementary Fig. 8), likely exhibit functional interdependence. Valine is most probably the ancestral state at both positions, but leucine at position 96 is also tolerated. Although rare, leucine at position 189 is generally found in combination with leucine at position 96, and tentatively, a V96L mutation might potentiate a subsequent V189L mutation.
The above analysis suggested that a V96L substitution might be sufficient to confer altered substrate specificity upon KAI2 proteins. To test this hypothesis, we generated transgenic Arabidopsis seedlings expressing GFP-BtKAI2b proteins with just a single amino-acid substitution from leucine to valine at either position 98 or 191 (i.e., KAI2pro:GFP-BtKAI2bV98 or KAI2pro:GFP-BtKAI2bV191), and examined their response to KAR1 and KAR2 in hypocotyl elongation assays. We found that the L98V substitution was sufficient to invert the karrikin preference of GFP-BtKAI2b in three independent homozygous transgenic lines (Fig. 5c; Supplementary Fig. 13). Although this result does not rule out an additional contribution towards ligand specificity from L191, it is consistent with the higher frequency of L96 compared to L189 observed among the angiosperms, and suggests that substitution at position 96 may have been selected relatively frequently in the evolution of KAI2 function.
Discussion
The plant α/β-hydrolases KAI2 and D14 are characterised by responsiveness towards butenolide compounds, including endogenous strigolactones and strigolactone-related compounds, abiotic karrikins derived from burnt vegetation, and synthetic strigolactone analogues with a wide array of functional groups. Direct evidence for a receptor-ligand relationship between karrikins and KAI2 homologues stems from crystallography and in vitro binding assays. Two crystal structures of KAR-responsive KAI2 proteins from Arabidopsis and the parasitic plant Striga hermonthica reveal largely similar overall protein structure, but surprisingly are non-congruent with respect to KAR1-binding position and orientation52,53. The affinity of AtKAI2 for KAR1 is imprecisely defined, with estimates of dissociation coefficients (Kd) ranging from 4.6 µM54 to 148 µM55 using isothermal calorimetry, and 9 µM using fluorescence microdialysis52. Although variability in affinity estimates can be explained in part by different experimental conditions and techniques, it should also be considered that, depending on the homologue under examination, KAR1 may not be the optimal ligand. Furthermore, binding of a hydrophobic compound to a hydrophobic pocket does not necessarily equate to ligand recognition or receptor activation. Our data, which are derived from clear and distinct biological responses, provide strong evidence that KAI2 is sufficient to determine ligand specificity, which, in turn, strengthens the case that KAI2 is the receptor by which karrikins are perceived.
The predominance of valine and leucine at position 96 (98 in BtKAI2 proteins), observed in 99.2% of our sample of 476 KAI2 sequences, suggests that this site is critical to protein function, which makes sense given its immediate proximity to the catalytic serine. This position not only contributes to ligand preference, but also to protein stability, given the negative effects of R98 in BtKAI2c. Compared with valine and leucine, arginine is relatively bulky with a charged side chain; indeed, in the few KAI2 sequences in which position 96 is neither leucine nor valine, the residue is nevertheless small and hydrophobic (either methionine or alanine; Supplementary Table 3). Interestingly, substitutions in AtKAI2 at or near the catalytic site, such as S95F, S119F and G245D, are not tolerated and render the protein unstable in plants43. The likely signalling mechanism for KAI2—by analogy of that for D14—involves ligand binding and/or hydrolysis, which triggers conformational changes in the lid domain and formation of a protein complex with MAX2 and SMAX1/SMXL2 protein partners30,31,56. As a result of signalling, KAI2 itself is degraded40,43. It is possible that KAI2 is an inherently metastable protein that is prone to conformational change and subsequent degradation by necessity; perhaps, this explains why it is so susceptible to instability through mutation.
Ligand specificity is a key contributor to the functional distinction between karrikin and strigolactone receptors, and elucidating the molecular mechanisms behind ligand specificity is a significant research challenge. In certain root-parasitic weeds in the Orobanchaceae, substitutions of bulky residues found in AtKAI2 with smaller hydrophobic amino acids that increase the ligand-binding pocket size have likely improved the affinity for host-derived strigolactone ligands, as opposed to the smaller-sized karrikin-type ligands57–59. Moreover, lid-loop residues that affect the rigidity and size of the ligand entry tunnel determine the ligand selectivity between KAR1 and ent-5-deoxystrigol (a strigolactone analogue with non-natural stereochemistry) among eleven KAI2 homologues in Physcomitrella patens60. Karrikin and strigolactone compounds also show chemical diversity within themselves, yet ligand discrimination among KAI2 and D14 receptors is not well characterised. Our data demonstrate that, in the case of BtKAI2 proteins, the two KAI2 homologues respond differently to subtly different KAR analogues, and that subtle changes in pocket residues likely account for preferences between these ligands. Position 96 appears to play a primary role in determining ligand specificity, whereas position 189 may improve ligand sensitivity, protein stability, or protein conformational dynamics following a prior mutation at position 96. Although the identified residues L98 and L191 are not predicted to change dramatically the pocket size of BtKAI2b in comparison with BtKAI2a, these residues would make the BtKAI2b pocket more hydrophobic, which is consistent with KAR1 being more hydrophobic than KAR24. Therefore, ligand specificity between highly similar chemical analogues can be achieved through fine-tuning of pocket hydrophobicity. Similarly, subtle changes have been reported for the rice gibberellin receptor GID1: changing Ile133 to leucine or valine increases the affinity for GA34 relative to the less-polar GA4, which lacks just one hydroxyl group61.
It is possible that what we learn about ligand specificity in KAI2 proteins may also be informative about strigolactone perception by D14. Different D14 proteins may show ligand specificity towards diverse natural strigolactones, resulting in part from multiplicity of biosynthetic enzymes, such as the cytochrome P450 enzymes in the MAX1 family45,62,63 and supplementary enzymes such as LBO64. Although the functional reasons underlying such strigolactone diversity are still unclear, it is possible that variation among D14 homologues yields varying affinities for different strigolactone analogues65. As an increasingly refined picture emerges of the features that determine ligand specificity for KAI2 and D14 receptors, we envision the rational design of synthetic receptor proteins with desirable ligand specificity in the future.
Gene duplication is a common feature in plant evolutionary histories as an initial step toward neofunctionalisation66. In obligate parasitic weeds, duplication and diversification of KAI2 genes (also referred to as HTL genes) have shifted ligand specificity towards strigolactones57,58. Karrikins, as abiotic molecules with limited natural occurrence, are unlikely to be the natural ligand for most KAI2 proteins, which are found throughout land plants. Instead, the evolutionary maintenance of a highly conserved receptor probably reflects the core KAI2 function of perceiving an endogenous ligand (KL) that regulates plant development34–36. This core function is presumably retained after gene duplication events through original, non-divergent copies that are under purifying selection57,58. KAI2 diversity may also reflect differing affinity for KL variants in different species and at different life stages. Our results are consistent with a scenario in which both BtKAI2a and BtKAI2b have retained the function of perceiving KL, because both copies complement the Arabidopsis kai2 phenotype, especially at the seedling and later stages. However, BtKAI2b has also acquired mutations that alter its ligand specificity, which in turn enhance sensitivity to the archetypal karrikin first discovered in smoke. The BtKAI2b gene is also more highly expressed than BtKAI2a in seeds and seedlings, consistent with an adaptation to post-fire seedling establishment. This diversity among KAI2 proteins may provide B. tournefortii with a selective advantage in fire-prone environments, contributing to its invasive nature.
We have shown evidence for the independent evolution of valine-to-leucine substitutions in KAI2 homologues throughout the dicots, albeit at low frequency, suggesting that these changes may have adaptive significance in various ecological contexts. It will be of interest to assess the function of such homologues in other species, and to validate and extend the specific conclusions we have made here. Recent findings indicate that KAI2 is an integrator that also modulates germination in response to other abiotic environmental signals, including temperature and salinity67. As germination is a critical life stage for seed plants, strategies for environmental conservation, restoration and weed control will benefit from specific knowledge of KAI2 sequence diversity and expression profiles.
Methods
Chemical synthesis
Karrikins (KAR1 and KAR2), 13[C]5-labelled karrikins and GR24 enantiomers (GR245DS and GR24ent-5DS) were prepared as previously described68,69.
Plant material and measurement of growth
Arabidopsis kai2-2 (Ler) and Atd14-1 (Col-0) mutants were previously described20. Arabidopsis plants were grown on a 6:1:1 mixture of peat-based compost (Seedling Substrate Plus; Bord Na Mona, Newbridge, Ireland), vermiculite and perlite, respectively. Light was provided by wide-spectrum white LED panels emitting 120–150 µmol photons m−2 s−1 with a 16 h light/8 h dark photoperiod, a 22 °C light/16 °C dark temperature cycle, and constant 60% relative humidity. The seeds of B. tournefortii used in this work were collected in November and December 2009 from two sites in Western Australia (Kings Park in Perth, and Merridin)70. B. tournefortii seeds were dried to 15% relative humidity for 1 month prior to storage in air-tight bags at −20 °C.
Arabidopsis rosettes were photographed at ~3–4 weeks after germination, prior to visible flowering, as specified in the figure legends. Plant height and shoot branching were determined once growth of the primary inflorescence had ceased. Height was measured using a ruler from the base of the primary inflorescence to the apex. Primary shoot branches were counted by inspecting the axils of each leaf; a bud was counted as a primary branch if it had elongated more than 5 mm.
Seed germination assays
Arabidopsis seeds were harvested from three pools of four plants per genotype, grown simultaneously under identical conditions (~120–150 μmol photons m−2 s−2 white light, 16 h light/8 h dark, 22 °C light/16 °C dark). After harvesting, seeds were dried under silica gel for four days, and stored at –80 °C. Seeds were sowed on 60 mm petri dishes containing 1% Phytagel solidified with 1.5 mM MgCl2 and supplemented with 0.1% v/v acetone (mock) or 1 µM KAR1/KAR2 (1:1000 dilution). Petri dishes were incubated at 25 °C under constant light for up to 1 week, and germination was scored using a dissecting microscope every 24 h. B. tournefortii seeds were sowed in triplicates (35–70 seeds each) on glass microfibre filter paper (Grade 393; Filtech, NSW Australia) held in 9-cm petri dishes and supplemented with mock or karrikin treatments (3 mL of aqueous treatment solution per petri dish). Treatments were prepared by diluting acetone (mock) or karrikin stocks (dissolved in acetone) 1:1000 with ultrapure water. Because germination of B. tournefortii is inhibited by light9, the seeds were imbibed in the dark at 22 °C. Numbers of germinated seeds were counted each day until the germination percentages remained unchanged.
Hypocotyl elongation assays
Approximately 50 Arabidopsis seeds were sowed on 60 mm petri dishes containing half-strength MS salts (pH 5.9) and 0.7% w/v agar. The plates were stratified in the dark at 4 °C for 72 h, transferred to a growth incubator and exposed to white light at 22 °C for 3 h, and then to darkness for a further 21 h at 22 °C. The plates were then exposed to continuous red light supplied by light-emitting diodes (LEDs) (Philips GreenPower LED Research module; λmax 660 nm, 5 µmol photons m−2 s−1) for 4 days at 22 °C. Seedlings were laid out horizontally on 0.7% w/v agar for photography, and hypocotyl lengths were measured using ImageJ software (https://imagej.nih.gov/ij/index.html). For B. tournefortii, assays were performed with the following modifications to the Arabidopsis protocol: B. tournefortii seeds were sowed on glass microfibre filter paper (Filtech) held in petri dishes supplemented with mock or karrikin treatments. The seeds were imbibed in the dark for 22 h at 24 °C before exposing to continuous red light (5 µmol photons m−2 s−1) for 4 days, at which point the hypocotyls were measured as above.
Transcript analysis
B. tournefortii seeds were imbibed on glass fibre filters in the dark at 22 °C and treated with karrikins using the same procedure as described under seed germination assays, above. For transcript quantification in B. tournefortii seeds, samples were taken at the indicated time points, blot-dried on paper towel, and frozen in liquid nitrogen. For B. tournefortii seedlings, two methods were used. For data shown in Fig. 1e, seeds were sowed and seedlings grown in triplicate under identical conditions to those described for hypocotyl elongation assays. For data shown in Fig. 2b, seeds were first imbibed for 24 h in the dark, and then transferred to the growth room (~120–150 μmol photons m−2 s−2 white light, 16 h light/8 h dark, 22 °C light/16 °C dark) for 3 days. A sample of ~20 seedlings was then transferred to a 250 mL conical flask containing 50 mL sterile ultrapure water supplemented with 1 µM KAR1, or an equivalent volume of acetone (0.1% v/v), with three replicate samples per treatment. The flasks were then shaken for 24 h before seedlings were harvested.
Arabidopsis seedlings were grown on 0.5 × MS agar under growth room conditions (wide-spectrum white LED panels emitting 120–150 µmol photons m−2 s−1 with a 16 h light/8 h dark photoperiod and a 22 °C light/16 °C dark temperature cycle) for 7 days. On the 7th day, seedlings were transferred to 3 mL liquid 0.5 × MS medium in 12-well culture plates (CORNING Costar 3513) and shaken at 70 rpm for a further 22 h under the same growth room conditions, after which the medium was removed by pipette and replaced with a fresh medium containing relevant compounds or an equivalent volume (0.1% v/v) of acetone. After further incubation with shaking (8 h), the seedlings were harvested, blotted dry, and frozen in liquid nitrogen. Dry Arabidopsis seeds (20 mg) were transferred to a 2-mL tube and imbibed in 1 mL of water, supplemented with karrikins or an equivalent volume of acetone, and incubated with end-over-end rotation for 72 h in the dark at 4 °C. Seeds were transferred to the light (16 h light/8 h dark, 120–150 µmol photons m−2 s−1) for at 22 °C for a further 24 h with rotation. Seeds were harvested by brief centrifugation, removal of excess water, and freezing in liquid nitrogen and were ground to a fine powder in a pestle and mortar.
Transcript quantification
RNA extraction was performed using the Spectrum Plant Total RNA kit (Sigma-Aldrich) with an on-column DNase digestion step. cDNA was generated from 500 ng total RNA using the iScript cDNA Synthesis kit (Bio-Rad), and reactions were subsequently diluted fourfold in water. Quantitative PCR was conducted in 5-microlitre reactions in a 384-well format using a Roche LightCycler 480. Reactions contained 0.4 µM oligonucleotides, 0.5 µl diluted cDNA and 1× Luna Universal qPCR Master Mix (New England Biolabs). Two technical replicates of each reaction were performed for each biological replicate, and the mean Cp value was used to normalise expression to CACS reference transcripts using the formula (1 + Egene)–Cp_gene/(1 + ECACS)–Cp_CACS, where E is the primer efficiency. Primer efficiencies (0.9 or greater) were determined in separate runs using serial dilutions of pooled cDNA. All oligonucleotides are listed in the Supplementary Table 3.
Cloning and mutagenesis of KAI2 and D14 homologues
Full-length BtKAI2-coding sequences (and unique 3′-UTR sequences) were amplified from B. tournefortii cDNA with Gateway-compatible attB sites using the universal forward primer BtKAI2_universal_F and homologue-specific reverse primers RACE_R (BtKAI2a), Contig1_R (BtKAI2b) and Contig5_R (BtKAI2c), before cloning into pDONR207 (Life Technologies). The pDONR207 clones were confirmed by Sanger sequencing and recombined with pKAI2pro-GFP-GW41 to generate the binary plant expression plasmids pKAI2pro-GFP-BtKAI2a, pKAI2pro-GFP-BtKAI2b and pKAI2pro-GFP-BtKAI2c. For transient overexpression in tobacco (Supplementary Fig. 5d), BtKAI2-coding sequences were transferred via Gateway-mediated recombination into pSKI106, which drives very high expression of coding sequences placed immediately downstream of a full-length cDNA clone of the tobacco mosaic virus U1 strain, all under control of the CaMV 35 S promoter in a standard T-DNA binary vector42. pSKI106 also encodes an N-terminal 3 × c-myc tag. For transient expression in tobacco using standard binary vectors (Supplementary Fig. 6b), the BtKAI2 coding sequences were transferred into pMDC43, which is identical to pKAI2pro-GFP-GW except that the AtKAI2 promoter & 5′-UTR sequences are replaced with a double CaMV 35 S promoter71. The BtD14a-coding sequence was amplified from cDNA using oligonucleotides BtD14_F and BtD14_R, cloned into pDONR207 as above, and transferred into pD14pro-GW41.
The full-length BtKAI2-coding sequences (excluding 3′-UTRs) were amplified from the pDONR207 clones and reconstituted with the pE-SUMO vector by Gibson Assembly to generate the heterologous expression plasmids pE-SUMO-BtKAI2a, -BtKAI2b and -BtKAI2c. Site-directed mutagenesis generated pE-SUMO-BtKAI2aL98;L191, pE-SUMO-BtKAI2bV98;V191 and the BtKAI2c variants. For expression of mutated versions of BtKAI2a, BtKAI2b, BtKAI2c, and AtKAI2 in tobacco and Arabidopsis, site-directed mutagenesis was performed on pDONR207 clones prior to recombination with pKAI2pro-GFP-GW or pMDC43. In the case of the double substitutions in BtKAI2a and BtKAI2b, both targeted residues were mutated simultaneously in one PCR product, whereas the remainder of the plasmid was amplified in a second PCR product. These double mutated plasmids were reconstituted by Gibson assembly. For the single mutants, the entire plasmid was amplified using a single pair of mutagenesis primers, and the PCR product was self-ligated. The BtKAI2cQUINT variant was generated by two successive rounds of mutagenesis. Coding regions were confirmed by Sanger sequencing.
Plant transformation
Homozygous kai2-2 and Atd14-1 plants were transformed by floral dip. Primary transgenic seedlings were selected on sterile 0.5 × MS medium supplemented with 20 µg/mL hygromycin B. T2 lines exhibiting a 3:1 ratio of hygromycin resistant-to-sensitive seedlings were propagated further to identify homozygous lines in the T3 generation. Experiments were performed from the T3 generation onwards.
For transient expression in tobacco, Agrobacterium (GV3101) carrying pSKI106 or pMDC43 variants was grown in LB medium (25 mL) supplemented with antibiotics and 20 µM acetosyringone until OD600 reached 1.0. The bacteria were then harvested by centrifugation (15 min, 5000 × g) and resuspended in 10 mM MgCl2, 10 mM MES (pH 5.6) and 100 µM acetosyringone. The optical density was adjusted to 0.4, and the suspension was left standing at 22 °C overnight (~14 h). Leaves of 3-week-old Nicotiana benthamiana were then infiltrated with a 5-mL syringe, through the abaxial leaf surface. After 4 days, the leaves were collected and frozen in liquid nitrogen.
Immunoblotting and antibodies
Seedlings were frozen in liquid nitrogen and ground to a fine powder using a bead mill. Soluble proteins were extracted with 150 µl of lysis buffer (50 mM TRIS-HCl pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% Tween-20, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol (DTT) and 1 × complete protease inhibitors (Roche)) per 100 mg of tissue. Lysates were clarified by two successive centrifugations at 20,000 × g for 10 minutes each. Approximately 40 µg protein was separated on a 12% SDS-PAGE gel and blotted onto PVDF membrane. Blots were blocked in 1 × TBS-T (pH 7.5) and 2% (w/v) bovine serum albumin (BSA) for 1 h at 22 °C. Primary antibodies were diluted in 1 × TBS-T + 0.2% (w/v) BSA as follows: 1:2500 (anti-KAI2 raised in rabbit40); 1:1000 (anti-GFP raised in rabbit, ThermoFisher Scientific A11122), 1:2500 (anti-actin raised in mouse, Sigma A0480) and 1:1000 (anti-c-myc raised in mouse, Genscript A00704). Primary antibodies were incubated with the blot either overnight at 4 °C, or for 1 h at 22 °C, with gentle rocking. Between antibody incubations, blots were washed four times (5 min per wash) with 1 × TBS-T. Secondary antibody (goat anti-rabbit IgG-HRP conjugate, ThermoFisher 32460; or goat anti-mouse IgG-AP conjugate, ThermoFisher 31321) was diluted 1:1000 (HRP) or 1:5000 (AP) in 1 × TBS-T and 0.2% (w/v) BSA, and incubated with the blot for 1 h at 22 °C. HRP activity was detected using Clarity Western ECL substrate (Bio-Rad), and AP activity was detected with ImmunStar AP substrate (Bio-Rad). Chemiluminescence images (16-bit) were collected using the ImageQuantRT ECL system (GE Healthcare).
Karrikin uptake measurements
Fifteen samples of seeds were sowed for each karrikin treatment (five time points, each in triplicate). In each sample, ~40 mg of B. tournefortii seeds were imbibed in 3 mL ultrapure water for 24 h in 5-mL tubes. After centrifugation (2 min at 3220 × g), excess water was removed by pipette and the volume of residual water (mostly absorbed into the seeds) was calculated by weighing the seeds before and after imbibition. Fresh ultrapure water was added to the fully imbibed seeds to reach a total volume of 980 μL. Then 20 μL of 100 μM KAR1 or KAR2 was added to a final concentration of 2 μM. The seeds were imbibed at 22 °C in darkness. At the indicated time point (0, 2, 4, 8 or 24 h post treatment), 500 μL of the imbibition solution was removed and combined with 100 ng of either13[C]5-KAR1 or 13[C]5-KAR2 (100 μL at 1 μg/mL) as an internal standard for quantification purposes. The sample was then extracted once with ethyl acetate (500 μL), and 1 μL of this organic layer was analysed using GC-MS in selective ion monitoring (SIM) mode as previously described72.
The amount of KAR1 in each sample was calculated by the formula × 100 ng and converted to moles, where A(Ion150) indicates the peak area of the ion 150 (KAR1 to be measured), A(Ion151) indicates the peak area of the ion 151 (13[C]5-KAR1), and 100 ng is the amount of 13[C]5-KAR1 spiked in before the ethyl acetate extraction. Similarly, the amount of KAR2 in each sample was calculated by the formula × 100 ng and converted to moles. The uptake percentage adjusted to 40 mg of B. tournefortii seeds was calculated by the formula: × , where N(0) indicates moles of karrikins at time 0, N(x) indicates moles of karrikins at time point x, and m(seed) indicates the dry weight (mg) of seeds tested in each replicate. For Arabidopsis seeds, the procedure was scaled down for a smaller mass of seeds (20 mg).
Transcriptome assembly and analysis
Twenty milligrams of dry B. tournefortii seeds were imbibed in water for 24 h and incubated at 22 °C in the dark. The seeds were collected by centrifugation, blotted dry and frozen in liquid nitrogen. A separate sample of seeds was sown on glass filter paper, imbibed for 24 h as above, and then incubated for 96 h under continuous red light (20 µmol m−2 s−1) with a 22 °C (16 h)/16 °C (8 h) temperature cycle. A single sample of seedlings (50 mg fresh weight) was harvested and frozen in liquid nitrogen. Total RNA was extracted from both seed and seedling samples using the Spectrum Plant RNA kit (Sigma-Aldrich), including an on-column DNase step. PolyA+ mRNA was purified using oligo(dT) magnetic beads (Illumina), and cDNA libraries for sequencing were generated as described73. Sequencing was performed on the Illumina HiSeq 2000 platform at the Beijing Genomic Institute, Shenzhen, China. Raw reads were filtered to remove adapters and low-quality reads (those with >5% unknown nucleotides, or those in which >20% of base calls had quality scores ≤10). After filtering, both libraries generated reads with >99.9% of nucleotides attaining Q20 quality score. Transcriptome de novo assembly was performed with Trinity74. For each library, contigs were assembled into Unigenes; Unigenes from both libraries were then combined, yielding a total of 45,553 predicted coding region sequences with a mean length of 1011 nt. The combined Unigenes were then interrogated for homology to AtKAI2, AtD14, AtDLK2, AtSTH7 and AtCACS using BLASTn searches.
Phylogenetic analysis and protein sequence analysis
KAI2 and D14 homologues in Brassica species were identified from BLAST searches using Arabidopsis coding sequences as a query. Additional sequences were sampled from an existing phylogenetic analysis33. Multiple sequence alignments were performed using MAFFT plugin implemented in Geneious R10 (Biomatters). The alignment was trimmed slightly at the 5′-end to remove non-aligned regions of monocot D14 sequences. Maximum likelihood phylogenies were generated using PHYML (GTR + G + I substitution model, combination of NNI and SPR search, and 100 bootstraps). The choice of substitution model was guided by Smart Model Selection in PhyML75 (http://www.atgc-montpellier.fr/sms). A list of all sequences, and their sources, is provided in Supplementary Table 1.
For analysis of BtKAI2c using PROVEAN, all 10 positions that were unique to BtKAI2c when compared with AtKAI2 and other Brassica sequences (Supplementary Fig. 3) were manually edited to the consensus identity at each position. This was the ‘reverted' sequence against which each of the native BtKAI2c amino-acid substitutions were tested using the PROVEAN server (http://provean.jcvi.org) using the default cutoff score of −2.5. The PROVEAN output is presented in Supplementary Table 2.
KAI2 homologues from The 1000 Plant Transcriptomes Project50 were identified by TBLASTN searches using A. thaliana KAI2 protein sequence as a query against ‘onekp database v5', which comprises 1328 individual samples (https://db.cngbdb.org). The search was configured to return a maximum of 500 target sequences with expect values <0.01, using a word size of six and the BLOSUM62 scoring matrix. Nucleotide sequences were collated, and open reading frames (ORFs) >700 nucleotides were predicted in Geneious software. A single ORF was selected for each sequence; those with multiple ORFs were screened manually for the one with homology to AtKAI2. The ORFs were translated and aligned using the MAFFT multiple alignment tool. A few misaligned or mis-translated sequences were removed from the alignment manually, resulting in a data set of 476 unique sequences. Extracted parts of this alignment centred on positions 96 and 189 were used to generate WebLogos (http://weblogo.threeplusone.com76). A list of identified KAI2 homologues and their positional analysis are presented in Supplementary Data 1.
Protein homology modelling
KAI2 structures were modelled using the SWISS-MODEL server (https://swissmodel.expasy.org) using the alignment mode77 and the A. thaliana KAI2 structure 3w06 as a template54. Figures of protein structure and homology models were generated using PyMOL v1.3 (Schrödinger LLC). Cavity surfaces were visualised using the ‘cavities & pockets (culled)' setting in PyMOL and a cavity detection cutoff value of four solvent radii. Cavity volumes were calculated using the CASTp server v3.078 (http://sts.bioe.uic.edu/castp) with a probe radius of 1.4 Å. Values indicate Connolly’s solvent-excluded volumes. Cavities were inspected visually using Chimera v1.12 (https://www.cgl.ucsf.edu/chimera/). For both BtKAI2a and BtKAI2b models, CASTp erroneously included surface residues near the primary pocket entrance in the calculation of the pocket volumes. This issue was resolved by the artificial placement of a free alanine residue adjacent to the cavity entrance, as described previously79.
Protein expression and purification
AtKAI2 and BtKAI2-coding sequences were cloned into pE-SUMO-Amp (LifeSensors) to generate N-terminal 6 × HIS-SUMO fusion proteins. All proteins were expressed in BL21 Rosetta DE3 pLysS cells (Merck-Millipore) and purified using IMAC as described in detail previously41. In brief, cultures were grown in Luria-Bertani medium at 30 °C until the optical density reached 0.8–1, at which point the cultures were chilled to 16 °C and induced with 0.1 mM isopropyl β-d-1 thiogalactopyranoside and allowed to grow for a further 14–16 h. Pellets were lysed in 20 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol, 10 mM imidazole and 1 × BugBuster reagent (Merck-Millipore). Proteins were purified from lysates in batch mode using gravity columns containing TALON cobalt affinity resin (Takara). Columns were washed with 10 mM imidazole and eluted with 200 mM imidazole in 1-mL fractions. Proteins were concentrated and imidazole removed by ultrafiltration and buffer exchange into 20 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol.
Differential scanning fluorimetry
DSF was performed in 384-well format using a LightCycler 480 instrument (Roche). Reactions (10 µL) contained 20 µM protein, 20 mM HEPES pH 7.5, 150 mM NaCl, 1.25% (v/v) glycerol, 5 × SYPRO Tangerine dye (ThermoFisher Scientific) and varying concentrations of ligand that resulted in a final concentration of 5% (v/v) acetone. The programme parameters were: excitation at 483 nm, emission at 640 nm, ramp rate of 0.2 °C/s from 20 to 80 °C. The first derivative of fluorescence over temperature was calculated using the ‘Tm calling' function of the LightCycler software. Raw data from technical replicates were averaged and plotted using GraphPad Prism 8.1 (GraphPad Software, graphpad.com).
Statistical analysis
Data were analysed using one- or two-way analysis of variance (ANOVA) (α = 0.05, with Tukey’s multiple comparisons test). For Figs. 4c and 5c, in which data from three experimental replicates were combined, data were analysed using a mixed effects model with experimental replicate as a random effect, and genotype and treatment as fixed effects. Prior to ANOVA, germination data were arcsine-transformed, and gene expression data were log-transformed. Tests were implemented in GraphPad Prism version 8.1.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by funding from the Australian Research Council (DP130103646 to S.M.S. and G.R.F.; DP140104567 to S.M.S.; FT150100162 to M.T.W., DP160102888 to G.R.F. and M.T.W.). Y.K.S was recipient of a Research Training Program Scholarship from the Australian Government. We thank Dr Rowena Long for the original provision of B. tournefortii seeds, Dr Rohan Bythell-Douglas for advice on homology modelling and Mr Yongjie Meng with assistance with RNA extractions. We are grateful to Dr Kevin Rozwadowski (Agriculture and Agri-Food Canada) for the provision of pSKI106.
Source Data
Author contributions
Y.K.S., S.M.S., C.S.B., G.R.F. and M.T.W. conceived and designed the research. Y.K.S. and M.T.W. performed the majority of experiments with assistance from J.Y. (hypocotyl elongation assays and screening of transgenic lines), A.S. (chemical synthesis and seed germination assays), K.T.M. and S.F.D. (protein expression and purification). Y.K.S. and M.T.W. analysed the data. Y.K.S., S.M.S. and M.T.W. wrote the manuscript.
Data availability
All relevant data and materials are available from the authors upon reasonable request. Sequence data are available at NCBI Genbank under the following accessions: BtKAI2a, MG783328; BtKAI2b, MG783329; BtKAI2c, MG783330; BtD14a, MG783331; BtD14b, MG783332; BtDLK2, MG783333; BtSTH7, MK756121; BtCACS, MK756122. Raw RNA sequence data from B. tournefortii seed and seedlings are available in the NCBI SRA database under accession SRP128835. The original data underlying the following figures are provided as a Source Data file: Fig. 1a, b, d, e; 2b, e; 3a–d; 4a–d; 5a–c; Supplementary Figs. 1b–f; 2; 3; 4c; 5b–d; 6b, c, e; 7; 8a–e; 9a–c; 10a; 11; 12; 13.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-14991-w.
References
- 1.Early R, et al. Global threats from invasive alien species in the twenty-first century and national response capacities. Nat. Commun. 2016;7:12485–12485. doi: 10.1038/ncomms12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flematti GR, et al. Karrikin and cyanohydrin smoke signals provide clues to new endogenous plant signaling compounds. Mol. Plant. 2013;6:29–37. doi: 10.1093/mp/sss132. [DOI] [PubMed] [Google Scholar]
- 3.Keeley JE, Pausas JG. Evolution of ‘smoke’ induced seed germination in pyroendemic plants. South Afr. J. Bot. 2018;115:251–255. doi: 10.1016/j.sajb.2016.07.012. [DOI] [Google Scholar]
- 4.Flematti, G. R., Ghisalberti, E. L., Dixon, K. W. & Trengove, R. D. Identification of Alkyl substituted 2H-Furo[2,3- c]pyran-2-ones as germination stimulants present in smoke. J. Agric. Food Chem.57, 9475–9480 (2009). [DOI] [PubMed]
- 5.Hrdlička J, et al. Quantification of karrikins in smoke water using ultra-high performance liquid chromatography-tandem mass spectrometry. Plant Methods. 2019;15:81. doi: 10.1186/s13007-019-0467-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long RL, et al. Detecting karrikinolide responses in seeds of the Poaceae. Aust. J. Bot. 2011;59:610–620. doi: 10.1071/BT11170. [DOI] [Google Scholar]
- 7.Milberg P, Lamont BB. Fire enhances weed invasion of roadside vegetation in southwestern Australia. Biol. Conserv. 1995;73:45–49. doi: 10.1016/0006-3207(95)90061-6. [DOI] [Google Scholar]
- 8.Flematti GR, et al. Preparation of 2H-furo[2,3-c]pyran-2-one derivatives and evaluation of their germination-promoting activity. J. Agric. Food Chem. 2007;55:2189–2194. doi: 10.1021/jf0633241. [DOI] [PubMed] [Google Scholar]
- 9.Long RL, et al. Seeds of Brassicaceae weeds have an inherent or inducible response to the germination stimulant karrikinolide. Ann. Bot. 2011;108:933–944. doi: 10.1093/aob/mcr198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stevens J, Merritt D, Flematti G, Ghisalberti E, Dixon K. Seed germination of agricultural weeds is promoted by the butenolide 3-methyl-2H-furo[2,3-c]pyran-2-one under laboratory and field conditions. Plant Soil. 2007;298:113–124. doi: 10.1007/s11104-007-9344-z. [DOI] [Google Scholar]
- 11.Nelson DC, et al. Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc. Natl Acad. Sci. 2010;107:7095–7100. doi: 10.1073/pnas.0911635107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulkarni MG, Sparg SG, Light ME, Staden JV. Stimulation of rice (Oryza sativa L.) seedling vigour by smoke‐water and butenolide. J. Agron. Crop Sci. 2006;192:395–398. doi: 10.1111/j.1439-037X.2006.00213.x. [DOI] [Google Scholar]
- 13.Staden Jv, Sparg SG, Kulkarni MG, Light ME. Post-germination effects of the smoke-derived compound 3-methyl-2H-furo[2,3-c]pyran-2-one, and its potential as a preconditioning agent. Field Crop Res. 2006;98:98–105. doi: 10.1016/j.fcr.2005.12.007. [DOI] [Google Scholar]
- 14.Bangle DN, Walker LR, Powell EA. Seed germination of the invasive plant Brassica tournefortii (Sahara mustard) in the Mojave Desert. West. North Am. Naturalist. 2008;68:334–342. doi: 10.3398/1527-0904(2008)68[334:SGOTIP]2.0.CO;2. [DOI] [Google Scholar]
- 15.Brooks ML, et al. Effects of invasive alien plants on fire regimes. BioScience. 2004;54:677. doi: 10.1641/0006-3568(2004)054[0677:EOIAPO]2.0.CO;2. [DOI] [Google Scholar]
- 16.Sanders R., Minnich R. Brassica tournefortii. In: Invasive Plants of California’s Wildlands (eds. Bossard C. C., Randall J. M., Hoshovsky M. C.). University of California Press (2000).
- 17.Schiermeier Q. Pall hangs over desert’s future as alien weeds fuel wildfires. Nature. 2005;435:724. doi: 10.1038/435724b. [DOI] [PubMed] [Google Scholar]
- 18.Steers R. J. Invasive Plants, Fire succession, and Restoration of Creosote Bush Scrub in Southern California. University of California Riverside (2008).
- 19.Sun X-D, Ni M. HYPOSENSITIVE TO LIGHT, an alpha/beta fold protein, acts downstream of ELONGATED HYPOCOTYL 5 to regulate seedling de-etiolation. Mol. Plant. 2011;4:116–126. doi: 10.1093/mp/ssq055. [DOI] [PubMed] [Google Scholar]
- 20.Waters MT, et al. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development. 2012;139:1285–1295. doi: 10.1242/dev.074567. [DOI] [PubMed] [Google Scholar]
- 21.Durvasula A, et al. African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana. Proc. Natl Acad. Sci. 2017;114:5213–5218. doi: 10.1073/pnas.1616736114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson DC, et al. Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol. 2009;149:863–873. doi: 10.1104/pp.108.131516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flematti GR, Ghisalberti EL, Dixon KW, Trengove RD. A compound from smoke that promotes seed germination. Science. 2004;305:977. doi: 10.1126/science.1099944. [DOI] [PubMed] [Google Scholar]
- 24.Waters MT, Gutjahr C, Bennett T, Nelson DC. Strigolactone sgnaling and evolution. Annu. Rev. Plant Biol. 2017;68:291–322. doi: 10.1146/annurev-arplant-042916-040925. [DOI] [PubMed] [Google Scholar]
- 25.Yao R, Chen L, Xie D. Irreversible strigolactone recognition: a non-canonical mechanism for hormone perception. Curr. Opin. Plant Biol. 2018;45:155–161. doi: 10.1016/j.pbi.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Scaffidi A, et al. Exploring the molecular mechanism of karrikins and strigolactones. Bioorg. Medicinal Chem. 2012;22:3743–3746. doi: 10.1016/j.bmcl.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 27.Zwanenburg B, Pospíšil T. Structure and activity of strigolactones: new plant hormones with a rich future. Mol. Plant. 2013;6:38–62. doi: 10.1093/mp/sss141. [DOI] [PubMed] [Google Scholar]
- 28.de Saint Germain A, et al. An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat. Chem. Biol. 2016;12:787–794. doi: 10.1038/nchembio.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seto Y, et al. Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat. Commun. 2019;10:191. doi: 10.1038/s41467-018-08124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shabek N, et al. Structural plasticity of D3–D14 ubiquitin ligase in strigolactone signalling. Nature. 2018;563:652–656. doi: 10.1038/s41586-018-0743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao R, et al. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature. 2016;536:469–473. doi: 10.1038/nature19073. [DOI] [PubMed] [Google Scholar]
- 32.Marzec M, Brewer P. Binding or hydrolysis? how does the strigolactone receptor work? Trends Plant Sci. 2019;24:571–574. doi: 10.1016/j.tplants.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Bythell-Douglas R, et al. Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues. BMC Biol. 2017;15:52. doi: 10.1186/s12915-017-0397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conn CE, Nelson DC. Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front. Plant Sci. 2015;6:1219. doi: 10.3389/fpls.2015.01219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W, et al. The karrikin receptor KAI2 promotes drought resistance in Arabidopsis thaliana. PLoS Genet. 2017;13:e1007076. doi: 10.1371/journal.pgen.1007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun YK, Flematti GR, Smith SM, Waters MT. Reporter gene-facilitated detection of compounds in arabidopsis leaf extracts that activate the karrikin signaling pathway. Front. Plant Sci. 2016;7:1799. doi: 10.3389/fpls.2016.01799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S, et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lysak MA, Koch MA, Pecinka A, Schubert I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005;15:516–525. doi: 10.1101/gr.3531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 40.Waters MT, Scaffidi A, Flematti G, Smith SM. Substrate-induced degradation of the α/β-fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Mol. Plant. 2015;8:814–817. doi: 10.1016/j.molp.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 41.Waters MT, et al. A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in arabidopsis development but cannot mediate responses to Karrikins or strigolactones. Plant Cell. 2015;27:1925–1944. doi: 10.1105/tpc.15.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kagale S, et al. TMV-gate vectors: gateway compatible tobacco mosaic virus based expression vectors for functional analysis of proteins. Sci. Rep. 2012;2:874. doi: 10.1038/srep00874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao J, et al. An allelic series at the KARRIKIN INSENSITIVE 2 locus of Arabidopsis thaliana decouples ligand hydrolysis and receptor degradation from downstream signalling. Plant J. 2018;96:75–89. doi: 10.1111/tpj.14017. [DOI] [PubMed] [Google Scholar]
- 44.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abe S, et al. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl Acad. Sci. USA. 2014;111:18084–18089. doi: 10.1073/pnas.1410801111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamiaux C, et al. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 2012;22:2032–2036. doi: 10.1016/j.cub.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 47.Hamiaux C, et al. Inhibition of strigolactone receptors by N-phenylanthranilic acid derivatives: structural and functional insights. J. Biol. Chem. 2018;293:6530–6543. doi: 10.1074/jbc.RA117.001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Végh A, et al. Comprehensive analysis of DWARF14-LIKE2 (DLK2) reveals its functional divergence from strigolactone-related paralogs. Front. Plant Sci. 2017;8:1641. doi: 10.3389/fpls.2017.01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scaffidi A, et al. Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 2014;165:1221–1232. doi: 10.1104/pp.114.240036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leebens-Mack JH, et al. One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019;574:679–685. doi: 10.1038/s41586-019-1693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richardson DM, Wilgen BWV, Mitchell DT. Aspects of the reproductive ecology of four australian Hakea species (Proteaceae) in South Africa. Oecologia. 1987;71:345–354. doi: 10.1007/BF00378706. [DOI] [PubMed] [Google Scholar]
- 52.Guo Y, Zheng Z, La Clair JJ, Chory J, Noel JP. Smoke-derived karrikin perception by the α/β-hydrolase KAI2 from Arabidopsis. Proc. Natl Acad. Sci. USA. 2013;110:8284–8289. doi: 10.1073/pnas.1306265110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu Y, et al. Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica. Sci. Rep. 2016;6:31386. doi: 10.1038/srep31386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kagiyama M, et al. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells. 2013;18:147–160. doi: 10.1111/gtc.12025. [DOI] [PubMed] [Google Scholar]
- 55.Lee I, et al. A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana. J. Exp. Bot. 2018;69:3609–3623. doi: 10.1093/jxb/ery164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao, J. & Waters, M. T. Perception of karrikins by plants: a continuing enigma. J. Exp. Bot.erz548, 10.1093/jxb/erz1548 (2019). [DOI] [PMC free article] [PubMed]
- 57.Conn CE, et al. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science. 2015;349:540–543. doi: 10.1126/science.aab1140. [DOI] [PubMed] [Google Scholar]
- 58.Toh S, et al. Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science. 2015;350:203–207. doi: 10.1126/science.aac9476. [DOI] [PubMed] [Google Scholar]
- 59.Xu Y, et al. Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga. Nat. Commun. 2018;9:3947. doi: 10.1038/s41467-018-06452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bürger M, et al. Structural basis of Karrikin and non-natural strigolactone perception in Physcomitrella patens. Cell Rep. 2019;26:855–865. doi: 10.1016/j.celrep.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shimada A, et al. Structural basis for gibberellin recognition by its receptor GID1. Nature. 2008;456:520–523. doi: 10.1038/nature07546. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, et al. Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat. Chem. Biol. 2014;10:1028–1033. doi: 10.1038/nchembio.1660. [DOI] [PubMed] [Google Scholar]
- 63.Challis RJ, Hepworth J, Mouchel C, Waites R, Leyser O. A role for MORE AXILLARY GROWTH1 (MAX1) in evolutionary diversity in strigolactone signaling upstream of MAX2. Plant Physiol. 2013;161:1885–1902. doi: 10.1104/pp.112.211383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brewer PB, et al. LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc. Natl Acad. Sci. 2016;113:6301–6306. doi: 10.1073/pnas.1601729113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoneyama K, et al. Which are the major players, canonical or non-canonical strigolactones? J. Exp. Bot. 2018;69:2231–2239. doi: 10.1093/jxb/ery090. [DOI] [PubMed] [Google Scholar]
- 66.Panchy N, Lehti-Shiu MD, Shiu S-H. Evolution of gene duplication in plants. Plant Physiol. 2016;171:2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang L, Waters MT, Smith SM. Karrikin-KAI2 signalling provides Arabidopsis seeds with tolerance to abiotic stress and inhibits germination under conditions unfavourable for seedling establishment. New Phytol. 2018;219:605–618. doi: 10.1111/nph.15192. [DOI] [PubMed] [Google Scholar]
- 68.Goddard-Borger E. D., Ghisalberti E. L., Stick R. V. Synthesis of the germination stimulant 3-methyl-2H-furo[2,3-c]pyran-2-one and analogous compounds from carbohydrates. Eur. J. Org. Chem.2007, 3925-3934 (2007).
- 69.Mangnus E, Vanvliet LA, Vadenput D, Zwanenburg B. Structural modifications of strigol analogs - influence of the B and C rings on the bioactivity of the germination stimulant GR24. J. Agric. Food Chem. 1992;40:1222–1229. doi: 10.1021/jf00019a030. [DOI] [Google Scholar]
- 70.Gorecki MJ, Long RL, Flematti GR, Stevens JC. Parental environment changes the dormancy state and karrikinolide response of Brassica tournefortii seeds. Ann. Bot. 2012;109:1369–1378. doi: 10.1093/aob/mcs067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Flematti GR, Scaffidi A, Dixon KW, Smith SM, Ghisalberti EL. Production of the seed germination stimulant karrikinolide from combustion of simple carbohydrates. J. Agric. Food Chem. 2011;59:1195–1198. doi: 10.1021/jf1041728. [DOI] [PubMed] [Google Scholar]
- 73.Liang C, Liu X, Yiu S-M, Lim BL. De novo assembly and characterization of Camelina sativa transcriptome by paired-end sequencing. BMC Genomics. 2013;14:146. doi: 10.1186/1471-2164-14-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lefort V, Longueville J-E, Gascuel O. SMS: smart model selection in PhyML. Mol. Biol. Evol. 2017;34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bordoli L, et al. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 78.Dundas J, et al. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lopez-Obando M, et al. Structural modelling and transcriptional responses highlight a clade of PpKAI2-LIKE genes as candidate receptors for strigolactones in Physcomitrella patens. Planta. 2016;243:1441–1453. doi: 10.1007/s00425-016-2481-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All relevant data and materials are available from the authors upon reasonable request. Sequence data are available at NCBI Genbank under the following accessions: BtKAI2a, MG783328; BtKAI2b, MG783329; BtKAI2c, MG783330; BtD14a, MG783331; BtD14b, MG783332; BtDLK2, MG783333; BtSTH7, MK756121; BtCACS, MK756122. Raw RNA sequence data from B. tournefortii seed and seedlings are available in the NCBI SRA database under accession SRP128835. The original data underlying the following figures are provided as a Source Data file: Fig. 1a, b, d, e; 2b, e; 3a–d; 4a–d; 5a–c; Supplementary Figs. 1b–f; 2; 3; 4c; 5b–d; 6b, c, e; 7; 8a–e; 9a–c; 10a; 11; 12; 13.