Abstract
Turtles demonstrate variability in sex determination and, hence, constitute an excellent model for the evolution of sex chromosomes. Notably, the sex determination of the freshwater turtles from the family Chelidae, a species-rich group with wide geographical distribution in the southern hemisphere, is still poorly explored. Here we documented the presence of an XX/XY sex determination system in seven species of the Australasian chelid genera Chelodina, Emydura, and Elseya by conventional (karyogram reconstruction, C-banding) and molecular cytogenetic methods (comparative genome hybridization, in situ hybridization with probes specific for GATA microsatellite motif, the rDNA loci, and the telomeric repeats). The sex chromosomes are microchromosomes in all examined species of the genus Chelodina. In contrast, the sex chromosomes are the 4th largest pair of macrochromosomes in the genera Emydura and Elseya. Their X chromosomes are submetacentric, while their Y chromosomes are metacentric. The chelid Y chromosomes contain a substantial male-specific genomic region with an accumulation of the GATA microsatellite motif, and occasionally, of the rDNA loci and telomeric repeats. Despite morphological differences between sex chromosomes, we conclude that male heterogamety was likely already present in the common ancestor of Chelodina, Emydura and Elseya in the Mesozoic period.
Subject terms: Evolutionary genetics, Herpetology
Introduction
Amniotes possess two major sex determination systems: genotypic sex determination (GSD) and environmental sex determination (ESD). In GSD, the sex of an individual is determined by its sex-specific genotype, i.e. the combination of sex chromosomes. On the contrary, in ESD, the sex of an individual is influenced by environmental conditions and there are no consistent genotypic differences between sexes. The most well studied type of ESD is the temperature-dependent sex determination (TSD), where the sex of the individual is influenced by the temperature during a sensitive period of embryonic development (the definitions follow Johnson Pokorná & Kratochvíl1). Three amniote lineages, the geckos (infraorder Gekkota), the dragon lizards (family Agamidae) and the turtles (order Testudines), show extensive variability of sex determination systems, and closely related species have either GSD or ESD1–4, making them excellent groups for exploring the evolution of sex determination.
Turtles include 361 currently recognized extant species5–7. Unfortunately, the sex determination system is known in only approximately 24% of all species, and sex chromosomes have been up to now reported for only 20 species4,8–10. Phylogenetic reconstruction of sex determination systems suggested that ESD is ancestral in turtles and sex chromosomes, and thus GSD, evolved at least five times independently. In the suborder Cryptodira, XX/XY sex chromosomes have been reported for Siebenrockiella crassicollis (family Geoemydidae)4,11,12 and for the genera Staurotypus (family Kinosternidae)13 and Glyptemys (family Emydidae)14,15. In contrast, ZZ/ZW sex chromosomes are widely shared in softshell turtles (family Trionychidae)9,16,17. Recently, we demonstrated that the report on ZZ/ZW sex chromosomes in Pangshura smithii (Geoemydidae)18 was based on the erroneous pairing of chromosomes in the karyogram, and that this species has either GSD with poorly differentiated sex chromosomes or ESD19.
In the suborder Pleurodira, GSD was previously described for a few freshwater turtles of the family Chelidae20–22, a group consisting of 58 currently recognized species5. Chelid turtles form two geographically distinct clades, one distributed in Australasia and the other in South America23–27. Members of the family Chelidae have generally high diploid chromosome numbers ranging from 2n = 48 to 2n = 6410,20. Stable sex ratios of hatchlings incubated across a range of constant temperatures suggest the presence of GSD in at least three chelid species (Mesoclemmys gibba, Phrynops geoffroanus, Phrynops hilarii)8. Cytogenetic studies reported differentiated XX/XY sex chromosomes in three additional species, namely in Acanthochelys radiolata, Chelodina longicollis, and Emydura macquarii20–22,28. In A. radiolata, the pair of sex chromosomes consists of a medium-sized metacentric and a small acrocentric chromosome20. However, McBee et al.20 examined just a single individual (male), and the authors could neither determine the X or the Y chromosome in the heteromorphic pair, nor test whether this heteromorphism is linked to sex. Therefore, we consider the report on sex chromosomes in A. radiolata dubious. The sex chromosomes in Chelodina longicollis were identified as a pair of small chromosomes with a subtelocentric X and a submetacentric Y chromosome21. However, based on the accumulation of the GATA microsatellite motif, Matsubara et al.28 identified the Y chromosome in the same species as another, notably smaller microchromosome. The sex chromosomes in Emydura macquarii form the fourth-largest pair in the karyogram, consisting of a metacentric X and a submetacentric Y chromosome22, with a prominent C-positive band in the telomeric region of the short (p) chromosome arm28. In contrast to turtles from the family Chelidae, temperature-dependent sex determination was previously reported in species from the pleurodiran families Pelomedusidae (Pelomedusa subrufa, Pelusius castaneus) and Podocnemididae (Podocnemis unifilis, Podocnemis expansa, and Podocnemis erythrocephala)8,29–32.
In the current study, we explored sex chromosomes and karyotypes in the side-necked turtles of the genera Chelodina (C. expansa, C. novaeguineae, C. mccordi, C. reimanni, C. rugosa), Emydura (Em. macquarii krefftii), Elseya (El. novaeguineae), and two sibling individuals of the intergeneric hybrid Em. subglobosa (♀) × El. novaeguineae (♂) by applying a combination of conventional and molecular cytogenetic methods. We reconstructed karyograms and examined the presence of differentiated sex chromosomes by C-banding, comparative genome hybridization (CGH), and fluorescence in situ hybridization (FISH) with probes specific for GATA motif, telomeric repeats and rDNA loci, i.e. repetitive elements which often accumulate on the sex chromosomes of reptiles15,28.
Results
Species verification
The 5′ end of the mitochondrial cytochrome c oxidase I gene (COI) and/or the mitochondrial cytochrome b gene (cytb) were successfully amplified and sequenced and whenever possible compared to sequences from type specimens recently published by Kehlmaier et al.33. All studied individuals showed distinctly less than 3% genetic p-distance from the respective type specimens of the species with which they were identified. However, C. novaeguineae and C. reimanni do not differ in their mitochondrial DNA and the validity of C. reimanni is doubtful33. Accordingly, the COI of our specimens of C. reimanni was identical with the type sequences of these two species and our material was identified based on morphology.
Karyotype reconstruction and heterochromatin distribution
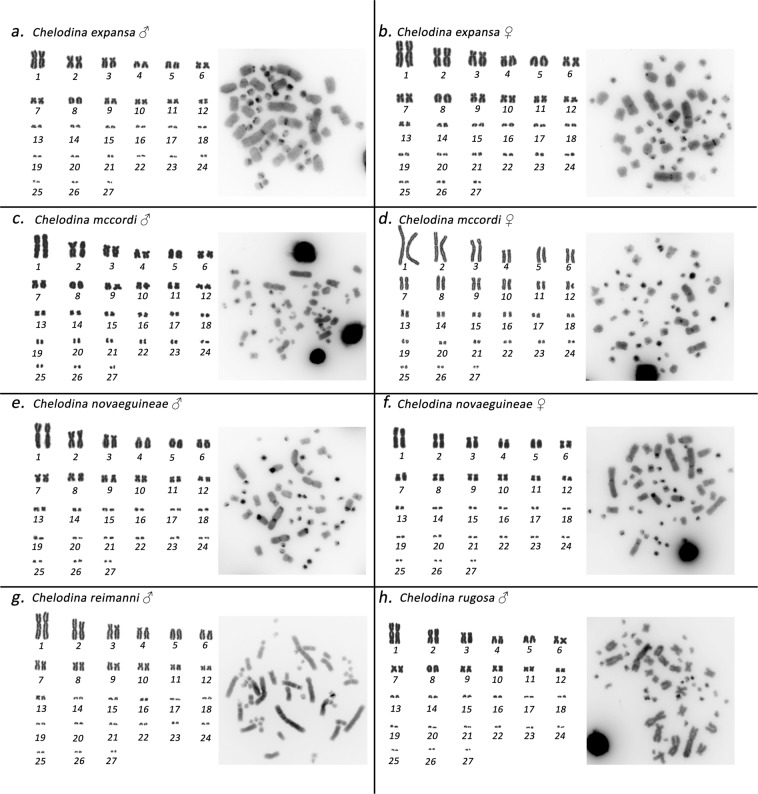
All examined individuals of the genus Chelodina had similar karyotypes with 2n = 54 chromosomes consisting of 12 pairs of macrochromosomes and 15 pairs of microchromosomes. All macrochromosomes were bi-armed, with the exception of the acrocentric chromosome pairs 5 and 8 in C. expansa, C. mccordi, and C. rugosa and of chromosome pair 5 in C. novaeguineae and C. reimanni (Fig. 1). C-banding stain revealed constitutive heterochromatin in the centromeric regions of all chromosomes. In addition, heterochromatic blocks were detected in up to four pairs of microchromosomes in all species as well as in the p-arms of the submetacentric chromosomes from the 4th pair in C. novaeguineae (Fig. 1).
Figure 1.
Giemsa-stained karyotype and C-banded metaphases in Chelodina expansa (a,b), Chelodina mccordi (c,d), Chelodina novaeguineae (e,f), Chelodina reimanni (g), and Chelodina rugosa (h). The pairing of microchromosomes does not indicate homology but morphological similarity.
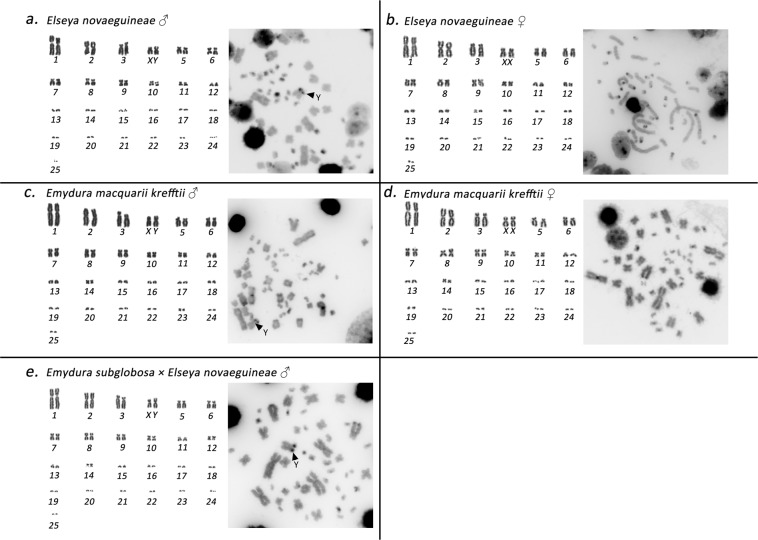
The individuals from the genera Emydura and Elseya possessed karyotypes with 2n = 50 chromosomes consisting of 12 pairs of macrochromosomes and 13 pairs of microchromosomes. All macrochromosomes were bi-armed. The 4th largest chromosome pair consisted of two submetacentric chromosomes in females, but a metacentric chromosome and a submetacentric chromosome in males (El. novaeguineae, Em. macquarii krefftii, and the two male hybrids Em. subglobosa × El. novaeguineae; Fig. 2). C-banding revealed constitutive heterochromatin in the centromeric regions of all chromosomes. In addition, heterochromatic blocks were observed in four pairs of microchromosomes and in the 4th largest pair (Fig. 2).
Figure 2.
Giemsa-stained karyotype and C-banded metaphases in Elseya novaeguineae (a,b), Emydura macquarii krefftii (c,d), and the hybrid Em. subglobosa × El. novaeguineae (e). The pairing of microchromosomes does not indicate homology but morphological similarity.
In situ hybridization with probes for GATA motif, telomeric repeats and rDNA loci
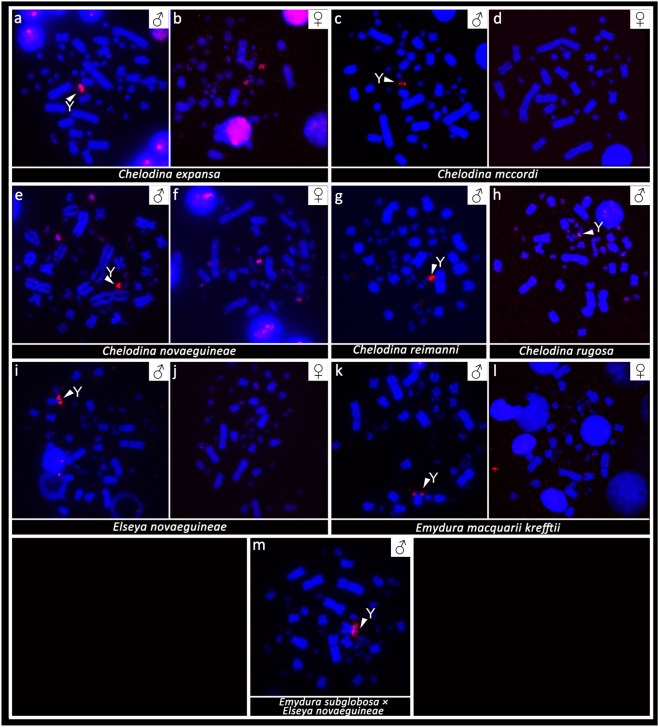
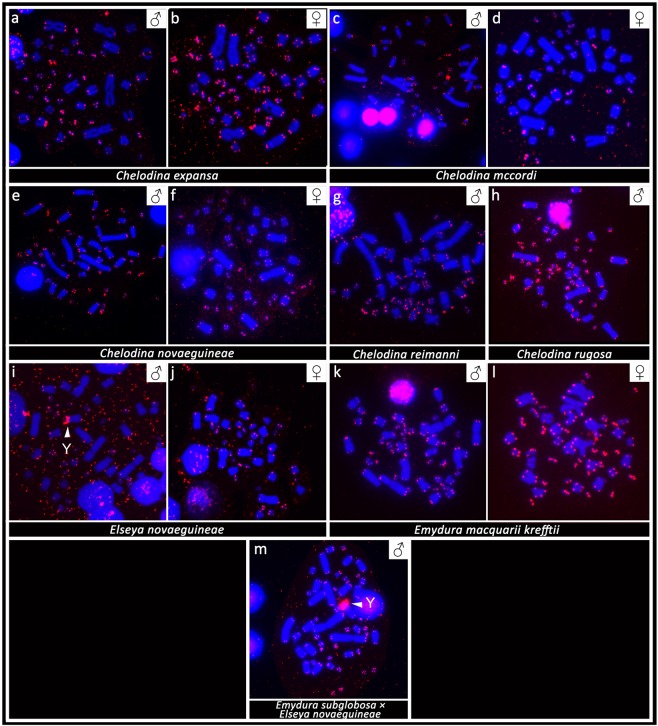
FISH with probes specific for the GATA microsatellite motif revealed a strong accumulation in a single microchromosome in the males of the genus Chelodina. Strong accumulations of this motif were revealed in all males of the genera Emydura and Elseya in the heterochromatic region in the terminal position of the p-arm of the metacentric chromosome from the 4th pair. No accumulation of the GATA motif was detected in females (Fig. 3) with the only exception of C. expansa, where the accumulation of the GATA microsatellite motif was identified in three microchromosomes in males but in only two microchromosomes in females.
Figure 3.
In situ hybridization with the probe specific for the (GATA)8 microsatellite motif in Chelodina expansa (a,b), Chelodina mccordi (c,d), Chelodina novaeguineae (e,f), Chelodina reimanni (g), Chelodina rugosa (h), Elseya novaeguineae (i,j), Emydura macquarii krefftii (k,l), and the hybrid Em. subglobosa × El. novaeguineae (m). The FITC signal of the GATA probe was pseudocolourized in red. All metaphases were counterstained with DAPI (blue). The Y chromosome is indicated with a white arrow.
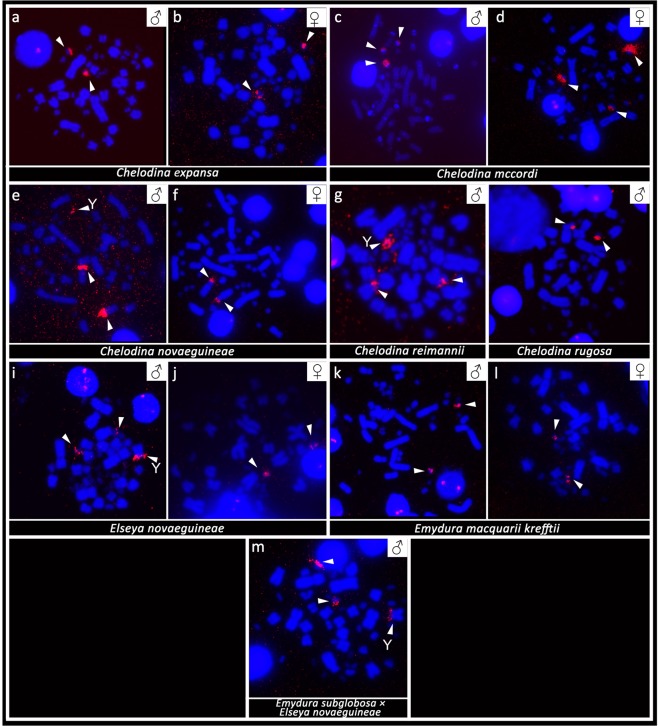
The probe specific for the telomeric repeats revealed the expected terminal topology. In addition, strong accumulation of telomeric-like motifs was detected in microchromosomes in all studied individuals as well as in the terminal position of the p-arm of the metacentric chromosome from the 4th pair in the hybrid Em. subglobosa × El. novaeguineae (Fig. 4).
Figure 4.
In situ hybridization with probe specific for the (TTAGGG)n telomeric motif in Chelodina expansa (a,b), Chelodina mccordi (c,d), Chelodina novaeguineae (e,f), Chelodina reimanni (g), Chelodina rugosa (h), Elseya novaeguineae (i,j), Emydura macquarii krefftii (k,l), and the hybrid Em. subglobosa × El. novaeguineae (m). The FITC signal was pseudocolourized in red. All metaphases were counterstained with DAPI (blue). The Y chromosome is indicated with a white arrow.
FISH with probes specific for the rDNA loci showed strong accumulation in two chromosomes in both sexes of C. expansa and C. rugosa. rDNA loci were accumulated in two chromosomes in females of C. novaeguineae, but in three chromosomes in males of C. novaeguineae and C. reimanni. Notably, an accumulation of rDNA loci was detected in three microchromosomes in both sexes of C. mccordi. rDNA loci accumulated in two microchromosomes in females of El. novaeguineae and Em. macquarii krefftii. In addition, rDNA loci accumulated also in the telomeric position of the p-arm of the metacentric chromosome from the 4th pair in male turtles of El. novaeguineae and the Em. subglobosa × El. novaeguineae hybrids but not in the homologous chromosome of Em. macquarii krefftii (Fig. 5).
Figure 5.
In situ hybridization with probe specific for the rDNA sequence in Chelodina expansa (a,b), Chelodina mccordi (c,d), Chelodina novaeguineae (e,f), Chelodina reimanni (g), Chelodina rugosa (h), Elseya novaeguineae (i,j), Emydura macquarii krefftii (k,l), and the hybrid Em. subglobosa × El. novaeguineae (m). The FITC signal was pseudocolourized in red. All metaphases were counterstained with DAPI (blue). The Y chromosome is indicated with a white arrow.
Comparative genome hybridization
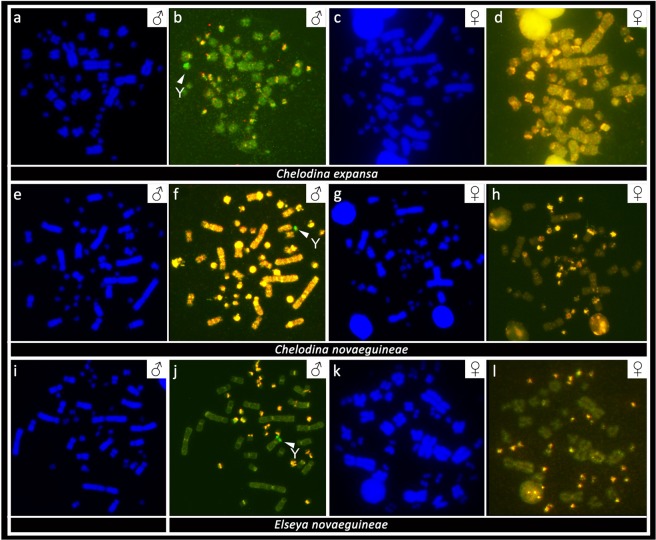
CGH revealed strong male-specific genomic content in a single microchromosome in metaphases from males of C. expansa and C. novaeguineae. Male-specific genomic content was detected at the terminal position of the p-arm of the metacentric chromosome from the 4th pair in metaphases from males of El. novaeguineae. No sex-specific content was found in metaphases of females in C. expansa, C. novaeguineae and El. novaeguineae (Fig. 6).
Figure 6.
Comparative genome hybridization with FITC-labelled probe specific for male (green) and rhodamine-labelled probe specific for female (red) genomic content in Chelodina expansa (a–d), Chelodina novaeguineae (e–h), and Elseya novaeguineae (i–l). Chromosomal regions with similar genomic content between sexes are visualized in yellow. The white arrow indicates male-specific region (green), corresponding to the Y chromosome.
Discussion
Freshwater turtles of the family Chelidae have karyotypes with 2n = 54 chromosomes in the genus Chelodina and 2n = 50 chromosomes in the genera Elseya and Emydura (Figs. 1 and 2). Our cytogenetic examination confirmed previously published karyotypes for C. expansa, C. novaeguineae, C. rugosa and Em. m. krefftii, with respect to chromosome numbers and morphology10, while karyotypes for C. mccordi, C. reimanni, and El. novaeguineae are presented here for the first time (Figs. 1 and 2). Within the genus Chelodina there was an evolutionary change in the shape of the chromosome pair 8, which is metacentric in C. novaeguineae and C. reimanni, but acrocentric in C. expansa, C. mccordi and C. rugosa. The metacentric shape in C. novaeguineae and C. reimanni can be a synapomorphy of these closely related or synonymous species33. The transitions between acrocentric and metacentric shape in a chromosome pair together with the conservation in chromosome numbers are often caused by intrachromosomal rearrangements in reptiles34–37. This hypothesis should be tested in the genus Chelodina by comparative cytogenetics in future, using whole chromosome painting or comparative BAC-FISH.
In situ hybridization with probes specific for repetitive elements that often accumulate on vertebrate sex chromosomes revealed an extensive accumulation of the GATA microsatellite motif in odd numbers of chromosomes in the metaphases of all male chelids. The relevant chromosome corresponds to a dot-like microchromosome in the genus Chelodina but to a single chromosome of the 4th pair of the complement in the genera Emydura and Elseya (Fig. 3). In addition, rDNA loci are amplified in odd numbers of chromosomes in males of C. novaeguineae, C. reimanni, El. novaeguineae and in the Em. subglobosa × El. novaeguineae hybrids (Fig. 4). Telomeric repeats seem to accumulate in a single chromosome of the 4th pair of the complement in El. novaeguineae and in the Em. subglobosa × El. novaeguineae hybrids (Fig. 5). We suggest that the chromosome with the amplification of the GATA microsatellite motif and in some species also of the rDNA loci and telomeric-like repeats in males is the Y chromosome. This conclusion is further supported by the results of CGH performed here for three species (C. expansa, C. novaeguineae, and El. novaeguineae) visualizing a male-specific genomic content within these chromosomes (Fig. 6). A metacentric Y chromosome was previously described for Em. m. macquarii by Martinez et al.22 with similar morphology as in El. novaeguineae, Em. macquarii krefftii and the two male Em. subglobosa × El. novaeguineae hybrids. In contrast to El. novaeguineae and the two male Em. subglobosa × El. novaeguineae hybrids, the accumulation of GATA microsatellite repeats was not detected here in the metacentric Y chromosome of Em. macquarii krefftii and previously also in Em. m. macquarii28. We assume that the GATA motif does not exist or accumulates in very low copy numbers in the Y chromosome of Em. macquarii krefftii and Em. m. macquarii, below the detection threshold of molecular cytogenetic methods. This situation likely reflects the extensive evolutionary dynamics of the heterochromatic content of degenerated sex chromosomes in sauropsids28,38,39.
As identified with our cytogenetic methods (i.e. karyotype reconstruction, C-banding, and FISH), the X chromosome is the submetacentric chromosome in the 4th largest pair of the complement in Elseya and Emydura (Fig. 2). We were not able to visualize the X chromosome in the genus Chelodina by our cytogenetic methods, yet, the chromosome pairing in karyograms suggests that it should be a microchromosome (Fig. 1). Ezaz et al.21 concluded that sex chromosomes in C. longicollis correspond to a pair of small-sized chromosomes with a prominent heterochromatic block. However, we assume that the sex chromosomes of C. longicollis were misidentified in the study of Ezaz et al.21. Our results agree with Matsubara et al.28 who showed the Y chromosome in Chelodina is a different, tiny microchromosome with a prominent amplification of microsatellite repeats.
All Australasian chelid species studied to date possess an XX/XY sex determination system (this study21,22,28). Homology between XX/XY sex chromosomes with dissimilar morphology in representatives from the genus Chelodina when compared with Elseya and Emydura might be supported by accumulation of the same repetitive motifs (GATA microsatellite, rDNA, telomeric-like sequences) in at least some members of these two clades (Figs. 3–5), but the accumulation of the same repetitive motifs in heterochromatic regions is generally a poor indicator of sex chromosome homology28,39. Matsubara et al.28 suggested that the sex chromosomes in the ancestor of Australasian chelids were a pair of microchromosomes, similar to the recent Chelodina, and a rearrangement occurred in the common ancestor of Elseya and Emydura. According to Matsubara et al.28, the ancestral sex chromosomes either (i) fused with a medium-sized pair of autosomes or (ii) a part of the ancestral sex chromosomes, including the sex determining region and a surrounding repetitive content, was translocated to a medium-sized autosome. However, the clades of the genera Chelodina and Elseya/Emydura show a sister group relationship (Fig. 7). Therefore, we assume that another scenario for sex chromosome homology is equally parsimonious, i.e. that the ancestral sex chromosomes were of the Elseya/Emydura type and a chromosomal rearrangement in the ancestor of the genus Chelodina transferred the sex-determining locus to a microchromosome.
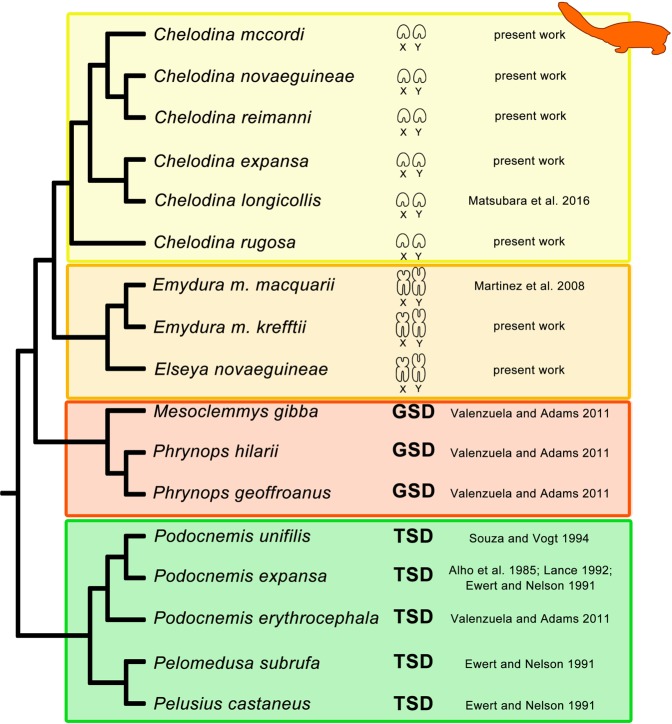
Figure 7.
Overview of current knowledge on sex determination in side-necked turtles. Phylogeny follows Valenzuela & Adams8 and Kehlmaier et al.33. Information on sex determination systems was compiled from this and previously published studies8,21,22,28–32. Sex chromosomes are microchromosomes in turtles from the genus Chelodina, but macrochromosomes in turtles from the genera Elseya and Emydura. GSD: genotypic sex determination, TSD: temperature dependent sex determination.
If sex determination is homologous between the two chelid clades, the XX/XY chromosomes in this group could date back to their last common ancestor living c. 50–120 million years ago27,40,41. In order to scrutinize the possible homology of sex chromosomes across Australasian chelids in future, it will be crucial to identify the gene content of their sex chromosomes using genomic methods, as recently applied in other reptilian lineages9,42–46.
Material and Methods
Studied material
We collected blood samples to establish cell cultures for chromosome preparations and for DNA isolation from side-necked turtles of the genera Chelodina (C. expansa, C. mccordi, C. novaeguineae, C. reimanni, C. rugosa), Emydura (Em. macquarii krefftii) and Elseya (El. novaeguineae), and two sibling hybrids Em. subglobosa (♀) × El. novaeguineae (♂). All turtles are either captive-bred or legally imported from the wild, and kept at Plzeň Zoo (Czech Republic), the Zoo Prague (Czech Republic), Turtle Island (Austria), or the Museum of Zoology, Senckenberg Dresden (Germany). A detailed list of specimens is provided in Table 1. Blood samples were collected by veterinarians primarily for diagnostic purposes, which is not considered as an experiment on animals according to Czech legislation (No. 46/1992). The owners of the turtles approved the use of blood samples for the current study. All methods were carried out in accordance with relevant guidelines and regulations, by researchers accredited for animal experimental design by the Ministry of Agriculture of the Czech Republic (Lukáš Kratochvíl: accreditation CZ02535; Michail Rovatsos: accreditation CZ03540), and with the approval of the Ethical Committee of Faculty of Science, Charles University.
Table 1.
List of individuals analyzed in this study. Diploid chromosome number (2n) and sex are indicated.
| Species | 2n | ♂ | ♀ |
|---|---|---|---|
| Chelodina expansa | 54 | 1 | 2 |
| Chelodina mccordi | 54 | 2 | 2 |
| Chelodina novaeguineae | 54 | 3 | 2 |
| Chelodina reimanni | 54 | 2 | — |
| Chelodina rugosa | 54 | 2 | — |
| Elseya novaeguineae | 50 | 1 | 1 |
| Emydura macquarii krefftii | 50 | 1 | 2 |
| Em. subglobosa (♀) × El. novaeguineae (♂) | 50 | 2 | — |
DNA isolation, chromosome preparation and staining
Genomic DNA was extracted from blood samples using a DNeasy Blood and Tissue Kit (Qiagen). Chromosome suspensions for cytogenetic analyses were obtained from whole-blood lymphocyte cultures following the protocol described in Mazzoleni et al.19. Chromosome spreads were stained with Giemsa for karyotype reconstruction (Figs. 1 and 2). The distribution of constitutive heterochromatin was detected by C-banding47, with slight modifications as described in Mazzoleni et al.19.
Species verification
Species identification is challenging in the genus Chelodina, as taxonomy is complicated and not fully resolved yet (for review see Kehlmaier et al.33). Therefore, we characterized our material and verified its taxonomy by sequencing the standard “DNA barcoding” region from the mitochondrial cytochrome c oxidase subunit I gene (COI) and/or the mitochondrial cytochrome b gene (cytb). This data is intended to genetically identify our cytogenetic material in future, regardless of potential taxonomic changes. The COI fragment was amplified by PCR using either the reptile-specific primers RepCOI-F and RepCOI-R48 or the universal primers LCO1490 and HCO219849. The cytb gene was amplified by PCR using the primers H16064 and L1491950. For both genes, we prepared the PCR reaction and cycling conditions according to Koubová et al.51. The PCR products were sequenced bi-directionally by Macrogen (Korea), and the obtained haplotype sequences were deposited in GenBank under the accession numbers MN757883-MN757886. The COI and cytb sequences were aligned using CLUSTALW52, as implemented in BioEdit v5.0.953, and subsequently analyzed in DnaSP v5.10.154. All sequences were compared with those from Kehlmaier et al.33, derived from type specimens, and Le et al.55. Genetic distances among haplotypes were calculated in MEGA v756.
Fluorescence in situ hybridization
The distributions of the GATA microsatellite motif was examined, as well as of the TTAGGG telomeric repeat and the rDNA loci, using FISH. The (GATA)8 probe was synthesized and labeled with biotin (Macrogen, Korea). The telomeric probe was synthesized and labeled with biotin by PCR according to a previously published protocol57. The probe for the rDNA loci was prepared from a plasmid (pDm r.a51#1) with an 11.5-kb insertion, encoding the 18S and 28S rRNA units of Drosophila melanogaster58; for the labeling protocol see Rovatsos et al.9. Hybridization conditions, post-hybridization washes, signal amplification and detection are explained in detail in Rovatsos et al.59.
Comparative genome hybridization
To detect sex-specific regions of the genome, CGH was performed using metaphase chromosomes of both male and female individuals of C. expansa, C. novaeguineae, and El. novaeguineae. The detailed protocol for probe and hybridization experiments is presented in Rovatsos et al.59.
Microscopy and image analysis
Giemsa-stained metaphase chromosomes were studied under a Carl Zeiss AxioImager.Z2 microscope, equipped with Metafer Scanning Platform (Metasystems) and a MetaSystems CoolCube digital camera. Images were processed for karyotype reconstruction with Ikaros karyotyping software (Metasystems). For C-banding, FISH and CGH methods, images from at least 20 metaphase chromosomes were analyzed using a Provis AX70 (Olympus) fluorescence microscope, equipped with a DP30BW digital camera (Olympus). All images were acquired in black and white, and later superimposed with colours in DP Manager imaging software (Olympus).
Acknowledgements
We would like to express our gratitude to Petr Ráb for providing laboratory space and constant support, and to Jana Thomayerová and Šárka Pelikánova for technical assistance. We thank the staff of Plzeň Zoo for providing blood samples. We also thank Tomáš Jirásek and Václav Burka. The project was supported by the Charles University project PRIMUS/SCI/46, the Grant Agency of Charles University (GAUK 444217) and the Charles University Research Centre program (204069).
Author contributions
S.M., B.A., L.C. and M.R. performed the experimental part, M.R., S.M. and L.K. developed the project, M.A., U.F., P.P., T.P. and P.V. contributed with material and consulting for the development of the project, M.R. and S.M. wrote the first draft, all authors read the manuscript and contributed to its final form.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Johnson Pokorná M, Kratochvíl L. What was the ancestral sex-determining mechanism in amniote vertebrates? Biol. Rev. 2016;91:1–12. doi: 10.1111/brv.12156. [DOI] [PubMed] [Google Scholar]
- 2.Pokorná M, Kratochvíl L. Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zool. J. Linn. Soc. 2009;156:168–183. doi: 10.1111/j.1096-3642.2008.00481.x. [DOI] [Google Scholar]
- 3.Gamble T, et al. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol. Biol. Evol. 2015;32:1296–1309. doi: 10.1093/molbev/msv023. [DOI] [PubMed] [Google Scholar]
- 4.Montiel EE, et al. Cytogenetic insights into the evolution of chromosomes and sex determination reveal striking homology of turtle sex chromosomes to amphibian autosomes. Cytogenet. Genome Res. 2016;148:292–304. doi: 10.1159/000447478. [DOI] [PubMed] [Google Scholar]
- 5.Turtle Taxonomy Working Group [Rhodin, A. G. J. et al.]. Turtles of the World: Annotated Checklist and Atlas of Taxonomy, Synonymy, Distribution, and Conservation Status (8th Ed.) In Conservation Biology of Freshwater Turtles and Tortoises: A Compilation Project of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group. (eds. Rhodin, A. G. J. et al.). Chelonian Res. Monogr. 7, 1–292 (2017).
- 6.Rhodin AGJ, et al. Global Conservation Status of Turtles and Tortoises (Order Testudines) Chelonian Conserv. Bi. 2018;17:135–161. doi: 10.2744/CCB-1348.1. [DOI] [Google Scholar]
- 7.Farkas, B., Ziegler, T., Pham, C. T., Ong, A. V. & Fritz, U. A new species of Pelodiscus from northeastern Indochina (Testudines, Trionychidae). ZooKeys824, 71–86 (2019). [DOI] [PMC free article] [PubMed]
- 8.Valenzuela N, Adams DC. Chromosome number and sex determination coevolve in turtles. Evolution. 2011;65:1808–1813. doi: 10.1111/j.1558-5646.2011.01258.x. [DOI] [PubMed] [Google Scholar]
- 9.Rovatsos M, Praschag P, Fritz U, Kratochvil L. Stable Cretaceous sex chromosomes enable molecular sexing in softshell turtles (Testudines: Trionychidae) Sci. Rep. 2017;7:42150. doi: 10.1038/srep42150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olmo, E. & Signorino, G. G. Chromorep: A reptile chromosomes database. http://chromorep.univpm.it Accessed 4 June 2019. (2005).
- 11.Carr JL, Bickham JW. Sex chromosomes of the Asian black pond turtle, Siebenrockiella crassicollis (Testudines: Emydidae) Cytogenet. Genome Res. 1981;31:178–183. doi: 10.1159/000131644. [DOI] [PubMed] [Google Scholar]
- 12.Kawagoshi T, Nishida C, Matsuda Y. The origin and differentiation process of X and Y chromosomes of the black marsh turtle (Siebenrockiella crassicollis, Geoemydidae, Testudines) Chromosome Res. 2012;20:95–110. doi: 10.1007/s10577-011-9267-7. [DOI] [PubMed] [Google Scholar]
- 13.Kawagoshi T, Uno Y, Nishida C, Matsuda Y. The Staurotypus turtles and aves share the same origin of sex chromosomes but evolved different types of heterogametic sex determination. PLoS One. 2014;9:105315. doi: 10.1371/journal.pone.0105315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montiel EE, Badenhorst D, Tamplin J, Burke RL, Valenzuela N. Discovery of the youngest sex chromosomes reveals first case of convergent co-option of ancestral autosomes in turtles. Chromosoma. 2017;126:105–113. doi: 10.1007/s00412-016-0576-7. [DOI] [PubMed] [Google Scholar]
- 15.Literman R, et al. Development of sexing primers in Glyptemys insculpta and Apalone spinifera turtles uncovers an XX/XY sex-determining system in the critically-endangered bog turtle Glyptemys muhlenbergii. Conserv. Genet. Resour. 2017;9:651–658. doi: 10.1007/s12686-017-0711-7. [DOI] [Google Scholar]
- 16.Kawagoshi T, Uno Y, Matsubara K, Matsuda Y, Nishida C. The ZW micro-sex chromosomes of the Chinese soft-shelled turtle (Pelodiscus sinensis, Trionychidae, Testudines) have the same origin as chicken chromosome 15. Cytogenet. Genome Res. 2009;125:125–131. doi: 10.1159/000227837. [DOI] [PubMed] [Google Scholar]
- 17.Badenhorst D, Stanyon R, Engstrom T, Valenzuela N. A ZZ/ZW microchromosome system in the spiny softshell turtle, Apalone spinifera, reveals an intriguing sex chromosome conservation in Trionychidae. Chromosome Res. 2013;21:137–147. doi: 10.1007/s10577-013-9343-2. [DOI] [PubMed] [Google Scholar]
- 18.Sharma, G. P., Kaur, P. & Nakhasi, U. Female heterogamety in the Indian cryptodiran chelonian, Kachuga smithi Gray. In Dr. B. S. Chuahah Commemoration Volume (eds. Tiwari K. K. & Srivistava C. B.) 359–368 (Orissa: Zoological Society of India) (1975).
- 19.Mazzoleni S, et al. Turtles of the genera Geoemyda and Pangshura (Testudines: Geoemydidae) lack differentiated sex chromosomes: the end of a 40-year error cascade for Pangshura. PeerJ. 2019;7:6241. doi: 10.7717/peerj.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McBee K, Bickham JW, Rhodin AG, Mittermeier RA. Karyotypic variation in the genus Platemys (Testudines: Pleurodira) Copeia. 1985;2:445–449. doi: 10.2307/1444856. [DOI] [Google Scholar]
- 21.Ezaz T, et al. An XX/XY sex microchromosome system in a freshwater turtle, Chelodina longicollis (Testudines: Chelidae) with genetic sex determination. Chromosome Res. 2006;14:139–150. doi: 10.1007/s10577-006-1029-6. [DOI] [PubMed] [Google Scholar]
- 22.Martinez PA, Ezaz T, Valenzuela N, Georges A, Marshall Graves JA. An XX/XY heteromorphic sex chromosome system in the Australian chelid turtle Emydura macquarii: a new piece in the puzzle of sex chromosome evolution in turtles. Chromosome Res. 2008;16:815–825. doi: 10.1007/s10577-008-1228-4. [DOI] [PubMed] [Google Scholar]
- 23.Seddon JM, Georges A, Baverstock PR, McCord W. Phylogenetic relationships of chelid turtles (Pleurodira: Chelidae) based on mitochondrial 12S rRNA gene sequence variation. Mol. Phylogenet. Evol. 1997;7:55–61. doi: 10.1006/mpev.1996.0372. [DOI] [PubMed] [Google Scholar]
- 24.Shaffer, H. B., Meylan, P. & McKnight, M. L. Tests of turtle phylogeny: Molecular, morphological, and paleontological approaches. Syst. Biol.46, 235–268 (1997). [DOI] [PubMed]
- 25.Georges A, Birrell J, Saint KM, McCord W, Donellan SC. A phylogeny of side-necked turtles (Chelonia: Pleurodira) based on mitochondrial and nuclear gene sequence variation. Biol. J. Linn. Soc. 1998;67:213–246. doi: 10.1111/j.1095-8312.1999.tb01862.x. [DOI] [Google Scholar]
- 26.Fujita MK, Engstrom TN, Starkey DE, Shaffer HB. Turtle phylogeny: insights from a novel nuclear intron. Mol. Phylogenet. Evol. 2004;31:1031–1040. doi: 10.1016/j.ympev.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 27.Pereira AG, Sterli J, Moreira FR, Schrago CG. Multilocus phylogeny and statistical biogeography clarify the evolutionary history of major lineages of turtles. Mol. Phylogenet. Evol. 2017;113:59–66. doi: 10.1016/j.ympev.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 28.Matsubara K, et al. Amplification of microsatellite repeat motifs is associated with the evolutionary differentiation and heterochromatinization of sex chromosomes in Sauropsida. Chromosoma. 2016;125:111–123. doi: 10.1007/s00412-015-0531-z. [DOI] [PubMed] [Google Scholar]
- 29.Alho CJ, Danni TM, Pádua LF. Temperature-dependent sex determination in Podocnemis expansa (Testudinata: Pelomedusidae) Biotropica. 1985;17:75–78. doi: 10.2307/2388383. [DOI] [Google Scholar]
- 30.Ewert, M. A. & Nelson, C. E. Sex determination in turtles: diverse patterns and some possible adaptive values. Copeia1991, 50–69 (1991).
- 31.Lance, V. A., Valenzuela, N. & Von Hildebrand, P. A hormonal method to determine sex of hatchling giant river turtles, Podocnemis expansa: application to endangered species. Am. Zool.32, 16A (1992).
- 32.de Souza RR, Vogt RC. Incubation temperature influences sex and hatchling size in the neotropical turtle Podocnemis unifilis. J. Herpetol. 1994;28:453–464. doi: 10.2307/1564958. [DOI] [Google Scholar]
- 33.Kehlmaier C, et al. Mitogenomics of historical type specimens of Australasian turtles: clarification of taxonomic confusion and old mitochondrial introgression. Sci. Rep. 2019;9:5841. doi: 10.1038/s41598-019-42310-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iannucci, A. et al. Conserved sex chromosomes and karyotype evolution in monitor lizards (Varanidae). Heredity123, 215–227 (2019). [DOI] [PMC free article] [PubMed]
- 35.King, M. Chromosome Change and Speciation in Lizards. In Essays on Evolution and Speciation (eds. Atchley, W. & Woodruff, D.) 262–285 (Cambridge University Press, Cambridge) (1981).
- 36.King M, Mengden GA, King D. A pericentric- inversion polymorphism and a ZZ/ZW sex- chromosome system in Varanus acanthurus Boulenger analyzed by G- and C-banding and Ag staining. Genetica. 1982;58:39–45. doi: 10.1007/BF00056001. [DOI] [Google Scholar]
- 37.King M. Species evolution: the role of chromosome change. (Cambridge University Press, Cambridge) (1993).
- 38.Rovatsos M, Johnson Pokorná M, Altmanová M, Kratochvíl L. Mixed-up sex chromosomes: identification of sex chromosomes in the X1X1X2X2/X1X2Y system of the legless lizards of the genus Lialis (Squamata: Gekkota: Pygopodidae) Cytogenet. Genome Res. 2016;149:282–289. doi: 10.1159/000450734. [DOI] [PubMed] [Google Scholar]
- 39.Augstenová B, Mazzoleni S, Kratochvíl L, Rovatsos M. Evolutionary dynamics of the W chromosome in caenophidian snakes. Genes. 2018;9:5. doi: 10.3390/genes9010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joyce WG, Parham JF, Lyson TR, Warnock RC, Donoghue PC. A divergence dating analysis of turtles using fossil calibrations: an example of best practices. J. Paleontol. 2013;87:612–634. doi: 10.1666/12-149. [DOI] [Google Scholar]
- 41.Lourenço JM, Claude J, Galtier N, Chiari Y. Dating cryptodiran nodes: origin and diversification of the turtle superfamily Testudinoidea. Mol. Phylogenet. Evol. 2013;62:496–507. doi: 10.1016/j.ympev.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 42.Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 2013;11:1001643. doi: 10.1371/journal.pbio.1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gamble T, et al. The discovery of XY sex chromosomes in a boa and python. Curr. Biol. 2017;27:2148–2153. doi: 10.1016/j.cub.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 44.Rovatsos M, Vukić J, Lymberakis P, Kratochvíl L. Evolutionary stability of sex chromosomes in snakes. Proc. Royal. Soc. B. 2015;282:20151992. doi: 10.1098/rspb.2015.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rovatsos M, et al. Conservation of sex chromosomes in lacertid lizards. Mol. Ecol. 2016;25:3120–3126. doi: 10.1111/mec.13635. [DOI] [PubMed] [Google Scholar]
- 46.Rovatsos, M., Rehák, I., Velenský, P. & Kratochvíl, L. Shared ancient sex chromosomes in varanids, beaded lizards and alligator lizards. Mol. Biol. Evol. 10.1093/molbev/msz024 (2019). [DOI] [PubMed]
- 47.Sumner AT. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell. Res. 1972;75:304–306. doi: 10.1016/0014-4827(72)90558-7. [DOI] [PubMed] [Google Scholar]
- 48.Nagy ZT, Sonet G, Glaw F, Vences M. First large-scale DNA barcoding assessment of reptiles in the biodiversity hotspot of Madagascar, based on newly designed COI primers. PLoS One. 2012;7:e34506. doi: 10.1371/journal.pone.0034506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 50.Burbrink FT, Lawson R, Slowinski JB. Mitochondrial DNA phylogeography of the polytypic North American ratsnake (Elaphe obsoleta): a critique of the subspecies concept. Evolution. 2000;54:2107–2118. doi: 10.1111/j.0014-3820.2000.tb01253.x. [DOI] [PubMed] [Google Scholar]
- 51.Koubová M, et al. Sex determination in Madagascar geckos of the genus Paroedura (Squamata: Gekkonidae): are differentiated sex chromosomes indeed so evolutionary stable? Chromosome Res. 2014;22:441–452. doi: 10.1007/s10577-014-9430-z. [DOI] [PubMed] [Google Scholar]
- 52.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 54.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 55.Le M, Reid BN, McCord WP, Naro-Maciel E, Raxworthy CJ, Amato G, Georges A. Resolving the phylogenetic history of the short–necked turtles, genera Elseya and Myuchelys (Testudines: Chelidae) from Australia and New Guinea. Mol. Phylogenet. Evol. 2013;68:251–258. doi: 10.1016/j.ympev.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 56.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ijdo JW, Wells RA, Baldini A, Reeders ST. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991;19:4780. doi: 10.1093/nar/19.17.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Endow SA. Polytenization of the ribosomal genes on the X and Y chromosomes of Drosophila melanogaster. Genetics. 1982;100:375–385. doi: 10.1093/genetics/100.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rovatsos M, Johnson Pokorná M, Altmanová M, Kratochvíl L. Female heterogamety in Madagascar chameleons (Squamata: Chamaeleonidae: Furcifer): differentiation of sex and neo-sex chromosomes. Sci. Rep. 2015;5:13196. doi: 10.1038/srep13196. [DOI] [PMC free article] [PubMed] [Google Scholar]