Abstract
Background
More than half of non-small cell lung cancer (NSCLC) patients present with metastatic disease at initial diagnosis with an estimated five-year survival rate of ~5%. Despite advances in understanding primary lung cancer oncogenesis metastatic disease remains poorly characterized. Recent studies demonstrate important roles of long non-coding RNAs (lncRNAs) in tumor physiology and as prognostic markers. Therefore, we present the first transcriptome analysis to identify lncRNAs altered in metastatic lung adenocarcinoma leading to the discovery and characterization of the lncRNA LCAL62 as a prognostic biomarker.
Patients and methods
RNA-Seq, microarray, nanoString expression, and clinical data from 1,116 LUAD patients across six independent cohorts and 83 LUAD cell lines were used to discover and evaluate the survival association of metastasis associated lncRNAs. Coexpression and gene set enrichment analyses were used to establish gene regulatory networks and implicate metastasis associated lncRNAs in specific biological processes.
Results
Our integrative analysis discovered LCAL62 as the most down-regulated lncRNA in metastasis. Further low LCAL62 expression promoted aggressive phenotypes and regulated genes associated with metastasis (such as metastasis repressor FOXA2). Low LCAL62 expression corresponded to poor overall patient survival across five independent lung adenocarcinoma cohorts (n = 881) including our own nanoString validation cohort.
Conclusion
We discovered that LCAL62 was down-regulated in lung cancer progression to promote invasive phenotypes, and lower expression was significantly associated with poor patient outcome and aggressive lung adenocarcinoma.
Keywords: Bioinformatics, Cancer research, Cell biology, Computational biology, Genetics, Molecular biology, Oncology, Long non-coding RNA, Transcriptome, Genomics, Lung cancer, Metastasis, Prognostic, Biomarker
Bioinformatics; Cancer research; Cell biology; Computational biology; Genetics; Molecular biology; Oncology; Long non-coding RNA; Transcriptome; Genomics; Lung cancer; Metastasis; Prognostic; Biomarker.
1. Introduction
Non-small cell lung cancer (NSCLC) accounts for ~85% of all lung cancers and is the leading cause of cancer-related deaths worldwide [1]. Greater than 50% of patients present with metastasis at initial diagnosis [2]. Furthermore, most patients with early stage and locally advanced NSCLC eventually recur with metastatic disease, despite undergoing potentially curative therapies. The five-year survival rate for metastatic disease is ~5% [1]. Current inadequacies for stratifying patients with aggressive disease presents a strong rationale to systematically identify biomarkers of aggressive lung cancer to improve patient diagnosis and care.
To date lung cancer treatment and research have primarily focused on the deregulation of protein-coding genes, thereby missing potentially important non-coding RNAs. Long non-coding RNAs have emerged as a biologically and clinically relevant class of transcripts contributing to tumorigenesis [3, 4]. We recently conducted a first-of-its-kind ab initio transcriptome assembly of publicly available RNA-Seq data from ~550 primary lung cancer patients to discover 111 lung cancer associated long non-coding RNAs (LCALs) altered in lung cancer [5]. Just as this study established the lung cancer lncRNA landscape in primary tumors for researchers to further explore potential mechanisms [6] and biomarker utility [7, 8], a similar systematic approach in lung cancer metastasis would improve our understanding of disease progression. Despite the availability of multiple RNA-Seq data collections derived from primary tumors [5], none of these cohorts include metastatic lung cancer specimens. While previous technologies such as microarrays were designed primarily to monitor protein-coding gene expression levels, some platforms can be exploited to monitor the expression of a subset of lncRNAs [9]. Therefore, our study leveraged RNA-Seq, microarray, and nanoString expression platforms to identify lncRNAs associated with lung cancer metastasis and their use as biomarkers for stratifying patients with poor outcome to ultimately improve patient care.
Through our integrative analysis we discovered 281 lncRNAs differentially expressed in lung adenocarcinoma (LUAD) metastasis. We found that low expression of LCAL62 (aliases LINC00261, onco-lncRNA-17 and DEANR1), the most down-regulated lncRNA in metastatic disease, promoted aggressive phenotypes in vitro. Furthermore, we found and validated that low expression of LCAL62 was associated with poor overall survival in LUAD patients from five independent cohorts totaling 881 patients. We also observed a strong coexpression between LCAL62 and FOXA2, a known tumor and metastasis suppressor of lung cancer [10]. Taken together, LCAL62 could serve as a biomarker of aggressive disease and patient outcome in LUAD.
2. Materials and methods
2.1. Datasets and patients
Six public gene expression datasets representing 5,320 tumor samples (1,035 LUAD patients, 83 LUAD cell lines, and 4,121 other cancer patients) were used for the discovery of metastasis associated lncRNAs, functional prediction, and survival analysis of LCAL62. An independent cohort of 81 LUAD patients were enrolled at Washington University School of Medicine in St. Louis, MO (WashU cohort). Primary tumor specimens were evaluated for LCAL62 and FOXA2 expression using the nanoString nCounter platform. Informed consent was obtained from all patients and the protocol was approved by the Washington University Institutional Review Board (IRB). Dataset details are provided in Table 1.
Table 1.
Summary of patient cohorts and cell line datasets used in this study.
| Dataset/cohort | GEO accession | Cancer | Patients | Patients w/usable clinical data | Samples (N-normal, P-primary, M-metastasis) | Platform | Ref. |
|---|---|---|---|---|---|---|---|
| Bittner | GSE2109 | LUAD | 82 | 0 | 82 (72P+10M) | Affymetrix HG U133 Plus 2.0 | |
| TCGA | NA | LUAD | 298 | 220 | 353 (55N + 298P) | RNA-Seq | [17] |
| Others | 4121 | Not evaluated | 4585 (464N + 4121P) | RNA-Seq | [17] | ||
| Schabath | GSE72094 | LUAD | 442 | 374 | 442P | Rosetta/Merck Human RSTA Custom Affymetrix 2.0 microarray | [18] |
| Der | GSE50081 | LUAD | 128 | 124 | 128P | Affymetrix HG U133 Plus 2.0 | [19] |
| Rousseaux | GSE30219 | LUAD | 85 | 82 | 85P | Affymetrix HG U133 Plus 2.0 | [20] |
| WashU | NA | LUAD | 81 | 81 | 81P | nanoString nCounter | |
| CCLE | NA | LUAD | 83 | 0 | 83 cell lines | RNA-Seq | |
| Total | all tissues | 5237 | |||||
| Total | LUAD tissue | 1116 | 881 | ||||
| Total | LUAD cell lines | 83 | |||||
| Total | all tissues & cell lines | 5320 | |||||
2.2. Bioinformatics analysis
Microarray probes were repurposed by realigning their sequences against the transcripts to identify probes uniquely mapped to a lncRNA. TCGA RNA-Seq pre-alignments and microarray pre-normalized probe expression were used for subsequent analyses. Differential expression analysis was performed using the negative binomial model for RNA-Seq raw counts and the two-sample t-test for microarray data. Digital transcript counts from nanoString nCounter assays were normalized according to nanoString nCounter data analysis manual. The Kaplan-Meier product limit method and Cox proportional hazard model was used to evaluate association between gene expression and patient overall survival. More details of the bioinformatics methods can be found in the Supplementary Methods.
2.3. Experimental characterization
Experimental methods are detailed in the Supplemental Methods.
3. Results
3.1. Discovery of down-regulated lncRNA LCAL62 in metastatic lung adenocarcinoma
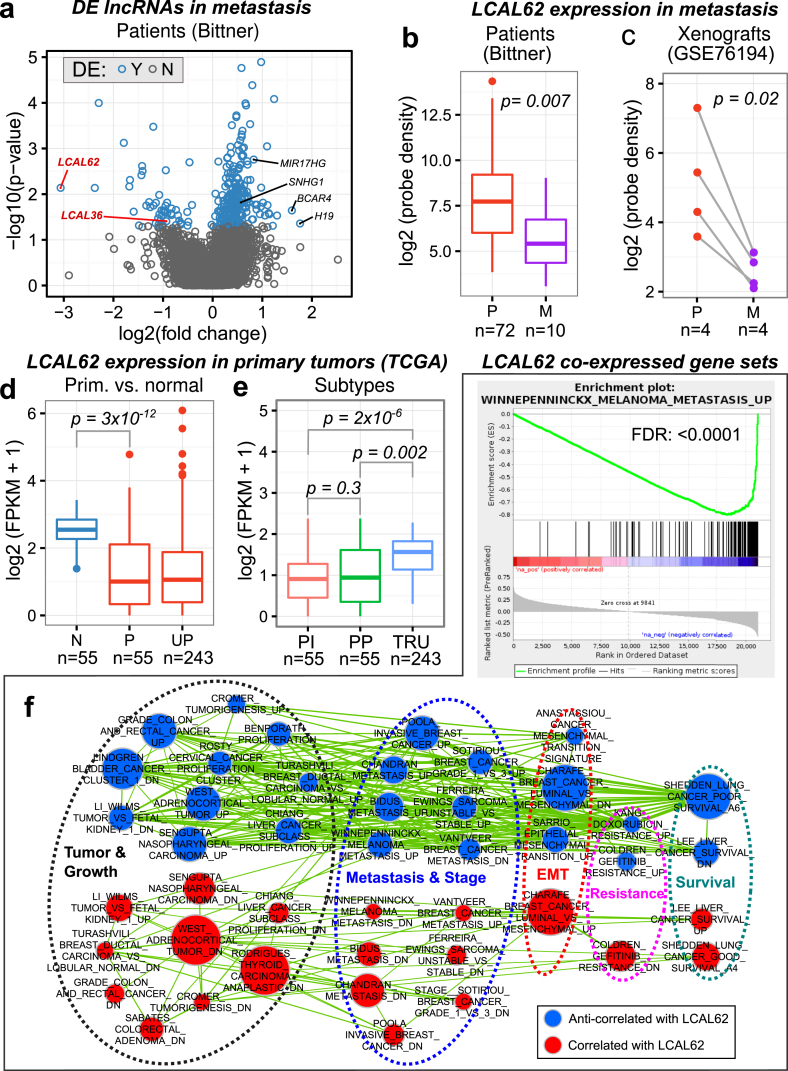
To identify lncRNAs altered in metastatic LUAD we leveraged a publicly available microarray gene expression dataset, referred to as the ‘Bittner cohort’ (GSE2109), that included 72 primary tumors and 10 unmatched metastatic tumors from brain (2), adrenal gland (3), cerebellar tissue (1), small bowel (1), soft tissue of the arm (1), and secondary lung tumor (2). We identified 5,357 probe sets in the microarray platforms that uniquely mapped to 4,443 lncRNA genes compiled from multiple annotation sources (Supplementary Methods). Comparison of primary and metastatic tumors from the Bittner cohort identified 281 lncRNA genes differentially expressed in metastasis including 235 up-regulated and 49 down-regulated lncRNAs (Figure 1a). We found up-regulated lncRNAs known to promote tumor proliferation and metastasis in lung and other cancers such as H19 [11], BCAR4 [12], SNHG1 [13], and MIR17HG [14]. Our previous study, using three independent RNA-Seq cohorts, characterized 111 lncRNAs dysregulated in non-small cell lung cancer primary tumors compared to matched normal tissues, termed LCALs, including 78 lncRNAs dysregulated in LUAD [5]. We hypothesized that a subset of LCALs may be dysregulated in metastasis compared to primary tumors. To select a candidate for subsequent characterization we prioritized lncRNAs that are altered throughout lung cancer progression by overlapping the metastasis-associated lncRNAs with our previously identified primary-associated lncRNAs, LCALs. A total of 43 LCALs were uniquely annotated with at least one probe set from the Bittner cohort microarray. We discovered LCAL62 had the most dramatic decreased expression in metastasis (Figure 1a,b). Additional analysis confirmed that LCAL62 was downregulated in bone metastasis of patient derived xenografts (Figure 1c). LCAL62 was also down-regulated in primary tumors compared with normal tissues (Figure 1d).
Figure 1.
Low expression of LCAL62 associates with metastasis and cancer aggressiveness. (a) Volcano plot of the lncRNAs in the metastasis versus primary tumor comparison from the Bittner cohort. LCAL62 is the most down-regulated lncRNA in metastasis; (b) Expression of LCAL62 in patients (Bittner) and (c) patient derived cell lines and xenografts (GSE76194, P - patient primary tumor derived cell lines; M - matched bone metastases developed in mouse xenograft model); (d) Expression of LCAL62 in primary tumors (TCGA RNA-Seq) compared with normal tissues (N - normal, P - matched primary, UP - unmatched primary) and (e) grouped by expression-based subtypes (PI: proximal-inflammatory/squamoid, PP: proximal-proliferative/magnoid, TRU: terminal respiratory unit/bronchioid); (f) Cancer related gene sets enriched in LCAL62 GSEA analysis (top - enrichment plot of a representative metastatic gene set in melanoma negatively co-expressed with LCAL62, bottom - network of enriched gene sets). Nodes represent gene sets that are significantly correlated (red) or anti-correlated (blue) with LCAL62 expression, grouped by manually curated categories (dashed circles). Node size represents the number of genes in the corresponding gene set. Edge width represents the number of genes shared between gene sets.
Initial characterization of the TCGA LUAD cohort identified lower LCAL62 expression in proximal-inflammatory and proximal-proliferative expression-based subtypes (Figure 1e). To further investigate the involvement of LCAL62 in tumor progression and metastasis, we employed a ‘guilt-by-association’ approach that assumes a lncRNA is likely involved in similar biological processes as coexpressing protein-coding genes [15, 16]. We found that low LCAL62 expression was correlated with genes up-regulated in tumor growth and proliferation, tumor stage and metastasis, epithelial mesenchymal transition (EMT), and drug resistance (Figure 1f; FDR adjusted p < 0.001).
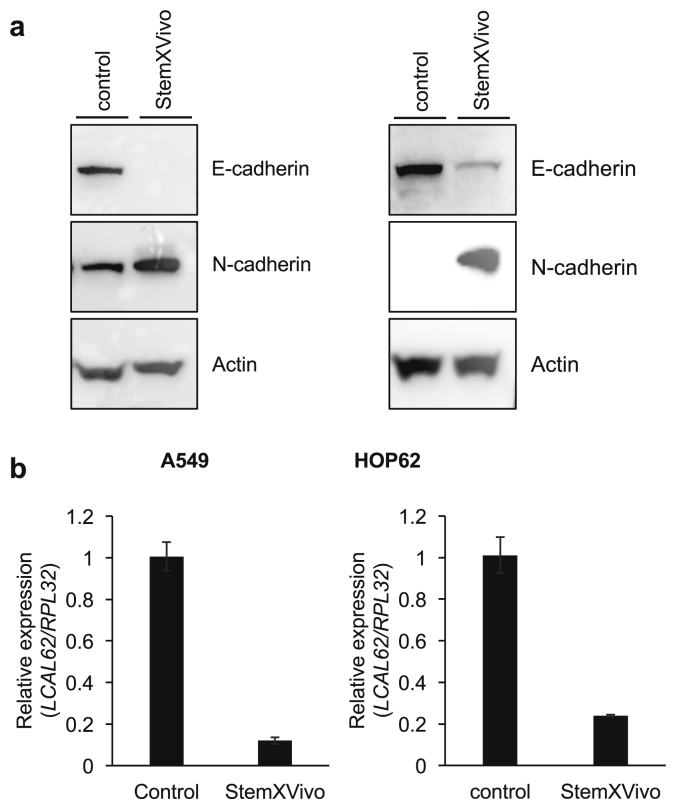
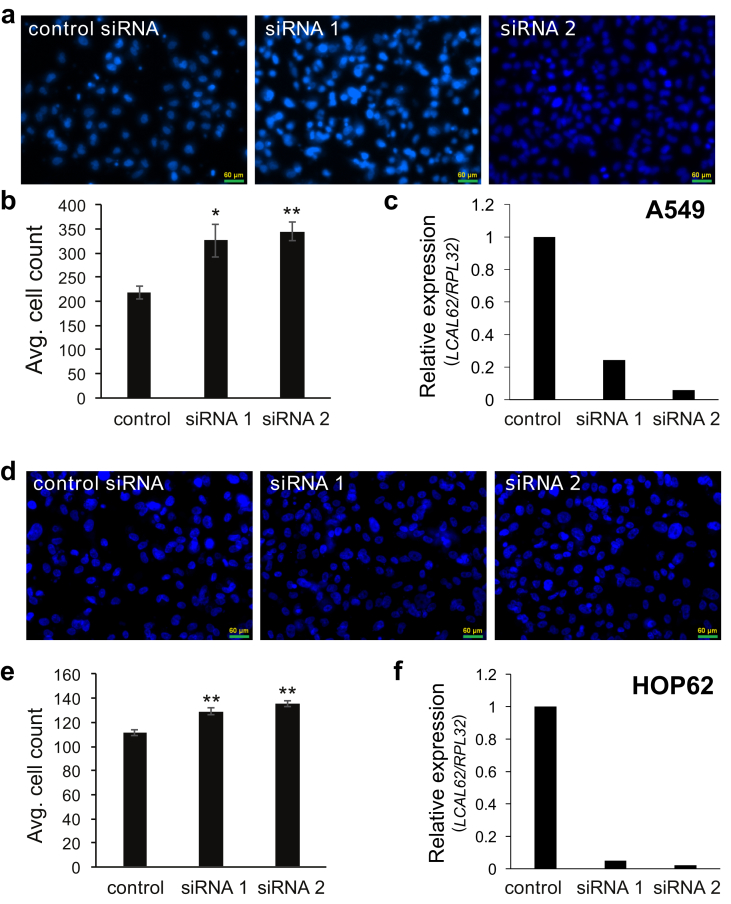
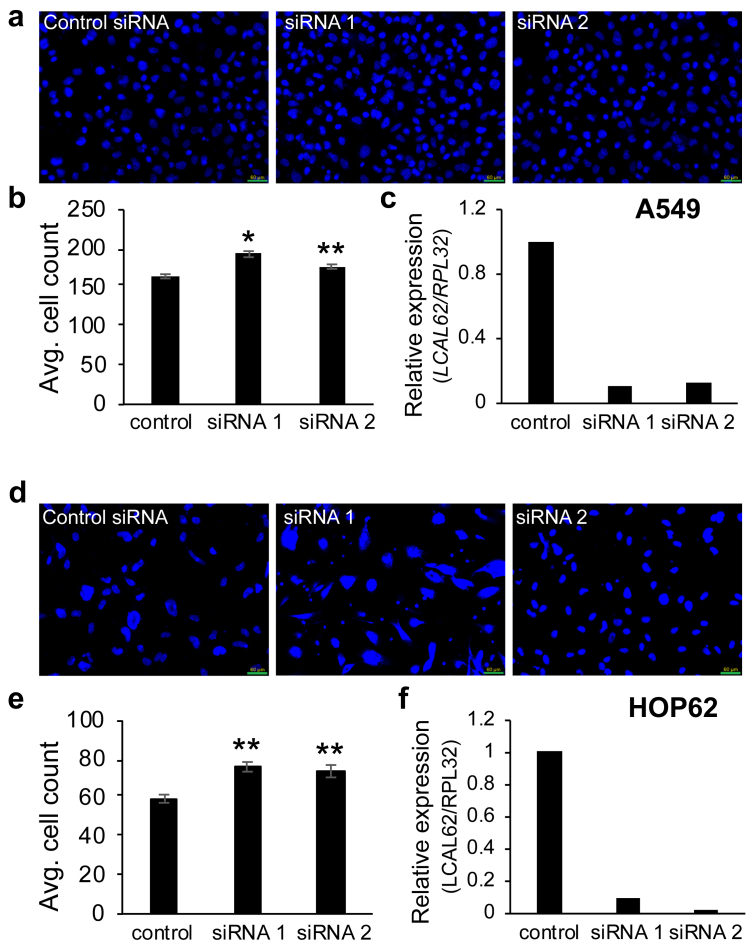
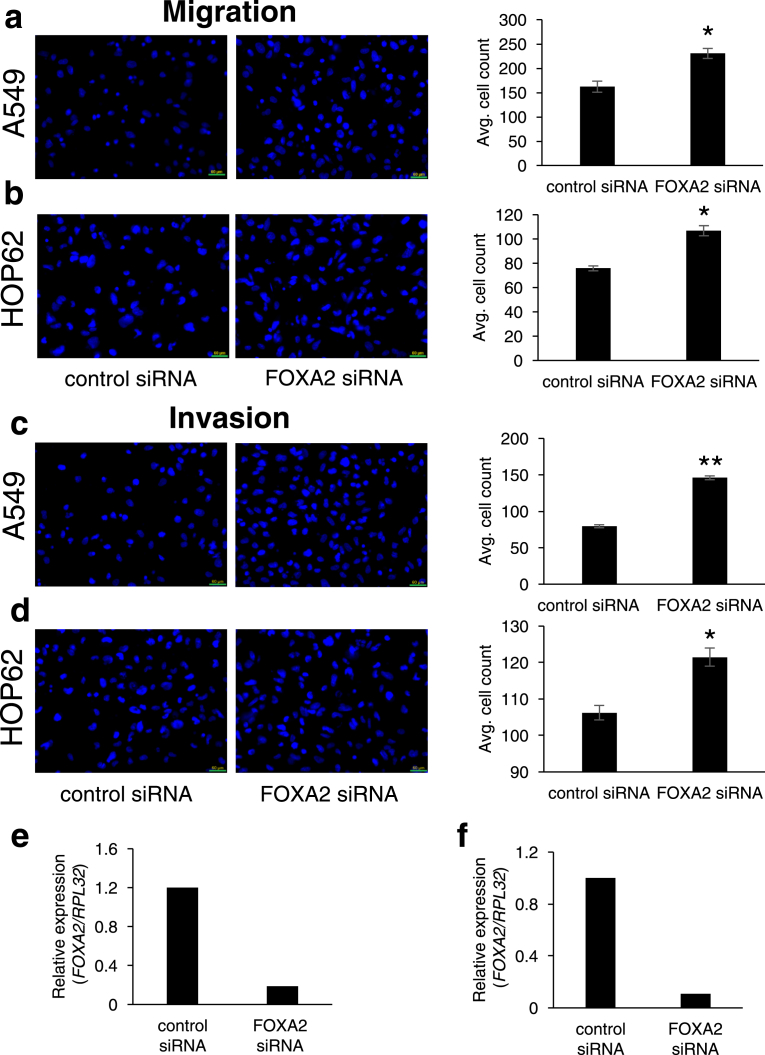
To further demonstrate if the lack of LCAL62 promotes aggressive disease, lung cancer cells were cultured with StemXVivo (R&D Systems) to induce an epithelial to mesenchymal transition (EMT) as determined by changes in E-cadherin and N-cadherin protein levels, hallmark events of EMT (Figure 2a). EMT-induced A549 and HOP62 cells showed a dramatic 80% decrease in LCAL62 expression compared to untreated cells (Figure 2b). Further, silencing LCAL62 in A549 cells increased cellular motility relative to control cells expressing LCAL62. After knockdown of LCAL62, with two independent siRNAs, cells were seeded on a transwell membrane in a modified Boyden Chamber assay. After 18 h there was a significant 49% and 57.5% increase (p < 0.02, p < 0.001) in migrated cells with siRNA 1 and siRNA 2 (at least 80% decreased LCAL62 expression), respectively, compared to control transfected cells (Figure 3a, b, c). In HOP62 cells, there was a 16.2% and 22.5% increase (p < 0.001) in migrated cells with siRNA 1 and siRNA 2 (at least 80% decreased LCAL62 expression) compared to control transfected cells (Figure 3d, e, f). Further silencing LCAL62 in both cell lines increased cellular invasion compared to LCAL62 expressing cells. There was a 17.3% (p < 0.02) and 7.6% (p < 0.001) increase in invaded cells with siRNA 1 and siRNA 2 in A549 cells (Figure 4a, b, c) and a 27.7% and 23.8% (p < 0.001) increase in invaded cells with siRNA 1 and siRNA 2 in HOP62 cells (Figure 4d, e, f). These data support the in silico predictions of decreased LCAL62 expression promoting aggressive phenotypes in LUAD.
Figure 2.
Low LCAL62 expression in metastatic cells. (a) Western blot of A549 (left) and HOP62 (right) protein lysates collected after 5-day treatment with StemXVivo showing EMT hallmark proteins (E-cadherin and N-cadherin) altered during epithelial to mesenchymal transition. (b) Inducing epithelial to mesenchymal transition in the presence of StemXVivo supplement decreased LCAL62 expression in A549 (left) and HOP62 (right) cell lines as measured by quantitative RT-PCR (qRT-PCR); n of 2. Full non-adjusted images of the Western blots in this figure are provided in Fig. S1.
Figure 3.
Low expression of LCAL62 increases cellular migration in lung cancer cell lines. (a,b) A549 cells were seeded on a transwell and DAPI-stained cells quantified; (c) qRT-PCR confirmed knockdown of LCAL62 in A549 using two independent siRNAs. ∗p < 0.02, ∗∗p < 0.001; n of 3. (d–f) Similar results for HOP62 cells.
Figure 4.
Low expression of LCAL62 increases cellular invasion in lung cancer cell lines. (a,b) A549 cells were seeded on a Matrigel-coated transwell and DAPI-stained cells quantified. (c) qRT-PCR confirmed knockdown of LCAL62 in A549 using two independent siRNAs. ∗p < 0.02, ∗∗p < 0.001; n of 3; (d–f) Similar results for HOP62 cells.
3.2. Low LCAL62 expression is associated with poor overall survival
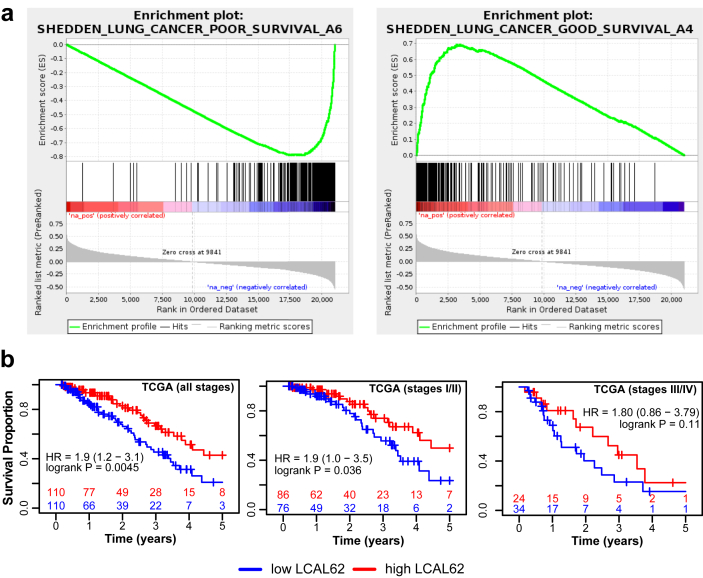
Our ‘guilt-by-association’ analysis revealed a striking enrichment of gene signatures for lung cancer survival (Figure 5a) suggesting that lower LCAL62 expression is associated with poor clinical outcome. Building upon this, we evaluated the clinical significance of LCAL62 on patient overall survival within the TCGA LUAD cohort (n = 220, RNA-Seq) [17]. Most patients in TCGA LUAD cohort presented with early stage disease. As expected, there was an association between stage and outcome (p < 0.001, Table 2). We performed survival analysis stratifying patients by median LCAL62 expression and stage and found that decreased expression of LCAL62 was significantly associated with poor overall survival in LUAD patients (p = 0.04, HR = 1.9; Figure 5b). The survival effect was similar in early and late stages although 5% significance was not reached in stage III/IV due to the small sample size.
Figure 5.
Discovery of the association of low LCAL62 expression with poor overall survival in TCGA cohort. (a) LCAL62 co-expressed with gene signature of lung cancer survival; (b) Low expression of LCAL62 correlated with poor survival in TCGA LUAD cohort.
Table 2.
Association of clinical variables with overall survival across cohorts.
| Variable | TCGA |
Schabath |
Der |
Rousseaux |
WashU |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | HR∗ | p | n | HR∗ | p | n | HR∗ | p | n | HR∗ | p | n | HR∗ | p | |
| Age | 0.83–2.14 | 0.24 | 0.87–2.17 | 0.17 | 0.77–2.73 | 0.25 | 1.1–3.8 | 0.02 | 0.7–1.9 | 0.57 | |||||
| <65 | 92 | 103 | 40 | 27 | 55 | ||||||||||
| ≥65 | 128 | 271 | 84 | 55 | 27 | ||||||||||
| Gender | 0.52–1.30 | 0.41 | 1.1–2.4 | 0.016 | 0.81–2.47 | 0.22 | 0.5–2.3 | 0.83 | 1.1–3.0 | 0.02 | |||||
| Male | 97 | 164 | 64 | 64 | 37 | ||||||||||
| Female | 123 | 210 | 60 | 18 | 44 | ||||||||||
| Stage | 1.7–4.3 | 1.6E-6 | 1.9–4.4 | 1.6E-7 | NA | NA | NA | NA | NA | NA | |||||
| I & II | 162 | 305 | 124 | 80 | 81 | ||||||||||
| III & IV | 58 | 69 | 0 | 2 | |||||||||||
95% confidence interval.
3.3. Microarray data across three independent cohorts confirms LCAL62 correlation with patient outcome
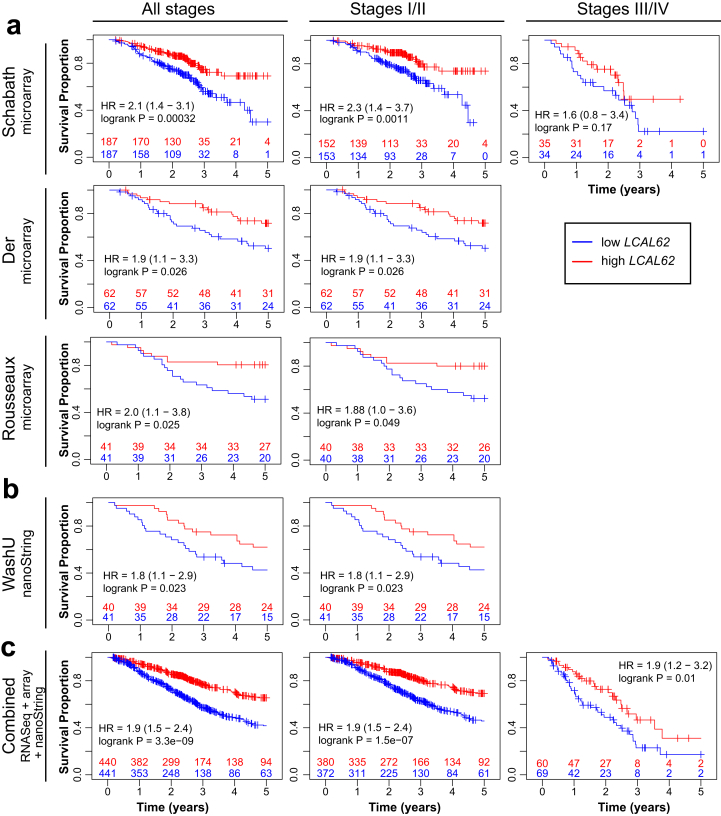
We confirmed association of LCAL62 expression with patient outcome by leveraging public microarray data from three independent LUAD cohorts totaling 583 patients (Figure 6a, Table 1). These cohorts included the Schabath cohort (n = 374) [18], Der cohort (n = 124) [19], and Rousseaux cohort (n = 85) [20] that leveraged two microarray platforms (Table 1). The majority of patients in these cohorts presented with early stage LUAD (Table 2). Low expression of LCAL62 was strongly and consistently associated with poor overall survival in all three cohorts (Schabath, p = 0.0003, HR = 1.7; Der, p = 0.026, HR = 1.9; Rousseaux, p = 0.025, HR = 2.0) (Figure 6a).
Figure 6.
Orthogonal and independent validation of the association of low LCAL62 expression with poor overall survival. (a) Public microarray datasets; (b) WashU nanoString cohort; (c) Combination of all cohorts including microarray, nanoString, and TCGA RNA-Seq. Kaplan-Meier curves for late stage (III/IV) are not generated for Der, WashU (no late stage patients), and Rousseaux (only 2 late stage patients) cohorts.
3.4. Orthogonal validation of LCAL62 correlation with patient outcome using nanoString
In addition to publicly available microarray datasets, we further validated the prognostic significance of LCAL62 in our independent WashU cohort (n = 81, stages I/II) using the nanoString nCounter platform. We confirmed low expression of LCAL62 was associated with poor overall survival in the WashU cohort (p = 0.023, HR = 1.9; Figure 6b). When patients from all cohorts were combined a significant association was observed between low LCAL62 expression and poor survival in both early and late stages (p = 3.3 × 10−9, HR = 1.9, Figure 6c). Overall, our analysis using three independent platforms (RNA-Seq, microarray, and nanoString) across five independent cohorts (n = 881) revealed a consistent association of LCAL62 expression with overall survival.
3.5. LCAL62 association with FOXA2
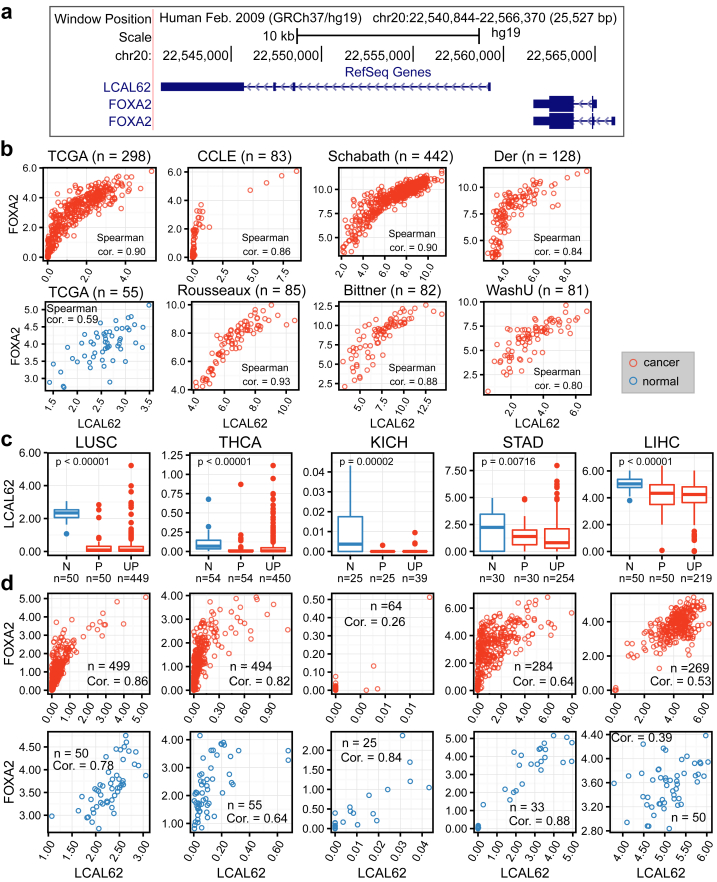
To identify LCAL62 target genes, we performed a coexpression analysis on six independent LUAD gene expression datasets (Table 1). The most highly coexpressed protein-coding gene with LCAL62 was FOXA2 (Forkhead Box A2), a gene encoding a transcription factor upstream of LCAL62 (Figure 7a) that is known to repress tumor growth and metastasis in multiple cancers including lung cancer [10]. Spearman correlation coefficient of expression between LCAL62 and FOXA2 ranged from 0.84 to 0.93 (Figure 7b). We further validated the strong correlation between LCAL62 and FOXA2 expression using the nanoString platform in our independent cohort (WashU cohort, Spearman correlation = 0.80, Figure 7b).
Figure 7.
LCAL62 down-regulation and correlation with FOXA2 are conserved across cancer types. (a) UCSC Genome browser screenshot of the LCAL62 and FOXA2 region; (b) Correlation of expression of LCAL62 and FOXA2 in lung adenocarcinoma across cohorts (TCGA, CCLE - RNA-Seq; Schabath, Der, Rousseaux; Bittner - microarray; WashU - nanoString); (c) Down-regulation of LCAL62 and (d) correlation with FOXA2 in other cancer types (TCGA RNA-Seq, top - tumor, bottom - normal). Expression is measured by log2(FPKM+1) in RNA-Seq data and log2 (probe intensity) in microarray data; N - normal, P - matched primary, UP - unmatched primary.
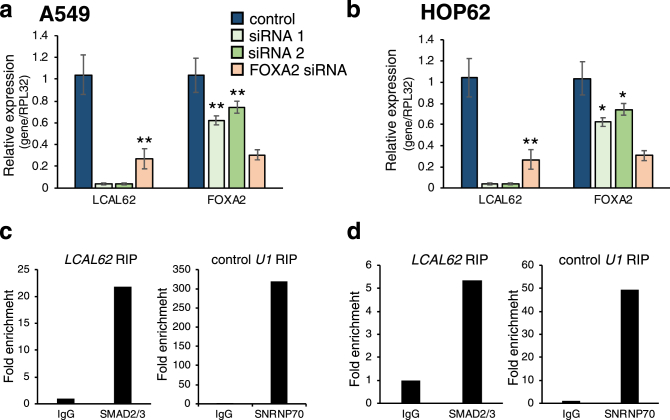
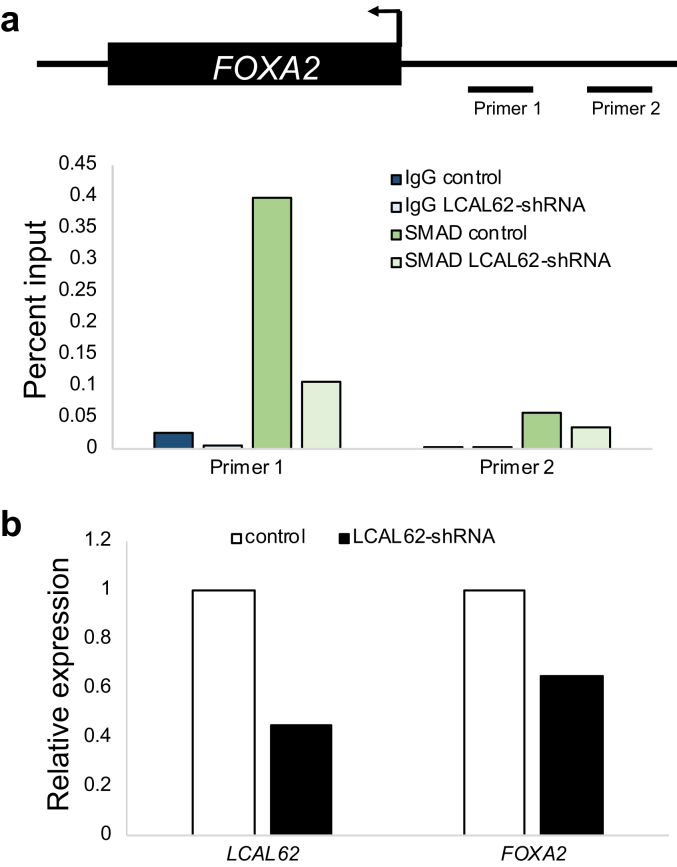
Since cis-regulation of nearby genes is a common mechanism of lncRNA gene regulation [21] we investigated the relationship between LCAL62 and FOXA2. After silencing LCAL62 (at least 90% knockdown) with two independent siRNAs there was a 37.8% and 25.8% (p < 0.05), respectively, decrease in FOXA2 expression in A549 cells (Figure 8a) and a 26.9% and 34.1% (p < 0.0005), respectively, decrease in FOXA2 expression in HOP62 cells (Figure 8b). Interestingly, silencing FOXA2 also caused a decrease in LCAL62 expression (p < 0.0005; Figure 8a,b). Moreover, silencing FOXA2 (with 80% knockdown) in our lung cancer cell models increased cellular migration and invasion similarly to phenotypes observed in LCAL62-silenced cells (Figure 9). A previous study found that LCAL62 positively regulated FOXA2 in endoderm differentiation by recruiting the SMAD2/3 transcription factor to the FOXA2 promoter [22]. We hypothesized this regulatory mechanism would be conserved in the lung which is endoderm derived. In lung cancer models, we determined that LCAL62 bound to SMAD2/3 with a 20-fold and 5-fold enrichment in the A549 and HOP62 cell lines, respectively, as determined by RNA immunoprecipitation (RIP) (Figure 8c,d). Further, A549 cells with stably silenced LCAL62 showed decreased occupancy of SMAD2/3 at the FOXA2 promoter (Figure 10). These data highlight that LCAL62 may control transcription of FOXA2 through its interaction with SMAD2/3.
Figure 8.
LCAL62 regulates FOXA2 expression in lung cancer cell lines. qRT-PCR determined a decrease in FOXA2 expression upon silencing of LCAL62 relative to a scrambled control siRNA and normalized to the control housekeeping gene, RPL32 in (a) A549 and (b) HOP62 cell lines. RNA immunoprecipitation (RIP) assay coupled to qRT-PCR showed LCAL62 binding to SMAD2/3 relative to IgG control in (c) A549 and (d) HOP62 cells. As a positive control, U1 was enriched for its binding partner, SNRNP70, in RIP assay. ∗p < 0.05, ∗∗p < 0.0005, n of 2.
Figure 9.
Low expression of FOXA2 increases aggressive phenotypes in lung cancer models. Increased cellular migration and invasion in (a,c) A549 and (b,d) HOP62 cells. qRT-PCR confirmed knockdown of FOXA2 in (e) A549 and (f) HOP62 cells for assays. ∗p < 0.0005 ∗∗p < 0.00001, n of 3.
Figure 10.
Knockdown of LCAL62 decreases SMAD2/3 binding at the FOXA2 promoter. (a) ChIP-qPCR shows SMAD2/3 occupancy at two genomic locations of the FOXA2 promoter in wild type and LCAL62-shRNA A549 cells. IgG was used as a negative control. Primer locations are depicted in the schematic above. n of 2 (b) qRT-PCR confirmation of expression of LCAL62 and FOXA2 in cell lines.
In contrast to the tissue-specific expression profiles commonly observed for lncRNAs [15, 23], we have observed that ~40% of LCALs were altered in multiple cancers suggestive of their conserved oncogenic roles across cancer types [5, 16]. In addition to LUAD, LCAL62 was down-regulated in lung squamous cell carcinoma (LUSC) under the gene alias onco-lncRNA-17 [5,16]. Therefore, we expanded our analysis to incorporate 12 cancer types using 4,938 TCGA RNA-Seq samples and found that the expression of LCAL62 was also decreased in thyroid cancer (THCA), kidney cancer (KICH), stomach cancer (STAD), and liver cancer (LIHC) (Figure 7c). Furthermore, coexpression analysis revealed that LCAL62 and FOXA2 were highly correlated (in both tumor and normal tissues) across multiple cancer types with LCAL62 differential expression (Figure 7d). LCAL62 and FOXA2 were the top correlated genes in LUAD, LUSC, STAD, and THCA. These results suggest LCAL62 has a conserved role regulating FOXA2 across cancers.
4. Discussion
Our comprehensive and systematic analysis of metastatic lung cancer is the first to show previously characterized non-coding RNAs in other cancers (i.e., BCAR4 and MIR17HG) [12, 14] are also up-regulated in metastatic lung cancer. Our systematic approach also revealed additional uncharacterized lncRNAs that could serve as the basis for subsequent mechanistic studies dissecting the role of lncRNAs in lung cancer metastasis. We prioritized LCAL62 since the lack of LCAL62 promoted aggressive phenotypes, associated with metastasis gene signatures, and correlated with patient outcome. We previously discovered LCAL62 as a down-regulated lncRNA in primary lung tumors via large-scale RNA-Seq analysis. While this current work was under review, several studies showed LCAL62 association with prognosis and function in vivo [24, 25, 26, 27]. However, these studies evaluated limited numbers of patients and utilized single transcriptomic platforms. Here we provide the most comprehensive and systematic evaluation of LCAL62 expression and association with prognosis in the largest compendium of patients to date (n = 1,116) from six multi-institutional and independent cohorts. Notably, low LCAL62 expression consistently correlated with patient outcome across 881 LUAD patients from five independent cohorts. Further supporting the robust nature of this association, we observed low LCAL62 expression correlated with overall survival using microarray, RNA-Seq, and nanoString expression platforms. Within individual cohorts we observed a stronger association within early stage compared with late stage patients, however this may be due to the limited number of late stage patients. However, despite having only 69 and 58 late stage patients in the Schabath and TCGA cohorts, respectively, we still observed an association between low LCAL62 expression and poor overall survival. When the late stage patients from all cohorts were combined, a significant association of low LCAL62 and poor overall survival was observed supporting that LCAL62 is associated with patient outcome independent of stage. Additionally, we showed that LCAL62 was associated with recurrence-free survival despite a limited number of patients (n = 387, 44%) with recurrence-free follow-up (p = 0.00045, HR = 1.8, combined cohort, Table 3).
Table 3.
Association of LCAL62 and FOXA2 expression with overall and recurrence-free survival across cohorts.
| Cohort | Overall survival |
Recurrence-free survival |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | LCAL62 |
FOXA2 |
n | LCAL62 |
FOXA2 |
|||||
| HR∗ | p | HR∗ | p | HR∗ | p | HR∗ | p | |||
| TCGA | 220 | 1.2–3.1 | 0.004 | 1.0–2.5 | 0.051 | 132 | 1.03–3.7 | 0.036 | 0.7–2.4 | 0.440 |
| Schabath | 374 | 1.4–3.1 | 0.0003 | 1.2–2.6 | 0.006 | 0 | NA | NA | NA | NA |
| Der | 124 | 1.1–3.3 | 0.026 | 1.1–3.3 | 0.023 | 124 | 0.8–3.0 | 0.164 | 0.8–2.8 | 0.261 |
| Rousseaux | 82 | 1.1–3.8 | 0.025 | 0.9–3.1 | 0.135 | 81 | 1.03–5.5 | 0.036 | 0.7–3.5 | 0.277 |
| WashU | 81 | 1.1–2.9 | 0.023 | 0.9–2.2 | 0.149 | 50 | 0.8–2.5 | 0.276 | 1.1–3.6 | 0.028 |
| Combined | 881 | 1.5–2.4 | 3.3E-09 | 1.3–2.0 | 1.6E-05 | 387 | 1.3–2.5 | 0.00045 | 1.03–2.0 | 0.031 |
95% confidence interval.
Through computational prediction of LCAL62 function, we found gene signatures supporting an association of LCAL62 with metastasis. Our in vitro findings support our clinical observations by demonstrating that initiating EMT in lung cancer models resulted in a dramatic decrease in LCAL62 expression, and silencing LCAL62 expression resulted in increased cellular motility and invasiveness. Our analysis also revealed the consistent coexpression across 1,116 lung cancer patients between LCAL62 and the protein-coding gene, FOXA2, which is implicated as a metastasis suppressor in LUAD [10]. FOXA2 expression also had an association (although not as strong as LCAL62) with overall survival (p = 1.6 × 10−5, HR = 1.6, combined cohort) and recurrence-free survival (p = 0.031, HR = 1.4, combined cohort, Table 3). The interplay between LCAL62 and FOXA2 was experimentally validated by silencing LCAL62 and observing decreased expression of FOXA2. Additionally, we determined that LCAL62 bound to SMAD2/3 to regulate FOXA2 expression in lung cancer, a relationship previously shown in definitive endoderm [22]. In contrast to the up-regulation of LCAL62 during endoderm differentiation, we observed a decrease in LCAL62 in metastatic samples potentially corresponding to dedifferentiation. However, subsequent mechanistic studies are necessary to determine how the loss of LCAL62 confers malignant phenotypes. We also believe this relationship extends beyond lung cancer. Our pan-cancer analysis revealed consistent coexpression between LCAL62 and FOXA2 in multiple cancer types suggesting a conserved regulatory interaction between these genes. Collectively, our in silico and in vitro results suggest that LCAL62 regulates FOXA2 to potentially help suppress metastasis across cancer types.
Overall, the poor patient outcome in advanced LUAD highlights a critical need for molecular biomarkers that can stratify patients with aggressive disease to improve patient care. Through our integrative analysis of metastatic lung cancer samples we have discovered that LCAL62 is down-regulated in lung cancer progression, loss of LCAL62 expression increases invasive phenotypes, and LCAL62 can serve as a prognostic biomarker of overall survival that could have potential clinical utility.
Declarations
Author contribution statement
Ha X. Dang, Nicole M. White, Christopher A. Maher: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Emily B. Rozycki, Brooke M. Felsheim: Performed the experiments; Analyzed and interpreted the data.
Mark A. Watson, Ramaswamy Govindan: Contributed reagents, materials, analysis tools or data.
Jingqin Luo: Analyzed and interpreted the data.
Funding statement
Christopher A. Maher was supported by a LUNGevity Career Development Award, an American Lung Association Biomedical Research Grant, and an NIH NCI R01 CA203995. Nicole M. White was supported by an American Lung Association/American Thoracic Society Senior Research Training Fellowship.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank Patricia Alldredge and Kalin Guebert for obtaining patient clinical data for the WashU cohort.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Molina J.R., Yang P., Cassivi S.D. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 2008;83(5):584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Canc. Discov. 2011;1(5):391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White N.M., Zhao S.G., Zhang J. Multi-institutional analysis shows that low PCAT-14 expression associates with poor outcomes in prostate cancer. Eur. Urol. 2017;71(2):257–266. doi: 10.1016/j.eururo.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 5.White N.M., Cabanski C.R., Silva-Fisher J.M. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15(8):429. doi: 10.1186/s13059-014-0429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang G., Chen H., Liu J. The long noncoding RNA LINC01207 promotes proliferation of lung adenocarcinoma. Am. J. Cancer Res. 2015;5(10):3162–3173. [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L., He Y., Liu W. Non-coding RNA LINC00857 is predictive of poor patient survival and promotes tumor progression via cell cycle regulation in lung cancer. Oncotarget. 2016;7(10):11487–11499. doi: 10.18632/oncotarget.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z., Jin Y., Ren H. Downregulation of the long non-coding RNA TUSC7 promotes NSCLC cell proliferation and correlates with poor prognosis. Am. J. Transl. Res. 2016;8(2):680–687. [PMC free article] [PubMed] [Google Scholar]
- 9.Du Z., Fei T., Verhaak R.G.W. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 2013;20(7):908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Y., Shu G., Yuan X. FOXA2 functions as a suppressor of tumor metastasis by inhibition of epithelial-to-mesenchymal transition in human lung cancers. Cell Res. 2011;21(2):316–326. doi: 10.1038/cr.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matouk I.J., Raveh E., Abu-lail R. Oncofetal H19 RNA promotes tumor metastasis. Biochim. Biophys. Acta. 2014;1843(7):1414–1426. doi: 10.1016/j.bbamcr.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 12.Xing Z., Lin A., Li C. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159(5):1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui Y., Zhang F., Zhu C. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/β-catenin signaling pathway. Oncotarget. 2017;8(11):17785–17794. doi: 10.18632/oncotarget.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mogilyansky E., Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20(12):1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttman M., Amit I., Garber M. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabanski C.R., White N.M., Dang H.X. Pan-cancer transcriptome analysis reveals long noncoding RNAs with conserved function. RNA Biol. 2015;12(6):628–642. doi: 10.1080/15476286.2015.1038012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research Network. Weinstein J.N., Collisson E.A. The cancer Genome atlas pan-cancer analysis project. Nat. Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schabath M.B., Welsh E.A., Fulp W.J. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene. 2016;35(24):3209–3216. doi: 10.1038/onc.2015.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Der S.D., Sykes J., Pintilie M. Validation of a histology-independent prognostic gene signature for early-stage, non-small-cell lung cancer including stage IA patients. J. Thorac. Oncol. 2014;9(1):59–64. doi: 10.1097/JTO.0000000000000042. [DOI] [PubMed] [Google Scholar]
- 20.Rousseaux S., Debernardi A., Jacquiau B. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci. Transl. Med. 2013;5(186) doi: 10.1126/scitranslmed.3005723. 186ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang W., Liu Y., Liu R. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep. 2015;11(1):137–148. doi: 10.1016/j.celrep.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer M.K., Niknafs Y.S., Malik R. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhamija S., Becker A.C., Sharma Y. LINC00261 and the adjacent gene FOXA2 are epithelial markers and are suppressed during lung cancer tumorigenesis and progression. Noncoding RNA. 2018;5(1) doi: 10.3390/ncrna5010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi J., Ma H., Wang H. Overexpression of LINC00261 inhibits non-small cell lung cancer cells progression by interacting with miR-522-3p and suppressing Wnt signaling. J. Cell. Biochem. 2019;120(10):18378–18387. doi: 10.1002/jcb.29149. [DOI] [PubMed] [Google Scholar]
- 26.Shahabi S., Kumaran V., Castillo J. LINC00261 is an epigenetically regulated tumor suppressor essential for activation of the DNA damage response. Cancer Res. 2019;79(12):3050–3062. doi: 10.1158/0008-5472.CAN-18-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao J., Dong L.-P. Linc00261 suppresses growth and metastasis of non-small cell lung cancer via repressing epithelial-mesenchymal transition. Eur. Rev. Med. Pharmacol. Sci. 2019;23(9):3829–3837. doi: 10.26355/eurrev_201905_17810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.