Abstract
Powdery mildew caused by Blumeria graminis f. sp. hordei (Bgh) is one of the main foliar diseases in barley (Hordeum vulgare L.; Hv). Naturally occurring resistance genes used in barley breeding are a cost effective and environmentally sustainable strategy to minimize the impact of pathogens, however, the primary gene pool of H. vulgare contains limited diversity owing to recent domestication bottlenecks. To ensure durable resistance against this pathogen, more genes are required that could be unraveled by investigation of secondary barley gene-pool. A large set of Hordeum bulbosum (Hb) introgression lines (ILs) harboring a diverse set of desirable resistance traits have been developed and are being routinely used as source of novel diversity in gene mapping studies. Nevertheless, this strategy is often compromised by a lack of recombination between the introgressed fragment and the orthologous chromosome of the barley genome. In this study, we fine-mapped a Hb gene conferring resistance to barley powdery mildew. The initial genotyping of two Hb ILs mapping populations with differently sized 2HS introgressions revealed severely reduced interspecific recombination in the region of the introgressed segment, preventing precise localization of the gene. To overcome this difficulty, we developed an alternative strategy, exploiting intraspecific recombination by crossing two Hv/Hb ILs with collinear Hb introgressions, one of which carries a powdery mildew resistance gene, while the other doesn’t. The intraspecific recombination rate in the Hb-introgressed fragment of 2HS was approximately 20 times higher than it was in the initial simple ILs mapping populations. Using high-throughput genotyping-by-sequencing (GBS), we allocated the resistance gene to a 1.4 Mb interval, based on an estimate using the Hv genome as reference, in populations of only 103 and 146 individuals, respectively, similar to what is expected at this locus in barley. The most likely candidate resistance gene within this interval is part of the coiled-coil nucleotide-binding-site leucine-rich-repeat (CC-NBS-LLR) gene family, which is over-represented among genes conferring strong dominant resistance to pathogens. The reported strategy can be applied as a general strategic approach for identifying genes underlying traits of interest in crop wild relatives.
Keywords: introgression lines, Hordeum bulbosum, recombination, mapping, powdery mildew, resistance, crop wild relative
Introduction
The increased genetic uniformity of cultivated crops, makes them highly vulnerable to various biotic and abiotic stresses, leading to crop yield losses and serious food security issues (Hoisington et al., 1999). Disease resistance breeding is a cost effective and environmentally sustainable strategy for minimizing the damage caused by plant pathogens. Hence, plant breeders are continuously working to discover novel sources of genetic resistance. Crop wild relatives (CWRs) are a reservoir of genetic variation providing an important source of novel alleles for the genetic improvement of cultivated species. Crosses between cultivars and CWRs have been carried out in several crop species to unlock this favorable genetic diversity (Tanksley and McCouch, 1997; Feuillet et al., 2008). Some prominent examples of the introgression of favorable disease resistance alleles from CWRs are the introductions of late blight resistance into potato from the wild potato Solanum demissum (Rao, 1979; Prescott-Allen and Prescott-Allen, 1986), and of stem rust resistance genes Sr21 (Chen et al., 2015) and Sr39 (Kerber and Dyck, 1990), both effective against the race Ug99, into bread wheat from Triticum monococcum and Aegilops speltoides, respectively.
Barley (Hordeum vulgare L.), the fourth most important cereal crop in the world, is affected each year by up to 30% potential yield loss due to pests and diseases (Savary et al., 2012). Limitations in the availability of novel resistance genes or alleles in the primary gene pool of barley, comprising the cultivated barley H. vulgare spp. vulgare and its wild progenitor H. vulgare spp. spontaneum, has directed the focus of research toward other barley gene pools. Bulbous barley, Hordeum bulbosum (Hb), a wild self-incompatible species and the only member of the secondary gene pool of cultivated barley (von Bothmer et al., 1995) is resistant to many barley pathogens (Xu and Kasha, 1992; Walther et al., 2000). A large panel of Hb introgression lines (ILs) harboring a diverse spectrum of resistance traits has been developed during recent years (Pickering et al., 1995; Johnston et al., 2009). This resource comprises ILs carrying, among others, the barley leaf rust resistance gene Rph26 on chromosome 1HbL (Yu et al., 2018); barley leaf rust gene Rph22 (Johnston et al., 2013) and barley mild mosaic virus gene Rym16Hb (Ruge-Wehling et al., 2006) both located on chromosome 2HbL; barley yellow dwarf virus resistance gene Ryd4Hb on chromosome 3HbL (Scholz et al., 2009) as well as loci conferring powdery mildew resistance located on chromosome 2HbS, 2HbL and 7HbL in barley/Hb introgression lines (Xu and Kasha, 1992; Pickering et al., 1995; Shtaya et al., 2007). These ILs represent a unique genetic resource for improving barley resistance to pathogens and for scientific investigation of resistance mechanisms as they provide access to further genetic diversity out of the primary gene pool of barley (Tanksley and Nelson, 1996; Zamir, 2001; Johnston et al., 2009). Genotyping-by-sequencing (GBS) of 145 Hv/Hb introgression lines (Wendler et al., 2014, 2015) has provided an extensive pool of molecular markers, sequence resources and single-nucleotide polymorphisms (SNPs) information, greatly improving the efficiency of mapping Hb loci.
Since the identification and the extensive use of the durable and complete mlo resistance gene in European germplasm (Jørgensen, 1992), the threat of powdery mildew to barley has been largely mitigated. However, mlo is associated with yield penalties (Kjær et al., 1990) and increased susceptibility to some hemibiothrophic and necrotrophic fungi (Jarosch et al., 1999; Kumar et al., 2001; McGrann et al., 2014). Thus, the search for alternative sources of resistance to powdery mildew remains important for barley breeding (Czembor, 2002; Corrion and Day, 2015). The wild progenitor H. vulgare spp. spontaneum proved to be a great source of diversity of resistance genes (Fischbeck et al., 1976; Dreiseitl and Bockelman, 2003). However, these genes are mostly race-specific, major resistance genes. They are particularly effective, but prone to be rapidly overcome by emerging new pathotypes. Therefore, the search for non-host resistance, from plant species to which the pathogen is not coevolutionary adapted, represent a great hope to achieve a complete and durable resistance. Several Hv/Hb introgression lines carrying a locus conferring powdery mildew resistance have been described (Xu and Kasha, 1992; Pickering et al., 1995, 1998; Shtaya et al., 2007). The Hb accession A42 displays a dominant high resistance to powdery mildew that has been localized on the short arm of chromosome 2Hb in preliminary studies (Szigat and Szigat, 1991; Michel, 1996). Interestingly, several significant QTLs and major genes associated with powdery mildew resistance have repeatedly been reported in this region in cultivated barley (Backes et al., 2003; von Korff et al., 2005; Řepková et al., 2009). Moreover, a resistance gene to powdery mildew has been reported in this region in various Hb accessions (Pickering et al., 1995; Zhang et al., 2001; Shtaya et al., 2007).
While the potential value of untapped genetic diversity of CWR is immense, their application in breeding programs through the use of ILs is hampered by negative linkage drag, mainly caused by severely repressed genetic recombination (Wijnker and de Jong, 2008; Prohens et al., 2017), which can confer yield penalties or other unfavorable characteristics (Hospital, 2001). The degree of drag is correlated with the size of introgressed CWR chromatin segments, and thus can be mitigated by reducing the size of respective introgressions through recombination (Frisch and Melchinger, 2001). However, the efficient utilization of Hb germplasm in barley crop improvement and the genetic mapping of loci contributed by Hb suffers from highly reduced frequency of recombination in introgressed intervals up to 14-fold compared to intraspecific barley crosses (Ruge et al., 2003; Ruge-Wehling et al., 2006; Kakeda et al., 2008; Johnston et al., 2013). Possible explanations for this phenomenon include excessive sequence diversity, structural variation among Hordeum genomes, and probably other unknown mechanisms (Pickering, 1991; Hoffmann et al., 2004; Wendler et al., 2017). To reduce the negative linkage drag, precise delimitation of the causal gene is required, which usually demands intensive screening of large segregating Hv/Hb ILs populations.
Canady et al. (2006) compared the recombination rate in crosses between cultivated tomato (Solanum lycopersicum) and ILs with Solanum lycopersicoides fragments in S. lycopersicum backgrounds with the recombination in crosses between ILs with S. lycopersicoides fragments and ILs with Solanum pennellii fragments in the same region. They showed that tomato ILs with overlapping fragments from closely related species exhibited increased recombination rates in those fragments. Similarly, in barley, Johnston et al. (2015) demonstrated the usefulness of intraspecific recombination between Hb ILs to overcome the negative linkage between genes conferring pathogen resistance and reduced yield. They crossed two recombinant ILs containing an Hb locus on chromosome 2HL comprising the genes Rph22 and Rym16Hb, together with the proximal region of the original introgression for one of them, and the distal region for the other. They obtained four lines with reduced introgressions around the locus of interest for which the yield was close to the one of the recurrent Hv genotype.
The current study aimed to map a dominant powdery mildew resistance locus on chromosome 2HbS introgressed from the tetraploid A42 powdery mildew resistant Hb accession into a susceptible barley cultivar “Borwina.” Mapping in populations of over 200 F7 and BC1F6 from crosses between a susceptible barley cultivar and an Hb IL showed severely repressed recombination between the introgressed segment and the Hv genome. To overcome this difficulty, we exploited intraspecific recombination instead of interspecific recombination by crossing the IL carrying the resistance locus with another Hv/Hb IL carrying a homologous Hb introgression without resistance loci.
Materials and Methods
Fungal Isolate and Phenotyping Test
The swiss powdery mildew field isolate CH4.8 was chosen based on its hypersensitive response it triggers on resistant plants and its capacity to overcome the resistance gene MlRu2 present in Borwina, allowing a clear discrimination between resistant and susceptible plants. The resistance test was carried out in two technical replicates by detached leaf assay on the second seedling leaf sampled 14 days after sowing, as described in Hoseinzadeh et al. (2019). The inoculated detached leaves were kept for 7 days in a growth cabinet (MLR-352H, Panasonic, Japan) with LED light sources (S2 20W matt nw, ARTEKO-LED, Germany) at 20°C with 60% humidity and a 16:8 h photoperiod. Powdery mildew resistance phenotype was scored macroscopically based on percentage infection area as described by Mains and Dietz (1930). Plants without mycelial growth and sporulation but presenting necrotic flecks (infection type 1)were classified as resistant, while those having infection types similar to the susceptible parent “Borwina,” with sporulation of well-developed colonies (infection types 3 and 4) were classified as susceptible.
Plant Material and Population Development
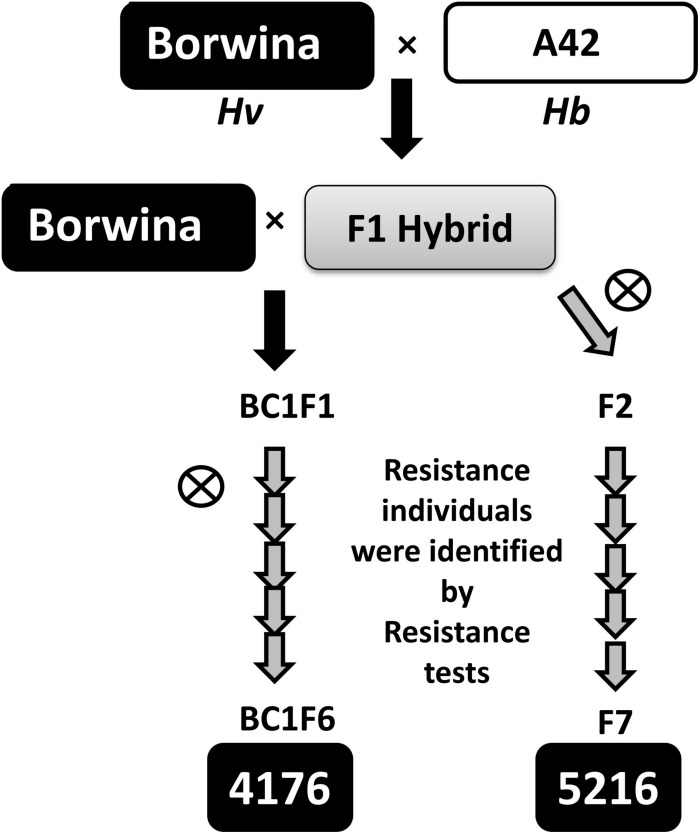
Initial genetic mapping was performed on the F7 and BC1F6 populations “5216” and “4176,” respectively, both derived from crossing a tetraploid derivative of the colchicine treated German winter barley cultivar “Borwina” and the tetraploid Hb accession A42, which is resistant to several European barley powdery mildew isolates. Both “5216” and “4176” populations were derived from selfing of a single plant from the previous generation established as both resistant and heterozygous at the resistance locus. The crossing scheme used to generate those populations is described in Figure 1.
FIGURE 1.
Schematic outline of the mapping population design. The tetraploid Hb accession A42, resistant to the isolate CH4.8, was crossed to the tetraploid derivative of the susceptible barley cultivar “Borwina.” From tetraploid F1 hybrids, two different introgression mapping populations were developed: the “4176” population was developed by backcrossing the F1 hybrid once to the parent “Borwina,” followed by five generations of selfing. A gray arrow indicates a selfing generation. The “5216” population was derived from six generations of selfing from the F1 hybrid. In the course of development of populations “4176” and “5216,” the generations were diploid from BC1F2 and F2, respectively. Through the population development, each new selfing generation was obtained by selecting and selfing a single resistant heterozygous plant identified by resistance test with the powdery mildew isolate CH4.8 and marker data analysis, in order to promote recombination.
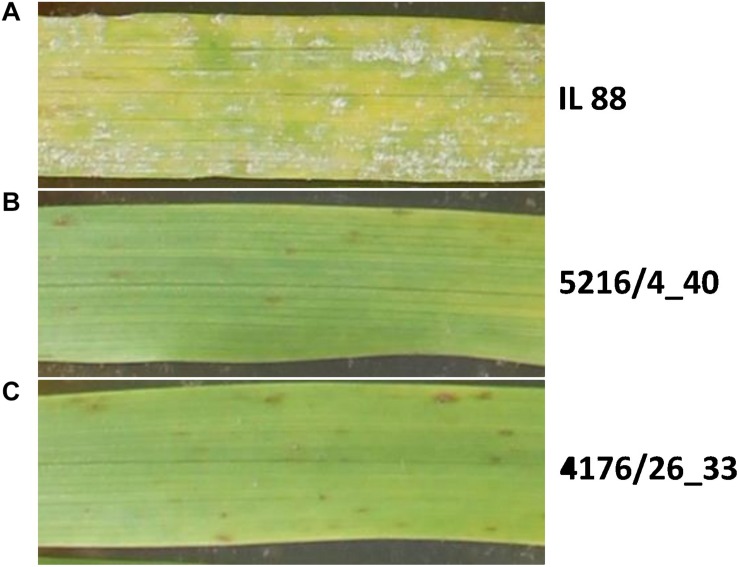
We hypothesized that differences in the sequence or organization between Hv and Hb orthologous genome regions would severely reduce meiotic recombination. To test this hypothesis, we analyzed intraspecific Hb/Hb recombination in a Hv background. We generated two F2 populations by crossing two independent Hv/Hb ILs carrying independent but overlapping Hb introgressions at the terminal 2HS chromosome, thus representing different Hb genotypes at the resistance locus. Three introgression lines “IL 88,” “IL 99,” and “IL 116,” developed in New Zealand Institute for Crop and Food Research, were selected that carry independent Hb introgressions at end of the short arm of barley chromosome 2H (Wendler et al., 2015; Table 1). These three ILs were phenotyped for resistance against isolate CH4.8 (Figure 2). Only “IL 88” displayed full susceptibility to CH4.8 and was used for further population development.
TABLE 1.
Hb introgression lines containing segments overlapping with IL “4176” and IL “5216” and their observed resistance phenotype to the Bgh isolate CH4.8.
| IPK ID | IL code (New Zealand) | Crossing scheme | Introgression location GBS | Start (Mb) | End (Mb) | Phenotype of ILs to isolate CH4.8 |
| 88 | 200A3/7/M1 | Emir x A17/1 | 2HS | 0 | 19,4 | Susceptible |
| 99 | 213G3/2/2/2/1 | Emir x (2920/4 x Tinos16/1) | 2HS, 6HS | 0 | 22,1 | Resistant |
| 116 | 230H24/5/M1/M1 | Morex x 2032 | 2HS | 0 | 19,4 | Resistant |
FIGURE 2.
Powdery mildew infection (isolate CH4.8) on the second leaf of seedling from the three independent Hv/Hb parental ILs, 7 days after inoculation. (A) Phenotype of the susceptible parental introgression line “IL 88” population “dIL_5216” and “dIL_4176.” (B) Phenotype of the resistant parental introgression line 5216/4_40 from population “dIL_5216.” (C) Phenotype of the resistant parental introgression line 4176/26_33 from population “dIL_4176.”
The homozygous resistant ILs 5216/4_40 and 4176/26_33 from the populations “5216” and “4176” were crossed to the susceptible “IL 88.” From each cross, a single F1 plant was selfed, resulting in 103 and 146 F2 seeds, respectively. These two F2 populations were named “dIL_5216” and “dIL_4176,” respectively.
Genomic DNA Extraction
Genomic DNA was extracted from third leaves of barley seedlings using a guanidine isothiocyanate-based DNA isolation method in 96-well plate format as described earlier (Milner et al., 2018). dsDNA concentration was measured by Qubit® 2.0 Fluorometer using the QubitTM dsDNA BR (Broad Range) Assay Kit (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s protocol.
Marker Development
To screen the initial IL mapping populations “4176” and “5216” for recombinants, nine CAPS markers (Supplementary Table 1) were designed to be evenly distributed over the distal 20 cM of barley chromosome 2HS, based on the conserved interspecific SNPs identified by targeted enrichment re-sequencing of 145 Hb ILs (Wendler et al., 2015). This set of ILs included the F4 homozygous 2HS IL (3026) from the mapping population “5216” and its associated donor lines, plus four additional Hv cultivars and four Hb accessions (Wendler et al., 2015). Only conserved Hv/Hb SNPs with a minimum of six-fold read coverage and located in the target region on the barley draft genome (International Barley Genome Sequencing Consortium., 2012) were selected and converted into CAPS markers using SNP2CAPS software (Thiel et al., 2004). Primer design was carried out using the default settings of Primer3 v.0.4.01 (Koressaar and Remm, 2007; O’Halloran, 2015) with minor modifications: The primer length was set between 19–21 bp. Primer melting temperature (Tm) was set to minimum Tm = 58°C, optimum Tm = 59°C and maximum Tm = 60°C. The product size was defined to be between 700 and 1,000 bp and Guanine-Cytosine content (GC-content) was set within the range of 50–55%.
CAPS Genotyping
Genotyping of populations “4176” and “5216” with the described CAPS markers was performed in a 20 μl PCR reaction volume including 20 ng genomic DNA, 0.1 U of HotStarTaq DNA Polymerase (Qiagen, Hilden, Germany), 1x PCR reaction buffer containing 15 mM MgCl2 (Qiagen, Hilden, Germany), 0.2 mM of each dNTP (Fermentas, Fermentas, St. Leon-Rot, Germany), and 0.5 mM of each primer. All fragments were amplified using the following touchdown PCR profile: an initial denaturing step of 15 min at 95°C was followed by four cycles with denaturation at 95°C for 30 s, annealing at 62°C for 30 s (decreasing by 1°C per cycle), and extension at 72°C for 30 s. A final extension step was performed at 72°C for 7 min. The enzymatic digestion of the amplicons was performed in a 10 μl volume containing 5 μl of PCR product, 1× of appropriate buffer (New England Biolabs, Hitchin, United Kingdom), 1 U of enzyme (New England Biolabs, Hitchin, United Kingdom) and adjusted to final volume by adding molecular biology grade pure water. The reaction mix was incubated for 1 h at recommended incubation temperature. The digested PCR products were resolved by 1.5–2.5% gel-electrophoresis depending on amplicon size.
Genotyping-by-Sequencing and Data Analysis
GBS was used, following published procedures (Mascher et al., 2013b), to check the genetic purity and state of heterozygosity of F1 hybrid seeds derived from crosses between overlapping 2HS introgression lines as well as to genotype the whole F2 populations derived from these crosses. DNA of the progeny and parental lines were pooled per Illumina HiSeq2500 lane in an equimolar manner and sequenced for 107 cycles, single read, using a custom sequencing primer as previously described (Hoseinzadeh et al., 2019). The reads were aligned to the TRITEX genome assembly of barley cultivar Morex (Monat et al., 2019) with BWA-MEM version 0.7.12a (Li and Durbin, 2009). Variants were filtered following the protocol of Milner et al. (2018) for a minimum depth of sequencing of four to accept a genotype call, a minimum mapping quality score of the SNPs (based on read depth ratios calculated from the total read depth and depth of the alternative allele) of six, a maximum fraction of heterozygous call of 70% and a maximum fraction of 25% of missing data. The resulting tables of polymorphisms are provided in Supplementary Tables 2, 3.
Results
Inheritance of the Resistance Contributed by Hb
The susceptible parental Hv genotype “Borwina” consistently displayed a leaf infection area of ≥80% in all experiments. The scoring rates of all susceptible individuals of both populations “4176” and “5216” was similar to the susceptible parent and did not significantly vary between phenotyping experiments, indicating high infection efficiency and reproducibility in all phenotyping experiments. The resistance phenotype to the CH4.8 powdery mildew isolate of plants carrying the Hb introgressed segment in a heterozygous state was identical to that of plants homozygous for the Hb fragment (Figure 3). The resistance phenotype was invariably accompanied with a hypersensitive response (HR) forming a necrotic lesion.
FIGURE 3.
Powdery mildew infection (isolate CH4.8) on the second leaf of seedling from two BC2F6 ILs from population “4176,” 7 days after inoculation. 4176/16_97 is homozygous at the resistance locus whereas 4176/16_51 is heterozygous. Their resistance phenotype is identical and present necrotic lesions characteristic of HR.
Phenotypic segregation for powdery mildew resistance against CH4.8 isolate was consistent with a 3:1 ratio (resistant/susceptible, R/S, P < 0.05) in all mapping populations, indicating the control of resistance by a single dominant resistance gene (Table 2). We propose the temporary name Mlhb.A42 for this locus, based on previous naming of Hb powdery mildew resistance genes (Pickering et al., 1995; Steffenson, 1998).
TABLE 2.
Phenotypic segregation of powdery mildew resistance in each of 2HS IL mapping populations.
| Mapping population | Number of resistant lines | Number of susceptible lines | χ2 |
| “4176” (BC1F6) | 199 | 67 | 0,005** |
| “5216” (F7) | 150 | 53 | 0,13** |
| “dIL_5216” (F2) | 81 | 22 | 0.73** |
| “dIL_4176” (F2) | 114 | 32 | 0.74* |
χ2 indicates the result of the Chi squared test performed to determine the goodness to fit a 3R:1S ratio, expected for a dominant monogenic inheritance. Asterisks indicate significance of the test with a p-value inferior to 1% (**) and 5% (*).
Recombination Frequency and Mapping of Mlhb.A42 in Single-Introgression Line Mapping Populations
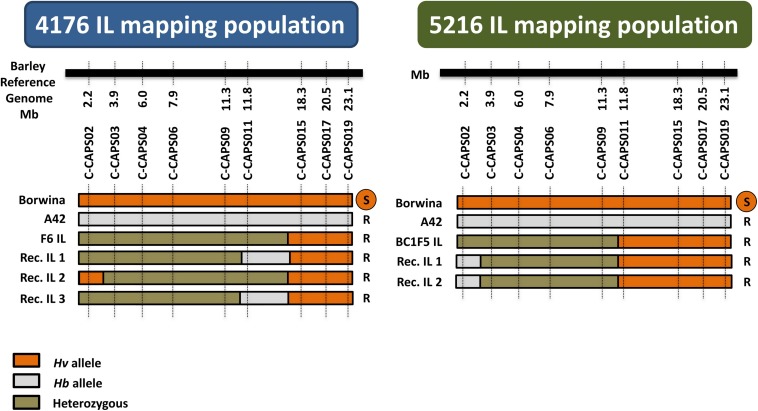
Genotyping of the mapping populations employed nine CAPS markers designed based on existing exome capture re-sequencing data (Supplementary Table 1). This showed that the BC1F6 population “4176” carried a longer introgressed segment compared to the F7 population “5612” (Figure 4). The genotyping results for population “5612” confirmed the extent of the introgressed segment, previously detected by exome capture in the F6 generation (Wendler et al., 2015).
FIGURE 4.
Graphical genotypes and phenotypes of the recombinant Hv/Hb ILs from the two IL mapping populations. “Borwina” and “A42” are the original susceptible and resistant parents of the populations. “F6 IL” and “BC1F5 IL” represent the plant that was selfed in the previous generation to obtain populations “4176” and “5216,” respectively. “Rec. IL” 1 to 3 are the identified recombinant plants. Nine CAPS markers named C-CAPS02 to C-CAPS019, were developed based on conserved Hv/Hb SNP loci described in Wendler et al. (2014), spanning the terminal 23 Mbp of barley chromosome 2HS. The black horizontal bars represent schematically the barley reference genome. The physical position of each selected SNPs on the barley reference genome is written below the black line. The homozygous state of the alleles from susceptible and resistant parents is shown as orange and gray colors, respectively, whereas heterozygous state is shown in green. The phenotype of each recombinant is indicated on the right of their genotype (R = resistant; S = susceptible). The number of recombinants identified through screening with the developed markers was only three and two in IL mapping population “4176” and “5216,” respectively.
Genotyping of 266 and 203 individuals in the initial IL mapping populations “4176” and “5612,” uncovered only three and two recombinants within the introgressed segments, respectively (Figure 4). The introgressed segment therefore has a genetic length of approximately 1 centiMorgan (cM). Yet, the segment of population “5612” represents 10 cM on the barley POPSEQ map (Mascher et al., 2013a; Wendler et al., 2015). This confirms the anticipated reduced recombination between the Hv genome and the introgressed Hb segment. The resulting genetic interval for Mlhb.A42 is flanked by markers CAPS02 and CAPS11, corresponding to a 9.5 Mbp interval between bp positions 2,269,761 and 11,819,688 on chromosome 2HS.
Recombination and Mapping of Mlhb.A42 in Double Introgression Lines
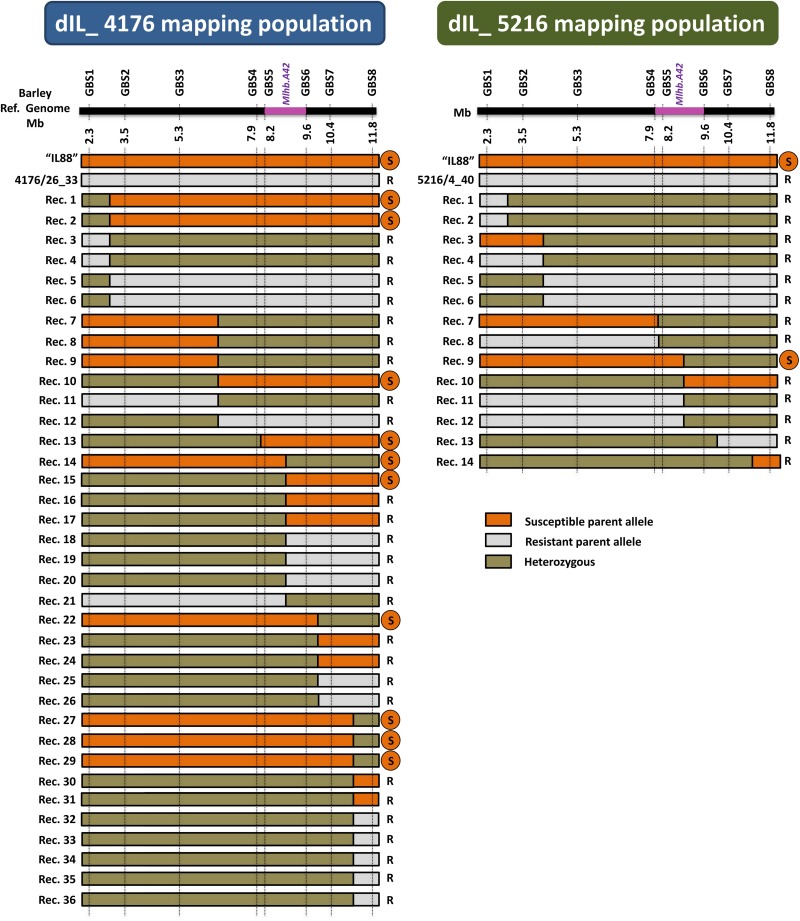
The 103 and 146 F2 plants from the two double introgression populations “dIL_5216” and “dIL_4176,” respectively, were genotyped by GBS and phenotyped for resistance against CH4.8 isolate. Based on obtained SNP variants across candidate interval for Mlhb.A42 defined in the initial mapping populations, 14 and 36 recombinants were obtained in “dIL_5216” and “dIL_4176,” respectively (Figure 5). The phenotypes of all non-recombinant plants corresponded to their genotype in this interval. Association between the phenotypes and genotypes of the recombinants defined overlapping 1.7 Mbp (between 7,943,409 and 9,595,691 bp) and 1.4 Mbp (between 8,193,151 and 9,595,691 bp) intervals on barley chromosome 2HS (Monat et al., 2019) in “dIL_5216” and “dIL_4176,” respectively (Figure 5).
FIGURE 5.
Intraspecific mapping of the powdery mildew resistance locus Mlhb.A42 in F2 populations derived from two different 2HS Hv/Hb introgression lines. The horizontal black bar schematically represents barley chromosome 2HS. The physical coordinates of each GBS marker are indicated below it. The genomic region containing the Mlhb.A42 locus on barley chromosome 2HS is shown in purple. The graphical genotypes of recombinants are indicated as horizontal bars. The genotype of the susceptible parent “IL88” is represented in orange, the one of the resistant parents 4176/26_33 and 5216/4_40 is represented in gray, and heterozygous state is shown in olive green. The phenotype of each recombinant plant is indicated on the right of its genotype (R = resistant; S = susceptible). The recombination rates in “dIL_4176” and “dIL_5216” are 24.7 and 13.6%, respectively, corresponding to a 20-fold increase compared to single introgression lines. The Mlhb.A42 gene was allocated to a 1.4 Mb interval, based on an estimate using the Hv genome as reference.
Candidate Genes Within the Target Interval
Based on the annotated barley reference genome sequence (Monat et al., 2019), 46 high confidence (HC) genes (Table 3) are located within the 1.4 Mbp Mlhb.A42 interval as delimited in the “dIL_4176” population. Those genes with a putative functional annotation included a nucleotide-binding-site leucine-rich-repeat class of gene (NBS-LLR), an HR-like lesion-inducing protein-coding gene, a lectin receptor kinase (LecRK) and a Heat shock protein 90, all gene functions that could be directly or indirectly implied with plant resistance to biotic and/or abiotic stresses and therefore, qualify as candidate genes for Mlhb.A42. HORVU.MOREX.r2.2HG0082250 is annotated as a LecRK. However, analysis of its protein sequence with InterPro (Mitchell et al., 2018), showed that, like LecRKs, it is composed of an extracellular legume (L-type) lectin domain and a transmembrane domain. However, its cytoplasmic domain is only 23 amino-acid long and does not bear a kinase domain, making this gene a probable lectin receptor-like protein (LecRLP).
TABLE 3.
High confidence (HC) genes based on automated annotation of barley reference genome.
| Gene name | Start1 | End1 | Annotation |
| HORVU.MOREX.r2.2HG0081650 | 8223803 | 8224706 | Serine/threonine-protein phosphatase 7 long form-like protein |
| HORVU.MOREX.r2.2HG0081660 | 8227095 | 8233246 | NBS-LRR-like resistance protein |
| HORVU.MOREX.r2.2HG0081670 | 8278638 | 8280547 | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein, putative |
| HORVU.MOREX.r2.2HG0081680 | 8302525 | 8304486 | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein, putative |
| HORVU.MOREX.r2.2HG0081690 | 8320263 | 8321485 | 12-oxophytodienoate reductase-like protein |
| HORVU.MOREX.r2.2HG0081700 | 8328715 | 8329968 | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein, putative |
| HORVU.MOREX.r2.2HG0081720 | 8364720 | 8365206 | NADH-ubiquinone oxidoreductase chain 1 |
| HORVU.MOREX.r2.2HG0081740 | 8381983 | 8383512 | ATP synthase subunit alpha |
| HORVU.MOREX.r2.2HG0081760 | 8470715 | 8473118 | HR-like lesion-inducing protein-related protein |
| HORVU.MOREX.r2.2HG0081770 | 8474573 | 8487616 | Actin-related protein |
| HORVU.MOREX.r2.2HG0081780 | 8511271 | 8512941 | Cytochrome P450 |
| HORVU.MOREX.r2.2HG0081810 | 8576680 | 8577147 | Cytochrome P450 |
| HORVU.MOREX.r2.2HG0081820 | 8607940 | 8608750 | Cytochrome P450 |
| HORVU.MOREX.r2.2HG0081830 | 8615323 | 8617046 | Cytochrome P450 |
| HORVU.MOREX.r2.2HG0081840 | 8664852 | 8679924 | Cytochrome P450 |
| HORVU.MOREX.r2.2HG0081850 | 8686994 | 8691803 | Kaurene synthase |
| HORVU.MOREX.r2.2HG0081860 | 8729062 | 8731108 | Cytochrome P450, putative |
| HORVU.MOREX.r2.2HG0081880 | 8896440 | 8898414 | Copalyl diphosphate synthase |
| HORVU.MOREX.r2.2HG0081890 | 8905788 | 8907365 | Cytochrome P450 |
| HORVU.MOREX.r2.2HG0081900 | 8930477 | 8930938 | Isoaspartyl peptidase/L-asparaginase |
| HORVU.MOREX.r2.2HG0081920 | 8961925 | 8964012 | Copalyl diphosphate synthase |
| HORVU.MOREX.r2.2HG0081930 | 8965503 | 8967761 | Cytochrome P450 |
| HORVU.MOREX.r2.2HG0081980 | 9008320 | 9009897 | Cytochrome P450 |
| HORVU.MOREX.r2.2HG0082000 | 9064624 | 9068933 | Copalyl diphosphate synthase |
| HORVU.MOREX.r2.2HG0082010 | 9095159 | 9096736 | Cytochrome P450 |
| HORVU.MOREX.r2.2HG0082020 | 9253167 | 9258032 | Agenet domain, putative |
| HORVU.MOREX.r2.2HG0082050 | 9310583 | 9312095 | Chalcone synthase |
| HORVU.MOREX.r2.2HG0082060 | 9337837 | 9339296 | O-methyltransferase family protein |
| HORVU.MOREX.r2.2HG0082080 | 9357711 | 9359256 | Glycosyltransferase |
| HORVU.MOREX.r2.2HG0082090 | 9360337 | 9378163 | ABC transporter B family protein |
| HORVU.MOREX.r2.2HG0082100 | 9379916 | 9381655 | Glycosyltransferase |
| HORVU.MOREX.r2.2HG0082140 | 9435844 | 9439807 | Transcription factor |
| HORVU.MOREX.r2.2HG0082150 | 9441094 | 9442595 | Serine/threonine-protein kinase |
| HORVU.MOREX.r2.2HG0082160 | 9462767 | 9463051 | TTF-type zinc finger protein with HAT dimerization domain-containing protein |
| HORVU.MOREX.r2.2HG0082180 | 9468781 | 9469636 | F-box protein PP2-A13 |
| HORVU.MOREX.r2.2HG0082190 | 9472177 | 9476645 | ATP sulfurylase (Sulfate adenylyltransferase) |
| HORVU.MOREX.r2.2HG0082200 | 9478069 | 9482961 | Zinc finger family protein |
| HORVU.MOREX.r2.2HG0082210 | 9488137 | 9491244 | carbohydrate esterase, putative (DUF303) |
| HORVU.MOREX.r2.2HG0082220 | 9498236 | 9499725 | Serpin |
| HORVU.MOREX.r2.2HG0082240 | 9505214 | 9505813 | Maternal effect embryo arrest protein |
| HORVU.MOREX.r2.2HG0082250 | 9538406 | 9539521 | Lectin receptor kinase |
| HORVU.MOREX.r2.2HG0082260 | 9543886 | 9546930 | Carboxymethylenebutenolidase-like protein |
| HORVU.MOREX.r2.2HG0082270 | 9550542 | 9550993 | NAD(P)-binding rossmann-fold protein |
| HORVU.MOREX.r2.2HG0082280 | 9555561 | 9557307 | Nicotianamine synthase |
| HORVU.MOREX.r2.2HG0082310 | 9591121 | 9593401 | Heat shock protein 90 |
| HORVU.MOREX.r2.2HG0082320 | 9595584 | 9597283 | caspase-6 protein |
1Coordinates based on TRITEX Morex assembly (Monat et al., 2019).
Discussion
We report the fine mapping of Mlhb.A42, a dominant powdery mildew resistance locus originating from Hb, using mapping populations derived by crossing two independent, resistant and susceptible, Hv/Hb ILs. The strong suppression of recombination between homeologous genomic segments in Hv/Hb introgression lines, which typically results in severe linkage drag, previously represented a barrier to the efficient utilization of Hb germplasm in barley crop improvement, and to the isolation of disease resistance genes introgressed from Hb.
The genomic resource created by the GBS study of Wendler et al. (2015) on 145 Hv/Hb ILs proved to be a useful tool to identify suitable partners for the development of double ILs populations. The exploitation of intraspecific recombination allowed us to overcome the barrier to recombination usually observed in IL populations. Recombination rates in the region carrying the introgressed fragment were estimated to be 24.7% (“dIL_4176”) and 13.6% (“dIL_5216”), comparable to the corresponding 10% rate observed in pure Hv/Hv mapping populations [e.g. POPSEQ map (Mascher et al., 2013a)]. The rate of recombination was exceeded by a factor of 20-fold as compared to the F7 and BC1F6 single IL populations “5216” and “4176” (approximately 1%). The polymorphisms and markers identified in this study can be converted into KASP assays which would enable for rapid and high-throughput screening of large breeding population for the purpose of introgression of Mlhb.A42 into barley cultivars.
The 1.4 Mbp identified target interval is containing 46 annotated HC genes on the most recent chromosome-scale genome assembly of cultivar “Morex” (Monat et al., 2019). The powdery mildew resistance conferred by this Hb locus is dominantly inherited, displaying chlorotic/necrotic flecks of HR with collapsed hyphae at inoculation sites, suggesting a salicylic acid (SA)-independent resistance pathway. Genes from the NBS-LLR or the receptor-like-kinase (RLK) families are over-represented among genes conferring this type of strong dominant resistance to pathogens (Kourelis and van der Hoorn, 2018), making genes from these families likely candidates in the context of this study. HORVU.MOREX.r2.2HG0081660, one of the annotated genes in the interval, is a coiled-coil (CC)-NBS-LRR gene and therefore the most likely candidate gene. However, HORVU.MOREX.r2.2HG0082250 is annotated as a LecRK and could also be a good candidate. LecRKs are a type of RLK characterized by an extracellular lectin motif (Wu and Zhou, 2013). They have been described as implicated in biotic stress resistance, mostly to bacteria and fungi (Singh and Zimmerli, 2013). This type of gene has been identified in resistance to oomycetes (Wang et al., 2015a, b; Balagué et al., 2017) and fungi (Huang et al., 2013; Wang et al., 2014a) in Arabidopsis thaliana and to wheat powdery mildew in Haynaldia villosa (Wang et al., 2018). According to InterPro (Mitchell et al., 2018), HORVU.MOREX.r2.2HG0082250 does not bear a kinase domain and is likely to be a L-type LecRLP. So far, the only LecRLPs described have a lectin-like Lysin-motif (LysM)-type lectin domain. Two LysM-LecRLPs from A. thaliana and three from rice have been identified reported in context of disease resistance through interaction with the LysM-LecRK CERK1. The rice LysM-LecRLP CEBiP recognizes chitin and, through a direct interaction with CERK1, confers pattern-triggered immunity against fungi (Shimizu et al., 2010). Similarly, LYP4 and LYP6 both perceive peptidoglycan and chitin and interact with CERK1 (Liu et al., 2012). In A. thaliana, LYM1 and LYM3 sense pectidoglycan and trigger immunity, through CERK1, to bacterial infection (Willmann et al., 2011). A. thaliana contains only four L-type LecRLPs (Bellande et al., 2017) but, so far, no L-type LecRLPs have been functionally described. However, a mode of action of HORVU.MOREX.r2.2HG0082250 similar to the one of LysM-LecRLPs is a possibility for further investigation.
Almost all the genes annotated in the delimited target interval might be directly or indirectly involved in resistance to biotic and abiotic stresses and should be considered tentative candidates. NADH-ubiquinone oxidoreductase is involved in intracellular ROS production that can prevent pathogen infection (van der Merwe and Dubery, 2006). Cytochrome P450 and O-methyltransferase proteins are responsible for production of several molecules that can play a role in resistance to pathogens or pests (He and Dixon, 2000; Dixon, 2001; Noordermeer et al., 2001). Some glycosyl-transferase genes have been identified as necessary for the HR (Langlois-Meurinne et al., 2005). The rice gene OsHRL encodes for a HR-like lesion inducing protein and has been shown to be associated with resistance to bacterial blight (Park et al., 2010). Copalyl-diphosphate synthases are implicated in the biosynthesis of phytohormones including gibberellic acid or phytoalexins, which contribute to defensive secondary metabolism (Prisic et al., 2004; Harris et al., 2005). Finally, HORVU.MOREX.r2.2HG0082310 is annotated as a Heat shock protein 90 (Hsp90), which are involved in stress resistance, in particular disease resistance, mediating signal transduction for HR (Xu et al., 2012). Notably, it has been shown by virus-induced gene silencing that an Hsp90 gene is required for Mla13 resistance of barley to powdery mildew (Hein et al., 2005).
In addition to the annotated genes, other genes, e.g. members of NBS-LRR and RLK families, might be present in the interval of the resistant Hb parent but missing in the respective interval of the “Morex” reference sequence. Indeed, Hv and Hb diverged 6 million years ago, accumulating structural variations since then (Jakob and Blattner, 2006). Therefore, this Hb resistance to powdery mildew could be due to a gene absent from the barley reference genome. In particular, NBS-LRR genes are subject to frequent duplication (Flagel and Wendel, 2009), and resistances conferred by NBS-LRRs genes are frequently due to presence/absence variation of such genes (Grant et al., 1998; Griffiths et al., 1999; Henk et al., 1999; Stahl et al., 1999; Tian et al., 2002). The wheat powdery mildew-resistance gene Pm21 (Xing et al., 2018) originates from the wheat/Dasypyrum villosum translocation line T6AL.6VS and is localized in a region presenting a high level of synteny with the Mlhb.A42 locus. This gene confers broad spectrum dominant resistance against wheat powdery mildew isolates and encodes a RPP13-like NBS-LRR gene (He et al., 2017; Xing et al., 2018). The protein sequence of Pm21 only shares 34% identity with the translated nucleotide sequence of HORVU.MOREX.r2.2HG0081660. However, its localization in a syntenic region to the resistance conferring Mlhb.A42 locus provides a tempting working hypothesis that Pm21 and Mlhb.A42 could represent orthologous genes or members of the same locally evolved gene family.
A resequencing strategy of the Mlhb.A42 locus in both resistant and susceptible Hb will be necessary to ascertain the structure of the locus and the number and nature of the candidate genes. With the development of new sequencing methods this could be achieved by Cas9-guided enrichment of the locus (Wang et al., 2014b) or sorting and sequencing for assembly of the chromosome 2H in one of the two introgression lines (Thind et al., 2017). Moreover, a locus of resistance to powdery mildew has been identified on chromosome 2HS in the Hb accessions S1 – where it was named Mlhb1.a – (Pickering et al., 1995; Shtaya et al., 2007), 2032 (Zhang et al., 2001; Shtaya et al., 2007), and A17 (Shtaya et al., 2007). Allelism tests are required to check whether the same locus is involved in the resistance from those four accessions or not. Xu and Kasha (1992) identified another Hb powdery mildew resistance gene in the accession GBC141. However, the causal dominant gene, designated as Mlhb2.b, was located by Kasha et al. (1996) on chromosome 2HL and therefore not allelic to Mlhb1.a or Mlhb.A42.
Durability and spectrum of a resistance gene are two major criteria to assess its application potential (Mundt, 2014). Durability of resistance genes can become a major concern as deployed resistance genes are experiencing a boom-and-bust phenomenon (Gladieux et al., 2015). The spectrum of Mlhb.A42 was not evaluated in this study and should be tested in order to ascertain its potential for field resistance. Durability is difficult to estimate under laboratory scenarios. The durability of orthologs genes can be used as a proxy, yet quite imperfect. As discussed earlier, the wheat resistance gene Pm21could be orthologous to Mlhb.A42. Varieties carrying Pm21 have increasingly been cultivated in China in the recent years (Bie et al., 2015) and its durability can therefore be evaluated in real conditions. Unfortunately, in some wheat fields, new Bgt isolates, virulent against Pm21 have been identified (Shi et al., 2009; Yang et al., 2009). However, resistance based on this gene persisted close to 40 years (Tang et al., 2018) and is still effective against more than a thousand of field isolates in China (Zeng et al., 2014) and Poland (Czembor et al., 2014). To counteract the risk of isolates breaking the resistance provided by a single locus it is of ongoing importance to identify new resistance genes and to pyramid new loci with existing sources of resistance to increase the durability of resistance in the system (Wu et al., 2019). Moreover, exploiting CWR resistances could be a way to unravel even more effective resistance genes. Indeed, Wang et al. (2019) showed that the Hb LecRLK gene of resistance to leaf rust Rph22 confers a stronger resistance to leaf rust adapted to Hv than its Hv ortholog Rphq2. The hypothesis is that crop receptors have a lower recognition of crop-adapted pathogens than the CWR receptors because of adaptation of the pathogens during centuries of coevolution with their host plant.
In the current study we report genetic mapping of Mlhb.A42, a dominant resistance locus introgressed to cultivated barley from Hb. This work is a proof-of-concept study for establishing the basic steps of map-based cloning of genes present in Hv/Hb IL collections by exploiting double ILs mapping populations. Using this strategy, we circumvented the limitation of repressed meiotic recombination which was frequently observed in attempts of genetic mapping employing populations derived between Hv/Hb introgression lines and pure barley cultivars. Here, we observed similar or even higher recombination rates as expected in Hv and thus providing a major step toward facilitated exploitation of secondary gene pool-derived resistance genes in barley crop improvement.
Data Availability Statement
The GBS datasets generated and analyzed in this study are deposited at e!DAL PGP repository (Arend et al., 2016) under the https://doi.org/10.5447/IPK/2020/2.
Author Contributions
PH performed the experimental work. PH and HP performed the data analysis and wrote the manuscript. BR-W provided seed material and sample information for two IL mapping populations “4176” and “5216.” PS supervised the phenotyping. NS designed the study, supervised the experimental work, and contributed to the writing of the manuscript. All authors read, corrected, and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully appreciate the excellent technical support by Mary Ziems in performing crossings and Susanne Koenig in GBS library preparation. We kindly acknowledge Dr. Axel Himmelbach for his valuable support in the sequencing, Dr. Martin Mascher for his invaluable guidance in data processing and analysis, Dr. Dimitar Douchkov for his input on the phenotyping test and fungal material, and Dr. Timothy Rabanus-Wallace for language editing. We are grateful to Dr. Neele Wendler for her support in the employment of the previously developed GBS and exome capture data presented in the current study.
Footnotes
Funding. This manuscript is part of a Ph.D. study of PH, which was financially supported by a grant from the German Research Foundation (DFG) to PS and NS (“DURESTrit,” STE 1102/5-1/608699) in frame of the ERACAPS initiative. This study was further supported by German Ministry of Food and Agriculture (BMEL, FKZ:2818201615) by reason to the decision of the German Bundestag.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00225/full#supplementary-material
References
- Arend D., Junker A., Scholz U., Schüler D., Wylie J., Lange M. (2016). PGP repository: a plant phenomics and genomics data publication infrastructure. Database 2016:baw033. 10.1093/database/baw033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes G., Madsen L., Jaiser H., Stougaard J., Herz M., Mohler V., et al. (2003). Localisation of genes for resistance against Blumeria graminis f. sp. hordei and Puccinia graminis in a cross between a barley cultivar and a wild barley (Hordeum vulgare ssp. spontaneum) line. Theor. Appl. Genet. 106 353–362. 10.1007/s00122-002-1148-1 [DOI] [PubMed] [Google Scholar]
- Balagué C., Gouget A., Bouchez O., Souriac C., Haget N., Boutet-Mercey S., et al. (2017). The Arabidopsis thaliana lectin receptor kinase LecRK-I.9 is required for full resistance to Pseudomonas syringae and affects jasmonate signalling. Mol. Plant Pathol. 18 937–948. 10.1111/mpp.12457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellande K., Bono J.-J., Savelli B., Jamet E., Canut H. (2017). Plant lectins and lectin receptor-like kinases: how do they sense the outside? Int. J. Mol. Sci. 18:1164. 10.3390/ijms18061164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bie T., Zhao R., Jiang Z., Gao D., Zhang B., He H. (2015). Efficient marker-assisted screening of structural changes involving Haynaldia villosa chromosome 6V using a double-distal-marker strategy. Mol. Breed. 35:34 10.1007/s11032-015-0211-y [DOI] [Google Scholar]
- Canady M. A., Ji Y., Chetelat R. T. (2006). Homeologous recombination in Solanum lycopersicoides introgression lines of cultivated tomato. Genetics 174 1775–1788. 10.1534/genetics.106.065144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Rouse M. N., Zhang W., Jin Y., Akhunov E., Wei Y., et al. (2015). Fine mapping and characterization of Sr21, a temperature-sensitive diploid wheat resistance gene effective against the Puccinia graminis f. sp. tritici Ug99 race group. Theor. Appl. Genet. 128 645–656. 10.1007/s00122-015-2460-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrion A., Day B. (2015). “Pathogen Resistance Signalling in Plants,” in eLS. New York, NY: American Cancer Society, 1–14. 10.1002/9780470015902.a0020119.pub2 [DOI] [Google Scholar]
- Czembor H. J., Domeradzka O., Czembor J. H., Mańkowski D. R. (2014). Virulence structure of the powdery mildew (Blumeria graminis) population occurring on triticale (x Triticosecale) in poland. J. Phytopathol. 162 499–512. 10.1111/jph.12225 [DOI] [Google Scholar]
- Czembor J. H. (2002). Resistance to powdery mildew in selections from Moroccan barley landraces. Euphytica 125 397–409. 10.1023/A:1016061508160 [DOI] [Google Scholar]
- Dixon R. A. (2001). Natural products and plant disease resistance. Nature 411 843–847. 10.1038/35081178 [DOI] [PubMed] [Google Scholar]
- Dreiseitl A., Bockelman H. E. (2003). Sources of powdery mildew resistance in a wild barley collection. Genet. Resour. Crop. Evol. 50 345–350. 10.1023/A:1023953819787 30812385 [DOI] [Google Scholar]
- Feuillet C., Langridge P., Waugh R. (2008). Cereal breeding takes a walk on the wild side. Trends Genet. 24 24–32. 10.1016/j.tig.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Fischbeck G., Schwarzbach E., Sobel Z., Wahl I. (1976). Mildew resistance in Israeli populations of 2-rowed wild barley (Hordeum spontaneum). Z. Pflanzenzüchtung 76 163–166. [Google Scholar]
- Flagel L. E., Wendel J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183 557–564. 10.1111/j.1469-8137.2009.02923.x [DOI] [PubMed] [Google Scholar]
- Frisch M., Melchinger A. E. (2001). The length of the intact donor chromosome segment around a target gene in marker-assisted backcrossing. Genetics 157 1343–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladieux P., Feurtey A., Hood M. E., Snirc A., Clavel J., Dutech C., et al. (2015). The population biology of fungal invasions. Mol. Ecol. 24 1969–1986. 10.1111/mec.13028 [DOI] [PubMed] [Google Scholar]
- Grant M. R., McDowell J. M., Sharpe A. G., de Torres Zabala M., Lydiate D. J., Dangl J. L. (1998). Independent deletions of a pathogen-resistance gene in Brassica and Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95 15843–15848. 10.1073/pnas.95.26.15843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths A. J. F., Gelbart W. M., Miller J. H., Lewontin R. C. (eds) (1999). “Chromosomal rearrangements,” in Modern Genetic Analysis (New York, NY: W. H. Freeman; ). [Google Scholar]
- Harris L. J., Saparno A., Johnston A., Prisic S., Xu M., Allard S., et al. (2005). The Maize An2 gene is induced by Fusarium attack and encodesan ent-copalyl diphosphate synthase. Plant Mol. Biol. 59 881–894. 10.1007/s11103-005-1674-8 [DOI] [PubMed] [Google Scholar]
- He H., Zhu S., Ji Y., Jiang Z., Zhao R., Bie T. (2017). Map-based cloning of the gene Pm21 that confers broad spectrum resistance to wheat powdery mildew. bioRxiv [preprint]. 10.1101/177857 [DOI] [Google Scholar]
- He X.-Z., Dixon R. A. (2000). Genetic manipulation of isoflavone 7-O-methyltransferase enhances biosynthesis of 4’-O-Methylated isoflavonoid phytoalexins and disease resistance in alfalfa. Plant Cell 12 1689–1702. 10.1105/tpc.12.9.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein I., Barciszewska-Pacak M., Hrubikova K., Williamson S., Dinesen M., Soenderby I. E., et al. (2005). Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol. 138 2155–2164. 10.1104/pp.105.062810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henk A. D., Warren R. F., Innes R. W. (1999). A new ac-like transposon of Arabidopsis Is associated with a deletion of the RPS5 disease resistance gene. Genetics 151 1581–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Sgrò C. M., Weeks A. R. (2004). Chromosomal inversion polymorphisms and adaptation. Trends Ecol. Evol. 19 482–488. 10.1016/j.tree.2004.06.013 [DOI] [PubMed] [Google Scholar]
- Hoisington D., Khairallah M., Reeves T., Ribaut J.-M., Skovmand B., Taba S., et al. (1999). Plant genetic resources: what can they contribute toward increased crop productivity? Proc. Natl. Acad. Sci. U.S.A. 96 5937–5943. 10.1073/pnas.96.11.5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoseinzadeh P., Zhou R., Mascher M., Himmelbach A., Niks R. E., Schweizer P., et al. (2019). High resolution genetic and physical mapping of a major powdery mildew resistance locus in barley. Front. Plant Sci. 10:146. 10.3389/fpls.2019.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital F. (2001). Size of donor chromosome segments around introgressed loci and reduction of linkage drag in marker-assisted backcross programs. Genetics 158 1363–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Ju H.-W., Min J.-H., Zhang X., Kim S.-H., Yang K.-Y., et al. (2013). Overexpression of L-type lectin-like protein kinase 1 confers pathogen resistance and regulates salinity response in Arabidopsis thaliana. Plant Sci. 20 98–106. 10.1016/j.plantsci.2012.12.019 [DOI] [PubMed] [Google Scholar]
- International Barley Genome Sequencing Consortium. (2012). A physical, genetic and functional sequence assembly of the barley genome. Nature 491 711–716. 10.1038/nature11543 [DOI] [PubMed] [Google Scholar]
- Jakob S. S., Blattner F. R. (2006). A chloroplast genealogy of Hordeum (Poaceae): long-term persisting haplotypes, incomplete lineage sorting, regional extinction, and the consequences for phylogenetic inference. Mol. Biol. Evol. 23 1602–1612. 10.1093/molbev/msl018 [DOI] [PubMed] [Google Scholar]
- Jarosch B., Kogel K.-H., Schaffrath U. (1999). The Ambivalence of the barley Mlo locus: mutations conferring resistance against powdery mildew (Blumeria graminis f. sp. hordei) Enhance Susceptibility to the Rice Blast Fungus Magnaporthe grisea. Mol. Plant-Microbe Interact. 12 508–514. 10.1094/MPMI.1999.12.6.508 [DOI] [Google Scholar]
- Johnston P. A., Meiyalaghan V., Forbes M. E., Habekuß A., Butler R. C., Pickering R. (2015). Marker assisted separation of resistance genes Rph22 and Rym16Hb from an associated yield penalty in a barley: Hordeum bulbosum introgression line. Theor. Appl. Genet. 128 1137–1149. 10.1007/s00122-015-2495-z [DOI] [PubMed] [Google Scholar]
- Johnston P. A., Niks R. E., Meiyalaghan V., Blanchet E., Pickering R. (2013). Rph22: mapping of a novel leaf rust resistance gene introgressed from the non-host Hordeum bulbosum L. into cultivated barley (Hordeum vulgare L.). Theor. Appl. Genet. 126 1613–1625. 10.1007/s00122-013-2078-9 [DOI] [PubMed] [Google Scholar]
- Johnston P. A., Timmerman-Vaughan G. M., Farnden K. J. F., Pickering R. (2009). Marker development and characterisation of Hordeum bulbosum introgression lines: a resource for barley improvement. Theor. Appl. Genet. 118 1429–1437. 10.1007/s00122-009-0992-7 [DOI] [PubMed] [Google Scholar]
- Jørgensen I. H. (1992). Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63 141–152. 10.1007/BF00023919 [DOI] [Google Scholar]
- Kakeda K., Ibuki T., Suzuki J., Tadano H., Kurita Y., Hanai Y., et al. (2008). Molecular and genetic characterization of the S locus in Hordeum bulbosum L., a wild self-incompatible species related to cultivated barley. Mol. Genet. Genomics 280 509–519. 10.1007/s00438-008-0383-9 [DOI] [PubMed] [Google Scholar]
- Kasha K. J., Pickering R. A., William H. M., Hill M. A., Oro R., Reader S., et al. (1996). “GISH and RFLP facilitated identification of a barley chromosome carrying powdery mildew resistance from Hordeum bulbosum,” in Proceedings 7th International Barley Genetics Symposium, Saskatoon, Canada. [Google Scholar]
- Kerber E. R., Dyck P. L. (1990). Transfer to hexaploid wheat of linked genes for adult-plant leaf rust and seedling stem rust resistance from an amphiploid of Aegilops speltoides × Triticum monococcum. Genome 33 530–537. 10.1139/g90-079 [DOI] [Google Scholar]
- Kjær B., Jensen H. P., Jensen J., Jørgensen J. H. (1990). Associations between three ml-o powdery mildew resistance genes and agronomic traits in barley. Euphytica 46 185–193. 10.1007/BF00027217 [DOI] [Google Scholar]
- Koressaar T., Remm M. (2007). Enhancements and modifications of primer design program Primer3. Bioinformatics 23 1289–1291. 10.1093/bioinformatics/btm091 [DOI] [PubMed] [Google Scholar]
- Kourelis J., van der Hoorn R. A. L. (2018). Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30 285–299. 10.1105/tpc.17.00579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J., Hückelhoven R., Beckhove U., Nagarajan S., Kogel K.-H. (2001). A compromised mlo pathway affects the response of barley to the necrotrophic fungus Bipolaris sorokiniana (Teleomorph: Cochliobolus sativus) and its toxins. Phytopathology 91 127–133. 10.1094/PHYTO.2001.91.2.127 [DOI] [PubMed] [Google Scholar]
- Langlois-Meurinne M., Gachon C. M. M., Saindrenan P. (2005). Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol. 139 1890–1901. 10.1104/pp.105.067223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Li J.-F., Ao Y., Qu J., Li Z., Su J., et al. (2012). Lysin Motif6Containing Proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 24 3406–3419. 10.1105/tpc.112.102475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains E. B., Dietz S. M. (1930). Physiologic forms of barley mildew. Erysiphe graminis hordei. Phytopathology 20 229–239. [Google Scholar]
- Mascher M., Muehlbauer G. J., Rokhsar D. S., Chapman J., Schmutz J., Barry K., et al. (2013a). Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J. 76 718–727. 10.1111/tpj.12319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M., Wu S., Amand P. St, Stein N., Poland J. (2013b). Application of genotyping-by-sequencing on semiconductor sequencing platforms: a comparison of genetic and reference-based marker ordering in barley. PLoS One 8:e076925. 10.1371/journal.pone.0076925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrann G. R. D., Stavrinides A., Russell J., Corbitt M. M., Booth A., Chartrain L., et al. (2014). A trade off between mlo resistance to powdery mildew and increased susceptibility of barley to a newly important disease. Ramularia leaf spot. J. Exp. Bot. 65 1025–1037. 10.1093/jxb/ert452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel M. (1996). Untersuchungen zur Übertragung von Resistenzgenen aus der Wildart Hordeum bulbosum L. in die Kulturgerste Hordeum vulgare L. Diss. Munich: TU München. [Google Scholar]
- Milner S., Jost M., Taketa S., Mazón E., Himmelbach A., Oppermann M., et al. (2018). Genebank genomics reveals the diversity of a global barley collection. Nat. Genet. Forthcomin. 51 319–326. 10.1038/s41588-018-0266-x [DOI] [PubMed] [Google Scholar]
- Mitchell A. L., Attwood T. K., Babbitt P. C., Blum M., Bork P., Bridge A., et al. (2018). InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47 D351–D360. 10.1093/nar/gky1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monat C., Padmarasu S., Lux T., Wicker T., Gundlach H., Himmelbach A., et al. (2019). TRITEX: chromosome-scale sequence assembly of Triticeae genomes with open-source tools. bioRxiv [preprint]. 10.1101/631648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt C. C. (2014). Durable resistance: a key to sustainable management of pathogens and pests. Infect. Genet. Evol. 27 446–455. 10.1016/j.meegid.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer M. A., Veldink G. A., Vliegenthart J. F. G. (2001). Fatty acid hydroperoxide lyase: a plant cytochrome P450 enzyme involved in wound healing and pest resistance. ChemBioChem 2 494–504. [DOI] [PubMed] [Google Scholar]
- O’Halloran D. M. (2015). PrimerView: high-throughput primer design and visualization. Source Code Biol. Med. 10:8. 10.1186/s13029-015-0038-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-R., Moon S.-J., Shin D.-J., Kim M.-G., Hwang D.-J., Bae S.-C., et al. (2010). Isolation and characterization of rice OsHRL gene related to bacterial blight resistance. Plant Pathol. J. 26 417–420. 10.5423/PPJ.2010.26.4.417 [DOI] [Google Scholar]
- Pickering R. A. (1991). Comparison of crossover frequencies in barley (Hordeum vulgare) and H. vulgare × H. bulbosum hybrids using a paracentric inversion. Genome 34 666–673. 10.1139/g91-102 [DOI] [Google Scholar]
- Pickering R. A., Hill A. M., Michel M., Timmerman-Vaughan G. M. (1995). The transfer of a powdery mildew resistance gene from Hordeum bulbosum L to barley (H. vulgare L.) chromosome 2 (2I). Theor. Appl. Genet. 91 1288–1292. 10.1007/BF00220943 [DOI] [PubMed] [Google Scholar]
- Pickering R. A., Steeffenson B. J., Hill A. M., Borovkova I. (1998). Association of leaf rust and powdery mildew resistance in a recombinant derived from a Hordeum vulgare × Hordeum bulbosum hybrid. Plant Breed 117 83–84. 10.1111/j.1439-0523.1998.tb01453.x [DOI] [Google Scholar]
- Prescott-Allen C., Prescott-Allen R. (1986). The First Resource. New Haven, CT: Yale University Press. [Google Scholar]
- Prisic S., Xu M., Wilderman P. R., Peters R. J. (2004). Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol. 136 4228–4236. 10.1104/pp.104.050567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prohens J., Gramazio P., Plazas M., Dempewolf H., Kilian B., Díez M. J., et al. (2017). Introgressiomics: a new approach for using crop wild relatives in breeding for adaptation to climate change. Euphytica 213:158. 10.1007/s10681-017-1938-9 27242876 [DOI] [Google Scholar]
- Rao M. V. P. (1979). The Transfer of Alien Genes for Stem rust Resistance to Durum Wheat, ed. Ramanujam S. (New Delhi: Indian Society of Genetics & Plant Breeding; ), 338–341. [Google Scholar]
- Řepková J., Teturová K., Dreiseitl A., Soldánová M. (2009). Characterization and chromosomal location of powdery mildew resistance genes from wild barley PI282605. J. Plant Dis. Prot. 116 257–259. 10.1007/BF03356319 [DOI] [Google Scholar]
- Ruge B., Linz A., Pickering R., Proeseler G., Greif P., Wehling P. (2003). Mapping of Rym14Hb, a gene introgressed from Hordeum bulbosum and conferring resistance to BaMMV and BaYMV in barley. Theor. Appl. Genet. 107 965–971. 10.1007/s00122-003-1339-4 [DOI] [PubMed] [Google Scholar]
- Ruge-Wehling B., Linz A., Habekuß A., Wehling P. (2006). Mapping of Rym16Hb, the second soil-borne virus-resistance gene introgressed from Hordeum bulbosum. Theor. Appl. Genet. 113 867–873. 10.1007/s00122-006-0345-8 [DOI] [PubMed] [Google Scholar]
- Savary S., Ficke A., Aubertot J.-N., Hollier C. (2012). Crop losses due to diseases and their implications for global food production losses and food security. Food Secur. 4 519–537. 10.1007/s12571-012-0200-5 28831526 [DOI] [Google Scholar]
- Scholz M., Ruge-Wehling B., Habekuß A., Schrader O., Pendinen G., Fischer K., et al. (2009). Ryd4Hb: a novel resistance gene introgressed from Hordeum bulbosum into barley and conferring complete and dominant resistance to the barley yellow dwarf virus. Theor. Appl. Genet. 119 837–849. 10.1007/s00122-009-1093-3 [DOI] [PubMed] [Google Scholar]
- Shi Y., Wang B., Li Q., Wu X., Wang F., Liu H., et al. (2009). Analysis of the virulent genes of Erysiphe graminis f. sp. tritici and the resistance genes of wheat commercial cultivars in Shaanxi Province. J. Triticeae Crop. 29 706–711. [Google Scholar]
- Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., et al. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64 204–214. 10.1111/j.1365-313X.2010.04324.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtaya M. J. Y., Sillero J. C., Flath K., Pickering R., Rubiales D. (2007). The resistance to leaf rust and powdery mildew of recombinant lines of barley (Hordeum vulgare L.) derived from H. vulgare × H. bulbosum crosses. Plant Breed 126 259–267. 10.1111/j.1439-0523.2007.01328.x [DOI] [Google Scholar]
- Singh P., Zimmerli L. (2013). Lectin receptor kinases in plant innate immunity. Front. Plant Sci. 4:124. 10.3389/fpls.2013.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl E. A., Dwyer G., Mauricio R., Kreitman M., Bergelson J. (1999). Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400 667–671. 10.1038/23260 [DOI] [PubMed] [Google Scholar]
- Steffenson B. J. (1998). Coordinators reports: disease and pest resistance genes. Barley Genet. newslette [Google Scholar]
- Szigat G., Szigat G. (1991). Amphidiploid hybrids between Hordeum vulgare and H. bulbosum-basis for the development of new initial material for winter barley breeding. Vor. Pflanzenzüchtg 20 34–39. [Google Scholar]
- Tang S., Hu Y., Zhong S., Luo P. (2018). The potential role of powdery mildew-resistance Gene Pm40 in Chinese wheat-breeding programs in the Post-Pm21 Era. Engineering 4 500–506. 10.1016/j.eng.2018.06.004 [DOI] [Google Scholar]
- Tanksley S. D., McCouch S. R. (1997). Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277 1063–1066. 10.1126/science.277.5329.1063 [DOI] [PubMed] [Google Scholar]
- Tanksley S. D., Nelson J. C. (1996). Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor. Appl. Genet. 92 191–203. 10.1007/BF00223376 [DOI] [PubMed] [Google Scholar]
- Thiel T., Kota R., Grosse I., Stein N., Graner A. (2004). SNP2CAPS: a SNP and INDEL analysis tool for CAPS marker development. Nucleic Acids Res. 32 e5–e5. 10.1093/nar/gnh006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thind A. K., Wicker T., Šimková H., Fossati D., Moullet O., Brabant C., et al. (2017). Rapid cloning of genes in hexaploid wheat using cultivar-specific long-range chromosome assembly. Nat. Biotechnol. 35 793–796. 10.1038/nbt.3877 [DOI] [PubMed] [Google Scholar]
- Tian D., Araki H., Stahl E., Bergelson J., Kreitman M. (2002). Signature of balancing selection in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99 11525–11530. 10.1073/pnas.172203599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe J. A., Dubery I. A. (2006). Benzothiadiazole inhibits mitochondrial NADH: ubiquinone oxidoreductase in tobacco. J. Plant Physiol. 163 877–882. 10.1016/j.jplph.2005.08.016 [DOI] [PubMed] [Google Scholar]
- von Bothmer R., Jacobsen N., Baden C., Jorgensen R. B., Linde-Laursen I. (1995). “An ecogeographical study of the genus Hor-deum,” in Systematic and Ecogeographic Studies on Crop Gene Pools 7, (Rome: International Plant Genetic Resources Institute; ). [Google Scholar]
- von Korff M., Wang H., Léon J., Pillen K. (2005). AB-QTL analysis in spring barley. I. Detection of resistance genes against powdery mildew, leaf rust and scald introgressed from wild barley. Theor. Appl. Genet. 111 583–590. 10.1007/s00122-005-2049-x [DOI] [PubMed] [Google Scholar]
- Walther U., Rapke H., Proeseler G., Szigat G. (2000). Hordeum bulbosum-a new source of disease resistance-transfer of resistance to leaf rust and mosaic viruses from H. bulbosum into winter barley. Plant Breed 119 215–218. 10.1046/j.1439-0523.2000.00475.x [DOI] [Google Scholar]
- Wang Y., Bouwmeester K., Beseh P., Shan W., Govers F. (2014a). Phenotypic analyses of Arabidopsis T-DNA insertion lines and expression profiling reveal that multiple L-Type lectin receptor kinases are involved in plant immunity. Mol. Plant-Microbe Interact. 27 1390–1402. 10.1094/MPMI-06-14-0191-R [DOI] [PubMed] [Google Scholar]
- Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., et al. (2014b). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32 947–951. 10.1038/nbt.2969 [DOI] [PubMed] [Google Scholar]
- Wang Y., Cordewener J. H. G., America A. H. P., Shan W., Bouwmeester K., Govers F. (2015a). Arabidopsis lectin receptor kinases LecRK-IX.1 and LecRK-IX.2 Are functional analogs in regulating phytophthora resistance and plant cell death. Mol. Plant-Microbe Interact 28 1032–1048. 10.1094/MPMI-02-15-0025-R [DOI] [PubMed] [Google Scholar]
- Wang Y., Subedi S., de Vries H., Doornenbal P., Vels A., Hensel G., et al. (2019). Orthologous receptor kinases quantitatively affect the host status of barley to leaf rust fungi. Nat. Plants 5 1129–1135. 10.1038/s41477-019-0545-2 [DOI] [PubMed] [Google Scholar]
- Wang Y., Weide R., Govers F., Bouwmeester K. (2015b). L-type lectin receptor kinases in Nicotiana benthamiana and tomato and their role in Phytophthora resistance. J. Exp. Bot. 66 6731–6743. 10.1093/jxb/erv379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Cheng J., Fan A., Zhao J., Yu Z., Li Y., et al. (2018). LecRK-V, an L-type lectin receptor kinase in Haynaldia villosa, plays positive role in resistance to wheat powdery mildew. Plant Biotechnol. J. 16 50–62. 10.1111/pbi.12748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler N., Mascher M., Himmelbach A., Johnston P., Pickering R., Stein N. (2015). Bulbosum to Go: a toolbox to utilize Hordeum vulgare/bulbosum Introgressions for Breeding and Beyond. Mol. Plant 8 1507–1519. 10.1016/j.molp.2015.05.004 [DOI] [PubMed] [Google Scholar]
- Wendler N., Mascher M., Nöh C., Himmelbach A., Scholz U., Ruge-Wehling B., et al. (2014). Unlocking the secondary gene-pool of barley with next-generation sequencing. Plant Biotechnol. J. 12 1122–1131. 10.1111/pbi.12219 [DOI] [PubMed] [Google Scholar]
- Wendler N., Mascher M., Himmelbach A., Bini F., Kumlehn J., Stein N. (2017). A High-density, sequence-enriched genetic map of Hordeum bulbosum and Its collinearity to H. vulgare. Plant Genome 10 1–11. 10.3835/plantgenome2017.06.0049 [DOI] [PubMed] [Google Scholar]
- Wijnker E., de Jong H. (2008). Managing meiotic recombination in plant breeding. Trends Plant Sci. 13 640–646. 10.1016/j.tplants.2008.09.004 [DOI] [PubMed] [Google Scholar]
- Willmann R., Lajunen H. M., Erbs G., Newman M.-A., Kolb D., Tsuda K., et al. (2011). Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 108 19824–19829. 10.1073/pnas.1112862108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. X., Xu X. F., Ma D. X., Chen R. Z., Li T. Y., Cao Y. Y. (2019). Virulence structure and its genetic diversity analyses of Blumeria graminis f. sp. tritici isolates in China. BMC Evol. Biol. 19:183. 10.1186/s12862-019-1511-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhou J.-M. (2013). Receptor-Like Kinases in Plant Innate Immunity. J. Integr. Plant Biol. 55 1271–1286. 10.1111/jipb.12123 [DOI] [PubMed] [Google Scholar]
- Xing L., Hu P., Liu J., Witek K., Zhou S., Xu J., et al. (2018). Pm21 from Haynaldia villosa Encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol. Plant 11 874–878. 10.1016/j.molp.2018.02.013 [DOI] [PubMed] [Google Scholar]
- Xu J., Kasha K. J. (1992). Transfer of a dominant gene for powdery mildew resistance and DNA from Hordeum bulbosum into cultivated barley (H. vulgare). Theor. Appl. Genet. 84 771–777. 10.1007/BF00227383 [DOI] [PubMed] [Google Scholar]
- Xu Z.-S., Li Z.-Y., Chen Y., Chen M., Li L.-C., Ma Y.-Z. (2012). Heat shock protein 90 in plants: molecular mechanisms and roles in stress responses. Int. J. Mol. Sci. 13 15706–15723. 10.3390/ijms131215706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Xiang L., Zeng F., Wang H., Shi W., Yu D. (2009). Virulence gene structure analysis of Blumeria graminis f. sp. tritici in Hubei. Plant Prot 35 76–79. [Google Scholar]
- Yu X., Kong H. Y., Meiyalaghan V., Casonato S., Chng S., Jones E. E., et al. (2018). Genetic mapping of a barley leaf rust resistance gene Rph26 introgressed from Hordeum bulbosum. Theor. Appl. Genet. 131 2567–2580. 10.1007/s00122-018-3173-8 [DOI] [PubMed] [Google Scholar]
- Zamir D. (2001). Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2 983–989. 10.1038/35103590 [DOI] [PubMed] [Google Scholar]
- Zeng F., Yang L., Gong S., Shi W., Zhang X., Wang H., et al. (2014). Virulence and Diversity of Blumeria graminis f. sp. tritici Populations in China. J. Integr. Agric. 13 2424–2437. 10.1016/S2095-3119(13)60669-3 [DOI] [Google Scholar]
- Zhang L., Pickering R. A., Murray B. G. (2001). Hordeum vulgare × H. bulbosum tetraploid hybrid provides useful agronomic introgression lines for breeders. New Zeal. J. Crop Hortic. Sci. 29 239–246. 10.1080/01140671.2001.9514185 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GBS datasets generated and analyzed in this study are deposited at e!DAL PGP repository (Arend et al., 2016) under the https://doi.org/10.5447/IPK/2020/2.