Abstract
Aflatoxins are secondary metabolites produced by soilborne saprophytic fungus Aspergillus flavus and closely related species that infect several agricultural commodities including groundnut and maize. The consumption of contaminated commodities adversely affects the health of humans and livestock. Aflatoxin contamination also causes significant economic and financial losses to producers. Research efforts and significant progress have been made in the past three decades to understand the genetic behavior, molecular mechanisms, as well as the detailed biology of host-pathogen interactions. A range of omics approaches have facilitated better understanding of the resistance mechanisms and identified pathways involved during host-pathogen interactions. Most of such studies were however undertaken in groundnut and maize. Current efforts are geared toward harnessing knowledge on host-pathogen interactions and crop resistant factors that control aflatoxin contamination. This study provides a summary of the recent progress made in enhancing the understanding of the functional biology and molecular mechanisms associated with host-pathogen interactions during aflatoxin contamination in groundnut and maize.
Keywords: Aspergillus flavus, aflatoxin contamination, host-pathogen interactions, molecular mechanisms, QTLs, groundnut, maize
Introduction
Aflatoxins are teratogenic, carcinogenic and immunosuppressive secondary metabolites produced by several Aspergillus section Flavi species (Frisvad et al., 2019). The most common aflatoxin-producing species is A. flavus (Amaike and Keller, 2011) but, A. parasiticus, A. nomius, and other species may be important causal agents of contamination in some areas/years (Diedhiou et al., 2011; Probst et al., 2014; Kachapulula et al., 2017; Kumar P. et al., 2017). Aflatoxin-producing fungi contaminate several agricultural commodities such as groundnut, maize, cottonseed, wheat, rice, tree nuts, and chili peppers (Doster et al., 2014; Khan et al., 2014; Kumar P. et al., 2017; Sarma et al., 2017; Ezekiel et al., 2019).
Aflatoxin remains in food and feed even after cooking and drying of the crop because of its heat and freeze stable nature. There are four major types of aflatoxins, namely, AFB1, AFB2, AFG1, and AFG2 which are discernible based on their blue and green fluorescence under UV light and migration rate. AFB1, the most potent and toxic, is associated with hepatocellular carcinoma (Liu and Wu, 2010). Consuming contaminated commodities may have chronic and/or acute effects that may lead to mortality (Sarma et al., 2017). In addition to the large array of negative health effects of the toxins, the contamination of crops results in large economic losses to farmers and to countries because of produce rejected by markets seeking aflatoxin-compliant crops (Wild and Gong, 2010; Bryden, 2012). For instance, India could export only 800,000 tons each year despite being 2nd largest groundnut producer in the world, and aflatoxin contamination being one of the major reason behind low export (Suneja, 2019). In semi-arid and arid regions of the United States, and tropical and sub-tropical Asia and Africa, aflatoxin contamination of agricultural products occurs frequently (Cotty et al., 2008; Razzaghi-Abyanehed, 2013; Bandyopadhyay et al., 2016). In such affected areas, mitigation of contamination is necessary to protect the health of consumers, maintain crop competitiveness, and to harness the full potential of crops to ensure food and nutritional security.
Deploying pre- and post-harvest genetic resistance in new crop varieties together with good agricultural practices may provide a permanent solution to this problem (Ayalew et al., 2017; Meseka et al., 2018). In this context, it is imperative to explore and deploy all possible resistance mechanisms/methods to control aflatoxin accumulation in the field followed by best practices in the entire value chain. In the case of groundnut, three different types of resistance mechanisms, namely in vitro seed colonization (IVSC), pre-harvest aflatoxin contamination (PAC), and aflatoxin production (AP) have been reported, which are inherited independently (Nigam et al., 2009). In addition, genetic resistance is modulated by high soil temperature and moisture stress which promote higher rates of fungal infection and contamination. To achieve stable genetic resistance against A. flavus infection, we believe all three mechanisms should be examined and integrated to effectively provide resistance under field conditions, during harvest, and throughout storage (see Pandey et al., 2019).
Groundnut and maize are among the most aflatoxin-prone crops. Both are commonly exposed to Aspergillus infection during pre- and post-harvest stages (Guo et al., 2008). For example in Ghana, these two crops that are considered as staples are frequently infected by Aspergillus species, with unsafe aflatoxin levels (Samson et al., 1981; MoFA, 2011; Agbetiameh et al., 2018). In Ghana, as in any other country, aflatoxin-resistant varieties are not commercially available. In addition, farmers typically do not follow good agricultural practices; so contamination begins in the field and may continue until the crops are consumed. Therefore, farmers and traders must receive training and information on good agricultural practices such as timely sowing and irrigation, ensuring adequate dry field conditions before harvest, timely harvesting, and post-harvest management strategies to limit aflatoxin contamination (Dorner, 2004; Jaime-Garcia and Cotty, 2004; Hell et al., 2008; Florkowski and Kolavalli, 2013; Bandyopadhyay et al., 2016). Although some success has been achieved, good management practices are neither very cost effective nor always practical for the resource-poor farmers, or are not effective in reducing aflatoxin content below tolerance thresholds if not used as part of a holistic aflatoxin management strategy. Climate change and frequent extreme weather events, hot and dry conditions, and erratic rainfall have become more pronounced, allowing aflatoxin-producing fungi to thrive, exacerbating the frequency and severity of contamination events (Chen et al., 2015). Heat and drought stresses are the most important abiotic stresses that predispose crops to Aspergillus infection and also affect crop productivity.
A promising strategy is the field application of atoxigenic A. flavus strains to reduce aflatoxin content in crops. In the United States and several African countries, driven primarily by USDA-ARS and IITA, respectively, the application of carefully selected atoxigenic A. flavus strains as biocontrol agents has consistently reduced aflatoxin contamination in commercially produced crops and allowed farmers to enter domestic and international premium markets (Cotty et al., 2007; Dorner, 2009; Mehl et al., 2012; Doster et al., 2014; Bandyopadhyay et al., 2019; Ortega-Beltran and Bandyopadhyay, 2019; Schreurs et al., 2019; Senghor et al., 2019). When applied at the right stage, treated crops accumulate over 80% less and sometimes even 100% less aflatoxin than non-treated adjacent crops. In addition, when biocontrol is used as a centerpiece of a holistic aflatoxin management strategy, lower aflatoxins accumulate in treated crops at harvest and throughout storage (Bandyopadhyay et al., 2019). Research groups in Italy, Argentina, China, Thailand, and Australia have conducted extensive work on biocontrol in addition to the United States and Africa (Alaniz Zanon et al., 2013, 2016; Mauro et al., 2015; Pitt et al., 2015). Although significant progress has been made, there are many countries where the biocontrol technology has not yet been developed and in the meantime other aflatoxin management strategies need to be employed.
In rainfed areas where farmers are subjected to unavoidable biotic and abiotic stresses that influence aflatoxin accumulation, it is paramount to conduct comprehensive genetics and genomics studies for a better understanding of the genetic behavior, genetic architecture, and molecular mechanisms that govern different types of aflatoxin resistance in groundnut and maize. Several genetic mapping studies conducted in both groundnut and maize have concluded that aflatoxin resistance is a quantitative trait and has complex genetic behavior with high G × E interaction (Chen et al., 2015; Pandey et al., 2019). Hence, by dissecting host-pathogen interactions during fungal infection by aflatoxin producers and aflatoxin contamination, important host-specific, resistance-related genes/proteins/pathways/resistant factors can be characterized in both groundnut and maize. This study focusses on the current status of resistance and molecular mechanisms in these two major crops using different omics approaches such as genetics, genomics, transcriptomics, and proteomics in addition to emphasizing on host-pathogen interactions. We also discuss the research gaps in global efforts to understand resistance mechanisms and translational genomics in developing aflatoxin-resistant groundnut and maize varieties to provide safe products to consumers as well as safeguard the multibillion-dollar industries associated with both crops.
General Characteristics of Aflatoxin-Producing Fungi
Aspergillus is a diverse genus of fungi that contains more than 200 species (Samson, 1992). Among those that produce aflatoxin, the agriculturally important species belong to section Flavi (Frisvad et al., 2019). Within section Flavi, A. flavus and A. parasiticus are the most common causal agents of aflatoxin contamination and are associated with a large number of crops (Pildain et al., 2008; Probst et al., 2014). A. flavus produces B aflatoxins and A. parasiticus produces both B and G aflatoxins. Some A. flavus strains cannot produce aflatoxin due to deletions or defects in the aflatoxin biosynthesis gene cluster (Chang et al., 2005; Adhikari et al., 2016). A. flavus strains may also produce other toxic compounds such as sterigmatocystin, cyclopiazonic acid, kojic acid, β-nitropropionic acid, aspertoxin, aflatrem, gliotoxin, and aspergillic acid (Hedayati et al., 2007); however, their incidence and frequency in field crops and toxicity to humans and animals are not clear.
Based on sclerotia size, A. flavus can be classified into two groups, L and S morphotypes. L morphotype produces few, large sclerotia (>400 μm), abundant conidia, and variable aflatoxin levels while S morphotype produces few conidia, abundant small sclerotia (<400 μm), and consistently high aflatoxin levels (Cotty, 1989). Some L morphotype strains do not produce aflatoxin due to lesions in the aflatoxin gene cluster and are known as atoxigenic (Chang et al., 2005; Adhikari et al., 2016). In nature, A. flavus produces primarily asexual spores (conidia) (Amaike and Keller, 2011). The fungus lives in the soil as conidia and the sclerotia, aggregates of hyphae that serve as survival structures that germinate to form saprophytically growing mycelia. Conidia are carried by wind or insects to host tissues, where they germinate and infect both aerial and subterraneanly grown organs of agronomically important crops (Cotty, 2001; Amaike and Keller, 2011); hence, insects may act as vectors during crop infection. Sclerotia allow aflatoxin producers to survive in extreme environmental conditions (Wicklow et al., 1993; Payne, 1998). Certain strains of A. flavus – both aflatoxin producers and atoxigenic strains – have higher adaptation and increased competitiveness in diverse cropping systems (Mehl and Cotty, 2011; Atehnkeng et al., 2016; Agbetiameh et al., 2019). Further, sexual reproduction has been reported to occur in A. flavus, A. parasiticus, and A. nomius under highly artificial laboratory conditions (Horn et al., 2009a, b) and also in the field after the release of A. flavus sclerotia incubated for 6 months (Horn et al., 2014). However, the significance of sexual reproduction in nature needs further studies.
Factors Affecting Toxigenicity and Aflatoxin Contamination
Different biotic factors such as fungal virulence, host susceptibility, insect damage, and abiotic factors such as soil moisture, temperature, high humidity, and mechanical damage while attempting inter-cultivation practices significantly influence A. flavus invasion and aflatoxin accumulation in groundnut (Asis et al., 2005). In maize, hot and dry environments (>32°C and >70% RH), drought conditions and damage to kernel seed coat compromise predispose the crop to aflatoxin contamination. Under drought conditions, drought-tolerant varieties accumulate lower aflatoxin levels compared to non-drought-tolerant varieties. High grain moisture increases post-harvest molding and aflatoxin contamination. Hence, proper drying of grains after harvest to 7% moisture level in groundnut and 12% moisture level in maize is ideal to prevent fungal growth (Liang et al., 2009). Temperature is also an important factor as A. flavus thrives well in a wide range of temperatures between 10 and 40°C. However, the optimum temperature range for high AP by A. flavus is 25–30°C (Gqaleni et al., 1997). Storage conditions largely influence aflatoxin in crops. Storing pods/grains in jute bags provides favorable conditions for A. flavus growth. Jute bags can easily absorb moisture because of high porosity which favors rapid growth and multiplication of molds. Purdue Improved Crop Storage (PICS) bags that rely on the principle of hermetic storage have been used to prevent A. flavus infestation and aflatoxin contamination during storage (Sudini et al., 2015; Danso et al., 2018, 2019; Walker et al., 2018). Although aflatoxin contamination is more severe in the field during pre-harvest stage, contamination may increase during post-harvest if management practices such as transportation and storage are deficient. Hence, integrated management of aflatoxin contamination during pre-harvest, post-harvest and storage is necessary to reduce aflatoxin contamination and aflatoxin exposure.
Genetics of Resistance Mechanisms
The mechanisms of resistance to infection and reduced AP are quantitative in nature (Warburton and Williams, 2014). In groundnut, the mechanisms include resistance to infection in the pod wall, resistance to seed invasion and colonization of seed coat, and resistance to AP in cotyledons. At the time of infection, aflatoxin producers have to penetrate the pod wall and then the seed coat to reach the cotyledons, from which they derive nutrients and produce aflatoxin. In groundnut, resistance to pod infection is attributed to pod shell structure, while resistance to seed invasion and colonization are mostly physical and attributed to seed coat thickness, density of palisade cell layers, and the presence of wax layers (Upadhyaya et al., 2002). In the case of maize, resistance mechanisms include good husk coverage, presence of proteins inhibiting fungal growth (Moore et al., 2004; Chen et al., 2010) wax, and cutin layers (Russin et al., 1997; Gembeh et al., 2001). Maize with kernel integrity intact and a living embryo typically accumulates less aflatoxin (Brown et al., 1993).
Generation mean analysis in maize has shown that additive and dominant gene action are important for resistance to AP (Campbell et al., 1997; Busboom and White, 2004). Diallele mating designs were used to study the inheritance of resistance to both Aspergillus ear rot and aflatoxin accumulation. These two studies reported that general combining ability had a greater effect on aflatoxin resistance in maize than specific combining ability, suggesting that additive gene effect is more important than dominant gene effect (Darrah et al., 1987; Gorman et al., 1992).
A resistant inbred of maize Oh516 was developed from the cross (B14 × L97) × B14 at Ohio State University and the hybrid derived from testcross Oh516 × B73 showed resistance to A. flavus infection and low aflatoxin concentration in grain (Campbell and White, 1995). The resistant inbred lines from testcross Oh516 × B73 were not significantly different from the inbred lines developed from the testcross Tex6 × B73 (Paul et al., 2003). F1 crosses developed with inbred lines Oh516 or Tex6 had lower aflatoxin concentration in grain than crosses without Oh516 or Tex6. The F1 cross Oh516 × Tex6 had the lowest aflatoxin concentration in grain of all F1 crosses. These findings indicate that the resistance mechanism is quantitative in nature and may be governed by multiple genes.
Types of Resistance Mechanisms
Groundnut has three types of resistance mechanisms, i.e., IVSC, PAC, and AP (Nigam et al., 2009; Figure 1). Similarly, in maize, the resistance is a sum of (1) prevention of fungal infection; (2) prevention of subsequent growth of the fungus after infection; and (3) inhibition of aflatoxin biosynthesis after infection (Williams et al., 2015). The extent of aflatoxin contamination varies with geographical location, cultural and agronomic practices, storage and processing period.
FIGURE 1.

Aflatoxin resistance mechanisms in groundnut. IVSC, in vitro seed colonization; PAC, pre-harvest aflatoxin contamination; AP, aflatoxin production.
In groundnut, the majority of contamination occurs in the field. Hence in the context of developing aflatoxin-resistant groundnut cultivars, host resistance for PAC is a preventive approach that is economical and easy to disseminate. Such strategy does not require extra resources for farmers, leaves no chemical residues as a result of fungicide usage, and is an alternative for areas/nations where atoxigenic biocontrol measures are not available (Garrido-Bazan et al., 2018). ICRISAT has been deploying genetics and genomics approaches to understand resistance mechanisms and identify resistant genes/haplotypes to amalgamate all the three resistance mechanisms into a single genetic background in groundnut using genomics-assisted breeding (GAB) (Pandey et al., 2019). In addition to genetic resistance in groundnut and maize, reduced aflatoxin accumulation will require multidisciplinary approaches such as the use of biocontrol agents, good harvesting practices, appropriate drying, and optimal post-harvest storage (Logrieco et al., 2018). In the long run, the development of new breeding lines using introgression of validated quantitative trait loci (QTLs), single nucleotide polymorphism (SNPs) associated with resistance at the pre-harvest and/or post-harvest stages, optimized markers for marker-assisted selection (MAS), marker-assisted recurrent selection (MARS), and genomic selection (GS), can help the farming community grow crop varieties that may accumulate less/minimal aflatoxin.
Physical and Chemical Barriers to Infection
In groundnut, seed coat thickness and its permeability confer resistance against A. flavus infection as a seed coat is the outermost layer that acts as a physical barrier (LaPrade et al., 1973). Smaller hila, a more compact arrangement of palisade-like layer of testa, and thicker waxy surface contribute to resistance against A. flavus infection (Taber et al., 1973). It has been reported that higher wax and cutin deposits in groundnut lead to resistance to A. flavus invasion and AP in resistant genotypes than in susceptible genotypes (Liang et al., 2003b). Hence, the seed coat, wax, and cutin are effective physical barriers to pathogen invasion and colonization. Groundnut testa is a rich source of tannins that inhibit A. flavus infection. 5-7-dimethoxyisoflavone (Turner et al., 1975) and tannins (Sanders and Mixon, 1979) have been reported as important inhibitors of A. flavus infection. In groundnut, tannins inhibit A. parasiticus growth by arresting mycelial growth and reducing AP (Sanders and Mixon, 1979). The basic composition of testa also contributes to the resistance to invasion. A study on protein profiling in a panel of 15 groundnut genotypes revealed that resistant genotypes had higher trypsin content and activity than susceptible genotypes (Liang et al., 2003a).
In maize, trypsin, ribosome-inactivating protein (RIP), and zeamatin act as inhibitors to the infection of A. flavus and A. parasiticus, and many other fungi (Chen et al., 1998). Resistance to colonization results from a variety of physiological, biochemical, and molecular factors at different levels of infection. Elevated levels of chitinases pCh2 and pCh11 were reported in the aleurone layer of maize in damaged grains colonized by A. flavus (Moore et al., 2004). Hence, breeding to strengthen physical features such as thick testa and chemical barriers such as thick cutin and lignin layers can inhibit A. flavus infection and aflatoxin contamination. Similarly, improving the aleurone layer of maize with high chitinase and trypsin inhibitor can reduce aflatoxin accumulation.
Constitutive and Induced Resistance Mechanisms
Host plant resistance to biotic stresses has been characterized into two categories, i.e., constitutive and induced resistance. Phytoanticipins confer constitutive resistance while phytoalexins contribute to induced resistance (VanEtten et al., 1994). Secondary metabolites are known to be involved in controlling several immune responses, e.g., callose deposition and programed cell death (Piasecka et al., 2015). Phytoanticipins are antimicrobial metabolites (Pedras and Yaya, 2015). For instance, the groundnut plant produces a variety of phenylpropanoids, such as p-coumaric acid, caffeic acid, ferulic acid, methoxycinnamic acid, and mucilagin A, a phenylpropanoid-polyketide-isoprenoid. These metabolites have been known to have antifungal activities against both A. flavus and A. parasiticus (Sobolev et al., 2006). These phenylpropanoids are likely to function as phytoanticipins in specific groundnut plant tissues (Pedras and Yaya, 2015). Phenylalanine ammonia lyase (PAL) which is a precursor of lignin and phytoalexins, has increased rapidly and reached maximum levels in resistant groundnut genotypes than in susceptible ones (Liang et al., 2001). In the case of membrane lipid peroxidation, the level of malondialdehyde (MDA) increased by 8-fold 2–3 days after inoculation (DAI). Moreover, the generation of O2–, H2O2, and lipoxygenase (LOX) also increased markedly at the early stage after infection in groundnut (Liang et al., 2002). Resveratrol is an antifungal secondary metabolite or phytoalexin compound found in groundnut seeds (Wang et al., 2015). In resistant genotypes, resveratrol levels increased by 30-fold on the third DAI (Liang et al., 2006). In contrast, the resveratrol level remained unchanged even on the 4-DAI in susceptible genotypes. Plants have several inducible defense responses to pathogens, such as lignification, cell wall cross-linking, phytoalexins, hypersensitive response, production of reactive oxygen species (ROS), and pathogenesis-related (PR) proteins (Liang et al., 2006).
In maize, the first line of defense in response to A. flavus results in the activation of expression of transcriptional factors such as WRKY that confer resistance against pathogens (Skriver and Mundy, 1990). WRKY transcription factors were found to be significantly upregulated by A. flavus infection in developing maize kernels of resistant maize line TZAR101 (Fountain et al., 2015). ZmWRKY53 is highly expressed in response to a necrotrophic pathogen and also regulates chitinase and peroxidase gene expression. Lignin cross-linking in the cell wall contributes to the resistance to A. flavus infection. For instance, less A. flavus growth was observed in Mp313E, a maize line that has high cross-linked lignin compared to the susceptible line SC212 (Magbanua et al., 2013). For breeding aflatoxin resistance, the genetic transformation or introgression of resistance genes and transcription factors such as WRKY, PAL, and LOX genes can improve groundnut and maize varieties and reduce the burden of aflatoxin contamination.
Genomic Regions Controlling Aflatoxin Resistance
Several QTL mapping studies have been performed leading to discovery of genomic regions for aflatoxin resistance in groundnut and maize (Table 1). Each QTL mapping experiment in groundnut has had at least one QTL with phenotypic variation explained (PVE) > 10% and reaching up to >20% in some cases. Interestingly in maize, some QTLs were mapped on same genomic regions in different mapping populations which indicated that there are some genes underlying similar function in different studies (Warburton and Williams, 2014; Parish et al., 2019).
TABLE 1.
Key bi-parental QTL mapping and GWAS studies for discovery of genomic regions controlling aflatoxin contamination in groundnut and maize.
| Population | Trait | No. of QTLs/MTAs | LOD/p-value range | PVE% range | References |
| Groundnut (Arachis hypogaea) | |||||
| Bi-parental QTL mapping | |||||
| Zhonghua 10 × ICG 12625 (RIL population) | PSII | 2 | 3.1–5.0 | 8.0–13.0 | Yu et al., 2019 |
| AFB1 | 7 | 3.1–6.4 | 7.3–17.9 | Yu et al., 2019 | |
| AFB2 | 5 | 3.5–8.8 | 8.3–21.0 | Yu et al., 2019 | |
| Yueyou 92 × Xinhuixiaoli (RIL population) | Resistance to A. flavus | 2 | 2.9–10.5 | 5.2–19.0 | W. Zhuang (personal communication) |
| Genome-wide association study (GWAS) | |||||
| ICRISAT Reference Set 300 | Resistance to A. flavus | 1 | 9.68 × 10−7 | 24.7 | Pandey et al., 2014 |
| Maize (Zea mays) | |||||
| Bi-parental QTL mapping | |||||
| M53 × RA (F8:9 RIL population) | Resistance to A. flavus | 8 | 2.2–5.4 | 3.6–9.9 | Yin et al., 2014 |
| Mp313E × Va35 (F2:3 population) | Aflatoxin content | 20 | 2.4–8.0 | 0.2–21.6 | Willcox et al., 2013 |
| Mp715 × T173 (F2:3 population) | Aflatoxin content | 12 | 1.8–11.5 | 2.7–18.5 | Warburton et al., 2011 |
| NC300 × Mp717 (F2:3 population) | Aflatoxin content | 12 | − | 1.0–11.0 | Warburton et al., 2009 |
| B73 × Mp313E (F2:3 population) | Aflatoxin content | 13 | 2.9–7.8 | 5.0–18.4 | Brooks et al., 2005 |
| Tex6 × B73 (BC1S1) | Aflatoxin content | 2 | 3.8–4.2 | 16.1–17.8 | Paul et al., 2003 |
| Tex6 × B73 (F2:3) | Aflatoxin content | 3 | 2.5–5.2 | 6.7–15.1 | Paul et al., 2003 |
| RA × M53 (RIL population) | Amount of Aflatoxin (AA) | 1 major QTL (qAA8) | 8.42 | 18.23 | Zhang et al., 2016 |
| 6 epistatic QTLs | 5.0–5.4 | 14.05–22.6 | Zhang et al., 2016 | ||
| B73 × CML322 (F2S5) RIL population | Afl, ICS, IFS, KSP, and SSP | 10 | 2.6–6.2 | 6.0–16.0 | Mideros et al., 2014 |
| B73o2/o2 × CML161 RIL population | Aflatoxin accumulation | 9 | 3.0–4.0 | 8.0–11.0 | Mayfield et al., 2011 |
| B73o2/o2 × CML161 RIL population | Aflatoxin accumulation | 9 | 2.7–3.9 | 7.8–11.3 | Bello, 2007 |
| Genome-wide association study (GWAS) | |||||
| Maize inbred lines (346 line) | Aflatoxin resistance | 6 | 5.1–5.5 | 4.8–6.1 | Farfan et al., 2015 |
| Inbred lines (300 line) | Resistance to aflatoxin accumulation (RAA) | 107 | 9.8 × 10–6 to 2.9 × 10–10 | 5.4–16.0 | Warburton et al., 2015 |
| Maize inbred lines (437 lines) | Amount of aflatoxin (AA) | 3 | 1.1 × 10−8 to 2.1 × 10−7 | 6.7–10.4 | Zhang et al., 2016 |
| Resistance to A. flavus infection (RAI) | 22 | 3.7 × 10−22 to 8.7 × 10−6 | 6.4–26.8 | Zhang et al., 2016 | |
| Maize inbred lines (287 lines) | Grain aflatoxin levels | 298 Maize Cyc pathways | 2.9 × 10–10 to 1.0 | 6.4 × 10–14 to 0.3 | Tang et al., 2015 |
BC1S1, selfed backcross population; PSII, percent seed infection index; AFB1, aflatoxin B1; AFB2, aflatoxin B2; IVSC, in vitro seed colonization; RIL, recombinant inbred lines; Chr, chromosome; LOD, logarithm of odds; ICS, infection on silk tissue; IFS, infection frequency on silk tissue; KSP, sporulation on developing kernels; SSP, sporulation on silk tissue; Afl, aflatoxin accumulation.
In groundnut, very few genetic mapping studies have been reported for aflatoxin resistance. Individual QTLs were identified for AFB1, AFB2, and (percent seed infection index; PSII) using a recombinant inbred line (RIL) population Zhonghua 10 × ICG 12625 by Yu et al. (2019). The study identified two QTLs for PSII, one on chromosome A03 with 8.0% PVE and another on chromosome A10 with 13.0% PVE. Seven QTLs were identified for AFB1 (Aflatoxin B1) resistance, of which two major QTLs were detected on chromosomes A07 and B06 with 17.9 and 16.3% PVE, respectively. Similarly, five QTLs were identified for resistance to AFB2, of which chromosomes A07, B05, B06, and B07 recorded higher PVEs of 12.2, 11.1, 21.0, and 14.5% PVE, respectively. Two consistent QTLs for AFB1 (Aflatoxin B1) and AFB2 (Aflatoxin B2) and one for PSII were identified (Yu et al., 2019). Genetic mapping using a groundnut RIL population Yueyou 92 × Xinhuixiaoli for IVSC identified two major QTLs on chromosomes A03 and B04 with LOD of 10.5 and 2.9 and 19.0 and 5.1% PVE, respectively (W. Zhuang, personal communication). Similarly, genome-wide association studies using a groundnut reference set identified a marker associated with IVSC explaining 24.7% PVE (Pandey et al., 2014). One groundnut MAGIC population using eight genotypes possessing resistance to Aspergillus infection and reduced aflatoxin accumulation has been developed at ICRISAT for genetic dissection of component traits.
In the case of maize, major effect QTLs were identified in crosses Tex6 × B73 (F2:3) and Tex6 × B73 (BC1S1) on chromosomes 3, 4, 5, and 10 with 6.7–17.8% PVE (Paul et al., 2003). Another study (Brooks et al., 2005) conducted in F2:3-derived maize populations reported two major effect QTLs for aflatoxin resistance in B73 × Mp313E population that were significant across environments. Other studies in maize have identified one stable QTL in NC300 × Mp717 population which was stable across years. Warburton et al. (2009), three major effect QTLs explaining PVE ranging from 12.1–21.6% in Mp313E × Va35 population (Willcox et al., 2013); small effect QTLs in M53 × Mo17 population (Yin et al., 2014), and single QTL explaining 18.5% PVE in Mp715 × T173 population (Warburton et al., 2011). Similarly, QTL for log aflatoxin accumulations were detected on chromosomes 1, 3, 4, and 9, explaining a total of 17% PVE; while QTL for aflatoxin were detected on chromosomes 3, 4, and 8, explaining a total of 15% PVE in RIL population B73o2/o2 × CML161 (Mayfield et al., 2011). In fact, the same population (B73o2/o2 × CML161) was used earlier (Bello, 2007). QTLs affecting aflatoxin from both parents; however, the favorable alleles for the QTL detected by Bello (2007) were derived mainly from CML161 (Mayfield et al., 2011). In earlier aflatoxin QTL studies, Brooks et al. (2005) evaluated their germplasm in four environments, Paul et al. (2003) used two environments, and Warburton et al. (2009) used four environments. All these studies reported few significant QTLs detected in more than one environment. Warburton et al. (2009) reported the most, with one QTL present in all four environments and one QTL detected in two environments. However, Mayfield et al. (2011) reported three QTLs one on each of chromosomes 1, 4, and 9, across multiple years and environments. In another study by using the B73 × CML322 population, ten QTLs with 6.0–16.0% PVE were found using two QTL mapping methods, six of which were located on the same chromosome segments using both approaches (Mideros et al., 2014). By using various sources of near-isogenic lines (NILs) for selected loci, the resistance QTL located in bin 4.08 was confirmed using a NIL pair. Furthermore, the meta-analysis of QTLs using data from 12 populations indicated that the QTL in bin 4.08 has been reported in four mapping populations. The study showed that the largest-effect QTL, located in bin 4.08, is a good candidate for further characterization and use.
In addition to bi-parental QTL mapping studies, many diverse association panels have been used for genome-wide association study (GWAS) leading to the identification of markers/genomic regions for aflatoxin resistance in maize. For instance, Farfan et al. (2015) identified 6 MTAs for aflatoxin resistance with 4.79–6.06% PVE. In another study (Warburton et al., 2015), GWAS analysis using 300 maize inbred lines identified 107 SNPs associated with aflatoxin accumulation in one or more environments in the association panel. Similarly, in another study using an association panel of 437 maize inbred lines, Zhang et al. (2016) identified 3 MTAs for AA and 22 MTAs for resistance to A. flavus infection (RAI). In a comprehensive GWAS analysis undertaken by Tang et al. (2015), 298 maize Cyc pathways were reported to be associated with resistance mechanisms, 17 of the pathways reported high enrichment scores of false discovery rate (FDR) < 0.2, of which the jasmonic acid biosynthesis pathway seems to be a major one for aflatoxin resistance. While these studies are informative, comprehensive efforts are required to perform high resolution GWAS in maize and especially in groundnut so that candidate genomic regions/genes can be identified and validated for breeding applications.
Molecular Basis of Aflatoxin Resistance Mechanisms
Identification of Resistance-Associated Proteins
Proteomics approaches have identified several plant proteins involved in host-pathogen interaction and in controlling resistance to fungal invasion and toxin production in both groundnut and maize. For instance, in groundnut, a 2D-based proteomics study identified pathways/proteins including resistance-associated proteins (RAPs) which were associated with pre-harvest aflatoxin resistance under drought stress conditions (Wang et al., 2010). That study highlighted the role of iso Ara-h3, oxalate oxidase, PII protein, trypsin inhibitor, SAP domain-containing protein, CDK1, L-ascorbate peroxidase, RIO kinase, and heat shock proteins in reducing aflatoxin accumulation at pre-harvest aflatoxin resistance. Later, Wang et al. (2012) identified several RAPs in groundnut which were key controllers of pathways such as immune signaling, PAMP perception, cell wall responses, and detoxification. The study on effect of H2O2-derived oxidative stress on A. flavus isolates discovered a sub-set of genes that control fungus pathogenicity, mycelial development, and manage ROS production (Fountain et al., 2018).
In maize, several proteomic approaches have been used to understand the molecular mechanisms involved in host-pathogen interaction and resistance to AP. For instance, RIP and zeamatin were present in higher concentrations in germinating maize kernels and led to decreased aflatoxin levels in susceptible maize kernels and thereby inhibited the growth of A. flavus under imbibed conditions (Guo et al., 1997). A similar study has indicated the importance of fungal cell wall degrading enzymes, particularly isoforms of beta-l,3-glucanase and chitinase, which are induced in maturing kernels in response to A. flavus infection and also in maturing uninfected kernels (Lozovaya et al., 1998; Ji et al., 2000). Importantly, antifungal proteins chitinase and zeamatin appear to be associated with the host first and second layer of resistance (Guo et al., 1997), and their constitutive expression in maize can provide resistance against A. flavus. Grains of resistant maize genotypes can accumulate inhibitory proteins such as 22 and 28kDa which restrict the growth of the fungus as they are associated key resistant proteins like PR-5 thaumatin-like proteins and zeamatin (Huang et al., 1997; Moore et al., 2004). In another study, the proteome analysis of resistant maize genotypes identified a constitutive expression of 14-kDa trypsin inhibitor that can cause spore rupture and abnormal hyphal development in A. flavus (Chen et al., 1998). Also, the trypsin inhibitor produced by maize can inhibit fungal-amylase activity that limits pathogen access to the host food resource (starch) which in turn restrict fungus mycelial growth and sclerotia development (Woloshuk et al., 1997; Chen et al., 1998, 1999).
A proteomic examination of maize seeds has identified several groups of proteins associated with the embryo and endosperm that were significantly upregulated upon A. flavus infection. These proteins were grouped into four categories: storage proteins, water stress-related proteins, PR proteins, and antifungal proteins (Chen et al., 2002, 2004b, 2006, 2007, 2012). Storage proteins globulin 1 and 2, water stress responsive related proteins WSI18, aldose reductase, late embryogenesis abundant (LEA; LEA3 and LEA14) and heat stress related proteins (HSP16.9) impart kernel resistance (Chen et al., 2002). Further, glyoxalase I (GLX-I; EC 4.4.1.5), a stress-related protein, directly controls methylglyoxal levels, an aflatoxin inducing substrate, thereby contributing to lower aflatoxin levels in resistant maize genotypes (Chen et al., 2004b). The RAP involves maize PR-10, which exhibits ribonucleolytic and antifungal activities (Chen et al., 2006, 2007); and the genes of encoding PR proteins are usually highly expressed in resistant genotypes (Chen et al., 2007). A United States–Africa collaborative project identified resistant maize inbred lines (Menkir et al., 2006, 2008; Meseka et al., 2018). The project reported the development of 52 BC1S4 lines from crosses between five African maize inbreds and five temperate aflatoxin-resistant lines followed by the identification of RAPs related to antifungal, stress-related, storage or regulatory protein categories (Chen et al., 2012). Resistant inbred lines of maize are known to express higher levels of chitinase and proteins associated with phenylpropanoid metabolism pathways (Peethambaran et al., 2009; Pechanova et al., 2011).
Using multiple approaches in groundnut and maize have led to the identification of several moderate/low/high resistant lines for A. flavus infection and reduced aflatoxin contamination. These advances have facilitated the development of aflatoxin-resistant transgenic groundnut (Sharma et al., 2018) and maize (Thakare et al., 2017); and it is expected that in the coming years, farmers may have access to superior and aflatoxin-resistant varieties. However, the release of transgenic cultivars is dependent on their acceptance by regulators in the target countries. To date, the use of transgenic maize is accepted only in South Africa and Sudan in Africa. A summary of different proteomic studies in maize and groundnut is provided in Table 2. Cumulatively, these studies enhance our knowledge of target proteins in order to identify protein encoding resistance genes in response to aflatoxin contamination in these crops.
TABLE 2.
List of key proteins and their functions associated with resistance to aflatoxin contamination in groundnut and maize.
| RAPs | Function | References |
| Groundnut | ||
| Oxalate oxidase | Seed storage protein | Wang et al., 2010 |
| Trypsin inhibitor | Antifungal compound | |
| SAP domain-containing protein | Abiotic stress tolerance protein | |
| L-ascorbate peroxidase | Regulates antioxidant metabolism | |
| Iso Ara-h3 | Seed storage protein | |
| Heat shock protein precursor | Regulates heat shock factors | |
| LRR receptor serine/threonine kinase | PAMPs perception | Wang et al., 2012 |
| Protein phosphatase 2A regulatory B subunit | Dephosphorylation | |
| Pentatricopeptide repeat-containing protein | RNA stabilization | |
| Esterase_lipase | Lipid metabolism | |
| Cytochrome P450 | Degrades toxins | |
| Maize | ||
| Zeamatin | Antimicrobial, fungicide | Guo et al., 1997; Huang et al., 1997; Chen et al., 2002 |
| Ribosome-inactivating protein (RIP) | Protein synthesis inhibitor | Guo et al., 1997 |
| Chitinase | Hydrolytic enzymes that degrade chitin | Guo et al., 1997; Ji et al., 2000; Chen et al., 2002 |
| Glucanase | Destroys cell wall of fungi | Guo et al., 1997 |
| Beta-1,3-glucanase | PR-2 family protein, antifungal | Lozovaya et al., 1998; Ji et al., 2000 |
| PR-5 thaumatin-like protein | PR protein | Huang et al., 1997 |
| Globulin-1,2 | Seed storage proteins | Chen et al., 2001, 2002, 2006, 2012 |
| Endochitinase | Degrades chitin molecule at random point | Huang et al., 1997 |
| 14-kDa trypsin inhibitor | Spores rupture and cause abnormal hyphal development | Chen et al., 1998, 1999 |
| LEA3,14 | Stress responsive proteins | Chen et al., 2002, 2006, 2012 |
| WSI18 and aldose reductase | Osmo-stress responsive and oxidative stress responsive proteins | Chen et al., 2002 |
| HSP16.9 (Heat stress related) | Stress responsive protein | Chen et al., 2002 |
| Glyoxalase I | Controls methylglyoxal level as it stimulates the expression of aflR, an aflatoxin regulatory gene | Chen et al., 2004a |
| PR-10 | Disease resistance | Chen et al., 2006 |
| Stress-related-peroxiredoxin antioxidant (PER1) | Antioxidants proteins that protect against oxygen species | |
| Heat shock proteins (HSP17.2) | Stress responsive proteins | |
| Antifungal trypsin inhibitor protein (TI) | Inhibits A. flavus growth | |
| Cold-regulated protein (COR) | Inhibits germination of A. flavus conidia and mycelial growth | Chen et al., 2006, 2012 |
| Superoxide dismutase | Enhances oxidative stress tolerance | Chen et al., 2012 |
| Peroxiredoxin | Enhances oxidative stress tolerance | |
| Cupindomain-containing proteins | Seed storage protein | |
| Putative lipid transfer protein | Stress responsive | |
| Eukaryotic translation initiation factor 5A | Plays a role in plant growth and development | |
| Abiotic stress responsive proteins | PR protein and stress responsive | Peethambaran et al., 2009 |
| PRm3 chitinase | Fungal cell wall degradation and stress resistance | |
| Chitinase 1 | Defense mechanism in response to biotic stress | |
| Chitinase A | Suppresses fungal growth | |
| Phenylpropanoid metabolism | Secondary metabolite production | Pechanova et al., 2011 |
Identification of Candidate Genes
Functional genomics provides new insights into a wide number of candidate genes associated with resistance to aflatoxin contamination in both groundnut and maize (Table 3). In the case of groundnut, transcriptomics studies have identified candidate genes, pathways, and the regulatory networks for the three resistance mechanisms of aflatoxin accumulation (IVSC, PAC, and AP). Earlier efforts to identify resistance/differentially expressed genes in groundnut were based on EST or microarray-based techniques (Luo et al., 2005; Guo et al., 2011; Wang et al., 2013). The gene expression profiling approach was deployed by Luo et al. (2005) in A13 drought-tolerant and pre-harvest aflatoxin-resistant groundnut genotypes in which a cDNA microarray containing 384 unigenes was selected from two cDNA libraries. Overall, the microarray-based screening approach identified defense responsive (Kunitz-type trypsin inhibitor, auxin repressed protein, cystatin-like protein), signaling component (ethylene-responsive protein, calcium-binding protein), ion-proton transporter (aquaporin 1), stress proteins, and secondary metabolites (lipoxygenase 1) resistance genes in groundnut in response to A. parasiticus infection under drought stress (Luo et al., 2005).
TABLE 3.
A summary of some transcriptomics studies to identify candidate genes involved in aflatoxin contamination in groundnut and maize.
| Candidate genes | Functions of candidate genes | References |
| Groundnut | ||
| Seed maturation protein LEA 4 | Stress responsive protein | Guo et al., 2008 |
| Serine protease inhibitor | Involved in inflammatory responses | |
| Cu/Zn superoxide dismutase II | Antioxidant defensive protein | |
| Serine protease inhibitor | Involved in inflammatory responses | |
| Lipoxygenase | Regulates jasmonic acid signaling pathway | Guo et al., 2011 |
| Proline-rich protein | Stress responsive protein | |
| Cupin//Oxalate oxidase | Seed storage protein | |
| LEA-protein 2 | Stress responsive protein | |
| Brassinosteroid Insensitive 1-associated Receptor kinase 1 | Defense response | Wang et al., 2013 |
| 3-ketoacyl-CoA synthase | Fatty acid biosynthetic process | |
| Em protein | Stress responsive | |
| TIR | Defense response | |
| Defensin | Defense response | |
| Mitogen-activated protein kinase | Signaling cascade gene | Wang et al., 2016 |
| PR proteins | Disease resistance | |
| Nucleotide-binding site-leucine-rich repeat proteins | PAMPs perception | |
| Polygalacturonase inhibitor proteins | Inhibit polygalactouronase produced by the fungal pathogen | |
| Abscisic acid insensitive5 | Participates in ABA signaling pathway | Clevenger et al., 2016 |
| BLH1 | Modulates seed development | |
| Respiratory burst oxidase homolog | Regulates numerous plant cell responses | |
| 13S-lipoxygenases | Lipid metabolism | |
| PR-2 | Disease resistance in plants | |
| Deoxy-chalcone synthase | Synthesizes phytoalexins | |
| Resveratrol synthase | Biosynthesis stilbene type-phytoalexins | Nayak et al., 2017 |
| Chalcone synthase | Involved in the flavonoid biosynthesis pathway | |
| Epoxide hydrolase | Detoxification of reactive epoxide | |
| Receptor-like kinases | Cell wall signaling | |
| 9s-LOX | Lipid metabolism | |
| WRKY genes | Transcriptional regulators; regulates plant development | Korani et al., 2018 |
| Toll/Interleukin-1 receptor-nucleotide-binding site leucine-rich repeat (TIR-NBS-LRR) | Defense responsive | |
| α-linolenic acid metabolism | Lipid metabolism | |
| Hevamine-A | Defense protein | Zhao et al., 2019 |
| PR proteins | Disease resistance | |
| Chitinase | Hydrolytic enzymes that degrade chitin | |
| Maize | ||
| Kunitz-type trypsin inhibitor | Serine protease inhibitor activity | Luo et al., 2005 |
| Auxin repressed protein | Regulates growth and disease resistance | |
| Cystatin-like protein | Defense mechanism | |
| Lipoxygenase 1 | Regulates the jasmonic acid pathway | |
| Ion-proton transporter (Aquaporin 1), | Accelerates oxidative stress and cell signaling | |
| Glutathione S-transferase | Antioxidant | |
| Heat shock protein | Defense mechanism; regulates heat shock factors | |
| PR protein 1 | Disease resistance | |
| ADP glucose pyrophosphorylase | Starch metabolism | Luo et al., 2009 |
| 1-acyl-glycerol-3-phosphate acyltransferase | Lipid metabolism | |
| Lipoxygenase | Regulates the jasmonic acid pathway | |
| Oleosin 17 | Oil body formation and storage protein | |
| Abscisic acid inducible gene | Defense-related genes | |
| Chalcone synthase C2 | Involved in the flavonoid biosynthesis pathway | |
| Glutathione transferase | Antioxidant gene | Luo et al., 2011 |
| Leucine-rich repeat-like protein | Biotic stress-related gene | |
| ABI3-interacting protein 2 | A transcription factor of the abscisic acid signal transduction pathway that plays a role in seed development | |
| Beta-1,3-glucanase | Classified in PR-2 family of PR proteins, antifungal | |
| Zeamatin-like protein | Antimicrobial, fungicide | |
| PR genes | PR genes | |
| Phosphoglycerate dehydratase 1 | Plays a role in catalysis | Luo et al., 2010 |
| Heat shock protein 90 | Signal transduction and stress responsive | |
| Glycine−rich protein | Stress responsive and signaling | |
| Cytochrome P450 | Degrades toxins | |
| Ethylene-responsive element binding factor | Regulates jasmonic acid signaling pathway | |
| 9-oxylipins | Suppresses aflatoxin biosynthesis pathway | Fountain et al., 2013 |
| Lipoxygenase-3 (LOX3) | Regulates jasmonic acid signaling pathway | |
| PR proteins | Disease resistance | |
| NUP85-like genes | Transports RNA, R-proteins and macromolecules from the nucleus to the cytoplasm | Kelley et al., 2012 |
| Heat shock protein (HSP101) | Molecular chaperone protein | |
| Molecular chaperones | Plays a role in protein folding | |
| Cinnamoyl-CoA | Synthesizes lignin compounds | |
| PR-4 | Antifungal proteins play a role in pathogenicity | Dhakal et al., 2017 |
| Leucine-rich repeat family protein | Highly conserved region for disease resistance genes | |
| DEAD-box RNA helicase | Defense-related signaling | |
| Fructose-1,6-bisphosphatase | Carbohydrate metabolism | |
| Plant receptor protein kinases (RPK) | Senses pathogen signals and accelerates defense | |
| Cysteine proteinase inhibitor | Stress responsive | |
| PR-1, PR-4, PR-5, PR-10 | Disease resistance-related genes | |
| CC-NBS-LRR | Conserves disease resistance genes | Shu et al., 2017 |
| LRR-RLK | Conserves disease resistance genes | |
| Thaumatin- like protein | Regulates host defense mechanism | |
| Chitinase | Hydrolytic enzymes that degrade chitin | |
To understand the molecular mechanism of host-mediated resistance, a separate study was conducted in Aspergillus resistant (GT-C20) and susceptible (Tifrunner) genotypes of groundnut which identified 52 highly and 126 moderately expressed genes (Guo et al., 2011). This study reported several important genes including lipoxygenase, lea-protein 2, proline-rich protein, cupin//Oxalate oxidase, among others, in response to A. flavus infection. Some studies have suggested the possible involvement of LOX pathway in the production of jasmonic acid which plays hormone-like regulatory and defense-related roles in plants (Royo et al., 1996; Kolomiets et al., 2001; Yan et al., 2013; Ogunola et al., 2017).
Studies have reported that LOX genes also play a major role in plant defense mechanisms, growth, and developmental processes (Kolomiets et al., 2001, 2018; Gao et al., 2008; Park and Kolomiets, 2010). In this emerging field, more investigations are needed on host-pathogen cross-talk communication that fungi use to exploit the plant host in order to meet their biological needs (Christensen and Kolomiets, 2011). Some LOX genes have been shown to play an important role in plant defense resistance and in mediating fungal colonization and toxin production (Battilani et al., 2018).
A microarray study representing 36,158 unigenes was used to identify genes associated with aflatoxin resistance in groundnut (Wang et al., 2013), providing insights into the co-regulation of multiple pathways such as host defensive responses including carbohydrate biosynthesis/metabolism, transmembrane transport, coenzyme A biosynthesis, oxidation-reduction, proteolysis metabolism, etc., during aflatoxin resistance. Modern approaches such as RNA-seq have been used to identify host resistance associated pathways in different crops including maize and groundnut. For instance, in case of groundnut, an integrated IVSC and RNA-seq approach that analyzed the four different stages of infected seed samples from J11 (resistant) and JL24 (susceptible) identified 4,445 differentially expressed unigenes (DEGs) that were involved in multiple pathways such as defense-related, PR or metabolic pathway targeting genes provided a more solid understanding of cross-talk between host-pathogen interactions (Nayak et al., 2017).
Likewise, an RNA-seq-based approach was deployed in groundnut to identify genes that confer resistance during PAC (Clevenger et al., 2016). The study was able to associate the role of abscisic acid (ABA) signaling pathway during drought stress-induced aflatoxin contamination and/or PAC, and also revealed the role of genes from the fatty acid metabolism, cell wall restructuring and morphology, sugar metabolism and nitrogen metabolism pathways during A. flavus contamination in soil. Recently, Zhao et al. (2019) suggested the role of hevamine-A protein in groundnut during PAC resistance. Hevamine-A protein is an enzyme with chitinase activity that is also coordinated with PR proteins and can directly inhibit the growth of A. flavus (Zhao et al., 2019).
Post-harvest aflatoxin contamination can take place during drying, storage or transportation due to increase in humidity and/or insect damage, thereby promoting A. flavus infection. To understand the post-harvest resistance mechanism, Wang et al. (2016) performed global transcriptome profiling in the grains of resistant (Zhonghua 6) and susceptible (Zhonghua 12) genotypes of groundnut and identified 30,143 DEGs, of which 842 were defense-related genes, including mitogen-activated protein kinase, PR proteins, leucine-rich repeat receptor-like kinases transcription factors, nucleotide-binding site-leucine-rich repeat proteins, polygalacturonase inhibitor proteins, and ADP-ribosylation factors in response to AP by A. flavus. A recent study by Korani et al. (2018) provides new insights into post-harvest resistance mechanism in response to A. flavus infection by comparing the seed transcriptome of resistant (ICG 1471) and susceptible (Florida-07) groundnut cultivars. The study identified 4,272 DEGs and showed the importance of WRKY TFs, heat shock proteins and TIR-NBS-LRR in providing resistance. Further, this study also showed the altered expression of genes associated with protein processing in the endoplasmic reticulum, spliceosome mediated protein degradation and α-linolenic acid metabolism.
In maize, gene expression analysis of inbred line Tex6 identified 8,497 positive array spots including genes related to disease resistance (chitinase, zeamatin-like protein, endochitinase B precursor, PR-1;4;5), stress responsive (heat shock proteins, auxin responsive factor-1, D-type cyclin), ROS scavenger (glutathione S-transferase, superoxide dismutase), and defense-related genes, as well as storage protein genes and lipid metabolism genes (Luo et al., 2009). Further, Luo et al. (2010) have shown that jasmonate and abscisic acid biosynthetic and signaling pathways play crucial roles in drought-induced A. flavus infection and accumulation of aflatoxin in maize. The transcriptomic study of resistant maize (Eyl25) with susceptible (Eyl31) lines identified 530 DEGs including defense-related genes; beta-1,3-glucanase, zeamatin-like protein, trypsin inhibitor, and PR genes (Luo et al., 2011). Fountain et al. (2013) have highlighted the role of WRKY TFs in conferring resistance to Aspergillus infection and subsequently in reduced PAC in maize genotype. The transcriptomic study of maize kernels in two resistant inbred lines (Mp313E and Mp04:86) and two susceptible inbred lines (Va35 and B73) under artificial inoculation conditions identified NUP85-like genes in resistance (Kelley et al., 2012). The NUP85-like protein is a major part of nuclear pore complex (NPCs) and is involved in the transportation of RNA, R-proteins, and other macromolecules from the nucleus to the cytoplasm (Cheng et al., 2009; Garcia and Parker, 2009). A few more genes like heat shock protein (HSP101), metallothionein-like protein (MTLP), lecithin cholesterol acyltransferase (LCAT)-like gene, Prenylated Rab PRA1 proteins, molecular chaperones, and detoxification proteins were found to be highly expressed in resistant maize inbred line Mp313E. Some genes including a nuclease-phosphatase domain superfamily protein, a cinnamoyl-CoA, a heat shock protein HSP18a, and few significantly mapped genes like lysine-rich RNA binding domains, large and small ribosomal units had significantly higher expression in susceptible line Va35 than in resistant line Mp313E (Kelley et al., 2012).
Climate change has a devastating impact on mycotoxin production and fungal infection. Functional genomics tools have shown the impact of elevated CO2 levels on aflR gene (an aflatoxin biosynthetic regulatory gene) in A. flavus (Gilbert et al., 2016). A cDNA library of Mp715 (resistant inbred) and B73 (susceptible inbred) was designed to differentiate expression patterns for aflatoxin accumulation in maize, and those cDNA clones were mapped onto the maize genome by in silico mapping (Dhakal et al., 2017). This study identified 267 unigenes related to stress tolerance, metabolism, disease resistance, PR-4, and leucine-rich repeat family protein. A comparative study of maize kernels infected with A. flavus and F. verticillioides identified several candidate genes such as PR-1, 10,4,5,10.1; chitinase, CC-NBS-LRR, LRR-RLK, and Thaumatin-like proteins that showed temporal expression patterns during infection/stress (Shu et al., 2017). Several environmental/external factors affect the expression of transcripts, thus influencing the colonization of A. flavus and subsequently toxin production. For instance, the antifungal fumigant benzenamine affects aflatoxin biosynthesis, development, and virulence in A. flavus by downregulating the LeaA regulatory factor, thus acting as a fumigant against A. flavus (Yang et al., 2019).
Transgenic Approaches for Resistance to A. flavus Infection and Aflatoxin Contamination
Several transgenic approaches including expressing protein/enzyme that can reduce fungal infection or degrade the toxin have been deployed in groundnut and maize to mitigate aflatoxin contamination (Table 4). In groundnut, very few reports on transgenic approaches are available substantiating the importance of host genes like PR and defensin (Xie et al., 2013; Arias et al., 2015). A study (Sharma et al., 2018) has shown that the overexpression of Medicago defensin genes- MsDef1 and MtDef4.2 reduced Aspergillus infection as well as AP in susceptible groundnut variety JL 24. The study also demonstrated a host-induced gene silencing (HIGS) mediated silencing of aflatoxin biosynthetic pathway regulatory genes aflM and aflP to inhibit AP. Notably, both OE−Def and HIGS lines showed remarkably reduced levels of aflatoxin B1 ranging from 1 to 20 ppb compared to the wild type cultivar that accumulates up to > 4,000 ppb.
TABLE 4.
A summary of some overexpression, RNAi and host-induced gene silencing studies in groundnut and maize.
| Gene | Source | Approach | Promoter | Outcome | References |
| Groundnut | |||||
| ARAhPR10 | A. hypogaea | Overexpression | CaMV35S | Transgenic lines showed both reduced infection and less aflatoxin production | Xie et al., 2013 |
| aflR; aflS; aflJ; aflep; aflC/pksA/pksL1, pes1 | A. flavus | RNA interference gene silencing technology | CaMV35S | Transgenic lines showed up to 100% reduction in aflatoxin content | Arias et al., 2015 |
| MsDef1; MtDef4 | M. sativa; M. truncatula | Overexpression | FMV35S | OE-Def lines showed a significant reduction in aflatoxin content (up to 99%) HIGS lines showed a significant reduction in aflatoxin content (up to 99.9%) | Sharma et al., 2018 |
| aflM; aflP | A. flavus | Host-induced-gene silencing approach | CaMV35S | ||
| Maize | |||||
| ZmPR10 | Z. mays | RNA interference gene silencing technology | CaMV35S promoter | Downregulation of PR-10 caused increased susceptibility and aflatoxin contamination | Chen et al., 2010 |
| Thanatin | Podisus maculiventris | Heterologous expression | Ubiquitin-1 promoter | Cloning of thanatin (an antimicrobial synthetic peptide) improved resistance and reduced aflatoxin content (up to 68%) | Schubert et al., 2015 |
| aflR | A. flavus | Host-induced-gene silencing approach | Ubiquitin promoter | Transgenic lines showed up to 14-fold less aflatoxin concentration compared to the wild type | Masanga et al., 2015 |
| aflC | A. flavus | RNA interference | γ-zein endosperm-specific promoter | Transgenic lines showed up to 100% reduction in aflatoxin content | Thakare et al., 2017 |
| ZmPRms | Z. mays | RNA interference based gene silencing | Zein promoter | Downregulation of ZmPRms gene caused increased susceptibility and aflatoxin contamination | Majumdar et al., 2017 |
| AGM182 | Tachypleus tridentatus | Overexpression | Ubiquitin-1 promoter | Overexpression of AGM182 (an antimicrobial peptide) caused suppression of A. flavus growth and subsequently aflatoxin production (up to 98%) | Rajasekaran et al., 2018 |
Various studies on maize provide insights into using transgenic approaches and the knowledge of precise engineering strategies to improve food safety. A key approach is RNA interference (RNAi), a technology that limits the transcription of a target gene. This approach has been deployed to silence RAP genes (PR-10, GLXI, TI) in maize to identify the key role of RAPs in host resistance mechanism against A. flavus infection (Chen et al., 2004a, 2010). RNAi Pr10 silencing construct was introduced in maize plants showing increased susceptibility to A. flavus colonization and aflatoxin accumulation (Chen et al., 2010). Notably, PR-10 was involved in enhancing plant stress tolerance and severe suppression of their PR protein encoding genes drastically increased susceptibility to A. flavus infection (Xie et al., 2010; Majumdar et al., 2017). Recently, aflC and aflR genes were targeted that encode the enzyme in Aspergillus aflatoxin biosynthetic pathway to develop aflatoxin-free transgenic kernels (Masanga et al., 2015; Thakare et al., 2017). Also, thanatin, a growth inhibitor of A. flavus, was overexpressed in maize, reducing aflatoxin contamination and increasing resistance by three to four-fold resistance (Schubert et al., 2015).
In a recent study, expression analyses of polyamine (PA) metabolism/transport genes during A. flavus-maize interaction showed significant increase in the expression of arginine decarboxylase (Adc) and S-adenosylmethionine decarboxylase (Samdc) genes in the maize host and PA uptake transporters in the fungus (Majumdar et al., 2018). This study suggested that future studies targeting spermidine biosynthesis in A. flavus, using RNAi-based host-induced gene silencing approaches, may be an effective strategy to reduce aflatoxin contamination in maize and possibly in other susceptible crops. In contrary, Gressel and Polturak (2018) report that RNAi technology can’t help post-harvest AP as it may have only limited utility when the grain has been dried. However, the dormant state of seeds is usually alleviated during post-harvest storage conditions or under low moisture conditions and cannot accelerate the production of hpRNAs/siRNAs (Majumdar et al., 2017). Even in the post-transcriptional state, RNAi negatively regulates gene expression and does not produce any protein or enzyme in the host plant (Majumdar et al., 2017). Fakhoury and Woloshuk (1999) produced a mutant strain (101) of A. flavus which was defective in the α-amylase activity. The α-amylase enzyme is crucial in A. flavus as it is involved in the degradation of the host’s carbohydrate reservoir which is an essential energy source for fungus growth and reproduction, as well as AP. Therefore, an α-amylase inhibitor protein (AILP) that inhibits α-amylase activity was expressed in the host; this reduced fungus growth and subsequent AP (Fakhoury and Woloshuk, 2001; see Chen et al., 2015). Recently, a transgenic maize line expressing AGM182 which encodes a tachyplesin1-derived synthetic peptide (an antimicrobial peptide) was developed that exhibited reduced fungal growth and a significant reduction in aflatoxin level (76–98%) compared to the control (Rajasekaran et al., 2018). Characterization of these candidate genes through a transgenic approach would be important in safeguarding food commodities.
Managing Aflatoxin Contamination: Similarities Between Groundnut and Maize
Pre- and post-harvest management strategies largely predict the extent to which Aspergillus fungi invade seeds and exacerbate AP (Hell et al., 2008). Most post-harvest management practices like rapid drying of groundnut in-shell and maize ears coupled with appropriate storage conditions are crucial for reducing infection and toxin accumulation. During initiation stage, host-pathogen interactions occur in the cell wall where NBS-LRR receptors, oxylipins, and elicitors play an important role. This is followed by a change in ion flux across the plasma membrane and the activation of a number of genes that lead to changes in the plant’s cell wall. It activates various PR-related proteins, phytoalexins-like compounds and TFs which play an important role in defense mechanism. In addition, at the environmental level, PAC is largely exacerbated by drought stress and insect damage in groundnut and maize (Guo et al., 2008; Hell et al., 2008). Attempts to characterize resistance due to the physical barriers suggested that pod shell may serve as a barrier to A. flavus infection when the kernels are stored in-shell in the case of groundnut (Liang et al., 2006; Nigam et al., 2009). Similarly, in maize, a tight husk and non-upright ear act as a barrier to the entry of spores and keep the ear dryer, resulting in an unfavorable environment for fungal growth (Warburton and Williams, 2014). Such physical barriers are considered non-desirable traits since they pose serious challenges while threshing or dehulling.
In groundnut and maize, cross-talk communication between the pathogen and host plant is the first critical step toward the rapid activation of defense mechanisms in host plants. Functional and biological composition of resistance mechanisms in maize and groundnut using integrated approaches have led to the elucidation of the roles of several genes, PR-10, chitinase, 14-kDa trypsin inhibitor, zeatin and beta-1,3-glucanase, lipoxygenase, ROS, and stress responsive proteins (such as late embryogenesis abundant protein (LEA14), catalase, glutathione S-transferase, superoxide dismutase, heat shock proteins) which play a vital role in regulating resistance and in cross-kingdom interactions between host plants and Aspergillus species in groundnut (Luo et al., 2005; Chadha and Das, 2006; Liang et al., 2006; Wang et al., 2010; Guo et al., 2011; Kumari et al., 2011; Nayak et al., 2017) and maize (Guo et al., 1997; Chen et al., 1998, 1999, 2001, 2002, 2004b, 2006, 2007, 2012; Lozovaya et al., 1998; Ji et al., 2000; Moore et al., 2004; Magbanua et al., 2007; Pechanova et al., 2011; Pegoraro et al., 2011; Roze et al., 2013; Fountain et al., 2014, 2016; Hawkins et al., 2015; Ogunola et al., 2017).
Metabolomics Under A. flavus Infection and Aflatoxin Resistance
Metabolomics is an emerging field that represents the complete set of metabolites in a biological cell, tissue, organ or organism. It provides an instantaneous snapshot of the “physiological state” of an organism (Ramalingam et al., 2015; Kumar R. et al., 2017). Metabolites are small molecules that are directly involved in growth, development, and reproduction processes.
To understand the aflatoxin resistance mechanism at the metabolite level, some metabolome studies in response to A. flavus infection have been conducted in maize. For instance, metabolome profile under A. flavus infection showed significant induction and higher expression of polyamine (PA) biosynthesis genes in maize-resistant lines TZAR102, MI82 than in susceptible line SC212. Higher expression of spermidine (Spd), spermine (Spm), and diamine putrescine (Put) along with their increased catabolism in the resistant lines than in the susceptible line indicate that polyamines play an important role in A. flavus resistance (Majumdar et al., 2019). In addition, higher concentrations of amino acids such as glutamate (Glu), glutamine (Gln), and γ-aminobutyric acid in susceptible maize line SC212 showed that these amino acids favor A. flavus infection. In a similar study by Falade et al. (2018), metabolites were analyzed at R3 (milk), R4 (dough), and R5 (dent) stages of cob development under A. flavus infection (4 doses). The study showed that grain colonization decreases with increasing kernel maturity from milk-, dough-, and dent-stage kernels, with approximately 100%, 60%, and 30% colonization, respectively. However, aflatoxin levels increase with increased doses at dough and dent stages. This shows that initial stages of cob development (milk and dough) are more susceptible than the maturity stage (Falade et al., 2018). A study on aflatoxin accumulation in grains of 120 maize hybrids showed that higher concentrations of beta-carotene (BC), beta-cryptoxanthin (BCX), and total provitamin A had significantly less aflatoxin accumulation compared to that in hybrids with lower carotenoid concentration. Hence, breeding for increased carotenoid concentration can increase aflatoxin resistance in maize to help combat aflatoxin contamination as well as malnutrition (Suwarno et al., 2019). In short, metabolites significantly influence A. flavus infection and can be used as biomarkers for screening resistant and susceptible maize genotypes.
Molecular Biology of A. flavus for Aflatoxin Production and Resistance
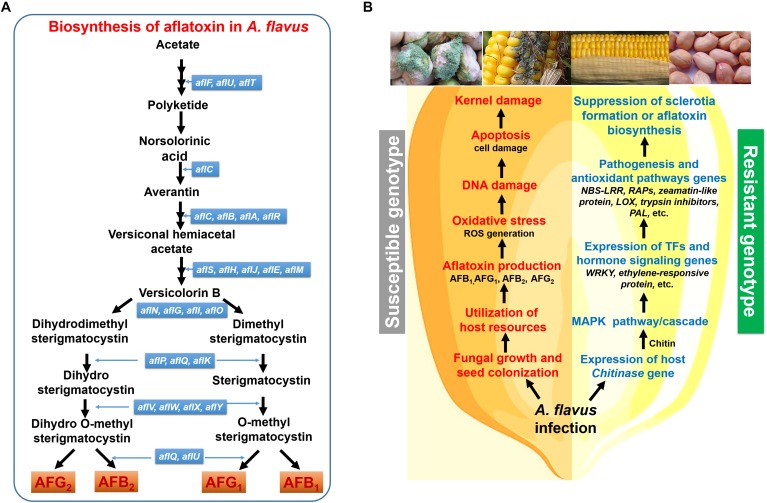
The genome of the toxigenic strain of A. flavus contains ∼12,000 genes involved in the synthesis of secondary metabolites, with more than 56 gene clusters contributing to the production of secondary metabolites, including aflatoxin (Rokas et al., 2007). The aflatoxin biosynthesis gene cluster includes 25 genes spanning approximately 70 kb of DNA (Yu et al., 2004). The aflatoxin gene cluster resides on chromosome 3, next to the telomeric region comprising of pathway-specific regulatory genes as well as surrounded by four sugar-utilization genes at the distal end (Yu et al., 2000). Some regulatory genes (e.g., aflR and aflS) are reported to be essential for the production of aflatoxin after infection, and they work in conjunction with several other regulators/factors such as VelB/VeA/LaeA complex, CreA transcription factor, among others. While the aflR gene encodes a DNA binding Zn-cluster protein that binds to DNA binding-domains of aflatoxin pathway genes, aflS is an aflatoxin pathway-specific regulatory gene required to mediate aflR transportation to/from the nucleus and assist in aflR localization (Figure 2; Ehrlich et al., 2012).
FIGURE 2.
A simplified representation of the aflatoxin biosynthesis pathway and the defense response mechanism in groundnut or maize. (A) Aflatoxin biosynthesis in A. flavus; (B) the aflatoxin biosynthesis pathway involve multiple genes which co-express together for the formation of toxin secondary metabolites. In the susceptible genotype infection leads to the A. flavus seed colonization and production of aflatoxin which causes suppression of host defense mechanism results in ROS generation and DNA damage causing cell death (apoptosis). In contrast, in resistant genotypes infection causes induction of host defense mechanism that include MAPK pathway which induces WRKY TF expression which is a key regulator of pathogenesis and antioxidant related genes involved in the suppression of aflatoxin biosynthesis pathway or detoxification of toxin.
Aspergillus flavus can hijack the host machinery to facilitate the uptake of resources required for AP. For instance, the fungus requires the spermidine synthase (a polyamine biosynthetic gene) for AP and can utilize the host substrate to enhance polyamine (PA) biosynthesis and AP (Majumdar et al., 2018). In susceptible maize kernel, the expression of the PA biosynthetic/metabolism genes S-adenosylmethionine decarboxylase (Samdc) and arginine decarboxylase (Adc) significantly increased; this was followed by the upregulation of PA transporters in the pathogen (Majumdar et al., 2018). Maize’s hypersensitivity and susceptibility to A. flavus involve a gene encoding glycine-rich RNA binding protein 2 which is associated with hormone and pathogen stress (Kelley et al., 2012), through salicylic-mediated defense signal transduction and HR reactions (Naqvi et al., 1998; Singh et al., 2011). The NPCs which transport RNA and other macromolecules are highly expressed in resistant maize cultivars and suppress A. flavus infection (Kelley et al., 2012). In Arabidopsis, a defect in MOS7 (an NPC encoding gene) suppresses the accumulation of R-protein in the nucleus that causes a defect in both basal and systemic acquired resistance and R-protein-mediated immunity (Cheng et al., 2009). The infection induces higher expression of ethylene-responsive protein (ETHRP) in resistant maize cultivars suggesting the role of the ethylene signaling pathway in aflatoxin accumulation resistance. ETHRP is a universal stress protein and a key regulator of stress responses, and confers stress survival (Kelley et al., 2012). Further, fungal infection induces the production of several antifungal proteins such as 14-kDa trypsin inhibitor, 18 kDa ribosome-inactivating-protein, 28, 38 and 100 kDa protein, non-specific lipid transfers proteins, 2 S storage proteins, and zeamatin (Liang et al., 2006). An infection can also induce lipid peroxidation, which facilitates resistance to AP in groundnut (Liang et al., 2002).
Aspergillus infection also involves a dynamic network of transcription factors that coordinate the expression of the target biosynthetic genes of the pathogen and the suppression of the host’s immune responses. This may involve the suppression of key gene WRKY, a transcription factor that modulates the expression of several genes involved in detoxification of ROS as well as aflatoxin (Korani et al., 2018), including NBS-LRR; its suppression is linked to aggravated accumulation of aflatoxin in plants such as groundnut (Nayak et al., 2017). Further, these TFs are also associated with PR proteins, which play a major role in resistance after infection (Pierpoint et al., 1981; Van Loon, 1985; Szerszen, 1990; Van Loon and Van Strien, 1999). In groundnut, WRKY and other key TFs such as ERF and NAC function in a coordinated fashion (Nayak et al., 2017; Korani et al., 2018); their modulation has a substantial impact on antioxidant biosynthetic, PR proteins, chitinase, and beta-1,3-glucanase genes. Modulation of these TFs in the host severely affects the transcription of ROS detoxifying genes such as catalases, superoxide dismutase, glutathione-S-transferase, and antioxidant biosynthesis genes like resveratrol synthase, PAL, chalcone synthase, chitinase, and beta-1,3-glucanase (Nayak et al., 2017; Korani et al., 2018). These genes protect host plants from oxidative damage, increase the levels of secondary metabolites involved in lignin biosynthesis, and restrict fungal invasion as well as its growth. In resistant groundnut genotypes, the activity of PAL enzyme that catalyzes the metabolism of phenolic compounds such as phytoalexin and lignin precursors, increases significantly (Nayak et al., 2017; Korani et al., 2018).
Resveratrol is a potent phytoalexin induced up to 30-fold in resistant genotypes of groundnut seeds upon infection (Liang et al., 2006). In wild groundnut species, the pod shell and seeds are rich in lignin content that prevents aflatoxin contamination (Guimarães et al., 2012). Notably, in maize, exposure to drought severely reduces PAL enzyme activity and phytoalexin production due to reduced moisture content in the kernel, resulting in fungal invasion and toxin production (Gholizadeh, 2011). Although, studies spanning 15 years have identified several gene clusters regulating host-pathogen interactions and AP, the characterization of individual genes is crucial to design strategies toward mitigation of aflatoxin contamination.
Challenges and Opportunities
Aspergillus flavus infection and subsequent aflatoxin contamination is highly influenced by environmental parameters such as high soil temperature, moisture stress, and relative humidity which often outsmart the low levels of genetic resistance available in groundnut and maize genotypes. This could be one of the key reasons in making this trait very complex and limited progress has been made under field conditions as compared to controlled environment. Even under controlled environmental conditions, most studies are targeted at understanding host-pathogen interactions using a single toxigenic A. flavus strain and its interaction with the host (groundnut or maize). However, under field conditions, the reality is different. Often, many species of Aspergillus group of fungi such as A. flavus and A. parasiticus are involved in causing aflatoxin contamination. The population dynamics of toxigenic Aspergillus in soils and possible shifts in toxigenic and non-toxigenic strains could be an important area to focus on while studying host-pathogen interactions. Also required is a knowledge of the soil composition of toxigenic A. flavus group of fungi and the ambient environment in a crop production region that drives Aspergillus population levels and other competing and co-existing pathogens. Similar conditions can be created/simulated under a controlled environment to facilitate the easy adoption and translation of results from laboratory conditions to the field. The lack of consistency in host-pathogen-toxin interactions inhibits the understanding of the precise genetic behavior of resistance in groundnut and maize. Despite a sequencing revolution in the last decade, genetic and gene discovery efforts have not led to solutions to aflatoxin reduction because of inconsistent phenotyping results. Devising novel phenotyping techniques to assay AP at different steps is a way forward. Dissecting components of resistance using known pre-harvest resistant sources of groundnut and maize may be an interesting area of research. In this context, studying the biochemical composition of the seed coat could lead to a better understanding of host-pathogen interactions.
Another key challenge as well as an opportunity would be to understand the impact of soil and its environment on AP. Plants growing in unhealthy soils are bound to be more stressed, and this might increase aflatoxin contamination. While most studies have concentrated on the physical and chemical components of soil, the biological component remains unexplored. An analysis of the phytobiome, the microbial component that surrounds the plant, from the leaves down to the roots, is another emerging area of research. A phytobiome that negatively impacts plant health would influence aflatoxin contamination. Insights into the phytobiomes of groundnut and maize would certainly influence our understanding of host-pathogen interactions, especially in complex traits such as aflatoxin contamination.
Summary
While discussing the progress made in understanding the resistance mechanisms of aflatoxin contamination in groundnut and maize using multidisciplinary approaches, the paper elaborates on several QTLs, genes, pathways and complex genetic architecture of the target trait. The paper has also reviewed the potential of different approaches in better understanding the complexities of candidate genes identified after the genome sequencing of host and pathogen. Various cultural and biological methods have been reported to prevent/sustainably manage aflatoxin contamination in groundnut and maize. The development of varieties/hybrids or transgenics with resistance to both fungal infection and aflatoxin contamination remains a challenge. To date, aflatoxin management strategies have centered around the use of good agricultural practices during pre-and post-harvest stages, including the use of biocontrol agents (particularly of non-toxigenic strains of A. flavus) in countries where they are available to farmers. Omics studies in the last couple of decades provide an array of genetic and genomic resources and expand the knowledge base on Aspergillus infection and aflatoxin reduction mechanisms, host-pathogen interactions, toxigenicity of the fungi, mechanism of aflatoxin biosynthesis, and inhibitors targeting the aflatoxin biosynthetic genes. Promising genomics and transgenic approaches have provided complimentary beneficial effects by integrating genes, peptides/antifungal proteins, and even silencing key genes for Aspergillus growth and aflatoxin biosynthesis in susceptible varieties to enhance resistance levels. These integrated approaches comprising of functional and structural genomics, together with NGS platform will provide more information on candidate genes to facilitate the development of molecular markers for use in molecular breeding. Conventional and modern breeding tools need to be deployed to develop aflatoxin-resistant maize and groundnut varieties that will lead to food safety, poverty reduction and boosting the industry and market.
Author Contributions
RV together with MP and PS conceptualized the idea after receiving the invitation from the journal. PS together with SG, AO-B, RK, SP, HS, YL, XN, DH, JF, SN, GM, TR, WZ, BG, BL, PSi, MP, and RB wrote the manuscript. PS, MP, and RV finalized the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The fellowship of PS and SP was supported by the Council of Scientific & Industrial Research (CSIR), Government of India. RK and RV are thankful to the Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Government of India for the N-Post Doctoral Fellowship and the JC Bose Fellowship, respectively, and SG acknowledges DST for the DST-INSPIRE Fellowship.
Footnotes
Funding. Thanks are due to the support from MARS-Wrigley, United States and CGIAR-National Natural Science Foundation of China (NSFC), China, and Bill & Melinda Gates Foundation. This work has been undertaken as part of the CGIAR Research Program on Grain Legumes and Dryland Cereals (CRP-GLDC, ICRISAT), MAIZE CGIAR Research Program (CRP-Maize, IITA) and Agriculture for Nutrition and Health (CRP-A4NH, IITA). ICRISAT and IITA are members of the CGIAR System Organization.
References
- Adhikari B. N., Bandyopadhyay R., Cotty P. J. (2016). Degeneration of aflatoxin gene clusters in Aspergillus flavus from Africa and North America. AMB Express 6:62. 10.1186/s13568-016-0228-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbetiameh D., Ortega-Beltran A., Awuah R. T., Atehnkeng J., Cotty P. J., Bandyopadhyay R. (2018). Prevalence of aflatoxin contamination in maize and groundnut in Ghana: population structure, distribution, and toxigenicity of the causal agents. Plant Dis. 102 764–772. 10.1094/PDIS-05-17-0749-RE [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbetiameh D., Ortega-Beltran A., Awuah R. T., Atehnkeng J., Islam M.-S., Callicott K. A., et al. (2019). Potential of atoxigenic strains of Aspergillus flavus associated with maize and groundnut in Ghana as biocontrol agents for aflatoxin management. Front. Microbiol. 10:2069 10.3389/fmicb.2019.02069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaniz Zanon M. S., Barros G. G., Chulze S. N. (2016). Non-aflatoxigenic Aspergillus flavus as potential biocontrol agents to reduce aflatoxin contamination in peanuts harvested in Northern Argentina. Int. J. Food Microbiol. 231 63–68. 10.1016/j.ijfoodmicro.2016.05.016 [DOI] [PubMed] [Google Scholar]
- Alaniz Zanon M. S., Chiotta M. L., Giaj-Merlera G., Barros G., Chulze S. (2013). Evaluation of potential biocontrol agent for aflatoxin in Argentinean peanuts. Int. J. Food Microbiol. 162 220–225. 10.1016/j.ijfoodmicro.2013.01.017 [DOI] [PubMed] [Google Scholar]
- Amaike S., Keller N. P. (2011). Aspergillus flavus. Annu. Rev. Phytopathol. 49 107–133. 10.1146/annurev-phyto-072910-095221 [DOI] [PubMed] [Google Scholar]
- Arias R. S., Dang P. M., Sobolev V. S. (2015). RNAi-mediated control of aflatoxins in peanut: method to analyze mycotoxin production and transgene expression in the peanut/Aspergillus pathosystem. J. Vis. Exp. 106 1–11. 10.3791/53398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asis R., Barrionuevo D. L., Giorda L. M., Nores M. L., Aldao M. A. (2005). Aflatoxin production in six peanut (Arachis hypogaea L.) genotypes infected with Aspergillus flavus and Aspergillus parasiticus, isolated from peanut production areas of Cordoba Argentina. J. Agric. Food Chem. 53 9274–9280. 10.1021/jf051259 [DOI] [PubMed] [Google Scholar]
- Atehnkeng J., Donner M., Ojiambo P. S., Ikotun B., Augusto J., Cotty P. J., et al. (2016). Environmental distribution and genetic diversity of vegetative compatibility groups determine biocontrol strategies to mitigate aflatoxin contamination of maize by Aspergillus flavus. Microb. Biotechnol. 9 75–88. 10.1111/1751-7915.12324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew A., Kimanya M., Matumba L., Bandyopadhyay R., Menkir A., Cotty P. (2017). “Controlling aflatoxins in maize in Africa: strategies, challenges and opportunities for improvement,” in Achieving Sustainable Cultivation of Maize. Cultivation Techniques, Pest and Disease Control, Vol. 2 ed. Watson D., (Cambridge: Burleigh Dodds Science Publishing; ), 1–24. [Google Scholar]
- Bandyopadhyay R., Atehnkeng J., Ortega-Beltran A., Akande A., Falade T. D. O., Cotty P. J. (2019). “Ground-truthing” efficacy of biological control for aflatoxin mitigation in farmers’ fields in Nigeria: from field trials to commercial usage. Front. Microbiol. 10:2528 10.3389/fmicb.2019.02528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay R., Ortega-Beltran A., Akande A., Mutegi C., Atehnkeng J., Kaptoge L., et al. (2016). Biological control of aflatoxins in Africa: current status and potential challenges in the face of climate change. World Mycotoxin J. 9 771–789. 10.3920/wmj2016.2130 [DOI] [Google Scholar]
- Battilani P., Lanubile A., Scala V., Reverberi M., Gregori R., Falavigna C., et al. (2018). Oxylipins from both pathogen and host antagonize jasmonic acid-mediated defence via the 9-lipoxygenase pathway in Fusarium verticillioides infection of maize. Mol Plant Pathol. 19 2162–2176. 10.1111/mpp.12690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello H. T. (2007). Phenotypic and Genotypic Evaluation of Generations and Recombinant Inbred Lines for Response to Aflatoxin. Ph.D. dissertation Texas A&M University, College Station, TX. [Google Scholar]
- Brooks T. D., Williams W. P., Windham G. L., Willcox M. C., Abbas H. K. (2005). Quantitative trait loci contributing resistance to aflatoxin accumulation in maize inbred Mp313E. Crop Sci. 45 171–174. 10.1371/journal.pone.0036892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R., Cotty P., Cleveland T., Widstrom N. (1993). Living maize embryo influences accumulation of aflatoxin in maize kernels. J. Food Prot. 56 967–971. 10.4315/0362-028X-56.11.967 [DOI] [PubMed] [Google Scholar]
- Bryden W. L. (2012). Mycotoxin contamination of the feed supply chain: implications for animal productivity and feed security. Anim. Feed Sci. Tech. 173 134–158. 10.1016/j.anifeedsci.2011.12.014 [DOI] [Google Scholar]
- Busboom K. N., White D. G. (2004). Inheritance of resistance to aflatoxin production and Aspergillus ear rot of corn from the cross of inbreds B73 and Oh516. Phytopathology 94 1107–1115. 10.1094/PHYTO.2004.94.10.1107 [DOI] [PubMed] [Google Scholar]
- Campbell K. W., Hamblin A. M., White D. G. (1997). Inheritance of resistance to aflatoxin production in the cross between corn inbreds B73 and LB31. Phytopathology 87 1144–1147. 10.1094/PHYTO.1997.87.11.1144 [DOI] [PubMed] [Google Scholar]
- Campbell K. W., White D. G. (1995). Evaluation of corn genotypes for resistance to Aspergillus ear rot, kernel infection, and aflatoxin production. Plant Dis. 79 1039–1045. [Google Scholar]
- Chadha P., Das R. H. (2006). A pathogenesis related protein, AhPR10 from peanut: an insight of its mode of antifungal activity. Planta 225 213–222. 10.1007/s00425-006-0344-7 [DOI] [PubMed] [Google Scholar]
- Chang P. K., Horn B. W., Dorner J. W. (2005). Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet. Biol. 42 914–923. 10.1016/j.fgb.2005.07.004 [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Cleveland T. E., Damann K. E. (2004a). Investigating the roles of an aflatoxin resistance-associated protein in maize using RNAi. Phytopathology 94 S18–S18. [Google Scholar]
- Chen Z. Y., Brown R. L., Cleveland T. E., Damann K. E., Russin J. S. (2001). Comparison of constitutive and inducible maize kernel proteins of genotypes resistant or susceptible to aflatoxin production. J. Food Protect. 64 1785–1792. 10.4315/0362-028x-64.11.1785 [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Damann K. E., Cleveland T. E. (2002). Identification of unique or elevated levels of kernel proteins in aflatoxin–resistant maize genotypes through proteome analysis. Phytopathology 92 1084–1094. 10.1094/PHYTO.2002.92.10.1084 [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Damann K. E., Cleveland T. E. (2004b). Identification of a maize kernel stress-related protein and its effect on aflatoxin accumulation. Phytopathology 94 938–945. 10.1094/PHYTO.2004.94.9.938 [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Damann K. E., Cleveland T. E. (2007). Identification of maize kernel endosperm proteins associated with resistance to aflatoxin contamination by Aspergillus flavus. Phytopathology 97 1094–1103. 10.1094/PHYTO-97-9-1094 [DOI] [PubMed] [Google Scholar]
- Horn B. W., Ramirez-Prado J. H., Carbone I. (2009b). Sexual reproduction and recombination in the aflatoxin-producing fungus Aspergillus parasiticus. Fungal Genet. Biol. 46 169–175. 10.1016/j.fgb.2008.11.004 [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Damann K. E., Cleveland T. E. (2010). PR10 expression in maize and its effect on host resistance against Aspergillus flavus infection and aflatoxin production. Mol. Plant Pathol. 11 69–81. 10.1111/j.1364-3703.2009.00574.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Lax A. R., Guo B. Z., Cleveland T. E., Russin J. S. (1998). Resistance to Aspergillus flavus in corn kernels is associated with a 14-kDa protein. Phytopathology 88 276–281. 10.1094/PHYTO.1998.88.4.276 [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Menkir A., Cleveland T. E. (2012). Identification of resistance-associated proteins in closely-related maize lines varying in aflatoxin accumulation. Mol. Breed. 30 53–68. 10.1007/s11032-011-9597-3 [DOI] [Google Scholar]
- Chen Z. Y., Brown R. L., Rajasekaran K., Damann K. E., Cleveland T. E. (2006). Identification of a maize kernel pathogenesis-related protein and evidence for its involvement in resistance to Aspergillus flavus infection and aflatoxin production. Phytopathology 96 87–95. 10.1094/PHYTO-96-0087 [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Russin J. S., Lax A. R., Cleveland T. E. (1999). A corn trypsin inhibitor with antifungal activity inhibits Aspergillus flavus α-amylase. Phytopathology 89 902–907. 10.1094/PHYTO.1999.89.10.902 [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., Rajasekaran K., Brown R. L., Sayler R. J., Bhatnagar D. (2015). Discovery and confirmation of genes/proteins associated with maize aflatoxin resistance. World Mycotoxin J. 8 211–224. 10.3920/wmj2014.1732 [DOI] [Google Scholar]
- Cheng Y. T., Germain H., Wiermer M., Bi D., Xu F., García A. V., et al. (2009). Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21 2503–2516. 10.1105/tpc.108.064519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S. A., Kolomiets M. V. (2011). The lipid language of plant–fungal interactions. Fungal Genet. Biol. 48 4–14. 10.1016/j.fgb.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Clevenger J., Marasigan K., Liakos V., Sobolev V., Vellidis G., Holbrook C., et al. (2016). RNA sequencing of contaminated seeds reveals the state of the seed permissive for pre-harvest aflatoxin contamination and points to a potential susceptibility factor. Toxins 8 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotty P. J. (1989). Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology 79 808–814. [Google Scholar]
- Cotty P. J. (2001). “Cotton seed losses and mycotoxins,” in Compendium of Cotton Diseases. Part 1, eds Kirkpatrick T. L., Rothrock C. S., (St Paul: APS Press; ), 9–13. [Google Scholar]
- Cotty P. J., Antilla L., Wakelyn P. J. (2007). “Competitive exclusion of aflatoxin producers: farmer-driven research and development,” in Biological Control a Global Perspective, eds Vincent C., Goettel M. S., Lazarovits G., (Wallingford: Centre for Agriculture and Bioscience International; ), 241–253. 10.1079/9781845932657.0241 [DOI] [Google Scholar]
- Cotty P. J., Probst C., Jaime-Garcia R. (2008). “Etiology and management of aflatoxin contamination,” in Mycotoxins: Detection Methods, Management, Public Health and Agricultural Trade, (Wallingford: Centre for Agriculture and Bioscience International; ), 287–299. 10.1079/9781845930820.0287 [DOI] [Google Scholar]
- Danso J. K., Osekre E. A., Opit G. P., Arthur F. H., Campbell J. F., Mbata G., et al. (2019). Impact of storage structures on moisture content, insect pests and mycotoxin levels of maize in Ghana. J. Stored Prod. Res. 81 114–120. 10.1016/j.jspr.2018.11.012 [DOI] [Google Scholar]
- Danso J. K., Osekre E. A., Opit G. P., Manu N., Armstrong P., Arthur F. H., et al. (2018). Post-harvest insect infestation and mycotoxin levels in maize markets in the Middle Belt of Ghana. J. Stored Prod. Res. 77 9–15. 10.1016/j.jspr.2018.02.004 [DOI] [Google Scholar]
- Darrah L. L., Lillehoj E. B., Zuber M. S., Scott G. E., Thompson D., West D. R., et al. (1987). Inheritance of aflatoxin B1 levels in maize kernels under modified natural inoculation with Aspergillus flavus. Crop Sci. 27 869–872. [Google Scholar]
- Dhakal R., Chai C., Karan R., Windham G. L., Williams W. P., Subudhi P. K. (2017). Expression profiling coupled with in-silico mapping identifies candidate genes for reducing aflatoxin accumulation in maize. Front. Plant. Sci. 8:503. 10.3389/fpls.2017.00503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diedhiou P. M., Bandyopadhyay R., Atehnkeng J., Ojiambo P. S. (2011). Aspergillus colonization and aflatoxin contamination of maize and sesame kernels in two agro–ecological zones in Senegal. J. Phytopathol. 159 268–275. 10.1111/j.1439-0434.2010.01761.x [DOI] [Google Scholar]
- Dorner J. W. (2004). Biological control of aflatoxin contamination of crops. J. Toxicol. Toxin Rev. 23 425–450. 10.1081/txr-200027877 [DOI] [Google Scholar]
- Dorner J. W. (2009). Development of biocontrol technology to manage aflatoxin contamination in peanuts. Peanut Sci. 36 60–67. 10.3146/at07-002.1 [DOI] [Google Scholar]
- Doster M. A., Cotty P. J., Michailides T. J. (2014). Evaluation of the atoxigenic Aspergillus flavus strain AF36 in pistachio orchards. Plant Dis. 98 948–956. 10.1094/PDIS-10-13-1053-RE [DOI] [PubMed] [Google Scholar]
- Ehrlich K., Mack B., Wei Q., Li P., Roze L., Dazzo F., et al. (2012). Association with AflR in endosomes reveals new functions for AflJ in aflatoxin biosynthesis. Toxins 4 1582–1600. 10.3390/toxins4121582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezekiel C. N., Ortega-Beltran A., Oyedeji E., Atehnkeng J., Kössler P., Tairu F., et al. (2019). Aflatoxin in chili peppers in Nigeria: extent of contamination and control using atoxigenic Aspergillus flavus genotypes as biocontrol agents. Toxins 11:429. 10.3390/toxins11070429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhoury A. M., Woloshuk C. P. (1999). Amy1, the α-amylase gene of Aspergillus flavus: involvement in aflatoxin biosynthesis in maize kernels. Phytopathology 89 908–914. 10.1094/PHYTO.1999.89.10.908 [DOI] [PubMed] [Google Scholar]
- Fakhoury A. M., Woloshuk C. P. (2001). Inhibition of growth of Aspergillus flavus and fungal α-amylases by a lectin-like protein from Lablab purpureus. Mol. Plant Microbe Interact. 14 955–961. 10.1094/mpmi.2001.14.8.955 [DOI] [PubMed] [Google Scholar]
- Falade T. D. O., Chrysanthopoulos P. K., Hodson M. P., Sultanbawa Y., Fletcher M., Darnell R., et al. (2018). Metabolites identified during varied doses of Aspergillus species in Zea mays grains, and their correlation with aflatoxin levels. Toxins 10:187. 10.3390/toxins10050187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfan I. D., De La Fuente GN, Murray S. C., Isakeit T., Huang P. C., Warburton M., et al. (2015). Genome Wide Association Study for Drought, Aflatoxin Resistance, and Important Agronomic Traits of Maize Hybrids in the Sub-Tropics. PLoS One 10:e0117737. 10.1371/journal.pone.0117737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florkowski W. J., Kolavalli S. (2013). “Aflatoxin control strategies in the groundnut value chain in Ghana,” in Proceedings of the IFPRI Ghana Strategy Support Program Working Paper, (Washington, DC: IFPRI; ), 33. [Google Scholar]
- Fountain J. C., Bajaj P., Nayak S. N., Yang L., Pandey M. K., Kumar V., et al. (2016). Responses of Aspergillus flavus to oxidative stress are related to fungal development regulator, antioxidant enzyme, and secondary metabolite biosynthetic gene expression. Front. Microbiol. 7:2048. 10.3389/fmicb.2016.02048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain J. C., Koh J., Yang L., Pandey M. K., Nayak S. N., Bajaj P., et al. (2018). Proteome analysis of Aspergillus flavus isolate-specific responses to oxidative stress in relationship to aflatoxin production capability. Sci. Rep. 8:3430. 10.1038/s41598-018-21653-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain J. C., Raruang Y., Luo M., Brown R. L., Chen Z. Y. (2013). The potential roles of WRKY transcription factors in regulating maize defense responses against Aspergillus flavus infection. Phytopathology 103:45. [Google Scholar]
- Fountain J. C., Raruang Y., Luo M., Brown R. L., Guo B. Z., Chen Z. Y. (2015). Potential roles of WRKY transcription factors in regulating host defense responses during Aspergillus flavus infection of immature maize kernels. Physiol. Mol. Plant P. 89 31–40. 10.1016/j.pmpp.2014.11.005 [DOI] [Google Scholar]
- Fountain J. C., Scully B. T., Ni X., Kemerait R. C., Lee R. D., Chen Z. Y., et al. (2014). Environmental influences on maize-Aspergillus flavus interactions and aflatoxin production. Front. Microbiol. 5:40. 10.3389/fmicb.2014.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J. C., Hubka V., Ezekiel C. N., Hong S. B., Novakova A., Chen A. J., et al. (2019). Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 93 1–63. 10.1016/j.simyco.2018.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Isakeit T., Brodhagen M., Keller N. P., Kolomiets M. V. (2008). Maize lipoxygenase ZmLOX3-mediated pathway suppresses seed colonization, production of spores and mycotoxins by Aspergilli spp. Phytopathology 98 S57–S57. [Google Scholar]
- Garcia A. V., Parker J. E. (2009). Heaven’s gate: nuclear accessibility and activities of plant immune regulators. Trends Plant Sci. 14 479–487. 10.1016/j.tplants.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Garrido-Bazan V., Mahuku G., Bibbins-Martinez M., Arroyo-Bacerra A., Villalobos-López M. Á. (2018). Dissection of mechanisms of resistance to Aspergillus flavus and aflatoxin using tropical maize germplasm. World Mycotoxin J. 11 215–224. 10.3920/wmj2017.2219 [DOI] [Google Scholar]
- Gembeh S. V., Brown R. L., Grimm C., Cleveland T. E. (2001). Identification of chemical components of corn kernel pericarp wax associated with resistance to Aspergillus flavus infection and aflatoxin production. J. Agric. Food Chem. 49 4635–4641. 10.1021/jf010450q [DOI] [PubMed] [Google Scholar]
- Gholizadeh A. (2011). Effects of drought on the activity of phenylalanine ammonia lyase in the leaves and roots of maize inbreds. Aust. J. Basic Appl. Sci. 5 952–956. [Google Scholar]
- Gilbert M. K., Mack B. M., Wei Q., Bland J. M., Bhatnagar D., Cary J. W. (2016). RNA sequencing of an nsdC mutant reveals global regulation of secondary metabolic gene clusters in Aspergillus flavus. Microbiol. Res. 182 150–161. 10.1016/j.micres.2015.08.007 [DOI] [PubMed] [Google Scholar]
- Gorman D. P., Kang M. S., Cleveland T., Hutchinson R. L. (1992). Combining ability for resistance to field aflatoxin accumulation in maize grain. Plant Breed. 109 296–303. 10.1111/j.1439-0523.1992.tb00188.x [DOI] [Google Scholar]
- Gqaleni N., Smith J. E., Lacey J., Gettinby G. (1997). Effects of temperature and water activity, and incubation time on production of aflatoxins and cyclopiazonic acid by an isolate of Aspergillus flavus in surface agar culture. Appl. Environ. Microbiol. 63 1048–1053. 10.1128/aem.63.3.1048-1053.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressel J., Polturak G. (2018). Suppressing aflatoxin biosynthesis is not a breakthrough if not useful. Pest Manag. Sci. 74 17–21. 10.1002/ps.4694 [DOI] [PubMed] [Google Scholar]
- Guimarães P. M., Brasileiro A. C., Morgante C. V., Martins A. C., Pappas G., Silva O. B., et al. (2012). Global transcriptome analysis of two wild relatives of peanut under drought and fungi infection. BMC Genom. 13:387. 10.1186/1471-2164-13-387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Chen X., Dang P., Scully B. T., Liang X., Holbrook C. C., et al. (2008). Peanut gene expression profiling in developing seeds at different reproduction stages during Aspergillus parasiticus infection. BMC Dev. Biol. 8:12. 10.1186/1471-213X-8-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Fedorova N. D., Chen X., Wan C. H., Wang W., Nierman W. C., et al. (2011). Gene expression profiling and identification of resistance genes to Aspergillus flavus infection in peanut through EST and microarray strategies. Toxins 3 737–753. 10.3390/toxins3070737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B. Z., Chen Z. Y., Brown R. L., Lax A. R., Cleveland T. E., Russin J. S., et al. (1997). Germination induces accumulation of specific proteins and antifungal activities in corn kernels. Phytopathology 87 1174–1178. 10.1094/PHYTO.1997.87.11.1174 [DOI] [PubMed] [Google Scholar]
- Hawkins L. K., Mylroie J. E., Oliveira D. A., Smith J. S., Ozkan S., Windham G. L., et al. (2015). Characterization of the maize chitinase genes and their effect on Aspergillus flavus and aflatoxin accumulation resistance. PLoS One 10:0126185. 10.1371/journal.pone.0126185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedayati M. T., Pasqualotto A. C., Warn P. A., Bowyer P., Denning D. W. (2007). Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiol. 153 1677–1692. 10.1099/mic.0.2007/007641-0 [DOI] [PubMed] [Google Scholar]
- Hell K., Fandohan P., Bandyopadhyay R., Kiewnick S., Sikora R., Cotty P. J. (2008). “Pre-and postharvest management of aflatoxin in maize: an African perspective,” in Mycotoxins: Detection Methods, Management, Public Health and Agricultural Trade, eds Bandyopadhyay R., Leslie J. F., (Wallingford: CABI; ), 219–229. 10.1079/9781845930820.0219 [DOI] [Google Scholar]
- Horn B. W., Moore G. G., Carbone I. (2009a). Sexual reproduction in Aspergillus flavus. Mycologia 101 423–429. 10.3852/09-011 [DOI] [PubMed] [Google Scholar]
- Horn B. W., Sorensen R. B., Lamb M. C., Sobolev V. S., Olarte R. A., Worthington C. J., et al. (2014). Sexual reproduction in Aspergillus flavus sclerotia naturally produced in corn. Phytopathology 104 75–85. 10.1094/PHYTO-05-13-0129-R [DOI] [PubMed] [Google Scholar]
- Huang Z., White D. G., Payne G. A. (1997). Corn seed proteins inhibitory to Aspergillus flavus and aflatoxin biosynthesis. Phytopathology 87 622–627. 10.1094/phyto.1997.87.6.622 [DOI] [PubMed] [Google Scholar]
- Jaime-Garcia R., Cotty P. J. (2004). Aspergillus flavus in soils and corncobs in south Texas: implications for management of aflatoxins in corn-cotton rotations. Plant Dis. 88 1366–1371. 10.1094/PDIS.2004.88.12.1366 [DOI] [PubMed] [Google Scholar]
- Ji C., Norton R. A., Wicklow D. T., Dowd P. F. (2000). Isoform patterns of chitinase and β-1, 3-glucanase in maturing corn kernels (Zea mays L.) associated with Aspergillus flavus milk stage infection. J. Agr. Food Chem. 48 507–511. 10.1021/jf9905119 [DOI] [PubMed] [Google Scholar]
- Kachapulula P. W., Akello J., Bandyopadhyay R., Cotty P. J. (2017). Aspergillus section Flavi community structure in Zambia influences aflatoxin contamination of maize and groundnut. Int. J Food Microbiol. 261 49–56. 10.1016/j.ijfoodmicro.2017.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R. Y., Williams W. P., Mylroie J. E., Boykin D. L., Harper J. W., Windham G. L. (2012). Identification of maize genes associated with host plant resistance or susceptibility to Aspergillus flavus infection and aflatoxin accumulation. PLoS One 7:e0036892. 10.1371/journal.pone.0036892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. A., Asghar M. A., Iqbal J., Ahmed A., Shamsuddin Z. A. (2014). Aflatoxins contamination and prevention in red chillies (Capsicum annuum L.) in Pakistan. Food Addit. Contaminants Part B Surveill. 7 1–6. 10.1080/19393210.2013.825330 [DOI] [PubMed] [Google Scholar]
- Kolomiets M., Battilani P., Borrego E., Reverberi M., Lanubile A., Scala V., et al. (2018). Mycotoxin contamination in maize is controlled by oxylipin signals. Phytopathology 108:1. [Google Scholar]
- Kolomiets M. V., Hannapel D. J., Chen H., Tymeson M., Gladon R. J. (2001). Lipoxygenase is involved in the control of potato tuber development. Plant Cell 13 613–626. 10.1105/tpc.13.3.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korani W., Chu Y., Holbrook C. C., Ozias-Akins P. (2018). Insight into genes regulating postharvest aflatoxin contamination of tetraploid peanut from transcriptional profiling. Genetic 209 143–156. 10.1534/genetics.118.300478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Mahato D. K., Kamle M., Mohanta T. K., Kang S. G. (2017). Aflatoxins: a global concern for food safety, human health and their management. Front. Microbiol. 7:2170 10.3389/fmicb.2016.02170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Bohra A., Pandey A. K., Pandey M. K., Kumar A. (2017). Metabolomics for plant improvement: status and prospects. Front. Plant Sci. 8:1302. 10.3389/fpls.2017.01302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A. M., Devi P. U., Sucharitha A. (2011). Differential effect of lipoxygenase on aflatoxin production by Aspergillus spp. Int J Plant Pathol. 4 153–164. 10.3923/ijpp.2011.153.164 [DOI] [Google Scholar]
- LaPrade J. C., Bartz J. A., Nordan A. J., DeMuynk T. J. (1973). Correlation of peanut seed coat surface wax accumulation with tolerance to colonization by Aspergillus flavus. J. Am. Peanut Res. Educ. Soc. 5 89–94. [Google Scholar]
- Liang X., Zhou G., Hong Y., Chen X., Liu H., Li S. (2009). Overview of research progress on peanut (Arachis hypogaea L.) host resistance to aflatoxin contamination and genomics at the Guangdong Academy of Agricultural Sciences. Peanut Sci. 369 29–34. 10.3146/at07-003.1 [DOI] [Google Scholar]
- Liang X. A., Luo M., Guo B. Z. (2006). Resistance mechanisms to Aspergillus flavus infection and aflatoxin contamination in peanut (Arachis hypogaea). Plant Pathol. J. 51 115–124. 28973784 [Google Scholar]
- Liang X. Q., Pan R. Z., Zhou G. (2002). Involvement of active oxygen generation and lipid peroxidation in the susceptibility/resistance of peanut seed infected by Aspergillus flavus. Chin. J. Oil Crops 24 19–23. [Google Scholar]
- Liang X. Q., Zhou G. Y., Pan R. (2001). Changes of some biochemical substances in peanut seeds under infection of Aspergillus flavus and their role in resistance to seed invasion. Chin. J. Oil Crop 23 26–30. [Google Scholar]
- Liang X. Q., Zhou G. Y., Pan R. Z. (2003b). Study on the relationship of wax and cutin layers in peanut seeds and resistance to invasion and aflatoxin production by Aspergillus flavus. J. Trop. Subtrop. Bot. 11 11–14. [Google Scholar]
- Liang X. Q., Pan R. C., Zhou G. Y. (2003a). Relationship of trypsin inhibitor in peanut seed and resistance to Aspergillus flavus invasion. Acta Agronom. Sin. 29 295–299. [Google Scholar]
- Liu Y., Wu F. (2010). Global burden of aflatoxin-induced hepatocellular carcinoma: a risk assessment. Environ. Health Persp. 118 818–824. 10.1289/ehp.0901388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logrieco A. F., Miller J. D., Eskola M., Krska R., Ayalew A., Bandyopadhyay R., et al. (2018). The mycotox charter: increasing awareness of, and concerted action for, minimizing mycotoxin exposure worldwide. Toxins 10:149. 10.3390/toxins10040149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozovaya V. V., Waranyuwat A., Widholm J. M. (1998). β-l, 3-glucanase and resistance to Aspergillus flavus infection in maize. Crop Sci. 38 1255–1260. 10.1111/jipb.12286 [DOI] [PubMed] [Google Scholar]
- Luo M., Brown R. L., Chen Z. Y., Cleveland T. E. (2009). Host genes involved in the interaction between Aspergillus flavus and maize. Toxin Rev. 28 118–128. 10.1080/15569540903089197 [DOI] [Google Scholar]
- Luo M., Brown R. L., Chen Z. Y., Menkir A., Yu J., Bhatnagar D. (2011). Transcriptional profiles uncover Aspergillus flavus-induced resistance in maize kernels. Toxins 3 766–786. 10.3390/toxins3070766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Liang X. Q., Dang P., Holbrook C. C., Bausher M. G., Lee R. D. (2005). Microarray-based screening of differentially expressed genes in peanut in response to Aspergillus parasiticus infection and drought stress. Plant Sci. J. 169 695–703. 10.1016/j.plantsci.2005.05.020 [DOI] [Google Scholar]
- Luo M., Liu J., Lee D., Scully B. T., Guo B. (2010). Monitoring the expression of maize genes in developing kernels under drought stress using oligo-microarray. J. Integr. Plant Biol. 52 1059–1074. 10.1111/j.1744-7909.2010.01000.x [DOI] [PubMed] [Google Scholar]
- Magbanua Z. V., De Moraes C. M., Brooks T. D., Williams W. P., Luthe D. S. (2007). Is catalase activity one of the factors associated with maize resistance to Aspergillus flavus? MPMI 20 697–706. 10.1094/mpmi-20-6-0697 [DOI] [PubMed] [Google Scholar]
- Magbanua Z. V., Williams P. W., Luthe D. S. (2013). The maize rachis affects Aspergillus flavus spread during ear development. Maydica 58 182–188. [Google Scholar]
- Majumdar R., Lebar M., Mack B., Minocha R., Minocha S., Wientjes C. (2018). The Aspergillus flavus Spermidine synthase (spds) gene, is required for normal development, aflatoxin production, and pathogenesis during infection of maize kernels. Front. Plant Sci. 9:317. 10.3389/fpls.2018.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar R., Minocha R., Lebar M. D., Rajasekaran K., Long S., Carter-Wientjes C., et al. (2019). Contribution of maize polyamine and amino acid metabolism towards resistance against Aspergillus flavus infection and aflatoxin production. Front. Plant Sci. 10:692. 10.3389/fpls.2019.00692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar R., Rajasekaran K., Sickler C., Lebar M., Musungu B. M., Fakhoury A. M., et al. (2017). The pathogenesis-related maize seed (PRms) gene plays a role in resistance to Aspergillus flavus infection and aflatoxin contamination. Front. Plant Sci. 8:1758. 10.3389/fpls.2017.01758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masanga J. O., Matheka J. M., Omer R. A., Ommeh S. C., Monda E. O., Alakonya A. E. (2015). Downregulation of transcription factor aflR in Aspergillus flavus confers reduction to aflatoxin accumulation in transgenic maize with alteration of host plant architecture. Plant Cell Rep. 34 1379–1387. 10.1007/s00299-015-1794-9 [DOI] [PubMed] [Google Scholar]
- Mauro A., Battilani P., Cotty P. J. (2015). Atoxigenic Aspergillus flavus endemic to Italy for biocontrol of aflatoxins in maize. Biocontrol 60 125–134. 10.1007/s10526-014-9624-5 [DOI] [Google Scholar]
- Mayfield K. L., Murray S. C., Rooney W. L., Isakeit T., Odvody G. A. (2011). Confirmation of QTL reducing aflatoxin in maize testcrosses. Crop Sci. 51 2489–2498. 10.2135/cropsci2011.02.0112 [DOI] [Google Scholar]
- Mehl H. L., Cotty P. J. (2011). Influence of the host contact sequence on the outcome of competition among Aspergillus flavus isolates during host tissue invasion. Appl. Environ. Microbiol. 77 1691–1697. 10.1128/AEM.02240-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehl H. L., Jaime R., Callicott K. A., Probst C., Garber N. P., Ortega-Beltran A., et al. (2012). Aspergillus flavus diversity on crops and in the environment can be exploited to reduce aflatoxin exposure and improve health. Ann. NY. Acad. Sci. 1273 7–17. 10.1111/j.1749-6632.2012.06800.x [DOI] [PubMed] [Google Scholar]
- Menkir A., Brown R. L., Bandyopadhyay R., Chen Z. Y., Cleveland T. E. (2006). A USA–Africa collaborative strategy for identifying, characterizing, and developing maize germplasm with resistance to aflatoxin contamination. Mycopathologia 162 225–232. 10.1007/s11046-006-0056-3 [DOI] [PubMed] [Google Scholar]
- Menkir A., Brown R. L., Bandyopadhyay R., Cleveland T. E. (2008). Registration of six tropical maize germplasm lines with resistance to aflatoxin contamination. J. Plant Regist. 2 246–250. 10.3198/jpr2008.01.0028crg [DOI] [Google Scholar]
- Meseka S., Williams W. P., Warburton M. L., Brown R. L., Augusto J., Ortega-Beltran A., et al. (2018). Heterotic affinity and combining ability of exotic maize inbred lines for resistance to aflatoxin accumulation. Euphytica 214:184. [Google Scholar]
- Mideros S. X., Warburton M. L., Jamann T. M., Windham G. L., Williams W. P., Nelson R. J. (2014). Quantitative trait loci influencing mycotoxin contamination of maize: analysis by linkage mapping, characterization of near-isogenic lines, and meta-analysis. Crop Sci. 54:127 10.2135/cropsci2013.04.0249 [DOI] [Google Scholar]
- MoFA, (2011). Agriculture in Ghana. Facts and figures (2010). Statistics, Research and 691 Information Directorate (SRID). Accra: MoFA. [Google Scholar]
- Moore K. G., Price M. S., Boston R. S., Weissinger A. K., Payne G. A. (2004). A chitinase from Tex6 maize kernels inhibits growth of Aspergillus flavus. Phytopathology 94 82–87. 10.1094/PHYTO.2004.94.1.82 [DOI] [PubMed] [Google Scholar]
- Naqvi S. S., Park K. S., Yi S. Y., Lee H. W., Bok S. H., Choi D. (1998). A glycine-rich RNA-binding protein gene is differentially expressed during acute hypersensitive response following Tobacco Mosaic Virus infection in tobacco. Plant Mol. Biol. 37 571–576. [DOI] [PubMed] [Google Scholar]
- Nayak S. N., Agarwal G., Pandey M. K., Sudini H. K., Jayale A. S., Purohit S., et al. (2017). Aspergillus flavus infection triggered immune responses and host-pathogen cross-talks in groundnut during in-vitro seed colonization. Sci. Rep. 7:9659. 10.1038/s41598-017-09260-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam S. N., Waliyar F., Aruna R., Reddy S. V., Kumar P. L., Craufurd P. Q., et al. (2009). Breeding peanut for resistance to aflatoxin contamination at ICRISAT. Peanut Sci. 36 42–49. 10.3146/at07-008.1 [DOI] [Google Scholar]
- Ogunola O. F., Hawkins L. K., Mylroie E., Kolomiets M. V., Borrego E., Tang J. D., et al. (2017). Characterization of the maize lipoxygenase gene family in relation to aflatoxin accumulation resistance. PLoS One 12:e0181265. 10.1371/journal.pone.0181265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Beltran A., Bandyopadhyay R. (2019). Comments on “Trial Summary on the Comparison of Various Non-Aflatoxigenic Strains of Aspergillus flavus on Mycotoxin Levels and Yield in Maize” by MS Molo, et al. Agron. J. 111 2625–2631. [Google Scholar]
- Pandey M. K., Kumar R., Pandey A. K., Soni P., Gangurde S. S., Sudini H. K., et al. (2019). Mitigating aflatoxin contamination in groundnut through a combination of genetic resistance and post-harvest management practices. Toxins 11:315. 10.3390/toxins11060315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey M. K., Upadhyaya H. D., Rathore A., Vadez V., Sheshshayee M. S., Sriswathi M., et al. (2014). Genome wide association studies for 50 agronomic traits in peanut using the ‘reference set’ comprising 300 genotypes from 48 countries of the semi-arid tropics of the world. PLoS One 9:e0105228. 10.1371/journal.pone.0105228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish F., Williams W. P., Windham G. L., Shan X. (2019). Differential expression of signaling pathway genes associated with aflatoxin reduction quantitative trait loci in maize (Zea mays L.). Front. Microbiol 10:2683. 10.3389/fmicb.2019.02683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y., Kolomiets M. (2010). Distinct roles of two 9-lipoxygenase paralogs in the regulation of aflatoxin accumulation in maize seed. Phytopathology 6 S96–S97. [Google Scholar]
- Paul C., Naidoo G., Forbes A., Mikkilineni V., White D., Rocheford T. (2003). Quantitative trait loci for low aflatoxin production in two related maize populations. Theor. Appl. Genet. 107 263–270. 10.1007/s00122-003-1241-0 [DOI] [PubMed] [Google Scholar]
- Payne G. A. (1998). “Process of contamination by aflatoxin-producing fungi and their impact on crops,” in Mycotoxin in Agriculture and Food safety, eds Sinha K. K., Bhatnagar D., (New York, NY: Marcel Decker Inc; ), 279–300. [Google Scholar]
- Pechanova O., Pechan T., Williams W. P., Luthe D. S. (2011). Proteomic analysis of the maize rachis: potential roles of constitutive and induced proteins in resistance to Aspergillus flavus infection and aflatoxin accumulation. Proteomics 11 114–127. 10.1002/pmic.201000368 [DOI] [PubMed] [Google Scholar]
- Pedras M. S. C., Yaya E. E. (2015). Plant chemical defenses: are all constitutive antimicrobial metabolites phytoanticipins? Nat. Prod. Commun. 10 209–218. [PubMed] [Google Scholar]
- Peethambaran B., Hawkins L., Windham G. L., Williams W. P., Luthe D. S. (2009). Anti-fungal activity of maize silk proteins and role of chitinases in Aspergillus flavus resistance. Toxin Rev. 29 27–39. 10.3109/15569540903402874 [DOI] [Google Scholar]
- Pegoraro C., Mertz L. M., da Maia L. C., Rombaldi C. V., de Oliveira A. C. (2011). Importance of heat shock proteins in maize. JCSB 14 85–95. [Google Scholar]
- Piasecka A., Jedrzejczak-Rey N., Bednarek P. (2015). Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 206 948–964. 10.1111/nph.13325 [DOI] [PubMed] [Google Scholar]
- Pierpoint W. S., Robinson N. P., Leason M. B. (1981). The pathogenesis-related proteins of tobacco: their induction by viruses in intact plants and their induction by chemicals in detached leaves. Physiol. Plant Pathol. 19 85–97. [Google Scholar]
- Pildain M. B., Frisvad J. C., Vaamonde G., Cabral D., Varga J., Samson R. A. (2008). Two novel aflatoxin-producing Aspergillus species from Argentinean peanuts. Int. J. Syst. Evol. Microbiol. 58 725–735. 10.1099/ijs.0.65123-0 [DOI] [PubMed] [Google Scholar]
- Pitt J., Manthong C., Siriacha P., Chotechaunmanirat S., Markwell P. (2015). Studies on the biocontrol of aflatoxin in maize in Thailand. Biocontrol Sci. Techn. 25 1070–1091. 10.1080/09583157.2015.1028893 [DOI] [Google Scholar]
- Probst C., Bandyopadhyay R., Cotty P. J. (2014). Diversity of aflatoxin-producing fungi and their impact on food safety in sub-Saharan Africa. Int. J. Food Microbiol. 174 113–122. 10.1016/j.ijfoodmicro.2013.12.010 [DOI] [PubMed] [Google Scholar]
- Rajasekaran K., Sayler R. J., Sickler C. M., Majumdar R., Jaynes J. M., Cary J. W. (2018). Control of Aspergillus flavus growth and aflatoxin production in transgenic maize kernels expressing a tachyplesin-derived synthetic peptide. AGM182. Plant Sci. 270 150–156. 10.1016/j.plantsci.2018.02.006 [DOI] [PubMed] [Google Scholar]
- Ramalingam A., Kudapa H., Pazhamala L. T., Weckwerth W., Varshney R. K. (2015). Proteomics and metabolomics: two emerging areas for legume improvement. Front. Plant Sci. 6:1116. 10.3389/fpls.2015.01116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzaghi-Abyaneh M. ed. (2013). Aflatoxins: Recent Advances and Future Prospects. Norderstedt: BoD–Books on Demand. [Google Scholar]
- Rokas A., Payne G., Fedorova N. D., Baker S. E., Machida M., Yu J., et al. (2007). What can comparative genomics tell us about species concepts in the genus Aspergillus? Stud. Mycol. 59 11–17. 10.3114/sim.2007.59.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo J., Vancanneyt G., Pérez A. G., Sanz C., Störmann K., Rosahl S., et al. (1996). Characterization of three potato lipoxygenases with distinct enzymatic activities and different organ-specific and wound-regulated expression patterns. J Biol Chem. 271 21012–21019. 10.1074/jbc.271.35.21012 [DOI] [PubMed] [Google Scholar]
- Roze L. V., Hong S. Y., Linz J. E. (2013). Aflatoxin biosynthesis: current frontiers. Annu. Rev. Food Sci. 4 293–311. 10.1146/annurev-food-083012-123702 [DOI] [PubMed] [Google Scholar]
- Russin J. S., Guo B. Z., Tubujika K. M., Brown R. L., Cleveland T. E., Widstrom N. W. (1997). Comparison of kernel wax from corn genotypes resistant or susceptible to Aspergillus flavus. Phytopathology 87 529–533. 10.1094/phyto.1997.87.5.529 [DOI] [PubMed] [Google Scholar]
- Samson R. A. (1992). Current taxonomic schemes of genus Aspergillus and its teleomorphs. Biotechnolgy 23:355. [PubMed] [Google Scholar]
- Samson R. A., Hoekstra E. S., Van Oorschot C. A. (1981). Introduction to Food-Borne Fungi. Centraalbureau voor Schimmelcultures. Available at: https://www.worldcat.org/title/introduction-to-food-borne-fungi/oclc/680002633 (accessed September 10, 2019). [Google Scholar]
- Sanders T. H., Mixon A. C. (1979). Effect of peanut tannin on percent seed colonization and in vitro growth by Aspergillus parasiticus. Mycopathologia 66 169–173. 10.1007/bf00683966 [DOI] [PubMed] [Google Scholar]
- Sarma U. P., Bhetaria P. J., Devi P., Varma A. (2017). Aflatoxins: implications on health. Indian J. Clin. Biochem. 32 124–133. 10.1007/s12291-017-0649-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreurs F., Bandyopadhyay R., Kooyman C., Ortega-Beltran A., Akande A., Konlambigue M., et al. (2019). Commercial Products Promoting Plant Health in African Agriculture. Cambridge: Burleigh Dodds Science Publishing Limited. [Google Scholar]
- Schubert M., Houdelet M., Kogel K. H., Fischer R., Schillberg S., Nölke G. (2015). Thanatin confers partial resistance against aflatoxigenic fungi in maize (Zea mays). Transgenic Res. 24 885–895. 10.1007/s11248-015-9888-2 [DOI] [PubMed] [Google Scholar]
- Senghor L., Ortega-Beltran A., Atehnkeng J., Callicott K., Cotty P., Bandyopadhyay R. (2019). The atoxigenic biocontrol product Aflasafe SN01 is a valuable tool to mitigate aflatoxin contamination of both maize and groundnut cultivated in Senegal. Plant Dis. 104 510–520. 10.1094/PDIS-03-19-0575-RE [DOI] [PubMed] [Google Scholar]
- Sharma K. K., Pothana A., Prasad K., Shah D., Kaur J., Bhatnagar D., et al. (2018). Peanuts that keep aflatoxin at bay: a threshold that matters. Plant Biotechnol. J. 16 1024–1033. 10.1111/pbi.12846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X., Livingston D. P., III, Woloshuk C. P., Payne G. A. (2017). Comparative histological and transcriptional analysis of maize kernels infected with Aspergillus flavus and Fusarium verticillioides. Front. Plant Sci. 8:2075. 10.3389/fpls.2017.02075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U., Deb D., Singh A., Grover A. (2011). Glycine-rich RNA binding protein of Oryza sativa inhibits growth of M15 E. coli cells. BMC Res. 4:18. 10.1186/1756-0500-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriver K., Mundy J. (1990). Gene expression in response to abscisic acid osmotic stress. Plant Cell 2:503 10.2307/3869112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev V. S., Potter T. L., Horn B. W. (2006). Prelylated stilbenes from peanut root mucilage. Phytochem. Anal. 17 312–322. 10.1002/pca.920 [DOI] [PubMed] [Google Scholar]
- Sudini H., Ranga Rao G. V., Gowda C. L. L., Chandrika R., Margam V., Rathore A., et al. (2015). Purdue Improved Crop Storage (PICS) bags for safe storage of groundnuts. J. Stored Prod. Res. 64 133–138. 10.1016/j.jspr.2014.09.002 [DOI] [Google Scholar]
- Suneja K. (2019). India Ropes in Global Institution to Curb Contamination in Spice Shipments. Geneva: World Trade Organization. [Google Scholar]
- Suwarno W. B., Hannok P., Palacios-Rojas N., Windham G., Crossa J., Pixley K. V. (2019). Provitamin A carotenoids in grain reduce aflatoxin contamination of maize while combating vitamin A deficiency. Front. Plant Sci. 10:30. 10.3389/fpls.2019.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerszen J. B. (1990). Change of isozymes of Aspergillus group infected peanut cotyledon from plants grown under drought stress. APRES 21:23. [Google Scholar]
- Taber R. A., Pettit R. E., Benedict C. R., Dieckert J. W., Kertring D. L. (1973). Comparison of Aspergillus flavus tolerant and susceptible lines. I. Light microscope investigation. Am. Peanut Res. Edu. Assoc. 5:206. [Google Scholar]
- Tang J. D., Perkins A., Williams W. P., Warburton M. L. (2015). Using genome-wide associations to identify metabolic pathways involved in maize aflatoxin accumulation resistance. BMC Genomics 16:673. 10.1186/s12864-015-1874-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakare D., Zhang J., Wing R. A., Cotty P. J., Schmidt M. A. (2017). Aflatoxin-free transgenic maize using host-induced gene silencing. Sci. Adv. 3:e1602382. 10.1126/sciadv.1602382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R. B., Linday D. L., Davis D. D., Bishop R. D. (1975). Isolation and identification of 5, 7-dimethoxyisoflavone, an inhibitor of Aspergillus flavus from peanut. Mycopathologia 57 39–40. 10.1007/bf00431177 [DOI] [PubMed] [Google Scholar]
- Upadhyaya H. D., Nigam S. N., Thakur R. P. (2002). “Genetic enhancement for resistance to aflatoxin contamination in groundnut,” in Proceedings of the Seventh ICRISAT Regional Groundnut Meeting for Western and Central Africa, 6–8 Dec. 2000, Cotonou, Benin. Patancheru 502 324, (Andhra Pradesh: International Crops Research Institute for the Semi-Arid Tropics; ), 29–36. [Google Scholar]
- Van Loon L. C. (1985). Pathogenesis-related proteins. Plant Mol. Biol. 4 111–116. [DOI] [PubMed] [Google Scholar]
- Van Loon L. C., Van Strien E. A. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55 85–97. 10.1006/pmpp.1999.0213 [DOI] [Google Scholar]
- VanEtten H. D., Mansfield J. W., Bailey J. A., Farmer E. E. (1994). Two classes of plant antibiotics: phytoalexins versus “phytoanticipins.”. Plant Cell 6 1191–1192. 10.1105/tpc.6.9.1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S., Jaime R., Kagot V., Probst C. (2018). Comparative effects of hermetic and traditional storage devices on maize grain: mycotoxin development, insect infestation and grain quality. J. Stored Prod. Res. 77 34–44. 10.1016/j.jspr.2018.02.002 [DOI] [Google Scholar]
- Wang H., Lei Y., Wan L., Yan L., Lv J., Dai X., et al. (2016). Comparative transcript profiling of resistant and susceptible peanut post-harvest seeds in response to aflatoxin production by Aspergillus flavus. BMC Plant Biol. 1:54. 10.1186/s12870-016-0738-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Lei Y., Yan L., Cheng K., Dai X., Wan L., et al. (2015). Deep sequencing analysis of transcriptomes in Aspergillus flavus in response to resveratrol. BMC Microbiol. 15:182. 10.1186/s12866-015-0513-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Chen X., Li H., Liu H., Hong Y., Yang Q., et al. (2013). Transcriptome identification of the resistance-associated genes (RAGs) to Aspergillus flavus infection in pre-harvested peanut (Arachis hypogaea). Funct. Plant Biol. 40 292–303. [DOI] [PubMed] [Google Scholar]
- Wang T., Zhang E., Chen X., Li L., Liang X. (2010). Identification of seed proteins associated with resistance to pre-harvested aflatoxin contamination in peanut (Arachis hypogaea L). BMC Plant Biol. 10:267. 10.1186/1471-2229-10-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yan S., Liu C., Chen F., Wang T. (2012). Proteomic analysis reveals an aflatoxin-triggered immune response in cotyledons of Arachis hypogaea infected with Aspergillus flavus. J. Proteome Res. 11 2739–2753. 10.1021/pr201105d [DOI] [PubMed] [Google Scholar]
- Warburton M. L., Brooks T. D., Krakowsky M. D., Shan X., Windham G. L., Williams W. P. (2009). Identification and mapping of new sources of resistance to aflatoxin accumulation in maize. Crop Sci. 49 1403–1408. 10.2135/cropsci2008.12.0696 [DOI] [Google Scholar]
- Warburton M. L., Tang J. D., Windham G. L., Hawkins L. K., Murray S. C., Xu W., et al. (2015). Genome-wide association mapping of Aspergillus flavus and aflatoxin accumulation resistance in maize. Crop Sci. 55 1857–1867. 27598199 [Google Scholar]
- Warburton M. L., Williams W. P. (2014). Aflatoxin resistance in maize: what have we learned lately? Adv. Bot. 2014:352831 10.1155/2014/352831 [DOI] [Google Scholar]
- Warburton M. L., Brooks T. D., Windham G. L., Williams W. P. (2011). Identification of novel QTL contributing resistance to aflatoxin accumulation in maize. Mol. Breeding 27 491–499. 10.1007/s11032-010-9446-9 [DOI] [Google Scholar]
- Wicklow D. T., Wilson D. M., Nelson T. C. (1993). Survival of Aspergillus flavus sclerotia and conidia buried in soil in Illinois or Georgia. Phytopathology 83 1141–1147. [Google Scholar]
- Wild C. P., Gong Y. Y. (2010). Mycotoxins and human disease: a largely ignored global health issue. Carcinogenesis 31 71–82. 10.1093/carcin/bgp264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox M. C., Davis G. L., Warburton M. L., Windham G. L., Abbas H. K., Betrán J., et al. (2013). Confirming quantitative trait loci for aflatoxin resistance from Mp313E in different genetic backgrounds. Mol. Breed. 32 15–26. 10.1007/s11032-012-9821-9 [DOI] [Google Scholar]
- Williams W. P., Krakowsky M. D., Scully B. T., Brown R. L., Menkir A., Warburton M. L., et al. (2015). Identifying and developing maize germplasm with resistance to accumulation of aflatoxins. World Mycotoxin J. 8 193–209. 10.3920/wmj2014.1751 16944289 [DOI] [Google Scholar]
- Woloshuk C. P., Cavaletto J. R., Cleveland T. E. (1997). Inducers of aflatoxin biosynthesis from colonized maize kernels are generated by an amylase activity from Aspergillus flavus. Phytopathology 87 164–169. 10.1094/phyto.1997.87.2.164 [DOI] [PubMed] [Google Scholar]
- Xie C., Wen S., Liu H., Chen X., Li H., Hong Y., et al. (2013). Overexpression of ARAhPR10, a member of the PR10 family, decreases levels of Aspergillus flavus infection in peanut seeds. Am. J. Plant Sci. 4:602 10.4236/ajps.2013.43079 [DOI] [Google Scholar]
- Xie Y. R., Chen Z. Y., Brown R. L., Bhatnagar D. (2010). Expression and functional characterization of two pathogenesis-related protein 10 genes from Zea mays. J. Plant Physiol. 167 121–130. 10.1016/j.jplph.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Yan Y., Borrego E., Kolomiets M. V. (2013). Jasmonate Biosynthesis, Perception and Function in Plant Development and Stress Responses Lipid metabolism. London: InTech, 393–442. [Google Scholar]
- Yang M., Lu L., Li S., Zhang J., Li Z., Wu S., et al. (2019). Transcriptomic insights into benzenamine effects on the development, aflatoxin biosynthesis, and virulence of Aspergillus flavus. Toxins 11:70. 10.3390/toxins11020070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Wang Y., Wu F., Gu X., Bian Y., Wang Y., et al. (2014). Quantitative trait locus mapping of resistance to Aspergillus flavus infection using a recombinant inbred line population in maize. Mol. Breed. 33 39–49. 10.1007/s11032-013-9932-y [DOI] [Google Scholar]
- Yu B., Huai D., Huang L., Kang Y., Ren X., Chen Y., et al. (2019). Identification of genomic regions and diagnostic markers for resistance to aflatoxin contamination in peanut (Arachis hypogaea L.). BMC Genet. 20:32. 10.1186/s12863-019-0734-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Chang P. K., Bhatnagar D., Cleveland T. E. (2000). Genes encoding cytochrome P450 and monooxygenase enzymes define one end of the aflatoxin pathway gene cluster in Aspergillus parasiticus. Appl. Microbiol. Biotechnol. 53 583–590. 10.1007/s002530051660 [DOI] [PubMed] [Google Scholar]
- Yu J., Chang P. K., Ehrlich K. C., Cary J. W., Bhatnagar D., Cleveland T. E., et al. (2004). Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 70 1253–1262. 10.1128/aem.70.3.1253-1262.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cui M., Zhang J., Zhang L., Li C., Kan X., et al. (2016). Confirmation and fine mapping of a major QTL for aflatoxin resistance in maize using a combination of linkage and association mapping. Toxins 8:258. 10.3390/toxins8090258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Li C., Yan C., Wang J., Yuan C., Zhang H., et al. (2019). Transcriptome and proteome analyses of resistant preharvest peanut seed coat in response to Aspergillus flavus infection. Electron. J. Biotechn. 39 82–90. 10.1016/j.ejbt.2019.03.003 [DOI] [Google Scholar]