Abstract
Background
Osteosarcoma (OS) is the most common primary malignant bone tumour. Unfortunately, no new treatments are approved and over the last 30 years the survival rate remains only 30% at 5 years for poor responders justifying an urgent need of new therapies. The Mutt homolog 1 (MTH1) enzyme prevents incorporation of oxidized nucleotides into DNA and recently developed MTH1 inhibitors may offer therapeutic potential as MTH1 is overexpressed in various cancers.
Methods
The aim of this study was to evaluate the therapeutic benefits of targeting MTH1 with two chemical inhibitors, TH588 and TH1579 on human osteosarcoma cells. Preclinical efficacy of TH1579 was assessed in human osteosarcoma xenograft model on tumour growth and development of pulmonary metastases.
Findings
MTH1 is overexpressed in OS patients and tumour cell lines, compared to mesenchymal stem cells. In vitro, chemical inhibition of MTH1 by TH588 and TH1579 decreases OS cells viability, impairs their cell cycle and increases apoptosis in OS cells. TH1579 was confirmed to bind MTH1 by CETSA in OS model. Moreover, 90 mg/kg of TH1579 reduces in vivo tumour growth by 80.5% compared to non-treated group at day 48. This result was associated with the increase in 8-oxo-dG integration into tumour cells DNA and the increase of apoptosis. Additionally, TH1579 also reduces the number of pulmonary metastases.
Interpretation
All these results strongly provide a pre-clinical proof-of-principle that TH1579 could be a therapeutic option for patients with osteosarcoma.
Funding
This study was supported by La Ligue Contre le Cancer, la SFCE and Enfants Cancers Santé.
Key words: Osteosarcoma, Bone tumours, MTH1, ROS, DNA damage
Research in context.
Evidence before this study
Osteosarcoma is the most prevalent primary malignant bone tumour with poor prognosis mainly affecting children and young adults. Since current therapies did not change in the last 30 years, there is a real need of new therapeutic strategies. Targeting MTH1 using TH1579 has shown to inhibit tumour growth in preclinical studies in various cancers.
Added value of this study
Using in vitro experiments and mouse models of osteosarcoma, we report that TH1579, MTH1 inhibitor, inhibits cell growth and induces apoptosis in osteosarcoma cells and, delays tumour growth and blocks the development of pulmonary metastases in preclinical model of osteosarcoma.
Implications of all the available evidence
In this study, we provide the new insight in the therapeutic efficacy of TH1579 on tumour growth and metastases development in preclinical model of osteosarcoma suggesting that TH1579 could be a therapeutic option in treatment of patient with osteosarcoma.
Alt-text: Unlabelled box
1. Introduction
Osteosarcoma (OS) is the most common primary malignant bone tumour. It mainly affects children and young adults, with an incidence peak at around 18 years old, and is mostly localized in long bones [1]. OS involves deregulation of the equilibrium between bone formation and bone resorption, which leads to ectopic bone formation and osteolysis. Current strategies combine surgical tumour excision and chemotherapy [2,3]. Unfortunately, 5 years survival rate drops from 75% to 25% for bad responders [4,5], hence the urgent necessity to develop new therapeutic strategies. In this context, DNA damaging compounds are particularly interesting candidates. Indeed, their antitumour effects have been known for over half a century [6,7]. In normal cells, DNA damages lead to cell cycle arrest in order to prevent transmission of genetic alterations during mitosis. Nevertheless, tumour cells have high tolerance to DNA damage, allowing greater proliferation rates [8]. However this fast proliferation also increases the chances of cell death through the transmission of fatal DNA damages to daughter cells. Therefore, several anticancer strategies are based on the induction of DNA damages to tumour cells. For example in OS cells, DNA damaging agents such as cisplatin, doxorubicin and methotrexate have been used in clinic for decades to treat the disease [1,8,9].
Recently, there has been a growing interest for antitumour capabilities of the reactive oxygen species (ROS), notably through their ability to induce DNA damage [10]. ROS can indeed interact with the pool of free nucleotides, creating oxidized nucleotides. These oxonucleotides can be integrated into the DNA double helix by a DNA polymerase, during replication, and disturb cellular processes [11]. Base Excision Repair (BER) mechanism then lead to excision of oxonucleotides from DNA by glycosylases such as the 8-oxoguanineglycosylase 1 (OGG1). However, this repair mechanism can be overwhelmed, leading to cell death. The Mutt homolog 1 (MTH1) or Nudix-Type 1 (NUDT1) protein is part of the nudix family of hydrolases. MTH1 acts as a nucleotide pool sanitation enzyme by hydrolysing oxonucleotides triphosphates such as 8-oxo-dGTP, 2-OH-dATP or 8-oxo-dATP, into their monophosphates counterparts. Its activity prevents the integration of oxidized nucleotides into DNA, since monophosphate nucleotides cannot be used by the DNA polymerase [12,13]. Thus, MTH1 protects cells against ROS effects on DNA integrity. Therefore, targeting MTH1 could imply impairing the genome integrity, i.e. through the targeting of DNA building blocks, nucleotides. Free nucleotides are also more likely to be oxidized by ROS than DNA macromolecules [12]. Furthermore, ROS are by-products of cell metabolism [14], and it has been shown that tumour cells produce high quantity of ROS compared to normal cells, due to their fast metabolism, their environment and mutations [15], [16], [17]. Consequently, tumour cells could strongly depend on MTH1 activity to protect their genome integrity, and thus increase its expression compared to normal cells. MTH1 has indeed been found to be overexpressed in several types of cancer such as breast and colorectal cancer [18,19], while MTH1-deficient mice developed spontaneous tumours 18 months after birth [20]. Since 2014, several studies have tested the effects of MTH1 inhibition in different cancer types including osteosarcoma, melanoma, breast, cervical or colorectal cancer. MTH1 inhibition reduced cell viability and colony formation, increased oxidized nucleotides integration in DNA, and induced DNA damage [21], [22], [23], [24]. In vivo, MTH1 inhibition decreased tumour growth and increased survival in mouse xenograft models [21], [22], [23], leading to an increased interest in MTH1 as therapeutic target in cancer [21,22]. But more recently, these results were challenged by questioning the role of MTH1 in tumour biology, as well as the benefits of targeting this protein in cancer [25], [26], [27], [28].
In this study we investigated the therapeutic efficacy of TH588 (first generation of MTH1 inhibitor) and TH1579 (optimized analogue of TH588), in OS. TH1579 (Karonudib) is currently in a phase I clinical trial. Using a panel of human OS cell lines and a xenograft model, we evaluated the effects of TH588 and TH1579 on several tumour processes such as proliferation, tumour growth or metastatic dissemination.
2. Materials and methods
2.1. Tumour cell lines
Human osteosarcoma cancer cell lines HOS-MNNG, KHOS, MG63, U2OS, SJSA-1, G292, Cal72 and 143B were purchased from the ATCC and maintained in DMEM (Invitrogen-Life Technologies, Inc.) supplemented with 10% FBS and 2 mmol/L l-glutamine. All cell lines were cultured in a humidified 5% CO2/air atmosphere at 37 °C.
2.2. Tissue specimens and mesenchymal stem cells
Patient tumour biopsy specimens were collected at Nantes University Hospital (Nantes, France). Samples were obtained following patient in formed consent, and after ethical approval by the Nantes University Hospital Ethics Committee.
Human mesenchymal stem cells (hMSCs) from healthy donors and osteoblasts derived from hMSCs on osteogenic differentiation are used as controls. Briefly, hMSCs are cultured in DMEM supplemented with vitamin D3 and dexamethasone. Then ascorbic acid and β-glycerophosphate was added to osteogenic medium. Before their use, the cells induced to osteogenic differentiation were tested for mineralization using alizarin red staining as described previously [29].
2.3. Therapeutic agent
MTH1 inhibitors, TH588 and TH1579 [21,22] was provided by the Karolinska Institutet and used for in vitro and in vivo studies. For the in vitro studies, the drugs were dissolved in dimethyl sulfoxide (DMSO) at 10 mmol/L stock solution and stored at −20 °C. For in vivo studies, TH1579 was dissolved in an acetate plus cyclodextrin buffer at the indicated concentrations.
2.4. Cell proliferation assay, clonogenic assay and apoptosis assay
OS cells were seeded in 96 wells plates. 24 h after seeding, OS cells were treated with the drugs at the indicated doses (from 78 nml/L to 20 µmol/L). After 72 h of treatment, the cells were fixed with 1% glutaraldehyde under agitation for 5 min, washed with water then stained with 0.1% crystal violet for 5 min. After drying, the dye was diluted in Sorensen's buffer (5 min in trisodium citrate, HCl 0,1N and ethanol) and the absorbance was measured at 570 nm using a spectrophotometer (Perkin Elmer Wallac 1420 Victor²). The proliferation rate is represented as a percentage of untreated cells.
Regarding the clonogenic assay, cells were treated for 48 h with 2 or 8 µmol/L of TH588 or TH1579. The cells were then trypsinated and plated at 1000 cells/well in 6 wells plate. 10 days later, the cells were fixed and stained with crystal violet as described above. The colonies were then counted.
2.5. Transfection and transduction
OS cells were transfected with 30 nmol/L of siRNA (sc-62647, sc-37007; Santa Cruz Biotechnology) using Lipofectamine RNAimax (thermofisher scientific) following the provider's recommendation. For the transduction, the shRNA (MOI20) targeting MTH1.pLKO.1shMTH1-1 and shMTH1-2 was a gift of Bob Weinberg (Addgene plasmids # 21297 and # 21298) [30]. The proliferation assay was performed as described above (without treatment), on cells trypsinated and re-seeded after 48 h of transfection or 72 h of transduction.
2.6. Apoptosis assay
Caspase activity was measured in protein extract using the Apo-ONE caspase 3/7 kit from Promega. The cells were treated for 48 h and the assay was performed according to the supplier's recommendation. The fluorescence was measured at 521 nm on the Tristar LB 941 (Berthold Technologies). The enzymatic activity was normalized by the protein quantity.
2.7. Cell-cycle analysis
Osteosarcoma cell were treated at indicated concentrations of TH588 or TH1579 for 48 h, trypsinized, washed twice, in cold PBS (5 min), and fixed with 70% ethanol at 4 °C for 30 min. After washing, cells were incubated for 10 min in 4 °C phospho-citrate buffer, washed and incubated in PBS containing 100 μg/mL ribonuclease A, for 30 min at 37 °C. Cells were then incubated in 50 µg/mL propidium iodide for 20 min at 4 °C. Cell-cycle distribution was analysed by flow cytometry (Beckman Coulter Epics Elite, Beckman Coulter), based on 2 N and 4 N DNA content.
2.8. Comet assay
OS cells were treated with TH588 or TH1579 for 24 h, then trypsinated, included in low melting agarose (37 °C) and put on Superfrost plus slides (ThermoFisher scientific). The cells were then lysated overnight at 4 °C, in a DMSO, NaOH, triton and Tris–HCl containing buffer. The cells were then washed in a digestion buffer (Hepes, KCl, 500 mmol/L EDTA and BSA in water; 3 × 20 min) and treated with 2µG OGG1 (diluted in the digestion buffer) when indicated, for 45 min at 37 °C. After 30 min in a denaturation buffer (EDTA, 10 N NaOH in water), an electrophoresis was performed at 300 mA, for 30 min in the denaturation buffer. The slides are then dipped in a pH 7.5 neutralization buffer for 45 min (Tris and HCL in water). DNA breaks are detected using the Thermofisher Sybr Gold dye and quantified using FIJI and the Open Comet plugin.
2.9. 8-oxo-dG assay
It is well known that Avidin binds with high specificity to 8-oxo-dG and used to evaluate the incorporation of 8-oxo-dG into DNA [31]. Tumour cells were treated with TH1579 at indicated concentrations for 24 h in slide 8 wells (Ibidi). Then, cells were fixed in methanol at −20 °C for 20 min following by incubation in TBS with 0.1% Triton X-100 for 15 min. Cells were incubated in blocking solution (15% FBS, 0.1% Triton X-100 in TBS) for 2 h at room temperature, and thereafter incubated with 10 μg/mL Alexa488-conjugated Avidin (Invitrogen) in blocking solution for 1 h at 37 °C. Then, the cells were washed in TBS 0.1% Triton X-100 for 2 × 5 min at room temperature, following by a quick rinse in distilled water. DNA was counterstained with DAPI for 15 min in dark at room temperature. The cells were rinsed in TBS and Ibidi mounting Medium were added in the wells of the slides. Images were acquired with the Olympus IX73 inverted microscope.
2.10. Quantitative reverse transcription PCR
Total RNA was extracted using the Macherey-Nagel Nucleospin RNA. Following a reverse transcription on 1 µg of total RNA, the cDNA were diluted to 10 ng/µL. A Real-time monitoring of PCR amplification of cDNA was performed using DNA primers (Supplementary Table I) on CFX96 Detector System (Bio-Rad) with SYBR PCR Master Mix (Bio-Rad). Target gene expression was normalized to GAPDH level in respective samples as an internal standard, and the comparative cycle threshold (Ct) method was used to calculate relative quantification of target mRNAs. Each assay was performed in triplicate.
2.11. Western blot
Samples containing equal amounts of protein from lysates of cultured osteosarcoma cells underwent electrophoresis on SDS-polyacrylamide gel and were transferred to PVDF membranes blocked in odyssey blocking buffer (LI-COR). Blots were probed overnight at 4 °C with primary antibodies (Supplementary Table II). After incubation, the membranes were washed in PBS containing 0.05% Tween. Membranes were then probed with a fluorescent antibody (Li-COR IRDye) diluted in the odyssey blocking buffer. Antibody binding was visualized using the LI-COR odyssey Fc.
2.12. Immunofluorescence
Cells were cultured in an imaging chamber (Ibidi) and treated as indicated in the figure. As control, cells were irradiated at 15 Gy, delivered at a rate of 1,67 Gy/min (Faxitron CP160 irradiator, Faxitron X-ray Corporation, IL, USA). Then, cells were fixed with 4% paraformaldehyde for 20 min at room temperature, then washed (washing buffer: PBS-Tween20 0.05%) and permeabilized 10 min at room temperature with PBS-0.1% triton X100. Cells were then blocked 10 min with goat serum (1/200) in washing buffer, then incubated overnight, at 4 °C, with primary antibody (Cell signalling Technology γH2AX antibody). Afterward, cells were washed and incubated with alexa Fluor 488 antibody (60 min at room temperature), then with DAPI.
2.13. Transwell motility and invasion
Cell migration and invasion assays were performed in Transwell chamber (8-μm pore size; Falcon). For cell invasion experiments, inserts were coated with 100 μL basement membrane Matrigel (3 μg/ml; Becton Dickinson) for 90 min at 37 °C. HOS-MNNG cells (1 × 105 cells/mL) were resuspended in DMEM containing 0.1% FBS, and 300 μL of this cell suspension was loaded into each insert (top chamber). DMEM 10% FBS was placed in the bottom chamber (750 μL/well). The plates were incubated for 6 h at 37 °C for migration and 24 h for invasion. Then, the inserts were carefully collected, the non-migrating cells were removed, and the migrating cells on the under-surface of the inserts were fixed with 1% glutaraldehyde under agitation for 5 min, washed with water, and then stained with 0.1% crystal violet for 5 min. The cells were then counted by bright-field microscopy.
2.14. Animal treatment
Female athymic nude mice were injected intramuscularly close to the tibia with 1.5 × 106 HOS-MNNG cells. Once their tumours were palpable, mice were randomly assigned to vehicle, TH1579 45 mg/kg or TH1579 90 mg/kg groups and treated orally b.i.d daily three times per week until day 48. Each experimental group consisted of 8 mice. Tumour volume was measured three times per week. Tumour volume was calculated by the formula: length × width × depth × 0.5432. Data points were expressed as average tumour volume ± SEM. Tumours and tibia were harvested for further analyses [micro computed tomography (microCT), histology]. Animal care and experimental protocols were approved by the French Ministry of Research and were done in accordance with the institutional guidelines of the French Ethical Committee protocol agreement number 1280.01 and under the supervision of authorized investigators.
2.15. MicroCT analysis
Tibias were scanned using high-resolution microcomputed tomography (Skyscan 1076) at 50 kV and 200 mA using a 0.5 mm aluminium filter and a detection pixel size of 9 mm. Images were captured every 0.8° through 180°rotation, and analysed using Skyscan software. After scanning, 3D microstructural image data was reconstructed using the Skyscan NRecon software. Bone parameters were calculated using the Skyscan CT Analyzer (CTAn) software. Using this software, the volume of interest was defined as cortical remodelling bone. Binarized images were performed: an upper threshold of 255 and a lower threshold of 55 was used to delineate each pixel as “bone” or “non-bone” Morphological traits of the mid-diaphyseal cortical shell were performed at a site starting 200 slices from the peronea insertion and then extending from this position for 700 slices (each slice = 9 µm). Then, total cross sectional area (Tt.Ar), cross sectional cortical area (Ct.Ar), cortical bone fraction (Ct.Ar/Tt.Ar), cortical thickness (Ct.Th), and cross sectional moment of inertia (polar) (CSMI) was measured in 2D.
2.16. Immunohistochemistry
IHC was performed on formalin-fixed, paraffin-embedded 3 μm sections of tumour samples. Immunostaining was conducted using the primary antibodies cleaved caspase-3 (1/400; Cell signalling Technology), CD146 (1/200; Abcam), gH2AX (1/750; Millipore), 8-oxo-dG (1/750; Millipore) and Sp7/Osterix (1/1000; Abcam) on tumour section.
Tibias were decalcified with 4.13% EDTA and 0.2% paraformaldehyde in PBS using the KOS microwave histostation (Milestone) before embedding in paraffin. Sections (Leica Microsystems) were analysed by Osterix and TRAP staining. Quantification of relative OC surface (TRAP+ cells) and OB number (Osterix+ cells) was evaluated by ImageJ (NIH, Bethesda, MD) software. All analyses were assessed by slide scanning using nanozoomer 2.0 RS (Hamamatsu).
2.17. Cellular thermal shift assay (CETSA)
CETSA is a target engagement assay detecting potential interaction between a specific target and studied compounds and has been described previously [32]. Briefly, frozen tumours from the TH1579 and vehicle treated animals were grinded in liquid nitrogen then resuspended in 600 µl TBS-T complete protease inhibitor cocktail (Roche). Aliquots were incubated for 3 min at a temperature gradient of 50–66 °C. Samples were frozen (−80 °C) and thawed (25 °C) three times to lyse the cells, followed by centrifugation at 17,000 g for 30 min. The supernatant (35 µl/sample) was transferred to a 96-well PCR plate (Agilent) and 12 µl of 4x Laemmli sample buffer were added to each sample. Proteins were denatured by heating the PCR plate at 95 °C for 10 min. Samples were run on SDS-PAGE and transferred to nitrocellulose membranes and incubated with rabbit anti MTH1 antibody (Novus Biological). Membranes were developed using Super Signal West Femto Substrate (Thermo Scientific), and measurements were performed using Odyssey Imaging System (LI-COR) and the bands signals were quantified using Odyssey image software.
2.18. Osteoclast differentiation assays
Osteoclasts were generated from human CD14+ monocytes of different donors. Purified CD14+ cells were cultured in α-MEM (Invitrogen Life Technologies) with 10% FBS and 25 ng/mL human macrophage colony stimulating factor (hM-CSF; R&D Systems). After 4 days, 100 ng/mL hRANKL +/- TH1579 were added. Multinucleated cells with more than three nuclei were counted after Tartrate-Resistant Acid Phosphatase (TRAP) staining (Sigma, France).
2.19. Short term in vivo study using HOS-MNNG-GFP cells
Procedures involving animal handling and care were approved by the Animal Care and Ethics Committee of the Home office in UK [PPL: 70/8967, Establishment license n°: 50/2509] and performed by trained, licensed personnel. To assess effects of short-term treatment with TH1579 on disseminated tumour cells (DTCs) to the lung and circulating tumour cells (CTCs) a paratibial osteosarcoma model was used. Briefly, 2.5 × 105 HOS-MNNG-GFP cells were injected partibially into anaesthetized female Balb/c nude mice (6–7 weeks old). Five days later animals received treatment of either vehicle or TH1579 at 90 mg/kg b.i.d three times per week for one week by oral gavage. Animals were culled 24 h (TH1579 group) or 48 h (vehicle group) after the last treatment. The tumour bearing leg was measured using callipers and the volume was calculated before assessing DTCs, CTCs.
2.20. Quantification of DTCs and CTCs using Parsortix
Animals were culled just before sample preparation for DTC analysis. Whole blood was collected via cardiac puncture into EDTA vacutainers (BD). Blood from all animals in one group was pooled. Lungs were excised, washed in PBS and minced using a scalpel blade before adding tumour dissociation solution (Tumour Dissociation Kit, Myltenyi) for 30 min at 37 °C. The single cell suspension was then filtered and centrifuged before resuspending in PBS and loading onto the Parsortix system (Angle) using the PX2_S50F separation program and a 6.5 μm separation cassette. Briefly, the Parsortix system is a microfluidic technology, which allows enrichment, isolation and quantification of small numbers of tumour cells based on cell size on deformability. The separation cassette containing the enriched sample was then vizualized using a fluorescent microscope and GFP-positive tumour cells trapped in the cassette were counted. As indicative experiment, the same procedure was performed for CTCs in whole blood (blood in each group was pooled before analysis; control N = 2; TH1579, N = 3) using PX2_S99F separation program.
2.21. Statistical analysis
All statistical tests were performed using the Graphpad Prism software. Statistical significativity was assessed by Kruskal–Wallis test or Anova. * p < 0.05; ** p < 0.01, ***p < 0.001, ****p < 0.0001.
3. Results
3.1. MTH1 mRNA level is overexpressed in osteosarcoma
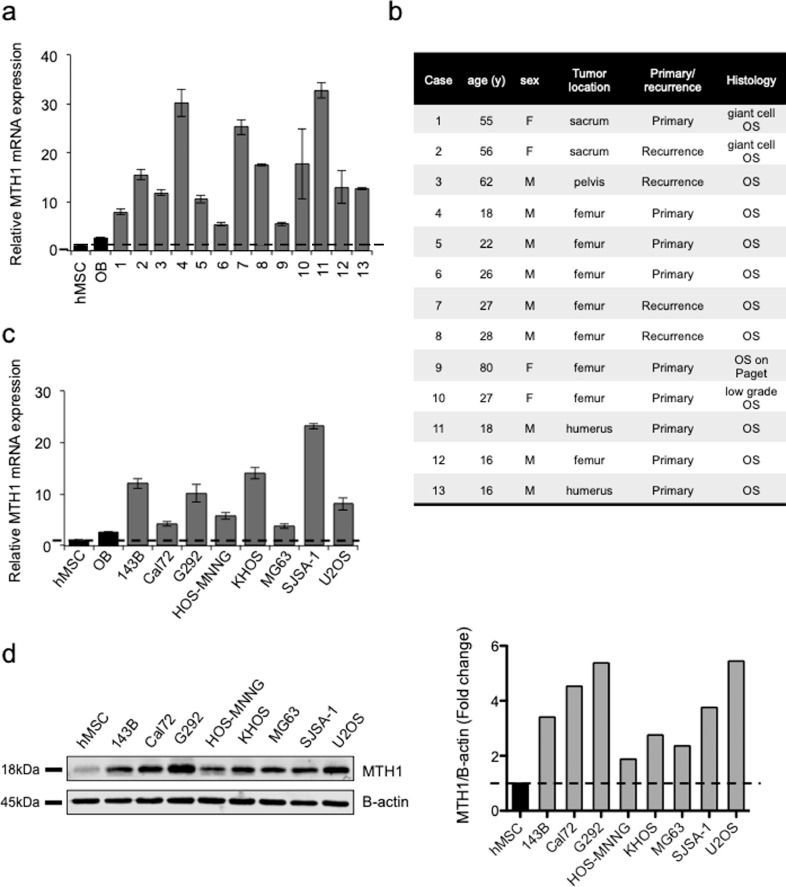
We first evaluated MTH1 mRNA expression level of in 13 human OS patient samples (Fig. 1a and 1b) and 7 OS cell lines (Fig. 1c and d). Compared to normal mesenchymal stem cells (Msc) or osteoblasts derived from hMSC (OB) from healthy donors, MTH1 was overexpressed in all patient samples at mRNA level, and in all OS cell lines tested at mRNA and protein levels.
Fig. 1.
MTH1 is overexpressed in osteosarcoma. (a) Expression of MTH1 was evaluated by RT-qPCR in 13 human osteosarcoma biopsies and compared to human mesenchymal stem cells (hMSC) or osteoblast derived from differentiation of MSCs (OB) from healthy donors. (b) Clinical features of osteosarcoma patients. (c) Expression of MTH1 in human osteosarcoma cell lines was also evaluated by RT-qPCR and compared to hMSC and OB from healthy donors. Errors bars show SD. (d) Expression of MTH1 in human osteosarcoma cell lines was also evaluated by Western blot and compared to hMSC from healthy donors and analysed using Empiria Studio software (Licor).
3.2. TH588 and TH1579 reduce osteosarcoma cell proliferation
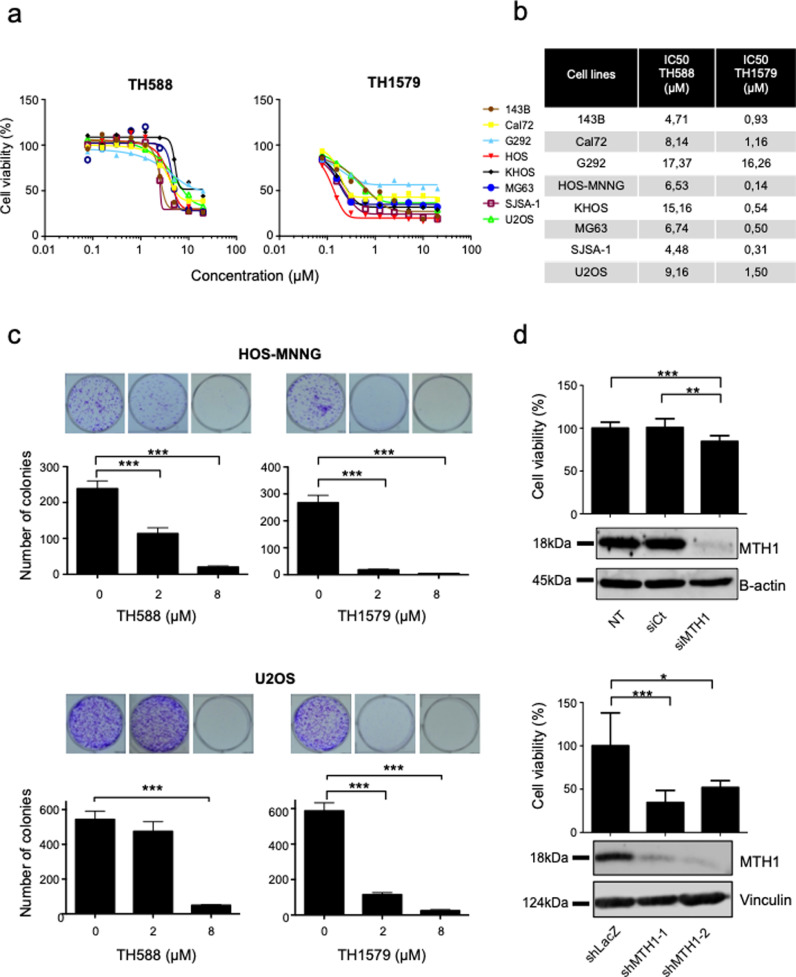
The effects of TH588 and TH1579 were evaluated on cell growth, in 8 OS cell lines (Fig. 2a). In all tested cell lines, TH588 and TH1579 inhibited cell growth in a dose dependent manner, with IC50 values ranging from 4.48 µmol/L to 17.37 µmol/L for TH588, and from 0.31 µmol/L to 16.26 µmol/L for TH1579 (Fig. 2b).
Fig. 2.
MTH1 inhibition decreases cell proliferation in human osteosarcoma cell lines. (a) Human osteosarcoma cell lines (143B, Cal72, G292, HOS-MNNG, KHOS, MG63, SJSA-1 and U2OS) were cultured for 72 h with TH588 or TH1579 at the indicated concentrations, and cell growth was determined by a crystal violet assay. The expression of an untreated population was equalled to 100%. (b) IC50 for TH588 and TH1579 in tumour cell lines was calculated. (c) HOS-MNNG and U2OS osteosarcoma cells were treated with 2 or 8 µM of the indicated drugs for 48 h then plated at clonal density for colony counts. (d) HOS-MNNG cells were transfected with siRNA (30 nM) or transduced with different shRNA targeting MTH1 (MOI 20). Cell viability was compared to controls after crystal violet staining (NT: non transfected, siCt: control siRNA, shLacZ: control shRNA). Error bars show SD Statistical significativity was assessed by Kruskal–Wallis test. * p < 0.05; ** p < 0.01, ***p < 0.001.
Colony formation assays was performed after 48 h of TH588 or TH1579 treatment and showed that both drugs significantly reduced tumour cells clonogenicity in HOS-MNNG and U2OS cell lines (Fig. 2c). Indeed, number of colonies was reduced by 53.4% in HOS-MNNG cells treated with 2 µmol/L of TH588 and by 91.5% when treated with 8 µmol/L of TH588 (p < 0.001). Likewise, number of U2OS colonies was respectively reduced by 13% and 91% (p < 0.001) when treated with 2 µmol/L and 8 µmol/L of TH588, respectively. For TH1579, number of colonies in both cell lines was reduced by over 90% for each concentration (p < 0.001). Similar results were observed 143B, Cal72, G292, KHOS, MG63 and SJSA-1 cell lines (Supplementary Fig. S1).
In order to evaluate whether these results were linked to specific MTH1 inhibition, we transfected or transduced HOS-MNNG cells with a mix of two specific siRNA or two different shRNA targeting MTH1. The decrease of MTH1 expression was validated by qPCR (Supplementary Fig.S2) and Western blot (Fig. 2d). Specific MTH1 inhibition reduced cell growth by 16% compared to control siRNA (p < 0.01), and by 66% (p < 0.001) and 48% (p < 0.01) respectively, for MTH1 shRNA-1 and 2 treatment (Fig 2d).
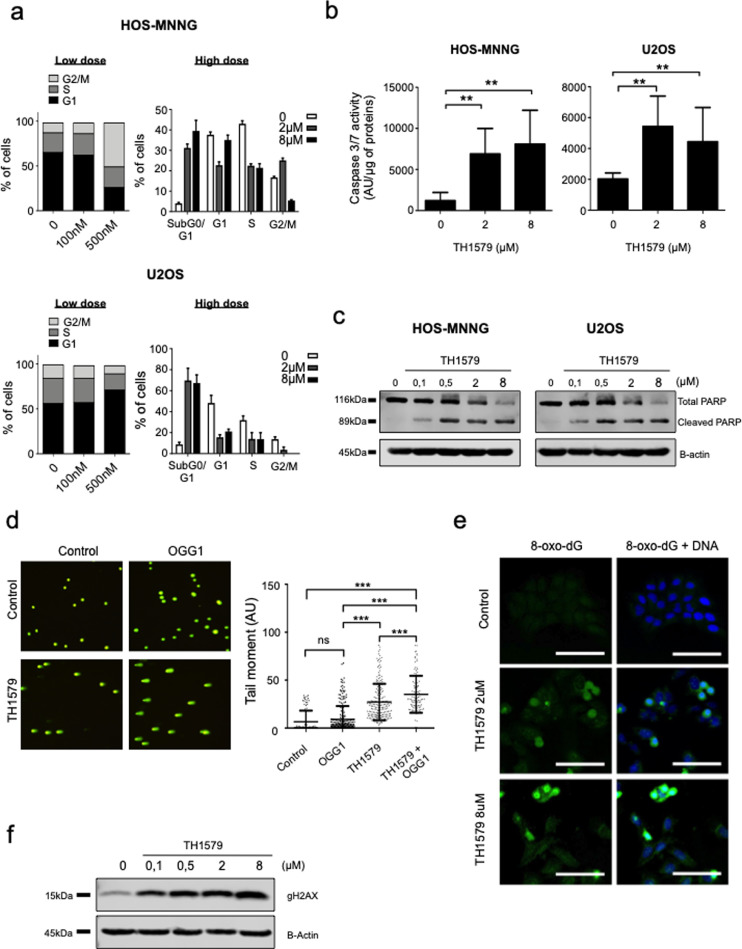
3.3. TH1579 induces apoptosis by increasing DNA strand breaks
Since the TH1579 compound being the most potent drug in this study, we focus the rest of the study on TH1579. Using this drug, we investigated the effect of TH1579 on cell cycle using propidium iodide staining after 48 h of treatment (Fig. 3a). At 500 nM, TH1579 induces G2/M arrest in HOS-MNNG cells as shown by 49% of cells in G2/M phase compared to 11% of cells in control condition. Moreover, in both HOS-MNNG and U2OS cells, TH1579 increased cells death (subG0/G1 fraction) in a dose-dependent manner compared to control. For HOS-MNNG cells, 4% of cells were in the subG0/G1 fraction in the control condition, while 31% and 39% of cells for the 2 µM and 8 µM TH1579 respectively. For U2OS cell line, 8%, 69% and 67% of cells were in the subG0/G1 fraction for control, 2 µM and 8 µM of TH1579 conditions, respectively. Indeed, TH1579 induced apoptosis as shown by a significant increase of caspase 3/7 activity compared to non-treated control (e.g. by 2-fold after 8 µM TH1579 treatment vs control in U2OS cells, p < 0.01; by 6-fold after 8 µM TH1579 treatment vs control in HOS-MNNG cells, p < 0.01; Fig. 3b). Same results were obtained for TH588 (Supplementary Fig. S3a and S3b). Induction of apoptosis was also confirmed by an increase in PARP cleavage in treated conditions compared to the control (Fig. 3c).
Fig. 3.
MTH1 inhibition disrupts cell cycle and induces apoptosis in osteosarcoma cells. (a) HOS-MNNG and U2OS cells were treated with 100 nM or 500 nM (low dose) and 2 or 8 µM (high dose) TH1579 for 48 h, and the proportion of cells in subG1, G0–G1, S, G2–M was determined by propidium iodide staining and flow cytometry. (b-c) Tumour cells were treated with TH1579 at the indicated concentrations and apoptosis was evaluated by a caspase 3/7 assay and cleaved poly (ADP-ribose) polymerase (PARP) expression using western blotting. (d) U2OS cells were treated with 8 µM TH1579 for 24 h, and modified comet assay was performed to evaluate DNA damage (+/- OGG1). The tail moment was calculated using FiJi and the Open comet plugin. (e) U2OS cells were treated with TH1579 at indicated concentrations for 24 h and 8-oxo-dG incorporation into DNA was evaluated by fluorescence using Alexa488-conjugated Avidin (scale bar 100 μm). (f) gH2AX expression was evaluated by western blot in U2OS cells, after 48 h of treatment with TH1579 at the indicated doses. Error bars show SD. Statistical significativity was assessed by Kruskal–Wallis or Anova. ** p < 0.01, ***p < 0.001.
Since MTH1 prevents oxonucleotides integration into DNA, we measured the DNA damage induced by TH1579 +/- OGG1 using comet assay (Fig. 3d). OGG1 is a glycosylase that excises the 8-oxo-dG nucleotide from the double helix, inducing DNA breaks and initiating the DNA repair process. In U2OS cells, combination TH1579+OGG1 treatment increased the tail moment compared to controls (Fig. 3d). The tail moment was increased by 3,93-fold in TH1579+OGG1 compared with untreated control condition (p < 0.001), by 3.88-fold compared with OGG1 condition (p < 0.001), and by 1.3-fold compared with TH1579 alone (p < 0.001), indicating that TH1579 induces DNA strand breaks probably by enhancing 8-oxo-dG integration into DNA. We confirmed the incorporation of 8-oxo-dG into DNA after treatment with TH1579 compared with control in OS cells as shown by an accumulation of 8-oxo-dG in the nucleus (counterstained with DAPI) observed by immunofluorescence (Fig. 3e).
Consequently, we next evaluated the DNA damage induced by TH1579. We analyzed the expression of phosphorylation on serine 319 of H2AX histone (γH2AX) known to be a marker of DNA double strand breaks [33,34]. TH1579 increased γH2AX expression in U2OS cells (Fig. 3f). However, we failed to show a significant impact of the treatment on γH2AX expression, using immunofluorescence, as γH2AX expression was very heterogeneous: some cells displayed a high number of foci expression while other showed a low number of foci (Supplementary Fig.S4).
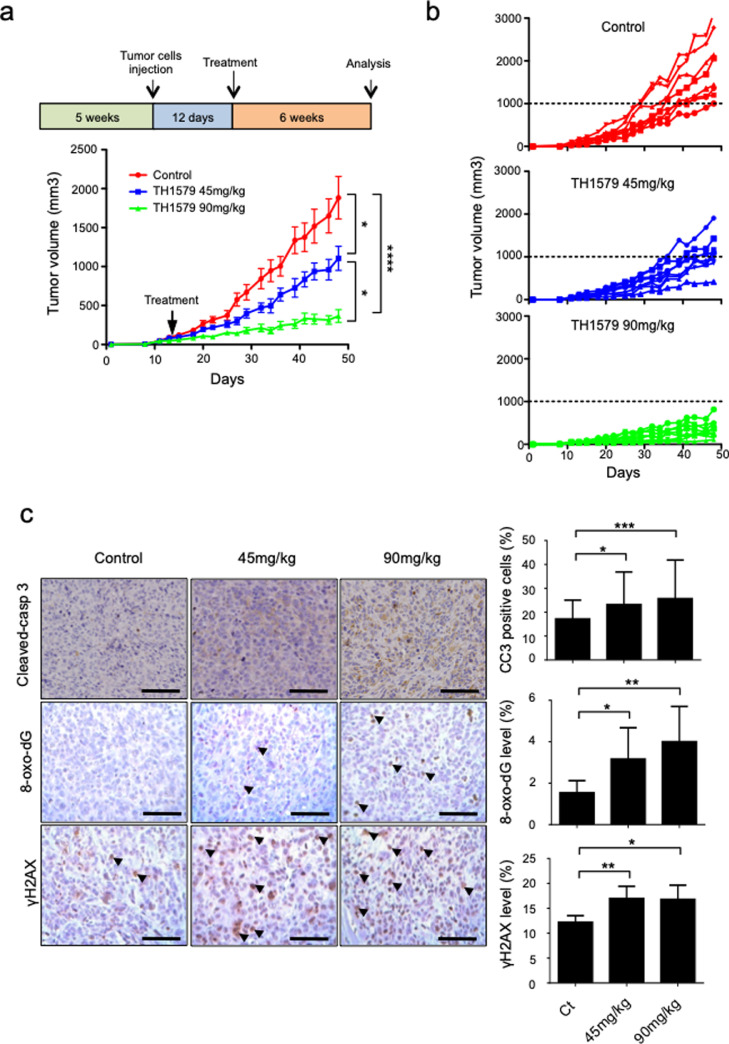
3.4. TH1579 delays tumour growth and increases DNA damages in HOS-MNNG xenograft model
Therapeutic potential of TH1579 was next evaluated in vivo in an OS xenograft model. Mice bearing HOS-MNNG tumours, which is an OS xenograft model reproducing the human pathology associated with development of pulmonary metastases, were randomly separated in three groups and orally treated 3 times per week with vehicle, 45 mg/kg of TH1579 or 90 mg/kg of TH1579 for 48 days. No significant change of body weight was observed at these concentrations. TH1579 treatment significantly decreased the tumour volume in both treated groups in a dose dependent-manner, compared to control group (Fig. 4a and b). At day 29, the mean tumour volume was diminished by 40.6% in the 45 mg/kg group (398 mm3± 57,2 mm3, p = 0,23), and 73.2% in the 90 mg/kg group (179.46 mm3 ± 39,4 mm3, p < 0.001), compared to control (670.3 mm3 ± 99 mm3). At day 48, the mean tumour volumes were reduced by 41.3% (1105.3 mm3 ± 153,7 mm3, p = 0,22) and 80.6% (365.96 mm3 ± 81,6 mm3, p < 0,001) in the 45 mg/kg and the 90 mg/kg groups respectively, compared to control (1883.09 mm3 +/- 272,3 mm3; Fig. 4A). At the individual level, the incidence of mice with a tumour volume of 1000 mm3 or greater was significantly reduced by day 48 in both TH1579 90 mg/kg-treated mice (0/8) and TH1579 45 mg/kg-treated mice (4/8) compared with control mice (8/8; Fig. 4b).
Fig. 4.
TH1579 significantly delays tumour growth in a HOS-MNNG xenograft model. Mice were orally treated with vehicle, 45 mg/kg or 90 mg/kg TH1579 twice a day, 3 days a week until day 48, when the tumour volume reached 100 mm3. The mean tumour volume (a) or the individual tumour volume (b) of the treated mice were compared to the control group +/- SEM (n = 8). (c) Tumours were collected after 48 days, 8-oxo-G, cleaved-caspase-3 and gH2AX were evaluated by immunohistochemical analysis (scale bar 100 μm). Statistical significativity was assessed by Anova (tumour growth) or Kruskal–Wallis. Error bars show SEM. * p < 0.05; ** p < 0.01, ***p < 0.001.
Moreover, the target engagement evaluated by Cellular Thermal Shift Assay (CETSA) is increased in a dose-dependent manner in tumour tissue following TH1579 treatment (Supplementary Fig.S5).
Immunohistochemistry staining carried out on tumour tissues indicated that TH1579 increased tumour cell death as shown by a significant increase of cleaved-caspase 3 expression by 6% and 8.5% in the 45 mg/kg (p < 0.05) and the 90 mg/kg (p < 0.001) groups respectively, compared to the control group (Fig. 4c). These data indicate that TH1579 induces apoptosis in vivo.
Added to this, effects of TH1579 treatment on 8-oxo-dG integration into DNA and DNA damage were evaluated in tumours by immunohistochemistry, using 8-oxo-dG and γH2AX antibodies. TH1579 significantly increased 8-oxo-dG integration and DNA damage in treated groups compared to control. Indeed, in TH1579 45 mg/kg group for example, 8-oxo-dG expression was increased by 2-fold (p < 0.05) and γH2AX expression was increased 1.4-fold (p < 0.01) compared to control. In TH1579 90 mg/kg group 8-oxo-dG and γH2AX were respectively increased by 2.6-fold (p < 0.01) and 1.38-fold (p < 0.05) compared to the control group (Fig. 4c).
3.5. TH1579 reduces pulmonary metastases by inhibiting migration and angiogenesis
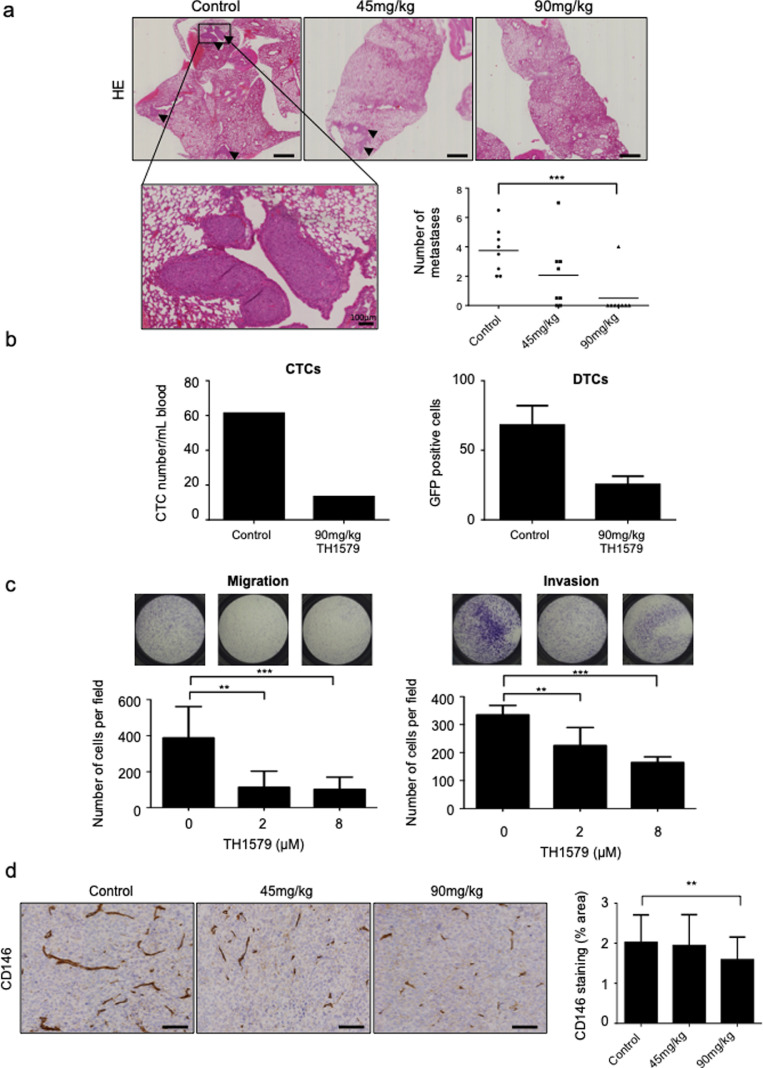
Thereafter, effects of TH1579 on metastases development were evaluated in HOS-MNNG xenograft model. Lung metastases were counted in each experimental group after H&E coloration. The average number of metastases was reduced by 45% (average of 2.06 metastases) in 45 mg/kg TH1579 group, and significantly reduced by 86.7% (average of 0.5 metastasis) in 90 mg/kg group (p < 0.001), compared to control (average of 3.75 metastases; Fig. 5a).
Fig. 5.
TH1579 decreases lung metastases in a HOS-MNNG xenograft model. (a) Lung metastases were counted after hematoxylin-eosin coloration (HE; scale bar 1 mm, except for the zoom: scale bar 100 μm as mentioned in the figure). (b) Circulating (CTC) and disseminated (DTC) tumour cells were counted in two groups of OS mice: non treated or treated with 90 mg/kg of TH1579 for one week. (c) TH1579 effect on invasion and migration was evaluated in vitro, on HOS-MNNG cells, by a migration assay and an invasion assay respectively. Cells were plated in non-coated boyden chambers (migration) or coated with 2 µg of matrigel (invasion) with the indicated TH1579 concentrations, for 6 h (migration) or 24 h (invasion). (d) Neo-angiogenesis was evaluated by CD146 staining, by immunohistochemical (scale bar 100 μm) and quantified using ImageJ. Error bars show s.d. Statistical significativity was assessed by Kruskal–Wallis or Anova. ** p < 0.01, ***p < 0.001.
Moreover, TH1579 also reduced the number of Circulating Tumour Cells (CTC) counted in blood and Disseminated Tumour Cells (DTC) counted in dissociated lungs of mice bearing HOS-MNNG-GFP tumours after one week treatment of 90 mg/kg TH1579 compared to control group, suggesting an effect of TH1579 on metastatic dissemination. Indeed, while the mean tumour volume of the two groups (90 mg/kg TH1579 vs vehicle) was equivalent (89.6 mm3 and 96.1 mm3 for the control and treated conditions respectively; Supplementary Fig. S6), the number was 61 CTC/mL of blood in control group, and 13 CTC/mL in 90 mg/kg TH1579 group. DTC number in lungs was also reduced by 2.68-fold after 90 mg/kg TH1579 (25 GFP-positive tumour cells) treatment compared to control group (68 GFP-positive tumour cells; Fig. 5b). One of the mice from the control group was excluded from the analysis since it did not have a visible GFP+ tumour.
In order to verify that TH1579 could affect processes involved in metastases, migration and invasion assays were performed on HOS-MNNG cells treated with TH1579. In the migration assay using transwells, number of migrating cells was significantly reduced by 70% (p < 0.01) and 73% (p < 0.001) after treatment with 2 µM TH1579 (113 cells) and with 8 µM TH1579 (102 cells) respectively, compared to control (388 cells). In the invasion assay, number of cells per field was significantly reduced by 32.8% (p < 0.01) after treatment with 2 µM TH1579, and 69.44% (p < 0.001) after treatment with 8 µM TH1579. TH1579 significantly reduced both migration and invasion capabilities of HOS-MNNG cells (Fig. 5c).
According to migration and invasion assays, TH1579 significantly reduced angiogenesis, as shown by a reduced CD146 expression in tumours treated with 90 mg/kg TH1579 (1.27 times lower) compared to control group (p < 0.01; Fig. 5d). No significant change was observed between 45 mg/kg TH1579 and control groups (Fig. 5d).
3.6. Effect of TH1579 on tumour-associated bone lesions
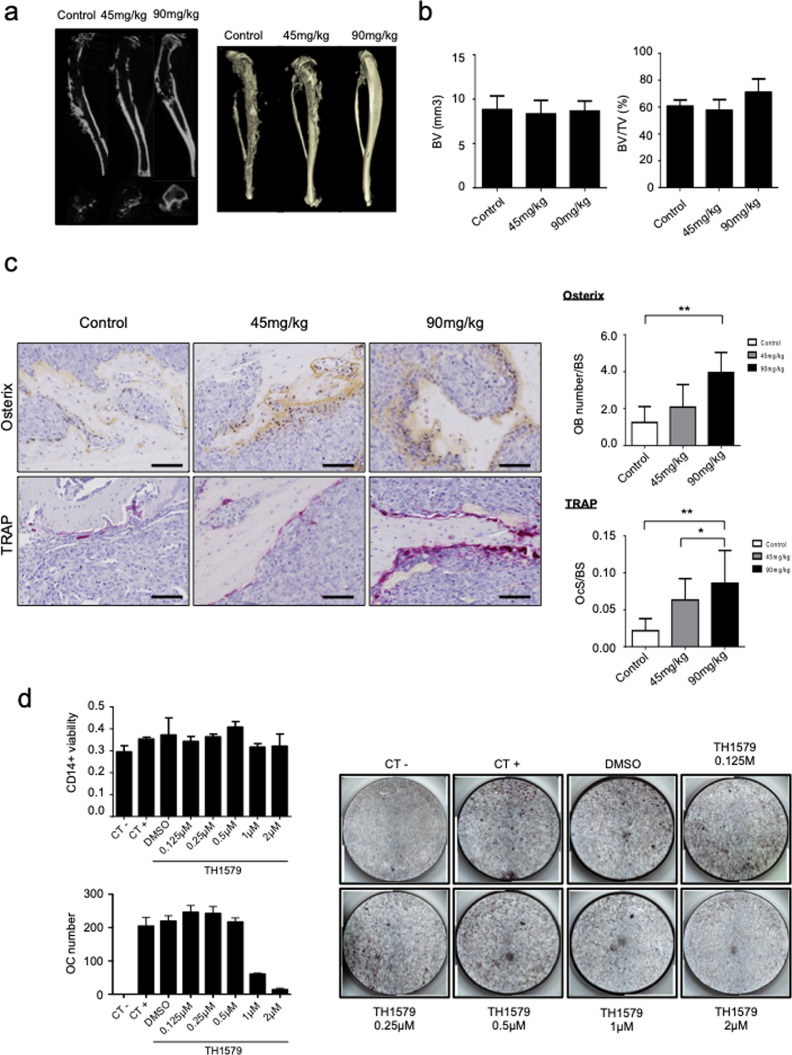
In order to evaluate the effect of TH1579 on tumour associated-osteolysis, cortical and trabecular bone parameters of tibia bearing tumours were examined (Fig. 6a and b). BV was identical between TH1579 treated (45 mg/kg and 90 mg/kg) and control groups. BV/TV was slightly but not significantly increased by 17% in 90 mg/kg TH1579 group compared to control group (p value = 0.0508).
Fig. 6.
TH1579 effect on bone lesions. (a) Representative microCt image of tibias bearing the tumour in the 3 groups. (b) Quantification of the Bone Volume (BV) and BV/TV (Total Volume) parameters. (c) Quantification of the IHC staining of TRAP and Osterix using ImageJ (scale bar 100 μm) and represented as number of osteoblasts on bone surface (OB number/BS) and number of osteoclasts on bone surface (OCs/BS). (d) CD14+ cells viability was assessed using crystal violet after TH1579 treatment as indicated concentration. Osteoclast differentiation was assessed after treatment of CD14+ cells with 25 ng/mL human macrophage colony stimulating factor (hM-CSF) and 100 ng/mL of hRANKL +/- TH1579 at the indicated concentrations. Osteoclasts number was evaluated after TRAP staining at day 11. Negative control (CT -) represents CD14+ cells treated with 25 ng/mL hM-CSF without human RANKL. Positive control (CT+) represents cells treated with 100 ng/mL of hRANKL. DMSO represents CT+ treated with DMSO. Error bars show s.d. Statistical significativity was assessed by Kruskal–Wallis. * p < 0.05; ** p < 0.01.
Immunohistochemical staining of osterix showed an increase of osteoblasts (OB) number over bone surface (BS) in both treated groups (2.08 and 3.95 OB/BS for the 45 mg/kg and 90 mg/kg conditions respectively), compared to control group (1.25 OB/BS). Likewise, TRAP staining also showed a significant increase of osteoclast number over bone surface in treated groups compared to control group (Fig. 6c). Ocs/BS was 2.9 times more important after treatment with 45 mg/kg TH1579 and 3.9 more important after treatment with 90 mg/kg TH1579 compared to control. Effects of TH1579 on bone cells were also evaluated in vitro on CD14+ osteoclast precursors as well as hMSC osteoblast precursors. TH1579 reduced osteoclasts differentiation, without any significant effect on their precursor's viability (Fig. 6d). However, TH1579 reduced both hMSC viability and mineralization (Supplementary Fig.S7).
4. Discussion
Osteosarcoma is a paediatric cancer with poor prognosis and high recurrence. Since 30 years, treatments are limited to chemotherapy associated with resistance development. Finding new therapeutic strategies for OS is urgently needed. In this context, the enzyme MTH1 is an attractive target since MTH1 is “druggable” by chemical compounds (TH588, TH1579, S-Crizotinib). MTH1 is described to support malignant transformation, proliferation and oncogenic ability through the degradation of oxidized guanine nucleotides from oxidative stress [21,23,35,36]. OS exhibits a high level of ROS suggesting a potential constitutive activation of MTH1 in tumour cells. Indeed, we observed an overexpression of MTH1 in OS biopsies as well as in a large panel of OS cell lines with various genetic alterations compared to hMSC.
Despite controversial opinions in literature about the dependence of MTH1 by cancer cells to survive and the efficiency of therapeutic drugs under development [25], [26], [27], [28], we aimed to evaluate the therapeutic efficacy of the MTH1 inhibitors, TH588 and TH1579 in OS model. In the present study, we clearly observed anti-tumour activity of both TH588 and TH1579 in OS cell lines with higher activity of TH1579 as shown by significant decrease of cell viability associated with increase of apoptosis as already described in the literature [21,22,37,38]. Indeed, TH1579 is more potent to inhibit MTH1 activity than TH588 [22], and is currently in phase 1 clinical trial in various advanced solid malignancies otherwise known as Karonudib (NCT03036228). TH1579 exhibits high anti-tumour activity through a dual mechanism of action, by inducing toxic 8-oxodG lesions into the DNA, that can be reversed by expression of the bacterial MutT enzyme [22], and also through arrest of cells in mitosis by inhibition of microtubule polymerisation. The detailed mechanism of action is complex as the MTH1 protein promotes tubulin polymerisation and also directly binds tubulin [39]. Furthermore, TH1579, but not other potent MTH1 inhibitors, breaks the tubulin-MTH1 interaction and arrest cells in mitosis. To complicate matters even further the TH1579 compound directly interfere with tubulin [39] which altogether make up the mechanism of action of Karonudib. Here, we want to evaluate the effect of TH1579 in OS and detailed understanding of the mechanism of action falls outside the scope of this report.
However, when MTH1 was specifically knock-down using siRNA or shRNA approaches, a reduction of cell viability of tumour cells was shown, but less dramatically than the effects observed with TH588 and TH1579, suggesting that MTH1 is probably not the only target of these drugs in OS and deserved to be deeply studied. Several studies reported that specific MTH1 inhibition does not affect tumour cell viability and/or apoptosis such as reported in non-small cell lung cancer [25,26,28,40]. While the cytotoxic effects of MTH1 inhibition in cancer are controversial in literature, TH588 has been shown to be cytotoxic in several studies [41,42]. We also found that TH588 and TH1579 induce apoptosis by increasing DNA damage as shown by comet assay and induction of H2AX phosphorylation in OS cells. These data are consistent with early cellular response to DNA strand breaks as already reported in the literature [33,43]. As reported in two studies TH588 induces a mitotic arrest with cells accumulation in G2/M phases in Hela cells or in colorectal cancer [25,41], we also observed mitotic arrest (G2/M blockage) in HOS-MNNG cell line after 48 h of treatment with TH1579 (500 nM). But we cannot exclude that the observed apoptosis associated with DNA damage could also be MTH1-dependent.
Our study further demonstrates that TH1579 strongly delayed tumour growth in vivo associated with an increase of apoptosis and DNA damage as shown by higher expression level of toxic 8-oxo-G and γH2AX after TH1579 treatment. By using CETSA, we could show that TH1579 binds to MTH1 inside the tumours of the treated mice in a dose dependent manner. The drug seems to be well tolerated with no toxicity observed. Additionally, TH1579 significantly reduced lung metastases in vivo in OS xenograft model associated with reduced-CD146 expression in primary tumour compared to control, suggesting less vascularization in treated-primary tumours. We also have shown that TH1579 decreased the number of Circulating Tumour Cells (CTC) and Disseminated Tumour Cells (DTC) in lung of treated mice bearing a non-advanced OS tumour, suggesting that TH1579 might have a direct effect on invasion and migration of tumour cells [44,45]. We effectively confirmed in vitro that TH1579 reduced invasion and migration of OS cells in line with the literature showing that MTH1 is required for invasion and migration [24,46]. Indeed, Arczewska et al. demonstrated that MTH1 depletion using siRNA significantly reduced invasion and migration of thyroid cancer cell lines by modulating Matrix Metalloproteinases (MMPs) activity and expression [46]. MTH1 inhibitor (S)-crizotinib was also described to reduce migratory ability of OS cells in vitro [24].
Furthermore, associated-bone lesions characterize OS development, and no significant change of bone parameters was observed after TH1579 treatment even if the bone integrity of the tibia is better than in the control group. These results can be explained by the smaller tumours in the treated groups compared to the control.
While targeting MTH1 is a contentious question in cancer research, our study demonstrates an anti-tumour activity of TH1579 both in vitro and in vivo in OS model associated with induction of apoptosis and DNA damage. All these data are the proof of principle that TH1579 could be considered as alternative therapy in metastatic osteosarcoma patient.
Declaration of Competing Interest
Helleday, T: A patent has been filed with TH588 and TH1579 where T.H., is listed as inventor. The Intellectual Property Right is owned by the non-profit Thomas Helleday Foundation for Medical Research (THF). T.H. and U.W.B are board members of the THF. U.W.B is chairman of Oxcia AB. THF is sponsor for on-going clinical trial with TH1579.
Acknowledgments
Acknowledgements
Drs Martin Scobie and Tobias Koolmeister are acknowledged for the identification and synthesis of TH588 and TH1579. The authors would like to thank the Therapeutic Experimental Unit (Nantes, France) for their technical assistance.
Funding sources
This study was supported by La Ligue Contre le Cancer (Ligue 44, 29 et 22 (F.L.)), la SFCE (Société Francaise des Cancers de l'Enfant; F.L.) et la fédération Enfants Cancers Santé (F.L.) which the authors gratefully acknowledge. The funders did not have any role in study design, data analysis, interpretation or writing the report.
Author's contributions
Conception and designs: Warpman Berglund, U; Heymann, D; Helleday, T; Ory, B; Lamoureux F
Acquisition of data: Moukengue, B; Brown HK, Charrier, C; Battaglia, S; Baud'huin, M; Pham T, Pateras IS; Gorgoulis VG (cetsa and tumour IHC); Lamoureux F
Analysis and interpretations of data: Moukengue, B; Brown HK, Charrier, C; Battaglia, S; Baud'huin, M; Quillard, T; Warpman Berglund, U; Heymann, D, Helleday, T; Ory, B; Lamoureux F
Writing, review, and/or revision of the manuscript: Moukengue, B; Brown HK, Quillard, T; Warpman Berglund, U; Heymann, D, Helleday, T; Ory, B; Lamoureux F
Study supervision: Warpman Berglund, U; Heymann, D; Helleday, T; Ory, B; Lamoureux F
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102704.
Appendix. Supplementary materials
References
- 1.Lamoureux F., Trichet V., Chipoy C., Blanchard F., Gouin F., Redini F. Recent advances in the management of osteosarcoma and forthcoming therapeutic strategies. Expert Rev Anticancer Ther. 2007;7(2):169–181. doi: 10.1586/14737140.7.2.169. [DOI] [PubMed] [Google Scholar]
- 2.Rosen G., Murphy M.L., Huvos A.G., Gutierrez M., Marcove R.C. Chemotherapy, en bloc resection, and prosthetic bone replacement in the treatment of osteogenic sarcoma. Cancer. 1976;37(1):1–11. doi: 10.1002/1097-0142(197601)37:1<1::aid-cncr2820370102>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 3.Lee J.A. Osteosarcoma in Korean children and adolescents. Korean J Pediatr. 2015;58(4):123–128. doi: 10.3345/kjp.2015.58.4.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He J.P., Hao Y., Wang X.L., Yang X.J., Shao J.F., Guo F.J. Review of the molecular pathogenesis of osteosarcoma. Asian Pac J Cancer Prev. 2014;15(15):5967–5976. doi: 10.7314/apjcp.2014.15.15.5967. [DOI] [PubMed] [Google Scholar]
- 5.Brown H.K., Schiavone K., Gouin F., Heymann M.F., Heymann D. Biology of bone sarcomas and new therapeutic developments. Calcif Tissue Int. 2018;102(2):174–195. doi: 10.1007/s00223-017-0372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman L.S., Wintrobe M.M. Nitrogen mustard therapy; use of methyl-bis (beta-chloroethyl) amine hydrochloride and tris (beta-chloroethyl) amine hydrochloride for Hodgkin's disease, lymphosarcoma, leukemia and certain allied and miscellaneous disorders. J Am Med Assoc. 1946;132:126–132. doi: 10.1001/jama.1946.02870380008004. [DOI] [PubMed] [Google Scholar]
- 7.Farber S. Some observations on the effect of folic acid antagonists on acute leukemia and other forms of incurable cancer. Blood. 1949;4(2):160–167. [PubMed] [Google Scholar]
- 8.Cheung-Ong K., Giaever G., Nislow C. DNA-damaging agents in cancer chemotherapy: serendipity and chemical biology. Chem Biol. 2013;20(5):648–659. doi: 10.1016/j.chembiol.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Park H.J., Bae J.S., Kim K.M., Moon Y.J., Park S.H., Ha S.H. The PARP inhibitor olaparib potentiates the effect of the DNA damaging agent doxorubicin in osteosarcoma. J Exp Clin Cancer Res. 2018;37(1):107. doi: 10.1186/s13046-018-0772-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou Z., Chang H., Li H., Wang S. Induction of reactive oxygen species: an emerging approach for cancer therapy. Apoptosis. 2017;22(11):1321–1335. doi: 10.1007/s10495-017-1424-9. [DOI] [PubMed] [Google Scholar]
- 11.Nakabeppu Y., Oka S., Sheng Z., Tsuchimoto D., Sakumi K. Programmed cell death triggered by nucleotide pool damage and its prevention by MutT homolog-1 (MTH1) with oxidized purine nucleoside triphosphatase. Mutat Res. 2010;703(1):51–58. doi: 10.1016/j.mrgentox.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Nakabeppu Y., Ohta E., Abolhassani N. MTH1 as a nucleotide pool sanitizing enzyme: friend or foe? Free Radic Biol Med. 2017;107:151–158. doi: 10.1016/j.freeradbiomed.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Fujikawa K., Kamiya H., Yakushiji H., Nakabeppu Y., Kasai H. Human MTH1 protein hydrolyzes the oxidized ribonucleotide, 2-hydroxy-ATP. Nucleic Acids Res. 2001;29(2):449–454. doi: 10.1093/nar/29.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pizzino G., Irrera N., Cucinotta M., Pallio G., Mannino F., Arcoraci V. Oxidative stress: harms and benefits for human health. Oxid Med Cell Longev. 2017;2017 doi: 10.1155/2017/8416763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liou G.Y., Storz P. Reactive oxygen species in cancer. Free Radic Res. 2010;44(5):479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moloney J.N., Cotter T.G. ROS signalling in the biology of cancer. Semin Cell Dev Biol. 2018;80:50–64. doi: 10.1016/j.semcdb.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 17.Storz P. Reactive oxygen species in tumor progression. Front Biosci. 2005;10:1881–1896. doi: 10.2741/1667. [DOI] [PubMed] [Google Scholar]
- 18.Coskun E., Jaruga P., Jemth A.S., Loseva O., Scanlan L.D., Tona A. Addiction to MTH1 protein results in intense expression in human breast cancer tissue as measured by liquid chromatography-isotope-dilution tandem mass spectrometry. DNA Repair. 2015;33:101–110. doi: 10.1016/j.dnarep.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Obtulowicz T., Swoboda M., Speina E., Gackowski D., Rozalski R., Siomek A. Oxidative stress and 8-oxoguanine repair are enhanced in colon adenoma and carcinoma patients. Mutagenesis. 2010;25(5):463–471. doi: 10.1093/mutage/geq028. [DOI] [PubMed] [Google Scholar]
- 20.Tsuzuki T., Egashira A., Igarashi H., Iwakuma T., Nakatsuru Y., Tominaga Y. Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase. Proc Natl Acad Sci USA. 2001;98(20):11456–11461. doi: 10.1073/pnas.191086798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gad H., Koolmeister T., Jemth A.S., Eshtad S., Jacques S.A., Strom C.E. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature. 2014;508(7495):215–221. doi: 10.1038/nature13181. [DOI] [PubMed] [Google Scholar]
- 22.Warpman Berglund U., Sanjiv K., Gad H., Kalderen C., Koolmeister T., Pham T. Validation and development of MTH1 inhibitors for treatment of cancer. Ann Oncol. 2016;27(12):2275–2283. doi: 10.1093/annonc/mdw429. [DOI] [PubMed] [Google Scholar]
- 23.Huber K.V., Salah E., Radic B., Gridling M., Elkins J.M., Stukalov A. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature. 2014;508(7495):222–227. doi: 10.1038/nature13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qing X., Shao Z., Lv X., Pu F., Gao F., Liu L. Anticancer effect of (S)-crizotinib on osteosarcoma cells by targeting MTH1 and activating reactive oxygen species. Anticancer Drugs. 2018;29(4):341–352. doi: 10.1097/CAD.0000000000000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawamura T., Kawatani M., Muroi M., Kondoh Y., Futamura Y., Aono H. Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival. Sci Rep. 2016;6:26521. doi: 10.1038/srep26521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kettle J.G., Alwan H., Bista M., Breed J., Davies N.L., Eckersley K. Potent and selective inhibitors of MTH1 probe its role in cancer cell survival. J Med Chem. 2016;59(6):2346–2361. doi: 10.1021/acs.jmedchem.5b01760. [DOI] [PubMed] [Google Scholar]
- 27.Samaranayake G.J., Huynh M., Rai P. MTH1 as a chemotherapeutic target: the elephant in the room. Cancers. 2017;9(5) doi: 10.3390/cancers9050047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrocchi A., Leo E., Reyna N.J., Hamilton M.M., Shi X., Parker C.A. Identification of potent and selective MTH1 inhibitors. Bioorg Med Chem Lett. 2016;26(6):1503–1507. doi: 10.1016/j.bmcl.2016.02.026. [DOI] [PubMed] [Google Scholar]
- 29.Baud'huin M., Lamoureux F., Jacques C., Rodriguez Calleja L., Quillard T., Charrier C. Inhibition of BET proteins and epigenetic signaling as a potential treatment for osteoporosis. Bone. 2017;94:10–21. doi: 10.1016/j.bone.2016.09.020. [DOI] [PubMed] [Google Scholar]
- 30.Rai P., Onder T.T., Young J.J., McFaline J.L., Pang B., Dedon P.C. Continuous elimination of oxidized nucleotides is necessary to prevent rapid onset of cellular senescence. Proc Natl Acad Sci USA. 2009;106(1):169–174. doi: 10.1073/pnas.0809834106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Struthers L., Patel R., Clark J., Thomas S. Direct detection of 8-oxodeoxyguanosine and 8-oxoguanine by avidin and its analogues. Anal Biochem. 1998;255(1):20–31. doi: 10.1006/abio.1997.2354. [DOI] [PubMed] [Google Scholar]
- 32.Martinez Molina D., Jafari R., Ignatushchenko M., Seki T., Larsson E.A., Dan C. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341(6141):84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 33.Burma S., Chen B.P., Murphy M., Kurimasa A., Chen D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276(45):42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 34.Georgoulis A., Vorgias C.E., Chrousos G.P., Rogakou E.P. Genome instability and gammaH2AX. Int J Mol Sci. 2017;18(9) doi: 10.3390/ijms18091979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rai P., Young J.J., Burton D.G., Giribaldi M.G., Onder T.T., Weinberg R.A. Enhanced elimination of oxidized guanine nucleotides inhibits oncogenic RAS-induced DNA damage and premature senescence. Oncogene. 2011;30(12):1489–1496. doi: 10.1038/onc.2010.520. [DOI] [PubMed] [Google Scholar]
- 36.Helleday T. Cancer phenotypic lethality, exemplified by the non-essential MTH1 enzyme being required for cancer survival. Ann Oncol. 2014;25(7):1253–1255. doi: 10.1093/annonc/mdu158. [DOI] [PubMed] [Google Scholar]
- 37.Pudelko L., Rouhi P., Sanjiv K., Gad H., Kalderen C., Hoglund A. Glioblastoma and glioblastoma stem cells are dependent on functional MTH1. Oncotarget. 2017;8(49):84671–84684. doi: 10.18632/oncotarget.19404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou W., Ma L., Yang J., Qiao H., Li L., Guo Q. Potent and specific MTH1 inhibitors targeting gastric cancer. Cell Death Dis. 2019;10(6):434. doi: 10.1038/s41419-019-1665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gad H., Mortusewicz O., Rudd S.G., Stolz A., Amaral N., Brautigam L. MTH1 promotes mitotic progression to avoid oxidative DNA damage in cancer cells. bioRxiv. 2019 [Google Scholar]
- 40.Abbas H.H.K., Alhamoudi K.M.H., Evans M.D., Jones G.D.D., Foster S.S. MTH1 deficiency selectively increases non-cytotoxic oxidative DNA damage in lung cancer cells: more bad news than good? BMC Cancer. 2018;18(1):423. doi: 10.1186/s12885-018-4332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Waals L.M., Laoukili J., Jongen J.M.J., Raats D.A., Borel Rinkes I.H.M., Kranenburg O. Differential anti-tumour effects of MTH1 inhibitors in patient-derived 3D colorectal cancer cultures. Sci Rep. 2019;9(1):819. doi: 10.1038/s41598-018-37316-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pompsch M., Vogel J., Classen F., Kranz P., Iliakis G., Riffkin H. The presumed MTH1-inhibitor TH588 sensitizes colorectal carcinoma cells to ionizing radiation in hypoxia. BMC Cancer. 2018;18(1):1190. doi: 10.1186/s12885-018-5095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Redon C., Pilch D., Rogakou E., Sedelnikova O., Newrock K., Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12(2):162–169. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 44.Chalopin A., Tellez-Gabriel M., Brown H.K., Vallette F., Heymann M.F., Gouin F. Isolation of circulating tumor cells in a preclinical model of osteosarcoma: effect of chemotherapy. J Bone Oncol. 2018;12:83–90. doi: 10.1016/j.jbo.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M., Lu Y., Long Z., Li M., Kong J., Chen G. Prognostic and clinicopathological significance of circulating tumor cells in osteosarcoma. J Bone Oncol. 2019;16 doi: 10.1016/j.jbo.2019.100236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arczewska K.D., Stachurska A., Wojewodzka M., Karpinska K., Kruszewski M., Nilsen H. hMTH1 is required for maintaining migration and invasion potential of human thyroid cancer cells. DNA Repair. 2018;69:53–62. doi: 10.1016/j.dnarep.2018.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.