Summary
The phenomenon of protein misfolding and aggregation is associated with a wide range of neurodegenerative conditions that cause progressive loss of function in specific regions of the human brain. To understand the causes of the selective cell and tissue vulnerability to the formation of these deposits, we analyzed the ability of different cell and tissue types to respond, in the absence of disease, to the presence of high levels of aggregation-prone proteins. By performing a transcriptional analysis, we found that the protein homeostasis system that regulates protein aggregation is weaker in neurons than in other cell types and in brain tissues than in other body tissues. These results suggest that the intrinsic level of regulation of protein aggregation in the healthy state is correlated with the selective vulnerability of cells and tissues to protein misfolding diseases.
Subject Areas: Molecular Physiology, Neuroscience, Transcriptomics
Graphical Abstract
Highlights
-
•
A branch of the protein homeostasis system regulates protein aggregation
-
•
This system is weaker in brain tissues than in other body tissues
-
•
This system is weaker in Braak regions than in other brain regions
-
•
This system is weaker in neurons than in other brain cell types
Molecular Physiology; Neuroscience; Transcriptomics
Introduction
Neurodegenerative disorders are multifactorial conditions characterized by extensive neuronal dysfunction associated with the misfolding and aggregation of a specific set of proteins that form aberrant deposits, including amyloid plaques and neurofibrillary tangles in Alzheimer's disease and Lewy bodies in Parkinson's disease (De Strooper and Karran, 2016, Eisenberg and Jucker, 2012, Holtzman et al., 2011, Selkoe and Hardy, 2016, Knowles et al., 2014). Increasing evidence also indicates that protein aggregation is a widespread phenomenon, as hundreds of proteins unrelated in sequence or structure have been found to form insoluble assemblies under conditions of stress, aging, or disease (Chapman et al., 2006, Ciryam et al., 2015, Ciryam et al., 2017, David et al., 2010, Gidalevitz et al., 2006, Tartaglia et al., 2007, Walther et al., 2015).
To rationalize these observations, it has recently been reported that a large fraction of the proteome is inherently metastable against aggregation within the cellular environment (Ciryam et al., 2013, Ciryam et al., 2015, Ciryam et al., 2016, Ciryam et al., 2017, Yerbury et al., 2019). Proteins are metastable in their native states when their cellular concentrations exceed their intrinsic solubilities (Tartaglia et al., 2007, Vecchi et al., 2020). Indeed, proteins that co-aggregate in plaques, tangles, and Lewy bodies have been found to be highly metastable against aggregation (Ciryam et al., 2013), and biochemical pathways associated with neurodegenerative diseases, including in particular oxidative phosphorylation, have been reported to be enriched in these proteins, hereafter referred to as metastable proteins, thus revealing a common feature for these otherwise very different neurodegenerative conditions (Ciryam et al., 2013). Moreover, a fraction of these metastable proteins were reported to be specifically transcriptionally downregulated in Alzheimer's disease (Ciryam et al., 2016). This metastable subproteome associated with Alzheimer's disease was found to be associated primarily with specific components of the protein trafficking and clearance mechanisms, in particular the endosomal-lysosomal and the ubiquitin-proteasome systems (Kundra et al., 2017). These results suggest that these quality control systems are crucial for the maintenance of the cellular homeostasis of a set of metastable proteins prone to aggregation in Alzheimer's disease.
In addition, the protein quality control mechanisms, which are collectively known as the protein homeostasis system, have been shown quite generally to become progressively impaired upon aging and in neurodegenerative diseases, resulting in increased accumulation of aggregated species (Balch et al., 2008, Freer et al., 2016, Freer et al., 2019, Fu et al., 2018, Fu et al., 2019, Hipp et al., 2014, Knowles et al., 2014). Hence, it is crucial to maintain a balance between the levels of aggregation-prone proteins and their associated protein homeostasis components in order to preserve overall cellular health. Further studying these effects may therefore help answer the question of why a large number of the diseases associated with protein aggregation involve the central nervous system. Although there have been a number of studies about the vulnerability of specific brain tissues (Bero et al., 2011, Mattson and Magnus, 2006, Miller et al., 2013), it is still not completely clear why the brain itself is more vulnerable to the aggregation phenomenon compared with other body tissues.
The aim of the present study is to address this question by investigating whether healthy brains are more susceptible to the effects of protein misfolding diseases than other organs, by examining at the transcriptional level the balance between the expression of aggregation-prone proteins and the efficacy of their corresponding protein homeostasis control system. To achieve this goal, we determined the expression levels of metastable proteins associated with Alzheimer's disease and their associated protein homeostasis components across nearly 80 different healthy human tissues, including several ones from the brain. Our expectation is that brain tissues, compared with those from other regions of the body, should have a higher level of expression of the set of metastable proteins but a lower expression of the associated homeostasis components. In this view, due to the inherent lower expression levels of the quality control components associated with metastable proteins, certain regions of the brain would not have the ability to mount a robust response to these highly aggregation-prone metastable proteins, suggesting an origin for the selective vulnerability of the brain to protein misfolding diseases.
Results
The Strength of the Protein Homeostasis System that Responds to Protein Aggregation Is Proportional to Level of Gene Expression of a Metastable Subproteome
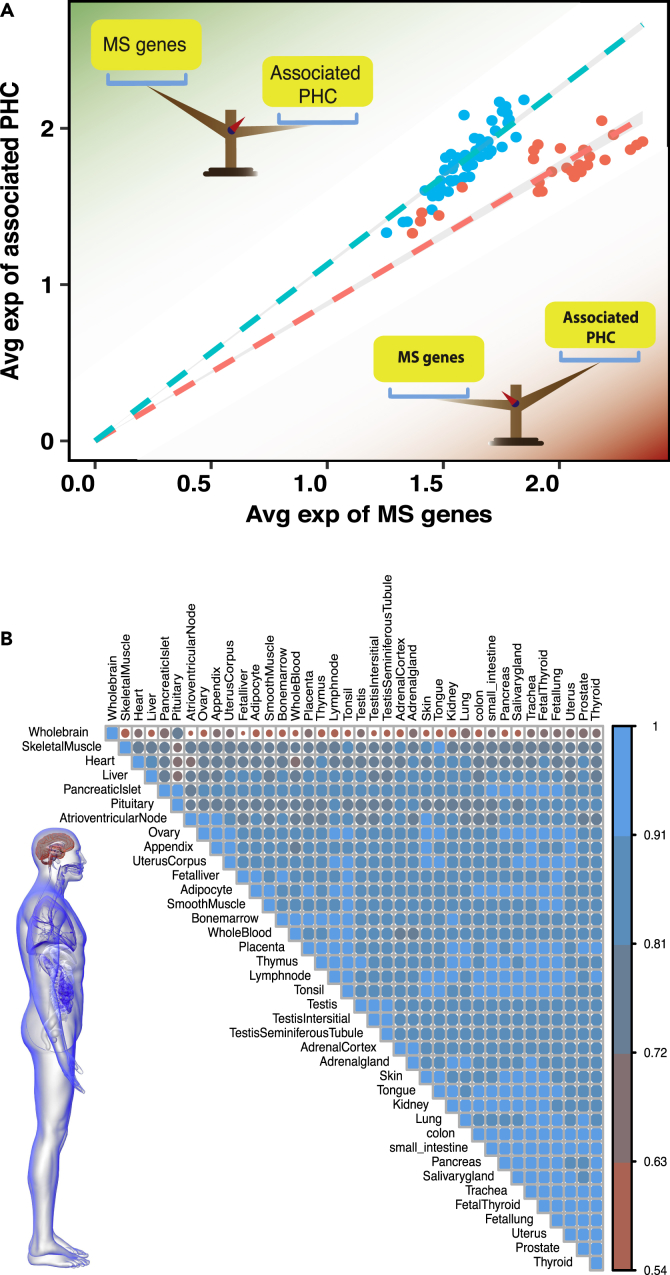
Because aggregation-prone proteins are intrinsically metastable even in the absence of disease, we studied healthy brain tissues to identify their associated protein homeostasis components using an approach based on coexpression network analysis (see Supplemental Information). We used a subset of these metastable proteins specifically associated with Alzheimer's disease (Ciryam et al., 2016), as our aim was to study the vulnerability of different tissues to this disease. In this way, using a weighted coexpression approach (Langfelder and Horvath, 2008), we identified the protein homeostasis components (PHC) associated with this metastable subproteome (MS) (Kundra et al., 2017). We then used here extensive microarray data obtained for 77 different types of healthy human tissues (Su et al., 2004) to study the vulnerability of various tissues based on the relative expression of these genes (Table S1). We carried out this analysis by defining a protection factor, s, as the correlation of the relative expression levels of the MS and its corresponding PHC. When s = 1 the response to protein aggregation in a given tissue matches exactly, at the transcript level, the requirement to prevent aggregation of the metastable proteins, whereas values below 1 indicate a lower, and possibly ineffective, response. We found from this analysis that the brain tissues have an elevated expression of genes corresponding to the MS, but not of their protein homeostasis counterpart, relative to other types of tissues in the body (Figure 1A). Although in body tissues the PHC expression level grows more rapidly than that of the MS (s = 1.33), in brain tissues the opposite is true (s = 0.88). Thus, although the protein homeostasis response in all tissues tends to be generally proportional to the pressure toward protein aggregation, brain tissues tend to behave differently from other tissues (Figure 1B) and have an overall weaker response.
Figure 1.
The Protein Homeostasis System that Responds to Protein Aggregation Is Weaker in Brain Tissues Compared with Other Tissues in the Human Body
(A) Comparison between the average expression of genes corresponding to the metastable subproteome (MS) and the average expression of the genes corresponding to the associated protein homeostasis components (PHC) in 77 different types of human tissue (Table S1). The protection factor s defines the strength of the protein homeostasis response to the presence of metastable proteins. Brain tissues have smaller values of s (orange, s = 0.88) than other tissues of the body (blue, s = 1.13), indicating that brain tissues have a reduced ability to protect themselves from protein aggregation relative to other tissues.
(B) Correlation matrix of gene expression levels in different tissues, where the correlation is calculated for the genes corresponding to the MS and their associated PHC. The intensity of the colors and the size of the circles are proportional to the correlation coefficients. Brain tissues (first row) are only weakly correlated with other body tissues, as also illustrated by the body image on the bottom left. The statistical significance was evaluated as explained in Methods and in Figure S1.
Vulnerable Brain Tissues Have a Weaker Response to the Pressure toward Protein Aggregation Than Non-vulnerable Ones
The greater vulnerability of brain tissues to the pressure toward protein aggregation observed above suggests the presence of a link between the levels of regulation of protein aggregation and the development of neurodegenerative processes in Alzheimer disease. In order to investigate this relationship in more detail, we differentiated between tissues that are vulnerable and those that are resistant to neurodegeneration in Alzheimer's disease, as assessed by the Braak staging (Braak and Braak, 1991). We mapped different types of brain tissue to the respective Braak stages as reported previously (Freer et al., 2016, Freer et al., 2019). Seven of these tissues corresponded to different Braak stages present in the microarray dataset used in the current study. We analyzed the levels of expression of the MS and the associated PHC in these types of tissue and compared them with the tissues not affected in Braak stages. We found that brain tissues corresponding to Braak regions have a lower value of s than do those of the non-Braak regions (Figure 2). These results reveal a close link between the regulation of the pressure toward protein aggregation with tissue vulnerability to Alzheimer's disease.
Figure 2.
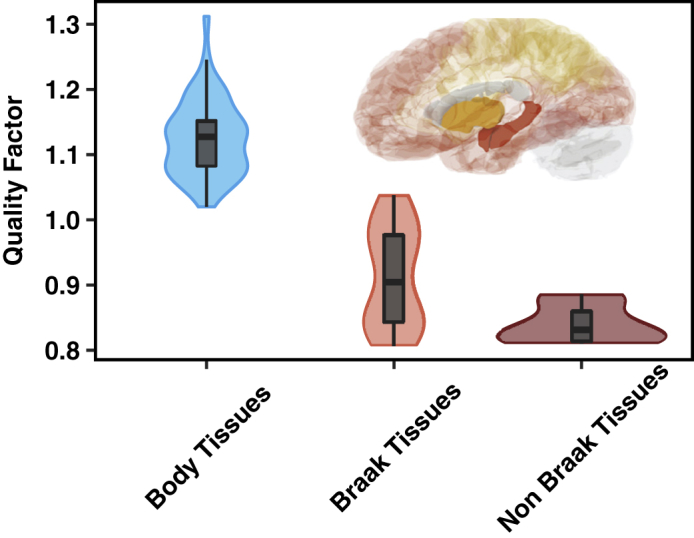
Brain Tissues Vulnerable to Alzheimer's Disease Have a Weaker Protein Homeostasis Response to Protein Aggregation than Non-vulnerable Brain Tissues
Brain tissues corresponding to different Braak regions have a lower value of the protection factor s compared with that of non-Braak regions; for comparison the protection factor of other body tissues is also shown. ∗∗∗p < 0.001.
Neurons Have a Lower Ability to Maintain Protein Homeostasis Than Other Types of Brain Cells
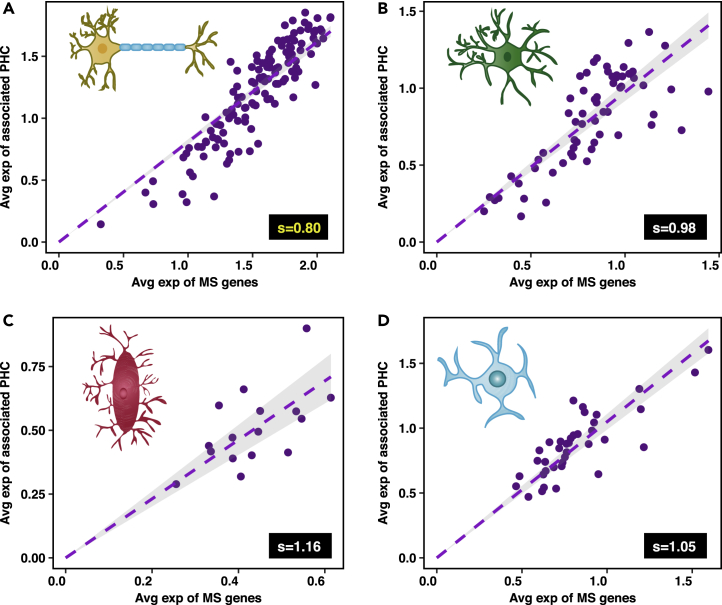
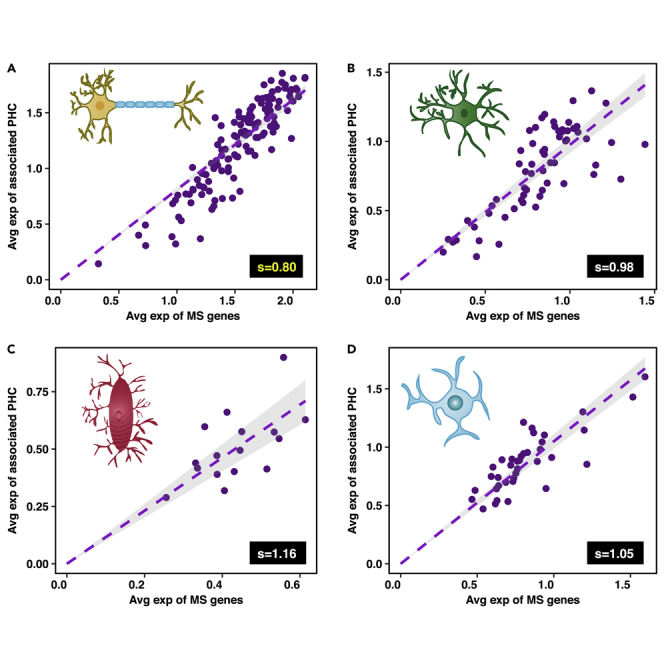
These results suggest a possible origin for the increased vulnerability of neurons to changes that lead to Alzheimer's disease relative to other non-neuronal brain cell types. In order to verify whether or not neurons are less able than other brain cell types to regulate protein aggregation, we compared the expression of genes corresponding to the MS and their associated protein homeostasis components (PHC) in neurons, astrocytes, microglia, and oligodendrocytes, using extensive single-cell RNA sequencing data (Habib et al., 2017, Lake et al., 2016). We found that neurons have the lowest value of the protection factor s among these cell types (Figure 3), indicating that neurons are indeed the cell types most vulnerable to protein aggregation in the brain.
Figure 3.
Neurons Have a Weaker Protein Homeostasis Response to Protein Aggregation than Non-neuronal Brain Cell Types
Scatterplots depicting the expression of genes corresponding to the metastable subproteome (MS) and their associated protein homeostasis components (PHC) in (A) neurons, (B) astrocytes, (C) microglia, and (D) oligodendrocytes. Each dot represents an individual cell. The level of significance between different values of s is shown in Figure S2.
Proteins in the Oxidative Phosphorylation Pathway Have a Reduced Ability to Maintain Protein Homeostasis in the Brain Than in Other Tissues
In order to understand in more detail the molecular origins of the increased vulnerability of certain types of brain cells and tissues to the effects leading to Alzheimer's disease, we considered the ability of the cells to regulate oxidative phosphorylation, because this is a specific process associated with metastable proteins (Ciryam et al., 2016, Kundra et al., 2017), and more generally with neurodegenerative diseases (Lin and Beal, 2006, Rhein et al., 2009). We divided the genes associated with the oxidative phosphorylation pathway into two parts, those that are metastable and those that are not (Table S2). We then calculated the value of protection factor s for the oxidative phosphorylation genes and their associated PHC for both groups across all tissues. The difference in s values between the body and brain tissues (Δs) is higher for the group of oxidative phosphorylation genes that are metastable, whereas there is almost no difference in the values of s for non-metastable oxidative phosphorylation genes. These results suggest that, even within the oxidative phosphorylation pathway, those genes that are also metastable pose a higher risk for the brain tissues, as these are less well prepared to face the challenge posed by the presence of these aggregation-prone proteins (Figure 4).
Figure 4.
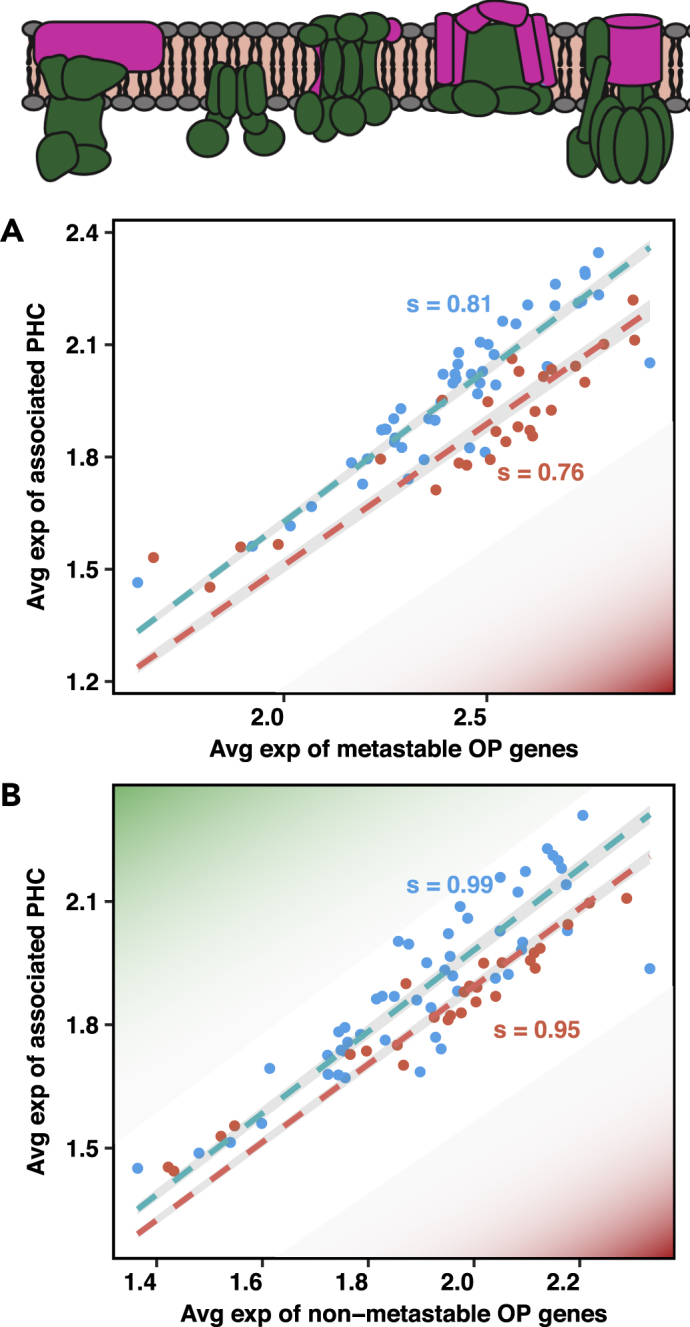
Proteins in the Oxidative Phosphorylation Pathway Have a Weaker Protein Homeostasis Response to Protein Aggregation in the Brain than in Other Tissues
(A) Average expression of the genes corresponding to the metastable subproteome (MS) within the KEGG oxidative phosphorylation (OP) pathway (inset) and their associated PHC.
(B) Average expression of all genes corresponding to non-metastable proteins within the OP pathway and their associated PHC. The value of s for these critical genes is lower (<1) throughout the body, not just in the brain, and the difference in the values of s between the body brain tissues is more pronounced for the genes encoding for metastable proteins in the oxidative phosphorylation pathway. The subunits in pink in the schematic show the location of the metastable genes within the oxidative phosphorylation pathway. The colour code is the same as in Figure 1.
Discussion
Taken together, the results of this study suggest that the strength of the protein homeostasis system that responds to protein aggregation is a central element in determining the vulnerability of cells and tissues to protein misfolding diseases. Our analysis shows that brain tissues are less effective than other body tissues in dealing with the problems posed by protein aggregation. The protection factor s serves as an effective measure of the vulnerability of different tissue types to protein aggregation, as it decreases progressively from body tissues to resistant brain tissues to highly vulnerable brain tissues (Figures 1 and 2). It also provides insights into the molecular origins of Alzheimer's disease, as neurons have the lowest value of s of all the types of cells found in the central nervous system (Figure 3).
As the human brain experienced a rapid cortical reorganization during the course of recent evolution from non-human species, one may speculate that the expression levels of critical genes could also have increased too rapidly for the simultaneous evolution of the regulatory components of the protein homeostasis system. In this view, when faced with the challenges posed by aggregation-prone proteins, brain tissues are more likely to suffer from protein aggregation diseases.
In conclusion, our results indicate that the specific vulnerability of certain brain tissues to protein aggregation is associated with their relative inability to regulate this process already in the healthy state. Furthermore, we have also provided evidence that this inability is characteristic of neurons, which are therefore observed to be the earliest affected by protein misfolding diseases, compared with other brain cell types. These observations link the molecular origins of this neurodegenerative disease to the specific branch of the protein homeostasis network that regulates the maintenance in their soluble state of the proteins that are metastable against aggregation.
Limitations of the Study
In this study, we investigated the cell- and tissue-specific capacity of the protein homeostasis system to respond to the pressure caused by the overall expression of aggregation-prone proteins. Although useful insights can be derived from a transcriptome-level analysis, as we have done here, a proteome-level analysis will in future studies provide more stringent conclusions.
It is possible that specific genetic traits of different individuals could influence the results in the transcriptional analysis carried out using just six control healthy brains from the Allen Brain Atlas. We also note that the transcriptional analyses of brain and non-brain tissues were carried out on different datasets, again leaving open the possibility that genetic traits of specific individuals could influence the results.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by the Center for Misfolding Diseases, University of Cambridge.
Authors Contributions
All the authors participated in the design of the study, its execution, the analysis of the results, and the writing of the manuscript.
Declaration of Interests
We declare no conflict of interests.
Published: March 27, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.100934.
Supplemental Information
References
- Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Bero A.W., Yan P., Roh J.H., Cirrito J.R., Stewart F.R., Raichle M.E., Lee J.-M., Holtzman D.M. Neuronal activity regulates the regional vulnerability to amyloid-β deposition. Nat. Neurosci. 2011;14:750. doi: 10.1038/nn.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H., Braak E. Neuropathologicalstageing of Alzheimer-related changes. Acta Neuropatholog. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Chapman E., Farr G.W., Usaite R., Furtak K., Fenton W.A., Chaudhuri T.K., Hondorp E.R., Matthews R.G., Wolf S.G., Yates J.R. Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperoninGroEL. Proc. Natl. Acad. Sci. U S A. 2006;103:15800–15805. doi: 10.1073/pnas.0607534103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P., Kundra R., Freer R., Morimoto R.I., Dobson C.M., Vendruscolo M. A transcriptional signature of Alzheimer’s disease is associated with a metastable subproteome at risk for aggregation. Proc. Natl. Acad. Sci. U S A. 2016;113:4753–4758. doi: 10.1073/pnas.1516604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P., Kundra R., Morimoto R.I., Dobson C.M., Vendruscolo M. Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol. Sci. 2015;36:72–77. doi: 10.1016/j.tips.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P., Lambert-Smith I.A., Bean D.M., Freer R., Cid F., Tartaglia G.G., Saunders D.N., Wilson M.R., Oliver S.G., Morimoto R.I. Spinal motor neuron protein supersaturation patterns are associated with inclusion body formation in ALS. Proc. Natl. Acad. Sci. U S A. 2017;114:E3935–E3943. doi: 10.1073/pnas.1613854114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P., Tartaglia G.G., Morimoto R.I., Dobson C.M., Vendruscolo M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 2013;5:781–790. doi: 10.1016/j.celrep.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David D.C., Ollikainen N., Trinidad J.C., Cary M.P., Burlingame A.L., Kenyon C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 2010;8:e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B., Karran E. The cellular phase of Alzheimer’s disease. Cell. 2016;164:603–615. doi: 10.1016/j.cell.2015.12.056. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freer R., Sormanni P., Vecchi G., Ciryam P., Dobson C.M., Vendruscolo M. A protein homeostasis signature in healthy brains recapitulates tissue vulnerability to Alzheimer’s disease. Sci. Adv. 2016;2:e1600947. doi: 10.1126/sciadv.1600947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freer R., Sormanni P., Ciryam P., Rammner B., Rizzoli S.O., Dobson C.M., Vendruscolo M. Supersaturated proteins are enriched at synapses and underlie cell and tissue vulnerability in Alzheimer's disease. Heliyon. 2019;5:e02589. doi: 10.1016/j.heliyon.2019.e02589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Hardy J., Duff K.E. Selective vulnerability in neurodegenerative diseases. Nat. Neurosci. 2018;21:1350–1358. doi: 10.1038/s41593-018-0221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Possenti A., Freer R., Nakano Y., Villegas N.C.H., Tang M., Cauhy P.V., Lassus B.A., Chen S., Fowler S.L. A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat. Neurosci. 2019;22:47. doi: 10.1038/s41593-018-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T., Ben-Zvi A., Ho K.H., Brignull H.R., Morimoto R.I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Habib N., Avraham-Davidi I., Basu A., Burks T., Shekhar K., Hofree M., Choudhury S.R., Aguet F., Gelfand E., Ardlie K. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods. 2017;14:955. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp M.S., Park S.-H., Hartl F.U. Proteostasis impairment in protein-misfolding and-aggregation diseases. Trends Cell Biol. 2014;24:506–514. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Holtzman D.M., Morris J.C., Goate A.M. Alzheimer’s disease: the challenge of the second century. Sci. Transl. Med. 2011;3:77sr71. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles T.P., Vendruscolo M., Dobson C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014;15:384. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- Kundra R., Ciryam P., Morimoto R.I., Dobson C.M., Vendruscolo M. Protein homeostasis of a metastable subproteome associated with Alzheimer’s disease. Proc. Natl. Acad. Sci. U S A. 2017;114:E5703. doi: 10.1073/pnas.1618417114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake B.B., Ai R., Kaeser G.E., Salathia N.S., Yung Y.C., Liu R., Wildberg A., Gao D., Fung H.-L., Chen S. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.T., Beal M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Mattson M.P., Magnus T. Ageing and neuronal vulnerability. Nat. Rev. Neurosci. 2006;7:278. doi: 10.1038/nrn1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.A., Woltjer R.L., Goodenbour J.M., Horvath S., Geschwind D.H. Genes and pathways underlying regional and cell type changes in Alzheimer's disease. Genome Med. 2013;5:48. doi: 10.1186/gm452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhein V., Song X., Wiesner A., Ittner L.M., Baysang G., Meier F., Ozmen L., Bluethmann H., Dröse S., Brandt U. Amyloid-β and tau synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer's disease mice. Proc. Natl. Acad. Sci. U S A. 2009;106:20057–20062. doi: 10.1073/pnas.0905529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe D.J., Hardy J. The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol. Med. 2016;8:595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia G.G., Pechmann S., Dobson C.M., Vendruscolo M. Life on the edge: a link between gene expression levels and aggregation rates of human proteins. Trends Biochem. Sci. 2007;32:204–205. doi: 10.1016/j.tibs.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Vecchi G., Sormanni P., Mannini B., Vandelli A., Tartaglia G.G., Dobson C.M., Hartl F.U., Vendruscolo M. Proteome-wide observation of the phenomenon of life on the edge of solubility. Proc. Natl. Acad. Sci. U S A. 2020;117:1015–1020. doi: 10.1073/pnas.1910444117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther D.M., Kasturi P., Zheng M., Pinkert S., Vecchi G., Ciryam P., Morimoto R.I., Dobson C.M., Vendruscolo M., Mann M. Widespread proteome remodeling and aggregation in aging C. elegans. Cell. 2015;161:919–932. doi: 10.1016/j.cell.2015.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerbury J.J., Ooi L., Blair I.P., Ciryam P., Dobson C.M., Vendruscolo M. The metastability of the proteome of spinal motor neurons underlies their selective vulnerability in ALS. Neurosci. Lett. 2019;704:89–94. doi: 10.1016/j.neulet.2019.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.