Abstract
Background
This article aims to develop and assess the radiomics paradigm for predicting colorectal cancer liver metastasis (CRLM) from the primary tumor.
Methods
This retrospective study included 100 patients from the First Hospital of Jilin University from June 2017 to December 2017. The 100 patients comprised 50 patients with and 50 without CRLM. The maximum-level enhanced computed tomography (CT) image of primary cancer in the portal venous phase of each patient was selected as the original image data. To automatically implement radiomics-related paradigms, we developed a toolkit called Radiomics Intelligent Analysis Toolkit (RIAT).
Results
With RIAT, the model based on logistic regression (LR) using both the radiomics and clinical information signatures showed the maximum net benefit. The area under the curve (AUC) value was 0.90±0.02 (sensitivity =0.85±0.02, specificity =0.79±0.04) for the training set, 0.86±0.11 (sensitivity =0.85±0.09, specificity =0.75±0.19) for the verification set, 0.906 (95% CI, 0.840–0.971; sensitivity =0.81, specificity =0.84) for the cross-validation set, and 0.899 (95% CI, 0.761–1.000; sensitivity =0.78, specificity =0.91) for the test set.
Conclusions
The radiomics nomogram-based LR with clinical risk and radiomics features allows for a more accurate classification of CRLM using CT images with RIAT.
Keywords: Radiomics, nomogram, colorectal cancer liver metastasis (CRLM), machine learning
Introduction
Colorectal cancer (CRC) is the third most diagnosed cancer and the second most common cause of cancer-related deaths worldwide (1). Approximately 50% of patients with CRC develop liver metastases either by the time of diagnosis or later during the disease course. Preoperative prediction of hepatic metastasis in patients with CRC is crucial to treatment decisions and patient prognosis (2,3). The liver is the most common metastatic organ of CRC. Thus, accurate diagnosis of liver metastases in patients with CRC before surgery is crucial to ensuring the appropriate treatment choice and for prognostic evaluation. Computed tomography (CT) is widely used to diagnose colorectal diseases because it can better show the primary tumor, local lymph node metastasis, and distant metastasis of the lesion, but the sensitivity and specificity of CT images are low when used to assess small or atypical liver metastases (4-8). Some studies have focused on colorectal cancer liver metastasis (CRLM) with magnetic resonance imaging (MRI) and achieved excellent results (9,10). For some CRLM patients with whom a diagnosis is difficult, liver MRI enhancement examination or needle biopsy is performed for diagnosis. However, MRI examination is expensive and time consuming, while biopsy is an invasive examination.
Radiomics (11) extracts mineable high-dimensional information from medical images (CT, MRI, etc.), and is a mature research field that aims to build models to improve diagnostic, prognostic, and prediction accuracy, which enhance precision medicine (12-15). Radiomics procedures can be divided into several steps, namely, region of interest (ROI) extraction, radiomics feature extraction, feature selection, and building models. Each step requires a reasonable assessment to build robust models that can be transferred to clinical practice for prognosis, noninvasive assessment, and tracking disease response to treatment. Scholars have also used radiomics in research on clinically relevant CRC issues (16-18). This study develops an individualized nomogram to predict CRC synchronous liver metastases based on radiomics data of primary CRC from CT images.
Methods
Framework for predicting colorectal liver metastases
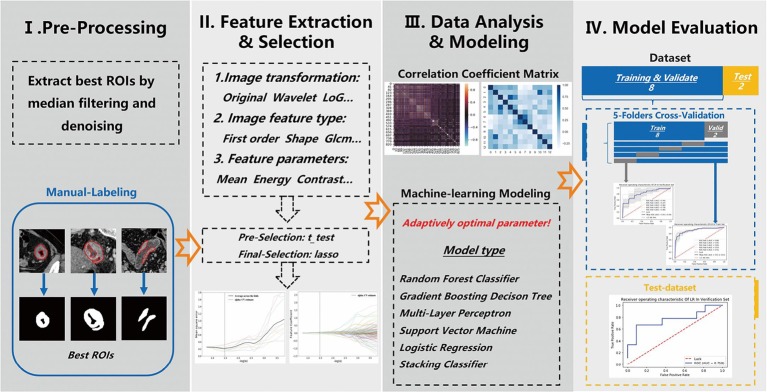
We built two models to predict colorectal liver metastases: one based on radiomics feature signatures, and the other one based on both radiomics features and clinical information signatures. The process for analyzing the radiomics features is shown in Figure 1.
Figure 1.
The process for analyzing radiomics features.
Materials
Subjects
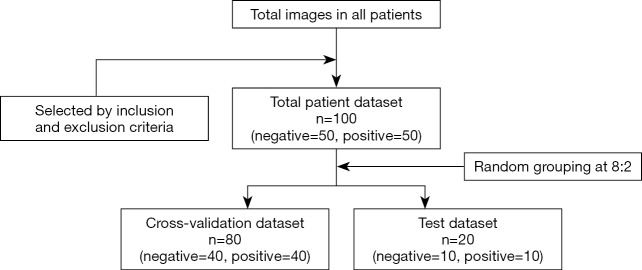
We performed a retrospective analysis of patients from the First Hospital of Jilin University from June 2017 to December 2017. The criteria for inclusion were (I) patients with CRC and liver metastases confirmed by MRI or pathology and (II) patients who received preoperative colon CT plain scan and enhanced examination. The criteria for exclusion were (I) patients who received new adjuvant chemotherapy before surgery and (II) patients with poor image quality that was unsuitable for quantitative analysis. One hundred patients were selected from the eligible patients; the selected patients included 50 with liver metastasis and 50 without liver metastasis.
The whole dataset comprising data of 100 patients was randomly divided into the cross-validation set and the test set at a ratio of 8:2; that is, 80 patients were assigned to the cross-validation set, and 20 were assigned to the test set.
All the primary CRC lesions were single tumors. Finally, we adopted the maximum-level enhanced CT image in the portal venous phase of each patient as the input image. The filtering process is shown in Figure 2.
Figure 2.
The patient selection process. Negative = patients without colorectal liver metastasis; positive = patients with colorectal liver metastasis.
CT scanning protocol
A Philips Brilliance 256 iCT scanner was used for plain and enhanced abdominal scanning. The layer thickness was 5 mm, the layer spacing was 5 mm, the frame rotation speed was 2 r/s, the pitch was 0.800, the tube voltage was 120 kV, the tube current was 150 mA, and the matrix size was 512×512. The output CT images were exported in DICOM format.
Data collection
Two radiology physicians with an average age of 37 years (radiologist 1, HM Zhang, chief physician, 25 years of experience; radiologist 2, Y Guo, 3 years of experience) read the CT images. In-house developed software was used to select the slice that included the largest of the venous image tumor lesions to place the ROIs along the contour of the lesion while removing cystic, necrotic, and transitional areas between lesions and normal intestine. An agreement between the two radiologists regarding ROI extraction was evaluated using the intraclass correlation coefficient (ICC). An ICC greater than 0.75 was considered good agreement; the remaining ROIs were placed by radiologist 1.
The patients’ clinical information included age, sex, carcinoembryonic antigen (CEA) (normal reference value ≤3.7 ng/mL), and carbohydrate antigen 19-9 (CA19-9) (normal reference value ≤27 U/mL).
Radiomics method
We developed a medical image quantitative analysis platform called the Radiomics Intelligent Analysis Toolkit (RIAT) to assist radiologists. RIAT consists of two main parts: (I) DICOM file preprocessing and (II) data processing. Its main functions include the following: (I) removing personal information from DICOM files and optimizing ROI extraction; (II) extracting either fewer or more radiomics features based on the radiologists’ needs; (III) acquiring the radiomics features signature using different algorithms; (IV) building and saving high-accuracy machine learning models by automatically selecting the optimal parameters; (V) using the receiver operating characteristic (ROC) curves of the cross-validation and test set to evaluate the models; and (VI) building models and performing predictions using an embedded device.
The graphical user interface (GUI)
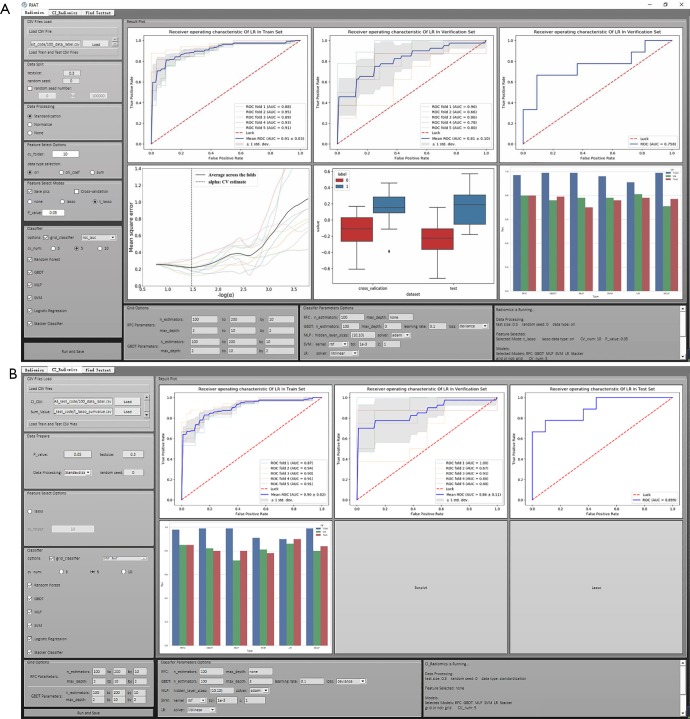
The GUI of RIAT supports DICOM file preprocessing and ROI extraction, radiomics feature extraction, data preprocessing, radiomics features and model parameter selection, model building and prediction, logging, etc. (see Figure 3A,B). The GUI was developed by QT® (Qtsoftware, Norway).
Figure 3.
The GUI: (A) for radiomics features in RIAT; (B) for radiomics features and clinical information in RIAT. GUI, graphical user interface; RIAT, Radiomics Intelligent Analysis Toolkit.
Optimized identification and extraction of ROI
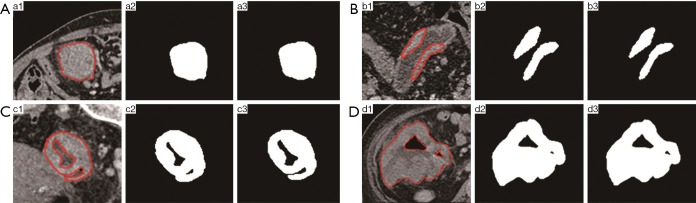
Some of the functions in RIAT include ROI optimized extraction and matching ROI binary images to DICOM files. RIAT can set the window widths and centers of DICOM files. The preprocessed DICOM and label files can be used directly for feature selection. A red line indicates the ROI of the CT image, and the optimization algorithm must extract the best ROI. RIAT supported the extraction of single ROIs, multiple ROIs, and nested ROIs. Some optimization results are shown in Figure 4.
Figure 4.
(a1,b1,c1,d1) are the ROIs of the original CT images marked by the red line; (a2,d2,c2,d2) are unoptimized ROI binary images; and (a3,b3,c3,d3) are optimized ROI binary images. ROI, region of interest.
Automated radiomics module
Radiomics consists of five parts: (I) ROI extraction; (II) feature extraction; (III) feature selection; (IV) machine learning model development; and (V) model evaluation and prediction. Recently, automatic and semiautomatic segmentation via deep learning have achieved excellent performance (19-21). However, most researchers still use manual extraction (22-24). Feature selection is conducted after feature extraction. Removing redundant features is conducive to building highly accurate models, which are based on hypothesis tests and regularization. Although statistics occupies a vital position in the study of radiomics (25), few studies have applied hypothesis tests to select radiomics features (26,27). Most studies use the least absolute shrinkage and selection operator (LASSO) regularization to select radiomics features because of its stability and effectiveness (23,24,28-30). In addition, minimum redundancy maximum relevance (mRMR) has proven to be effective in some studies (31).
Support vector machine (SVM) (32,33) and the random forest classifier (RFC) have been used (27,34) in radiomics. The nomogram is widely used with the radiomics feature and clinical information because it is more interpretable (23,35). ROC and area under the receiver operating characteristic (AUROC) curves are regarded as the most used evaluation standards. The results of cross-validation in large datasets are representative and enable objective validation of the data distribution of the whole dataset (36). The combination of radiomics features and clinical information can improve model accuracy for some diseases (33,35). Machine learning has the potential to improve the ability and accuracy of radiomics (37-41) and can improve radiologists’ efficiency (42-44).
RIAT uses pyradiomics, which is highly accurate and written in Python, to extract the following radiomics features (19): (I) up to 187 first-order and shape features based on the original image and (II) up to 1,470 first-order and higher-order features based on the original image and other image transforms (such as a wavelet transform). Using the ROIs annotated by a radiologist, RIAT extracts 841 features for each CT image on the complete dataset. These features include the following: (I) wavelet transform; (II) first-order texture feature; (III) shape features (surface area, sphericity, flatness, etc.); (IV) gray-level co-occurrence matrix (GLCM) (contrast, correlation, joint entropy, etc.); (V) gray-level size-zone matrix (GLSZM) [gray-level nonuniformity (GLN), size-zone nonuniformity (SZN), gray-level variance (GLV), etc.]; (VI) gray-level run-length matrix (GLRLM) [run variance (RV), run entropy (RE), short-run emphasis (SRE), etc.]; (VII) neighborhood gray-tone difference matrix (NGTDM) (coarseness, contrast, complexity, etc.); and (VIII) gray-level dependence matrix (GLDM) (gray level, GLV, GLN, and level dependence). Figure 5 shows an example of an original ROI image and feature matrix visualizations of the same ROI under different image transformations.
Figure 5.
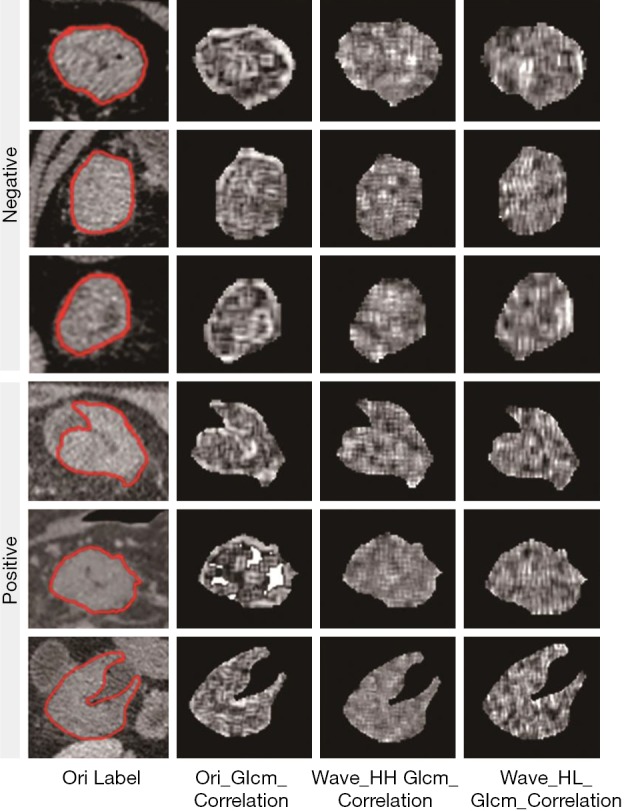
Original ROI images and feature visualizations under different radiomics features. ROI, region of interest.
LASSO regularization and gradient feature selection are implemented in RIAT. In each method, different numbers of cross-validation folds can be set by the user. RIAT automatically plots the convergence and feature selection curves after performing LASSO regularization. RIAT provides several stable machine learning models: RFC, gradient decision tree (GBDT), SVM, LR, multilayer perceptron (MLP), and stacking classifier (SCLF).
Evaluation module
Model evaluation methods have always been a focus of research. As RIAT randomly divides the dataset into a cross-validation set and a test set, we can see whether the feature selection method is effective across the entire cross-validation set. This procedure helps to understand data distribution characteristics and assess model generalizability. The ROC can effectively reduce accuracy deviations caused by imbalanced data. RIAT provides cross-validation for the training and verification sets and plots every ROC curve, mean ROC curve, and the standard deviations under different folds. Some diseases require high sensitivity, specificity, positive predictive value, or negative predictive value; this aspect forms a vital part of the model effectiveness evaluation. Based on the number of folds selected for cross-validation, RIAT calculates the average value and standard deviation for sensitivity, specificity, positive predictive value, and negative predictive value for each training and verification set.
Data standardization and gradient feature selection
Because the absolute values of the radiomics feature are quite different, data standardization is the first step before data processing. We obtain the mean and variance from the cross-validation set; these values are used to standardize the test set in RIAT.
Feature selection is required before modeling. We independently select the radiomics feature signature from the cross-validation set only to ensure that the test set is unaffected. We use gradient feature selection to evaluate the 841 features of each CT image. Features are selected when the t-test results are P<0.05 based on the hypothesis. Then, these features are further filtered by LASSO with 10-fold cross-validation. We select the α value in which the mean square error is the smallest. Using the α value, we obtain the radiomics feature signature and feature coefficients.
Models and performance for CRLM prediction
Radiomics feature signature
We built six models using RIAT on the radiomics feature signature for CRLM prediction. The optimal parameters for all the models were set by automatic selection except for the SCLF model, which consists of RFC, GBDT, and LR. The optimized model parameters were selected when the mean AUROC of the cross-validation set is the highest. The initial parameter range of each model is reported in Table 1; the three values in ‘n_estimators’ and ‘max_depth’ are the start value, end value, and step size, respectively.
Table 1. The parameter search range of models and selected model parameters.
| Parameter | RFC | GBDT | MLP | SVM | LR |
|---|---|---|---|---|---|
| n_estimators | (100, 200, 10)a, (130)b, (150)c | (100, 200, 10)a, (110)b, (160)c | |||
| max_depth | (2, 10, 2)a, (2)b, (2)c | (2, 10, 2)a, (2)b, (4)c | |||
| hidden_layer_size | [(10, 10),(50, 50),(100, 100)]a, (50, 50)b, (50, 50)c | ||||
| solver | (lbfgs, sgd, adam)a, (lbfgs)b, (lbfgs)c | (newton-cg, lbfgs, liblinear, sag, saga)a, (newton-cg)b, (newton-cg)c | |||
| kernel | (rbf, linear)a, (rbf)b, (rbf)c | ||||
| C | (1, 10, 100, 1000)a, (1)b, (1)c | ||||
| cv_folder | 5 | 5 | 5 | 5 | 5 |
The first column presents the parameter names of all models except ‘cv_folder’. The first row presents the names of all models. a, represents the parameter search range of each model. In addition, the third place of ‘n_estimators’ or ‘max_depth’ means the step size of the corresponding parameter change. The models automatically search for the optimal parameters from the parameter search range according to the cross-validation set. b, represents the optimal parameters of each model with the radiomics feature signature. c, represents the optimal parameters of each model with the radiomics feature signature and clinical information signature. ‘cv_folder’ represents the cross-validation folder.
After selecting the optimal parameters for each model, the ROC curve is plotted as 5-fold cross-validation on the cross-validation set. The proportion of negative and positive patients in each training and verification set is to 1:1. The ROC of the training set and verification set contains the independent ROC for each fold, the mean ROC of the 5-fold cross-validation, the mean AUROC, and its standard deviation. Thus, we can see the overall data distribution of the cross-validation set and the ROC fluctuation in each dataset. Using this approach not only serves to verify the efficacy and generalizability of feature selection on the entire cross-validation set but also functions as a reference for the test set ROC. Consequently, it increases the fairness and objectivity of the model evaluation.
The radiomics feature signature of the entire cross-validation set is used as a training set when building a new model to perform predictions based on the radiomics feature signature of the test set. Among all the models, the best model is found by comparing the mean AUROC and standard deviation of the cross-validation set with the AUROC of the test set.
In addition to using ROC and AUROC as a reference for model performance evaluation, RIAT also provides sensitivity, specificity, positive predictive value, and negative predictive value scores. RIAT only calculates the average value and standard deviation of the cross-validation set, and not those of the test set.
Radiomics feature signature and clinical information signature
We transformed the multidimensional radiomics feature signature into a one-dimensional radiomics feature signature using Eq. [1]. By applying a chi-square test to sex and lesion location clinical information and a t-test to the other clinical information, if we can obtain a clinical information signature of P<0.05, we use it to build a model with the one-dimensional radiomics feature signature. The model-building process is the same as that used to build the model for the radiomics feature signature:
| [1] |
where R* is the one-dimensional radiomics feature signature, vi is the multidimensional radiomics feature signature, and ci is the feature coefficient.
Results
Gradient feature selection and the heat map of the feature correlation coefficient matrix
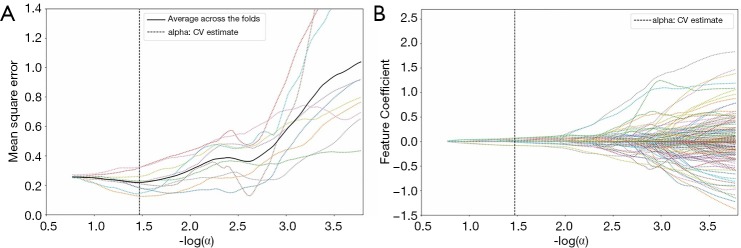
We first selected 210 radiomics features of P<0.05 from 841 radiomics features using a t-test. Then, the 12 radiomics feature signatures were selected by LASSO in a 10-fold cross-validation from the 210 radiomics features. An example LASSO regularized diagram is shown in Figure 6. The feature names, feature values, and feature coefficients of the 12 radiomics feature signatures are listed in Table 2.
Figure 6.
(A) The relationship between the mean square of the LASSO 10-fold cross-validation and the value of -log(α); (B) the relationship between the feature coefficient and the value of -log(α). The thick solid line is the mean curve of the LASSO 10-fold cross-validation curve, and the thin dotted line is the LASSO regularized curve of each fold in (A). The thin dotted line is the feature coefficient value of each fold at different -log(α) values in (B). The thick dotted line is the value of -log(α) at the minimum mean square error. LASSO, least absolute shrinkage and selection operator.
Table 2. Feature name and feature coefficient of radiomics feature signatures.
| Feature name | Coefficients |
|---|---|
| original_firstorder_Median | 0.0258837 |
| wavelet-LLL_gldm_DependenceVariance | 0.00187731 |
| wavelet-LLH_firstorder_Skewness | 0.0850055 |
| wavelet-LLH_glcm_MaximumProbability | 0.056486 |
| wavelet-LHL_glcm_Idmn | 0.0664622 |
| wavelet-HLL_glrlm_ShortRunLowGrayLevelEmphasis | −0.0743506 |
| wavelet-HLL_glszm_ZoneEntropy | −0.0415202 |
| wavelet-HLH_firstorder_Median | −0.10654 |
| wavelet-HLH_glrlm_RunLengthNonUniformityNormalized | 0.0606122 |
| wavelet-HHL_firstorder_Maximum | 0.0436455 |
| wavelet-HHL_glszm_SmallAreaEmphasis | 0.0410796 |
| wavelet-HHH_glcm_Contrast | 0.0380088 |
Original: the original image; wavelet: wavelet transform; LLL, LLH, LHL, HLL, HLH, HHL, and HHH: subbands of the wavelet transform; firstorder: first-order feature; gldm: gray-level dependence matrix; glcm: gray-level co-occurrence matrix; glrlm: gray-level run-length matrix; glszm: gray-level size-zone matrix; Idmn: inverse difference moment normalized.
The heat map of the radiomics feature correlation coefficient matrix and the heat map of the radiomics feature signature correlation coefficient matrix are shown in Figure 7. As shown in Figure 7, the gradient feature selection removed redundant features but retained the feature correlations.
Figure 7.
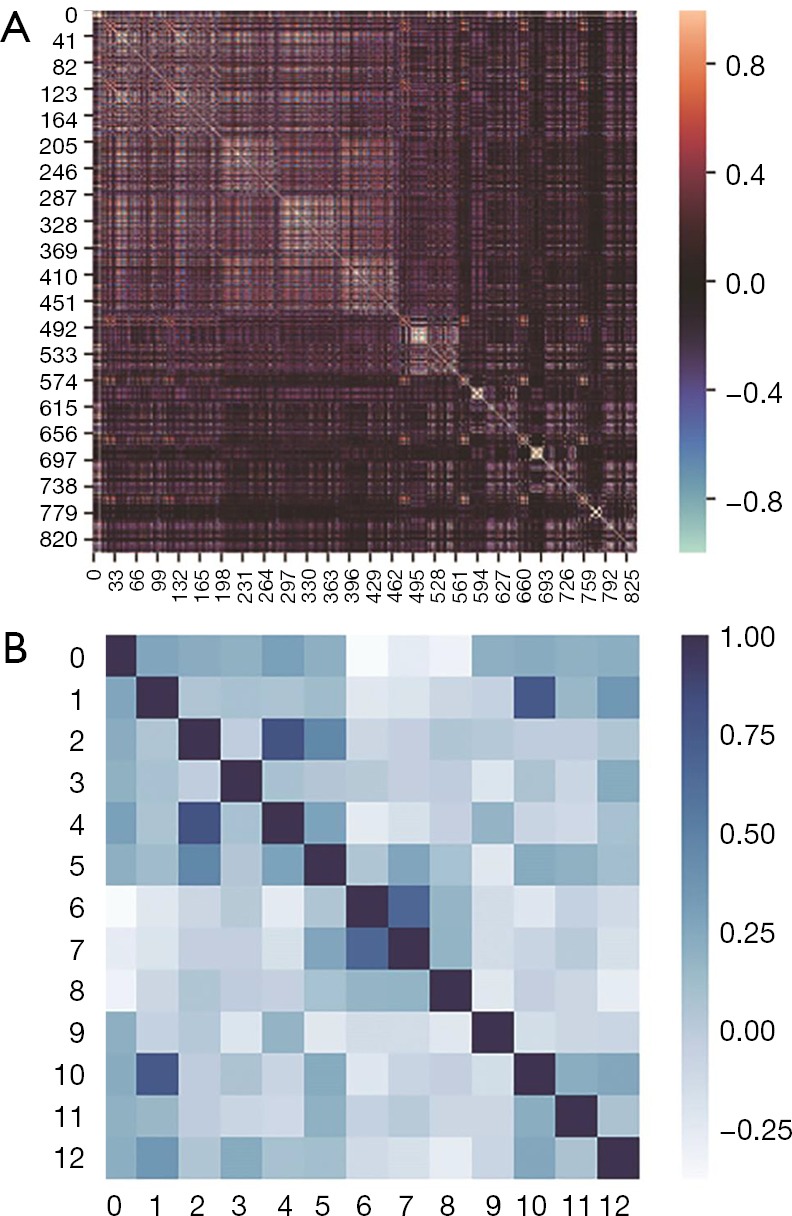
The abscissa and ordinate in (A) are the 841 standardized radiomics features. The abscissa and ordinate in (B) are the 12 radiomics feature signatures. When the correlation coefficient value is closer to 1, the two features are more correlated, and when the correlation coefficient value is closer to −1, the two features are less correlated.
Evaluation of radiomics feature signature models
According to the radiomics feature signature, we traversed all the initialized parameters using five models (e.g., RFC, GBDT, MLP, SVM, and LR) to select optimal parameters. The selection criterion is to select the largest mean AUROC of the 5-fold cross-validation. The final optimal parameters of each model are reported in Table 1.
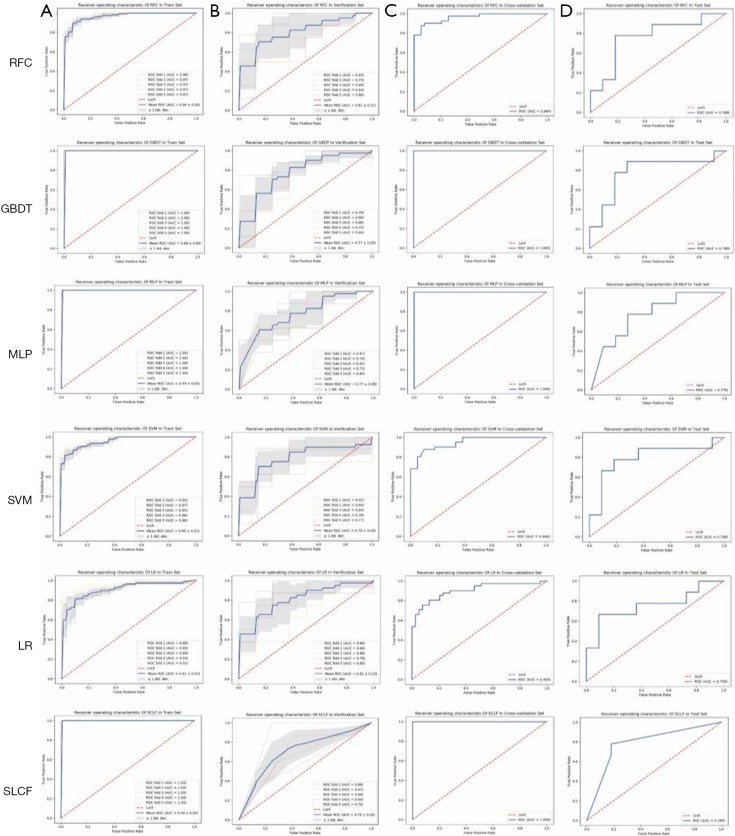
We plotted the ROCs of the training and verification sets in RIAT using the model with optimal parameters from the 5-fold cross-validation. Then, we applied the entire cross-validation set as a training set to build a model and use it to predict the test set. All the ROCs of the different models are shown in Figure 8.
Figure 8.
The training ROC curve, verification ROC curve, cross-validation ROC curve, and test ROC curve of the radiomics features signatures from different models. The solid blue line denotes the mean ROC curve or the ROC curve, while the red dotted line is the diagonal of the ROC curve. The gray shaded area is the standard deviation range of the ROC curve on the cross-validation set, and the thin solid line is the ROC curve for each fold in cross-validation. (A) The ROC of the training set on the cross-validation set; (B) the ROC of the verification set on the cross-validation set; (C) the ROC of the cross-validation; and (D) the ROC of the test set. ROC, receiver operating characteristic.
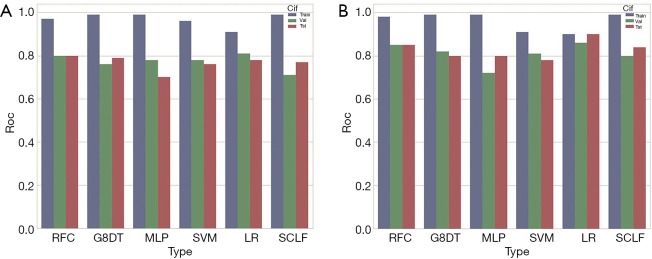
The AUROC scores for each model on the training set, verification set, cross-validation set, and test set are reported in Table 3. A histogram of the AUROC scores for each model on the training set, verification set, and test set is shown in Figure 9. The sensitivity, specificity, positive predictive value, and negative predictive value of the training set, verification set, cross-validation set and test set for each model with the radiomics feature signature are reported in Table 4.
Table 3. The AUROC scores for each model.
| Variable | Training set | Verification set | Cross-validation set (95% CI) | Test set (95% CI) |
|---|---|---|---|---|
| A: The AUROC scores for each model with the radiomics signature on the training set, verification set, cross-validation set and test set | ||||
| RFC | 0.97±0.00 | 0.79±0.10 | 0.964 (0.929–0.998) | 0.768 (0.542–0.994) |
| GBDT | 0.99±0.00 | 0.76±0.05 | 1.000 (*) | 0.788 (0.560–1.000) |
| MLP | 0.99±0.00 | 0.78±0.09 | 1.000 (*) | 0.778 (0.570–0.986) |
| SVM | 0.96±0.00 | 0.78±0.07 | 0.948 (0.906–0.990) | 0.798 (0.573–1.000) |
| LR | 0.91±0.03 | 0.79±0.04 | 0.904 (0.836–0.971) | 0.758 (0.521–0.994) |
| SCLF | 0.99±0.00 | 0.71±0.09 | 1.000 (*) | 0.788 (0.594–0.982) |
| B: The AUROC scores for each model with radiomics signature and clinical information in training, verification, cross-validation, and test set | ||||
| RFC | 0.98±0.01 | 0.85±0.09 | 0.967 (0.931–1.000) | 0.848 (0.640–1.000) |
| GBDT | 0.99±0.00 | 0.81±0.12 | 1.000 (*) | 0.848 (0.640–1.000) |
| MLP | 0.99±0.00 | 0.72±0.09 | 1.000 (*) | 0.768 (0.542–0.994) |
| SVM | 0.91±0.03 | 0.81±0.12 | 0.909 (0.847–0.972) | 0.778 (0.515–1.000) |
| LR | 0.90±0.02 | 0.86±0.11 | 0.906 (0.840–0.971) | 0.899 (0.761–1.000) |
| SCLF | 0.99±0.00 | 0.79±0.10 | 1.000 (*) | 0.843 (0.674–1.000) |
*, means that no 95% confidence interval exists. AUROC, area under the receiver operating characteristic.
Figure 9.
Histogram of the mean AUROC. (A) The AUROC of the training set, verification set, and test set of the radiomics feature signatures from six models; (B) the AUROC of the training set, verification set, and test set of radiomics feature signatures and clinical information signatures. AUROC, area under the receiver operating characteristic.
Table 4. The sensitivity, specificity, positive predictive value, and negative predictive value of different models with the radiomics feature signature in the cross-validation dataset and test set.
| Variable | The training set of cross-validation | The validation set of cross-validation | Cross-validation set | Test set | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sen | Spe | Ppv | Npv | Sen | Spe | Ppv | Npv | Sen | Spe | Ppv | Npv | Sen | Spe | Ppv | Npv | ||||
| RFC | 0.91±0.02 | 0.87±0.02 | 0.88±0.02 | 0.91±0.02 | 0.70±0.20 | 0.67±0.12 | 0.69±0.09 | 0.73±0.16 | 0.90 | 0.90 | 0.90 | 0.90 | 0.56 | 0.73 | 0.62 | 0.67 | |||
| GBDT | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 0.70±0.19 | 0.63±0.20 | 0.68±0.09 | 0.69±0.09 | 1.00 | 1.00 | 1.00 | 1.00 | 0.78 | 0.82 | 0.78 | 0.82 | |||
| MLP | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 0.70±0.13 | 0.67±0.17 | 0.71±0.09 | 0.69±0.08 | 1.00 | 1.00 | 1.00 | 1.00 | 0.67 | 0.55 | 0.55 | 0.67 | |||
| SVM | 0.87±0.03 | 0.90±0.05 | 0.90±0.04 | 0.87±0.03 | 0.70±0.17 | 0.66±0.15 | 0.70±0.06 | 0.71±0.08 | 0.92 | 0.88 | 0.92 | 0.88 | 0.67 | 0.82 | 0.75 | 0.75 | |||
| LR | 0.87±0.02 | 0.79±0.04 | 0.81±0.03 | 0.85±0.02 | 0.72±0.22 | 0.66±0.19 | 0.72±0.10 | 0.74±0.15 | 0.81 | 0.84 | 0.85 | 0.79 | 0.67 | 0.91 | 0.86 | 0.77 | |||
| SCLF | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 0.68±0.21 | 0.61±0.20 | 0.66±0.09 | 0.67±0.09 | 1.00 | 1.00 | 1.00 | 1.00 | 0.78 | 0.82 | 0.78 | 0.82 | |||
As Figure 9A and Table 3 (part A) show, the mean AUROC of the LR model on the verification set is 0.79, which is the highest of the six models. The absolute value of the standard deviation of the LR model is 0.04, which is the smallest of the six models. The AUROC of the RFC model on the test set is 0.798, which is the highest, but it is similar to the AUROC of the LR model. The sensitivity, specificity, positive predictive value, and negative predictive value of the LR model are all the highest among all the models, whether on the verification set or the test set. Therefore, the LR model has better classification efficiency for the radiomics feature signature.
Evaluation of the radiomics feature signature and clinical information signature models
The clinical information in which the chi-square test and the t-test result were P<0.05 was selected as the clinical information signature from all the patients’ clinical information. All the clinical information and the P values are reported in Table 5. CEA and CA19-9 can be used as clinical information signatures (P<0.01). We obtained the one-dimensional radiomics feature signature of all the datasets using Equation 4. Then, we plotted a box plot of the cross-validation set and test set, as shown in Figure 10.
Table 5. All clinical information of patients with colorectal cancer in the liver metastasis group and nonliver metastasis group.
| Label | Number of cases | Age (years) | Sex | Lesion location | CEA (ng/mL) | CA19-9 (U/mL) | |||
|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Right colon | Left colon | ||||||
| Liver metastasis | 50 | 59.5 (52.0–68.5) | 31 | 19 | 24 | 26 | 22.88 (5.15–67.17) | 31 (9.24–161.03) | |
| Nonliver metastasis | 50 | 63.5 (56.2–69.7) | 26 | 24 | 28 | 22 | 6.33 (2.79–19.40) | 15.45 (9.47–42.45) | |
| P value | 0.38 a | 0.31 b | 0.423 b | <0.01 a | 0.01 a | ||||
Age, CEA, CA19-9 are shown as medians (upper and lower quartiles). Sex and lesion locations are shown as the number of patients. a, means t-test; b, means chi-square test.
Figure 10.
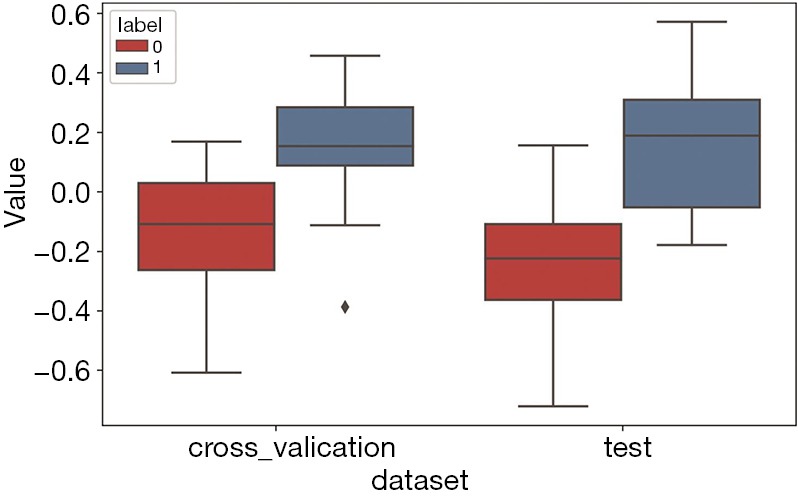
Box plot of the one-dimensional radiomics feature signature on the cross-validation and test sets. A label of 0 indicates colorectal cancer without liver metastasis, while a label of 1 indicates colorectal cancer with liver metastasis. As the figure shows, the two labels have obvious differences in both the cross-validation and test sets. This result indicates that the one-dimensional radiomics feature signature results in better classification performance.
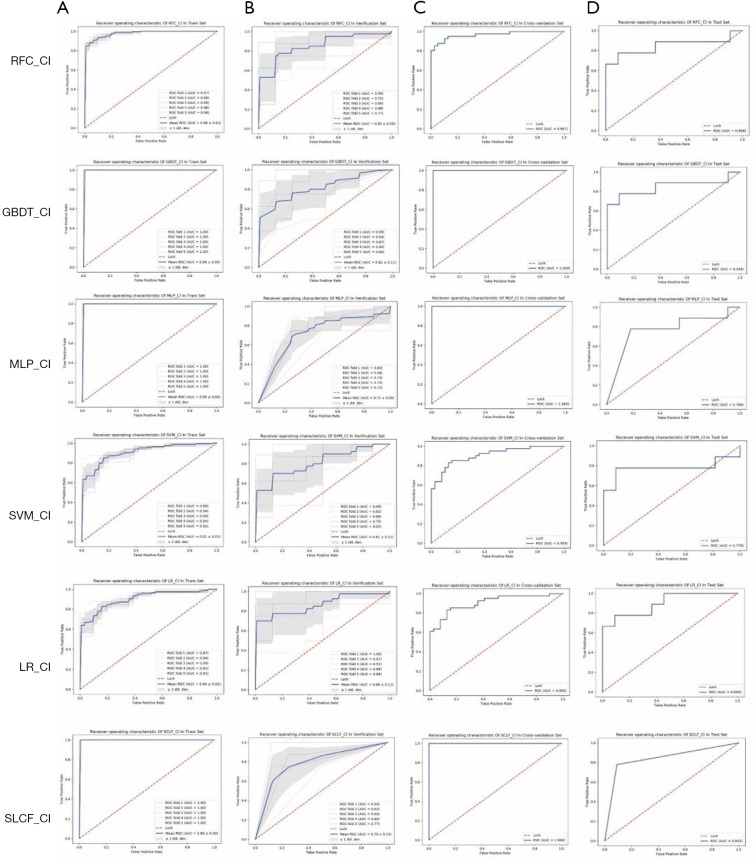
The one-dimensional radiomics feature signature and the clinical information signature were used to build different models as above. The best parameters of the various models are reported in Table 1. The ROC of the cross-validation set and the test set are plotted in Figure 11. The specific AUROC values are reported in Table 3 (part B), and a histogram of the AUROC of each model on the training, verification, and test sets are shown in Figure 9B. Figure 9 indicates that the highest mean AUROC of the verification set is 0.86, and the lowest standard deviation is 0.11; all are from the LR model. The AUROC of the test set is 0.899 (95% CI, 0.761–1.000), which is also the highest.
Figure 11.
ROCs of the training set, verification set, cross-validation set, and test set in different models. ROC, receiver operating characteristic.
The sensitivity, specificity, positive predictive value, and negative predictive value of the training, verification, cross-validation, and test sets for each model with the radiomics feature signature and clinical information signature are reported in Table 6, which shows that the LR model is the best among all models.
Table 6. The sensitivity, specificity, positive predictive value, and negative predictive value of different models with the radiomics feature signature and clinical information signature in the cross-validation dataset and test set.
| Variable | The training set of cross-validation | The validation set of cross-validation | Cross-validation set | Test set | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sen | Spe | Ppv | Npv | Sen | Spe | Ppv | Npv | Sen | Spe | Ppv | Npv | Sen | Spe | Ppv | Npv | ||||
| RFC | 0.88±0.01 | 0.94±0.04 | 0.94±0.03 | 0.91±0.01 | 0.80±0.13 | 0.70±0.15 | 0.75±0.09 | 0.79±0.12 | 0.95 | 0.86 | 0.85 | 0.95 | 0.78 | 0.91 | 0.88 | 0.83 | |||
| GBDT | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 0.82±0.10 | 0.66±0.11 | 0.72±0.08 | 0.79±0.13 | 1.00 | 1.00 | 1.00 | 1.00 | 0.78 | 0.91 | 0.88 | 0.83 | |||
| MLP | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 0.78±0.12 | 0.59±0.14 | 0.67±0.05 | 0.75±0.13 | 1.00 | 1.00 | 1.00 | 1.00 | 0.78 | 0.82 | 0.78 | 0.82 | |||
| SVM | 0.87±0.03 | 0.78±0.06 | 0.81±0.05 | 0.85±0.03 | 0.82±0.13 | 0.65±0.21 | 0.73±0.12 | 0.77±0.17 | 0.81 | 0.84 | 0.85 | 0.79 | 0.78 | 0.82 | 0.78 | 0.82 | |||
| LR | 0.85±0.02 | 0.79±0.04 | 0.81±0.04 | 0.84±0.03 | 0.85±0.09 | 0.75±0.19 | 0.79±0.13 | 0.82±0.13 | 0.81 | 0.84 | 0.85 | 0.79 | 0.78 | 0.91 | 0.88 | 0.83 | |||
| SCLF | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 0.82±0.10 | 0.69±0.10 | 0.74±0.07 | 0.80±0.12 | 1.00 | 1.00 | 1.00 | 1.00 | 0.78 | 0.91 | 0.88 | 0.83 | |||
Model selection and nomogram
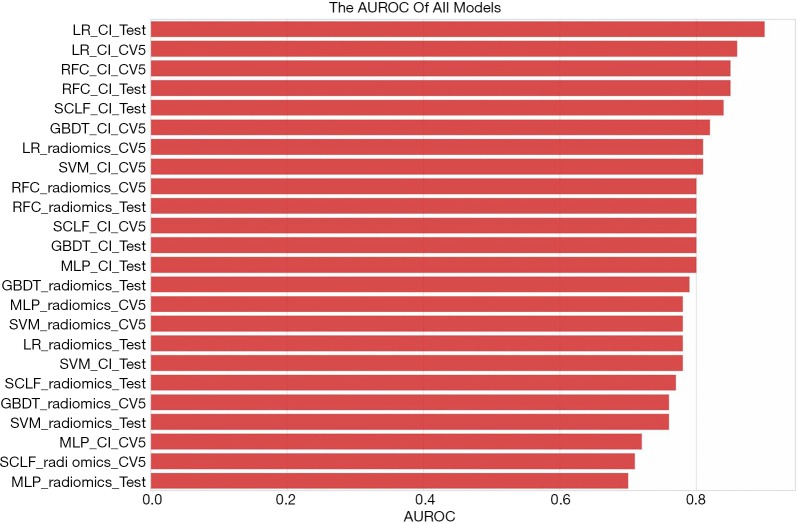
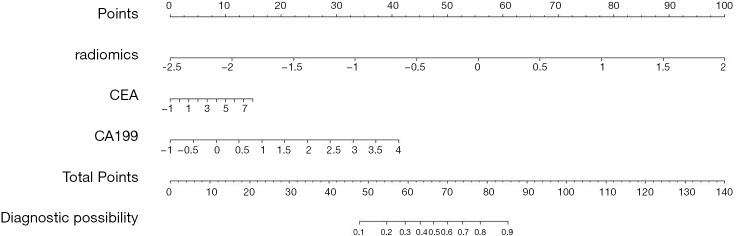
The mean AUROC scores on the verification sets and the AUROC scores on the test sets of all models are shown in Figure 12. As indicated in the figure, the LR model performed the best, followed by the RFC model. The LR_CI model with a radiomics feature signature and clinical information signature achieved the highest AUROC on the test set. The LR_CI model performed significantly better than the LR model only with the radiomics feature signature on the test set (P=0.0239). The nomogram drawn based on the LR_CI model is shown in Figure 13.
Figure 12.
The total AUROCs of different models. Radiomics: models with the radiomics feature signature; CI: models with the radiomics feature signature and clinical information signature; CV5: the AUROC in verification set of 5-fold cross-validation. AUROC, area under the receiver operating characteristic.
Figure 13.
The nomogram based on the LR_CI model. The nomogram critical score is 66; that is, the probability of liver metastasis of colorectal cancer is greater than 50% when the total score is greater than 66. The higher the score is, the higher the probability of colorectal cancer liver metastasis.
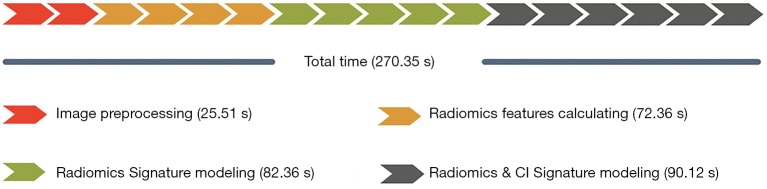
Execution time
We measured the execution time required for the four parts of RIAT; the results are shown in Figure 14. The total execution time was 270.35 s. The entire experiment was performed on a PC with a Windows 10 64-bit operating system equipped with an Intel i7 3.6 GHz CPU (Intel Core i7-7700), 16 GB of memory, and an NVIDIA GeForce GTX 1060 6G graphics card.
Figure 14.
The execution time of the four parts of RIAT described in this paper. RIAT, Radiomics Intelligent Analysis Toolkit.
Discussion
This study developed an individualized nomogram created using RIAT to predict CRLM in patients with CRC based on radiomics applied to CT images. This study provides the following two contributions. (I) To predict the risk of CRLM in CT images, we applied statistics to select features, for instance, using a t-test to preselect radiomics features, and the CRLM nomogram was used to show the prediction of each variable, including radiomics and clinical information signatures. (II) We designed a smart medical platform called RIAT, which can help radiologists conveniently analyze data and build models automatically. RIAT is the first radiomics software to predict CRLM. The main innovations in RIAT include the following: (I) RIAT minimizes the radiomics feature error by optimizing the ROI through image analysis and threshold tuning; (II) RIAT proposes gradient feature selection, which involves first performing preselection using a t-test to evaluate a hypothesis and then using LASSO to select the features. This method effectively retains more highly correlated features; therefore, it can help in constructing a highly accurate model. Benefiting from statistical methods, the selected features have good interpretability. (III) RIAT sets the range of initialized parameters for different models and saves models with high accuracy and strong generalization ability by selecting the best parameters. (IV) RIAT plots the mean ROC of the cross-validation set and calculates the mean accuracy and standard deviation to assess the ROC fluctuation. Finally, it builds a model by training on the complete cross-validation set and plotting the ROC results of the test set to evaluate the generalization ability of the model.
By using RIAT, we developed an individualized nomogram to predict CRLM in patients with CRC, and we have achieved the goal of measuring the radiomics feature signature and clinical information signature according to the presence of CRLM.
At present, radiomics computing methods mainly use MATLAB® (MathWorks Inc., USA), Python, and commercial software such as the Artificial Intelligence Kit, A.K. (General Electric Company) (35). MATLAB is a semi-open-source software package in which viewing and changing the base functions is neither easy nor convenient for researchers. Pyradiomics offers a more comprehensive method for calculating feature values. Some open-source projects (e.g., IBEX and QIFE) have been developed for radiomics (44,45) but offer only a few functions, such as image processing and simple model building.
In this study, we developed RIAT quantitative medical imaging for the prediction of CRLM based on maximum-level enhanced CT images in the portal venous phase. In contrast, RIAT not only provides basic radiomics functions but also enables a new feature selection method and statistics while outputting the evaluation results of models intelligently. In addition to the above advantages, RIAT uses contour recognition and edge acquisition algorithms, median filtering denoising, and binary image transformation to extract the main ROI contour. Then, it determines the best edge through dynamic threshold processing of the RGB channel. In addition, RIAT can independently identify and optimize multiple ROIs in one image at the same time.
In this paper, we introduced some of the functions and algorithms of RIAT for analysis of CRLM. From Figure 12, it can be observed that the LR_CI model has the best performance in the test set and verification set compared with other models. According to Figure 9 and Table 3, the model using both the radiomics features signature and the clinical information signature is better than the model using only the radiomics feature signature. By comparing the mean AUROC (5-fold) on the cross-validation set with the AUROC on the test set in all models, along with the sensitivity, specificity, positive predictive value, and negative predictive value, we found that the LR_CI model achieved the best performance. The improvement is reflected not only by the AUROC score but also by the sensitivity, specificity, positive predictive value, and negative predictive value results. The Delong test was performed on the ROC curves of the nomogram to assess possible overfitting and revealed that the differences were not statistically significant among the AUCs of the cross-validation set and test set in LR_CI, with a P value of 0.9330. From the boxplot and nomogram, we can see that the radiomics feature signature is the most critical weight among all the signatures. Nomograms can effectively help physicians understand the probability of CRC liver metastasis by combining one-dimensional radiomics feature signatures with clinical information signatures.
Although the AUROC scores of the six models provided by RIAT differ, even the lowest AUROC of the model exceeded 0.70. Moreover, the AUROC of each model was significantly improved after adding the clinical information signature. Thus, the data drive the models. Of course, there are no specific models. In RIAT, we included a process that automatically tunes each model parameter. To find the initially optimal parameters for the models, it is necessary to adopt the values when the mean AUROC on the cross-validation set is highest.
To remain objective when evaluating the models, we proposed a double test in RIAT. The double-test approach involves first obtaining the mean AUROC from the cross-validation set and then obtaining the AUROC from the test set using the entire cross-validation set as a training set. The mean AUROC shows the model’s generalizability after performing features selection on the cross-validation set and helps us observe the distribution of data. The standard deviation of the mean AUROC shows the range of fluctuations in the curve. The ROC of the test set further reveals the generalizability and robustness of the final model, and it is tested on data that are not involved in training. The double-test method can conclusively reveal the phenomenon that large fluctuations of ROC are common because of training data limitations. It can also explain the real data distribution and avoid the limitation of having only a single ROC of the test set.
Our study has some limitations. First, only 100 cases met the inclusion criteria, and the sample size was not large. Second, this was retrospective, single-center study, and more center patients are expected to participate in the future. Radiologists have different requirements for different research purposes, such as higher sensitivity or specificity. Therefore, a key problem is how to make the model smarter in order to not only achieve high AUROC scores but also to achieve higher sensitivity and specificity. Sometimes large ROC fluctuations are unavoidable due to the limited amounts of data in radiomics. How to more effectively build optimal models and reduce the ROC fluctuations is an issue that should be addressed by future work.
Conclusions
For predicting CRLM, we found that the LR model with both the radiomics feature signature and the clinical information signature has good predictive ability. The AUROC, sensitivity, specificity, positive predictive values, and negative predictive values of the LR_CI model all exceed those of the other models.
Acknowledgments
Funding: We would like to acknowledge the support of the following organizations: the National Health Commission of the People’s Republic of China (No. 131025000000170001)), the Natural Science Foundation of Jilin Province (No. 20180101038JC), the Science and Technology Development Plan of Jilin Province (No. 20170622009JC), the Provincial and School Joint Construction Project of Jilin University (No. SXGJXX2017-8), the National Natural Science Foundation of China (No. 81871406), the Foundation of Jilin Provincial Department of Finance (No. 2018SCZWSZX-026), the Foundation of Jilin Provincial Department of Finance, Establishment of standardized database for colorectal cancer and exploration of new diagnosis and treatment model based on big data analysis, the Foundation of Health Commission of Jilin Province (No. 2017J073), and Jilin Province Science and Technology Department Science and Technology Innovation Talents Cultivation Program (No. 20180519008JH).
Ethical Statement: This retrospective study has been approved by the ethics committee at the corresponding author’s institution.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018 GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg 2006;244:254-59. 10.1097/01.sla.0000217629.94941.cf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones RP, Brudvik KW, Franklin JM, Poston GJ. Precision surgery for colorectal liver metastases: Opportunities and challenges of omics-based decision making. EJSO 2017;43:875-83. 10.1016/j.ejso.2017.02.014 [DOI] [PubMed] [Google Scholar]
- 4.Nakai H, Arizono S, Isoda H, Togashi K. Imaging Characteristics of Liver Metastases Overlooked at Contrast-Enhanced CT. AJR 2019;212:782-87. 10.2214/AJR.18.20526 [DOI] [PubMed] [Google Scholar]
- 5.Ludwig DR, Mintz AJ, Sanders VR, Fowler KJ. Liver Imaging for Colorectal Cancer Metastases. Curr Colorectal Cancer Rep 2017;13:470-80. 10.1007/s11888-017-0391-4 [DOI] [Google Scholar]
- 6.Sivesgaard K, Larsen LP, Sørensen M, Kramer S, Schlander S, Amanavicius N, Bharadwaz A, Tønner Nielsen D, Viborg Mortensen F, Morre Pedersen E. Diagnostic accuracy of CE-CT, MRI and FDG PET/CT for detecting colorectal cancer liver metastases in patients considered eligible for hepatic resection and/or local ablation. Eur Radiol 2018;28:4735-47. 10.1007/s00330-018-5469-0 [DOI] [PubMed] [Google Scholar]
- 7.Chan A, Hodgson D, Barker J. Liver diffusion magnetic resonance imaging for detecting liver metastasis in rectal and anal cancers. Int J Colorectal Dis 2016;31:1573-75. 10.1007/s00384-016-2591-9 [DOI] [PubMed] [Google Scholar]
- 8.Seo N, Park MS, Han K, Lee KH, Kim MJ. Magnetic resonance imaging for colorectal cancer metastasis to the liver: comparative effectiveness research for the choice of contrast agents. Cancer Res Treat 2018;50:60-70. 10.4143/crt.2016.533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, Aerts HJWL. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE T Syst Man CY-S 1973;SMC3:610-21.
- 12.Yip SSF, Aerts HJWL. Applications and limitations of radiomics. Phys Med Biol 2016;61:R150-66. 10.1088/0031-9155/61/13/R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lubner MG. Reflections on radiogenomics and oncologic radiomics. Abdom Radiol (NY) 2019;44:1959. 10.1007/s00261-019-02047-7 [DOI] [PubMed] [Google Scholar]
- 14.Liang C, Huang Y, He L, Chen X, Ma Z, Dong D, Tian J, Liang C, Liu Z. The development and validation of a CT-based radiomics signature for the preoperative discrimination of stage I-II and stage III-IV colorectal cancer. Oncotarget 2016;7:31401-12. 10.18632/oncotarget.8919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shu Z, Fang S, Ding Z, Mao D, Cai R, Chen Y, Pang P, Gong X. MRI-based Radiomics nomogram to detect primary rectal cancer with synchronous liver metastases. Sci Rep 2019;9:3374. 10.1038/s41598-019-39651-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou Z, Yang Y, Li S, Yan J, Ren W, Liu J, Wang K, Liu B, Wan S. Radiomic analysis using contrast-enhanced CT: predict treatment response to pulsed low dose rate radiotherapy in gastric carcinoma with abdominal cavity metastasis. Quant Imaging Med Surg 2018;8:410-20. 10.21037/qims.2018.05.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Chen H, Qi X, Dou Q, Fu C, Heng P. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation from CT Volumes. IEEE Trans Med Imaging 2018;37:2663-74. 10.1109/TMI.2018.2845918 [DOI] [PubMed] [Google Scholar]
- 18.Roth HR, Lu L, Lay N, Harrison AP, Farag A, Sohn A, Summers RM. Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation. Med Image Anal 2018;45:94-107. 10.1016/j.media.2018.01.006 [DOI] [PubMed] [Google Scholar]
- 19.Avendi MR, Kheradvar A, Jafarkhani H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal 2016;30:108-19. 10.1016/j.media.2016.01.005 [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Zhou M, Liu ZY, Liu ZY, Gu DS, Zang YL, Dong D, Gevaert O, Tian J. Central focused convolutional neural networks: Developing a data-driven model for lung nodule segmentation. Med Image Anal 2017;40:172-83. 10.1016/j.media.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parmar C, Velazquez ER, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, Mitra S, Shankar BU, Kikinis R, Haibe-Kains B, Lambin P, Aerts HJWL. Robust Radiomics Feature Quantification Using Semiautomatic Volumetric Segmentation. PLoS One 2014;9:e102107. 10.1371/journal.pone.0102107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SE, Han K, Kwak JY, Lee E, Kim EK. Radiomics of US texture features in differential diagnosis between triple-negative breast cancer and fibroadenoma. Sci Rep 2018;8:13546. 10.1038/s41598-018-31906-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren J, Tian J, Yuan Y, Dong D, Li X, Shi Y, Tao X. Magnetic resonance imaging based radiomics signature for the preoperative discrimination of stage I-II and III-IV head and neck squamous cell carcinoma. Eur J Radiol 2018;106:1-6. 10.1016/j.ejrad.2018.07.002 [DOI] [PubMed] [Google Scholar]
- 24.Wu L, Wang C, Tan XZ, Cheng ZX, Zhao K, Yan LF, Liang YL, Liu ZY, Liang CH. Radiomics approach for preoperative identification of stages I−II and III−IV of esophageal cancer. Chin J Cancer Res 2018;30:396-405. 10.21147/j.issn.1000-9604.2018.04.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruppert David. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. J Am Stat Assoc 2004;99:567 10.1198/jasa.2004.s339 [DOI] [Google Scholar]
- 26.Fan TW, Malhi H, Varghese B, Cen S, Hwang D, Aron M, Rajarubendra N, Desai M, Duddalwar V. Computed tomography-based texture analysis of bladder cancer: differentiating urothelial carcinoma from micropapillary carcinoma. Abdom Radiol (NY) 2019;44:201-08. 10.1007/s00261-018-1694-x [DOI] [PubMed] [Google Scholar]
- 27.Suh HB, Choi YS, Bae S, Ahn SS, Chang JH, Kang SG, Kim EH, Kim SH, Lee SK. Primary central nervous system lymphoma and atypical glioblastoma: Differentiation using radiomics approach. Eur Radiol 2018;28:3832-39. 10.1007/s00330-018-5368-4 [DOI] [PubMed] [Google Scholar]
- 28.Huang X, Cheng Z, Huang Y, Liang C, He L, Ma Z, Chen X, Wu X, Li Y, Liang C, Liu Z. CT-based Radiomics Signature to Discriminate High-grade From Low-grade Colorectal Adenocarcinoma. Acad Radiol 2018;25:1285-97. 10.1016/j.acra.2018.01.020 [DOI] [PubMed] [Google Scholar]
- 29.Cui LB, Liu L, Wang HN, Wang LX, Guo F, Xi YB, Liu TT, Li C, Tian P, Liu K, Wu WJ, Chen YH, Qin W, Yin H. Disease Definition for Schizophrenia by Functional Connectivity Using Radiomics Strategy. Schizophr Bull 2018;44:1053-59. 10.1093/schbul/sby007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao L, Chen H, Liang M, Li K, Gao J, Qin P, Ding X, Li X, Liu X. Quantitative radiomic model for predicting malignancy of small solid pulmonary nodules detected by low-dose CT screening. Quant Imaging Med Surg 2019;9:263-72. 10.21037/qims.2019.02.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braman NM, Etesami M, Prasanna P, Dubchuk C, Gilmore H, Tiwari P, Pletcha D, Madabhushi A. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res 2017;19:57. 10.1186/s13058-017-0846-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu LC, Park S, Kawamoto S, Fouladi DF, Shayesteh S, Zinreich ES, Graves JS, Horton KM, Hruban RH, Yuille AL, Kinzler KW, Vogelstein B, Fishman EK. Utility of CT Radiomics Features in Differentiation of Pancreatic Ductal Adenocarcinoma From Normal Pancreatic Tissue. AJR 2019;213:349-57. 10.2214/AJR.18.20901 [DOI] [PubMed] [Google Scholar]
- 33.Ren Y, Zhang X, Rui WT, Pang HP, Qiu TM, Wang J, Xie Q, Jin T, Zhang H, Chen H, Zhang Y, Lu HB, Yao ZW, Zhang JH, Feng XY. Noninvasive Prediction of IDH1 Mutation and ATRX Expression Loss in Low-Grade Gliomas Using Multiparametric MR Radiomic Features. J Magn Reson Imaging 2019;49:808-17. 10.1002/jmri.26240 [DOI] [PubMed] [Google Scholar]
- 34.Li W, Huang Y, Zhuang BW, Liu GJ, Hu HT, Li X, Liang JY, Wang Z, Huang XW, Zhang CQ, Ruan SM, Xie XY, Kuang M, Lu MD, Chen LD, Wang W. Multiparametric ultrasomics of significant liver fibrosis: A machine learning-based analysis. Eur Radiol 2019;29:1496-506. 10.1007/s00330-018-5680-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen LD, Liang JY, Wu H, Wang Z, Li SR, Li W, Zhang XH, Chen JH, Ye JN, Li X, Xie XY, Lu MD, Kuang M, Xu JB, Wang W. Multiparametric radiomics improve prediction of lymph node metastasis of rectal cancer compared with conventional radiomics. Life Sci 2018;208:55-63. 10.1016/j.lfs.2018.07.007 [DOI] [PubMed] [Google Scholar]
- 36.Simon RM, Subramanian J, Li MC, Menezes S. Using cross-validation to evaluate predictive accuracy of survival risk classifiers based on high-dimensional data. Brief Bioinform 2011;12:203-14. 10.1093/bib/bbr001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kai C, Uchiyama Y, Shiraishi J, Fujita H, Doi K. Computer-aided diagnosis with radiogenomics: analysis of the relationship between genotype and morphological changes of the brain magnetic resonance images. Radiol Phys Technol 2018;11:265-73. 10.1007/s12194-018-0462-5 [DOI] [PubMed] [Google Scholar]
- 38.Doi K. Overview on research and development of computer-aided diagnostic schemes. Semin Ultrasound CT MR 2004;25:404-10. 10.1053/j.sult.2004.02.006 [DOI] [PubMed] [Google Scholar]
- 39.Doi K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Comput Med Imaging Graph 2007;31:198-211. 10.1016/j.compmedimag.2007.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinto Dos Santos D, Giese D, Brodehl S, Chon SH, Staab W, Kleinert R, Maintz D, Baessler B. Medical students’ attitude towards artificial intelligence: a multicentre survey. Eur Radiol 2019;29:1640-46. 10.1007/s00330-018-5601-1 [DOI] [PubMed] [Google Scholar]
- 41.Liew C. The future of radiology augmented with Artificial Intelligence: A strategy for success. Eur J Radiol 2018;102:152-56. 10.1016/j.ejrad.2018.03.019 [DOI] [PubMed] [Google Scholar]
- 42.Obermeyer Z, Emanuel EJ. Predicting the Future - Big Data, Machine Learning, and Clinical Medicine. New Engl J Med 2016;375:1216-19. 10.1056/NEJMp1606181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collado-Mesa F, Alvarez E, Arheart K. The Role of Artificial Intelligence in Diagnostic Radiology: A Survey at a Single Radiology Residency Training Program. J Am Coll Radiol 2018;15:1753-57. 10.1016/j.jacr.2017.12.021 [DOI] [PubMed] [Google Scholar]
- 44.Zhang Lifei, Fried David V, Fave Xenia J, Hunter Luke A, Yang Jinzhong, Court Laurence E. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys 2015;42:1341-53. 10.1118/1.4908210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Echegaray S, Bakr S, Rubin DL, Napel S. Quantitative Image Feature Engine (QIFE): an Open-Source, Modular Engine for 3D Quantitative Feature Extraction from Volumetric Medical Images. J Digit Imaging 2018;31:403-14. 10.1007/s10278-017-0019-x [DOI] [PMC free article] [PubMed] [Google Scholar]