Abstract
Highly charged, single α-helical (SAH) domains contain a high percentage of Arg, Lys, and Glu residues. Their dynamic salt bridge pairing creates the exceptional stiffness of these helical rods, with a persistence length of more than 200 Å for the myosin VI SAH domain. With the aim of modulating the stiffness of the helical structure, we investigated the effect, using NMR spectroscopy, of substituting key charged Arg, Lys, Glu, and Asp residues by Gly or His. Results indicate that such mutations result in the transient breaking of the helix at the site of mutation but with noticeable impact on amide hydrogen exchange rates extending as far as ±2 helical turns, pointing to a substantial degree of cooperativity in SAH stability. Whereas a single Gly substitution caused transient breaks ∼20% of the time, two consecutive Gly substitutions break the helix ∼65% of the time. NMR relaxation measurements indicate that the exchange rate between an intact and a broken helix is fast (>300,000 s−1) and that for the wild-type sequence, the finite persistence length is dominated by thermal fluctuations of backbone torsion angles and H-bond lengths, not by transient helix breaking. The double mutation D27H/E28H causes a pH-dependent fraction of helix disruption, in which the helix breakage increases from 26% at pH 7.5 to 53% at pH 5.5. The ability to modulate helical integrity by pH may enable incorporation of externally tunable dynamic components in the design of molecular machines.
Significance
Charged single α-helical domains are remarkably robust molecular rods that have been shown to have spring-like properties. They also hold considerable potential for protein engineering. Their dynamic properties, including their persistence length, can be studied at residue-specific resolution by NMR spectroscopy. Whereas helical integrity is found to be rather insensitive to single residue substitutions, double mutations cause transient breaks. By introducing His residues, the helical stiffness can be tuned through pH adjustment in the physiological range.
Introduction
Single α-helical (SAH) domains, stabilized by intramolecular salt bridges between their numerous, positively charged Lys/Arg (K/R) and negatively charged Glu (E) residues, are remarkable structural entities predicted to be present in a significant fraction (∼4%) of proteins (1, 2, 3). Their discovery as independent structural entities was delayed by a dearth of early x-ray crystallographic structural information. In recent years, however, their remarkable structural and spring-like properties, which potentially can make them powerful building blocks in protein engineering, have generated considerable interest in the thermodynamic components stabilizing such entities (4, 5, 6, 7, 8). The sequence of SAH domains typically consists of repeating clusters of four like-charged residues followed by four residues of the opposite charge, referred to as E4(R/K)4 clusters. These four-residue repeats place the oppositely charged side chains one helical turn apart, making it possible for a side chain to form a salt bridge in either the C-terminal or N-terminal direction, adding an entropic component to its stabilization (7,8). Indeed, compelling computational and experimental evidence, supported by the absence of high electron density for most of the SAH salt bridge interactions in x-ray structures, points to the entropy associated with the dynamic network of salt bridges as an important component of the free energy of these helices (1,8). A circular dichroism study of a large array of synthetic peptides containing E4K4 clusters revealed that a minimum of two such eight-residue clusters was needed to approach the maximal molar ellipticity per residue (7). However, insertion of four Ala residues between such eight-residue clusters had no adverse effect on helicity (7), despite restricting the dynamic side chain interactions to intracluster salt bridges, i.e., imposing a modest entropic penalty (8).
As in the synthetic peptide study, natural SAH domains can also include substantial deviations from the canonical E4(R/K)4 sequence pattern. Specifically, Ala, Leu, Met, and Gln residues are also rather common, whereas, intriguingly, the myosin 7a (m7a) SAH domain contains a single His residue at the position (residue 896 for Homo sapiens; residue 897 for Sus scrofa) where a positively charged residue would be expected (9,10). With the protonation state of His residues being a function of the physiological state, this raises the question of whether cellular pH can impact the stiffness of such His-containing SAH helices. In other words, could nature have placed this residue to impart tunable flexibility in the m7a SAH domain for a functional purpose? If pH sensitive, this could confer the ability to easily tune the dynamic properties of otherwise quite rigid helical SAH rods, thereby extending their utility in protein engineering. Furthermore, with the ability to change the His protonation state by the application of hydrostatic pressure (11), SAH stiffness then could become tunable without changing the sample composition.
Charged SAH domains are particularly prevalent in myosins, in which they have been shown to function as extensions of the lever arm (12,13). Detailed studies by a wide variety of methods have been performed on their structural and biophysical characterization. These include small angle x-ray scattering, x-ray crystallography, single molecule pulling, atomic force microscopy, cryo-electron microscopy, circular dichroism, and NMR spectroscopy (2). The latter was long considered too challenging for full-length SAH domains because of their exceptionally poor resonance dispersion (14). This poor dispersion is generally associated with α-helices but strongly exacerbated by the virtual absence of aromatic residues and the dominant presence of only three residue types, Glu, Lys, and Arg, which have nearly indistinguishable 13Cα and 13Cβ α-helical chemical shifts, arranged in a highly repetitive sequence, thereby making resonance assignment by traditional three-dimensional triple resonance technology particularly challenging. A very recent NMR study by Peckham and co-workers focused on m7a, a system reportedly chosen for its slightly less repetitive sequence compared to other myosins but nevertheless posing a formidable challenge for analysis of data collected at high magnetic fields (750 and 950 MHz) (8).
Our study concerns the 68-residue SAH domain of myosin VI. Instead of relying on traditional three-dimensional triple resonance NMR spectroscopy, we previously took advantage of the helical nature of the SAH domain to derive backbone amide assignments by recording four-dimensional (4D) nuclear Overhauser enhancement (NOE) spectra (15). Relying on perdeuteration and transverse relaxation-optimized spectroscopy (TROSY) versions (16) of traditional NMR experiments for line narrowing, as well as advances in nonuniform sampling (NUS) (17,18), such spectra can be recorded relatively rapidly (<24 h) and provide exceptional spectral resolution, directly linking the (1H, 15N) frequencies of one amide, i, with those of its two immediate neighbors, i ± 1, and frequently showing weak crosspeaks to the amides of i ± 2 and i ± 3 as well (15).
The stiffness of this SAH domain can be evaluated in a site-specific manner by attaching a paramagnetic tag near the N- or C-terminal end of the helix (15). This tag weakly aligns the SAH domain relative to an external magnetic field, and the measurement of the residual dipolar couplings (RDCs) then provides a highly sensitive, site-specific probe for the local SAH alignment as a function of distance from the tagging site (19,20).
Here, we use this RDC technology to evaluate the effect of two types of substitutions on the stability of the S. scrofa SAH domain of myosin VI. First, we measure the effect of substituting Arg-32 and Lys-33 residues by Gly, often considered a “helix-breaking” residue. Whereas the introduction of two adjacent Gly residues indeed significantly disrupts the SAH domain, a single R32G mutation only introduces minimal transient breaks in the SAH domain despite giving rise to large increases in the hydrogen exchange (HX) rates between backbone amides and solvent. Second, we explore the effect of mutating two adjacent, negatively charged residues to His and the effect on helix stability when the sample pH is above and below the pKa of these His residues.
Methods
Protein expression and purification
The cloning details and design of a fusion protein comprising the IgG-binding domain of protein G (GB1) and a Tobacco Etch Virus protease (TEV) cleavage site, followed by the myosin VI SAH domain, were described previously (15). In brief, the complementary DNA of GB1-TEV-SAH was cloned with its N-terminal end in frame with the native 6xHis tag of the pET24a vector, supplied by Genscript (Piscataway, NJ). This plasmid, as well as a New England Biolabs Site-Directed Mutagenesis kit (Ipswich, MA), was then used to prepare the I13C, I13C/R32G, I13C/R32G/K33G, and I13C/D27H/E28H mutants of the SAH domain (Fig. 1 A). Expression and purification of these GB1-TEV-SAH domains was performed as described previously (15), except for the use of high-performance liquid chromatography as the final purification step immediately after the TEV cleavage reaction. For this high-performance liquid chromatography step, an Agilent Prep-C18 Scalar 5-μm 4.6 × 250 mm column (Santa Clara, CA), along with solvent A (H2O + 0.1% trifluoroacetic acid) and solvent B (acetonitrile + 0.1% trifluoroacetic acid), was used to separate and isolate the SAH domain using a 1.5% per min gradient of solvent B from 0 to 70%. The SAH-containing fractions were then flash frozen in liquid N2 and lyophilized. All samples were perdeuterated and 15N labeled by using 99% D2O (Cambridge Isotope Laboratories, Tewksbury, MA) in place of H2O in the M9 expression media with 15N-labeled ammonium chloride and 2H-labeled D-glucose as the sole sources of nitrogen and carbon, respectively.
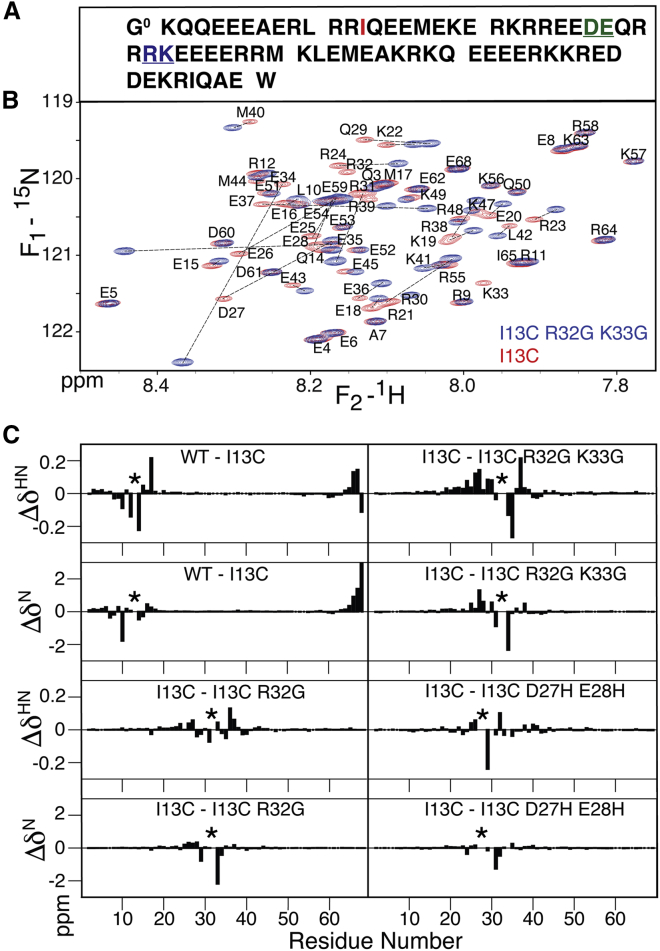
Figure 1.
NMR analysis of myosin VI SAH domain. (A) A sequence with color marking for mutated sites is shown here. Residue K1 corresponds to K918 in S. scrofa myosin VI. (B) Superposition of 900 MHz 1H-15N TROSY-HSQC spectra of paramagnetically tagged WT (red) and R32G/K33G (blue) spectra is shown here. (C) 1HN and 15N chemical shift differences, Δδ, of WT − I13C, I13C − I13C/R32G, I13C − I13C/R32G/K33G, and I13C − I13C/D27H/E28H are shown here (Table S1). Here, “WT” refers to the WT construct that lacked W69, and all I13C-containing constructs include the C-terminal W69 addition. Locations where the compared sequences differ are marked by asterisks. To see this figure in color, go online.
For tagging of the SAH domain with 4R,4S-1,4,7,10 tetraaza-cyclododecane-tetraacetic acid-M8-Tm (DOTA-M8-Tm) or S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL), each lyophilized I13C mutant of SAH was fully dissolved in 100 mM Tris-HCl (pH = 7.5) at a concentration of ∼200–400 μM. Then, a molar excess (10×) of dithiothreitol to SAH was added to the solution, followed by a 30-min incubation at room temperature to ensure full reduction of Cys. Next, an appropriate aliquot of this solution, sufficient to yield a 100- to 200-μM NMR sample of DOTA-M8-Tm/MTSL-tagged SAH, was buffer exchanged into 100 mM Tris-HCl (pH 7.5) using a PD10 G25 column (GE Healthcare, Chicago, IL). Specifically, the protein-containing eluent from the PD10 column, at a concentration of ∼15–20 μM, was collected into a 10-mL Falcon tube containing sufficient stock DOTA-M8-Tm/MTSL at a concentration of ∼100 or 200 μM to create a reaction mixture with ∼5- to 10-fold molar excess of DOTA-M8-Tm/MTSL to SAH. This reaction solution was then incubated at room temperature in the dark, overnight, before being concentrated using a 3-kDa cutoff Amicon filter (Millipore, Burlington, MA) and added to another PD10 G25 column to simultaneously buffer exchange into the desired NMR buffer and remove excess DOTA/MTSL tag. The NMR buffer contained 20 mM sodium phosphate, 2 mM EDTA, 2% D2O at pH 5.5, 6.3, or 7.5 for the I13C D27H E28H-tagged samples and pH 6.3 for the I13C- and R32/K33G-tagged samples. Liquid chromatography-mass spectrometry was used to verify the identity and purity of the MTSL/DOTA-M8-Tm-tagged SAH. No unlabeled SAH or unbound DOTA/MTSL was observed in this analysis. To make the diamagnetic MTSL sample, a 10-fold molar excess of sodium ascorbate (at pH 6.3) was added to the pH 6.3 sample of I13C-MTSL SAH (or I13C-MTSL R32G/K33G SAH) and incubated for 30 min at room temperature before recording the diamagnetic 1H transverse relaxation rates.
NMR spectroscopy
All NMR experiments were performed on a 900-MHz 1H frequency Bruker Avance-III spectrometer (Billerica, MA) equipped with a z-gradient TCI cryogenic probe. For resonance assignment of the mutant SAH domains (I13C/R32G, I13C/K33G/R32G, I13C/D27H/E28H), 4D 1H-15N-15N-1H HMQC-NOESY-TROSY-heteronuclear single quantum coherence (HSQC) spectra with NUS, as previously described (15), were acquired on each mutant in 20 mM NaPi, 2 mM EDTA, 1 mM tris(2-carboxyethyl)phosphine, and 2% D2O (pH 6.3) at 20°C. Each 4D spectrum required 12–24 h of data collection. Reconstruction of the NUS spectra was performed using the SMILE program (18). Stepwise titration with pH and/or temperature was then used to assign resonances at pH values other than 6.3 and at temperatures other than 20°C. All NMR data were extensively zero filled, processed, and initially analyzed with NMRPipe software (21), whereas Sparky (22) and nmrglue (23) were used for further analysis.
RDC measurements of DOTA-M8-Tm and bacterial phage (Pf1)-aligned samples (100–200 μM) were carried out at 900 MHz 1H frequency using the two-dimensional TROSY-based ARTSY experiment (24). The DOTA-M8-Tm tag was kindly synthesized and provided to us by Dr. D. Haussinger (Biozentrum, Basel, Switzerland) and linked to residue C13 of the various SAH mutants, as described above. The Pf1 phage was purchased as a 50 ± 4 mg/mL stock solution from ASLA Biotech (Riga, Latvia). Because of the small variability in the noted concentration and volume errors introduced by pipetting the viscous Pf1 solution, the reported Pf1 concentrations are taken from the deuterium 2H lock splitting in the aligned sample, i.e., p Hz splitting corresponds to p mg/mL Pf1 (25).
Paramagnetic relaxation enhancement (PRE) rates of the 1HN signals were measured as described by Clore and Iwahara (26) from the difference in Hahn-echo 1H TROSY transverse relaxation rates between the paramagnetic and reduced states of the MTSL spin label of the perdeuterated sample.
HX and 15N TROSY T2 experiments were performed on 100–200 μM samples as previously described (15). The sample buffer contained 20 sodium phosphate, 2 mM EDTA, 1 mM tris(2-carboxyethyl)phosphine, and 2% D2O. The 15N TROSY T2 experiments were performed at pH 6.3. For the HX experiments, the pH meter readings introduced small uncertainties in the actual sample pH. Because of the high sensitivity of HX rates to pH, the 1J(31P-1H) splitting of a small amount of added sodium phosphite (1 mM) was used as an internal pH reference (27).
Results and Discussion
The previously introduced I13C mutation in the medial tail domain of myosin VI, comprising its SAH, was used for this study, in which C13 serves as the site of attachment for a thulium-chelating DOTA-M8 tag (20). Here, we use the residue numbering of the expressed construct (Fig. 1 A), with G0 being a non-native remnant of the TEV cleavage site and W69 a non-native C-terminal residue added to facilitate concentration measurements. The 900 MHz 1H-15N TROSY-HSQC spectrum of the perdeuterated construct shows the narrow resonance dispersion associated with SAH domains but also highlights that, at such high magnetic fields, the vast majority of resonances can be resolved (Fig. 1 B). Use of high-field NMR, therefore, provides access to detailed studies of the structure and dynamics of such SAH domains (8,15).
Effect of I13C on SAH structure
Comparison of chemical shifts between the wild-type (WT) sequence and the I13C mutant (Fig. 1 C) shows that, with the exception of L10, the 15N chemical shift differences are very small. Similarly, 1HN shift differences are modest and localized around the site of mutation. This result indicates that, except for residues immediately surrounding the mutation site, the local structure of the helix remains unchanged. The relatively large change in L10 15N shift may be caused by a decrease in the H-bond strength between mutated residue I13 and its acceptor carbonyl of R9, which is known to have an inductive effect on the 15N shift of the same peptide bond moiety. Weakening of the H-bonds at the site of mutation is also reflected in the amide HX rates with solvent, which increase by more than an order of magnitude at the mutated site relative to the WT sequence (open black symbols in Fig. 2 A). As discussed previously, it is not straightforward to interpret HX protection factors of the SAH domain quantitatively in terms of the fraction of broken H-bonds because HX in a transiently broken helix is impacted by the electric dipole moments of its N- and C-terminal halves (15). However, they clearly point to the transient, local disruption of the α-helical H-bond network in the immediate vicinity of the mutation site, leading to some increased mobility of the N-terminal 13-residue segment relative to the remainder of the SAH domain, a conclusion supported by 15N TROSY transverse relaxation measurements (vide infra) and 4D NOESY data on the I13C/R32G/K33C construct (Fig. S1).
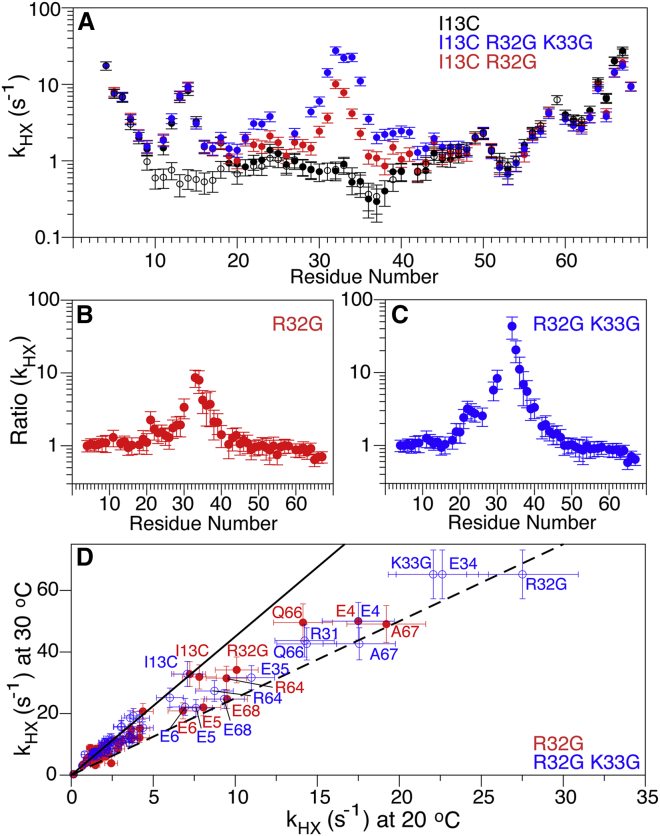
Figure 2.
Comparison of amide HX rates in the Ψ-WT SAH domain of myosin VI and its R32G and R32G/K33G mutants. Values measured at pH values close to 7.4 (Table S2) were normalized to pH 7.4 by multiplying the measured values by 10(7.4 − Q), where Q is the actual sample pH (|7.4 − Q| < 0.15) at 20°C. (A) HX rates in the Ψ-WT (black) and R32G (red) and R32G/K33G (blue) mutants of the Ψ-WT are shown here. Open symbols refer to WT lacking the I13C substitution and W69. (B and C) Ratios of HX rates in R32G (B) and R32G/K33G (C) mutants over those in the Ψ-WT are shown here. (D) Temperature dependence of HX rates in the R32G (red) and R32G/K33G (blue) mutants of the Ψ-WT is shown here. The solid black line (slope of 4.5) corresponds to the previously fitted activation energy for HX of 113 kJ/mole in WT-SAH (15); the dashed line corresponds to the activation energy for HX of non-H-bonded amides in a random coil (71 kJ/mole; slope 2.5) (30). HX rates are listed in Table S3. Acquisition parameters are listed in Table S5. To see this figure in color, go online.
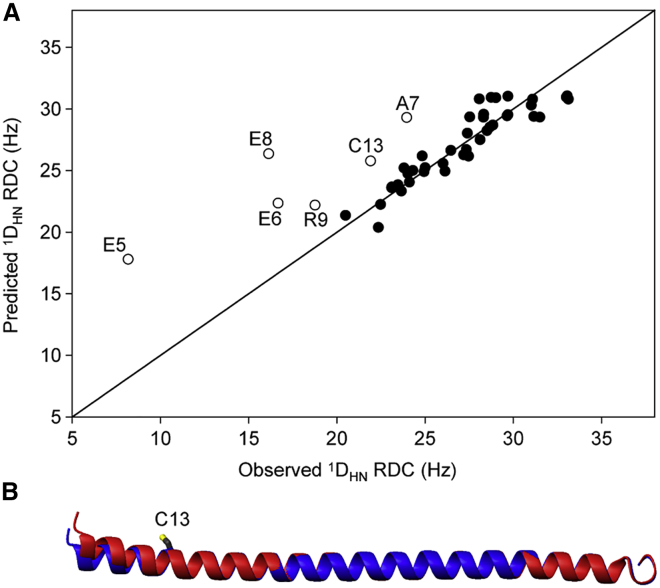
The impact of the I13C mutation on the ensemble-averaged helical structure can be visualized by comparing 1DHN RDCs measured in Pf1 alignment medium for the WT sequence and the I13C mutant, which we refer to as pseudo-WT (Ψ-WT). The largest differences are observed for the N-terminal residues (Fig. 3 A). Although a single set of 1DHN RDCs is insufficient to calculate a de novo structure for the Ψ-WT, the RDC data can readily be used to carry out a rigid body refinement. This approach keeps the structures of the N-terminal (residues K1–R11) and the C-terminal (E15–R64) segments frozen to those of the original WT coordinates (Protein Data Bank, PDB: 6OBI) while harmonically restraining the ϕ and ψ angles of the intervening residues to those of the 6OBI structure using a weak force constant. Such a rigid body refinement procedure reveals a small, ∼6° change in average orientation of the N-terminal segment helix axis relative to the remainder of the SAH domain, in the direction of the missing, sterically bulky side chain of I13 (Fig. 3 B).
Figure 3.
Effect of I13C mutation on the average structure of the S. scrofa myosin VI SAH domain. (A) Predicted versus observed 1DNH RDCs for the I13C mutant are shown here, using singular value decomposition fitting of the experimental RDCs of residues E15–R64 to the previously determined structure for the WT sequence (Protein Data Bank, PDB: 6OBI) to obtain the alignment tensor. Predicted RDCs for the N-terminal residues are depicted by open symbols. RDCs were measured at 20°C, 900 MHz, in a solution containing ∼11 mg/mL Pf1 and, as mentioned previously (15), were scaled by (R2,max/R2)1/2, where the scaling accounts for the increased dynamics (decreased generalized order parameter, S) when moving toward the termini of the helix. With a Q factor of 9%, the RDCs of residues E15–R64 fit well to those of the 6OBI coordinates. (B) Superposition of the backbone ribbon of the previously determined WT sequence (6OBI, blue) and the I13C mutant (red) is shown, obtained by rigid body refinement, leaving the backbone torsion angles of R12-Q14 harmonically restrained by a weak force constant to their WT values. To see this figure in color, go online.
Impact of R32G and K33G mutations on SAH structure
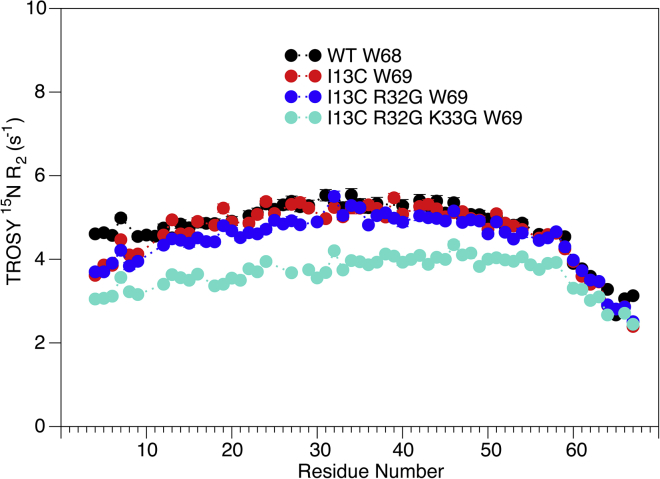
The effect on resonance assignments caused by two Gly substitutions (R32G/K33G) or the effect of the I13C substitution relative to the NMR assignments of the WT spectrum (15) is not easily tracked by simple inspection of the HSQC spectrum (blue resonances in Fig. 1 B) and required the recording of a separate 4D HMQC-NOESY-TROSY-HSQC spectrum (Fig. S1). For the N-terminal half of the triply mutated (I13C/R32G/K33G) SAH domain, the NOE crosspeak/diagonal peak intensity ratios in this 4D spectrum are systematically ∼20–30% weaker than previously observed for the WT sequence (Fig. S1). This decrease correlates with lower 15N transverse relaxation rates (Fig. 4) and is attributed to the flexibility introduced by the I13C mutation. Indeed, even for sequences with only a single or no Gly substitutions near the middle of the domain, a decrease in 15N transverse relaxation rates is observed for the residues preceding the I13C mutation (Fig. 4). For the second half of the triply mutated helix, NOE crosspeak/diagonal peak ratios are also lower than those of the WT sequence, but by a smaller amount, indicating that the structural integrity of this C-terminal half is little impacted by the R32G/K33G mutations, as foreshadowed by the similarity of its chemical shifts to those of the WT sequence (Fig. 1 C). As shown by simulations, the buildup of the ratio of crosspeak/diagonal peak intensity as a function of mixing time for NOEs between sequential amides remains linear for much longer than the buildup of NOE crosspeak intensity (15). This ratio scales with the rHH−6 and, therefore, represents a quite precise measure for interproton distances, with a 25% change in the ratio corresponding to a change of only ∼0.1 Å in interproton distance. The gradual decrease in the crosspeak/diagonal peak ratio when moving toward the termini of the helix mirrors the decrease in 15N transverse relaxation rate, therefore reflecting a decrease in J(0) spectral density and not an increase in rHH.
Figure 4.
TROSY Hahn-echo R2 values versus residue number for WT (black), Ψ-WT (I13C/W69; red), R32G (blue), and R32G/K33G (aqua) mutants of the myosin VI SAH domain. R2 values are listed in Table S2. To see this figure in color, go online.
A key question is whether the Gly substitutions simply introduce flexibility in the helical rod, i.e., reduce the local stiffness, or whether they cause transient breaks in the helix where the Gly residues simply act as a very short disordered linker. In the first scenario, the absence of a Gly side chain removes steric clashing, thereby allowing increased bending (i.e., lower helical axis rigidity) in the direction of the original side chain(s) of R32 and K33. This would result in a small kink in the average helical axis, in addition to the increased flexibility. In the second scenario, the SAH domain rapidly switches between two states: (A) intact and straight and (B) broken with a highly flexible, short-linker segment and no significant force to restore the helical H-bonding pattern. As discussed below, our experimental NMR data, and in particular the RDCs, HX, and PRE data, indicate that this latter case applies.
We first focus on the I13C SAH domain with a single internal Gly (R32G) mutation. By attaching a paramagnetic lanthanide tag at the I13C mutation site using a stereospecific linker (20), the N-terminal half of this SAH domain becomes oriented identically relative to the magnetic field, as the corresponding half of the paramagnetically aligned Ψ-WT sequence. We note that the alignment force exerted by the paramagnetic tag is about three orders of magnitude below thermal energy (kT) and, therefore, does not detectably alter the structure of the domain. In the vicinity of the tagging site, residues A7–R24 are paramagnetically broadened beyond detection by the Tm metal ion but, to within experimental uncertainty, residues immediately preceding and after this region indeed show the same RDCs for the mutated WT and the Ψ-WT SAH domain (Fig. 5 A). Comparison of RDCs between the Ψ-WT and R32G mutant for residues C-terminal of the mutation site yields a near-perfect linear correlation with a slope of 0.80 (Fig. 5 B). The latter confirms that the phase and amplitude of the dipolar wave pattern (28), exponentially decaying with the previously determined persistence length decay constant of 49 residues (15), is not significantly altered by the R32G mutation. Therefore, the time-averaged orientation of this segment is essentially the same as in the Ψ-WT structure but with an alignment strength that is reduced by 20%. Qualitatively, this result is consistent with the sharp increase in HX rates relative to the Ψ-WT sequence for residues in the immediate vicinity of the mutation site. The amide HX rate for G32 is ∼10-fold faster than in the WT SAH domain and approximately sevenfold slower than its random coil value (29,30). The latter suggests that its H-bond to the carbonyl oxygen of E28 is broken for an ∼15% fraction of time. However, as mentioned previously (15), quantitative interpretation of HX protection factors in SAH domains appears adversely impacted by the observation that the HX rate at the position of a helix break may differ from its canonical random coil HX rate, adding substantial uncertainty to this 15% fraction.
Figure 5.
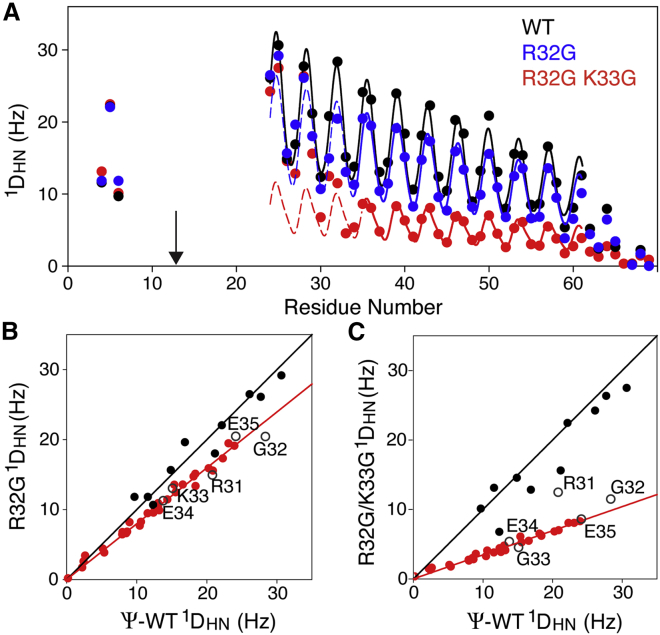
Effect of Gly mutations on 1DNH RDCs in the myosin VI SAH domain, which is paramagnetically aligned by 4R,4S-DOTA-M8 and loaded with Tm (20). The measurements used are pH 6.3, 20°C, and 900 MHz. (A) RDCs of the (I13C, W69)Ψ-WT (black), with corresponding couplings for its R32G mutant in blue and R32G/K33G mutant in red, are shown here. Solid lines correspond to the best-fitted dipolar wave patterns (28) expected for an idealized α-helix, multiplied by a decaying exponential function with a decay constant of 49.75 residues, previously derived from the persistence length of ∼149 residues. The arrow marks the location of the paramagnetic tag. (B) RDCs of the R32G mutant versus those of the Ψ-WT are shown here. RDCs of observable amides E4–R30 are shown as solid black symbols (R2 = 0.93), E36–E68 are shown as red symbols (R2 = 0.99), and R31–E35 are shown as open black symbols. (C) RDCs of the R32G/K33G mutant versus those of the Ψ-WT (R2 = 0.88 for E4–R30; R2 = 0.97 for E36–E68) are shown here. In (B) and (C), the slope for the RDCs of the N-terminal residues is 1.0; the slopes for residues E36–E38 are 0.80 (B) and 0.35 (C). All RDCs are included in Table S4. To see this figure in color, go online.
Whereas the single R32G mutation only decreases the alignment of the C-terminal half by ∼20% relative to the Ψ-WT, the double Gly mutation (R32G/K33G) causes a much larger disruption and decreases alignment of the C-terminal half by ∼65% (Fig. 5 C). Again, the correlation between RDCs of residues E36–E62 in the Ψ-WT and R32G/K33G mutant remains very tight, indicating that for an ∼35% fraction of time, the helix remains intact, without significant kinking. HX rates at the R32G/K33G sites are about threefold higher than in the R32G mutant (Fig. 2, A–C), consistent with a higher fraction of broken α-helical H-bonds. Interestingly, experimentally measured HX rates for E34 and E35 substantially exceed their intrinsic random coil values (29) (Table S3), confirming our prior conclusion (15) that care needs to be exercised with the quantitative interpretation of these SAH domain’s HX protection factors.
For a solvent-exposed, not intramolecular H-bonded amide, the activation energy for OH−-catalyzed HX with solvent is ∼71 kJ/mole. The ratio of HX rates measured at 30°C and at 20°C then should be ∼2.5, as is indeed observed for the most N- and C-terminal residues (Fig. 2 D). The activation energy for breaking H-bonds in the well-ordered part of an SAH helix was previously measured to be ∼42 kJ/mole (15), and correspondingly, their HX rates at 30°C are ∼4.5 times faster than at 20°C (solid line in Fig. 2 D). The HX rate for G32 in the R32G mutant falls midway in between the solid and dashed lines in Fig. 2 D, indicating that it is H-bonded ∼50% of the time. For the R32G/K33G mutant, HX rates of residues R31–E35 show a temperature dependence that approaches that of a random coil (dashed line in Fig. 2 D), indicating that they are engaged in α-helical H-bonds for only a modest fraction of time, which is in agreement with the above conclusion derived from the RDC analysis.
PRE of R32G/K33G helix
Attachment of a nitroxide spin label (MTSL) to C13 of the Ψ-WT sequence can be used to measure PRE for any hydrogens that transiently come into close proximity of the MTSL label (26). The enhancement of transverse relaxation scales with the inverse sixth power of the distance between the unpaired electron and nuclei of interest, allowing static effects to be measured for distances of up to ∼25 Å in perdeuterated proteins. As a result of the r−6 scaling of the PRE effect, even a fraction as low as 0.4% with a transient electron-1H distance of 10 Å should become detectable by PRE.
Although the PRE effect is often measured qualitatively from the intensity of resonances in a 1H-15N HSQC spectrum, quantitative measurement can be carried out by encoding the PRE rates in the initial magnetization transfer delay of the HSQC experiment (26). The difference in 1H transverse relaxation rates measured in the presence of paramagnetic and reduced MTSL then corresponds to the PRE. For the Ψ-WT SAH domain, this PRE steeply decreases with distance from the tagging site at C13 (Fig. 6). The absence of any significant PRE effect in the C-terminal half of the Ψ-WT SAH domain is consistent with its large persistence length and excludes the presence of even a small fraction of species with a transient break that would give rise to a detectable PRE.
Figure 6.
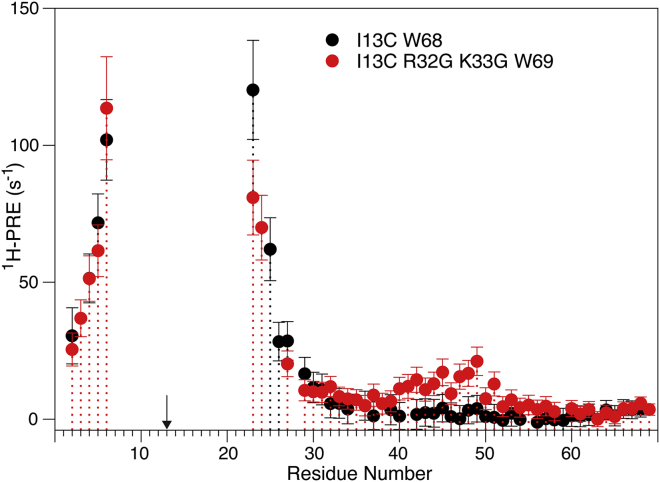
Comparison of amide 1H PRE in MTSL-tagged Ψ-WT SAH domain of myosin VI (black) and its R32G/K33G double mutant (red) at 900 MHz, 20°C. The enhancement seen in the region of residues M40–E51 indicates transient proximity to the MTSL label, consistent with transient breaks in the helix at G32/G33. PRE rates were measured as described by Clore and Iwahara (26) and represent the difference in Hahn-echo 1H T2 between the paramagnetic and reduced states of the spin label. The arrow marks the location of the paramagnetic tag. To see this figure in color, go online.
By contrast, substantial long-range PREs are seen for the R32G/K33G mutant (Fig. 6); whereas for residues preceding the mutation site, the PRE in the double mutant is the same as for the Ψ-WT sequence, a noticeable PRE is seen for residues M40–E51. The diffuse distribution of the PRE and the absence of any helical periodicity in the PRE magnitude for M40-E51 points to random transient positioning of the MTSL in the vicinity of these amides rather than a distinct, well-defined kink that would have brought sections of the helix transiently together in a specific manner. These PRE data therefore support the model of transient breaks in the SAH domain at or near the site of the R32G/K33G mutation, where the break acts as a short highly flexible covalent linker.
Kinetics of helix disruption
The timescale of the H-bond breaking process can be accessed by transverse relaxation (R2) measurement of the TROSY component of the 15N spin magnetization. Measurement of the R2 of the slowly relaxing TROSY component by simple Hahn-echo experiments provides a very sensitive probe for the presence of conformational exchange contributions, Rex, to the 15N R2 rate (31) and can be used to probe perturbations of the 15N chemical shift resulting from transient H-bond breaks. The presence of a single sharp resonance for each backbone amide in the TROSY-HSQC spectrum of the R32G/K33G mutant indicates that such processes are in the fast exchange limit, i.e., Δω τex « 1. Here, Δω is the angular 15N chemical shift difference (in units of radians per second) between open and H-bonded states, and τex is the timescale of the exchange process: τex = 1 / (ko→c + kc→o), with ko→c and kc→o the rate constants at which H-bonds close and open, respectively. The Rex contribution to the 15N R2 relaxation rate equals
| (1) |
where p = kc→o/(ko→c + kc→o) denotes the open-state fractional population.
Measurements of the TROSY Hahn-echo 15N R2 relaxation rates, which includes the Rex contribution, were carried out at high field (900 MHz 1H frequency) to maximize Rex, whereas the intrinsic TROSY R2 rate is close to its minimum at this magnetic field strength of 21 T (16), thereby maximizing the fractional contribution of Rex to the measured relaxation rates. Considering the large variations in intrinsic HX rates, which apply for an unfolded, coil-like structure of the same sequence (30), 15N R2 rates measured for the WT, Ψ-WT, and Ψ-WT R32G mutants show remarkably little variation over the entire range from Q14-K63 (Fig. 6). Compared to the WT sequence, SAH domains that contain the I13C mutation show a small decrease in TROSY R2 for the N-terminal segment preceding this mutation, an effect attributed to flexibility in the helix axis caused by the I13C mutation. Small differences seen for the two C-terminal residues are caused by the Ψ-WT constructs extending the sequence by one residue relative to the WT sequence. For the center part of the SAH domain, Q14-K63, TROSY R2 rates are quite homogeneous. At an average value of ∼5 s−1, these rates are approximately sevenfold lower than the previously reported HSQC 15N R2 rates (15).
For the Ψ-WT R32G/K33G construct, a modest decrease in R2,TROSY values is observed, attributed, as discussed above, to the destabilization of the helical integrity by the double mutation. The homogeneous nature of the R2,TROSY values excludes the presence of conformational exchange contributions, Rex, larger than ∼0.5 s−1. Rex contributions (Eq. 1) scale with the square of the chemical shift difference between intact and broken helical states and can be estimated from the difference in chemical shifts between the Ψ-WT and the Ψ-WT R32G/K33G constructs. The largest changes are ∼1 ppm and correspond to the difference between a fully helical state and a state that is rapidly averaging between a one third fraction of intact helix and a two thirds fraction of broken helix. The differences in 15N chemical shift between intact and broken helical states, therefore, reach values of ∼1.5 ppm or 850 radians per second. With p ≈ 0.65 and Rex ≤ 0.5 s−1, Eq. 1 yields an upper limit τex ≤ 3 μs.
pH sensitivity of helical integrity by D27H/E28H mutation
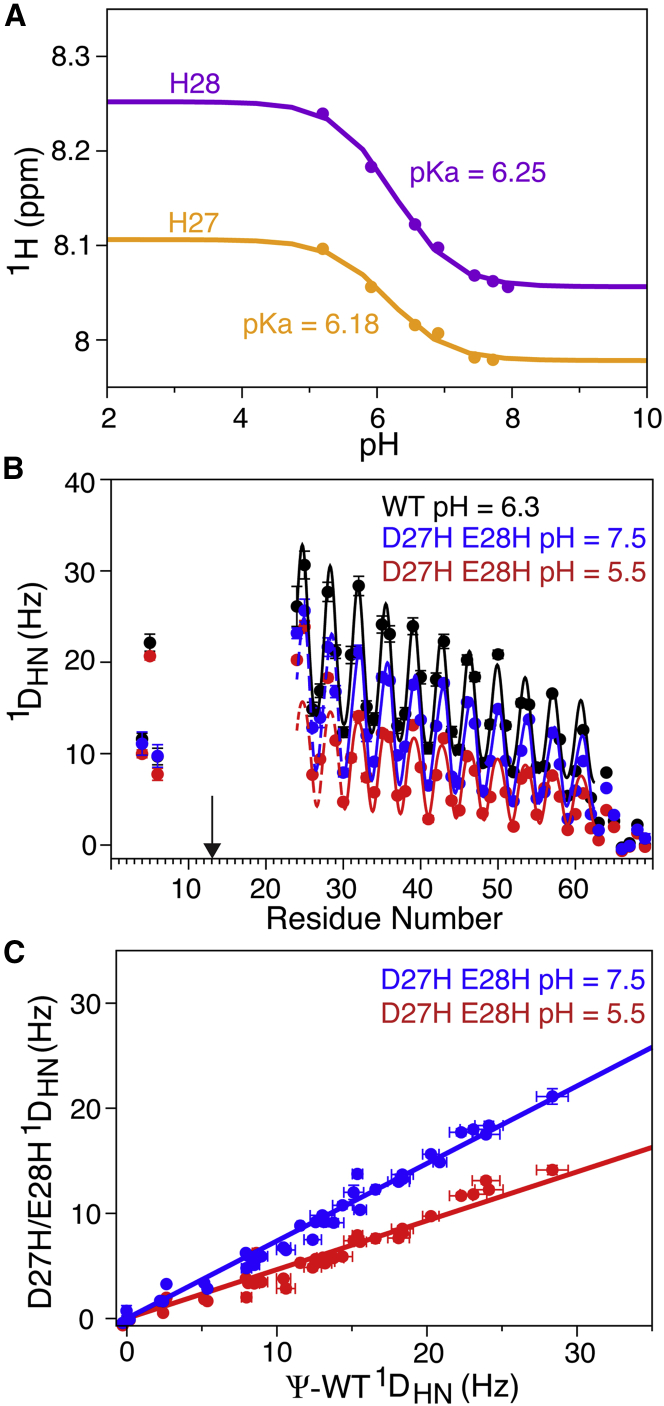
With the aim of controlling the SAH domain integrity by external parameters such as pH or hydrostatic pressure, we substituted two adjacent, negatively charged residues by His. The protonation state of His can be modulated by pH and slightly depressed pKa values are observed for H27 and H28 (Fig. 7 A). Elevated pKa values are observed when introducing R → H or K → H mutations, but these proved less effective at modulating helical integrity (data not shown). Protonation of His can also be promoted by hydrostatic pressure, in particular when combined with a pressure-sensitive buffer such as phosphate, which decreases solvent pH by nearly one unit when raising the sample from atmospheric pressure to 3 kbar (11).
Figure 7.
pH dependence of the helical stiffness of the D27H/E28H double mutant of the Ψ-WT SAH domain of myosin VI, paramagnetically tagged at I13C. (A) Shown here is the 1HN chemical shift titration of H27 (yellow) and H28 (magenta), with the solid lines representing the best fits to the Henderson-Hasselbalch equation. (B) RDC versus residue number in the Ψ-WT (black) and D27H/E28H mutants at pH 7.5 (blue) and pH 5.5 (red) is shown here. Solid lines correspond to the best-fitted dipolar wave patterns (28) for residues R24–E62, multiplied by a decaying exponential function with a decay constant of 49.75 residues. For the D27H/E28H mutants, only RDCs for residues R30–E62 were included in the dipolar wave fit; continuation of the dipolar wave in the N-terminal direction is depicted by dashed lines. The arrow marks the location of the paramagnetic tag. (C) The correlation between 1DNH RDCs of residues R30–E68 in the D27H/E28H double mutant and Ψ-WT at pH 7.5 (blue, slope 0.74; R2 = 0.985) and pH 5.5 (slope 0.47; R2 = 0.976) is shown here. To see this figure in color, go online.
Again, helical persistence is evaluated by comparing alignment of the C-terminal half of the SAH domain with that of the Ψ-WT, where for both samples, the N-terminal segment is aligned identically by the DOTA-M8-Tm paramagnetic tag attached to C13. As can be seen, RDCs for residues C-terminal of the mutation site are uniformly smaller than in the WT by a factor of 0.74 at pH 7.5 and by a factor of 0.47 at pH 5.5, at which both His residues are largely protonated.
Clearly, at alkaline pH, the His substitutions are less disruptive than the above discussed mutations to Gly. Counter to expectations, however, when lowering the sample pH to 5.5, at which both His residues become positively charged, the SAH helix remains intact nearly 50% of the time. So, even though in this case, two sequential, canonical negatively charged residues are substituted by positively charged His, the SAH helical integrity is only partially destroyed.
Conclusions
Our results confirm that SAH domains are remarkably resilient and not easily disrupted by even quite drastic substitutions. Introduction of a single Gly residue, often considered a helix breaker, had minimal impact on the helical integrity of the SAH domain and caused a transient disruption for at most a 20% fraction of time. Even with two adjacent Gly substitutions, the SAH domain remained intact for about one-third of the time. Somewhat unexpected was the relatively large effect of substituting Ile-13 by Cys, a mutation needed to introduce an anchoring site for the paramagnetic alignment and MTSL tags. Whereas β-branched residues are normally entropically disfavored in α-helices where, for steric reasons, they can adopt only a single χ1 rotameric state (32), their presence in SAH domains apparently can play a stabilizing role. In the myosin VI SAH domain, hydrophobic interactions between the side chains of L10, I13, and M17 are plausible contributing factors for such stabilization. The I13C mutation caused chemical shift changes and increases in backbone HX rates by amounts that are comparable to those of the R32G mutation. In this context, it may be noted that Arg has been recognized as a key residue in SAH domains, having an even higher entropic ability to engage in salt bridges than Lys (1). The R32G substitution by Gly, therefore, was expected to be particularly disruptive, and our finding that I13C results in a comparable perturbation is perhaps surprising. We note that the structural effect caused by I13C impacts the paramagnetic alignment of all constructs tagged at Cys-13 equally. The differences in alignment and the dynamic disorder observed for constructs with substitutions in the D27–K33 range, therefore, are solely caused by these latter substitutions, not by I13C.
The distance over which the effect of mutations extends, revealed by our study, is also quite remarkable. Elevated HX rates resulting from mutations spread in both directions by more than two full helical turns from the mutation site (Fig. 2, A–C), indicative of considerable cooperativity in the stability of the SAH structure. On the other hand, in contrast to small globular proteins, the structure does not behave like a two-state folder, and the helix can be disrupted by mutations without any detectable consequence on the chemical shifts or HX properties of residues that are more than three to four helical turns removed.
From the functional requirement that the myosin VI SAH domain serves as an extension of the lever arm, Sivaramakrishnan and Spudich concluded from their optical pulling experiments that the time between helical breaks must be larger than 100 μs (33). Considering that there are many sites where the helical break can occur, the rate at which the SAH is disrupted at any individual site must be more than an order of magnitude slower, i.e., kc→o ≤ 1000 s−1. Our 15N TROSY R2 measurements for the R32G/K33G mutant yielded kex = 1/(1/ko→c + 1/kc→o) ≥ 3.3 × 105 s−1. With the experimentally determined fraction p = kc→o/(ko→c + kc→o) ≈ 0.66, this yields ko→c ≥ 5 × 105 s−1. Whereas the rate at which the α-helical H-bond breaks at the GG mutation site, kc→o, clearly is much higher than for the WT sequence, ko→c for WT may be assumed to be comparable to or faster than in the R32G/K33G mutant, i.e., ko→c ≥ 5 × 105 s−1. Together with the lower limit, kc→o ≤ 1000 s−1, from the optical pulling experiments, this yields a population of p ≤ 0.002 for broken H-bonds at any given site. This value is too low to account for the persistence length of ∼150 residues measured for the myosin VI SAH domain (15). Therefore, a dominant fraction of the finite persistence length of the SAH domains must be attributed to the thermal fluctuations of the backbone torsion angles and H-bond lengths rather than to the breaking of helical H-bonds that would result in transient kinking of the SAH domain. This distinction is important when aiming to use the SAH domain to exert a weak, spring-like force on a substrate protein. In such applications, a kinked helix will only have a very weak restoring force, whereas an intact but bent SAH domain will be able to exert higher force.
The SAH domain is small enough to potentially be used in NMR experiments to probe the effect of its applied force on a substrate protein covalently linked between the N- and C-termini of the SAH domain. Modulation of the spring-like force exerted by the SAH domain on such a protein by a simple experimental variable, such as pH or hydrostatic pressure, may open an avenue to probe the effect of mechanical force at atomic resolution by NMR spectroscopy. The effect of pH on the D27H/E28H double mutant suggests that such experiments will become feasible, but a steeper dependence of the integrity of the helical rod on pH is clearly desirable for studies of this type. A number of analogous systems are currently being investigated for such purposes.
Author Contributions
A.B. and C.A.B. designed research. C.A.B. and J.Y. performed research. C.A.B. and Y.S. analyzed data. A.B. and C.A.B. wrote the article.
Acknowledgments
We thank Daniel Haussinger for his generous gift of the DOTA-M8-Tm tags used in our study; Angus J. Robertson for suggesting the use of sodium phosphate as an internal pH reference; James L. Baber for technical support; Duck-Yeon Lee at the National Heart, Lung, and Blood Institute Biochemistry Core Facility for providing access and use of his mass spectrometers; and Dennis A. Torchia, G. Marius Clore, and Robert Best for valuable discussions.
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Editor: Wendy Shaw.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.01.003.
Supporting Material
References
- 1.Wolny M., Batchelor M., Peckham M. Characterization of long and stable de novo single alpha-helix domains provides novel insight into their stability. Sci. Rep. 2017;7:44341. doi: 10.1038/srep44341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swanson C.J., Sivaramakrishnan S. Harnessing the unique structural properties of isolated α-helices. J. Biol. Chem. 2014;289:25460–25467. doi: 10.1074/jbc.R114.583906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gáspári Z., Süveges D., Tóth G. Charged single alpha-helices in proteomes revealed by a consensus prediction approach. Biochim. Biophys. Acta. 2012;1824:637–646. doi: 10.1016/j.bbapap.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Spink B.J., Sivaramakrishnan S., Spudich J.A. Long single alpha-helical tail domains bridge the gap between structure and function of myosin VI. Nat. Struct. Mol. Biol. 2008;15:591–597. doi: 10.1038/nsmb.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sivaramakrishnan S., Spink B.J., Spudich J.A. Dynamic charge interactions create surprising rigidity in the ER/K alpha-helical protein motif. Proc. Natl. Acad. Sci. USA. 2008;105:13356–13361. doi: 10.1073/pnas.0806256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolny M., Batchelor M., Peckham M. Stable single α-helices are constant force springs in proteins. J. Biol. Chem. 2014;289:27825–27835. doi: 10.1074/jbc.M114.585679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker E.G., Bartlett G.J., Woolfson D.N. Local and macroscopic electrostatic interactions in single α-helices. Nat. Chem. Biol. 2015;11:221–228. doi: 10.1038/nchembio.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batchelor M., Wolny M., Peckham M. Dynamic ion pair behavior stabilizes single α-helices in proteins. J. Biol. Chem. 2019;294:3219–3234. doi: 10.1074/jbc.RA118.006752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sivaramakrishnan S., Sung J., Spudich J.A. Combining single-molecule optical trapping and small-angle x-ray scattering measurements to compute the persistence length of a protein ER/K alpha-helix. Biophys. J. 2009;97:2993–2999. doi: 10.1016/j.bpj.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J., Chen Y., Zhang M. Ca2+-induced rigidity change of the myosin VIIa IQ motif-single α helix lever arm extension. Structure. 2017;25:579–591.e4. doi: 10.1016/j.str.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Quinlan R.J., Reinhart G.D. Baroresistant buffer mixtures for biochemical analyses. Anal. Biochem. 2005;341:69–76. doi: 10.1016/j.ab.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Baboolal T.G., Sakamoto T., Peckham M. The SAH domain extends the functional length of the myosin lever. Proc. Natl. Acad. Sci. USA. 2009;106:22193–22198. doi: 10.1073/pnas.0909851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn A.R., Chuan P., Spudich J.A. Contribution of the myosin VI tail domain to processive stepping and intramolecular tension sensing. Proc. Natl. Acad. Sci. USA. 2010;107:7746–7750. doi: 10.1073/pnas.1002430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knight P.J., Thirumurugan K., Peckham M. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J. Biol. Chem. 2005;280:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 15.Barnes C.A., Shen Y., Bax A. Remarkable rigidity of the single α-helical domain of myosin-VI as revealed by NMR spectroscopy. J. Am. Chem. Soc. 2019;141:9004–9017. doi: 10.1021/jacs.9b03116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pervushin K., Riek R., Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mobli M., Hoch J.C. Nonuniform sampling and non-Fourier signal processing methods in multidimensional NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2014;83:21–41. doi: 10.1016/j.pnmrs.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ying J., Delaglio F., Bax A. Sparse multidimensional iterative lineshape-enhanced (SMILE) reconstruction of both non-uniformly sampled and conventional NMR data. J. Biomol. NMR. 2017;68:101–118. doi: 10.1007/s10858-016-0072-7. Published online November 19, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertini I., Del Bianco C., Zoroddu M.A. Experimentally exploring the conformational space sampled by domain reorientation in calmodulin. Proc. Natl. Acad. Sci. USA. 2004;101:6841–6846. doi: 10.1073/pnas.0308641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Häussinger D., Huang J.R., Grzesiek S. DOTA-M8: an extremely rigid, high-affinity lanthanide chelating tag for PCS NMR spectroscopy. J. Am. Chem. Soc. 2009;131:14761–14767. doi: 10.1021/ja903233w. [DOI] [PubMed] [Google Scholar]
- 21.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 22.Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helmus J.J., Jaroniec C.P. Nmrglue: an open source Python package for the analysis of multidimensional NMR data. J. Biomol. NMR. 2013;55:355–367. doi: 10.1007/s10858-013-9718-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fitzkee N.C., Bax A. Facile measurement of 1H-15N residual dipolar couplings in larger perdeuterated proteins. J. Biomol. NMR. 2010;48:65–70. doi: 10.1007/s10858-010-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen M.R., Mueller L., Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 26.Clore G.M., Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eykyn T.R., Kuchel P.W. Scalar couplings as pH probes in compartmentalized biological systems: 31P NMR of phosphite. Magn. Reson. Med. 2003;50:693–696. doi: 10.1002/mrm.10580. [DOI] [PubMed] [Google Scholar]
- 28.Mesleh M.F., Opella S.J. Dipolar Waves as NMR maps of helices in proteins. J. Magn. Reson. 2003;163:288–299. doi: 10.1016/s1090-7807(03)00119-8. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y.Z. University of Pennsylvania; Philadelphia: 1995. Protein and Peptide Structure and Interactions Studied by Hydrogen Exchange and NMR. [Google Scholar]
- 30.Bai Y., Milne J.S., Englander S.W. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C., Rance M., Palmer A.G., III Mapping chemical exchange in proteins with MW > 50 kD. J. Am. Chem. Soc. 2003;125:8968–8969. doi: 10.1021/ja035139z. [DOI] [PubMed] [Google Scholar]
- 32.Zuiderweg E.R.P., Boelens R., Kaptein R. Stereospecific assignments of 1H NMR methyl lines and conformation of valyl residues in Lac repressor headpiece. Biopolymers. 1985;24:601–611. [Google Scholar]
- 33.Sivaramakrishnan S., Spudich J.A. Systematic control of protein interaction using a modular ER/K α-helix linker. Proc. Natl. Acad. Sci. USA. 2011;108:20467–20472. doi: 10.1073/pnas.1116066108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.