Abstract
Accumulating evidence indicates that RIPK1 is associated with inflammation and apoptotic. RIPK1 deficiency leads to proinflammatory signaling impaired. However, only few patients with homozygous loss-of-function mutation in RIPK1 gene had been reported until now. Here, we report a Chinese combined immunodeficiency patient. He had recurrent infection, diarrhea after 3 months old. Immune function indicated that T, B and NK cells decreased significantly but immunoglobulins approximately remained normal. Whole-exome sequencing indicated that he had novel compound heterozygous mutations (c.998 C > A from his mother and c.1934 C > T from his father) in RIPK1 gene, which were confirmed by Sanger sequencing. Our study reports novel mutations in RIPK1 gene and new phenotype of patient with RIPK1 deficiency.
Keywords: Combined immunodeficiency, Inflammatory bowel disease, Mutation, RIPK1
Introduction
RIPK1 (receptor-interacting serine/threonine kinase 1) is an essential element of multiple signaling pathways that responsible for development, antiviral immunity, inflammation, necroptotic cell death and homeostasis.1, 2, 3, 4, 5 Specifically, ubiquitination of RIPK1 is associated with TNF-α-induced NF-κB-mediated inflammation.6,7 The research of RIPK1 has been substantially implicated in mouse model, suggesting that mice lacking RIPK1 (RIPK1−/−) showed postnatal lethality,8 severe gut disease,9 skin inflammation and reduced body weight.10
In 2018, Cuchet-Lourenco et al11 first reported four patients with RIPK1 homozygous mutation, who suffered from lymphopenia, recurrent viral, bacterial and fungal infection. Patients had decreased T, B and NK cells in different degrees. Those patients also existed reduced Naïve CD4 and CD8 T cells and effector CD8 T cells. Impaired MAPK signaling and dysregulated cytokine production was found in RIPK1-dificient immune cells. In 2019, another study reported eight RIPK1 deficiency patients suffered from immune and intestinal epithelial cell dysfunctions. Study showed a decreased frequency of central memory and effector memory CD4 and CD8 T cells, regulatory T cells, Th1 cells, Th17 cells and switched memory B cells in their patients. Consent with previous report, their research revealed that RIPK1 deficiency was associated with high levels of inflammasome activity upon LPS stimulation and led to impaired TNF-α induced NF-κB signaling and necroptosis.12 However, there are only the two studies reported this disease until now.
Here, we report a Chinese boy with novel compound mutation in RIPK1, who had combined immunodeficiency and intestinal inflammation. This study is helpful for expanding the phenotype and genetic profile of this disease.
Materials and methods
This study was approved by the Ethics Committee of the Children's Hospital of Fudan University. Written informed consent was obtained from the patient's parents.
Patient
The patient enrolled in this study was a 3 years old boy. The disease was diagnosed by clinical manifestation, immune phenotype, and genetic analysis.
Serum immunoglobulins detection
As previously reported,13, 14, 15 serum immunoglobulins (IgG, IgA, IgM) were detected by nephelometry. The immunoglobulin kit was purchased from Orion Diagnostica Oy (Espoo, Finland). IgE was assessed by UniCAP (Pharmacia, Uppsala, Sweden).
Lymphocyte subset detection
The peripheral blood was collected from the patient. Staining for lymphocyte surface markers was performed after red blood cell lysis, according to a standard flow cytometric multicolor protocol with the appropriate fluorochrome-conjugated antibodies. Briefly, after washing with phosphate buffer solution (PBS) two times, 1–5 × 104 live cells were analyzed by flow cytometry (FACS Canto II) using Diva software (BD Biosciences). B cells, Total T cells, CD4 T cells, CD8 T cells and CD56+/CD16 + natural killer (NK) cells were detected by the BD Multitest IMK Kit. The following validated antibodies were used to define T-cell subsets: anti-human CD3 (PerCP-Cy5.5), anti-CD8 (BV510), anti-CD4 (FITC; fluorescein isothiocyanate), anti-CD27 (APC; allophycocyanin), anti-CD45RA (PE-Cy7), anti-TCRαβ (PE; phycoerythrin) and anti-TCRγδ (BV421). The following were used define B-cell subsets: anti-CD19 (APC), anti-human CD24 (PE), anti-CD27 (BV450), anti-CD38 (PerCP-Cy5.5) and anti-IgD (BV510) (BD Biosciences). The following isotype control reagents were used: BV510 Mouse IgG2a, κ Isotype Control; APC Mouse IgG1, κ Isotype Control; PE-Cy™7 Mouse IgG2b, κ Isotype Control; BV421 Mouse IgG1, k Isotype Control; PE Mouse IgM, κ Isotype Control; V450 Mouse IgG1, κ Isotype Control.
Genetics analysis
Whole exome sequencing (WES) was used for genetic analysis. Briefly, genomic DNA was extracted from the PBMC of patient and his parents. Then, genomic DNA fragments were enriched for the target region of the consensus coding sequence (CCDS) exons and subsequently sequenced on the HiSeq 2000 sequencer (Illumina, San Diego, CA). An average of 11.8 Gb of raw sequence data was generated with 92.65X depth of exome target regions for each individual as paired-end 150 base pair reads. The raw data was mapped to the human genome reference sequence (hg19). 91.2% of the raw date sequencing quality was above Q30. The average sequencing depth ranged from 35.73X-38.45X. The mapping rate of clear data ranged from 97.03 to 97.27%, and the genome coverage ranged from 99.83% to 99.85%. A nucleotide changes observed in more than 5% of aligned reads were called and reviewed by using NextGENe software (SoftGenetics, State College, PA).
The variants in RIPK1 were confirmed by using Sanger sequencing. Primers for PCR are listed in supplemental Table A. After an initial denaturation for 5 min at 95 °C, 35 cycles of amplification were carried out as follows: 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 40 s. Final extension was performed at 72 °C for 7 min. Sanger sequencing of PCR products was performed in both directions.
Results
Case presentation
The patient was a 41-month old boy from unrelated and healthy parents. At the age of 3–6 months, he suffered from anemia, thrush mycotic stomatitis, acute bronchopneumonia, enteritis and jaundice. From 13 months old, he had recurrent diarrhea, watery stool with mucous and then developed into hematochezia. Stool culture appeared that Clostridium difficile was positive while Salmonella and Shigella was negative. He had unexplained recurrent fever from 14 months year old. The maximum temperature was 40.3 °C and displayed a form of prolonged, intermittent, irregular fever. He was treated at local hospital. However, diarrhea and fever had no improvement. During his malady progress, hepatosplenomegaly was always existed with abnormal elevated serum glutamic oxalacetic transaminase (AST, the maximum value was 104 IU/L) and glutamic-pyruvic transaminase (ALT, the maximum value was 102 IU/L). As the patient got close to 24 months years old, perianal abscess and anal fistula was diagnosed. He underwent exploratory laparotomy, enterectomy, entero-anastomosis and subsequent sigmoid colostomy due to gradually aggravated stenosis in proximal colon and intestinal obstruction with the help of performing erect abdominal plain radiograph when he was 20- and 41-months old.
At 41-month old, he was transferred to Children's Hospital of Fudan University. The boy had severe malnutrition. All pathogens detection is normal. T-spot, PPD test, Epstein–Barr virus (EBV)-DNA, Cytomegalovirus (CMV)-DNA, and GM test were all negative. All of the autoantibody relevant examinations were also normal, including antinuclear antibody (ANA), dsDNA, and Coomb's test, et al. However, the patient had high level of serum interleukin-6 (IL-6) (the highest value was 1209 pg/ml while normal range should<7 pg/ml) and calcitonin zymogen (48.12 ng/ml, normal range <0.05 ng/ml), which suggested that the existence of systemic inflammatory response.
Immune phenotype
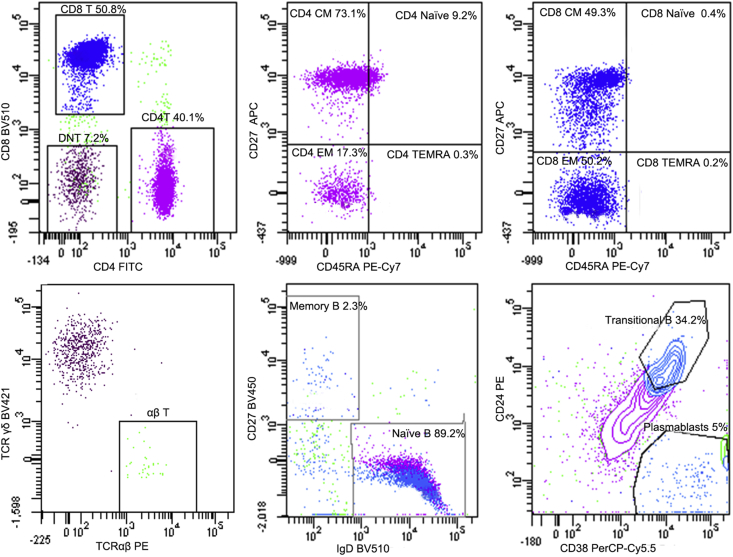
The patient's serum immunoglobulin levels were roughly normal in different periods (Table 1). We speculated one of factors which may lead to his relatively elevated IgG was intravenous immunoglobulin (IVIG). A small amount of IgA consisted in IVIG might lead to the mildly increase in IgA. To analyze the number of lymphocytes, we measured lymphocyte subpopulation of this patient. CD4 T lymphocytes, NK cells and B cells were all gradually decreased while CD8 T cells increased (Table 2). We performed more detailed detection for T cells and B cells when he was 41-month year old. For T lymphocytes, the percentage of memory CD4 T cells, including central memory CD4 T cells (CD4 CM) and effector memory CD4 T cells (CD4 EM) were dramatically rising while Naïve CD4 T cells obviously dwindled. A similar change was found in the CD8 CM, CD8 EM and Naïve CD8 T cells. In addition, TCRαβ+ double-negative T (DNT) cell was double increased. For B lymphocytes, transitional B cells rose markedly and memory B reduced slightly (Table 3, Fig. 1). In all, the abnormal lymphocyte subpopulation indicated the RIPK1-deficient patient suffered from disorder of immune system.
Table 1.
Serum immunoglobulin level of patient in different age.
| Age (month) | 2 | 4 | 15 | 17 | 24 | 29 | 41 | Reference value |
|---|---|---|---|---|---|---|---|---|
| IgG | 4.8 | 11.2 | 15.77 ↑ | 16.32 ↑ | 11.25 | 12.15 | 14.2 ↑ | 4.95–12.74 (g/L) |
| IgA | 0.53 | 0.53 | 0.55 | 0.75 | 0.63 | 0.37 | 2.25 ↑ | 0.33–1.89 (g/L) |
| IgM | 0.34 | 0.87 | 1.09 | 0.72 | 0.66 | 0.36 | 0.43 | 0.65–2.01 (g/L) |
| IgE | 58.7 | 4.1 | 13.1 | <17.8 | <17.9 | 23.3 | 8 | <100 (KU/L) |
Table 2.
The numbers and percent of T, B and NK cells of the patient.
| Age (month) | 15 | 17 | 24 | 29 | 41 | Reference value |
|---|---|---|---|---|---|---|
| Total T cell (cells/μL) | 4735.62 | – | – | 2942.50 | 1631.30 | 1794–4247 |
| Total T cell (%) | 83.65 ↑ | 87.21 ↑ | 89.75 ↑ | 87.65 ↑ | 83.4 ↑ | 53.88–72.87 |
| CD4 T cell (cells/μL) | 1286.59 | – | – | 628.33 ↓ | 449.42 ↓ | 902–2253 |
| CD4 T cell (%) | 22.73 | 14.89 ↓ | 20.60 | 18.72 ↓ | 22.98 | 24.08–42.52 |
| CD8 T cell (cells/μL) | 3229.16 ↑ | – | – | 2012.29 ↑ | 1159.54 | 580–1735 |
| CD8 T cell (%) | 57.04 ↑ | 63.12 ↑ | 62.45 ↑ | 59.94 ↑ | 59.28 ↑ | 19.00–31.51 |
| Total B cell (cells/μL) | 398.66 ↓ | – | – | 218.69 ↓ | 158.34 ↓ | 461–1456 |
| Total B cell (%) | 7.04 ↓ | 4.13 ↓ | 4.15 ↓ | 6.51 ↓ | 8.09 ↓ | 13.23–26.39 |
| NK cell (cells/μL) | 508.60 | – | – | 179.52 | 124.75 ↓ | 270–1053 |
| NK cell (%) | 8.98 | 7.30 | 4.6 ↓ | 5.53 ↓ | 6.38 ↓ | 7.21–20.90 |
| CD4/CD8 | 0.4 ↓ | 0.23 ↓ | 0.33 ↓ | 0.31 ↓ | 0.39 ↓ | 0.9–2.13 |
Table 3.
Lymphocyte subpopulation of the patient at 41 months old.
| Value (%) | Reference value (%) (1–4 year old) | |
|---|---|---|
| Cytotoxic T cell (CD8, CD45 + CD3 + CD8 + ) | 50.8 ↑ | 19.00–32.51 |
| Central memory cytotoxic T cells (CD8 CM, CD3 + CD8 + CD45RA - CD27 + ) | 49.3 ↑ | 5.18–31.66 |
| Naive differentiated cytotoxic T cells (CD8 Naive, CD3 + CD8 + CD45RA + CD27 + ) | 0.4 ↓ | 36.8–83.16 |
| Effector memory cytotoxic T cells (CD8 EM, CD3 + CD8 + CD45RA - CD27 - ) | 50.2 ↑ | 0.7–11.22 |
| Terminally differentiated effector memory cytotoxic T cells (CD8 TEMRA, CD3 + CD8 + CD45RA + CD27 - ) | 0.2 ↓ | 0.84–33.02 |
| CD4 T cell (CD45 + CD3 + CD4 + ) | 40.10 | 24.08–42.52 |
| Central memory helper T cells (CD4 CM, CD3 + CD4 + CD45RA - CD27 + ) | 73.1 ↑ | 13.88–48.12 |
| Naive differentiated helper T cells (CD4 Naive, CD3 + CD4 + CD45RA + CD27 + ) | 9.2 ↓ | 46.14–84.4 |
| Effector memory helper T cells (CD4 EM, CD3 + CD4 + CD45RA - CD27 - ) | 17.3 ↑ | 0.94–6.46 |
| Terminally differentiated effector memory helper T cells (CD4 TEMRA, CD3 + CD4 + CD45RA + CD27 - ) | 0.30 | 0–1.36 |
| γδ T cells (γδ T, CD3 + TCRγδ + ) | 7.70 | 4.94–17.98 |
| TCRαβ+ double-negative T (DNT) cells (CD3 + TCRαβ + CD4 - CD8 - ) | 7.2 ↑ | 0.37–1.80 |
| B Cells (CD45 + CD19 + ) | 10.6 ↓ | 13.23–26.39 |
| Memory B cells (CD19 + CD27 + IgD - ) | 2.3 ↓ | 2.98–14.18 |
| Naïve B cells (CD19 + CD27 - IgD + ) | 89.20 | 65.54–86.62 |
| Transitional B cells (CD19 + CD24 + + CD38 + +v) | 34.2 ↑ | 5.24–17.22 |
| Plasmablasts (CD19 + CD24 - CD38 + + ) | 5.00 | 0.5–7.06 |
Figure 1.
Lymphocyte subpopulation of the patient at 41 months old. For T lymphocytes, the percentage of central memory helper T cells (CD4 CM, CD3 + CD4 + CD45RA - CD27V+ ) and effector memory helper T cells (CD4 EM, CD3 + CD4 + CD45RA - CD27 - ) were dramatically rising while naive differentiated helper T cells (CD4 Naive, CD3 + CD4 + CD45RA + CD27 + ) obviously dwindled. A similar change was found in the central memory cytotoxic T cells (CD8 CM, CD3 + CD8 + CD45RA - CD27 + ), effector memory cytotoxic T cells (CD8 EM, CD3 + CD8 + CD45RA - CD27 - ) and naive differentiated cytotoxic T cells (CD8 Naive, CD3 + CD8 + CD45RA + CD27 + ), respectively. In addition, TCRαβ+ double-negative T (DNT) cell was double increased. For B lymphocytes, transitional B cells (CD19 + CD24 + + CD38 + + ) rose markedly while memory B cells (CD19 + CD27 + IgD - ) reduced slightly.
Genetics analysis
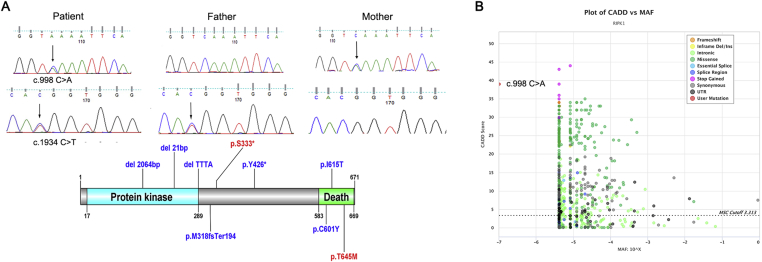
WES revealed that he carried a RIPK1 gene compound heterozygous mutation, c.998C > A (p. S333X) from his mother and c.1934 C > T (p. T645M) from his father. These two mutations were confirmed by Sanger sequencing. c.998 C > A in RIPK1 was a novel mutation which has not been reported in database, including the ClinVar database16 and the Human Gene Mutation Database.17 (Fig. 2A). c.998C > A in the patient was supposed to result in an early stop codon in the protein at position 333, leading to nonsense-mediated decay or creating a premature truncated protein in one of the prediction tools—Mutationtaster. Besides, this variant was also predicated to damage in the software PopViz.18 (Fig. 2B).
Figure 2.
The mutation in RIPK1 gene of the patient (A) The above figure illustrates that patient carries RIPK1 compound heterozygous mutation in c.998 C > A (p. S333X, from his mother) and c.1934 C > T (p. T645M, from his father). The black arrow shows the mutation. All RIPK1 mutation sites from literature are indicated in blue while mutations in this study are shown in red in the lower image. X represents stop. The schematic representation of RIPK1 mutation is plotted with DOG.21 (B) CADD vs MAF plot of RIPK1 by PopViz. The horizontal and vertical axis shows MAF and CADD scores, respectively. The MAF score is −7 and the CADD score is 39, which indicates that the novel mutation c.998 C > A in our patients is malignant.
Discussion
Here, we report a novel mutation of RIPK1 gene in immunodeficiency disease. It has been showed that typical features in RIPK1 deficiency patient include mouth ulcers, recurrent respiratory infections, inflammatory bowel disease, inflammatory polyarthritis, skin rash, bacterial, viral, and fungal infections, failure to thrive.11,12 Cuchet-Lourenco D et al11 concluded that the effects of RIPK1 deficiency was primarily to the immune system, because their patients mainly suffered from decreased T, B and NK cells, dysregulated cytokine production and impaired MAPK signaling. Consistent with literature, our patient has decreased T, B and NK cells in different degrees, as well as Naïve CD4 T and CD8 T cells, and memory B cells. However, memory T cells show opposite tendency in our patient, which maybe is attributed to the different inflammatory conditions in patient. Different from previous reports, our patient did not exhibit polyarthritis, skin lesions (just slight partial interspersed macule could be found) and no evidence of viral and fungal infection but obvious recurrent fever and intestinal obstruction in his early life stage. During hospitalization, the patient was treated by improvement pulmonary hypertension, drugs protecting liver, intravenous injection of globulin and antibiotics to resist the bacterial infection. Unfortunately, routine follow up shown that this boy had died at 4 years old.
Limited by obtaining of samples, our data did not provide functional experiment evidence that if the heterozygous mutation was pathogenic. However, c.1934 C > T is a common spot mutation in the previous report,12 confirmed by missense mutation prediction software (PolyPhen19 and SIFT20) and specifically functional studies. Meanwhile, c.998 C > A is predicated to be deleterious in both Mutationtaster and PopViz. Further study about this rare deficiency deserves to be conducted both in potential mechanism and therapeutic means. In all, those findings suggest that RIPK1 actually works in multi-system including immunity, digestion and respiration system.
Our study reports novel mutations in RIPK1 gene and new phenotype of patient with RIPK1 deficiency. It is helpful for understanding the phenotype of this disease and expanding the mutation database.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81471482). We thank the patient and his parents for their cooperation.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2019.10.008.
Contributor Information
Jinqiao Sun, Email: jinqiaosun@fudan.edu.cn.
Xiaochuan Wang, Email: xchwang@shmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sun L., Wang H., Wang Z. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148(1–2):213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 2.Saleh D., Degterev A. Emerging roles for RIPK1 and RIPK3 in pathogen-induced cell death and host immunity. Curr Top Microbiol Immunol. 2017;403:37–75. doi: 10.1007/82_2015_449. [DOI] [PubMed] [Google Scholar]
- 3.He S., Wang X. RIP kinases as modulators of inflammation and immunity. Nat Immunol. 2018;19(9):912–922. doi: 10.1038/s41590-018-0188-x. [DOI] [PubMed] [Google Scholar]
- 4.Kondylis V., Pasparakis M. RIP kinases in liver cell death, inflammation and cancer. Trends Mol Med. 2019;25(1):47–63. doi: 10.1016/j.molmed.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 5.O'Donnell J.A., Lehman J., Roderick J.E. Dendritic cell RIPK1 maintains immune homeostasis by preventing inflammation and autoimmunity. J Immunol. 2018;200(2):737–748. doi: 10.4049/jimmunol.1701229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wegner K.W., Saleh D., Degterev A. Complex pathologic roles of RIPK1 and RIPK3: moving beyond necroptosis. Trends Pharmacol Sci. 2017;38(3):202–225. doi: 10.1016/j.tips.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondylis V., Kumari S., Vlantis K., Pasparakis M. The interplay of IKK, NF-kappaB and RIPK1 signaling in the regulation of cell death, tissue homeostasis and inflammation. Immunol Rev. 2017;277(1):113–127. doi: 10.1111/imr.12550. [DOI] [PubMed] [Google Scholar]
- 8.Dillon C.P., Weinlich R., Rodriguez D.A. RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell. 2014;157(5):1189–1202. doi: 10.1016/j.cell.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dannappel M., Vlantis K., Kumari S. RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature. 2014;513(7516):90–94. doi: 10.1038/nature13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J., Kumari S., Kim C. RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nature. 2016;540(7631):124–128. doi: 10.1038/nature20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuchet-Lourenco D., Eletto D., Wu C. Biallelic RIPK1 mutations in humans cause severe immunodeficiency, arthritis, and intestinal inflammation. Science. 2018;361(6404):810–813. doi: 10.1126/science.aar2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y., Fuhrer M., Bahrami E. Human RIPK1 deficiency causes combined immunodeficiency and inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2019;116(3):970–975. doi: 10.1073/pnas.1813582116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L., Wang W., Wang Y. A Chinese DADA2 patient: report of two novel mutations and successful HSCT. Immunogenetics. 2019;71(4):299–305. doi: 10.1007/s00251-018-01101-w. [DOI] [PubMed] [Google Scholar]
- 14.Dong X., Liu L., Wang Y. Novel heterogeneous mutation of TNFAIP3 in a Chinese patient with behcet-like phenotype and persistent EBV viremia. J Clin Immunol. 2019;39(2):188–194. doi: 10.1007/s10875-019-00604-9. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y., Wang W., Liu L. Report of a Chinese cohort with activated phosphoinositide 3-kinase delta syndrome. J Clin Immunol. 2018;38(8):854–863. doi: 10.1007/s10875-018-0568-x. [DOI] [PubMed] [Google Scholar]
- 16.Landrum M.J., Lee J.M., Benson M. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenson P.D., Mort M., Ball E.V. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136(6):665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang P., Bigio B., Rapaport F. PopViz: a webserver for visualizing minor allele frequencies and damage prediction scores of human genetic variations. Bioinformatics. 2018;34(24):4307–4309. doi: 10.1093/bioinformatics/bty536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013 doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adzhubei I.A., Schmidt S., Peshkin L. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. Dog 1.0: illustrator of protein domain structures. Cell Res. 2009;19(2):271–273. doi: 10.1038/cr.2009.6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.