Abstract
Common variable immunodeficiency disorders (CVID), a heterogeneous group of inborn errors of immunity, is the most common symptomatic primary immunodeficiency disorder. Patients with CVID have highly variable clinical presentation. With the advent of whole genome sequencing and genome wide association studies (GWAS), there has been a remarkable improvement in understanding the genetics of CVID. This has also helped in understanding the pathogenesis of CVID and has drastically improved the management of these patients. A multi-omics approach integrating the DNA sequencing along with RNA sequencing, proteomics, epigenetic and metabolomics profile is the need of the hour to unravel specific CVID associated disease pathways and novel therapeutic targets. In this review, we elaborate various techniques that have helped in understanding the genetics of CVID.
Keywords: Common variable immunodeficiency (CVID), Epigenome, Genetics, Next generation sequencing (NGS), Transcriptome
Introduction
Inborn errors of immunity, a heterogeneous group of uncommon genetic disorders are characterized by impairment of immune system leading to varied clinical manifestations such as infections, autoimmunity, immune dysregulation, inflammation and malignancies.1 Nearly 400 genetic defects have been identified in patients with various PIDs till date.1., 2, 3 CVID is the most common symptomatic PID with predominant antibody deficiency which manifests in older children and adults.4 The prevalence of CVID ranges from 1/10,000 to 1/50,000 in the Middle East and Caucasian population; and is less frequently described in African and Asian population.4, 5, 6 The age of onset of symptoms is highly variable ranging from childhood to second and third decade of life.4,7 There is marked genetic and phenotypic heterogeneity in this disease. Several monogenic forms of CVID have been described and have paralleled rapid advancements and development of high-throughput sequencing technologies.8, 9, 10, 11, 12, 13 However, monogenic genetic variants still constitute only 2–10% of all CVID patients in different cohorts.4,5,7 The majority of CVID patients lack a monogenic basis and the disease is probably polygenic in origin in most cases. Understanding the genetic basis of CVID will help in developing personalized treatment approaches for better management of patients. This review attempts to illustrate the importance of omics-based technology and integration of next generation platforms in CVID genetics.
Clinical manifestations of CVID
CVID comprises a heterogeneous group of PIDs that present with decreased to undetectable levels of immunoglobulins and an increased susceptibility to develop infections.1,2 A large proportion of patients with CVID also have autoimmune manifestations that may sometimes be the presenting or the sole clinical manifestation of disease. Patients with CVID also have predilection to develop malignancies, commonly lymphomas.14 In addition, patients with CVID may also present with polyclonal lymphoproliferation (lymphoid interstitial pneumonia, persistent lymphadenopathy, splenomegaly, hepatomegaly), chronic inflammatory disorders like granulomas, and colitis.4,14,15 CVID is diagnosed by diminished IgG levels, and either low IgM or IgA levels along with absent or reduced antibody specific responses to infection or vaccine, along with exclusion of secondary forms of hypogammaglobulinemia. The clinical profile of patients with monogenic forms of CVID is different and several defects lead to a unique phenotype,1 in a given patient it may be extremely difficult based on clinical profile alone to determine the causative gene.
Can the causative gene be identified based on clinical profile of patient and what are the therapeutic implications of identifying the genetic defects in patients with CVID?
Although hypogammaglobulinemia is central to all forms of CVID and intravenous immunoglobulin (IVIg) replacement therapy is the cornerstone of management, specific treatment modalities are now being employed depending on the underlying genetic defect. Identification of the causative gene and its functional significance in patients with CVID may significantly alter the management options from IVIg replacement to hematopoietic stem cell transplant or specific targeted therapy. Clinical manifestations may occasionally suggest an underlying genetic etiology.
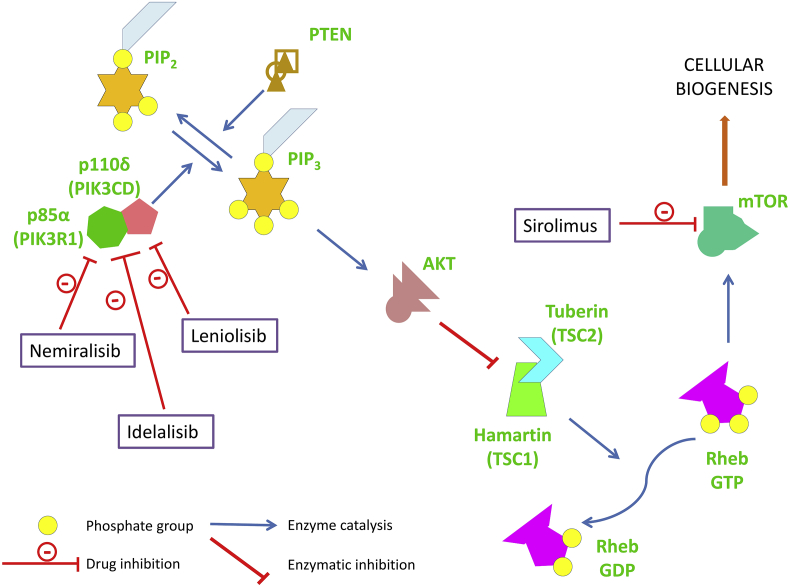
Mutations in the phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta (PIK3CD), phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1), and phosphatase and tensin homolog (PTEN) enzymes lead to a similar clinical phenotype characterized by hypogammaglobulinemia, lymphoproliferation, autoimmunity, and a combined immunodeficiency.16 PIK3CD phosphorylates phophatidyl inositol bisphosphate and activates the downstream signalling cascade. Gain of function mutations result in increased level of rheb GTP (Ras homolog, mTORC1 binding guanosine triphosphate) that leads to blockage of tuberin hamartin complex and increased activation of mTOR (mammalian target of rapamycin) thereby causing cell proliferation. PTEN and PIK3R1 act as regulators of this signalling cascade by decreasing the levels of metabolically active phosphatidyl inositol trisphosphate. This leads to de-phosphorylation of rheb GTP leading to decreased activation of mTOR. Hence, loss of function mutations in PTEN or PI3KR1 lead to a similar clinical profile as is seen in patients with gain of function mutations in PIK3CD gene. Patients with activated PI3K delta syndrome (APDS) may have low/normal IgG and IgA with normal to high IgM. The immunoglobulin profile may also suggest a clinical possibility of hyperIgM syndrome. Flow cytometry may reveal increased proportion of senescent T cells. Identification of these defects is very important in patients presenting with CVID phenotype because specific targeted therapies with PI3Kδ inhibitors or mTOR inhibitors are the treatment of choice17 (Fig. 1). LRBA (Lipopolysaccharide responsive beige-like anchor protein), and CTLA4 (cytotoxic T-lymphocyte associated protein 4) expression is also closely coordinated as they are present together in the Golgi bodies. Mutations in these genes may cause enteropathy, lymphoproliferation, and autoimmunity in addition to infections. Flow cytometry may reveal decreased expression of CTLA4. Management includes use of CTLA-4 agonists such as abatacept, and belatacept.18 Hematopoietic stem cell transplantation is also an effective treatment modality in these patients. Mutations in the members of the TNF (tumor necrosis factor) receptor superfamily; Transmembrane activator, and CAML interactor (TACI), TNF receptor superfamily member 13B (TNFRSF13B), B-cell activating factor receptor (BAFF-R), TNF receptor superfamily member 13C (TNFRSF13C) resulting in the CVID phenotype have no specific clinical features. However, flow cytometry analysis may suggest decreased expression of these proteins. Mutations in the ATPase H+ transporting accessory protein 1 (ATP6AP1) gene can also lead to a CVID phenotype, however differentiating features includes an X-linked inheritance, and liver involvement with low copper levels.19 Mutations in the genes encoding for nuclear factor kappa light chain enhancer of activated B cells (NF-κB), Nuclear factor kappa B subunit 1 (NFKB1), and Nuclear factor kappa B subunit 2 (NFKB2) have prominent endocrinopathies in addition to other autoimmune manifestations and recurrent sinopulmonary infections. Gain of function mutations in the NFKB2 may lead to similar manifestations as that of NFKB1 loss of function mutations because of physiological antagonism. Therapy with calcineurin inhibitors which modulate the NFκB signalling may be employed as a treatment modality.20 Similarly, mutations in other genes associated with the CVID phenotype may have suggestive clinical features such as congenital sideroblastic anemia with hearing loss and auto-inflammation in TRNT1 (TRNA nucleotidyl transferase 1) defects; neutropenia in TNF related weak inducer of apoptosis (TWEAK) (TNF superfamily member 12; TNFSF12) defect21, abnormal hair, facies, severe diarrhea with villous atrophy, and possible liver involvement in tetratricopeptide repeat domain 37 (TTC37) gene defect (trichohepatoenteric disease).22 Treatment with TNF inhibitors has been recently reported to be efficacious in patients with TRNT1 defects.23
Figure 1.
The effect of mutations in Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta (PIK3CD), Phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1), phosphatase and tensin homolog (PTEN) genes leading to lymphoproliferation in common variable immunodeficiency disease and their therapeutic modulators. [AKT (Serine/Threonine specific protein kinase); mTOR (Mammalian target of rapamycin); PIP2 (Plasma membrane intrinsic protein 2); PIP3 (Plasma membrane intrinsic protein 3); Rheb GTP (Ras homolog, mTORC1 binding guanosine triphosphate); Rheb GDP (Ras homolog, mTORC1 binding guanosine diphosphate); TSC1(Tuberous Sclerosis Complex 1); TSC2 (Tuberous Sclerosis Complex 2)].
Some of these clinical manifestations may guide clinicians regarding an underlying genetic defect. However, clinical manifestations often overlap and in a given case it may be difficult to identify a candidate gene based only on the available clinical information.
Genetic heterogeneity of CVID has made deciphering the genetic etiologies a challenging task. Over the past decade, CVID has been explored to delineate the monogenic variants, however, besides the genetic variants, epigenetic and environmental factors also play an important role in biology of complex diseases. This needs to be studied extensively to unravel the precise genetic predisposition in these patients. Various genetic defects that have been reported in patients with CVID include mutations in PIK3CD, PIK3R1, PTEN, CD19 (cluster of differentiation 19), CD81 (cluster of differentiation 81), CD20 (cluster of differentiation 20), CD21 (cluster of differentiation 21), TACI (TNFRSF13B), BAFF (TNFRSF13C), TWEAK, TRNT1, TTC37, NFKB1, NFKB2, IKZF1 (IKAROS family Zinc finger 1), IRF2BP2 (Interferon regulatory factor 2 binding protein 2), and ATP6AP1 genes.2 Illustrating the genetic architecture in patients with CVID phenotype has lead to identification of recessive autosomal inheritance with biallelic mutations in CD19, CD20, CD81, ICOS (inducible T cell costimulator), PRKCD (protein kinase C delta), and LRBA24, 25, 26 along with autosomal dominant inheritance with monoallelic mutations in NFKB1, NFKB2, PIK3CD, PIK3R1,27,28 and IKZF1.29 Also, the rare hypomorphic mutations associated with severe combined immunodeficiency (SCID) have also been identified in CVID. All of these genes have been reported to play significant roles in immune regulatory pathways such as T-cell signaling, B-cell signaling, and isotype switching.25 However, in majority (approximately 90%) of CVID patients, no underlying genetic defect has ever been identified till date. Hence, unraveling the genetic basis of sporadic CVID will provide opportunity for patient stratification on the basis of genetic profile and thereby provide insights into therapeutic management of patients.
Technological advancements in elucidating the genetics in CVID
Unlike most PIDs where in a single gene defect has been identified, CVID has a heterogeneous genetic etiology. While several genes have been identified to cause CVID like phenotype in approximately 10% of all patients, no genetic defect has ever been identified in remaining patients.5,30 The recent GWAS and next generation sequencing (NGS) platforms have highlighted the alternative theory of polygenic disorders as the underlying cause of CVID pathogenesis. These polygenic determinants are driven by complex gene-gene interactions, incomplete penetrance, and variations in non-coding regions, which are in sharp contrast to monogenic defects with defined genotype-phenotype correlation.5,25 Furthermore, it has also been highlighted that the majority of CVID cases are reportedly sporadic which emphasizes the fact that CVID genetics does not follow the classical Mendelian inheritance pattern. Various techniques that have been used in recent time to elucidate the genetic etiology of CVID are as follows:
Next generation screening of CVID
The advent of next generation techniques has revolutionized the identification of genetic basis of diseases and its use has also been extended to patients with CVID.30 The first attempt to elicit pathogenic genetic variants in inherited antibody deficiencies was made in 2010 by Hong-Ying and colleagues who used a customized 300 kb; 148-gene (implicated in immunoglobulin isotype switching and B cell development) re-sequencing Hyper-IgM/CVID chip and reported mutations in TNFRSF13B, AICDA (activation induced cytidine deaminase), CD40LG (cluster of differentiation 40 ligand), and IKBKG (inhibitor of nuclear factor kappa B kinase regulatory subunit gamma), along with novel mutations in TNF receptor associated factor 3 interacting protein 2 (TRAF3IP2) (rs13190932:C > T and rs33980500:G > A).31 This was followed by GWAS study in 363 CVID patients whereby authors quoted the association of CVID pathogenesis and copy number variations (CNVs) with ADAM (A Disintegrin and Metalloproteinase) and MHC (Major Histocompatibility Complex) genes.32 Following the initial reports on mutation profile, in 2015 two studies documented decreased diversity in VDJ gene rearrangement, and abnormal complementarity determining region 3 (CDR3) formation which may explain immunodeficiency and increased auto-reactivity in CVID patients.33, 34, 35, 36 Another study published in 2015 documented RAG1 (recombination activating 1) mutations in two CVID patients and the group anticipated that the judicious application of NGS will help in elucidating the single gene defects in solving challenging cases.37 The group further emphasized that the early identification of RAG1 mutations may help in expediting hematopoietic cell transplantation in CVID patients and thereby resorting to preventing translation of autoimmune disease into fatal infections. This was further supported by findings of RAG1 mutations (c.1871G > A, c.2182T > C and p. H249R) in a CVID patient38 and another study reporting with a novel RAG1 missense variant (c.1123C > G) and frameshift deletion (c.1430delC, p. F478Sfs*14) along with a known missense mutation (rs199474678) in CVID patient.39 Another study extended the use of next generation technology to illustrate reduction of TCR (T cell receptor) repertoire diversity is driven by reduction of naïve T cells and was found to be associated with reduction in class-switch memory B cells and elevated expression of CD21lo B cells.40 A novel mutation in IRF2BP2 gene (c.1652G > A) was identified in three family members of CVID patients which was reported to be linked to monogenic phenotype of CVID and lead to production of mutant protein which was reported to have effect on B-cell differentiation leading to decline in in vitro plasmablast production.41 Another novel heterozygous frameshift mutation was documented in NFKB1 which lead to a premature stop codon thereby resulting in reduced p105 phosphorylation and decrease p50 protein expression.42,43 This loss-of-function mutation attributed to NFKB1 haplo-insufficiency and has been documented to be related to immunodeficiency. Two other studies reported mutations in CVID patients in CECR1 (cat eye syndrome chromosome region, candidate 1) gene, which encodes for adenosine deaminase 2 (ADA2)44 and seventeen probable monoallelic mutations in STAT3 (signal transducer and activator of transcription 3), PIK3CD, NFKB1, CTLA4, and IKZF1 and biallelic mutations in STXBP2 (syntaxin binding protein 2), and, LRBA while screening 50 CVID patients.25 Jan et al (2017) reported several known mutations in STAT1 (signal transducer and activator of transcription 1), NLRP3 (NLR family pyrin domain containing 3), MEFV (Mediterranean fever), TNSFR13B, and novel mutations in LIG1 (DNA Ligase 1), MX1 (MX domain like GTPase 1), FBN1(Fibrillin-1), DSG1 (Desmoglein 1), UNC13D (Unc-13 homolog D), TLR1 (Toll like receptor 1), NBPF15 (NBPF member 15), FASN (Fatty acid synthase), IL1A (Interleukin 1 alpha), LAX1 (Lymphocyte transmembrane adaptor 1), SF3B1 (Splicing factor 3b subunit 1), CHD7 (Chromodomain helicase DNA binding protein 7), TUBB1 (Tubulin beta 1 class VI), ATM (ATM Serine/Threonine kinase), TYK2 (Tyrosine kinase 2), LRRC8A (Leucine rich repeat containing 8 VRAC subunit A), PRKCD, CFHR5 (Complement factor H related 5), LRBA, EPG5 (Ectopic P-granules autophagy protein 5), RAG2 (Recombination activating 2), RAG1, NCF2 (Neutrophil cytosolic factor 2), PTPRC (Protein tyrosine phosphatase receptor type C), and MASP2 (Mannan binding lectin serine peptidase 2) in seven CVID patients.45 A small subset of CVID patients, have been identified with monogenic defects, however, with introduction of whole genome sequencing and GWAS, polygenic basis of CVID disease etiology has also surfaced.46 In a recent study published by Ran et al (2018), three CVID patients were screened using NGS platform which reported with differential mutation profile (NFKB1 and LRBA) in each of the patient.47 Two other studies accomplished in 2018 reported genetic mutations in LRBA, ZBTB24 (Zinc finger and BTB domain containing 24), DNMT3B (DNA methyltransferase 3 beta), CTLA4, NFKB1, PIK3R1, PRKCD, MAPK8 (Mitogen-activated protein kinase 8), and DOCK8 (Dedicator of cytokinesis 8).48,49 Despite use of next generation technologies for unraveling the underlying genetic defects in CVID patients, different studies have reported differential variants which are attributed to genetic diversity among populations (Table 1). An amalgamation of sequencing data from different cohorts will be useful to identify a panel of genes which are responsible for pre-disposition of patients to CVID phenotype. Hence, in view of recent technological advancements, the genetic screening need to be further elaborated for effective disease management at early stage and targeting these altered genes may present as potential candidate for treatment on therapeutic front.
Table 1.
Genetic alterations identified through different next generation platforms in CVID samples. A Disintegrin and Metalloproteinase (ADAM); Activation induced cytidine deaminase (AICDA); Adenosine deaminase 2 (ADA2); AKT serine/threonine kinase 1 (AKT1); ATM Serine/Threonine kinase (ATM); B-cell lymphoma 2 like 1 (BCL2L1); B-cell lymphoma 6 (BCL6); C–C chemokine receptor type 7 (CCR7); CD40 ligand (CD40LG); CD 81 molecule (CD81); Cat eye syndrome chromosome region, candidate 1 (CECR1); Complement factor H related 5 (CFHR5); Chromodomain helicase DNA binding protein 7 (CHD7); Cytotoxic T-lymphocyte associated protein 4 (CTLA4); DNA methyltransferase 3 beta (DNMT3B); Dedicator of cytokinesis 8 (DOCK8); Desmoglein 1 (DSG1); Ectopic P-granules autophagy protein 5 (EPG5); Forkhead Box O(FOXO); Intercellular adhesion molecule 1 (ICAM1); Interferon (IFN); DNA Ligase 1 (LIG1); Fatty acid synthase (FASN); Fibrillin-1 (FBN1); Inhibitor of nuclear factor kappa B kinase regulatory subunit gamma (IKBKG); IKAROS family zinc finger 1 (IKZF1); Interleukin 1 alpha (IL1A); Interferon regulatory factor 2 binding protein 2 (IRF2BP2); Lymphocyte transmembrane adaptor 1 (LAX1); Lipopolysaccharide-responsive beige-like anchor protein (LRBA); Leucine rich repeat containing 8 VRAC subunit A (LRRC8A); Mitogen-activated protein kinase 8 (MAPK8); Mannan binding lectin serine peptidase 2 (MASP2); Mediterranean fever (MEFV); MX domain like GTPase 1 (MX1); NBPF member 15 (NBPF15); Neutrophil cytosolic factor 2 (NCF2); Nuclear factor kappa B subunit 1 (NFKB1); Nuclear factor kappa B subunit 2 (NFKB2); NLR family pyrin domain containing 3 (NLRP3); NLR family pyrin domain containing 12 (NLRP12); Paired Box 5 (PAX5); Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta (PIK3CD); Phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1); Phospholipase C, gamma (PLCG); Protein kinase C delta (PRKCD); Protein tyrosine phosphatase receptor type C (PTPRC); Recombination activating 1 (RAG1); Recombination activating 2 (RAG2); Retinoblastoma associated (RBA); Potassium calcium-activated channel subfamily N member 4 (KCNN4); Ribosomal protein S6 kinase beta-2 (RPS6KB2); Splicing factor 3b subunit 1 (SF3B1); Signal transducer and activator of transcription 1 (STAT1); Signal transducer and activator of transcription 3 (STAT3); Syntaxin binding protein 2 (STXBP2); Transcription factor 3 (TCF3); Toll like receptor 1 (TLR1); TNF receptor superfamily member 13B (TNFRSF13B); TRAF3 interacting protein 2 (TRAF3IP2); Tubulin beta 1 class VI (TUBB1); Tyrosine kinase 2 (TYK2); Unc-13 homolog D (UNC13D); Zinc finger and BTB domain containing 24 (ZBTB24).
| Technique | Patients Enrolled in study | Genes Alterations/Mutation | Year | Reference |
|---|---|---|---|---|
| Next Generation Sequencing Technology | ||||
| Hyper-IgM/CVID custom 148 gene Re-sequencing chip | 34 CVID patients | TNFRSF13B,AICDA,CD40LG, IKBKG, TRAF3IP2 | 2010 | 31 |
| Genome-wide SNP genotyping (InfiniumII HumanHap610 BeadChip) | 363 CVID patients | CVID association with ADAM and MHC genes. | 2011 | 32 |
| IGH rearrangements (Roche 454 sequencing) | 18 CVID patients | VDJ rearrangement and abnormal formation of complementarity determining region 3 (CDR3) | 2015 | 33 |
| High-throughput DNA sequencing of immunoglobulin heavy chain gene rearrangements (Roche 454 DNA sequencing) | 93 CVID patients | VDJ rearrangement and abnormal formation of complementarity determining region 3 (CDR3) | 2015 | 35 |
| Whole exome sequencing (1st patient) and Targeted Gene Panel (2nd patient) | 2 CVID patients | 1st Patient: RAG1 (c256_257delAA, c1835A > G) 2nd Patient: RAG1(c.1566G > T, c.2689C > T) |
2015 | 37 |
| Targeted PID gene sequencing (Ion PGM) | 1CVID patient | RAG1 mutation (c.1871G > A, c.2182T > C and p.H249R) | 2016 | 38 |
| Next-generation sequencing of TCRbrepertoire (ClonoSIGHT platform (Sequenta) | 42 CVID patients | Decrease in TCR repertoire diversity, naive T cells, and thymic volume was consistent with orthogonal evidence supporting thymic failure in CVID patients. | 2016 | 40 |
| Whole exome sequencing | A family with 3 affected individuals with CVID and unaffected family members | Novel mutation IRF2BP2 (c.1652G > A:p(S551N)) | 2016 | 41 |
| Targeted exome sequencing (Illumnia HiSeq 3000) | 1 patient | Novel frameshift mutation in NFKB1c.491delG (p.G165A*31) | 2016 | 42 |
| Whole exome sequencing | 5 family members of CVID patient | Two Heterozygous mutation in CECR1 (encoding ADA2) rs77563738 and novel Chr22:17,684,478 A > C) | 2016 | 44 |
| Whole exome sequencing (IlluminaHiSeq 2500) | 50 CVID patients | Monoallelic mutations (NFKB1, STAT3, CTLA4, PIK3CDand IKZF1) Biallelic mutations (LRBA and STXBP2) | 2016 | 25 |
| Targeted Sequencing (MiSeq (Illumina)) | 1 patient | RAG1: Three mutations. Two Novel:A missense variant (c.1123C > G) and frameshift deletion (c.1430delC, p.F478Sfs*14) and a known missense mutation (c.1420C > T, rs199474678) | 2017 | 39 |
| Whole exome sequencing (HiSeq 2000 or NextSeq 500 (Illumina)) | 7 CVID patients | STAT1, NLRP3, MEFV, TNSFR13B, LIG1, MX1, FBN1, DSG1, UNC13D, TLR1, NBPF15, FASN, IL1A, LAX1, SF3B1, CHD7, TUBB1, ATM, TYK2, LRRC8A, PRKDC, CFHR5, LRBA, EPG5, RAG2, RAG1, NCF2, PTPRC and MASP2 mutations. | 2017 | 45 |
| TCRβ High-throughput sequencing (Adaptive Biotechnologies) | 44 CVID patients | CVID TCRs had reduced junctional diversity and CVID CD3 sequence had increased clonality. | 2017 | 34 |
| Targeted Sequencing (MiSeq (Illumina)) | 1 patient | NFKB1frameshift mutation (c.1149delT, p.Gly384Glu ∗ 48) | 2018 | 43 |
| TCR Repertoire sequencing (Roche 454 sequencing) | 33 CVID patients | CVID patients had defective V(D)J recombination along with somatic Hyper-mutation (SHM). | 2018 | 36 |
| Whole exome sequencing (IlluminaHiseq 4000) | 3 CVID patients | 1st patient:RBA (c.8436G > C and c.4089A > T) and TNFRSF13B (c.226G > A) 2nd patient: LRBA (c.3764G > C) 3rd patient: LRBA (c.5084T > C) and NFKB1 (c.666dupG). |
2018 | 47 |
| Whole exome sequencing (IlluminaHiSeq 2000) | 36 CVID patients | LRBA, CTLA4, NFKB1,PIK3R1, PRKCD, MAPK8and DOCK8 | 2018 | 48 |
| Whole exome sequencing | 550 patients (HIgM, CVID and Agammaglobulinemia) | LRBA, ZBTB24andDNMT3Bmutations in CVID | 2018 | 49 |
| Transcriptome Expression | ||||
| GeneChip Human Genome U133A Array (Affymetrix) | 23 CVID patients | Enhanced cytotoxic effector functions, Predominance of CCR7-T cells, and Antigen activated T cells | 2004 | 50 |
| HT-12 V4 BeadChip (Illumina) | 91 CVID patients | Up-regulation of IFN responsive genes. | 2013 | 51 |
| Whole transcriptome sequencing (IlluminaHiSeq 2000) | 1 patient | NLRP12 (Heterozygous mutations) encoding NALP12 protein (p.H304Y and p.A629D) | 2014 | 52 |
| RNA sequencing (IlluminaHiSeq 2500) | 3 CVID patients | TNFRSF13B, LRBA, NLRP12and TNFRSF13Cvariants and up-regulation of NLRP12, ICAM1, CD81 and PLCG. | 2015 | 7 |
| RNA sequencing (IlluminaHiSeq 2000) | 7 equine CVID patients | Down-regulation of pro-B cell differentiation genes specifically PAX5 | 2015 | 56 |
| Epigenetic Alterations | ||||
| High-Throughput DNA methylation and Bisulfite-modified DNA pyrosequencing | 23 CVID patients | Impaired demethylation in AICDA, BCL6, STAT3, FOXO, AKT1 and NFKB2 | 2019 | 60 |
| High-Throughput DNA methylation | Monozygotic twins discordant for CVID | PIK3CD, BCL2L1, RPS6KB2, TCF3 and KCNN4 | 2015 | 55 |
| Genome wide bisulfite sequencing | Seven equine CVID patients | PAX5 gene silencing | 2015 | 56 |
Transcriptional regulation of CVID
The recent genetic platforms have tried to unravel the novel genetic mutations corroborated with patient's predisposition to PIDs. However, enormous data generated from NGS platforms have not been able to uncover the probable causes of PIDs which is in turn attributed to 70% of the human genome transcribed into non coding RNAs. The advent of RNA sequencing surmounted the lacunae as it provides quantitative determination of both coding and non-coding RNAs which will help in characterizing and identifying genetic basis of PIDs. Holm et al (2004) documented the predominance of CCR7-T (C–C chemokine receptor type 7) cells in subgroup of CVID patients.50 In another study conceptualized in 2013 on whole blood transcriptome analysis of CVID patients, the group reported up-regulation of IFN (Interferon) responsive genes manifested pertaining to impaired adaptive immunity which lead to aberrant activation of innate IFN downstream pathway.51 In another study; RNA sequencing in 20-year-old female patient revealed a novel heterozygous NLRP12 (NLR family pyrin domain containing 12) mutation along with down-regulation of CCR3 (CC chemokine receptor 3), IFN-γ (Interferon-gamma), and CCR4 (CC chemokine receptor 4) expression.52 In another study, van Schouwenburg and colleagues illustrated TNFRSF13B, LRBA, NLRP12, and TNFRSF13C variants were associated with up-regulation of NLRP12, ICAM1 (Intercellular adhesion molecule 1), CD81, and PLCG (Phospholipase C, gamma) upon corroborated analysis of whole genome and RNA sequencing data.7 The transcriptome profile of CVID patients still needs to be illustrated extensively to resolve the unanswered dimensions of CVID pathogenesis. In view of limited literature concerning the transcriptome expression of CVID patients, this needs to be further elucidated to unravel the underlying mystery of major portion of the genome transcribed into non-coding RNAs.
Epigenetic dysfunction of CVID
Despite extensive research in CVID patients, epigenome of CVID patients still remains a dimension unexplored. The relevance of epigenetic changes in CVID pathogenesis is illustrated by the role differential DNA methylation plays in the development and activation of B cells.53,54 This is attributed to CVID disease etiology which is not associated with a specific genetic defect and varies among patients with identical genetic alterations either because of polygenic defects or environmental factors which influence disease susceptibility in CVID patients. The first evidence of epigenetic pre-disposition of CVID was reported in 2015, when researchers of the Chromatin and Disease Group from the Bellvitge Biomedical Research Institute (IDIBELL) and La Paz Hospital (IDIPAZ) reported epigenetic modifications in identical monozygotic twins discordant for the disease.55 The group reported hyper-methylation of RPS6KB2 (Ribosomal protein S6 kinase beta-2), BCL2L1 (B-cell lymphoma 2 like 1), PIK3CD, KCNN4 (Potassium calcium-activated channel subfamily N member 4), and TCF3 (Transcription factor 3) genes, associated with B lymphocytes, led to defective memory cell generation, and aberrant presentation of CVID. In 2015, another report by Rebecca and colleagues documented increase in methylation of PAX5 (Paired Box 5) enhancer region leading to PAX5 gene silencing in bone marrow of equine CVID patients.56 In another recent study Kienzler and colleagues, emphasized on the role of epigenetic modifications in CVID and its importance in understanding the altered gene expression in pathogenesis of CVID.46 The B cells in CVID cells have an impaired knack to up-regulate and de-methylate genes associated with naive to memory B cells transition.57,58 In a study published in 2018 insights into cross-talk mechanism between immune activation, gut microbiome and epigenetic alterations in CVID for administration of personalized medicine based on clinical phenotype and molecular genotype.59 Lucia et al (2019) in a recent study suggested that the impaired de-methylation in CVID patients while undergoing transition from the naive to memory B cells in genes of CpG islands has been implicated in B cell signaling (STAT3, FOXO (Forkhead Box O), AKT1 (AKT serine/threonine kinase 1), and NFKB2), and at GC reaction in BCL6 (B-cell lymphoma 6), and AICDA, these may contribute to the defects reported in terminal B cell of CVID patients.60 In view of available scientific evidence, epigenetic treatment may hold as new paradigm of treatment avenues in CVID patient cohort.61
Advantages and limitations of genetic detection methods
Single gene defects in PIDs were initially identified using Sanger sequencing which involves dye-terminator methodology for DNA fragments which is coupled with capillary electrophoresis . Sanger sequencing till date remains the gold standard for DNA sequencing and is best carried out for known DNA fragments for mutational analysis. However, the major limitation of the technique includes more cost and time along with limited scalability, low throughput and resolution. Later on, sequencing-by-synthesis technique, pyro-sequencing was developed. This technology was based on nucleotide synthesis with release of pyrophosphate. The pyrophosphate is enzymatically converted to ATP, which comes in contact with enzyme luciferase leading to light production and addition of dNTPs individually to the growing DNA molecules. The major challenge encountered with this technique is the addition of new enzyme with every addition of nucleotide, laborious washing of sample between each nucleotide addition and correlation of signal intensity to incorporation of number of bases which is problematic at times for sequencing the homopolymer stretches. The major breakthrough in the sequencing technology, came with 13-years long, Human Genome Project (HGP) which was completed in 2003.62,63 With the transition of sequencing technology to development of next generation sequencing based technology (Illumina, Roche 454, Ion Torrent/PGM sequencing, Oxford Nanopore) for rapid amplification of larger stretch of human genome, the sequencing platforms allowed for sequencing with 30× coverage or more. NGS allows for massive parallel sequencing, wherein millions of DNA fragments per sample are sequenced with high precision and accuracy. The main advantages of NGS has been the feasibility to sequence whole genome and whole exome which have in turn helped decipher the underlying genetic basis of many diseases which remained unanswered till date. Of the NGS technologies; high throughput sequencing of human genome unravels genetic basis of diseases both in the coding and regulatory pathways. Transcriptome sequencing transitioned from microarray to RNA-seq and have helped decipher the entire transcriptome map associated with disease patients and provides information about the RNA expression profiles. DNA methylation profiling additionally helps study the expression profile of high and low methylated CpG islands intensity to study the role of epigenetic modifications in disease biology. With continuous technological advancements the cost incurred to sequence whole exome/genome or transcriptome have drastically been reduced. However, the major limitation of the advanced technologies is the huge amount of data generated from NGS runs with massive parallel sequencing of which data analysis is complex and need high end computational back-up and set-up for bioinformatics analysis. The advantages and limitations of various sequencing technologies (Sanger sequencing, Pyro-sequencing) and NGS platforms (whole exome sequencing, whole genome sequencing, Transcriptome profiling, epigenome profiling and proteome profiling) have been detailed in Table 2. Although, NGS is cost effective and fast in comparison to first generation sequencing technologies, it still remains expensive for small scale laboratories and individuals. The small read lengths leads to highly repetitive sequences which is a major shortcoming. Further, NGS data is complex, time consuming and need good bioinformatics expertise to comprehend the huge data generated. In view of these limitations, third generation sequencing technologies have been instated to address the gap between NGS and conventional sequencing technologies which involves direct sequencing of single molecules with long read lengths, low cost and time whereby maintaining quality of genome assembly.
Table 2.
Advantages and limitations of different genomic, transcriptomic and proteomics based platforms in Common variable immunodeficiency disease. ELISA, Enzyme-linked immunosorbent assay; SNP, Single nucleotide polymorphism.
| Technology | Basic technique | Advantages | Limitations | Ref |
|---|---|---|---|---|
| Genome-wide Association Studies |
|
|
|
32,64, 65, 66, 67 |
| Sanger Sequencing |
|
|
|
37,44,49,68, 69, 70 |
| Pyro-sequencing |
|
|
|
27,35,55,60 |
| Next Generation Sequencing |
|
|
|
|
| Targeted Sequencing |
|
|
31,38,39,42,43 | |
| Whole Exome Sequencing |
|
|
25,37,41,44,45,47,48 | |
| Whole Genome Sequencing |
|
|
30,48 | |
| Microarray |
|
|
|
50,51 |
| RNA-Sequencing |
|
|
|
7,52,56 |
| Epigenome profiling |
|
|
|
55,56,60 |
| Fourier-transform infrared (FTIR) spectroscopy |
|
|
|
71 |
Future perspective
The management of CVID patients has transitioned from the initial era of clinical diagnosis to molecular diagnosis. However, despite the technical advancements the precise genetic basis of CVID still remains enigmatic. The genetic diversity among different patient cohorts of diverse ethnic background is another challenge in CVID genetics. The advancements in the field of next generation has brought has many steps closer to treat and manage CVID patients; which remained enigmatic and static over the past decade. The muti-omics approach integrating the findings from NGS platform along with expression profiling, proteomics, metabolomics and methylation patterns concurrently will enhance our understanding of the underlying complexities of CVID genetics. The strength of coalescing complementary technologies together will help comprehend the CVID genetic architecture, which still remains an unanswered paradox. The complex challenges of CVID need to be deciphered for individualization and moderation of therapeutic treatments on individual basis.
Conclusion
The heterogeneous paradigm of CVID still remains a dimension to be explored. Recently, NGS has been actively deployed to comprehend the pathogenic mutations responsible for CVID clinical manifestations. However, pertaining to CVID disease heterogeneity, studies have reported different genetic profile in different cohort of CVID patients which is attributed to ethnic diversity, different risk factors and diverse epigenetic profile. Despite the recent studies to explore the genetic basis of CVID, the scope of translation of genome-wide association studies in patient care is limited in view of small sample size pertaining to relatively low prevalence of CVID in comparison to other primary immunodeficiency disorders. Thus, the available scientific evidence warrants an insight into multi-omics approach which would conjugate next generation sequencing technology, gene expression studies, epigenetics, proteomics and metabolomics together, with an aim to unleash the underlying molecular complexities in CVID pathogenesis. The implementations of next generation techniques to elucidate genetic alterations in health care have helped us realize the potential of precision medicine. The dawn of NGS and ‘omic’ technologies in molecular research has transformed the perception of looking through genetic and epigenetic diaspora of disease etiology and its associated underlying pathogenesis. Hence, personalized treatment based on phenotypic and genotypic alterations is the need of the hour for CVID patients. In summary, sustained efforts of multi-omics on integrative platform will pave the way ahead for better clinical characterization of CVID which can be translated into therapeutic targets for precision medicine.
Conflict of interest
The authors declare no conflict of interest.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Authorship
Contributions: A.V, B.A, J. AK and R.A conceptualized and designed the review; A.V, A.B and D. J performed literature search; A.V and A.B prepared manuscript; J. AK and R.A directed the review; A.V, A.B, J. AK and R.A edited the manuscript and all authors approved the final versions of the review.
Acknowledgments
The authors would like to acknowledge Department of Pediatrics, Advanced Pediatrics Center, Post Graduate Institute of Medical Education and Research (PGIMER), Chandigarh for providing a platform to complete this review.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Seidel M.G., Kindle G., Gathmann B. The European society for immunodeficiencies (ESID) registry working definitions for the clinical diagnosis of inborn errors of immunity. J Allergy Clin Immunol In Pract. 2019;7(6):1763–1770. doi: 10.1016/j.jaip.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Bousfiha A., Jeddane L., Picard C. The 2017 IUIS phenotypic classification for primary immunodeficiencies. J Clin Immunol. 2018;38(1):129–143. doi: 10.1007/s10875-017-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimbacher B., Party E.R.W. The European society for immunodeficiencies (ESID) registry 2014. Clin Exp Immunol. 2014;178(Suppl 1):18–20. doi: 10.1111/cei.12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonilla F.A., Barlan I., Chapel H. International consensus document (ICON): common variable immunodeficiency disorders. J Allergy Clin Immunol In Pract. 2016;4(1):38–59. doi: 10.1016/j.jaip.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogaert D.J., Dullaers M., Lambrecht B.N., Vermaelen K.Y., De Baere E., Haerynck F. Genes associated with common variable immunodeficiency: one diagnosis to rule them all? J Med Genet. 2016;53(9):575–590. doi: 10.1136/jmedgenet-2015-103690. [DOI] [PubMed] [Google Scholar]
- 6.Tseng C.W., Lai K.L., Chen D.Y., Lin C.H., Chen H.H. The incidence and prevalence of common variable immunodeficiency disease in taiwan, a population-based study. PLoS One. 2015;10(10):e0140473. doi: 10.1371/journal.pone.0140473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Schouwenburg P.A., Davenport E.E., Kienzler A.K. Application of whole genome and RNA sequencing to investigate the genomic landscape of common variable immunodeficiency disorders. Clin Immunol. 2015;160(2):301–314. doi: 10.1016/j.clim.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimbacher B., Hutloff A., Schlesier M. Homozygous loss of ICOS is associated with adult-onset common variable immunodeficiency. Nat Immunol. 2003;4(3):261–268. doi: 10.1038/ni902. [DOI] [PubMed] [Google Scholar]
- 9.Kuijpers T.W., Bende R.J., Baars P.A. CD20 deficiency in humans results in impaired T cell-independent antibody responses. J Clin Investig. 2010;120(1):214–222. doi: 10.1172/JCI40231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiel J., Kimmig L., Salzer U. Genetic CD21 deficiency is associated with hypogammaglobulinemia. J Allergy Clin Immunol. 2012;129(3):801–810. doi: 10.1016/j.jaci.2011.09.027. e806. [DOI] [PubMed] [Google Scholar]
- 11.van Zelm M.C., Reisli I., van der Burg M. An antibody-deficiency syndrome due to mutations in the CD19 gene. N Engl J Med. 2006;354(18):1901–1912. doi: 10.1056/NEJMoa051568. [DOI] [PubMed] [Google Scholar]
- 12.van Zelm M.C., Smet J., Adams B. CD81 gene defect in humans disrupts CD19 complex formation and leads to antibody deficiency. J Clin Investig. 2010;120(4):1265–1274. doi: 10.1172/JCI39748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warnatz K., Salzer U., Rizzi M. B-cell activating factor receptor deficiency is associated with an adult-onset antibody deficiency syndrome in humans. Proc Natl Acad Sci U S A. 2009;106(33):13945–13950. doi: 10.1073/pnas.0903543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Resnick E.S., Cunningham-Rundles C. The many faces of the clinical picture of common variable immune deficiency. Curr Opin Allergy Clin Immunol. 2012;12(6):595–601. doi: 10.1097/ACI.0b013e32835914b9. [DOI] [PubMed] [Google Scholar]
- 15.Chapel H., Lucas M., Lee M. Common variable immunodeficiency disorders: division into distinct clinical phenotypes. Blood. 2008;112(2):277–286. doi: 10.1182/blood-2007-11-124545. [DOI] [PubMed] [Google Scholar]
- 16.Lucas C.L., Chandra A., Nejentsev S., Condliffe A.M., Okkenhaug K. PI3Kdelta and primary immunodeficiencies. Nat Rev Immunol. 2016;16(11):702–714. doi: 10.1038/nri.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coulter T.I., Cant A.J. The treatment of activated PI3Kdelta syndrome. Front Immunol. 2018;9:2043. doi: 10.3389/fimmu.2018.02043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alkhairy O.K., Abolhassani H., Rezaei N. Spectrum of phenotypes associated with mutations in LRBA. J Clin Immunol. 2016;36(1):33–45. doi: 10.1007/s10875-015-0224-7. [DOI] [PubMed] [Google Scholar]
- 19.Jansen E.J., Timal S., Ryan M. ATP6AP1 deficiency causes an immunodeficiency with hepatopathy, cognitive impairment and abnormal protein glycosylation. Nat Commun. 2016;7:11600. doi: 10.1038/ncomms11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoeger B., Serwas N.K., Boztug K. Human NF-kappaB1 haploinsufficiency and Epstein-Barr virus-induced disease-molecular mechanisms and consequences. Front Immunol. 2017;8:1978. doi: 10.3389/fimmu.2017.01978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Picard C., Bobby Gaspar H., Al-Herz W. International union of immunological societies: 2017 primary immunodeficiency diseases committee report on inborn errors of immunity. J Clin Immunol. 2018;38(1):96–128. doi: 10.1007/s10875-017-0464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartley J.L., Zachos N.C., Dawood B. Mutations in TTC37 cause trichohepatoenteric syndrome (phenotypic diarrhea of infancy) Gastroenterology. 2010;138(7):2388–2398. doi: 10.1053/j.gastro.2010.02.010. 2398 e2381-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giannelou A., Wang H., Zhou Q. Aberrant tRNA processing causes an autoinflammatory syndrome responsive to TNF inhibitors. Ann Rheum Dis. 2018;77(4):612–619. doi: 10.1136/annrheumdis-2017-212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelsen J.R., Dawany N., Moran C.J. Exome sequencing analysis reveals variants in primary immunodeficiency genes in patients with very early onset inflammatory bowel disease. Gastroenterology. 2015;149(6):1415–1424. doi: 10.1053/j.gastro.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maffucci P., Filion C.A., Boisson B. Genetic diagnosis using whole exome sequencing in common variable immunodeficiency. Front Immunol. 2016;7:220. doi: 10.3389/fimmu.2016.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picard C., Al-Herz W., Bousfiha A. Primary immunodeficiency diseases: an update on the classification from the international union of immunological societies Expert committee for primary immunodeficiency 2015. J Clin Immunol. 2015;35(8):696–726. doi: 10.1007/s10875-015-0201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elgizouli M., Lowe D.M., Speckmann C. Activating PI3Kdelta mutations in a cohort of 669 patients with primary immunodeficiency. Clin Exp Immunol. 2016;183(2):221–229. doi: 10.1111/cei.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fliegauf M., Bryant V.L., Frede N. Haploinsufficiency of the NF-kappaB1 subunit p50 in common variable immunodeficiency. Am J Hum Genet. 2015;97(3):389–403. doi: 10.1016/j.ajhg.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuehn H.S., Boisson B., Cunningham-Rundles C. Loss of B Cells in patients with heterozygous mutations in IKAROS. N Engl J Med. 2016;374(11):1032–1043. doi: 10.1056/NEJMoa1512234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ameratunga R., Lehnert K., Woon S.T. Review: diagnosing common variable immunodeficiency disorder in the era of genome sequencing. Clin Rev Allergy Immunol. 2018;54(2):261–268. doi: 10.1007/s12016-017-8645-0. [DOI] [PubMed] [Google Scholar]
- 31.Wang H.Y., Gopalan V., Aksentijevich I. A custom 148 gene-based resequencing chip and the SNP explorer software: new tools to study antibody deficiency. Hum Mutat. 2010;31(9):1080–1088. doi: 10.1002/humu.21322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orange J.S., Glessner J.T., Resnick E. Genome-wide association identifies diverse causes of common variable immunodeficiency. J Allergy Clin Immunol. 2011;127(6):1360–1367. doi: 10.1016/j.jaci.2011.02.039. e1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.H IJ, Wentink M., van Zessen D. Strategies for B-cell receptor repertoire analysis in primary immunodeficiencies: from severe combined immunodeficiency to common variable immunodeficiency. Front Immunol. 2015;6:157. doi: 10.3389/fimmu.2015.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramesh M., Hamm D., Simchoni N., Cunningham-Rundles C. Clonal and constricted T cell repertoire in common variable immune deficiency. Clin Immunol. 2017;178:1–9. doi: 10.1016/j.clim.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roskin K.M., Simchoni N., Liu Y. IgH sequences in common variable immune deficiency reveal altered B cell development and selection. Sci Transl Med. 2015;7(302):302ra135. doi: 10.1126/scitranslmed.aab1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Schouwenburg P.A., H IJ, Pico-Knijnenburg I. Identification of CVID patients with defects in immune repertoire formation or specification. Front Immunol. 2018;9:2545. doi: 10.3389/fimmu.2018.02545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchbinder D., Baker R., Lee Y.N. Identification of patients with RAG mutations previously diagnosed with common variable immunodeficiency disorders. J Clin Immunol. 2015;35(2):119–124. doi: 10.1007/s10875-014-0121-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cifaldi C., Scarselli A., Petricone D. Agammaglobulinemia associated to nasal polyposis due to a hypomorphic RAG1 mutation in a 12 years old boy. Clin Immunol. 2016;173:121–123. doi: 10.1016/j.clim.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 39.Schroder C., Baerlecken N.T., Pannicke U. Evaluation of RAG1 mutations in an adult with combined immunodeficiency and progressive multifocal leukoencephalopathy. Clin Immunol. 2017;179:1–7. doi: 10.1016/j.clim.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 40.Wong G.K., Millar D., Penny S. Accelerated loss of TCR repertoire diversity in common variable immunodeficiency. J Immunol. 2016;197(5):1642–1649. doi: 10.4049/jimmunol.1600526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keller M.D., Pandey R., Li D. Mutation in IRF2BP2 is responsible for a familial form of common variable immunodeficiency disorder. J Allergy Clin Immunol. 2016;138(2):544–550. doi: 10.1016/j.jaci.2016.01.018. e544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boztug H., Hirschmugl T., Holter W. NF-kappaB1 haploinsufficiency causing immunodeficiency and EBV-driven lymphoproliferation. J Clin Immunol. 2016;36(6):533–540. doi: 10.1007/s10875-016-0306-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dieli-Crimi R., Martinez-Gallo M., Franco-Jarava C. Th1-skewed profile and excessive production of proinflammatory cytokines in a NFKB1-deficient patient with CVID and severe gastrointestinal manifestations. Clin Immunol. 2018;195:49–58. doi: 10.1016/j.clim.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 44.Schepp J., Bulashevska A., Mannhardt-Laakmann W. Deficiency of adenosine deaminase 2 causes antibody deficiency. J Clin Immunol. 2016;36(3):179–186. doi: 10.1007/s10875-016-0245-x. [DOI] [PubMed] [Google Scholar]
- 45.Stuchly J., Kanderova V., Vlkova M. Common Variable Immunodeficiency patients with a phenotypic profile of immunosenescence present with thrombocytopenia. Sci Rep. 2017;7:39710. doi: 10.1038/srep39710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kienzler A.K., Hargreaves C.E., Patel S.Y. The role of genomics in common variable immunodeficiency disorders. Clin Exp Immunol. 2017;188(3):326–332. doi: 10.1111/cei.12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li R., Zheng Y., Li Y. Common variable immunodeficiency with genetic defects identified by whole exome sequencing. BioMed Res Int. 2018;2018:3724630. doi: 10.1155/2018/3724630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Valles-Ibanez G., Esteve-Sole A., Piquer M. Evaluating the genetics of common variable immunodeficiency: monogenetic model and beyond. Front Immunol. 2018;9:636. doi: 10.3389/fimmu.2018.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yazdani R., Abolhassani H., Kiaee F. Comparison of common monogenic defects in a large predominantly antibody deficiency cohort. J Allergy Clin Immunol In Pract. 2019;7(3):864–878. doi: 10.1016/j.jaip.2018.09.004. e869. [DOI] [PubMed] [Google Scholar]
- 50.Holm A.M., Sivertsen E.A., Tunheim S.H. Gene expression analysis of peripheral T cells in a subgroup of common variable immunodeficiency shows predominance of CCR7(-) effector-memory T cells. Clin Exp Immunol. 2004;138(2):278–289. doi: 10.1111/j.1365-2249.2004.02630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park J., Munagala I., Xu H. Interferon signature in the blood in inflammatory common variable immune deficiency. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0074893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borte S., Celiksoy M.H., Menzel V. Novel NLRP12 mutations associated with intestinal amyloidosis in a patient diagnosed with common variable immunodeficiency. Clin Immunol. 2014;154(2):105–111. doi: 10.1016/j.clim.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 53.Lai A.Y., Mav D., Shah R. DNA methylation profiling in human B cells reveals immune regulatory elements and epigenetic plasticity at Alu elements during B-cell activation. Genome Res. 2013;23(12):2030–2041. doi: 10.1101/gr.155473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee S.T., Xiao Y., Muench M.O. A global DNA methylation and gene expression analysis of early human B-cell development reveals a demethylation signature and transcription factor network. Nucleic Acids Res. 2012;40(22):11339–11351. doi: 10.1093/nar/gks957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodriguez-Cortez V.C., Del Pino-Molina L., Rodriguez-Ubreva J. Monozygotic twins discordant for common variable immunodeficiency reveal impaired DNA demethylation during naive-to-memory B-cell transition. Nat Commun. 2015;6:7335. doi: 10.1038/ncomms8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tallmadge R.L., Shen L., Tseng C.T., Miller S.C., Barry J., Felippe M.J. Bone marrow transcriptome and epigenome profiles of equine common variable immunodeficiency patients unveil block of B lymphocyte differentiation. Clin Immunol. 2015;160(2):261–276. doi: 10.1016/j.clim.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J., Wei Z., Li Y.R. Understanding the genetic and epigenetic basis of common variable immunodeficiency disorder through omics approaches. Biochim Biophys Acta. 2016;1860(11 Pt B):2656–2663. doi: 10.1016/j.bbagen.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 58.Salzer U., Unger S., Warnatz K. Common variable immunodeficiency (CVID): exploring the multiple dimensions of a heterogeneous disease. Ann N Y Acad Sci. 2012;1250:41–49. doi: 10.1111/j.1749-6632.2011.06377.x. [DOI] [PubMed] [Google Scholar]
- 59.Jorgensen S.F., Fevang B., Aukrust P. Autoimmunity and inflammation in CVID: a possible crosstalk between immune activation, gut microbiota, and epigenetic modifications. J Clin Immunol. 2019;39(1):30–36. doi: 10.1007/s10875-018-0574-z. [DOI] [PubMed] [Google Scholar]
- 60.Del Pino-Molina L., Rodriguez-Ubreva J., Torres Canizales J. Impaired CpG demethylation in common variable immunodeficiency associates with B cell phenotype and proliferation rate. Front Immunol. 2019;10:878. doi: 10.3389/fimmu.2019.00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rae W. Indications to epigenetic dysfunction in the pathogenesis of common variable immunodeficiency. Arch Immunol Ther Exp. 2017;65(2):101–110. doi: 10.1007/s00005-016-0414-x. [DOI] [PubMed] [Google Scholar]
- 62.Collins F.S., Morgan M., Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003;300(5617):286–290. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 63.International Human Genome Sequencing C. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 64.Kralovicova J., Hammarstrom L., Plebani A., Webster A.D., Vorechovsky I. Fine-scale mapping at IGAD1 and genome-wide genetic linkage analysis implicate HLA-DQ/DR as a major susceptibility locus in selective IgA deficiency and common variable immunodeficiency. J Immunol. 2003;170(5):2765–2775. doi: 10.4049/jimmunol.170.5.2765. [DOI] [PubMed] [Google Scholar]
- 65.Li J., Jorgensen S.F., Maggadottir S.M. Association of CLEC16A with human common variable immunodeficiency disorder and role in murine B cells. Nat Commun. 2015;6:6804. doi: 10.1038/ncomms7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y.R., Zhao S.D., Li J. Genetic sharing and heritability of paediatric age of onset autoimmune diseases. Nat Commun. 2015;6:8442. doi: 10.1038/ncomms9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maggadottir S.M., Li J., Glessner J.T. Rare variants at 16p11.2 are associated with common variable immunodeficiency. J Allergy Clin Immunol. 2015;135(6):1569–1577. doi: 10.1016/j.jaci.2014.12.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abolhassani H., Wang N., Aghamohammadi A. A hypomorphic recombination-activating gene 1 (RAG1) mutation resulting in a phenotype resembling common variable immunodeficiency. J Allergy Clin Immunol. 2014;134(6):1375–1380. doi: 10.1016/j.jaci.2014.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aird A., Lagos M., Vargas-Hernandez A. Novel heterozygous mutation in NFKB2 is associated with early onset CVID and a functional defect in NK cells complicated by disseminated CMV infection and severe nephrotic syndrome. Front Pediatr. 2019;7:303. doi: 10.3389/fped.2019.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kotlinowski J., Bukowska-Strakova K., Koppolu A. A novel monoallelic nonsense mutation in the NFKB2 gene does not cause a clinical manifestation. Front Genet. 2019;10:140. doi: 10.3389/fgene.2019.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Callery E.L., Morais C.L.M., Paraskevaidi M. New approach to investigate Common Variable Immunodeficiency patients using spectrochemical analysis of blood. Sci Rep. 2019;9(1):7239. doi: 10.1038/s41598-019-43196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]