Abstract
Phosphoenolpyruvate carboxykinase 1 (PCK1), a step limiting enzyme of gluconeogenesis, is downregulated in hepatocellular carcinoma (HCC). Overexpression of PCK1 has been shown to suppress hepatoma cell growth, but the underlying mechanism remains unclear. We used recombinant adenovirus overexpressing PCK1 or GFP in Huh7 cells, and the differentially expressed genes (DEGs) were identified by RNA-Seq. 180 were upregulated by PCK1 overexpression, whereas 316 were downregulated. Pathway analysis illustrated that PCK1 was closely correlated with Wnt signaling pathway and TGF-beta signaling pathway. Hence, Wnt signaling pathway and its downstream component, FZD2, FZD6, FZD7 and β-catenin were confirmed by qRT-PCR and Western blot. In vivo we also observed that PCK1 had restrained tumor growth as a result of decreasing expression of β-catenin. Whole-transcriptomic profile analysis discovered that overexpression of PCK1 downregulates several oncogenic signaling pathways in HCC, providing potential therapeutic targets for improving HCC therapy.
Keywords: Hepatocellular carcinoma cells, Oncogene, Phosphoenolpyruvate carboxylase kinase1 (PCK1), RNA sequencing, Wnt signaling pathway
Introduction
Liver cancer is the fourth cause of cancer death worldwide in 2018, with about 841,000 new cases and 782,000 deaths annually. Hepatocellular carcinoma (HCC) comprises 75%–85% of primary liver cancer and lacks effective treatment measures.1 In recent years, the role of metabolism in tumorigenesis received extensive attention. A tumor is not only considered to be a genetic disease but also a metabolic disease.2 Tumor cells are achieved by reprogramming their own metabolic patterns to balance between energy and synthesis.3
Gluconeogenesis is a process by which noncarbohydrate precursor molecules are converted to glucose and glycogen. Nowadays, scientists believe that activating gluconeogenesis will disrupt metabolic reprogramming, and leads to an imbalance of cancer energy.4 Therefore, researchers have made great efforts to promote the development of anti-tumor treatment based on gluconeogenesis.5 Phosphoenolpyruvate carboxykinase (PEPCK also known as PCK, EC number 4.1.1.32) is capable of catalyzing the reaction from oxaloacetate to phosphoenolpyruvate, and is one of the limiting enzymes in gluconeogenesis. PEPCK has two isozymes, one of which is cytoplasmic phosphoenolpyruvate carboxykinase (PEPCK-C), encoded by the PCK1 gene; the other is located in the mitochondria, encoded by the PCK2 gene. In the mammalian liver, PCK1 accounts for over 95% of the activity.6
Early studies found that PCK1 was downregulated in early stage HCC.4, 7 p53 was found to downregulate PCK1 and G6PC in the colon cancer cells, and may exert anti-cancer function by inhibiting gluconeogenesis.8 In the liver and kidney, PCK1 also acts as a tumor suppressor.9, 10 Taken together, these studies indicate that PCK1 is closely related to oncogenesis, and the molecular mechanism remains to be further studied. In the presented work, we used RNA-seq analysis in an attempt to explore the differentially expressed genes (DEGs) in PCK-overexpression hepatoma cells. Several oncogenic signaling pathways were downregulated upon overexpression of PCK1 in HCC cell lines. Our results provide more profound insights into the underlying molecular mechanisms of PCK1 deficiency and HCC progression.
Materials and methods
Adenovirus production
The full-length cDNA of PCK1 (coding sequence of NM_002591) was cloned from plasmid pOTB7-PCK1 (Cat# FL07339; GeneCopoeia, Guangzhou, China) and cloned into the vector pAdTrack-TO4 (kindly gifted by Dr. Tong-Chuan He, University of Chicago, USA). Adenoviral recombinant Ad-PCK1 was generated using the AdEasy system. Green fluorescent protein-expressing analogous adenovirus (Ad-GFP) was applied as a control.
CRISPR/Cas9-mediated knockout of PCK1
The CRISPR/Cas9 plasmids lentiCRISPR v2, pMD2.G, and psPAX2 were kindly provided by Prof. Ding Xue from the School of Life Sciences, Tsinghua University (Beijing, China). Single-guide RNAs targeting human PCK1 were designed using the E-CRISP online tool (http://www.e-crisp.org/E-CRISP/designcrispr.html). The PCK1 targeting sequences were synthesized by TsingKe Biological Technology (Chongqing, China) and cloned into lentiCRISPR v2 vector. PCK1-knockout efficiency was confirmed by western blotting. PCK1-knockout and control cells are referred to as PCK1-KO and parental cells. All primers are shown in Table S1.
Western blot analysis
Cell proteins were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis and electrotransferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). The membranes were incubated with primary antibodies against PCK1 (1:1000; Cat#BS6870; Bioworld), β-catenin (1:5000; Cat#66379-1-Ig; Proteintech). Then, the membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibody (Abcam). Protein bands were detected using Super Signal West Pico Chemiluminescent Substrate Kit (Millipore). β-actin (Cat# BL005B; Biosharp) were used as an internal control. All experiments were repeated three times independently.
Cell culture
Huh7, PRF/PLC/5 and MHCC97H cells were acquired from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Hyclone, Logan, UT, USA) added with 10% fetal bovine serum (FBS; Gibco, Rockville, MD, USA), 100 U/mL penicillin, and 100 mg/mL streptomycin at 37 °C in an atmosphere containing 5% CO2.
RNA-sequencing (RNA-seq)
For RNA sequencing, Huh7 cells were infected with Ad-GFP or Ad-PCK1 for 36 h. Total RNA was extracted using TRIzol (Invitrogen), according to the manufacturer's instructions. RNA-seq experiments were performed by Shanghai Novel Bio Ltd. Briefly, strand-specific RNA-seq libraries were prepared using the Total RNA-seq (H/M/R) Library Prep Kit (Vazyme Biotech, Nanjing, China) and were sequenced on Ion Torrent Proton. Sequences have been deposited in the NCBI GEO database, https://www.ncbi.nlm.nih.gov/geo (accession number GSE117822).
Differential gene expression analysis
Based on the data, we performed differential gene expression analysis in the overexpressing vs Control group. We used the internationally recognized algorithm DESeq to screen differential genes for Counts. Screening was performed under the threshold of Log2FC > 0.585 or < -0.585, and FDR<0.05.
Gene ontology (GO) analysis
We acquired GO annotations from NCBI, UniProt (http://www.uniprot.org/), and the Gene Ontology Consortium (http://www.geneontology.org/). Fisher's exact test was used to select significant GO categories, and false discovery rate (FDR) was applied to rectify the P-values.11
Pathway analysis
Pathway analysis was applied to decide the significant pathways of the DEGs in the light of the KEGG database. Fisher's exact test was employed to identify significant pathways, and the threshold of significance was used on the basis of the P-value and FDR.12
Path-act-network and gene-act-network analysis
The inter-regulation associated with all pathways was organized into a database. In this analysis, we used pathway analysis to select the upregulated and downregulated pathway term with P-value < 0.05.13
Reverse transcription (RT)-PCR, and real-time PCR
RNA was reverse transcribed using moloney murine leukemia virus reverse transcriptase (MMuLV-RT, Promega). Quantification of target genes was used by SYBR Green qPCR on a CFX Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). All primer sequences are listed in the Table S1. Relative expression was counted as a ratio of the expression of the transcript to GAPDH.
Histological and immunohistochemistry (IHC) analysis
Liver samples were fixed in fresh 4% paraformaldehyde and subjected to routine histological procedures for embedding in paraffin. Then, the samples were cut in to 4.5-μm sections, which were processed for hematoxylin and eosin (HE) staining or IHC staining with antibodies targeting PCK1(1:500), and β-catenin (1:500). For IHC assay, the sections were incubated with secondary anti-rabbit IgG (ZSGB-BIO, Beijing, China) and stained with 3,3′-diaminobenzidine (ZSGB-BIO). Stained slides were scanned with a Pannoramic Scan 250 Flash or MIDI system and images were acquired using Pannoramic Viewer 1.15.2 (3DHistech, Budapest, Hungary).
Animal models
For the orthotopic implantation model, 12 BALB/c nude mice were randomly divided into AdGFP and AdPCK1 groups (6 mice per group). The MHCC97H cells infected by AdGFP or AdPCK1 (1 × 105 cells/injection) were collected and implanted into the left lobes of the livers of nude mice. At 7 weeks after implantation, the mice were sacrificed and liver tissues were collected for histological examination. Animal experiments were carried out according to the guidelines of the Institutional Animal Care and Use Committee at Chongqing Medical University (The project license number: 2017012), and protocols of animal care and use adhere to National Regulations for the administration of laboratory animals.
Statistical analysis
All values are showed as means standard deviations (SDs). Student's t-tests were applied to compare two groups. Differences with P-values<0.05 were deemed statistically significant.
Results
RNA-Seq data analysis from Huh7 overexpressing PCK1
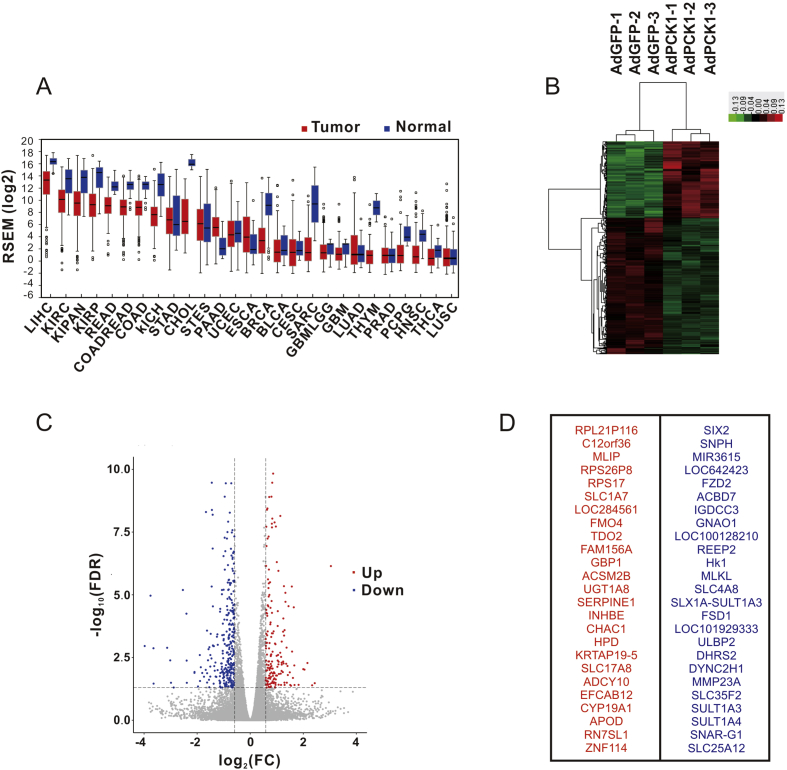
The TCGA database was queried for PCK1 mRNA expression levels across different human tumor types (Fig. 1A). PCK1 is significantly low expressed in most tumor tissues, especially in liver hepatocellular carcinoma, kidney renal papillary cell carcinoma, cholangiocarcinoma, breast invasive carcinoma, and thymoma. However, some tumor types, such as pancreatic adenocarcinoma, esophageal carcinoma, and stomach adenocarcinoma, have high expression level of PCK1. This observation indicated that PCK1 might have contrasting roles in tumorigenesis in different tumor types.
Figure 1.
Identification of DEGs between Huh7 cells overexpressing PCK1 and the control. (A) The levels of PCK1 expression in tumor tissues are shown with normal tissues. The colorized bars represent tumor nd normal issues. The analysis data is derived from Firebrowse (http://firebrowse.org/) for study details. (B) Heat map visualization of DEGs between samples of different data sets. Transcript enrichment is encoded in the heat map from low o high. (C) Volcano plot of genes differentially expressed in DEGs. The log2 fold change difference associating with the Ad-GFP and Ad-PCK1 samples is showed on the x-axis, and negative log of P-values is showed on the y-axis. Each point shows a gene with detectable expression in both sample. (D) The top 50 differential genes affected by PCK1, red for up-regulation and blue for down-regulation.
A major function of the liver is to provide fuel to other organs in the human body through glycogenolysis or gluconeogenesis. Recent studies found that in more than 200 pairs of HCC and surrounding normal tissues, the expression of PCK1 and PCK2 were both significantly downregulated in HCC.14 We firstly generated the PCK1-overexpression model in Huh7 cells infected by the Ad-PCK1 and Ad-GFP. Total RNA was extracted to apply RNA-Seq analysis and then screen DEGs. According to DEGs Painted into a heat map, there were 180 upregulated genes and 316 downregulated genes in the PCK1-overexpression cells (Fig. 1B). To acquire an overview of the differential status of DEGs, in the volcano map, the gene expression rate is displayed on the x-axis, and the differences in gene expression between the groups are exhibited on the y-axis, as shown in Fig. 1C. In the PCK1-overexpression model, we screened the top 50 DEGs (Fig. 1D), in which the up-regulated significant genes contained MLIP, SLC1A7, FMO4, ACSM2B, INHBE, CHAC1, etc., and the down-regulated significant genes included SLC25A12, DHRS2, ULBP2, MLKL, FZD2, SIX2, etc. These genes regulated by PCK1 are closely related to biological function and also significantly correlated with tumorigenesis.
Gene ontology analysis in PCK1-overexpression cells
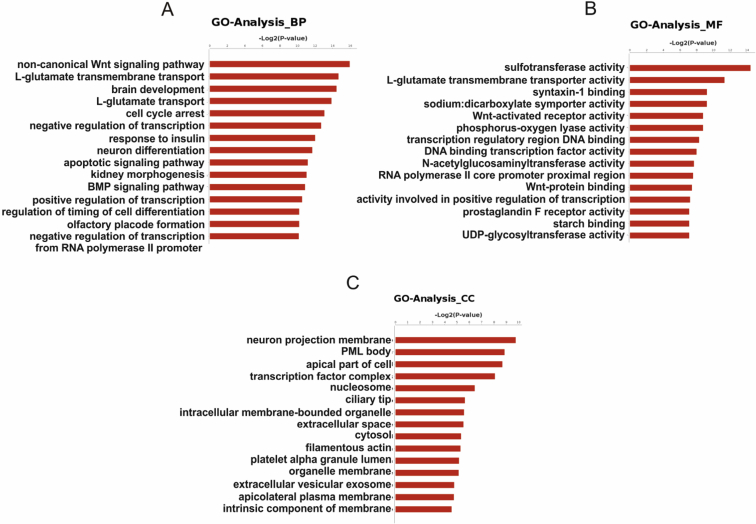
An overview analysis of the significant functions of these DEGs might further deepen our understanding of the relationship between PCK1 and HCC. Therefore, Enriched GO terms are exhibited in Figure 2, Figure 3C and arranged based on biological processes, molecular functions and cellular components. The significant biological processes for DEGs were mainly enriched in Wnt signaling pathway, l-glutamate transport, cell cycle arrest, negative regulation of transcription, and response to insulin. Referring to molecular function, both upregulated and downregulated genes were mainly mapped to “activity” (sulfotransferase activity, l-glutamate transmembrane transporter activity, Wnt-activated receptor activity, and phosphorus-oxygen lyase activity) and “binding” (syntaxin-1 binding, transcription regulatory region DNA binding and Wnt-protein binding). For cellular components annotation classification, most DEGs mapped to the membrane, cytosol and extracellular space.
Figure 2.
Significant GO terms from the overexpressing PCK1 cell line. (A) BP, Biological Process; (B) MF, Molecular Function; (C) CC, Cellular Component. P values < 0.01 for all significant GO terms.
Figure 3.
Pathway enrichment and Gene act network analysis. (A) Pathway interaction network for overexpressing PCK1 cells. (B) Gene act network analysis in huh7 cells overexpressing PCK1.
Pathway analysis and differentially expressed gene interaction network
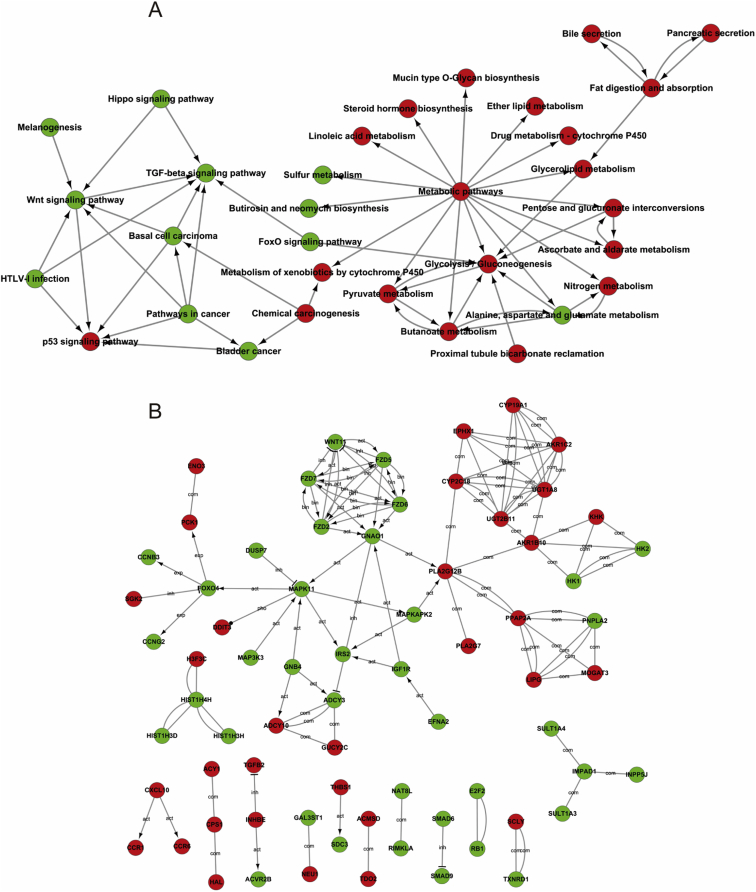
Pathway terms with significant enrichment were screened in PCK1 overexpression cells. The pathway terms that showed important enrichment were applied to acquire a pathway interaction network for further analysis (Fig. 3A). Pathway terms that exhibited significant enrichment included several major processes, such as P53 signaling pathway, TGF-beta signaling pathway, Hippo signaling pathway, FoxO signaling pathway, etc. Moreover, we found that most pathways were associated with the metabolic pathway interacted with glycolysis/gluconeogenesis, pyruvate metabolism, type II diabetes mellitus, nitrogen metabolism, glycerolipid metabolism, steroid hormone biosynthesis, etc. Pathway interaction network analysis confirmed that the metabolic pathway was the principal core pathway due to PCK1 is a key enzyme in gluconeogenesis. The Wnt signaling pathway, TGF-beta signaling pathway and pathways involved in cancer also interacted with many pathways indicated by the analysis. Thus, these pathways were also core networks regulated by PCK1 in HCC.
To further investigate the relationship between PCK1 and HCC, we presented a gene interaction network to decide the associations between the significant DEGs (Fig. 3B). The possible associations between DEGs contained activation, binding, expression, inhibition, and compound. Combined with the pathway analysis results, MAPK11 and FOXO4 appeared to be significant genes based on the gene interaction network analysis because these genes showed a strong centrality. UGT1A8, CYP2C18, EPHX1 and AKR1B10 showed interactive effects with many upregulated tumor-associated genes. We observed that the Wnt signaling pathway-related genes WNT1, FZD2, FZD5, FZD6, FZD7, and GNAO1 were downregulated and that they interacted with each other. SMAD6 and SMAD9 in the TGF-beta signaling pathway were also downregulated and interacted with each other.
Validation of the relationship between PCK1 and Wnt signaling pathway
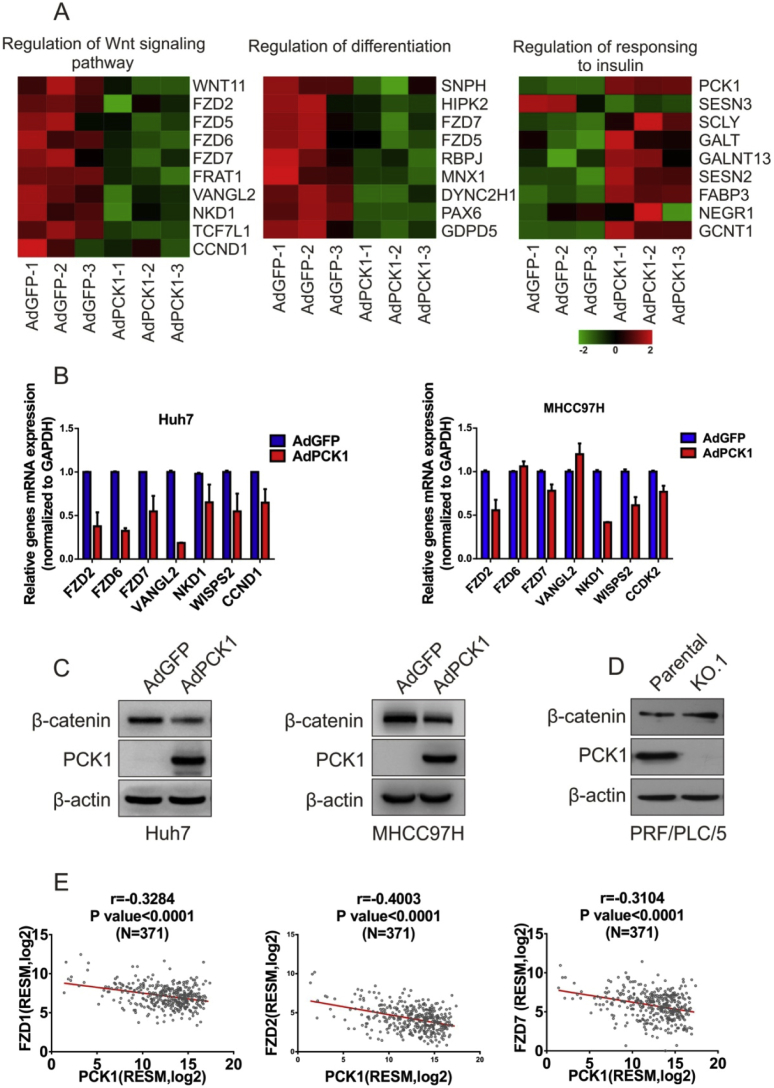
According to heat map, we found that DEGs were closely interrelated with the Wnt signaling pathway, regulation of differentiation, and regulation of response to insulin (Fig. 4A). The Wnt signaling pathway is closely associated with HCC. Our results, for the first time, indicated that PCK1 could regulate the Wnt signaling pathway in hepatoma cells. Specifically, PCK1 was negative-correlated with down expression of FZD1, FZD2 and FZD7 in patient HCC data from The Cancer Genome Atlas (TCGA) (Fig. 4E), which further confirmed the relationship between PCK1 and the Wnt signaling pathway. Further, we found that several genes of Wnt signaling pathway, such as FZD2, FZD6, FZD7, etc, were downregulated as detection of qRT-PCR after overexpression of PCK1 in Huh7 cells and MHCC97H (Fig. 4B). Consistently, immunoblot showed that PCK1 also downregulated β-catenin, the key downstream component of Wnt signaling pathway (Fig. 4C and D).
Figure 4.
Validation of the relationship between PCK1 and Wnt signaling pathway. (A) Heat map representations of differentially expressed genes belonging to the “Wnt signaling pathway”, “regulation of differentiation”, and “regulation of responsing to insulin”. (B) Target genes of Wnt pathway were analyzed by qRT-PCR. (C) and (D) Western blot analysis of β-catenin in PCK1-OE and PCK1-KO cells. (E) Correlation of PCK1 and Wnt signaling pathway gene FZD1, FZD2, FZD7 in HCC patient samples from TCGA database.
PCK1 suppresses tumor growth in vivo
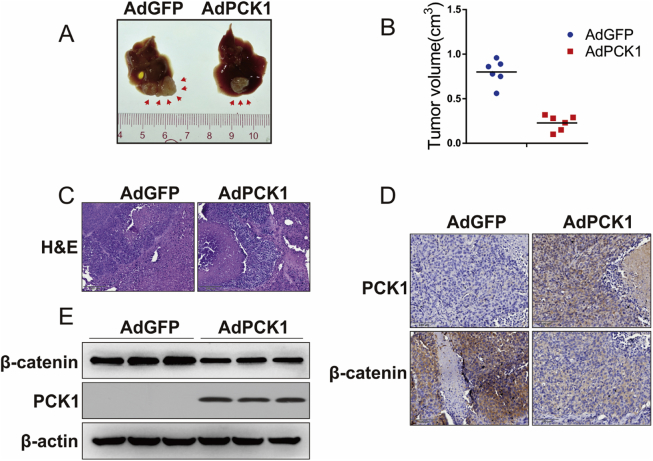
We next examined the effects of PCK1 overexpression on tumor growth in a murine orthotopic HCC model. MHCC97H infected AdGFP or AdPCK1 were transplanted into the livers of nude mice. Compared with AdGFP, AdPCK1 significantly inhibited tumor formation in orthotopic tumor model mice (Fig. 5A and B). HE staining revealed that livers in the AdPCK1 group displayed plenty of pleomorphic and atypical hepatocytes and a remarkably altered nodular liver structure (Fig. 5C). Furthermore, β-catenin expression was significantly lower in AdPCK1 groups (Fig. 5D and E), which was consistent with the in-vitro findings. Taken together, these results demonstrated that PCK1 efficiently inhibits the tumorigenesis of hepatoma cells in nude mice through Wnt signaling pathway.
Figure 5.
PCK1 suppresses tumor growth in orthotopic implantation tumor model established with MHCC97H. (A) Representative gross appearances of murine livers. Red arrows: tumor foci. (B) Tumor foci number in the liver of each mouse. (C) and (D) Representative HE staining and IHC as detection of PCK1 and β-catenin in liver tumor tissues. Magnification: 100×, 200×. (E) PCK1 and β-catenin protein expression in tumor tissue samples as detected by western blotting.
Discussion
Development and progression of HCC are correlative with lost control of cell proliferation, tissue invasion, angiogenesis, and metastasis. HCC frequently originates as a result of the dysregulation of critical genes and signaling pathways.15 In mammals, the liver is the main organ of gluconeogenesis. PCK1 is a regulator of liver energy metabolism and gluconeogenesis, and its abnormal regulation is associated with metabolic diseases such as diabetes, obesity, insulin resistance, and tumorigenesis.16, 17, 18 However, PCK1 is expressed differently in different tumor tissue. It has been reported that increased expression of PCK1 and PCK2 was discovered in tumor of several organs, such as lung, colon and skin and motivated anabolic metabolism and cell proliferation.19, 20 As one of the rate-limiting enzyme of gluconeogenesis, PCK1 was observed to be downregulated in HCC and restored its expression to inhibit tumors.9 Liu et al14 and our previous work16 showed PCK1 might participate in the regulation of energy metabolism, reactive oxygen species (ROS), and apoptosis in HCC to inhibit HCC. In the currently study, using a high throughput RNA-seq approach, we aimed to search for important signaling pathways in PCK1-overexpression hepatoma cells.
In the PCK1-overexpression cell model, we identified several significant upregulated genes containing MLIP, SLC1A7, FMO4, ACSM2B, INHBE, CHAC1, etc., and the vital downregulated genes including SLC25A12, DHRS2, ULBP2, MLKL, FZD2, SIX2, etc. These genes, such as SLC1A7, ACSM2B and SLC25A12, were shown to be involved in metabolic pathways, Acyl-CoA Synthesis, glucose and glutamate transporting.21, 22, 23 Previous studies showed that PCK1 promotes anabolic metabolism from glucose and glutamine to support tumor growth.17 FMO4, DHRS2 and MLKL are associated with oxidative stress, apoptosis and autophagy.24, 25, 26 PCK1 can induced the increase of mitochondrial NADH/NAD + ratio,14 decreased viability, enhanced apoptosis,18 and suppressed tumor progression in HCC cell lines. These DEGs might make up a mechanistic link between PCK1 and oxidative stress and apoptosis. Further, these genes were involved in significant signaling pathways. MLIP is a factor of regulation of the AKT/mTOR pathways and FOXO1.19 Other data suggested that regulation of mTORC1 activity by PEPCK was at the level of glutamine levels.17 INHBE encodes a member of the TGF-beta super family of proteins, and FZD2, SIX2 are associated with Wnt signaling pathway. In addition, CHAC120 and ULBP227 had been reported to be associated with tumorigenesis. Together, these varieties suggested that the DEGs caused by PCK1 are extremely importantly related to HCC. Next, we will design other experiments to explore how PCK1 inhibit HCC through signal transduction pathways.
Furthermore, the analysis of the functional terms was helpful for acquiring the relationships between PCK1 and HCC; therefore, the identified DEGs were subjected to GO analysis. The results revealed that the DEGs in significant biological processes were mainly associated with the Wnt signaling pathway, l-glutamate transport, cell cycle arrest, negative regulation of transcription, and response to insulin. Studies have shown that deletion of PCK1 in mice resulted in increased plasma free fatty acids, thus contributing to the occurrence of insulin resistance.28 From another perspective, pathway analyses were used to explore the molecular mechanism by which PCK1 suppresses HCC. Pathway terms that showed significant enrichment represented several vital pathway, including P53 signaling pathway, TGF-beta signaling pathway, Hippo signaling pathway and FoxO signaling pathway, and closely associated with HCC occurrence.21 Pathway interactions exhibited that many metabolic pathways were obvious activated (or inhibited) either directly or indirectly. These metabolic pathways mainly appears in glucose, amino, glycerolipid and steroid metabolism,22, 23, 26 and diseases associated with type II diabetes mellitus,22 suggesting that PCK1 might inhibit the development of HCC through rectifying metabolic disturbance.18, 24, 25 Previous reports also explained that overexpression of PCK1 represses HCC with inhibition of glycolysis and induction of gluconeogenesis, confirming that increased gluconeogenesis in HCC resisted cancer cell survival.26 Our results suggested that PCK1 not only affected metabolic pathways, but also altered signaling pathways, including the Wnt signaling pathway and TGF-beta signaling pathway, thus, playing much broader roles in HCC than currently understood.
Besides GO and pathway analyses, we provided a DEGs interaction network to explore genes-network communication regulated by PCK1 in HCC. In the DEGs interaction network, we found MAPK11 and FOXO4 showed a stronger degree of centrality, and UGT1A8, CYP2C18, EPHX1 and AKR1B10 showed interactive effects with many upregulated tumor-associated genes. These genes also suggest that PCK1 is associated with MAPK pathway and drug metabolism. Meanwhile, the DEGs in the Wnt signaling pathway and TGF-beta signaling pathway are respectively interacted with each other.
To validate the reliability of the sequencing results, qRT-PCR and Western blot was used to measure the expression of the crucial genes of Wnt signaling pathway and its downstream component β-catenin in PCK1-overexpressing cells. Consistently, we observed that PCK1 could suppress tumor growth though downregulation the expression of β-catenin in vivo.
Wnt/β-catenin signaling pathway is involved in tumorigenesis.29 The activation of Wnt/β-catenin signaling pathway promotes the growth and metastasis of multiple tumors. Wnt/β-catenin signaling is activated in HCC, and the inhibition of Wnt/β-catenin signaling suppresses proliferation.30 Wnt signaling is known to regulate gluconeogenesis by the Wnt signaling pathway effector TCF7L2.31 Interestingly, a recent study reported that the gluconeogenesis enzymes, Fructose-1, 6-bisphosphatase, is a novel regulator of Wnt/β-catenin pathway in breast cancer.32 Our findings, for the first time, showed that PCK1 suppressed Wnt/β-catenin signaling, which is critical in development and progression of HCC. In cancer cells, aberrant glucose metabolism-related enzymes, such as PKM and FBP1, caused mitochondrial dysfunction to participate in the regulation of reactive oxygen species (ROS) production.33, 34 Our previously study showed that PCK1 overexpression increases the ratio of NADPH/NADP+, and further decreases ROS production.16 As previously reported, a thioredoxin-related protein, nucleoredoxin (NRX), usually interacts with Dishevelled (Dvl), an essential adaptor protein for Wnt signalling. ROS causes dissociation of NRX from Dvl, which enhances Dvl to activate Wnt signalling pathway.35 On the one hand, PCK1 might regulate Wnt signalling pathway owing to ROS. On the other hand, Overexpression of PCK1 decreased cellular ATP levels and enabled AMPK phosphorylation.36 p-AMPK attenuates Wnt/β-catenin signaling by reducing β-catenin protein levels.37 Further insights into the PCK1 will clarify how dysregulation of gluconeogenesis promote HCC development, and how metabolic reprogramming regulate signaling pathways in cancer cells.
In summary, PCK1 is significantly low expressed in most tumor tissues, especially in hepatocellular carcinoma, kidney renal papillary cell carcinoma and cholangiocarcinoma. We also have a validation of the relationship between PCK1 and insulin resistance, oxidative stress, apoptosis, mTOR pathways, etc. Further, we discovery that overexpression of PCK1 downregulates Wnt/β-catenin signaling pathway in HCC. Our results may be helpful in further elucidating the underlying molecular mechanism of PCK1 deficiency and HCC progression, providing potential therapeutic targets for improving HCC therapy.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
We would like to thank Dr. Tong-Chuan He (University of Chicago, USA) for providing the AdEasy system. This study was supported by research grants from China National Natural Science Foundation (grant nos. 81602417 to KW, and 81872270 and 81572683 to NT), the Major National S&T program (2017ZX10202203-004 to NT), Natural Science Foundation Project of CQ CSTC (grant no. cstc2018jcyjAX0254 to NT), and The Program for Innovation Team of Higher Education in Chongqing (grant no.CXTDX201601015).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2019.04.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Masoudi-Nejad A., Asgari Y. Metabolic cancer biology: structural-based analysis of cancer as a metabolic disease, new sights and opportunities for disease treatment. Semin Canc Biol. 2015;30:21–29. doi: 10.1016/j.semcancer.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Ward P.S., Thompson C.B. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma R., Zhang W., Tang K. Switch of glycolysis to gluconeogenesis by dexamethasone for treatment of hepatocarcinoma. Nat Commun. 2013;4:2508. doi: 10.1038/ncomms3508. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein I., Yizhak K., Madar S., Goldfinger N., Ruppin E., Rotter V. p53 promotes the expression of gluconeogenesis-related genes and enhances hepatic glucose production. Cancer Metabol. 2013;1(1):9. doi: 10.1186/2049-3002-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendez-Lucas A., Duarte J.A.G., Sunny N.E. PEPCK-M expression in mouse liver potentiates, not replaces, PEPCK-C mediated gluconeogenesis. J Hepatol. 2013;59(1):105–113. doi: 10.1016/j.jhep.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wurmbach E., Chen Y., Khitrov G. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45(4):938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 8.Zhang P., Tu B., Wang H. Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proc Natl Acad Sci U S A. 2014;111(29):10684–10689. doi: 10.1073/pnas.1411026111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y., Wang X., Sang Z. Quantitative proteomics by SWATH-MS reveals sophisticated metabolic reprogramming in hepatocellular carcinoma tissues. Sci Rep. 2017;7:45913. doi: 10.1038/srep45913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders E., Diehl S. Analysis and interpretation of transcriptomic data obtained from extended Warburg effect genes in patients with clear cell renal cell carcinoma. Oncoscience. 2015;2(2):151–186. doi: 10.18632/oncoscience.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupuy D., Bertin N., Hidalgo C.A. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat Biotechnol. 2007;25(6):663–668. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- 12.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J.D., Wiemann S. KEGGgraph: a graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics. 2009;25(11):1470–1471. doi: 10.1093/bioinformatics/btp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu M.-X., Jin L., Sun S.-J. Metabolic reprogramming by PCK1 promotes TCA cataplerosis, oxidative stress and apoptosis in liver cancer cells and suppresses hepatocellular carcinoma. Oncogene. 2018;37(12):1637–1653. doi: 10.1038/s41388-017-0070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto S., Tomita Y., Hoshida Y. Expression of hepatoma-derived growth factor is correlated with lymph node metastasis and prognosis of gastric carcinoma. Clin Cancer Res. 2006;12(1):117–122. doi: 10.1158/1078-0432.CCR-05-1347. [DOI] [PubMed] [Google Scholar]
- 16.Tuo L., Xiang J., Pan X. PCK1 downregulation promotes TXNRD1 expression and hepatoma cell growth via the Nrf2/Keap1 pathway. Front Oncol. 2018;8:611. doi: 10.3389/fonc.2018.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montal E.D., Dewi R., Bhalla K. PEPCK coordinates the regulation of central carbon metabolism to promote cancer cell growth. Mol Cell. 2015;60(4):571–583. doi: 10.1016/j.molcel.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Y., Zhang Y., Wang C. Overexpression of PCK1 gene antagonizes hepatocellular carcinoma through the activation of gluconeogenesis and suppression of glycolysis pathways. Cell Physiol Biochem. 2018;47(1):344–355. doi: 10.1159/000489811. [DOI] [PubMed] [Google Scholar]
- 19.Cattin M.-E., Wang J., Weldrick J.J. Deletion of MLIP (muscle-enriched A-type lamin-interacting protein) leads to cardiac hyperactivation of Akt/mammalian target of rapamycin (mTOR) and impaired cardiac adaptation. J Biol Chem. 2015;290(44):26699–26714. doi: 10.1074/jbc.M115.678433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goebel G., Berger R., Strasak A.M. Elevated mRNA expression of CHAC1 splicing variants is associated with poor outcome for breast and ovarian cancer patients. Br J Canc. 2012;106(1):189–198. doi: 10.1038/bjc.2011.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michelotti G.A., Machado M.V., Diehl A.M. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol. 2013;10(11):656–665. doi: 10.1038/nrgastro.2013.183. [DOI] [PubMed] [Google Scholar]
- 22.Beale E.G., Hammer R.E., Antoine B., Forest C. Disregulated glyceroneogenesis: PCK1 as a candidate diabetes and obesity gene. Trends Endocrinol Metabol. 2004;15(3):129–135. doi: 10.1016/j.tem.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Gomez-Valades A.G., Mendez-Lucas A., Vidal-Alabro A. Pck1 gene silencing in the liver improves glycemia control, insulin sensitivity, and dyslipidemia in db/db mice. Diabetes. 2008;57(8):2199–2210. doi: 10.2337/db07-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho P.-C., Bihuniak J.D., Macintyre A.N. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell. 2015;162(6):1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma R., Ji T., Zhang H. A Pck1-directed glycogen metabolic program regulates formation and maintenance of memory CD8(+) T cells. Nat Cell Biol. 2018;20(1):21–27. doi: 10.1038/s41556-017-0002-2. [DOI] [PubMed] [Google Scholar]
- 26.Khan M.W., Biswas D., Ghosh M., Mandloi S., Chakrabarti S., Chakrabarti P. mTORC2 controls cancer cell survival by modulating gluconeogenesis. Cell Death Dis. 2015;1:15016. doi: 10.1038/cddiscovery.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothe A., Jachimowicz R.D., Borchmann S. The bispecific immunoligand ULBP2-aCEA redirects natural killer cells to tumor cells and reveals potent anti-tumor activity against colon carcinoma. Int J Cancer. 2014;134(12):2829–2840. doi: 10.1002/ijc.28609. [DOI] [PubMed] [Google Scholar]
- 28.Millward C.A., Desantis D., Hsieh C.-W. Phosphoenolpyruvate carboxykinase (Pck1) helps regulate the triglyceride/fatty acid cycle and development of insulin resistance in mice. J Lipid Res. 2010;51(6):1452–1463. doi: 10.1194/jlr.M005363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klaus A., Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Canc. 2008;8(5):387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 30.Wang W., Pan Q., Fuhler G.M., Smits R., Peppelenbosch M.P. Action and function of Wnt/beta-catenin signaling in the progression from chronic hepatitis C to hepatocellular carcinoma. J Gastroenterol. 2017;52(4):419–431. doi: 10.1007/s00535-016-1299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin T. Current understanding and dispute on the function of the Wnt signaling pathway effector TCF7L2 in hepatic gluconeogenesis. Genes Dis. 2016;3(1):48–55. doi: 10.1016/j.gendis.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li K., Ying M., Feng D. Fructose-1,6-bisphosphatase is a novel regulator of Wnt/beta-Catenin pathway in breast cancer. Biomed Pharmacother. 2016;84:1144–1149. doi: 10.1016/j.biopha.2016.10.050. [DOI] [PubMed] [Google Scholar]
- 33.Shinohara H., Taniguchi K., Kumazaki M. Anti-cancer fatty-acid derivative induces autophagic cell death through modulation of PKM isoform expression profile mediated by bcr-abl in chronic myeloid leukemia. Cancer Lett. 2015;360(1):28–38. doi: 10.1016/j.canlet.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 34.Dai J., Ji Y., Wang W. Loss of fructose-1,6-bisphosphatase induces glycolysis and promotes apoptosis resistance of cancer stem-like cells: an important role in hexavalent chromium-induced carcinogenesis. Toxicol Appl Pharmacol. 2017;331:164–173. doi: 10.1016/j.taap.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Funato Y., Michiue T., Asashima M., Miki H. The thioredoxin-related redox-regulating protein nucleoredoxin inhibits Wnt-beta-catenin signalling through dishevelled. Nat Cell Biol. 2006;8(5):501–508. doi: 10.1038/ncb1405. [DOI] [PubMed] [Google Scholar]
- 36.Tuo L., Xiang J., Pan X. PCK1 negatively regulates cell cycle progression and hepatoma cell proliferation via the AMPK/p27(Kip1) axis. J Exp Clin Cancer Res. 2019;38(1):50. doi: 10.1186/s13046-019-1029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takatani T., Minagawa M., Takatani R., Kinoshita K., Kohno Y. AMP-activated protein kinase attenuates Wnt/beta-catenin signaling in human osteoblastic Saos-2 cells. Mol Cell Endocrinol. 2011;339(1–2):114–119. doi: 10.1016/j.mce.2011.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.