Abstract
Background
Inhibitors of proprotein convertase subtilisin kexin 9 are indicated in Canada for treatment of patients with familial hypercholesterolemia (FH). Classically, FH is considered to be a monogenic condition caused by rare pathogenic mutations; however, some patients have hypercholesterolemia on a polygenic basis. Whether the effect of proprotein convertase subtilisin kexin 9 inhibitor treatment differs between patients with monogenic hypercholesterolemia and patients with polygenic hypercholesterolemia is unclear.
Methods
We performed retrospective chart reviews on patients treated with evolocumab 140 mg subcutaneously biweekly from the Lipid Genetics Clinic, London Health Sciences Centre. Evolocumab-treated patients with hypercholesterolemia were grouped into monogenic or polygenic categories on the basis of their genotype determined by targeted next-generation sequencing. Absolute and relative changes in low-density lipoprotein cholesterol (LDL-C) levels before and after evolocumab treatment were studied.
Results
In 32 patients with monogenic heterozygous FH and 7 patients with polygenic hypercholesterolemia treated with evolocumab, absolute incremental reductions in LDL-C were 2.94 ± 1.22 mmol/L and 3.15 ± 0.90 mmol/L, respectively (P = not significant), whereas percent reductions in LDL-C were 63.9% ± 16.0% and 67.7% ± 20.7%, respectively (P = not significant).
Conclusion
Although the sample size is small, the findings suggest comparable biochemical responsiveness to evolocumab in both monogenic (heterozygous) and polygenic hypercholesterolemia.
Résumé
Introduction
Au Canada, les inhibiteurs de la proprotéine convertase subtilisine/kexine de type 9 sont indiqués dans le traitement des patients atteints d’hypercholestérolémie familiale (HF). Traditionnellement, la HF est considérée comme une maladie monogénique causée par des mutations pathogènes rares. Toutefois, certains patients ont une hypercholestérolémie de forme polygénique. On ignore si les effets du traitement par inhibiteurs de la proprotéine convertase subtilisine/kexine de type 9 diffèrent entre les patients ayant une hypercholestérolémie monogénique et les patients ayant une hypercholestérolémie polygénique.
Méthodes
Nous avons réalisé une revue rétrospective de dossiers de patients traités par évolocumab à raison de 140 mg par voie sous-cutanée 2 fois par semaine à la Lipid Genetics Clinic du London Health Sciences Centre. Les patients atteints d’hypercholestérolémie qui étaient traités par évolocumab ont été regroupés dans la catégorie de la forme monogénique et la catégorie de la forme polygénique en fonction de leur génotype déterminé par le séquençage ciblé de nouvelle génération. Nous avons étudié les changements absolus et relatifs des concentrations de cholestérol à lipoprotéines de faible densité (LDL-C) avant et après le traitement par évolocumab.
Résultats
Chez 32 patients ayant une HF hétérozygote monogénique et 7 patients ayant une hypercholestérolémie polygénique traités par évolocumab, les réductions progressives absolues du LDL-C étaient respectivement de 2,94 ± 1,22 mmol/l et de 3,15 ± 0,90 mmol/l (P = non significatif), alors que les réductions du LDL-C en pourcentage étaient respectivement de 63,9 % ± 16,0 % et de 67,7 % ± 20,7 % (P = non significatif).
Conclusion
Bien que la taille de l’échantillon soit petite, les résultats montrent une capacité de réponse biochimique comparable à l’évolocumab lors d’hypercholestérolémie (hétérozygote) monogénique et d’hypercholestérolémie polygénique.
Familial hypercholesterolemia (FH) is a relatively common genetic disorder leading to elevated plasma concentrations of low-density lipoprotein cholesterol (LDL-C).1, 2 Approximately half of patients referred to the lipid clinic with suspected heterozygous FH have a rare mutation in 1 of 3 FH-related genes, namely, LDLR, APOB, or PCSK9.3 An additional 20% to 30% of such referred patients instead have polygenic hypercholesterolemia.3, 4 The latter condition features an accumulation of numerous common polymorphisms that cumulatively act to increase LDL-C greater than 5 mmol/L;5 this is commonly considered to be a threshold value to diagnose FH.6 When compared with individuals with normal LDL-C levels, the risk of atherosclerotic cardiovascular disease is high for patients with LDL-C > 5 mmol/L, irrespective of whether their hypercholesterolemia is on a monogenic or polygenic basis. In one study of individuals with LDL-C > 5 mmol/L compared with those with LDL-C < 3.4 mmol/L, the odds of a clinical atherosclerotic event were increased 22-fold if a monogenic mutation was present versus 6-fold increased if a mutation was absent.7
Although DNA sequencing to detect rare monogenic mutations in typical heterozygous FH is being performed with increasing frequency,8 concurrent polygenic risk scoring is not yet routinely performed.5 Among patients referred to the Lipid Genetics Clinic, London Health Sciences Centre, both types of genetic susceptibility are screened simultaneously using a targeted next-generation sequencing panel.9 Because we can diagnose patients unequivocally as having monogenic or polygenic hypercholesterolemia, we can explore hypotheses related to genetic differences between individuals. In the current study, we asked whether LDL-C response to the proprotein convertase subtilisin kexin 9 (PCSK9) inhibitor evolocumab differed according to the genetic basis for the patients’ hypercholesterolemia.
Methods
Study subjects
Charts of patients who had been treated with evolocumab, from the Lipid Genetics Clinic, London Health Sciences Centre (London, Ontario), were retrospectively reviewed. Patients provided written informed consent, and the study was approved by the Western University Research Ethics board (protocol 07290E). A database of the patients’ demographics, diagnosis, other treatments, and lipid laboratory values was created.
Genetic characterization
Patients were genotyped using LipidSeq, a targeted next-generation sequencing panel,9 at the London Regional Genomics Centre using standard protocols (www.lrgc.ca). Patients were classified as monogenic (heterozygous) FH or polygenic FH. Patients with monogenic FH were defined as those with mutation(s) classified as “pathogenic” or “likely pathogenic” according to the American College of Medical Genetics and Genomics classification guidelines.10 Weighted genetic risk scores (wGRS) for LDL-C were calculated for all patients as previously described.11 Patients with wGRS >90th percentile were classified as having polygenic hypercholesterolemia.
Lipid values
To calculate LDL-C reduction with evolocumab treatment, pretreatment and post-treatment lipid laboratory measurements were selected using the following criteria: (1) Pretreatment values were chosen from the most recent laboratory results before evolocumab enrolment, and (2) post-treatment values were chosen from laboratory results obtained 12 weeks after initiation of evolocumab 140 mg subcutaneously biweekly. Statin therapy dose or intensity was categorized as high, moderate, or low according to previously published criteria.12, 13 There was no need to impute lipid profiles in any patient.14
Statistical analysis
Statistical analyses were conducted with SAS version 9.4 (SAS Institute, Inc., Cary, NC). Differences between discrete and quantitative traits between molecularly characterized patient groups were assessed using chi-square analysis, unpaired Student t tests, and Kruskal–Wallis test, as appropriate. All tests were performed assuming unequal variances and are reported as the mean ± standard deviation (SD). Power and sample size calculations were made using the PS Power and Sample Size Calculation program.15
Results
All patients studied could be unequivocally classified as having monogenic (heterozygous) or polygenic hypercholesterolemia. Table 1 shows the clinical and demographic features of the study patients. There were no differences between the genotypic classes for these variables. As expected, the wGRS of patients with polygenic hypercholesterolemia was significantly higher than of patients with monogenic hypercholesterolemia. No patient had monogenic hypercholesterolemia together with a high polygenic score. None of the other demographic values were significantly different between the 2 groups. Table 1 also shows information on the use of statin and ezetimibe therapies.
Table 1.
Patient demographics and biochemical variables before and after evolocumab treatment
| Monogenic (He) hypercholesterolemia | Polygenic hypercholesterolemia | P value | |
|---|---|---|---|
| Number | 32 | 7 | - |
| Female N (%) | 12 (37.5%) | 2 (28.5%) | NS (0.6555) |
| Age (y) | 51.4 ± 11.0 | 57.7 ± 8.44 | NS (0.1230) |
| Body mass index (kg/m2) | 29.6 ± 4.30 | 31.2 ± 6.08 | NS (0.5217) |
| Total cholesterol (mmol/L) | |||
| Pretreatment | 6.73 ± 2.32 | 6.88 ± 1.54 | NS (0.8477) |
| Post-treatment | 3.75 ± 1.71 | 3.73 ± 1.34 | NS (0.9740) |
| Triglyceride (mmol/L) | |||
| Pretreatment | 1.62 ± 0.66 | 1.74 ± 0.61 | NS (0.6393) |
| Post-treatment | 1.49 ± 0.70 | 1.91 ± 0.66 | NS (0.1624) |
| HDL-C (mmol/L) | |||
| Pretreatment | 1.22 ± 0.36 | 1.26 ± 0.44 | NS (0.8230) |
| Post-treatment | 1.24 ± 0.36 | 1.33 ± 0.52 | NS (0.6814) |
| LDL-C (mmol/L) | |||
| Pretreatment | 4.77 ± 2.21 | 4.82 ± 1.44 | NS (0.9424) |
| Post-treatment | 1.83 ± 1.52 | 1.67 ± 1.11 | NS (0.7561) |
| Absolute LDL-C reduction (mmol/L) | 2.94 ± 1.22 | 3.15 ± 0.90 | NS (0.6174) |
| Percent change in LDL-C (%) | 63.9 ± 16.0 | 67.7 ± 20.7 | NS (0.6603) |
| Mean wGRS | 1.64 ± 0.18 | 1.95 ± 0.17 | 0.0020 |
| Baseline statin therapy N (%) | |||
| No statin | 3 (9.38) | 3 (42.9) | 0.0261 |
| Low intensity | 1 (3.13) | 0 | NS (0.6356) |
| Moderate intensity | 6 (18.8) | 2 (28.6) | NS (0.5600) |
| High intensity | 22 (68.8) | 2 (28.6) | 0.0478 |
| Ezetimibe therapy N (%) | 22 (68.8) | 4 (57.1) | NS (0.5551) |
Means and standard deviations (SDs) for quantitative variables are shown.
HDL-C, high-density lipoprotein cholesterol; He, heterozygous; LDL-C, low-density lipoprotein cholesterol; N, number of individuals; NS, not significant; wGRS, weighted genetic risk score.
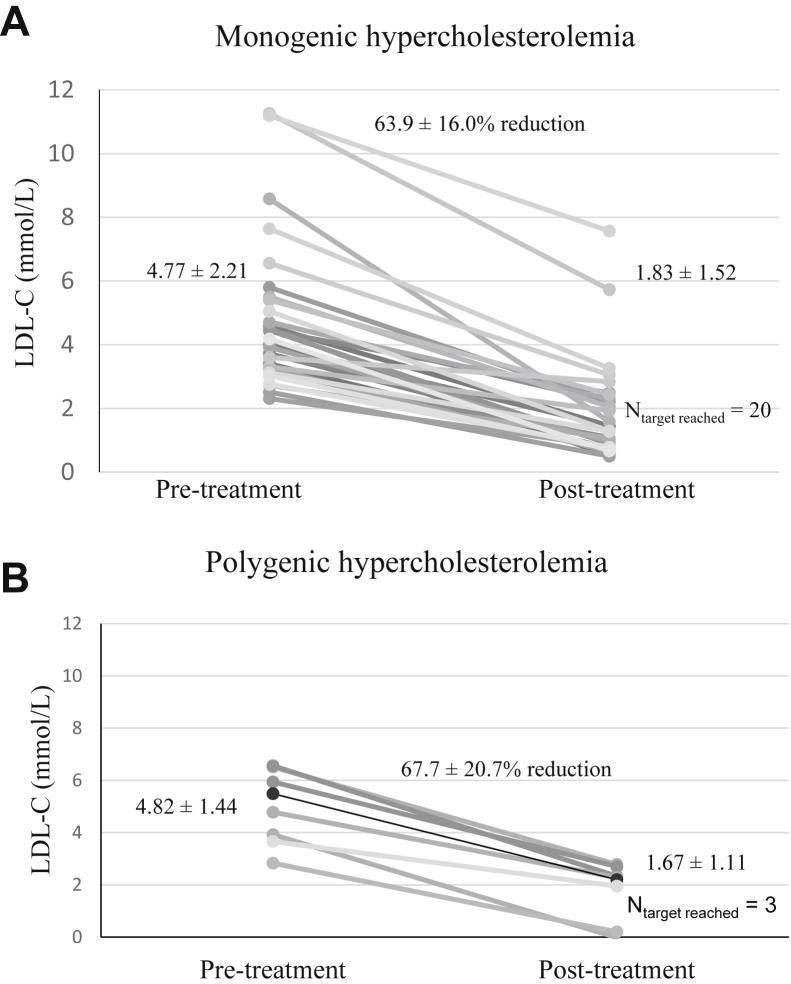
Figure 1 shows LDL-C reduction of each patient in each genetic category. The mean ± SD absolute LDL-C reduction achieved with evolocumab after 12 weeks of treatment was 2.94 ± 1.22 mmol/L and 3.15 ± 0.90 mmol/L in the monogenic and polygenic hypercholesterolemia groups, respectively. The mean ± SD percent LDL-C reduction achieved with evolocumab was 63.9% ± 16.0% and 67.7% ± 20.7% in the monogenic and polygenic hypercholesterolemia groups, respectively. These absolute and percent LDL-C reductions were not significantly different between the 2 groups.
Figure 1.
Low-density lipoprotein cholesterol (LDL-C) response to evolocumab according to genotype. LDL-C levels before and after evolocumab treatment in each patient, grouped into (A) monogenic (N = 32; all heterozygotes) and (B) polygenic (N = 7) hypercholesterolemia, where pretreatment levels indicate the most recent lipid panel result before evolocumab initiation, and post-treatment levels indicate results 12 weeks after first injection. Means ± standard deviations (SDs) are shown, as are the numbers of treated individuals who attained target LDL-C <2 mmol/L.
Twenty of 32 patients (62.5%) with monogenic hypercholesterolemia treated with evolocumab reached an LDL-C target < 2.0 mmol/L after treatment compared with 3 of 7 treated patients with polygenic hypercholesterolemia (42.9%). Twenty-seven of 32 patients (84.4%) with monogenic hypercholesterolemia treated with evolocumab achieved 50% reduction in LDL-C compared with 6 of 7 treated patients with polygenic hypercholesterolemia (85.7%). None of these differences were significant.
Discussion
In 32 patients with monogenic heterozygous FH and 7 patients with polygenic hypercholesterolemia who were treated with evolocumab, we observed no statistical differences in absolute incremental reductions or percent reduction in LDL-C. There were also no differences in the proportions of patients in each group who attained target LDL-C < 2.0 mmol/L or LDL-C reduction < 50%. Although the sample size is small, the findings suggest at least comparable biochemical responsiveness irrespective of the genetic basis of the hypercholesterolemia. This observed efficacy is consistent with observations in patients with heterozygous FH whose hypercholesterolemia had not been genetically sub-stratified as monogenic versus polygenic.16
We note that for each of 4 biochemical read-outs related to LDL-C, namely, absolute and relative reductions, percent attaining target, and percent with reduction >50%, the polygenic hypercholesterolemia group tended to have nonsignificantly greater efficacy benefit with evolocumab treatment. It is possible that with a larger sample size, between-group differences of the magnitude observed would become nominally significant. For instance, a sample size of 2282 individuals in each group would be required to observe a significant between-group LDL-C difference of 0.22 mmol/L with a power of 0.8 and alpha of 0.05. Going forward, it would be of interest to repeat this type of study in a large controlled sample. However, the relatively small nonsignificant differences observed would not necessarily be clinically relevant, nor would they likely have any impact on clinical decision-making with respect to initiating evolocumab.
Conclusion
We could detect no clinical difference in PCSK9 inhibitor treatment whether or not the hypercholesterolemic patients carried a single gene mutation or had polygenic predisposition. Whether or not there is a difference between these genotype strata in responsiveness to statin therapy remains to determined. This study confirms the general effectiveness of PCSK9 inhibitor therapy in lowering LDL-C level in patients diagnosed clinically with phenotypic heterozygous FH.
Funding Sources
Dr Hegele is supported by the Jacob J. Wolfe Distinguished Medical Research Chair, the Edith Schulich Vinet Research Chair in Human Genetics, and the Martha G. Blackburn Chair in Cardiovascular Research, and has received operating grants from the Canadian Institutes of Health Research (Foundation Grant) and the Heart and Stroke Foundation of Ontario (HSF G-18-0022147).
Disclosures
Dr Hegele has received honoraria for membership on advisory boards and speakers’ bureaus for Aegerion, Amgen, Gemphire, Lilly, Merck, Pfizer, Regeneron, Sanofi, and Valeant, all unrelated to the topic of this manuscript. The rest of the authors have no conflicts of interest to disclose.
Footnotes
Ethics Statement: The reported research has adhered to the relevant ethical guidelines.
See page 118 for disclosure information.
References
- 1.Brunham L.R., Ruel I., Aljenedil S. Canadian Cardiovascular Society position statement on familial hypercholesterolemia: update 2018. Can J Cardiol. 2018;34:1553–1563. doi: 10.1016/j.cjca.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Defesche J.C., Gidding S.S., Harada-Shiba M. Familial hypercholesterolaemia. Nat Rev Dis Primers. 2017;7:17093. doi: 10.1038/nrdp.2017.93. [DOI] [PubMed] [Google Scholar]
- 3.Berberich A.J., Hegele R.A. The complex molecular genetics of familial hypercholesterolaemia. Nat Rev Cardiol. 2019;16:9–20. doi: 10.1038/s41569-018-0052-6. [DOI] [PubMed] [Google Scholar]
- 4.Futema M., Shah S., Cooper J.A., Li K. Refinement of variant selection for the LDL cholesterol genetic risk score in the diagnosis of the polygenic form of clinical familial hypercholesterolemia and replication in samples from 6 countries. Clin Chem. 2015;61:231–238. doi: 10.1373/clinchem.2014.231365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dron J.S., Hegele R.A. Polygenic influences on dyslipidemias. Curr Opin Lipidol. 2018;29:133–143. doi: 10.1097/MOL.0000000000000482. [DOI] [PubMed] [Google Scholar]
- 6.Ruel I., Brisson D., Aljenedil S. Simplified Canadian definition for familial hypercholesterolemia. Can J Cardiol. 2018;34:1210–1214. doi: 10.1016/j.cjca.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Khera A.V., Won H.H., Peloso G.M. Diagnostic yield and clinical utility of sequencing familial hypercholesterolemia genes in patients with severe hypercholesterolemia. J Am Coll Cardiol. 2016;67:2578–2589. doi: 10.1016/j.jacc.2016.03.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham C.A., Latten M.J., Hart P.J. Molecular diagnosis of familial hypercholesterolaemia. Curr Opin Lipidol. 2017;28:313–320. doi: 10.1097/MOL.0000000000000430. [DOI] [PubMed] [Google Scholar]
- 9.Hegele R.A., Ban M.R., Cao H. Targeted next-generation sequencing in monogenic dyslipidemias. Curr Opin Lipidol. 2015;26:103–113. doi: 10.1097/MOL.0000000000000163. [DOI] [PubMed] [Google Scholar]
- 10.Iacocca M.A., Chora J.R., Carrié A. ClinVar database of global familial hypercholesterolemia-associated DNA variants. Hum Mutat. 2018;39:1631–1640. doi: 10.1002/humu.23634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J., Dron J.S., Ban M.R. Polygenic versus monogenic causes of hypercholesterolemia ascertained clinically. Arterioscler Thromb Vasc Biol. 2016;36:2439–2445. doi: 10.1161/ATVBAHA.116.308027. [DOI] [PubMed] [Google Scholar]
- 12.Lloyd-Jones D.M., Morris P.B., Ballantyne C.M. 2016 ACC Expert Consensus Decision Pathway on the role of non-statin therapies for LDL-cholesterol lowering in the management of atherosclerotic cardiovascular disease Risk: a report of the American College of Cardiology Task Force on Clinical Expert Consensus Documents. J Am Coll Cardiol. 2016;68:92–125. doi: 10.1016/j.jacc.2016.03.519. [DOI] [PubMed] [Google Scholar]
- 13.Clemens K.K., Shariff S.Z., McArthur E., Hegele R.A. Ezetimibe prescriptions in older Canadian adults after an acute myocardial infarction: a population-based cohort study. Lipids Health Dis. 2018;17:8. doi: 10.1186/s12944-017-0649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruel I., Aljenedil S., Sadri I. Imputation of baseline LDL cholesterol concentration in patients with familial hypercholesterolemia on statins or ezetimibe. Clin Chem. 2018;64:355–362. doi: 10.1373/clinchem.2017.279422. [DOI] [PubMed] [Google Scholar]
- 15.Dupont W.D., Plummer W.D., Jr. Power and sample size calculations for studies involving linear regression. Control Clin Trials. 1998;19:589–601. doi: 10.1016/s0197-2456(98)00037-3. [DOI] [PubMed] [Google Scholar]
- 16.Razek O., Cermakova L., Armani H. Attainment of recommended lipid targets in patients with familial hypercholesterolemia: real-world experience with PCSK9 inhibitors. Can J Cardiol. 2018;34:1004–1009. doi: 10.1016/j.cjca.2018.04.014. [DOI] [PubMed] [Google Scholar]