ABSTRACT
Vertebrate heart development requires the integration of temporally distinct differentiating progenitors. However, few signals are understood that restrict the size of the later-differentiating outflow tract (OFT). We show that improper specification and proliferation of second heart field (SHF) progenitors in zebrafish lazarus (lzr) mutants, which lack the transcription factor Pbx4, produces enlarged hearts owing to an increase in ventricular and smooth muscle cells. Specifically, Pbx4 initially promotes the partitioning of the SHF into anterior progenitors, which contribute to the OFT, and adjacent endothelial cell progenitors, which contribute to posterior pharyngeal arches. Subsequently, Pbx4 limits SHF progenitor (SHFP) proliferation. Single cell RNA sequencing of nkx2.5+ cells revealed previously unappreciated distinct differentiation states and progenitor subpopulations that normally reside within the SHF and arterial pole of the heart. Specifically, the transcriptional profiles of Pbx4-deficient nkx2.5+ SHFPs are less distinct and display characteristics of normally discrete proliferative progenitor and anterior, differentiated cardiomyocyte populations. Therefore, our data indicate that the generation of proper OFT size and arch arteries requires Pbx-dependent stratification of unique differentiation states to facilitate both homeotic-like transformations and limit progenitor production within the SHF.
KEY WORDS: Heart development, Pbx transcription factor, Second heart field, Zebrafish, Pharyngeal arch arteries, Outflow tract
Summary: The transcription factor Pbx4 restricts outflow tract size in the zebrafish heart by reinforcing distinct cardiac progenitor differentiation states.
INTRODUCTION
Appropriate vertebrate heart size is determined through inductive and repressive signals that intricately coordinate the assembly of multiple progenitor fields into a unified organ. At least two temporally distinguishable sources of progenitors have been identified in all vertebrate hearts: earlier-differentiating progenitors, termed the first heart field (FHF), which contribute to the formation of the nascent linear heart tube; and later-differentiating progenitors, termed the second heart field (SHF), which allow growth of the heart through addition to the outflow tract (OFT) and venous pole (Staudt and Stainier, 2012). A failure to determine proper size of the vertebrate heart, or its individual chambers, can lead to congenital heart defects (CHDs), which are the most common congenital malformations observed in humans (Hoffman and Kaplan, 2002; Hoffman et al., 2004). Furthermore, OFT defects comprise nearly 30% of diagnosed CHDs, implying that these are derived from improper development of the SHF (Neeb et al., 2013). Despite the high incidence of OFT-related CHDs and significant gains in our understanding of conserved factors that regulate SHF development, the molecular etiologies of most OFT defects are still largely unknown.
Recent work has indicated that pre-B-cell leukemia (Pbx) transcription factors are conserved regulators of vertebrate heart development (Chang et al., 2008; Kao et al., 2015; Stankunas et al., 2008). PBX3 mutations in humans are associated with an increased risk of OFT defects, including persistent truncus arteriosus (PTA), tetralogy of Fallot (TOF) and atrioventricular septal defects (ASVDs) (Arrington et al., 2012). Although a direct association of PBX gene mutations causing CHDs in humans is currently limited, analysis of Pbx1 knockout (KO) mice supports the idea that there is a conserved requirement for Pbx factors in establishing proper development of the mammalian OFT. Specifically, Pbx1 KO mice have a spectrum of OFT defects, including PTA, TOF, overriding aorta with ventricular septal defect, and bicuspid aortic valves (Chang et al., 2008; Stankunas et al., 2008), as well as malformations of the adjacent great arteries, which exit from the aorta. Importantly, although septation defects observed in the distal OFT are attributable to impaired cardiac neural crest cell (cNCC) migration, septation defects in the proximal conus and malformations of the great arteries are not due to inappropriate cNCC migration (Chang et al., 2008). Zebrafish Pbx4 is the functional equivalent of murine Pbx1 in multiple developmental contexts (Arenkiel et al., 2004; Choe et al., 2002; McClintock et al., 2002; Pöpperl et al., 2000; Studer et al., 1998). Zebrafish pbx4 (lazarus, lzr) mutants have distinctive abnormally shaped atria and ventricles (Kao et al., 2015). It has been proposed that Pbx4 acts in a biphasic manner to restrict heart size in zebrafish embryos: during earlier somitogenesis, Pbx4 is required for myocardial differentiation; at later stages of cardiogenesis, Pbx4 transitions to a role in restricting the number of both atrial and ventricular cardiomyocytes (CMs) (Kao et al., 2015). However, the mechanisms underlying these proposed differential roles of Pbx4 in zebrafish have not been interrogated. Furthermore, it is not clear how the reported defects in lzr hearts relate to the OFT defects found in Pbx1 KO mice and associated with PBX3 mutations in humans.
Here, we show that zebrafish lzr mutant hearts have a specific increase in SHF-derived ventricular CMs and smooth muscle cells within the OFT. Furthermore, we find that Pbx4 limits the number of SHF-derived OFT progenitors through two mechanisms: initially, Pbx4 promotes the specification of posterior endothelial cell (EC) progenitors at the expense of anterior progenitors within the anterior lateral plate mesoderm (ALPM), and, subsequently, it limits SHFP proliferation as differentiating cells add to the elongating arterial pole of the heart tube. Interrogation of nkx2.5+ cells with single cell RNA sequencing (scRNA-seq) supports the idea that Pbx4 is required to define previously unappreciated CM progenitor and differentiation states within the SHF and arterial pole of the nascent heart tube. Altogether, our data support a mechanism by which Pbx factors have conserved roles in coordinating OFT size and great artery development in vertebrates through reinforcement of SHFP states at distinct stages of cardiogenesis.
RESULTS
lzr mutants have a specific increase in ventricular CMs
The conserved requirements for Pbx transcription factors in vertebrate heart and great artery development remain unclear, despite previous studies in mice and zebrafish (Chang et al., 2008; Kao et al., 2015; Stankunas et al., 2008). Examining the hearts of lzr mutants at 48 h post-fertilization (hpf), we found that their ventricles appeared elongated and contained a variable number of protrusions, whereas their atria were more bulbous (Fig. 1A-D, Fig. S1), consistent with a previous report (Kao et al., 2015). However, in contrast to the previous study that suggested both atrial and ventricular CMs are increased in lzr mutant hearts (Kao et al., 2015), our quantification of CMs using the myl7:DsRed2-NLS transgene (Mably et al., 2003) revealed that lzr mutants only have an increase in ventricular CMs (Fig. 1E). Furthermore, a specific increase in ventricular CMs within the heart was also found when Pbx4 was depleted using a previously validated cocktail of morpholinos (Maves et al., 2009; Pöpperl et al., 2000; Waskiewicz et al., 2002) (Fig. S1). Therefore, in contrast to the global effect on CMs that has been proposed previously (Kao et al., 2015), our analysis suggests that Pbx4 is required to specifically limit ventricular CM number within the heart.
Fig. 1.
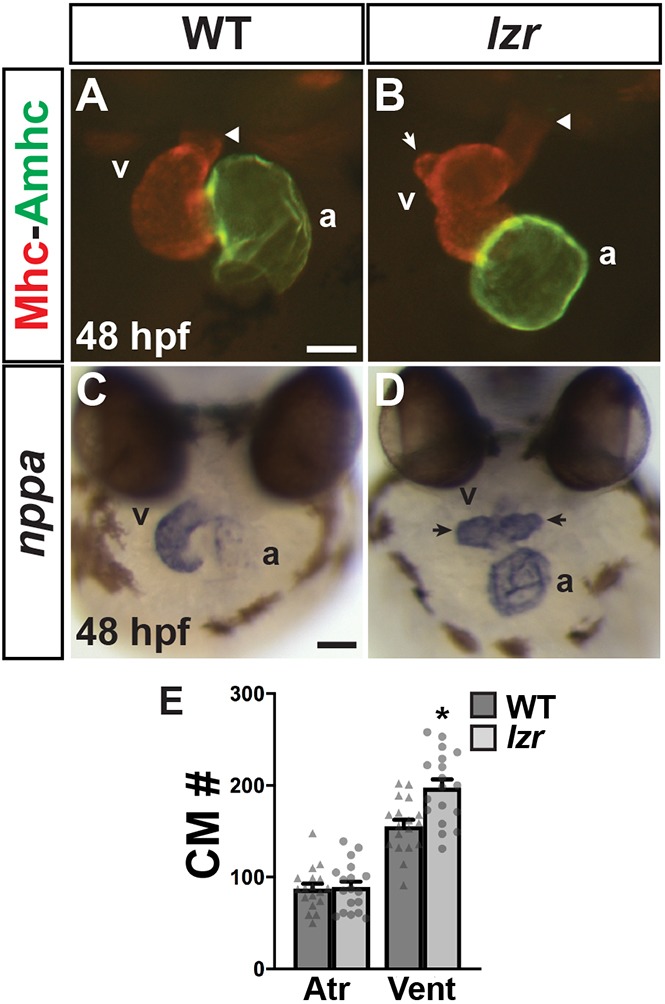
lzr mutants have an increase in ventricular CMs. (A,B) Hearts of WT and lzr embryos at 48 hpf. IHC for Sarcomeric myosin heavy chain (Mhc, red) and Atrial myosin heavy chain (Amhc, green). Images are frontal views. Arrowheads indicate the arterial pole of the heart. White arrow indicates a protrusion from the elongated OFT in a heart from a lzr embryo. Scale bar: 50 µm. (C,D) ISH for natriuretic peptide A (nppa). Images are frontal views. Arrows indicate ventricular protrusions. At least 30 embryos were examined for each condition. Scale bar: 100 µm. a, atrium; v, ventricle. (E) Number of CMs in WT (n=18) and lzr mutant (n=18) hearts with the myl7:DsRed2-NLS transgene. Atr, atrium; Vent, ventricle. Error bars indicate s.e.m. *P<0.05.
We next wanted to determine when the increased CMs in lzr mutants could first be observed. We did not observe a delay in CM differentiation, as was suggested previously (Kao et al., 2015), through late somitogenesis [16 and 20 somite (s) stages] using in situ hybridization (ISH) for the pan-cardiac differentiation marker myl7 (Fig. S2) or the ventricular CM-specific differentiation marker ventricular myosin heavy chain (vmhc; also known as myh7) (Fig. 2A,B) (Yelon et al., 1999). Furthermore, quantification of differentiating CMs with the myl7:EGFP transgene at the 20 s stage and 24 hpf did not show a difference in the number of CMs in lzr mutant hearts compared with their wild-type (WT) siblings (Fig. S3). Despite the lack of effect on earlier-differentiating CMs, we did observe morphological differences between lzr mutants and WT embryos in the forming hearts by the 20 s stage. Although vmhc was not overtly increased in lzr mutants, the cardiac cone had a greater aggregation of CMs more anteriorly and sometimes initiated fusion in the anterior (Fig. 2A,B, Fig. S2). Importantly, real-time quantitative PCR (RT-qPCR) on Pbx4-depleted embryos demonstrated that vmhc expression was not increased at the 20 s stage (Fig. 2G), corroborating the quantification of CMs with the myl7:EGFP transgene. By 24 hpf, the lzr mutant hearts were overtly shorter and wider compared with WT (Fig. 2C,D), despite not having a difference in differentiated CMs (Fig. S3). Although defects in the endoderm underlying differentiating CMs can lead to impaired migration, fusion and morphology (Fukushima et al., 2001; Maceyka et al., 2002), we did not observe overt defects in the endodermal marker sox32 (Muto et al., 2011) during gastrulation (Fig. S4) or anterior endodermal markers nkx2.7, gata4 or foxa2 (also known as axial) (Piotrowski and Nüsslein-Volhard, 2000) during later somitogenesis stages when CMs are migrating toward the midline (Fig. S4) (Grant et al., 2017).
Fig. 2.
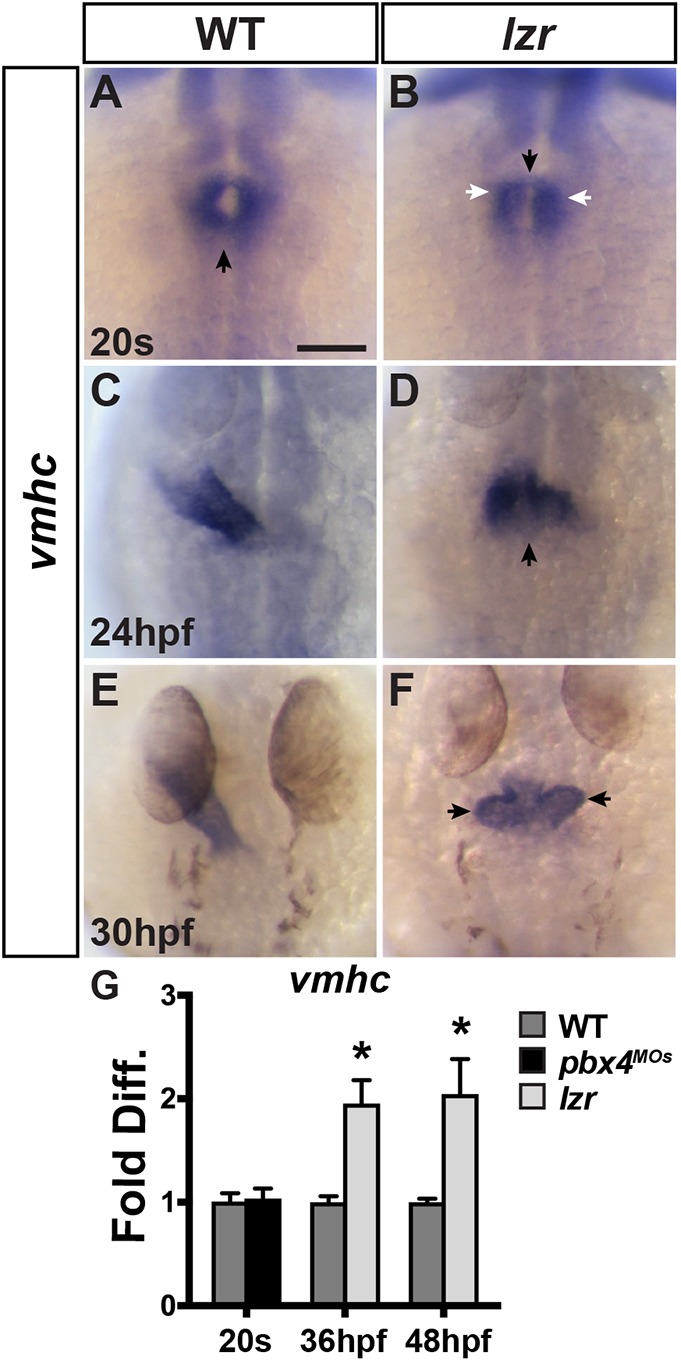
Cardiac fusion and elongation are abnormal in lzr mutants. (A-F) ISH for vmhc in WT and lzr mutant hearts at the 20 s stage, 24 hpf and 30 hpf. Views are dorsal with anterior up. Black arrows in A and B indicate the location of cardiac fusion when forming the cone. White arrows in B indicate anterior aggregates of CMs. Arrow in D indicates the larger ventricular pole. Arrows in F indicate already visible ventricular protrusions. At least 48 embryos per developmental stage were examined and genotyped. Scale bar: 100 µm. (G) RT-qPCR for vmhc expression in WT and Pbx4-depleted embryos at 20 s and in WT and lzr mutants at 36 hpf and 48 hpf. Error bars indicate s.e.m. *P<0.05.
Because of the lack of effect on early CM differentiation in lzr mutants, we sought to determine whether the increase in ventricular CMs derived from an increase in the number of CM progenitors at these earlier stages. However, we did not observe an expansion in myocyte enhancer factor 2cb (mef2cb), a marker of CM progenitors (Hinits et al., 2012; Lazic and Scott, 2011), at the 20 s stage (Fig. S2) nor a difference in the number of Nkx2.5+ progenitor cells at the 16 s stage (Fig. S5) or 24 hpf (Fig. S3). Therefore, although the location of CMs during cardiac cone formation is perturbed in lzr mutants, our data indicate overt specification of the initial Nkx2.5+ CM progenitor field and that early differentiation of FHF CMs is not affected.
lzr mutants have surplus SHF derivatives
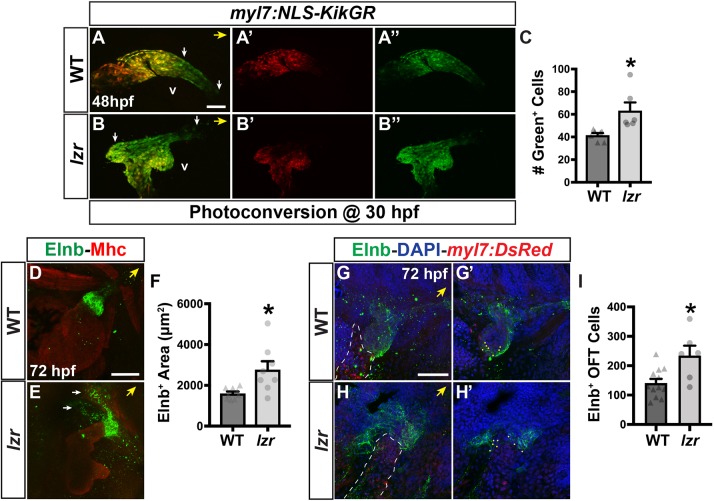
By 30 hpf, bilateral ventricular protrusions in the lzr mutant hearts were often evident (Fig. 2E,F). By 36 hpf, RT-qPCR indicated that vmhc expression was significantly increased in lzr mutants (Fig. 2G), which, together with the exacerbated morphology after 24 hpf, suggests that the increase in ventricular CMs may in part be derived from later-differentiating CMs. In contrast to vmhc, expression of the atrial-specific differentiation marker myh6 [also known as atrial myosin heavy chain (amhc)] (Berdougo, 2003) was not affected in lzr mutants at these stages (Fig. S6), again corroborating a specific defect in ventricular CMs. To assess whether later-differentiating ventricular CMs are increased in lzr mutants, we performed a temporal differentiation assay with the myl7:NLS-KikGR transgene (Lazic and Scott, 2011). CMs in transgenic embryos were photoconverted from green to red at 30 hpf. At 48 hpf, WT sibling and lzr mutant embryos were sorted and imaged (Fig. 3A-B″). We found that lzr mutants have a significant increase in these later-differentiating ventricular CMs (Fig. 3C). Moreover, this observation was corroborated using the myl7:Kaede transgene and photoconversion at 36 hpf (Fig. S7) (de Pater et al., 2009). Because both these results imply that the surplus ventricular CMs are derived from later-differentiating progenitors, we next wanted to determine whether there is a general expansion of SHF-derived cells in lzr mutants. Quantifying the amount of Elastin b (Elnb), a marker for SHF-derived smooth muscle (Miao et al., 2007; Moriyama et al., 2016), and the number of Elnb-surrounded cells within the OFT, we observed that there is also a surplus of SHF-derived smooth muscle within the OFT in lzr embryos at 72 hpf (Fig. 3D-I). Therefore, these results indicate that the enlarged OFTs of lzr mutant hearts are composed of surplus SHF-derived ventricular CMs and smooth muscle.
Fig. 3.
lzr mutants have an increase in later-differentiating ventricular CMs and SHF-derived smooth muscle. (A-B″) Confocal images of hearts from WT and lzr mutant myl7:NLS-KikGR embryos at 48 hpf following photoconversion at 30 hpf. White arrows indicate the area encompassing green-only cells within the OFT that was quantified. v, ventricle. (C) Number of green-only cells in WT (n=7) and lzr (n=6) myl7:NLS-KikGR hearts at 48 hpf following photoconversion. (D,E) IHC for Elnb (green) and Mhc (red) on hearts of WT and lzr embryos at 72 hpf. White arrows indicate extensions of Elnb in lzr mutant embryos. (F) Area (µm2) of Elnb expression in the OFT of WT (n=7) and lzr mutant (n=8) embryos. (G-H′) The OFT from hearts of WT and lzr mutant myl7:DsRed2-NLS embryos at 72 hpf. Elnb (green), DAPI (blue). Dashed line outlines the area containing CMs (red). G′ and H′ show the sections used to count Elnb-surrounded nuclei. Dots indicate representative nuclei. (I) Number of Elnb-surrounded cells in the OFT of WT (n=12) and lzr mutant (n=6) embryos. Yellow arrows indicate the direction of the arterial pole of the heart. Scale bars: 50 µm. Error bars indicate s.e.m. *P<0.05.
In addition to the SHF, cNCCs contribute to the OFT in vertebrate hearts and impaired cNCC migration manifests as distal OFT septation defects in Pbx1 KO mice (Chang et al., 2008). Furthermore, an early-migrating wave of cNCCs in zebrafish embryos differentiate as CMs (Abdul-Wajid et al., 2018; Cavanaugh et al., 2015) and zebrafish lzr mutants have neural crest (NC) defects (Pöpperl et al., 2000). Therefore, to assess NC contribution to the CMs within the hearts of Pbx4-depleted embryos, we used the established NC lineage-tracing lines sox10:GAL4, UAS:Cre; ubb:loxP-EGFP-loxP-mCherry (abbreviated as NC:mCherry) (Cavanaugh et al., 2015). We found that Pbx4-depletion does not affect the number of NC-derived CMs (Fig. S8), supporting the idea that cNCC defects do not contribute to the increased number of ventricular CMs within the hearts of lzr mutants.
Pbx4 limits proliferation within SHFPs
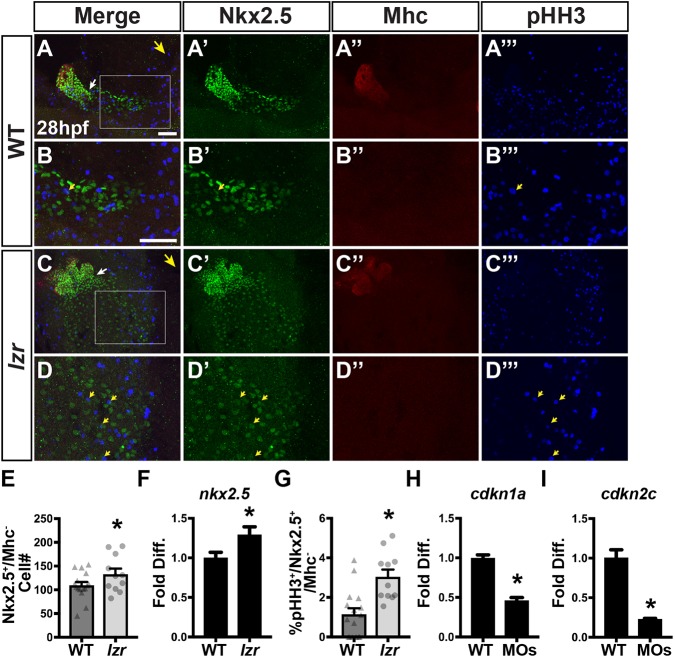
To determine whether the excess SHF-derived cells within the heart are from an enlarged population of SHFPs at later stages, we performed immunohistochemistry (IHC) for Nkx2.5 and sarcomeric myosin heavy chain (Mhc), which marks differentiated CMs, at 28 hpf. In contrast to quantification at the earlier stages, we found an ∼30% increase in the number of Nkx2.5+/Mhc− cells in lzr mutants by 28 hpf (Fig. 4A-E). Moreover, a similar increase in nkx2.5 expression was found using RT-qPCR on cDNA from lzr mutant embryos at 36 hpf (Fig. 4F). To determine whether excess proliferation can, at least in part, explain the increase in Nkx2.5+ progenitors of lzr mutants, the IHC for Nkx2.5 and Mhc also incorporated phospho-histone H3 (pHH3) labeling. We found that at 28 hpf lzr mutants have an increased percentage of pHH3+/Nkx2.5+/Mhc− progenitor cells (Fig. 4G). Furthermore, an increase in SHFP proliferation was also observed at 28 hpf following 5-ethynyl-2′deoxyuridine (EdU) incorporation at 26 hpf (Fig. S9). We did not find an increase in pHH3+/Nkx2.5+ progenitor cells within the ALPM of lzr mutants at the 16 s stages (Fig. S5) nor an increased rate of proliferation in differentiated Nkx2.5+/Mhc+ CMs of lzr mutants at 28 hpf (Fig. S10). In many contexts, increased cellular proliferation is associated with decreased expression of the cell cycle repressors cdkn1a (p21) and cdkn2c (p18) (Franklin et al., 2000; Jalili et al., 2012; Kreis et al., 2015; Robitaille et al., 2016). Therefore, we used fluorescence-activated cell sorting (FACS) to isolate nkx2.5:ZsYellow+ cells from transgenic control and Pbx4-depleted embryos at 28 hpf and assayed the expression of these genes with RT-qPCR. Consistent with increased proliferation of Nkx2.5+ progenitors after 24 hpf, Pbx4-depleted nkx2.5:ZsYellow+ cells had decreased cdkn1a and cdkn2c expression (Fig. 4H,I). Collectively, these results indicate that one mechanism by which Pbx4 restricts the number of cells within the OFT is through limiting proliferation of later-differentiating SHFPs.
Fig. 4.
lzr mutants have an increase in proliferating SHFPs. (A-D‴) IHC of Nkx2.5, Mhc and pHH3 in WT and lzr mutant embryos at 28 hpf. B and D are higher magnification images of the boxed areas in A and C, respectively. Large yellow arrows in A and C indicate direction of the arterial pole of the heart. White arrows indicate the border between Nkx2.5+/Mhc+ and Nkx2.5+/Mhc− cells. Small yellow arrows in B, B′, B‴, D, D′ and D‴ denote cells co-expressing Nkx2.5 and pHH3. Scale bars: 50 µm (in A for A-A‴,C-C‴; in B for B-B‴,D-D‴). (E) Number of Nkx2.5+/Mhc− cells at 28 hpf in WT (n=16) and lzr (n=11) embryos. (F) RT-qPCR for nkx2.5 at 36 hpf in WT (n=4) and lzr mutant (n=4) embryos. (G) Percentage of Nkx2.5+/Mhc− cells co-expressing pHH3. WT (n=16), lzr (n=11). (H,I) RT-qPCR for cdkn1a and cdkn2c from sorted nkx2.5:ZsYellow+ cells isolated at 28 hpf (n=3). Error bars indicate s.e.m. *P<0.05.
Pbx4 partitions subpopulations of Nkx2.5+ progenitors within the SHF
Despite the increased proliferation of SHFPs, it was not clear whether this was the sole mechanism that could explain the ∼50% increase in OFT ventricular CMs of lzr mutants. An indication that the increased OFT size in lzr mutants may also be the result of improper CM progenitor specification first came from analysis of markers mef2cb and ltbp3, which have enriched expression in SHFPs adjacent to the OFT (Hinits et al., 2012; Lazic and Scott, 2011; Zhou et al., 2011). Both ISH in lzr mutant embryos and RT-qPCR from the sorted Pbx4-depleted nkx2.5:ZsYellow+ cells indicated that the expression of mef2cb and ltbp3 was increased at 28 hpf (Fig. 5A-F), which is right at the onset of when we observe increased proliferation of Nkx2.5+ cells. Importantly, in contrast to these markers, we did not observe an increase in nkx2.5 expression in sorted nkx2.5:ZsYellow+ cells from Pbx4-depleted embryos (Fig. 5G), suggesting that the nkx2.5 transcript level per cell was not increased. Thus, the increased expression of SHF markers at the arterial pole in lzr mutants at 28 hpf, prior to when the increased CM proliferation could have significantly affected SHF size, suggests that the specification of SHFPs contributing to the OFT may be expanded at the expense of another Nkx2.5+ population at earlier stages.
Fig. 5.
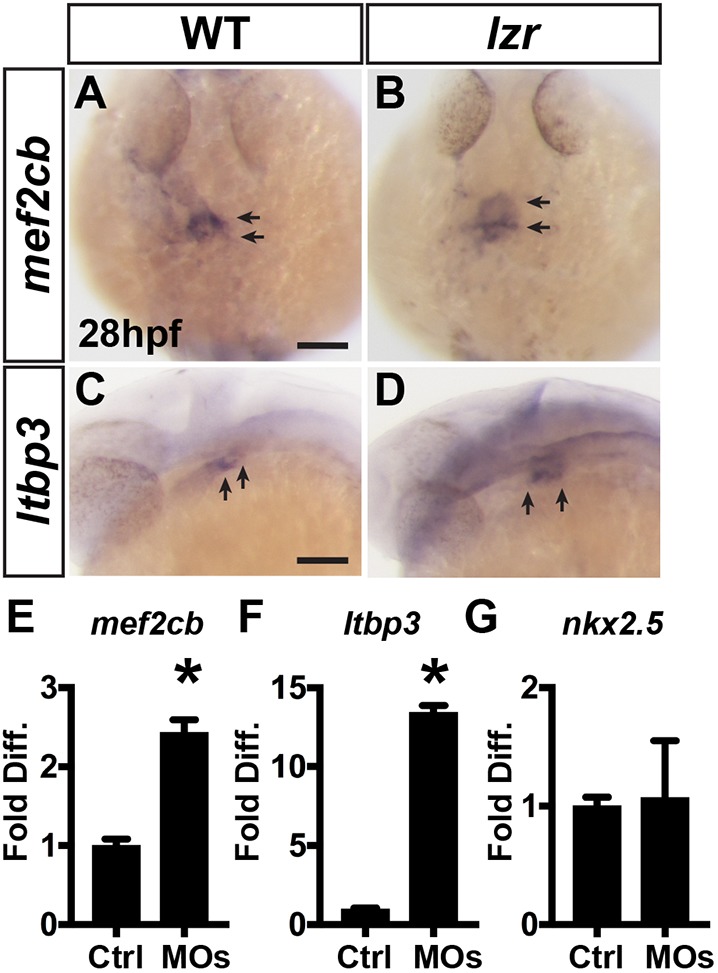
SHFPs adjacent to the arterial pole of the ventricle are expanded in lzr mutants. (A,B) ISH for mef2cb at 28 hpf. WT (n=7) and lzr (n=6). (C,D) ISH for ltbp3 at 28 hpf. WT (n=7), lzr (n=4). Views are dorsal with anterior up in A and B and lateral with anterior left in C and D. Arrows indicate expression at the arteriole pole of the ventricle. Scale bars: 100 µm. (E-G) RT-qPCR for meb2cb, ltbp3 and nkx2.5 from sorted nkx2.5:ZsYellow+ cells isolated at 28 hpf. Error bars indicate s.e.m. *P<0.05.
To test this hypothesis, we investigated posterior pharyngeal arch artery (pPAA) development in lzr mutants because Nkx2.5+ progenitors contribute to both the heart and pPAAs (3-6) (Abrial et al., 2017; Guner-Ataman et al., 2018; Paffett-Lugassy et al., 2013, 2017). Furthermore, similar to Nkx2.5 in zebrafish, recent lineage tracing has demonstrated that Isl1+ SHFPs in mice give rise to ECs within the posterior 3-6 PAAs in addition to the OFT (Wang et al., 2017), implying that a conserved aspect of vertebrate SHFPs is that they may contribute to the OFT of the heart and ECs of the pPAAs. Additionally, although the pPAAs are lost in lzr mutants (Pöpperl et al., 2000) and Pbx1 KO mice have great artery malformations (Chang et al., 2008), the mechanisms underlying these conserved defects are not understood. Concurrently examining the pPAAs and hearts using kdrl:mCherry; myl7:EGP transgenes at 48 hpf, we found that lzr mutants lack ECs of the pPAAs (Fig. 6A-D), confirming what has been reported (Pöpperl et al., 2000). However, despite the loss of pPAAs, lzr mutants retained anterior ECs within malformed, putative first arch arteries (AAs; mandibular arteries) and opercular arteries (ORAs) (Isogai et al., 2001) (Fig. 6A-D). To determine whether aggregates of ECs that give rise to the pPAAs develop in lzr mutants, we examined the EC markers tie1, fli1 and kdrl at 36 hpf (Anderson et al., 2008). Aggregates of EC precursors within that pharyngeal region did not form in lzr mutants. Instead, lzr mutants had a thin, broken line of differentiating ECs within this region connecting to the dorsal aorta (Fig. 6E,F, Fig. S11). Therefore, lzr mutants lack the pPAAs that are derived from Nkx2.5+ progenitors.
Fig. 6.
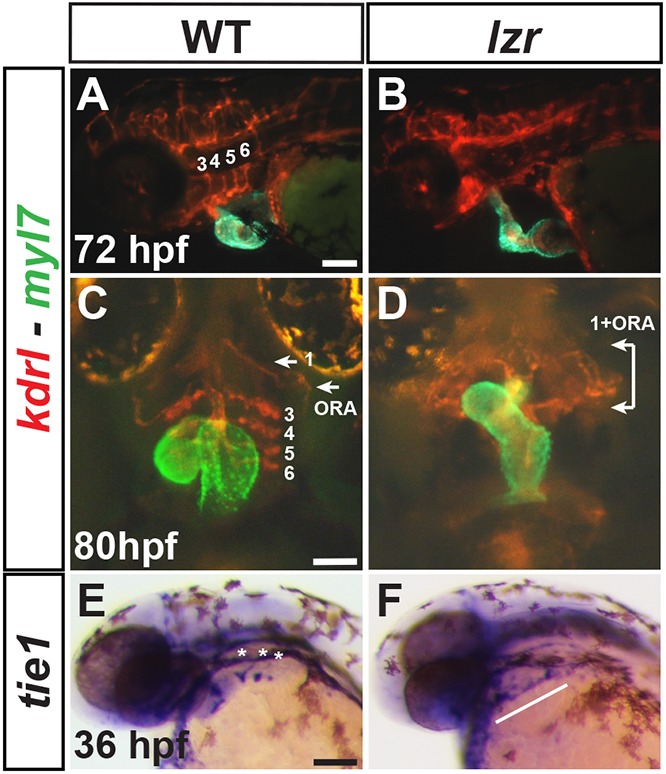
ECs in pPAAs are absent in lzr mutants. (A-D) IHC for mCherry (red) and EGFP (green) in WT and lzr mutant embryos with the kdrl:mCherry (ECs) and myl7:EGFP (CMs) transgenes at 72 and 80 hpf. Views in A and B are lateral with anterior left. Views in C and D are ventral with anterior up. Numbers designate the PAAs. ORA, opercular artery. White arrows and brackets indicate anterior arches. (E,F) ISH for tie1 at 36 hpf. Views are lateral with anterior left. WT (n=17), lzr (n=9). Asterisks denote aggregates of EC progenitors within developing posterior arches. White line marks tie1+ cells that extend to the dorsal aorta in lzr mutants. Scale bars: 100 µm.
Although one possibility is that the failure to form pPAAs is secondary at later stages due to NC segmentation defects, a prominent role of Pbx4 during early development is to promote homeotic transformations through the specification of more posterior fates (Erickson et al., 2007; Kimmel et al., 2001; Waskiewicz et al., 2001, 2002). Therefore, we used the kdrl:NLS-EGFP transgene to determine whether the number of EC progenitors in the anterior first AA and ORA is changed in lzr embryos. We did not find a difference between the number of ECs within the anterior first AA and ORA of WT sibling and the remaining, malformed first AA and ORA in lzr mutant embryos (Fig. S12). Additionally, we did not find an increase in the number of endocardial cells within the hearts of lzr mutants using the kdrl:NLS-EGFP transgene (Fig. S13). We also examined the anterior craniofacial muscles, which have been shown to derive partially from anterior nkx2.5+ cells (Nagelberg et al., 2015; Paffett-Lugassy et al., 2017). These muscles were present in lzr mutants and not discernibly different in size than those of WT siblings (Fig. S14). Therefore, our data suggested that there is not a conversion of pPAA progenitors into anterior ECs, endocardial cells or craniofacial muscles.
Within the ALPM, anterior nkx2.5+ progenitors will predominantly contribute to the heart, but also to the first AA, hypobranchial artery and craniofacial muscles (Nagelberg et al., 2015; Paffett-Lugassy et al., 2017), whereas smaller posterior nkx2.5+ progenitor populations will remain lateral and contribute to ECs of the pPAAs (Paffett-Lugassy et al., 2013). Thus, we next examined whether these nkx2.5+ progenitor subpopulations segregate during late somitogenesis stages. We used the nkx2.5:Kaede line to mark nkx2.5+ cells because this transgene has been shown to recapitulate most aspects of endogenous nkx2.5 expression (Paffett-Lugassy et al., 2013) and to provide continuity with subsequent lineage-tracing experiments described below. ISH of nkx2.5:Kaede expression demonstrated that nkx2.5+ progenitors through the 14-16 s stages were overtly unaffected in lzr mutants compared with siblings (Fig. 7A-D), consistent with what was found with cardiac specification markers at the 8 s stage (Kao et al., 2015). However, by the 18 s stage, aggregates of nkx2.5+ cells that go on to form the ECs of the PAAs were found separating from the more anterior population in WT embryos (Fig. 7E,G). These posterior nkx2.5+ aggregates were never observed in lzr mutants (Fig. 7E-H). Therefore, lzr mutants demonstrate an early failure to partition nkx2.5+ progenitor populations that contribute to the heart and pPAAs.
Fig. 7.
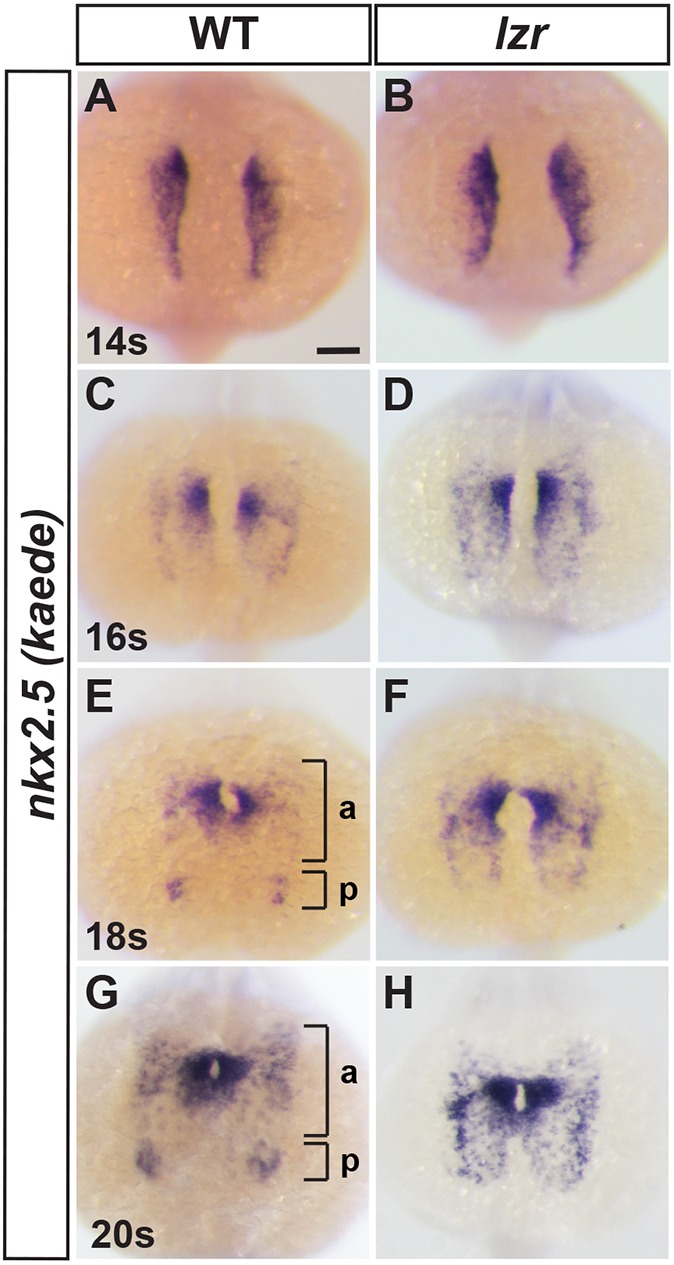
Nkx2.5+ progenitors fail to segregate into cardiac and endothelial populations in lzr mutants. (A-H) ISH for nkx2.5:Kaede in WT and lzr embryos at 14 s, 16 s, 18 s and 20 s. Views are dorsal with anterior up. Brackets in E and G denote anterior (a) and posterior (p) nkx2.5:Kaede+ clusters. 14 s: WT (n=47), lzr (n=5); 16 s: WT (n=26), lzr (n=6); 18 s: WT (n=29), lzr (n=6); 20 s: WT (n=58), lzr (n=6). Scale bar: 100 µm.
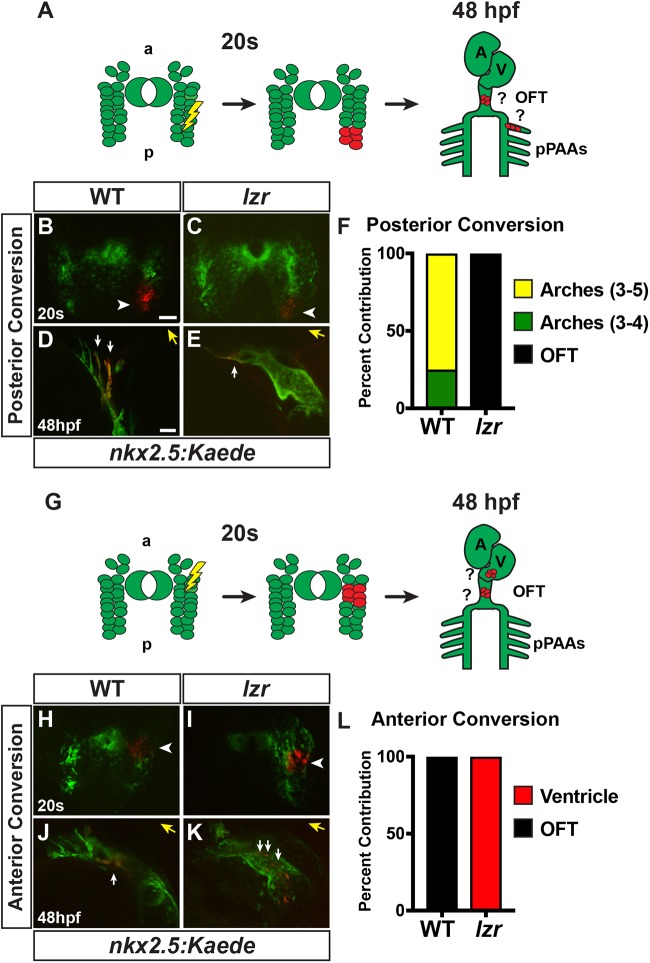
Given that we did not observe an effect on Nkx2.5+ progenitors within the ALPM of lzr mutants and that there is a requirement for Pbx proteins in posteriorizing tissues, a failure to form the posterior nkx2.5+ aggregates in lzr mutants indicated that these progenitors may be improperly specified, and instead contribute to the more anterior nkx2.5+ progenitors that form the OFT of the heart. To test this hypothesis, we performed optogenetic lineage tracing using the aforementioned nkx2.5:Kaede transgene. At the 20 s stage, clusters of nkx2.5:Kaede+ cells were converted from green to red in the most posterior region of expression in embryos from lzr heterozygous adults (Fig. 8A-C). Despite the clear distinction that was observed with ISH, there was not a clear separation of posterior nkx2.5:Kaede progenitors when examining fluorescence, potentially owing to a persistence of Kaede in these expressing cells. However, these clusters were also not visible when detecting endogenous Nkx2.5 expression (Figs S3 and S5). At 48 hpf, the embryos were sorted into WT and lzr mutant embryos and imaged to determine the contributions of the labeled cells to the ventricle, OFT and pPAAs (Fig. 8A). In WT sibling embryos, labeled nkx2.5:Kaede+ cells contributed to the ECs of the pPAAs (Fig. 8B,D,F), as would be expected (Guner-Ataman et al., 2013, 2018; Paffett-Lugassy et al., 2013; Rydeen and Waxman, 2016; Zhou et al., 2011). Conversely, in lzr mutants, the labeled nkx2.5:Kaede+ cells contributed to the tip of the OFT (Fig. 8C,E,F). A caveat of this experiment is that lzr mutants never form pPAAs. Therefore, we also photoconverted cells in more anterior positions within the nkx2.5:Kaede+ field, which are closer to the developing arterial pole of the heart (Fig. 8G). We found that labeled progenitors from this region contributed to the tip of the OFT and aorta before the third PAA in WT embryos (Fig. 8H,J,L). However, in lzr mutants, these more anterior-medial nkx2.5+ populations contributed to ventricular CMs that extended within the ventricle almost to the atrioventricular canal (Fig. 8I,K,L). Collectively, these results support that in lzr mutants the identity of nkx2.5+ progenitors within the ALPM is shifted anteriorly, with nkx2.5+ progenitors that normally segregate into ECs of the pPAAs instead integrating into the anterior SHF pool that contributes to the arterial ventricle and OFT.
Fig. 8.
Posterior nkx2.5+ progenitors contribute to the OFT and ventricle in lzr mutants. (A,G) Approaches for photoconversion and analysis of posterior and anterior nkx2.5+ cells. At the 20 s stage, small clusters of nkx2.5:Kaede cells were converted from green to red. At 48 hpf, they were analyzed for contributions to the ventricle, OFT, and pPAAs. a, anterior; A, atrium; p, posterior; V, ventricle. (B,C,H,I) Confocal images of photoconverted nkx2.5:Kaede clusters (red; white arrowheads) from transgenic WT and lzr mutant embryos at the 20 s stage. Views are dorsal with anterior up. (D,E,J,K) Confocal images of the photoconverted cells from B, C, H and I at 48 hpf in the pPAA, OFT and ventricle. White arrows in D indicate photoconverted Kaede+ cells (red) in PAAs 3 and 4. Views are lateral with anterior right. White arrows in E and J indicate photoconverted Kaede+ cells (red) within the OFTs. White arrows in K indicate photoconverted Kaede+ cells within the ventricle. Yellow arrows indicate direction of the OFT. Scale bars: 50 µm. (F) Percentage of cells converted in the posterior that labeled pPAAs and OFT. WT (n=8), lzr mutants (n=3). (L) Percentage of cells converted in the anterior-medial region that labeled the ventricular CMs and OFT. WT (n=9), lzr (n=3).
scRNA-seq reveals unappreciated stratification of differentiation states within nkx2.5+ cells
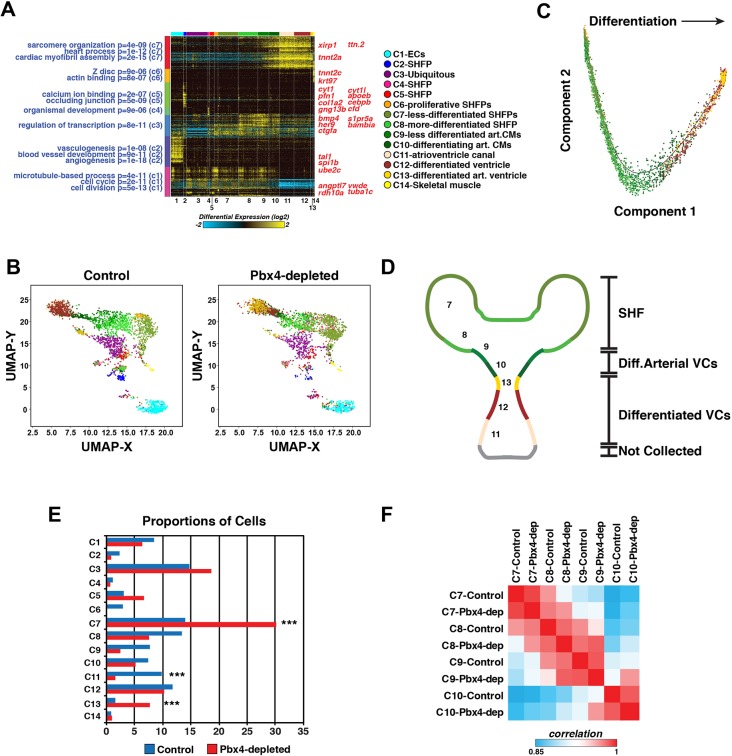
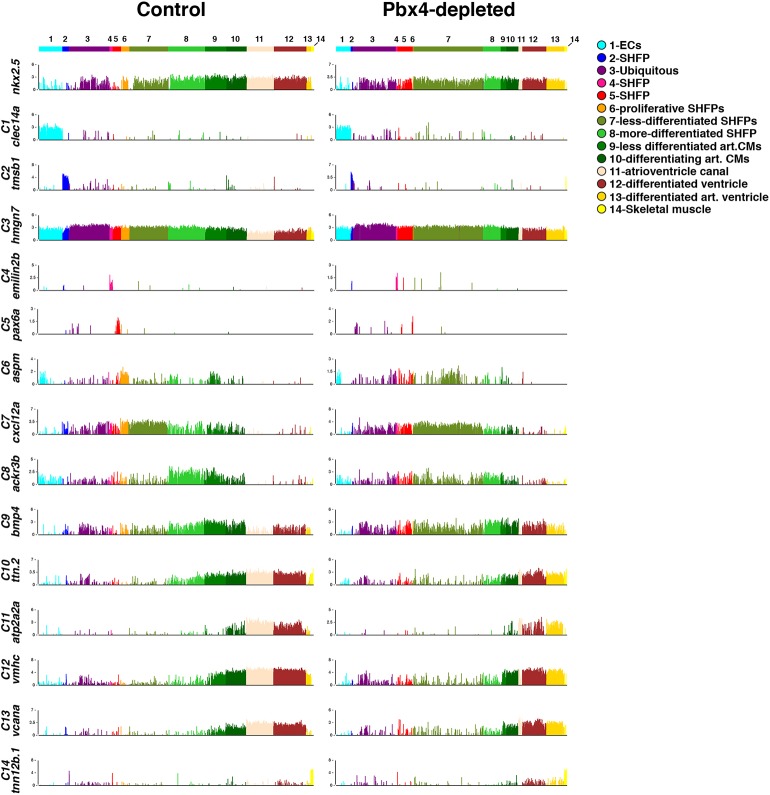
We sought to garner a better understanding of how Pbx4 affects identity states within the differentiating heart and SHF. Therefore, we used scRNA-sequencing to profile ∼2900 control and 2400 Pbx4-depleted nkx2.5:ZsYellow+ cells from 28 hpf embryos using the 10x Genomics platform. Unsupervised analyses of the control embryos with the ‘iterative clustering and guide-gene selection’ (ICGS) algorithm in AltAnalyze (Olsson et al., 2016) organized the control nkx2.5:ZsYellow+ cells into 14 transcriptionally distinct clusters (Fig. 9A), which were further evaluated using uniform manifold approximation and projection (UMAP) (Fig. 9B). The expression of population-discriminating genes was evaluated with the AltAnalyze ICGS viewer (http://altanalyze.org/ICGS/Public/NKX2-5/User.php) and in the UMAP plot. The majority of the sequenced cells expressed nkx2.5 (Figs 10 and 11), validating our isolation of nkx2.5+ cells. Smaller clusters that had lower levels or below detectable levels of nkx2.5 expression were indicated to be ECs (C1) and skeletal muscle progenitors (C14) (Figs 9A,B, 10 and 11), populations that transiently express endogenous nkx2.5 (Guner-Ataman et al., 2018; Nagelberg et al., 2015; Paffett-Lugassy et al., 2013). Hence, some discordance between expression from the nkx2.5:ZsYellow transgene used for sorting and endogenous nkx2.5 was not unexpected due to the persistence of ZsYellow (Zhou et al., 2011). Furthermore, searches using the ICGS viewer for endodermal markers (sox17, sox32, nkx2.3) indicated there were not any endodermal cells isolated, supporting the conclusion that all the Nkx2.5+ cells collected and analyzed were predominantly of mesodermal origin or transcriptional signature.
Fig. 9.
scRNA-seq reveals distinct differentiation states within the SHF and heart tube. (A) Heat map of predicted cell clusters from ICGS and identified guide-genes (right). Statistically enriched biological pathways from the software GO-Elite are shown on the left-hand side. (B) UMAP plots of nkx2.5:ZsYellow+ cells from control and Pbx4-depleted embryos. The Pbx4-depleted cells were aligned to the control cells in AltAnalyze. Colors correspond to the clusters shown in A. (C) Pseudo-time differentiation projection of the cardiac lineage clusters 6-13. (D) Schematic of the location of predicted cell types reflected in the cardiac lineage clusters 7-13. (E) Relative proportions of cells within each cluster from control and Pbx4-depleted nkx2.5:ZsYellow+ cells. Asterisks indicate the most significant differences using Fisher's exact test with P<10−28. (F) Pairwise comparison of gene expression within clusters 7-10 from control and Pbx4-depleted nkx2.5:ZsYellow+ cells using cellHarmony.
Fig. 10.
Gene expression within each cluster. Bar chart comparisons of gene expression in individual cells for genes found to be enriched in each cellHarmony assigned cluster. Each of the ∼2900 and ∼2400 cells from control and Pbx4-depleted embryos, respectively, is represented by a line on the x-axis of the graph. Y-axis indicates fold difference (log2) for the expression of each gene within a cell.
Fig. 11.
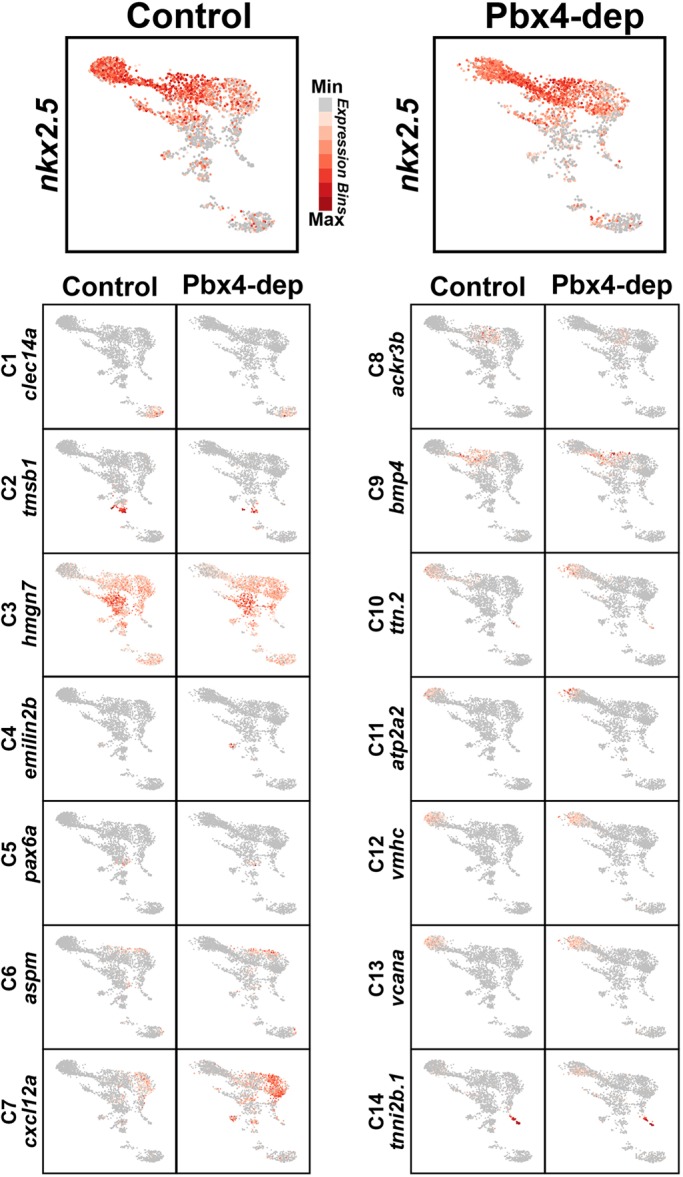
UMAP plots showing expression of genes enriched in each cluster. UMAP plots for control and Pbx4-depleted nkx2.5:ZsYellow+ cells. Expression of the genes shown in Fig. 10 is shown.
Gene expression within the clusters of nkx2.5:ZsYellow+ cells from control embryos revealed they were predominantly SHFPs and differentiated CMs. However, there was not a bimodal distinction between SHFPs and differentiated ventricular CMs in the nkx2.5+ cells. Instead, there was a greater spectrum of differentiation states coupled with unexpected, distinct expression boundaries, particularly within the SHFPs and arterial pole of the ventricle. The main developmental progression of differentiation states within the nkx2.5+ cells was revealed in clusters 7-13. C7 represents the least-differentiated, most-distal SHFPs relative to differentiated ventricular CMs, with genes expressed in this cluster including tbx1 (Liao et al., 2008; Nevis et al., 2013; Rana et al., 2014) and tcf21 (Nagelberg et al., 2015) (Figs 9A,B, 10 and 11, Fig. S15). Furthermore, expression of genes enriched in C7 progressively waned in clusters C8 through C10, and were predominantly absent in cells from clusters 11-13. Interestingly, although gene expression within C8 suggests that these cells are also SHFPs, genes enriched in this cluster, which includes ackr3b, rdh10a, meis1a and meis3, are largely excluded from cells in C7 (Figs 10 and 11, Fig. S15). Therefore, we posit that C8 is a specific subset of more-differentiated SHFPs residing closer to the arterial pole of the ventricle.
C9 was enriched for genes that promote CM differentiation, including bmp4 and hand2 (Reifers et al., 2000; Schindler et al., 2014), whereas C10 showed an increase in CM differentiation markers and concomitant decrease in SHFP marker expression (Figs 10 and 11, Fig. S16). These trends suggest that these two clusters contain differentiating ventricular CMs adjacent to or within the arterial pole of the ventricle. Clusters 12 and 13 were enriched for genes in differentiated ventricular CMs, including vmhc, suggesting that they are part of the arterial ventricular chamber (Figs 10 and 11, Fig. S16). The gene myh6, which is predominantly expressed in atrial CMs, was most highly expressed in C11 (Fig. S16). However, in the nascent heart tube, myh6 is also expressed in ventricular CMs at the venous pole of the chamber and the nascent atrioventricular canal (Foglia et al., 2016; Zhang et al., 2013), suggesting that these populations make up C11. No clusters were identified that exclusively expressed myh6 and did not express vmhc, implying that atrial CMs at the venous pole were not collected. Testing the progression of these differentiation states with pseudo-temporal ordering using Monocle2 (Qiu et al., 2017a,b; Trapnell et al., 2014) further corroborated this developmental progression, with clusters 7 and 11 being at opposite ends of a differentiation spectrum (Fig. 9C). Therefore, scRNA-seq of nkx2.5:ZsYellow+ cells reveals previously unappreciated stratification of SHFPs and differentiating ventricular CMs at the arterial pole in zebrafish embryos (Fig. 9D).
Although a progression of differentiation states within the developing heart was identified by gene expression in clusters 7-13, clusters 2-6 did not fit clearly into this progression. Clusters 2, 4, 5 and 6 are likely to be SHFPs, based on shared expression with markers enriched in C7, but also C8 and C9 (Fig. 10). Interestingly, these smaller clusters suggest they may be specialized, previously unanticipated subpopulations that reside within the SHF. For instance, gene ontology analysis indicated that C2 had broad signaling regulators of development, whereas C4 was characterized by numerous ‘posterior’ Hox genes (Table S1). C5 had genes associated with neuronal signature, including sox10, suggesting it may be cNCCs (Abdul-Wajid et al., 2018; Cavanaugh et al., 2015). C6 was enriched for genes associated with proliferation and the mitotic complex (Fig. 10, Fig. S15, Table S1). Furthermore, this cluster overlapped with SHFPs of C7 in the UMAP (Figs 9B and 11) and pseudo-temporal ordering (Fig. 9C). Although it was anticipated that SHFPs would be proliferative, it was not expected that proliferating SHF cells would be so distinct that they would form a unique cluster. In contrast to these SHF clusters, C3 was defined by enrichment for genes that are otherwise ubiquitously expressed (Figs 10 and 11, Table S1). Therefore, the scRNA-seq analysis also identified unanticipated smaller subpopulations of cells within the SHF that potentially may serve unique functions.
Pbx4 is necessary to stratify SHFP states
Next, we compared the nkx2.5:ZsYellow+ cells of Pbx4-depleted embryos with those in control embryos using the scRNA-Seq alignment tool cellHarmony (Hay et al., 2018). The major difference we found is that the relative number of SHFP cells within C7 (less-differentiated SHFPs) was increased in Pbx4-depleted embryos (Figs 9B,E, 10 and 11), which corroborates our quantification of nkx2.5+ SHFPs at 28 hpf by IHC in lzr mutant embryos (Fig. 4E). Additionally, the number of EC progenitors (C1) in Pbx4-depleted embryos was not reduced compared with controls, which likely reflects they are nkx2.5-derived ECs within the anterior AAs (Guner-Ataman et al., 2018; Nagelberg et al., 2015) and corroborates our quantification of ECs in the anterior AAs at 48 hpf (Fig. S12). Interestingly, although we find Nkx2.5+ SHFPs have increased proliferation in lzr mutants, the proliferative C6 cell population was dramatically reduced in Pbx4-depleted embryos. However, the proliferation-associated genes that defined this cluster in control embryos were expressed within the expanded C7 population of Pbx4-depleted embryos (Fig. 10, Fig. S15). This result supports that either less-differentiated SHFPs are more highly proliferative and/or this proliferative population has been incorporated into C7 of Pbx4-depleted embryos because it does not have as distinct a transcriptional signature.
In addition to the indication that the C7 of Pbx4-depleted embryos is more proliferative, genes normally restricted to the anterior clusters were expanded and/or had increased expression in posterior clusters. For instance, bmp4 and hand2, which are enriched in C9 of control cells, were highly expressed throughout C7 in Pbx4-depleted embryos (Fig. 10, Fig. S16). The expression of these genes in different cardiac clusters and their expanded expression was corroborated in WT and lzr mutant embryos using fluorescent ISH (FISH) (Fig. S17). Furthermore, mef2cb, which marks differentiating cells in the arterial pole of the ventricle and has a ‘posterior’ expression boundary between C8 and C9 of control embryos, was highly expressed within C8 cells of Pbx4-depleted embryos (Fig. S16), corroborating the expansion of mef2cb at the arterial pole with ISH in lzr mutant embryos (Fig. 5A,B). Note that ltbp3 could not be examined as its expression was too low to be detected in the isolated nkx2.5+ cells. Pairwise comparison of overall expression within clusters confirmed the anterior clusters were more similar to the posterior clusters in Pbx4-depleted embryos compared with controls (Fig. 9F). Additionally, the C13 population (arterial ventricular CMs) appeared to be expanded in the Pbx4-depleted embryos relative to control embryos, at the expense of C11 (venous ventricle and AVC), potentially reflecting the enlarged and elongating lzr mutant ventricles at 28 hpf (Fig. 9E). Comparison of differential gene expression (DGE) in the clusters from control and Pbx-depleted embryos showed that the greatest extent of DGE occurred in presumptive OFT progenitors (C13) (Fig. S18, Table S2). Indeed, ISH confirmed that genes associated with ventricular differentiation, including crip2, vcana, myl10 and lft2, were improperly expressed in lzr mutant hearts (Fig. S19). Overall, our scRNA-seq analysis indicates that Pbx4 restricts proliferation and anteriorization by reinforcing the stratification of distinct cellular identities within the SHF and arterial pole of the heart.
DISCUSSION
We still have limited knowledge of the mechanisms that restrict the size of the vertebrate heart. At earlier stages of cardiogenesis, retinoic acid and Wnt signaling restrict atrial and ventricular CM differentiation within the ALPM (Dohn and Waxman, 2012; Keegan et al., 2005; Ryckebusch et al., 2008; Sirbu et al., 2008; Sorrell and Waxman, 2011; Waxman et al., 2008), and the transcription factor Hey2 limits atrial and ventricular CM proliferation (Gibb et al., 2018; Jia et al., 2007). At later stages, cellular adhesion molecule 4 (Cadm4) limits the size of the OFT by restricting the deployment of SHFPs in zebrafish embryos (Zeng and Yelon, 2014). We find that Pbx4 has a unique role in specifically limiting the size of the SHF, and consequently the OFT and ventricle, which contrasts with a previously proposed role for Pbx4 in controlling the temporal differentiation of CMs that affects both the first and second heart fields (Kao et al., 2015). Using similar standard assays as Kao et al. (2015) to examine cardiomyocyte differentiation (vmhc), SHFPs (ltbp3) and SHF-derived smooth muscle (elnb), as well as additional complementary and quantitative assays, we did not observe the reported significant delays in CM differentiation during somitogenesis and later general increase in both atrial and ventricular CMs. Additionally, the OFT defects reported by Kao et al. (2015) were ambiguous, including both weaker ltbp3 expression and highly variable effects on elnb expression in lzr mutants (Kao et al., 2015). We presently do not understand why we observe these differences. Altogether, our extensive use of complementary assays, quantitative analyses, lineage tracing and scRNA-seq support a model whereby Pbx4 first partitions OFT and pPAA progenitors within the posterior ALPM and subsequently restricts proliferation of the SHFPs that will differentiate at the arterial pole of the heart (Fig. 12).
Fig. 12.
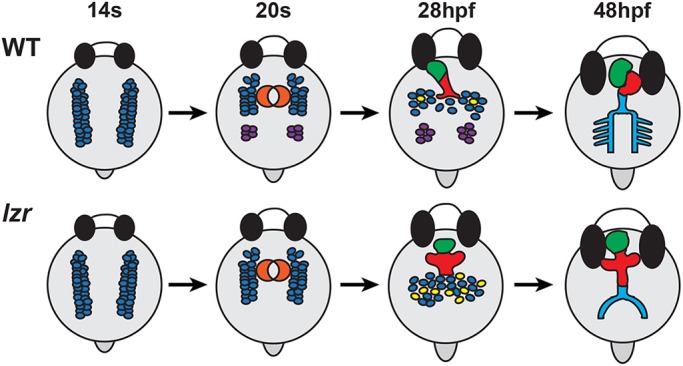
Schematic of the requirements of Pbx4 in OFT and great artery development. In WT embryos, nkx2.5+ cells within the ALPM separate into anterior populations that will primarily contribute to the heart tube and later to differentiating proliferative SHFPs, and posterior populations that will contribute to the pPAAs. In lzr mutants, the anterior and posterior nkx2.5+ populations initially fail to separate and later the SHFPs are more proliferative, leading to hearts with enlarged OFTs coupled with loss of the pPAAs. Blue, nkx2.5+ progenitors; purple, pPAA progenitors; orange, FHF progenitors forming a cone; red, ventricular CMs; green, atrial CMs; yellow, proliferating cells; light blue, aorta and pPAAs.
Previous work has shown that Pbx4 is expressed ubiquitously (Kao et al., 2015; Pöpperl et al., 2000). Our scRNA-seq data corroborated that pbx4 is expressed throughout the SHFPs and differentiated CMs (Fig. S15). Thus, other factors must work in conjunction with Pbx4 to promote the specific requirements in the OFT and greater arteries. Pbx factors often function in a complex with Hox and Meis transcription factors (Ferretti et al., 2000). In mice, Hoxa1 and Hoxb1 are expressed in the posterior SHF and are required for OFT development (Bertrand et al., 2011; Roux et al., 2015). Reminiscent of lzr mutants, Hoxb1 mouse mutants have an increased mitotic index of SHFPs and premature myocardial differentiation, the latter of which is enhanced in Hoxa1; Hoxb1 double mutants (Roux et al., 2015). However, a distinction may be that our scRNA-seq data indicate nkx2.5+ SHFPs in Pbx4-depleted embryos actually have a less-defined, mixed cell state identity, rather than precocious differentiation. Whereas Hoxa1; Hoxb1 double mutants also have PAA defects, PAA EC specification is not perturbed, suggesting that, unlike lzr mutants, they are not functioning to partition these progenitors. Therefore, the similarity of SHF defects in Hoxa1; Hoxb1 KO mice, although less severe, implies that Hox proteins may be co-factors that act with Pbx proteins to limit OFT size at later stages of its formation.
Mutations in MEIS1 and MEIS2 genes have been associated with CHDs in humans (Crowley et al., 2010; Pfeufer et al., 2010). Meis proteins can function with core cardiogenic transcription factors, including Tbx5 and Nkx2.5, during different stages of cardiogenesis (Luna-Zurita et al., 2016; Wamstad et al., 2017). Meis1 has been shown to compete with Nkx2.5 for binding sites to allow progression from SHFPs to differentiated CMs (Dupays et al., 2015). Meis1 KO mice have similar OFT septation defects as Pbx1 KO mice (González-Lázaro et al., 2014; Stankunas et al., 2008), implying that they may function with Pbx genes in cardiac development (Roux and Zaffran, 2016). However, the specific mechanisms underlying the OFT defects in Meis1 and Pbx1 KO mice remain to be elucidated. Although Meis1 represses proliferation as a direct regulator of Cdkn1a in postnatal hearts (Mahmoud et al., 2013), we have not yet examined whether Pbx4 directly regulates expression of cell cycle inhibitors in this context. Regardless of the transcriptional mechanism, our work supports the hypothesis that repression of proliferation may be a broad requirement of a Meis-Pbx complex in cardiac tissues.
Impaired cNCC contribution underlies the distal OFT septation defects in Pbx1 KO mice. However, the mechanisms underlying proximal septation of the conus and arch arteries have remained unexplored (Chang et al., 2008; Stankunas et al., 2008). Importantly, our data show that specific OFT and PAA (great artery) defects in Pbx1 KO mice and zebrafish lzr mutants are conserved consequences of Pbx loss in vertebrates. Furthermore, our data support that the SHF-derived OFT and pPAA defects in these Pbx-deficient embryos are directly related. Recent reports have greatly expanded upon the lineages derived from SHFPs (Wang et al., 2017). Although the SHF typically has not been proposed to include the pPAAs, lineage tracing of the Isl1+ SHFPs in mice indicates that they give rise to ECs within the posterior 3-6 PAAs (Wang et al., 2017). While there is currently not an equivalent SHFP marker in zebrafish, lineage tracing of nkx2.5+ progenitors within the ALPM demonstrates that they overtly start as a single field that then separates into an anterior population, which predominantly contributes to the heart, and a posterior population, which contributes to the ECs of the pPAAs (Paffett-Lugassy et al., 2013). Similarly, lineage tracing of tbx1 and tcf21 in zebrafish shows contribution to both the arterial pole of the ventricle and PAAs (Abrial et al., 2017; Guner-Ataman et al., 2018; Nagelberg et al., 2015). Thus, the contribution of cell lineages within the ALPM to what has traditionally been considered the derivatives of the SHF and ECs of the pPAAs appears to be conserved in vertebrates. Our data further illustrate the proximity of these progenitor populations within an initial organ field, as lzr mutants fail to partition the nkx2.5+ progenitors into their respective anterior (OFT) and posterior (pPAA) populations.
Our scRNA-seq of nkx2.5+ cells in control and Pbx4-depleted embryos provides insight into previously unappreciated stratification of differentiation states and subpopulations that occur during the development of the SHF, arterial pole of the heart, and ventricular CMs in zebrafish embryos. We find that normally within the broader nkx2.5+ population at 28 hpf, there are surprisingly distinct boundaries between less-differentiated (posterior) and more-differentiated (anterior) SHFP populations, as well as at least four definable differentiation states within cells at the arterial pole of the heart. Importantly, the transcriptional resolution of the scRNA-seq data bolster our experimental analysis and model, as well as add depth to our understanding of the cardiac defects in lzr mutants. The stratification of normally distinct progenitor populations within the SHF is lost in Pbx4-deficient embryos, with the enlarged SHFP population acquiring gene expression associated with both proliferation and more anterior, differentiated progenitors. Interestingly, the role of Pbx4 in generating posterior cell types within the SHF of the ALPM is reminiscent of homeotic transformations observed in defined metameric tissues, like the hindbrain (Arenkiel et al., 2004; Choe et al., 2002; Erickson et al., 2007; McClintock et al., 2002; Pöpperl et al., 2000; Studer et al., 1998; Waskiewicz et al., 2001, 2002). Thus, altogether, our scRNA-seq data provide additional mechanistic insights into previously unappreciated SHFP populations in both normal and Pbx-deficient conditions and will serve as a resource for the interrogation of novel hypotheses regarding mechanisms of congenital heart disease caused by improper SHF development.
Overall, our study establishes new models for the conserved requirements of Pbx transcription factors in the development of the OFT and pPAAs, as well as providing maps of distinct progenitor populations within the vertebrate SHF and ventricle of normal and Pbx4-depleted embryos. Future studies will allow us to decipher the specific transcriptional mechanisms and gene regulatory networks by which Pbx proteins regulate allocation and proliferation of SHFPs, which may illuminate the molecular origins of CHDs in humans.
MATERIALS AND METHODS
Ethics statement
Zebrafish husbandry and experiments were performed as outlined in approved IACUC protocols at the Cincinnati Children's Hospital Medical Center.
Zebrafish husbandry and lines used
Adult wild-type and transgenic zebrafish (Danio rerio) lines were maintained under standard laboratory conditions (Westerfield, 2000). The following transgenic lines were used: Tg(-5.1myl7:DsRed2-NLS)f2 (Mably et al., 2003), TgBAC(-36nkx2.5:ZsYellow)fb7 (Zhou et al., 2011), TgBAC(-36nkx2.5:Kaede)fb9 (Zhou et al., 2011), Tg(myl7:Kaede)sd22 (de Pater et al., 2009), Tg(myl7:NLS-KikGR)hsc6 (Lazic and Scott, 2011), Tg(myl7:EGFP)twu (Huang et al., 2003), Tg(kdrl:NLS-EGFP)ubs1 (Blum et al., 2008), Tg(-6.5kdrl:mCherry)ci5 (Proulx et al., 2010), Tg(-5sox10:GAL4, UAS:Cre)la2326 (Cavanaugh et al., 2015) and Tg(-3.5ubb:loxP-EGFP-loxP-mCherry)cz1701. The lzrb557 allele was used (Pöpperl et al., 2000). Genotyping of lzr mutants was performed with XbaI digest, which cuts the mutant allele, following PCR using previously reported primers (Table S3) (Kao et al., 2015).
Morpholino injections
Embryos were injected at the one-cell stage with 0.5 ng of both pbx4-MO1 (5′-AATACTTTTGAGCCGAATCTCTCCG-3′) and pbx4-MO2 (5′-CGCCGCAAACCAATGAAAGCGTGTT-3′) (Gene Tools) (Maves et al., 2007). All injections also included 0.5 ng of zebrafish p53 (tp53) morpholino to minimize nonspecific cell death (Robu et al., 2007).
ISH
Standard (NBT/BCIP color reaction) ISH and fluorescent (tyramide conjugated) ISH (FISH) were performed as previously reported (Oxtoby and Jowett, 1993; Row and Martin, 2017). Probes were used for the following: bmp4 (ZDB-GENE-980528), crip2 (ZDB-GENE-040426-2889), fli1a (ZDB-GENE-980526-426), foxa2 (ZDB-GENE-980526-404), gata4 (ZDB-GENE-980526-476), hand2 (ZDB-GENE-000511-1), kaede (ZDB-EFG-080131-2), kdrl1 (ZDB-GENE-000705-1), lft2 (ZDB-GENE-990630-11), ltbp3 (ZDB-GENE-060526-130), mef2cb (ZDB-GENE-040901-7), myl7 (ZDB-GENE-991019-3), myl10 (ZDB-GENE-050417-421), myoD (ZDB-GENE-980526-561), nkx2.7 (ZDB-GENE-990415-179), nppa (ZDB-GENE-030131-95), sox32 (ZDB-GENE-011026-1), tie1 (ZDB-GENE-990415-55), vcana (ZDB-GENE-011023-1) and vmhc (ZDB-GENE-991123-5). Embryos from standard ISH experiments were imaged using Zeiss M2Bio V8 or Zeiss M2BioV12 stereomicroscopes. Embryos from FISH experiments were imaged using a Nikon A1 confocal microscope. All embryos were genotyped following imaging. Area measurements for elnb were performed using Fiji (Schindelin et al., 2012).
IHC
IHC for quantification of Nkx2.5+ cells, myl7:DsRed2-NLS+ CMs and kdrl:NLS-EGFP+ ECs and endocardial cells was performed as described previously (Waxman et al., 2008). Embryos were fixed for 1 h in 1% formaldehyde/1×PBS. Following fixation, embryos were repeatedly washed in 0.2% Saponin in 1×PBS. Blocking was performed for 1 h at room temperature (RT) in a Saponin Block solution (SBS) [1×PBS, 10% sheep serum, 2 mg/ml bovine serum albumin (BSA) and 0.2% Saponin]. Embryos were then incubated with primary antibodies diluted in SBS overnight at 4°C. Embryos were washed 3× in 0.2% Saponin/1×PBS before incubation for 2 h at RT with secondary antibodies diluted in SBS. Embryos were washed multiple times in 0.2% Saponin/1×PBS prior to imaging. For quantification of myl7:DsRed2-NLS+ CMs and kdrl:NLS-EGFP+ ECs and endocardial cells, embryos were gently compressed with a coverslip and imaged using a Zeiss M2BioV12 or V8 stereomicroscope. Cells with DsRed+ or GFP+ nuclei were counted using Adobe Photoshop. To quantify Nkx2.5+ and pHH3+ co-labeled cells, embryos were mounted in µ-Slide 2 Well chamber slides (Ibidi, 80286). To quantify myl7:EGFP+ CMs and Nkx2.5+ cells, embryos were flat-mounted on a coverslip ventral side down. These embryos were imaged using a Nikon A1 confocal microscope. For myl7:EGFP+ cells where the nuclei were not obvious, Nkx2.5+ nuclear localization was also used to aid counting. Imaris software (Bitplane) was used to assist in determining colocalization of Nkx2.5 and pHH3 and to assist with cell counting.
The affinity-purified Elnb antibody was custom generated by YenZym to the peptide PGAGYQQQYPGFGGPGAGGPGS, which corresponds to amino acids 1992-2013 of the protein, and validated by comparison to the established expression of Elnb in the OFT (Miao et al., 2007). IHC with Mhc (MF20) was performed and imaged as indicated above. Elnb-surrounded smooth muscle cell nuclei were quantified with Imaris software (Bitplane) by using a ‘masked channel’ for Elnb. Subsequently, the ‘spots’ function was used to identify DAPI+ nuclei within our masked region of interest. Area measurements for Elnb were performed using Fiji (Schindelin et al., 2012).
For detection of NC cells, embryos were prepared as described previously (Rydeen and Waxman, 2016). NC:mCherry embryos were fixed in 4% paraformaldehyde and stored at 4°C. Subsequently, embryos were gradually dehydrated in a PBS with 0.1% Tween (PBST)/methanol series and stored at −20°C. Embryos were gradually rehydrated in PBST and incubated for 20 min in PBST supplemented with 1% DMSO and 0.3% Triton X-100 at RT. Embryos were blocked for 30 min at RT in 0.1% Tween20/10% sheep serum/1×PBS, and incubated overnight with primary antibodies at 4°C. Subsequently, embryos were washed three times for 5 min with PBST, blocked for 30 min, and incubated with secondary antibody for 2 h at RT. Embryos were then washed in PBST and stored at 4°C. Prior to imaging, the heads of the embryos were removed using forceps. Embryos were mounted and imaged as above.
To determine EC contributions to the anterior PAAs, IHC was performed on WT and lzr embryos harboring the kdrl:NLS-EGFP transgenes. Embryos were fixed at 48 hpf and IHC was performed as described above. Following IHC, embryos were mounted in µ-Slide 4 Well chamber slides (Ibidi, 80426) and imaged using a Nikon A1 confocal microscope. Cells with GFP+ nuclei within the anterior first and second PAAs were counted from images in Photoshop.
All antibody concentrations and supplier information are provided in Table S4.
EdU labeling
EdU labeling was performed using the Click-iT EdU Alexa Fluor 594 Imaging Kit (Life Technologies) similar to a previous report (Mahler et al., 2010). Briefly, nkx2.5:ZsYellow transgenic embryos were dechorionated at 26 hpf and incubated in 10 mM EdU for 20 min on ice. Following EdU incorporation, embryos were rinsed once with cold 0.3× Danieau's solution and then three times with RT 0.3× Danieau's for 5 min. Embryos were chased for 2 h at 28°C and subsequently fixed in 1% formaldehyde for IHC, as described above. Following IHC, embryos were mounted in µ-Slide 4 Well chamber slides (Ibidi, 80426) and imaged using a Nikon A1 confocal microscope. Imaris software (Bitplane) was used to assist in determining colocalization of Nkx2.5 and Edu.
Photoconversion and lineage-tracing assay
To assess the temporal differentiation of CMs with the myl7:NLS-KikGR and myl7:Kaede transgenes, photoconversion was performed as previously reported (Rydeen and Waxman, 2016). Briefly, embryos resulting from adult lzr+/−;myl7:NLS-KikGR fish or adult lzr+/−;myl7:Kaede fish were exposed to light from a DAPI filter for 20 min at 30 and 36 hpf, respectively, with a Zeiss M2BioV12 fluorescent stereomicroscope. At 48 hpf, transgenic lzr mutants and their WT sibling embryos were sorted, anesthetized with Tricaine, and prepped for confocal imaging by gentle compression with a coverslip. Newly differentiated CMs (green-only cells) from photoconverted myl7:NLS-KikGR embryos were counted from images in Adobe Photoshop. The contribution of green-only Kaede+ cells to the OFT was determined by measuring a region of interest using ImageJ software (Rueden et al., 2017).
To perform lineage tracing with the nkx2.5:Kaede transgene, photoconversion of nkx2.5:Kaede+ cell clusters was performed in a manner similar to that previously described (Rydeen and Waxman, 2016). Two-well μ-slides (Ibidi, 80286) were covered in a thin layer of 2% agarose and wells for embryos were created by puncturing the agarose with a capillary tube. Embryos were mounted dorsal side up in these wells and specific populations of nkx2.5:Kaede+ cells in either the most posterior or anterior-medial regions of expression were photoconverted from green to red using the 20× objective with 10× zoom and the DAPI filter on a Nikon A1 confocal microscope. Embryos were then incubated at 28.5°C until 48 hpf, at which point they were sorted into WT sibling and lzr mutant embryos, mounted on their side in embryo water, and imaged using a Nikon A1 confocal microscope. The contributions of converted red cells to the ventricle, OFT and pPAAs were scored.
RT-qPCR
RNA isolation, cDNA synthesis and qPCR were performed as previously described (D'Aniello et al., 2013). Total RNA was isolated from 30 embryos using the TRIzol reagent (Life Technologies, Invitrogen) protocol. The Pure link RNA Micro Kit (Invitrogen) was used to collect RNA. cDNA synthesis was performed using the ThermoScript Reverse Transcriptase kit (Invitrogen). RT-qPCR was performed using SYBR green PCR master mix (Applied Biosystems) in a Bio-Rad CFX PCR machine followed by melt curve analysis. Relative changes in expression with RT-qPCR were calculated using actb1 levels for normalization with the Livak method (Schmittgen and Livak, 2008). Primers used for RT-qPCR are listed in Table S3.
FACS and scRNA-seq
nkx2.5:ZsYellow+ cells from control and Pbx4-depleted embryos were isolated by FACS at 28 hpf. Dissociation of embryos was performed as described previously (Stachura and Traver, 2016) with the following modifications. Embryos (∼180 per condition) were manually dechorionated and rinsed in ice cold Hank's Balance Salt Solution (HBSS) containing Ca2+ and Mg2+. Embryos were subsequently transferred to 1.5 ml tubes in 500 µl HBSS and dissociation was initiated by adding 60 µl Liberase (Roche). Embryos were deyolked by gently pipetting up and down, first with a p1000 tip and subsequently with a p100 tip, as described previously (Samsa et al., 2016). Following yolk removal, embryos were incubated at 32.5°C. To facilitate dissociation, embryos were gently homogenized using a handheld pestle homogenizer (5-10 s) and returned to 32.5°C. This process was repeated until complete dissociation was achieved, which took approximately 15 min. Dissociation of embryos was monitored using a dissecting microscope. Following dissociation, cell suspensions were centrifuged for 5 min at 13,000 rpm (14,549 g) at 4°C. The supernatant was discarded and the resulting pellets were re-suspended in 500 µl of FACS medium consisting of 0.9× PBS and 2% fetal bovine serum. Samples were filtered twice through a 40 µm strainer and collected in a FACS tube. FACS sorting was performed by the CCHMC Research Flow Cytometry Core using a 70 µm nozzle and a gating strategy as described by Samsa et al. (2016). After FACS, cDNA for RT-qPCR was generated from mRNA isolated using the Single Cell RNA Purification Kit (Norgen Biotek) following the manufacturer's instructions. cDNA was synthesized using the Ovation RNA Amplification System V2 (NuGEN Technologies). For scRNA-seq, sorted cells were transferred to the CCHMC genomics core for processing and library generation with the 10x Genomics platform, followed by high-throughput sequencing in the CCHMC DNA core on an Illumina 2500 system.
Analysis of scRNA-seq data
Alignment and quantification of the scRNA-Seq data was performed using version 2.1.1 of Cell Ranger (10x Genomics) against the Zebrafish genome (Ensembl version 91); 2885 cellular barcodes with 52,617 mean reads/cell and 2039 mean genes/cell were obtained for the control nkx2.5:ZsYellow+ cells; 2417 cellular barcodes with 63,042 mean reads/cell and 2162 mean genes/cell were obtained for Pbx4-depleted nkx2.5:ZsYellow+ cells; 10,745 and 10,634 UMIs per barcode were obtained from the default filtered HDF5 outputs, respectively. The wild-type HDF5 file was used as input for unsupervised population detection in ICGS version 1.0, available through the software AltAnalyze and the Ensembl version 91 zebrafish database (Olsson et al., 2016). Using the ICGS-identified cell populations and associated population variable genes (Guide 3 output), label projection of the Pbx4-filtered HDF5 sample was performed using the cellHarmony software in AltAnalyze version 2.1.4 (DePasquale et al., 2019). For cellHarmony, population labels from the wild-type ICGS results were inferred using the built-in BioMarker database, with zebrafish orthologs from human tissue and cell-type makers (default ICGS output). Genes with a fold-difference of 1.5 and an empirical Bayes-moderated t-test P-value were reported for all wild-type aligned cell populations. Associated UMAP projections were produced by cellHarmony with gene-level expression UMAP plots produced in AltAnalyze.
Statistical analysis
Statistical significance comparing quantitative data for two conditions was determined using a two-tailed Student's t-test. For categorical data, Fisher's exact test was used. Statistical tests were performed using GraphPad Prism software. P<0.05 was considered statistically significant. Error bars in all graphs indicate s.e.m. Other than the scRNA-seq data, experiments shown are representative of at least two biological replicates.
Supplementary Material
Acknowledgements
The authors thank members of the Waxman lab for insightful discussions and Danielle Fritsch for excellent care of the zebrafish.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.H., J.S.W.; Methodology: A.H., N.S., J.S.W.; Software: K.C., N.S.; Validation: A.H., K.L., P.R., J.S.W.; Formal analysis: A.H., K.L., P.R., K.C., N.S., J.S.W.; Investigation: A.H., K.L., P.R., J.S.W.; Resources: A.H., J.S.W.; Data curation: K.C., N.S.; Writing - original draft: A.H., N.S., J.S.W.; Writing - review & editing: A.H., N.S., J.S.W.; Visualization: A.H., K.L., P.R.; Supervision: N.S.; Project administration: J.S.W.; Funding acquisition: A.H., J.S.W.
Funding
This work was funded by the National Institutes of Health [R01 HL112893, R01 HL137766, R01 HL141186 to J.S.W.] and the American Heart Association [18POST34050034 to A.H.]. Deposited in PMC for release after 12 months.
Data availability
The sequencing data used were deposited in Gene Expression Omnibus under accession number GSE126647. The gene level-normalized expression data for nkx2.5:ZsYellow+ cells from control and Pbx4-depleted embryos can be navigated at http://altanalyze.org/ICGS/Public/NKX2-5/User.php.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.185652.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.185652.reviewer-comments.pdf
References
- Abdul-Wajid S., Demarest B. L. and Yost H. J. (2018). Loss of embryonic neural crest derived cardiomyocytes causes adult onset hypertrophic cardiomyopathy in zebrafish. Nat. Commun. 9, 4603 10.1038/s41467-018-07054-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrial M., Paffett-Lugassy N., Jeffrey S., Jordan D., O'Loughlin E., Frederick C. J., Burns C. G. and Burns C. E. (2017). TGF-β signaling is necessary and sufficient for pharyngeal arch artery angioblast formation. Cell Rep. 20, 973-983. 10.1016/j.celrep.2017.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. J., Pham V. N., Vogel A. M., Weinstein B. M. and Roman B. L. (2008). Loss of unc45a precipitates arteriovenous shunting in the aortic arches. Dev. Biol. 318, 258-267. 10.1016/j.ydbio.2008.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenkiel B. R., Tvrdik P., Gaufo G. O. and Capecchi M. R. (2004). Hoxb1 functions in both motoneurons and in tissues of the periphery to establish and maintain the proper neuronal circuitry. Genes Dev. 18, 1539-1552. 10.1101/gad.1207204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrington C. B., Dowse B. R., Bleyl S. B. and Bowles N. E. (2012). Non-synonymous variants in pre-B cell leukemia homeobox (PBX) genes are associated with congenital heart defects. Eur. J. Med. Genet. 55, 235-237. 10.1016/j.ejmg.2012.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdougo E. (2003). Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development 130, 6121-6129. 10.1242/dev.00838 [DOI] [PubMed] [Google Scholar]
- Bertrand N., Roux M., Ryckebüsch L., Niederreither K., Dollé P., Moon A., Capecchi M. and Zaffran S. (2011). Hox genes define distinct progenitor sub-domains within the second heart field. Dev. Biol. 353, 266-274. 10.1016/j.ydbio.2011.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum Y., Belting H.-G., Ellertsdottir E., Herwig L., Lüders F. and Affolter M. (2008). Complex cell rearrangements during intersegmental vessel sprouting and vessel fusion in the zebrafish embryo. Dev. Biol. 316, 312-322. 10.1016/j.ydbio.2008.01.038 [DOI] [PubMed] [Google Scholar]
- Cavanaugh A. M., Huang J. and Chen J.-N. (2015). Two developmentally distinct populations of neural crest cells contribute to the zebrafish heart. Dev. Biol. 404, 103-112. 10.1016/j.ydbio.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-P., Stankunas K., Shang C., Kao S.-C., Twu K. Y. and Cleary M. L. (2008). Pbx1 functions in distinct regulatory networks to pattern the great arteries and cardiac outflow tract. Development 135, 3577-3586. 10.1242/dev.022350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S.-K., Vlachakis N. and Sagerström C. G. (2002). Meis family proteins are required for hindbrain development in the zebrafish. Development 129, 585-595. [DOI] [PubMed] [Google Scholar]
- Crowley M. A., Conlin L. K., Zackai E. H., Deardorff M. A., Thiel B. D. and Spinner N. B. (2010). Further evidence for the possible role of MEIS2 in the development of cleft palate and cardiac septum. Am. J. Med. Genet. A 152A, 1326-1327. 10.1002/ajmg.a.33375 [DOI] [PubMed] [Google Scholar]
- D'Aniello E., Rydeen A. B., Anderson J. L., Mandal A. and Waxman J. S. (2013). Depletion of retinoic acid receptors initiates a novel positive feedback mechanism that promotes teratogenic increases in retinoic acid. PLoS Genet. 9, e1003689 10.1371/journal.pgen.1003689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pater E., Clijsters L., Marques S. R., Lin Y.-F., Garavito-Aguilar Z. V., Yelon D. and Bakkers J. (2009). Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development 136, 1633-1641. 10.1242/dev.030924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePasquale E. A. K., Dexheimer P., Schnell D., Ferchen K., Hay S., Chetal K., Valiente-Alandí Í., Blaxall B. C., Grimes H. L. and Salomonis N. (2019). cellHarmony: cell-level matching and holistic comparison of single-cell transcriptomes. Nucleic Acids Res. 47, e138 10.1093/nar/gkz789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohn T. E. and Waxman J. S. (2012). Distinct phases of Wnt/beta-catenin signaling direct cardiomyocyte formation in zebrafish. Dev. Biol. 361, 364-376. 10.1016/j.ydbio.2011.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupays L., Shang C., Wilson R., Kotecha S., Wood S., Towers N. and Mohun T. (2015). Sequential binding of MEIS1 and NKX2-5 on the Popdc2 gene: a mechanism for spatiotemporal regulation of enhancers during cardiogenesis. Cell Rep. 13, 183-195. 10.1016/j.celrep.2015.08.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson T., Scholpp S., Brand M., Moens C. B. and Waskiewicz A. J. (2007). Pbx proteins cooperate with engrailed to pattern the midbrain-hindbrain and diencephalic-mesencephalic boundaries. Dev. Biol. 301, 504-517. 10.1016/j.ydbio.2006.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E., Marshall H., Pöpperl H., Maconochie M., Krumlauf R. and Blasi F. (2000). Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development 127, 155-166. [DOI] [PubMed] [Google Scholar]
- Foglia M. J., Cao J., Tornini V. A. and Poss K. D. (2016). Multicolor mapping of the cardiomyocyte proliferation dynamics that construct the atrium. Development 143, 1688-1696. 10.1242/dev.136606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin D. S., Godfrey V. L., O'Brien D. A., Deng C. and Xiong Y. (2000). Functional collaboration between different cyclin-dependent kinase inhibitors suppresses tumor growth with distinct tissue specificity. Mol. Cell. Biol. 20, 6147-6158. 10.1128/MCB.20.16.6147-6158.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima N., Ishii I., Contos J. J. A., Weiner J. A. and Chun J. (2001). Lysophospholipid receptors. Annu. Rev. Pharmacol. Toxicol. 41, 507-534. 10.1146/annurev.pharmtox.41.1.507 [DOI] [PubMed] [Google Scholar]
- Gibb N., Lazic S., Yuan X., Deshwar A. R., Leslie M., Wilson M. D. and Scott I. C. (2018). Hey2 regulates the size of the cardiac progenitor pool during vertebrate heart development. Development 145, dev167510 10.1242/dev.167510 [DOI] [PubMed] [Google Scholar]
- González-Lázaro M., Roselló-Díez A., Delgado I., Carramolino L., Sanguino M. A., Giovinazzo G. and Torres M. (2014). Two new targeted alleles for the comprehensive analysis of Meis1 functions in the mouse. Genesis 52, 967-975. 10.1002/dvg.22833 [DOI] [PubMed] [Google Scholar]
- Grant M. G., Patterson V. L., Grimes D. T. and Burdine R. D. (2017). Modeling syndromic congenital heart defects in zebrafish. Curr. Top. Dev. Biol. 124, 1-40. 10.1016/bs.ctdb.2016.11.010 [DOI] [PubMed] [Google Scholar]
- Guner-Ataman B., Paffett-Lugassy N., Adams M. S., Nevis K. R., Jahangiri L., Obregon P., Kikuchi K., Poss K. D., Burns C. E. and Burns C. G. (2013). Zebrafish second heart field development relies on progenitor specification in anterior lateral plate mesoderm and nkx2.5 function. Development 140, 1353-1363. 10.1242/dev.088351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guner-Ataman B., González-Rosa J. M., Shah H. N., Butty V. L., Jeffrey S., Abrial M., Boyer L. A., Burns C. G. and Burns C. E. (2018). Failed progenitor specification underlies the cardiopharyngeal phenotypes in a zebrafish model of 22q11.2 deletion syndrome. Cell Rep. 24, 1342-1354.e5. 10.1016/j.celrep.2018.06.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay S. B., Ferchen K., Chetal K., Grimes H. L. and Salomonis N. (2018). The human cell atlas bone marrow single-cell interactive web portal. Exp. Hematol. 68, 51-61. 10.1016/j.exphem.2018.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y., Pan L., Walker C., Dowd J., Moens C. B. and Hughes S. M. (2012). Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation. Dev. Biol. 369, 199-210. 10.1016/j.ydbio.2012.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman J. I. E. and Kaplan S. (2002). The incidence of congenital heart disease. J. Am. Coll. Cardiol. 39, 1890-1900. 10.1016/S0735-1097(02)01886-7 [DOI] [PubMed] [Google Scholar]
- Hoffman J. I. E., Kaplan S. and Liberthson R. R. (2004). Prevalence of congenital heart disease. Am. Heart J. 147, 425-439. 10.1016/j.ahj.2003.05.003 [DOI] [PubMed] [Google Scholar]
- Huang C.-J., Tu C.-T., Hsiao C.-D., Hsieh F.-J. and Tsai H.-J. (2003). Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 228, 30-40. 10.1002/dvdy.10356 [DOI] [PubMed] [Google Scholar]
- Isogai S., Horiguchi M. and Weinstein B. M. (2001). The vascular anatomy of the developing zebrafish: an atlas of embryonic and early larval development. Dev. Biol. 230, 278-301. 10.1006/dbio.2000.9995 [DOI] [PubMed] [Google Scholar]
- Jalili A., Wagner C., Pashenkov M., Pathria G., Mertz K. D., Widlund H. R., Lupien M., Brunet J.-P., Golub T. R., Stingl G. et al. (2012). Dual suppression of the cyclin-dependent kinase inhibitors CDKN2C and CDKN1A in human melanoma. J. Natl. Cancer Inst. 104, 1673-1679. 10.1093/jnci/djs373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., King I. N., Chopra S. S., Wan H., Ni T. T., Jiang C., Guan X., Wells S., Srivastava D. and Zhong T. P. (2007). Vertebrate heart growth is regulated by functional antagonism between Gridlock and Gata5. Proc. Natl. Acad. Sci. USA 104, 14008-14013. 10.1073/pnas.0702240104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao R. M., Rurik J. G., Farr G. H. III, Dong X. R., Majesky M. W. and Maves L. (2015). Pbx4 is required for the temporal onset of zebrafish myocardial differentiation. J. Dev. Biol. 3, 93-111. 10.3390/jdb3040093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan B. R., Feldman J. L., Begemann G., Ingham P. W. and Yelon D. (2005). Retinoic acid signaling restricts the cardiac progenitor pool. Science 307, 247-249. 10.1126/science.1101573 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Miller C. T. and Moens C. B. (2001). Specification and morphogenesis of the zebrafish larval head skeleton. Dev. Biol. 233, 239-257. 10.1006/dbio.2001.0201 [DOI] [PubMed] [Google Scholar]
- Kreis N.-N., Louwen F., Zimmer B. and Yuan J. (2015). Loss of p21Cip1/CDKN1A renders cancer cells susceptible to Polo-like kinase 1 inhibition. Oncotarget 6, 6611-6626. 10.18632/oncotarget.2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazic S. and Scott I. C. (2011). Mef2cb regulates late myocardial cell addition from a second heart field-like population of progenitors in zebrafish. Dev. Biol. 354, 123-133. 10.1016/j.ydbio.2011.03.028 [DOI] [PubMed] [Google Scholar]
- Liao J., Aggarwal V. S., Nowotschin S., Bondarev A., Lipner S. and Morrow B. E. (2008). Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev. Biol. 316, 524-537. 10.1016/j.ydbio.2008.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Zurita L., Stirnimann C. U., Glatt S., Kaynak B. L., Thomas S., Baudin F., Samee M. A. H., He D., Small E. M., Mileikovsky M. et al. (2016). Complex interdependence regulates heterotypic transcription factor distribution and coordinates cardiogenesis. Cell 164, 999-1014. 10.1016/j.cell.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mably J. D., Mohideen M.-A. P. K., Burns C. G., Chen J.-N. and Fishman M. C. (2003). heart of glass regulates the concentric growth of the heart in zebrafish. Curr. Biol. 13, 2138-2147. 10.1016/j.cub.2003.11.055 [DOI] [PubMed] [Google Scholar]
- Maceyka M., Payne S. G., Milstien S. and Spiegel S. (2002). Sphingosine kinase, sphingosine-1-phosphate, and apoptosis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1585, 193-201. 10.1016/S1388-1981(02)00341-4 [DOI] [PubMed] [Google Scholar]
- Mahler J., Filippi A. and Driever W. (2010). DeltaA/DeltaD regulate multiple and temporally distinct phases of notch signaling during dopaminergic neurogenesis in zebrafish. J. Neurosci. 30, 16621-16635. 10.1523/JNEUROSCI.4769-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud A. I., Kocabas F., Muralidhar S. A., Kimura W., Koura A. S., Thet S., Porrello E. R. and Sadek H. A. (2013). Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature 497, 249-253. 10.1038/nature12054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L., Waskiewicz A. J., Paul B., Cao Y., Tyler A., Moens C. B. and Tapscott S. J. (2007). Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development 134, 3371-3382. 10.1242/dev.003905 [DOI] [PubMed] [Google Scholar]
- Maves L., Tyler A., Moens C. B. and Tapscott S. J. (2009). Pbx acts with Hand2 in early myocardial differentiation. Dev. Biol. 333, 409-418. 10.1016/j.ydbio.2009.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock J. M., Kheirbek M. A. and Prince V. E. (2002). Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development 129, 2339-2354. [DOI] [PubMed] [Google Scholar]
- Miao M., Bruce A. E. E., Bhanji T., Davis E. C. and Keeley F. W. (2007). Differential expression of two tropoelastin genes in zebrafish. Matrix Biol. 26, 115-124. 10.1016/j.matbio.2006.09.011 [DOI] [PubMed] [Google Scholar]
- Moriyama Y., Ito F., Takeda H., Yano T., Okabe M., Kuraku S., Keeley F. W. and Koshiba-Takeuchi K. (2016). Evolution of the fish heart by sub/neofunctionalization of an elastin gene. Nat. Commun. 7, 10397 10.1038/ncomms10397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Calof A. L., Lander A. D. and Schilling T. F. (2011). Multifactorial origins of heart and gut defects in nipbl-deficient zebrafish, a model of cornelia de lange syndrome. PLoS Biol. 9, e1001181 10.1371/journal.pbio.1001181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagelberg D., Wang J., Su R., Torres-Vázquez J., Targoff K. L., Poss K. D. and Knaut H. (2015). Origin, specification, and plasticity of the great vessels of the heart. Curr. Biol. 25, 2099-2110. 10.1016/j.cub.2015.06.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeb Z., Lajiness J. D., Bolanis E. and Conway S. J. (2013). Cardiac outflow tract anomalies. Wiley Interdiscip. Rev. Dev. Biol. 2, 499-530. 10.1002/wdev.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevis K., Obregon P., Walsh C., Guner-Ataman B., Burns C. G. and Burns C. E. (2013). Tbx1 is required for second heart field proliferation in zebrafish. Dev. Dyn. 242, 550-559. 10.1002/dvdy.23928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A., Venkatasubramanian M., Chaudhri V. K., Aronow B. J., Salomonis N., Singh H. and Grimes H. L. (2016). Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537, 698-702. 10.1038/nature19348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxtoby E. and Jowett T. (1993). Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 21, 1087-1095. 10.1093/nar/21.5.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffett-Lugassy N., Singh R., Nevis K. R., Guner-Ataman B., O'Loughlin E., Jahangiri L., Harvey R. P., Burns C. G. and Burns C. E. (2013). Heart field origin of great vessel precursors relies on nkx2.5-mediated vasculogenesis. Nat. Cell Biol. 15, 1362-1369. 10.1038/ncb2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffett-Lugassy N., Novikov N., Jeffrey S., Abrial M., Guner-Ataman B., Sakthivel S., Burns C. E. and Burns C. G. (2017). Unique developmental trajectories and genetic regulation of ventricular and outflow tract progenitors in the zebrafish second heart field. Development 144, 4616-4624. 10.1242/dev.153411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeufer A., van Noord C., Marciante K. D., Arking D. E., Larson M. G., Smith A. V., Tarasov K. V., Müller M., Sotoodehnia N., Sinner M. F. et al. (2010). Genome-wide association study of PR interval. Nat. Genet. 42, 153-159. 10.1038/ng.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski T. and Nüsslein-Volhard C. (2000). The endoderm plays an important role in patterning the segmented pharyngeal region in zebrafish (Danio rerio). Dev. Biol. 225, 339-356. 10.1006/dbio.2000.9842 [DOI] [PubMed] [Google Scholar]
- Pöpperl H., Rikhof H., Cheng H., Haffter P., Kimmel C. B. and Moens C. B. (2000). lazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol. Cell 6, 255-267. 10.1016/S1097-2765(00)00027-7 [DOI] [PubMed] [Google Scholar]
- Proulx K., Lu A. and Sumanas S. (2010). Cranial vasculature in zebrafish forms by angioblast cluster-derived angiogenesis. Dev. Biol. 348, 34-46. 10.1016/j.ydbio.2010.08.036 [DOI] [PubMed] [Google Scholar]
- Qiu X., Hill A., Packer J., Lin D., Ma Y.-A. and Trapnell C. (2017a). Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309 10.1038/nmeth.4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H. A. and Trapnell C. (2017b). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana M. S., Théveniau-Ruissy M., De Bono C., Mesbah K., Francou A., Rammah M., Domínguez J. N., Roux M., Laforest B., Anderson R. H. et al. (2014). Tbx1 coordinates addition of posterior second heart field progenitor cells to the arterial and venous poles of the heart. Circ. Res. 115, 790-799. 10.1161/CIRCRESAHA.115.305020 [DOI] [PubMed] [Google Scholar]
- Reifers F., Walsh E. C., Léger S., Stainier D. Y. and Brand M. (2000). Induction and differentiation of the zebrafish heart requires fibroblast growth factor 8 (fgf8/acerebellar). Development 127, 225-235. [DOI] [PubMed] [Google Scholar]
- Robitaille K., Rourke J. L., McBane J. E., Fu A., Baird S., Du Q., Kin T., Shapiro A. M. J. and Screaton R. A. (2016). High-throughput functional genomics identifies regulators of primary human beta cell proliferation. J. Biol. Chem. 291, 4614-4625. 10.1074/jbc.M115.683912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A. and Ekker S. C. (2007). p53 Activation by Knockdown Technologies. PLoS Genet. 3, e78 10.1371/journal.pgen.0030078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M. and Zaffran S. (2016). Hox genes in cardiovascular development and diseases. J. Dev. Biol. 4, 14 10.3390/jdb4020014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M., Laforest B., Capecchi M., Bertrand N. and Zaffran S. (2015). Hoxb1 regulates proliferation and differentiation of second heart field progenitors in pharyngeal mesoderm and genetically interacts with Hoxa1 during cardiac outflow tract development. Dev. Biol. 406, 247-258. 10.1016/j.ydbio.2015.08.015 [DOI] [PubMed] [Google Scholar]
- Row R. H. and Martin B. L. (2017). itFISH: enhanced staining by iterative fluorescent in situ hybridization. Zebrafish 14, 578-580. 10.1089/zeb.2016.1413 [DOI] [PubMed] [Google Scholar]
- Rueden C. T., Schindelin J., Hiner M. C., DeZonia B. E., Walter A. E., Arena E. T. and Eliceiri K. W. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 1-26. 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckebusch L., Wang Z., Bertrand N., Lin S.-C., Chi X., Schwartz R., Zaffran S. and Niederreither K. (2008). Retinoic acid deficiency alters second heart field formation. Proc. Natl. Acad. Sci. USA 105, 2913-2918. 10.1073/pnas.0712344105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydeen A. B. and Waxman J. S. (2016). Cyp26 enzymes facilitate second heart field progenitor addition and maintenance of ventricular integrity. PLoS Biol. 14, e2000504 10.1371/journal.pbio.2000504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsa L. A., Fleming N., Magness S., Qian L. and Liu J. (2016). Isolation and characterization of single cells from zebrafish embryos. J. Vis. Exp., e53877 10.3791/53877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler Y. L., Garske K. M., Wang J., Firulli B. A., Firulli A. B., Poss K. D. and Yelon D. (2014). Hand2 elevates cardiomyocyte production during zebrafish heart development and regeneration. Development 141, 3112-3122. 10.1242/dev.106336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D. and Livak K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101-1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Sirbu I. O., Zhao X. and Duester G. (2008). Retinoic acid controls heart anteroposterior patterning by down-regulating Isl1 through the Fgf8 pathway. Dev. Dyn. 237, 1627-1635. 10.1002/dvdy.21570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell M. R. J. and Waxman J. S. (2011). Restraint of Fgf8 signaling by retinoic acid signaling is required for proper heart and forelimb formation. Dev. Biol. 358, 44-55. 10.1016/j.ydbio.2011.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachura D. L. and Traver D. (2016). Cellular dissection of zebrafish hematopoiesis. Methods Cell Biol. 133, 11-53. 10.1016/bs.mcb.2016.03.022 [DOI] [PubMed] [Google Scholar]
- Stankunas K., Shang C., Twu K. Y., Kao S.-C., Jenkins N. A., Copeland N. G., Sanyal M., Selleri L., Cleary M. L. and Chang C.-P. (2008). Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circ. Res. 103, 702-709. 10.1161/CIRCRESAHA.108.175489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt D. and Stainier D. (2012). Uncovering the molecular and cellular mechanisms of heart development using the zebrafish. Annu. Rev. Genet. 46, 397-418. 10.1146/annurev-genet-110711-155646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer M., Gavalas A., Marshall H., Ariza-McNaughton L., Rijli F. M., Chambon P. and Krumlauf R. (1998). Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development 125, 1025-1036. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N. J., Livak K. J., Mikkelsen T. S. and Rinn J. L. (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad J. A., Alexander J. M., Truty R. M., Shrikumar A., Li F., Eilertson K. E., Ding H., Wylie J. N., Pico A. R., Capra J. A. et al. (2017). Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 151, 206-220. 10.1016/j.cell.2012.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen D., Chen K., Jubran A., Ramirez A. J. and Astrof S. (2017). Endothelium in the pharyngeal arches 3, 4 and 6 is derived from the second heart field. Dev. Biol. 421, 108-117. 10.1016/j.ydbio.2016.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz A. J., Rikhof H. A., Hernandez R. E. and Moens C. B. (2001). Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development 128, 4139-4151. 10.3410/f.1004495.19155 [DOI] [PubMed] [Google Scholar]
- Waskiewicz A. J., Rikhof H. A. and Moens C. B. (2002). Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev. Cell 3, 723-733. 10.1016/S1534-5807(02)00319-2 [DOI] [PubMed] [Google Scholar]
- Waxman J. S., Keegan B. R., Roberts R. W., Poss K. D. and Yelon D. (2008). Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev. Cell 15, 923-934. 10.1016/j.devcel.2008.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). Eugene, OR: University of Oregon Press. [Google Scholar]
- Yelon D., Horne S. A. and Stainier D. Y. R. (1999). Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 214, 23-37. 10.1006/dbio.1999.9406 [DOI] [PubMed] [Google Scholar]
- Zeng X.-X. I. and Yelon D. (2014). Cadm4 restricts the production of cardiac outflow tract progenitor cells. Cell Rep. 7, 951-960. 10.1016/j.celrep.2014.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Han P., Yang H., Ouyang K., Lee D., Lin Y.-F., Ocorr K., Kang G., Chen J., Stainier D. Y. R. et al. (2013). In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 498, 497-501. 10.1038/nature12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Cashman T. J., Nevis K. R., Obregon P., Carney S. A., Liu Y., Gu A., Mosimann C., Sondalle S., Peterson R. E. et al. (2011). Latent TGF-beta binding protein 3 identifies a second heart field in zebrafish. Nature 474, 645-648. 10.1038/nature10094 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.